Abstract
The glutamate receptor δ2 (GluRδ2) subunit is selectively expressed in cerebellar Purkinje cells and plays an important role in cerebellar long-term depression, motor learning, motor coordination, and synapse development. We identified a novel GluRδ2-interacting protein, named Delphilin, that contains a single PDZ domain and formin homology (FH) domains FH1 and FH2 plus coiled-coil structure. As far as we know, this is the first reported protein that contains both PDZ and FH domains. Yeast two-hybrid and surface plasmon resonance (SPR) analyses indicated that Delphilin interacts with the GluRδ2 C terminus via its PDZ domain. This was also supported by coimmunoprecipitation experiments using a heterologous expression system in mammalian cells. Yeast cell and SPR analyses also demonstrated the possibility that the FH1 proline-rich region of Delphilin interacts with profilin, an actin-binding protein, and with the Src homology 3 domain of neuronal Src protein tyrosine kinase. In situ hybridization demonstrated the highest expression of Delphilin mRNA in Purkinje cells. Delphilin polypeptide was highly enriched in the synaptosomal membrane fraction of the cerebellum and coimmunoprecipitated with the GluRδ2 subunit. The post-embedding immunogold technique demonstrated that Delphilin is selectively localized at the postsynaptic junction site of the parallel fiber–Purkinje cell synapse and colocalized with GluRδ2. Thus, Delphilin is a postsynaptic scaffolding protein at the parallel fiber–Purkinje cell synapse, where it may serve to link GluRδ2 with actin cytoskeleton and various signaling molecules.
Keywords: glutamate receptor δ2 subunit, cerebellum, Purkinje cell, parallel fiber synapses, yeast two-hybrid, surface plasmon resonance, PDZ domain, FH domain, post-embedding immunogold labeling
Glutamate receptor (GluR) channels play major roles in fast excitatory synaptic transmission. The δ subfamily of GluR channel subunits consists of two (δ1 and δ2) subunits and is positioned between the NMDA and non-NMDA receptor channel subunits with respect to the amino acid sequence (Yamazaki et al., 1992; Araki et al., 1993; Lomeli et al., 1993). Thus far, the GluRδ2 subunit has not been shown to bind glutamatergic ligands, and no GluR channel activity was detected when it was expressed in Xenopus oocytes or mammalian cells, even in combination with other GluR channel subunits.
The GluRδ2 subunit is selectively expressed in Purkinje cells (Araki et al., 1993), which receive two excitatory inputs, one from the parallel fibers of cerebellar granular cells and the other from the climbing fibers of the inferior olivary nucleus. Early in development, the GluRδ2 is distributed both in parallel fiber– and climbing fiber–Purkinje cell synapses but becomes restricted to the parallel fiber synapses according to postnatal maturation (Takayama et al., 1996; Landsend et al., 1997; Zhao et al., 1998). Cerebellar long-term depression (LTD) in parallel fiber–Purkinje cell neurotransmission is produced by combined stimulation (simultaneous or costimulation) of both parallel and climbing fiber synapses on the Purkinje cells (Ito, 1989; Linden and Connor, 1995). Knock-out mice lacking the GluRδ2 gene showed perturbation in the parallel fiber synapse formation and in the elimination of surplus climbing fibers (Kashiwabuchi et al., 1995). Analyses of the gene knock-out mice further revealed that Gluδ2 plays an important role in motor learning, motor coordination, and induction of LTD in parallel fiber–Purkinje cell neurotransmission (Kashiwabuchi et al., 1995; Hirano, 1996; Kurihara et al., 1997). Studies using antisense oligonucleotides (Hirano et al., 1994; Jeromin et al., 1996) also showed involvement of GluRδ2 in the LTD. Recently, neurodegeneration in Lurcher mice was reported to be caused by a gain-of-function mutation in the GluRδ2 gene (Zuo et al., 1997), and channel properties of receptors with the Lurcher mutation have been investigated (Kohda at al., 2000; Taverna et al., 2000; Wollmuth et al., 2000).
Effective neurotransmission requires the precise localization of the neurotransmitter receptors on the proper postsynaptic membranes (Scannervin and Huganir, 2000). Postsynaptic density (PSD) is a dense thickening of submembranous cytoskeleton on the postsynaptic membrane, at which various ionotropic receptors are localized with their interacting molecules. PSD-95/synapse-associated protein-90 (SAP-90) and its family proteins, which interact with NMDA receptor channel subunits and Shaker-type potassium channels, have been identified as molecules that function in the accumulation, clustering, and signaling of the receptors. The common feature of these postsynaptic proteins is that they have PSD-95/Dlg/ZO-1 (PDZ) modular domains for protein–protein interaction. PDZ domains of these proteins directly recognize the C termini of their target proteins and are believed to play a central role in targeting and clustering of receptors to proper synaptic membranes (Sheng and Pak, 1999). Quite recently, it has been reported that PDZ domain-containing proteins, such as PSD-93 protein (Roche et al., 1999) and protein–tyrosine phosphatase PTPMEG (Hironaka et al., 2000), interact with GluRδ2 subunit. We report here a novel PDZ protein, named Delphilin, that interacts with GluRδ2 and is selectively targeted to a postsynaptic site of parallel fiber–Purkinje cell synapses, where it is colocalized with GluRδ2.
MATERIALS AND METHODS
Yeast two-hybrid experiments and library screening. Yeast two-hybrid studies were generally performed using the MATCHMAKER Two-Hybrid System 2 (Clontech Laboratories, Palo Alto, CA). Briefly, the bait polypeptides were fused to the yeast GAL4 DNA-binding domain in the vector pAS2–1 and introduced into Y190 yeast cells harboring the reporter genes HIS3 and β-galactosidase under control of the upstream GAL4 recognition sites. The bait used for library screening consisted of amino acid (aa) residues from 963 to the C terminus of the mouse GluRδ2 subunit. Construction was performed by PCR with proof reading polymerase pfu (Stratagene, La Jolla, CA), and the nucleotide sequence was confirmed by sequencing. Primers for PCR amplification were agccatgggcggcggcccttttaggcacagggctcc and gcgaattctcatatggacgtgcctcggtc. A mouse brain cDNA library (Clontech Laboratories) constructed in the pACT2, which adds GAL4 transcription activation domain to the polypeptide encoded by the insert, was subsequently introduced into the Y190 already harboring the bait plasmid. The number of independent clones that were screened was ∼2 × 106. Isolated clones were verified by yeast two-hybrid system to activate reporter genes specifically with the bait plasmid and not with the pAS2–1 empty vector or a recombinant pAS2–1 containing unrelated human protein, lamin C (Bartel et al., 1993). It was also confirmed that the isolated library plasmid alone does not directly activate the reporter genes. Yeast growth selection on synthetic dropout media and β-galactosidase assays with filter membranes were performed in strict accord with the manufacturer's protocol (Clontech Laboratories).
cDNA cloning and Northern blot analysis. Poly(A) RNA was isolated from adult BALB/c mouse cerebellum, and both oligo-dT-primed and random hexanucleotide-primed cDNA libraries were constructed in λgt10 or λgt11 phages. The insert of the original two hybrid clone MB2 (see details in Results) was labeled with P32 and used for conventional screening as a probe. Each cDNA library was screened for ∼8 × 105 plaques. The longest clones from each screening, clone 10-1 from the random-primed library and clone 9-2 from the dT-primed library, were subjected to nucleotide sequencing for both strands. These two clones overlapped and together covered the putative entire protein coding region and the 3′ terminus with a poly(A) tail.
The transcriptional start site of the gene was determined by the cap site hunting method (Murata and Yamaguchi, 1999) with mouse brain cap site cDNA (Nippon Gene, Toyama, Japan) according to the manufacturer's instructions. Briefly, cap site cDNA of mouse brain was constructed by removing the mRNA cap with tobacco acid pyrophosphatase and ligating oligoribonucleotides (r-oligos) of a known sequence to the decapped mRNA end with T4 RNA ligase. The first-strand cDNA was synthesized with random primers, followed by the gene-specific PCR amplification with the primer of the sequence identical to the ligated r-oligo and a gene specific downward primer (gaccctgcgacaccacctcgtcgaa).
Total RNAs were isolated from various tissues of adult BALB/c mice and subjected to Northern blot analysis. An 850 bp fragment corresponding to the entire sequence of the MB2 clone was labeled with P32 and used as a probe.
Plasmids for yeast two-hybrid studies. cDNA fragments encoding C-terminals of the GluRδ2 mutants and a wild-type GluRδ1 in pAS2–1 DNA-binding domain plasmid were prepared by PCR as described above. PCR primers for the C-terminal tail of the GluRδ1 subunit were agccatgggcggcattcagtgcaaacacaggtcg and gcgaattctcagatggaggtgccatgagag. The common 5′-side primer for the GluRδ2 mutants was agccatgggcggcggcccttttaggcacagggctcc. The 3′ primers were (1) for the mutant without the C-terminal isoleucine, gcgaattctcaggacgtgcctcggtcggggt; (2) for the mutant without isoleucine and serine, gcgaattctcacgtgcctcggtcggggtcatt; (3) for the mutant without isoleucine, serine, and threonine, gcgaattctcagcctcggtcggggtcattgc; (4) for the mutant without isoleucine, serine, threonine, and glycine, gcgaattctcatcggtcggggtcattgccca; and (5) for the mutant having an alanine residue instead of the C-terminal isoleucine, gcgaattctcaagcggacgtgcctcggtcggggt. cDNA fragments encoding the mouse neuronal Src (n-Src) SH3 domain and the rat PSD-93 Src homology 3 (SH3) domain in pAS2–1 were amplified by reverse transcriptase (RT)-PCR using mouse and rat RNAs of brain as templates, respectively. Primers for n-Src SH3 were ctagaattcccggtggggtgaccaccttt and gatctcgagttaggagtcggagggcgccacat. Primers for PSD-93 SH3 were ctagaattcagaaacgctccctgtat and gatctcgagttaggcacgctcctttctttcca. For the mouse profilin I entire coding region in pAS2–1, a PCR was done with primers cagaattcaggggcggcatggccgggtggaacgcctac and gactcgagtcagtactgggaacgccgcagg using a mouse expression sequence tagged (EST) clone 1095742 plasmid (GenBank accession number AA869045) as a template. A corresponding pAS2–1 plasmid for the mouse profilin II was also made by a PCR with primers cagaattcagggcggcatggccggttggcagagctac and gactcgagctagaacccagagtctctcaag using a mouse EST clone 464995 plasmid (GenBank accession number AA032658) as a template.
Biosensor measurements. Surface plasmon resonance measurements using analytes and ligands were performed on a BIAcore 2000 (BIAcore AB, Uppsala, Sweden). Analytes for the measurements were prepared as glutathione S-transferase (GST) fusion proteins using the Escherichia coli expression vector pGEX-4T-2 (Amersham Biosciences, Arlington, IL). The coding region of the Delphilin PDZ domain was prepared by PCR with primers containing the sites for EcoRI or XhoI restriction endonucleases (cgagaattccggcaacctcgctgctg and gctctcgagtcacagagaatctgaatc, respectively) using the clone MB2 as the template. After digestion withEcoRI and XhoI, the PCR product was subcloned into the corresponding position of the pGEX-4T-2 multiple cloning site in frame. The entire coding region for mouse profilin II was obtained from the corresponding pAct2 plasmid, prepared for the yeast two-hybrid assay as an EcoRI–XhoI fragment, and subcloned into the pGEX-4T-2 in frame. E. coli DH5 or BL21 were transformed with the pGEX-4T-2 plasmids, and the soluble fraction of each GST fusion protein was affinity-purified with the glutathione Sepharose 4B (Amersham Biosciences) according to the manufacturer's protocol. Ligands for the measurements were prepared as synthetic peptides. The GluRδ1 and GluRδ2 C-terminal eight-residue peptides (DTSHGTSI and DPDRGTSI, respectively) were synthesized (Peptide Synthetizer PSSM-8; Shimadzu Corporation, Kyoto, Japan), purified by reversed-phase HPLC, and immobilized on a CM5 research grade sensor chip with amine coupling to a density of 472 and 496 resonance units (RUs), respectively. The Delphilin FH1 domain peptide (aa residues 563–627), with a cysteine residue elongated to the N terminus, was also synthesized, purified by HPLC, confirmed by amino acid sequencing (Peptide Sequencer PPSQ-21, Shimadzu Corp.) and immobilized on a CM5 research grade sensor chip with ligand thiol coupling to a density of 1402 RUs. The sensor chip was equilibrated with running buffer [10 mm HEPES, pH 7.4, 150 mmNaCl, 3 mm EDTA, 0.005% (v/v) Surfactant P20]. GST and GST fusion proteins were diluted in the running buffer at different concentrations and injected at a 20 μl/min flow rate. The surface regeneration was performed by 20 μl injections of 10 mm NaOH at a 20 μl/min flow rate. All experiments were performed at 25°C. Data were analyzed with the BIAevaluation program 3.0 (BIAcore).
