Abstract
Numerous transcription factors recruit C-terminal binding protein (CtBP) corepressors. We show that the large zinc finger protein ZNF217 contacts CtBP. ZNF217 is encoded by an oncogene frequently amplified in tumors. ZNF217 contains a typical Pro-X-Asp-Leu-Ser (PXDLS) motif that binds in CtBP's PXDLS-binding cleft. However, ZNF217 also contains a second motif, Arg-Arg-Thr (RRT), that binds a separate surface on CtBP. The crystal structure of CtBP bound to an RRTGAPPAL peptide shows that it contacts a surface crevice distinct from the PXDLS binding cleft. Interestingly, both PXDLS and RRT motifs are also found in other zinc finger proteins, such as RIZ. Finally, we show that ZNF217 represses several promoters, including one from a known CtBP target gene, and mutations preventing ZNF217's contact with CtBP reduce repression. These results identify a new CtBP interaction motif and establish ZNF217 as a transcriptional repressor protein that functions, at least in part, by associating with CtBP.
The C-terminal binding proteins (CtBPs) are multifunctional proteins implicated in gene regulation, Golgi maintenance, and synaptic ribbon formation (3, 7, 41, 43). They function in gene regulation as transcriptional corepressors. CtBPs interact with the repression domains of sequence-specific DNA-binding proteins (transcription factors) and recruit a repressor complex that contains histone modifying enzymes, such as histone deacetylases 1 and 2, the histone methyltransferase G9a, and the histone demethylase LSD1 (38-40). Approximately 30 transcription factors that recruit CtBP to gene regulatory elements have been identified. These transcription factors come from diverse families and include proteins with the zinc finger, homeodomain, Ets, and Sox types of DNA-binding domains. They are united, however, by the fact that they typically contain a Pro-X-Asp-Leu-Ser (PXDLS) or related motif in their repression domains through which they contact CtBP (3, 43).
Crystallographic studies have shown that CtBP is composed of a nucleotide-binding domain that exhibits homology to dehydrogenase enzymes and includes an extensive dimerization interface and NADH binding motif, and a substrate-binding domain, formed by the N terminus and part of the C-terminal region (22, 30). The X-ray crystal structure shows that the substrate-binding domain forms CtBP's PXDLS-peptide binding cleft (30). In addition, CtBP contains 80 C-terminal residues recently shown to be intrinsically unstructured (31).
Although the mechanism through which CtBP is recruited by PXDLS partners is well understood, the other CtBP protein contacts remain to be characterized. In an effort to identify other important contact sites on CtBP, we constructed a CtBP protein with a “filled” PXDLS cleft. This protein was generated from a fusion gene encoding the well-characterized PXDLS motif found in the transcription factor basic Krüppel-like factor (BKLF/KLF3) (42) linked to the 3′ end of the murine CtBP2 gene. The resulting fusion protein thereby contains a C-terminal tail carrying a PXDLS motif, and since the C terminus of CtBP is flexible and is structurally located near CtBP's PXDLS-binding cleft, we expect this tail to be able to fill the cleft. Indeed, we have found that the linked PXDLS tail does block the binding of additional PXDLS motif partners (data not shown). Importantly, a similar fusion protein, incorporating a point mutation in the PXDLS sequence, does not interfere with the binding of exogenous PXDLS motif partners, arguing against the possibility that the fusion tail is nonspecifically impeding access to the PXDLS binding cleft (data not shown).
We used this fusion protein in yeast two-hybrid screens and identified murine Znf217 as a protein partner of CtBP2 that does not depend on the PXDLS cleft for association. Murine Znf217 has not previously been described, but based on homology (see Fig. 1) and synteny (10), it appears to be the orthologue of human ZNF217, a recognized oncogene implicated in numerous cancers, most notably breast and colon cancers (47). The human ZNF217 gene resides on the long arm of chromosome 20 at position q13.2 (5). This region is amplified in up to 40% of breast and 60% of colon cancers (35, 47). The amplification has been shown to correlate with increased ZNF217 protein and a poor prognosis. Furthermore, it has been found that overexpression of ZNF217 promotes the immortalization of breast epithelial cells (32), although the precise mechanism through which ZNF217 drives immortalization is not known. Interestingly, human ZNF217 has been found to be present in a number of repression complexes (14, 24, 48), including the CtBP-associated repression complex (39). However, the mechanism through which ZNF217 functions remains unknown.
FIG. 1.
The human and murine ZNF217 protein sequences show significant homology. The sequences of full-length human ZNF217 (NM_006526) and murine Znf217 (NM_001033299) proteins are shown. The zinc finger regions (1 to 8) are underlined with solid gray lines. The conserved PXDLS motifs and RRT motifs are underlined with gray dashed lines. Residues 530 (lysine) and 932 (glycine) of mZnf217, the first and last amino acids of the yeast two-hybrid screen isolate, are indicated with asterisks. Identical and similar residues are boxed in black and gray, respectively. Dashes indicate gaps introduced to maintain alignment.
Here we show that murine and human ZNF217 directly interact with CtBP. We find that ZNF217 contains a PXDLS motif that binds the CtBP cleft but also contains a motif that binds elsewhere on CtBP. We map this second motif within ZNF217 to the sequence RRTGCPPAL. Cocrystallization of CtBP with an RRTGAPPAL peptide reveals the location of the second peptide binding site on CtBP.
We demonstrate that ZNF217 represses transcription driven by a number of promoters, and mutations that prevent it from contacting CtBP impair its ability to repress transcription. This suggests that ZNF217 functions in gene repression by recruiting CtBP and its associated repression complex.
Our results further indicate that other zinc finger proteins, such as RIZ and ZNF516, which also contain a PXDLS and the novel RRT motif, may play direct roles in gene repression through contacting CtBP. We suggest that overexpression of ZNF217 may contribute to tumorigenesis through initiating changes in gene expression profiles.
MATERIALS AND METHODS
Plasmid constructs.
Full-length murine CtBP2 (mCtBP2) was amplified by PCR, and the product was cloned into an XmaI/SalI pGBT9 (Clontech) vector. The resulting construct, pGBT9-mCtBP2, was used as wild-type CtBP2 in the yeast two-hybrid experiments described throughout this report. Secondly, murine BKLF 30-75, containing the 61PVDLT65 motif, was amplified by PCR and cloned into the NotI/SalI sites of pGBT9-mCtBP2. The resulting yeast two-hybrid constructs expressed BKLF 30-75 fused to the C terminus of CtBP2. The pGBT9-mCtBP2-BKLF 30-75 construct is referred to as “cleft-filled” CtBP2 throughout this report. The “control ΔDL fusion,” which contains a DL-to-AS mutation in the PVDLT motif in the BKLF 30-75 portion of the fusion, was generated from the pGBT9-mCtBP2-BKLF 30-75 construct by overlap PCR mutagenesis. The mCtBP2, mCtBP2-BKLF 30-75, and mCtBP2-BKLF 30-75 ΔDL inserts were subcloned into the XmaI/SalI sites of the pGAD10(new) (derived from pGAD10 from Clontech) vector to allow them to be expressed in the yeast two-hybrid system as both Gal4 activation domain (Gal4AD) and Gal4-DNA-binding domain (Gal4DBD) fusions.
The A58E, V72R, E181A, and D237A mutations were introduced into mCtBP2 by overlap PCR mutagenesis. BglII/SalI-digested mutant inserts were ligated into the BamHI/SalI sites of the pGBT9 and pGAD10(new) vectors to generate pGBT9-mCtBP2-A58E, pGBT9-mCtBP2-V72R, pGAD10(new)-mCtBP2-A58E, pGAD10(new)-mCtBP2-V72R, pGBT9-mCtBP2-E181A/D237A, and pGBT9-mCtBP2 A58E/E181A/D237A.
Gal4DBD (147 amino acids) was amplified by PCR using appropriate primers and was ligated into the PstI/NotI sites of the pMT3 (derived from pMT2) vector to generate pMT3-Gal4 without a stop codon. A separate pMT3-Gal4 construct with a stop codon was generated to act as a control in mammalian repression assays. Secondly, wild-type mCtBP2, mCtBP2-A58E, mCtBP2-E181A/D237A, and mCtBP2-A58E/E181A/D237A mutant inserts were reamplified by PCR using appropriate primers and cloned into the NotI/SalI sites of pMT3-Gal4 without stop, 3′ of the Gal4 gene, to generate pMT3-Gal4-mCtBP2, pMT3-Gal4-mCtBP2-A58E, pMT3-Gal4-mCtBP2-E181A/D237A, and pMT3-Gal4-mCtBP2-A58E/E181A/D237A.
pMT3-YFP was generated by ligating a NsiI/NotI yellow fluorescent protein (YFP) PCR fragment from the pEYFP-C1 vector (Clontech) into PstI/NotI sites of the pMT3 vector (derived from pMT2). mCtBP2, mCtBP2-A58E, mCtBP2-E181A/D237A, and mCtBP2-A58E/E181A/D237A were reamplified by PCR using appropriate primers and cloned into NotI/SalI sites of pMT3-YFP. pMT2-HA-mCtBP2 has been previously described (44).
pGAD10-mZnf217 530-932 was isolated from a murine erythroleukemia cell (MEL) cDNA library with the pGBT9-mCtBP2-BKLF 30-75 bait protein. The NL→AS mutation (ΔDL) was introduced into the putative PXDLS motif, 680PLNLS684, in the pGAD10-mZnf217 530-932 construct using overlap PCR site-directed mutagenesis. The mZnf217 530-932 and mZnf217 530-932 ΔDL inserts were liberated from the pGAD10 vector by digestion with BamHI and BglII and were ligated into the BamHI site of the pGBT9(new) vector.
