Abstract
The emergence of the severe acute respiratory syndrome (SARS) that resulted in a pandemic in 2003 spurred a flurry of interest in the development of vaccines to prevent and treat the potentially deadly viral infection. Researchers around the world pooled their scientific resources and shared early data in an unprecedented manner in light of the impending public health crisis. There are still large gaps in knowledge about the pathogenesis of this virus. While significant advances have been made in the development of animal models, the practicality of their use may be hampered by a lack of pathological similarity with human disease. Described here are issues related to progress in vaccine development and the obstacles that lie ahead for both researchers and regulatory agencies.
Keywords: Severe acute respiratory syndrome, Coronavirus, SARS-CoV, Animal models, Vaccine
1. Introduction
Severe acute respiratory syndrome (SARS) was first reported as a disease of unknown etiology in Guangdong province, China, in November of 2002. According to the World Health Organization (WHO), the outbreak had spread and eventually 29 countries were treating infected individuals [1]. By the end of the outbreak in the summer of 2003, the number of SARS infected individuals exceeded 8096 and resulted in 774 deaths, a fatality rate of 9.6% [1].
SARS is characterized by fever, cough and flu-like symptoms. Severe cases resulted in alveolar damage, interstitial mononuclear cells and heavy fibrin deposition in the lungs. Respiratory distress resulted in atypical pneumonia, requiring ventilation for approximately 20% of patients [2]. The aim of this review is to describe the advances made in the development of animal models for SARS and to identify gaps in scientific understanding that need to be filled. By addressing and possibly overcoming these challenges and making use of the advances made, a safe and effective vaccine may be attainable. The information here may provide the scientific basis for facilitating future regulatory decisions related to the licensing of a SARS vaccine.
1.1. Molecular biology of the SARS coronavirus
The causative agent for SARS is a novel member of the coronavirus family, termed SARS-CoV [3]. Coronaviruses are large enveloped RNA viruses, so named for the radiating spike proteins found at the surface of the virion (Fig. 1 ; [4]). There are three groups of related coronaviruses and SARS may be a member of a new fourth group [4]. Classification of the SARS coronavirus has been controversial, although phylogenetic similarities may place the virus in a subgroup of the group 2 coronaviruses [5]. Human coronaviruses include three members that cause common respiratory infections [6]. Nonhuman coronaviruses include those that cause respiratory infections in birds, and enteric infections in cattle, pigs, dogs and cats [7]. Some of these viruses affect multiple organs, for example, both mouse hepatitis virus (MHV) and feline infectious peritonitis (FIP) virus are capable of causing respiratory, enteric, neurologic and hepatic infections [7].
Fig. 1.
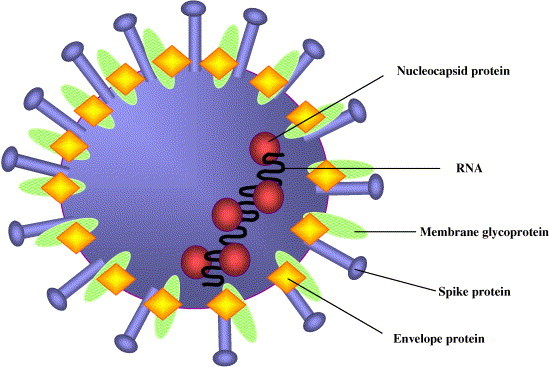
Diagram of the SARS coronavirus structure. The outer virion proteins are marked by the spike proteins that form the corona-like projections. The envelope and membrane glycoproteins are embedded in a lipid bilayer obtained from host-cell membranes. The viral RNA is coated with the nucleocapsid protein [4].
Coronaviruses are positive-sense RNA viruses that replicate by a unique mechanism whereby the structural genes are expressed as a nested set of subgenomic mRNAs, characterized by shared common 3′ ends and a conserved, capped 5′ leader sequence [4]. The nonstructural genes are transcribed from the 5′ end as a polyprotein that is processed by viral proteases (Fig. 2 ; [4]). Proteins are translated from the 5′ open reading frame of each mRNA [4].
Fig. 2.
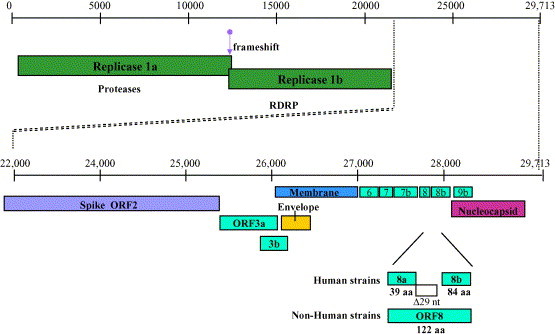
Genome organization of SARS-CoV. Coronaviruses are among the largest RNA viruses. The full-length positive sense SARS CoV is approximately 30 kb long. Nonstructural genes are encoded by ORFs 1a and 1b in the 5′ end of the virus. The structural genes (spike, envelope, membrane, and nucleocapsid) are located in the 3′ end, which includes as many as eight genes of unknown function [4].
SARS CoV has eight open reading frames of unknown function, but has structural proteins found in all coronaviruses that include the envelope (E), the matrix or membrane protein (M), spike (S) and nucleocapsid (N) [4]. The S protein is glycosylated and required for viral attachment and possibly entry. The nucleocapsid protein coats the viral genomic RNA. Viruses that belong to group 2, such as MHV, also contain a hemagglutinin-esterase (HE) protein, which is not present in other groups [4].
