Abstract
To achieve full ripening, climacteric fruits, such as tomato require synthesis, perception and signal transduction of the plant hormone ethylene. The nonripening phenotype of the dominant Green-ripe (Gr) and Never-ripe 2 (Nr-2) mutants of tomato is the result of reduced ethylene responsiveness in fruit tissues. In addition, a subset of ethylene responses associated with floral senescence, abscission, and root elongation are also impacted in mutant plants, but to a lesser extent. Using positional cloning, we have identified an identical 334-bp deletion in a gene of unknown biochemical function at the Gr/Nr-2 locus. Consistent with a dominant gain of function mutation, this deletion causes ectopic expression of Gr/Nr-2, which in turn leads to ripening inhibition. A CaMV35::GR transgene recreates the Gr/Nr-2 mutant phenotype but does not lead to a global reduction in ethylene responsiveness, suggesting tissue-specific modulation of ethylene responses in tomato. Gr/Nr-2 encodes an evolutionary conserved protein of unknown biochemical function that we associate here with ethylene signaling. Because Gr/Nr-2 has no sequence homology with the previously described Nr (Never-ripe) ethylene receptor of tomato we now refer to this gene only as GR. Identification of GR expands the current repertoire of ethylene signaling components in plants and provides a tool for further elucidation of ethylene response mechanisms and for controlling ethylene signal specificity in crop plants.
Keywords: fruit development, hormonal regulation, positional cloning
Altered ethylene responsiveness in plant tissues influences development and can compromise the plants ability to respond to environmental stimuli (1–4). The mechanisms by which the ethylene signal is perceived and transduced to mediate phenotypic changes is not fully understood, although many elegant studies exploiting the triple response screen in Arabidopsis have led to the identification of critical components of this signaling pathway (5).
Ethylene is perceived by a family of receptors that share homology to bacterial two-component regulators (5). Loss-of-function analysis indicates that the receptors act in a semiredundant manner to negatively regulate ethylene responses (6). At least two receptors interact with CONSTITUTIVE TRIPLE RESPONSE 1, a serine threonine mitogen-activated protein kinase kinase (MAPKKK) that acts as a negative regulator of the pathway (7–9). An integral membrane protein, EIN2, with homology to the NRAMP family of metal ion transporters acts downstream of the receptors and CTR1 (10, 11). The biochemical function of EIN2 remains unknown, but genetic studies have indicated that all ethylene responses described to date are transduced through this signaling intermediate (12). A family of transcription factors encoded by EIN3 and EIL (EIN3-like) act downstream of EIN2 (13, 14). Homodimers of EIN3, EIL1, and EIL2 bind to a defined target in the promoter region of the transcription factor, ETHYLENE RESPONSE FACTOR1 (ERF-1) (14). ERF1 is a member of a multigene family of transcription factors and is important in the regulation of downstream ethylene responsive genes via binding to the “GCC” box promoter element (15, 16). Ethylene responses are regulated at the level of EIN3 via ubiquitin/proteasome-dependent proteolysis mediated by the F-box proteins, EBF1 and EBF2 (17, 18).
The importance of ethylene in regulating traits of agronomic importance, particularly fruit ripening and floral senescence, has driven research on the identification and functional characterization of components of the ethylene signaling pathway in crop species (19, 20). Studies using tomato and petunia have been at the forefront of this comparative analysis and have revealed structural and functional conservation of the ethylene signaling pathway (21–25). However, there is an expansion of gene families encoding the receptors and CTR components in tomato and other crop plants, revealing an added layer of complexity to the ethylene response pathway (19–21).
We have recently reported that inhibition of fruit ripening in the Green-ripe (Gr) and Never-ripe 2 (Nr-2) mutants of tomato is the result of ethylene insensitivity (26). Gr and Nr-2 plants also display subdued ethylene responses associated with floral senescence, abscission, and root elongation during the triple response. However, ethylene-mediated inhibition of hypocotyl elongation and petiole epinasty remain normal, suggesting that these loci affect a subset of ethylene responses in tomato, with the strongest phenotypes observed in fruit (26).
Here we report the isolation of the GR/NR-2 locus by positional cloning. DNA sequencing revealed the presence of an identical 334-bp deletion in Gr/Gr and Nr-2/Nr-2 genotypes, indicating that these mutations are allelic. The deletion resides in the 5′-flanking region of a gene encoding an evolutionary conserved protein of unknown function that is predicted to be membrane localized. Molecular analysis revealed that the Gr/Nr-2 deletion causes ectopic expression of GR, a phenomenon consistent with a dominant gain of function mutation. Constitutive overexpression of GR in transgenic plants recreates the Gr mutant phenotype but does not result in plants that display whole plant ethylene insensitivity. The ability of GR to selectively inhibit ethylene responses suggests that tissue-specific signaling mechanisms operate in tomato.
Results
High-Resolution Genetic and Physical Mapping of the Nr-2 and Gr Loci.