Construction of expression vectors, transfection into human embryonic kidney 293T cells, and immunoprecipitation. The 3.1 kbNaeI–EcoRI fragment of pKF3-MB2, which contains the entire coding region of Delphilin, and the 5.4 kbKpnI–EcoRI fragment of pcDNA3.1-Myc-His (Invitrogen) were ligated to yield pcDNA3.1-Delph-Myc-His. The 0.1 kbEcoRI–SmaI fragment of pZeo-SV2-MB2 fused to hemagglutinin (HA)-tag sequence by PCR using primers 5′-CTGCAGCGAGAGGCCATG GAGG-3′ and 5′-TTTCCCGGGTTAAGCGTAGTCTGGGACGTCGTATGGGTACCAGGCCAGGGGTGACACCAT-3′ was inserted into the 8.4 kb EcoRI–PmeI fragment of pcDNA3-Delph-Myc-His to yield pcDNA3-Delph-HA, the expression vector for C-terminally HA epitope-tagged Delphilin. pFLAG-δ2 (expression vector for amino-terminally FLAG-tagged GluRδ2) and pFLAG-δ2Δ1 (expression vector for amino-terminally FLAG-tagged GluRδ2Δ1) were constructed previously (Matsuda and Mishina, 2000).
Human embryonic kidney (HEK)293T cells were maintained in DMEM supplemented with 10% fetal calf serum, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C under 5% CO2. HEK293T cells (2 × 106 cells per 10 cm dish) were transfected with pcDNA3-Delph-HA and either pFLAG-δ2 or pFLAG-δ2Δ1 by the standard calcium phosphate method. Immunoprecipitation studies were performed essentially as described (Hironaka et al., 2000). Two days after transfection, cells were lysed in TNE buffer (50 mm Tris-HCl, pH 7.5, 1% Nonidet P-40, 5 mm EDTA, 145 mm NaCl) containing 100 μm phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 1 μg/ml leupetin, and 1 μg/ml pepstatin A. Insoluble fraction was excluded by centrifugation at 15,000 rpm for 10 min, and the supernatants were precleared by protein G Sepharose 4 Fast Flow (Amersham Biosciences) for 1 hr at 4°C. The precleared lysates were used for coimmunoprecipitation by incubation with anti-FLAG M2 antibodies (Sigma, St. Louis, MO), rabbit anti-HA antibodies (Santa Cruz Biotechnology, Santa Cruz, CA), normal mouse IgG or normal rabbit IgG (Santa Cruz Biotechnology) for 1 hr at 4°C. The complexes were precipitated by incubation with protein G Sepharose for 1 hr at 4°C. The immunoprecipitates were washed six times with TNE buffer, eluted with sample buffer. Proteins in the cell lysates and immunoprecipitates were separated on SDS-polyacrylamide gels and transferred to nitrocellulose membranes. After blocking with 5% skim milk in TBST (0.1% Tween 20 in Tris-buffered saline, pH 7.6), the blots were probed with anti-FLAG antibody or anti-HA antibody in TBST for 1 hr. The membranes were then washed with TBST and incubated with horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG (Amersham Biosciences) for 1 hr. The proteins were imaged with an ECL chemoluminescence detection system (Amersham Biosciences).
In situ hybridization. Under pentobarbital anesthesia at a lethal dose, brains of the adult C57BL mouse were freshly removed from the skull and frozen in powdered dry ice. Fresh-frozen sections were prepared and mounted on glass slides precoated with 3-aminopropyltriethoxisilane (Sigma). Two nonoverlapping antisense oligonucleotides were synthesized as a hybridization probe. The sequence was ACGGCACAGCTTGCCCAGCAGACCCTGCGACACCACCTCGTCGAA (complementary to nucleotide residues 100–144) and GCTGCTGGCTGAAGGTCCTCCCCTGGACTCGGATACTGGAAGGCA (624–668). They were labeled with 35S-dATP to a specific activity of 0.5 × 109 dpm/μg DNA, using terminal deoxyribonucleotidyl transferase (BRL, Bethesda, MD). Procedures for in situ hybridization have been reported previously (Watanabe et al., 1993). Sections were exposed to x-ray film (Hyperfilm-β max; Amersham Biosciences) for 2 weeks, or to nuclear track emulsion (NTB-2, Kodak, Rochester, NY) for 1 month.
Antibodies. Rabbit anti-GluRδ2 antibody has been described previously (Araki et al., 1993). Guinea pig anti-mouse Delphilin antibody was produced against aa residues 8–227. The antigen polypeptide was expressed as a GST fusion protein, using pGEX-4T-2 plasmid vector (Amersham Biosciences, Uppsala, Sweden) and E. coli BL21. After removal of the GST-tag with thrombin, antigen polypeptide (100 μg) emulsified with Freund's complete adjuvant (Difco Laboratories, Detroit, MI) was injected subcutaneously into female guinea pigs at intervals of 2 weeks. From antisera sampled 2 weeks after the sixth injection, immunoglobulins specific to Delphilin were affinity purified using the fusion protein coupled to cyanogen bromide-activated Sepharose 4B (Amersham Biosciences).
Synaptosomal plasma membrane preparation, immunoprecipitation, and Western blotting. Synaptosomal plasma membranes were prepared essentially according to the procedures described previously (Blackstone et al., 1992; Lau et al., 1996). All procedures were performed at 4°C, and all buffers included protease inhibitors of 1 mm methanesulfonyl fluoride hydrochloride, 1 μg/ml pepstatin A, 2 μg/ml aprotinin, and 0.5 μg/ml leupeptin. Briefly, adult BALB/c mouse brain tissue was homogenized in HEPES-buffered 0.32 m sucrose (0.32m sucrose, 4 mm HEPES, pH 7.4), and after removal of the nuclear fraction the homogenate was centrifuged at 9000 × g for 15 min at 4°C. The supernatant was kept as the soluble fraction. The pellet was washed once with HEPES-buffered 0.32 m sucrose and then lysed by hypo-osmotic shock in water. The resultant lysate was centrifuged at 25,000 × g for 20 min, and the pellet was suspended in HEPES-buffered 0.25 m sucrose. The suspension was then fractionated in a discontinuous gradient containing 0.8 m/1.0 m/1.2m sucrose. The resulting 1.0m/1.2 m sucrose interface was collected as the synaptosomal plasma membrane fraction. Immunoprecipitation studies were performed as described by Lau et al. (1996), with minor modifications. In brief, 300 μg of synaptosomal plasma membrane was first solubilized in 2% SDS in immunoprecipitation buffer (150 mm NaCl, 20 mmHEPES, pH 7.4, 5 mm EDTA, 5 mm EGTA, and protease inhibitors described above) for 10 min at room temperature and then diluted with 5 vol of cold 2% Triton X-100 in immunoprecipitation buffer. The solubilized membranes were then incubated with protein A-Sepharose (Amersham Biosciences) and 5 μl of the anti-GluRδ2 antiserum or preimmune serum for 3 hr at 10°C. The reaction mixtures were washed once with 1% Triton X-100, twice with 1% Triton X-100 containing 500 mmNaCl, and finally three times with immunoprecipitation buffer. The proteins were eluted from protein A-Sepharose by Laemmli sample buffer, separated by SDS-PAGE, and transferred to a polyvinylidene difluoride membrane. The membrane was probed with guinea pig anti-Delphilin antiserum. An HRP-conjugated secondary antibody and ECL chemoluminescence (Amersham Biosciences) were used for visualization of the signals.
Immunohistochemistry for brain sections. Using adult mouse brains (2 months of age) fixed transcardially with 4% paraformaldehyde in 0.1 m sodium phosphate buffer, pH 7.2, paraffin (5 μm) and microslicer (50 μm) sections were prepared for immunoperoxidase or immunofluorescence. Before immunohistochemical incubations, all sections were subjected to protease pretreatment, because we found that this method was very effective for immunohistochemical detection for Delphilin and GluRδ2 subunit as well as for NMDA receptor subunits (Watanabe et al., 1998). Briefly, both paraffin and microslicer sections on glass slides were dipped in 1 mg/ml of pepsin (Dako, Carpinteria, CA)/0.2N HCl at 37°C for 10 min. Without the pepsin treatment, no immunoreactivity appeared at the antibody concentration adopted in the present study. For immunoperoxidase, sections were incubated successively with the primary antibody (0.3–0.5 μg/ml) overnight, biotinylated secondary antibodies for 2 hr, and avidin–biotin–peroxidase complex for 30 min, using a Histofine SAB-PO kit (Nichirei Corporation, Tokyo, Japan). The immunoreaction was visualized with 3,3′-diaminobenzidine. For immunofluorescence, microslicer sections were incubated with a mixture of guinea pig anti-Delphilin antibody and rabbit anti-GluRδ2 antibody (1 μg/ml for each). Immunoreaction was visualized by a 2 hr incubation with Cy3- or FITC-labeled secondary antibody (1:200) (Jackson ImmunoResearch, West Grove, PA).
Electron microscope immunocytochemistry using post-embedding immunogold. For post-embedding immunogold analysis, adult mice (2 months of age) were perfused transcardially with 4% paraformaldehyde/0.1% glutaraldehyde in 0.1 mphosphate buffer (PB), pH 7.2. Parasagittal cerebellar sections (400 μm in thickness) were prepared on a microslicer (VT1000S; Leica, Nussloch, Germany), cryoprotected with 30% glycerol in 0.1m PB, and frozen rapidly with liquid propane in a Leica EM CPC unit (Vienna, Austria). Frozen sections were immersed in 0.5% uranyl acetate in methanol at −90°C in a Leica AFS freeze-substitution unit (Vienna, Austria), infiltrated at −45°C with Lowicryl HM-20 resin (Lowi, Waldkraiburg, Germany), and polymerized with UV light. Ultrathin sections were mounted on nickel grids precoated with neoprene W (Nisshin EM, Tokyo, Japan). After etching with saturated sodium ethanolate solution for 3 sec, they were treated successively with 1% human serum albumin (Wako, Osaka, Japan) in TBS (HTBS), pH 7.5, for 1 hr, guinea pig anti-Delphilin antibody (10 μg/ml) in HTBS for 3 hr, and colloidal gold (10 nm)-conjugated anti-guinea pig IgG (1:100; British Bio Cell International, Cardiff, UK) in HTBS for 1 hr. For double immunogold labeling, grids were incubated first with a mixture of guinea pig anti-Delphilin antibody and rabbit anti-GluRδ2 antibody (10 μg/ml for each), and then with a mixture of colloidal gold (15 nm)-conjugated anti-guinea pig IgG and colloidal gold (10 nm)-conjugated anti-rabbit IgG (1:100 for each; British Bio Cell International). Finally, grids were stained with uranyl acetate for 15 min and examined with an H-7100 electron microscope (Hitachi, Tokyo, Japan). For quantitative analysis, immunogold particles at Purkinje cell synapses were counted on electron micrographs. In addition, subsynaptic distribution was examined in the perpendicular and tangential axes of Purkinje cell synapses, as reported previously (Landsend et al., 1997). Using images digitized on a scanner and NIH Image software (Version 1.61), perpendicular synaptic localization of gold particles was evaluated by measuring the distance between the midline of the synaptic cleft and the center of each gold particle. The tangential synaptic localization was evaluated by measuring the distances from the center of postsynaptic density to the center of each gold particle and to the edge of the postsynaptic density.
