Abstract
Sphingomyelin (SM) is a major component of animal plasma membranes. Its production involves the transfer of phosphocholine from phosphatidylcholine onto ceramide, yielding diacylglycerol as a side product. This reaction is catalysed by SM synthase, an enzyme whose biological potential can be judged from the roles of diacylglycerol and ceramide as anti- and proapoptotic stimuli, respectively. SM synthesis occurs in the lumen of the Golgi as well as on the cell surface. As no gene for SM synthase has been cloned so far, it is unclear whether different enzymes are present at these locations. Using a functional cloning strategy in yeast, we identified a novel family of integral membrane proteins exhibiting all enzymatic features previously attributed to animal SM synthase. Strikingly, human, mouse and Caenorhabditis elegans genomes each contain at least two different SM synthase (SMS) genes. Whereas human SMS1 is localised to the Golgi, SMS2 resides primarily at the plasma membrane. Collectively, these findings open up important new avenues for studying sphingolipid function in animals.
Keywords: apoptosis, ceramide, Golgi, lipid phosphate phosphatase, sphingomyelin synthase
Introduction
Sphingomyelin (SM) is an abundant constituent of cellular membranes in a wide range of organisms, from mammals (Ullman and Radin, 1974) and nematodes (Satouchi et al, 1993) to protozoa like the human malaria parasite Plasmodium falciparum (Elmendorf and Haldar, 1994). SM is preferentially concentrated in the outer leaflet of the plasma membrane. Its high packing density and affinity for sterols help provide a rigid barrier to the extracellular environment and play a role in the formation of lipid rafts, specialised areas in cellular membranes with important functions in signal transduction and membrane trafficking (Simons and Toomre, 2000; Holthuis et al, 2001). Since the discovery of the ‘SM cycle' as a putative signalling system analogous to well-known second messenger systems like the phosphoinositide pathway, SM has emerged to the focus of interest in many research laboratories (Kolesnick and Hannun, 1999; Andrieu-Abadie and Levade, 2002).
SM synthesis is mediated by a phosphatidylcholine:ceramide cholinephosphotransferase, or SM synthase, which transfers the phosphorylcholine moiety from phosphatidylcholine (PC) onto the primary hydroxyl of ceramide, thus generating SM and diacylglycerol (DAG; Ullman and Radin, 1974; Voelker and Kennedy, 1982; Marggraf and Kanfer, 1984). Since the enzyme is also able to catalyse the reverse reaction, namely the formation of PC from SM and DAG (Marggraf and Kanfer, 1984; van Helvoort et al, 1994), it may regulate, in opposite directions, the cellular levels of the bioactive lipids ceramide and DAG. The latter two are important regulators of membrane trafficking, cell proliferation and apoptosis (Kolesnick and Hannun, 1999; Bankaitis, 2002; Brose and Rosenmund, 2002; Pettus et al, 2002). Hence, one may anticipate that the physiological significance of SM synthase goes beyond the formation of SM. The subcellular localisation of the enzyme has been the subject of numerous studies. After the initial debate had focused on whether SM synthase is located in the Golgi or at the plasma membrane (Marggraf et al, 1981; Voelker and Kennedy, 1982; Lipsky and Pagano, 1985), subsequent work revealed evidence for the presence of SM synthase activity in both membranes (Futerman et al, 1990; Jeckel et al, 1990; van Helvoort et al, 1994). Whether SM synthesis detected at these locations is due to the presence of more than one isoform of the enzyme has remained an open issue.
Progress in understanding the biological roles of SM synthesis and its regulation is hampered by the fact that no successful purification of the responsible enzyme has been achieved. Recent work has identified a bacterial SM synthase released from Pseudomonas aeruginosa (Luberto et al, 2003). However, this activity is a soluble protein, hence in contrast to the animal enzyme that is tightly associated with the membrane (Ullman and Radin, 1974; Voelker and Kennedy, 1982). Other efforts focused on the isolation of SM synthase mutants by screening Chinese hamster ovary cells for resistance to an SM-directed cytolysin (Hanada et al, 1998). Instead of mutants with a primary defect in SM synthase, this method yielded mutants defective in serine palmitoyltransferase activity or blocked in ceramide transport to the site of SM synthesis (Fukasawa et al, 1999).
Here, we pursued a complementary approach for the identification of animal SM synthase that takes advantage of structural information available for enzymes catalysing analogous reactions. In contrast to most animal cells, plants, fungi and yeast do not produce SM. Instead, these organisms add phosphoinositol to phytoceramide to generate inositolphosphorylceramide (IPC) (Dickson, 1998). IPC production in the yeast Saccharomyces cerevisiae requires the product of the AUR1 gene (Nagiec et al, 1997). Sequence analysis of Aur1p proteins from different fungi revealed four conserved motifs (Heidler and Radding, 2000), two of which are similar to the C2 and C3 domains present in members of the lipid phosphate phosphatase (LPP) family (Waggoner et al, 1999). LPPs play a critical role in cell signalling by controlling the conversion of bioactive lipid phosphate esters such as (lyso)phosphatidic acid and sphingosine-1-phosphate to their dephosphorylated counterparts. The conserved C1, C2 and C3 domains in LPPs likely constitute the active site for cleavage of the bond between the lipid hydroxyl and phosphate groups (Neuwald, 1997). In the case of Aur1p, this reaction would represent the first step in the transfer of inositolphosphate from PI, with the resulting enzyme-phosphate intermediate being subjected to nucleophilic attack by the oxygen of ceramide rather than the oxygen of water used by LPPs. The presence of LPP-like motifs, together with the IPC synthase activity found associated with affinity-purified Aur1p (van den Dikkenberg and Holthuis, in preparation), suggests that Aur1p is directly responsible for IPC synthesis.
The above considerations led us to develop a bioinformatics and functional cloning strategy to identify the enzyme responsible for SM synthesis in animals. By searching the database using a sequence motif shared by LPPs and Aur1p homologues, a set of 27 candidate SM synthase sequences from humans, mice and Caenorhabditis elegans was collected and assembled into different protein families. Several members from each family were cloned and analysed for their ability to mediate SM synthesis upon heterologous expression in S. cerevisiae, an organism lacking SM synthase activity. This approach resulted in the identification of multiple animal SM synthases. We find that animal genomes each contain at least two SM synthase genes and provide evidence that this multiplicity in enzymes is used to interconvert PC and ceramide to DAG and SM at different cellular locations.
Results
Identification of candidate SM synthases from the animal database
Figure 1A shows an outline of the bioinformatics approach used to identify candidate sequences for SM synthases from the animal database. CSSs were identified based on the following criteria: (1) presence of a sequence motif, H-[YFWH]-X2-D-[VLI]-X2-[GA]-X3-[GSTA], shared by previously characterised LPPs and Aur1p homologues; (2) biochemical function should be unknown; (3) no structural homologues in S. cerevisiae, since this organism lacks SM; and (4) presence of multiple (>2) transmembrane domains, since the enzyme mechanism is intramembranous and because LPPs and Aur1p proteins have six predicted membrane spans. Sequences conforming to all four criteria were subsequently used as queries in a BLAST search to track down homologous sequences that were missed in the initial search due to deviations in the LPP/Aur1p motif. This approach yielded nine human, nine mouse and nine C. elegans sequences that could be grouped into three major protein families, designated CSS1, CSS2 and CSS3 (for candidate SM synthase family 1–3; Figure 1B and C). Except for the presence of a common sequence motif, CSS proteins displayed no significant sequence similarity to Aur1p proteins or to LPP family members with known biochemical functions. However, human CSS1β1 is identical to PRG1, a neuron-specific candidate phosphatidic acid phosphatase with a role in axon growth and regenerative sprouting (Brauer et al, 2003). Database accession numbers of CSS proteins are listed in the legend of Figure 1.
Figure 1.