1.2. Immunological features associated with SARS-CoV
The flu-like symptoms and atypical pneumonia, characteristic of SARS-CoV infection, was also frequently accompanied by lymphopenia [8]. Alveolar macrophages were prevalent in patients with fatal SARS [8] and contributed to the immune-mediated nature of the disease. In situ hybridization showed that viral infection was present in alveolar epithelial cells and viral RNA could also be detected in alveolar macrophages and bronchiolar epithelial cells [9].
Several studies have suggested that the immune system may be impaired by SARS CoV. T-cell lymphopenia was observed in 94% of patients observed, with a decline in CD4+ and CD8+ cell types [10]. Two weeks after disease onset, Th1 cell-mediated immunity and inflammatory response was noted by the marked elevation of cytokines, IFN gamma, the neutrophil chemokine IL-8, IL-1, -6 and -12, but not TNF, IL-2, -4, or -10. Accumulation of monocytes/macrophages and neutrophils was also observed [11]. Li et al. [12] noted that a rapid decline of T-cell subsets in the periphery was observed in patients during the acute phase of SARS infection, but they observed restoration of T cells during recovery. The presence of proinflammatory cytokines may result from activated alveolar macrophages, suggesting that they may play a role in the pathogenesis of SARS [8].
Antibodies to the SARS coronavirus were found retrospectively in 1.8% of samples collected in 2001, indicating that the 2003 outbreak was not the first time that SARS had entered the human population [13]. Most infected patients developed a humoral response to SARS-CoV and antiviral antibodies (IgG and IgM) were detected at 14 days post-onset of symptoms [14]. IgM antibodies declined after 30 days and IgG antibodies persisted up to day 210 [14] and antiviral neutralizing antibodies were obtained from convalescent patients [14]. Morbidity rates were greater for older individuals, while children under 12 years of age did not develop the severe disease that was seen in adults [15]. Taken together these data may suggest that the quality of the immune response may play a role in the outcome of virus infection.
1.3. Animal models
An additional challenge related to the containment of SARS-CoV is the lack in identification of its natural host. Virus has been detected in wild and domestic animals [16]. In 2003, the first people to be infected were animal handlers in a food market in Guangdong Province, China, suggesting a role for zoonotic transmission [17]. The SARS strain observed in animals varies only slightly from the human virus and may represent a recent jump across species. The development of good animal models will not only be useful for identifying the natural host, but will be invaluable for determining correlates of immunity, for testing therapeutics and vaccine development.
A remarkable advance in SARS research came with the discovery that mice were susceptible to infection with SARS-CoV [18]. Balb/c mice were infected with 103 or 105 50% tissue culture infective doses (TCID50) and by day 2 after infection, yielded 106 and 107 TCID50 per gram, respectively, from lung tissue. Although no clinical disease was observed, mild and focal peribronchiolar mononuclear inflammatory infiltrates were observed upon microscopic examination of the respiratory tract on day 2 [18] after infection. The presence of these infiltrates may suggest some mimicry with human clinical features, although much milder. The respiratory tracts of the mice were cleared of the virus by day 7 after infection. Wentworth et al. found SARS-CoV in the stomach, intestine, and duodenum, in addition to the respiratory tracts of infected mice [19]. Subbarao et al. also protected mice from infection by passive administration of SARS-CoV neutralizing antibody from previously infected mice, suggesting that neutralization in vivo is possible [18]. Mice clear the virus by day 7 post-infection, while humans begin to clear the infection by days 9–14 [8]. A small animal model will allow researchers to test therapeutics and vaccines, and because the mice recover from virus infection so efficiently, also identify host factors that contribute to virus resolution. Hamsters have also been shown to be a good model for SARS-CoV infection, reaching similar titers to those seen in mice [20].
Surprisingly, immunodeficient mice can clear a SARS-CoV infection. Mice (C57BL/6 background) that lack NK-T cells (CD1−/−), NK cells (beige) or those that lack T and B cells (Ragl−/−) cleared the virus by day 9 after infection [21]. The mice displayed high induction of proinflammatory cytokines, suggesting that the adaptive immune response and NK cells were not required for viral clearance in mice. Furthermore, it indicates that the involvement of the innate immune response is important in controlling the virus. It is interesting to speculate that interferon pathways may be important in viral clearance.
More evidence for the importance of innate immunity was provided through the infection of Stat1-deficient mice with SARS-CoV [22]. Stat1 is important to the regulation of interferons and Stat1-deficient mice produced a two log increase in viral titer over control mice. Additionally, the mutant mice developed interstitial pneumonia, not seen in the control mice [22] but not alveolar damage as seen in the lungs of human patients. It is unclear at this time if the observed pathological differences between human and Stat1-deficient mouse lungs were due to time of sampling or differences in host responses [22].
Domestic cats and ferrets have also been tested for use as a SARS-CoV animal model. Cats and ferrets were inoculated with 106 TCID50 SARS-CoV. Cats showed no clinical symptoms [23], while three out of six ferrets became lethargic and one died. Virus was recovered from the lungs of infected cats (103 TCID50/1 gram of tissue) and ferrets (106 TCID50/1 gram of tissue) [23]. Experiments also showed that horizontal transmission of the virus may have occurred between cats that were housed together or ferrets that were housed together, although the kinetics and mode of transmission are still unknown. The non-inoculated infected ferrets became lethargic, developed conjunctivitis and died at 16 and 21 days post-infection. While the ferrets did not show evidence of pneumonia, they did exhibit hepatic lipidosis and emaciation [23]. ter Meulen et al. showed that ferrets that were infected with SARS-CoV showed signs of multifocal pulmonary lesions affecting about 5–10% of the lung [24]. Alveolar damage and lymphocyte infiltration was also observed upon histological examination of infected ferret lung tissue.