RESULTS
Isolation of Delphilin, which binds the cytoplasmic tail of the GluRδ2 subunit and contains a PDZ domain and formin homology domains
Various studies to demonstrate ion channel activity or ligands for the GluRδ2 subunit have been performed, but without success. We speculated that identification of proteins interacting with the cytoplasmic region of the subunit might provide good clues to understanding its physiological functions. We screened a mouse brain cDNA library by the yeast two-hybrid system, using the C-terminal 45 aa residues of the subunit as bait. From 2 × 106 independent clones, 14 clones were finally graduated from the verification experiments described in Materials and Methods. The nucleotide sequence of the longest clone (designated clone MB2) among four overlapping ones was determined. A BLAST search against the databases demonstrated that the clone could encode 350 aa residues of a novel sequence containing a PDZ domain resembling those of PSD-95/SAP-90 family proteins of membrane-associated guanylate kinases, such as PSD-95 or Chapsyn110/PSD-93. The insert of the clone MB2 was used as a probe for conventional screening of a phage cDNA library made of adult mouse cerebellum. Nucleotide sequencing analyses of two overlapping clones (designated clone 10-1 and 9-2) and the cap site cDNA clone obtained by the cap site hunting method (Murata and Yamaguchi, 1999) together disclosed a cDNA sequence of a 3495 base pair (Fig.1A). The nucleotide sequence around an ATG at nucleotide 55 corresponded well to Kozak's consensus sequence for translation initiation (Kozak, 1987) and was followed by a long open reading frame for 1024 aa residues (Fig.1B). Although the nucleotide sequence upstream of the ATG does not contain in-frame stop codons, we considered it as the translation initiation codon because of our analysis of its transcriptional start site by the cap site hunting method (see details in Materials and Methods). The putative 3′ untranslated region has an AATAAA polyadenylation signal followed by a poly(A) tail at 50 bp downstream from the signal. The encoded protein was designated as Delphilin (for Delta2- philic-prote in) with a predicted molecular weight of 112,577. The nucleotide sequence of Delphilin cDNA has been deposited in GenBank under the accession number AF099933. The predicted PDZ domain of Delphilin is located from aa residue 88 to residue 163 (Figs.1B,C). The PDZ domain of Delphilin was most homologous to that of regulator of G-protein signaling 12 (Snow et al., 1997), with 36% identity (Fig.2A). Roche et al. (1999) reported that the PDZ domains of PSD-93/Chapsyn-110 or PSD-95 bind to the C terminus of GluRδ2, and the PDZ domains -1, -2, and -3 of rat PSD-93 (Brenman et al., 1996) (GenBank accession number U50717) showed 28, 24, and 24% identity to that of Delphilin, respectively (Fig. 2A). The Delphilin PDZ domain is considered a class I PDZ domain (Daniels et al., 1998) because it has a polar residue histidine as the first residue of the α-helix B (Fig.2A). In addition to a PDZ domain, BLAST searches further identified a region in Delphilin showing homology to proteins containing the FH domain, such as formins (Woychik et al., 1990;Jackson-Grusby et al., 1992), Diaphanous (Castrillon and Wasserman, 1994), or Bni1p (Evangelista et al., 1997) (for review, see Frazier and Field, 1997). By multiple alignment with amino acid sequences of these proteins, residues 777–907 of Delphilin was recognized as FH2, first described by Castrillon and Wasserman (1994) (Fig.2B). There was a proline-rich region of 63 aa residues at 151 residues N-terminal to the FH2 domain (Figs.1C, 2C). This region was considered to be FH1, usually accompanied by the FH2 domain. Although most FH domains are known to have two independent coiled-coil structures in the regions flanking the FH1 and FH2 domains, Delphilin demonstrated a single coiled-coil only at the C-terminal to the FH2 domain. A recently identified FH3 domain (Petersen et al., 1998) was not found in Delphilin. The schematic domain structure of Delphilin is presented in Figure 1C. In addition to the coiled-coil structure, Delphilin has a cysteine residue at the N-terminal third aa position as another possible tool for homomultimerization. The surrounding sequence of the cysteine residue resembles that of a palmitoylated cysteine residue of Gαs protein (Milligan et al., 1995). In the case of PSD-95, cysteine residues at the third and fifth aa positions are reported to be involved in palmitoylation (Topinka and Bredt, 1998) or disulfide bond formation (Hsueh et al., 1997; Hsueh and Sheng, 1999). These modifications may be required for cell membrane association or head-to-head homomultimerization of PSD-95. The cysteine residue of Delphilin is a potential candidate for palmitoylation or may form a disulfide bond for homodimerization.
Fig. 1.
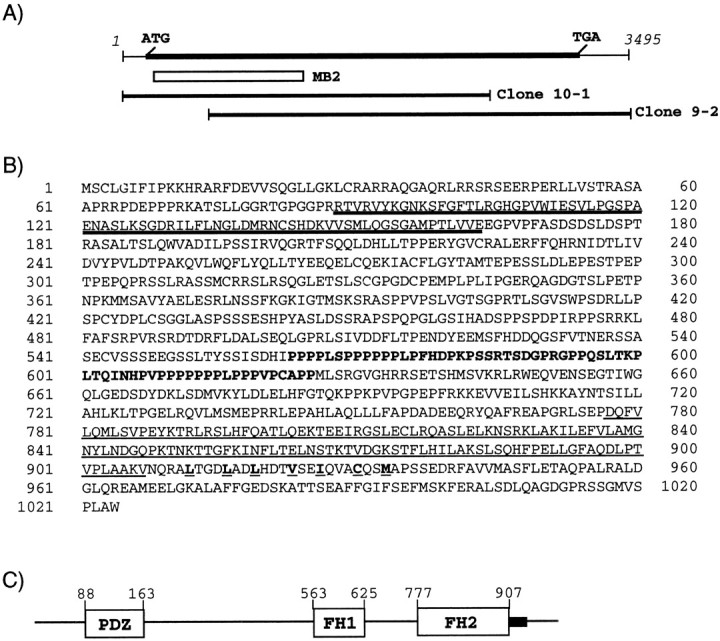
Schematic presentations of the cDNA cloning, deduced amino acid sequence, and domain structure of Delphilin.A, The Delphilin cDNA is presented at thetop with a putative protein coding region as aheavy line associated with start (ATG) and termination (TGA) codons. The open box below designates the location of the clone MB2 [corresponding to nucleotide (nt) 76–1125], the original Delphilin clone obtained by yeast two-hybrid library screening. The locations of the clone 10-1 (nt 1–2526) and clone 9-2 (nt 426–3495), which were cloned from conventional screening of a phage cDNA library, are also presented as horizontal bars, respectively. B, Deduced amino acid sequence of Delphilin. The putative PDZ domain is marked bydouble underlining. Amino acid residues of the putative FH1 domain are in boldface, and the putative FH2 domain is underlined. Hydrophobic residues involved in the coiled-coil structure after the FH2 domain are boldfacewith underlining. C, A schematic presentation of the Delphilin domain structure. Each domain is shown as an open box with amino acid positions of the N and C terminals of the domain. The coiled-coil structure is presented as ablack box after the FH2 domain.
Fig. 2.
The PDZ and FH domains of Delphilin.A, Alignment of the PDZ domain of Delphilin with the known PDZ domains. The secondary structure elements of six β sheets and two α helices are boxed and indicated as βA–F and αA and αB, respectively, according to Doyle et al. (1996). The sequences used for the alignment are as follows: PDZ domain ofDelphilin (residues 88–165), RGS12(residues 20–97 from rat regulator of G-protein signaling 12),93PDZ-1, -2, and -3(residues 97–182, 192–297, and 420–501 from rat PSD-93 corresponding to the PDZ-1, -2, and -3 domains, respectively), and95PDZ-1 and -3 (residues 64–151 and 312–393 from mouse PSD-95 corresponding to the PDZ-1 and PDZ-3, respectively). Histidine residues at the first position of the α-helix B are boldface. B, Alignment of selected FH2 domains from Diaphanous (Castrillon and Wasserman, 1994), Bni1p (Sen-Gupta et al., 1996), mouseForminVI (Jackson-Grusby et al., 1992), cell division control protein 12 (CCP12) (Chang et al., 1997), and cell fusion protein (FUS1) (Petersen et al., 1995) with the putative FH2 domain of Delphilin. Amino acid residues conserved in more than three proteins are shown inboldface. Dots at the topof the alignment demonstrate invariant residues. C, FH1 domain of Delphilin. Prolines are shown in boldface, and two PPLP motifs are boxed.
Specific interaction of Delphilin with GluRδ2 protein at its C terminus through the unique PDZ domain
The clone MB2, the original clone picked up by our yeast two-hybrid screening, contains the single PDZ domain of Delphilin. Because a PDZ domain is a binding motif generally recognizing the C terminus of its target proteins (Saras and Heldin, 1996), we assayed Delphilin interaction with various C-terminal mutants of the GluRδ2 subunit by using the clone MB2. The results of yeast two-hybrid studies are summarized in Table 1. All of the GluRδ2 mutants lacking their C-terminal -GTSI, -TSI, -SI or -I residues completely lost the ability to activate reporter genes, HIS3 and β-galactosidase. A mutant, which has an alanine as the C-terminal residue replaced for a isoleucine, also completely lost the interaction in yeast cells. These results indicate that the PDZ domain of Delphilin is the binding motif for the C terminus of GluRδ2. Although the GluRδ1 subunit, the other subunit of the GluRδ subfamily, has four C-terminal residues GTSI-COOH completely identical to those of GluRδ2, the Delphilin PDZ appeared not to interact with the GluRδ1 C terminus (Table 1).
Table 1.
Interaction between Delphilin and wild type or mutants of the δ family subunit C-terminal 45-residue peptide
| C terminal | HIS selection | β-Galactosidase |
|---|---|---|
| δ2 subunit | ||
| DPDRGTSI-COOH (wt) | + | + |
| DPDR-COOH | − | − |
| DPDRG-COOH | − | − |
| DPDRGT-COOH | − | − |
| DPDRGTS-COOH | − | − |
| DPDRGTSA-COOH | − | − |
| δ1 subunit | ||
| DTSHGTSI-COOH (wt) | − | − |
Yeast two-hybrid experiments using Delphilin clone MB2 as a bait and various constructs related to δ family subunit C-terminal 45 residues as preys (see Materials and Methods) were performed. + indicates that yeast growth on the plate without histidine (HIS selection assay) or blue color (β-galactosidase assay) was observed; − indicates failure to detect them. wt, Wild type.
