Abstract
Viruses are devastating pathogens of humans, animals, and plants. To further our understanding of how viruses use the resources of infected cells, we systematically tested the yeast single-gene-knockout library for the effect of each host gene on the replication of tomato bushy stunt virus (TBSV), a positive-strand RNA virus of plants. The genome-wide screen identified 96 host genes whose absence either reduced or increased the accumulation of the TBSV replicon. The identified genes are involved in the metabolism of nucleic acids, lipids, proteins, and other compounds and in protein targeting/transport. Comparison with published genome-wide screens reveals that the replication of TBSV and brome mosaic virus (BMV), which belongs to a different supergroup among plus-strand RNA viruses, is affected by vastly different yeast genes. Moreover, a set of yeast genes involved in vacuolar targeting of proteins and vesicle-mediated transport both affected replication of the TBSV replicon and enhanced the cytotoxicity of the Parkinson's disease-related α-synuclein when this protein was expressed in yeast. In addition, a set of host genes involved in ubiquitin-dependent protein catabolism affected both TBSV replication and the cytotoxicity of a mutant huntingtin protein, a candidate agent in Huntington's disease. This finding suggests that virus infection and disease-causing proteins might use or alter similar host pathways and may suggest connections between chronic diseases and prior virus infection.
Keywords: host factors, plus-stranded RNA virus, tomato bushy stunt tombusvirus, virus replication, yeast-knockout strains
The success of viruses as pathogens of humans, animals, and plants depends on the viruses' ability to reprogram the host-cell metabolism to support the infection. The virus-host interaction is more complex than the term “reprogramming” suggests because host cells have antiviral defense mechanisms. Identifying host genes that can affect virus replication and the infection process is central to understanding the complex role of the host in viral infections. The largest group of viruses, the positive-strand RNA viruses, which include the severe acute respiratory syndrome (SARS) coronavirus and hepatitis C and West Nile viruses, has virion RNAs that are directly translated in the infected cell. Synthesized viral proteins and recruited host proteins mediate processes that lead to efficient multiplication of the viral RNA (1, 2). RNA viruses are important not only as infectious agents but also as tools in biotechnology and gene therapy for expressing selected proteins in cells (3-5).
Yeast is a model eukaryotic cell that has been used extensively to study the roles of individual genes in cellular processes based on genome-wide screens (6-9). Many screens have been based on the yeast single-gene-knockout (YKO) library, because the role of each nonessential yeast gene can be tested for selected functions (10). We and others have developed systems for inducing yeast cells to support the replication of certain positive-strand RNA viruses or their surrogates (11-14). Here, we apply our previously developed system for robust replication of a small RNA replicon of the tomato bushy stunt virus (TBSV) in yeast (13, 15) to screen the entire YKO library for genes influencing the efficiency of viral replication. A total of 96 YKO strains were identified. The identified host genes are either involved in many cellular processes, including nucleic acid, protein, and lipid metabolism, protein targeting/transport, and general and stress metabolism or have unknown functions. Our results show that the replication of positive-strand RNA viruses of different supergroups is influenced by distinct groups of genes and that TBSV replication is associated with sets of genes that also have been associated with certain human disease states.
Materials and Methods
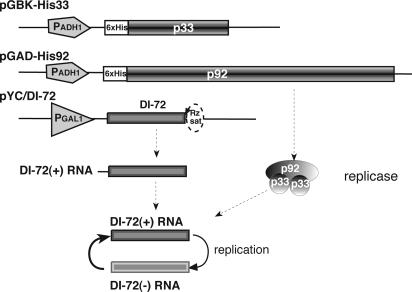
Yeast Strains and Expression Plasmids. Yeast strain 4741 and the YKO deletion series (10) were obtained from Open Biosystems (Huntsville, AL). Yeast transformation was modified from the standard lithium acetate (LiOAc)/polyethylene glycol (PEG) protocol (16) to facilitate a 96-well plate format. Briefly, yeast strains were grown overnight in yeast extract/peptone/dextrose medium supplemented with 200 mg/liter geneticin G418. Cultures were then diluted to ≈0.3 OD600 in fresh medium (0.25 ml per well) and cultured for an additional 4 h at 30°C. The cells were pelleted, washed with sterile water, and resuspended in 0.1 M LiOAc. This procedure was followed by repelleting and resuspending the cells in 100 μl of transformation mixture (34% PEG 3350, 0.1 M LiOAc, 0.5 mg/ml single-stranded salmon sperm DNA, and 10 mg/ml each of three plasmids) (Fig. 1). This mixture was incubated for 30 min at 30°C and then for 40 min at 42°C. The yeast cells were then pelleted and plated on minimal medium supplemented with G418.
Fig. 1.
A schematic presentation of expression of tombusvirus replicase genes and an RNA replicon (DI-72 RNA) in the YKO library. p33 and p92 replicase proteins are expressed constitutively from separate plasmids by using the ADH1 promoter (PADH1), whereas the precursor for DI-72 RNA is under the control of inducible GAL1 promoter (PGAL1). 6× His, 6× His-tag at the amino terminus of p33 and p92. The self-cleaving tobacco ringspot virus satellite ribozyme (Rz sat) produces an authentic 3′end in DI-72 RNA after the transcription of DI-72 precursor from pYC/DI-72 plasmid.
Each strain in the YKO library was cotransformed with plasmids pGBK-His 33 and pGAD-His 92, which express p33 and p92 replicase proteins separately from the constitutive ADH1 promoter (15), and pYC/defective interfering (DI)-72, which expresses the TBSV-derived DI-72 RNA replicon from an inducible GAL1 promoter (13).
