Abstract
Hsp90 is a ubiquitous molecular chaperone responsible for assembly and regulation of many eukaryotic signalling systems, and an emerging target for rational chemotherapy of many cancers. Although the structures of isolated domains of Hsp90 have been determined, the arrangement and ATP-dependent dynamics of these in the full Hsp90 dimer have been elusive and contentious. Here we present the crystal structure of full-length yeast Hsp90 in complex with an ATP analogue and the co-chaperone p23/Sba1. The structure reveals the complex architecture of the ‘closed’ state of the Hsp90 chaperone, the extensive inter-domain and inter-strand interactions, the detailed conformational changes in the N-terminal domain that accompany ATP binding, and the structural basis for stabilisation of the closed state by p23/Sba1. Contrary to expectations, the closed Hsp90 would not enclose its client proteins but provides a bipartite binding surface whose formation and disruption is coupled to the chaperone ATPase cycle.
Hsp90 is an essential molecular chaperone in eukaryotes, required for activation of many regulatory and signalling ‘client’ proteins. Hsp90 function depends on its ability to bind and hydrolyse ATP1, 2, and pharmacological inhibition by ATP-competitors promotes client degradation3, 4. The requirement of Hsp90 for the function of oncogenic protein kinases such as ErbB2, Cdk4, B-Raf, Akt/PKB etc (reviewed in5) makes it an attractive target for novel cancer therapeutics6. Hsp90 associates with a plethora of co-chaperones, several of which regulate progress through its ATPase cycle7–9. The p23 co-chaperone and its S.cerevisiae homologue Sba1, preferentially bind Hsp90 in the presence of ATP or ATP-mimetics10–12. As p23/Sba1 binds ~70-fold more tightly to ATP-bound Hsp90{Siligardi, 2004 #658}, it stabilises the state required for client-protein activation, slowing ATP turnover7, 13, 14. This regulatory property explains why, although not essential for Hsp90-dependent activation of client proteins, p23/Sba1 makes it more efficient15, 16.
Previous studies suggested that the dimeric Hsp90 operates a ‘molecular clamp’ mechanism coupled to its ATPase cycle, involving closure of a ‘lid’ segment and transient dimerisation of the N-terminal nucleotide-binding domain in the ATP-bound state17, 18. However, this model has recently been challenged19–21. We have now determined the crystal structure of yeast Hsp90 trapped in a closed conformation, in complex with a non-hydrolysable ATP analogue and p23/Sba1. The structure provides a first view of Hsp90 in the ATP-bound state, defining the conformational changes in the N-domain that accompany closure, and revealing how p23/Sba1 recognises and stabilises the ATP-bound conformation of the Hsp90 dimer. The structure confirms the ATPase coupled molecular clamp mechanism, and provides a structural basis from which to understand ATP-dependent activation of Hsp90 client proteins.
Architecture of the Hsp90-p23/Sba1 Complex
Yeast Hsp90, with an Ala107Asn mutation shown to activate Hsp90s ATPase cycle18, and with truncation of the dispensable charged-linker connecting the N-domain and middle segments22, was co-crystallised with the non-hydrolysable ATP analogue AMP-PNP and Sba1, the yeast homologue of p2311. Crystals were phased by molecular replacement with the isolated N-terminal domain23 and middle segment of yeast Hsp9024, and the globular core of human p2325. The structure of the yeast Hsp90 C-terminal domain was determined independently from crystals of a middle-and C-terminal Hsp90 construct (M-C) consisting of residues 273-709 refined at 3.0Å, and completed the molecular replacement solution, allowing refinement of the full ~ 220 kDa (Hsp90)2 - (Sba1)2 complex against anisotropic data to a maximum resolution of 3.1Å (3.5Å isotropic) (see Supplementary Information).