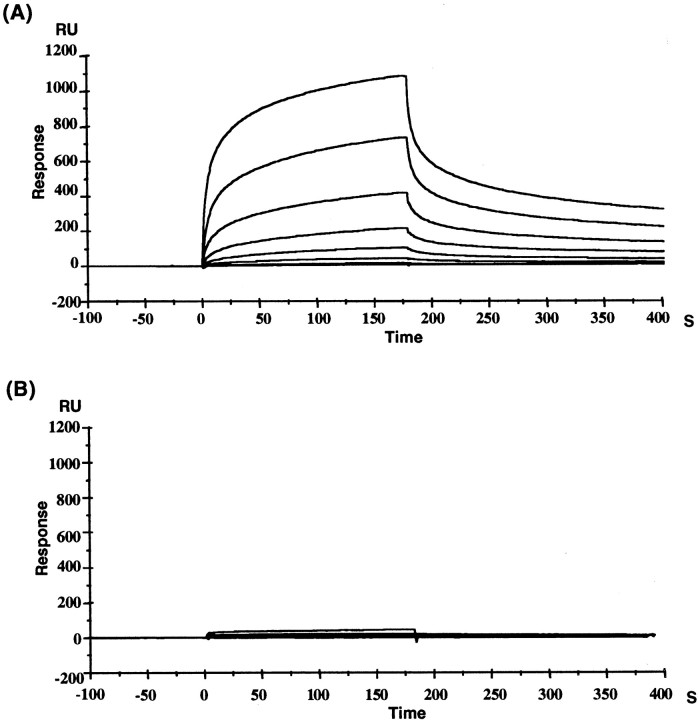
We further confirmed this by surface plasmon resonance analyses, which effectively monitor interactions between molecules in real time. In these experiments, a solution of a GST fusion protein of Delphilin PDZ was passed over two flow cells: one immobilized with the GluR δ2 C-terminal eight-residue peptide (DPDRGTSI) and one with the GluRδ1 corresponding peptide (DTSHGTSI). Binding of the control GST protein itself to the GluRδ2 peptide or to the GluRδ1 peptide was not detected (data not shown). The Delphilin PDZ exhibited a specific binding to the GluR δ2 C-terminal peptide but showed little, if any, binding to the GluRδ1 C-terminal peptide. From the data in Figure3, the dissociation constant (KD) value of the Delphilin PDZ for the GluR δ2 C-terminal peptide was calculated to be 22.6 μm (ka = 3.2 × 103m−1sec−1 andkd = 7.25 × 10−2sec−1). The binding of Delphilin PDZ to the GluRδ2 peptide analog lacking C-terminal isoleucine or having an alanine replacing an isoleucine as the C-terminal residue was not detected (data not shown), which is consistent with the results (Table 1) in yeast two-hybrid analyses. From these results, it appears that the PDZ domain of Delphilin interacts directly with the C terminus of GluRδ2 and that amino acids −6 to −4 of the C terminus may critically contribute to the binding.
Fig. 3.
Surface plasmon resonance analyses of the interaction between the Delphilin PDZ domain and C-terminal eight-residue peptides of the GluRδ family subunits. The binding of the Delphilin PDZ domain fused with GST, at different concentrations (0.025–3.2 μm), to the GluRδ2 C-terminal peptide (A) or to the GluRδ1 corresponding peptide (B) immobilized on the surface of flow cells on a sensor chip was detected by changes in RUs over time. The sensorgrams shown were obtained after the blank sensorgram of the control nonimmobilized flow cell was subtracted from the sensorgrams of the peptide-immobilized cells.
Interaction of Delphilin with GluRδ2 in HEK293T cells
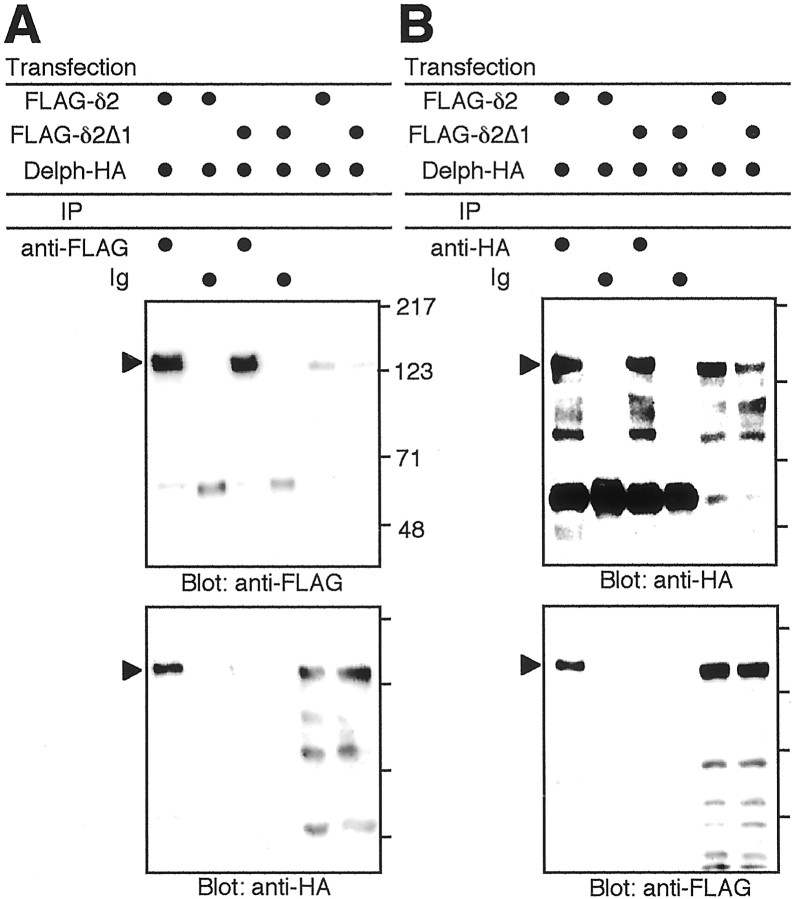
We examined the interaction of Delphilin and GluRδ2 proteins using a heterologous expression system. Immunofluorescence staining showed that HA-tagged Delphilin expressed in HEK293T cells distributed both on the cell surface and in the cytoplasm (data not shown). The distribution of FLAG-tagged GluRδ2 in HEK293T cells was similar to that of HA-tagged Delphilin. We observed no significant effects of coexpression on the intracellular distribution of the two proteins. Because the two proteins colocalized on the cell surface when expressed together, we examined the interaction of Delphilin and GluRδ2 proteins by immunoprecipitation. As shown in Figure4, immunoprecipitates from cell lysates with anti-HA antibody contained FLAG-tagged GluRδ2 proteins as well as HA-tagged Delphilin proteins (Fig. 4B). None of these proteins were present in immunoprecipitates with nonimmune control antibody. Consistently, immunoprecipitates from cell lysates with anti-FLAG antibody contained HA-tagged Delphilin proteins as well as FLAG-tagged GluRδ2 proteins (Fig. 4A). These results showed that Delphilin and GluRδ2 proteins interacted with each other in HEK293T cells.
Fig. 4.
Interaction of Delphilin with GluRδ2 in HEK293T cells. HA-tagged Delphilin (Delph-HA) together with FLAG-tagged GluRδ2 (FLAG-δ2) or FLAG-tagged GluRδ2Δ1 (FLAG-δ2Δ1) was expressed in HEK293T cells by transfection of respective expression vectors. A, Lysates of the transfected cells were immunoprecipitated with anti-FLAG antibody (anti-FLAG) or nonimmune control IgG (Ig). The immunoprecipitates were electrophoresed and immunoblotted with anti-FLAG (top panel) or anti-HA (bottom panel) antibodies. B, Lysates of the transfected cells were immunoprecipitated with anti-HA antibody or nonimmune control IgG. The immunoprecipitates were electrophoresed and immunoblotted with anti-HA (top panel) or anti-FLAG (bottom panel) antibodies. The input lanes (right two lanes of each panel) represent ∼1% of the cell lysates used for the immunoprecipitation experiments. The positions of precipitated HA-tagged Delphilin [bottom (A) and top (B)panels] and FLAG-tagged GluRδ2 [top(A) and bottom(B) panels] proteins are indicated by arrowheads. Positions of molecular weight markers are also indicated in kilodaltons on the right side of the top panel of A. In other panels, only corresponding bars are shown.IP, Immunoprecipitation.
Deletion of the C-terminal seven amino acids of GluRδ2 did not affect the distribution pattern in HEK293T cells, as reported in Madin-Darby canine kidney (MDCK) cells (Matsuda and Mishina, 2000). FLAG-tagged GluRδ2Δ1 (GluRδ2 mutant lacking C-terminal seven amino acids) proteins similarly distributed on the cell surface and in the cytoplasm when expressed together with HA-tagged Delphilin. However, immunoprecipitates from cell lysates with anti-HA antibody contained HA-tagged Delphilin proteins but no FLAG-tagged GluRδ2Δ1 proteins (Fig. 4B). Consistently, immunoprecipitates from cell lysates with anti-FLAG antibody contained FLAG-tagged GluRδ2Δ1 proteins but no HA-tagged Delphilin proteins (Fig.4A). Thus, the C-terminal seven amino acids of GluRδ2 were indispensable for the interaction of Delphilin and GluRδ2 proteins in HEK293T cells.
Possible interaction of the Delphilin FH1 domain with profilins I and II, and with the SH3 domain of neuronal type Src
In addition to a PDZ domain, Delphilin harbors an FH1 domain followed by an FH2 domain and a coiled-coil structure. Although the functions of FH2 domains have not yet been elucidated, proline-rich sequences of the FH1 domain are known as binding sites for the WW/WWP domain, the SH3 domain, or profilins that function in actine assembly (for review, see Frazier and Field, 1997). There is a possibility that Delphilin has some physiological function via this domain in addition to the interaction with the GluRδ2 subunit through the PDZ domain. Using the yeast two-hybrid system, we tested whether the Delphilin FH1 domain interacts with profilins I and II and with the SH3 domains of PSD-93 and n-Src (Table2). N-Src has been reported to be expressed in Purkinje cells (Sugrue et al., 1990). We first performed the following control experiments. The plasmid expressing the Delphilin FH1 domain fused to the Gal4 DNA-binding domain did not activate reporter genes by itself or by coexpression with the plasmid expressing only the Gal4 transcription activation domain. However, the Delphilin FH1-expressing plasmid specifically activated the reporter genes when it was cotransfected with the plasmids expressing profilins I and II or the n-Src SH3 domain fused to Gal4 transcription activation domain, indicating that the Delphilin FH1 domain interacted with profilins I, II, and the n-Src SH3 domain in yeast cells.
Table 2.
Binding profile of Delphilin FH1 domain to profilins and SH3 domains of PSD-93 and neuronal Src
| HIS selection | β-Galactosidase | |
|---|---|---|
| Profilin-I | + | + |
| Profilin-II | + | + |
| PSD-93 SH3 | − | − |
| N-Src SH3 | + | + |
Yeast two-hybrid experiments using Delphilin FH1 domain as a bait and profilin-1, profilin-2, and SH3 domains of PSD-93 and n-Src as preys (see Materials and Methods) were performed. See Table 1 legend for + and −. wt, Wild type.
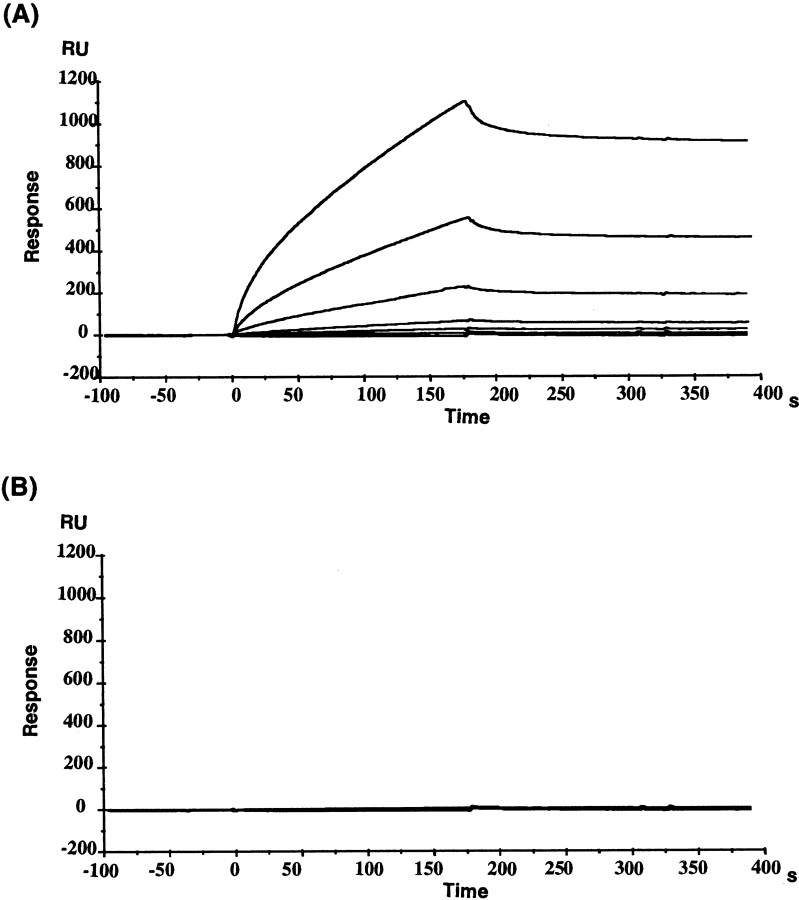
We further confirmed the interaction of the Delphilin FH1 domain with profilin II, which has been reported to be expressed at high levels only in the brain (Witke et al., 2001), using surface plasmon resonance analyses to measure molecular interaction directly in real time (Fig.5). In these experiments, a solution of GST fusion protein of mouse profilin II was passed over the flow cell immobilized with the Delphilin FH1 domain peptide (Fig. 5A). Binding of the control GST protein itself to the peptide was not detected (Fig. 5B). From the data in Figure 5A, the dissociation constant (KD) value of the profilin II protein for the Delphilin FH1 was calculated to be 39.5 nm (ka = 1.04 × 104m−1sec−1 andkd = 4.09 × 10−4sec−1). From these results, it appears that the FH1 domain of Delphilin interacts directly and specifically with profilin II.
