Abstract
The positive transcription elongation factor b (P-TEFb) stimulates transcriptional elongation by phosphorylating the carboxy-terminal domain of RNA polymerase II and antagonizing the effects of negative elongation factors. Not only is P-TEFb essential for transcription of the vast majority of cellular genes, but it is also a critical host cellular cofactor for the expression of the human immunodeficiency virus (HIV) type 1 genome. Given its important role in globally affecting transcription, P-TEFb's activity is dynamically controlled by both positive and negative regulators in order to achieve a functional equilibrium in sync with the overall transcriptional demand as well as the proliferative state of cells. Notably, this equilibrium can be shifted toward either the active or inactive state in response to diverse physiological stimuli that can ultimately affect the cellular decision between growth and differentiation. In this review, we examine the mechanisms by which the recently identified positive (the bromodomain protein Brd4) and negative (the noncoding 7SK small nuclear RNA and the HEXIM1 protein) regulators of P-TEFb affect the P-TEFb-dependent transcriptional elongation. We also discuss the consequences of perturbations of the dynamic associations of these regulators with P-TEFb in relation to the pathogenesis and progression of several major human diseases, such as cardiac hypertrophy, breast cancer, and HIV infection.
INTRODUCTION
RNA polymerase II (Pol II)-mediated transcription can be subdivided into several stages, designated preinitiation, initiation, promoter clearance, elongation, and termination (85, 91). Over the past two decades or so, major efforts have been devoted to the elucidation of the mechanisms governing the assembly of preinitiation complexes at promoters and to exploration of the intricacies of transcriptional initiation and promoter clearance. This has resulted in the identification of many basal transcription factors as well as gene-specific transcriptional regulators that are important for these early stages of the transcription cycle (85, 91). In contrast, the elongation phase of transcription, in which Pol II spends the bulk of its time during pre-mRNA synthesis, has received relatively little attention. In fact, there has been a long-held misconception that elongation is merely an unregulated event involving repetitive additions of ribonucleotides to the growing RNA chain.
Only recently, it has become increasingly clear that the elongation stage of eukaryotic transcription is a highly regulated process critical not only for generating the full-length mRNA transcripts but also for the coupling of transcription with other major gene expression events, such as pre-mRNA capping, splicing, and polyadenylation (91). Moreover, recent years have seen major advances in the biochemical analysis of mechanisms and factors that control the elongation process. One of the most significant achievements has been the identification and characterization of P-TEFb (positive transcription elongation factor b) as a factor that can specifically stimulate the processivity of Pol II elongation and antagonize the actions of negative elongation factors. In addition, our understanding of the general mechanisms controlling elongation has also benefited tremendously from studies of human immunodeficiency virus type 1 (HIV-1), which uses P-TEFb as a specific cofactor for efficient transcriptional elongation along the integrated proviral DNA template (4, 42, 45).
Besides being a cellular cofactor for HIV-1, P-TEFb is also a general transcription factor required for efficient expression of the vast majority of cellular genes (10, 86), and therefore, its activity must be carefully regulated to respond to changes in global transcriptional demand at different stages of cell growth or differentiation. In support of this notion, recent studies have indicated that the activity of P-TEFb is dynamically controlled by both positive and negative regulators in order to maintain a functional equilibrium appropriate for the growth or differentiation state of a cell. We will discuss how these novel regulators of P-TEFb, namely, the Brd4 and HEXIM1 proteins and the 7SK small nuclear RNA (snRNA), can reciprocally affect P-TEFb's activity to globally control transcriptional elongation and the physiological consequences resulting from this dynamic regulation.
P-TEFb STIMULATES GENERAL AND HIV-1-SPECIFIC TRANSCRIPTIONAL ELONGATION AND MEDIATES THE COUPLING OF TRANSCRIPTION WITH RNA PROCESSING
P-TEFb Is a General Transcriptional Elongation Factor
P-TEFb is composed of cyclin-dependent kinase 9 (CDK9) and its regulatory partner cyclin T1 (CycT1) (74, 99). Besides the 42-kDa form of CDK9, termed CDK9(42), there is also a second isoform of CDK9 called CDK9(55), with a 55-kDa molecular mass resulting from a 117-residue amino-terminal extension that is not present in the 42-kDa form (88). In addition to CycT1, minor CDK9-associated CycT2a, T2b, and K molecules are also present at much lower concentrations than CycT1 in many cell types (27, 75). For this review, only the P-TEFb complex containing CDK9(42) and CycT1 will be discussed, while complexes containing CDK9(55) and other minor cyclin molecules behave similarly in most cases. As expected of any CDK-cyclin pairs, CDK9 exerts its kinase activity only when associated with its cyclin partner. Importantly, this kinase activity is absolutely essential for P-TEFb to stimulate HIV-1 transcriptional elongation in vitro and in vivo (31, 55, 119). As for cellular genes, studies employing RNA interference (RNAi) in Caenorhabditis elegans reveal a broad requirement for P-TEFb during early embryonic gene expression (86). Consistent with this, treatment of human HeLa cells with flavopiridol, a potent inhibitor of CDK9, implicates P-TEFb as being essential for the expression of most protein-coding genes (10). However, apparently not all class II genes depend on P-TEFb for transcription. A recent report suggests that certain genes within the p53 pathway are able to bypass the requirement for P-TEFb (32). Furthermore, P-TEFb is not an essential elongation factor for the intronless histone H2b and the U2 snRNA genes (61).
Key Substrates of P-TEFb during Transcriptional Elongation
A primary target/substrate of the P-TEFb kinase during transcriptional elongation is the carboxy-terminal domain (CTD) of the largest subunit (RPB1) of RNA Pol II, which exists in two major forms depending on the phosphorylation status of the CTD. The hypophosphorylated form is called Pol IIa, and the hyperphosphorylated form, where the CTD is extensively phosphorylated, is termed Pol IIo. The CTD contains many repeats of the heptapeptide sequence YSPTSPS. Although this sequence is conserved throughout the eukaryotic kingdom, the numbers of repeats vary among different species. The CTD plays a critical role during the transcription cycle, as it interacts with a variety of transcription factors modulating different stages of transcription. It also serves as a scaffold for the assembly and operation of other machineries involved in the cotranscriptional processing of pre-mRNAs (48, 59, 60, 101).
The CTD undergoes dynamic phosphorylation and dephosphorylation throughout each transcription cycle (14), and P-TEFb plays a key part in this process (Fig. 1). To form the preinitiation complexes, the hypophosphorylated Pol IIa is recruited to promoters (91). Shortly after transcription initiation, the CTD becomes heavily phosphorylated at Ser5 positions in the heptapeptide repeats by CDK7, a component of the general transcription factor TFIIH (70). This event is essential for Pol II to escape the promoter and shift into an early elongation mode (71, 79). However, soon after the transition into early elongation, Pol II is arrested at a checkpoint imposed by the concerted actions of two negative elongation factors, i.e., 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole sensitivity-inducing factor (DSIF) and negative elongation factor (NELF) (97, 105, 106). This checkpoint serves to ensure the proper capping of pre-mRNA, and the Ser5-phosphorylated CTD helps recruit capping enzymes to the 5′ end of the nascent transcript (49, 79, 82).
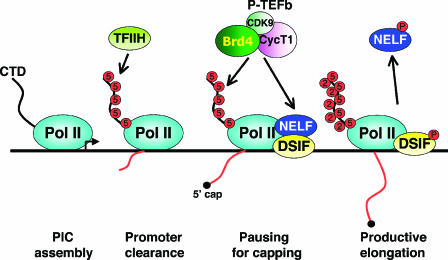
FIG. 1.
P-TEFb phosphorylates the Pol II CTD and negative elongation factors to stimulate processive elongation. At the start of the transcription cycle, Pol II with the hypophosphorylated CTD is assembled into the preinitiation complex (PIC) at the promoter. To facilitate promoter clearance and jump-start initiation, Ser5 residues of the CTD heptapeptide repeats are phosphorylated by CDK7 (a component of the TFIIH complex). However, shortly after initiation, the progression of Pol II is stalled by the concerted actions of two negative elongation factors, DSIF and NELF. This checkpoint facilitates the recruitment of capping enzymes to ensure proper capping of the nascent pre-mRNA. To overcome this checkpoint, P-TEFb, which is recruited by Brd4 to transcription template in the vicinity of the stalled Pol II, phosphorylates DISF, NELF, and the CTD repeats at the Ser2 positions. These phosphorylation events promote the dissociation of NELF and convert DSIF into a positive elongation factor, thereby allowing the Pol II to engage in productive elongation to produce full-length transcripts.
Without further stimulation at this stage, the paused Pol II will slip into abortive elongation, generating only short mRNA transcripts. Releasing the stalled Pol II from this checkpoint requires the enzymatic action of P-TEFb, which phosphorylates CTD at the Ser2 positions of the heptapeptide repeats, yielding heavily phosphorylated Pol IIo. Moreover, the Spt5 subunit of DSIF (38) and the RD subunit of NELF (28) are also phosphorylated by P-TEFb. In the case of Spt5, Thr4 residues within an evolutionarily conserved repetitive motif (consensus, G-S-R/Q-T-P) in the C-terminal region are, like the Ser2 residues in the Pol II CTD, highly phosphorylated by P-TEFb (104). These phosphorylation events lead to the dissociation of NELF and conversion of DSIF into a positive elongation factor, which, together with the hyperphosphorylation of Ser2 in the Pol II CTD, allow Pol II to shift into the productive phase of transcriptional elongation (Fig. 1) (78, 97, 106). Finally, at the end of the transcription cycle, the phosphorylated Pol IIo is recycled by the action of CTD phosphatases (e.g., FCP1) into the hypophosphorylated IIa form, allowing it to participate in a fresh round of transcription (12, 56).
P-TEFb as a Host Cellular Cofactor for HIV-1 Transcription
The observations that P-TEFb functions as a general transcription factor by impinging upon the basal RNA Pol II elongation machinery do not rule out the possibility that P-TEFb could be particularly rate limiting for the expression of certain genes under specific conditions. Indeed, there are no other cellular or viral genes that are more sensitive to the availability of P-TEFb than those carried by HIV-1 (10, 109), which uses a sophisticated scheme to capture and recruit host cellular P-TEFb to the integrated proviral DNA template. Thus, for transcription from the HIV-1 long terminal repeat (LTR), P-TEFb assumes an essential role as a specific cofactor. In fact, our understanding of the general mechanism of P-TEFb stimulation of Pol II elongation has benefited tremendously from studies of the role of P-TEFb in regulating HIV-1 transcription.
