Abstract
RNA silencing is conserved in a broad range of eukaryotes and operates in the development and maintenance of genome integrity in many organisms. Plants have adapted this system for antiviral defense, and plant viruses have in turn developed mechanisms to suppress RNA silencing. RNA silencing-related RNA inactivation is likely based on target RNA cleavage or translational arrest. Although it is widely assumed that virus-induced gene silencing (VIGS) promotes the endonucleolytic cleavage of the viral RNA genome, this popular assumption has never been tested experimentally. Here we analyzed the viral RNA targeting by VIGS in tombusvirus-infected plants, and we show evidence that antiviral response of VIGS is based on viral RNA cleavage by RNA-induced silencing effector complex (RISC) programmed by virus-specific small interfering RNAs (siRNAs). In addition, we found that the RISC-mediated cleavages do not occur randomly on the viral genome. Indeed, sequence analysis of cloned cleavage products identified hot spots for target RNA cleavage, and the regions of specific RISC-mediated cleavages are asymmetrically distributed along the positive- and negative-sense viral RNA strands. In addition, we identified viral siRNAs containing high-molecular-mass protein complexes purified from the recovery leaves of the silencing suppressor mutant virus-infected plants. Strikingly, these large nucleoproteins cofractionated with microRNA-containing complexes, suggesting that these nucleoproteins are silencing related effector complexes.
RNA silencing is an ancient self-defense mechanism induced by double-stranded RNA (dsRNA) leading to a homology-dependent degradation of a target RNA. RNA silencing is conserved across kingdoms and is known as quelling in fungi, RNA interference in animals, and posttranscriptional gene silencing in plants.
The unifying feature of RNA silencing relies on a set of highly conserved reactions that are triggered by dsRNA, which is processed into 21- to 24-nucleotide (nt) small interfering RNAs (siRNAs) (13, 14, 33) by the RNase III enzyme Dicer and its homologues (4, 30). These siRNAs program the multisubunit RNA-induced silencing complex (RISC) (15) and guide the complex to degrade cellular RNA molecules having perfect or near perfect base pairing between mRNA and the guide siRNA, while they mediate translation repression in case of partial complementarity (2, 8, 11, 18).
Replicating plant viruses are strong inducers as well as targets of virus-induced RNA silencing (VIGS). Indeed, the accumulation of virus-derived siRNAs were reported in local and systemic tissues of virus-infected plants (14, 41) demonstrating the activation of VIGS, which resulted in lowering the accumulation of the invading virus and the recovery phenotype in upper noninoculated leaves (41). Thus, VIGS emerged as an RNA-mediated defense response to protect plants against viral infection (27, 46, 47, 49). The finding that many plant viruses have evolved proteins that suppress at different stages of the silencing pathway (20, 23, 39, 40, 48) provides the most compelling evidence for the function of VIGS as a natural antiviral defense mechanism (46). Although several virus-encoded RNA silencing suppressor proteins have been described to date, only a few of them are characterized at the molecular level. The best-characterized silencing suppressor is the p19 protein of tombusviruses, which specifically binds 21-nt double-stranded siRNAs in vitro and in vivo, preventing siRNA incorporation into effector complexes such as RISC (21, 40, 45, 50). Similarly, the role and mechanism of VIGS in natural virus infection were studied in detail in tombusvirus-infected plants (16, 17, 40-42). However, the mode of inactivation of viral RNAs was not investigated.
Cymbidium ringspot virus (CymRSV) is a member of the genus Tombusvirus consisting on a positive single-stranded RNA genome with five open reading frames (ORFs) (36). The viral RNA of CymRSV, similar to most of the positive-strand RNA viruses, replicates its genome via dsRNA intermediates, which are believed to trigger the siRNA-generating machinery (1). However, recent findings demonstrated that folded viral RNA is preferentially processed into siRNAs in virus-infected plants (19, 28, 41).
CymRSV infection of Nicotiana benthamiana causes necrotic symptoms in systemic leaves that lead to plant death within 10 to 14 days. In contrast, plants infected with the silencing suppressor mutant virus (Cym19stop) produced different symptoms. New leaves of Cym19stop-infected plants, which develop after the first symptomatic leaves, are symptomless (recovered) and contained very low or nondetectable levels of viral RNA (41, 42). However, protoplast studies and in situ analysis of virus-infected plants revealed that accumulation of virus RNA in a single cell infected by Cym19stop was similar to that of CymRSV, suggesting that the replication of Cym19stop mutant is not impaired (16, 40). Instead, the spread of mutant virus is confined in and around the veins of recovered leaves of Cym19stop-infected plants (16). Moreover, the recovered leaves of Cym19stop-infected plants were resistant to challenge of virus infection carrying CymRSV sequences (41), suggesting an activated VIGS in the recovered plant tissues. This unique feature of the recovered leaves of Cym19stop-infected plants allows us to analyze the mechanism of VIGS in detail and in natural circumstances. VIGS was discovered many years ago; however, the major steps of the antiviral response are still not known. Indeed, we do not know (it is theoretically possible) that the DICER activity alone is responsible for viral RNA degradation or the viral siRNA-containing RISC is also required for the antiviral effect. Moreover, it is also not clear that viral siRNA-programmed RISC mediates the cleavage of viral RNA or inhibits its translation. Indeed, the high level of viral siRNAs generated from folded viral RNA, displaying only partial complementarity with viral RNA, raises the possibility that translational inhibition is also a possible mode of viral RNA inactivation in addition to the well-accepted (but never tested) target RNA cleavage-based mechanism.
This paper reports the use of recently developed in vivo system based on Agrobacterium-mediated delivery of sensor sequences sensitive to the VIGS in the recovered leaves of Cym19stop-infected plants (20). By using this approach, we demonstrated that VIGS-mediated viral RNA inactivation by the virus-induced RNA silencing is likely based on a sequence-specific cleavage of the viral target sequence rather than translational inhibition. Our finding also showed that viral sequences having either plus or minus polarity are good targets of VIGS, although viral RNA with minus polarity was cleaved more efficiently than the plus-sense viral RNA. Moreover, no cause-effect links were found between the cleavage site hot spots and the virus-generated siRNA profile. In addition, we identified high-molecular-mass nucleoprotein complexes containing virus-derived siRNAs.
MATERIALS AND METHODS
Plant material, virus infection, and Agrobacterium tumefaciens infiltration.
Six-leaf-stage Nicotiana benthamiana plants were inoculated with Cym19stop or PoLVΔp14 purified virions (42) and grown at a constant temperature of 22°C. The leaves showing the recovery phenotype developed 10 to 15 days postinoculation (dpi), and the sensor constructs were expressed in these recovered leaves by Agrobacterium-mediated transient expression as described earlier (32).
Preparation of sensor constructs.