Fig. 5.
Surface plasmon resonance analyses of the interaction between the Delphilin FH1 domain and profilin II. The binding of the GST fusion protein of mouse profilin II (A), or the control GST protein itself (B) at different concentrations (0.001–1.0 μm), to the Delphilin FH1 domain peptide immobilized on the surface of a flow cell on a sensor chip was detected by changes in RUs over time. The sensorgrams shown were obtained after the blank sensorgram of the control cystein-immobilized flow cell was subtracted from the sensorgrams of the peptide-immobilized cells.
Tissue distribution of the Delphilin gene transcripts and in situ hybridization analysis for brain sections
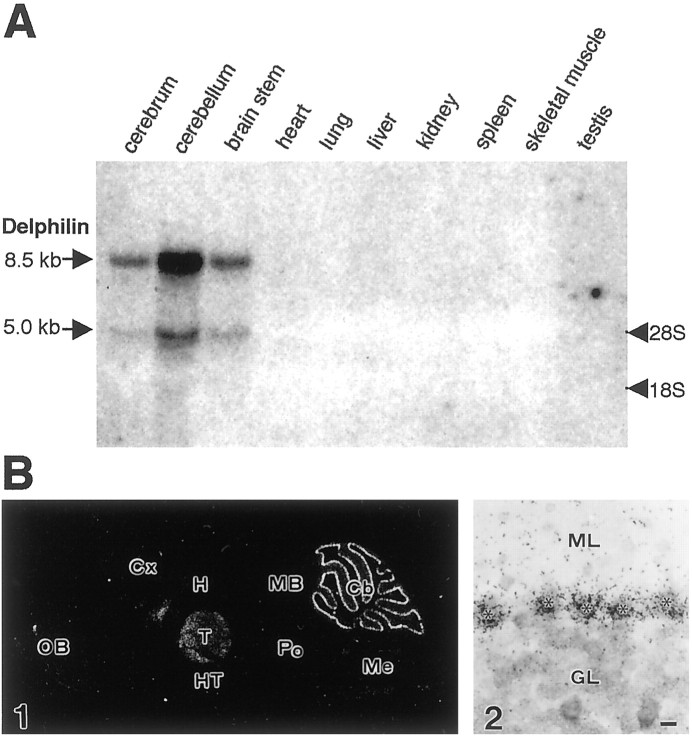
Northern blot analysis for 8-week-old mouse tissues demonstrated a major (∼8.5 kb) and a minor (∼5.0 kb) transcript of Delphilin in the cerebellum (Fig.6A). Weak but detectable expression was also observed in the cerebrum and the brainstem with both major and minor bands. Other organs including heart, lung, liver, kidney, spleen, skeletal muscle, and testis did not show any detectable signals even after a long exposure. To further localize the Delphilin gene transcripts in the brain, we performedin situ hybridization analysis. Using35S-labeled antisense oligonucleotide probe, distinct distribution of the Delphilin mRNA was revealed in the adult mouse brain (Fig. 6B1). The highest expression was detected in the cerebellum, where the cell bodies of Purkinje cells were strongly labeled (Fig. 6B2). Moderate levels were noted in various nuclei of the thalamus (Fig.6B1). A longer exposure showed low transcription levels in several other regions: the olfactory granular layer, the lateral septal nuclei, and the brainstem motor nuclei, such as the facial nucleus, the hypoglossal nucleus, and the ambiguus nucleus (data not shown). These results correlated well with Northern blot analysis.
Fig. 6.
mRNA expression of Delphilin. A, Northern blot analysis of Delphilin gene transcripts. Each 20 μg of total RNA, extracted from the various mouse tissues designated above the panel, was subjected to Northern analysis. Staining of the gel with ethidium bromide after electrophoresis revealed the integrity and an equally applied amount of each RNA (data not shown). The locations of 28S and 18S ribosomal RNAs were indicated on the right side of the panel and used as size markers.B, In situ hybridization of Delphilin mRNA in the adult mouse brain. B1 demonstrates a negative image of an x-ray film autoradiogram. B2 is a bright-field micrograph of emulsion-dipped cerebellar cortex. Note that Delphilin mRNA is concentrated in the cell body of cerebellar Purkinje cells (asterisks). Cb, Cerebellum;Cx, cerebral cortex; GL, granular layer;H, hippocampus; HT, hypothalamus;MB, midbrain; Me, medulla oblongata;ML, molecular layer; OB, olfactory bulb;Po, pons; T, thalamus. Scale bars:B1, 1 mm; B2, 10 μm.
Western blot analyses and coimmunoprecipitation of Delphilin with GluRδ2 protein from cerebellar synaptic membrane fractions
Polyclonal guinea pig antibody was raised against a partial Delphilin polypeptide (corresponding to aa residues 8–227), expressed in E. coli as a GST fusion protein. Western blot analysis with the antibody demonstrated a single ∼135 kDa band from cerebellar homogenate (Fig. 7A). An identical band was also obtained from homogenate prepared from COS-7 cells transfected with a Delphilin expression vector, which confirmed that the antibody actually recognizes Delphilin. The difference between the molecular size of Delphilin estimated from the data on the Western blotting and that calculated from the deduced amino acid sequence may be attributable to posttranslational modification of the protein, such as phosphorylation. The deduced Delphilin polypeptide sequence actually consists of multiple phosphorylation sites for protein kinase C and casein kinase II (data not shown). Delphilin protein in the cerebellum was highly enriched in the synaptosomal membrane fraction, but detectable immunoreactivity was also found in the soluble fraction. Samples from the cerebrum showed a very faint band of the same size as the cerebellar band in the synaptosomal membrane fraction (Fig. 7A). Like proteins localized at PSD such as PSD-95/SAP90, SAP102, or various GluR subunits, Delphilin in cerebellar synaptosomal membrane fraction was resistant to solubilization by the nondenaturing detergents Triton X-100 and 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate. A part of Delphilin was solubilized by 1% deoxycholate and totally solubilized by 2% SDS solution (Fig. 7B). Because the GluRδ2 subunit itself showed a similar solubility profile, Delphilin seemed to be localized at PSD together with GluRδ2. To confirm the colocalization of Delphilin and GluRδ2 in vivo, cerebellar synaptosomal membrane fraction was solubilized by 2% SDS and subjected to a coimmunoprecipitation experiment with a rabbit anti-GluRδ2 antiserum. Immunoprecipitation without the serum or with the preimmune serum did not precipitate Delphilin, but the anti-GluRδ2 antiserum coprecipitated Delphilin with a molecular size identical to that obtained in the Western blot analysis for a cerebellar synaptosomal membrane fraction (Fig.7C).
Fig. 7.
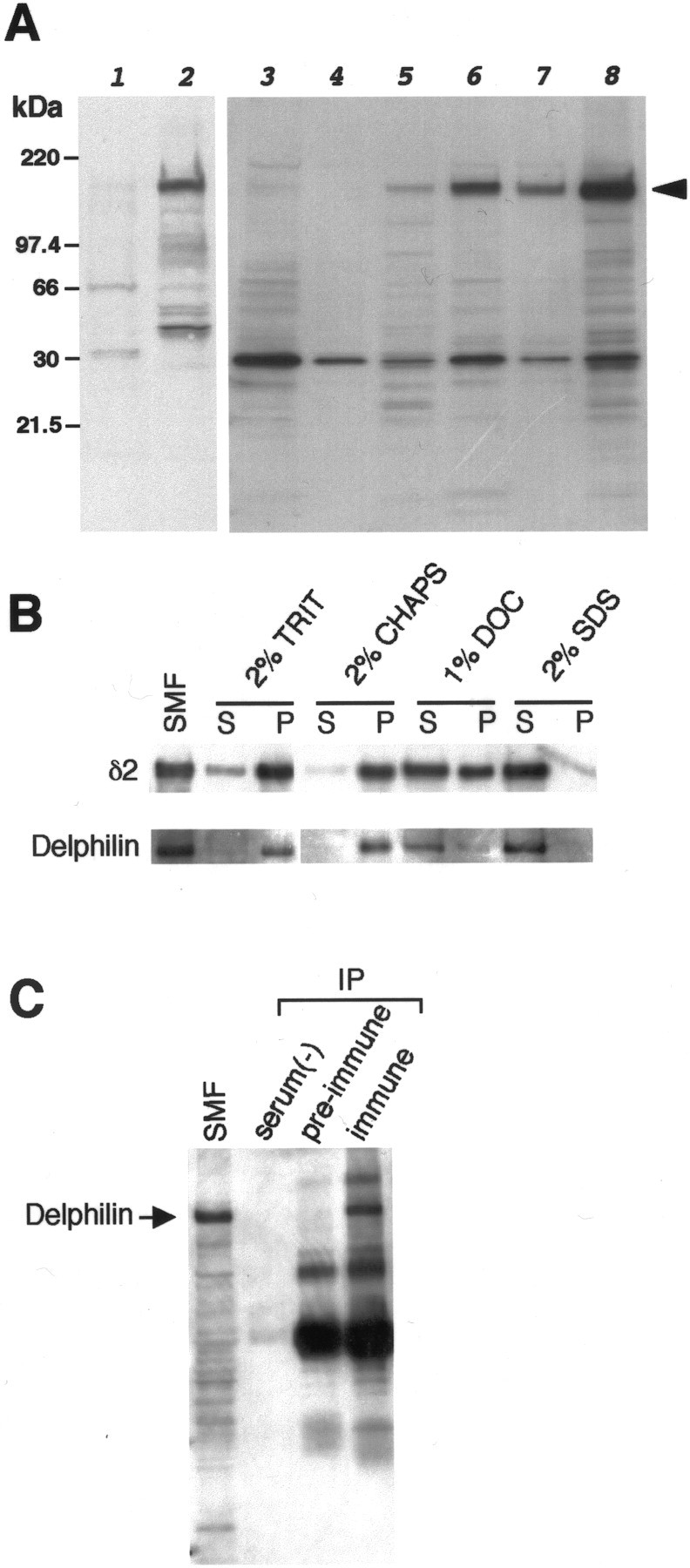
Identification and characterization of the Delphilin protein by Western blots. All blots were probed with guinea pig anti-Delphilin antibody or rabbit anti-GluRδ2 antibody.A, Delphilin expression in COS-7 cells and brain tissues. Five micrograms of each protein from transfected COS-7 cells with pZeoSV2 empty vector (lane 1) or pZeoSV2-Delphilin (lane 2) were analyzed on the left panel. Note the single 135 kDa band specific for the pZeoSV2-Delphilin transfectant. Ten micrograms of each cerebral (lanes 3,4, 5) or cerebellar (lanes 6, 7, 8) total homogenate, soluble fraction, or synaptosomal membrane fraction were also analyzed. Immunoreactive bands of the same size as the pZeoSV2-Delphilin transfectant were also identified in brain specimens except for the soluble fraction of cerebrum. B, Detergent solubility of Delphilin and GluRδ2. Ten micrograms of each cerebellar synaptosomal membrane were solubilized in the detergent presented at thetop of the panel, separated into soluble and insoluble fractions, and subjected to Western analyses.C, Immunoprecipitation of Delphilin from cerebellar synaptosomal membrane by anti-GluRδ2 antibody. Fifteen micrograms of synaptosomal membrane and each immunoprecipitate from 300 μg of membrane without serum [serum(−)] and with preimmune serum (pre-immune) or anti-GluRδ2 immune serum (immune) were analyzed by Western blotting with anti-Delphilin antibody. SMF, Synaptosomal membrane fraction; TRIT, Triton X-100; DOC, deoxycholate; S, supernatant; P, precipitate; IP, immunoprecipitation.