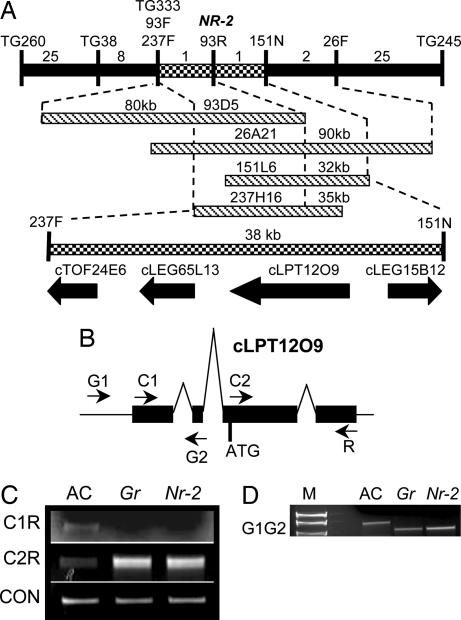
Using F2 populations segregating for normal and nonripening fruit between Solanum lycopersicum (Gr/Gr) × Solanum cheesmaniae (gr/gr) and S. lycopersicum (Nr-2/Nr-2) × S. cheesmaniae (nr-2/nr-2), we positioned the Gr and Nr-2 loci to overlapping regions of the long arm of chromosome 1 with tight linkage to the RFLP marker TG333 (26). The mapping resolution of each locus was increased to ≈0.03 cM per recombination event by screening for recombinants between TG260 and TG245 in 1,810 and 1,856 F2 individuals segregating for Nr-2 and Gr, respectively. TG333 was used to screen BAC libraries derived from S. lycopersicum and S. cheesmaniae (27, 28). A total of 13 BAC clones were recovered (data not shown). The ends from clone 93D5 (Fig. 1A) were isolated by sequencing and converted into the RFLP markers 93F and 93R. 93R was used to rescreen the BAC libraries and an S. lycopersicum cosmid library. Three additional clones 26A21, 237H16, and 151L6 were recovered. The ends of these clones were isolated and converted to either RFLP- or PCR-based markers. Mapping of these markers indicated that the Nr-2 locus cosegregated with 93R between the flanking loci 237F and 151N (Fig. 1A). The Gr locus was positioned within the same interval (data not shown). Using a combination of BAC subclones and primer walking, this interval was sequenced and found to be 38-kb in length. A blast search of the resulting sequence against build three of the Solanaceous Genomics Network unigene set (www.sgn.cornell.edu) identified four predicted genes (Fig. 1A). ESTs were identified corresponding to each of these genes and the longest corresponding cDNA clone was sequenced and aligned to the genomic sequence. blastx searches of GenBank (www.ncbi.nlm.nih.gov/blast) and the predicted proteins of Arabidopsis (www.arabidopsis.org) failed to reveal any additional genes residing in the 38-kb interval, suggesting that one or more of the four genes is altered in the Gr and Nr-2 mutations.
Fig. 1.
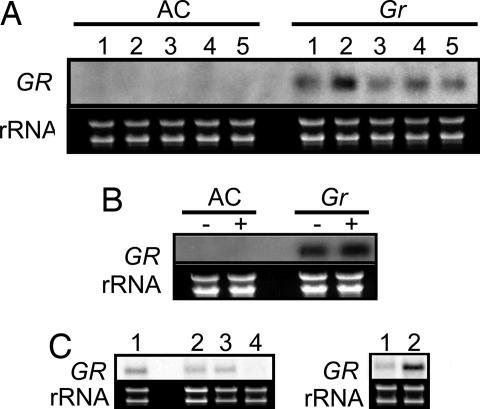
Structure of the Gr and Nr-2 loci. (A) Genetic and physical map of the Nr-2 locus based on 1,810 F2 plants. The number of recombinant individuals between adjacent markers is shown. BAC and cosmid clones are represented as horizontal bars, and approximate sizes are indicated (in kilobases). Four candidate genes at the Nr-2 locus are defined by their corresponding EST identifier. Arrows indicate the predicted direction of transcription. (B) Genomic structure corresponding to the EST clone cLPT12O9. Positions of primers used for RT-PCR (C1, C2, R) and amplification of genomic DNA (G1, G2) are shown. (C) RT-PCR amplification of cLPT12O9 from AC, Gr, and Nr-2 genotypes. SGN-U239539 was used as an equal loading control. (D) Identification of a deletion at the Gr and Nr-2 loci. Amplification of genomic DNA using the primers G1 and G2 is shown. The amplicon from control (AC) DNA is ≈1,400 bp. M refers to a DNA ladder.
An Identical Deletion in the Gr and Nr-2 5′-Flanking Region Causes a Dominant Gain of Function Phenotype.
Using primers designed to amplify full-length cDNA clones for each of the four candidate genes, RT-PCR analysis was performed on cDNA made from RNA of mixed stages of fruit development from three genotypes: AC (normal nearly isogenic control), Gr/Gr, and Nr-2/Nr-2. Amplifying the predicted full length cDNA corresponding to cLPT12O9 (Fig. 1A) using the primers C1 and R (Fig. 1 B and C) resulted in a faint band of the correct size from the AC control template, but no product was obtained from mutant samples. An independent RT-PCR using a nested forward primer, C2, and the reverse primer, R, on the same RNA samples amplified a product of the predicted size from each genotype (Fig. 1 B and C). Furthermore, a greater yield of PCR product was obtained from Gr and Nr-2 backgrounds, implying that cLPT12O9 is more highly expressed than in the control sample. Lack of amplification of cLPT12O9 in RT-PCR experiments using the C1 primer with mutant-derived RNAs suggested the presence of a deletion covering, or a mutation at, the C1 primer site in the Gr and Nr-2 mutants; this was confirmed by PCR amplification from genomic DNA with primers G1 and G2 (Fig. 1D). A smaller fragment was amplified from Gr and Nr-2 mutant plants compared to that obtained from AC control plants. These fragments were cloned and sequenced, and a single identical deletion of 334 bp was confirmed in both Gr and Nr-2. Sequence alignment of the deletion to the S. lycopersicum genomic and full-length cDNA sequences, obtained by 5′ RACE, revealed that the deletion results in the removal of 278 bp of the first exon (within the 5′ UTR) and 56 bp of the putative promoter; however, the predicted protein-coding region of cLPT12O9 remains unchanged in both the Gr and Nr-2 alleles. The deletion begins just 10 bp downstream of the predicted TATA box, suggesting that transcription initiation may be disrupted in Gr/Gr and Nr-2/Nr-2 genotypes. This disruption was confirmed by performing 5′ RACE amplification on cDNA synthesized from wild-type and Gr RNA, which indicated that transcription is initiated at a point 372 bp downstream of the wild-type site of transcription initiation of the normal, nondeleted genomic sequence, in mutant compared to wild-type plants. Alignment of cDNA and genomic sequences from wild-type plants reveals a 1,451-base transcript derived from a genomic clone comprised of four exons. Two of the three introns reside within the 5′ UTR, the first being 101 bases and the second being particularly large at 8,880 bases. This gene structure is conserved in Gr, although the first two introns are 1,655 bases and 7,172 bases, respectively, and occur at different positions to those in the wild-type gene. Additionally, the processed transcript size is 1,235 bases in mutant plants. An additional 694 bases upstream of the normal GR start of transcription was recovered through sequencing of the mutant allele and no additional sequence alterations were identified as compared the normal near isoline. This result suggests that the 334 deletion observed in Gr is responsible for elevated expression in mutant lines and this in turn confers the ethylene insensitive phenotypes observed in mutant plants.