Three species of monkeys have been tested for infectivity with SARS-CoV: cynomolgus, rhesus and African green. African green monkeys supported the highest level of viral replication, yielding a viral titer of approximately 104 TCID50/ml from nasal swabs [25]. Some researchers working with cynomolgus macaques reported signs of disease resembling SARS after the monkeys were infected with SARS-CoV [26], [27]. From day 3 post-infection, macaques became lethargic, had a temporary skin rash, and one animal showed signs of respiratory distress [26]. Two of the macaques had interstitial pneumonia with lesions present, similar to autopsy tissue obtained from SARS patients [26]. Other groups have reported that cynomolgous macaques show limited pathology, mild disease and upper respiratory symptoms [28], [29]. There is no apparent explanation for the discrepancy between these groups, possibly virus strain differences or monkey subspecies differences may account for the differences in outcome, but to conclude that non-human primates most similarly mimic human disease is still controversial.
1.4. Considerations in vaccine development
Vaccine efficacy is measured by the ability of the antigen to raise a protective immunologic response from B and T cells after exposure to the viral agent. Ideally, by creating memory within the immune system, individuals will be protected from infection for decades. Several veterinary coronavirus vaccines are currently available, but their efficacy is variable. The vaccine for prevention of infectious bronchitis virus (IBV), which infects chickens, is effective [30], but the canine and porcine vaccines are only partially effective [31]. The feline infectious peritonitis (FIP) vaccine is actually deleterious to the health of the animal and is discussed in further detail below [32].
Vaccines can be produced by inactivation of the virus, by using an attenuated or weak form of the virus, or by using recombinant forms of viral components. Inactivated virus vaccines are relatively safe because they cannot revert back to the live form. They are also relatively stable and may not even require refrigeration. This is important in developing countries and for ease in mobilization during outbreak or emergency situations. However, there are limitations to their use. Inactivated vaccines usually require several doses and some are weakly effective at stimulating an immune response. The vaccine to prevent hepatitis A is an example of an inactivated viral vaccine [33].
Live attenuated viral vaccines may require special laboratory development and cannot always be obtained. To reach effective levels, the virus must be capable of robust replication, but must have lost the ability to cause disease. Several problems are associated with the use of a live attenuated vaccine. These vaccines must be kept refrigerated or frozen, and have safety issues related to the possibility of reversion to the wild-type form. Additionally, they are almost never given to immunocompromised individuals for fears that the attenuated form may cause disease in the absence of an effective immune response [34].
Recombinant DNA or viral vectors have been constructed in the lab for use as potential vaccines or to study the tissue tropism of the SARS virus [25], [35]. The vectors can be used to deliver foreign antigens using attenuated or nonpathogenic organisms. The safety of these types of vaccines [36] centers around persistence of expression in vivo, possible genomic integration of the foreign DNA and possible evolutionary changes that may cause instability of the viral vector. The potential transmission of the viral vector, including its introduction into the environment should also be evaluated. An ideal recombinant vaccine might be engineered to include the inherent ability of the foreign substance to be cleared by drug treatments that are proven safe, such as an antibiotic. Antibiotic sensitivity introduced into a recombinant vector may allay fears of future adverse events although these designs may raise additional safety concerns. Recombinant proteins can also be used to stimulate the immune response. These proteins are purified from yeast or bacteria and currently used in the manufacture of a licensed hepatitis B vaccine [36].
The cell substrate used to manufacture all of these vaccines is also a concern so vaccine production must be performed in a well-characterized cell substrate. Vero E6 cells have been used to produce the licensed poliovirus vaccine [37] and may be appropriate for use in the development of a SARS vaccine as SARS grows well in Vero cells. The FDA Center for Biologics Evaluation and Research (CBER) issued a letter to sponsors using Vero cells as a cell substrate for investigational vaccines which can be found on the CBER website [38]. Another consideration is the use of fetal bovine serum and bovine derivatives in the growth of cells. Bovine tissues may contain the bovine spongiform encephalitis (BSE) agent. Guidelines on the use of BSE-free blood products appear on the FDA website [39] as do guidelines on the use of BSE-free bovine derivatives in the production of vaccines [40].
For viruses that have variable strains, a combination vaccine may be effective. A combination vaccine is one that consists of two or more live organisms, inactivated organisms or purified antigens combined [41]. This type of vaccine may be useful to prevent multiple organisms or strains of 1 organism. Each component must make a contribution to the whole and compatibility of the components is necessary.
1.5. RNA viruses and challenges for vaccine development
RNA viruses replicate through RNA-dependent RNA polymerase encoded by the virus. This type of polymerase has no proof-reading mechanism associated with it, which results in a high rate of uncorrected mutations. These mutations may or may not be lethal to virus replication and may even persist, resulting in rapid evolution of the virus. For this reason, many RNA viruses have multiple genomic strains, or quasispecies, present at one time in an individual. Quasispecies may arise in response to selective immune pressure, thus allowing for escape mutants. The existence of quasispecies during SARS-CoV infection is just coming to light [42] and their importance in escape from immune surveillance is still unknown.