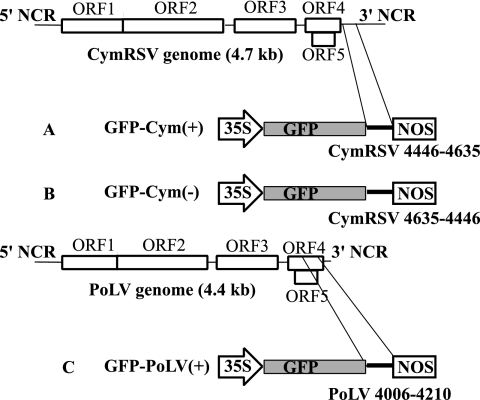
All sensor constructs were prepared from the previously reported 35S-green fluorescent protein (GFP) binary plasmid (6). First, a SmaI restriction site was inserted downstream of the GFP ORF by using the QuikChange site-directed mutagenesis kit (Stratagene) by following the instruction manual. The PCR fragment of 190 bp, corresponding to positions 4446 to 4635 of the CymRSV genome (accession no. NC003532), was amplified by using appropriate 5′-phosphorylated oligonucleotides and placed into the unique SmaI site of the modified 35S-GFP plasmid. The plus and minus orientations of the insert were selected, thus generating GFP-Cym(+) and GFP-Cym(−) sensor constructs (Fig. 1A and 1B, respectively). By analogy, the PCR fragment 205 bp in length, corresponding to positions 4006 to 4210 of the Pothos latent virus (PoLV) genome (accession no. X87115), was cloned as described above, thus generating the GFP-PoLV(+) plasmid (Fig. 1C). Constructs GFP 171.1 and GFP 171.2 were kindly provided by O. Voinnet and were described previously (32).
FIG. 1.
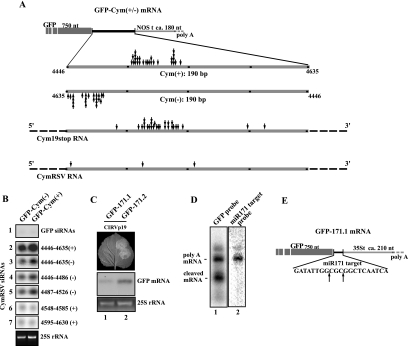
Schematic representation of the cloning strategy used to generate sensor sequences. The 190-bp-long amplified product corresponding to the positions 4446 to 4635 (referred to as the CymRSV genome) was placed downstream of the GFP reporter ORF in both orientations, thus generating GFP-Cym(+) (A) and GFP-Cym(−) (B) sensor sequences. By analogy, the 205-bp-long amplified product corresponding to positions 4006 to 4210 of the PoLV genome was cloned in the same way, thus generating the sensor GFP-PoLV(+) (C). The cloned PoLV and CymRSV fragment showed a level of similarity below 15% (data not shown). Sensor sequences were under the control of the 35S promoter and Nopaline synthase terminator in a pBIN-based binary vector. NCR, noncoding region; NOS, nopalin synthase.
Protein and RNA analysis.
Agro-infiltrated spots were excised, and ca. 100 mg in weight of leaf tissues were homogenized as previously described (9). Equal volumes of the grounded plant material were then submitted to RNA extraction and protein extraction.
Protein extraction was performed as described previously (37), separated in a 12% volume of sodium dodecyl sulfate-polyacrylamide gel, and then transferred onto a Hybond P filter (Amersham-Pharmacia, United Kingdom). GFP was probed with mouse monoclonal antibody (Amersham Biosciences, United Kingdom) and Carnation Italian ringspot virus (CIRV) p19 with rabbit antisera (45). Immunodetection was performed by using an enhanced chemiluminescence kit (ECL; Amersham-Biosciences, United Kingdom). Ponceau staining of total proteins provided a control for protein loading.
RNA extraction was performed by Tri-reagent (Sigma) according to the manufacturer's directions. The same total RNA extract was used for high- and low-molecular-mass RNA analysis as described previously (41). RNAs were detected by using 32P-labeled probes generated by in vitro transcription with the StripEZ RNA kit (Ambion). The GFP from position 608 to 750 was PCR amplified with a reverse oligonucleotide flanked with T7 RNA polymerase promoter and was used to detect GFP mRNA, whereas GFP mRNA-deriving siRNAs were detected by a riboprobe against the entire GFP mRNA. CymRSV-deriving siRNAs were detected by riboprobes obtained from the in vitro transcription of PCR-amplified products of the CymRSV regions, as detailed in the text and figures. MicroRNAs (miRNAs) were detected by using 32P-labeled locked nucleic acid (LNA) oligonucleotide probes described previously (44).
3′ rapid amplification of cDNA ends sequence analysis of the cleaved sensor and viral RNAs.
Total RNA (5 μg) extracted as previously described was ligated with 3′-end adapter oligonucleotide (28). The ligated RNA was submitted to reverse transcription with the specific oligonucleotide complementary to the 3′-end adapter and then submitted to PCR amplification using the same oligonucleotide used for the reverse transcription as a reverse primer and an oligonucleotide homologue to the region 608 to 630 of the GFP ORF (in the case of the 3′-end analysis of sensor sequences) or an oligonucleotide homologue to the region 4230 to 4253 of the CymRSV genome (in the case of the 3′-end analysis CymRSV and Cym19stop cleavage product) as a forward primer. The amplified products were then cloned into pUC18 SmaI and sequenced.
Fractionation of virus-derived siRNA- and miRNA-containing complexes from virus-infected plants.
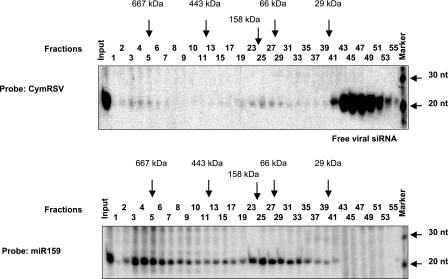
The protein extract was prepared at 7 dpi from systemically infected leaves of Cym19stop-infected N. benthamiana plants and size separated by a Superdex-200 gel filtration column (21). RNA was extracted from each fraction and analyzed on a gel blot. The gel blot was hybridized first with CymRSV probe, and then the membrane was stripped and reprobed with [γ-32P]ATP-labeled miR159 LNA oligonucleotide. The elution positions of protein molecular markers are as follows: 669 kDa, thyroglobulin; 443 kDa, ferritin, 150 kDa, aldolase; 66 kDa, bovine serum albumin; 29 kDa, carbonic anhydrase.
RESULTS
Tombusvirus-induced gene silencing is based on a sequence-specific RNA degradation mediated by viral siRNA-programmed RISC.