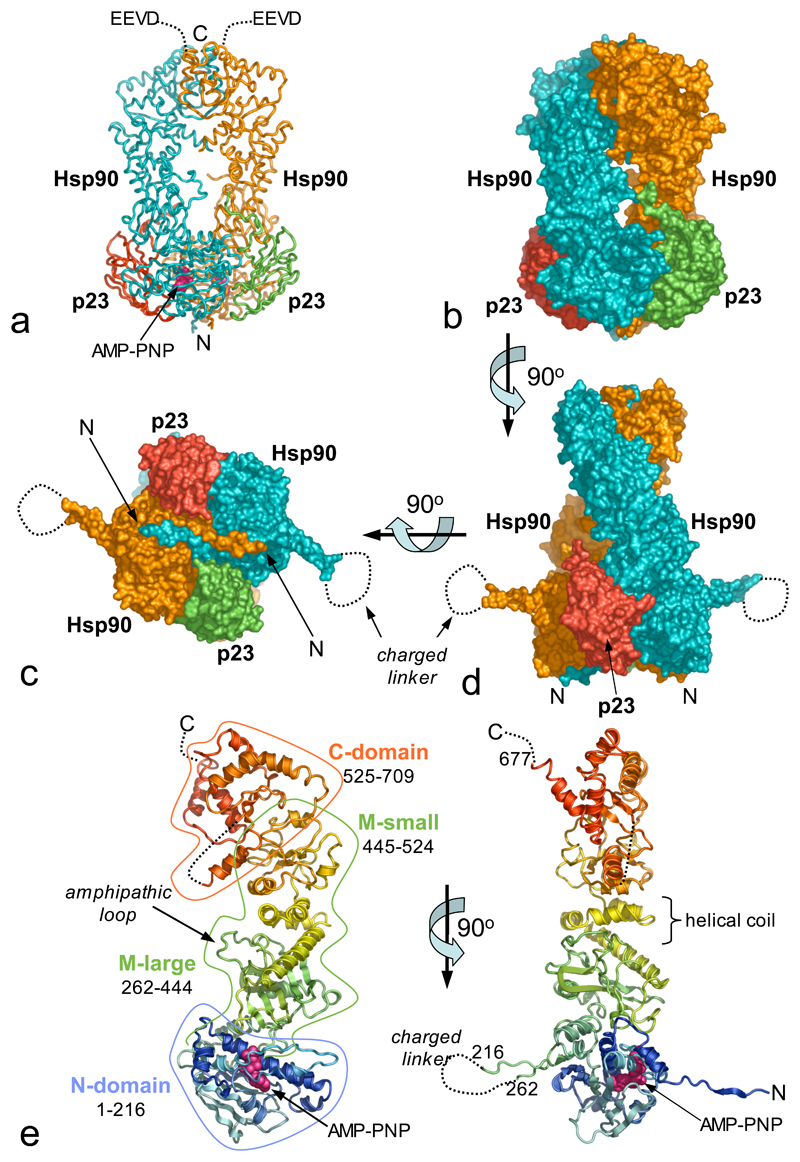
The Hsp90-p23/Sba1 complex contains a central Hsp90 dimer with symmetrically disposed p23/Sba1 molecules on either side (Figure 1a,b,c,d). The Hsp90 protomers have a parallel arrangement, with domains arranged in linear order such that the N-terminal domains are at one end of the dimer, and the C-terminal domains at the other. The protomers have a left-handed helical twist around the long axis of the dimer. Each Hsp90 protomer consists of four domains (Figure 1e). The N-domain (residues 1-216) is a two-layer sandwich, formed by a twisted β-sheet on one face, and a cluster of α-helices on the other, delimiting the binding pocket for ATP and a range of inhibitors26–30. The N-domain connects to the middle segment via an antiparallel β-strand involving residues 206-213 and 262-269. The loop connecting these strands formed by the truncated charged-linker, is disordered.
Figure 1. Architecture of Hsp90-p23/Sba1 Complex.
a) Backbone tracing of the (Hsp90)2-(p23/Sba1)2 complex; Hsp90 - blue and orange, p23/Sba1 - red and green. All molecular graphics were produced with MacPyMOL (www.pymol.org)
b) Molecular surface equivalent of a). The left-handed twist of the Hsp90 molecules is evident.
c) As b) the projecting β-ribbon is connected by the truncated charged-linker, which is disordered in the crystals.
d) As c) but rotated by 90° around the horizontal so that the exchange of the N-terminal strands is evident.
e) Orthogonal view of Hsp90 protomer, rainbow coloured from the N-terminus (blue) to the C-terminus (red).
The middle segment consists of a large three-layer α-β-α domain (residues 273-409) connected to a small α-β-α domain (435-525) via a helical coil (411-443)24. The relative orientation of the large and small domains is little changed from the isolated middle segment structure. An amphipathic loop (residues 329-339) implicated in client protein interactions24, projects from the inner face of the larger domain towards its equivalent in the other monomer. An extended loop connects the middle segment to the beginning of a curved α-helix (534-559) at the start of the C-domain, which consists of a three stranded β-sheet, packed beneath the curved helix and capping one face of a three-helix coil, whose other face forms the constitutive dimerisation interface. A helix-strand segment (residues 587-610) projects from the core of the C-domain towards the N-terminal end of the dimer and is involved in dimeric interactions with its equivalent in the other protomer. The yeast C-domain is similar in structure to the E.coli HtpG C-domain31, but with a significantly different orientation for this projecting helix-strand segment. Residues 678-709, which provide the C-terminal EEVD binding sequence for TPR-domain co-chaperones32, are disordered.
Sba1 has the same two-layer β-sandwich structure as human p2325, with some differences in the loops connecting the β-strands. Unlike unbound p23, ordered structure is evident beyond the core, with a segment of the charged C-terminal tail (residues 129-139) snaking up towards the middle of the complex.