HIV-1 encodes a small regulatory protein called Tat that is essential for activating transcriptional elongation from the HIV-1 LTR and producing the full-length viral transcripts (4, 42, 45). In the absence of Tat, transcription from the HIV-1 LTR generates only short transcripts; however, the presence of Tat dramatically stimulates the efficiency of elongation and results in a large increase in the level of full-length transcripts that extend through the more-than-9-kb HIV-1 genome. Tat is able to accomplish this by recruiting P-TEFb to the 5′ end of the nascent viral transcript (nucleotides +1 to +59), which forms a stem-loop RNA structure called the trans-acting response (TAR) RNA element (Fig. 2). Tat specifically binds to the TAR RNA at the three-nucleotide bulge and several flanking nucleotides in the double-stranded RNA stem just below the apical loop. On the other hand, CycT1 of P-TEFb recognizes specific residues in the apical loop of the TAR RNA, while Tat also interacts directly with CycT1 near its cyclin box region. These simultaneous and cooperative interactions among Tat, TAR, and P-TEFb allow the formation of a stable ternary complex that results in the recruitment of P-TEFb to the vicinity of the stalled Pol II (Fig. 2), where it can phosphorylate the CTD and negative elongation factors DSIF and NELF, leading to stimulation of transcriptional elongation as described above (4, 42, 45).
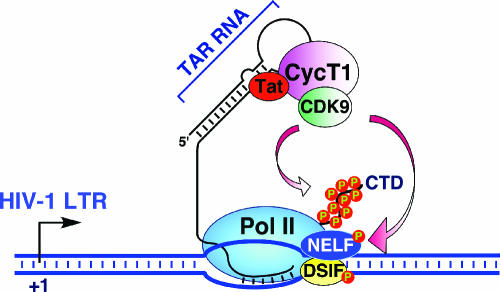
FIG. 2.
P-TEFb is essential for Tat transactivation of HIV-1 transcription. Shortly after transcription is initiated from the HIV-1 promoter, the progression of Pol II is stalled by the concerted actions of negative elongation factors DSIF and NELF. For Pol II to escape from this promoter-proximal pausing, the HIV-1-encoded Tat protein binds to host cellular P-TEFb and recruits it to the stalled Pol II through forming a stable ternary complex involving the TAR RNA stem-loop structure located near the 5′ end of the nascent viral transcript. Subsequently, P-TEFb phosphorylates the Pol II CTD as well as the negative elongation factors to stimulate processive elongation.
Role of P-TEFb in RNA Processing
In addition to P-TEFb having a role in stimulating transcriptional elongation, several lines of evidences have suggested additional roles of P-TEFb in affecting pre-mRNA processing. For example, P-TEFb has been proposed to mediate the coupling of transcriptional elongation with pre-mRNA splicing. The CycT1 subunit of P-TEFb is known to interact with Tat-SF1 (24), a nuclear protein implicated in stimulating general and HIV-1 transcriptional elongation (12, 46, 52, 73, 119, 120). On the other hand, Tat-SF1, which is a homologue of the yeast splicing factor CUS2 (107), can also interact with the spliceosomal U snRNPs to form a multisubunit complex, which strongly stimulates Pol II elongation when directed to an intron-free DNA template (25). This effect is mediated through the binding of Tat-SF1 to the C-terminal domain of CycT1, which, as part of P-TEFb, travels along with the elongating Pol II (76, 117, 118). Notably, inclusion of splicing signals in the nascent transcript further stimulates transcription, supporting the notion that the recruitment of U snRNPs near the elongating polymerase is important for transcription. Since the Tat-SF1-U snRNP complex stimulates both transcription and splicing, recruitment of this dual-function factor to Pol II by P-TEFb is believed to mediate an efficient coupling of transcriptional elongation with splicing.
Recently, yet another human splicing factor, called splicing-associated c-Ski-interacting protein (SKIP), has also been shown to bind to P-TEFb as well as the Tat-TAR-P-TEFb ternary complex (7). SKIP, an essential component of activated spliceosomes (66), is required for Tat transactivation. Moreover, through its association with U5 snRNP proteins and tri-snRNP110K protein, SKIP also facilitates recognition of an alternative Tat-specific splice site. However, unlike Tat-SF1 and its associated U snRNPs, which are able to stimulate both elongation and splicing, SKIP appears to play independent roles in pre-mRNA splicing and P-TEFb-mediated transcriptional elongation (7). Future studies are necessary to determine whether the bindings of P-TEFb to SKIP and Tat-SF1 allow the latter two proteins to function in the same or different pathways to couple transcription with pre-mRNA splicing.
Besides splicing, Ser2 phosphorylation of the Pol II CTD mediated by Drosophila P-TEFb or Saccharomyces cerevisiae Ctk1, a kinase similar to mammalian CDK9, has also been shown to contribute to the recruitment of RNA-processing factors involved in the 3′ end formation of a number of genes. Surprisingly, the depletion of Drosophila CDK9 or yeast Ctk1 activity causes defective 3′ polyadenylation but has no major effect on the distribution of elongation factors and transcribing Pol II along the transcription unit (1, 68). Consistent with these observations, a recent report indicates that for the relatively short, intronless genes, such as the human U2 snRNA gene, P-TEFb is not an essential elongation factor (61). Rather, it functions exclusively as an RNA-processing factor for recognition of the 3′ box RNA 3′ end processing signal. These results are intriguing, as they imply that the role of P-TEFb as an activator of transcriptional elongation can be separated from its role in RNA 3′ end processing. It remains to be tested whether this notion is also applicable to many mammalian protein-coding genes that are known to depend on P-TEFb for elongation. It would be very interesting if P-TEFb turns out to be dispensable for 3′ end formation for some of these genes.
NEGATIVE REGULATION OF P-TEFb ACTIVITY
The 7SK snRNA and HEXIM1 Protein Sequester P-TEFb into an Inactive Complex
It is well known that for proper progression of the cell cycle, the activities of CDKs involved in cell cycle regulation must be kept in check by specific cyclin-dependent kinase inhibitors (CKIs) that associate with either CDKs or CDK-cyclin complexes to inhibit their kinase activities (84). As a member of the CDK superfamily, CDK9 is no exception. In fact, recent studies have indicated that in the nucleus, not every CDK9-CycT1 heterodimer displays the P-TEFb kinase and transcriptional activities. Two independent efforts to isolate nuclear factors that can bind to and control the activity of human P-TEFb have led to the revelation that about half of the nuclear P-TEFb in log-phase HeLa cells is sequestered in a kinase-inactive complex that also contains the 7SK snRNA (67, 109). Transcribed by RNA Pol III, 7SK is an abundant (2 × 105 copies/cell) snRNA of 331 nucleotides that is highly conserved in vertebrates (22, 65, 121). After the first description of this RNA species in the 1970s, little was known about its physiological function for the next quarter of a century. Through its interaction with P-TEFb, it is now clear that 7SK plays a key role in suppressing the kinase and transcriptional activities of P-TEFb (67, 109). Moreover, the 7SK-bound P-TEFb cannot be recruited to the HIV-1 promoter/template either in vivo or in vitro (109).
While investigating the functional significance of a reconstituted interaction between 7SK and P-TEFb, it was subsequently found that this interaction alone was necessary but not sufficient to inactivate P-TEFb, indicating the presence of another factor in the 7SK snRNP for effective P-TEFb inactivation (110). Indeed, through affinity purification, a nuclear protein called HEXIM1 has been identified as a novel component of the 7SK-P-TEFb snRNP formed in vivo (63, 110). Originally identified as a hexamethylene bisacetamide (HMBA)-inducible protein in human vascular smooth muscle cells (72), the expression of HEXIM1 can also be induced in many other cell types (112). HMBA, a hybrid bipolar compound, is known to be a potent suppressor of cell growth and inducer of cell differentiation (57). Reflecting these effects of HMBA, HEXIM1 has been shown to inhibit the growth of many cell types and tissues (discussed in detail below). An explanation for HEXIM1's growth-inhibitory effect comes from the observations that this protein potently and specifically inhibits the kinase and transcriptional activities of the general transcription factor P-TEFb (62, 110).
It is important to point out that HEXIM1 exerts its inhibitory effect on P-TEFb in vivo and in vitro in a 7SK snRNA-dependent fashion (62, 110). This is due to the fact that 7SK plays a scaffolding role in mediating the interaction between HEXIM1 and P-TEFb (62, 63, 110). A recent study has revealed two structurally and functionally distinct protein-binding elements located in the 5′- and 3′-terminal hairpins of 7SK, which supports the in vivo recruitment of HEXIM1 and P-TEFb (22). While HEXIM1 binds independently and specifically to the G24-C48/G60-C87 distal segment of the 5′ hairpin of 7SK, the association of P-TEFb with the G302-C324 apical region of the 3′ hairpin of 7SK occurs only after HEXIM1 is already in complex with the 7SK RNA (22), implying that cooperative interactions among 7SK, HEXIM1, and P-TEFb are involved in forming the 7SK snRNP.
Similar Architectural Plans for Forming the Tat-TAR-P-TEFb and HEXIM1-7SK-P-TEFb Ribonucleoprotein Complexes
The HEXIM1 protein can be subdivided into three major domains: an N-terminal proline-rich domain that acts as a self-inhibitory domain (110), a C-terminal domain that contains many acidic amino acid residues and functions as the P-TEFb-targeting and inhibitory domain (2, 62, 63, 110), and a centrally located nuclear localization signal that largely overlaps with a 7SK-binding domain (111). The 7SK-binding domain (amino acids 150 to 166) (Fig. 3) is composed of bipartite arginine-rich motifs, which are often found in other RNA-interacting proteins (100). Moreover, this domain can interact with the adjacent acidic region in the absence of 7SK (2). The interaction of 7SK with the arginine-rich basic region appears to break up the intramolecular interaction between the oppositely charged regions in HEXIM1 and permit HEXIM1's specific binding to P-TEFb and proper nuclear localization (exclusion from nuclear speckles), which culminates in the inhibition of transcription (2).
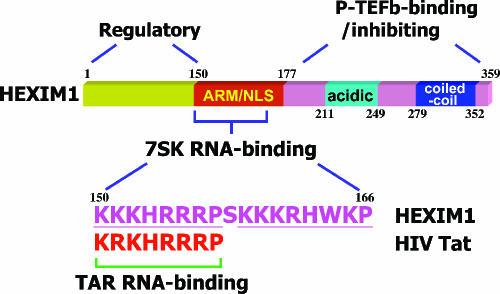
FIG. 3.