RNA Analysis. Transformed yeast strains were incubated in 96-well deep-well plates containing 0.5 ml per well minimal synthetic complete medium lacking uracil, leucine, and histidine and supplemented with 200 mg/liter geneticin G418 (SC-ULH) containing galactose (2%) for 48 h at 23°C or for 40 h at 30°C, with shaking at 525 rpm. After the cells were pelleted, the RNA was extracted by using a modified hot-phenol method (17). Briefly, 200 μl of sodium acetate (NaOAc)-EDTA+SDS buffer (50 mM NaOAc, pH 5.3/10 mM EDTA/1% SDS) and 200 μl of water-equilibrated phenol were added to each well, briefly vortexed, and incubated at 65°C for 4 min, followed by incubation in ice-cold water. After centrifugation, the total RNA samples (water phase) were transferred to a new plate and precipitated by using NaOAc and ethanol. Total RNA samples were visualized on 1.5% agarose gels. For the Northern blot analysis, the total RNA samples were diluted 100 times (except for the detection of DI-72 transcripts, for which undiluted samples were used) before electrophoresis, followed by transfer of RNA to membranes (18). RNA hybridization was done with a mixture of two probes to detect DI-72(+) RNA and 18S yeast ribosomal RNA. The probes were tested for crosshybridization but none was detected (data not shown). The RNA probes were prepared by in vitro transcription with T7 RNA polymerase from appropriate PCR products (18). The DI-72 probe was made by using primers 15 (GTAATACGACTCACTATAGGGCATGTCGCTTGTTTGTTGG) and 20 (GGAAATTCTCCAGGATTTCTC) and DI-72 as a template (13). The 18S rRNA probe was amplified from the yeast genome with primers 1251 (GGTGGAGTGATTTGTCTGCTT) and 1252 (TAATACGACTCACTATAGGTTTGTCCAAATTCTCCGCTCT). The template for the probe to detect transcription of DI-72 transcript from pYC/DI-72 was obtained by PCR with primers 1289 (AAGTATCAACAAAAAATTGTTAATATACCT) and 1290 (TAATACGACTCACTATAGGAAGCTTAATATTCCCTATAGT).
Protein Analysis. For protein analysis, yeast strains were grown as for RNA extraction. Pelleted cells were resuspended in 200 μl of 0.1M NaOH and incubated at 23°C for 10 min. NaOH was aspirated after a short centrifugation, and the samples were resuspended in SDS/PAGE buffer and boiled for 5 min. The supernatant was used for SDS/PAGE and Western blot analysis as described in refs. 13 and 15. The primary antibodies were anti-6× His (Invitrogen), and the secondary antibodies were alkaline-phosphatase-conjugated anti-mouse IgG (Sigma) (15).
Results and Discussion
Tests of Single Yeast Genes for Effects on Accumulation of a TBSV Replicon. Each strain in the YKO library was transformed with three plasmids simultaneously to induce replication of a TBSV replicon (termed DI-72 RNA). As shown in Fig. 1, two of the plasmids expressed the essential tombusvirus replicase proteins, p33 and p92, constitutively from ADH1 promoters, whereas the third plasmid directed transcription of DI-72 RNA replicon from the GAL1 promoter. After induction with galactose, the transcribed DI-72 RNA was cleaved by a cis-ribozyme to generate an authentic 3′ terminus in DI-72 RNA in yeast cells (Fig. 1). We found that the autonomously replicating DI-72 RNA accumulated to ribosomal RNA levels in yeast cells, comparable with the virus replication level in plant cells (15, 19). We also confirmed that, similar to plant infections, replication of DI-72 RNA replicon in yeast depends on cis-acting regulatory sequences in the RNA template and the transacting p33 and p92 proteins, which are part of the tombusvirus replicase (13, 15). The DI-72 RNA replicon, which is derived naturally from TBSV infections of plants, does not encode proteins, and high levels of DI-72 RNA were achieved without artificial selection pressure to maintain this RNA in yeast cells. Yeast-based replication of DI-72 RNA replicon mimics its replication in plant cells (13, 15).