Colocalization of Delphilin with GluRδ2 protein in the cerebellum
The distribution of the Delphilin protein was examined by immunohistochemistry with the same polyclonal guinea pig antibody used in the Western analyses. Immunoperoxidase using parasagittal brain sections showed the highest immunoreactivity of Delphilin in the cerebellar molecular layer, with low levels in the thalamus (Fig.8A). The spatial patterns were consistent with those obtained by in situhybridization (Fig. 6B) and were also similar to those of the GluRδ2 subunit specific to Purkinje cells (Fig.8B). Double immunofluorescence with a confocal laser scanning microscope was adopted to compare their distribution in Purkinje cells (Fig. 8C–E). The Delphilin antibody labeled tiny punctate structures in the neuropil of the cerebellar molecular layer (Fig. 8C). In these structures, Delphilin was extensively colocalized with GluRδ2 (Fig.8D), yielding numerous yellowish puncta (Fig.8E). Weak Delphilin immunoreactivity was also detected inside the cell body and, very rarely, in stem dendrites of Purkinje cells. GluRδ2 was hardly detected at all in these two sites. These results strongly suggest that Delphilin is distributed in Purkinje cell synapses together with GluRδ2.
Fig. 8.
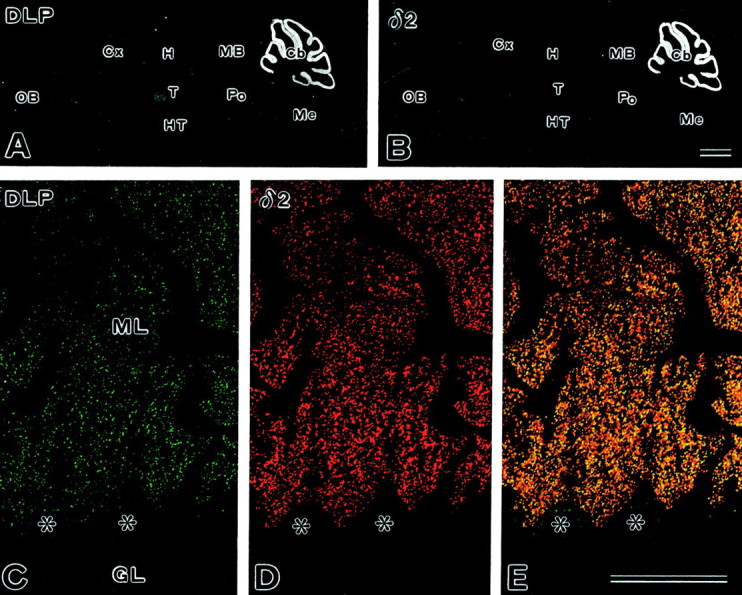
Immunohistochemistry of Delphilin on brain sections. A, B, Immunoperoxidase for Delphilin (A) and GluRδ2 (B). C–E, Double immunofluorescence for Delphilin (C) and GluRδ2 (D). E is a merged view ofC and D.Asterisks mark the cell bodies of Purkinje cells. Cb, Cerebellum;Cx, cerebral cortex; GL, granular layer;H, hippocampus; HT, hypothalamus;MB, midbrain; Me, medulla oblongata;ML, molecular layer; OB, olfactory bulb;Po, pons; T, thalamus. Scale bars:B, 1 mm; E, 50 μm.
Delphilin is selectively targeted to the postsynaptic site of parallel fiber-Purkinje cell synapses
Synaptic distribution and localization of Delphilin were examined by post-embedding immunogold in the molecular layer of the adult mouse cerebellum (Fig. 9). Immunogold labeling was concentrated at axospinous synapses in the molecular layer, suggesting the presence at the parallel fiber–Purkinje cell synapse or climbing fiber–Purkinje cell synapse, or both. Of the two synapses, Delphilin was detected at the parallel fiber synapse (Fig.9A), which was judged from a small terminal swelling (<1 μm in diameter) that usually forms synaptic contact with a single spine. Double labeling with different gold particle diameters showed that Delphilin was colocalized with GluRδ2 at individual parallel fiber–Purkinje cell synapses (Fig. 9B). In contrast, Delphilin was hardly detected at the climbing fiber synapse, which was judged by large terminal swelling that contains numerous synaptic vesicles and forms synaptic contact with multiple spines (Fig.9C). No specific labeling for Delphilin was found at axodendritic synapses, i.e., interneuron–Purkinje cell synapses. The distinct synaptic distribution of Delphilin was confirmed by counting gold particles (Fig. 9D). The parallel fiber synapse had 1.490 ± 1.070 particles per synapse (723 particles on 486 parallel fiber synapses), whereas the climbing fiber and interneuron synapses had 0.070 ± 0.256 particles per synapse (6 particles on 85 climbing fiber synapses) or 0.024 ± 0.154 particles per synapse (1 particle on 42 interneuron synapses), respectively. For the control experiment, we replaced anti-Delphilin antibody with normal guinea pig immunoglobulin, resulting in almost blank labeling. These results indicate that Delphilin is targeted selectively to Purkinje cell spines in contact with parallel fiber terminals, as is the case for GluRδ2 in the adult rat cerebellum (Landsend et al., 1997).
Fig. 9.
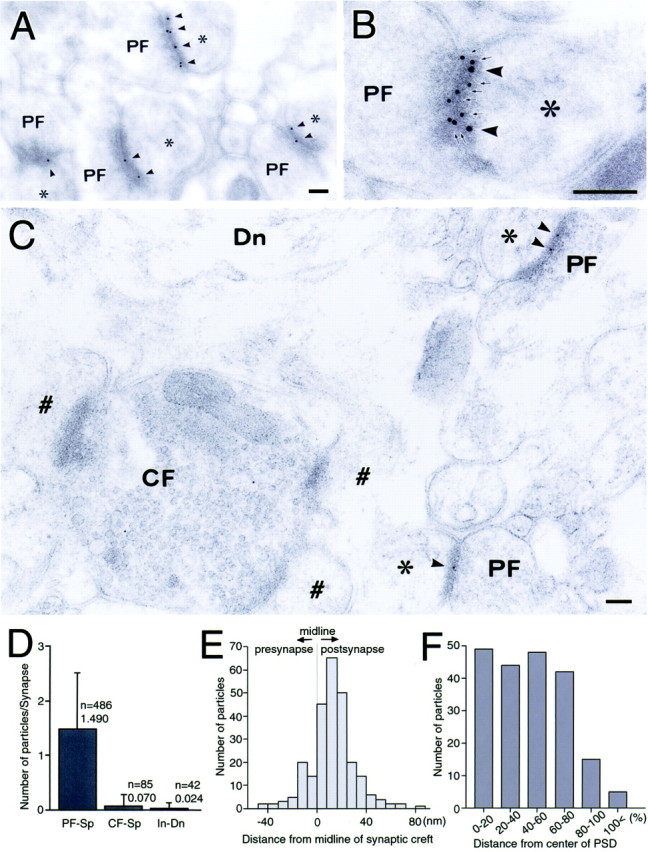
Ultrastructural localization of Delphilin (A–C, arrowheads) at Purkinje cell synapses. A, Delphilin labeling at parallel fiber synapses. Purkinje cell spines forming synaptic contact with parallel fiber terminals (PF) are indicated by asterisks. B, Colocalization of Delphilin (arrowheads) and GluRδ2 (arrows) at a PF synapse. C, Lack of Delphilin labeling at climbing fiber synapses. Purkinje cell spines forming synaptic contact with climbing fiber terminals (CF) are indicated by #. Dn, Dendrite. D, Quantitative analysis showing selective Delphilin labeling at synapses between parallel fiber and Purkinje cell spine (PF-Sp). CF-Sp, Synapses between climbing fiber and Purkinje cell spine; In-Dn, synapses between interneuron axon and Purkinje cell dendrite. Error bars indicate SD. E, Perpendicular distribution of Delphilin from the midline of synaptic cleft. A total of 123 parallel fiber–Purkinje cell synapses were analyzed. The distances from the midline of synaptic cleft to the center of gold particles were grouped into 8 nm bins. F, Tangential distribution of Delphilin in the postsynapse. A total of 86 parallel fiber–Purkinje cell synapses were analyzed. Relative mediolateral position of gold particles is indicated as the percentage of the distance from the center (0%) to the edge (100%) of the postsynaptic density. Scale bars: A–C, 100 nm.
To clarify subsynaptic distribution of Delphilin, perpendicular and tangential synaptic localization were examined at parallel fiber–Purkinje cell synapses (Fig.9E,F). The quantitative analysis revealed a normal distribution of gold particles from presynapse to postsynapse, with the peak at 8–16 nm postsynaptic from the midline of the synaptic cleft (Fig. 9E). In the tangential distribution on the parallel fiber–Purkinje cell synapse, gold particles were deposited uniformly along the postsynaptic membrane decorated with the PSD, except for a marginal 20% with reduced labeling rate (Fig. 9F). The postsynaptic membrane outside the PSD was associated with very low immunogold labeling (Fig.9F). Therefore, Delphilin is localized at the postsynaptic junction site of the parallel fiber–Purkinje cell synapse.
DISCUSSION
In this paper, we report a novel protein, Delphilin, that interacts with GluRδ2 subunit and contains multiple putative protein–protein interaction motifs, a single PDZ domain, and FH domains FH1 and FH2, followed by coiled-coil structure (Figs. 1, 2). Delphilin appears to be a good candidate as a postsynaptic scaffold protein that links GluRδ2 with multiple signaling molecules, including actin cytoskeleton.
Delphilin colocalization and interaction with GluRδ2
The GluRδ2 subunit is the indispensable molecule for cerebellar LTD and for proper development of both parallel fiber– and climbing fiber–Purkinje cell synapses (Kashiwabuchi et al., 1995). The GluRδ2 subunit is distributed more widely in Purkinje cells early in development, but it is selectively localized in the parallel fiber–Purkinje cell synapses in the adult. Delphilin, which interacts with the GluRδ2 protein via its PDZ domain, was distributed predominantly to the cerebellar molecular layer and was colocalized with GluRδ2 at the postsynaptic site of parallel fiber–Purkinje cell synapses (Figs. 6, 8, 9), the locus of LTD expression (Linden and Connor, 1995). Moreover, perpendicular and tangential subsynaptic localizations at parallel fiber synapses are quite similar to those of GluRδ2 (Landsend et al., 1997). Biochemically, Delphilin was localized almost exclusively in the insoluble synaptosomal membrane fraction (Fig. 7) or in the PSD (Fig. 9), like several PDZ domain-containing proteins involved in neurotransmitter receptor targeting, clustering, and signaling (for review, see Kornau et al., 1997; Sheng and Wyszynski, 1997; Craven and Bredt, 1998; Sheng and Pak, 1999). The colocalization of Delphilin with GluRδ2 in the postsynaptic membrane supports anatomically their interaction at given synapses. Furthermore, the interaction between Delphlin and GluRδ2 seems to be highly specific, because the Delphilin PDZ domain showed little, if any, interaction with the C terminus of GluRδ1, which has four C-terminal aa residues completely identical to those of GluRδ2 (Table 1, Fig. 3).
