Abstract
Alzheimer’s disease-associated β-amyloid peptides (Aβ) are generated by the sequential proteolytic processing of amyloid precursor protein (APP) by β- and γ-secretases. There is growing evidence that cholesterol- and sphingolipid-rich membrane microdomains are involved in regulating trafficking and processing of APP. BACE1, the major γ-secretase in neurons is a palmi-toylated transmembrane protein that resides in lipid rafts. A subset of APP is subject to amyloidogenic processing by BACE1 in lipid rafts, and this process depends on the integrity of lipid rafts. Here we describe the association of all four components of the γ-secretase complex, namely presenilin 1 (PS1)-derived fragments, mature nicastrin, APH-1, and PEN-2, with cholesterol-rich detergent insoluble membrane (DIM) domains of non-neuronal cells and neurons that fulfill the criteria of lipid rafts. In PS1−/−/PS2−/− and NCT−/− fibroblasts, γ-secretase components that still remain fail to become detergent-resistant, suggesting that raft association requires γ-secretase complex assembly. Biochemical evidence shows that subunits of the γ-secretase complex and three TGN/endosome-resident SNAREs cofractionate in sucrose density gradients, and show similar solubility or insolubility characteristics in distinct non-ionic and zwitterionic detergents, indicative of their co-residence in membrane microdomains with similar protein-lipid composition. This notion is confirmed using magnetic immunoisolation of PS1- or syntaxin 6-positive membrane patches from a mixture of membranes with similar buoyant densities following Lubrol WX extraction or sonication, and gradient centrifugation. These findings are consistent with the localization of γ-secretase in lipid raft microdomains of post-Golgi and endosomes, organelles previously implicated in amyloidogenic processing of APP.
Alzheimer’s disease, a neurodegenerative dementing disorder, is pathologically characterized by the cerebral deposition of 39–42 amino acid peptides termed β-amyloid (Aβ)1 peptides. Aβ is generated by the sequential processing of amyloid precursor protein (APP) by β- and γ-secretases (1, 2). BACE1, a transmembrane aspartyl protease is the major β -secretase, which cleaves APP within the extracellular/luminal domain, generating the N terminus of Aβ (3). A multimeric complex made of four transmembrane proteins is responsible for executing intramembraneous γ-secretase cleavage of APP C-terminal fragments (CTF) and releasing Aβ (4). Genetic mutations in the APP, PSEN1, and PSEN2 genes are responsible for familial early-onset Alzheimer’s disease (5, 6). PSEN1 and PSEN2 encode polytopic membrane proteins presenilin 1 (PS1) and PS2, respectively, which as components of the γ-secretase complex play essential role in Aβ production (7, 8). Considerable evidence suggests that familial Alzheimer’s disease-linked PS1 and PS2 (PS) variants exert their pathogenic influence by selectively elevating the levels of highly fibrillogenic Aβ42 peptides (2, 4). In addition to APP proteolysis, PS play a crucial role in the intramembraneous γ-secretase cleavage of select type I membrane proteins including the homologues of APP (APLP1 and APLP2), Notch1, and homologues, Notch ligands Delta and Jagged, ErbB-4, CD44, low density lipoprotein receptor-related protein, N- and E-cadherins, nectin-1α, DCC, p75 neurotrophin receptor, etc. (see Ref. 9). In addition to the growing number of γ-secretase substrates, accumulating evidence from protein interaction and loss of function studies suggest that, aside from Aβ production, PS regulate diverse physiological functions (reviewed in Ref. 9).
It is evident from recent studies that γ-secretase is a multi-protein complex comprised of PS1 (or PS2)-derived N- and C-terminal fragments (NTF and CTF), nicastrin, APH-1, and PEN-2 (10–12). Nicastrin is a type I membrane protein, and PEN-2 and APH-1 are predicted to span the membrane two and eight times, respectively. Results from gene knockout and knockdown studies reveal that γ-secretase components assist each other during biogenesis and exit out of the endoplasmic reticulum (ER), and all four components are required to undergo proper post-translational maturation and achieve stability (reviewed in Ref. 13). APH-1 is presumed to have a role in the initial assembly and maturation of PS-nicastrin complexes (14), while PEN-2 is required for endoproteolytic processing of PS (15). The subcellular site(s) of γ-secretase cleavage of APP is a subject of considerable interest. Available data indicate the presence of γ-secretase activity in multiple compartments including the ER, late-Golgi/TGN, endosomes and plasma membrane (16–19). PS1 has been localized to multiple intracellular membranes including the ER, ER/Golgi intermediate compartments, Golgi apparatus, endosomes and the plasma membrane by immunogold-electron microscopy (20–23). Although localization of the other components have not been investigated by electron microscopy, confocal microscopy, and subcellular fractionation studies reveal significant co-localization of all four γ-secretase components in the ER, Golgi, and the TGN (14, 15, 24, 25). Recent biochemical evidence suggests that γ-secretase components assemble into the proteolytically active complex in the Golgi/TGN compartments (25).
There has been considerable epidemiological interest in the relationship between cholesterol and susceptibility to Alzheimer’s disease (26, 27). Cholesterol is the major sterol component in most mammalian membranes. Growing evidence implicates specialized cellular membrane microdomains rich in cholesterol and sphingolipids, termed lipid rafts, in a number of important biological functions (28). DIM lipid rafts contribute to trafficking of proteins and lipids in the secretory and endocytic pathways by regulating vesicle sorting and formation (29). Several lines of evidence indicate that amyloidogenic processing of APP occurs in cholesterol- and sphingolipid-enriched DIM domains. First, DIMs were found to be the principal compartment containing monomeric and oligomeric Aβ in brain (30, 31). Second, experimental manipulations that result in cholesterol loading and depletion, or affect intracellular cholesterol transport, modulate Aβ production in cultured cells and animal models (26, 32–35). Third, BACE1 is localized and cleaves APP in lipid rafts (33, 36). Fourth, buoyant cholesterol-rich DIMs contain high levels of γ-secretase activity (33, 37), and a subset of PS1 and nicastrin partition into these membrane domains (30, 31, 33, 37). Interestingly, a recent study reported that γ-secretase cleavage occurs in lipid rafts, but γ-secretase catalytic activity is independent of the presence of cholesterol (38). Thus, it appears that APP processing within cholesterol-rich lipid rafts by secretases, and not the cholesterol levels per se, determines the levels of Aβ production.
In this study, we have provided the first detailed characterization of lipid raft association of each of the components of γ-secretase, the enzyme, which plays an essential role in Alzheimer’s disease-associated Aβ production. We report cholesterol-dependent association of significant amounts of each of the γ-secretase components to lipid rafts. Moreover, by subcellular fractionation, confocal microscopy, and immunoisolation methods, we show that endogenous PS1, nicastrin, APH-1, and PEN-2 associate with raft microdomains in post-Golgi and endosome membranes enriched in syntaxin 6, syntaxin 13, and VAMP4. These findings provide novel insights into the subcellular localization of γ-secretase, which has important implications in understanding the regulation of amyloidogenic processing of APP.
EXPERIMENTAL PROCEDURES
Cell Culture
Mouse N2a neuroblastoma cells stably transfected with human PS1 (N2aWt.11) (39) were cultured in 45% Dulbecco’s modified Eagle’s medium and 50% Opti-MEM (Invitrogen) supplemented with 5% fetal bovine serum and 200 μg/ml of G418. Wt, PS1−/−/PS2−/−, and NCT−/− mouse embryonic fibroblasts (40, 41), and HeLa cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. NCT−/− fibroblasts were transfected with human wild-type nicastrin expression plasmid and a pool of stable transfectants (NCTwt) were selected in 400 μg/ml hygromycin. Nicastrin expression in NCTwt pool was confirmed by Western blotting and immunofluorescence labeling. Primary cultures of cortical neurons were prepared from 17-day-old mouse embryos by dissociation with trypsin. Cultures were maintained in 60-mm polyethyleneimine-coated dishes in Neurobasal medium with B27 supplement (Invitrogen) for 11 days in vitro. For the MβCD treatment, cells grown to confluency were washed once with warm phosphate-buffered saline and incubated with or without 5 mm methyl-β-cyclodextrin in serum-free medium for 2 h at 37 °C (42).
Antibodies
The following antibodies were used in this study: PS1NT was raised against residues 1–65 of PS1 (43); Ab14 was raised against residues 1–25 of PS1 (44); αPS1Loop was raised against residues 263–407 of PS1 (44); SP716 was raised against residues 62–93 of NCT (24); polyclonal antibody 3925 recognizes C-terminal 19 amino acids of NCT (40); PNT2 was raised against residues 1–26 of PEN-2 (15); polyclonal antibody O2C2 recognizes C-terminal 20 amino acids of long isoform of human APH-1a was provided by Drs. Paul Fraser and Peter St George-Hyslop (Univ. of Toronto) (14); antibody H2D2 raised against the same epitope of APH-1aL was used in some experiments (provided by Dr. Gang Yu, Univ. of Texas Southwestern Medical Center) (45); polyclonal antibody STC261 was raised against stanniocalcin 2 (STC2);2 polyclonal PrP antibody D13 was generously provided by Dr. James A. Mastrianni (University of Chicago). Anti-PS1loop (Chemicon), mono-clonal anti-nicastrin (BD Transduction Laboratories), and polyclonal PrP (46) antibodies were used in primary neuronal culture studies. Antibodies against organelle markers and lipid raft-associated proteins were purchased from commercial sources: anti-KDEL and calnexin (Stressgen); caveolin-1, (Santa Cruz Biotechnology); GM130, p115, γ-adaptin, syntaxin 6, EEA1, flotillin-1, and flotillin-2 (BD Transduction Laboratories); cholera toxin (Calbiochem); syntaxin 13, SNAP-23, and VAMP4 (Synaptic Systems, Göttingen, Germany); Na+/K+-ATPase (Upstate Biotechnology, Inc.); LAMP-1 (Developmental Studies Hybridoma Bank); OKT8 (ATCC). Cy2-, or Cy3-conjugated secondary antibodies were purchased from Jackson Immunoresearch Laboratories.
