Abstract
ESCRT (endosomal sorting complex required for transport) complexes orchestrate efficient sorting of ubiquitinated transmembrane receptors to lysosomes via multivesicular bodies (MVBs). Yeast ESCRT-I and ESCRT-II interact directly in vitro; however, this association is not detected in yeast cytosol. To gain understanding of the molecular mechanisms of this link, we have characterised the ESCRT-I/-II supercomplex and determined the crystal structure of its interface. The link is formed by the vacuolar protein sorting (Vps)28 C-terminus (ESCRT-I) binding with nanomolar affinity to the Vps36-NZF-N zinc-finger domain (ESCRT-II). A hydrophobic patch on the Vps28-CT four-helix bundle contacts the hydrophobic knuckles of Vps36-NZF-N. Mutation of the ESCRT-I/-II link results in a cargo-sorting defect in yeast. Interestingly, the two Vps36 NZF domains, NZF-N and NZF-C, despite having the same core fold, use distinct surfaces to bind ESCRT-I or ubiquitinated cargo. We also show that a new component of ESCRT-I, Mvb12 (YGR206W), engages ESCRT-I directly with nanomolar affinity to form a 1:1:1:1 heterotetramer. Mvb12 does not affect the affinity of ESCRT-I for ESCRT-II in vitro. Our data suggest a complex regulatory mechanism for the ESCRT-I/-II link in yeast.
Keywords: CHMP, ESCRT, HIV budding, MVB, NZF finger
Introduction
Transport of protein cargo through maturing endosomes to lysosomes/vacuoles via the multivesicular body (MVB) pathway is dependent on a network of interactions formed between the MVB sorting machinery, protein cargo and endosomal membranes. ESCRT (endosomal sorting complex required for transport)-I, -II and -III complexes are an ancient component of eukaryotic endomembrane machinery and are found in all six major supergroups of eukaryotes (Field et al, 2006). In concert with protein cargo sorting, the ESCRT machinery remodels the protein and lipid content of the limiting endosomal membrane, initially driving formation of internal vesicles and subsequently promoting fusion of the MVB with the lysosome/vacuole. Components of the ESCRT machinery also play a critical role in retroviral budding.
Ten of the 18 class E MVB sorting proteins assemble into the ESCRT complexes, ESCRT-I, -II and -III (reviewed in Hurley and Emr, 2006). A model of sequential recruitment of these ESCRT complexes on endosomal membranes has been proposed for their function in cargo sorting. ESCRT-I (vacuolar protein sorting (Vps)23, Vps28 and Vps37) (Katzmann et al, 2001) and ESCRT-II (Vps22, Vps25 and Vps36) (Babst et al, 2002b) assemble into discrete heterotrimeric and heterotetrameric complexes, respectively (Hierro et al, 2004; Teo et al, 2004, 2006; Kostelansky et al, 2006). Unlike ESCRT-I and -II, ESCRT-III (Vps2, Vps20, Vps24 and Snf7) appears to form an oligomeric scaffold on endosomal membranes, functioning in conjunction with several ESCRT-III-like accessory proteins (Babst et al, 2002a; Lin et al, 2005). Additional isoforms of human ESCRT-I and -III subunits have been identified, suggesting an expanding set of ESCRT regulatory mechanisms evolving from primitive eukaryotes to mammals (see for a review Slagsvold et al, 2006).
Efficient recruitment of ESCRT-I to endosomal membranes is dependent on the upstream Vps27/Hrs complex (Bilodeau et al, 2003; Katzmann et al, 2003). Vps27/Hrs is a recent evolutionary addition specific to fungal and metazoan MVB sorting pathways, suggesting that the efficiency of ESCRT-I recruitment to endosomal membranes is important for MVB sorting in these eukaryotes (Field et al, 2006). ESCRT-II (Vps36 subunit) and ESCRT-III (Vps24 subunit) have Vps27-independent mechanisms of recruitment to endosomes through direct binding to 3-phosphoinositides (Whitley et al, 2003; Slagsvold et al, 2005; Teo et al, 2006). Direct binding of ESCRT-I to -II (Kostelansky et al, 2006; Teo et al, 2006) and ESCRT-II to -III (Teo et al, 2004; Yorikawa et al, 2005) in vitro suggests that an ESCRT-I/-II/-III network may be stabilised once it assembles on endosomal membranes. Much progress has been made in determining the isolated structures of the ESCRT-I and -II complexes and, most recently, the ESCRT-III hVps24 subunit (Muziol et al, 2006). However, there is a critical need to find out how the ESCRT complexes are linked and how they regulate cargo sorting, internal vesicle formation and fusion of the MVB with the lysosome.
Several components of MVB sorting machinery bind to ubiquitinated cargo and all of these interactions are essential for efficient sorting of protein cargo into internal vesicles. It has yet to be resolved whether all these ubiquitin (Ub)-binding domains act in an orchestrated sequential hand-off or whether they are needed to generate a critical density of clustered ubiquitinated cargo before cargo sorting into internal vesicles. Recent work has shown that Hrs and clathrin colocalise in distinct microdomains on early endosomes that exclude EEA1 (an early endosomal marker) but are enriched in Ub (Raiborg et al, 2006). Flat clathrin lattices assembling on early endosomes through binding to Hrs may represent an initial mechanism for clustering of ubiquitinated cargo, which is subsequently maintained by the ESCRT-I and -II complexes. Interestingly, recent studies have shown that at least some subunits of human ESCRT-III are not required for formation of internal vesicles. ESCRT-III is required downstream for regulation of fusion of the MVB with the lysosome (Bache et al, 2006; Shim et al, 2006).
The role of the ESCRT complexes is not limited to sorting of ubiquitinated cargo. Silencing the Tsg101 (tumour susceptibility gene) subunit of ESCRT-I with siRNA results in abnormal endosomes. These structures are either tubular or contain abundant cisternae, but with an almost complete lack of internal vesicles, suggesting a role in regulating the morphology and dynamics of the limiting endosomal membrane (Doyotte et al, 2005; Razi and Futter, 2006).
The functional importance of links between the components of the ESCRT machinery is becoming increasingly apparent. We have previously characterised the direct interaction between ESCRT-I and ESCRT-II in vitro and shown that this requires the C-terminus of the Vps28 subunit of ESCRT-I and the N-terminal Npl4 zinc-finger (NZF) domain within the Vps36-GLUE domain of ESCRT-II (Teo et al, 2006). Here, we report the high-resolution crystal structure of the C-terminus of Vps28 (ESCRT-I) in isolation and in complex with the N-terminal NZF domain of Vps36 (ESCRT-II). We also demonstrate that the full-length yeast ESCRT-I and ESCRT-II form a supercomplex of affinity similar to that of isolated domains. Importantly, we find that mutation of residues involved in the ESCRT-I/-II interface affects cargo sorting and generates a class E phenotype in yeast. In addition, we find that a previously uncharacterised 12 kDa protein, Mvb12 (YGR206W), forms a nanomolar affinity 1:1:1:1 heterotetrameric complex in vitro with Vps23, Vps28 and Vps37 to complete the functional unit of the yeast ESCRT-I complex.
Results
Yeast ESCRT-I binds directly to ESCRT-II with nanomolar affinity
To determine the affinity of the ESCRT-I/-II interaction, we used an N-terminally FlAsH-tagged Vps28 C-terminus (residues 148–242) (FlAsH-Vps28-CT) (Adams et al, 2002). The isolated FlAsH-Vps28-CT, a small 12 kDa construct, has a fluorescence anisotropy of 0.19. Upon binding of FlAsH-Vps28-CT to the 140 kDa full-length ESCRT-II complex, the fluorescence anisotropy increases to 0.27. Direct titration with intact ESCRT-II shows that the isolated FlAsH-Vps28-CT binds ESCRT-II with a Kd of approximately 27 nM (Figure 1A). A similar Kd (57 nM) was determined for the direct titration of a Vps36 fragment containing the double NZF insertion (residues 110–205, encompassing both the N- and C-terminal NZF domains, Vps36-NZF-NC) (Figure 1A). This is in good agreement with an affinity of 10 nM for Vps28-CT binding to GST-Vps36-NZF-NC as determined using BIAcore analysis (data not shown). Therefore, the yeast-specific insertion present within Vps36 of ESCRT-II is sufficient for the high-affinity interaction we observe with the C-terminus of Vps28 in ESCRT-I.
Figure 1.