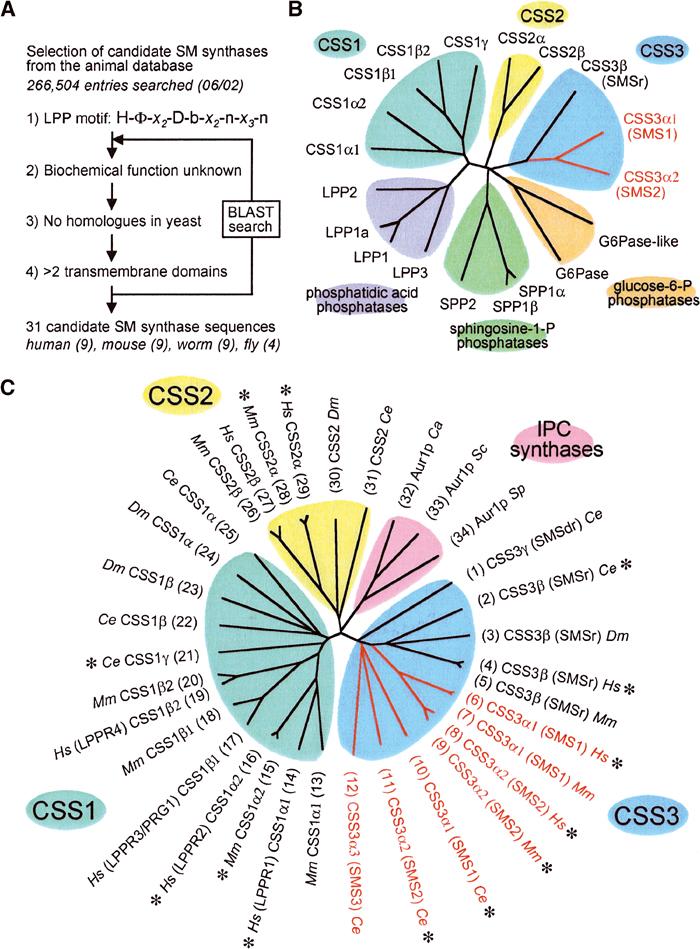
Selection and phylogenetic analysis of CSSs. (A) Animal entries in SwissProt/TrEMBL were searched for the presence of a sequence motif shared by LPPs and Aur1p proteins and then further selected on the basis of three additional criteria, as indicated. (B) Phylogenetic tree of human CSSs and previously characterised members of the human LPP superfamily. (C) Phylogenetic tree of CSS proteins from human (Hs), mouse (Mm), C. elegans (Ce) and D. melanogaster (Dm), and of Aur1p proteins from S. cerevisiae (Sc), Schizosaccharomyces pombe (Sp) and Candida albicans (Ca). Asterisks denote CSS proteins expressed in S. cerevisiae and tested for SM synthase activity. SM synthases (SMS) are marked in red. SwissProt/TrEMBL accession numbers of CSS proteins are: (1) Q9TYV2; (2) Q20696; (3) Q9VS60/Q9VS61; (4) Q96LT4; (5) Q9DA37; (6) Q86VZ5; (7) Q8VCQ6; (8) Q8NHU3; (9) Q9D4B1; (10) Q9XTV2; (11) Q20735; (12) Q965Q4; (13) Q9D606; (14) Q9NXE2; (15) Q8VCY8; (16) Q96GM1; (17) Q96MP0; (18) AAP57768; (19) Q9BQF9; (20) AAP57767; (21) Q22250; (22) Q9TXU1; (23) Q9VNT9; (24) Q9VNU1; (25) Q10022; (26) Q9D4F2; (27) Q8IY26; (28) Q91WB2; (29) Q96SS7; (30) Q8T8T9; (31) Q22461. Note that Q96LT4 contains a partial protein sequence and that the complete ORF was deduced from a corresponding EST clone (Table I).
A subset of CSS3 family members displays SM synthase activity
To investigate whether any of the three CSS families indeed contained SM synthases, the open reading frames of human, mouse and C. elegans members for which full-length cDNAs could be obtained were cloned into a yeast multicopy, GAL1 promoter plasmid in frame with a carboxy-terminal V5 epitope. The resulting plasmids were used to transform wild-type yeast and the transformants were shifted to galactose-containing medium to induce expression of recombinant proteins. Expression of proteins was verified by Western blot analysis using anti-V5 antibodies (Figure 2A). Thus, 13 of the 27 selected CSS sequences were expressed and analysed for SM synthase activity (marked by asterisks, Figure 1C). To this end, yeast cells expressing CSS protein were lysed and incubated with fluorescent C6-NBD-ceramide (NBD-Cer), a known substrate of mammalian SM synthase. Incubations were performed in the presence of N-ethylmaleimide (NEM), a potent inhibitor of yeast-associated SMases that does not affect mammalian SM synthase (see Supplementary Figure 1). Reaction mixtures were next subjected to one-phase lipid extraction and the lipids separated by one-dimensional thin layer chromatography (TLC).
Figure 2.
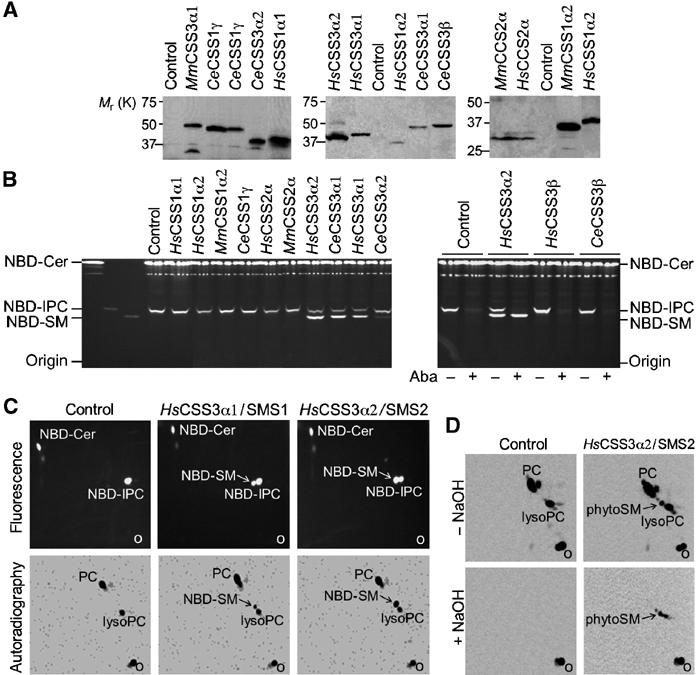
A subset of CSS3 family members displays SM synthase activity upon expression in yeast. (A) Immunoblots of cells expressing various CSS proteins were stained with antibodies recognising the V5 epitope-tagged carboxy termini of CSS proteins. Control denotes cells transformed with empty vector. (B) TLC separation of reaction products generated when NBD-ceramide (NBD-Cer) was incubated with lysates of control or CSS-expressing cells in the presence (+) or absence (−) of the IPC synthase inhibitor aureobasidin A (Aba). (C) Metabolic labelling of cells expressing human CSS3α1/SMS1 or CSS3α2/SMS2 with [14C]-choline and NBD-ceramide. The lipids were extracted, separated by two-dimensional TLC and analysed for fluorescence and radioactivity. Note that SMS1- and SMS2-expressing cells, but not control cells, synthesised NBD-SM, and that this NBD-SM was labelled with [14C]-choline. (D) Metabolic labelling of cells expressing human CSS3α2/SMS2 with [14C]-choline. The lipids were extracted, deacylated by mild alkaline hydrolysis (+NaOH) or control incubated (−NaOH) and separated by two-dimensional TLC before autoradiography. Note that SMS2-expressing cells synthesised alkaline-resistant species of [14C]-choline-labelled lipids (phytoSM) that were absent in control cells.
Figure 2B shows that in lysates of control cells, NBD-Cer was converted exclusively into NBD-IPC. The same was true for lysates of cells expressing members of the CSS1 and CSS2 protein families. However, in lysates of cells expressing human CSS3α1, human CSS3α2, C. elegans CSS3α1 or C. elegans CSS3α2, NBD-Cer was converted to a second product with an Rf value identical to that of NBD-SM. The identity of this product as NBD-SM was confirmed by electrospray ionisation tandem mass spectrometry (ESI-MS/MS; data not shown). Moreover, metabolic labelling of cells expressing human CSS3α1 or CSS3α2 with [14C]-choline in the presence of NBD-Cer resulted in the production of radiolabelled NBD-SM (Figure 2C). When NBD-Cer was omitted, we noticed that CSS3α2-expressing cells generated small amounts of [14C]-choline-labelled lipids that were absent in control cells and distinct from PC and lysoPC. In addition, these lipids were resistant to mild alkaline hydrolysis, suggesting that they represent phytoceramide-based SMs (phytoSM; Figure 2D). Importantly, ESI-MS revealed that bovine brain ceramides incubated with detergent extracts of CSS3α2-expressing cells, but not control cells, are converted to SM (Figure 3). Together, these results indicate that CSS3α proteins not only use NBD-Cer but also recognise naturally occurring ceramides as substrates for SM synthesis.
Figure 3.
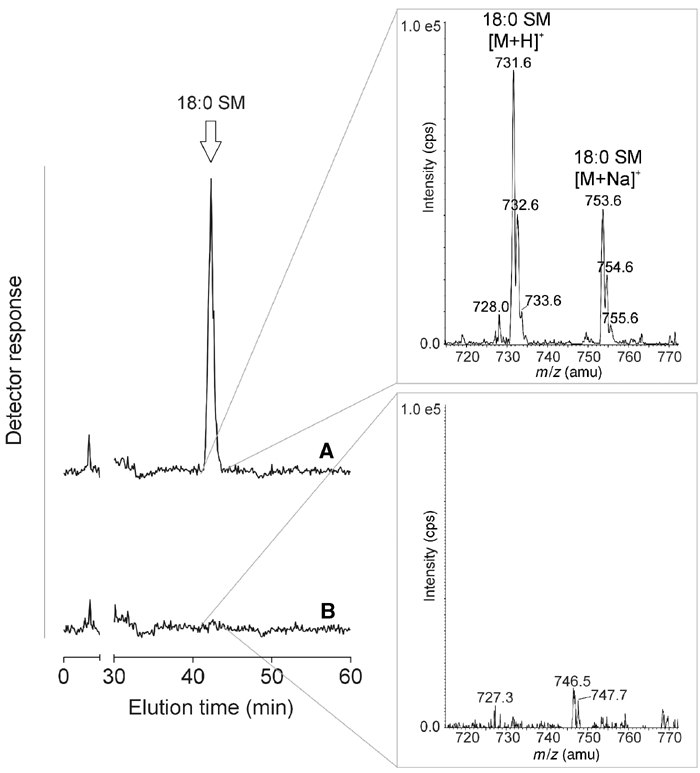
Detergent extracts of human CSS3α2/SMS2-expressing yeast cells support SM formation from bovine brain ceramides. Lines A and B show the reconstituted ion chromatograms of m/z 731.6, corresponding to the m/z ratio of protonated 18:0 SM, during the separation of molecular species of choline-containing phospholipids after solid-phase extraction of membrane extracts that had been incubated with bovine brain ceramides and egg PC. The elution time of an authentic standard of 18:0 SM is indicated in the chromatogram by an arrow (42.3 min). Line A was derived from detergent extracts of human CSS3α2/SMS2-expressing cells and line B from detergent extracts of control cells. Mass spectra recorded at the elution time of 18:0 SM confirmed that formation of 18:0 SM occurred in CSS3α2/SMS2-containing extracts (top right panel) but not in control extracts (bottom right panel).