Lipid Raft Isolation
Lipid rafts were isolated from nearly confluent cells grown in two 100-mm dishes as described previously (47–49) with some modifications. Briefly, cells were washed twice in phosphate-buffered saline, scraped into 1.5 ml of lysis buffer containing different detergents: 0.5% Lubrol WX (Lubrol 17A17; Serva) or 0.5% Brij-96V (Fluka) in 25 mm Tris-HCl (pH 7.4), 150 mm NaCl, and 5 mm EDTA; 1% CHAPSO (Amresco) in 25 mm MES (pH 6.5) and 150 mm NaCl (MBS buffer); or 1% Triton X-100 in 25 mm Tris-HCl (pH 8) and 140 mm NaCl. All the lysis buffers were supplemented with a protease inhibitor mixture (Sigma) and 1 mm phenylmethylsulfonyl fluoride. Cells were homogenized by five passages through a 25-gauge needle. Lysates were adjusted to 45% final concentration of sucrose (final volume, 4 ml) and transferred to a 12-ml ultracentrifuge tube. A discontinuous sucrose gradient is then formed by sequentially layering 35% sucrose (4 ml) and 5% sucrose (4 ml), and the tubes were subject to ultracentrifugation at 39,000 rpm for 19 h in Beckman SW41 rotor at 4 °C. Twelve 1-ml fractions were collected from the top of the gradient using a fraction collector and equal volume of each fraction was analyzed by Western blotting. In some cases samples were concentrated by methanol/chloroform precipitation, or subject to immunoprecipitation before analysis. Mouse primary neurons were lysed in buffer containing 1% Lubrol WX in MES-buffered saline containing Complete protease inhibitor mixture (Roche Applied Science) at 4 °C and DIM were prepared as previously described (47).
Lipid rafts were also isolated using a non-detergent method as described previously (50). Briefly, cells grown to confluency were washed in cold phosphate-buffered saline and scraped into 2 ml of ice-cold MBS buffer containing 0.5 m sodium carbonate and protease inhibitors. Cells were disrupted on ice by Dounce homogenization (15 strokes) followed by sonication using 130 watt Ultrasonic Processor (Model GE130) set at amplitude 50 (twelve 5-s bursts). This homogenate was then subjected to fractionation as described above in sucrose gradient containing 250 mm sodium carbonate.
Subcellular Fractionation
Cells from two 100-mm dishes were homogenized using a ball-bearing homogenizer with a 12 μm clearance and postnuclear supernatants were fractionated on sucrose density gradients as described previously (18). Twelve 1-ml fractions were collected from the top of the gradient using a fractionator, and 60 μl of each fraction were analyzed by Western blotting. To isolate lipid rafts from post-Golgi and endosome membranes, fraction 6 (enriched in Golgi/TGN/endosome/lysosome markers) from four gradients were pooled, diluted with 25 mm Tris-HCl (pH 7.4), and 5 mm EDTA, and subject to centrifugation at 27,000 rpm for 1 h at 4 °C in a SW41Ti rotor (Beckman Instruments). The resulting membrane pellet was resuspended in 0.5% Lubrol WX in 25 mm Tris-HCl (pH 7.4), 150 mm NaCl and 5 mm EDTA, homogenized by five passages through a 25-gauge needle, and subject to flotation density gradient centrifugation as described above.
Immunoisolation of Raft Patches Using Magnetic Beads
Mono-PS1Loop clonal antibody syntaxin 6, and affinity-purified PS1NT and α antibodies were bound to Dynabeads M-280 beads pre-coated with sheep anti-rabbit or anti-mouse IgG (Dynal) according to the manufacturer’s instructions. Antibody OKT8 (recognizes CD8α) and affinity-purified polyclonal antibody STC261 (recognizes a luminal protein that is processed through the secretory pathway) were used as negative controls to establish the specificity of the immunoisolation procedure. Pooled lipid raft fractions 4 and 5 generated from Lubrol WX or non-detergent lysates were incubated with primary antibody-coated magnetic beads for 5 h with continuous slow rotation at 4 °C. Bound immunocomplexes were captured using a magnetic device and washed four times in 20 mm Tris-HCl (pH 7.4) and 150 mm NaCl, and eluted in Laemmli buffer. Bound immunocomplexes and an aliquot of the input were analyzed by Western blotting.
Immunofluorescence Labeling
Mouse embryonic fibroblasts or HeLa cells cultured on glass coverslips were fixed as described previously (51) and blocked with 3% bovine serum albumin. Primary and secondary antibodies were diluted in phosphate-buffered saline containing 0.2% Tween 20 and 3% bovine serum albumin. Confocal images were acquired with a 100× oil immersion objective using a Zeiss laser scanning microscope (Pascal 5) and processed using MetaMorph software.
RESULTS
PS1 and Mature Nicastrin Are Present in Lubrol WX-insoluble Lipid Rafts
In order to gain mechanistic insights into the amyloidogenic processing of APP in detergent insoluble buoyant membrane domains, we undertook studies to determine the localization of endogenous γ-secretase components within lipid rafts of cultured cells. Previous studies have reported the presence of PS1 derivatives in membrane domains resistant to extraction with certain detergents, indicative of raft localization (30, 33, 37, 38). The choice of detergent is a critical step in isolation of lipid rafts as it is becoming clear that raft domains that differ in their protein composition and properties can be subclassified based on their differential solubility in select detergents (48, 49, 52, 53). For example, Thy-1 and PrP (both anchored by glycosylphosphatidylinositol anchor) localize to distinct membrane domains with variable detergent solubility due to differences in glycolipid and lipid composition (48, 54). Hence, we first evaluated partitioning of PS1 and nicastrin into DIMs in detergents that are routinely used to isolate lipid rafts from cultured cells: non-ionic detergents Brij-96 (0.5%), Triton X-100 (1%), and Lubrol WX (0.5%). We also used CHAPSO (1%), a zwitterionic detergent commonly used to examine association between γ-secretase components (55, 56), and DIM association of PS1 (38).
Membrane rafts from mouse neuroblastoma N2a cells stably expressing human PS1 were isolated on the basis of their enriched content of glycosphingolipids and cholesterol and the resulting insolubility in detergents at low temperature, and their low buoyant density. DIM prepared with different detergents have been observed to have different densities (53), and fractions enriched in DIM were identified by immunoblotting with antibodies against flotillin-2, an abundant raft-associated protein (57). Distribution of calnexin, a non-raft transmembrane protein localized in the ER, was also examined to monitor the contamination of DIM lipid rafts with ER membrane. In all four cases flotillin-2 was enriched in fractions 4 and 5 (the interphase between 5 and 35% sucrose in the gradient); unlike the case of Triton X-100 and Lubrol WX, flotillin-2 was broadly distributed in raft and non-raft fractions in cells solubilized in Brij 96 and CHAPSO (Fig. 1). Although it has been previously employed to separate raft-associated PrP (54), 0.5% Brij 96 only provided poor separation of DIM from non-raft proteins as calnexin was found distributed in all but the topmost fraction. DIM fractionated using 1% CHAPSO also contained detectable levels of calnexin, indicating contamination with ER membrane proteins. Extraction with either Lubrol WX or Triton X-100 yielded good separation of the raft marker flotillin-2 from the ER-resident protein calnexin (Fig. 1, C and D).
Fig. 1. Analysis of the detergent solubility characteristics of PS1 and nicastrin.
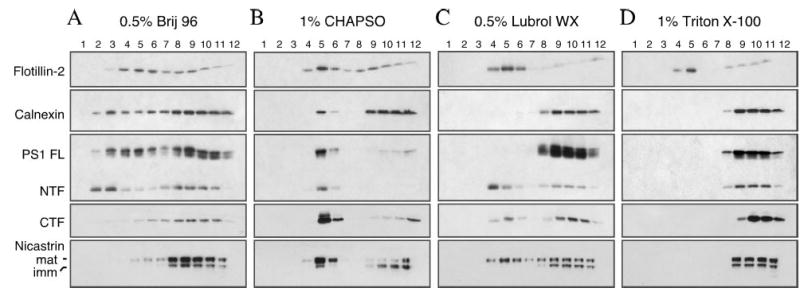
N2aWt.11 cells were solubilized either in 0.5% Brij 96 (A), 1% CHAPSO (B), 0.5% Lubrol WX (C), or 1% Triton X-100 (D) at 4 °C for 30 min. The lysates were then subject to flotation centrifugation on discontinuous sucrose gradients as described under “Experimental Procedures.” The gradients were harvested from the top, and the distribution of PS1 and nicastrin were determined by fractionating 60-μl aliquots of gradient fractions (1–12; top to bottom) on SDS gels followed by Western blot analysis. Flotillin-2 and calnexin were analyzed in parallel as a marker to indicate DRM and detergent-soluble fractions, respectively.