Most coronaviruses are thought to have the ability to recombine due to homologous sequences in the 5′ and 3′ ends of the mRNAs. Evidence suggests that the SARS-CoV originated by recombination between coronaviruses [43], [44] and that there was an additional host-species drift [43].
1.6. Challenges in the development of a SARS-CoV vaccine
Both large and small animals can be infected with SARS-CoV, a giant advance for vaccine research. Examining infected animal models will provide information that will lead to an understanding of the correlates of immunity. Mice have been used to further the understanding of virus neutralization, cytokine upregulation and the minimum requirements for viral clearance, but have yet to show disease that mimics the atypical pneumonia seen in adult humans. While there have been some reports of disease in cynomolgous macaques [26], [27], many groups have not reproduced these findings [28]. A promising animal model may be the domestic ferret. Ferrets show elevated liver enzymes, lymphocytic infiltration and alveolar damage, which has also been observed in humans [8].
Despite the usefulness of these animal models, many challenges lie ahead. First, animal models of SARS-CoV infection do not mimic human disease. In mice, the virus is cleared in less than 1 week and minor pathology in the lung is observed [18]. Histopathology performed on necropsy samples suggests that lung epithelial cells are involved, although the absence of pneumonia and infiltrating macrophages [18] is disappointing. Stat1-deficient mice may prove promising as an animal model that most similarly mimics human disease [22]. Second, in order to test efficacy, large human populations must be tested in areas where the virus is endemic. Finally, if SARS fails to return, how will vaccine manufacturers test candidate vaccines for efficacy? While the animal rule has been provided for just this type of case, an animal model should mimic human disease in order to be applicable. The final rule was published in the federal register and can be found on the federal register website [45].
Additionally, coronaviruses may induce a short-lived immunity. This may be the reason that humans are subject to multiple infections with coronaviruses that cause the common cold. Long-term immunity studies for SARS-CoV are currently underway.
Antibody-dependent enhancement (ADE) has been observed in vaccinated and wild-type infections of FIP. ADE is thought to potentiate viral infection through the infection of macrophages. Viral entry into macrophages occurs when antibodies bind the virus and attach to macrophages via the Fc region of the antibody and its interaction with cell surface expressed Fc receptors [46]. Neutralizing antibodies can also be enhancing antibodies if antibody titer is low or is of the IgG class [47], [48]. Because macrophages increase with viral disease, this cell type may provide an abundant reservoir for the virus and thus expansion of the virus in the host. Some similarities between FIP and SARS exist. First, in both cases macrophages can be infected with the virus [9], [49], and in the case of SARS, the etiology of disease is contributed by infiltrating alveolar macrophages leading to pneumonitis [8]. Second, the treatment with corticosteroids and/or interferon alpha ameliorates SARS disease [50], suggesting an inflammatory, immune-mediated disease. While there has been no observation of ADE during SARS infection, it is worth noting that one coronavirus, FIPV, is capable of eliciting ADE and in the evaluation of vaccines, we may want to consider this possible outcome. However, the difficulty in testing animal models for ADE bears the caveat that if ADE is not observed; it has not proved that vaccines are safe with regard to ADE in humans. In contrast, if an animal model for ADE is developed, we may learn more about the mechanism of SARS-induced ADE, which may help form the basis for developing guidelines for safe vaccine development.
1.7. Potential vaccine candidates and prototypes
1.7.1. Therapeutic vaccines and neutralizing antibodies
A potential SARS vaccine might target the virus specifically through humoral or cell-mediated immune responses. Alternatively, therapeutic vaccines may be useful in the treatment of viral infection. Spike-specific monoclonal and polyclonal antibodies that neutralize the virus have been developed [51], [52] and passive transfer of immune serum into naive mice protected them from infection with SARS-CoV [18]. This suggests that neutralizing antibody alone can prevent viral infection. Neutralization may not require host recognition of the Fc region of the antibody, but the need to develop humanized forms of these types of antibodies may be critical if they are to be considered for use as a treatment. A human monoclonal antibody, derived from a phage display library, was administered to ferrets and protected the ferrets from lung disease and the shedding of virus in pharyngeal secretions [24].
Neutralizing antibodies from convalescent patients have been identified and characterized. Usually neutralizing epitopes are located in the spike protein of the virus [20], [52]. Recent evidence has determined that virus neutralization is sensitive to deglycosylation of the spike protein, suggesting that conformational epitopes are important in antibody recognition [53].
1.7.2. Inactivated SARS-Co V
SARS-CoV can be efficiently inactivated by ultraviolet (UV) irradiation [54]. Mice immunized with UV-inactivated SARS-CoV develop humoral and cell-mediated immune responses [55]. Both T cell proliferation and cytokine upregulation was observed after boosting with the inactivated virus. Beta-propiolactone-inactivated virus also elicited neutralizing antibodies when administered to Balb/c mice [56] Formalin-inactivated SARS-CoV yielded potent humoral responses in Balb/c mice as well [57].
1.7.3. Recombinant viruses and virus-like particles
Recombinant viruses may be used to elicit responses to introduced SARS-CoV genes. The first type of recombinant virus is a defective or non-pathogenic vector that expresses SARS-CoV proteins. The second type is one that is stimulated to assemble virus-like particles (VLP) in vitro. VLPs containing the structural envelope proteins including spike (S), envelope (E) and membrane protein (M) have been assembled by coinfecting insect cells with three baculoviruses expressing one of the three structural proteins [58].