Human CSS3α1 and CSS3α2 share 57% sequence identity and are highly conserved in mammals (human–mouse: >90%). Based on the results presented herein, we propose to rename these proteins as SMS1 and SMS2, respectively. C. elegans CSS3α1 (ceSMS1) and CSS3α2 (ceSMS2) share 22–27% sequence identity with human SMS1 and SMS2, and apparently represent their functional counterparts in the nematode (Figure 2B). C. elegans contains a third, CSS3α-related protein, termed ceCSS3α3. Although we have not tested this protein for SM synthase activity, the finding implies that C. elegans is equipped with three independent SM synthases (Figure 1C). BLAST searches for orthologous sequences identified two proteins in the human malaria parasite P. falciparum (PlasmoDB accession numbers MAL6P1.177 and MAL6P1.178; also see below). Hence, a multiplicity of SM synthase genes appears to be a general feature of organisms generating SM.
Apart from CSS3α/SMS proteins, the CSS3 family contains a second cluster of CSS3β or SMS-related (SMSr) proteins with members in humans, mice, C. elegans and Drosophila (Figure 1C). Human SMSr is highly conserved in mammals (human–mouse: 95%), shares >40% sequence identity with its orthologues in C. elegans and Drosophila, and is about 34% identical to human SMS1 and SMS2. Heterologous expression of human or C. elegans SMSr did not yield any detectable SM synthase activity (Figure 2B, right panel). The C. elegans CSS3γ protein, renamed SMSdr, is only distantly related to SMS and SMSr proteins (<22% identical; see also Figure 1C) and its ability to mediate SM synthesis was not tested.
Human SMS1 and SMS2 function as PC:ceramide cholinephosphotransferases with reverse activity
SM synthesis in animals proceeds by the liberation of phosphorylcholine from PC and its subsequent transfer onto the primary hydroxyl of ceramide. To establish whether human SMS1 and SMS2 function as PC:ceramide cholinephosphotransferases, their enzymatic characteristics were analysed in more detail. Hence, different donors of choline-P were tested as substrates for SM synthesis. To this end, detergent extracts of cells expressing human SMS1 or SMS2 were incubated with NBD-Cer and the formation of NBD-SM was monitored by TLC. A dilution of extracts proved necessary to render SMS-mediated synthesis of NBD-SM dependent on externally added head group donors (see Supplementary Figure 2). Under these conditions, free phosphorylcholine (Ch-P) and CDP-choline (CDP-Ch) did not support SMS1- or SMS2-mediated SM synthesis (Figure 4A). PC, on the other hand, was efficiently recognised as a substrate. SM itself was also used as a donor of the phosphorylcholine group. LysoPC was a very poor substrate. These results suggest that human SMS1 and SMS2 are transferases that require two fatty chains on the choline-P donor molecule in order to be recognised efficiently as a substrate.
Figure 4.
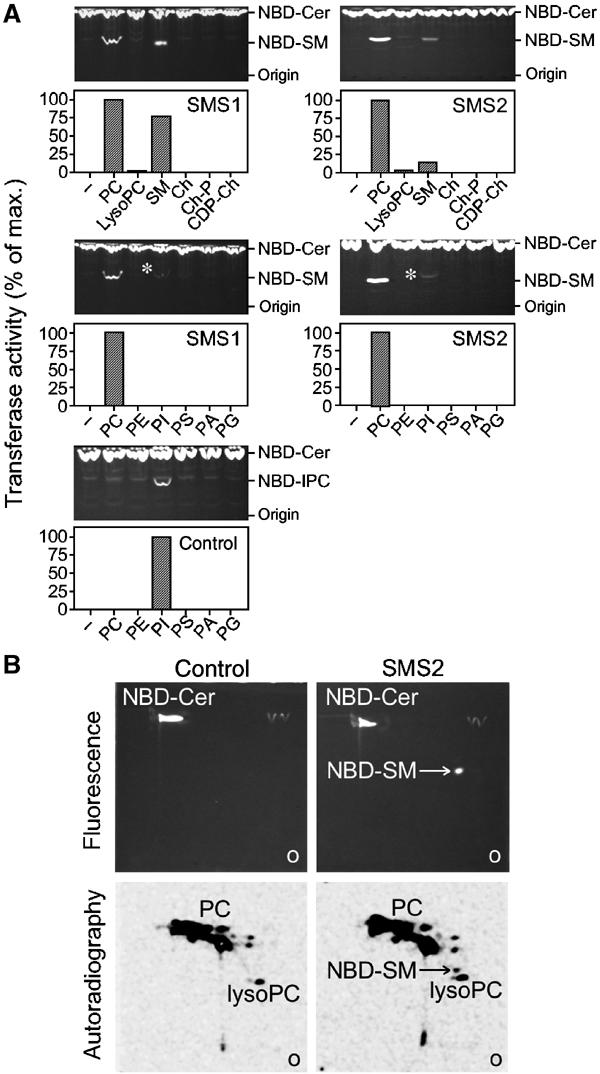
Human SMS1 and SMS2 function as PC:ceramide cholinephosphotransferases. (A) Detergent extracts of yeast cells expressing human SMS1, SMS2 or transformed with empty vector (control) were incubated with NBD-ceramide (18 μM) in the presence or absence of different potential head group donors (220 μM), as indicated. Formation of NBD-SM or NBD-IPC was monitored by one-dimensional TLC and quantified as described in Materials and methods. Note that addition of PI stimulated formation of NBD-IPC in all three extracts (asterisks). PC, phosphatidylcholine; SM, sphingomyelin; Ch, choline; Ch-P, phosphorylcholine; CDP-Ch, cytidine-5′ diphosphocholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PS, phosphatidylserine; PA, phosphatidic acid; PG, phosphatidylglycerol. (B) Detergent extracts of SMS2-expressing and control cells were incubated with NBD-ceramide and [3H]-choline-labelled PC. The lipids were extracted, separated by two-dimensional TLC and analysed for fluorescence and radioactivity. Note that only SMS2-expressing cells synthesised NBD-SM, and that this NBD-SM was labelled with [3H]-choline.
We next investigated the possibility that human SMS1 or SMS2 would use non-choline phospholipids as substrates. None of the phospholipids tested (PE, PI, PS, PA, PG) other than PC supported SM formation (Figure 4A). Moreover, when extracts of cells expressing SMS2 were incubated with PC containing [3H]-choline, the formation of radiolabelled NBD-SM was observed (Figure 4B). Hence, it appears that SMS1 and SMS2 directly and specifically recognise the choline head group on their substrates.
Previous work suggested that mammalian SM synthase is also capable of catalysing the reverse reaction, namely the formation of PC from SM and DAG (Marggraf and Kanfer, 1984; van Helvoort et al, 1994). To investigate whether this was also the case for human SMS1 and SMS2, extracts of cells expressing these proteins were incubated with NBD-DAG and the formation of NBD-PC monitored by TLC. Addition of SM induced NBD-PC formation in extracts of SMS1- or SMS2-expressing cells, but not in control cell extracts (Figure 5). Strikingly, PC itself proved more efficient than SM in stimulating SMS1- or SMS2-dependent NBD-PC formation. In contrast, addition of non-choline phospholipids (e.g. PI) had no effect. These data suggest that human SMS1 and SMS2, rather than functioning strictly as SM synthases, are transferases capable of using PC and SM as phosphocholine donors to produce PC or SM, dependent on the relative concentrations of DAG and ceramide as phosphocholine acceptors, respectively.
Figure 5.
