Abstract
Human cytomegalovirus (HCMV) major immediate-early protein IE1 is an abundant 72-kDa nuclear phosphoprotein that is thought to play an important role in efficient triggering of the lytic cycle, especially at low multiplicity of infection. The best-known properties of IE1 at present are its transient targeting to punctate promyelocytic leukemia protein (PML)-associated nuclear bodies (PML oncogenic domains [PODs] or nuclear domain 10 [ND10]), with associated displacement of the cellular PML tumor suppressor protein into a diffuse nucleoplasmic form and its association with metaphase chromosomes. Recent studies have shown that the targeting of PML (and associated proteins such as hDaxx) to PODs is dependent on modification of PML by ubiquitin-like protein SUMO-1. In this study, we provide direct evidence that IE1 is also covalently modified by SUMO-1 in both infected and cotransfected cells, as well as in in vitro assays, with up to 30% of the protein representing the covalently conjugated 90-kDa form in stable U373/IE1 cell lines. Lysine 450 was mapped as the major SUMO-1 conjugation site, but a point mutation of this lysine residue in IE1 did not interfere with its targeting to and disruption of the PODs. Surprisingly, unlike PML or IE2, IE1 did not interact with either Ubc9 or SUMO-1 in yeast two-hybrid assays, suggesting that some additional unknown intranuclear cofactors must play a role in IE1 sumoylation. Interestingly, stable expression of either exogenous PML or exogenous Flag-SUMO-1 in U373 cell lines greatly enhanced both the levels and rate of in vivo IE1 sumoylation during HCMV infection. Unlike the disruption of PODs by the herpes simplex virus type 1 IE110(ICP0) protein, the disruption of PODs by HCMV IE1 proved not to involve proteasome-dependent degradation of PML. We also demonstrate here that the 560-amino-acid PML1 isoform functions as a transcriptional repressor when fused to the GAL4 DNA-binding domain and that wild-type IE1 inhibits the repressor function of PML1 in transient cotransfection assays. Furthermore, both IE1(1-346) and IE1(L174P) mutants, which are defective in displacing PML from PODs, failed to inhibit the repression activity of PML1, whereas the sumoylation-negative IE1(K450R) mutant derepressed as efficiently as wild-type IE1. Taken together, our results suggest that proteasome-independent disruption of PODs, but not IE1 sumoylation, is required for efficient IE1 inhibition of PML-mediated transcriptional repression.
The human cytomegalovirus (HCMV) major immediate-early (MIE) locus encodes two nuclear phosphoproteins, namely, IE1 (UL123 or IE72) and IE2 (UL122 or IE86) (55). IE1 is the most abundantly expressed viral protein at very early times or under cycloheximide-restricted conditions in HCMV-infected permissive HF or U373 cells (32, 72, 78), and both MIE proteins are also the only HCMV proteins known to be expressed at any stages after infection in nonpermissive rodent fibroblasts (39). The IE2 protein is a powerful nonspecific transcriptional activator and is thought to be essential for all subsequent lytic-cycle HCMV gene expression even in cell culture. IE2 is also a specific DNA-binding protein with negative autoregulatory activity (13, 41, 43, 62, 63, 77) and interacts with numerous cellular proteins. IE2 is also efficiently incorporated into viral DNA replication compartments (6, 68). However, the exact role of the 491-amino-acid (aa) acidic IE1 protein, which is important for efficient lytic-cycle infection in cell culture at low multiplicity of infection (MOI) but which is not essential under high-MOI infection conditions (4, 21), has remained obscure.
The IE1 protein colocalizes with metaphase chromosomes in both stable cell lines and HCMV-infected cells (40), and it can also inhibit apoptosis induced by tumor necrosis factor alpha in HeLa cells (85). It has also been suggested that IE1 up-regulates the MIE promoter through upstream NF-κB sites (14), but in transient cotransfection assays IE1 is not a significant transactivator on its own (63). Under certain conditions, a synergistic transactivation of some target promoters when IE1 is introduced into cells together with IE2 has been reported (11, 37, 45, 49, 71). The IE1 protein has also been reported to interact with components of TFIID in vitro and to coimmunoprecipitate with purified TFIID complexes from HCMV-infected nuclear extracts (44) and may physically interact with cellular transcription factors SP-1, E2F1, and CTF-1 (24, 45, 50). Overall, these diverse activities have not formed a clear picture of the precise functional role of IE1 in the context of the HCMV life cycle.