Regions of mZnf217 corresponding to amino acids 548 to 617, 660 to 715, 753 to 794, 869 to 911, and 686 to 737 were amplified from the pGAD10-mZnf217 530-932 template. Regions of mZnf217 corresponding to amino acids 660 to 715 ΔDL, 548 to 715 ΔDL, 753 to 911, 660 to 794 ΔDL, 548 to 794 ΔDL, 660 to 911 ΔDL, 548 to 911 ΔDL, 548 to 775 ΔDL, 548 to 750 ΔDL, 548 to 725 ΔDL, 700 to 911, 725 to 911, and 730 to 760 were amplified from the pGAD10-mZnf217 ΔDL template. PCR products were cloned into the XmaI/BamHI sites of the pGBT9 vector to allow expression of Gal4DBD fusions in yeast. The region of mZnf217 encoding amino acids 700 to 790 was amplified from the pGAD10-mZnf217 530-932 template, and the PCR product was cloned into the BamHI/PstI sites of the pGBT9 vector.
Triple and single alanine scanning mutations (shown in Fig. 2D) were introduced into pGBT9-mZnf217 700-790 using overlap PCR mutagenesis.
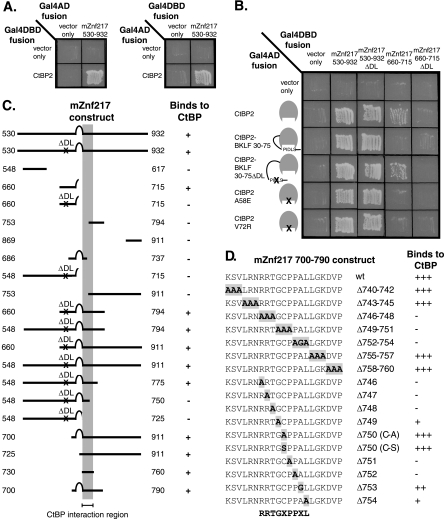
FIG. 2.
Znf217 interacts with CtBP2, and the non-PXDLS interaction site was mapped to the motif RRTGXPPXL. A. Yeast two-hybrid assays were performed to examine the interactions between mZnf217 530-932 and CtBP2. These assays were performed with each of the two test proteins fused to the C terminus of either Gal4AD or Gal4DBD. Growth on -His-Leu-Trp plates (pictured) indicates that the two test proteins interact. B. The PXDLS motifs in Znf217 530-932 and Znf217 660-715 were mutated, and both wild-type and mutant proteins were tested for binding to wild-type CtBP and CtBP with mutations in the PXDLS cleft. Interaction with only wild-type CtBP (and the BKLF 30-75 ΔDL control protein) indicates binding which is dependent on a PXDLS motif. Interaction with both wild-type and mutant CtBPs indicates binding which is not dependent on a PXDLS motif. C. Deletion mapping was performed to determine the minimal portion of murine Znf217 capable of interacting with CtBP in a PXDLS motif-independent manner. The Gal4DBD-Znf217 proteins are depicted schematically, and results of yeast two-hybrid assays with these proteins and Gal4AD-CtBP are shown as either plus (for growth of yeast) or minus. A ΔDL mutation (NL-AS in the PLNLS motif) was introduced into many of the mZnf217 proteins so that only non-PXDLS binding was being examined, and this is indicated. The minimal region of mZnf217 required for interaction with CtBP, amino acids 730 to 760, is indicated by a gray column. D. Both single and triple mutations were introduced into amino acids 740 to 760 of Gal4DBD-Znf217 700-790. The mutations in each of the constructs are highlighted within the sequence of Znf217 amino acids 740 to 760. The results of yeast two-hybrid assays with these mutant Gal4DBD-mZnf217 proteins and Gal4AD-CtBP are shown as either pluses (for relative growth of yeast) or minus. The consensus motif suggested, RRTGXPPXL, is shown below.
Full-length human ZNF217 (hZNF217) (1 to 1048) was amplified by PCR from pLXSN-ZNF217 (32) and cloned into the EcoRI site of the pGAD10 vector to produce pGAD10-hZNF217 1-1048. An NL→AS mutation (ΔDL) was introduced into the 686PLNLS690 motif of pGAD10-hZNF217 1-1048 using overlap PCR mutagenesis to generate pGAD10-hZNF217 1-1048 ΔDL. An RRT→AAA mutation (ΔRRT) was introduced into the 752RRTGCPPAL760 motif of pGAD10-hZNF217 1-1048 and pGAD10-hZNF217 1-1048 ΔDL by overlap PCR mutagenesis to produce pGAD10-hZNF217 1-1048 ΔRRT and pGAD10-hZNF217 1-1048 ΔDL ΔRRT.
hZNF217 1-1048, 1-1048 ΔDL, 1-1048 ΔRRT, and 1-1048 ΔDL ΔRRT inserts were subcloned into the EcoRI site of the pMT3-FLAGb vector to generate hZNF217 expression constructs with the FLAG sequence fused to the N terminus.
Segments of hRIZ1 (amino acids 661 to 820, containing 735RRTSSPPSS743), mRiz1 (amino acids 772 to 931, containing 858RRTSSPPSS866), and hZNF516 (amino acids 2381 to 2580, containing 2442GRTGPPPAL2450) were amplified from human genomic DNA, murine genomic DNA, the K562 (human) cDNA library, and the MEL (murine) cDNA library by PCR and were cloned into the EcoRI/BamHI sites of the pGBT9 vector. RRT/GRT→AAA mutations were introduced into the putative RRT motifs of hRIZ1 661-820, mRiz1 772-931, and hZNF516 2381-2580 using overlap PCR mutagenesis to generate pGBT9-hRIZ1 661-820 ΔRRT, pGBT9-mRiz1 772-931 ΔRRT, and pGBT9-hZNF516 ΔGRT.
Details of primers used in plasmid construction are available on request. The identities of the inserts in each construct were confirmed by automated DNA sequencing.
The firefly luciferase reporter vector pGL2-(Gal4)5-(LexA)2-E1B-Luc and the LexA-VP16 mammalian expression plasmid pCMV-LexA (1-202)-VP16 (410-490) were generous gifts from Luke Gaudreau and Mark Ptashne (The Sloan-Kettering Institute, New York, NY). A second firefly luciferase reporter vector containing five Gal4 binding sites and the thymidine kinase (TK) promoter, pGL2-(Gal4)5-TK-Luc, has been described previously (33). The pGL3-human E-cadherin (−427/+53)-luciferase reporter vector was a gift from Stephen Sugrue (Department of Anatomy and Cell Biology, Harvard Medical School) and has been previously described (1).
Yeast two-hybrid screen and assays.
Yeast two-hybrid screens were performed with pGBT9-mCtBP2 BKLF 30-75 as bait and MEL and human K562 cell cDNA libraries as described previously (42). For yeast two-hybrid assays, test proteins were expressed in yeast strain HF7c as either Gal4DBD or Gal4AD fusions. Transformant colonies were selected on Leu/Trp-deficient plates and patched onto His/Leu/Trp-deficient plates. Growth was scored following 72 h of incubation.
Mammalian cell culture.
COS-1 and HEK293 cells were cultured as described previously (1, 34) and transfected, using the transfection reagent FuGENE6 (Roche Diagnostics), following the manufacturer's instructions. CtBP1+/− CtBP2+/− (CtBP+/−) and CtBP1−/− CtBP2−/− (CtBP−/−) cells were a gift from J. Hildebrand and were cultured and transfected as described previously (16).
Coimmunoprecipitation experiments.