Next, we examined the distribution of PS1 and nicastrin across sucrose gradient fractions isolated using different detergents. Similar to calnexin, full-length PS1, and NTF were also found in all except the first fraction in Brij 96 gradients, although PS1 NTF was slightly enriched in light density fractions 2 and 3. However, PS1 CTF and nicastrin were recovered mainly in the heavier fractions, which contain detergent soluble proteins (Fig. 1A). On the other hand, nearly all of full-length PS1, PS1 NTF/CTF, and mature nicastrin were recovered in DIM fractions 5 and 6 when cells were lysed in 1% CHAPSO (Fig. 1B). When cells were lysed in Lubrol WX, the majority of PS1 NTF and substantial amount of PS1 CTF and mature nicastrin were associated with DIM, which floated to the low density fractions, whereas full-length PS1 and immature nicastrin along with a fraction of mature nicastrin co-sedimented with calnexin in heavier fractions (Fig. 1C). Finally, full-length PS1, PS1 NTF/CTF or nicastrin were undetectable in membranes insoluble in 1% Triton X-100 at 4 °C (Fig. 1D). Interestingly, it is worth noting that greater amount of PS1 derivatives and mature nicastrin is recovered in DIM when we examined naïve cells not overexpressing PS1 were solubilized in Lubrol WX (see Figs. 3 and 4A). Based on the above results, we conclude that mature components of the γ-secretase are soluble in Triton X-100, but remain insoluble in Lubrol WX-resistant cholesterol-rich membranes consistent with their recruitment into biochemically distinct lipid rafts, as it has been previously described for microvilli resident protein prominin, copper-transporting P-type ATPase ATP7B, and multidrug resistance protein 1 (47, 58).
Fig. 3. DIM localization of endogenous γ-secretase components in neuronal and non-neuronal cells.
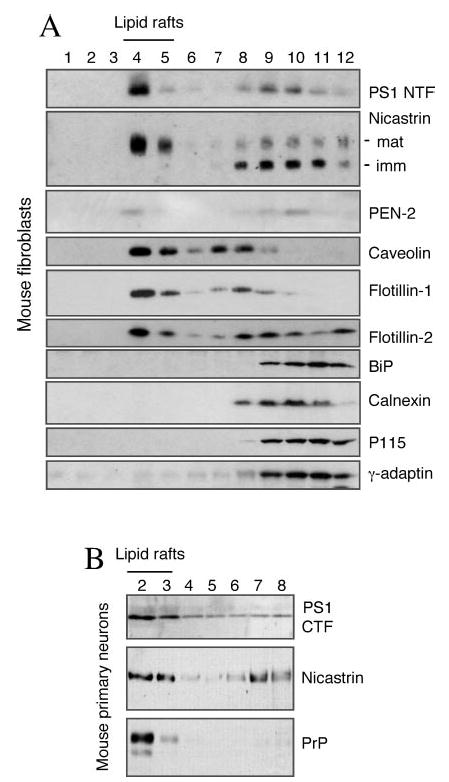
A, mouse embryonic fibroblasts were lysed in 0.5% Lubrol WX and subjected to flotation sucrose density gradient centrifugation. Equal volumes of each fraction was analyzed by Western blotting with antibodies against PS1, nicastrin and PEN-2. Caveolin, flotillin-1, and flotillin-2 mark low buoyant density lipid raft fractions. Non-raft proteins residing in the ER (BiP and calnexin), Golgi (P115), and the TGN (γ-adaptin) (TGN) are recovered in heavier fractions. B, mouse primary neurons were subject to sucrose density gradient fractionation and analyzed as above using anti-PS1 loop, mAb anti-nicastrin, and PrP antibodies. Lipid raft fractions were identified by the presence of GPI-anchored PrP protein as the marker.
Fig. 4. γ-Secretase components are recruited in Lubrol WX DIM following their assembly.
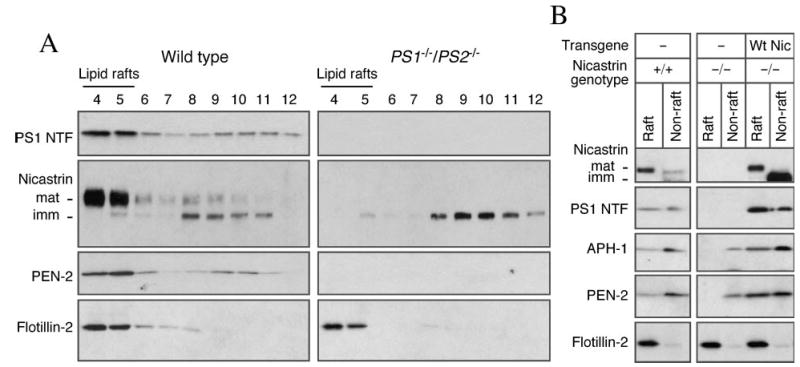
A, PS1−/−/PS2−/− fibroblasts were solubilized in 0.5% Lubrol WX and subject to sucrose gradient fractionation. In the absence of PS1 and PS2, immature nicastrin fails to become Lubrol WX-resistant and were recovered in non-raft fractions. B, wild-type and NCT−/− fibroblasts stably transfected with an empty vector (Vec) or human wild-type nicastrin cDNA were lysed and fractionated on a sucrose gradients. Raft (fractions 4 and 5) and non-raft fractions 8–12 were pooled, methanol/chloroform-precipitated, and analyzed by Western blotting. In the absence of nicastrin, APH-1, and PEN-2 remain in the non-raft fractions. Introduction of nicastrin in NCT−/− fibroblasts restores raft association of γ-secretase components.
The Lubrol WX-resistant DIM-containing γ-Secretase Components Are Sensitive to Cholesterol Depletion
The above results indicate that a subset of mature PS1 and nicastrin associate with Lubrol WX-insoluble membranes that float in a sucrose density gradient. Next, we sought to establish the role of cholesterol in the maintenance of γ-secretase components at Lubrol WX-insoluble membranes. Cholesterol, which is present in both leaflets of cell membranes, plays an important role in stabilizing “liquid-ordered” microdomains enriched in cholesterol and sphingolipids, and depletion of cholesterol disrupts lipid raft integrity. Treatment of cells at 37 °C with methyl-β-cyclodextrin (MβCD), a drug known to selectively deplete biological membranes of cholesterol and render certain raft-associated proteins detergent soluble, leading to their dissociation from rafts and disappearance from the floating fraction in density gradients (59, 60). Hence, we reasoned that cholesterol-dependent association of γ-secretase components with DIM will fulfill an important criterion for their association with lipid rafts. Relative to control cells, cholesterol depletion by exposure of N2a cells to MβCD caused displacement of PS1 NTF and mature nicastrin from the low density fractions upon gradient analysis of Lubrol WX lysates (Fig. 2). In contrast, MβCD treatment did not affect sedimentation of PS1 holoprotein or immature nicastrin in Lubrol WX-soluble heavier fractions. We also examined the distribution of PEN-2 and show that DIM association of PEN-2 is also disrupted by MβCD treatment (Fig. 2). Similarly, we found that lipid raft association of PrP, a GPI-linked protein was sensitive to MβCD treatment. In agreement with a previous report, MβCD treatment did not affect flotillin-2 association with DIM (61). Thus we conclude that mature γ-secretase associates with lipid rafts in a cholesterol-dependent manner.
Fig. 2. DIM association of γ-secretase components is sensitive to cholesterol depletion.
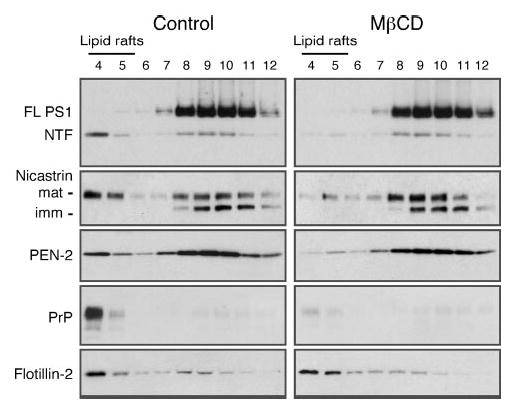
N2aWt.11 cells were treated or not with 5 mm MβCD for 2 h at 37 °C, lysed in 0.5% Lubrol WX, and analyzed by flotation on sucrose gradients. DIM localization of γ-secretase components, and raft-associated proteins PrP and flotillin-2, were assessed by Western blotting.
Endogenous Components of the γ-Secretase Are Enriched in Lipid Rafts in Neuronal and Non-neuronal Cells
Since the studies described above were performed in N2a neuroblastoma cells stably transfected with human PS1, we undertook studies to determine the localization of endogenous PS1 in mouse fibroblasts. Sucrose density fractionation revealed that the majority of endogenous PS1 and mature nicastrin are associated with Lubrol WX-insoluble rafts that float to low-density fractions (Fig. 3A). About 30% of PEN-2 is also associated with Lubrol WX-insoluble rafts. As expected, these fractions are enriched in lipid raft markers caveolin, flotillin-1 and flotillin-2. Furthermore, immature nicastrin fractionates with Lubrol WX-soluble heavier membrane fractions that contain luminal, integral membrane or membrane-associated protein markers of the ER (BiP and calnexin), Golgi (P115) and the TGN (γ-adaptin).