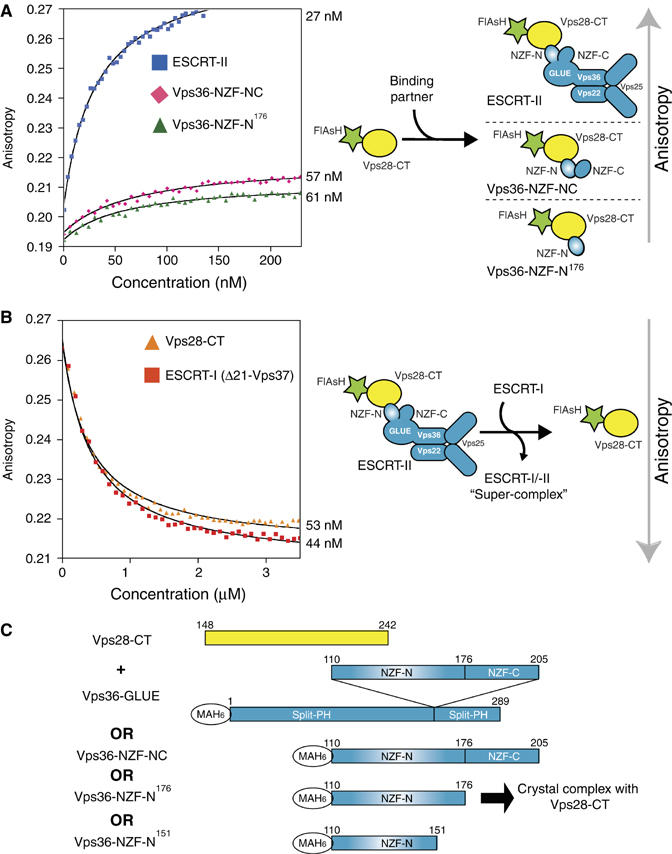
Yeast ESCRT-I binds to ESCRT-II with nanomolar affinity. (A) The fluorescence anisotropy of the isolated recombinant FlAsH-Vps28-CT increases in accordance with the size of the titrated recombinant ESCRT-II binding partner: 9.5 kDa extended Vps36-NZF-N (residues 110–176), 12.5 kDa Vps36-NZF-NC (residues 110–205) and 140 kDa full-length ESCRT-II. Direct fitting of the titration curves shows similar Kd values of Vps36-NZF-N (61 nM), Vps36-NZF-NC (57 nM) and full-length ESCRT-II (27 nM) for FlAsH-Vps28-CT. (B) In a fluorescence anisotropy competition assay, various ESCRT-I proteins are titrated into a solution containing a preformed complex of the FlAsH-Vps28-CT/full-length ESCRT-II. Titration of either ESCRT-I(Δ21-Vps37) or non-labelled Vps28-CT results in a displacement of fluorescent FlAsH-Vps28-CT from ESCRT-II and consequently, in a decrease of anisotropy. Fitting of the titration curves allows determination of Kd values for Vps28-CT (53 nM) and ESCRT-I(Δ21-Vps37) (44 nM) for ESCRT-II. (C) Constructs used to generate ESCRT-I/-II subcomplexes used in this study.
To measure the affinity of intact ESCRT-I for intact ESCRT-II, we used a competition assay, where we monitored dissociation of FlAsH-Vps28-CT from ESCRT-II by titration of heterotrimeric ESCRT-I(Δ21-Vps37) (an engineered complex missing 21 residues in the N-terminus of the Vps37 subunit) (Teo et al, 2006). Competition experiments show that non-labelled ESCRT-I(Δ21-Vps37) and non-labelled Vps28-CT have similar Kd values (44 and 53 nM, respectively) for ESCRT-II (Figure 1B). In addition, a competition experiment showed that a non-labelled Vps36-NZF-NC also gives a similar Kd (32 nM) (data not shown). These results show that the yeast Vps28-CT in ESCRT-I and the yeast-specific insertion of Vps36 within ESCRT-II are both necessary and sufficient for high-affinity ESCRT-I and ESCRT-II complex formation in vitro.
Crystallisation of the yeast ESCRT-I/-II link
To structurally characterise the yeast ESCRT-I/-II ‘supercomplex', we focused on the minimal domains required for these two large complexes to form a stable interaction in vitro. We generated subcomplexes of Vps28-CT (residues 148–242) with various Vps36-GLUE constructs (Vps36-GLUE domain (residues 1–289), Vps36 yeast-specific insertion (residues 110–205) and a minimal Vps36-NZF-N domain (residues 110–151)) (Figure 1C). For crystallisation, all the subcomplexes tested could be copurified by gel filtration except for a Vps28-CT/minimal Vps36-NZF-N complex. The interaction between these two fragments could be easily detected by GST-pulldown analysis, but the two proteins did not copurify on gel filtration, and therefore were mixed for crystallisation trials.
Because initial co-crystallisation experiments using the Vps28-CT/minimal Vps36-NZF-N complex failed, we re-examined the domain boundaries for the Vps36-NZF-N that were originally derived from the structure of the homologous NZF domain from Np14 (Wang et al, 2003). As the minimal Vps36-NZF-N domain showed only low-affinity binding to ESCRT-I, in comparison to full-length ESCRT-II, we tested whether a C-terminally extended construct (residues 110–176, including conserved hydrophobic residues in the linker region between NZF-N and NZF-C) would bind with higher affinity. Coexpression of Vps28-CT with this extended Vps36-NZF-N resulted in a stable complex that could be copurified by gel filtration. The extended Vps36-NZF-N binds to FlAsH-Vps28-CT with a Kd of 61 nM, confirming that only the extended Vps36-NZF-N and not the Vps36-NZF-C domain interacts with ESCRT-I (Figure 1A).
This Vps28-CT/extended Vps36-NZF-N subcomplex crystallised readily in space group C2221 with unit cell dimensions a=66.6 Å, b=100.0 Å and c=115.6 Å. The 2.0 Å resolution structure was determined with a single anomalous dispersion experiment using the intrinsic Zn2+ bound to the protein. The data collection, phasing and refinement statistics are given in Table I.
Table 1.
Data collection, structure determination and refinement statistics
| ScVps28-CT/NZF-N complex (HR)a | ScVps28-CT/NZF-N complex (Zn SAD)b | Sc Vps28-CT (HR)c | Xl Vps28-CT (HR)d | Xl Vps28-CT (peak)e | Xl Vps28-CT (remote)f | |
|---|---|---|---|---|---|---|
| Data collection | ||||||
| Resolution | 2.0 Å | 2.5 Å | 2.0 Å | 1.3 Å | 1.9 Å | 2.3 |
| Completeness (last shell) | 99.9 (99.9) | 99.9 (100.0) | 99.5 (99.5) | 88.3 (88.3) | 100.0 (99.9) | 63.0(67) |
| Rmerge g (last shell) | 0.10 (0.49) | 0.088 (0.46) | 0.097 (0.41) | 0.099 (0.33) | 0.17 (0.44) | 0.15 (0.52) |
| Multiplicity (last shell) | 7.0 (6.2) | 14.3 (14.4) | 6.1 (4.6) | 4.3 (1.2) | 10.2 (3.7) | 1.9 (1.8) |
| 〈I/σ〉 (last shell) | 14.4 (2.5) | 25.6 (5.3) | 14.1 (3.1) | 9.0 (0.9) | 13.8 (1.8) | 4.6 (0.9) |
| Phasing | ||||||
| Resolution for phasing | — | 2.5 | — | — | 2.3 Å | 2.3 Å |
| Phasing power (iso)h | — | — | 0.4 | |||
| Phasing power (anom)h | — | 1.1 | — | — | 1.3 | 0.9 |
| Sites found (expected) | — | 2 Zn (2) | — | — | 17 Se (18) | — |
| FOM after SHARP | — | 0.31 | — | — | 0.40 | — |
| FOM after SOLOMON (% solvent) | — | 0.91 (49.7%) | — | — | 0.93 (40.1%) | — |
| Refinement | ||||||
| Resolution range | 32–2.0 Å | — | 42.2–2.0 Å | 44.9–1.3 Å | — | — |
| Number of reflections | 25 086 | — | 9619 | 37 809 | — | — |
| Cutoff (F/s) | None | — | None | None | — | — |
| Completeness | 99.8% | — | 99.4% | 88.2 | — | — |
| Protein atoms | 2242 | — | 821 | 1654 | — | — |
| Number of TLS groups | 35 | — | — | 9 | — | — |
| Average B factor (Wilson B factor) | 32 Å2 (29 Å2) | (48 Å2) | 30 Å2 (25 Å2) | 16.5 Å2 (13.1 Å2) | (13.5 Å2) | (19.6 Å2) |
| Waters | 51 | 53 | 134 | |||
| Rcryst i | 0.22 | 0.23 | 0.20 | |||
| Rfree i (% data used) | 0.25 (5) | 0.26 (5) | 0.23 (5) | |||
| RMSD from idealityj | ||||||
| Bonds | 0.021 Å | 0.017 Å | 0.011 Å | |||
| Angles | 1.7° | 1.7° | 1.3° | |||
| Dihedrals | 5.1° | 6.1° | 4.3° | |||
| High-resolution (HR) native data set, ESRF beamline ID23-2 at λ=0.873 Å. | ||||||
| ESRF beamline BM16 at λ=1.28 Å. | ||||||
| ESRF beamline ID14-4 at λ=0.9795 Å. | ||||||
| ESRF beamline ID14-4 at λ=0.9789 Å. | ||||||
| ESRF beamline ID14-4 at λ=0.9788 Å. | ||||||
| ESRF beamline ID14.4 at λ=0.9393 Å. | ||||||
| Rmerge=∑hkl∑i∣Ii(hkl)−〈I(hkl)〉∣/∑hkl∑iIi(hkl). | ||||||
| The ratio of the heavy atom structure factor amplitudes to the lack-of-closure error. | ||||||
| Rcryst and Rfree=∑∥Fobs∣−∣Fcalc∥/∑∣Fobs∣; Rfree calculated with the percentage of the data shown in parentheses. | ||||||
| RMSD for bond angles and lengths with regard to Engh and Huber parameters. | ||||||
The structure of the yeast ESCRT-I/-II link
There are two copies of Vps28-CT/extended Vps36-NZF-N complex in the asymmetric unit. The entire Vps28-CT domain is a well-ordered four-helix bundle with an up-down-up-down topology with each helix packing in an antiparallel fashion with the helix immediately preceding it in the sequence (Figure 2). The first two helices have a similar length and tightly associate to form a straight helical hairpin connected by a short loop. The second two helices form a more splayed helical hairpin, where the shorter helix α3 packs against the longer helix α4 with an angle of −154°. The α3/α4 loop is the longest loop in the four-helix bundle and is well ordered, in part owing to hydrogen bonding with the adjacent α1/α2 loop. The two helical hairpins associate with each other around an extensive hydrophobic core. There are essentially no differences between the 2.0 Å resolution Vps28-CT and the 3.05 Å resolution structure that was recently reported for unliganded yeast Vps28-CT (Pineda-Molina et al, 2006). Sequence alignment of fungal and metazoan ESCRT-I Vps28-CT domains is shown in Figure 3A.