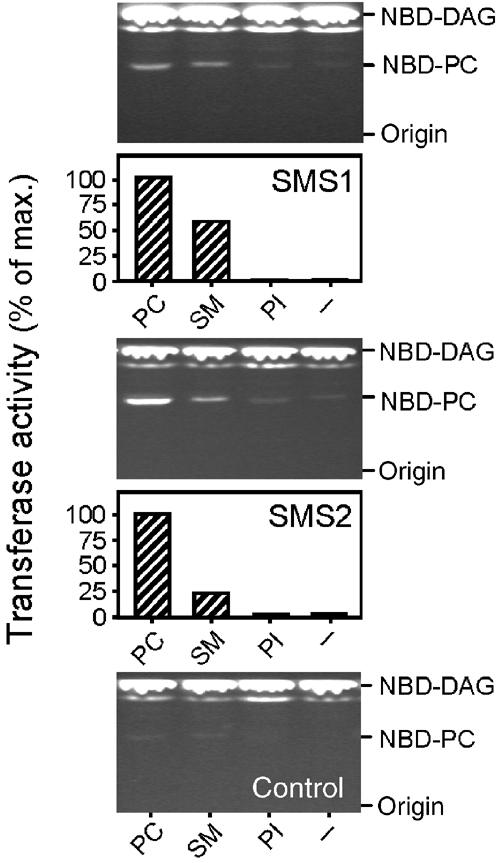
Human SMS1 and SMS2 exhibit reverse activity. Detergent extracts of yeast cells expressing human SMS1, SMS2 or transformed with empty vector (control) were incubated with NBD-diacylglycerol (NBD-DAG; 18 μM) in the presence or absence of different head group donors (220 μM), as indicated. Formation of NBD-PC was monitored by one-dimensional TLC and quantified as described in Materials and methods.
Mammalian SM synthase was found to be sensitive to the bacterial PC-phospholipase C inhibitor, D609 (Luberto and Hannun, 1998). In lysates of SMS1- or SMS2-expressing yeast cells, D609 inhibited SM synthesis in a dose-dependent manner (data not shown). The extent of inhibition observed for SMS1-mediated SM synthesis (50% at 100 μg/ml D609) was comparable to that reported for mammalian SM synthase (Luberto and Hannun, 1998). SMS2 proved two-fold less sensitive to the drug.
Hence, human SMS1, SMS2 and the mammalian SM synthase activities described in the literature share many enzymatic characteristics.
Structure and expression of human SMS1 and SMS2
Figure 6A shows a sequence alignment of human SMS1, SMS2 and SMSr. Hydrophobicity analysis predicted six membrane-spanning alpha helices connected by hydrophilic regions that would form extramembrane loops. A comparative analysis with SMS and SMSr sequences from mice, C. elegans, P. falciparum and Drosophila revealed that the number and spacing properties of transmembrane helices are well conserved. In addition, SMS proteins contain four highly conserved sequence motifs, designated D1, D2, D3 and D4 (Figure 6A and B, residues highlighted in blue). Motifs D3 (C-G-D-X3-S-G-H-T) and D4 (H-Y-T-X-D-V-X3-Y-X6-F-X2-Y-H) are similar to the C2 and C3 motifs in LPPs (shared residues highlighted in red) and include the histidine and aspartate residues (underlined) that form a catalytic triad mediating the nucleophilic attack on the lipid phosphate ester bond (Neuwald, 1997). As in LPPs, these residues are juxtaposed to transmembrane segments 4 and 6 of the SMS proteins and consequently would be oriented towards the same side of the membrane. This would suggest that motifs D3 and D4 are part of the catalytic site responsible for liberating cholinephosphate from PC during SM synthesis. Motifs D1 (P-L-P-D) and D2 (R-R-X8-Y-X2-R-X6-T), on the other hand, appear entirely unique to SMS proteins and are located in the first extramembrane loop and third transmembrane helix, respectively (Figure 6A). The SMSr proteins found in humans, mouse, Drosophila and C. elegans each contain exact copies of the D1 and D3 motifs, yet exhibit one or more conserved amino-acid substitutions in motifs D2 and D4 (Figure 6A and B, residues highlighted in green).
Figure 6.
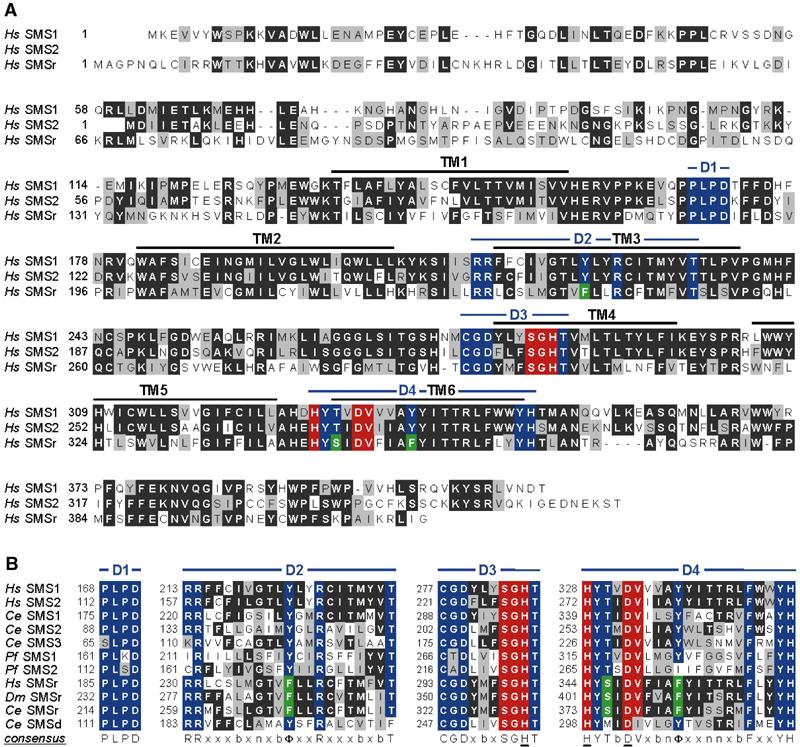
Conserved sequence motifs in SMS1, SMS2 and SMSr proteins. (A) Alignment of human SMS1, SMS2 and SMSr amino-acid sequences. Identical residues are highlighted in black and conservative amino-acid substitutions in grey. Conserved residues within four homology motifs, designated D1–D4, are highlighted in blue and conservative amino-acid substitutions in the D2 and D4 motifs of SMSr proteins in green. Note that motifs D3 and D4 display similarity to the C2 and C3 domains in LPPs, with identical residues highlighted in red. Regions predicted to form transmembrane domains, TM1–TM6, are marked by a black line. (B) Alignment of the four homology motifs in SMS and SMSr proteins from humans (Hs), C. elegans (Ce), P. falciparum (Pf) and D. melanogaster (Dm). PlasmoDB accession numbers of PfSMS1 and PfSMS2 are MAL6P1.178 and MAL6P1.177, respectively. Putative active site residues in consensus sequences are underlined. Symbols used are: n, small neutral amino acid; Φ, aromatic amino acid; b, branched amino acid; x, any amino acid.
Previous work showed that some members of the mammalian LPP superfamily are expressed in only a limited set of tissues (Waggoner et al, 1999; Brauer et al, 2003). Since differences in tissue distribution would provide a possible explanation for the existence of two different SM synthase isoforms in mammals, we investigated the expression profiles of human SMS1 and SMS2. As shown in Figure 7, northern blot analysis detected the presence of a low abundant 3.8 kb transcript for SMS1 in human brain, heart, kidney, liver, muscle and stomach. A major 1.9 kb transcript for SMS2 was expressed to a similar level in all of the above human tissues. These results suggest that human SMS1 and SMS2 are encoded by ubiquitously expressed genes.
Figure 7.
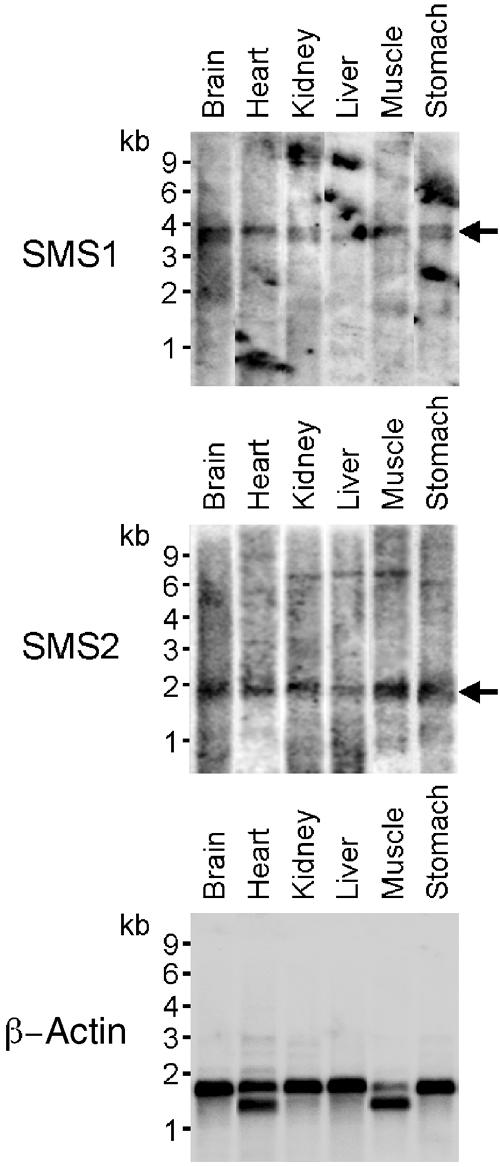
Human SMS1 and SMS2 are encoded by ubiquitously expressed genes. Northern blot analysis of SMS1 and SMS2 transcripts (arrows) in various human tissues. Random prime-labelled human SMS1 or SMS2 cDNA was hybridised to a human poly(A)+ RNA blot (Origene, Rockville, MD). As a control for loading, the RNA blot was stripped and rehybridised with a human β-actin cDNA probe. Mobilities of RNA size markers are indicated.