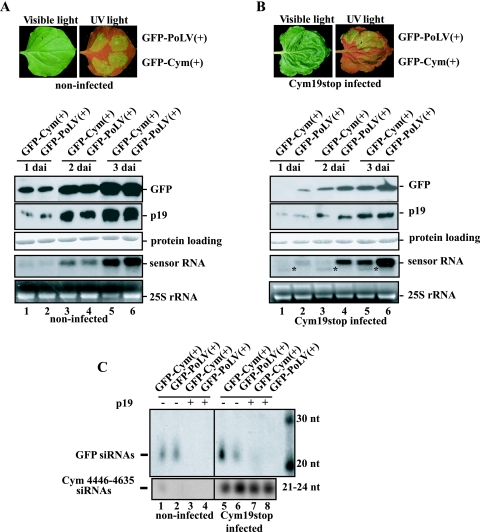
To get better insight into the molecular mechanism of RNA silencing in natural virus infection, a new experimental system was further developed. This new approach is based on Cym19stop virus-infected plant. Previously, we have shown that the recovery phenotype of Cym19stop-infected Nicotiana benthamiana plants is a consequence of virus-induced gene silencing. The recovered leaves of Cym19stop plants were susceptible to Potato virus X (PVX) infection but resistant to PVX-Cym constructs containing sequences homologous with CymRSV, suggesting that the PVX-Cym viral genome was efficiently targeted by viral siRNA-programmed RISC (41). Indeed, these virus-infected plants contained virus-derived siRNAs, which predominantly originated from folded regions of the positive strand of the viral RNA (28, 41). Importantly, these virus-derived siRNAs show partial complementarity to the plus-sense viral RNAs and perfect complementarity to the minus-sense viral RNAs. Considering that in plants siRNA-guided RISC can mediate cleavage of target mRNA when there is perfect or near perfect base pairing between target mRNAs and the short guide RNA (24) and translation repression when there is partial complementarity (2, 8), it is difficult to predict how viral RNA is inactivated by VIGS. To clear up the meaning of RISC-mediated “viral RNA inactivation” and to distinguish between viral inhibition of translation and/or viral RNA degradation, we set up the following experiment. N. benthamiana plants at the six-leaf stage were infected with the Cym19stop virus (41). The infected plants showed the typical recovery phenotype in 10 to 15 dpi at a standard temperature of 22°C. We expected that these recovered leaves contain viral siRNA-programmed RISC, which can be sensed unambiguously by the virus-specific sensor construct. To do so, we developed sensor constructs containing GFP-encoding ORF fused to virus-specific target sequences. A 190-nt-long region of CymRSV corresponding to positions 4446 to 4635 of the viral genome was used to develop the GFP-Cym(+) sensor (Fig. 1A). This target region was chosen because it is particularly sensitive for targeting by VIGS (41). A 205-nt-long region (similar in size and position) of PoLV corresponding to positions 4006 to 4210 of the viral genome was also used to construct the GFP-PoLV(+) sensor (Fig. 1C). The two constructs GFP-Cym(+) and GFP-PoLV(+) were delivered by agro-infiltration in plant leaves recovering from Cym19stop infection. Then a time course analysis of protein and mRNAs was carried out on samples extracted from the agro-infiltrated spots. Since the low level of homology between the PoLV target sequence and the corresponding region in CymRSV (data not shown), we expected that CymRSV-derived siRNA-activated RISC would target only the homologous sensor construct (GFP-Cym) but not the GFP-PoLV sensor containing the heterologous target sequence. As a control, both sensors were also expressed by agro-infiltration in the noninfected plants (Fig. 2A). The expression of a sensor construct by agro-infiltration leads to the activation of gene silencing against the expressed sensors mRNAs, which is demonstrated by accumulation of sensor-specific siRNAs (Fig. 2C, lanes 1 and 2). To avoid the expressed sensors activating transgene-induced silencing against themselves, p19 silencing suppressor protein of CIRV was coexpressed with each of the two sensors, which indeed blocked the accumulation of sensor-specific siRNAs (Fig. 2C, lanes 3 and 4). The use of p19 is not expected to interfere with sensor RNA targeting by preassembled RISC, since we previously demonstrated that p19 prevents the assembly of active RISC by siRNA sequestration (20, 21, 40) but does not inhibit the RNA targeting by preassembled RISC (20). Therefore, we predicted that the coexpression of p19 will not compromise the cleavage of sensor RNAs by the preassembled RISCs, which contain virus specific siRNAs. To monitor the expression of the two sensors, GFP-Cym(+) and GFP-PoLV(+), we monitored the accumulations of GFPs and the corresponding sensor mRNAs in a time course experiment. Both GFP-Cym(+) and GFP-PoLV(+) expressed the same amount of GFP mRNA and protein in noninfected plants, confirming that the expression levels of the two sensor molecules were very similar during the first 3 days after agro-infiltration (dai) (Fig. 2A, lanes 1 to 2, 3 to 4, and 5 to 6). However, remarkable differences were observed in the accumulation of GFP-Cym(+) and GFP-PoLV(+) when the sensors were delivered in the recovering leaves of Cym19stop-infected plants (Fig. 2B). At 1 dai, we could detect the expression of GFP-PoLV(+) construct at either protein or RNA level, while the accumulation of GFP-Cym(+) was hardly detectable (Fig. 2B, lane 2 versus 1). At 2 dai, the GFP expression of GFP-PoLV(+) was significantly higher than that of GFP-Cym(+), and this difference was more pronounced at the RNA level (Fig. 2B, lane 3 versus 4). The accumulation of GFP-Cym(+) mRNA was just above the detection level and the accumulation of a new RNA species shorter than the sensor mRNA, which hybridized with GFP-specific probe, could be observed (Fig. 2B, mRNA panel, lanes 1, 3, and 5). This shorter RNA band was identified as the 5′ cleavage product of GFP-Cym(+) sensor mRNA (see later). The level of GFP-PoLV(+) mRNA remained intact and accumulated at a significantly higher level than the GFP-Cym(+) mRNA, indicating that the Cym19stop-specific siRNA-programmed RISC can cleave only the homologous sequence containing sensor mRNA (Fig. 2B, lanes 1, 3, and 5). At 3 dai, the accumulations of the two sensors were more similar to each other than at 2 dai, and the GFP expression from GFP-PoLV(+) was slightly higher than the GFP level from GFP-Cym(+), while the mRNA accumulation from GFP-PoLV(+) was much higher than from GFP-Cym(+) (Fig. 2B, lane 6 against lane 5). It is worthy of note that the accumulation of the uncleaved GFP-Cym(+) mRNA increased at 3 dai (Fig. 2B, lanes 1, 3, and 5). This suggests that the activity of preassembled antiviral RISC lasts for approximately 2 to 3 days, since the coexpressed p19 suppressor with the sensor sequences prevents the assembly of new siRNA-containing RISC from the time point of agro-infiltration. These results also clearly suggest that the viral RNA targeting by VIGS is mediated mainly by RNA cleavage.
FIG. 2.