Domain Interactions and Conformational Changes
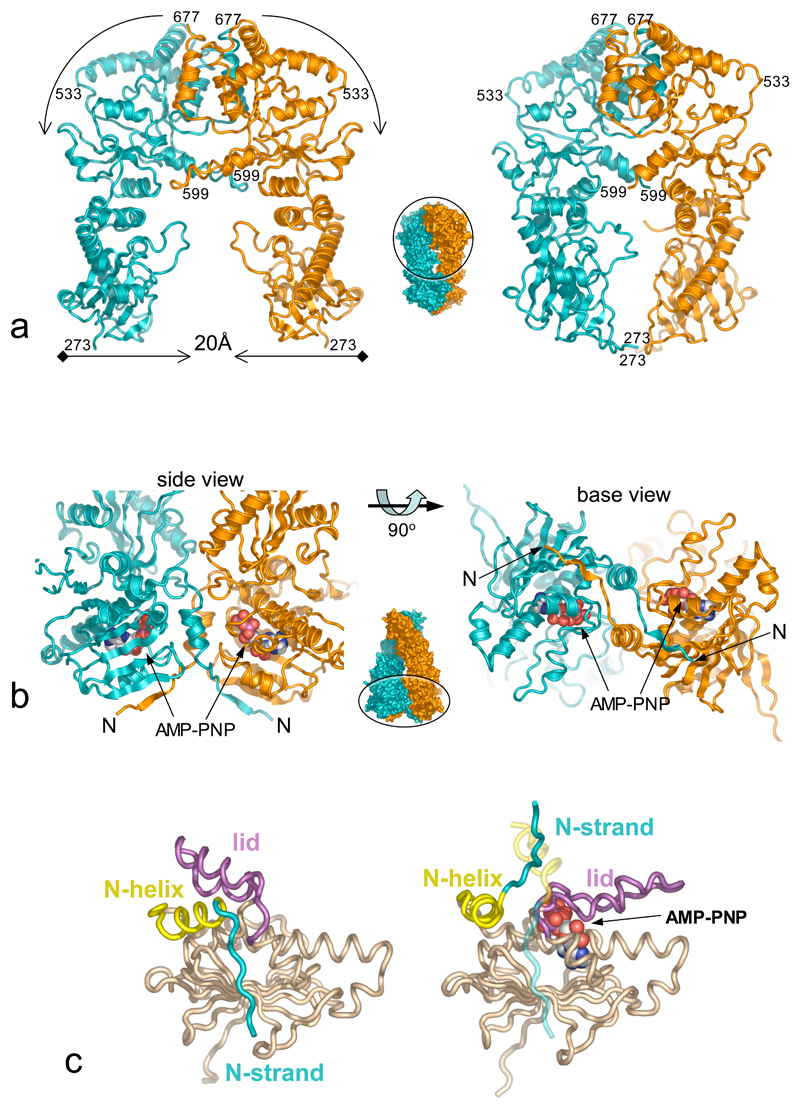
Extensive interfaces occur between the domains of the two Hsp90s, and the p23/Sba1 molecules. The constitutive dimerisation interface between the C-domains is essentially identical in the M-C structure (residues 273-709) and the full complex, suggesting little change on ATP-binding. The interface between the C-domain and small middle segment domain, flexes on going from the unconstrained M-C structure to the ATP-bound conformation, bringing the small middle domains ~10Å closer together. This displaces the projecting helix-strand segments, which tilt downwards and become less well ordered. As several temperature sensitive mutations of yeast Hsp90 map in this regions33, 34, this conformational change may have functional significance. The middle segment large domains move even more in the ATP-bound conformation, coming ~20Å closer together. Although the middle segments do not actually come into contact, the gap remaining between them is too small to accommodate a folded client protein (Figure 2a).
Figure 2. ATP-Dependent Conformational Changes.
a) Comparison of the dimerised M-C construct (left), and equivalent regions from the Hsp90-p23/Sba1 complex (right). ATP-dependent association of the N-domains brings the the middle segments ~20Å closer together. The central ‘thumbnail’ of the Hsp90 dimer shows the direction of view and location of the M-C region in the overall structure.
b) Secondary structure cartoon of the strand-swapped interface between the N-domains of Hsp90, viewed from the base (left) and side (right).
c) Comparison of the isolated N-domain (left), and the AMP-PNP-bound state (right). In addition to lid closure, there is considerable movement of the N-terminal helix and complete detachment of the N-terminal strand, which swaps into the other N-domain of the dimer (shown as a ghost).
As previously shown17, 18, and in contradiction to recent suggestions19–21 the N-domains form an intimate dimeric interaction, involving significant changes in conformation relative to their structure in isolation. Firstly, the N-terminal β-strand (residues 1-9) swaps over to hydrogen bond to the edge of the main β-sheet in the N-domain of the other monomer, with concomitant movement of the first α-helix (residues 13-22) – an arrangement also seen in dimers of MutL and GyrB35, 36 (Figure 2b). Secondly, the ‘lid’ segment (residues 94-125), swings through nearly 180° from its ‘open’ position in the isolated N-domain, hinged at Gly 94 and Gly121, to fold over the nucleotide-binding pocket. The lid movement exposes a hydrophobic patch centred on Leu15 and Leu18, which along with Gln 14, and Thr 95, Ile 96, Ala 97 and Phe 120 from the hinge-points of the lid, form a substantial interface with their equivalents in the other monomer. Thus, as we previously suggested, lid closure in the ATP-bound state reveals a hydrophobic patch whose burial stabilises N-domain association18. The lid movement explains the effects of Thr101Ile and Ala107Asn mutations. Thr101 is buried in the open conformation of the lid, becoming exposed in the ATP-bound complex; its mutation to the more hydrophobic isoleucine stabilises the open conformation, decreasing ATPase activity. Ala107 comes close to Tyr 47 in the closed conformation; mutation to asparagine allows a polar interaction with Tyr47, stabilising the closed conformation and enhancing ATPase activity (Figure 2c).