Subcellular localisation and membrane topology
SM synthesis occurs in the Golgi complex as well as at the plasma membrane of mammalian cells. Endosomes have been put forward as another major site of SM synthesis (Kallen et al, 1994), but this has been disputed (van Helvoort et al, 1997). It is not known whether SM synthase activity detected at these locations is due to the presence of more than one isoenzyme in the cell. This led us to examine the subcellular distribution of V5-tagged versions of human SMS1 and SMS2 in transfected HeLa cells using immunofluorescence microscopy. As shown in Figure 8A, SMS1 was concentrated in the perinuclear region where it displayed extensive colocalisation with sialyltransferase, a marker of trans Golgi cisternae. This colocalisation was also observed in cells treated with nocodazole, a drug causing fragmentation of the Golgi by disrupting the microtubular network, hence confirming the association of SMS1 with the Golgi apparatus. SMS2 displayed a different localisation pattern and was primarily concentrated at the plasma membrane (Figure 8B). A portion of SMS2 was also found in the perinuclear region where it colocalised with sialyltransferase (see Supplementary Figure 3). This Golgi-associated pool of SMS2 unlikely represents newly synthesised material en route to the cell surface, since it was also observed in cells after a 4-h chase with cycloheximide. There was no substantial colocalisation of SMS2 with markers of the endosomal/lysosomal system (e.g. EEA1, CD63, internalised transferrin; data not shown). Whether SMS2 cycles between the Golgi and the plasma membrane, and thereby passes through endosomes remains to be established.
Figure 8.
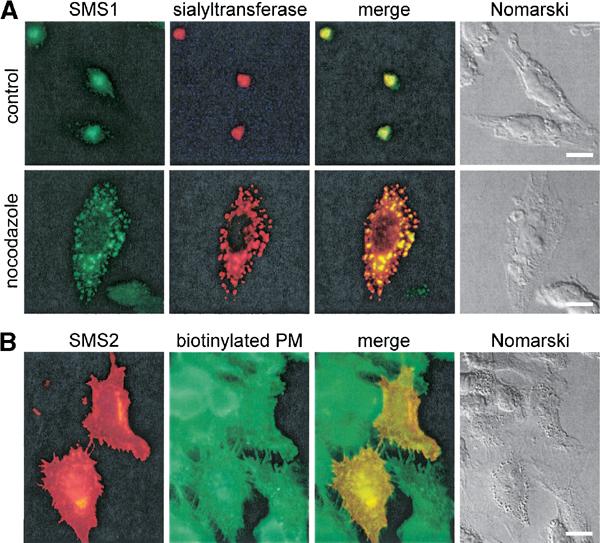
Human SMS1 and SMS2 localise to different cellular organelles. (A) HeLa cells co-transfected with V5-tagged SMS1 and myc-tagged sialyltransferase were incubated in the presence or absence of 10 μM nocodazole for 60 min at 37°C, fixed and then co-stained with rabbit anti-V5 and mouse anti-myc antibodies. Counterstaining was with FITC-conjugated goat anti-rabbit and Texas red-conjugated goat anti-mouse antibodies. (B) HeLa cells transfected with V5-tagged SMS2 were biotinylated on ice, fixed and then co-stained with mouse anti-V5 and rabbit anti-biotin antibodies. Counterstaining was with Texas red-conjugated goat-anti mouse and FITC-conjugated goat anti-rabbit antibodies. Bar, 10 μm.
Since SM synthesis takes place in the exoplasmic leaflet of the Golgi and the plasma membrane (Futerman et al, 1990; van Helvoort et al, 1994), the putative catalytic residues in motifs D3 and D4 of SMS1 and SMS2 would be oriented towards the Golgi lumen and cell surface, respectively, whereas their termini would be located on the opposite, cytosolic side of the membrane (Figure 9A). To test this prediction, we investigated the sidedness of the V5-tagged carboxy termini of human SMS1 and SMS2 by protease protection analysis. When HeLa cells transfected with the SMS2-V5 construct were trypsinised, the V5 tag remained intact, unless cells were lysed or treated with detergent prior to incubation with the protease (Figure 9B). Trypsinisation of lysates from SMS1-V5-expressing cells in the absence of detergent resulted in a complete removal of the V5 tag. Under these conditions, the Golgi-associated type I membrane protein, p24 (Gommel et al, 1999), was largely protected. Collectively, these results indicate that the carboxy termini of SMS1 and SMS2 are cytosolic. Consequently, the putative active site residues in these proteins would be positioned on the exoplasmic leaflet (Figure 9A), hence where SM synthesis was found to occur. Together, these findings demonstrate yet another level of similarity between SMS1, SMS2 and the SM synthase activity previously described in mammalian cells.
Figure 9.
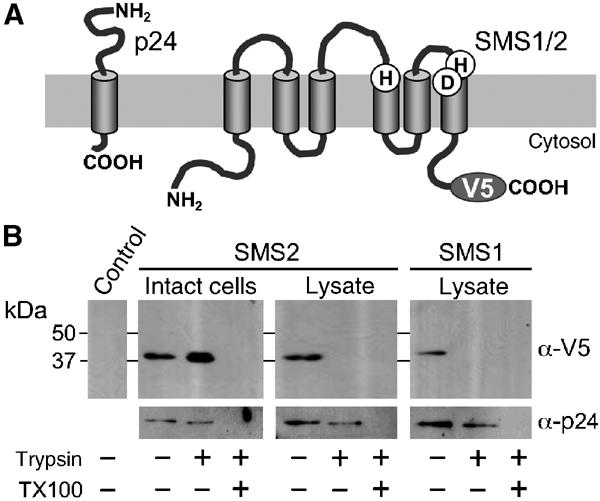
Membrane topology of human SMS1 and SMS2 proteins. (A) Schematic view of the predicted membrane topologies of V5-tagged SMS1 and SMS2, and the Golgi-associated type I membrane protein, p24. (B) Immunoblot of intact or lysed HeLa cells expressing V5-tagged SMS1 or SMS2 and pretreated with 8 mM trypsin for 30 min at 30°C in the presence or absence of 0.4% Triton X-100, as indicated. Immunoblots were stained with antibodies against the V5 epitope (α-V5) or against p24 (α-p24).
Discussion
Identification of a conserved family of SM synthases
In this study, we identified a novel family of integral membrane proteins likely responsible for SM synthesis in animal cells. The two human members of this family, SMS1 and SMS2, were subjected to a detailed biochemical analysis, and we believe that their functional assignment as SM synthases is justified on the following grounds.
First, heterologous expression of SMS1 or SMS2 proves sufficient to support SM synthesis in the yeast S. cerevisiae, an organism lacking endogenous SM synthase activity. Second, in keeping with the enzymatic properties of mammalian SM synthase reported in the literature, SMS1 and SMS2 function as bidirectional lipid cholinephosphotransferases capable of converting PC and ceramide to SM and DAG and vice versa. Third, SMS1 and SMS2 localise to cellular organelles previously established as the principal sites of SM synthesis, namely the Golgi (SMS1) and the plasma membrane (SMS2). Fourth, SMS1 and SMS2 share sequence motifs containing putative active site residues with lipid phosphate phosphatases and candidate PI:ceramide inositolphosphotransferases. Finally, SMS1 and SMS2 adopt a membrane topology whereby the putative active site residues are facing the exoplasmic leaflet, hence the side of the membrane where SM synthesis was found to occur.
A multiplicity of SM synthase genes appears widely spread among organisms generating SM. Human SMS1 and SMS2 are highly conserved in mammals, and we found evidence for the existence of up to three functional homologues in the nematode C. elegans. Moreover, we identified two orthologous sequences in the human malaria parasite P. falciparum, suggesting that this lower eukaryote too contains different SM synthases encoded by separate genes. SM synthesis in P. falciparum occurs in the Golgi apparatus as well as in the network of tubovesicular membranes (TVM) emerging from the parasitophorous vacuolar membrane during intraerythrocytic development (Elmendorf and Haldar, 1994). A differential sensitivity to 1-phenyl-2-acylamino-3-morpholino-1-propanol has been taken as evidence that the Golgi- and TVM-associated activities correspond to different enzymes (Lauer et al, 1995). Our current findings provide a novel opportunity to test this prediction and may facilitate the development of new drugs against malaria.