More-recent studies of the intracellular localization of IE1 have revealed that it is one of several key regulatory proteins encoded by DNA viruses that target subnuclear punctate domains at very early times after infection. In most cells, the promyelocytic leukemia protein (PML)-associated nuclear bodies known as PML oncogenic domains (PODs) or nuclear domain 10 (ND10) form several dozen small spherical structures with an average size of 0.3 to 0.5 μm. Electron microscopy shows that the PML protein surrounds an electron-dense core associated with the nuclear matrix. Several other cellular proteins have been found to be present in the PODs, including Sp100, Daxx, and recently characterized ubiquitin-like protein SUMO-1. Intriguingly, both PML and Sp100 are posttranslationally modified by covalent conjugation with SUMO-1. The unmodified form of PML is found in the NP-40-soluble nucleoplasmic fraction, whereas the SUMO-1-conjugated forms are compartmentalized in the NP-40-insoluble PODs (59). Recent studies further demonstrated that SUMO-1 modification of PML is required for its recruitment of Daxx into the PODs (30, 83) and for its own punctate nuclear localization pattern (83). The peripheries of PODs have also been implicated as sites for deposition of input viral DNA, as well as being associated with initiation of both immediate-early transcription and viral DNA replication in several DNA viruses (6, 29, 31). We and others have shown that, within the first 1 to 2 h of infection and even before IE2 is synthesized, the IE1 protein transiently localizes to PODs, followed by displacement of both IE1 and PML into a diffuse nucleoplasmic form (4–6, 38, 79). This disruption of the PODs appears to be mediated through a specific interaction of IE1 with PML in a RING finger-dependent manner (2).
PML is a member of a growing family of proteins characterized by the presence of a RING finger domain. RING finger proteins are implicated in transcriptional regulation (69) and in some cases as E3 ligases that mediate the transfer of ubiquitin both to heterologous substrates and to the RING finger proteins themselves (33). PML exists in several alternatively spliced isoforms that differ in their C-terminal sequences (19). All isoforms described so far contain the RING finger, two additonal Cys- and His-rich regions (B1 and B2 boxes), and an α-helical coiled-coil domain (collectively known as the RBCC domain) and a nuclear localization signal. The three most common PML splice variants are PML1 (560 aa) (35), PML-L (641 aa) (16), and PML3 (633 aa) (19). PML-L was reported to be a coactivator for c-Jun and for the progesterone receptor through its RBCC domain (22, 74). Unlike bona fide transcriptional coactivators such as p300 and CBP (60, 76), PML does not have intrinsic histone acetyltransferase activity. Although it does not bind to DNA directly, we found that PML1 displayed a RING finger-dependent cryptic transactivating activity when its RBCC domain was tethered to DNA and the C terminus was removed (2).
On the other hand, PML also acts as a transcriptional corepressor, inhibiting the transactivation of Sp1 on the epidermal growth factor receptor promoter (73) and abolishing the activation of glucocorticoid receptor-regulated transcription by the retinablastoma protein (Rb) (8). When fused to the GAL4 DNA-binding domain, PML-L acts as a transcriptional repressor, inhibiting transcription from a GAL4-responsive promoter (75, 80). Deletion analysis of PML-L revealed that the repressive effects depended on an intact coiled-coil (dimerization) domain (75). Recent studies further demonstrated that PML-L represses basal transcription by physically interacting with all three isoforms of histone deacetylases (HDAC) (80). In contrast, PML1 did not bind to any of the three HDAC isoforms directly (42). Nevertheless, recent studies showed that PML1 inhibits transcriptional repression mediated by Tax and Daxx (17, 42).