Overexpression of cLPT12O9 Recreates the Gr Phenotype.
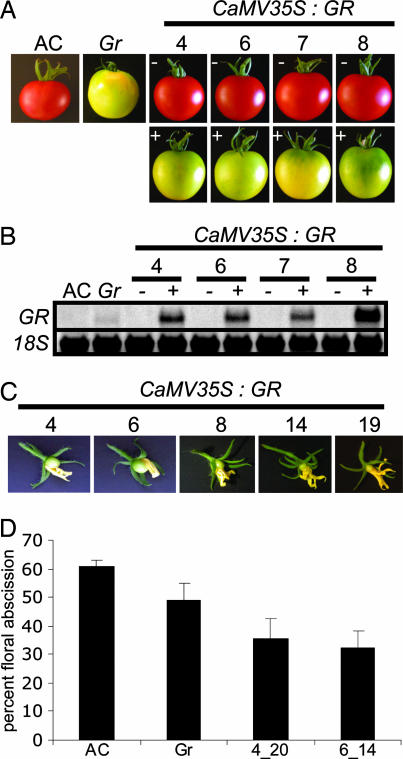
Confirmation that ectopic expression of cLPT12O9 is responsible for conferring the nonripening phenotype of Gr and Nr-2 fruit was achieved through overexpression of the full-length cDNA, derived from Gr/Gr RNA, under the control of the cauliflower mosaic virus (CaMV) 35S promoter in transgenic tomato plants. Seventeen of 18 primary transformants regenerated from tissue culture displayed a nonripening phenotype characteristic of the Gr mutant (data not shown). T1 progeny derived from four independently transformed lines, segregating for the NPTII marker gene and GR overexpression, clearly demonstrated a link between the transgene and the nonripening phenotype (Fig. 2 A and B). We have previously documented a weak inhibition of petal senescence associated with the Gr mutant (26). In support of this observation, several of the primary transgenic lines displayed petal retention on developing fruits (Fig. 2C). In addition, ethylene-induced floral abscission is reduced in CaMV35S::GR lines compared to wild-type. This response was stronger in the transgenic lines than in flowers of the Gr mutant (Fig. 2D).
Fig. 2.
CaMV35S::GR expression recreates the Gr phenotype. (A) Segregation of ripening inhibition in T1 progeny of four CaMV35S::GR independent transgenic lines (4, 6, 7, and 8). Normal ripening fruit (−) have segregated out the transgene, whereas nonripening fruit (+) have retained the transgene. Wild-type (AC) and Gr fruit of identical age are shown for comparison. (B) GR expression in fruit samples shown in A. Total RNA (20 μg) extracted from normal ripening fruit was hybridized to a 32P-labeled GR probe. The filter was stripped and reprobed with an 18S rRNA probe. (C) Petal retention on developing fruits of multiple CaMV35S::GR transgenic lines. (D) Frequency of ethylene-induced floral abscission. Floral abscission was monitored in wild type (AC), Gr, and two homozygous CaMV35S::GR lines (4-20 and 6-14) 72 h after ethylene treatment. The mean of three independent experiments derived from at least 134 flowers is presented. Vertical bars represent SE.
CaMV35S::GR Does Not Lead to Whole-Plant Ethylene Insensitivity in Tomato.
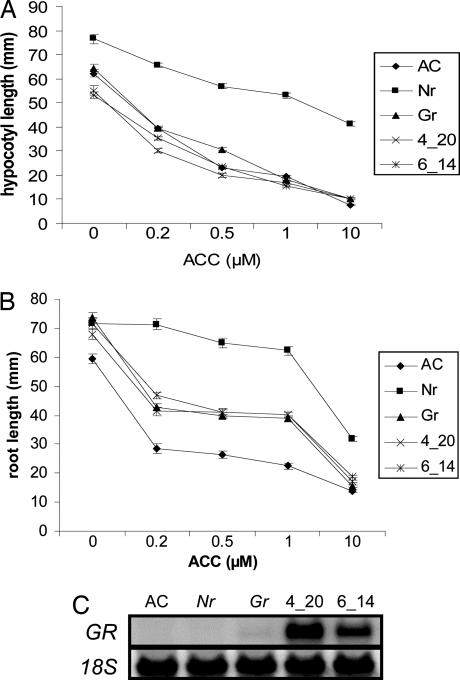
Dark grown hypocotyls of Gr undergo normal inhibition of cell elongation in response to ethylene; however, roots display a slightly reduced response (26). The triple response phenotype was monitored in two homozygous CaMV35S::GR transgenic lines to determine whether ethylene-insensitivity could be induced in dark grown hypocotyls (Fig. 3 A and B). A dose–response curve of hypocotyl length in response to increasing concentrations of the ethylene precursor ACC revealed that CaMV35S::GR lines 4-20 and 6-14 showed a similar pattern of growth inhibition as wild-type (AC) and Gr. In contrast, hypocotyls of the partial ethylene-insensitive Nr mutant (3) display reduced inhibition of growth (Fig. 3A). In roots, three phenotypic classes were observed with respect to ethylene-induced growth inhibition (Fig. 3B). Wild-type (AC) was most responsive; Gr, 4-20, and 6-14 had an intermediate ethylene response; and Nr was least responsive. Given the wild-type response of Gr and CaMV35S::GR hypocotyls to ethylene, we monitored GR transcript levels in hypocotyls (Fig. 3C). GR transcripts were undetectable in AC and Nr hypocotyls, but were detectable in Gr and were abundant in lines 4-20 and 6-14. Similar to the response observed in hypocotyls, ethylene-induced petiole epinasty was observed in AC, Gr, 4-20 and 6-14 plants but was greatly reduced in Nr (data not shown), again demonstrating that GR effects are limited to a subset of tissues.