Figure 2.

The structure of the Vps28 C-terminus (ESCRT-I) bound to the Vps36-NZF-N domain (ESCRT-II). (A) Ribbon diagrams of Vps28-CT/extended Vps36-NZF-N complex (generated by PyMOL). Two orthogonal views are shown illustrating the ‘fist'-shaped NZF-N contacting the splayed α2/α3 helices on the ‘punchbag'-shaped Vps28-CT four-helix bundle. (B) A schematic of the Vps28-CT/Vps36-NZF-N binding interface highlighting residues important for this interaction. (C) Two close-up representations of the Vps28-CT/Vps36-NZF-N binding interface. Important residues on the Vps28-CT binding interface are coloured green (hydrophobic), blue (Arg190 and Arg193) and orange (Asn210). Upper panel, critical residues in Vps36-NZF-N comprising the Vps28-CT binding motif (Ile122Vps36/Val148Vps36/Leu154Vps36) are shown as pink sticks. Lower panel, flanking polar interactions are shown. Ordered waters at the interface are shown as yellow spheres. (D) The full-length Vps36 with an I122D/V148D double mutation or an I122D/V148D/D151R/L154R quadruple mutation in the NZF-N domain exhibits a strong defect in GFP-CPS cargo sorting in the context of vps36Δ strain. The GFP-CPS cargo is mis-sorted to the limiting membrane of prevacuolar endosomes (class E compartment, arrowheads) and the limiting membrane of the vacuole (both labelled by FM4-64) upon shift to 37°C in the double mutant or at 30°C in the vps36Δ and quadruple mutant.
Figure 3.

The two Vps36 NZF domains (NZF-N and NZF-C) have the same core fold with distinct binding sites for either ESCRT-I (in NZF-N) or Ub (in NZF-C). (A) Structure-based sequence alignment of the Vps28 C-terminus in ESCRT-I. Species used in this alignment are fungi S. cerevisiae (Sc), Yarrowia lipolytica (Yl) and Schizosaccharomyces pombe (Sp) and metazoa Caenorhabditis elegans (Ce), Xenopus laevis (Xl) and Homo sapiens isoforms 1 (Hs-1) and 2 (Hs-2). Invariant (red), conserved (black), similar (grey) and non-conserved residues (white) are coloured with BOXSHADE using a sequence identity cutoff of 50%. Secondary structure is shown schematically above the Sc sequence. Important functional residues in the Vps28-CT/Vps36-NZF-N binding interface are labelled with asterisks (hydrophobic green, positively charged as blue and polar as orange). The conserved surface patch on Vps28 is marked with a maroon bar. Residues implicated in binding to ScVps20 (Pineda-Molina et al, 2006) are labelled with red asterisks. Polar residues in X. laevis Vps28-CT spatially equivalent to the ScVps28-CT residues that are involved in the Vps36-NZF-N binding site are shown as red circles. (B) Structure-based sequence alignment of the Vps28-CT-binding and Ub-binding NZF domains: Vps36-NZF-N domains of S. cerevisiae (Sc), Candida glabrata (Cg), Aspergillus nidulans (An), Y. lipolytica (Yl), S. pombe (Sp) and Ustilago maydis (Um) against the Ub-binding Sc Vps36-NZF-C domain (NZF-C Sc) and the rat Npl4-NZF domain (NZF Npl4). Important hydrophobic residues in the Vps36-NZF-N and Npl4-NZF domains for binding Vps28-CT and Ub are marked with green and brown bars, respectively. Asp151 in Vps36-NZF-N important for binding Vps28-CT is marked with a red bar. Secondary structure elements of Vps36-NZF-N and rat Npl4-NZF domains are shown above and below the sequence alignment. (C) Structural alignment of the Vps36-NZF-N (coloured ribbon diagram) and Npl4-NZF (black worm) (1Q5W) domains reveal a core common fold with additional features in Vps36-NZF-N (C-terminal extension in pink and extended β2/β3 loop in cyan). Cysteine residues binding to the Zn2+ ion in the Vps36-NZF-N (yellow sticks) and Npl4-NZF domains (black sticks) are shown. (D) Vps36-NZF-N and Vps36-NZF-C domains use distinct binding sites on the rubredoxin knuckles for binding to ESCRT-I and Ub. The NZF domains from the crystal structures of Vps36-NZF-N bound to ESCRT-I and Npl4-NZF bound to Ub (1Q5W) were superimposed, and the Vps28-CT (yellow) and Ub (silver) ligands are shown on the Vps36-NZF-N domain (blue).
The Vps36-NZF-N domain binds to the α2/α3 face of the Vps28-CT helical bundle (Figure 2A). Residues 114–162 of the Vps36-NZF-N domain are ordered. The fold of the Vps36-NZF-N domain is a classical zinc-ribbon fold with a left-handed coordination of the Zn2+ ion between two loops containing CXXC motifs that are often referred to as rubredoxin knuckles (Krishna et al, 2003). The Vps36-NZF-N is a β-sandwich consisting of a two-stranded β-sheet and a three-stranded β-sheet. The three-stranded β-sheet (β1/β2/β5 sheet, lower sheet in Figure 2B) packs orthogonally against the two-stranded β-sheet (β3/β4 sheet, upper sheet). The β1/β2 and β3/β4 loops form the primary contact with the Vps28-CT four-helix bundle. The interaction between the β1/β2 and β3/β4 loops is stabilised by a tetrahedral coordination with a single Zn2+ ion involving two Cys residues from each of the loops. These Cys residues are part of two rubredoxin knuckles (120–123 and 143–146). Residues 110–151 of the Vps36-NZF-N domain have a core fold similar to the NZF domain from Npl4 (Wang et al, 2003); however, additional residues in the Vps36-NZF-N domain form a single-turn α-helix and an additional β-strand, which pairs in an antiparallel fashion to strand β1 making a three-stranded β-sheet to complete the overall fold of the Vps36-NZF-N domain. The resulting five-stranded β-sandwich has a fold that is similar to the fold of the iron-sulfur protein rubredoxin.
A schematic of the binding interface is shown in Figure 2B and detailed interactions are highlighted in Figure 2C. The primary interaction site is hydrophobic; the invariant Ile122Vps36 and highly conserved Val148Vps36 of the two hairpin loops in the Vps36-NZF-N domain slot into a hydrophobic pocket (Figure 2B and C) formed between the splayed helices α2/α3 on the Vps28-CT. Flanking this primary hydrophobic interface are additional polar and hydrophobic contacts. His178Vps28 and Asn210Vps28 form hydrogen bonds with the main-chain carbonyls of Cys146Vps36 and Asn145Vps36, respectively. On the other flanking side, Arg190Vps28 forms a hydrogen bond with Asp151Vps36. A secondary hydrophobic interface is formed by the highly conserved Leu154Vps36 slotting into a small pocket lined by the aliphatic portions of the side chains of Arg190Vps28 and Arg193Vps28.