To examine interactions between mCtBP2 and hZNF217 mutants, duplicate 100-mm plates of COS-1 cells were transfected with combinations of 1 μg of pMT2-HA-mCtBP2 and 3 μg of pMT3-FLAGb-hZNF217 wild type, ΔDL mutant, ΔRRT mutant, and double ΔDL ΔRRT mutant DNA. Forty-eight hours following transfection, cells were harvested, duplicates were pooled, and whole-cell protein extracts were prepared (in 50 mM Tris, pH 8.0, 150 mM NaCl, 1% NP-40, 0.2 mM phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, and 1 μg/ml leupeptin; total volume, 500 μl). For the input lanes, 20 μl of each extract (10% of the amount used in immunoprecipitation) was mixed with sodium dodecyl sulfate (SDS) loading dye, boiled, and run on an 8% SDS-polyacrylamide gel electrophoresis (PAGE) gel. Immunoprecipitation was performed with 200 μl of each extract, 10 μl of protein G beads, and 7.5 μg of either mouse monoclonal antihemagglutinin (anti-HA) (12CA5; Roche Corporation) or mouse monoclonal anti-FLAG (Sigma) antibody (Ab) to immunoprecipitate HA-mCtBP2 or FLAG-hZNF217, respectively. Following washes, beads were mixed with SDS loading dye, boiled, and run on 8% SDS-PAGE gels. Proteins in SDS-PAGE gels were blotted onto nitrocellulose membranes (Western blot) and immunodetected with 10 μg of both anti-HA Ab and anti-FLAG Ab in 10 ml Tris-buffered saline-Tween 20 to detect HA-mCtBP2 and FLAG-hZNF217, respectively. A sheep antimouse horseradish peroxidase-conjugated secondary Ab (Amersham Bioscience) was used, and bands were detected with Western Lightning Chemiluminescence Reagent Plus (Perkin Elmer Life Sciences) and X-ray film (Eastman Kodak Company). The exposures show the results of a representative experiment.
To examine interactions between hZNF217 and mCtBP2 mutants, 100-mm petri dishes of COS-1 cells were transiently transfected with combinations of 3 μg pMT3-FLAGb-hZNF217 and 250 ng of either pMT3-YFP-mCtBP2, pMT3-YFP-mCtBP2 A58E, pMT3-YFP-mCtBP2 E181A/D237A, or pMT3-YFP-mCtBP2 A58E/E181A/D237A. Protein extracts were prepared and immunoprecipitation and Western blotting were performed as described in the above coimmunoprecipitation methods except that immunoprecipitations were conducted with 10 μg of anti-HA antibody only and Western blots were immunodetected using anti-HA and monoclonal mouse anti-YFP (BD Living Colors, JL-8; Clontech) antibodies.
Western blots for assessment of protein expression levels.
Western blots were performed to confirm equivalent expression of the Gal4-mCtBP2 and FLAG-hZNF217 proteins. One-hundred-millimeter petri dishes of COS-1 cells were transiently transfected with 4 μg pMT3 alone, pMT3-Gal4-mCtBP2, pMT3-Gal4-mCtBP2 A58E, pMT3-Gal4-mCtBP2 E181A/D237A, or pMT3-Gal4-mCtBP2 A58E/E181A/D237A or 3 μg pMT3 alone, pMT3-FLAGb-hZNF217, or pMT3-FLAGb-hZNF217 ΔDL ΔRRT. Cells were incubated for 48 h following the transfection before cells were harvested and nuclear extracts prepared. Equal amounts of each nuclear extract were run on a 12% SDS-PAGE gel, and Western blotting was performed as described above. Gal4-mCtBP2 was visualized using a mouse monoclonal anti-CtBP2 antibody (BD Biosciences). FLAG-hZNF217 was visualized using a mouse monoclonal anti-FLAG antibody.
Mammalian cell repression assays.
To examine CtBP repression of reporter gene expression, six-well plates of COS-1 cells or CtBP+/− and CtBP−/− cells were transiently transfected. To examine repression of basal expression, the following plasmids were used: 3 μg pGL2-(Gal4)5-TK-Luc reporter and 50 ng of either pMT3-Gal4-mCtBP2, pMT3-Gal4-mCtBP2-A58E, pMT3-Gal4-mCtBP2-E181A/D237A, or pMT3-Gal4-mCtBP2-A58E/E181A/D237A. To examine repression of activated expression, the following plasmids were used: 3 μg pGL2-(Gal4)5-(LexA)2-E1B-Luc reporter, 1 μg pCMV-LexA (1-202)-VP16 (410-490) expression vector, and 50 ng of either pMT3-Gal4-mCtBP2, pMT3-Gal4-mCtBP2-A58E, pMT3-Gal4-mCtBP2-E181A/D237A, or pMT3-Gal4-mCtBP2-A58E/E181A/D237A. In both experiments, 10 ng of the Renilla (R) luciferase vector pRL-Luc (Promega) was cotransfected to allow the firefly (FF) luciferase measurements to be corrected to control for transfection efficiency. Luciferase activity was measured 48 h posttransfection in a Turner Designs model TD 20/20 luminometer using the dual-luciferase reporter assay system (Promega). Results shown are averaged FF/R luciferase ratios from four replicates of a representative experiment.
To examine ZNF217 repression of reporter gene expression, six-well plates of COS-1 cells were transiently transfected. To examine repression of basal expression, the following plasmids were used: 3 μg pGL2-(Gal4)5-TK-Luc reporter and 150 ng of either pMT3-FLAGb-hZNF217 or pMT3-FLAGb-hZNF217 ΔDL ΔRRT. To examine repression of activated expression, the following plasmids were used: 3 μg pGL2-(Gal4)5-(LexA)2-E1B-Luc reporter, 1 μg pCMV-LexA (1-202)-VP16 (410-490) expression vector, and 150 ng of either pMT3-FLAGb-hZNF217 or pMT3-FLAGb-hZNF217 ΔDL ΔRRT. FF luciferase activity was measured as described above. Results shown are averaged FF luciferase ratios from four replicates of a representative experiment.
To examine ZNF217 repression of the E-cadherin promoter, six-well plates of HEK293 cells or CtBP+/− and CtBP−/− cells were transiently transfected with 1 μg of pGL3-E-cad-Luc, 1 μg of various pMT3-FLAG-hZNF217 wild-type or mutant derivatives, and 10 ng of pRL-Luc. Forty-eight hours posttransfection, FF and R luciferase activities were quantified as described above. Results shown are averaged FF/R luciferase ratios from two replicates of a representative experiment.
Crystallization, structure determination, and refinement.
t-CtBP1-S, bearing a His tag at its N terminus, was expressed in Escherichia coli and purified as described previously (29). Vapor diffusion cocrystallization experiments on the protein-peptide complex were performed after overnight incubation of t-CtBP1-S (at a 10-mg/ml concentration) with 10 mM RRTGAPPAL peptide. Bipyramidally shaped crystals of the t-CtBP1-S/peptide complex grew in a few days using a crystallization solution containing 1.8 to 2.1 M ammonium formate, 100 mM HEPES, pH 7.5. The crystals belong to the space group P6422, with the following unit cell parameters: a = b = 89.3 Å; c = 162.7 Å, one molecule per asymmetric unit. A full diffraction data set was collected at a 2.85-Å resolution using synchrotron radiation (ID14-EH3 beamline; ESRF, Grenoble, France). All diffraction data were processed using MOSFLM and SCALA (9, 25) (see Table 1).
TABLE 1.
Data collection and refinement statistics
| Parameter | Value(s) for t-CtBP1-S: NAD(H)/RRTGAPPAL |
|---|---|
| Data collection statisticsa | |
| Space group | P6422 |
| Unit cell dimensions (Å) | a = b = 89.3; c = 162.7 |
| Resolution (Å) | 2.85 |
| Completeness (%) | 99.8 (100) |
| Multiplicity | 9.3 (9.5) |
| Rmergeb(%) | 9.4 (39.9) |
| I/σ(I) | 21.1 (4.6) |
| Refinement statistics and model quality | |
| Rfactor (%) | 22.7 |
| Rfreec (%) | 27.5 |
| No. of residues | 331 + 9 (peptide) |
| No. of waters | 19 |
| No. of formate anions | 1 |
| rmsdd bond lengthse (Å) | 0.006 |
| rmsd bond angles (°) | 0.95 |
Values in parentheses are for the highest-resolution shell.
Rmerge = ΣhΣi |Ihi − < Ih >|/ΣhΣi Ihi.
Rfree estimation is based on 10% of data withheld for cross-validation.
rmsd, root mean square deviation.
The quality of the final model was assessed using the program PROCHECK (23).
The RRTGAPPAL peptide was prepared on an Applied Biosystems model 433A synthesizer according to standard 9-fluorenylmethoxycarbonyl solid-phase synthesis. After purification by preparative reverse-phase high-performance liquid chromatography, it was shown to be >95% homogeneous by analytical reverse-phase high-performance liquid chromatography. Its identity and molecular weight were confirmed by electrospray ionization mass spectrometry (Finnigan LCQ Advantage) (m/z, found, 938.3; calculated for C40H71N15O11, 938.097).
The structure of the t-CtBP1-S/peptide complex was determined by molecular replacement using the program MolRep (4, 45). The crystal structure of t-CtBP1-S (Protein Data Bank entry code 1HKU) (30) was used as a search model. The structure was then refined using the program REFMAC (28) (rigid body and restrained refinement). After a few cycles of refinement, 2Fo-Fc electron density maps showed structural details that allowed unambiguous modeling of the peptide, with the exception of the C-terminal A-L residues, for which poor density was available. As in the case of the t-CtBP1-S structure (30), a NAD(H) molecule was found, likely the result of specific uploading during t-CtBP1-S expression/purification (30), tightly bound at the nucleotide-binding domain. The final model contains 331 t-CtBP1-S residues (15 to 345), 19 water molecules, 1 formate molecule, 1 NAD(H), and 1 RRTGAPPAL peptide molecule (Rfactor = 22.7% and Rfree = 27.5%, respectively), with ideal stereochemical parameters (see Table 1) (8, 23).