Because it appears likely that lipid rafts have different properties depending on the cell type, it was important to determine whether γ-secretase components are associated with neuronal lipid rafts. To this end, we performed sucrose density gradient analysis of mouse primary neurons lysed in Lubrol WX. To accommodate analysis of smaller number of cells present in neuronal cultures, we scaled down the size of the gradients and collected 8 fractions; DIMs were recovered in fractions 2 and 3. Consistent with results from analysis of mouse neuroblastoma and fibroblasts, nicastrin, and PS1-derived fragments are predominantly associated with Lubrol WX-insoluble lipid raft fractions (Fig. 3B). As expected, GPI-anchored prion protein was exclusively localized in low-density lipid raft fractions of primary neurons.
Assembly of γ-Secretase Components Precedes Raft Association
It is now well established that formation of high molecular weight γ-secretase complex involves co-operative maturation and ER exit of PS1, nicastrin, APH-1, and PEN-2 (10, 11, 13). Previous reports also indicate that when one of the core components of the γ-secretase complex is not present, the others fail to exit the ER. In the case of PS1−/−/PS2−/− cells, nicastrin remains in the core glycosylated immature form, whereas the steady-state levels of PEN-2 are markedly diminished (24, 62). Under these circumstances, we expected not to recover substantial amounts of the unassembled γ-secretase subunits in lipid raft fractions. This prediction is based on the absence of lipid raft microdomains in the ER (63). Fractionation of Lubrol WX lysates confirmed our prediction and showed that greater than 90% of immature nicastrin remains detergent soluble in PS1−/−/PS2−/− cells (Fig. 4A). To confirm these findings, we also analyzed raft association of γ-secretase components in NCT−/− fibroblasts. As reported previously, NCT−/− cells have nearly undetectable levels of PS1 NTF and CTF. On the other hand, the levels of APH-1 and PEN-2 are greatly reduced, but still detectable, in NCT−/− cells as compared with wild-type cells. Fractionation analysis revealed that substantial amounts of endogenous nicastrin, PS1 fragments, APH-1 and PEN-2 partitioned into lipid rafts in wild-type fibroblasts (Fig. 4B, lane 1). On the other hand, APH-1 and PEN-2 in NCT−/− cells are present only in the non-raft fractions (Fig. 4B, lane 4). Stable expression of human nicastrin in NCT−/− cells fully restored the steady-state levels of PS1, APH-1, and PEN-2, as well as promoted their efficient partitioning into Lubrol WX-resistant lipid raft domains (Fig. 4B, lane 5). Together, these results indicate that the subunits that still remain in cells lacking one or more components of γ-secretase complex, reside exclusively in detergent-soluble non-raft membranes.
Subcellular Localization of γ-Secretase Complex in Lubrol WX-resistant Membrane Microdomains
Components of the γ-secretase complex have been found to reside in the ER, as well as in secretory and endocytic compartments (20, 21, 64, 65). Following exit from the ER, the core components of γ-secretase assemble into a functional complex in the Golgi/TGN compartments (25). Cholesterol and sphingolipids get organized into detergent-insoluble raft domains in the secretory pathway as early as in the Golgi membranes, and play a role in apical trafficking of certain raft-associated proteins in TGN-derived secretory vesicles (63, 66). With the exception of nicastrin, endogenous γ-secretase complex components are poorly labeled by cell surface biotinylation in our hands, and are not readily detectable by live staining of non-permeabilized cells using antibodies raised against luminal epitopes. Double immunofluorescence confocal microscopy analysis of PS1 and organelle markers reveals considerable overlap in the subcellular localization of endogenous PS1 with resident proteins of the ER (BiP and GRP94), cis-Golgi (GM130), TGN and TGN-derived vesicles (γ-adaptin and syntaxin 6) and only slight overlap with markers of the plasma membrane (Na+/K+-ATPase) early endosomes (EEA1) (Fig. 5). Hence, we postulated that the majority of raft-associated γ-secretase likely resides in intracellular membranes. To experimentally test this idea, we carried out a two-step subcellular fractionation method. First, cells were disrupted without the use of detergents using a ball-bearing homogenizer and lysates were fractionated through a sucrose step gradient to yield a fraction (fraction 6) highly enriched in mature components of the γ-secretase complex (Fig. 6). Using antibodies against organelle markers we determined that fraction 6 contains Golgi (GM130), TGN (γ-adaptin, syntaxin 6, and VAMP4), late endosomes (syntaxin 13), and lysosomes (LAMP-1). In addition, the majority of raft marker flotillin-2 and the raft component ganglioside GM1 were also recovered in fraction 6. Notably, fraction 6 was devoid of plasma membrane resident protein Na+/K+-ATPase (recovered in fractions 1 and 2), and ER-resident protein calnexin (enriched in fractions 10 and 11).
Fig. 5. Subcellular localization of endogenous PS1 by confocal microscopy.
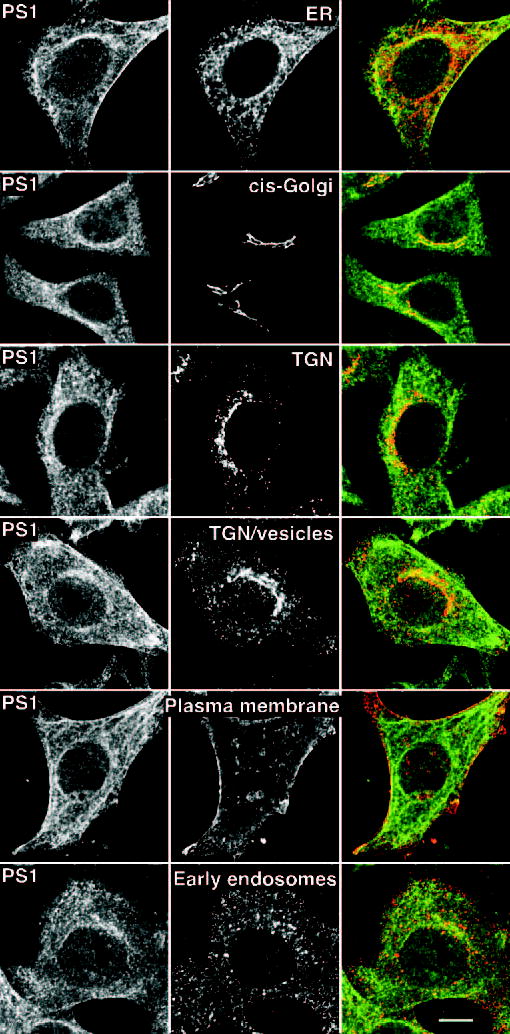
HeLa cells were grown on coverslips were fixed and analyzed by double immunofluorescence staining with PS1 N-terminal antiserum Ab14, and antibodies raised against resident proteins of the ER (anti-KDEL), cis-Golgi (GM130), TGN (γ-adaptin), TGN/TGN-derived vesicles (syntaxin 6), plasma membrane (Na+/K+-ATPase), or early endosomes (EEA1). Note the colocalization between PS1 (green) and Golgi/TGN markers (red). Scale bar, 10 μm.
Fig. 6. Subcellular fractionation of γ-secretase components in sucrose gradients.
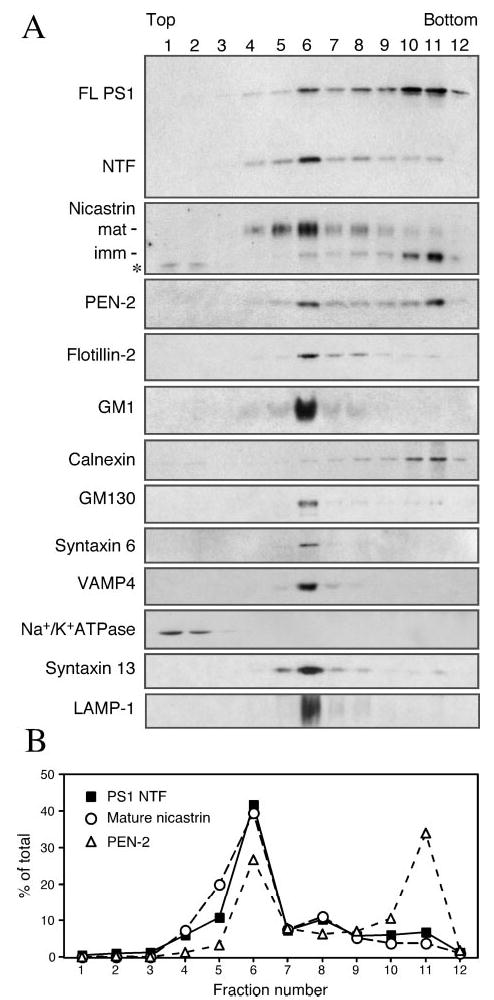
N2aWt.11 cells were grown to confluency, and cells were lysed using a ball-bearing homogenizer and fractionated by velocity sedimentation. A, equal volume aliquots of fractions were analyzed by immunoblotting using antibodies against the following organelle-specific markers: calnexin (ER), GM130 (Golgi), syntaxin 6, and VAMP4 (TGN/TGN-derived vesicles), Na+/K+-ATPase (plasma membrane), syntaxin 13 (late endosome), and LAMP-1 (lysosome). Ganglioside GM1 distribution in the gradient fractions was detected by binding of cholera toxin followed by incubation with anti-cholera toxin antiserum. Note that significant amounts of PS1 NTF, nicastrin, and PEN-2 are present in fraction 6, which is enriched in Golgi, TGN, endosome, and lysosome markers. An asterisk indicates nonspecific protein (migrating faster than immature nicastrin polypeptide) in fractions 1 and 2 that is reactive with nicastrin antibody. B, signal intensities of PS1 NTF, mature nicastrin and PEN-2 were quantified, and the relative distribution in each of the sucrose density fractions is plotted as % of total intensity for each protein. Note that the second peak for PEN-2 (fraction 11) likely represents nascent PEN-2 that is in the process of assembly with full-length PS1 and immature nicastrin (also abundant in fraction 11) within the ER.