Domain structure of HEXIM1. The N-terminal domain of HEXIM1 functions as a self-inhibiting regulatory domain. The centrally located arginine-rich motif (ARM) that largely overlaps with the nuclear localization signal (NLS) also serves as the 7SK RNA-binding domain. The 7SK-binding domain in HEXIM1 has a bipartite structure, with the first half showing a near-perfect match with the TAR RNA-binding motif found in HIV-1 Tat. The P-TEFb-binding/inhibiting function of HEXIM1 resides in the HEXIM1 C-terminal domain. Within this domain, the coiled-coil region mediates the dimerization of HEXIM1, whereas a region enriched in acidic residues is proposed to interact with the adjacent basic residues in the ARM/NLS domain, thus preventing the premature binding of HEXIM1 to P-TEFb in the absence of the 7SK snRNA (2). All numbers refer to amino acid positions in HEXIM1.
It is worth noting that the 7SK-binding motif in HEXIM1 is highly homologous to, and in fact functionally interchangeable with, the arginine-rich TAR-binding motif in the HIV-1 Tat protein (Fig. 3) (reference 111 and our unpublished data). In addition, the G302-C324 apical region of the 3′ hairpin of 7SK also displays remarkable sequence similarity to the bulge-loop region of the HIV-1 TAR RNA (positions +20 to +42), both of which also make direct contacts with CycT1 (Fig. 4) (22, 45). Finally, both HEXIM1 and Tat can contact P-TEFb through a region near the cyclin box in CycT1 (11, 45, 63). These similarities observed at multiple levels are unlikely to be purely coincidental. Rather, similar RNA-dependent architectural plans may exist to form both the Tat-TAR-P-TEFb and HEXIM1-7SK-P-TEFb ribonucleoprotein complexes (Fig. 4). Since the Tat-TAR-P-TEFb complex emerges much later during evolution, it is possible that HIV-1 simply borrows an existing and effective theme from the host cell to efficiently recruit P-TEFb to the HIV-1 LTR for activated transcription.
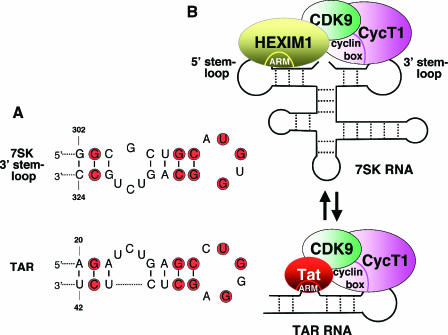
FIG. 4.
Architectural resemblance between the Tat-TAR-P-TEFb and HEXIM1-7SK-P-TEFb ribonucleoprotein complexes. (A) Comparison of nucleotide sequences between the G302-C324 apical region of the 3′ hairpin of 7SK RNA and the bulge-loop region of the HIV-1 TAR RNA (positions +20 to +42). The conserved nucleotides are shaded in red. Reprinted from reference 22 with permission.) (B) The Tat-TAR-P-TEFb and HEXIM1-7SK-P-TEFb complexes display many similar features and may share a common evolutionary origin. In addition to the sequence similarity between the HIV-1 TAR RNA and the 3′ hairpin of the 7SK RNA, both sequences also make direct contacts with CycT1. Moreover, both Tat and HEXIM1 directly interact with P-TEFb through binding to a region near the cyclin box in CycT1. Lastly, both HEXIM1 and Tat utilize a highly homologous and functionally interchangeable arginine-rich motif (ARM) for binding to their respective RNA partners. Given the multiple similarities shared between the two P-TEFb-containing ribonucleoprotein complexes, Tat has been shown to efficiently compete with HEXIM1 for binding to the same P-TEFb molecule. Tat can also disrupt the 7SK snRNP in an effort to convert the inactive P-TEFb into the active Tat/TAR-bound form for stimulating HIV-1 transcription. The secondary structure of 7SK is a modified version based on two previous reports (77, 92). For simplicity, only one copy each of HEXIM1, CDK9, and CycT1 is shown, although two copies of each are believed to interact with one 7SK molecule to form one HEXIM1-7SK-P-TEFb snRNP.
This hypothesis, although yet to be further substantiated through structural analysis of the two P-TEFb complexes, raises an intriguing possibility that the nuclear level of HEXIM1 can be therapeutically manipulated to effectively modulate the amount of P-TEFb in the Tat-TAR-P-TEFb complex, which in turn would affect HIV-1 transcription. In support of this notion, overexpression of HEXIM1 has been shown to potently inhibit HIV-1 transcription both in vitro and in vivo (26, 110). Furthermore, a similar structural assembly of P-TEFb with either Tat/TAR or HEXIM1/7SK suggests that the binding of Tat/TAR might preclude or disrupt the binding of HEXIM1/7SK to the same P-TEFb molecule, thus converting an inactive form of P-TEFb into an active form for Tat transactivation. Preliminary studies have indicated that this may indeed be the case. Tat has been shown to efficiently compete with HEXIM1 for binding to the CycT1 subunit of P-TEFb in vitro (63, 83). In transfected HeLa cells, Tat can also release P-TEFb from the sequestration by HEXIM1 and 7SK via its cysteine-rich activation region, which is known to interact with CycT1 (our unpublished data). Thus, it appears that HIV-1 has evolved a mechanism that can alleviate the negative regulation of P-TEFb by hijacking it from the inactive HEXIM1-7SK-P-TEFb complex for activated HIV-1 transcription.
Homo- and Hetero-Oligomer Formation by HEXIM1 and Its Homologue HEXIM2
Several recent studies have demonstrated that HEXIM1 can form oligomers (most likely dimers) (20, 51, 112) that are mediated by the centrally located 7SK-binding motif and the coiled-coil domain at the C terminus of HEXIM1 (Fig. 3) (2, 6). Although the precise reason for HEXIM1 to do this is not clear, it is suspected that the oligomerization may serve to nucleate the formation of the 7SK snRNP, as the HEXIM1 oligomers can be detected both inside and outside the 7SK complex (112). It has been determined by a number of techniques that the 7SK snRNP most likely contains a single molecule of the 7SK snRNA and at least two copies each of HEXIM1 and P-TEFb (20, 51, 112). However, given the wide range of the estimated molecular sizes of the 7SK snRNP (∼300 to 600 kDa) (67, 112), it is possible that heterogeneous complexes may exist with different copy numbers of HEXIM1 and/or P-TEFb under different conditions.
In addition to HEXIM1, a smaller HEXIM1 homologue, termed HEXIM2, has recently been described (8, 112). HEXIM2 is shorter than HEXIM1 by 73 amino acids. Sequence alignment between the two showed that the area of homology (>50% identical) begins at the N-terminal boundary of the central arginine-rich 7SK-binding/nuclear localization signal domain (amino acids 150 to 177 of HEXIM1) and extends into the C-terminal domain of HEXIM1 to position 343 (8, 112). In contrast, sequences in their N-terminal regions are mostly unrelated. The unique N-terminal half of HEXIM1 has been shown to play a regulatory role, as the deletion of this region results in a mutant HEXIM1 that targets and inactivates P-TEFb with higher efficiency than does the wild-type protein (110). These observations suggest that while the two homologous HEXIM proteins are likely to have similar physiological functions and/or mechanisms of action, they may be regulated differently through their regions of unique sequences. Furthermore, the two HEXIM homologues show distinct expression patterns in different mammalian tissues or cell lines (8, 112). For instance, HEXIM1 was found to be expressed at ∼4- and ∼6-times-higher levels than HEXIM2 in two established human cell lines, HeLa and TK6, respectively (112). Importantly, like HEXIM1, HEXIM2 can also form homo-oligomers. In addition, it can hetero-oligomerize with HEXIM1 within the 7SK snRNP (20, 51, 112). Thus, there are potentially three distinct populations of the 7SK snRNP containing homo-oligomers of HEXIM1 or HEXIM2, or their hetero-oligomers, which explains to some extent the observed heterogeneity of the sizes of the 7SK complexes.
CDK9 with a Phosphorylated T-Loop Is Incorporated into the 7SK snRNP
Although the precise mechanism and signaling pathway that regulate the assembly of the 7SK snRNP are largely unknown at this time, phosphorylation of CDK9 appears to play a critical role in this process. Several lines of evidence support this view. First, the 7SK snRNP is sensitive to conditions that lead to the dephosphorylation of proteins, as the treatment of this complex with phosphatases (such as protein phosphatase 1) causes the dissociation of HEXIM1 and 7SK from P-TEFb in vitro (11). Second, site-directed mutagenesis has revealed that the threonine residue at position 186 (T186) in CDK9 is important for CDK9's association with HEXIM1 and 7SK (11). This residue is phosphorylated as revealed by mass spectrometry (51). Moreover, immunoblotting with a phospho-specific antibody indicates that T186 is indeed phosphorylated when CDK9 is sequestered into the 7SK snRNP (our unpublished data).
The T186 residue, located at the tip of a flexible loop (the so-called T-loop) in CDK9, is absolutely conserved among all members of the CDK superfamily. Its phosphorylation is known to induce a major conformational change of the T-loop that allows the entry of the substrate and ATP into the kinase catalytic pocket (80). When T186 in CDK9 is mutated into Ala or Glu, P-TEFb can no longer interact with HEXIM1 and 7SK (11). Since the phosphorylation of this threonine residue is a hallmark for an activated CDK (64, 80), the 7SK/HEXIM1 regulatory circuit apparently targets active P-TEFb molecules that are poised to carry out kinase reactions and mediate transcription (11). By this means, 7SK and HEXIM1 are able to keep the nuclear level of active P-TEFb in check to prevent excessive transcriptional activity when demand is low. More importantly, this mechanism renders the regulation of P-TEFb activity fast and efficient, as the dissociation of 7SK/HEXIM1 from P-TEFb in response to environmental stimuli (discussed below) can lead to the generation of P-TEFb molecules that are ready to take action without the need to be reactivated through the phosphorylation of the T-loop.
Association of HEXIM1 with Nuclear Hormone Receptors
7SK and P-TEFb are the principal cellular factors associated with HEXIM1 (110), and hence regulating the transcriptional activity of P-TEFb could be a major function of HEXIM1. However, recent reports suggest that HEXIM1 may have other ways to regulate gene expression. For example, HEXIM1 has been shown to modulate the activities of certain nuclear hormone receptors, such as estrogen receptor alpha (ERα) and the glucocorticoid receptor (GR), which function as transcription factors to regulate a diverse set of genes in response to their respective lipid-soluble ligands (98). Both ERα and GR have been independently identified as HEXIM1-associated factors. In the case of GR, it forms a separate complex with HEXIM1 that is distinct from the HEXIM1-7SK-P-TEFb complex (87). Overexpression of HEXIM1 decreases ligand-dependent association between GR and the p160 coactivator transcriptional intermediary factor 2, resulting in suppression of glucocorticoid-responsive gene activation (87). For ERα, it appears that an interaction between ERα and CycT1 is modulated by HEXIM1. An increased HEXIM1 expression prevents the ERα-CycT1 interaction, resulting in inhibition of the recruitment of CycT1 as well as the binding of ERα to the promoter (102). Although more details of the functional consequences of the interactions of HEXIM1 with these two nuclear hormone receptors remain to be investigated, one fact that has become abundantly clear based on the published results is the negative role of HEXIM1 in repressing transactivation mediated by ERα and GR (87, 102). However, since HEXIM1 also inhibits general transcription through targeting P-TEFb, future studies are necessary to distinguish between the inhibitory effects of HEXIM1 on P-TEFb-dependent elongation and the nuclear receptor-mediated transactivation with regard to those GR- and ER-responsive genes.