Protein structure accession numbers.
Coordinates and structure factors have been deposited with the Protein Data Bank (2) with accession codes 2HU2 and r2HU2sf, respectively.
RESULTS
Identification of Znf217 as a CtBP partner protein.
The CtBP2/BKLF fusion protein, containing residues 30 to 75 of murine BKLF (encompassing its well-characterized PVDLT CtBP contact site) extending from the CtBP2 C terminus, was used as a bait in yeast two-hybrid screens. Several positive clones were isolated, including previously reported CtBP partners, Ubc9 (18, 26), HIPK2 (49), and the related protein HIPK1. One clone encoding residues 530 to 932 of murine Znf217 was recovered. This isolate was tested for its ability to interact with normal full-length CtBP2 in yeast two-hybrid assays as both prey and bait (see Fig. 2A). Yeast growth was observed in both experiments, suggesting that Znf217 is a direct binding partner of CtBP2.
Defining the contact regions in Znf217.
We next mapped the domains of Znf217 that contact CtBP2. Znf217 contains eight classical zinc fingers. The original cDNA fragment we recovered encodes amino acids 530 to 932 and includes zinc finger 7. Inspection of this fragment revealed that it also contained the motif PLNLS just upstream of zinc finger 7. The PLNLS sequence fits the general consensus for PXDLS motifs (3, 43) and is conserved in human ZNF217 (Fig. 1).
First, experiments were carried out to confirm that this PLNLS motif was functional and could slot into the CtBP PXDLS peptide binding cleft. Residues 660 to 715 of Znf217 were amplified and tested for their ability to bind CtBP2 in the yeast two-hybrid assay system. PLNLS was mutated to PLASS, since the substitution of the central two residues, often DL (but here NL) and referred to as the ΔDL mutation, is known to disrupt binding to the CtBP PXDLS peptide binding cleft (36, 37). In addition, CtBP derivatives that contain defective PXDLS peptide binding clefts were also tested. Two previously described mutations in the cleft, A58E and V72R (30), as well as the “cleft-filled” mutant, were also tested for their ability to bind the Znf217 PXDLS motif (Fig. 2B). The fragment containing the PLNLS motif was able to interact with wild-type CtBP2, the mutation in this motif prevented binding, and the CtBP2 derivatives with defective clefts could not bind this fragment. In summary, Znf217 contains a functional PXDLS motif, as shown in Fig. 1.
Our screen was designed to identify CtBP partners that did not rely on PXDLS motifs for associating with CtBP. To determine if Znf217 did require the PXDLS motif for binding to CtBP, its PLNLS motif was mutated in the context of a longer fragment of Znf217, residues 530 to 932. We found that this Znf217 fragment retained the ability to interact with CtBP (Fig. 2B). This fragment also retained the ability to bind to the A58E and V72R CtBP cleft mutants and the “cleft-filled” derivative in both orientations in yeast (only one orientation is shown). This result confirmed our expectation that Znf217 was a partner protein that did not rely solely on the CtBP PXDLS peptide binding cleft for contact and suggested that Znf217 contained a second CtBP contact motif.
We next used deletion analyses with Znf217 fragments containing a mutated PLNLS motif to define the second contact motif (Fig. 2C) and localized it to the region downstream of zinc finger 7. Further alanine scanning experiments demonstrate that the second contact surface in Znf217 comprises the motif RRTGCPPAL (Fig. 2D). We term this an RRT motif.
We next carried out experiments with full-length human ZNF217 and full-length CtBP2 to verify the interaction. Full-length ZNF217 mutants with defective PXDLS (ΔDL) or RRT (RRTGCPPAL→AAAGCPPAL; ΔRRT) motifs were generated, as well as a double mutant that contained mutations in both motifs. These three mutants were first tested in the yeast two-hybrid assay system. As expected, wild-type full-length ZNF217 interacted with wild type CtBP2. Additionally, both the single mutants retained the ability to interact. However, the double mutant showed very little CtBP binding (Fig. 3A). This result suggests that the PXDLS and RRT motifs in ZNF217 are the major determinants through which it contacts CtBP2.
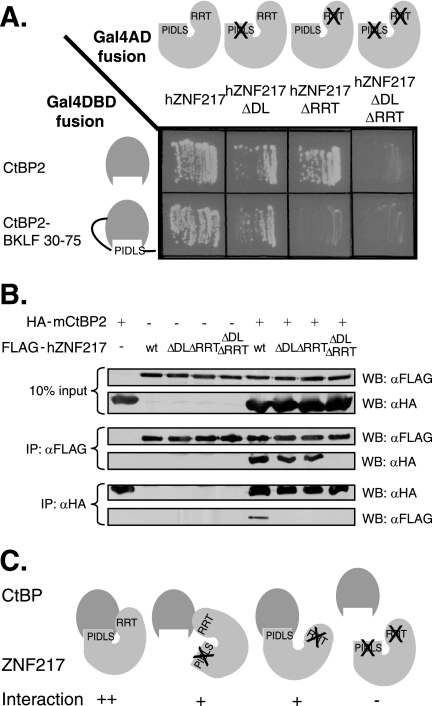
FIG. 3.
Mutation of the PXDLS and RRT motifs of ZNF217 reduce the ability to bind to CtBP, and the double mutant has a severe reduction in binding. A. Gal4AD-fused wild-type hZNF217 or hZNF217 with mutations in the PLNLS motif (ΔDL), RRTGCPPAL motif (ΔRRT), or both motifs (ΔDL ΔRRT) was examined for its ability to interact with Gal4DBD-fused wild-type or cleft-filled (CtBP2-BKLF 30-75) CtBP in yeast two-hybrid assays. B. Fusion of FLAG and wild-type hZNF217 or hZNF217 with mutations in the PLNLS motif (ΔDL), RRTGCPPAL motif (ΔRRT), or both motifs (ΔDL ΔRRT) was examined for its ability to interact with HA-CtBP2 in coimmunoprecipitation experiments. COS-1 cells were transfected with the expression vectors indicated, and whole-cell extracts were immunoprecipitated (IP) separately with both the anti-FLAG (αFLAG) and anti-HA (αHA) antibodies. Expression of each of the FLAG-fused and HA-fused proteins is shown in the top two panels (10% input). FLAG-ZNF217 immunoprecipitated by the anti-FLAG antibody and the resulting coimmunoprecipitated HA-CtBP2 is shown in the middle two panels (IP: αFLAG). HA-CtBP2 immunoprecipitated by the anti-HA antibody and the resulting coimmunoprecipitated FLAG-hZNF217 is shown in the bottom two panels (IP: αHA). C. A summary diagram combining the results of interaction studies between CtBP and wild-type or mutant ZNF217.
We then sought to test whether the protein interactions also occurred in the context of mammalian cells. Epitope-tagged FLAG-ZNF217 and HA-CtBP2 were transfected into COS-1 cells, and their interaction was monitored using immunoprecipitation. As shown in Fig. 3B, when ZNF217 was recovered with Flag antibody, CtBP2 was efficiently retained, as revealed by Western blotting against HA. The PXDLS and RRT ZNF217 mutants and the double mutants were also tested. Both the single ZNF217 mutants bound some CtBP2 (though slightly less than the wild type), but the double mutant did not associate with detectable CtBP2. The converse experiment (immunoprecipitating with anti-HA and Western blotting with anti-Flag) was also carried out with similar results, except that in this orientation the reduction in binding brought about by the single mutations was more striking, possibly because the immunoprecipitation or detection of associated proteins by Western blotting was somewhat less efficient in this orientation. Nevertheless, these results confirm the inferences from the yeast two-hybrid assays that full-length CtBP2 associates with full-length ZNF217 and that the two motifs, the PXDLS and the RRT motifs, are primarily responsible for the association (Fig. 3C).
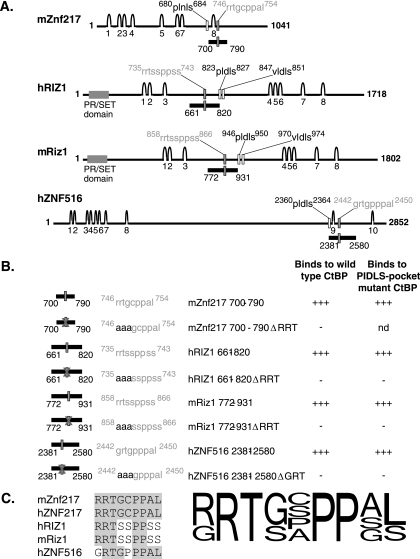
RRT motifs occur in several CtBP partner proteins.