Given the 61 nM affinity for this interaction, the total surface area buried by complex formation is surprisingly low, 1008 Å2. To verify the functional importance of the interactions observed in the structure of the complex, we mutated a set of interacting residues in the Vps36-NZF-N domain and determined the affinity of the mutants for FlAsH-Vps28-CT (Table II). Mutations of residues within the central hydrophobic interface in Vps36-NZF-N have the greatest impact on binding to Vps28-CT. The I122D mutation reduced the binding over 8000-fold, and the double mutant I122D/V148D has no detectable binding to FlAsH-Vps28-CT. However, single mutations of flanking residues Asp151 or Leu154 resulted in Vps36 NZF-N domains that still retained moderate binding affinity (0.4 and 3.9 μM, respectively).
Table 2.
Affinities of ESCRT-I for ESCRT-II and for Mvb12
| ESCRT-I binding to ESCRT-II | |
| Direct binding assays (binding to FlAsH-Vps28-CT) | Kd (nM) |
| Full-length ESCRT-II wt | 27±4.0 |
| Vps36 NZF-NC wt | 57±4.9 |
| Vps36 NZF-N (110–176) wt | 61±25.3 |
| Vps36 NZF-N (110–176) I122D | >188 μM |
| Vps36 NZF-N (110–176) I122D/V148D | No binding |
| Vps36 NZF-N (110–176) D151R | 403±8 |
| Vps36 NZF-N (110–176) L154R | 2582±969 |
| Competition assays (competition with FlAsH-Vps28-CT/E-II) | |
| ESCRT-I (Δ21-Vps37) | 44±1.8 |
| ESCRT-I (Δ21-Vps37)/Mvb12 | 30±13.2 |
| ESCRT-I/Mvb12 | 23±0.3 |
| Vps28-CT | 53±11.8 |
| Vps36 NZF-NC | 32±1.1 |
| Direct binding assays (binding to FlAsH-Mvb12/ESCRT-I) | |
| Full-length ESCRT-II | 34±10.8 |
| ESCRT-I binding to Mvb12 | |
| Direct binding assays (binding to FlAsH-Mvb12) | |
| ESCRT-I (Δ21-Vps37) | 28±9.2 |
| ESCRT-I (28-NT/Δ21-Vps37) | 22±2.9 |
| ESCRT-I (Δ251-Vps23/Δ21-Vps37) | 20±0.9 |
| ESCRT-I (28-NT/Δ304-Vps23/Δ21-Vps37) | 735±43 |
| ESCRT-I (28-NT/Δ304-Vps23/Δ129-Vps37) | 3000±1697 |
ESCRT-I/-II coupling is essential for efficient MVB cargo sorting
The functional effect of mutations that abolish the ESCRT-I/-II link in vitro was investigated in yeast using a GFP-CPS vacuolar-sorting assay. Both a double mutant (Vps36I122D/V148D) and quadruple mutant (Vps36I122D/V148D/D151R/L154R) when expressed in a yeast strain lacking Vps36 (vps36Δ) resulted in GFP-CPS localisation to the limiting membrane of the vacuole and to a class E compartment rather than localisation in the vacuolar lumen as observed for the wild-type Vps36 (Figure 2D). This class E phenotype observed in the ESCRT-I/-II link mutants is identical to the vps36Δ mutant. Therefore, the ESCRT-I/-II link is essential for efficient cargo sorting.
Structural alignment of Vps36-NZF-N and Npl4-NZF domains reveals a core fold with non-overlapping binding sites
NZF are a widespread family of Zn-binding modules, many of which function in recognising Ub using their rubredoxin knuckles. The NMR structure of the rat Npl4-NZF domain in a complex with Ub identified a critical Ub-binding motif, C-X2-CTF-X8-C-X-φ-C, where φ is a bulky hydrophobic residue (Alam et al, 2004). ESCRT-II Vps36 subunit contains two tandem NZF domains: Vps36-NZF-N, which binds to Vps28-CT with nanomolar affinity, and Vps36-NZF-C, which binds Ub with micromolar affinity (Alam et al, 2004). As the Vps36-NZF-N and Vps36-NZF-C domains appear to be highly specific for their respective binding partners despite having similar folds, we wanted to examine the structural basis for this selectivity.
Alignment and superimposition of the Vps36-NZF-N structure on the Npl4-NZF is shown in Figure 3B and C. The hydrophobic core formed by the packing of two orthogonal β-sheets and the conformation of the rubredoxin knuckles are essentially identical in both the Vps36-NZF-N and Npl4 NZF structures (Figure 3C). However, two major structural differences are apparent. Firstly, the NZF-N domain contains an additional α-helix (α1) and a β-strand (β5), with the strand completing the lower β1/β2/β5 sheet (Figure 3C). The extended C-terminus is essential for generating nanomolar binding affinity (mutation of Leu154Vps36 in this region decreases the affinity more than 60-fold) (Table II) as opposed to the weaker interaction formed by Vps36-NZF-C (encompassing only the NZF core fold) for Ub (Kd 180 μM) (Alam et al, 2004). The second striking difference between the Vps36-NZF-N and other Npl4-family zinc-fingers is that the β2/β3 loop in Vps36-NZF-N is much longer (Figure 3C). It is unlikely that this extended loop in Vps36-NZF-N has a functional role, as it is distant from the Vps28-CT-binding site and not conserved among fungi (Figure 3B).
Mapping the binding interfaces from the Vps36-NZF-C/Ub and Vps36-NZF-N/Vps28-CT complexes on the Vps36-NZF-N domain shows clearly that the binding selectivity arises from two distinct binding sites on the same fold (Figure 3D). It is apparent from sequence alignment of Vps36-NZF-N and Vps36-NZF-C with the Npl4-NZF domain that each NZF domain in the yeast Vps36 subunit of ESCRT-II is adapted for only one of these two potential binding partners (Figure 3B). We have identified a critical motif in Vps36-NZF-N (Ile122/Val148/Leu154) that is required for Vps28-CT binding. The corresponding residues in the Saccharomyces cerevisiae Vps36-NZF-C domain are Ala185, His202 and Lys209. The presence of the charged His149 in the place of valine is a substitution similar to the V148D mutation that reduced the affinity of Vps36-NZF-N for Vps28-CT by more than 8000-fold (Table II), suggesting why Vps36-NZF-C does not bind to Vps28-CT. Conversely, Vps36-NZF-N lacks the critical Ub-binding motif, with the 187-TF-X10-M-199 of Vps36-NZF-C aligning to 124-MV-X10-N-145 in Vps36-NZF-N. The NZF Ub-binding interface has been shown to be sensitive to such minor changes, where mutation of residues 141-TF-142 in Vps36-NZF-C into 141-GS-142 completely abolishes Ub binding (Alam et al, 2004).
The effect of Vps36-NZF-N binding on the structure of Vps28-CT
The isolated yeast Vps28-CT crystallised readily in a high-salt crystallisation condition that dissociated the Vps28-CT/Vps36-NZF-NC complex. This enabled us to compare the unliganded Vps28-CT with the Vps36-bound form. Overall, the 2.0 Å resolution structure of the isolated Vps28-CT domain is essentially identical to Vps28-CT in a complex with Vps36-NZF-N (0.5 Å RMSD for 94 Cα atoms) (Figure 4A, left panel; see Table I for crystallographic data collection and refinement statistics). Conformational change is restricted to side chains involved in the Vps36-NZF-N binding interface. The orientation of Arg193Vps28 changes and Arg190Vps28 becomes ordered on Vps28-CT binding to the Vps36-NZF-N domain, and this minor conformational change acts to provide flanking interactions that are important for generating the nanomolar binding affinity. The rest of the binding interface is rigid and remains unchanged in the non-bound and bound crystal structures, suggesting that the high surface complementarity with the Vps36-NZF-N knuckles is important for generating the high-affinity interaction despite the low contact surface area.
Figure 4.
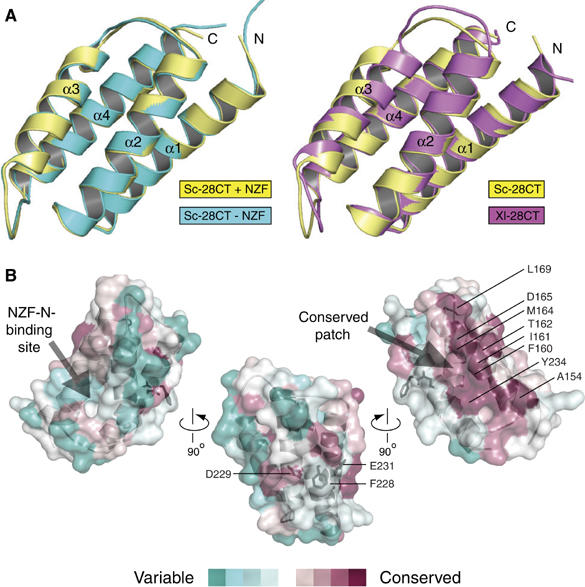
The structural comparison of the isolated S. cerevisiae and X. laevis ESCRT-I Vps28 C-terminus reveals that the four-helix bundle fold is rigid and highly conserved throughout evolution. (A) Alignment of the isolated (cyan) and Vps36-NZF-N-bound (yellow) S. cerevisiae Vps28-CT domains (left panel) and the S. cerevisiae (yellow) and X. laevis (purple) Vps28-CT domains (right panel). (B) Three orthogonal views of conserved surface residues calculated using CONSURF (Landau et al, 2005) displayed on the semi-transparent surface of the isolated S. cerevisiae Vps28-CT structure. Sequence conservation is scored by colour (from most conserved (maroon) to most variable (green)) and displayed by PyMOL. The 238-FDxE-241 sequence motif is shown as black sticks.