Structural proteins expressed by the live attenuated bovine parainfluenza virus type 3 (BHPIV3) were evaluated for efficacy in hamsters [20] and African green monkeys [25]. High titer neutralizing antibodies were obtained after only one intranasal immunization with this vector. Single immunization with BHPIV3 expressing S alone provided complete protection upon challenge with SARS-CoV [20], [25].
Recombinant live attenuated modified vaccinia virus Ankara (MVA) was used to deliver the SARS spike protein (rMVA-S) into Balb/c mice [59]. Neutralizing antibodies were obtained and a reduction in the viral titer was observed after challenge with live SARS-CoV [59]. Only ferrets that were challenged with SARS-CoV after vaccination with rMVA-S showed enhanced liver disease as demonstrated by increases in ALT values and the presence of mononuclear hepatitis upon histological examination [60]. These data suggest enhanced disease due to vaccination with a SARS protein.
Adenoviruses expressing the S, M or N proteins were used in combination to vaccinate rhesus macaques [61]. The immunized animals all had antibody responses to the S protein and T-cell responses to the N protein [61].
Highly infectious HIV particles expressing the S protein have been made, primarily to study the host-cell distribution of the putative SARS-CoV receptor [35]. Additionally, investigating the requirements for viral receptor binding and entry will also enhance our understanding of the requirements for viral control. Recombinant HIV particles that express the SARS spike protein may provide insight into cell tropism and receptor expression profiles [35]. Another retrovirus, murine leukemia virus, was used to generate infectious particles containing most of the S protein. Convalescent serum was able to neutralize infection of the recombinant virus in Vero cells [62].
1.7.4. DNA vaccines
High cytotoxic T-lymphocyte (CTL) and antibody responses were observed after mice were injected three times with a recombinant plasmid vector expressing the N protein [63]. Mice immunized with a plasmid containing the S protein produced anti-SARS-CoV IgG [64] and developed neutralizing antibodies and a T-cell mediated response resulting in a six-fold reduction in viral titer in the lungs [65]. Plasmids encoding either the S1 or S2 regions of the spike protein elicited antibody production in mice [66]. Neither the S1 or S2 antibodies alone were capable of neutralizing the virus; however, cooperatively they enabled neutralization of the virus, suggesting that both regions of the spike protein are important for host-cell viral entry [66]. The nucleocapsid protein may also stimulate an effective immune response. DNA vaccination with calreticulin fused to the N protein generated SARS-specific humoral and cellular immunity in C57BL/6 mice [67]. Calreticulin was used because it was found to enhance major histocompatibility complex class I presentation of fusion proteins to CD8 (+) T cells [67].
Recombinant viruses may be generated from the full-length infectious cDNA clone of SARS-CoV [68]. This clone may provide a source for genetic manipulation of the genome [68]. Once the viral virulence factors are understood, attenuated strains may be obtained by engineered mutation of the virus.
Vaccine development may proceed through the undertaking of a systematic approach to understanding the correlates of immunity raised by SARS-CoV. Much of the focus has centered towards the humoral response and neutralizing epitopes, but cell-mediated immunity may also be important. CTL epitopes within SARS-CoV that may be presented by 99% of the human leukocyte antigen supertypes were identified by advanced bioinformatics [69]. Further characterization of these epitopes, including their recognition by convalescent serum, should advance the understanding of important immunological features in the control of SARS-CoV.
1.8. Summary
While there has been intense study of SARS, there is still much that is unknown about the pathology of the virus. Many research questions will need to be answered, thus providing the resources necessary to develop an effective and safe vaccine. To be sure that a vaccine will provide coverage for a potentially variable virus, we need to know what the occurrences of viral quasispecies are and how variable are the important epitopes. What are the viral virulence factors and can we use this information to develop an attenuated strain? Can the virus establish persistent infection and can humans be repeatedly infected? We also need to determine the innate, humoral and cell-mediated immune responses in recovered SARS patients in order to understand how viral clearance comes about. To understand viral pathogenesis we must know how the immune system mediates disease. What is the role of lymphocytic infiltrates and do alveolar macrophages enhance disease or control infection? If we define the ability of the virus to replicate in monocytes/macrophages can we be assured that antibody-dependent enhancement will not be a problem for a SARS vaccine? Most importantly, identification of the natural animal host may launch the further development of large and small animal models for correlates of immunity and drug and vaccine screening. Development of an animal model that mimics human disease will be the single most important advance in the development of a SARS vaccine. Development of a vaccine for SARS-CoV is imperative and research headed in a forward direction will enable the public health community to be ready.
Acknowledgments
Kanta Subbarao, Edward Tabor, Miriam Darnell, Robin Levis and Hira Nakhasi are gratefully acknowledged for comments on the manuscript.
References
- 1.http://www.who.int/csr/sars/country/table2004_04_21/en/index.html
- 2.Fowler R.A., Lapinsky S.E., Hallett D., Detsky A.S., Sibbald W.J., Slutsky A.S., Toronto SARS Critical Care Group Critically ill patients with severe acute respiratory syndrome. JAMA. 2003;290(3):367–373. doi: 10.1001/jama.290.3.367. [DOI] [PubMed] [Google Scholar]
- 3.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., SARS Working Group A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 4.Stadler K., Masignani V., Eickmann M., Becker S., Abrignani S., Klenk H.D. SARS–beginning to understand a new virus. Nat Rev Microbiol. 2003;1(3):209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorbalenya A.E., Snijder E.J., Spaan W.J. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J Virol. 2004;78(15):7863–7866. doi: 10.1128/JVI.78.15.7863-7866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C. Identification of a new human coronavirus. Nat Med. 2004;10(4):368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holmes KV, Lai MMC. Coronaviridae: the viruses and their replication. In: Fundamental Virology, 3rd ed., 1996; p. 541–60.