We searched protein databases to determine whether RRT motifs occur in other proteins. Similar motifs were identified in ZNF516 and RIZ. Both proteins also contained recognizable PXDLS motifs within their sequences (Fig. 4A). We did not identify proteins that contained clear RRT motifs in the absence of the PXDLS motif. Although little is known about ZNF516, it is notable that it was found to copurify in the repression complex that associates with CtBP in HeLa cells (reported under the name KIA0222) (39). RIZ is a well-studied eight-zinc-finger protein that contains a PR/SET domain and has been reported to possess histone methyltransferase activity (17). It contains two PXDLS motifs and has previously been inferred to be a CtBP partner, although the sites and functional effects of CtBP contact have not been described (15). RIZ also contains two potential RRT motifs. These motifs are conserved in the human and murine forms of RIZ.
FIG. 4.
RRT motifs are also found in RIZ and ZNF516 and are capable of mediating binding to CtBP. A. mZnf217, hRIZ1, mRiz1, and hZNF516 are large zinc finger proteins which possess both PXDLS motifs and putative RRT motifs. The features of each protein are shown. The predicted zinc fingers are shown as arches and numbered, the consensus PXDLS motifs are indicated by hollow rectangles with the motifs outlined above in black, and the putative RRT motifs are indicated by gray filled rectangles with the motifs outlined above in gray. The PR/SET domains in the RIZ proteins are indicated by wide gray rectangles. The portion of each protein containing the putative RRT motif which was tested for interaction with CtBP is indicated below each sequence as a black bar with the numbers of the flanking amino acids indicated. B. Segments of ZNF516 and both murine and human RIZ1 with and without mutations in the putative RRT motifs fused to Gal4DBD were tested for their ability to bind to wild-type and cleft mutant CtBP fused to Gal4AD in yeast two-hybrid assays. nd, not determined. C. An alignment is shown of the RRT motifs that have been shown to mediate binding to CtBP. Amino acids within the sequences which are identical to the amino acids in the mZnf217 RRT motif are boxed in gray. The consensus combines information obtained from validated natural RRT motifs and also from mutagenesis studies. The height of each amino acid at each position is representative of the relative frequency in the naturally occurring RRT motif proteins and the tolerance for various amino acids as determined by mutational analysis.
In order to test whether the RRT motifs that had been identified by bioinformatics screening were able to physically interact with CtBP, segments of RIZ and ZNF516 were tested for binding to CtBP2 using the yeast two-hybrid system. It was found that the ZNF516 motif and one (but not the other; data not shown) of the RRT motifs in RIZ were able to interact with CtBP (Fig. 4A). Mutations of the RRT sequence abolished the interaction (Fig. 4B), as summarized in Fig. 4C.
Defining the regions in CtBP that contact the RRT motif using X-ray crystallography.
To shed more light on the structural bases of the CtBP-ZNF217 interaction, crystallographic evidence was sought on the location of the RRTGCPPAL peptide recognition site on CtBP. To avoid aggregation during crystallization, mutants with the peptide's C residue altered to A or S were tested in yeast two-hybrid assays for binding to CtBP2 (Fig. 2D). Both A and S are tolerated at this amino acid position, so an RRTGAPPAL peptide was synthesized. Cocrystallization experiments were performed by incubating the synthetic peptide with a truncated form of the CtBP1-S isoform (or the short-CtBP1 splice isoform, previously known as CtBP3/BARS). This truncated CtBP1-S (t-CtBP1-S), devoid of 80 C-terminal residues, was successfully used in the past to identify the PXDLS consensus binding site (30). The protein-RRTGAPPAL complex three-dimensional (3D) structure was solved by molecular replacement methods using the t-CtBP1-S structure as a starting model (PDB entrycode 1HKU) and refined to a 2.85-Å resolution(Rfactor = 22.7% and Rfree = 27.5%, respectively) (Table 1) (8). Crystallized t-CtBP1-S appears as a tight dimer, built across a twofold crystallographic symmetry axis, with major packing interactions based on pairing of two nucleotide-binding domains of each monomer, as observed for other t-CtBP1-S crystal forms (Fig. 5A and B) (30).
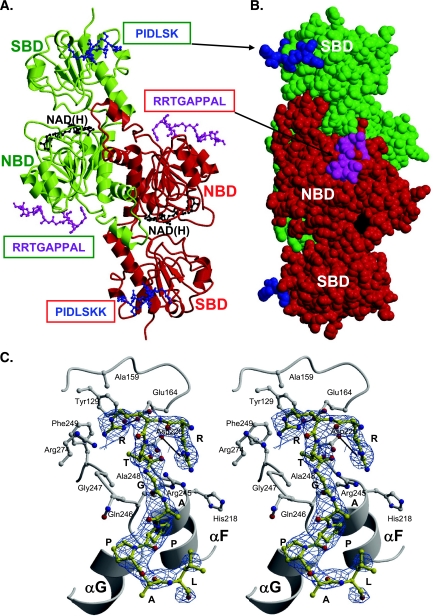
FIG. 5.
X-ray crystal structure of the RRTGAPPAL peptide bound to t-CtBP1-S. A. Ribbon diagram of the t-CtBP1-S dimer. The protein subunits composing the dimer are shown in green and red. The substrate- and nucleotide-binding domains of each subunit are labeled as SBD and NBD, respectively. The bound NAD(H) and RRTGAPPAL peptide molecules are shown in ball-and-stick representations (black and magenta, respectively). The PXDLS binding site is reported from the crystal structure of the complex formed by t-CtBP1-S and the PIDLSKK peptide, shown in blue (PDB entry code 1HL3) (prepared with MOLSCRIPT [21] and Raster3D [27]). B. CPK representation of the t-CtBP1-S dimer. In this space-filling representation, the molecular complex displayed in panel A has been rotated by about 90° around the vertical axis. In this view the location of the PXDLS and RRTGAPPAL binding sites belonging to different subunits that fall on the same face of the dimeric assembly are clearly depicted. C. Consensus peptide binding site. Stereo view of the consensus RRTGAPPAL peptide (yellow) bound to the t-CtBP1-S nucleotide-binding domain. Salt bridges (black lines) between R1 and D220 and between R2 and E164 are highlighted. The 2Fo-Fc electron density map at 2.85-Å resolution is shown as a blue grid.
The crystal structure of the protein-RRTGAPPAL complex shows that the consensus peptide binds at a surface cleft mainly defined by the loop connecting helix αC to strand βA, and by helices αF and αG, of the nucleotide-binding domain (Fig. 5A and C). The bound peptide adopts an extended conformation, antiparallel to the αG helix, burying 146 Å2 of protein surface. Binding of the exogenous peptide is supported by docking of its R1, R2, T3 side chains into a surface groove lined by CtBP residues Y129, A159, E164, H218, D220, R245, Q246, G247, A248, F249, and R274 (Fig. 5C). The rest of the peptide lies at the protein surface and shows a kink at residues P6-P7 that locates the peptide C-terminal part next to the last turn of helix αG. The main stabilizing peptide-protein interactions involve two salt bridges (R1-D220 and R2-E164), hydrogen bonds in the residue pairs R1-H217, R2-G247, T3-D220, T3-R245, G4-Q246, and G4-Q246, and intermolecular van der Waals contacts at the P6 and P7 residues (Fig. 5C). Interestingly, all protein residues involved in peptide binding/recognition are conserved within the CtBP family, except for the conservative substitution of H218→Q in the CtBP2 sequence.
An overlay of t-CtBP1-S and t-CtBP1-S/peptide complex 3D structures yields a root-means-square deviation of 0.45 Å, indicating that binding of the consensus peptide RRTGAPPAL is not associated with significant tertiary/quaternary structure modifications. Only local side chain conformational changes are induced by peptide binding. Among these, we notice the substitution of the R245 guanidino group of t-CtBP1-S with the guanidino head of R1, from the peptide, which thus replaces the intramolecular salt bridge R245-D220 with the intermolecular R1-D220 ion pair. The consensus RRTGAPPAL binding site has no direct contact with the NAD(H) binding region (about 27 Å apart), although both are hosted in the nucleotide-binding domain, nor with the previously identified PXDLS binding site (about 53 Å apart). The latter is localized at the N-terminal region of the substrate-binding domain and on the opposite face of the t-CtBP1-S subunit (Fig. 5A and B). It is worth noting, however, that in the t-CtBP1-S dimeric assembly, where the two substrate-binding domains lie at opposite poles, the PXDLS binding site of one subunit is located on the same dimer face of the RRT binding site of the opposite subunit (about 30 Å apart). Considering the close proximity of the 686PLNLS690 and 752RRTGCPPAL760 motifs in the ZNF217 sequence (only 61 amino acids apart), it is possible for ZNF217 to bind across the CtBP dimer, accessing the PXDLS and RRT binding sites on distinct CtBP subunits, respectively (Fig. 5A and B).
Confirmation of structural results using mutagenesis.