POSITIVE REGULATION OF P-TEFb ACTIVITY
CDK9 and CycT1 Interact with the Bromodomain Protein Brd4 To Form the Transcriptionally Active P-TEFb Complex
Compared to the case for the population of CDK9-CycT1 heterodimers sequestered in the 7SK snRNP, where the CDK9 kinase activity is suppressed, considerably less had been known about the state of active P-TEFb in vivo, especially whether there may exist an unknown factor(s) that can bind to CDK9-CycT1 and mediate its transcriptional activity. Despite this uncertainty, it is generally presumed that the transcriptionally active P-TEFb exists and functions in the form of a free CDK9-CycT1 heterodimer (69, 74, 75). Two recent reports (39, 108), however, have provided biochemical evidence challenging this conventional view. In these two studies, an interaction of the bromodomain protein Brd4 with a major portion of cellular P-TEFb has been identified. This interaction was shown to be required for P-TEFb to stimulate transcriptional elongation in general.
Brd4, a ubiquitously expressed nuclear protein of ∼200 kDa, belongs to the conserved BET protein family (19), whose members contain two tandem bromodomains and a single extra terminal domain (40). The bromodomain has been recognized as a functional module to help decipher the histone code through interacting with acetylated histone tails (114). Consistent with this view, Brd4 has been shown to bind to actively transcribed chromatin regions through a direct interaction with acetylated histone H3 and H4 (18). Recently, Brd4 was also found to be a component of the Mediator complex (36, 39, 41), which is a multisubunit protein complex responsible for bridging the interactions of sequence-specific transcription activators with the basal transcriptional machinery (13, 54). Another salient feature of Brd4 is that it remains bound to chromatin during mitosis, suggesting a possible role for this protein in transmitting epigenetic memory across cell division (18, 19) (see discussion below).
A surprising link between Brd4 and P-TEFb was revealed when CDK9 and CycT1 were identified as specific Brd4-associated factors during affinity purification (39). In fact, roughly half of nuclear P-TEFb in HeLa cells cofractionates with Brd4 as revealed by density gradient analysis. Furthermore, depletion of Brd4 by small interfering RNA (siRNA) results in the concomitant disappearance of the population of P-TEFb comigrating with Brd4 in the same gradient fractions (39). Importantly, functional analyses have demonstrated that the Brd4-bound P-TEFb represents the transcriptionally active form of P-TEFb that is capable of phosphorylating the Pol II CTD and stimulating elongation in vivo and in vitro (39, 108). Since the other half of nuclear P-TEFb is sequestered in the kinase-inactive 7SK snRNP, there is probably not much free P-TEFb existing off transcriptional templates and in the nucleoplasm (the P-TEFb that is comigrating with the elongating Pol II at a distance from the promoter regions could be free of Brd4 [see below]). This is contrary to the traditional view that the core P-TEFb, consisting of only the CDK9-CycT1 heterodimer, is fully functional as a free entity throughout the transcription cycle (69, 74, 75).
Domain analysis indicates that the two bromodomains of Brd4, which are known to bind to acetylated histones, are also required for the Brd4-P-TEFb interaction in vivo (39). On the P-TEFb side, CycT1, but not CDK9, is able to interact with Brd4 in vitro as a glutathione S-transferase fusion protein (39). However, a serine residue at position 175 (S175) in CDK9, which is located within the flexible T-loop and conserved between CDK9 and CDK7, is also important for the Brd4-P-TEFb interaction in vivo, as the mutation of this residue to Ala (S175A) or Asp (S175D) completely abolishes the interaction (108). Reflecting the fact that Brd4 and HEXIM1/7SK exist in two mutually exclusive P-TEFb complexes, mutation of S175 does not affect the interactions of 7SK and HEXIM1 with P-TEFb (108). It is speculated that the S175A and S175D mutations in the T-loop, which is well known to undergo major conformational changes in a phosphorylation-dependent manner, may induce an altered conformation in the CDK9-CycT1 heterodimer that blocks a direct interaction between Brd4 and CycT1 (108).
Recruitment of P-TEFb for Stimulation of General Transcriptional Elongation by Brd4
Neither CDK9 nor CycT1 is known to have any sequence-specific DNA-binding activity. How, then, is P-TEFb recruited to diverse class II promoters for stimulation of transcriptional elongation in general? For activated transcription from the HIV-1 LTR, the answer is already clear. The recruitment of P-TEFb to the HIV-1 promoter is mediated through the formation of a ternary complex containing Tat, P-TEFb, and the TAR RNA element, a stem-loop structure located at the 5′ end of the nascent viral transcript (Fig. 2) (45). Besides HIV-1 Tat, a group of DNA sequence-specific transcription factors, such as CIITA (43), NF-κB (3), Myc (21, 44), STAT3 (30), the androgen receptor (50), the aryl hydrocarbon receptor (94), MyoD (90), HIC (113), B-Myb (15), GRIP1 (47), and MCEF (23), have also been identified as P-TEFb-associated factors that can potentially recruit P-TEFb to their respective promoter targets. Since these are gene-specific transcription factors, their recruitment of P-TEFb is expected to affect only a limited group of genes that bear their specific recognition sites in the promoter or enhancer regions. However, given that P-TEFb is globally required for the transcription of the vast majority of cellular genes (10, 86), it is conceivable that there should exist a common mechanism to recruit P-TEFb to generic cellular promoters that do not contain any binding sites for the above-mentioned P-TEFb-associated gene-specific transcription factors.
Indeed, using approaches that include chromatin immunoprecipitation, immobilized DNA template-binding assay, and RNAi, it has been demonstrated that Brd4 may well be this long-sought P-TEFb recruitment factor that delivers P-TEFb to generic transcription templates (39, 108). The Brd4-mediated recruitment leads to the stimulation of Ser2 phosphorylation on the Pol II CTD and transcriptional elongation from a variety of promoters in vivo and in vitro (39, 108). As for the target on transcriptional templates for the Brd4-P-TEFb complex, it has been proposed that the recruitment is achieved through Brd4's direct interactions with both acetylated histones and the Mediator complex (39, 108). The latter interaction may explain why the stimulatory effect of Brd4 on elongation was detected not only on chromatin templates in vivo but also on chromatin-free DNA template in vitro (108).
Since the distribution of Brd4 along the entire transcription unit is similar but not completely identical to that of P-TEFb as revealed by the chromatin immunoprecipitation assay (39), it is not clear whether Brd4 is released from P-TEFb or whether it tracks along with P-TEFb and the elongating Pol II once it has recruited P-TEFb to the template. So far, several observations suggest that it is the Brd4-free P-TEFb that is probably involved in the actual elongation process. First, there is no compelling evidence indicating that Brd4 can modulate P-TEFb's catalytic activity once it recruits P-TEFb to the template (reference 108 and our unpublished data). Second, although the Brd4-mediated recruitment of P-TEFb is important for general transcription, for Tat-activated HIV-1 transcription, the P-TEFb recruitment role of Brd4 can be functionally fulfilled by Tat (108), which directly recruits P-TEFb to the HIV-1 LTR through formation of the Tat-TAR-P-TEFb ternary complex (Fig. 2). In addition, overexpression of Brd4 inhibits Tat transactivation, due to the competition between Tat and Brd4 for binding to the same P-TEFb heterodimer (108). These observations suggest that, at least for transcription from the HIV-1 LTR, the Brd4-P-TEFb interaction must be disrupted to allow the released P-TEFb to travel with Pol II along the template. In support of this view, an earlier study has shown that the C terminus of CycT1 can directly interact with the Pol II CTD (93), suggesting that P-TEFb may not require other factors to mediate its interaction with the elongating Pol II, once it has been recruited to transcription templates by Brd4. The observation that the PIE-1 transcriptional repressor of C. elegans can inhibit transcriptional elongation by blocking the binding of CycT1 to the Pol II CTD through an alanine-heptapeptide repeat (115) is consistent with the notion that Brd4 may not be needed for the P-TEFb-CTD binding after the initial recruitment step.
Possible Cooperation between Brd4 and Gene-Specific Transcriptional Activators in Recruitment of P-TEFb
It should be pointed out that although Brd4 recruits P-TEFb for general transcription, it is not necessarily the only or most efficient way to deliver P-TEFb to a particular template. Given that a rather large number of gene-specific transcription activators have been shown to interact with P-TEFb, these interactions could potentially contribute to the P-TEFb recruitment process by supplementing or facilitating the function of Brd4. In fact, it is possible that for any genes whose transcription is preferentially regulated at the elongation stage and thus likely to be strongly P-TEFb dependent, multiple mechanisms are employed to maximize the chance of getting P-TEFb to the promoters. An excellent example to illustrate this point concerns the transcription from the HIV-1 LTR, where P-TEFb could be recruited through different mechanisms depending on when the Tat protein is produced. It is known that with the integrated provirus, the DNA sequence-specific transcriptional activator NF-κB, which binds to two enhancer elements located within the 5′ LTR, is required to get the early transcription going. Due to its ability to bind to P-TEFb (3), NF-κB may function in conjunction with Brd4 to recruit P-TEFb to the HIV-1 promoter. In addition, NF-κB can bring in a histone acetylase and cause histone acetylation (116), which in turn may further facilitate the recruitment of P-TEFb to the viral promoter by Brd4. These events can be envisioned to occur before the HIV-1 Tat protein is produced. After the initial round of transcription is completed and a small amount of Tat is synthesized, Tat clearly takes over and directly recruits P-TEFb to the TAR RNA element (108). This establishes a positive feed-forward loop, and optimal replication of the virus ensues.