Fig. 3.
Seedling triple response phenotype in CaMV35S::GR transgenic lines. Seeds of five different genotypes: wild type (AC), Nr, Gr, and CaMV35S::GR lines 4-20 and 6-14 were surface sterilized and sown on 0.8% water agar containing ACC at the indicated concentrations. Hypocotyl (A) and root (B) lengths were determined 8 days after sowing. Each data point is the mean of at least 31 seedlings. Vertical bars represent SE. (C) GR expression in hypocotyls of etiolated seedlings. Total RNA (20 μg) extracted from the genotypes described in A was hybridized to a 32P-labeled GR probe. The filter was stripped and reprobed with an 18S rRNA probe.
GR Is a Member of a Small Gene Family in Higher Plants That Is Conserved in Eukaryotes.
GR encodes a protein of 243 aa with a molecular mass of ≈27.9 kDa and a pI of 6.92. A search of several transmembrane domain prediction programs with GR gave variable results with either two or three transmembrane spanning domains predicted depending on the program. A blastp search of the predicted GR protein sequence against the Conserved Domain Database (www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (29) revealed that GR contains a domain of unknown function (DUF778) that is conserved in several eukaryotic proteins of undetermined biochemical function. A blastp search of the GenBank nonredundant coding sequence (CDS) database (www.ncbi.nlm.nih.gov/blast) identified five homologous proteins in plants, two from Arabidopsis (At2G26070 and At3G51040), and three from rice (GenBank accession nos. NP_916598, AAV59409, and AAO37528). Numerous related proteins are present in metazoan genomes having e values in the range of 1e-17 to 2e-23. No homology was observed to proteins from fungal or bacterial genomes. A tblastn search of build three of the solanaceous unigene set (www.sgn.cornell.edu) revealed the existence of two additional tomato genes represented by the unigene numbers SGN-U225677 and SGN-U219847 that we have designated GREEN RIPE LIKE 1 and 2 (GRL1 and GRL2), respectively, and two potato genes represented by the unigenes SGN-U292599 and SGN-U276841. The TIGR gene indices (www.tigr.org/tdb/tgi) contain several predicted full-length GR-like ESTs from multiple plant species. GR shares 53%, 51%, and 37% amino acid identity with GRL1, At2G26070, and GRL2, respectively.
Alignment of GR with several homologous proteins reveals divergence at the N termini followed by two blocks of ≈60 aa that are highly conserved. An interesting feature of these proteins is that they contain a relatively large number of conserved cysteine and histidine residues throughout the protein (Fig. 6, which is published as supporting information on the PNAS web site). These are residues that possess high affinity for divalent cations. In addition, GR possesses a single copy of the motif MXCXXC at the C terminus and an MXXXM motif (where X is any hydrophobic residue) in a predicted membrane-spanning domain. These motifs have been shown to participate in the binding of copper ions in the Atx1 protein of yeast and the copper transport activity of the high-affinity copper uptake proteins (Ctr proteins) of human and yeast, respectively (30, 31). However, these putative motifs are not conserved in any of the homologous proteins, including GRL1 and GRL2, identified to date.
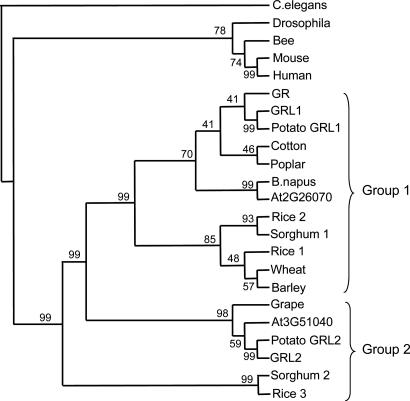
Phylogenetic analysis indicates that the plant derived proteins form two distinct clades designated group 1 and group 2 (Fig. 4). Distribution of the proteins indicates that higher plants seem likely to possess at least a single protein in each group; however, rice and tomato each possess a second protein within group 1. In the case of tomato, GR and GRL1 both reside in group 1, and GRL1 has higher homology than GR to At2g26070 (60% versus 51%), indicating that GR is the most divergent of the three proteins.
Fig. 4.
Phylogenetic analysis of GR related proteins. A nonrooted phylogenetic tree was generated by using the phylip 3.5c suite of programs (http://evolution.genetics.washington.edu/phylip.html) with the C. elegans protein as the out-group. The single most parsimonious tree obtained in a heuristic search following 100 random sequence addition replicates is shown. Bootstrap percentage supports are indicated. Sequence identifiers are available in Supporting Text.
GR Expression Is Associated with Seed Development.
Our results indicate that ripening inhibition of Gr is caused by a dominant gain of function mutation that causes elevated expression of GR in mutant fruit (Figs. 1C and 2A). As a first step to address where GR may be functioning under normal conditions, we examined the expression pattern of GR in various organs and tissues of tomato using RNA gel blot analysis. GR transcripts were undetectable during wild-type fruit ripening but, as suggested by RT-PCR data (Fig. 1B), expression was readily detectable in fruits of the Gr mutant at all stages examined (Fig. 5A). Treatment of mature green fruit with ethylene failed to result in a change of GR expression in either wild-type or the Gr mutant (Fig. 5B). Furthermore, expression was undetectable in leaves of different ages, roots, flowers, and trichomes (data not shown). However, GR transcripts were readily detectable in developing seeds extracted from immature and mature green fruit, but not in seeds from ripe fruit (Fig. 5C). Concomitant with increased GR expression in Gr fruit, elevated transcript levels were also observed in immature Gr seed (Fig. 5D). In agreement with data obtained from 5′ RACE, PCR amplification, and DNA sequence analysis of the GR locus (Fig. 1D), a slight reduction in transcript size was observed in Gr tissues (Fig. 5 C and D).