SM synthases and signalling
The uniform tissue distributions of transcripts for SMS1 and SMS2 in humans suggest that most mammalian cell types would contain both SM synthase isoforms. Biochemical analysis of human SMS1 and SMS2 thus far revealed no fundamental differences in enzymatic properties, even though we cannot exclude the possibility that substrate preferences are lost when enzymes are assayed in a heterologous, detergent-containing system. For now, the most striking difference between SMS1 and SMS2 concerns their subcellular localisation. While SMS1 seems to represent the well-known Golgi-associated SM synthase, SMS2 primarily resides at the plasma membrane. Hence, SMS1 would be proximal to SMS2 with respect to receiving newly synthesised ceramide from the endoplasmic reticulum. A challenging prospect is that, whereas the Golgi enzyme would be responsible for generating the bulk of cellular SM, the second enzyme may serve a principal role in signal transduction at the plasma membrane. The activation of a sphingomyelinase (SMase) and subsequent liberation of ceramide at the plasma membrane has been recognised as an important signalling event in the regulation of fundamental cellular processes that include cell cycle arrest, differentiation and apoptosis (Kolesnick and Hannun, 1999; Pettus et al, 2002; Andrieu-Abadie and Levade, 2002). By converting ceramide back to SM, plasma membrane-associated SM synthase may attenuate SMase-induced signalling. This reaction would produce DAG as a side product, which is a signalling molecule in its own right (Brose and Rosenmund, 2002). Hence, the presence of an SM synthase at the plasma membrane complicates a proper understanding of the role of ceramide as a specific signal transducer.
A current bottleneck for elucidating the role of SM hydrolysis in cellular signalling is the unambiguous identification of the agonist-stimulated SMase. Both neutral and acid SMases have been implicated, but their precise cellular roles in SM hydrolysis and signalling remain to be clarified (Andrieu-Abadie and Levade, 2002). Since the plasma membrane-associated SM synthase is capable of catalysing the reverse reaction of SM synthesis (van Helvoort et al, 1994), it is tempting to speculate that it may execute part of the signalling events currently attributed to ligand-induced SMase. The observation that both plasma membrane- and Golgi-associated SM synthases exhibit reverse activity brings up the question of how the directionality of these enzymes is regulated. One possibility is that the direction of the reaction is primarily determined by the relative concentrations of ceramide and DAG in the membrane. On the other hand, given the biological importance attributed to DAG and ceramide, one may expect the interconversion of these molecules to be subjected to a more elaborate form of control. Indeed, it has been shown that activation of SM synthesis in primary astrocytes is an early event associated with the mitogenic activity of basic fibroblast growth factor (Riboni et al, 2001) and that the onset of TNFα-induced apoptosis in rhabdomyosarcoma cells is preceded by TNF-dependent inhibition of SM synthesis (Bourteele et al, 1998). Mammalian SMS1 contains a predicted SAM (sterile alpha motif) domain at the amino terminus (residues 7–63 in human SMS1) that might provide a means for the enzyme to interact with regulatory proteins (Schultz et al, 1997). The present identification of a family of animal SM synthases offers unprecedented opportunities to further clarify the biological significance of SM metabolism and its regulation.
Mechanism of action
SM synthases contain four highly conserved sequence motifs. Two of these, D3 and D4, are similar to the C2 and C3 phosphatase domains in LPPs and include the putative catalytic histidine and aspartate residues implicated in LPP-mediated hydrolysis of lipid phosphate esters (Neuwald, 1997). This suggests a working mechanism for SM synthases in which the catalytic triad previously described for LPPs would function in a similar fashion. The reaction would proceed via the following steps: (1) binding of a two-chain choline phospholipid (PC or SM) to a unique binding site; (2) nucleophilic attack on the lipid-phosphate ester bond by the histidine in D4 assisted by the conserved aspartate in this motif; (3) formation of a cholinephospho-histidine intermediate and release of DAG or ceramide, facilitated by the histidine in D3 acting as a base; (4) nucleophilic attack of the C1-hydroxyl of ceramide or DAG on the cholinephospho-histidine intermediate, assisted by the histidine in D3; and (5) release of SM or PC from the active site to allow another round of catalysis. This model provides a framework for site-directed mutagenesis and kinetic studies to reveal the working mechanism of SM synthases.
There is a striking parallel between animal SM synthase and IPC synthase in yeast. Like human SMS1, the yeast enzyme is Golgi localised and contains the key D4 motif in the Golgi lumen at the start of the last membrane span (Levine et al, 2000). This suggests that animal SM synthase and yeast IPC synthase may have evolved from a common ancestor. In contrast, the SM synthase recently identified from P. aeruginosa (Luberto et al, 2003) lacks the key motifs of animal SM synthase and is a soluble rather than an integral membrane protein. Hence, it appears that SM synthases arose at least twice during evolution.
Ethanolaminephosphotransferases
Database searches using the catalytic site sequences from LPPs yielded, in addition to SM synthases, numerous other proteins containing partially conserved phosphatase motifs. In many cases, the biochemical functions of these proteins are not known. Given our present findings, the possibility that some of these LPP-like proteins do not serve as phospholipases but instead catalyse novel kinds of synthetic or transphosphatidylation reactions deserves consideration. For example, mammals and insects synthesise ethanolamine phosphorylceramide, a sphingolipid analogous to SM (Malgat et al, 1986; Rietveld et al, 1999). Whether production of EPC and SM involves a single transferase that accepts both PC and PE as substrates or requires two separate enzymes has remained an open issue. However, Drosophila lacks SM and generates only EPC (Rietveld et al, 1999). Our finding that Drosophila contains an SMS-related protein, but no SMS proteins, suggests that the SMS-related proteins described in this study may function as dedicated EPC synthases. This possibility is currently under investigation.
Materials and methods
Chemicals
Sphingosyl-(NBD-hexanoyl)-phosphocholine (NBD-SM) and (NBD-hexanoyl)-ceramide (NBD-Cer) were from Molecular Probes (Eugene, OR), and oleoyl-(NBD-hexanoyl)-phosphocholine (NBD-PC) was from Avanti Polar Lipids (Alabaster, AL). [Methyl-14C]-choline was from ICN Biomedicals (Irvine, CA) and L-3-phosphatidyl [N-methyl-3H]-choline,1,2-dipalmitoyl from Amersham Pharmacia (Piscataway, NJ). NBD-DAG was produced from NBD-PC by treatment with phospholipase C from Bacillus cereus as described (Trotter, 2000). All other lipids and chemicals were from Sigma Aldrich (St Louis, MO), as listed in Supplementary data.
Selection, cloning and expression of CSS sequences
Animal entries in the Swiss-Prot/TrEMBL protein sequence database were searched using Prosite (http://www.expasy.org/tools/scanprosite/) against sequence motif H-[YFWH]-X2-D-[VLI]-X2-[GA]-X3-[GSTA] and then further selected based on three additional criteria as described under Results. CSSs were assembled into different groups by multiple sequence alignments and phylogenetic trees generated with ClustalW and PHYLIP. Open reading frames (ORFs) in selected CSS sequences were PCR amplified using Taq polymerase (MBI Fermentas, Hanover, MD) according to information provided in Table I. PCR products were cloned into yeast expression vector pYES2.1/V5-His-TOPO® (Invitrogen Corporation, Carlsbad, CA) and the resulting plasmids were used to transform S. cerevisiae strain IAY11 (MATα ura3-52 his3-Δ200 leu2-3,-112 trp-Δ901 ade2-101 ade3-Δ853, provided by Ian Adams, MRC-LMB, Cambridge, UK). Transformants were grown in complete minimal uracil dropout medium containing 2% (w/v) galactose. Expression of CSS proteins was verified by Western blot analysis with mouse anti-V5 antibodies (Invitrogen). PCR-amplified ORFs of human SMS1 and SMS2 were cloned into mammalian expression vector pcDNA3.1/V5-His-TOPO® (Invitrogen) and the resulting plasmids (SMS1-V5/pcDNA3.1 and SMS2-V5/pcDNA3.1) were used to transfect HeLa cells.
Table 1.