To confirm the inferred location of the RRTGAPPAL binding site, two CtBP1-S residues building up the peptide recognition cleft, E164 and D220, were selected for mutation to alanine. Mutations were also made in the corresponding residues in CtBP2: E181A and D237A. CtBP derivatives containing each mutation or the two mutations together were generated. In addition, CtBP proteins containing defective PXDLS-binding clefts were further mutated so that they also carried these additional mutations in the putative RRT binding sites. This panel of CtBP mutants was first tested for interaction with ZNF217 using the yeast two-hybrid system (Fig. 6A). Each of the mutations in the CtBP2 and CtBP1-S RRT motif binding clefts was individually sufficient to abrogate binding (only the results from double mutation of two of the amino acids in this cleft for CtBP2 are shown). As expected, the CtBP mutants bearing mutations either in the RRT motif contact region or in the PXDLS binding cleft retained the ability to contact ZNF217; however, when both regions were mutated, binding was abrogated. Each of the CtBP mutants retained the ability to dimerize with wild-type CtBP, indicating that these proteins are expressed and properly folded in yeast. This result is consistent with the structural data and confirms the inference that ZNF217 does contact residues E181 and D237 of CtBP2 through its RRT motif (Fig. 6A). Coimmunoprecipitation experiments were then performed and validated the yeast two-hybrid assay results (Fig. 6B). A summary of the interactions is shown in Fig. 6C.
FIG. 6.
Mutagenesis confirms the RRT contact residues of CtBP. A. Gal4AD-hZNF217 was examined for its ability to interact with the Gal4DBD-CtBP2 wild type or with a PXDLS motif binding cleft (A58E) mutant, the newly identified RRT motif binding cleft (E181A D237A) mutant, or with CtBP2 with mutations in both clefts in yeast two-hybrid assays. Interactions between the Gal4DBD CtBP2 mutants and with Gal4AD wild-type CtBP were also examined as a positive control for the expression and folding of the CtBP2 mutants in yeast. B. The ability of FLAG-hZNF217 to interact with YFP-CtBP2 wild type or with a PXDLS motif binding cleft (A58E) or RRT motif binding cleft (E181A D237A) mutant or with a construct with mutations in both clefts in coimmunoprecipitation experiments. COS-1 cells were transfected with the expression vectors indicated above each lane, and whole-cell extracts of those cells were immunoprecipitated (IP) with anti-FLAg (αFLAG) antibody. Expression of each of the FLAG-fused or YFP-fused proteins is shown in the top two panels (5% input). FLAG-ZNF217 immunoprecipitated by the anti-FLAG antibody and the resulting coimmunopreciptated YFP-CtBP2 are shown in the bottom two panels (IP: αFLAG). C. A summary diagram combining the results of interaction studies between ZNF217 and wild-type or mutant CtBP. αYFP, anti-YFP antibody; WB, Western blot.
Mutations in the PXDLS and RRT motif binding clefts of CtBP have little effect on its ability to repress transcription.
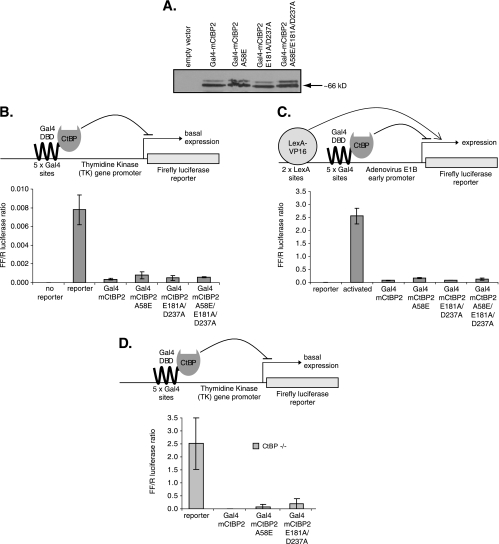
Having generated a CtBP mutant (A58E E181A D237A) that was unable to bind to ZNF217, we sought to assess the effect of this mutation on the ability of CtBP to repress transcription. Since CtBP cannot bind to DNA directly, the conventional Gal4DBD fusion strategy that is widely used to assess CtBP repression activity was employed (11, 20, 42). The cDNA encoding wild-type CtBP or CtBP with a mutation(s) in thePXDLS motif binding cleft (A58E), in the RRT motif binding cleft (E181A D237A), or at both clefts was fused to a cDNA encoding Gal4DBD. These constructs were transfected into COS-1 cells and were shown to be expressed at equivalent levels (Fig. 7A). Their ability to repress transcription of the firefly luciferase reporter driven by a core TK promoter with five Gal4 binding sites or a LexA-VP16 activated E1B promoter with five Gal4 binding sites and two LexA binding sites was examined (Fig. 7B and C). The mutation in the PXDLS binding cleft (A58E) had a modest effect on the ability of CtBP to repress, but additional mutations in the RRT binding cleft (E181A D237A) or the RRT cleft mutations alone had no discernible effect on activity. To exclude the possibility that the mutant proteins were retaining repression activity by virtue of their ability to dimerize with wild-type endogenous CtBP, we repeated the experiments with murine embryonic fibroblasts derived from CtBP1−/− CtBP2−/− double-knockout murine embryos (16). Wild-type CtBP2 and A58E and E181A D237A mutants all exhibited strong repression activity in these CtBP−/− cells (Fig. 7D), although the mutants showed a slight reduction in repression. In summary, these results suggest that ZNF217 contact does not make a major contribution to repression by CtBP.
FIG. 7.
CtBP repression activity does not depend on its ability to bind ZNF217. A. Western blotting was performed to examine the expression level of Gal4DBD-fused CtBP2 wild type or A58E, E181A D237A, or A58E E181A D237A mutant in transiently transfected COS-1 cells. B and C. Gal4DBD-CtBP2 constructs were tested for their abilities to repress (B) basal firefly luciferase reporter gene expression from the TK promoter or (C) LexA-VP16 activated firefly luciferase reporter gene expression from the E1B promoter in COS-1 cells following transient transfection (n = 4; ± standard deviation; representative experiment). D. Gal4DBD-CtBP2 constructs were tested for their abilities to repress basal firefly luciferase reporter gene expression from the TK promoter in CtBP−/− cells following transient transfection (n = 2; ± standard deviation; representative experiment).
ZNF217 is a transcriptional repressor, and mutations in the PXDLS and RRT motifs of ZNF217 reduce this activity.
We next examined whether ZNF217 is able to repress transcription. We also tested ZNF217 mutants that cannot bind to CtBP in these assays. The molecular mechanism through which ZNF217 operates has not been determined, but the finding that it associates directly with CtBP2 suggested that it may play a role in gene repression. We therefore examined its activity on a number of test promoters.
We first tested the ability of ZNF217 to repress the Gal4 site-linked TK promoter. Cotransfection of a plasmid encoding wild-type ZNF217 resulted in significant repression. We then tested the mutant derivatives of ZNF217. Mutation of both the PXDLS and RRT motifs in ZNF217 significantly reduced repression (Fig. 8B). Taken together, these results suggest that ZNF217 can act as a repressor of transcription and that it in part utilizes CtBP to mediate repression. The residual repression observed indicates that it may also have additional mechanisms through which it can repress gene expression. Similar results were obtained when ZNF217 and the mutant were tested against a second promoter containing LexA and Gal4 sites upstream of the adenovirus E1B promoter driving a luciferase reporter gene (Fig. 8C).
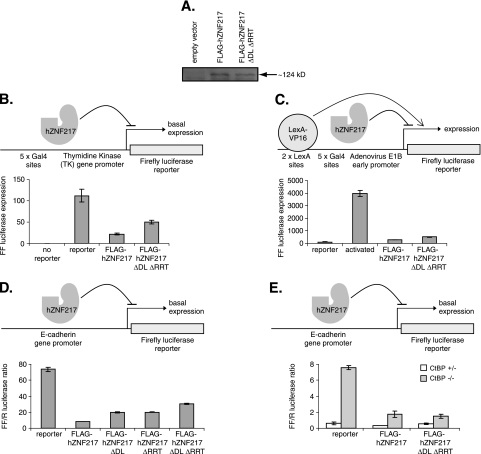
FIG. 8.
ZNF217 represses gene transcription, and this effect is partially dependent on its ability to bind to CtBP. A. A Western blot was performed to examine the expression levels of the FLAG-fused hZNF217 wild type and an ΔDL ΔRRT mutant in transiently transfected COS-1 cells. B to E. FLAG-hZNF217 constructs were tested for their ability to repress (B) firefly luciferase reporter gene expression from the TK promoter in COS-1 cells (n = 4; ± standard deviation; representative experiment), (C) LexA-VP16 activated firefly luciferase reporter gene expression from the E1B promoter in COS-1 cells (n = 4; ± standard deviation; representative experiment), (D) basal firefly luciferase reporter gene expression from the E-cadherin promoter in HEK293 cells (n = 2; range of values; representative experiment), or (E) basal firefly luciferase reporter gene expression from the E-cadherin promoter in CtBP+/− and CtBP−/− cells (n = 2; range of values; representative experiment), following transient transfection.