Several families of ubiquitin-like proteins have recently been described; these include aforementioned small ubiquitin-like modifier SUMO-1, which can be covalently conjugated to proteins such as RanGAP1, IκBα, PML, Sp100, p53, c-Jun, and MDM2 through a pathway distinct from but analogous to the ubiquitin conjugation system (25). Three mammalian SUMO proteins have been identified so far; SUMO-2 and SUMO-3 have 87% amino acid sequence identity but are only 47% identical to SUMO-1. SUMO precursors are proteolytically processed at their C-terminal double-glycine motifs before being activated by the formation of a thioester bond with an E1 SUMO activation enzyme in an ATP-dependent reaction. Then the SUMO moiety is transferred to E2 SUMO conjugation enzyme Ubc9. While there are several families of E3 ubiquitin ligases that provide substrate specificity for ubiquitination, the existence of E3 SUMO ligases has not yet been reported. SUMO-1 modification of RanGAP1 was shown to be critical for its intracellular targeting to nuclear pore complexes as well as for its interaction with RanBP2 (48, 52). Addition of SUMO-1 to IκBα and MDM2 antagonizes both their ubiquitination and down-regulation by modifying the same lysine residues used for ubiquitination (10, 15). SUMO-1 conjugation to p53 enhances transactivation activity on p53 target promoters (20, 65), whereas SUMO-1 modification of c-Jun down-regulates its transactivation (57). Therefore, the functional consequences of SUMO-1 modification appear to be diverse among the different known substrates.
Our earlier colocalization mapping and yeast two-hybrid assays suggested that the disruption of PODs by IE1 may be mediated through a specific physical interaction of IE1 and PML (2). Furthermore, a truncated IE1(1-346) mutant could still colocalize with PODs but failed to displace PML from the PODs, thus uncoupling the requirements for targeting from subsequent disruption. Importantly, this IE1(1-346) mutant still interacted with PML in the yeast two-hybrid assay (2). In agreement with our report, Muller and Dejean also detected a specific interaction between PML and wild-type IE1 in Saccharomyces cerevisiae two-hybrid assays (58). In addition, they found than an L174P IE1 mutant did not interact with PML in yeast and that the same mutant protein also failed to target PODs. These observations suggest that the interaction with PML may be responsible for the targeting of IE1 to PODs but that additional features are necessary for the displacement of PML from PODs. Muller and Dejean first detected a 90-kDa IE1 isoform in addition to the regular 72-kDa band in HeLa cells transiently transfected with an IE1 expression plasmid (58). They were able to precipitate the 90-kDa isoform with Ni-agarose after cotransfecting HeLa cells with His-tagged SUMO-1, indicating that the IE1 protein might also be a substrate for SUMO-1 modification. They also suggested that cotransfection with either herpes simplex virus type 1 (HSV-1) IE110 or HCMV IE1 either prevented or removed the SUMO modification of PML.
In this study, we demonstrate that IE1 is covalently modified by SUMO-1 both in HCMV-infected permissive cells and in stable DNA-transfected cell lines. We also identify the lysine residue at position 450 as the SUMO-1 conjugation site in IE1 and provide evidence that stable expression of either PML1 or SUMO-1 in HCMV-permissive U373 cells enhances IE1 sumoylation during HCMV infection. Finally, we show that wild-type IE1 can inhibit PML1-mediated repression of basal transcription and that this novel functional aspect of IE1 requires its ability to disrupt PODs but not covalent modification by SUMO-1.
MATERIALS AND METHODS
Plasmids.
Wild-type HCMV IE1 cDNA expression plasmid pJHA303 and truncation mutant pJHA304 encoding IE1(1-346) were described previously (2). Mutant forms encoding IE1(K163R) (pYX108), -(K197R) (pYX109), -(K331R) (pYX102), -K450R (pYX118), and -(L174P) (pYX145) were generated from pJHA303 with the Stratagene QuikChange site-directed mutagenesis protocol. Expression plasmid pJHA312, encoding Flag-SUMO-1, was described elsewhere (7). The GST-PML-L plasmid was a generous gift from Kun-Sang Chang (University of Texas MD Anderson Cancer Center, Houston, Tex.). Plasmid pYX143 encoding glutathione S-transferase (GST)-PML(1-96) was constructed by placing the NcoI (blunt ended)-SalI fragment from pCMX-PML1 between the SmaI and XhoI sites of pGEX-4T-1. Plasmids pGH250 (pSV2-GAL4-DB vector), pJHA258 (pSV2-GAL4-DB/PML1) (2), pJH93 (pSV2-GAL4-DB/CBF1) (27), (GAL4)5-TK-CAT (70), and (GAL4)5-TK-LUC (67) were previously described. A full-length PCR fragment of human Daxx cDNA was isolated from pQE38 (a gift from Jerome F. Strauss III, University of Pennsylvania, Philadelphia) and digested with BamHI. Then the BamHI insert was placed into the BglII site of pGH251 (another version of the pSV2-GAL4-DB vector) to make the pJHA347 (pSV2-GAL4-DB/Daxx) expression plasmid.