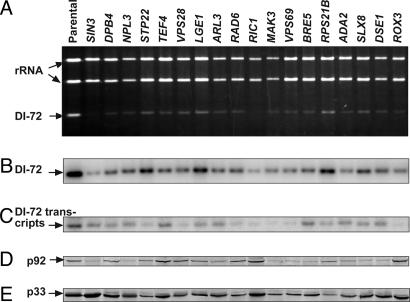
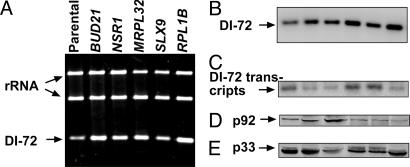
Identification of 96 Yeast Genes Affecting Accumulation of TBSV Replicon. We found that, of the 4,848 strains present in the YKO library (which covers ≈80% of all of the predicted yeast genes), we were unable to transform 71 strains, and an additional 229 strains did not grow on galactose-containing media. The remaining 4,548 YKO strains were tested individually (four samples per strain) for their ability to support DI-72 RNA replication by measuring the accumulation level of DI-72 RNA in total yeast RNA extracts, as described in Materials and Methods. Measurements of DI-72 RNA replication products in samples should give easily interpretable data on replication when compared with more complex virus-based reporter-protein assays, which depend not only on replication but on translation of the reporter gene (20). We found that 90 YKO strains (<2% of total genes) supported the accumulation of DI-72 RNA replicon at <50% of the level observed in the parental yeast strain (Table 1 and Fig. 2). On the contrary, six YKO strains (0.1% of total genes) supported the accumulation of DI-72 RNA replicon at >150% of the parental yeast level (Fig. 3). The low number of yeast strains with an increased level of DI-72 RNA replication might be due to the originally high level of DI-72 RNA accumulation in the parental strain, which could make further increase of replication difficult. Analysis of transcription levels of DI-72 RNA in the identified YKO strains revealed the lack of direct correlation between transcription and replication levels (see Fig. 5, which is published as supporting information on the PNAS web site). For example, in SIN3 and RPL1B strains (Fig. 2, lane 2, and Fig. 3, lane 6), transcription of DI-72 RNA was ≈50%, but replication of DI-72 RNA was ≈5% and 200%, respectively, when compared with the parental strain. Thus, it seems that the initial transcription of DI-72 might have only a limited effect on the subsequent robust replication process. The amounts of p92 and p33 replicase proteins also varied in the identified YKO strains, and we did not find close correlation between the amounts of p33/p92 and the level of DI-72 RNA replication (Fig. 2, and see Fig. 6, which is published as supporting information on the PNAS web site). We should point out, however, that almost half (43 of 96) of the YKO strains identified showed reduced amounts (≤50%) of p33 and/or p92 proteins (Table 1). Based on this observation, we predict that ≈45% of the identified host genes, which reduced DI-72 RNA accumulation, might affect virus replication by reducing accumulation levels of the replicase proteins in cells. The identified genes, based on their known cellular functions, were placed into 12 groups (Table 1), albeit several genes might have multiple direct and indirect functions in both TBSV replication and cellular processes.
Table 1. Functional grouping of identified host genes affecting TBSV DI-72 RNA replication.
| Gene* | Replication† | p33‡ | Transcription† | Function |
|---|---|---|---|---|
| Group 1: Protein biosynthesis | ||||
| MRPL32§¶ | 162 | 100 | 111 | Protein biosynthesis |
| RPL1B§¶ | 192 | 100 | 39 | Protein biosynthesis |
| RPL7A§¶ | 192 | 120 | 97 | Protein biosynthesis |
| RPS21B§¶ | 33 | 50 | 70 | Protein biosynthesis |
| TEF4 | 15 | 100 | 68 | Translation elongation factor |
| Group 2: Protein metabolism, posttranslation modification | ||||
| ARO1∥ | 38 | 40 | 50 | Aromatic amino acid synthesis |
| BRE1§ | 20 | 30 | 20 | Ubiquitin-protein ligase |
| DOA4** | 22 | 30 | 50 | Protein deubiquitination |
| LGE1** | 30 | 80 | 48 | Protein monoubiquitination |
| MAK3 | 17 | 50 | 10 | Protein amino acid acetylation |
| MET1§ | 33 | 100 | 95 | Uroporphyrin-methyltransferase |
| RAD6§ | 23 | 70 | 22 | Ubiquitin conjugating enzyme |
| SIW14** | 50 | 100 | 61 | Protein tyrosine phosphatase |
| Group 3: RNA metabolism | ||||
| BUD21 | 184 | 150 | 51 | snoRNA binding |
| CCR4 | 45 | 100 | 46 | 3′-5′ exoribonuclease |
| KEM1 | 40 | 80 | 65 | 5′-3′ exoribonuclease |
| NPL3 | 21 | 80 | 49 | mRNA binding |
| MSR1 | 153 | 100 | 43 | RNA binding rRNA processing |
| Group 4: Lipid metabolism | ||||
| ERG4 | 18 | 30 | 55 | †24 (24-1) sterol reductase |
| INO2¶ | 29 | 100 | 70 | Phospholipid biosynthesis |
| MCT1 | 49 | 60 | 51 | S-malonyltransferase/fatty acid metabolism |
| POX1∥ | 29 | 30 | 55 | Acyl-CoA oxidase/fatty acid beta-oxidation |
| TGL2 | 20 | 50 | 105 | Triacylglycerol lipase/lipid metabolism |