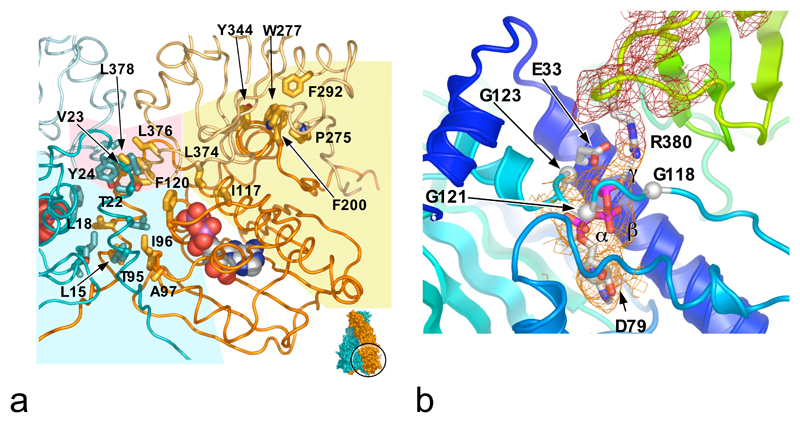
While the middle segments move together, they do not make contact. However each one interacts with the N-domain of the other protomer, in addition to the interface with its own N-domain (see below). This inter-molecular inter-domain interface involves Thr22, Val23 and Tyr24 from the N-domain, in a hydrophobic interaction with Leu376 and Leu378 in the middle segment loop bearing the catalytic residue Arg380. The involvement of Thr22 in this interface is particularly interesting, as Thr22Ile had been isolated as a ts allele in vivo34 and activated Hsp90s ATPase in vitro18, although the mechanism of this was not clear. Mutation to the more hydrophobic isoleucine would reinforce the interface with the catalytic loop, stabilising its active conformation (Figure 3a).
Figure 3. Domain Interfaces and Active Site Formation.
a) Interfaces between the Hsp90 N-domains and middle segments: the inter-molecular N-domain (pale blue background), inter-molecular N-domain – middle (pale pink), and intra-molecular N-domain – middle segment (pale yellow) that assembles the catalytic apparatus.
b) Details of the split catalytic apparatus. The γ-phosphate of AMP-PNP is orientated for attack by a water activated by Glu33. Arg380 polarises the γ-β phosphodiester bond and neutralises the transition state. Mutation of Glu33 or Arg380 kills catalysis; mutation of Asp79 abolishes ATP binding 24, 1. The mesh shows Fo-Fc ‘omit’ electron-density for the catalytic loop (red) and ATP-analogue (orange).
Nucleotide Interactions and active site formation
The adenine, ribose and α and β phosphate groups of AMP-PNP occupy the same position as ADP in high-resolution studies of the isolated N-terminal domain26, 2. The γ-phosphate is cradled in a glycine-rich loop at the C-terminal hinge of the lid, hydrogen bonding with the peptide backbone of Gln119, Gly121, Val122 and Gly123. The β-phosphate hydrogen bonds with the peptides of Phe 124, and Gly100 from the N-terminal hinge (Figure 3b).
The nucleotide makes a single contact outside the N-domain – a polar interaction between its γ-phosphate and the head group of Arg380, which projects into the nucleotide-binding pocket from the middle segment, close to the catalytically essential Glu331. The N-domain makes extensive interactions with the middle segment of the same protomer, involving the side chain of Phe200 from the N-domain, packing into a hydrophobic pocket formed by Thr273, Pro275, Trp277, Phe292 and Tyr344 in the middle segment. Residues 114-120 from the lid in the N-domain make polar and hydrophobic interactions with residues 372-379 from the middle segment catalytic loop. Both these segments have very different conformations in the isolated domains, so that their conformations in the ATP-bound complex are mutually interdependent (Figure 3a).
Previous studies suggested close interactions between the N-domain and middle segment of the protein in the ATP-bound state, and identified Arg380 as an essential catalytic residue 37, 24. The juxtaposition of the N-domain and middle segments, and the proximity of Arg380 to the bound nucleotide in the structure presented here, completely vindicate that mechanistic proposal. However, involvement of Arg380 has recently been questioned on the basis of structures of the E.coli Hsp90 HtpG, lacking the C-terminal dimerisation domain (residues 1-559), and in the presence of ADP19. These structures show radically different conformations to the present study, with little contact between the middle segment and the nucleotide-binding pocket, and with the equivalent of Arg380 too far from the nucleotide to play a catalytic role. While the bacterial homologue could operate by a different mechanism, the strong conservation of the residues involved in the N-domain - middle segment interface, and the sequence identity of almost all residues involved in nucleotide interactions, make this highly improbable. Most likely the HtpG conformation is an artefact, stabilised by favourable crystal lattice contacts, and reflecting the considerable flexibility of the protein in the ATP-free state.