Fig. 5.
GR expression is associated with seed development. (A) GR expression during fruit ripening in wild type (AC) and Gr. Stages 1–5 are defined in ref. 26. (B) GR expression in mature green fruit with (+) or without (−) ethylene treatment. (C) GR expression during tomato seed development. RNA was extracted from seeds of wild-type tomato fruit at the immature green (2), mature green (3), and red-ripe (4) stages of development. RNA extracted from Gr mature green fruit pericarp tissue was included as a positive control (1). (D) GR expression in wild-type and mutant seeds. Seeds were extracted from immature wild-type (1) and Gr (2) fruit. All blots contained 15 μg of total RNA and were hybridized to a 32P-labeled GR probe. Images of the rRNA are shown as a guide to equal loading.
Discussion
Gr and Nr-2 Are Identical Mutations.
The phenotypic similarity between Gr and Nr-2 mutants coupled with their close physical proximity within the genome led us to speculate that they may represent allelic mutations (26). The presence of an identical deletion in both mutants (Fig. 1D) was unexpected given that both arose spontaneously (32, 33). To exclude any possibility that our seed stocks were contaminated we confirmed the presence of the deletion in the accessions LA2453 and LA2455 (data not shown). These accessions are homozygous for the Gr and Nr-2 mutation, respectively, possess distinct plant and fruit morphology, and were used as donors in the generation of our nearly isogenic lines and F2 mapping populations (26). To our knowledge, no other accessions carrying these mutations exist and, therefore, two possibilities remain: (i) an identical spontaneous mutation arose independently on two occasions or (ii) Nr-2 is the result of a pollen or seed contaminant derived from Gr. We are proposing to name the gene at the Gr/Nr-2 locus GREEN RIPE (GR) after the accession LA2453 that was first described (33). This name will avoid any confusion regarding comparisons between Never-ripe and Never-ripe 2, the former encoding an ethylene receptor (25).
Ethylene Signaling Specificity.
The Gr mutant displays reduced ethylene responsiveness in fruit, during floral senescence and abscission and root elongation but not in hypocotyls or petioles (26). Our data indicate that these phenotypes are caused by ectopic expression of GR resulting from a deletion of 5′ UTR and upstream regulatory sequences (Figs. 1C, 2, and 3). We hypothesized that CaMV35S::GR expression might lead to reduced ethylene responsiveness throughout the plant, resulting in reduced ethylene sensitivity in dark-grown hypocotyls. However, our data do not support this hypothesis despite high accumulation of GR transcripts in transgenic lines (Fig. 3 A and C). These data suggest that GR can modulate ethylene responses in a tissue-specific manner and, given that this effect is brought about by a dominant gain of function mutation, indicates that components of the ethylene signaling pathway differ in tomato between hypocotyls and the other tissues examined in this study (i.e., fruit, petals, abscission zones, and roots). The factors responsible for these differences remain unknown; however, it is possible that GR may function to disrupt ethylene signaling from specific receptors. Analysis of the expression patterns of the different ethylene receptor genes suggests that different tissues likely contain different pools of receptor proteins (19). For example, LeETR4 and LeETR5 are predominantly expressed in floral and fruit tissues with very low or no expression detected in etiolated hypocotyls (34). Therefore, a correlation exists between the expression of these receptor isoforms and GR function. Identification of GR provides a tool to assess apparent tissue-specific ethylene signal transduction.
Tissue-specific perturbation of ethylene responses has been previously documented in several Arabidopsis mutants, namely hookless 1 (hls 1), ethylene-insensitive root 1 (eir 1), enhanced ethylene response 1 (eer1), and weak ethylene insensitive 2 and 3 (wei2, wei3) (2, 10, 35, 36). These mutants display ethylene insensitivity in a single aspect of seedling morphology, and molecular characterization has revealed that all function to regulate synthesis, transport or responsiveness to auxin (37–40) or, in the case of eer1, which encodes the protein phosphatase 2A A regulatory subunit, RCN1 (41), to participate in the function of multiple hormonal signaling pathways (42, 43). The specificity of the Gr mutant phenotype differs from that of the Arabidopsis tissue-specific mutants in that ethylene responsiveness was reduced notably, although only moderately in the majority of the tissues examined but with a dramatic impact on fruit ripening. At present, we cannot rule out that GR participates in the signaling of multiple hormone response pathways.
GR as a Potential Regulator of Ethylene Responses.
Because Gr confers a dominant gain-of-function mutation, a fundamental question regarding GR function is whether the protein is an integral component of the ethylene signaling pathway or whether, through ectopic expression, GR is able to mediate cellular changes that lead to altered ethylene responsiveness yet may have a different function under normal expression conditions and levels. RNA interference-mediated repression of GR in transgenic tomatoes should address these possibilities.
One hypothesis to explain Gr phenotypes that is consistent with the second scenario is that deregulated expression of GR in mutant or transgenic fruit is able to inhibit normal functioning of GRL1 or GRL2 via a currently undetermined mechanism that may involve competing for binding partners or disrupting protein complexes. This hypothesis assumes that either or both GRL1 and GRL2 normally function as part of the ethylene signaling pathway in tomato. Consistent with this hypothesis, primary transformants overexpressing GRL1 and GRL2 do not display ethylene insensitive phenotypes as do the GR overexpression lines described in this study. For example, fruit from CaMV35S::GRL1 and CaMV35S::GRL2 lines ripen normally and show no signs of delayed petal senescence despite high transgene expression (unpublished data).