Structural comparison of the isolated yeast and metazoan Vps28-CT
We also determined a 1.3 Å resolution structure of the Xenopus laevis Vps28-CT domain. The isolated X. laevis Vps28-CT domain forms a four-helix bundle that is very similar to the yeast domain (1.2 Å RMSD for 90 Cα atoms) (Figure 4A, right panel). Despite this structural conservation, the X. laevis Vps28-CT does not bind to the yeast ESCRT-II (data not shown). Examining the X. laevis Vps28-CT surface corresponding to the Vps36-NZF-N interface highlights that the central hydrophobic pocket in yeast Vps28 has been disrupted by the protrusion of two charged residues that are highly conserved in metazoans (Arg158 and Glu162 in X. laevis, corresponding to Ala182 and Ile186 in yeast Vps28) (Figure 3A). These charged residues would disrupt the hydrophobic patch essential for Vps36-NZF-N binding.
Interestingly, a highly conserved surface patch is seen on the opposite face of the Vps28-CT four-helix bundle as compared to the yeast Vps36-NZF-N-binding site (Figure 4B). Adjacent to this most conserved region is a solvent-exposed phenylalanine (Phe228) flanked on either side by acidic residues, Asp229 and Glu231. Recently, this region was implicated in binding the ESCRT-III subunit Vps20, as mutation of the yeast sequence 228-FDLE-231 to 228-ASLA-231 abolishes the interaction of Vps28-CT with yeast Vps20 in vitro (Pineda-Molina et al, 2006). Further investigation of the functional role of this larger conserved patch in ESCRT-I is required.
ESCRT-I binds directly to a new subunit, Mvb12, forming a stable 1:1:1:1 heterotetrameric complex
We have previously shown that yeast ESCRT-I(Δ21-Vps37), a heterotrimeric complex composed of Vps28, Vps23 and Δ21-Vps37, forms a 1:1:1 complex in vitro (Teo et al, 2006) (Δ21-Vps37 is a deletion variant of S. cerevisiae Vps37, missing residues 1–21 that are prone to proteolysis in Escherichia coli and which significantly promote ESCRT-I aggregation). However, a recent S. cerevisiae proteome-wide screen of protein complexes identified a new putative ESCRT-I-binding protein, YGR206W (systematic yeast ORF designation) (Krogan et al, 2006). This 12 kDa protein of unknown function localises to endosomes in vivo, making it an enticing target for further investigation (Huh et al, 2003) (http://yeastgfp.ucsf/getOrf.php?orf=YGR206W). Independently, YGR206W was recently identified as a component of ESCRT-I and renamed Mvb12 (multivesicular body sorting factor of 12 kDa), a nomenclature we have adopted (Chu et al, 2006).
Gel filtration analysis shows that purified recombinant Mvb12 binds directly to recombinant ESCRT-I(Δ21-Vps37) to form a stable heterotetrameric complex (Figure 5A). Similarly, Mvb12 (with an N-terminal MAH6 tag) and full-length ESCRT-I (untagged) can be readily copurified when the four components are coexpressed in E. coli (Supplementary Figure S1). The most striking result of full-length ESCRT-I coexpression/purification in the presence of Mvb12 was that no aggregation of full-length ESCRT-I was observed (Figure 5B). This is in contrast to full-length ESCRT-I behaviour when purified without Mvb12, which elutes on gel filtration as an aggregate. Despite its small 12 kDa size, Mvb12 changes the apparent size of ESCRT-I(Δ21-Vps37) from 233 to 313 kDa as judged by gel filtration (Figure 5B).
Figure 5.
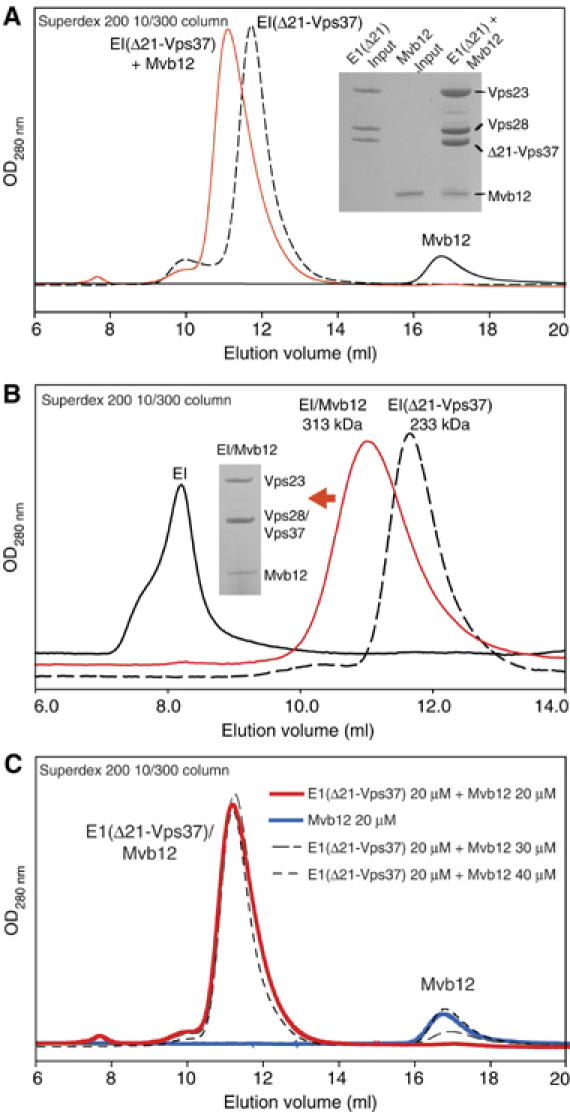
A new ESCRT-I component, Mvb12, forms a stable heterotetrameric ESCRT-I complex in vitro. (A) Gel filtration of recombinant Mvb12 (20 μM), ESCRT-I(Δ21-Vps37) (20 μM) or their mixture (20 μM each) on a Superdex 200 10/300 column. SDS analysis of input ESCRT-I(Δ21-Vps37) and Mvb12 proteins (each diluted to 0.4 mg/ml) is shown alongside a peak fraction from the ESCRT-I(Δ21-Vps37)/Mvb12 complex. (B) Elution profiles of recombinant aggregated heterotrimeric ESCRT-I (≫670 kDa), heterotetrameric ESCRT-I/Mvb12 (313 kDa) and heterotrimeric ESCRT-I(Δ21-Vps37) (233 kDa) (all 6 μM) on a Superdex 200 10/300 column. SDS analysis of a peak fraction from the heterotetrameric ESCRT-I/Mvb12 complex is shown as an inset. (C) Gel filtration of pre-mixtures of ESCRT-I(Δ21-Vps37) (20 μM) with Mvb12 (20 μM: red solid line; 30 μM: grey dashed line; 40 μM: black dashed line) and Mvb12 alone (20 μM: blue solid line) on a Superdex 200 10/300 column.
To determine the stoichiometry of the ESCRT-I/Mvb12 complex, we carried out gel filtration and ultracentrifugation analyses. Gel filtration indicated that a single copy of Mvb12 binds to a single ESCRT-I(Δ21-Vps37) heterotrimer. An equimolar mixture of Mvb12 and ESCRT-I(Δ21-Vps37) yields a single species containing four components; however, even 50% molar excess of Mvb12 results in an additional peak containing free Mvb12 (Figure 5C). Furthermore, we used equilibrium analytical ultracentrifugation to determine accurately the size and stoichiometry of the ESCRT-I/Mvb12 complex. This gave an apparent molecular weight of 104 407±379 Da for the ESCRT-I/Mvb12 heterotetramer (data not shown). This is in agreement with the expected size of ESCRT-I/Mvb12 complex with a 1:1:1:1 stoichiometry (109 072 Da).
Mvb12 binds heterotrimeric ESCRT-I with nanomolar affinity
We measured the affinity of Mvb12 for ESCRT-I using a fluorescence anisotropy assay with an FlAsH-tagged Mvb12 (FlAsH-Mvb12). Direct titration of ESCRT-I(Δ21-Vps37) (the most intact soluble, non-aggregated complex that can be purified without Mvb12) into FlAsH-Mvb12 gave a Kd of 28 nM (Figure 6A). To map the Mvb12-binding site in the ESCRT-I complex, we carried out the fluorescence assay with various truncations of heterotrimeric ESCRT-I (Table II). These results indicate that the coiled-coil region of Vps23 (residues 252–304) represents the primary binding site for Mvb12. Deletion of this region from the ESCRT-I complex reduces the affinity for Mvb12 more than 30-fold. However, the 3 μM Kd of Mvb12 for the ESCRT-I core suggests that there are additional interactions with ESCRT-I outside the coiled-coil region of Vps23. In addition, we find that the C-terminal domain of Vps28 does not participate in binding Mvb12. A construct consisting of only Vps23 residues 200–304 encompassing the coiled-coil region required for high-affinity Mvb12 binding was not soluble when expressed in E. coli (data not shown), and therefore we could not measure its affinity for Mvb12 directly.