A FUNCTIONAL P-TEFb EQUILIBRIUM IS KEY FOR CELLULAR CONTROL OF GROWTH AND DIFFERENTIATION
Dynamic Exchanges between Positive and Negative Regulators in Association with P-TEFb
Global regulation of transcription requires fast responses to ever-changing physiological conditions. The HEXIM1 and 7SK regulatory circuit can temporarily sequester P-TEFb into an inactive complex when transcriptional demand is low. Since CDK9 in the 7SK snRNP is already phosphorylated on its T-loop and thus is ready to perform, the release of P-TEFb and its subsequent association with Brd4 should then allow a nearly instantaneous response to various signals for increased transcription. Thus, the opposing effects of Brd4 and 7SK/HEXIM1 on P-TEFb could theoretically maintain the activity of this general transcription factor in a dynamic equilibrium, which may serve as a key signal integration point whereby diverse physiological stimuli converge and globally affect gene expression. Although the signaling pathway(s) governing the formation/disruption of the 7SK snRNP and the Brd4-P-TEFb complex is still poorly understood, it has indeed been demonstrated that the population of the inactive, HEXIM1/7SK-bound P-TEFb does not remain static in the cell (11, 62, 63, 67, 109, 110). Rather, it can be readily and quantitatively converted into the active, Brd4-bound complexes under a variety of conditions (108).
For example, stressful events that globally interrupt gene expression, particularly transcription, such as exposure of cells to the DNA-damaging agents actinomycin D and UV irradiation and the kinase inhibitors 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole, staurosporine, and 1-(5-isoquinolinesulfonyl)-2-methylpiperazine, can all lead to disruption of the 7SK snRNP and enhanced formation of the Brd4-P-TEFb complex (references 11, 62, 63, 67, and 108 to 110 and our unpublished data). Although these agents usually cause a global inhibition of transcription, earlier observations have indicated that at least for actinomycin D and UV, a low dosage can induce the phosphorylation of the Pol II CTD and activation of HIV-1 transcription (9, 96). Moreover, UV irradiation of human T cells prior to HIV infection also significantly shortens the viral growth cycle (96). The ability of these agents to dissociate HEXIM1 and 7SK and activate P-TEFb provides a mechanistic explanation for their effects on HIV-1 transcription and replication. As for cellular genes, although the activation of P-TEFb could conceivably be important for the expression of many or all stress-induced genes, the induced shift of the P-TEFb equilibrium toward the activated, Brd4-bound state may also simply be an indication of a natural cellular response to those stressful events that suppress transcription and cell growth.
Besides stress responses in HeLa and many other cell types, treatment of cardiac myocytes with conditions that cause cardiac hypertrophy has also been shown to induce the disruption of the 7SK snRNP and activation of P-TEFb (81). Because P-TEFb activity is limiting in normal cardiac myocytes, the activation of P-TEFb by hypertrophic signals can lead to a global increase in cellular RNA and protein contents and consequently the enlargement of heart cells, which is the cause of hypertrophy (81). Thus, the release of HEXIM1/7SK from P-TEFb provides a necessary means to deal with an increased transcriptional demand under these highly proliferative conditions. Consistent with the notion that HEXIM1 has a growth-inhibitory function during normal heart growth and development through its inactivation of P-TEFb, ablation of the HEXIM1 (CLP-1) gene in mice leads to fetal death that is caused by conditions reminiscent of cardiac hypertrophy (37).
In contrast to the above-mentioned stress and hypertrophic signals that cause the disruption of the 7SK snRNP, treatment of murine erythroleukemia cells (MELC) with HMBA can significantly up-regulate the HEXIM1 gene expression and sequester more P-TEFb into the inactive 7SK snRNP (N. He, A. C. Pezda, and Q. Zhou, submitted for publication). Interestingly, the initial response to HMBA is a disruption of the 7SK snRNP, which peaks at around 2 h after the start of the treatment, an effect very similar to that caused by the above-mentioned stress-inducing agents. However, this disruption is transient (lasts for 1 to 2 h), and the long-term effect of HMBA is a permanent shift of the P-TEFb equilibrium toward the inactive HEXIM1/7SK-bound state. The hybrid bipolar compound HMBA is a highly effective (and the best characterized) inducer of terminal differentiation of MELC, which serves as a model for examining the regulation of erythroid differentiation (58). In contrast to the case for MELC, treatment of HeLa cells with HMBA, which causes only a small increase in HEXIM1 expression and a minor, transient growth retardation but not differentiation, produces no long-lasting effect on the P-TEFb equilibrium, and the ratio between the active and inactive P-TEFb complexes remains the same as that prior to the treatment (He et al., submitted).
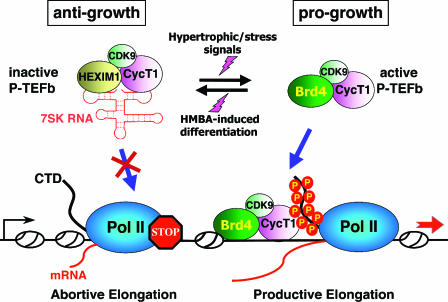
It is not clear at this time whether the observed shift of the P-TEFb equilibrium toward the inactive state in MELC is actually required for the commitment and then establishment of the differentiated state. Nonetheless, the data have so far revealed a highly controlled and dynamic process in which P-TEFb is kept in a delicate balance between two distinctive states: the HEXIM1/7SK-bound inactive state and the Brd4-associated active state ready for elongation (Fig. 5). Perturbations of this equilibrium result in dynamic exchanges of binding partners between the two P-TEFb subpopulations.
FIG. 5.
P-TEFb is maintained in a functional equilibrium by dynamic associations with its positive and negative regulators. In the nucleus, a major portion of P-TEFb is sequestered into the 7SK/HEXIM1 snRNP, where P-TEFb's kinase activity is inhibited by HEXIM1 in a 7SK-dependent manner. Because P-TEFb within the 7SK snRNP is unable to phosphorylate the Pol II CTD or associate with promoters, Pol II goes into the abortive elongation mode. Treatment of HeLa cells with stress-inducing agents or cardiac myocytes with hypertrophic signals can cause a rapid disruption of the 7SK snRNP and quantitative conversion of the released P-TEFb into the Brd4-bound form. This results in the increased recruitment of P-TEFb by Brd4 to transcriptional templates and stimulation of productive elongation by Pol II. On the other hand, when murine erythroleukemia cells are induced to differentiate by the treatment with HMBA, the P-TEFb equilibrium is shifted to the inactive, HEXIM1/7SK-bound state. Thus, the dynamic associations of P-TEFb with its positive and negative regulators are kept under tight cellular control in response to ever-changing transcriptional demand in the cell. Since HEXIM1 is known to display an antigrowth effect in a number of cell types, whereas Brd4 is progrowth during mouse development, their targeting of the general transcription factor P-TEFb is expected to affect the global control of cell growth and differentiation. For simplicity, only a monomer each of P-TEFb and HEXIM1 is depicted in the 7SK snRNP.
In addition to the proliferation of cardiac myocytes and differentiation of erytholeukemia cells mentioned above, P-TEFb has also been implicated in playing a role in regulating the differentiation programs of several other cell types, such as skeletal muscle cells, monocytes, lymphocytes, and neurons (5, 16, 17). However, only the expression of CDK9/CycT1 has been monitored in these studies; therefore, the regulatory effects of Brd4 and 7SK/HEXIM1 on P-TEFb in relation to the differentiation control of these cell types still remain to be examined.
Cells Strive To Maintain a Delicate Balance between the Active and Inactive P-TEFb Subpopulations
Several pieces of evidence have indicated that under normal growth conditions, cells strive hard to maintain a delicate balance between the active and inactive P-TEFb subpopulations, underscoring the importance of maintaining a proper P-TEFb equilibrium for critical cellular functions. Although P-TEFb is the major binding partner and target of HEXIM1 in vivo, only about 20% of total nuclear HEXIM1 is actually in association with P-TEFb in log-phase HeLa cells (63, 112). Surprisingly, when the total cellular level of HEXIM1 is being gradually reduced by RNAi, the population of free HEXIM1 is the first to disappear, whereas the level of HEXIM1 associated with 7SK/P-TEFb is only minimally affected (8; He et al., submitted). In these HEXIM1-depleted cells, there apparently is an effort to compensate for the loss of HEXIM1 by mobilizing virtually all the remaining free HEXIM1 into the 7SK snRNP so as to avoid a major perturbation of the P-TEFb equilibrium. In addition to HEXIM1, the HEXIM1 homologue HEXIM2 is also increasingly recruited into the 7SK snRNP in cells expressing the HEXIM1-specific siRNA (8, 112). Normally, HEXIM1 is the preferred component of the 7SK snRNP in HeLa cells, whereas most of the HEXIM2 exists in free forms not associated with this complex (8, 112). The mobilization of both free HEXIM1 and HEXIM2 into the 7SK snRNP to compensate for the siRNA-mediated HEXIM1 reduction can be explained only by the assumption that the functional P-TEFb equilibrium plays an extremely important role in globally regulating transcription and that its disruption would therefore be detrimental to many key cellular functions.
As mentioned above, in HeLa cells, roughly half of the P-TEFb heterodimers are sequestered into the 7SK snRNP, while the remaining half are associated with Brd4. Although it has not been tested extensively in other cell types, the ratio of these two P-TEFb populations is likely to be different among different cell lines or tissues. Moreover, the P-TEFb distribution in a given cell type may also change according to its growth and differentiation status. For example, in quiescent peripheral blood lymphocytes (PBL), where transcriptional demand is low, the level of functional P-TEFb is kept very low through the suppression of CycT1 expression (33). Under these conditions, the expression of HEXIM1 is also very low (34), as there is no excessive P-TEFb activity that must be suppressed at this moment. However, upon the activation of PBL, the level of CycT1 is rapidly induced, which results in an accumulation of functional P-TEFb. At the same time, the levels of HEIXM1 and 7SK are also up-regulated to allow formation of the 7SK snRNP (33). The simultaneous up-regulation of 7SK and HEXIM1 as the P-TEFb level increases in activated PBL suggests that once a fine balance between supply and demand for active P-TEFb is established in the cell, the P-TEFb activity is immediately kept under tight control by the opposing effects of 7SK/HEXIM1 and Brd4 to avoid excessive transcriptional activity.
Targeting the Functional P-TEFb Equilibrium for the Global Control of Cell Growth and Differentiation
What could be the reason for cells striving to maintain a delicate balance between the active and inactive P-TEFb populations? The growth-regulatory functions of the P-TEFb-associated factors very likely hold the key to this important question. As a negative regulator of P-TEFb activity, HEXIM1's expression is elevated in cells that are induced to differentiate by HMBA (72), suggesting that this protein plays an important role during the critical cellular decision to transit from proliferative growth to terminal differentiation. Consistent with this view, an antigrowth function of HEXIM1 has also been demonstrated in cardiac myocytes, where the absence of HEXIM1 causes the enlargement of heart cells in a pathological condition known as hypertrophy (37). In breast epithelial cells, HEXIM1 has also been recognized as an inhibitor of cell proliferation, as its expression is down-regulated by estrogens and decreased in breast tumors (103). Finally, a recent report indicates that ectopic expression of HEXIM1 causes growth inhibition and promotes neuronal differentiation (95).