We also sought to test a natural CtBP-dependent promoter and chose the E-cadherin promoter, since it has previously been shown to be a CtBP target gene (12, 13, 39).We transfected a ZNF217-encoding plasmid together with the E-cadherin promoter driving a luciferase reporter gene and observed that ZNF217 significantly repressed expression of the reporter gene. We also tested the PXDLS mutant, the RRT mutant, and the double mutant. We found that each single mutation modestly reduced repression and that the double mutation more significantly reduced repression (Fig. 8D). These results were similar to those obtained with the viral promoters used above. We also noted that no repression was observed with other promoters, such as the cytomegalovirus promoter-driven Renilla reporter plasmid, showing that ZNF217 does not nonspecifically repress all promoters (data not shown). Wild-type ZNF217 and ZNF217 with mutations in both the PXDLS motif (ΔDL) and the RRT motif (ΔRRT) were expressed at equivalent levels in COS-1 cells (Fig. 8A).
To further assess the contribution of CtBP to ZNF217 repression activity, we repeated the experiments with CtBP+/− and double-knockout cells (Fig. 8E). When transfected into CtBP+/− control cells, ZNF217 represses the E-cadherin promoter. Again, this repression activity appears to be mediated in part by CtBP, since the double mutant (ΔDL ΔRTT) shows reduced repression. When tested with CtBP−/− cells, the E-cadherin reporter is derepressed and shows high activity (Fig. 8E). Significantly, cotransfection of ZNF217 leads to significant repression even in the CtBP−/− cells. Double-mutant ZNF217 retains equivalent repression activity in these cells (Fig. 8E). Taken together, the results suggest that recruitment of CtBP enhances repression but that ZNF217 contacts additional partners that can mediate repression in the absence of CtBP.
DISCUSSION
We have shown that the zinc finger oncoprotein ZNF217 interacts with CtBP, utilizing both a conventional PXDLS motif (localizing to a binding cleft in the CtBP substrate-binding domain (30) and a distinct RRT motif, which binds at a newly defined surface cleft in the CtBP nucleotide-binding domain (Fig. 5). The two peptide recognition sites are physically well separated (53 Å), being roughly at opposite poles of the CtBP subunit. Moreover, based on geometrical considerations, the two sites do not appear to support simultaneous contacts with the same ZNF217 molecule within one CtBP subunit. Rather, ZNF217 may bind across the CtBP dimer, contacting surfaces of both substrate- and nucleotide-binding domains from the two protein subunits, thus recognizing both PXDLS and RRT binding sites on distinct CtBP subunits.
ZNF217 is implicated in human cancers and has been shown to contribute to the immortalization of breast epithelial cells in culture (32). Our work suggests that one mechanism by which an increased copy number of ZNF217 contributes to tumorigenesis could be through altering gene expression, for example, via increased repression of tumor suppressor gene promoters. We also show that the ability of ZNF217 to repress transcription is partially dependent on its ability to bind to CtBP.
Having established that ZNF217 represses transcription, future research will focus on the full mechanisms by which it mediates repression. It is known that the protein RIZ, which also contains zinc fingers and PXDLS and RRT motifs, can bind GC-rich sites in DNA through its zinc fingers 1 to 3 (46) and also has been reported to possess histone methyltransferase activity (19). By analogy, ZNF217 may be a sequence-specific DNA-binding protein that recruits CtBP to silence specific genes, and the residual repression activity observed when it is unable to recruit CtBP may reflect an additional repression mechanism. However, to date we have not detected direct DNA binding by ZNF217 (data not shown). The relationship between ZNF217 and the E-cadherin promoter and the mechanism through which it may be recruited to the promoter in vivo are still under investigation.
Interestingly, ZNF217 and ZNF516 (recorded as KIA0222) have been found to be present in a number of repression complexes (14, 24, 48), including the CtBP-associated repression complex that exists in HeLa cells (39), consistent with our data that these proteins directly contact CtBP. Relatively few typical sequence-specific transcription factors have been found in these repression complexes. One known DNA-binding protein that has been found in the CtBP repressor complex is the large zinc finger homeodomain transcription factor ZEB (39). It is possible that ZNF217, RIZ, and ZEB can function as conventional transcription factors and also display additional activities allowing them to contribute directly to gene repression (6). However, our observation that mutations in CtBP that prevent it from binding ZNF217 had little effect on its ability to repress transcription argues against ZNF217 being an essential effector protein in the CtBP repression complex. It should be noted that a slight loss of repression was apparent when the CtBP mutants were tested in CtBP−/− cells (Fig. 7D), so it is possible that ZNF217 makes some contribution to repression. But taken together, the results indicate that ZNF217 is not a critical effector of CtBP activity, at least in the promoter and cellular contexts tested.
In summary, we have shown that ZNF217 is a direct partner protein contacting CtBP through the known PXDLS motif but also through a second RRT motif that binds a novel peptide recognition groove. Other large zinc finger proteins also contain PXDLS and RRT motifs. We have shown that mutation of these motifs in ZNF217 reduces its ability to repress transcription. These results suggest that one mechanism by which the ZNF217 oncogene may contribute to tumorigenesis is through CtBP-associated repression of transcription.
Acknowledgments
We are grateful to L. Gaudreau and M. Ptashne for the pGL2-(Gal4)5-(LexA)2-E1B-Luc and LexA-VP16 mammalian expression plasmids, S. Sugrue for the E-cad-Luc reporter construct, J. Hildebrand for the CtBP knockout cells, and C. Cericola and A. Colanzi for their skillful assistance. We thank the staff of the ID14-EH3 beamline at ESRF, Grenoble (France), for data collection facilities and assistance.
K.Q. is supported by an Australian Postgraduate Award. This work was supported by NIH grant NHLBI HL073443 and grants from the Australian ARC and NHMRC to M.C., by Italian Ministry of University FIRB grants, by AIRC (Italy) grants to M.B. and D.C., and by a Telethon (Italy) grant toD.C. M.B. is grateful to CIMAINA (University of Milano) and to Fondazione CARIPLO (Milano, Italy) for continuous support.
Footnotes
Published ahead of print on 28 August 2006.
REFERENCES
- 1.Alpatov, R., G. C. Munguba, P. Caton, J. H. Joo, Y. Shi, Y. Shi, M. E. Hunt, and S. P. Sugrue. 2004. Nuclear speckle-associated protein Pnn/DRS binds to the transcriptional corepressor CtBP and relieves CtBP-mediated repression of the E-cadherin gene. Mol. Cell. Biol. 24:10223-10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chinnadurai, G. 2002. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell 9:213-224. [DOI] [PubMed] [Google Scholar]
- 4.Collaborative Computational Project, no. 4. 1994. The CCP4 suite. Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 5.Collins, C., J. M. Rommens, D. Kowbel, T. Godfrey, M. Tanner, S. I. Hwang, D. Polikoff, G. Nonet, J. Cochran, K. Myambo, K. E. Jay, J. Froula, T. Cloutier, W. L. Kuo, P. Yaswen, S. Dairkee, J. Giovanola, G. B. Hutchinson, J. Isola, O. P. Kallioniemi, M. Palazzolo, C. Martin, C. Ericsson, D. Pinkel, D. Albertson, W. B. Li, and J. W. Gray. 1998. Positional cloning of ZNF217 and NABC1: genes amplified at 20q13.2 and overexpressed in breast carcinoma. Proc. Natl. Acad. Sci. USA 95:8703-8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Comijn, J., G. Berx, P. Vermassen, K. Verschueren, L. van Grunsven, E. Bruyneel, M. Mareel, D. Huylebroeck, and F. van Roy. 2001. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol. Cell 7:1267-1278. [DOI] [PubMed] [Google Scholar]
- 7.Corda, D., A. Colanzi, and A. Luini. 2006. The multiple activities of CtBP/BARS proteins: the Golgi view. Trends Cell Biol. 16:167-173. [DOI] [PubMed] [Google Scholar]
- 8.Engh, R. A., and R. Huber. 1991. Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr. A 47:392-400. [Google Scholar]
- 9.Evans, P. R. 1993. Proceedings of the CCP4 study weekend on data collection and processing, p. 114-122. CLRC Daresbury Laboratory, Warrington, United Kingdom.