- 8.Nicholls J.M., Poon L.L., Lee K.C., Ng W.F., Lai S.T., Leung C.Y. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361(9371):1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakajima N., Asahi-Ozaki Y., Nagata N., Sato Y., Dizon F., Paladin F.J. SARS coronavirus-infected cells in lung detected by new in situ hybridization technique. Jpn J Infect Dis. 2003;56(3):139–141. [PubMed] [Google Scholar]
- 10.Cui W., Fan Y., Wu W., Zhang F., Wang J.Y., Ni A.P. Expression of lymphocytes and lymphocyte subsets in patients with severe acute respiratory syndrome. Clin Infect Dis. 2003;37(6):857–859. doi: 10.1086/378587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong C.K., Lam C.W., Wu A.K., Ip W.K., Lee N.L., Chan I.H. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol. 2004;136(1):95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li T., Qiu Z., Zhang L., Han Y., He W., Liu Z. Significant changes of peripheral T lymphocyte subsets in patients with severe acute respiratory syndrome. J Infect Dis. 2004;189(4):648–651. doi: 10.1086/381535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng B.J., Wong K.H., Zhou J., Wong K.L., Young B.W., Lu L.W. SARS-related virus predating SARS outbreak, Hong Kong. Emerg Infect Dis. 2004;10(2):176–178. doi: 10.3201/eid1002.030533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi J., Wei Z., Song J. Dissection study on the severe acute respiratory syndrome 3C-like protease reveals the critical role of the extra domain in dimerization of the enzyme: defining the extra domain as a new target for design of highly specific protease inhibitors. J Biol Chem. 2004;279(23):24765–24773. doi: 10.1074/jbc.M311744200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng P.C., Leung C.W., Chiu W.K., Wong S.F., Hon E.K. SARS in newborns and children. Biol Neonate. 2004;85(4):293–298. doi: 10.1159/000078174. [DOI] [PubMed] [Google Scholar]
- 16.Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302(5643):276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 17.Breiman R.F., Evans M.R., Preiser W., Maguire J., Schnur A., Li A. Role of China in the quest to define and control severe acute respiratory syndrome. Emerg Infect Dis. 2003;9(9):1037–1041. doi: 10.3201/eid0909.030390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subbarao K., McAuliffe J., Vogel L., Fahle G., Fischer S., Tatti K. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J Virol. 2004;78(7):3572–3577. doi: 10.1128/JVI.78.7.3572-3577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wentworth D.E., Gillim-Ross L., Espina N., Bernard K.A. Mice susceptible to SARS coronavirus. Emerg Infect Dis. 2004;10(7):1293–1296. doi: 10.3201/eid1007.031119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buchholz U.J., Bukreyev A., Yang L., Lamirande E.W., Murphy B.R., Subbarao K. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc Natl Acad Sci USA. 2004;101(26):9804–9809. doi: 10.1073/pnas.0403492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glass W.G., Subbarao K., Murphy B., Murphy P.M. Mechanisms of host defense following severe acute respiratory syndrome-coronavirus (SARS-CoV) pulmonary infection of mice. J Immunol. 2004;173(6):4030–4039. doi: 10.4049/jimmunol.173.6.4030. [DOI] [PubMed] [Google Scholar]
- 22.Hogan R.J., Gao G., Rowe T., Bell P., Flieder D., Paragas J. Resolution of primary severe acute respiratory syndrome-associated coronavirus infection requires Stat1. J Virol. 2004;78(20):11416–11421. doi: 10.1128/JVI.78.20.11416-11421.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martina B.E., Haagmans B.L., Kuiken T., Fouchier R.A., Rimmelzwaan G.F., Van Amerongen G. Virology: SARS virus infection of cats and ferrets. Nature. 2003;425(6961):915. doi: 10.1038/425915a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.ter Meulen J., Bakker A.B., van den Brink E.N., Weverling G.J., Martina B.E., Haagmans B.L. Human monoclonal antibody as prophylaxis for SARS coronavirus infection in ferrets. Lancet. 2004;363(9427):2139–2141. doi: 10.1016/S0140-6736(04)16506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bukreyev A., Lamirande E.W., Buchholz U.J., Vogel L.N., Elkins W.R., St Claire M. Mucosal immunisation of African green monkeys (Cercopithecus aethiops) with an attenuated parainfluenza virus expressing the SARS coronavirus spike protein for the prevention of SARS. Lancet. 