In vivo analysis of GFP-Cym(+) and GFP-PoLV(+) sensor sequences. Spots in half leaves were infiltrated with Agrobacterium containing the indicated sensor sequences and CIRV p19 into noninfected and Cym19stop-recovering plants (A and B, respectively). Leaves were viewed at 3 dai under visible and long-wavelength UV illumination (A and B, upper panels). Time course of protein- and RNA-normalized extracts at 1, 2, and 3 dai subjected to immunoblot analysis or to RNA blot analysis using a radiolabeled GFP sequence probe. Total protein loading control was stained by Ponceau reagent, whereas the 25S RNA was used as a loading control for total RNA (A and B, lower panels). Total RNA extracts at 3 dai were submitted to a low-molecular-mass RNA analysis (C). 32P-labeled RNA probes raised against GFP and CymRSV (positions 4446 to 4635) were used to detect siRNAs deriving from the GFP part and the viral sequences, respectively. Decade markers (Ambion) were used as small RNA molecular markers. *, position of 5′ cleavage product target RNA.
Sequence-specific cleavage of sensor RNAs by VIGS.
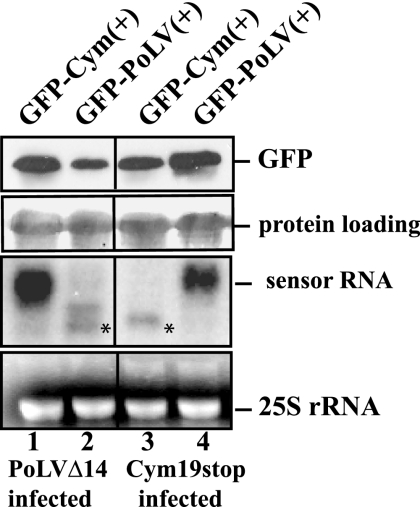
Although the obtained data suggest that the inactivation of GFP-Cym(+) sensor and the appearance of its low-molecular-mass form is due to a sequence-specific phenomenon, we designed experiments to confirm this hypothesis and to further analyze the sequence-specific nature of VIGS. To this end, we used the previously described PoLV p14 silencing suppressor-deficient mutant (PoLVΔ14) virus (26, 35) along with the Cym19stop mutant virus to preinoculate N. benthamiana plants. Both silencing suppressor mutant virus infections resulted in the development of recovery phenotype 12 to 15 days after inoculation. The recovered leaves of both Cym19stop and PoLVΔ14 were agro-infiltrated either with GFP-PoLV(+) or with GFP-Cym(+), and the expression of GFP and the sensor mRNAs were monitored. Both sensor molecules were efficiently targeted by VIGS when expressed by agro-infiltration in the recovered leaf of corresponding homologous mutant virus-preinoculated plant [e.g., GFP-PoLV(+) agro-infiltrated PoLVΔ14-infected plant] (Fig. 3, lane 2). At 2 dai, the sensor RNA 5′ cleavage products were clearly detectable in the homologous sensor-virus combination, indicating that the viral siRNA programmed RISC activity (Fig. 3, lanes 2 and 3). In contrast, the sensor RNAs remained intact and accumulated at a high level when they were expressed in heterologous virus-preinoculated plant [e.g., GFP-PoLV(+) agro-infiltrated Cym19stop-infected plant] (Fig. 3, lane 4). These results definitely confirmed the presence of preassembled cleavage-compatible RISC in the recovered leaves of preinoculated plants and also demonstrated that the target recognition and cleavage of viral siRNA-programmed RISC are highly sequence specific.
FIG. 3.
In vivo analysis of the accumulation of GFP-Cym(+) and GFP-PoLV(+) in homologous and heterologous virus-infected N. benthamiana. Total proteins and RNAs were extracted at 3 dai from Cym19stop-recovering (lanes 3 and 4) or PolVΔ14-recovering (lanes 1 and 2) N. benthamiana leaves agro-infiltrated with GFP-Cym(+) and GFP-PoLV(+) sensors. Protein extracts were subjected to immunoblot analysis using GFP-specific antibody, and RNA samples were analyzed by Northern blotting using a radiolabeled GFP sequence probe. The extracted total RNAs are visualized in an ethidium bromide staining gel at the bottom panels; the 25S ribosomal RNAs were used as loading controls. *, position of 5′ cleavage product target RNA.
Strand-specific degradation of sensor RNA.
It was reported previously that viral siRNAs originated predominantly from plus-stranded viral RNA (28, 41), and we showed in this report that sensor RNA targeting occurs mostly if not exclusively via RNA cleavage. These two observations predict that the virus-derived siRNA-programmed RISC may contain predominantly plus-stranded siRNAs; thus, this preassembled RISC will cleave sensor RNA with negative viral sense sequence more efficiently than the sensor RNAs with plus-stranded sequence. To this end, an additional sensor construct was also prepared [GFP-Cym(−)], containing the complementary viral target sequence which was inserted in GFP-Cym(+) sensor (Fig. 1A, B).
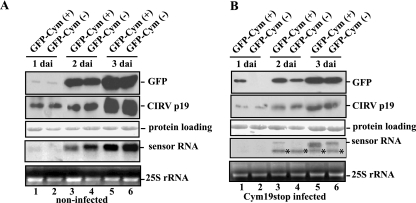
GFP-Cym(+) and GFP-Cym(−) constructs were coexpressed with p19 protein in the control nonpreinoculated and the Cym19stop-infected N. benthamiana plants. Both constructs expressed GFP very efficiently and equally in the control nonpreinoculated plants (Fig. 4A). In contrast, when the recovering leaves of Cym19stop-preinoculated plants were infiltrated with both GFP-Cym(+) and GFP-Cym(−), the Western analysis showed an evident difference in the accumulation of GFP. While GFP accumulation was clearly detectable at 1 dai in the leaf tissue agro-infiltrated with GFP-Cym(+), no GFP was detected in the protein sample derived from GFP-Cym(−)-infiltrated tissue. This difference was not detectable at the level of mRNA, since we were not able to detect sensor RNA at 1 dai. It is worthy to note that we compared the accumulation of two sensor RNAs in the two halves of the same leaf (Fig. 4A, B) to avoid the effect of the existing slight variability of sensor RNA expression in different plant leaves. The differences in the accumulation of either GFP or sensor RNA levels between the two constructs were more pronounced at 2 dai (Fig. 4B, lanes 3, 4). At the RNA level, the presence of 5′ cleavage product of both sensor RNAs confirmed the activity of RISC programmed by either plus-sense or minus-sense siRNAs derived from Cym19stop. Although both GFP-Cym(+) and GFP-Cym(−) sensor RNAs were cleaved by viral siRNA-preassembled RISC, the levels of GFP-Cym(+) RNA and the GFP derived from this sensor were higher than that from the GFP-Cym(−) sensor, suggesting that more RISC complexes contain siRNA from plus-sense viral RNA than from minus sense. However, this asymmetry is not as pronounced as we expected from the previously observed 3:1 ratio between the plus and minus siRNAs (28).
FIG. 4.