- 10.Ewart-Toland, A., P. Briassouli, J. P. de Koning, J. H. Mao, J. Yuan, F. Chan, L. MacCarthy-Morrogh, B. A. Ponder, H. Nagase, J. Burn, S. Ball, M. Almeida, S. Linardopoulos, and A. Balmain. 2003. Identification of Stk6/STK15 as a candidate low-penetrance tumor-susceptibility gene in mouse and human. Nat. Genet. 34:403-412. [DOI] [PubMed] [Google Scholar]
- 11.Furusawa, T., H. Moribe, H. Kondoh, and Y. Higashi. 1999. Identification of CtBP1 and CtBP2 as corepressors of zinc finger-homeodomain factor δEF1. Mol. Cell. Biol. 19:8581-8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grooteclaes, M., Q. Deveraux, J. Hildebrand, Q. Zhang, R. H. Goodman, and S. M. Frisch. 2003. C-terminal-binding protein corepresses epithelial and proapoptotic gene expression programs. Proc. Natl. Acad. Sci. USA 100:4568-4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grooteclaes, M. L., and S. M. Frisch. 2000. Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene 19:3823-3828. [DOI] [PubMed] [Google Scholar]
- 14.Hakimi, M. A., Y. Dong, W. S. Lane, D. W. Speicher, and R. Shiekhattar. 2003. A candidate X-linked mental retardation gene is a component of a new family of histone deacetylase-containing complexes. J. Biol. Chem. 278:7234-7239. [DOI] [PubMed] [Google Scholar]
- 15.Hickabottom, M., G. A. Parker, P. Freemont, T. Crook, and M. J. Allday. 2002. Two nonconsensus sites in the Epstein-Barr virus oncoprotein EBNA3A cooperate to bind the co-repressor carboxyl-terminal-binding protein (CtBP). J. Biol. Chem. 277:47197-47204. [DOI] [PubMed] [Google Scholar]
- 16.Hildebrand, J. D., and P. Soriano. 2002. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol. Cell. Biol. 22:5296-5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, S., G. Shao, and L. Liu. 1998. The PR domain of the Rb-binding zinc finger protein RIZ1 is a protein binding interface and is related to the SET domain functioning in chromatin-mediated gene expression. J. Biol. Chem. 273:15933-15939. [DOI] [PubMed] [Google Scholar]
- 18.Kagey, M. H., T. A. Melhuish, and D. Wotton. 2003. The polycomb protein Pc2 is a SUMO E3. Cell 113:127-137. [DOI] [PubMed] [Google Scholar]
- 19.Kim, K. C., L. Geng, and S. Huang. 2003. Inactivation of a histone methyltransferase by mutations in human cancers. Cancer Res. 63:7619-7623. [PubMed] [Google Scholar]
- 20.Koipally, J., and K. Georgopoulos. 2000. Ikaros interactions with CtBP reveal a repression mechanism that is independent of histone deacetylase activity. J. Biol. Chem. 275:19594-19602. [DOI] [PubMed] [Google Scholar]
- 21.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 22.Kumar, V., J. E. Carlson, K. A. Ohgi, T. A. Edwards, D. W. Rose, C. R. Escalante, M. G. Rosenfeld, and A. K. Aggarwal. 2002. Transcription corepressor CtBP is an NAD(+)-regulated dehydrogenase. Mol. Cell 10:857-869. [DOI] [PubMed] [Google Scholar]
- 23.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK, a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 24.Lee, M. G., C. Wynder, N. Cooch, and R. Shiekhattar. 2005. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 437:432-435. [DOI] [PubMed] [Google Scholar]
- 25.Leslie, A. G. M. 2003. MOSFLM user guide, Mosflm version 6.2.3. MRC Laboratory of Molecular Biology, Cambridge, United Kingdom.
- 26.Lin, X., B. Sun, M. Liang, Y. Y. Liang, A. Gast, J. Hildebrand, F. C. Brunicardi, F. Melchior, and X. H. Feng. 2003. Opposed regulation of corepressor CtBP by SUMOylation and PDZ binding. Mol. Cell 11:1389-1396. [DOI] [PubMed] [Google Scholar]
- 27.Merritt, E. A., and M. E. Murphy. 1994. Raster3D version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 50:869-873. [DOI] [PubMed] [Google Scholar]
- 28.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 29.Nardini, M., S. Spano, C. Cericola, A. Pesce, G. Damonte, A. Luini, D. Corda, and M. Bolognesi. 2002. Crystallization and preliminary X-ray diffraction analysis of brefeldin A-ADP ribosylated substrate (BARS). Acta Crystallogr. D Biol. Crystallogr. 58:1068-1070. [DOI] [PubMed] [Google Scholar]
- 30.Nardini, M., S. Spano, C. Cericola, A. Pesce, A. Massaro, E. Millo, A. Luini, D. Corda, and M. Bolognesi. 2003. CtBP/BARS: a dual-function protein involved in transcription co-repression and Golgi membrane fission. EMBO J. 22:3122-3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nardini, M., D. Svergun, P. V. Konarev, S. Spano, M. Fasano, C. Bracco, A. Pesce, A. Donadini, C. Cericola, F. Secundo, A. Luini, D. Corda, and M. Bolognesi. 2006. The C-terminal domain of the transcriptional co-repressor CtBP is intrinsically unstructured. Protein Sci. 15:1042-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nonet, G. H., M. R. Stampfer, K. Chin, J. W. Gray, C. C. Collins, and P. Yaswen. 2001. The ZNF217 gene amplified in breast cancers promotes immortalization of human mammary epithelial cells. Cancer Res. 61:1250-1254. [PubMed] [Google Scholar]
- 33.Perdomo, J., and M. Crossley. 2002. The Ikaros family protein Eos associates with C-terminal-binding protein corepressors. Eur. J. Biochem. 269:5885-5892. [DOI] [PubMed] [Google Scholar]
- 34.Perdomo, J., A. Verger, J. Turner, and M. Crossley. 2005. Role for SUMO modification in facilitating transcriptional repression by BKLF. Mol. Cell. Biol. 25:1549-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rooney, P. H., A. Boonsong, M. C. McFadyen, H. L. McLeod, J. Cassidy, S. Curran, and G. I. Murray. 2004. The candidate oncogene ZNF217 is frequently amplified in colon cancer. J. Pathol. 204:282-288. [DOI] [PubMed] [Google Scholar]
- 36.Schaeper, U., J. M. Boyd, S. Verma, E. Uhlmann, T. Subramanian, and G. Chinnadurai. 1995. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc. Natl. Acad. Sci. USA 92:10467-10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaeper, U., T. Subramanian, L. Lim, J. M. Boyd, and G. Chinnadurai. 1998. Interaction between a cellular protein that binds to the C-terminal region of adenovirus E1A (CtBP) and a novel cellular protein is disrupted by E1A through a conserved PLDLS motif. J. Biol. Chem. 273:8549-8552. [DOI] [PubMed] [Google Scholar]
- 38.Shi, Y., F. Lan, C. Matson, P. Mulligan, J. R. Whetstine, P. A. Cole, R. A. Casero, and Y. Shi. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941-953. [DOI] [PubMed] [Google Scholar]
- 39.Shi, Y., J. Sawada, G. Sui, E. B. Affar, J. R. Whetstine, F. Lan, H. Ogawa, M. P. Luke, Y. Nakatani, and Y. Shi. 2003. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422:735-738. [DOI] [PubMed] [Google Scholar]
- 40.Shi, Y., and Y. Shi. 2005. CtBP corepressor complex—a multi-enzyme machinery that coordinates chromatin modifications. In G. Chinnadurai (ed.), CtBP family proteins. LANDES BIOSCIENCES, Georgetown, Tex. [Online.] http://eurekah.com/abstract.php?chapid=2751&bookid=198&catid=30.
- 41.tom Dieck, S., F. Schmitz, and J. H. Brandstatter. 2005. CtBPs as synaptic proteins. In G. Chinnadurai (ed.), CtBP family proteins. LANDES BIOSCIENCES, Georgetown, Tex. [Online.] http://eurekah.com/abstract.php?chapid=2646&bookid=198&catid=30.
- 42.Turner, J., and M. Crossley. 1998. Cloning and characterization of mCtBP2, a co-repressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. EMBO J. 17:5129-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner, J., and M. Crossley. 2001. The CtBP family: enigmatic and enzymatic transcriptional co-repressors. Bioessays 23:683-690. [DOI] [PubMed] [Google Scholar]
- 44.Turner, J., H. Nicholas, D. Bishop, J. M. Matthews, and M. Crossley. 2003. The LIM protein FHL3 binds basic Kruppel-like factor/Kruppel-like factor 3 and its co-repressor C-terminal-binding protein 2. J. Biol. Chem. 278:12786-12795. [DOI] [PubMed] [Google Scholar]
- 45.Vagin, A., and A. Teplyakov. 1997. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30:1022-1025. [Google Scholar]
- 46.Xie, M., G. Shao, I. M. Buyse, and S. Huang. 1997. Transcriptional repression mediated by the PR domain zinc finger gene RIZ. J. Biol. Chem. 272:26360-26366. [DOI] [PubMed] [Google Scholar]
- 47.Yaswen, P., and M. R. Stampfer. 2002. Molecular changes accompanying senescence and immortalization of cultured human mammary epithelial cells. Int. J. Biochem. Cell Biol. 34:1382-1394. [DOI] [PubMed] [Google Scholar]
- 48.You, A., J. K. Tong, C. M. Grozinger, and S. L. Schreiber. 2001. CoREST is an integral component of the CoREST-human histone deacetylase complex. Proc. Natl. Acad. Sci. USA 98:1454-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, Q., Y. Yoshimatsu, J. Hildebrand, S. M. Frisch, and R. H. Goodman. 2003. Homeodomain interacting protein kinase 2 promotes apoptosis by downregulating the transcriptional corepressor CtBP. Cell 115:177-186. [DOI] [PubMed] [Google Scholar]