In contrast to HEXIM1, Brd4, the positive regulator and recruitment factor for P-TEFb, has been implicated in playing a growth-stimulatory role. While mice lacking both alleles of the Brd4 gene are lethal to embryos, Brd4-heterozygotic mice display pre- and postnatal growth defects associated with a reduced proliferation rate (36). In primary cell cultures, heterozygous cells also display reduced proliferation rates. In addition, Brd4 is required for the proper progression of the cell cycle, as heterozygotes display a significantly reduced number of mitotic cells compared to that in wild-type tissues (36). Because of Brd4's ability to remain bound to chromosomes during mitosis, when both specific and general transcription factors are released from chromatin (19), the Brd4-P-TEFb interaction may help recruit P-TEFb to chromatin templates at the end of mitosis and beginning of G1, when transcription restarts. This may allow Brd4 to play a key role in transmitting transcriptional memory from one generation of cells to the next (18, 53).
The opposing effects on cellular growth exerted by Brd4 and HEXIM1, both of which target P-TEFb but produce antagonizing results, support the idea that controlling the activity of the general transcription factor P-TEFb, which affects the expression of a vast array of genes, is central to the global regulation of cell growth and differentiation. With this in mind, it is not difficult to understand that for normal cell growth, a well-controlled equilibrium must be maintained between the two P-TEFb subpopulations, so that cells would not accidentally slip into either an overproliferative state or growth arrest/terminal differentiation.
CONCLUSIONS AND PERSPECTIVES
The elucidation of a functional P-TEFb equilibrium maintained by dynamic and opposing actions of its positive and negative regulators holds important implications for the control of HIV-1 gene expression and the cellular decision between growth and differentiation. The exciting discoveries described in this review suggest numerous important future experiments. For example, structure-function analyses of the 7SK-HEXIM1-P-TEFb snRNP by methods such as X-ray crystallography or cryoelectron microscopy will determine the precise mechanism by which HEXIM1 functions as a CKI to inhibit CDK9. Different from the traditional CKIs that are important for cell cycle control, HEXIM1 requires an RNA cofactor, 7SK, for its inhibitory action. In addition to its demonstrated role in mediating the HEXIM1-P-TEFb interaction, future structural studies will reveal whether 7SK also directly participates in the inhibition of CDK9 kinase activity. Another area worth exploring is the unraveling of the signaling pathways that can shift the P-TEFb equilibrium toward either the active or inactive state in response to different physiological stimuli. Studies of these pathways will not only help elucidate the mechanisms governing the fundamental cellular processes of growth versus differentiation but also facilitate the understanding of diverse disease processes ranging from cardiac hypertrophy to breast cancer to HIV infection.
On the Brd4 front, it will be interesting to determine whether this bromodomain protein may bring P-TEFb to the core of cell cycle control. Unlike the case for many other members of the CDK superfamily, the function of P-TEFb has been thought to be rather independent of the cell cycle progression. For example, CDK9 is known to be constitutively expressed throughout the cell cycle. Moreover, the kinase activity associated with the isolated CDK9-CycT1 heterodimer (Brd4-free P-TEFb) also remains unchanged as the cell cycle progresses (29, 89). However, in light of the recently revealed interaction of P-TEFb with Brd4, which is implicated in playing an important role in cell cycle progression, the activity of P-TEFb may actually be regulated throughout the cell cycle and be important for the cell cycle-dependent regulation of transcription. Compared to the mature and much broader field of eukaryotic transcriptional control or even the relatively young subfield of transcriptional elongation, the study of P-TEFb regulation is a focused area that is still in early infancy and will undoubtedly yield many more exciting findings in the future.
Acknowledgments
This work is supported by grants from the National Institutes of Health (AI41757) and the American Cancer Society (RSG-01-171-01-MBC) to Q. Zhou and by a Ruth L. Kirschstein NRSA postdoctoral fellowship from the National Institutes of Health (AI058400) to J. Yik.
REFERENCES
- 1.Ahn, S. H., M. Kim, and S. Buratowski. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 13:67-76. [DOI] [PubMed] [Google Scholar]
- 2.Barboric, M., J. Kohoutek, J. P. Price, D. Blazek, D. H. Price, and B. M. Peterlin. 2005. Interplay between 7SK snRNA and oppositely charged regions in HEXIM1 direct the inhibition of P-TEFb. EMBO J. 24:4291-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barboric, M., R. M. Nissen, S. Kanazawa, N. Jabrane-Ferrat, and B. M. Peterlin. 2001. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 8:327-337. [DOI] [PubMed] [Google Scholar]
- 4.Barboric, M., and B. M. Peterlin. 2005. A new paradigm in eukaryotic biology: HIV Tat and the control of transcriptional elongation. PLoS Biol. 3:e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellan, C., G. De Falco, S. Lazzi, P. Micheli, S. Vicidomini, K. Schurfeld, T. Amato, A. Palumbo, L. Bagella, E. Sabattini, S. Bartolommei, M. Hummel, S. Pileri, P. Tosi, L. Leoncini, and A. Giordano. 2004. CDK9/CYCLIN T1 expression during normal lymphoid differentiation and malignant transformation. J. Pathol. 203:946-952. [DOI] [PubMed] [Google Scholar]
- 6.Blazek, D., M. Barboric, J. Kohoutek, I. Oven, and B. M. Peterlin. 2005. Oligomerization of HEXIM1 via 7SK snRNA and coiled-coil region directs the inhibition of P-TEFb. Nucleic Acids Res. 33:7000-7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bres, V., N. Gomes, L. Pickle, and K. A. Jones. 2005. A human splicing factor, SKIP, associates with P-TEFb and enhances transcription elongation by HIV-1 Tat. Genes Dev. 19:1211-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byers, S. A., J. P. Price, J. J. Cooper, Q. Li, and D. H. Price. 2005. HEXIM2, a HEXIM1-related protein, regulates positive transcription elongation factor b through association with 7SK. J. Biol. Chem. 280:16360-16367. [DOI] [PubMed] [Google Scholar]
- 9.Casse, C., F. Giannoni, V. T. Nguyen, M.-F. Dubois, and O. Bensaude. 1999. The transcriptional inhibitors, actinomycin D and alpha-amanitin, activate the HIV-1 promoter and favor phosphorylation of the RNA polymerase II C-terminal domain. J. Biol. Chem. 274:16097-16106. [DOI] [PubMed] [Google Scholar]
- 10.Chao, S.-H., and D. H. Price. 2001. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J. Biol. Chem. 276:31793-31799. [DOI] [PubMed] [Google Scholar]
- 11.Chen, R., Z. Yang, and Q. Zhou. 2004. Phosphorylated positive transcription elongation factor b (P-TEFb) is tagged for inhibition through association with 7SK snRNA. J. Biol. Chem. 279:4153-4160. [DOI] [PubMed] [Google Scholar]
- 12.Cho, H., T. K. Kim, H. Mancebo, W. S. Lane, O. Flores, and D. Reinberg. 1999. A protein phosphatase functions to recycle RNA polymerase II. Genes Dev. 13:1540-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conaway, R. C., S. Sato, C. Tomomori-Sato, T. Yao, and J. W. Conaway. 2005. The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem. Sci. 30:250-255. [DOI] [PubMed] [Google Scholar]
- 14.Dahmus, M. E. 1996. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J. Biol. Chem. 271:19009-19012. [DOI] [PubMed] [Google Scholar]
- 15.De Falco, G., L. Bagella, P. P. Claudio, A. De Luca, Y. Fu, B. Calabretta, A. Sala, and A. Giordano. 2000. Physical interaction between CDK9 and B-Myb results in suppression of B-Myb gene autoregulation. Oncogene 19:373-379. [DOI] [PubMed] [Google Scholar]
- 16.De Falco, G., C. Bellan, A. D'Amuri, G. Angeloni, E. Leucci, A. Giordano, and L. Leoncini. 2005. Cdk9 regulates neural differentiation and its expression correlates with the differentiation grade of neuroblastoma and PNET tumors. Cancer Biol. Ther. 4:277-281. [DOI] [PubMed] [Google Scholar]
- 17.De Falco, G., and A. Giordano. 2002. CDK9: from basal transcription to cancer and AIDS. Cancer Biol. Ther. 1:342-347. [PubMed] [Google Scholar]
- 18.Dey, A., F. Chitsaz, A. Abbasi, T. Misteli, and K. Ozato. 2003. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. USA 100:8758-8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dey, A., J. Ellenberg, A. Farina, A. E. Coleman, T. Maruyama, S. Sciortino, J. Lippincott-Schwartz, and K. Ozato. 2000. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G2-to-M transition. Mol. Cell. Biol. 20:6537-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dulac, C., A. A. Michels, A. Fraldi, F. Bonnet, V. T. Nguyen, G. Napolitano, L. Lania, and O. Bensaude. 2005. Transcription-dependent association of multiple positive transcription elongation factor units to a HEXIM multimer. J. Biol. Chem. 280:30619-30629. [DOI] [PubMed] [Google Scholar]
- 21.Eberhardy, S. R., and P. J. Farnham. 2001. c-Myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. J. Biol. Chem. 276:48562-48571. [DOI] [PubMed] [Google Scholar]
- 22.Egloff, S., E. Van Herreweghe, and T. Kiss. 2006. Regulation of polymerase II transcription by 7SK snRNA: two distinct RNA elements direct P-TEFb and HEXIM1 binding. Mol. Cell. Biol. 26:630-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Estable, M. C., M. H. Naghavi, H. Kato, H. Xiao, J. Qin, A. Vahlne, and R. G. Roeder. 2002. MCEF, the newest member of the AF4 family of transcription factors involved in leukemia, is a positive transcription elongation factor-b-associated protein. J. Biomed. Sci. 9:234-245. [DOI] [PubMed] [Google Scholar]
- 24.Fong, Y. W., and Q. Zhou. 2000. Relief of two built-in autoinhibitory mechanisms in P-TEFb is required for assembly of a multicomponent transcription elongation complex at the human immunodeficiency virus type 1 promoter. Mol. Cell. Biol. 20:5897-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fong, Y. W., and Q. Zhou. 2001. Stimulatory effect of splicing factors on transcriptional elongation. Nature (London) 414:929-933. [DOI] [PubMed] [Google Scholar]
- 26.Fraldi, A., F. Varrone, G. Napolitano, A. A. Michels, B. Majello, O. Bensaude, and L. Lania. 2005. Inhibition of Tat activity by the HEXIM1 protein. Retrovirology 2:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu, T.-J., J. Peng, G. Lee, D. H. Price, and O. Flores. 1999. Cyclin K functions as a CDK9 regulatory subunit and participates in RNA polymerase II transcription. J. Biol. Chem. 274:34527-34530. [DOI] [PubMed] [Google Scholar]
- 28.Fujinaga, K., D. Irwin, Y. Huang, R. Taube, T. Kurosu, and B. M. Peterlin. 2004. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol. Cell. Biol. 24:787-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garriga, J., S. Bhattacharya, J. Calbo, R. M. Marshall, M. Truongcao, D. S. Haines, and X. Grana. 2003. CDK9 is constitutively expressed throughout the cell cycle, and its steady-state expression is independent of SKP2. Mol. Cell. Biol. 23:5165-5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giraud, S., A. Hurlstone, S. Avril, and O. Coqueret. 2004. Implication of BRG1 and cdk9 in the STAT3-mediated activation of the p21waf1 gene. Oncogene 23:7391-7398. [DOI] [PubMed] [Google Scholar]
- 31.Gold, M. O., X. Yang, C. H. Herrmann, and A. P. Rice. 1998. PITALRE, the catalytic subunit of TAK, is required for human immunodeficiency virus Tat transactivation in vivo. J. Virol. 72:4448-4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gomes, N. P., G. Bjerke, B. Llorente, S. A. Szostek, B. M. Emerson, and J. M. Espinosa. 2006. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 20:601-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haaland, R. E., C. H. Herrmann, and A. P. Rice. 2003. Increased association of 7SK snRNA with Tat cofactor P-TEFb following activation of peripheral blood lymphocytes. AIDS 17:2429-2436. [DOI] [PubMed] [Google Scholar]
- 34.Haaland, R. E., C. H. Herrmann, and A. P. Rice. 2005. siRNA depletion of 7SK snRNA induces apoptosis but does not affect expression of the HIV-1 LTR or P-TEFb-dependent cellular genes. J. Cell Physiol. 205:463-470. [DOI] [PubMed] [Google Scholar]
- 35.Reference deleted.