To ascertain whether components of the γ-secretase complex in this Golgi/TGN/endosomes/lysosome (GTEL)-enriched membranes are present within detergent-insoluble lipid raft microdomains, we solubilized fraction 6 in Lubrol WX and subject to flotation density gradient centrifugation. Western blot analysis revealed that about 30% of mature PS1 fragments and nicastrin and about 20% PEN-2 in the GTEL membrane fraction partition into Lubrol DIM, which is recovered in low-density raft fractions (Fig. 7). On the other hand, PS1 holoprotein remained Lubrol WX soluble and located in the high density fractions 8–12, indicating that only proteolytically processed PS1 derivatives reside in DIM domains of GTEL membranes. Similarly, immature nicastrin was soluble in Lubrol WX, further providing evidence that only the mature components of the γ-secretase complex acquire resistance to solubilization in Lubrol WX. Raft-associated flotillin-2 and GM1 were found enriched in low-density DIM fractions 4 and 5, suggesting that the integrity of lipid rafts are maintained during cell homogenization and subsequent centrifugation steps. As expected, Golgi and TGN-resident non-raft proteins GM130 and γ-adaptin, respectively, were recovered in Lubrol soluble heavier fractions 8–12 (Fig. 7). Similarly, one of the major lysosome associated transmembrane proteins, LAMP-1, was also found in Lubrol WX-soluble heavier fractions 8–12.
Fig. 7. Raft association of γ-secretase components in GTEL-enriched membranes.
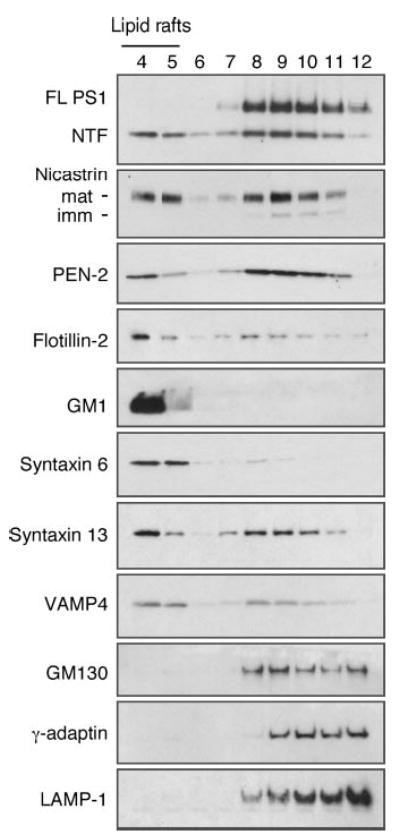
N2aWt.11 cells grown to confluency were lysed and fractionated on sucrose gradients as described under legend to Fig. 5. Membranes in fraction 6 were pooled from four gradients. Lipid rafts from these GTEL-enriched membranes were isolated by solubilization in 0.5% Lubrol WX and subject to sucrose gradient fractionation. Aliquot of each fraction were subject to SDS-PAGE and Western blot analyses. Raft fractions were identified by the presence of lipid raft markers flotillin-2 and GM1. Mature components of γ-secretase complex, syntaxin 6, syntaxin 13, and VAMP4 partition into DIM fractions, whereas GM130, γ-adaptin, and LAMP-1 remain Lubrol WX-soluble.
We then searched for additional vesicle/organelle markers that cofractionated with γ-secretase components in light density DIMs derived from GTEL membranes. Virtually all of syntaxin 6, a t-SNARE implicated in the delivery of TGN-derived AP-1/clathrin-coated vesicles to endosomes (67), was recovered in light density Lubrol WX-resistant fractions (Fig. 7). Next we turned our attention to syntaxin 13, a t-SNARE that is localized in tubular extensions of early endosomes as well as recycling endosomes, and thought to mediate recycling of plasma membrane proteins (68). Western blot analysis revealed that 30% of syntaxin 13 partitioned into DIM (Fig. 7). We further examined the distribution of VAMP4, a protein that colocalizes with syntaxin 6 in tubular and vesicular membranes of the TGN (69) and found that about one-third of VAMP4 is present in DIM.
Co-residence of PS1 with SNAREs in Lipid Rafts of TGN and Endosomal Compartments
The results described above suggest that a fraction of γ-secretase residing in TGN as well as early and recycling endosome compartments becomes insoluble in Lubrol WX. However, it is still possible that syntaxin 6, syntaxin 13, and VAMP4 are not co-resident with components of the γ-secretase on the same lipid raft patches or DIM microdomains. We performed two sets of experiments to address this issue. First, we asked whether syntaxin 6, syntaxin 13 and VAMP4 show detergent-resistant characteristics similar to what was observed for mature PS1 and nicastrin (see Fig. 1). Flotation gradient centrifugation analysis of cell lysates prepared with different detergents show high degree of similarity in the distribution of syntaxin 6, syntaxin 13, VAMP4 (Fig. 8) and PS1 NTF/CTF (Fig. 1) across the light and heavy density gradient fractions. Furthermore, treatment of cells with MβCD prior to solubilization in Lubrol WX also revealed that similar to components of the γ-secretase complex, DIM association of syntaxin 6, syntaxin 13 and VAMP4 is cholesterol-dependent (data not shown). Second, to obtain direct evidence for co-residence of syntaxin 6, syntaxin 13, and VAMP4 with PS1, we carried out antibody-mediated immunoisolation of syntaxin 6 and PS1-containing lipid raft microdomains from a pool of Lubrol WX-resistant DIM isolated using flotation density gradients. Aliquots of DIM were incubated with magnetic beads coated with affinity-purified antibodies raised against syntaxin 6. This enabled gentle separation of Lubrol-resistant membrane patches containing syntaxin 6 away from DIM microdomains devoid of this t-SNARE. Subsequent Western blot analysis revealed the presence of PS1 NTF/CTF, syntaxin 13, and VAMP4 co-residing with syntaxin 6 in immunoisolated Lubrol-resistant membrane patches (Fig. 9A). More importantly, we demonstrate the absence of SNAP-23 in DIM isolated using magnetic beads coated with syntaxin 6 antibody, which strongly suggests that DIM domains originating from the plasma membrane (containing SNAP-23 in cholesterol-rich rafts (70)) do not coalesce with DIM originating from TGN and TGN-derived vesicles (containing syntaxin 6) during lysates preparation or subsequent flotation gradient centrifugation steps. We then immunoisolated PS1-containing DIM using antibodies raised against the N terminus or the cytoplasmic loop domain of PS1 and confirmed the co-residence of syntaxin 6, syntaxin 13, and VAMP4 with PS1 (Fig. 9B). As expected, PS1 positive DIM also contained nicastrin and PEN-2, confirming the presence of other γ-secretase components, likely in a functional complex. Moreover, absence of SNAP-23 in PS1-containing DIM indicates that significant amount of PS1 is not localized in lipid rafts originating from the plasma membrane.
Fig. 8. Syntaxin 6, syntaxin 13, and VAMP4 display detergent solubility characteristics similar to γ-secretase components.
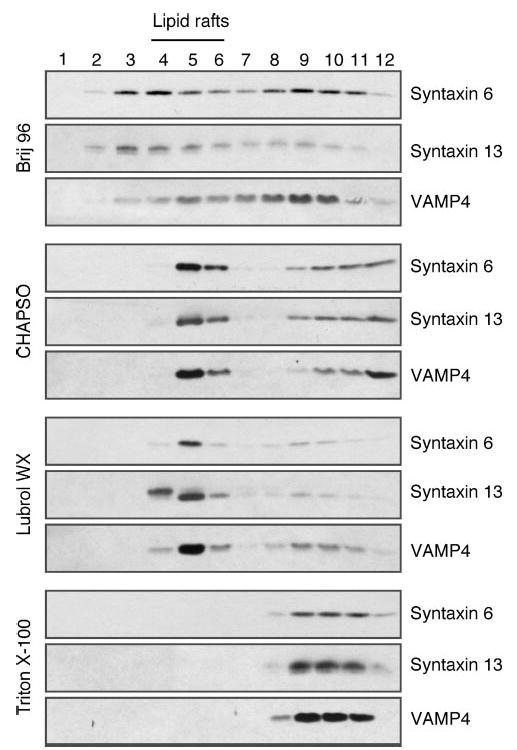
N2aWt.11 cells were lysed either in 0.5% Brij 96, 1% CHAPSO, 0.5% Lubrol WX, or 1% Triton X-100 at 4 °C for 30 min and fractionated on discontinuous sucrose gradients. Aliquots of fractions (1–12; from top to bottom) were resolved on SDS-PAGE and analyzed by Western blotting.