In vivo analysis of GFP-Cym(+) and GFP-Cym(−) sensor sequences. Noninfected (A) and Cym19stop-recovering (B) N. benthamiana plants were agro-infiltrated with the indicated sensor constructs, and the GFP, p19 proteins, and sensor mRNAs were analyzed at the indicated time points. Normalized extracts were prepared at 1, 2, and 3 dai and subjected to immunoblot analysis with anti-GFP antibody and anti-CIRV p19 antiserum or subjected to RNA blot analysis using a radiolabeled GFP sequence probe. The extracted total RNAs are visualized in an ethidium bromide staining gel at the bottom panels; the 25S ribosomal RNAs were used as loading controls. *, position of 5′ cleavage product target RNA.
Mapping the RISC-mediated cleavage sites on viral sequences in vivo.
The analysis of sensor mRNAs expressed in the recovered leaves of Cym19stop-inoculated plants revealed the presence of distinct low-molecular-mass RNA species supposed to be the cleaved sensor mRNAs (Fig. 2B, lanes 1, 3, and 5, and Fig. 4B, lanes 3, 4, 5, and 6). This suggests that the RISC-mediated cleavage does not occur randomly in the virus-specific target sequence. To better characterize these shorter RNA species shown in Fig. 2B and 4B, they were cloned and sequenced. The higher-molecular-mass species was identified as the entire not cleaved sensor sequence containing the GFP ORF followed by the nopalin synthase terminator (ca. 180 nt in length) and poly(A) tail variable in length from 20 to 80 nt, according to the previous observation (5). Identification of the lower-molecular-mass species revealed that the cleavage occurred in the regions containing CymRSV sequences either in plus [Cym(+)] or minus [Cym(−)] orientations (Fig. 5A). Strikingly, the identified cleavage sites were not distributed uniformly along the viral target sequence between nucleotide positions 4446 to 4635 (Fig. 5A and data not shown). Instead, all cleavage sites, identified by sequencing of the 5′ cleavage product of the GFP-Cym(+) sensor, were localized in a 40-nt-long region between base positions 4493 to 4533 referred to in the CymRSV genome. Similar results were observed when we localized the cleavage sites on the minus-sense viral target sequence of GFP-Cym(−). The cleavage sites were in the last 40-base-long region of viral target sequence (positions 4592 to 4635, referred to in the CymRSV genome) (Fig. 5A and data not shown). To test whether the presence of target hot spots in the virus sequence is a particular characteristic of these artificial sensor constructs, 3′ rapid amplification of cDNA ends cloning and sequencing of the expected cleavage products of natural viral genomic RNAs were also performed. The cloned and sequenced region contained the corresponding viral sequence with some flanking sequences, which was inserted into the GFP-Cym(+) sensor construct. The distribution of RISC-mediated cleavage sites of the natural viral genome occurred in the same hot spot (in the region corresponding to the sensor sequence), which was identified by sensor RNAs (Fig. 5A and data not shown). The same approach performed on the natural minus-strand viral RNA did not reproduce any amplified product, probably due to the very low amount of the minus-strand viral RNA in natural infections (data not shown). Indeed, we observed previously that the ratio of plus to minus viral RNA strands is 100:1 (J. Burgyan, unpublished data). An alternative explanation for the lack of cleavage in the minus viral strand is that the minus viral RNA strand is likely present only in replication complexes and the viral siRNA-programmed RISC has no access to it. The identified hot spots for the cleavage in the viral target sequence could be explained by the preferential generation of viral siRNA from these regions. However, the analysis of siRNA origin did not support this hypothesis. We found a similar amount of viral siRNAs from the cleavage hot spot region as well as from regions where no cleavage was identified (Fig. 5B, panels 4, 5, 6, and 7). The cleavage sites in this region (the sequence between nt 4446 to 4635) of viral genomes were also searched in wild-type (wt) p19 suppressor-expressing virus (CymRSV)-infected plants. The obtained results convincingly demonstrated the lack of cleavage hot spot in the analyzed region (Fig. 5A), which likely suggests that presence of p19 suppressor protects the viral genome against RNA silencing-mediated degradation by sequestering siRNAs.
FIG. 5.
In vivo analysis of VIGS and miR171-mediated sensor sequences cleavage. Schematic illustration of the GFP-Cym(+) and GFP-Cym(−) deriving from the in vivo 3′-end analysis of the uncleaved and cleaved mRNAs. The GFP ORF is represented by a gray open box, and the noncoding regions are indicated by thick lines (black for viral target sequence and pale gray for nopalin synthase [NOS] terminator-deriving sequence). Arrows indicate the sites of cleavage on the schematic enlargement of viral target sequences (+ and −) in the sensor sequences and in the viral Cym19stop and CymRSV context (gray lines) (A). The position and the frequency of cleavage sites are not shown. (B) RNA gel blot analysis of siRNAs deriving from Cym19stop. Total RNA extracted from Cym19stop-infected plants were separated on 12% acrylamide gels, blotted, and hybridized with specific riboprobes to detect siRNAs deriving from the sensor sequences (referred to as GFP) and from specific viral regions, as indicated in each panel of the picture. (C) An agro-infiltrated N. benthamiana leaf viewed at 3 dai under long-wavelength UV illumination and the corresponding RNA gel blot analysis of RNA extracted from tissues infiltrated with GFP 171.1 and GFP 171.2 constructs are shown. A riboprobe for the GFP ORF was used to detect sensor mRNAs. (D) Southern blot analysis of the PCR products obtained from the 3′-end analysis performed on the sensor mRNAs. Two blots of the same sample were hybridized with a riboprobe raised against the GFP ORF or with 32P-oligonucleotide complementary to the miR171 target sequence (lanes 1 and 2, respectively). (E) Schematic illustration of the GFP 171.1 deriving from the in vivo 3′-end analysis of the uncleaved and the cleaved mRNAs. The GFP ORF is represented by a gray open box, and the noncoding regions are indicated by thick lines (black for miRNA target sequence and pale gray for 35S terminator-deriving sequence). Arrows indicate the sites of cleavage on the miR171 target sequence.