EST clones and primers used in this study
| Protein name | Accession numbera | EST clone ID | Primer pairs N- and C-terminal (5′–3′) |
|---|---|---|---|
| HsCSS1α1 | Q9NXE2 | IRAKp961F1432b | AAGCCATGGCTGTAGGAAACAACACGGTAACTTCGGTCA TGGAC |
| HsCSS1α2 | Q96GM1 | IRALp962F0526b | AAGCCATGGCGGGAGGGAGAGGTGGCCACGGCGGGC |
| HsCSS2α | Q96SS7 | IRALp962I0417b | AAGCCATGCCAGCTTCCCAGAGCCAGGCAGAGATGAGCA TC |
| HsCSS3α1 (SMS1) | Q86VZ5 | LGN01891c | AAGCCATGATCCTTGTAGGACTCTGTGTGTCATTCACCA GCCGG |
| HsCSS3α2 (SMS2) | Q8NHU3 | IRAKp961F1433b | AAGCCATGAATATGTTTGATGCTGACAATCCTTATAAGC CCGTGTGG |
| HsCSS3β | Q96LT4 | IMAGp958I131239Qb | AAGCCATGGCAGGTCCTAATCAACTCTCCAATTAGTCTT TTCATTATTGC |
| MmCSS1α2 | Q8VCY8 | IRAKp961L1531b | AAGCCATGGCTGGAGGGAGGGTGGCCACGGCGGGC |
| MmCSS2α | Q91WB2 | IMAGp998B0810812b | AAGCCATGCCAGCTTCCCAGACCAGGCAGAGATGAGCATC |
| MmCSS3α1 (SMS1) | Q8VCQ6 | IRAKp961L2141b | AAGCCATGAAGGAAGTGGTTTACTGTGTCGTTTACCAGCC GG |
| CeCSS1γ | Q22250 | YK289g7 /YK572h5d | AAGCCATGTCCGTGCCAGCTTCGGTACCGATATCCATCAT TTTG |
| CeCSS3α1 (SMS1) | Q9XTV2 | YK559h9/YK517e10d | AAGCCATGAAAATGTCTTGGAATCATCAATTTTGGCAGA GACATGGTAG |
| CeCSS3α2 (SMS2) | Q20735 | YK428e6/YK109h8d | AAGCCATGACAAACAGTTCGGAGTTCTTGCAATTTGTAGT TGATACGA |
|
CeCSS3β |
Q20696 |
YK524b8d |
AAGCCATGCTGGATAACAGACCTATACATTGTGCTTTTT GGTATGATTTTTG |
| a Swiss-Prot/TrEMBL. | |||
| b Obtained from RZPD Deutsches Ressourcenzentrum für Genomforschung, Berlin, Germany. | |||
| c Provided by Dr Sumio Sugano, Institute of Medical Science, University of Tokyo, Japan. | |||
| d Provided by Dr Yuji Kohara, National Institute of Genetics, Mishima, Japan. | |||
SM synthase assay on cell lysates
CSS-expressing yeast cells were lysed at 100 OD600/ml by bead bashing at 4°C in RB1 buffer (120 mM K-glutamate, 15 mM KCl, 5 mM NaCl, 2 mM MnCl2, 2 mM MgCl2, 20 mM Hepes-KOH, pH 7.2) containing freshly added protease inhibitors (Holthuis et al, 1998). Lysates were centrifuged at 700 g for 10 min at 4°C and supernatants (0.25 ml per reaction) preincubated with NEM (1 mM) for 10 min at 37°C. The reaction was initiated by addition of 5 nmol of NBD-Cer in 0.1 ml RB1 buffer and incubation continued for 15 min. Lipids were extracted by addition of 1.7 ml chloroform:methanol (1:2.2), dried under N2 and subjected to butanol/water partitioning. Lipids recovered from the butanol phase were separated by one-dimensional TLC for 30 min in solvent I (chloroform/methanol/4.2 M NH4OH (9:7:3, v/v)) followed by 30 min in solvent II (chloroform/methanol/acetic acid (45:30:7, v/v)). Fluorescent images were recorded on a STORM 860 Imaging Analysis System (Molecular Dynamics, Sunnyvale, CA) and analysed with ImageQuant software.
Metabolic labelling
CSS-expressing yeast cells (0.5 OD600) were inoculated in 5 ml medium containing 10 μCi [methyl-14C]-choline and 10 nmol NBD-Cer. Cells were grown for 16 h at 30°C, washed in water, and lipids were extracted by bead bashing in H2O/methanol/chloroform (5:16:16, v/v). The organic extracts were dried under N2, subjected to butanol/water partitioning, and lipids recovered from the butanol phase were deacylated by mild base treatment using 0.2 N NaOH in methanol (60 min, 30°C). After neutralising with 1 M acetic acid, lipids were extracted with chloroform and separated by two-dimensional TLC using solvent I in the first and solvent II in the second dimension. Radiolabelled lipids were detected by exposure to BAS-MS imaging screens (Fuji Photo Film Co., Japan) and read out on a BIO-RAD Personal Molecular Imager.
Preparation of detergent extracts
SMS1/SMS2-expressing yeast cells were lysed at 100 OD600/ml by bead bashing at 4°C in lysis buffer (50 mM Tris–HCl, pH 7.0, 1 mM EDTA, 0.3 M sucrose) containing protease inhibitors. Post nuclear supernatants were prepared as above and loaded onto a 60% (w/w) sucrose cushion and then centrifuged at 100 000 g for 60 min at 4°C. Membranes derived from ±500 OD600 of cells were resuspended in 1 ml ice-cold extraction buffer (50 mM Tris–HCl, pH 7.0, 10% glycerol, 1% Triton X-100, 1 mM MnCl2) containing protease inhibitors, incubated for 60 min at 4°C while rotating and then centrifuged at 100 000 g for 60 min at 4°C. Detergent extracts were diluted to a final protein concentration of 1.5 mg/ml in extraction buffer and 100 μl aliquots were snap frozen in liquid N2 and stored at −80°C.
SM synthase and reverse transferase assays on detergent extracts
Detergent extracts were diluted five- to eight-fold in extraction buffer, and 50 μl mixed with 5 μl of a 12 mM head group donor stock (e.g. CDP-choline, PC, PI) or with 15 μCi (350–700 fmol) of [3H]-choline-containing PC prepared in extraction buffer. Following a 15 min preincubation at 37°C, reactions were started by addition of 220 μl RB2 buffer (50 mM Tris–HCl, pH 7.0, 5 mM MgCl2, 1 mM MnCl2, 1 mM NEM) containing 5 nmol NBD-ceramide (SM synthase assay) or 5 nmol NBD-DAG (reverse transferase assay) and incubated for 60 min at 37°C. Lipids were extracted, separated by TLC, and analysed as above.
Mass spectrometry
Detergent extracts (400 μl) were mixed with bovine brain ceramide (180 μM) and egg PC (500 μM) in a total volume of 500 μl. After 15 min at 37°C, reactions were diluted four-fold in RB2 buffer and then incubated for another 60 min at 37°C. Lipids were extracted in chloroform:methanol (1:2.2) and subjected to butanol/water partitioning. Residual Triton X-100 was removed by solid-phase extraction on Si-60 columns (500 mg stationary phase) in acetone. Phospholipids were eluted in methanol/chloroform (19:1, v/v), dried under N2, dissolved in methanol/chloroform (2:1, v/v) and then separated by HPLC on two 250 × 4.6 mm Lichrosphere RP-18 end-capped columns (Merck, Darmstadt, Germany) in series (Brouwers et al, 1998). The column effluent was split in a 10:1 ratio, with the smaller fraction going to the mass spectrometer. Mass spectrometry was performed on a Sciex API-365 triple quadrupole mass spectrometer (Applied Biosystems, Nieuwerkerk a/d IJssel, The Netherlands). Positive ions were generated by a turbo-ion spray ionisation source operating at +5.5 kV ionisation potential. N2 (2 l/min) was used as drying gas at a temperature of 350°C. The declustering potential (cone voltage) was set to 45 V and the focus potential to 220 V. Mass spectra from mass to charge ratio (m/z) 650–950 amu were recorded at a speed of 120 amu/s.
Mammalian cell transfection and immunofluorescence microscopy
HeLa cells were cultured in DMEM medium containing 10% fetal calf serum. Cells were grown on glass coverslips to 40% confluence and then transfected with human SMS1-V5/pcDNA3.1, SMS2-V5/pcDNA3.1 and myc-tagged sialyltransferase/pCB7 constructs using Lipofectamine 2000 (Invitrogen). After 48 h, cells were fixed in 3% paraformaldehyde/PBS and processed for immunofluorescence microscopy (see Supplementary data for details). For cell surface biotinylation, cells were incubated twice with 0.5 mg/ml sulpho-NHS-SS-biotin (Pierce) in PBS for 20 min on ice, quenched in 10 mM glycine/PBS for 20 min and then washed in PBS prior to fixation. Immunostaining was with rabbit anti-V5 antibodies (Sigma), mouse anti-V5 antibodies (Invitrogen), mouse 9E10 anti-myc antibody (SanverTech, CA), rabbit anti-biotin antibodies (Rockland, Gilbertsville, PA), FITC- or Texas red-conjugated goat anti-rabbit and goat anti-mouse antibodies (Jackson Laboratories, West Grove, PA). Images were obtained using a Nikon D-ɛclipse C1 confocal microscope.
Protease protection assay
HeLa cells transfected with human SMS1-V5 or SMS2-V5 constructs were washed, scraped and then lysed in ice-cold PNS buffer (120 mM K-glutamate, 15 mM KCl, 5 mM NaCl, 2 mM MnCl2, 0.8 mM CaCl2, 2 mM MgCl2, 1.6 mM EDTA and 20 mM Hepes-KOH, pH 7.2) by 20 passages through a 26¾-gauge needle. The homogenate was treated with 8 mM trypsin (Sigma Aldrich) in the presence or absence of 0.4% Triton X-100 for 30 min at 37°C. Alternatively, cells were only washed and then trypsin-treated in PNS buffer as above. Homogenates and cell suspensions were transferred on ice, trypsin inhibitor (Sigma Aldrich) was added to 160 mM and samples were processed for Western blot analysis using mouse anti-V5 antibodies and rabbit anti-p24 protein (Gommel et al, 1999; provided by BJ Helms, Faculty of Veterinary Medicine, Utrecht).
Supplementary Material
Supplementary Fig. 1.
Supplementary Fig. 2.
Supplementary Fig. 3.
Supplementary materials
Acknowledgments
We are indebted to Drs Sumio Sugano and Yuji Kohara for providing cDNA clones, to Bernd Helms for anti-p24 antibodies, and to Maarten Egmond and Gerrit van Meer for many helpful discussions. This work was supported by a grant from the Royal Netherlands Academy of Arts and Sciences (to JCMH), by EC grant HPRN-CT-2000-00077 and by SENTER, the Dutch Ministry of Economic Affairs.