Quite recently it has been reported that PSD-93 (Roche et al., 1999) and PTPMEG (Hironaka et al., 2000) interact with the C terminus of the GluRδ2 subunit. Against our expectation, PDZ domains of PSD-93 and PTPMEG are not homologous to that of Delphilin, even in the carboxylated-loop region, which is known to directly associate with the C terminus of the target protein. The C terminus of GluRδ2 is essential for interaction with PDZ proteins such as Delphilin (Table 1, Figs. 3, 4), PTPMEG (Hironaka et al., 2000), and PSD-93 (Roche et al., 1999) but was dispensable for the plasma membrane targeting in MDCK (Matsuda and Mishina, 2000) and HEK293T (this study) cells. Very recently, it has been reported that PSD-93 knock-out mice reveal that PSD-93 is not required for development or function of parallel fiber synapses in cerebellum (McGee et al., 2001). In contrast to PSD-93, which is broadly distributed in the brain (Aoki et al., 2001), Delphilin was highly concentrated in the Purkinje cells, especially at the postsynpatic site of parallel fiber–Purkinje cell synapses, where it resided with GluRδ2. Precise studies of developmental changes in expression and distribution of Delphilin, PSD-93, PTPMEG, and GluRδ2 in Purkinje cells, and also studies comparing the interacting profiles, such as binding affinity, of GluRδ2 to these proteins may provide clues to help discriminate the role difference among these three GluRδ2-interacting PDZ proteins in the GluRδ2 function or signaling.
Possible implications of Delphilin protein–protein interaction motifs
Recent works demonstrated that multiplicity of the PDZ domain of scaffolding proteins has been considered to be important for receptor clustering (for review, see Gomperts, 1996). On the other hand, a protein interacting with C kinase 1 (PICK1), which contains a single PDZ domain and a coiled-coil structure (Staudinger et al., 1995) just like Delphilin, has been shown to bind and cluster AMPA receptor subunits (Xia et al., 1999). In this case, the coiled-coil structure serves as a protein multimerization motif (for review, see Adamson et al., 1993; Blake et al., 1995). Delphilin may multimerize via its coiled-coil structure to function as a molecule involved in the GluRδ2 function or signaling.
We further demonstrated that Delphilin, through its FH1 proline-rich domain, interacted with profilins I and II and the SH3 domain of n-Src protein tyrosine kinase (PTK) in a yeast two-hybrid system (Table 2) and surface plasmon resonance analyses (Fig. 5). FH proteins, proteins containing FH domains (Castrillon and Wasserman, 1994), have been implicated in cytokinesis and the establishment of cell polarity in yeast and limb formation in vertebrates (for review, see Frazier and Field, 1997). Although the function of the FH2 domain is unknown, the FH1 proline-rich domains of these proteins have been shown to directly interact with profilin (Watanabe et al., 1997). Profilin is a small actin monomer binding protein, facilitating actin multimerization, and is considered as a key molecule that mediates between signal transduction and the actin cytoskeleton (for review, see Machesky and Pollard, 1993; Schlüter et al., 1997). We could speculate that through the FH1 domain, Delphilin directly regulates the PSD–actin cytoskeleton reorganization.
The FH1 proline-rich domain is also known to interact with the WW/WWP and SH3 domains of various proteins (Chan et al., 1996). Our data demonstrating the interaction of the Delphilin FH1 domain with the SH3 domain of n-Src PTK in yeast cells (Table 1) raised a possibility that Delphilin may contribute to signal transduction by modulating the signaling pathway involving Src PTKs. A recent work by Hayashi et al. (1999) showed involvement of Lyn, a PTK, in the AMPA receptor-mediated signal transduction leading to synaptic plasticity (Lo, 1995; Thoenen, 1995). Furthermore, Boxall et al. (1996) demonstrated that PTK activity is necessary for the LTD at the parallel fiber–Purkinje cell synapse in the cerebellum. Taking these works into consideration, Delphilin might be involved in the formation of the cerebellar LTD by transducing stimuli from GluRδ2 to the n-Src PTK signaling pathway.
Conclusions
We identified a novel PDZ protein that synaptically (at the postsynpatic site of parallel fiber–cerebellar Purkinje cell synpases) colocalizes with and interacts with the GluRδ2 subunit. The protein, designated Delphilin, has FH domains in addition to a PDZ domain. As far as we know, this is the first reported protein that contains both PDZ and FH domains. Delphilin may directly connect GluRδ2 via its FH1 domain both to actin cytoskeleton and to various signaling molecules containing SH3 domains or WW/WWP domains. Thus, although the ligand of the GluRδ2 protein is still unknown, the discovery of Delphilin should help us to understand the GluRδ2 function and the induction mechanism of cerebellar LTD.
Footnotes
This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan, and by a grant from the Naito Foundation. We thank Drs. H. Mori and I. Matsuda (University of Tokyo) for providing a mouse random-primed cDNA library and the expression construct pcDNA3-GluRδ2, respectively. We also thank Dr. F. Yazama (Yokohama City University) for amino acid sequencing.
Correspondence should be addressed to Dr. Susumu Kawamoto, Department of Bacteriology, Yokohama City University School of Medicine, 3-9 Fukuura, Kanazawa-ku, Yokohama 236-0004, Japan. E-mail:skawamot@med.yokohama-cu.ac.jp.
Y. Miyagi's present address: Pathology Division, Kanagawa Cancer Center Research Institute, Yokohama 241-0815, Japan.
REFERENCES
- 1.Adamson JG, Zhou NE, Hodges RS. Structure, function and application of the coiled-coil protein folding motif. Curr Opin Biotechnol. 1993;4:428–437. doi: 10.1016/0958-1669(93)90008-k. [DOI] [PubMed] [Google Scholar]
- 2.Aoki C, Miko I, Oviedo H, Mikelandze-Dvali T, Alexandre L, Sweeney N, Bredt DS. Electron microscopic immunocytochemical detection of PSD-95, PSD-93, SAP-102, and SAP-97 at postsynaptic, presynaptic, and nonsynaptic sites of adult and neonatal rat visual cortex. Synapse. 2001;40:239–257. doi: 10.1002/syn.1047. [DOI] [PubMed] [Google Scholar]
- 3.Araki K, Meguro H, Kushiya E, Takayama C, Inoue Y, Mishina M. Selective expression of the glutamate receptor channel δ2 subunit in cerebellar Purkinje cells. Biochem Biophys Res Commun. 1993;197:1267–1276. doi: 10.1006/bbrc.1993.2614. [DOI] [PubMed] [Google Scholar]
- 4.Bartel P, Chien CT, Sternglanz R, Fields S. Elimination of false positives that arise in using the two-hybrid system. Biotechniques. 1993;14:920–924. [PubMed] [Google Scholar]
- 5.Blackstone CD, Moss SJ, Martin LJ, Levey AI, Price DL, Hugani RL. Biochemical characterization and localization of a non-N-methyl-d-aspartate glutamate receptor in rat brain. J Neurochem. 1992;58:1118–1126. doi: 10.1111/j.1471-4159.1992.tb09370.x. [DOI] [PubMed] [Google Scholar]
- 6.Blake DJ, Tinsley JM, Davies K, Knight AE, Winder SJ, Kendrick-Jones J. Coiled-coil regions in the carboxy-terminal domains of dystrophin and related proteins: potentials for protein-protein interactions. Trends Biochem Sci. 1995;20:133–135. doi: 10.1016/s0968-0004(00)88986-0. [DOI] [PubMed] [Google Scholar]
- 7.Boxall AR, Lancaster B, Garthwaite J. Tyrosine kinase is required for long-term depression in the cerebellum. Neuron. 1996;16:805–813. doi: 10.1016/s0896-6273(00)80100-2. [DOI] [PubMed] [Google Scholar]
- 8.Brenman JE, Christopherson KS, Craven SE, McGee AW, Bredt DS. Cloning and characterization of postsynaptic density 93, a nitric oxide synthase interacting protein. J Neurosci. 1996;16:7407–7415. doi: 10.1523/JNEUROSCI.16-23-07407.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castrillon DH, Wasserman SA. Diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development. 1994;120:3367–3377. doi: 10.1242/dev.120.12.3367. [DOI] [PubMed] [Google Scholar]
- 10.Chan DC, Bedford MT, Leder P. Formin binding proteins bear WWP/WW domains that bind proline-rich peptides and functionally resemble SH3 domains. EMBO J. 1996;15:1045–1054. [PMC free article] [PubMed] [Google Scholar]
- 11.Chang F, Drubin D, Nurse P. cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J Cell Biol. 1997;137:169–182. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craven SE, Bredt DS. PDZ proteins organize synaptic signaling pathways. Cell. 1998;93:495–498. doi: 10.1016/s0092-8674(00)81179-4. [DOI] [PubMed] [Google Scholar]
- 13.Daniels DL, Cohen AR, Anderson JM, Brunger AT. Crystal structure of the hCASK PDZ domain reveals the structural basis of class II PDZ domain target recognition. Nat Struct Biol. 1998;5:317–325. doi: 10.1038/nsb0498-317. [DOI] [PubMed] [Google Scholar]
- 14.Doyle DA, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 15.Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- 16.Frazier JA, Field CM. Actin cytoskeleton: are FH proteins local organizers? Curr Biol. 1997;7:414–417. doi: 10.1016/s0960-9822(06)00205-3. [DOI] [PubMed] [Google Scholar]
- 17.Gomperts SN. Clustering membrane proteins: it's all coming together with the PSD-95/SAP90 protein family. Cell. 1996;84:659–662. doi: 10.1016/s0092-8674(00)81043-0. [DOI] [PubMed] [Google Scholar]
- 18.Hayashi T, Umemori H, Mishina M, Yamamoto T. The AMPA receptor interacts with and signals through the protein tyrosine kinase Lyn. Nature. 1999;397:72–76. doi: 10.1038/16269. [DOI] [PubMed] [Google Scholar]
- 19.Hirano T. Molecular basis of the cerebellar long-term depression. In: Ishikawa K, McGaugh JL, Sakata H, editors. Brain processes and memory. Elsevier; Amsterdam: 1996. pp. 269–277. [Google Scholar]
- 20.Hirano T, Kasono K, Araki K, Shinozuka K, Mishina M. Involvement of the glutamate receptor δ2 subunit in the long-term depression of glutamate responsiveness in cultured rat Purkinje cells. Neurosci Lett. 1994;182:172–176. doi: 10.1016/0304-3940(94)90790-0. [DOI] [PubMed] [Google Scholar]
- 21.Hironaka K, Umemori H, Tezuka T, Mishina M, Yamamoto T. The protein-tyrosine phosphatase PTPMEG interacts with glutamate receptor δ2 and ε subunits. J Biol Chem. 2000;275:16167–16173. doi: 10.1074/jbc.M909302199. [DOI] [PubMed] [Google Scholar]
- 22.Hsueh YP, Sheng M. Requirement of N-terminal cysteines of PSD-95 for PSD-95 multimerization and ternary complex formation, but not for binding to potassium channel Kv1.4. J Biol Chem. 1999;274:532–536. doi: 10.1074/jbc.274.1.532. [DOI] [PubMed] [Google Scholar]
- 23.Hsueh YP, Kim E, Sheng M. Disulfide-linked head-to-head multimerization in the mechanism of ion channel clustering by PSD-95. Neuron. 1997;18:803–814. doi: 10.1016/s0896-6273(00)80319-0. [DOI] [PubMed] [Google Scholar]