2004;363(9427):2122–21227. doi: 10.1016/S0140-6736(04)16501-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fouchier R.A., Kuiken T., Schutten M., van Amerongen G., van Doornum G.J., van den Hoogen B.G. Aetiology: Koch's postulates fulfilled for SARS virus. Nature. 2003;423(6937):240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuiken T., Fouchier R.A., Schutten M., Rimmelzwaan G.F., van Amerongen G., van Riel D. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362(9380):263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rowe T., Gao G., Hogan R.J., Crystal R.G., Voss T.G., Grant R.L. Macaque model for severe acute respiratory syndrome. Virology. 2004;78(20):11401–11404. doi: 10.1128/JVI.78.20.11401-11404.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McAuliffe J., Vogel L., Roberts A., Fahle G., Fischer S., Shieh W.J. Replication of SARS coronavirus administered into the respiratory tract of African Green, rhesus and cynomolgus monkeys. Virology. 2004;330:8–15. doi: 10.1016/j.virol.2004.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ladman B.S., Pope C.R., Ziegler A.F., Swieczkowski T., Callahan C.J., Davison S. Protection of chickens after live and inactivated virus vaccination against challenge with nephropathologenic infectious bronchitis virus. Avian Dis. 2002;46:938–944. doi: 10.1637/0005-2086(2002)046[0938:POCALA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 31.Pratelli A., Tinelli A., Decaro N., Martella V., Camera M., Tempesta M. Safety and efficacy of a modified-live canine coronavirus vaccine in dogs. Vet Microbiol. 2004;99:43–49. doi: 10.1016/j.vetmic.2003.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiss R.C., Scott F.W. Antibody-mediated enhancement of disease in feline infectious peritonitis: comparisons with dengue hemorrhagic fever. Comp Immunol Microbiol Infect Dis. 1981;4(2):175–189. doi: 10.1016/0147-9571(81)90003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purcell R.H., D’Hondt E., Bradbury R., Emerson S.U., Govindarajan S., Binn L. Inactivated hepatitis A vaccine: active and passive immunoprophylaxis in chimpanzees. Vaccine. 1992;10(Suppl. 1):S148–S451. doi: 10.1016/0264-410x(92)90572-2. [DOI] [PubMed] [Google Scholar]
- 34.Arvin A.M., Gershon A.A. Live attenuated varicella vaccine. Annu Rev Microbiol. 1996;50:59–100. doi: 10.1146/annurev.micro.50.1.59. [DOI] [PubMed] [Google Scholar]
- 35.Nie Y., Wang P., Shi X., Wang G., Chen J., Zheng A. Highly infectious SARS-CoV pseudotyped virus reveals the cell tropism and its correlation with receptor expression. Biochem Biophys Res Commun. 2004;321(4):994–1000. doi: 10.1016/j.bbrc.2004.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dertzbaugh M.T. Genetically engineered vaccines: an overview. Plasmid. 1998;39(2):100–113. doi: 10.1006/plas.1997.1329. [DOI] [PubMed] [Google Scholar]
- 37.Rosenthal S.R., Merchlinsky M., Kleppinger C., Goldenthal K.L. Developing new smallpox vaccines. Emerg Infect Dis. 2001;7(6):920–926. doi: 10.3201/eid0706.010602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.http://www.fda.gov/cber/ltr/vero031301.htm
- 39.http://www.fda.gov/cber/gdlns/cjdvcjdq&a.htm
- 40.http://www.fda.gov/cber/bse/bseqa.htm#a3
- 41.Falk L.A., Ball L.K. Current status and future trends in vaccine regulation--USA. Vaccine. 2001;19(13/14):1567–1572. doi: 10.1016/s0264-410x(00)00353-4. [DOI] [PubMed] [Google Scholar]
- 42.Xu D., Zhang Z., Wang F.S. SARS-associated coronavirus quasispecies in individual patients. N Engl J Med. 2004;350(13):1366–1367. doi: 10.1056/NEJMc032421. [DOI] [PubMed] [Google Scholar]
- 43.Rest J.S., Mindell D.P. SARS associated coronavirus has a recombinant polymerase and coronaviruses have a history of host-shifting. Infect Genet Evol. 2003;3(3):219–225. doi: 10.1016/j.meegid.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stavrinides J., Guttman D.S. Mosaic evolution of the severe acute respiratory syndrome coronavirus. J Virol. 2004;78(1):76–82. doi: 10.1128/JVI.78.1.76-82.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.FR 67:37988-98; May 31, 2002; http://www.gpoaccess.gov/fr/index.html.