References
- Andrieu-Abadie N, Levade T (2002) Sphingomyelin hydrolysis during apoptosis. Biochim Biophys Acta 1585: 126–134 [DOI] [PubMed] [Google Scholar]
- Bankaitis VA (2002) Cell biology Slick recruitment to the Golgi. Science 295: 290–291 [DOI] [PubMed] [Google Scholar]
- Bourteele S, Hausser A, Doppler H, Horn-Muller J, Ropke C, Schwarzmann G, Pfizenmaier K, Muller G (1998) Tumor necrosis factor induces ceramide oscillations and negatively controls sphingolipid synthases by caspases in apoptotic Kym-1 cells. J Biol Chem 273: 31245–31251 [DOI] [PubMed] [Google Scholar]
- Brauer AU, Savaskan NE, Kuhn H, Prehn S, Ninnemann O, Nitsch R (2003) A new phospholipid phosphatase, PRG-1, is involved in axon growth and regenerative sprouting. Nat Neurosci 6: 572–578 [DOI] [PubMed] [Google Scholar]
- Brose N, Rosenmund C (2002) Move over protein kinase C, you've got company. J Cell Sci 115: 4399–4411 [DOI] [PubMed] [Google Scholar]
- Brouwers JF, Gadella BM, van Golde LM, Tielens AG (1998) Quantitative analysis of phosphatidylcholine molecular species using HPLC and light scattering detection. J Lipid Res 39: 344–353 [PubMed] [Google Scholar]
- Dickson RC (1998) Sphingolipid functions in Saccharomyces cerevisiae: comparison to mammals. Annu Rev Biochem 67: 27–48 [DOI] [PubMed] [Google Scholar]
- Elmendorf HG, Haldar K (1994) Plasmodium falciparum exports the Golgi marker sphingomyelin synthase into a tubovesicular network in the cytoplasm of mature erythrocytes. J Cell Biol 124: 449–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa M, Nishijima M, Hanada K (1999) Genetic evidence for ATP-dependent ER-to-Golgi apparatus trafficking of ceramide for sphingomyelin synthesis in CHO cells. J Cell Biol 144: 673–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futerman AH, Stieger B, Hubbard AL, Pagano RE (1990) Sphingomyelin synthesis in rat liver occurs predominantly at the cis and medial cisternae of the Golgi apparatus. J Biol Chem 265: 8650–8657 [PubMed] [Google Scholar]
- Gommel D, Orci L, Emig EM, Hannah MJ, Ravazzola M, Nickel W, Helms JB, Wieland FT, Sohn K (1999) p24 and p23, the major transmembrane proteins of COPI-coated transport vesicles, form hetero-oligomeric complexes and cycle between the organelles of the early secretory pathway. FEBS Lett 447: 179–185 [DOI] [PubMed] [Google Scholar]
- Hanada K, Hara T, Fukasawa M, Yamaji A, Umeda M, Nishijima M (1998) Mammalian cell mutants resistant to a sphingomyelin-directed cytolysin. J Biol Chem 273: 33787–33794 [DOI] [PubMed] [Google Scholar]
- Heidler SA, Radding JA (2000) Inositol phosphoryl transferases from human pathogenic fungi. Biochim Biophys Acta 1500: 147–152 [DOI] [PubMed] [Google Scholar]
- Holthuis JCM, Nichols BJ, Dhruvakumar S, Pelham HR (1998) Two syntaxin homologues in the TGN/endosomal system of yeast. EMBO J 17: 113–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis JCM, Pomorski T, Raggers RJ, Sprong H, Van Meer G (2001) The organizing potential of sphingolipids in intracellular membrane transport. Physiol Rev 81: 1689–1723 [DOI] [PubMed] [Google Scholar]
- Jeckel D, Karrenbauer A, Birk R, Schmidt RR, Wieland F (1990) Sphingomyelin is synthesized in the cis Golgi. FEBS Lett 261: 155–157 [DOI] [PubMed] [Google Scholar]
- Kallen K-J, Allan D, Whatmore J, Quinn P (1994) Synthesis of surface sphingomyelin in the plasma membrane recycling pathway of BHK Cells. Biochim Biophys Acta 1191: 52–58 [DOI] [PubMed] [Google Scholar]
- Kolesnick R, Hannun YA (1999) Ceramide and apoptosis. Trends Biochem Sci 24: 224–225 [DOI] [PubMed] [Google Scholar]
- Lauer SA, Ghori N, Haldar K (1995) Sphingolipid synthesis as a target for chemotherapy against malaria parasites. Proc Natl Acad Sci USA 92: 9181–9185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine TP, Wiggins CA, Munro S (2000) Inositol phosphorylceramide synthase is located in the Golgi apparatus of Saccharomyces cerevisiae. Mol Biol Cell 11: 2267–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsky NG, Pagano RE (1985) Intracellular translocation of fluorescent sphingolipids in cultured fibroblasts. J Cell Biol 100: 27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luberto C, Hannun YA (1998) Sphingomyelin synthase, a potential regulator of intracellular levels of ceramide and diacylglycerol during SV40 transformation. J Biol Chem 273: 14550–14559 [DOI] [PubMed] [Google Scholar]
- Luberto C, Stonehouse MJ, Collins EA, Marchesini N, El-Bawab S, Vasil AI, Vasil ML, Hannun YA (2003) Purification, characterization, and identification of a sphingomyelin synthase from Pseudomonas aeruginosa. J Biol Chem 278: 32733–32743 [DOI] [PubMed] [Google Scholar]
- Malgat M, Maurice A, Baraud J (1986) Sphingomyelin and ceramide-phosphoethanolamine synthesis by microsomes and plasma membranes from rat liver and brain. J Lipid Res 27: 251–260 [PubMed] [Google Scholar]
- Marggraf WD, Anderer FA, Kanfer JN (1981) The formation of sphingomyelin from phosphatidylcholine in plasma membrane preparations from mouse fibroblasts. Biochim Biophys Acta 664: 61–73 [DOI] [PubMed] [Google Scholar]
- Marggraf W-D, Kanfer JN (1984) The phosphorylcholine receptor in the phosphatidylcholine:ceramide cholinephosphotransferase reaction. Biochim Biophys Acta 793: 346–353 [DOI] [PubMed] [Google Scholar]
- Nagiec MM, Nagiec EE, Baltisberger JA, Wells GB, Lester RL, Dickson RC (1997) Sphingolipid synthesis as a target for antifungal drugs. J Biol Chem 272: 9809–9817 [DOI] [PubMed] [Google Scholar]
- Neuwald AF (1997) An unexpected structural relationship between integral membrane phosphatases and soluble haloperoxidases. Protein Sci 6: 1764–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettus BJ, Chalfant CE, Hannun YA (2002) Ceramide in apoptosis: an overview and current perspectives. Biochim Biophys Acta 1585: 114–125 [DOI] [PubMed] [Google Scholar]
- Riboni L, Viani P, Bassi R, Giussani P, Tettamanti G (2001) Basic fibroblast growth factor-induced proliferation of primary astrocytes. J Biol Chem 276: 12797–12804 [DOI] [PubMed] [Google Scholar]
- Rietveld A, Neutz S, Simons K, Eaton S (1999) Association of sterol- and glycosylphosphatidylinositol-linked proteins with Drosophila raft lipid microdomains. J Biol Chem 274: 12049–12054 [DOI] [PubMed] [Google Scholar]
- Satouchi K, Hirano K, Sakaguchi M, Takehara H, Matsuura F (1993) Phospholipids from the free-living nematode Caenorhabditis elegans. Lipids 28: 837–840 [DOI] [PubMed] [Google Scholar]
- Schultz J, Ponting CP, Hofmann K, Bork P (1997) SAM as a protein interaction domain involved in developmental regulation. Protein Sci 6: 249–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K, Toomre D (2000) Lipid rafts and signal transduction. Nat Rev Mol Cell Biol 1: 31–39 [DOI] [PubMed] [Google Scholar]
- Trotter PJ (2000) A novel pathway for transport and metabolism of a fluorescent phosphatidic acid analog in yeast. Traffic 1: 425–434 [DOI] [PubMed] [Google Scholar]
- Ullman MD, Radin NS (1974) The enzymatic formation of sphingomyelin from ceramide and lecithin in mouse liver. J Biol Chem 249: 1506–1512 [PubMed] [Google Scholar]
- van Helvoort A, Stoorvogel W, van Meer G, Burger KNJ (1997) Sphingomyelin synthase is absent from endosomes. J Cell Sci 110: 781–788 [DOI] [PubMed] [Google Scholar]
- van Helvoort A, van't Hof W, Ritsema T, Sandra A, van Meer G (1994) Conversion of diacylglycerol to phosphatidylcholine on the basolateral surface of epithelial MDCK cells. J Biol Chem 269: 1763–1769 [PubMed] [Google Scholar]
- Voelker DR, Kennedy EP (1982) Cellular and enzymic synthesis of sphingomyelin. Biochemistry 21: 2753–2759 [DOI] [PubMed] [Google Scholar]
- Waggoner DW, Xu J, Singh I, Jasinska R, Zhang QX, Brindley DN (1999) Structural organization of mammalian lipid phosphate phosphatases: implications for signal transduction. Biochim Biophys Acta 1439: 299–316 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1.
Supplementary Fig. 2.
Supplementary Fig. 3.
Supplementary materials