To confirm that the performed 3′-end analysis is accurate enough to detect the virus-derived siRNA-programmed RISC-mediated cleavage sites, we identified the natural miRNA171-mediated cleavage position in the previously described GFP 171.1 sensor RNA (32) using the same cloning and sequencing methodology. The GFP 171.1 sensors and the uncleavable control GFP 171.2 sensors (32) were coexpressed with CIRV p19 in N. benthamiana plants by agro-infiltration to avoid the activation of transgene silencing against the sensor itself (20). As expected, at 2 to 3 dai, a bright green fluorescence of GFP was observed under UV light in the control GFP 171.2 sensor but not at the wt GFP 171.1 sensor-infiltrated area (Fig. 5C). The lack of GFP expression in the wt GFP 171.1-infiltrated area was the consequence of endogenous miR171-mediated degradation of GFP 171.1 sensor RNA (Fig. 5C, lane 1 versus 2) (32). Although the expected miR171-mediated cleavage product was not visible by RNA gel blot analysis, the 5′ part of the cleaved mRNA was revealed by reverse transcription-PCR amplification of the region of GFP-miR171.1 sensor containing the miR171.1 target site, and the amplified products were analyzed by Southern blotting. Figure 5D shows that the low-molecular-mass amplified product was detected only with a GFP-specific probe and not with a probe containing miR171 sequence. The miR171 probe detects only the full-length not cleaved sensor, while the GFP-specific probe detects both full-length and the 5′ cleavage products of GFP 171.1 sensor RNA (Fig. 5D). By cloning and sequencing of the low-molecular-mass species, we identified the miR171-mediated cleavage sites in the GFP 171.1 sensor RNA, which coincided with that observed on natural miR171 targets (24), confirming the accuracy of our methodology (Fig. 5D, E).
Viral siRNA-containing protein complexes cofractionate with miRNA-containing complexes.
Recent studies identified plant miRNA-Argonaute1 (AGO1) protein complexes in plants, and the purified miRNA-AGO1 complex was able to slice the target mRNA containing the corresponding miRNA target sequences (3, 34). Moreover, it was also reported that AGO1 is physically associated with miRNAs, trans-acting siRNAs, and transgene-derived siRNAs but excludes virus-derived siRNAs and 24-nt siRNAs involved in chromatin silencing (3). It is possible that another plant AGO protein specific for viral siRNA is responsible for antiviral function. Moreover, the involvement of RISC-mediated cleavage of viral RNA in the silencing-based antiviral response is still a matter of debate (46). Since we described a slicer activity in the recovered leaves of Cym19stop-infected plants and this slicing activity was shown to be sequence specific and likely mediated by virus-derived siRNA, we expected that these recovered leaves contain viral siRNA-guided slicer complexes, similar to miRNA-containing AGO1. To identify viral siRNA-containing protein complexes, a protein extract was prepared at 7 dpi from systemically infected leaves of Cym19stop-inoculated N. benthamiana plants and size separated by a Superdex-200 gel filtration column. Then RNA was extracted from each fraction and analyzed on an RNA gel blot. The gel blot was hybridized first with CymRSV probe, and then the membrane was stripped and reprobed with [γ-32P]ATP-labeled miR159 LNA oligonucleotide. Figure 6 demonstrates that we were able to separate three viral siRNA-containing “complexes.” The majority of viral siRNA was eluted in the low-molecular-mass fraction corresponding to the protein-free siRNA size as reported earlier (21) (Fig. 6A, fractions from 41 to 55). More importantly, we identified two other viral siRNA-containing protein complexes characterized by a different molecular mass of 150 kDa (fractions from 19 to 29) and 670 kDa (fractions from 3 to 6), respectively. Strikingly, these two complexes cofractionated with miRNA (miR159)-containing proteins (Fig. 6B). The miRNA-containing protein fraction of 150 kDa in size may correspond to the recently identified AGO1 slicer (3), and we suggest that the viral siRNA-containing counterpart (Fig. 6A, fractions from 19 to 29) is another slicer, probably 1 of the 10 family members of plant AGO proteins that is specifically adapted to retain viral siRNAs and for antiviral response. The large (670 kDa) siRNA and miRNA complexes (Fig. 6A and B, fractions from 3 to 6) may correspond to the DICER complex identified in animal systems or, alternatively, to the multi-protein-subunit-containing holo RISC complexes (15, 43).
FIG. 6.
Cofractionation of viral siRNAs and plant miRNAs containing complexes derived from virus-infected N. benthamiana plants. The protein extract was prepared at 7 dpi from systemic leaves of Cym19stop-infected N. benthamiana plants and size separated by a Superdex-200 gel filtration column. RNA was extracted from each fraction and analyzed on a gel blot. The gel blot was hybridized first with CymRSV probe, and then the membrane was stripped and reprobed with [γ-32P]ATP-labeled oligonucleotide complementary to miR159. The elution positions of protein molecular markers are shown above the panels and are as follows: 669 kDa, thyroglobulin; 443 kDa, ferritin, 150 kDa, aldolase; 66 kDa, bovine serum albumin; 29 kDa, carbonic anhydrase. RNA size markers are shown at the right side.
DISCUSSION
Although our knowledge about the different RNA silencing-based pathways in plants was extended significantly in recent years, still little is known about the molecular bases of RNA silencing-based antiviral response. We previously demonstrated that a plant-encoded DICER-like enzyme(s) is(are) able to recognize highly structured regions within the single-stranded viral RNA and process them into siRNAs. This process resembled the DICER-like activity involved in the generation of miRNAs from short-hairpin precursors (28), although it has been shown very recently using genetic approaches that both DCL2 and DCL4 are responsible for the generation of viral siRNAs (10). However, the molecular bases of the following steps in silencing-based antiviral pathway were never analyzed. Even the mere existence of a viral siRNA-activated antiviral RISC was a matter of debate because Dicer-mediated processing of viral RNA with a dsRNA feature could be, in principle, sufficient to dampen virus infections (22, 46). In addition, the recent analysis of short RNA content of Arabidopsis AGO1, which was identified as a plant slicer (containing miRNA, trans-acting siRNA, and transgene-derived siRNA) resulted in the lack of virus-derived siRNA (3). In this study, using different approaches, we demonstrated the existence of an antiviral RISC and we showed that this putative viral siRNA-programmed RISC cleaves the target viral RNA with high sequence specificity.
Activation of antiviral RISC by tombusvirus infection resulted in the cleavage of the viral target RNA rather than its translational inhibition.
The particular feature of Cym19stop silencing suppressor-deficient mutant virus is that, in this mutant, the basic viral functions, replication and spreading to short and long distance, are not impaired (41). This unique feature allowed us to address questions of how the silencing-based plant antiviral response targets viral RNA. In the recovered leaves of Cym19stop-infected N. benthamiana plants, which is resistant to secondary virus infection carrying homologous sequence, we were able to analyze separately the targeting step of viral sequences from the activation step of the antiviral silencing response. The coexpression of sequence-specific sensors with the p19 silencing suppressor inhibited the new assembly of virus-specific RISC complexes by the sequestration of viral siRNA. In this way, we were able to demonstrate the endonucleotic cleavage activity of the preassembled antiviral RISC in trans on the transiently expressed virus-specific sensor RNAs. Furthermore, the expression of the two virus-specific sensors [GFP-PoLV(+) and GFP-Cym(+)] in the recovered leaves of plants infected with one of two suppressor mutant viruses demonstrated a highly sequence-specific endonuclease activity of an antiviral RISC, which is an important feature of siRNA- or miRNA-activated RISC-mediated target cleavage in plants. The analysis of protein and RNA accumulation expressed from the sensor construct clearly showed that the sequence-specific targeting of sensor RNAs is mediated by cleavage, although our data cannot exclude the possibility of translational inhibition for some extent.