Figure 6.
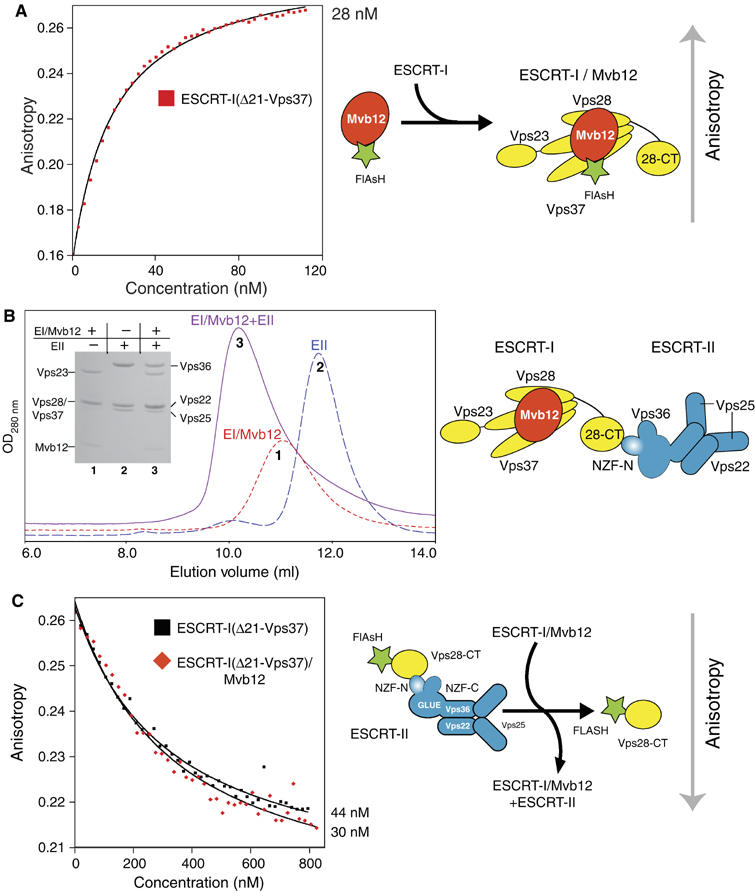
Mvb12 binds to ESCRT-I with nanomolar affinity and does not affect the affinity of ESCRT-I for recombinant ESCRT-II in vitro. (A) The fluorescence anisotropy of the isolated recombinant FlAsH-Mvb12 increases when titrated with recombinant ESCRT-I complexes. Direct fitting of the titration curves shows that ESCRT-I(Δ21-Vps37) binds to FlAsH-Mvb12 with a Kd of approximately 28 nM. (B) Elution profiles of ESCRT-I/Mvb12 alone (6 μM, peak 1), ESCRT-II alone (6 μM, peak 2) and their mixture (6 μM each, peak 3) on a Superdex 200 10/300 column showing formation of an ESCRT-I/Mvb12/ESCRT-II supercomplex. SDS–PAGE of samples from each gel filtration is shown as an inset. (C) In a fluorescence anisotropy competition assay, both heterotrimeric ESCRT-I(Δ21-Vps37) and heterotetrameric ESCRT-I(Δ21-Vps37)/Mvb12 are equally efficient competitors of an FlAsH-Vps28-CT/ESCRT-II complex (Kd 44 and 30 nM, respectively).
The large footprint of Mvb12 on ESCRT-I suggests an important structural role in stabilising the ESCRT-I complex. Indeed, coexpression of Mvb12 with ESCRT-I prevents aggregation (Figure 5B) and greatly enhances the homogeneity of purified recombinant ESCRT-I complexes as Mvb12 protects regions of ESCRT-I that are prone to proteolysis (data not shown).
Mvb12 does not regulate the ESCRT-I/-II supercomplex in vitro
Gel filtration analysis was carried out using purified recombinant ESCRT-I/Mvb12 and ESCRT-II. The addition of Mvb12 to ESCRT-I did not affect the formation of an ESCRT-I/-II supercomplex in vitro (Figure 6B). Using a competition fluorescence anisotropy assay similar to that previously outlined (Figure 1B), we measured the affinity of the heterotetrameric ESCRT-I(Δ21-Vps37)/Mvb12 complex for ESCRT-II. ESCRT-I(Δ21-Vps37) either with or without Mvb12 is equally efficient at competing with FlAsH-Vps28-CT for ESCRT-II (Kd=44 and 30 nM, respectively) (Figure 6C). Therefore, binding of Mvb12 does not affect ESCRT-I binding to ESCRT-II in a recombinant system in vitro. Full-length ESCRT-I/Mvb12 complex is also an equally efficient competitor (Kd=23 nM) of an FlAsH-Vps28-CT/ESCRT-II complex, demonstrating that the N-terminus of the ESCRT-I Vps37 subunit also does not affect the affinity of the ESCRT-I/-II supercomplex formation in vitro (data not shown). Using direct titration, a FlAsH-Mvb12/ESCRT-I complex binds to ESCRT-II with a similar affinity (Kd=34 nM) as determined using FlAsH-Vps28-CT (data not shown).
Discussion
Studies of the ESCRT complexes have revealed extended networks of ESCRT interactions that ensure efficient sorting of mono-ubiquitinated cargo into lysosomes. We have recently identified a direct interaction between recombinant yeast ESCRT-I and ESCRT-II. Consistent with in vitro observations, yeast two-hybrid analyses show ESCRT-I/-II interactions (Bowers, 2004). However, an ESCRT-I/-II complex has not been detected in yeast cell extracts (Katzmann et al, 2001; Babst et al, 2002b). To help reconcile these observations, we undertook a detailed, quantitative analysis of yeast ESCRT-I/-II interactions by structural and fluorescence methods. Here, we have measured the affinity of recombinant ESCRT-I for ESCRT-II and we demonstrated that both the intact complexes and isolated Vps28-CT (from ESCRT-I)/extended Vps36-NZF-N (from ESCRT-II) domains associate with similar nanomolar affinities. Therefore, the Vps28-CT domain and the Vps36-NZF-N domain are both necessary and sufficient for the high-affinity interaction we observe between recombinant full-length yeast ESCRT-I and -II. The Vps28-CT and Vps36-NZF-N are conserved among all fungi. Mutation of residues observed crystallographically in the interface between these two domains critically affects complex formation in vitro and has a strong effect on MVB cargo sorting in vivo, suggesting that a high-affinity ESCRT-I/-II link has evolved in yeast to coregulate the effect of the ESCRT-I and ESCRT-II complexes on endosomal membranes.
The role of the Vps28 (ESCRT-I) C-terminal domain
The Vps28 subunit of ESCRT-I is composed of two discrete domains, the N-terminal core domain, which is essential for the formation of a stable ESCRT-I complex, and the C-terminal domain, which is not necessary for the stability of the ESCRT-I complex. Deletion of the C-terminal domain in yeast in vivo results in a strong class E phenotype (Kostelansky et al, 2006), showing that this domain has an important function in the MVB pathway. The region of the Vps28 core immediately upstream of this domain folds into three small helices and packs differently in the two different crystal forms of the ESCRT-I core (Kostelansky et al, 2006; Teo et al, 2006). Therefore, it appears that the Vps28-CT domain is connected to the rigid ESCRT-I core by a flexible tether. The use of small flexibly connected domains (Vps28-CT and Vps36-NZF-N) in the ESCRT-I/-II interaction may be important to prevent steric clashes between the two large multi-subunit ESCRT-I and -II complexes.
Structural comparison of the yeast Vps28-CT and X. laevis Vps28-CT domains highlights that the rigid four-helix bundle is highly conserved in evolution, but despite this, the NZF-N-binding surface is not conserved. As both plants and metazoa lack an NZF insertion in their Vps36 subunits (ESCRT-II), it appears that the NZF is a relatively recent elaboration of the ancient ESCRT complexes. If metazoan ESCRT-I and -II do associate, it would involve a different mechanism. Surface analysis of the ESCRT-I Vps28-CT domain reveals a highly conserved patch centred on residues 160-FITxxDxx(K/R)L-169. Surface residues near this patch (228-FDxE-231) have been recently linked to binding the yeast Vps20 subunit of ESCRT-III in vitro (Pineda-Molina et al, 2006). Further experiments are necessary to explore the role of this conserved Vps28 patch in yeast and mammalian MVB pathways.