Fig. 9. Co-residence of γ-secretase components in syntaxin 6, syntaxin 13 and VAMP4 containing raft domains revealed by immunoisolation.
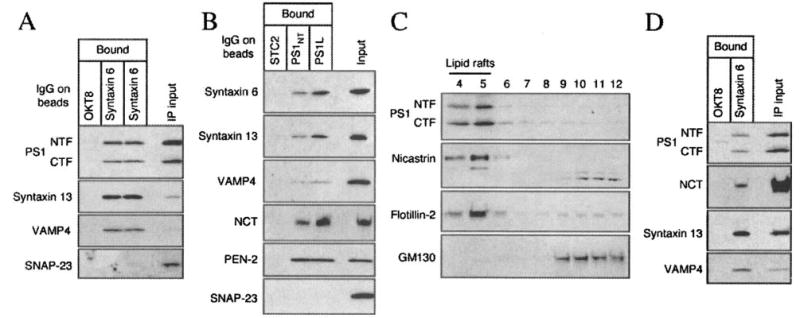
A and B, N2aWt.11 cells were solubilized in 0.5% Lubrol WX and fractionated for raft isolation on sucrose gradients. Raft fractions (4 and 5) were pooled and an aliquot was incubated with magnetic beads coated with monoclonal syntaxin 6 antibody, OKT8 (negative control), affinity-purified polyclonal PS1 antibodies, or STC2 antibody (negative control). Bound DIM were analyzed by Western blotting using indicated antibodies. An aliquot of the input (1/30th volume) was also separated in the same gel for comparison. C, N2a cells were lysed by a non-detergent method (0.5 m sodium carbonate/sonication) and analyzed by flotation on sucrose gradients. DIM localization of PS1 NTF/CTF and nicastrin was assessed by Western blotting. Raft and non-raft fractions were identified by blotting with flotillin-2 and GM130 antibodies, respectively. D, non-detergent raft fractions were incubated with magnetic beads coated with syntaxin 6 or OKT8 antibody and immunoisolated membranes were analyzed by Western blotting using indicated antibodies.
While the fractionation studies described above are consistent with the presence of PS1 with syntaxin 6, syntaxin 13, and VAMP4 in Lubrol WX-resistant DIM, these results are insufficient to formally establish their spatial organization with the same microdomain in native TGN/endosome membranes before cell lysis using Lubrol WX. Therefore, we employed an alternate detergent-free strategy previously used to characterize lipid raft membranes (50). In this method, non-detergent cell homogenates are disrupted by sonication followed by flotation gradient analysis. Western blot analysis revealed that the vast majority of PS1 NTF/CTF, nicastrin, and the raft marker flotillin-2 are recovered in low-density lipid raft fractions (Fig. 9C). Aliquots of non-detergent lipid raft fractions (4 and 5) were then used to immunoisolate membrane patches containing syntaxin 6 using magnetic beads as described above. Subsequent analysis of bound DIM confirmed the co-residence of syntaxin 6 with PS1 NTF/CTF, nicastrin, syntaxin 13, and VAMP4, validating the results from detergent lipid raft analyses (Fig. 9D).
DISCUSSION
Our study reveals the presence of all four components of the γ-secretase complex within cholesterol and sphingolipid-rich microdomains in intracellular membranes. In PS1−/−/PS2−/− and NCT−/− fibroblasts we found that γ-secretase components that still remain fail to become detergent-resistant. Consistent with results from immunofluorescence labeling studies, subcellular fractionation in sucrose density gradients shows that the majority of γ-secretase components reside in intracellular membranes enriched in organelle markers for Golgi, TGN and endosomes. When we further characterized the peak fraction containing γ-secretase components, we found that a subset of these proteins partitions into DIM domains enriched in t-SNAREs syntaxin 6 and syntaxin 13, and v-SNARE VAMP4. Immunoisolation using magnetic beads coated with antibodies confirm the co-residence of syntaxin 6, syntaxin 13, and VAMP4 with components of the γ-secretase complex in Lubrol WX-resistant DIM as well as in low buoyant density lipid rafts isolated by a non-detergent method. Plasma membrane raft-associated t-SNARE SNAP-23 was not present in DIM immunoisolated using PS1 or syntaxin 6 antibodies. Based on the above results we conclude that a significant fraction of mature γ-secretase resides in lipid raft microdomains of post-Golgi and endosome membranes.
Although cholesterol is synthesized in the ER, raft assembly can occur first in the Golgi complex, where sphingolipids are synthesized (71). As such, GPI-anchored proteins first become Triton-insoluble in the medial Golgi during biosynthetic transport (63). We provide biochemical evidence from cultured primary neurons, neuroblastoma cells and fibroblasts that significant levels of individual γ-secretase components are associated with DIM and non-detergent lipid rafts. Furthermore, we show that full-length PS1 and immature nicastrin are virtually excluded from lipid rafts. Thus, partitioning into lipid rafts likely signifies complete assembly of the mature γ-secretase complex and successful progression through the secretory pathway. It has been previously reported by independent groups that subunits of the γ-secretase complex cooperatively mature and proceed through the secretory pathway, and incomplete assemblies are incapable of exit from the ER, and are targeted for degradation (14, 15, 24, 40, 62, 72). For example, in the absence of PS expression nicastrin fails to undergo complex glycosylation, a post-translational modification that usually occurs when transmembrane and luminal proteins reach the medial Golgi compartment (24). Consistent with these findings, analysis of γ-secretase components in PS1−/−/PS2−/− and NCT−/− fibroblasts showed that subunits that still remain do not partition into DIM microdomains. In the future, it will be interesting to determine the mechanisms regulating γ-secretase localization in lipid rafts.
We have employed a combination of organelle fractionation, immunofluorescence localization, flotation sucrose density fractionation and immunoisolation of lipid rafts, to arrive at the conclusion that a significant fraction of mature PS1 and other γ-secretase components resides in membrane microdomains containing the t-SNAREs syntaxin 6 and 13, and the v-SNARE VAMP4. Although confocal microscopy reveals significant colocalization between endogenous PS1 and markers of the TGN and TGN-derived vesicles, the spatial resolution achieved using immunofluorescence labeling is insufficient to demonstrate the presence of γ-secretase components in close proximity to the SNAREs within microdomains in native membranes. Such a level of resolution is feasible by immunogold labeling and high resolution electron microscopy, and has been reported to date for only abundant raft-resident proteins such as GPI-linked Thy-1 and palmitoylated LAT (73). Plasma membrane lipid raft proteins such as placental alkaline phosphatase have been successfully examined by another technique involving antibody-mediated cross-linking of cell surface proteins in live cells (74). Unfortunately, this technique is not applicable to the analysis of raft-resident proteins in intracellular organelles. Flotation gradient analysis of DIM is the most commonly used method to assess raft association of proteins. While it is clear that well characterized raft markers such as flotillins and caveolin readily partition into low buoyant density fractions, results from gradient centrifugation experiments alone is insufficient to support raft association of a given protein. For example, it has been suggested that the use of detergents for lipid raft isolation may facilitate merger of detergent-resistant proteins localized in spatially distinct membrane domains in intact cells. Results of our magnetic immunoisolation experiments rule out this possibility as we show that syntaxin 6 and SNAP-23, two t-SNAREs associated with cholesterol-rich lipid rafts present in the TGN/TGN vesicles (this study) and plasma membrane (70), respectively, do not form mixed raft patches containing both these proteins following extraction of cells in Lubrol WX. We have further addressed this issue by the use of a non-detergent method to examine raft association of γ-secretase components and corroborated our major conclusions.
Lipid rafts participate in a number of important biological functions including the trafficking of proteins and lipids in the secretory and endocytic pathways in epithelial cells and neurons (28). Multiple lines of evidence converge on the role of lipid rafts in facilitating amyloidogenic processing of APP. BACE1 is targeted to lipid rafts via cysteine palmitoylation within trans-membrane/cytoplasmic domains (42, 75). Addition of GPI anchor to localize BACE1 exclusively to lipid rafts increases APP processing at the β-cleavage site (36). Conversely, disruption of raft assemblies by cholesterol depletion leads to marked diminution in Aβ secretion (32), presumably as a result of displacement of both β-secretase (42) and γ-secretase (Fig. 2). Although the present work is the first detailed investigation on the localization of each of the four essential γ-secretase component in lipid rafts, presence of γ-secretase activity has been previously demonstrated in low buoyant density membranes and cholesterol-rich microdomains affinity isolated by the use of perfringolysin O (37, 38). Further evidence for cholesterol-dependent localization and function of γ-secretase results from investigations on amyloidogenic processing under conditions where intracellular cholesterol transport was inhibited by Niemann-Pick C1 protein deficiency or exposure to selective inhibitors. Cholesterol is non-uniformly distributed within the endosomallysosomal system, with relative enrichment in early endosomes and recycling endosomes and lower levels in late endosomes and lysosomes (76). However, cholesterol accumulates in late endosome-like storage organelles when intracellular cholesterol transport is inhibited. Interestingly, PS1 and PS2 also redistribute to these organelles and consequently result in enhanced γ-secretase processing of APP (35). In agreement with these data, we show that components of the γ-secretase complex reside in cholesterol-rich domains of TGN and TGN-derived vesicles (containing syntaxin 6 and VAMP4), and recycling endosomes (containing syntaxin 13) and are not readily detectable in plasma membrane rafts (containing SNAP-23). Endosomes are the major site for processing internalized surface receptors for recycling to plasma membrane or degradation in lysosomes. Hence, localization of γ-secretase components within this organelle has functional significance not only for amyloidogenic processing of endocytosed APP, but also for the facilitation or down-regulation of signaling by intramembraneous cleavage of diverse γ-secretase substrates.