A second hypothesis to explain GR function that is consistent with a normal role in ethylene signaling is that the plant controls levels of GR to selectively repress ethylene signaling in tissues where this may be detrimental to plant or cell survival. The levels of GR expression are low or undetectable in all tissues that we examined with the exception of developing seeds (Fig. 5C). It is possible that GR expression may be increased in developing seeds to inhibit ethylene signaling and protect the developing embryo. The function of ethylene in developing tomato seeds is unclear, but studies in Arabidopsis, petunia, and maize implicate ethylene in regulating the levels of multiple hormones and ABA signaling to control dormancy, seed weight, and cell death in the endosperm, respectively (44–47).
GR Homologues Are Conserved in Plants and Animals.
Comparison of the deduced amino acid sequence of GR with various sequence repositories identified a number of homologous proteins in a range of eukaryotes (Fig. 4). The biochemical function of this family remains a mystery but all possess a conserved domain of unknown function (DUF778). GR is currently the only member of this family to which a mutant phenotype has been assigned. The tomato members of the GR family are diverse in the composition of their primary sequence. For example, the two most closely related proteins, GR and GRL1, share only 52% amino acid identity. In addition, GR, but not other family members, contains an MXCXXC at the C terminus of the protein and an MXXXM motif within one of the predicted transmembrane-spanning domains. These motifs have been shown to participate in the binding of copper ions in the Atx1 protein of yeast and the copper transport activity of the high-affinity copper uptake proteins (Ctr proteins) of human and yeast, respectively (30, 31). The significance of these motifs requires further investigation, but metal ion homeostasis is fundamental for ethylene signaling. The receptors contain copper that mediates ethylene binding (48). Furthermore, mutations at the response to antagonist 1 (ran1) locus, which encodes a conserved copper transporting P-type ATPase disrupt ethylene signaling (49, 50), the ein2 locus encodes a protein of unknown function that shares homology with the NRAMP family of metal ion transporters (11), and pharmacological studies implicate calcium in transduction of the ethylene signal (51).
Potential for GR in Crop Improvement.
Control of ethylene responsiveness in crops is of commercial importance to reduce senescence, overripening, and postharvest deterioration of fruit, vegetable, and floral crops. Previous research has led to the generation of transgenic horticultural crops with altered ethylene responsiveness to counteract the negative impacts of ethylene on ripening and floral senescence (52, 53). These studies have successfully achieved their aims, but subsequent evaluation of horticultural performance has revealed that constitutive ethylene insensitivity mediated by a dominant gain of function receptor mutation has deleterious effects on seed germination, seedling vigor, and adventitious rooting in tomato and petunia (46, 54, 55). The CaMV35S::GR transgene has a range of phenotypic penetrance in different tissues (i.e., a strong influence in fruit, moderate impact on floral senescence and abscission, a weak effect on root growth and no discernable changes in hypocotyl or shoot growth; Figs. 2 and 3). This differential mediation of ethylene responsiveness by GR may be useful for reducing the impact of the less desirable consequences of ethylene on tissues such as ripe fruit whilst maintaining normal plant vigor. In the accompanying paper, loss of function of an Arabidopsis GR homolog, RTE1/At2G26070, suppresses Etr 1-2-mediated ethylene insensitivity (56).
Materials and Methods
Plant Material and Treatments.
Plant growth conditions, mutant lines, and mapping populations used in this study have been described (26). Fruit were harvested at five developmental stages (termed 1–5) representing mature green through to fully ripe or equivalence for mutant fruit. Ethylene treatments were performed as described (26). Experiments to evaluate the triple response and floral abscission were performed as described (26), except that seedlings were measured 8 days after sowing, and flowers were induced to abscise by treatment with 2 μl·l−1 ethylene.
Genetic and Physical Mapping.
Genomic DNA isolation and genetic mapping were performed as described (26). A physical contig spanning the Gr locus was obtained via screening and characterization of ordered BAC and cosmid libraries derived from S. lycopersicum and S. cheesmaniae (27, 28). Clones 93D5, 26A21, and 237H16 were isolated from an S. cheesmaniae BAC library, and 151L6 was isolated from an S. lycopersicum cosmid library. BAC and cosmid ends were isolated by DNA sequencing and converted to RFLP or CAPS markers. Details of marker polymorphisms can be found in Supporting Text, which is published as supporting information on the PNAS web site.
Molecular Analysis of the Gr Locus.
Total RNA was extracted from plant tissues and fractionated through 1% denaturing agarose gels as described (26). RT-PCR amplification of GR from AC, Gr, and Nr-2 genotypes was achieved through use of primers designed from the sequence of the EST clone cLPT12O9 (GenBank accession no. AW618118): C1, 5′-GAATCATGAATGCTCCACCGCATGA-3′; C2, 5′-TGCTGAGAAGACACATTAAGGTAAC-3′; CR, 5′-TAACATTGCATTACAACACTGGACA-3′. cDNA was synthesized from 500 ng of total RNA extracted from a pool of mixed fruit stages by using Superscript II reverse transcriptase (Invitrogen). PCR amplification of genomic DNA spanning the deletion in Gr and Nr-2 was achieved by using the primers G1 (5′-CATGAATGCTCCACCGCATGACGTA-3′) and G2 (5′-TTCACTGGCACGCCCTAACA-3′). The 5′ ends of cDNAs for GR, GRL1, and GRL2 were obtained by RACE using the BD Smart RACE cDNA kit (Clontech) (see Supporting Text).
Vector Construction and Plant Transformation.
The full-length cDNA sequence of GR derived from Gr/Gr was cloned downstream of the CaMV35S promoter in a modified form of the binary vector pBI121 (see online supporting information for details). Transgenic tomato plants were generated by Agrobacterium tumefaciens-mediated transformation (strain GV3101).