The role of the Vps36 (ESCRT-II) NZF insertion
The Vps36 subunit of ESCRT-II contains two tandem NZF domains, Vps36-NZF-N and Vps36-NZF-C, which enable ESCRT-II to bind simultaneously to the Vps28 subunit of ESCRT-I and Ub, respectively (Teo et al, 2006). The structure of the Vps36-NZF-N domain offers an insight into the molecular mechanisms of a non-Ub-binding class of NZF domains. Despite an identical conformation of the rubredoxin knuckles, NZF zinc-fingers that bind Ub or Vps28-CT use two different surfaces to achieve binding partner specificity. A simple predictive tool for identifying Ub-binding NZF domains was characterised by the highly conserved sequence motif 187-TF-188/φ-199 (where φ is a bulky hydrophobic residue; numbers corresponding to the yeast Vps36-NZF-C, which binds Ub) (Alam et al, 2004). Here, we show that the Vps36-NZF-N domain, which binds to Vps28-CT, uses a distinct binding surface characterised by the hydrophobic signature motif Ile-122/Val-148/Leu-154 in S. cerevisiae.
The binding interface in Vps36-NZF-N for Vps28-CT is more extensive than the Ub-binding interface in Npl4-NZF. This is due to a C-terminal extension in Vps36-NZF-N, which is important for achieving the nanomolar affinity of yeast ESCRT-I for ESCRT-II. It is likely that this extension also stabilises the overall NZF fold, as it forms the third strand of the three-stranded β-sheet in the Vps36-NZF-N domain. A C-terminal extension may be a feature that is shared by at least some other NZF domains, but in the subset of NZF domains that bind Ub, such an extension would not directly participate in binding because Ub interacts with the NZF surface distal to the extension.
Mvb12 and its role in the ESCRT-I/-II link
We have demonstrated that ESCRT-I, which was previously thought to be a heterotrimer, binds directly with nanomolar affinity to a new interacting protein, Mvb12, identified in a recent proteome-wide purification of yeast complexes (Krogan et al, 2006). Deletion of the gene for Mvb12 is associated with partial mis-sorting of carboxypeptidase-Y (Bonangelino et al, 2002) and CPS (Chu et al, 2006), suggesting Mvb12 has a positive role in the MVB pathway. Despite the similar nanomolar affinities of the Mvb12/ESCRT-I and ESCRT-I/-II interactions (Kd of 28 and 27 nM, respectively), only the Mvb12/ESCRT-I heterotetramer and not the ESCRT-I/-II supercomplex copurifies from wild-type yeast, implying that the ESCRT-I/-II interaction is regulated in yeast (Chu et al, 2006). However, an ESCRT-I/-II supercomplex readily copurifies from an Mvb12Δ yeast strain (Chu et al, 2006), suggesting that Mvb12 either directly or indirectly regulates an ESCRT-I/-II link in vivo. Furthermore, this study shows the formation of an Mvb12-dependent ESCRT-I oligomer in yeast. Our in vitro measurements with recombinant proteins demonstrate that Mvb12 does not result in ESCRT-I oligomerisation, nor does Mvb12 affect the ESCRT-I interaction with ESCRT-II, implying a more complex regulatory mechanism in yeast cytosol. This regulatory mechanism could involve either a covalent modification of ESCRT-I or ESCRT-II components or the recruitment of another component that prevents a constitutive ESCRT-I/-II supercomplex in yeast.
We find that Mvb12 considerably reduces proteolysis during purification of the ESCRT-I complex expressed in E. coli and also completely eliminates the aggregation of full-length ESCRT-I heterotetramer. This is likely to be a simple consequence of Mvb12 binding to a significant portion of the ESCRT-I complex, which encompasses the central coiled-coil region of Vps23 (residues 252–304) and the ESCRT-I core. Such a stabilising role of Mvb12 for ESCRT-I we observe in vitro may also be important for ESCRT-I in vivo.
The ESCRT interaction network
Structural information on the ESCRT complexes has offered glimpses into an extended network formed between ESCRT machinery, ubiquitinated cargo and endosomal membranes. The ESCRT complexes use multifunctional domains to efficiently sort mono-ubiquitinated cargo into the lysosomes. These multifunctional binding domains can be thought of as hubs or branches regulating a network of interactions (Praefcke et al, 2004). The GLUE domain of ESCRT-II is one such hub that can recognise endosomal membranes, ubiquitinated cargo and ESCRT-I simultaneously. The yeast Vps28-CT domain could act as a branch point by its ability to bind to both ESCRT-II and ESCRT-III complexes. Structural information on the extended networks formed by the ESCRT machinery with cargo on endosomes and how this might influence the morphology of the endosomes is needed to understand fully the MVB pathway. Critical questions remain to be answered. What are the molecular mechanisms for triggering lumenal vesicle formation at the start of MVB biogenesis? What is the functional significance of the differences between yeast and metazoan ESCRT complexes and their links? The discovery of a new component of ESCRT-I underscores that there is still much to be learned about the MVB pathway.
Materials and methods
Plasmids
The S. cerevisiae Vps28 C-terminus (Vps28-CT, residues 148–242), Vps23 C-terminus (residues 252–385), Vps36-GLUE (residues 1–289), Vps36 yeast-specific insertion (residues 110–205), minimal Vps36-NZF-N (residues 110–151) and extended Vps36-NZF-N (residues 110–176) were PCR amplified from full-length clones described previously (Teo et al, 2006). Vps36 constructs and Vps28-CT were cloned into the first and second cassettes of the polycistronic, coexpression vector pOPC with an N-terminal MAH6 tag on Vps36 or on Vps28-CT, as indicated in the figures, following the general cloning strategy described by Tan (2001). The Vps28-CT and the minimal Vps36-NZF-N were also expressed individually with an N-terminal MAH6 tag using the pOPTH vector. All heterotrimeric ESCRT-I complexes were cloned into the pOPCH vector where Vps28 is in the first cassette (with an N-terminal MAH6 tag), Vps23 in the second cassette and Vps37 in the third cassette. The S. cerevisiae Mvb12 (YGR206W) was amplified from genomic DNA (ATCC 9763D) and cloned into the pOPTH vector. Subcloning of His6-tagged Mvb12 into the fourth cassette of the pOPC vector (Vps28, Vps23 and Vps37 in the first, second and third cassettes) generated coexpression vectors for the heterotetrameric complexes. Mutations in Vps36 were generated using the QuikChange Mutagenesis Kit (Stratagene). A new vector (pOPTHFlash) was constructed for expression of proteins in E. coli and subsequent labelling with Lumio Green reagent (FlAsH, Invitrogen). This vector introduces an MAHHHHHHSSGSENLYFQGSCCPGCCGHM tag (His-rsTEV-Flash tag) to the N-terminus of a protein. The N-terminal His-tag can be cleaved by the addition of TEV protease leaving a small 11 amino-acid tag for labelling with FlAsH (Adams et al, 2002). Similarly, another vector (pOPTHLFlash) was constructed to introduce a His6-tagged lipoyl domain (followed by the TEV cleavage site and Flash tag) to the N-terminus of a protein (for expression of proteins poorly expressed in E. coli using pOPTHFlash). All constructs were verified by sequencing.
Protein expression and purification
See Supplementary data.
Fluorescence binding assays
In all direct binding assays, analyte protein was titrated into a cuvette containing 15 nM Vps28-CT or 25 nM full-length Mvb12 labelled at its N-terminus with Lumio Green (FlAsH-Vps28-CT/FlAsH-Mvb12) in 2.5 ml binding buffer (20 mM Tris (pH 7.4, 20°C), 100 mM NaCl, 10 mM ammonium sulphate and 10mM β-mercaptoethanol). In all competition assays, test proteins were titrated into a cuvette containing preincubated 15 nM FlAsH-Vps28-CT and 120 nM full-length yeast ESCRT-II. Fluorescence was measured using a Perkin-Elmer LS-55 spectrophotometer with λex=490 nm and λem=530 nm. Excitation and emission slits were 15 and 20 nm, respectively. Anisotropy was measured with an integration time of 5 s. Protein was titrated into the cuvette with a Hamilton-MicroLab titrator, allowing 50 s stirring after each titration step and a wait of 10 s before the anisotropy was recorded. Titration of binding buffer alone in either a direct or competition binding assay results in no overall change in anisotropy. The Kd values were calculated from direct fitting of the curves obtained from direct binding assays and competition assays in the program Datafitter (DB Veprintsev). At least two independent experiments were conducted to determine Kd values.
Crystallisation, data collection and structure determination
See Supplementary data.
ESCRT-I analytical gel filtration and ultracentrifugation
See Supplementary data.
Microscopy
Living yeast cells expressing GFP-CPS were harvested at an OD600 of 0.4–0.6 and labelled with FM4-64 for vacuolar membrane staining and resuspended in a medium for visualisation. Visualisation of cells was performed on a fluorescence microscope (Axiovert S1002TV; Carl Zeiss Microimaging Inc.) equipped with fluorescein isothiocyanate and rhodamine filters, captured with a digital camera (Photometrix) and deconvoluted using Delta Vision software (Applied Precision Inc.). Results presented were based on observations of >120 cells at 30°C for vps36Δ and Vps36I122D/V148D/D151R/L154R and at 37°C for Vps36I122D/V148D.