- 36.Houzelstein, D., S. L. Bullock, D. E. Lynch, E. F. Grigorieva, V. A. Wilson, and R. S. Beddington. 2002. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol. Cell. Biol. 22:3794-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang, F., M. Wagner, and M. A. Siddiqui. 2004. Ablation of the CLP-1 gene leads to down-regulation of the HAND1 gene and abnormality of the left ventricle of the heart and fetal death. Mech. Dev. 121:559-572. [DOI] [PubMed] [Google Scholar]
- 38.Ivanov, D., Y. T. Kwak, J. Guo, and R. B. Gaynor. 2000. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol. Cell. Biol. 20:2970-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jang, M. K., K. Mochizuki, M. Zhou, H. S. Jeong, J. N. Brady, and K. Ozato. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19:523-534. [DOI] [PubMed] [Google Scholar]
- 40.Jeanmougin, F., J. M. Wurtz, B. Le Douarin, P. Chambon, and R. Losson. 1997. The bromodomain revisited. Trends Biochem. Sci. 22:151-153. [DOI] [PubMed] [Google Scholar]
- 41.Jiang, Y. W., P. Veschambre, H. Erdjument-Bromage, P. Tempst, J. W. Conaway, R. C. Conaway, and R. D. Kornberg. 1998. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc. Natl. Acad. Sci. USA 95:8538-8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones, K. A. 1997. Taking a new TAK on tat transactivation. Genes Dev. 11:2593-2599. [DOI] [PubMed] [Google Scholar]
- 43.Kanazawa, S., T. Okamoto, and B. M. Peterlin. 2000. Tat competes with CIITA for the binding to P-TEFb and blocks the expression of MHC class II genes in HIV infection. Immunity 12:61-70. [DOI] [PubMed] [Google Scholar]
- 44.Kanazawa, S., L. Soucek, G. Evan, T. Okamoto, and B. M. Peterlin. 2003. c-Myc recruits P-TEFb for transcription, cellular proliferation and apoptosis. Oncogene 22:5707-5711. [DOI] [PubMed] [Google Scholar]
- 45.Karn, J. 1999. Tackling Tat. J. Mol. Biol. 293:235-254. [DOI] [PubMed] [Google Scholar]
- 46.Kim, J. B., Y. Yamaguchi, T. Wada, H. Handa, and P. A. Sharp. 1999. Tat-SF1 protein associates with RAP30 and human SPT5 proteins. Mol. Cell. Biol. 19:5960-5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kino, T., O. Slobodskaya, G. N. Pavlakis, and G. P. Chrousos. 2002. Nuclear receptor coactivator p160 proteins enhance the HIV-1 long terminal repeat promoter by bridging promoter-bound factors and the Tat-P-TEFb complex. J. Biol. Chem. 277:2396-2405. [DOI] [PubMed] [Google Scholar]
- 48.Kobor, M. S., and J. Greenblatt. 2002. Regulation of transcription elongation by phosphorylation. Biochim. Biophys. Acta 1577:261-275. [DOI] [PubMed] [Google Scholar]
- 49.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee, D. K., H. O. Duan, and C. Chang. 2001. Androgen receptor interacts with the positive elongation factor P-TEFb and enhances the efficiency of transcriptional elongation. J. Biol. Chem. 276:9978-9984. [DOI] [PubMed] [Google Scholar]
- 51.Li, Q., J. P. Price, S. A. Byers, D. Cheng, J. Peng, and D. H. Price. 2005. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J. Biol. Chem. 280:28819-28826. [DOI] [PubMed] [Google Scholar]
- 52.Li, X. Y., and M. R. Green. 1998. The HIV-1 Tat cellular coactivator Tat-SF1 is a general transcription elongation factor. Genes Dev. 12:2992-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loyola, A., and G. Almouzni. 2004. Bromodomains in living cells participate in deciphering the histone code. Trends Cell Biol. 14:279-281. [DOI] [PubMed] [Google Scholar]
- 54.Malik, S., and R. G. Roeder. 2005. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 30:256-263. [DOI] [PubMed] [Google Scholar]
- 55.Mancebo, H. S., G. Lee, J. Flygare, J. Tomassini, P. Luu, Y. Zhu, J. Peng, C. Blau, D. Hazuda, D. Price, and O. Flores. 1997. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 11:2633-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mandal, S. S., H. Cho, S. Kim, K. Cabane, and D. Reinberg. 2002. FCP1, a phosphatase specific for the heptapeptide repeat of the largest subunit of RNA polymerase II, stimulates transcription elongation. Mol. Cell. Biol. 22:7543-7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marks, P. A., V. M. Richon, H. Kiyokawa, and R. A. Rifkind. 1994. Inducing differentiation of transformed cells with hybrid polar compounds: a cell cycle-dependent process. Proc. Natl. Acad. Sci. USA 91:10251-10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marks, P. A., and R. A. Rifkind. 1989. Induced differentiation of erythroleukemia cells by hexamethylene bisacetamide: a model for cytodifferentiation of transformed cells. Environ. Health Perspect. 80:181-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McCracken, S., N. Fong, E. Rosonina, K. Yankulov, G. Brothers, D. Siderovski, A. Hessel, S. Foster, S. Shuman, and D. L. Bentley. 1997. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 11:3306-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCracken, S., N. Fong, K. Yankulov, S. Ballantyne, G. Pan, J. Greenblatt, S. D. Patterson, M. Wickens, and D. L. Bentley. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385:357-361. [DOI] [PubMed] [Google Scholar]
- 61.Medlin, J., A. Scurry, A. Taylor, F. Zhang, B. M. Peterlin, and S. Murphy. 2005. P-TEFb is not an essential elongation factor for the intronless human U2 snRNA and histone H2b genes. EMBO J. 24:4154-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Michels, A. A., A. Fraldi, Q. Li, T. E. Adamson, F. Bonnet, V. T. Nguyen, S. C. Sedore, J. P. Price, D. H. Price, L. Lania, and O. Bensaude. 2004. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 23:2608-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Michels, A. A., V. T. Nguyen, A. Fraldi, V. Labas, M. Edwards, F. Bonnet, L. Lania, and O. Bensaude. 2003. MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol. Cell. Biol. 23:4859-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morgan, D. O. 1995. Principles of CDK regulation. Nature 374:131-134. [DOI] [PubMed] [Google Scholar]
- 65.Murphy, S., C. Di Liegro, and M. Melli. 1987. The in vitro transcription of the 7SK RNA gene by RNA polymerase III is dependent only on the presence of an upstream promoter. Cell 51:81-87. [DOI] [PubMed] [Google Scholar]
- 66.Neubauer, G., A. King, J. Rappsilber, C. Calvio, M. Watson, P. Ajuh, J. Sleeman, A. Lamond, and M. Mann. 1998. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat. Genet. 20:46-50. [DOI] [PubMed] [Google Scholar]
- 67.Nguyen, V. T., T. Kiss, A. A. Michels, and O. Bensaude. 2001. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414:322-325. [DOI] [PubMed] [Google Scholar]
- 68.Ni, Z., B. E. Schwartz, J. Werner, J. R. Suarez, and J. T. Lis. 2004. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol. Cell 13:55-65. [DOI] [PubMed] [Google Scholar]
- 69.O'Keeffe, B., Y. Fong, D. Chen, S. Zhou, and Q. Zhou. 2000. Requirement for a kinase-specific chaperone pathway in the production of a Cdk9/cyclin T1 heterodimer responsible for P-TEFb-mediated Tat stimulation of HIV-1 transcription. J. Biol. Chem. 275:279-287. [DOI] [PubMed] [Google Scholar]
- 70.Orphanides, G., T. Lagrange, and D. Reinberg. 1996. The general transcription factors of RNA polymerase II. Genes Dev. 10:2657-2683. [DOI] [PubMed] [Google Scholar]
- 71.Orphanides, G., and D. Reinberg. 2002. A unified theory of gene expression. Cell 108:439-451. [DOI] [PubMed] [Google Scholar]
- 72.Ouchida, R., M. Kusuhara, N. Shimizu, T. Hisada, Y. Makino, C. Morimoto, H. Handa, F. Ohsuzu, and H. Tanaka. 2003. Suppression of NF-kappaB-dependent gene expression by a hexamethylene bisacetamide-inducible protein HEXIM1 in human vascular smooth muscle cells. Genes Cells 8:95-107. [DOI] [PubMed] [Google Scholar]
- 73.Parada, C. A., and R. G. Roeder. 1999. A novel RNA polymerase II-containing complex potentiates Tat-enhanced HIV-1 transcription. EMBO J. 18:3688-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peng, J., N. F. Marshall, and D. H. Price. 1998. Identification of a cyclin subunit required for the function of Drosophila P-TEFb. J. Biol. Chem. 273:13855-13860. [DOI] [PubMed] [Google Scholar]
- 75.Peng, J., Y. Zhu, J. T. Milton, and D. H. Price. 1998. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 12:755-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ping, Y. H., and T. M. Rana. 1999. Tat-associated kinase (P-TEFb): a component of transcription preinitiation and elongation complexes. J. Biol. Chem. 274:7399-7404. [DOI] [PubMed] [Google Scholar]
- 77.Reddy, R., D. Henning, C. S. Subrahmanyam, and H. Busch. 1984. Primary and secondary structure of 7-3 (K) RNA of Novikoff hepatoma. J. Biol. Chem. 259:12265-12270. [PubMed] [Google Scholar]
- 78.Renner, D. B., Y. Yamaguchi, T. Wada, H. Handa, and D. H. Price. 2001. A highly purified RNA polymerase II elongation control system. J. Biol. Chem. 276:42601-42609. [DOI] [PubMed] [Google Scholar]