Structural basis for conformation-dependent recruitment of p23/Sba1
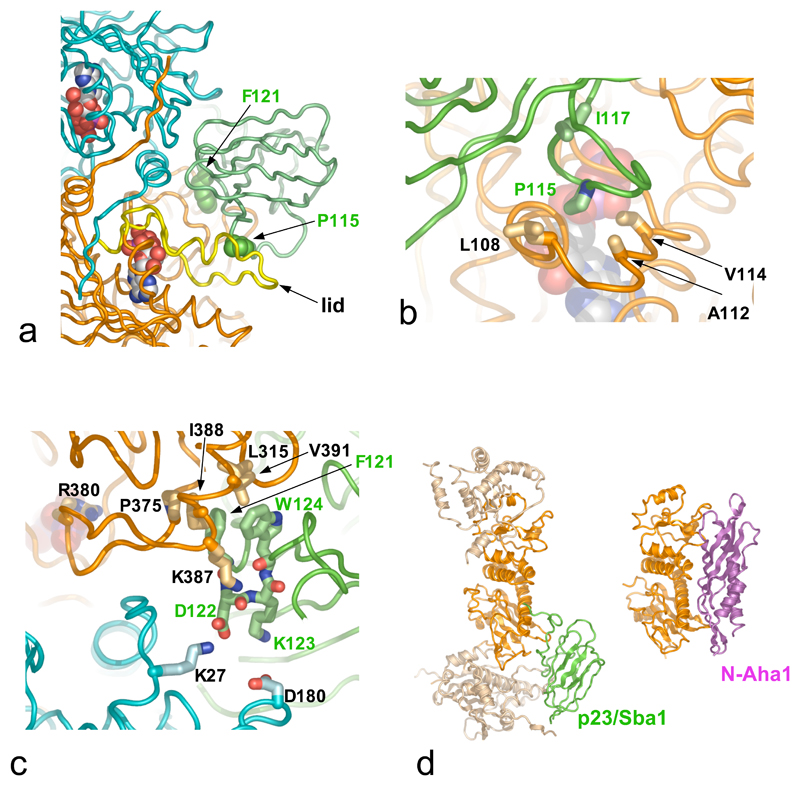
Each p23/Sba1 molecule lies in a depression at the junction of the two Hsp90 N-domains, contacting each via distinct surface patches (Figure 4a). The smaller interface involves residues 31-37 and 85-91 at the tips of β-hairpins in p23/Sba1 and residues 12-21 and 151-155 from one Hsp90 N-domain. The larger interface involves residues 13-16 from the N-terminal strand of p23/Sba1 along with a broad loop formed by residues 113-118, which packs against the surface of the lid segment of the other N-domain (Figure 4b). Formation of this interface requires lid closure, so that binding of p23/Sba1 would stabilise the ATP-bound conformation. Consistent with this, mutation of Ile117 in this loop causes significant loss of Sba1 function in vivo38.
Figure 4. Mechanism of ATPase Regulation by p23/Sba1.
a) Overall view of one p23/Sba1 molecule (green) packed against the Hsp90 dimer (cyan and orange), and interacting with the ‘lid’ (yellow) in its ATP-dependent closed conformation. Details are shown in b) and c).
b) Residues 113-118 of p23/Sba1 (green) pack onto the ‘lid’ (orange). Mutation of Ile117 causes severe Sba1 loss of function in yeast38.
c) The conserved DFxxW motif of p23/Sba1 binds across the inter-molecular inter-domain interface, reinforcing the activated conformation of the catalytic loop carrying Arg380.
d) Binding sites for p23/Sba1 (left) and Aha1 (right) overlap, so that their recruitment to Hsp90 is mutually exclusive39.
A third interface involves residues 120-125 of p23/Sba1 binding across the inter-molecular inter-domain junction (see above). One side of this bridging interaction involves Lys27 and Lys387 of Hsp90, and Asp122 and the peptide carbonyls of Phe121, Asp122 and Trp124, of p23/Sba1. On the other side, Phe121 and Trp124 from p23/Sba1 pack into a hydrophobic recess formed by Leu315, Pro375, Ile388 and Val391 from Hsp90 (Figure 4c). Remarkably, Lys387, Ile388 and Val391 from the catalytic loop make very similar interactions with Aha137, an Hsp90 co-chaperone that significantly activates Hsp90s inherent ATPase{Panaretou, 2002 #520; Lotz, 2003 #579; Siligardi, 2004 #658} (Figure 4d). Binding of Aha1 to Hsp90 remodels the catalytic loop towards the conformation seen in the AMP-PNP - bound Hsp90-p23/Sba1 complex. Although unrelated, Aha1 and p23/Sba1 share an ability to stabilise the catalytic loop in an active conformation, promoting ATP-hydrolysis. However, with a common binding site, their interaction with Hsp90 is mutually exclusive39.