DNA Sequence Analysis and Bioinformatics Resources.
Details of DNA sequence analysis, amino acid alignments, phylogenetic analysis, and membrane prediction programs can be found in Supporting Text.
Supplementary Material
Acknowledgments
We thank Ryan McQuinn for assistance with genetic mapping, Sherry Roof for plant transformation, Julia Vrebalov (Boyce Thompson Institute for Plant Research) and Ruth White [U.S. Department of Agriculture (USDA)/Agriculture Research Service] for BAC resources, and Ken Manning and Graham Seymour (Warwick-HRI, Warwick, U.K.) for the S. lycopersicum cosmid library. This work was supported by USDA/National Research Initiative 2002-35304-12530, the USDA/Agricultural Research Service, and National Science Foundation Grants DBI-0116076 and DBI-0501778.
Abbreviation
- CaMV
cauliflower mosaic virus
See Commentary on page 7537.
References
- 1.Bleecker A. B., Estelle M. A., Somerville C., Kende H. Science. 1988;241:1086–1089. doi: 10.1126/science.241.4869.1086. [DOI] [PubMed] [Google Scholar]
- 2.Guzman P., Ecker J. R. Plant Cell. 1990;2:513–524. doi: 10.1105/tpc.2.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lanahan M. B., Yen H. C., Giovannoni J. J., Klee H. J. Plant Cell. 1994;6:521–530. doi: 10.1105/tpc.6.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang K. L. C., Li H., Ecker J. R. Plant Cell. 2002;14:S131–S151. doi: 10.1105/tpc.001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo H., Ecker J. R. Curr. Opin. Plant Biol. 2004;7:40–49. doi: 10.1016/j.pbi.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 6.Hua J., Meyerowitz E. M. Cell. 1998;94:261–271. doi: 10.1016/s0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- 7.Kieber J. J., Rothenberg M., Roman G., Feldmann K. A., Ecker J. R. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- 8.Clark K. L., Larsen P. B., Wang X., Chang C. Proc. Natl. Acad. Sci. USA. 1998;95:5401–5406. doi: 10.1073/pnas.95.9.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao Z., Chen Y.-F., Randlett M. D., Zhao X.-C., Findell J. L., Kieber J. J., Schaller G. E. J. Biol. Chem. 2003;278:34725–34732. doi: 10.1074/jbc.M305548200. [DOI] [PubMed] [Google Scholar]
- 10.Roman G., Lubarsky B., Kieber J. J., Rothenberg M., Ecker J. R. Genetics. 1995;139:1393–1409. doi: 10.1093/genetics/139.3.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alonso J. M., Hirayama T., Roman G., Nourizadeh S., Ecker J. R. Science. 1999;284:2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- 12.Hall A. E., Bleecker A. B. Plant Cell. 2003;15:2032–2041. doi: 10.1105/tpc.013060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chao Q., Rothenberg M., Solano R., Roman G., Terzaghi W., Ecker J. R. Cell. 1997;89:1133–1144. doi: 10.1016/s0092-8674(00)80300-1. [DOI] [PubMed] [Google Scholar]
- 14.Solano R., Stepanova A., Chao Q., Ecker J. R. Genes Dev. 1998;12:3703–3714. doi: 10.1101/gad.12.23.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohme-Takagi M., Suzuki K., Shinshi H. Plant Cell Physiol. 2000;41:1187–1192. doi: 10.1093/pcp/pcd057. [DOI] [PubMed] [Google Scholar]
- 16.Fujimoto S. Y., Ohta M., Usui A., Shinshi H., Ohme-Takagi M. Plant Cell. 2000;12:393–404. doi: 10.1105/tpc.12.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo H., Ecker J. R. Cell. 2003;115:667–677. doi: 10.1016/s0092-8674(03)00969-3. [DOI] [PubMed] [Google Scholar]
- 18.Potuschak T., Lechner E., Parmentier Y., Yanagisawa S., Grava S., Koncz C., Genschik P. Cell. 2003;115:679–689. doi: 10.1016/s0092-8674(03)00968-1. [DOI] [PubMed] [Google Scholar]
- 19.Klee H. J. Plant Physiol. 2004;135:660–667. doi: 10.1104/pp.104.040998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams-Phillips L., Barry C., Giovannoni J. Trends Plant Sci. 2004;9:331–338. doi: 10.1016/j.tplants.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Adams-Phillips L., Barry C., Kannan P., Leclercq J., Bouzayen M., Giovannoni J. Plant Mol. Biol. 2004;54:387–404. doi: 10.1023/B:PLAN.0000036371.30528.26. [DOI] [PubMed] [Google Scholar]
- 22.Shibuya K., Barry K. G., Ciardi J. A., Loucas H. M., Underwood B. A., Nourizadeh S., Ecker J. R., Klee H. J., Clark D. G. Plant Physiol. 2004;136:2900–2912. doi: 10.1104/pp.104.046979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tieman D. M., Ciardi J. A., Taylor M. G., Klee H. J. Plant J. 2001;26:47–58. doi: 10.1046/j.1365-313x.2001.01006.x. [DOI] [PubMed] [Google Scholar]
- 24.Tieman D. M., Taylor M. G., Ciardi J. A., Klee H. J. Proc. Natl. Acad. Sci. USA. 2000;97:5663–5668. doi: 10.1073/pnas.090550597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilkinson J. Q., Lanahan M. B., Yen H. C., Giovannoni J. J., Klee H. J. Science. 1995;270:1807–1809. doi: 10.1126/science.270.5243.1807. [DOI] [PubMed] [Google Scholar]