- 24.Ito M. Long-term depression. Annu Rev Neurosci. 1989;12:85–102. doi: 10.1146/annurev.ne.12.030189.000505. [DOI] [PubMed] [Google Scholar]
- 25.Jackson-Grusby L, Kuo A, Leder P. A variant limb deformity transcript expressed in the embryonic mouse limb defines a novel formin. Genes Dev. 1992;6:29–37. doi: 10.1101/gad.6.1.29. [DOI] [PubMed] [Google Scholar]
- 26.Jeromin A, Huganir RL, Linden DJ. Suppression of the glutamate receptor δ2 subunit produces a specific impairment in cerebellar long-term depression. J Neurophysiol. 1996;76:3578–3583. doi: 10.1152/jn.1996.76.5.3578. [DOI] [PubMed] [Google Scholar]
- 27.Kashiwabuchi N, Ikeda K, Araki K, Hirano T, Shibuki K, Takayama C, Inoue Y, Kutsuwada T, Yagi T, Kang Y, Aizawa S, Mishina M. Impairment of motor coordination, Purkinje cell synapse formation, and cerebellar long-term depression in GluR δ2 mutant mice. Cell. 1995;81:245–252. doi: 10.1016/0092-8674(95)90334-8. [DOI] [PubMed] [Google Scholar]
- 28.Kohda K, Wang Y, Yuzaki M. Mutation of a glutamate receptor motif reveals its role in gating and δ2 receptor channel properties. Nat Neurosci. 2000;3:315–322. doi: 10.1038/73877. [DOI] [PubMed] [Google Scholar]
- 29.Kornau HC, Seeburg PH, Kennedy MB. Interaction of ion channels and receptors with PDZ domain proteins. Curr Opin Neurobiol. 1997;7:368–373. doi: 10.1016/s0959-4388(97)80064-5. [DOI] [PubMed] [Google Scholar]
- 30.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurihara H, Hashimoto K, Kano M, Takayama C, Sakimura K, Mishina M, Inoue Y, Watanabe M. Impaired parallel fiber → Purkinje cell synapse stabilization during cerebellar development of mutant mice lacking the glutamate receptor δ2 subunit. J Neurosci. 1997;17:9613–9623. doi: 10.1523/JNEUROSCI.17-24-09613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landsend AS, Amiry-Moghaddam M, Matsubara A, Bergersen L, Usami S, Wenthold RJ, Ottersen OP. Differential localization of δ glutamate receptors in the rat cerebellum: coexpression with AMPA receptors in parallel fiber-spine synapses and absence from climbing fiber-spine synapses. J Neurosci. 1997;17:834–842. doi: 10.1523/JNEUROSCI.17-02-00834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lau LF, Mammen A, Ehlers MD, Kindler S, Chung WJ, Garner CC, Huganir RL. Interaction of the N-methyl-d-aspartate receptor complex with a novel synapse-associated protein, SAP102. J Biol Chem. 1996;271:21622–21628. doi: 10.1074/jbc.271.35.21622. [DOI] [PubMed] [Google Scholar]
- 34.Linden DJ, Connor JA. Long-term synaptic depression. Annu Rev Neurosci. 1995;18:319–357. doi: 10.1146/annurev.ne.18.030195.001535. [DOI] [PubMed] [Google Scholar]
- 35.Lo DC. Neurotrophic factors and synaptic plasticity. Neuron. 1995;15:979–981. doi: 10.1016/0896-6273(95)90085-3. [DOI] [PubMed] [Google Scholar]
- 36.Lomeli H, Sprengel R, Laurie DJ, Kohr G, Herb A, Seeburg PH, Wisden W. The rat δ1 and δ2 subunits extend the excitatory amino acid receptor family. FEBS Lett. 1993;315:318–322. doi: 10.1016/0014-5793(93)81186-4. [DOI] [PubMed] [Google Scholar]
- 37.Machesky LM, Pollard TD. Profilin as a potential mediator of membrane-cytoskeleton communication. Trends Cell Biol. 1993;3:381–385. doi: 10.1016/0962-8924(93)90087-h. [DOI] [PubMed] [Google Scholar]
- 38.Matsuda I, Mishina M. Identification of a juxtamembrane segment of the glutamate receptor δ2 subunit required for the plasma membrane localization. Biochem Biophys Res Commun. 2000;275:565–571. doi: 10.1006/bbrc.2000.3351. [DOI] [PubMed] [Google Scholar]
- 39.McGee AW, Topinka JR, Hashimoto K, Petralia RS, Kakizawa S, Kauer F, Aguilera-Moreno A, Wenthold RJ, Kano M, Bredt DS. PSD-93 knock-out mice reveal that neuronal MAGUKs are not required for development or function of parallel fiber synapses in cerebellum. J Neurosci. 2001;21:3085–3091. doi: 10.1523/JNEUROSCI.21-09-03085.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milligan G, Grassie MA, Wise A, MacEwan DJ, Magee AI, Parenti M. G-protein palmitoylation: regulation and functional significance. Biochem Soc Trans. 1995;23:583–587. doi: 10.1042/bst0230583. [DOI] [PubMed] [Google Scholar]
- 41.Murata T, Yamaguchi M. Promoter characterization of the rat gene for Ca2+-binding protein regucalcin: transcriptional regulation by signaling factors. J Biol Chem. 1999;274:1277–1285. doi: 10.1074/jbc.274.3.1277. [DOI] [PubMed] [Google Scholar]
- 42.Petersen J, Weilguny D, Egel R, Nielsen O. Characterization of fus1 of Schizosaccharomyces pombe: a developmentally controlled function needed for conjugation. Mol Cell Biol. 1995;15:3697–3707. doi: 10.1128/mcb.15.7.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petersen J, Nielsen O, Egel R, Hagan IM. FH3, a domain found in formins, targets the fission yeast formin Fus1 to the projection tip during conjugation. J Cell Biol. 1998;141:1217–1228. doi: 10.1083/jcb.141.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roche KW, Ly CD, Petralia RS, Wang YX, McGee AW, Bredt DS, Wenthold RJ. Postsynaptic density-93 interacts with the δ2 glutamate receptor subunit at parallel fiber synapses. J Neurosci. 1999;19:3926–3934. doi: 10.1523/JNEUROSCI.19-10-03926.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saras J, Heldin CH. PDZ domains bind carboxy-terminal sequences of target proteins. Trends Biochem Sci. 1996;21:455–458. doi: 10.1016/s0968-0004(96)30044-3. [DOI] [PubMed] [Google Scholar]
- 46.Scannervin RH, Huganir RL. Postsynaptic organization and regulation of excitatory synapses. Nat Rev Neurosci. 2000;1:133–141. doi: 10.1038/35039075. [DOI] [PubMed] [Google Scholar]
- 47.Schlüter K, Jockusch BM, Rothkegel M. Profilins as regulators of actin dynamics. Biochim Biophys Acta. 1997;1359:97–109. doi: 10.1016/s0167-4889(97)00100-6. [DOI] [PubMed] [Google Scholar]
- 48.Sen-Gupta M, Lyck R, Fleig U, Niedenthal RK, Hegemann JH. The sequence of a 24,152 bp segment from the left arm of chromosome XIV from Saccharomyces cerevisiae between the BNI1 and the POL2 genes. Yeast. 1996;12:505–514. doi: 10.1002/(SICI)1097-0061(199604)12:5%3C505::AID-YEA932%3E3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 49.Sheng M, Pak DT. Glutamate receptor anchoring proteins and the molecular organization of excitatory synapses. Ann NY Acad Sci. 1999;868:483–493. doi: 10.1111/j.1749-6632.1999.tb11317.x. [DOI] [PubMed] [Google Scholar]
- 50.Sheng M, Wyszynski M. Ion channel targeting in neurons. BioEssays. 1997;19:847–853. doi: 10.1002/bies.950191004. [DOI] [PubMed] [Google Scholar]
- 51.Snow BE, Antonio L, Suggs S, Gutstein HB, Siderovski DP. Molecular cloning and expression analysis of rat Rgs12 and Rgs14. Biochem Biophys Res Commun. 1997;233:770–777. doi: 10.1006/bbrc.1997.6537. [DOI] [PubMed] [Google Scholar]
- 52.Staudinger J, Zhou J, Burgess R, Elledge SJ, Olson EN. PICK1: a perinuclear binding protein and substrate for protein kinase C isolated by the yeast two-hybrid system. J Cell Biol. 1995;128:263–271. doi: 10.1083/jcb.128.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sugrue MM, Brugge JS, Marshak DR, Greengard P, Gustafson EL. Immunocytochemical localization of the neuron-specific form of the c-src gene product, pp60c-src(+), in rat brain. J Neurosci. 1990;10:2513–2527. doi: 10.1523/JNEUROSCI.10-08-02513.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takayama C, Nakagawa S, Watanabe M, Mishina M, Inoue Y. Developmental changes in expression and distribution of the glutamate receptor channel δ2 subunit according to the Purkinje cell maturation. Dev Brain Res. 1996;92:147–155. doi: 10.1016/0165-3806(95)00212-x. [DOI] [PubMed] [Google Scholar]
- 55.Taverna F, Xiong Z-G, Brandes L, Roder JC, Salter MW, MacDonald JF. The Lurcher mutation of an α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor subunit enhances potency of glutamate and converts an antagonist to an agonist. J Biol Chem. 2000;275:8475–8479. doi: 10.1074/jbc.275.12.8475. [DOI] [PubMed] [Google Scholar]
- 56.Thoenen H. Neurotrophins and neuronal plasticity. Science. 1995;270:593–598. doi: 10.1126/science.270.5236.593. [DOI] [PubMed] [Google Scholar]
- 57.Topinka JR, Bredt DS. N-terminal palmitoylation of PSD-95 regulates association with cell membranes and interaction with K+ channel Kv1.4. Neuron. 1998;20:125–134. doi: 10.1016/s0896-6273(00)80440-7. [DOI] [PubMed] [Google Scholar]
- 58.Watanabe M, Inoue Y, Sakimura K, Mishina M. Distinct distributions of five N-methyl-d-aspartate receptor channel subunit mRNAs in the forebrain. J Comp Neurol. 1993;338:377–390. doi: 10.1002/cne.903380305. [DOI] [PubMed] [Google Scholar]
- 59.Watanabe M, Fukaya M, Sakimura K, Manabe T, Mishina M, Inoue Y. Selective scarcity of NMDA receptor channel subunits in the stratum lucidum (mossy fibre-recipient layer) of the mouse hippocampal CA3 subfield. Eur J Neurosci. 1998;10:478–487. doi: 10.1046/j.1460-9568.1998.00063.x. [DOI] [PubMed] [Google Scholar]
- 60.Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, Saito Y, Nakao K, Jockusch BM, Narumiya S. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 1997;16:3044–3056. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Witke W, Sutherland JD, Sharpe A, Arai M, Kwiatkkowski DJ. Profilin I is essential for cell survival and cell division in early mouse development. Proc Natl Acad Sci USA. 2001;98:3832–3836. doi: 10.1073/pnas.051515498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wollmuth LF, Kuner T, Jatzke C, Seeburg PH, Heintz N, Zuo J. The Lurcher mutation identifies δ2 as an AMPA/kainate receptor-like channel that is potentiated by Ca2+. J Neurosci. 2000;20:5973–5980. doi: 10.1523/JNEUROSCI.20-16-05973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Woychik RP, Maas RL, Zeller R, Vogt TF, Leder P. “Formins”: proteins deduced from the alternative transcripts of the limb deformity gene. Nature. 1990;346:850–853. doi: 10.1038/346850a0. [DOI] [PubMed] [Google Scholar]
- 64.Xia J, Zhang X, Staudinger J, Huganir RL. Clustering of AMPA receptors by the synaptic PDZ domain-containing protein PICK1. Neuron. 1999;22:179–187. doi: 10.1016/s0896-6273(00)80689-3. [DOI] [PubMed] [Google Scholar]
- 65.Yamazaki M, Mori H, Araki K, Mori KJ, Mishina M. Cloning, expression and modulation of a mouse NMDA receptor subunit. FEBS Lett. 1992;300:39–45. doi: 10.1016/0014-5793(92)80160-i. [DOI] [PubMed] [Google Scholar]
- 66.Zhao H-M, Wenthold RJ, Petralia RS. Glutamate receptor targeting to synaptic populations on Purkinje cells is developmentally regulated. J Neurosci. 1998;18:5517–5528. doi: 10.1523/JNEUROSCI.18-14-05517.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zuo J, De Jager PL, Takahashi KA, Jiang W, Linden DJ, Heinz N. Neurodegeneration in Lurcher mice caused by mutation in δ2 glutamate receptor. Nature. 1997;388:769–773. doi: 10.1038/42009. [DOI] [PubMed] [Google Scholar]