None of the Hsp90 - p23/Sba1 interfaces have significant affinity individually, but in combination add to a substantial degree of interaction. However the p23/Sba1 binding sites are only presented in the correct three-dimensional configuration when Hsp90 is in the ATP-bound state. Thus, p23/Sba1 recruitment is ATP dependent, but once bound provides a scaffold, reinforcing the ATP-bound conformation and extending the lifetime of the state of the chaperone required for client protein activation.
ATPase coupled conformational cycle and client-protein activation
The structure presented here defines the ‘tense’ state of the chaperone and confirms the proposed dimerisation-coupled ‘split’ ATPase mechanism 18, 17, uniting Hsp90 with other dimeric GHKL ATPases, such as MutL and GyrB35, 36. As the key residues involved are highly conserved between yeast and human Hsp90s, it is difficult to rationalise suggestions that human Hsp90 has a different mechanism21. Hsp90 in solution does not have a single ‘relaxed’ conformation, but exists as a continuum of C-terminally dimerised conformations in which the rest of the molecule is unconstrained40. The ATP-bound state by contrast is a highly constrained structure whose formation involves coupled conformational switches in the N-terminus and lid of the N-domain, and in the catalytic loop of the middle segment, which together position the two halves of the catalytic apparatus for ATP hydrolysis. These switches rather than ATP hydrolysis itself, govern the rate of the chaperone cycle, and mutations that affect them alter Hsp90 ATPase activity in vitro and client protein activation in vivo33, 34, 18, 41. Furthermore, Hsp90 co-chaperones such as p23/Sba1, Aha1 and Cdc37, that regulate the chaperone ATPase cycle, do so by interacting with these switch components{Meyer, 2004 #619; Roe, 2004 #618; Siligardi, 2004 #658}.
Mutational analysis 1, 2, and the effect of ATP-competitive inhibitors6, show that ATP-binding to Hsp90 is crucial for client protein activation in vivo. Now the structural consequences of ATP binding to Hsp90 are understood, the key question remains – what property of this conformation contributes to client protein activation. At least for protein kinases some insight comes from an EM reconstruction of an Hsp90-Cdc37-Cdk4 complex (C.K. Vaughan, U.Gohlke, F. Sobott, V.M. Good, M.M.U. Ali, C. Prodromou, C.V. Robinson, H.R. Saibil, L.H. Pearl, manuscript submitted). This suggests that the two lobes of the kinase interact with different domains of Hsp90, so that the client conformation is directly coupled to the changes in their relative position that accompany the chaperone ATPase cycle. While this may provide a general mechanism for coupling chaperone and client conformations, how such changes in a client facilitate its activation remains to be defined. The structural data presented here provide a firm basis from which this key mechanistic question can be addressed.
Methods Overview
A full-length construct of yeast Hsp90 harbouring the mutation A107N and with residues 221-255 in the charged linker region deleted and replaced by LQHMASVD; an N-terminal and middle-segment (M-C) construct of yeast Hsp90 (residues 273-709); and full-length yeast p23 (Sba1) were inserted with addition of N-terminal His6-tag and PreScission protease cleavage site into pRSETA, and expressed in E.coli BL21(DE3) pLysS. The expressed proteins were purified to homogeneity by column chromatography and used in crystallisation experiments.
Crystals of the Hsp90-p23/Sba1 AMP-PNP complex and of the M-C construct were grown in hanging-drop vapour diffusion experiments, snap-frozen in liquid nitrogen, and used to collect X-ray diffraction data at ESRF Grenoble. The M-C structure was solved by molecular replacement with the yeast middle segment structure, and the C-terminal domain built from difference Fourier maps. The complex structure was determined by molecular replacement with structures of the separate N- middle and C-domains of yeast Hsp90, and human p23. Structures were refined using automated procedures interspersed with manual rebuilding.
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Acknowledgments
We are very grateful to Matt Gold, Paul Wan, Andrew Doré and Mairi Kilkenny for assistance with synchrotron data collection. This work was supported by a Programme Grant from The Wellcome Trust and infrastructural support for structural biology at The Institute of Cancer Research, from Cancer Research UK.
Footnotes
Author Information Coordinates and structure factors for the Hsp90-p23/Sba1/AMP-PNP complex and M-C construct have been deposited in the Protein Databank with accession codes : 2CGE and 2CG9. Reprints and permissions information is available at npg.nature.com/reprintsandpermissions.
The authors declare no competing financial interests.