Accession numbers
The coordinates for the structures described in this work have been deposited in the Protein Data Bank with ID codes 2J9U (ScVps28-CT/Vps36-NZF-N), 2J9V (ScVps28-CT) and 2J9W (Xenopus Vps28-CT).
Supplementary Material
Supplementary data
Supplementary Figure S1
Acknowledgments
We thank Gordon Leonard, Andrew McCarthy, Raimond Ravelli, Ana Labrador, Didier Nurizzo and David Flot for assistance at ESRF beamlines ID14-4, BM16 and ID23-2 (Grenoble, France). DJG is supported by a studentship from the Medical Research Council, HLT by the Agency for Science, Technology and Research of Singapore and JS as an HHMI Research Associate. SDE is supported as an investigator of the HHMI and RLW by the Medical Research Council.
References
- Adams SR, Campbell RE, Gross LA, Martin BR, Walkup GK, Yao Y, Llopis J, Tsien RY (2002) New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. J Am Chem Soc 124: 6063–6076 [DOI] [PubMed] [Google Scholar]
- Alam SL, Sun J, Payne M, Welch BD, Blake BK, Davis DR, Meyer HH, Emr SD, Sundquist WI (2004) Ubiquitin interactions of NZF zinc fingers. EMBO J 23: 1411–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD (2002a) ESCRT-III: an endosome-associated heterooligomeric protein complex required for MVB sorting. Dev Cell 3: 271–282 [DOI] [PubMed] [Google Scholar]
- Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD (2002b) Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell 3: 283–289 [DOI] [PubMed] [Google Scholar]
- Bache KG, Stuffers S, Malerod L, Slagsvold T, Raiborg C, Lechardeur D, Walchli S, Lukacs GL, Brech A, Stenmark H (2006) The ESCRT-III subunit hVps24 is required for degradation but not silencing of the epidermal growth factor receptor. Mol Biol Cell 17: 2513–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau PS, Winistorfer SC, Kearney WR, Robertson AD, Piper RC (2003) Vps27-Hse1 and ESCRT-I complexes cooperate to increase efficiency of sorting ubiquitinated proteins at the endosome. J Cell Biol 163: 237–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonangelino CJ, Chavez EM, Bonifacino JS (2002) Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol Biol Cell 13: 2486–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers K, Lottridge J, Helliwell SB, Goldthwaite LM, Luzio JP, Stevens TH (2004) Protein–protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic 5: 194–210 [DOI] [PubMed] [Google Scholar]
- Chu T, Sun J, Saksena S, Emr SD (2006) New component of ESCRT-I regulates endosomal sorting complex assembly. J Cell Biol 175: 815–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyotte A, Russell MR, Hopkins CR, Woodman PG (2005) Depletion of TSG101 forms a mammalian ‘Class E' compartment: a multicisternal early endosome with multiple sorting defects. J Cell Sci 118: 3003–3017 [DOI] [PubMed] [Google Scholar]
- Field MC, Gabernet-Castello C, Dacks JB (2006) Reconstructing the evolution of the endocytic system: insights from genomics and molecular cell biology In Origins and Evolution of Eukaryotic Endomembranes and Cytoskeleton, Jékely G (ed) pp. 309–334. Austin, TX: Eurekah/Landes Bioscience Press [DOI] [PubMed] [Google Scholar]
- Hierro A, Sun J, Rusnak AS, Kim J, Prag G, Emr SD, Hurley JH (2004) Structure of the ESCRT-II endosomal trafficking complex. Nature 431: 221–225 [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Hurley JH, Emr SD (2006) The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu Rev Biophys Biomol Struct 35: 277–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann DJ, Babst M, Emr SD (2001) Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106: 145–155 [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Stefan CJ, Babst M, Emr SD (2003) Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J Cell Biol 162: 413–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostelansky MS, Sun J, Lee S, Kim J, Ghirlando R, Hierro A, Emr SD, Hurley JH (2006) Structural and functional organization of the ESCRT-I trafficking complex. Cell 125: 113–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna SS, Majumdar I, Grishin NV (2003) Structural classification of zinc fingers: survey and summary. Nucleic Acids Res 31: 532–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B, Richards DP, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete MM, Vlasblom J, Wu S, Orsi C, Collins SR, Chandran S, Haw R, Rilstone JJ, Gandi K, Thompson NJ, Musso G, St Onge P, Ghanny S, Lam MH, Butland G, Altaf-Ul AM, Kanaya S, Shilatifard A, O'Shea E, Weissman JS, Ingles CJ, Hughes TR, Parkinson J, Gerstein M, Wodak SJ, Emili A, Greenblatt JF (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440: 637–643 [DOI] [PubMed] [Google Scholar]
- Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N (2005) ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res 33: W299–W302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Kimpler LA, Naismith TV, Lauer JM, Hanson PI (2005) Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7-1 with itself, membranes, and the AAA+ ATPase SKD1. J Biol Chem 280: 12799–12809 [DOI] [PubMed] [Google Scholar]
- Muziol T, Pineda-Molina E, Ravelli RB, Zamborlini A, Usami Y, Gottlinger H, Weissenhorn W (2006) Structural basis for budding by the ESCRT-III factor CHMP3. Dev Cell 10: 821–830 [DOI] [PubMed] [Google Scholar]
- Pineda-Molina E, Belrhali H, Piefer AJ, Akula I, Bates P, Weissenhorn W (2006) The crystal structure of the C-terminal domain of Vps28 reveals a conserved surface required for Vps20 recruitment. Traffic 7: 1007–1016 [DOI] [PubMed] [Google Scholar]
- Praefcke GJ, Ford MG, Schmid EM, Olesen LE, Gallop JL, Peak-Chew SY, Vallis Y, Babu MM, Mills IG, McMahon HT (2004) Evolving nature of the AP2 alpha-appendage hub during clathrin-coated vesicle endocytosis. EMBO J 23: 4371–4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C, Wesche J, Malerod L, Stenmark H (2006) Flat clathrin coats on endosomes mediate degradative protein sorting by scaffolding Hrs in dynamic microdomains. J Cell Sci 119: 2414–2424 [DOI] [PubMed] [Google Scholar]
- Razi M, Futter CE (2006) Distinct roles for Tsg101 and Hrs in multivesicular body formation and inward vesiculation. Mol Biol Cell 17: 3469–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim JH, Xiao C, Hayden MS, Lee KY, Trombetta ES, Pypaert M, Nara A, Yoshimori T, Wilm B, Erdjument-Bromage H, Tempst P, Hogan BL, Mellman I, Ghosh S (2006) CHMP5 is essential for late endosome function and down-regulation of receptor signaling during mouse embryogenesis. J Cell Biol 172: 1045–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slagsvold T, Aasland R, Hirano S, Bache KG, Raiborg C, Trambaiolo D, Wakatsuki S, Stenmark H (2005) Eap45 in mammalian ESCRT-II binds ubiquitin via a phosphoinositide-interacting GLUE domain. J Biol Chem 280: 19600–19606 [DOI] [PubMed] [Google Scholar]
- Slagsvold T, Pattni K, Malerod L, Stenmark H (2006) Endosomal and non-endosomal functions of ESCRT proteins. Trends Cell Biol 16: 317–326 [DOI] [PubMed] [Google Scholar]
- Tan S (2001) A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Protein Expr Purif 21: 224–234 [DOI] [PubMed] [Google Scholar]
- Teo H, Gill DJ, Sun J, Perisic O, Veprintsev DB, Vallis Y, Emr SD, Williams RL (2006) ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell 125: 99–111 [DOI] [PubMed] [Google Scholar]
- Teo H, Perisic O, Gonzalez B, Williams RL (2004) ESCRT-II, an endosome-associated complex required for protein sorting; crystal structure and interactions with ESCRT-III and membranes. Dev Cell 7: 559–569 [DOI] [PubMed] [Google Scholar]
- Wang B, Alam SL, Meyer HH, Payne M, Stemmler TL, Davis DR, Sundquist WI (2003) Structure and ubiquitin interactions of the conserved zinc finger domain of Npl4. J Biol Chem 278: 20225–20234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitley P, Reaves BJ, Hashimoto M, Riley AM, Potter BVL, Holman GD (2003) Identification of mammalian Vps24p as an effector of phosphatidylinositol 3,5-bisphosphate-dependent endosome compartmentalization. J Biol Chem 278: 38786–38795 [DOI] [PubMed] [Google Scholar]
- Yorikawa C, Shibata H, Waguri S, Hatta K, Horii M, Katoh K, Kobayashi T, Uchiyama Y, Maki M (2005) Human CHMP6, a myristoylated ESCRT-III protein, interacts directly with an ESCRT-II component EAP20 and regulates endosomal cargo sorting. Biochem J 387: 17–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary Figure S1