- 26.Barry C. S., McQuinn R. P., Thompson A. J., Seymour G. B., Grierson D., Giovannoni J. J. Plant Physiol. 2005;138:267–275. doi: 10.1104/pp.104.057745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li C., Schilmiller A. L., Liu G., Lee G. I., Jayanty S., Sageman C., Vrebalov J., Giovannoni J. J., Yagi K., Kobayashi Y., Howe G. A. Plant Cell. 2005;17:971–986. doi: 10.1105/tpc.104.029108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Budiman M. A., Mao L., Wood T. C., Wing R. A. Genome Res. 2000;10:129–136. [PMC free article] [PubMed] [Google Scholar]
- 29.Marchler-Bauer A., Bryant S. H. Nucleic Acids Res. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pufahl R. A., Singer C. P., Peariso K. L., Lin S. J., Schmidt P. J., Fahrni C. J., Cizewski-Culotta V., Penner-Hahn J. E., O’Halloran T. V. Science. 1997;278:853–856. doi: 10.1126/science.278.5339.853. [DOI] [PubMed] [Google Scholar]
- 31.Puig S., Lee J., Lau M., Thiele D. J. J. Biol. Chem. 2002;277:26021–26030. doi: 10.1074/jbc.M202547200. [DOI] [PubMed] [Google Scholar]
- 32.Kerr E. A. Rep. Tomato Genetics Coop. 1982;32:33. [Google Scholar]
- 33.Kerr E. A. Rep. Tomato Genetics Coop. 1958;8:22. [Google Scholar]
- 34.Tieman D. M., Klee H. J. Plant Physiol. 1999;120:165–172. doi: 10.1104/pp.120.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larsen P. B., Chang C. Plant Physiol. 2001;125:1061–1073. doi: 10.1104/pp.125.2.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alonso J. M., Stepanova A. N., Solano R., Wisman E., Ferrari S., Ausubel F. M., Ecker J. R. Proc. Natl. Acad. Sci. USA. 2003;100:2992–2997. doi: 10.1073/pnas.0438070100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lehman A., Black R., Ecker J. R. Cell. 1996;85:183–194. doi: 10.1016/s0092-8674(00)81095-8. [DOI] [PubMed] [Google Scholar]
- 38.Li H., Johnson P., Stepanova A., Alonso J. M., Ecker J. R. Dev. Cell. 2004;7:193–204. doi: 10.1016/j.devcel.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 39.Luschnig C., Gaxiola R. A., Grisafi P., Fink G. R. Genes Dev. 1998;12:2175–2187. doi: 10.1101/gad.12.14.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stepanova A. N., Hoyt J. M., Hamilton A. A., Alonso J. M. Plant Cell. 2005;17:2230–2242. doi: 10.1105/tpc.105.033365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larsen P. B., Cancel J. D. Plant J. 2003;34:709–718. doi: 10.1046/j.1365-313x.2003.01762.x. [DOI] [PubMed] [Google Scholar]
- 42.Kwak J. M., Moon J.-H., Murata Y., Kuchitsu K., Leonhardt N., DeLong A., Schroeder J. I. Plant Cell. 2002;14:2849–2861. doi: 10.1105/tpc.003335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garbers C., Delong A., Deruere J., Bernasconi P., Soll D. EMBO J. 1996;15:2115–2124. [PMC free article] [PubMed] [Google Scholar]
- 44.Beaudoin N., Serizet C., Gosti F., Giraudat J. Plant Cell. 2000;12:1103–1115. doi: 10.1105/tpc.12.7.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiwocha S. D. S., Cutler A. J., Abrams S. R., Ambrose S. J., Yang J., Ross A. R. S., Kermode A. R. Plant J. 2005;42:35–48. doi: 10.1111/j.1365-313X.2005.02359.x. [DOI] [PubMed] [Google Scholar]
- 46.Clevenger D. J., Barrett J. E., Klee H. J., Clark D. G. J. Am. Soc. Hortic. Sci. 2004;129:401–406. [Google Scholar]
- 47.Young T. E., Gallie D. R. Plant Mol. Biol. 2000;44:283–301. doi: 10.1023/a:1026588408152. [DOI] [PubMed] [Google Scholar]
- 48.Rodriguez F. I., Esch J. J., Hall A. E., Binder B. M., Schaller G. E., Bleecker A. B. Science. 1999;283:996–998. doi: 10.1126/science.283.5404.996. [DOI] [PubMed] [Google Scholar]
- 49.Woeste K. E., Kieber J. J. Plant Cell. 2000;12:443–455. doi: 10.1105/tpc.12.3.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hirayama T., Kieber J. J., Hirayama N., Kogan M., Guzman P., Nourizadeh S., Alonso J. M., Dailey W. P., Dancis A., Ecker J. R. Cell. 1999;97:383–393. doi: 10.1016/s0092-8674(00)80747-3. [DOI] [PubMed] [Google Scholar]
- 51.Raz V., Fluhr R. Plant Cell. 1992;4:1123–1130. doi: 10.1105/tpc.4.9.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilkinson J. Q., Lanahan M. B., Clark D. G., Bleecker A. B., Chang C., Meyerowitz E. M., Klee H. J. Nat. Biotechnol. 1997;15:444–447. doi: 10.1038/nbt0597-444. [DOI] [PubMed] [Google Scholar]
- 53.Bovy A. G., Angenent G. C., Dons H. J. M., van-Altvorst A. C. Mol. Breeding. 1999;5:301–308. [Google Scholar]
- 54.Clark D. G., Gubrium E. K., Barrett J. E., Nell T. A., Klee H. J. Plant Physiol. 1999;121:53–59. doi: 10.1104/pp.121.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gubrium E. K., Clevenger D. J., Clark D. G., Barrett J. E., Nell T. A. J. Am. Soc. Hort. Sci. 2000;125:277–281. [Google Scholar]
- 56.Resnick J. S., Wen C.-K., Shockey J. A., Chang C. Proc. Natl. Acad. Sci. USA. 2006;103:7917–7922. doi: 10.1073/pnas.0602239103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.