| Group 5: Vesicle-mediated transport | ||||
| ARL3∥ | 19 | 100 | 82 | Small monomeric GTPase |
| BRE5 | 17 | 70 | 90 | Vesicle-mediated transport |
| GOS1 | 23 | 30 | 59 | v-SNARE activity/intra-Golgi transport |
| MCH5 | 40 | 100 | 96 | Transporter/membrane associated |
| PEP3 | 17 | 60 | 30 | Transporter/vacuolar membrane associated |
| RIC1 | 5 | 100 | 22 | Guanyl-nucleotide exchange factor |
| SNF7 | 16 | 100 | 59 | Late endosome to vacuole transport |
| TLG2 | 14 | 50 | 105 | t-SNARE, v-SNARE/nonselective vesicle fusion |
| VPS24∥ | 44 | 50 | 38 | Late endosome to vacuole transport |
| VPS29 | 19 | 30 | 100 | Retrograde (endosome to Golgi) transport |
| VPS4¶ | 46 | 70 | 50 | ATPase/late endosome to vacuole transport |
| VPS41 | 14 | 50 | 90 | Rab guanyl-nucleotide exchange factor |
| VOS9 | 30 | 30 | 130 | Protein transporter/ER to Golgi transport |
| Group 6: Protein-vacuolar targeting | ||||
| DID2¶†‡ | 30 | 50 | 144 | Protein-vacuolar targeting |
| MON1¶†‡ | 42 | 80 | 54 | Protein-vacuolar targeting |
| STP22¶†‡ | 33 | 30 | 22 | Protein-vacuolar targeting |
| VPS28∥‡‡ | 20 | 70 | 10 | Protein-vacuolar targeting |
| VPS51¶‡‡ | 24 | 70 | 59 | Protein-vacuolar targeting |
| VPS61¶‡‡ | 31 | 100 | 58 | Protein-vacuolar targeting |
| VPS69¶‡‡ | 17 | 50 | 10 | Protein-vacuolar targeting |
| Group 7: Membrane associated | ||||
| MSP1 | 33 | 30 | 105 | ATPase/mitochondrial translocation |
| OPT1 | 25 | 30 | 106 | Oligopeptide transporter |
| SAC1 | 23 | 100 | 101 | Inositol/phosphatidylinositol phosphatase |
| SNF4 | 40 | 60 | 70 | Protein kinase activator |
| STE14 | 48 | 50 | 147 | Isoprenylcysteine-methyltransferase |
| STV1 | 16 | 50 | 90 | Hydrogen-transporting ATPase |
| TOK1 | 15 | 40 | 80 | Potassium channel |
| Group 8: Stress-related | ||||
| GRE3‡‡ | 32 | 40 | 138 | Aldehyde reductase |
| GTT1∥ | 29 | 60 | 106 | Glutathione transferase |
| IRA2 | 24 | 50 | 70 | Ras GTPase activator |
| UGA2 | 24 | 30 | 88 | Glutamate catabolism |
| WHI3 | 29 | 40 | 90 | Phosphatase activator |
| Group 9: General metabolism | ||||
| BEM4 | 15 | 100 | 22 | Rho protein signal transduction |
| COX12 | 25 | 100 | 41 | Cytochrome-c oxidase |
| DSE1 | 25 | 100 | 94 | Cell-wall organization and biogenesis |
| GLO2††¶ | 28 | 100 | 91 | Hydroxyacylglutathione hydrolase |
| GPH1 | 37 | 50 | 69 | Glycogen phosphorylase |
| HAP3 | 12 | 40 | 123 | Regulation of carbohydrate metabolism |
| MSB1 | 27 | 50 | 67 | Establishment of cell polarity |
| PHD1 | 33 | 100 | 75 | Pseudohyphal growth |
| RMD7 | 37 | 100 | 19 | Cell-wall organization and biogenesis |
| THI3 | 31 | 100 | 89 | Carboxy-lyase/thiamin biosynthesis |
| YIL064W | 20 | 50 | 93 | S-adenosylmethionine-methyltransferase |
| Group 10: RNA transcription | ||||
| CDC50** | 42 | 30 | 50 | Transcription regulator |
| ROX3 | 9 | 100 | 10 | RNA polymerase II transcription mediator |
| SRB8 | 35 | 50 | 68 | RNA polymerase II transcription mediator |
| SWI3 | 24 | 100 | 40 | General RNA polymerase II transcription factor |
| TEA1†† | 25 | 100 | 140 | Transcription regulator |
| UME6 | 39 | 100 | 41 | Transcription regulator |
| Group 11: DNA remodeling, metabolism | ||||
| ADA2§ | 8 | 70 | 62 | Chromatin modification, histone acetylation |
| DPB4 | 18 | 100 | 53 | Epsilon DNA polymerase |
| HEX3 | 26 | 30 | 44 | DNA recombination |
| HUR1 | 32 | 100 | 55 | DNA replication |
| NGG1§ | 27 | 100 | 18 | Chromatin modification, histone acetylation |
| SAS3†† | 27 | 80 | 158 | Acetyltransferase/chromatin silencing |
| SIN3§ | 4 | 100 | 50 | Histone deacetylase |
| SLX8 | 22 | 80 | 71 | DNA metabolism |
| SLX9 | 151 | 100 | 107 | DNA metabolism |
| SNF6 | 16 | 30 | 70 | Chromatin modeling/SWI-SNF complex |
| Group 12: Function unknown | ||||
| BSC2 | 49 | 50 | 90 | Unknown |
| LDB7 | 25 | 50 | 10 | Unknown |
| YBR007C | 26 | 100 | 106 | Unknown |
| YBR032W | 32 | 50 | 36 | Unknown |
| YCR099C | 37 | 80 | 91 | Unknown |
| YFL043C | 31 | 50 | 87 | Unknown |
| YGL140C | 17 | 60 | 90 | Unknown |
| YGR064W | 16 | 30 | 10 | Unknown |
| YHR029C | 31 | 50 | 122 | Unknown |
| YIL090W | 24 | 50 | 100 | Unknown |
| YJL175W | 13 | 30 | 69 | Unknown |
| YLR358C | 39 | 100 | 24 | Unknown |
| YNL321W | 29 | 100 | 150 | Unknown |
| YPR050C | 10 | 30 | 53 | Unknown |
A plant orthologue of the given gene (underlined) has been found by BLAST search.
Relative level of DI-72 RNA (100% in parental strain).
Relative p33 level.
Similar in BMV.
Similar in α-synuclein.
Same in α-synuclein.
Same in BMV.
Same in huntingtin.
Similar in huntingtin.
Fig. 2.