Raft-associated proteins cycle between cell-surface and Golgi by raft-mediated endocytosis characterized by clathrin independence and dynamin dependence. Inhibition of this process by expression of Rab5 GTPase-activating protein RN-tre or dominant negative K44A dynamin II mutant markedly diminishes Aβ production (33). Moreover, amyloidogenic processing is restored by antibody cross-linking of cell surface APP and BACE1, suggesting that APP is delivered to BACE1 containing lipid raft domains upon endocytosis from the cell surface (33). Together, these findings provide a glimpse of the dynamic regulation of amyloidogenic processing by protein trafficking of APP as well as underscore the functional significance of residence of secretases in lipid rafts of post-TGN and endosome membranes containing syntaxins 6 and 13, and VAMP4.
Acknowledgments
We thank Drs. Sangram S. Sisodia, Theodore L. Steck, and Benjamin S. Glick for helpful discussions. We thank Drs. Wim Annaert and Bart DeStrooper for generously providing wild-type and PS1−/−/PS2−/− fibroblasts, Paul Fraser, Peter St George-Hyslop, Gang Yu, and James A. Mastrianni for providing antibodies.
Footnotes
This work was supported in part by National Institutes of Health Grants AG21495 and AG 19070, Alzheimer’s Association, and American Health Assistance Foundation.
The abbreviations used are: Aβ, β-amyloid; APP, amyloid precursor protein; PS, presenilin(s); NTF, N-terminal fragment; CTF, C-terminal fragment; ER, endoplasmic reticulum; TGN, trans-Golgi network; CHAPSO, 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonic acid; DIM, detergent-insoluble membrane; GTEL, Golgi/TGN/endosomes/lysosome-enriched membranes; MES, 4-morpholine-ethanesulfonic acid; SNARE, soluble NSF attachment protein receptors; VAMP, vesicle-associated membrane protein; MβCD, methyl-β-cyclodextrin; GPI, glycosylphosphatidylinositol.
D. Ito and G. Thinakaran, unpublished data.
References
- 1.Selkoe DJ. Physiol Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 2.Sisodia SS, St George-Hyslop PH. Nat Rev Neurosci. 2002;3:281–290. doi: 10.1038/nrn785. [DOI] [PubMed] [Google Scholar]
- 3.Vassar R. J Mol Neurosci. 2004;23:105–114. doi: 10.1385/JMN:23:1-2:105. [DOI] [PubMed] [Google Scholar]
- 4.Iwatsubo T. Curr Opin Neurobiol. 2004;14:379–383. doi: 10.1016/j.conb.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Price DL, Tanzi RE, Borchelt DR, Sisodia SS. Annu Rev Genet. 1998;32:461–493. doi: 10.1146/annurev.genet.32.1.461. [DOI] [PubMed] [Google Scholar]
- 6.Tanzi RE, Bertram L. Neuron. 2001;32:181–184. doi: 10.1016/s0896-6273(01)00476-7. [DOI] [PubMed] [Google Scholar]
- 7.De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, Von Figura K, Van Leuven F. Nature. 1998;391:387–390. doi: 10.1038/34910. [DOI] [PubMed] [Google Scholar]
- 8.Naruse S, Thinakaran G, Luo JJ, Kusiak JW, Tomiata T, Iwatsubo T, Qian X, Ginty DD, Price DL, Borchelt DR, Wong PC. Neuron. 1998;21:1213–1221. doi: 10.1016/s0896-6273(00)80637-6. [DOI] [PubMed] [Google Scholar]
- 9.Thinakaran G, Parent AT. Pharmacol Res. 2004;50:411–418. doi: 10.1016/j.phrs.2003.12.026. [DOI] [PubMed] [Google Scholar]
- 10.Takasugi N, Tomita T, Hayashi I, Tsuruoka M, Niimura M, Takahashi Y, Thinakaran G, Iwatsubo T. Nature. 2003;422:438–441. doi: 10.1038/nature01506. [DOI] [PubMed] [Google Scholar]
- 11.Kimberly WT, LaVoie MJ, Ostaszewski BL, Ye W, Wolfe MS, Selkoe DJ. Proc Natl Acad Sci U S A. 2003;100:6382–6387. doi: 10.1073/pnas.1037392100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim SH, Ikeuchi T, Yu C, Sisodia SS. J Biol Chem. 2003;278:33992–34002. doi: 10.1074/jbc.M305834200. [DOI] [PubMed] [Google Scholar]
- 13.De Strooper B. Neuron. 2003;38:9–12. doi: 10.1016/s0896-6273(03)00205-8. [DOI] [PubMed] [Google Scholar]
- 14.Gu Y, Chen F, Sanjo N, Kawarai T, Hasegawa H, Duthie M, Li W, Ruan X, Luthra A, Mount HT, Tandon A, Fraser PE, St. George-Hyslop P. J Biol Chem. 2003;278:7374–7380. doi: 10.1074/jbc.M209499200. [DOI] [PubMed] [Google Scholar]
- 15.Luo WJ, Wang H, Li H, Kim BS, Shah S, Lee HJ, Thinakaran G, Kim TW, Yu G, Xu H. J Biol Chem. 2003;278:7850–7854. doi: 10.1074/jbc.C200648200. [DOI] [PubMed] [Google Scholar]
- 16.Xu H, Sweeney D, Wang R, Thinakaran G, Lo AC, Sisodia SS, Greengard P, Gandy S. Proc Natl Acad Sci U S A. 1997;94:3748–3752. doi: 10.1073/pnas.94.8.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cook DG, Forman MS, Sung JC, Leight S, Kolson DL, Iwatsubo T, Lee VM, Doms RW. Nat Med. 1997;3:1021–1023. doi: 10.1038/nm0997-1021. [DOI] [PubMed] [Google Scholar]
- 18.Greenfield JP, Tsai J, Gouras GK, Hai B, Thinakaran G, Checler F, Sisodia SS, Greengard P, Xu H. Proc Natl Acad Sci U S A. 1999;96:742–747. doi: 10.1073/pnas.96.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi RH, Milner TA, Li F, Nam EE, Edgar MA, Yamaguchi H, Beal MF, Xu H, Greengard P, Gouras GK. Am J Pathol. 2002;161:1869–1879. doi: 10.1016/s0002-9440(10)64463-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lah JJ, Heilman CJ, Nash NR, Rees HD, Yi H, Counts SE, Levey AI. J Neurosci. 1997;17:1971–1980. doi: 10.1523/JNEUROSCI.17-06-01971.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lah JJ, Levey AI. Mol Cell Neurosci. 2000;16:111–126. doi: 10.1006/mcne.2000.0861. [DOI] [PubMed] [Google Scholar]
- 22.Torp R, Ottersen OP, Cotman CW, Head E. Neuroscience. 2003;120:291–300. doi: 10.1016/s0306-4522(03)00320-8. [DOI] [PubMed] [Google Scholar]
- 23.Rechards M, Xia W, Oorschot VM, Selkoe DJ, Klumperman J. Traffic. 2003;4:553–565. doi: 10.1034/j.1600-0854.2003.t01-1-00114.x. [DOI] [PubMed] [Google Scholar]
- 24.Leem JY, Vijayan S, Han P, Cai D, Machura M, Lopes KO, Veselits ML, Xu H, Thinakaran G. J Biol Chem. 2002;277:19236–19240. doi: 10.1074/jbc.C200148200. [DOI] [PubMed] [Google Scholar]
- 25.Baulac S, LaVoie MJ, Kimberly WT, Strahle J, Wolfe MS, Selkoe DJ, Xia W. Neurobiol Dis. 2003;14:194–204. doi: 10.1016/s0969-9961(03)00123-2. [DOI] [PubMed] [Google Scholar]
- 26.Puglielli L, Tanzi RE, Kovacs DM. Nat Neurosci. 2003;6:345–351. doi: 10.1038/nn0403-345. [DOI] [PubMed] [Google Scholar]
- 27.Wolozin B. Neuron. 2004;41:7–10. doi: 10.1016/s0896-6273(03)00840-7. [DOI] [PubMed] [Google Scholar]
- 28.Simons K, Toomre D. Nat Rev Mol Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 29.Ikonen E. Curr Opin Cell Biol. 2001;13:470–477. doi: 10.1016/s0955-0674(00)00238-6. [DOI] [PubMed] [Google Scholar]
- 30.Lee SJ, Liyanage U, Bickel PE, Xia W, Lansbury PT, Jr, Kosik KS. Nat Med. 1998;4:730–734. doi: 10.1038/nm0698-730. [DOI] [PubMed] [Google Scholar]