- 46.Tirado S.M., Yoon K.J. Antibody-dependent enhancement of virus infection and disease. Viral Immunol. 2003;16(1):69–86. doi: 10.1089/088282403763635465. [DOI] [PubMed] [Google Scholar]
- 47.Hohdatsu T., Tokunaga J., Koyama H. The role of IgG subclass of mouse monoclonal antibodies in antibody-dependent enhancement of feline infectious peritonitis virus infection of feline macrophages. Arch Virol. 1994;139(3/4):273–285. doi: 10.1007/BF01310791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corapi W.V., Darteil R.J., Audonnet J.C., Chappuis G.E. Localization of antigenic sites of the S glycoprotein of feline infectious peritonitis virus involved in neutralization and antibody-dependent enhancement. J Virol. 1995;69(5):2858–2862. doi: 10.1128/jvi.69.5.2858-2862.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hohdatsu T., Yamada M., Tominaga R., Makino K., Kida K., Koyama H. Antibody-dependent enhancement of feline infectious peritonitis virus infection in feline alveolar macrophages and human monocyte cell line U937 by serum of cats experimentally or naturally infected with feline coronavirus. J Vet Med Sci. 1998;60(1):49–55. doi: 10.1292/jvms.60.49. [DOI] [PubMed] [Google Scholar]
- 50.Loutfy M.R., Blatt L.M., Siminovitch K.A., Ward S., Wolff B., Lho H. Interferon alfacon-1 plus corticosteroids in severe acute respiratory syndrome: a preliminary study. JAMA. 2003;290(24):3222–3228. doi: 10.1001/jama.290.24.3222. [DOI] [PubMed] [Google Scholar]
- 51.Berry J.D., Jones S., Drebot M.A., Andonov A., Sahara M., Yuan X.Y. Development and characterisation of neutralising monoclonal antibody to the SARS-coronavirus. J Virol Methods. 2004;120(1):87–96. doi: 10.1016/j.jviromet.2004.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang H., Wang G., Li J., Nie Y., Shi X., Lian G. Identification of an antigenie determinant on the S2 domain of the severe acute respiratory syndrome coronavirus spike glycoprotein capable of inducing neutralizing antibodies. J Virol. 2004;78(13):6938–6945. doi: 10.1128/JVI.78.13.6938-6945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song H.C., Seo M.Y., Stadler K., Yoo B.J., Choo Q.L., Coates S.R. Synthesis and characterization of a native, oligomeric form of recombinant severe acute respiratory syndrome coronavirus spike glycoprotein. J Virol. 2004;78(19):10328–10335. doi: 10.1128/JVI.78.19.10328-10335.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Darnell M.E., Subbarao K., Feinstone S.M., Taylor D.R. Inactivation of the coronavirus that induces severe acute respiratory syndrome, SARS-CoV. J Virol Methods. 2004;121(1):85–91. doi: 10.1016/j.jviromet.2004.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takasuka N., Fujii H., Takahashi Y., Kasai M., Morikawa S., Itamura S. Subcutaneously injected UV-inactivated SARS coronavirus vaccine elicits systemic humoral immunity in mice. Int Immunol. 2004;16(10):1423–1430. doi: 10.1093/intimm/dxh143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang L., Zhu Q., Qin E., Yu M., Ding Z., Shi H. Inactivated SARS-CoV vaccine prepared from whole virus induces a high level of neutralizing antibodies in BALB/c mice. DNA Cell Biol. 2004;23(6):391–394. doi: 10.1089/104454904323145272. [DOI] [PubMed] [Google Scholar]
- 57.Zhang C.H., Lu J.H., Wang Y.F., Zheng H.Y., Xiong S., Zhang M.Y. Immune responses in Balb/c mice induced by a candidate SARS-CoV inactivated vaccine prepared from F69 strain. Vaccine. 2005;23(24):3196–3201. doi: 10.1016/j.vaccine.2004.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ho Y., Lin P.H., Liu C.Y., Lee S.P., Chao Y.C. Assembly of human severe acute respiratory syndrome coronavirus-like particles. Biochem Biophys Res Commun. 2004;318(4):833–838. doi: 10.1016/j.bbrc.2004.04.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bisht H., Roberts A., Vogel L., Bukreyev A., Collins P.L., Murphy B.R. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc Natl Acad Sci USA. 2004;101(17):6641–6646. doi: 10.1073/pnas.0401939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weingartl H., Czub M., Czub S., Neufeld J., Marszal P., Gren J. Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J Virol. 2004;78:12672–12676. doi: 10.1128/JVI.78.22.12672-12676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao W., Tamin A., Soloff A., D’Aiuto L., Nwanegbo E., Robbins P.D. Effects of a SARS-associated coronavirus vaccine in monkeys. Lancet. 2003;362(9399):1895–1896. doi: 10.1016/S0140-6736(03)14962-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Giroglou T., Cinatl J., Jr., Rabenau H., Drosten C., Schwalbe H., Doerr H.W. Retroviral vectors pseudotyped with severe acute respiratory syndrome coronavirus S protein. J Virol. 2004;78(17):9007–9015. doi: 10.1128/JVI.78.17.9007-9015.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu M.S., Pan Y., Chen H.Q., Shen Y., Wang X.C., Sun Y.J. Induction of SARS-nucleoprotein-specific immune response by use of DNA vaccine. Immunol Lett. 2004;92(3):237–243. doi: 10.1016/j.imlet.2004.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao P., Ke J.S., Qin Z.L., Ren H., Zhao L.J., Yu J.G. DNA vaccine of SARS-CoV S gene induces antibody response in mice. Acta Biochim Biophys Sin (Shanghai) 2004;36(1):37–41. doi: 10.1093/abbs/36.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang Z.Y., Kong W.P., Huang Y., Roberts A., Murphy B.R., Subbarao K. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428(6982):561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zeng F., Chow K.Y., Hon C.C., Law K.M., Yip C.W., Chan K.H. Characterization of humoral responses in mice immunized with plasmid DNAs encoding SARS-CoV spike gene fragments. Biochem Biophys Res Commun. 2004;315(4):1134–1139. doi: 10.1016/j.bbrc.2004.01.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim T.W., Lee J.H., Hung C.F., Peng S., Roden R., Wang M.C. Generation and characterization of DNA vaccines targeting the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J Virol. 2004;78(9):4638–4645. doi: 10.1128/JVI.78.9.4638-4645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yount B., Curtis K.M., Fritz E.A., Hensley L.E., Jahrling P.B., Prentice E. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci USA. 2003;100(22):12995–13000. doi: 10.1073/pnas.1735582100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sylvester-Hvid C., Nielsen M., Lamberth K., Roder G., Justesen S., Lundegaard C. SARS CTL vaccine candidates; HLA supertype-, genome-wide scanning and biochemical validation. Tissue Antigens. 2004;63(5):395–400. doi: 10.1111/j.0001-2815.2004.00221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