Comparison of viral RNA and replicase protein levels in selected YKO strains with the parental yeast strain. (A) Ethidium-bromide-stained agarose gel of total RNA obtained from selected YKO strains showing a reduced level of DI-72 RNA accumulation. The ribosomal rRNA and DI-72 RNA bands are indicated with arrows. Each strain was transformed with three plasmids (see Fig. 1). (B and C) Northern blot analysis of total RNA extracts for DI-72 RNA (B) with the 3′ end of DI-72 as a probe and for DI-72 RNA transcript (C) with a probe for the 5′ plasmid-borne leader sequence, which is deleted in the replicating DI-72 RNA (13). (D and E) Western analysis of p33 and p92 replicase proteins in total protein samples with anti-His-tag antibody.
Fig. 3.
Increased viral RNA replication in six YKO strains. Northern blot and Western analyses were performed as described in the legend to Fig. 2.
Functional Grouping of the Identified Host Genes. The first group of host genes that affected DI-72 RNA replication includes five genes involved in protein biosynthesis either by being part of the ribosome (MRPL32, RPL1B, RPL7A, and RPS21B) or by acting as a translation-elongation factor (TEF4) (Saccharomyces Genome Database, www.yeastgenome.org). Interestingly, DI-72 RNA accumulation increased in the absence of three ribosomal proteins, suggesting that these proteins might bind to a host protein that is also needed for DI-72 RNA replication, resulting in competition between regular cellular processes and viral replication. These genes do not seem to inhibit p33/p92 translation, because the amounts of p33/p92 did not increase in the absence of these ribosomal proteins (Figs. 2 and 3). An interesting gene in this group is TEF4, which codes for translation-elongation factor EF-1γ, which binds to the L replicase protein of vesicular stomatitis virus (21). In addition, the bacterial homologue of EF-1 was shown to play a direct role in replication of Q beta bacteriophage RNA (22, 23), suggesting possible evolutionary conservation of replication strategies in these viruses. Several other RNA viruses are proposed to use EF-1α for replication (24-29), which might play a role comparable to that of EF-1γ for the above viruses.
The second group includes eight genes involved in protein metabolism and posttranslational modification (Table 1). The lack of these genes decreased replication of DI-72 RNA in yeast. Four genes (BRE1, DOA4, RAD6, and LGE1) are part of the ubiquitination pathway, whereas one gene (SIW14) codes for a protein tyrosine phosphatase, and another (MAK3) is responsible for N-terminal protein amino acid acetylation, suggesting that posttranslational modification and/or protein turnover is important for tombusvirus replication. Interestingly, three of these genes (DOA4, LGE1, and SIW14) also affected the replication of the distantly related brome mosaic virus (BMV), another plus-strand RNA virus, in yeast (20), suggesting that similar host genes are involved in posttranslational modification of either the viral replicase proteins or shared host factors of BMV and TBSV. Two genes (ARO1 and MET1) play a role in aromatic amino acid biosynthesis and methionine metabolism, respectively. Deletion of ARO1 and MET1 genes might affect virus replication indirectly through a 50-80% reduction of p33 replicase protein levels in these YKO strains when compared with the parental strain (Table 1).
The third group contains five RNA-metabolism genes with known RNA-binding (BUD21, NPL3, and NSR1) and/or ribonuclease activities (CCR4 and KEM1) (Table 1). Deletion of two of these genes increased the accumulation of DI-72 RNA, whereas deletion of three decreased it. Based on their cellular functions, these genes might affect the transport/turnover of DI-72 RNA and/or host mRNAs, which code for host factors critical for DI-72 RNA replication.
The fourth group of genes are involved in lipid metabolism. Deletion of each of the five genes in this group inhibited the accumulation of DI-72 RNA (Table 1). It is possible that the lack of these genes affected membrane composition used by TBSV for replication, i.e., peroxisomal membrane in both plants (30) and yeast (T.P. and P.D.N., unpublished data). These changes could inhibit the assembly/functionality of the replicase complex.
The fifth group includes 13 genes implicated in vesicle-mediated transport, the deletion of which reduced accumulation of DI-72 RNA (Table 1). These proteins are proposed to affect transport to endoplasmic reticulum (ARL3), Golgi (GOS1, RIC1, VSP29, and YOS9), or vacuole (SNF7, VSP24, and VPS4), or they act in membrane fusions (TLG2 and VSP41) or as transporters across membranes (MCH5, PEP3, and YOS9). Although BRE5 has been annotated as a protein with unknown function in the Saccharomyces Genome Database, BRE5 has been implicated in intracellular transport because of the sensitivity of the bre5 strain to brefeldin A, which affects intracellular transport (31). All of these 13 host genes might be involved in transporting either the viral replicase proteins/RNA or important host factors to the site of replication or posttranslational modifications. The large number of genes in this group (>10% of the total genes affecting DI-72 RNA replication) suggests that protein/RNA transport in the cytoplasm has significant influence on TBSV replication.
The sixth group contains seven genes with known functions in the vacuolar targeting of proteins (Table 1). Deletion of these genes inhibited TBSV accumulation, a finding that is unexpected because the viral replicase proteins are targeted to the peroxisome and not to the vacuoles in TBSV infections of plants (T.P. and P.D.N., unpublished data). Four strains in this group (STP22, DID2, VPS51, and VPS69) (Table 1 and Fig. 2, lanes 5 and 13) showed decreased steady-state levels of p33 in cells, suggesting that these genes could be involved in maintaining high levels of the replicase proteins.
The seventh group consists of seven gene products broadly defined as membrane-associated (Table 1). SNF4 is a protein kinase activator involved in peroxisome biogenesis, which could be important for assembling the replicase complexes on peroxisomal membranes. The other six genes code for integral membrane proteins, which might indirectly influence DI-72 RNA replication.