- 79.Rodriguez, C. R., E. J. Cho, M. C. Keogh, C. L. Moore, A. L. Greenleaf, and S. Buratowski. 2000. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol. Cell. Biol. 20:104-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Russo, A. A., P. D. Jeffrey, and N. P. Pavletich. 1996. Structural basis of cyclin-dependent kinase activation by phosphorylation. Nat. Struct. Biol. 3:696-700. [DOI] [PubMed] [Google Scholar]
- 81.Sano, M., M. Abdellatif, H. Oh, M. Xie, L. Bagella, A. Giordano, L. H. Michael, F. J. DeMayo, and M. D. Schneider. 2002. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat. Med. 8:1310-1317. [DOI] [PubMed] [Google Scholar]
- 82.Schroeder, S. C., B. Schwer, S. Shuman, and D. Bentley. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schulte, A., N. Czudnochowski, M. Barboric, A. Schonichen, D. Blazek, B. M. Peterlin, and M. Geyer. 2005. Identification of a cyclin T-binding domain in Hexim1 and biochemical analysis of its binding competition with HIV-1 Tat. J. Biol. Chem. 280:24968-24977. [DOI] [PubMed] [Google Scholar]
- 84.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501-1512. [DOI] [PubMed] [Google Scholar]
- 85.Shilatifard, A. 2004. Transcriptional elongation control by RNA polymerase II: a new frontier. Biochim. Biophys. Acta 1677:79-86. [DOI] [PubMed] [Google Scholar]
- 86.Shim, E. Y., A. K. Walker, Y. Shi, and T. K. Blackwell. 2002. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev. 16:2135-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shimizu, N., R. Ouchida, N. Yoshikawa, T. Hisada, H. Watanabe, K. Okamoto, M. Kusuhara, H. Handa, C. Morimoto, and H. Tanaka. 2005. HEXIM1 forms a transcriptionally abortive complex with glucocorticoid receptor without involving 7SK RNA and positive transcription elongation factor b. Proc. Natl. Acad. Sci. USA 102:8555-8560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shore, S. M., S. A. Byers, W. Maury, and D. H. Price. 2003. Identification of a novel isoform of Cdk9. Gene 307:175-182. [DOI] [PubMed] [Google Scholar]
- 89.Simone, C., and A. Giordano. 2001. New insight in cdk9 function: from Tat to MyoD. Front. Biosci. 6:D1073-D1082. [DOI] [PubMed] [Google Scholar]
- 90.Simone, C., P. Stiegler, L. Bagella, B. Pucci, C. Bellan, G. De Falco, A. De Luca, G. Guanti, P. L. Puri, and A. Giordano. 2002. Activation of MyoD-dependent transcription by cdk9/cyclin T2. Oncogene 21:4137-4148. [DOI] [PubMed] [Google Scholar]
- 91.Sims, R. J., III, R. Belotserkovskaya, and D. Reinberg. 2004. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 18:2437-2468. [DOI] [PubMed] [Google Scholar]
- 92.Suh, D., Y. Yuan, D. Henning, and R. Reddy. 1989. Secondary structure of 7SK and 7-2 small RNAs. Possible origin of some 7SK pseudogenes from cDNA formed through self-priming by 7SK RNA. Eur. J. Biochem. 186:221-226. [DOI] [PubMed] [Google Scholar]
- 93.Taube, R., X. Lin, D. Irwin, K. Fujinaga, and B. M. Peterlin. 2002. Interaction between P-TEFb and the C-terminal domain of RNA polymerase II activates transcriptional elongation from sites upstream or downstream of target genes. Mol. Cell. Biol. 22:321-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tian, Y., S. Ke, M. Chen, and T. Sheng. 2003. Interactions between the aryl hydrocarbon receptor and P-TEFb. Sequential recruitment of transcription factors and differential phosphorylation of C-terminal domain of RNA polymerase II at cyp1a1 promoter. J. Biol. Chem. 278:44041-44048. [DOI] [PubMed] [Google Scholar]
- 95.Turano, M., G. Napolitano, C. Dulac, B. Majello, O. Bensaude, and L. Lania. 2006. Increased HEXIM1 expression during erythroleukemia and neuroblastoma cell differentiation. J. Cell Physiol. 206:603-610. [DOI] [PubMed] [Google Scholar]
- 96.Valerie, K., A. Delers, C. Bruck, C. Thiriart, H. Rosenberg, C. Debouck, and M. Rosenberg. 1988. Activation of human immunodeficiency virus type 1 by DNA damage in human cells. Nature 333:78-81. [DOI] [PubMed] [Google Scholar]
- 97.Wada, T., T. Takagi, Y. Yamaguchi, D. Watanabe, and H. Handa. 1998. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17:7395-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Watson, C. S., and B. Gametchu. 2001. Membrane estrogen and glucocorticoid receptors—implications for hormonal control of immune function and autoimmunity. Int. Immunopharmacol. 1:1049-1063. [DOI] [PubMed] [Google Scholar]
- 99.Wei, P., M. E. Garber, S. M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451-462. [DOI] [PubMed] [Google Scholar]
- 100.Weiss, M. A., and N. Narayana. 1998. RNA recognition by arginine-rich peptide motifs. Biopolymers 48:167-180. [DOI] [PubMed] [Google Scholar]
- 101.West, M. L., and J. L. Corden. 1995. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics 140:1223-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wittmann, B. M., K. Fujinaga, H. Deng, N. Ogba, and M. M. Montano. 2005. The breast cell growth inhibitor, estrogen down regulated gene 1, modulates a novel functional interaction between estrogen receptor alpha and transcriptional elongation factor cyclin T1. Oncogene 24:5576-5588. [DOI] [PubMed] [Google Scholar]
- 103.Wittmann, B. M., N. Wang, and M. M. Montano. 2003. Identification of a novel inhibitor of breast cell growth that is down-regulated by estrogens and decreased in breast tumors. Cancer Res. 63:5151-5158. [PubMed] [Google Scholar]
- 104.Yamada, T., Y. Yamaguchi, N. Inukai, S. Okamoto, T. Mura, and H. Handa. 2006. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell 21:227-237. [DOI] [PubMed] [Google Scholar]
- 105.Yamaguchi, Y., N. Inukai, T. Narita, T. Wada, and H. Handa. 2002. Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol. Cell. Biol. 22:2918-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41-51. [DOI] [PubMed] [Google Scholar]
- 107.Yan, D., R. Perriman, H. Igel, K. J. Howe, M. Neville, and M. Ares, Jr. 1998. CUS2, a yeast homolog of human Tat-SF1, rescues function of misfolded U2 through an unusual RNA recognition motif. Mol. Cell. Biol. 18:5000-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yang, Z., J. H. Yik, R. Chen, N. He, M. K. Jang, K. Ozato, and Q. Zhou. 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 19:535-545. [DOI] [PubMed] [Google Scholar]
- 109.Yang, Z., Q. Zhu, K. Luo, and Q. Zhou. 2001. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414:317-322. [DOI] [PubMed] [Google Scholar]
- 110.Yik, J. H., R. Chen, R. Nishimura, J. L. Jennings, A. J. Link, and Q. Zhou. 2003. Inhibition of P-TEFb (CDK9/cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell 12:971-982. [DOI] [PubMed] [Google Scholar]
- 111.Yik, J. H., R. Chen, A. C. Pezda, C. S. Samford, and Q. Zhou. 2004. A human immunodeficiency virus type 1 Tat-like arginine-rich RNA-binding domain is essential for HEXIM1 to inhibit RNA polymerase II transcription through 7SK snRNA-mediated inactivation of P-TEFb. Mol. Cell. Biol. 24:5094-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yik, J. H., R. Chen, A. C. Pezda, and Q. Zhou. 2005. Compensatory contributions of HEXIM1 and HEXIM2 in maintaining the balance of active and inactive positive transcription elongation factor b complexes for control of transcription. J. Biol. Chem. 280:16368-16376. [DOI] [PubMed] [Google Scholar]
- 113.Young, T. M., Q. Wang, T. Pe'ery, and M. B. Mathews. 2003. The human I-mfa domain-containing protein, HIC, interacts with cyclin T1 and modulates P-TEFb-dependent transcription. Mol. Cell. Biol. 23:6373-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zeng, L., and M. M. Zhou. 2002. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 513:124-128. [DOI] [PubMed] [Google Scholar]
- 115.Zhang, F., M. Barboric, T. K. Blackwell, and B. M. Peterlin. 2003. A model of repression: CTD analogs and PIE-1 inhibit transcriptional elongation by P-TEFb. Genes Dev. 17:748-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhong, H., M. J. May, E. Jimi, and S. Ghosh. 2002. The phosphorylation status of nuclear NF-kappa B determines its association with CBP/p300 or HDAC-1. Mol. Cell 9:625-636. [DOI] [PubMed] [Google Scholar]
- 117.Zhou, M., M. A. Halanski, M. F. Radonovich, F. Kashanchi, J. Peng, D. H. Price, and J. N. Brady. 2000. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 20:5077-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhou, M., F. Kashanchi, H. Jiang, H. Ge, and J. N. Brady. 2000. Phosphorylation of the RAP74 subunit of TFIIF correlates with Tat-activated transcription of the HIV-1 long terminal repeat. Virology 268:452-460. [DOI] [PubMed] [Google Scholar]
- 119.Zhou, Q., D. Chen, E. Pierstorff, and K. Luo. 1998. Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J. 17:3681-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhou, Q., and P. A. Sharp. 1996. Tat-SF1: cofactor for stimulation of transcriptional elongation by HIV-1 Tat. Science 274:605-610. [DOI] [PubMed] [Google Scholar]
- 121.Zieve, G., B. J. Benecke, and S. Penman. 1977. Synthesis of two classes of small RNA species in vivo and in vitro. Biochemistry 16:4520-4525. [DOI] [PubMed] [Google Scholar]