Antiviral RISC cleaves viral RNAs in hot spots and shows preference for minus-stranded viral sequence.
It has been demonstrated recently that there is an asymmetry in the strandedness of the origin of virus-derived siRNAs, showing that the majority of viral siRNAs have plus-stranded viral sequences (28, 41). This finding predicts that viral siRNA-guided RISC targets more frequently the viral strand having negative polarity than the plus-stranded viral RNA. Indeed, our analysis using strand-specific sensors [GFP-Cym(+) and GFP-Cym(−)] for antiviral RISC-mediated cleavage confirmed this prediction. It is worthy of note that the amount of negative-strand viral RNA is a rate-limiting factor of viral replication; therefore, the preferential targeting of the negative viral strand makes the antiviral silencing response very efficient and very attractive for the plant defense.
The analyzed region (190 nt long) of the antiviral RISC-mediated cleavages was previously identified as a well-targeted sequence in the defective interfering RNA genome associated with CymRSV, and this region was highly represented in the pool of virus-specific siRNAs (41). Surprisingly, the identification of cleavage sites by sequencing the cloned 5′ cleavage products of GFP-Cym(+/−) sensors revealed that the cleavage sites were not distributed homogeneously along the analyzed target sequence but occurred in a short 40-nt-long hot spot. Comparison of the level of virus-derived siRNAs specific for the cleavage hot spot with the siRNA level specific for a nontargeted region did not show any difference. Thus, we hypothesized that the presence of a hot spot on the target RNA depends on the local RNA structure. Supporting this idea, it was illustrated recently that the local secondary structure of a target RNA interferes with the accessibility of target RNA for RISC-mediated cleavage (31, 51); however, we were not able to identify a distinct RNA structure for the hot spots identified. Importantly, the same hot spot for viral RNA cleavage was found in the natural viral genome derived from Cym19stop suppressor-deficient virus infection. In contrast, we found very few cleavages in the viral genome when the plants were infected with the wt virus. This finding fits well our previous model that p19 proteins prevent the RNA silencing-mediated viral RNA degradation by sequestering virus-specific siRNAs (16, 40, 45). Interestingly, the sequence analysis of cleaved viral and sensor RNAs also revealed the presence of nontemplated U residues at the site of cleavages (data not shown), which were previously shown as a signature of RISC-mediated cleavage (38).
Viral siRNA-containing protein complexes cofractionate with miRNA-containing complexes.
In this paper, we demonstrated a virus-specific slicer activity in plant leaves recovered from virus infection, thus strongly suggesting the existence of viral siRNA-programmed RISC. This antiviral RISC cleaves viral target RNA in trans with high sequence specificity. Moreover, we demonstrated that protein complexes containing viral siRNA cofractionated with complexes containing miRNA. We identified two major peaks which contained both viral siRNAs and miRNA. The smaller complex with a molecular mass of approximately 150 kDa, probably corresponds to the recently identified miRNA-containing AGO1 slicer (3), although that report also showed that no virus-derived siRNA has been found in AGO1 immunoprecipitate derived from virus-infected plant (3), suggesting that AGO1 is not involved as a slicer in antiviral response. In plants, AGO proteins are encoded by multigene families, allowing an extensive diversification of RNA interference pathways (7, 12); thus, it is a reasonable possibility that a specific AGO other than AGO1 is developed for the antiviral defense. Alternatively, the silencing suppressor proteins expressed by the viruses used in that experiment may inhibit the assembly of virus-specific siRNAs into AGO1 (3). Indeed, recently reported data demonstrated that many of the viral silencing suppressors act via siRNA sequestration (20, 25). Moreover, the observation that Arabidopsis ago1 hypomorphic mutant is hypersensitive to virus infection suggests that AGO1 is involved in antiviral defense (29). Further experiments analyzing the siRNA content of AGO1 in suppressor-deficient virus-infected plants may help to clarify this point. The other high-molecular-mass complex containing both miRNA and viral siRNAs identified in the fractionation experiment has a molecular mass of 650 kDa. It could be one of the other RNA silencing complexes associated with the RNA silencing pathways, and further studies need to explore the composition and the functions of these small-RNA-containing complexes.
ADDENDUM IN PROOF
Virus-derived siRNA containing AGO1 was recently identified in virus-infected plants by X. Zhang et al. (Genes Dev. 20:3255-3268, 2006.)
Acknowledgments
We are grateful to Olivier Voinnet for providing the miR171.1 and miR171.2 sensor constructs.
This research was supported by grants from the Hungarian Scientific Research Fund (OTKA T046728 and OTKA NK60352) and the “RIBOREG” EU project (LSHG-CT-2003503022). V.P. was a recipient of short-term EMBO fellowship ASTF 13-2005, and G.S. was supported by a Bolyai János Fellowship.
Footnotes
Published ahead of print on 31 January 2007.