References
- 1.Panaretou B, et al. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 1998;17:4829–4836. doi: 10.1093/emboj/17.16.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Obermann WMJ, Sondermann H, Russo AA, Pavletich NP, Hartl FU. In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J Cell Biol. 1998;143:901–910. doi: 10.1083/jcb.143.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mimnaugh EG, Chavany C, Neckers L. Polyubiquitination and proteasomal degradation of the p185c-erbB-2 receptor protein-tyrosine kinase induced by geldanamycin. J Biol Chem. 1996;271:22796–801. doi: 10.1074/jbc.271.37.22796. [DOI] [PubMed] [Google Scholar]
- 4.Schneider C, et al. Pharmacologic shifting of a balance between protein folding and degradation mediated by Hsp90. Proc natl Acad Sci USA. 1996;93:14536–14541. doi: 10.1073/pnas.93.25.14536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pearl LH. Hsp90 and Cdc37 -- a chaperone cancer conspiracy. Curr Opin Genet Dev. 2005;15:55–61. doi: 10.1016/j.gde.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 6.Workman P. Combinatorial attack on multistep oncogenesis by inhibiting the Hsp90 molecular chaperone. Cancer Lett. 2004;206:149–57. doi: 10.1016/j.canlet.2003.08.032. [DOI] [PubMed] [Google Scholar]
- 7.Panaretou B, et al. Activation of the ATPase activity of Hsp90 by the stress-regulated co-chaperone Aha1. Mol Cell. 2002;10:1307–1318. doi: 10.1016/s1097-2765(02)00785-2. [DOI] [PubMed] [Google Scholar]
- 8.Prodromou C, et al. Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J. 1999;18:754–762. doi: 10.1093/emboj/18.3.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siligardi G, et al. Regulation of Hsp90 ATPase activity by the co-chaperone Cdc37p/p50cdc37. J Biol Chem. 2002;277:20151–20159. doi: 10.1074/jbc.M201287200. [DOI] [PubMed] [Google Scholar]
- 10.Johnson JL, Toft DO. Binding of p23 and hsp90 during assembly with the progesterone receptor. Mol Endocrinol. 1995;9:670–8. doi: 10.1210/mend.9.6.8592513. [DOI] [PubMed] [Google Scholar]
- 11.Fang Y, Fliss AE, Rao J, Caplan AJ. SBA1 encodes a yeast hsp90 cochaperone that is homologous to vertebrate p23 proteins. Mol Cell Biol. 1998;18:3727–34. doi: 10.1128/mcb.18.7.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sullivan W, et al. Nucleotides and two functional states of hsp90. J Biol Chem. 1997;272:8007–12. doi: 10.1074/jbc.272.12.8007. [DOI] [PubMed] [Google Scholar]
- 13.McLaughlin SH, Smith HW, Jackson SE. Stimulation of the weak ATPase activity of human Hsp90 by a client protein. J Mol Biol. 2002;315:787–798. doi: 10.1006/jmbi.2001.5245. [DOI] [PubMed] [Google Scholar]
- 14.Richter K, Walter S, Buchner J. The Co-chaperone Sba1 connects the ATPase reaction of Hsp90 to the progression of the chaperone cycle. J Mol Biol. 2004;342:1403–13. doi: 10.1016/j.jmb.2004.07.064. [DOI] [PubMed] [Google Scholar]
- 15.Pratt WB, Dittmar KD. Studies with purified chaperones advance the understanding of the mechanism of glucocorticoid receptor hsp90 heterocomplex assembly. Trends Endocrinol Metab. 1998;9:244–252. doi: 10.1016/s1043-2760(98)00059-9. [DOI] [PubMed] [Google Scholar]
- 16.Young JC, Hartl FU. Polypeptide release by Hsp90 involves ATP hydrolysis and is enhanced by the co-chaperone p23. EMBO J. 2000;19:5930–5940. doi: 10.1093/emboj/19.21.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chadli A, et al. Dimerization and N-terminal domain proximity underlie the function of the molecular chaperone heat shock protein 90. Proc Natl Acad Sci USA. 2000;97:12524–12529. doi: 10.1073/pnas.220430297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prodromou C, et al. The ATPase cycle of Hsp90 drives a molecular 'clamp' via transient dimerization of the N-terminal domains. EMBO J. 2000;19:4383–4392. doi: 10.1093/emboj/19.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huai Q, et al. Structures of the N-terminal and middle domains of E. coli Hsp90 and conformation changes upon ADP binding. Structure (Camb) 2005;13:579–90. doi: 10.1016/j.str.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 20.Immormino RM, et al. Ligand-induced conformational shift in the N-terminal domain of GRP94, an Hsp90 chaperone. J Biol Chem. 2004;279:46162–71. doi: 10.1074/jbc.M405253200. [DOI] [PubMed] [Google Scholar]
- 21.McLaughlin SH, Ventouras LA, Lobbezoo B, Jackson SE. Independent ATPase activity of Hsp90 subunits creates a flexible assembly platform. J Mol Biol. 2004;344:813–26. doi: 10.1016/j.jmb.2004.09.055. [DOI] [PubMed] [Google Scholar]