The eighth group contains five genes involved in response to stress. Whether the effect of these genes on DI-72 RNA accumulation is direct or indirect is unknown. Because the robust replication of DI-72 RNA might cause stress in yeast, stress-induced genes might be important to help the cells cope with virus replication. It is possible that the number of stress-induced genes involved in DI-72 RNA replication is underestimated in this work because of the redundant nature of these genes in yeast and most other eukaryotes.
The ninth group includes 11 genes with variable functions in general metabolism (Table 1). Despite their diverse functions, deletion of these genes decreased DI-72 RNA accumulation. This group of genes affects glycogen metabolism (GPH1), thiamin biosynthesis (THI3), cytochrome c oxidase biogenesis (COX12), cell wall biogenesis (DSE1 and RMD7), cell polarity (MSB1), and hyphal growth (PHD1), all of which might have an indirect affect on tombusvirus replication. Two genes involved in carbohydrate metabolism (GLO2 and HAP3) might affect DI-72 RNA replication indirectly by affecting yeast growth on medium containing galactose. Deletion of YIL064W, a methyltransferase, might reduce the efficiency of the capping of mRNAs, which could affect the translation of p33/p92 or a critical host factor(s). The BEM4 signal-transduction gene is involved in actin cytoskeleton organization, which might affect protein/RNA trafficking and DI-72 RNA replication.
The 10th and 11th groups contain genes involved in transcription and DNA remodeling. The deletion of these genes, except SLX9, reduced DI-72 RNA accumulation (Table 1). In general, deletions of these genes are expected to affect the amount of mRNAs, which, in turn, should influence the levels of p33/p92 and/or a critical host factor(s) in cells. Not surprisingly, the genome-wide screen for BMV replication also found four related genes (20) belonging to this group, as indicated in Table 1. Because many of the host proteins in this group bind to nucleic acids and/or modify the functions of other proteins, we cannot exclude the possibility that several of these genes might be directly involved in DI-72 RNA replication. The 12th group contains 14 genes and hypothetical ORFs. All of these genes inhibited DI-72 RNA replication (Table 1). The genes of unknown function represent only ≈15% of the genes identified in this screen.
Factors Influencing the Number of Identified Host Genes. The systematic genome-wide screen of yeast genes revealed that ≈2% of the genes represented in the YKO library affected tombusvirus replication. However, it is possible that the number of genes involved in TBSV DI-72 RNA replication is underestimated in this work because of the redundant nature of some genes in yeast. The 96 host genes identified might have direct or indirect roles in tombusvirus replication. For example, some host proteins could directly interact with viral replicase proteins and/or the viral RNA during replication. Other host proteins, although not interacting directly with p33/p92 or the viral RNA, could be part of protein complexes, such as those involved in p33/p92 translation, metabolism, and intracellular transport, which could affect TBSV replication by altering functional complexes and, thus, changing their contribution to virus replication. Moreover, the nature of the defects, either replication- or RNA-degradation-related, is unknown. Our approach likely minimized the number of host factors affecting TBSV DI-72 RNA replication indirectly because (i) we measured DI-72 RNA replication levels in yeast cells that constitutively expressed the replicase proteins, (ii) we did not use selection markers in the viral RNA, and (iii) we normalized DI-72 RNA accumulation in each sample based on ribosomal RNA levels, which reflect on the availability of general factors for cell growth. YKO cells sickened by particular gene deletions that contained small amount of ribosomal RNAs were not included in Table 1.
Possible Functions of Yeast Genes in RNA Replication. Based on the known functions of the identified host genes, we predict that identified host genes might affect the accumulation of DI-72 RNA by altering (i) the amount of p33/p92 and/or a critical host factor(s) by altering the efficiency of their translation, (ii) cellular transport/targeting of replication factors, (iii) posttranslational modifications of replication factors, (iv) viral RNA/protein turnover, (v) membrane structures/biogenesis, and (vi) antiviral responses. Accordingly, purification of tombusvirus replicase complexes, followed by in vitro replicase assays (15), revealed that the replicase showed either reduced template activity (from arl3Δ, ric1Δ, and yos9Δ strains) or alteration in the ratio between plus-versus minus-strand synthesis (from ric1Δ and did2Δ strains) (S. Serva, X. Lu, and P.D.N., unpublished work). However, other events could also be affected. For example, in the absence of a specific yeast protein, the vacuoles or important membrane-containing vesicles might be damaged by the viral replicase or host proteins. If peroxisome membranes (the sites of TBSV replication) are altered, TBSV replication may be reduced. The host genes might also affect other general factors such as the availability of ribonucleotides and amino acids.
Connections of TBSV-Affecting Yeast Genes to Other Phenomena. TBSV and BMV, which are members of two distinct supergroups of positive-strand RNA viruses, were each affected in their accumulation in yeast by ≈100 host genes (20). However, we found that only four of the yeast genes were in both of these sets. Three of these genes are in the protein metabolism group (ubiquitin pathway) (Table 1), and the fourth is a transcription regulator. If we consider host genes with similar functions, the number of similar and identical host genes identified by both the BMV and TBSV systems is 14. These genes mainly belong to three groups: (i) protein biosynthesis, (ii) protein metabolism, and (iii) transcription/DNA remodeling (Table 1). Host genes present in the first two groups indicate that the dependence of BMV and TBSV on the protein translation and modification machinery of the host is somewhat similar. On the contrary, none of the host genes that affected the replication of the TBSV replicon and are involved in protein targeting, membrane association, vesicle-mediated transport, or lipid metabolism were found in the BMV set (20), suggesting important differences for BMV and TBSV, which replicate in different cellular compartments [peroxisome and perinuclear endoplasmic reticulum for TBSV and BMV (32), respectively].