REFERENCES
- 1.Ahlquist, P. 2002. RNA-dependent RNA polymerases, viruses, and RNA silencing. Science 296:1270-1273. [DOI] [PubMed] [Google Scholar]
- 2.Aukerman, M. J., and H. Sakai. 2003. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15:2730-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumberger, N., and D. C. Baulcombe. 2005. Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 102:11928-11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363-366. [DOI] [PubMed] [Google Scholar]
- 5.Bevan, M., W. M. Barnes, and M. D. Chilton. 1983. Structure and transcription of the nopaline synthase gene region of T-DNA. Nucleic Acids Res. 11:369-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brigneti, G., O. Voinnet, W. X. Li, L. H. Ji, S. W. Ding, and D. C. Baulcombe. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739-6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Carmell, M. A., Z. Xuan, M. Q. Zhang, and G. J. Hannon. 2002. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 16:2733-2742. [DOI] [PubMed] [Google Scholar]
- 8.Chen, X. 2003. A MicroRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 11:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalmay, T., L. Rubino, J. Burgyan, A. Kollar, and M. Russo. 1993. Functional analysis of cymbidium ringspot virus genome. Virology 194:697-704. [DOI] [PubMed] [Google Scholar]
- 10.Deleris, A., J. Gallego-Bartolome, J. Bao, K. D. Kasschau, J. C. Carrington, and O. Voinnet. 2006. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313:68-71. [DOI] [PubMed] [Google Scholar]
- 11.Doench, J. G., C. P. Petersen, and P. A. Sharp. 2003. siRNAs can function as miRNAs. Genes Dev. 17:438-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fagard, M., S. Boutet, J. B. Morel, C. Bellini, and H. Vaucheret. 2000. AGO1, QDE-2, and RDE-1 are related proteins required for transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. USA 97:11650-11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamilton, A., O. Voinnet, L. Chappell, and D. Baulcombe. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21:4671-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamilton, A. J., and D. C. Baulcombe. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286:950-952. [DOI] [PubMed] [Google Scholar]
- 15.Hammond, S. M., E. Bernstein, D. Beach, and G. J. Hannon. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293-296. [DOI] [PubMed] [Google Scholar]
- 16.Havelda, Z., C. Hornyik, A. Crescenzi, and J. Burgyan. 2003. In situ characterization of cymbidium ringspot tombusvirus infection-induced posttranscriptional gene silencing in Nicotiana benthamiana. J. Virol. 77:6082-6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Havelda, Z., C. Hornyik, A. Valoczi, and J. Burgyan. 2005. Defective interfering RNA hinders the activity of a tombusvirus-encoded posttranscriptional gene silencing suppressor. J. Virol. 79:450-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutvagner, G., and P. D. Zamore. 2002. RNAi: nature abhors a double-strand. Curr. Opin. Genet. Dev. 12:225-232. [DOI] [PubMed] [Google Scholar]
- 19.Lacomme, C., K. Hrubikova, and I. Hein. 2003. Enhancement of virus-induced gene silencing through viral-based production of inverted-repeats. Plant J. 34:543-553. [DOI] [PubMed] [Google Scholar]
- 20.Lakatos, L., T. Csorba, V. Pantaleo, E. J. Chapman, J. C. Carrington, Y. P. Liu, V. V. Dolja, L. F. Calvino, J. J. Lopez-Moya, and J. Burgyan. 2006. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 25:2768-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lakatos, L., G. Szittya, D. Silhavy, and J. Burgyan. 2004. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 23:876-884. [Epub 19 Feb 2004.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, F., and S. W. Ding. 2006. Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 60:503-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, W. X., and S. W. Ding. 2001. Viral suppressors of RNA silencing. Curr. Opin. Biotechnol. 12:150-154. [DOI] [PubMed] [Google Scholar]
- 24.Llave, C., Z. Xie, K. D. Kasschau, and J. C. Carrington. 2002. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297:2053-2056. [DOI] [PubMed] [Google Scholar]
- 25.Mérai, Z., Z. Kerenyi, S. Kertesz, M. Magna, L. Lakatos, and D. Silhavy. 2006. Double-stranded RNA binding may be a general plant RNA viral strategy to suppress RNA silencing. J. Virol. 80:5747-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mérai, Z., Z. Kerenyi, A. Molnar, E. Barta, A. Valoczi, G. Bisztray, Z. Havelda, J. Burgyan, and D. Silhavy. 2005. Aureusvirus P14 is an efficient RNA silencing suppressor that binds double-stranded RNAs without size specificity. J. Virol. 79:7217-7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moissiard, G., and O. Voinnet. 2004. Viral suppression of RNA silencing in plants. Mol. Plant Pathol. 5:71-82. [DOI] [PubMed] [Google Scholar]
- 28.Molnár, A., T. Csorba, L. Lakatos, E. Varallyay, C. Lacomme, and J. Burgyan. 2005. Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J. Virol. 79:7812-7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morel, J. B., C. Godon, P. Mourrain, C. Beclin, S. Boutet, F. Feuerbach, F. Proux, and H. Vaucheret. 2002. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14:629-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nykanen, A., B. Haley, and P. D. Zamore. 2001. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107:309-321. [DOI] [PubMed] [Google Scholar]
- 31.Overhoff, M., M. Alken, R. K. Far, M. Lemaitre, B. Lebleu, G. Sczakiel, and I. Robbins. 2005. Local RNA target structure influences siRNA efficacy: a systematic global analysis. J. Mol. Biol. 348:871-881. [DOI] [PubMed] [Google Scholar]
- 32.Parizotto, E. A., P. Dunoyer, N. Rahm, C. Himber, and O. Voinnet. 2004. In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 18:2237-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plasterk, R. H. 2002. RNA silencing: the genome's immune system. Science 296:1263-1265. [DOI] [PubMed] [Google Scholar]
- 34.Qi, Y., A. M. Denli, and G. J. Hannon. 2005. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell 19:421-428. [DOI] [PubMed] [Google Scholar]
- 35.Rubino, L., and M. Russo. 1997. Molecular analysis of the pothos latent virus genome. J. Gen. Virol. 78(Pt 6):1219-1226. [DOI] [PubMed] [Google Scholar]
- 36.Russo, M., J. Burgyan, and G. P. Martelli. 1994. Molecular biology of tombusviridae. Adv. Virus Res. 44:381-428. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Shen, B., and H. M. Goodman. 2004. Uridine addition after microRNA-directed cleavage. Science 306:997. [DOI] [PubMed] [Google Scholar]
- 39.Silhavy, D., and J. Burgyan. 2004. Effects and side-effects of viral RNA silencing suppressors on short RNAs. Trends Plant Sci. 9:76-83. [DOI] [PubMed] [Google Scholar]
- 40.Silhavy, D., A. Molnar, A. Lucioli, G. Szittya, C. Hornyik, M. Tavazza, and J. Burgyan. 2002. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 21:3070-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szittya, G., A. Molnar, D. Silhavy, C. Hornyik, and J. Burgyan. 2002. Short defective interfering RNAs of tombusviruses are not targeted but trigger post-transcriptional gene silencing against their helper virus. Plant Cell 14:359-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szittya, G., D. Silhavy, A. Molnar, Z. Havelda, A. Lovas, L. Lakatos, Z. Banfalvi, and J. Burgyan. 2003. Low temperature inhibits RNA silencing-mediated defence by the control of siRNA generation. EMBO J. 22:633-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomari, Y., and P. D. Zamore. 2005. Perspective: machines for RNAi. Genes Dev. 19:517-529. [DOI] [PubMed] [Google Scholar]
- 44.Valoczi, A., C. Hornyik, N. Varga, J. Burgyan, S. Kauppinen, and Z. Havelda. 2004. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 32:e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vargason, J., G. Szittya, J. Burgyan, and T. M. Hall. 2003. Size selective recognition of siRNA by an RNA silencing suppressor. Cell 115:799-811. [DOI] [PubMed] [Google Scholar]
- 46.Voinnet, O. 2005. Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6:206-220. [DOI] [PubMed] [Google Scholar]
- 47.Voinnet, O. 2001. RNA silencing as a plant immune system against viruses. Trends Genet. 17:449-459. [DOI] [PubMed] [Google Scholar]
- 48.Voinnet, O., Y. M. Pinto, and D. C. Baulcombe. 1999. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 96:14147-14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waterhouse, P. M., M. B. Wang, and T. Lough. 2001. Gene silencing as an adaptive defence against viruses. Nature 411:834-842. [DOI] [PubMed] [Google Scholar]
- 50.Ye, K., L. Malinina, and D. J. Patel. 2003. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature 3:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao, Y., E. Samal, and D. Srivastava. 2005. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 436:214-220. [DOI] [PubMed] [Google Scholar]