- 22.Louvion JF, Warth R, Picard D. Two eukaryote-specific regions of Hsp82 are dispensable for its viability and signal transduction functions in yeast. Proc Natl Acad Sci U S A. 1996;93:13937–42. doi: 10.1073/pnas.93.24.13937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prodromou C, Roe SM, Piper PW, Pearl LH. A molecular clamp in the crystal structure of the N-terminal domain of the yeast Hsp90 chaperone. Nature Struct Biol. 1997;4:477–482. doi: 10.1038/nsb0697-477. [DOI] [PubMed] [Google Scholar]
- 24.Meyer P, et al. Structural and functional analysis of the middle segment of Hsp90: Implications for ATP hydrolysis and client-protein and co-chaperone interactions. Mol Cell. 2003;11:647–658. doi: 10.1016/s1097-2765(03)00065-0. [DOI] [PubMed] [Google Scholar]
- 25.Weaver AJ, Sullivan WP, Felts SJ, Owen BA, Toft DO. Crystal structure and activity of human p23, a heat shock protein 90 co-chaperone. J Biol Chem. 2000;275:23045–52. doi: 10.1074/jbc.M003410200. [DOI] [PubMed] [Google Scholar]
- 26.Prodromou C, et al. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell. 1997;90:65–75. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 27.Stebbins CE, et al. Crystal structure of an Hsp90-geldanamycin complex: Targetting of a protein chaperone by an antitumor agent. Cell. 1997;89:239–250. doi: 10.1016/s0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- 28.Roe SM, et al. The Structural Basis for Inhibition of the Hsp90 Molecular Chaperone, by the Anti-tumour Antibiotics Radicicol and Geldanamycin. J Med Chem. 1999;42:260–266. doi: 10.1021/jm980403y. [DOI] [PubMed] [Google Scholar]
- 29.Cheung KM, et al. The identification, synthesis, protein crystal structure and in vitro biochemical evaluation of a new 3,4-diarylpyrazole class of Hsp90 inhibitors. Bioorg Med Chem Lett. 2005;15:3338–43. doi: 10.1016/j.bmcl.2005.05.046. [DOI] [PubMed] [Google Scholar]
- 30.Wright L, et al. Structure-activity relationships in purine-based inhibitor binding to HSP90 isoforms. Chem Biol. 2004;11:775–85. doi: 10.1016/j.chembiol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 31.Harris SF, Shiau AK, Agard DA. The crystal structure of the carboxy-terminal dimerization domain of htpG, the Escherichia coli Hsp90, reveals a potential substrate binding site. Structure (Camb) 2004;12:1087–97. doi: 10.1016/j.str.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 32.Scheufler C, et al. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 33.Bohen SP, Yamamoto KR. Isolation of Hsp90 mutants by screening for decreased steroid receptor function. Proc Natl Acad Sci U S A. 1993;90:11424–8. doi: 10.1073/pnas.90.23.11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nathan DF, Lindquist S. Mutational analysis of Hsp90 function: interactions with a steroid receptor and a protein kinase. Mol Cell Biol. 1995;15:3917–3925. doi: 10.1128/mcb.15.7.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ban C, Junop M, Yang W. Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair. Cell. 1999;97:85–97. doi: 10.1016/s0092-8674(00)80717-5. [DOI] [PubMed] [Google Scholar]
- 36.Wigley DB, Davies GJ, Dodson EJ, Maxwell A, Dodson G. Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature. 1991;351:624–629. doi: 10.1038/351624a0. [DOI] [PubMed] [Google Scholar]
- 37.Meyer P, et al. Structural basis for recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery. EMBO J. 2004;23:511–9. doi: 10.1038/sj.emboj.7600060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oxelmark E, et al. Genetic dissection of p23, an Hsp90 cochaperone, reveals a distinct surface involved in estrogen receptor signaling. J Biol Chem. 2003;278:36547–55. doi: 10.1074/jbc.M305960200. [DOI] [PubMed] [Google Scholar]
- 39.Harst A, Lin H, Obermann WM. Aha1 competes with Hop, p50 and p23 for binding to the molecular chaperone Hsp90 and contributes to kinase and hormone receptor activation. Biochem J. 2005;387:789–96. doi: 10.1042/BJ20041283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang W, et al. Biochemical and structural studies of the interaction of Cdc37 with Hsp90. J Mol Biol. 2004;340:891–907. doi: 10.1016/j.jmb.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 41.Richter K, Reinstein J, Buchner J. N-terminal residues regulate the catalytic efficiency of the Hsp90 ATPase cycle. J Biol Chem. 2002;277:44905–44910. doi: 10.1074/jbc.M208457200. [DOI] [PubMed] [Google Scholar]
- 42.Vaughan CK, et al. Structure and assembly of Hsp90 - Cdc37 - protein kinase complexes. EMBO J. 2006 submitted. [Google Scholar]
- 43.Roe SM, et al. The Mechanism of Hsp90 regulation by the protein kinase-specific cochaperone p50(cdc37) Cell. 2004;116:87–98. doi: 10.1016/s0092-8674(03)01027-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.