The largest group (≈20%) of host genes identified in the TBSV system consists of those genes likely to be involved in intracellular transport/targeting. Comparison of yeast genes, identified based on their sensitivity to brefeldin A with genes involved in tombusvirus replication, revealed six common genes, TLG2, MON1, ERG4, BRE1, BRE5, and BRE3 (similar to CDC50), that represent 30% of genes implicated for intracellular transport (31).
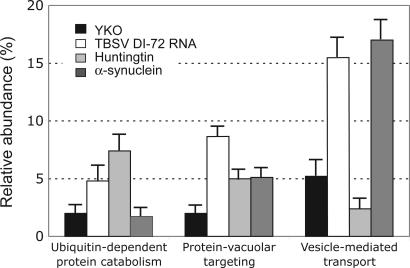
We compared our TBSV results with published genome-wide yeast screens for genes involved in vesicle-mediated transport and protein-vacuolar targeting, resulting in an intriguingly similar pair of profiles (Fig. 4). α-Synuclein has been observed to be a major component of inclusion bodies in the brains of Parkinson's disease victims, where it assembles into fibrillar protein aggregates (33). Because both replicase proteins and α-synuclein are membrane-associated in cells, they might use common protein-trafficking pathways. In the absence of these host factors, the replicase proteins and α-synuclein may not be transported efficiently to their usual locations, causing reduced viral replication and increased cytotoxicity, respectively. An alternative explanation is that both α-synuclein and the viral replicase proteins are sufficiently cytotoxic that factors such as GTT1 could protect the cell by facilitating export from cytoplasm to vacuoles (34).
Fig. 4.
Comparison of relative abundance of yeast genes with known functions within selected functional categories known to enhance the cytotoxicity of α-synuclein (35) or mutated huntingtin and affect tombusvirus replication. We included those genes from the three genome-wide screens, which matched or were known to have similar functions. The data for comparison are taken from ref. 35 and Table 1. YKO, the relative abundance of genes with given functions among nonessential genes in the YKO collection. The standard error is shown.
Additional comparison with previously published genome-wide screens revealed that the deletion of host genes involved in ubiquitin-mediated protein catabolism and protein-vacuolar targeting affected the replication of TBSV replicon and was reported to enhance the cytotoxicity of mutant huntingtin (35) (Fig. 4). Truncated huntingtin forms protein aggregates and is a major component of cytoplasmic and nuclear inclusion bodies found in Huntington's disease (35, 36). Ubiquitin-mediated protein catabolism and protein-vacuolar targeting might be part of a protein quality control system to prevent aggregation of mutated forms of huntingtin (35). Therefore, it is possible that this efficient protein quality control system, which reduces the cytotoxicity of the mutated huntingtin, is also important during the replication of the TBSV replicon in the parental yeast. A protein quality control system might help in maintaining correctly folded p33/p92 replicase proteins or critical host factors, whereas in this set of deletion strains, the protein quality control system is debilitated, leading to cytotoxicity by the mutated huntingtin and reduced efficiency of replication by the TBSV replicon.
Our results advance the possibility of further characterization of the roles of host genes in tombusvirus replication and for testing the relationships between pathways leading to virus replication and cytotoxicity of disease-causing proteins, possibly in the context of relationships between chronic diseases and prior virus infection as suggested for the Japanese encephalitis (37), influenza A (38), HIV-1, and simian immunodeficiency (39-41) viruses.
Supplementary Material
Acknowledgments
We thank Drs. Judit Pogany, Saulius Serva, and John Shaw for valuable comments. This work was supported by the U.S. Department of Agriclture National Research Initiative 2003-35319-13878 and the Kentucky Tobacco Research and Development Center.
Author contributions: T.P. and P.D.N. designed research; T.P., E.S., and J.B. performed research; T.P. and E.S. contributed new reagents/analytic tools; T.P., E.S., and P.D.N. analyzed data; and P.D.N. wrote the paper.
Abbreviations: BMV, brome mosaic virus; DI, defective interfering; TBSV, tomato bushy stunt virus; YKO, yeast knockout.