- 31.Kawarabayashi T, Shoji M, Younkin LH, Wen-Lang L, Dickson DW, Murakami T, Matsubara E, Abe K, Ashe KH, Younkin SG. J Neurosci. 2004;24:3801–3809. doi: 10.1523/JNEUROSCI.5543-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simons M, Keller P, De Strooper B, Beyreuther K, Dotti CG, Simons K. Proc Natl Acad Sci U S A. 1998;95:6460–6464. doi: 10.1073/pnas.95.11.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ehehalt R, Keller P, Haass C, Thiele C, Simons K. J Cell Biol. 2003;160:113–123. doi: 10.1083/jcb.200207113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burns M, Gaynor K, Olm V, Mercken M, LaFrancois J, Wang L, Mathews PM, Noble W, Matsuoka Y, Duff K. J Neurosci. 2003;23:5645–5649. doi: 10.1523/JNEUROSCI.23-13-05645.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Runz H, Rietdorf J, Tomic I, de Bernard M, Beyreuther K, Pepperkok R, Hartmann T. J Neurosci. 2002;22:1679–1689. doi: 10.1523/JNEUROSCI.22-05-01679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cordy JM, Hussain I, Dingwall C, Hooper NM, Turner AJ. Proc Natl Acad Sci U S A. 2003;100:11735–11740. doi: 10.1073/pnas.1635130100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wahrle S, Das P, Nyborg AC, McLendon C, Shoji M, Kawarabayashi T, Younkin LH, Younkin SG, Golde TE. Neurobiol Dis. 2002;9:11–23. doi: 10.1006/nbdi.2001.0470. [DOI] [PubMed] [Google Scholar]
- 38.Wada S, Morishima-Kawashima M, Qi Y, Misono H, Shimada Y, Ohno-Iwashita Y, Ihara Y. Biochemistry. 2003;42:13977–13986. doi: 10.1021/bi034904j. [DOI] [PubMed] [Google Scholar]
- 39.Thinakaran G, Harris CL, Ratovitski T, Davenport F, Slunt HH, Price DL, Borchelt DR, Sisodia SS. J Biol Chem. 1997;272:28415–28422. doi: 10.1074/jbc.272.45.28415. [DOI] [PubMed] [Google Scholar]
- 40.Li T, Ma G, Cai H, Price DL, Wong PC. J Neurosci. 2003;23:3272–3277. doi: 10.1523/JNEUROSCI.23-08-03272.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herreman A, Serneels L, Annaert W, Collen D, Schoonjans L, De Strooper B. Nat Cell Biol. 2000;2:461–462. doi: 10.1038/35017105. [DOI] [PubMed] [Google Scholar]
- 42.Riddell DR, Christie G, Hussain I, Dingwall C. Curr Biol. 2001;11:1288–1293. doi: 10.1016/s0960-9822(01)00394-3. [DOI] [PubMed] [Google Scholar]
- 43.Thinakaran G, Regard JB, Bouton CML, Harris CL, Price DL, Borchelt DR, Sisodia SS. Neurobiol Dis. 1998;4:438–453. doi: 10.1006/nbdi.1998.0171. [DOI] [PubMed] [Google Scholar]
- 44.Thinakaran G, Borchelt DR, Lee MK, Slunt HH, Spitzer L, Kim G, Ratovitsky T, Davenport F, Nordstedt C, Seeger M, Hardy J, Levey AI, Gandy SE, Jenkins NA, Copeland NG, Price DL, Sisodia SS. Neuron. 1996;17:181–190. doi: 10.1016/s0896-6273(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 45.Lee SF, Shah S, Li H, Yu C, Han W, Yu G. J Biol Chem. 2002;277:45013–45019. doi: 10.1074/jbc.M208164200. [DOI] [PubMed] [Google Scholar]
- 46.Hashimoto K, Mannen T, Nukina N. Acta Neuropathol (Berl) 1992;83:613–617. doi: 10.1007/BF00299410. [DOI] [PubMed] [Google Scholar]
- 47.Roper K, Corbeil D, Huttner WB. Nat Cell Biol. 2000;2:582–592. doi: 10.1038/35023524. [DOI] [PubMed] [Google Scholar]
- 48.Brugger B, Graham C, Leibrecht I, Mombelli E, Jen A, Wieland F, Morris R. J Biol Chem. 2004;279:7530–7536. doi: 10.1074/jbc.M310207200. [DOI] [PubMed] [Google Scholar]
- 49.Radeva G, Sharom FJ. Biochem J. 2004;380:219–230. doi: 10.1042/BJ20031348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waugh MG, Lawson D, Hsuan JJ. Biochem J. 1999;337:591–597. [PMC free article] [PubMed] [Google Scholar]
- 51.Hammond AT, Glick BS. Mol Biol Cell. 2000;11:3013–3030. doi: 10.1091/mbc.11.9.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chamberlain LH. FEBS Lett. 2004;559:1–5. doi: 10.1016/s0014-5793(04)00050-x. [DOI] [PubMed] [Google Scholar]
- 53.Schuck S, Honsho M, Ekroos K, Shevchenko A, Simons K. Proc Natl Acad Sci U S A. 2003;100:5795–5800. doi: 10.1073/pnas.0631579100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Madore N, Smith KL, Graham CH, Jen A, Brady K, Hall S, Morris R. EMBO J. 1999;18:6917–6926. doi: 10.1093/emboj/18.24.6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li YM, Lai MT, Xu M, Huang Q, DiMuzio-Mower J, Sardana MK, Shi XP, Yin KC, Shafer JA, Gardell SJ. Proc Natl Acad Sci U S A. 2000;97:6138–6143. doi: 10.1073/pnas.110126897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.LaVoie MJ, Fraering PC, Ostaszewski BL, Ye W, Kimberly WT, Wolfe MS, Selkoe DJ. J Biol Chem. 2003;278:37213–37222. doi: 10.1074/jbc.M303941200. [DOI] [PubMed] [Google Scholar]
- 57.Lang DM, Lommel S, Jung M, Ankerhold R, Petrausch B, Laessing U, Wiechers MF, Plattner H, Stuermer CA. J Neurobiol. 1998;37:502–523. doi: 10.1002/(sici)1097-4695(199812)37:4<502::aid-neu2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 58.Ahnert-Hilger G, Bader MF, Bhakdi S, Gratzl M. J Neurochem. 1989;52:1751–1758. doi: 10.1111/j.1471-4159.1989.tb07253.x. [DOI] [PubMed] [Google Scholar]
- 59.Klein U, Gimpl G, Fahrenholz F. Biochemistry. 1995;34:13784–13793. doi: 10.1021/bi00042a009. [DOI] [PubMed] [Google Scholar]
- 60.Harder T, Simons K. Curr Opin Cell Biol. 1997;9:534–542. doi: 10.1016/s0955-0674(97)80030-0. [DOI] [PubMed] [Google Scholar]
- 61.Rajendran L, Masilamani M, Solomon S, Tikkanen R, Stuermer CA, Plattner H, Illges H. Proc Natl Acad Sci U S A. 2003;100:8241–8246. doi: 10.1073/pnas.1331629100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steiner H, Winkler E, Edbauer D, Prokop S, Basset G, Yamasaki A, Kostka M, Haass C. J Biol Chem. 2002;277:39062–39065. doi: 10.1074/jbc.C200469200. [DOI] [PubMed] [Google Scholar]
- 63.Brown DA, Rose JK. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- 64.Kim SH, Lah JJ, Thinakaran G, Levey A, Sisodia SS. Neurobiol Dis. 2000;7:99–117. doi: 10.1006/nbdi.1999.0280. [DOI] [PubMed] [Google Scholar]
- 65.Pasternak SH, Bagshaw RD, Guiral M, Zhang S, Ackerley CA, Pak BJ, Callahan JW, Mahuran DJ. J Biol Chem. 2003;278:26687–26694. doi: 10.1074/jbc.m304009200. [DOI] [PubMed] [Google Scholar]
- 66.Keller P, Simons K. J Cell Biol. 1998;140:1357–1367. doi: 10.1083/jcb.140.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bock JB, Klumperman J, Davanger S, Scheller RH. Mol Biol Cell. 1997;8:1261–1271. doi: 10.1091/mbc.8.7.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prekeris R, Klumperman J, Chen YA, Scheller RH. J Cell Biol. 1998;143:957–971. doi: 10.1083/jcb.143.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Steegmaier M, Klumperman J, Foletti DL, Yoo JS, Scheller RH. Mol Biol Cell. 1999;10:1957–1972. doi: 10.1091/mbc.10.6.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chamberlain LH, Gould GW. J Biol Chem. 2002;277:49750–49754. doi: 10.1074/jbc.M206936200. [DOI] [PubMed] [Google Scholar]
- 71.Simons K, Ikonen E. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 72.Hu Y, Fortini ME. J Cell Biol. 2003;161:685–690. doi: 10.1083/jcb.200304014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wilson BS, Steinberg SL, Liederman K, Pfeiffer JR, Surviladze Z, Zhang J, Samelson LE, Yang LH, Kotula PG, Oliver JM. Mol Biol Cell. 2004;15:2580–2592. doi: 10.1091/mbc.E03-08-0574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harder T, Scheiffele P, Verkade P, Simons K. J Cell Biol. 1998;141:929–942. doi: 10.1083/jcb.141.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Benjannet S, Elagoz A, Wickham L, Mamarbachi M, Munzer JS, Basak A, Lazure C, Cromlish JA, Sisodia S, Checler F, Chretien M, Seidah NG. J Biol Chem. 2001;276:10879–10887. doi: 10.1074/jbc.M009899200. [DOI] [PubMed] [Google Scholar]
- 76.Maxfield FR, Wustner D. J Clin Investig. 2002;110:891–898. doi: 10.1172/JCI16500. [DOI] [PMC free article] [PubMed] [Google Scholar]