References
- 1.Ahlquist, P., Noueiry, A. O., Lee, W. M., Kushner, D. B. & Dye, B. T. (2003) J. Virol. 77, 8181-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buck, K. W. (1996) Adv. Virus Res. 47, 159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gleba, Y., Marillonnet, S. & Klimyuk, V. (2004) Curr. Opin. Plant Biol. 7, 182-188. [DOI] [PubMed] [Google Scholar]
- 4.Figlerowicz, M., Alejska, M. & Kurzynska-Kokorniak, A. (2003) Med. Res. Rev. 23, 488-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scholthof, K. B., Mirkov, T. E. & Scholthof, H. B. (2002) Genet. Eng. (N.Y.) 24, 67-85. [DOI] [PubMed] [Google Scholar]
- 6.Birrell, G. W., Giaever, G., Chu, A. M., Davis, R. W. & Brown, J. M. (2001) Proc. Natl. Acad. Sci. USA 98, 12608-12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edmonds, D., Breitkreutz, B. J. & Harrington, L. (2004) Proc. Natl. Acad. Sci. USA 101, 9515-9516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marston, A. L., Tham, W. H., Shah, H. & Amon, A. (2004) Science 303, 1367-1370. [DOI] [PubMed] [Google Scholar]
- 9.Page, N., Gerard-Vincent, M., Menard, P., Beaulieu, M., Azuma, M., Dijkgraaf, G. J., Li, H., Marcoux, J., Nguyen, T., Dowse, T., et al. (2003) Genetics 163, 875-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winzeler, E. A., Shoemaker, D. D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J. D., Bussey, H., et al. (1999) Science 285, 901-906. [DOI] [PubMed] [Google Scholar]
- 11.Noueiry, A. O. & Ahlquist, P. (2003) Annu. Rev. Phytopathol. 41, 77-98. [DOI] [PubMed] [Google Scholar]
- 12.Price, B. D., Roeder, M. & Ahlquist, P. (2000) J. Virol. 74, 11724-11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panavas, T. & Nagy, P. D. (2003) Virology 314, 315-325. [DOI] [PubMed] [Google Scholar]
- 14.Pantaleo, V., Rubino, L. & Russo, M. (2003) J. Virol. 77, 2116-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Panaviene, Z., Panavas, T., Serva, S. & Nagy, P. D. (2004) J. Virol. 78, 8254-8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gietz, R. D. & Woods, R. A. (2002) Methods Enzymol. 350, 87-96. [DOI] [PubMed] [Google Scholar]
- 17.Schmitt, M. E., Brown, T. A. & Trumpower, B. L. (1990) Nucleic Acids Res. 18, 3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pogany, J., Fabian, M. R., White, K. A. & Nagy, P. D. (2003) EMBO J. 22, 5602-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White, K. A. & Nagy, P. D. (2004) Prog. Nucleic Acid Res. Mol. Biol. 78, 187-226. [DOI] [PubMed] [Google Scholar]
- 20.Kushner, D. B., Lindenbach, B. D., Grdzelishvili, V. Z., Noueiry, A. O., Paul, S. M. & Ahlquist, P. (2003) Proc. Natl. Acad. Sci. USA 100, 15764-15769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das, T., Mathur, M., Gupta, A. K., Janssen, G. M. & Banerjee, A. K. (1998) Proc. Natl. Acad. Sci. USA 95, 1449-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blumenthal, T. (1980) Proc. R. Soc. London Ser. B 210, 321-335. [DOI] [PubMed] [Google Scholar]
- 23.Brown, D. & Gold, L. (1996) Proc. Natl. Acad. Sci. USA 93, 11558-11562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cimarelli, A. & Luban, J. (1999) J. Virol. 73, 5388-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blackwell, J. L. & Brinton, M. A. (1997) J. Virol. 71, 6433-6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, C. M., Perez, D. R., French, R., Merrick, W. C. & Donis, R. O. (2001) J. Gen. Virol. 82, 2935-2943. [DOI] [PubMed] [Google Scholar]
- 27.De Nova-Ocampo, M., Villegas-Sepulveda, N. & del Angel, R. M. (2002) Virology 295, 337-347. [DOI] [PubMed] [Google Scholar]
- 28.Matsuda, D., Yoshinari, S. & Dreher, T. W. (2004) Virology 321, 47-56. [DOI] [PubMed] [Google Scholar]
- 29.Zeenko, V. V., Ryabova, L. A., Spirin, A. S., Rothnie, H. M., Hess, D., Browning, K. S. & Hohn, T. (2002) J. Virol. 76, 5678-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rubino, L. & Russo, M. (1998) Virology 252, 431-437. [DOI] [PubMed] [Google Scholar]
- 31.Muren, E., Oyen, M., Barmark, G. & Ronne, H. (2001) Yeast 18, 163-172. [DOI] [PubMed] [Google Scholar]
- 32.Restrepo-Hartwig, M. A. & Ahlquist, P. (1996) J. Virol. 70, 8908-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conway, K. A., Lee, S. J., Rochet, J. C., Ding, T. T., Harper, J. D., Williamson, R. E. & Lansbury, P. T., Jr. (2000) Ann. N.Y. Acad. Sci. 920, 42-45. [DOI] [PubMed] [Google Scholar]
- 34.Choi, J. H., Lou, W. & Vancura, A. (1998) J. Biol. Chem. 273, 29915-29922. [DOI] [PubMed] [Google Scholar]
- 35.Willingham, S., Outeiro, T. F., DeVit, M. J., Lindquist, S. L. & Muchowski, P. J. (2003) Science 302, 1769-1772. [DOI] [PubMed] [Google Scholar]
- 36.Scherzinger, E., Lurz, R., Turmaine, M., Mangiarini, L., Hollenbach, B., Hasenbank, R., Bates, G. P., Davies, S. W., Lehrach, H. & Wanker, E. E. (1997) Cell 90, 549-558. [DOI] [PubMed] [Google Scholar]
- 37.Ogata, A., Tashiro, K., Nukuzuma, S., Nagashima, K. & Hall, W. W. (1997) J. Neurovirol. 3, 141-147. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi, M. & Yamada, T. (2001) Adv. Neurol. 86, 91-104. [PubMed] [Google Scholar]
- 39.Jenuwein, M., Scheller, C., Neuen-Jacob, E., Sopper, S., Tatschner, T., ter Meulen, V., Riederer, P. & Koutsilieri, E. (2004) J. Neurovirol. 10, 163-170. [DOI] [PubMed] [Google Scholar]
- 40.Berger, J. R. & Arendt, G. (2000) J. Psychopharmacol. 14, 214-221. [DOI] [PubMed] [Google Scholar]
- 41.Mirsattari, S. M., Power, C. & Nath, A. (1998) Movement Disorders 13, 684-689. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.