Cell lines and cell culture.
Permissive human diploid fibroblast (HF) cells, semipermissive U373-MG cells, and nonpermissive 293T and HeLa cells were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum. The U373-AN and U373-A45 cell lines, obtained from Susan Michelson (Institute Pasteur, Paris, France),were described previously (54, 81). The U373-A3 cell line was established by cotransfecting U373-MG cells with pRL103 plasmid DNA containing the entire MIE transcription unit (40). The U373-PML and the control U373-Neo cell lines were described elsewhere (4); the U373-SUMO-1 cell line was established by stable transfection of U373-MG cells with the expression plasmids pJHA312 encoding Flag-SUMO-1 and pSV2-Neo.
Virus preparation and infection.
Virus stocks for wild-type HCMV(Towne) were prepared as previously described (2, 40). Recombinant adenoviruses expressing either wild-type or K450R mutant IE1 proteins [Ad-IE1 and Ad-IE1(K450R)] were generated with the Cre-lox recombination system as described elsewhere (23). Briefly, a BamHI fragment from pJHA303 or pYX118 was subcloned into the BamHI site of recombinant transfer vector pAdlox to make recombinant transfer plasmids pYX137 and pYX138, respectively. pAdlox vector, Ψ5 virus, and CRE8 cells were all generous gifts from Stephen Hardy (Somatix Therapy Corporation, Alameda, Calif.). Recombinant adenoviruses were made by cotransfecting Ψ5 viral DNA with pYX137 or with pYX138 into CRE8 cells. The viruses were passed sequentially through CRE8 cells four times. Viral lysates were harvested by three freeze-thaw cycles and cleared by centrifugation.
For immunoblot analysis, U373-MG cells were seeded into six-well plates at 2 × 105 cells per well. Next day, cells were mock infected or infected with wild-type HCMV(Towne) at an MOI of 5 or with Ad-IE1 at an MOI of 20. Cell lysates were then harvested at different time points after infection. For immunofluorescence experiments, HF cells were infected with HCMV(Towne) at an MOI of 5 and Vero cells were infected with HSV-1(KOS) at an MOI of 20. When MG132 (Calbiochem) was used, the cells were preincubated with medium containing 5 μM MG132 for 30 min before infection and all subsequent incubations were performed in the presence of the drug. Slides for HSV-1-infected Vero cells were fixed at 4 h after infection, and slides for HCMV-infected HF cells were fixed at 6 h after infection.
Yeast two-hybrid interaction assays.
Yeast strain Y190 (MATa gal4Δ gal80Δ his3-200 trp1-901 ade2-101 ura3-52 leu2-3,112 URA3::GAL1-lacZ LYS::GAL-HIS3 cyhR) was the host for color development filter assays for lacZ expression using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and for a quantitative assay using o-nitrophenyl-β-d-galactopyranoside (ONPG). Media for yeast growth and the method for yeast transformation were described elsewhere (66). Both the X-Gal filter and ONPG assays were described previously (2, 3).
Transient DNA transfection.
For immunoblot analysis, 293T cells were seeded into six-well plates at 4 × 105 cells per well and DNA mixtures were introduced into subconfluent cells with the N,N-bis-(2-hydroxyethyl)-2-aminoethanesulfonic acid-buffered saline (BBS) version of the calcium phosphate procedure as described previously (62). For indirect immunofluorescence assays (IFA), HF cells were seeded into two-well slide chambers at 0.4 × 105 cells per well and transfected with Effectene reagent from Qiagen (Chatsworth, Calif.) according to the manufacturer's protocol. For chloramphenicol acetyltransferase (CAT) and luciferase reporter assays, HeLa cells were seeded into six-well plates at 2 × 105 cells per well and transfected by the calcium phosphate procedure.
Antibodies.
Mouse monoclonal antibody (MAb) 6E1 against HCMV IE1 (exon 4) was purchased from Vancouver Biotech (Vancouver, British Columbia, Canada). MAb CH810, which detects epitopes present in both IE1 and IE2 (exons 2 and 3), was purchased from Chemicon (Temecula, Calif.). Rabbit anti-SUMO-1 polyclonal antibody (PAb) FL101 was purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.). Rabbit antipeptide PAbs PML(C) and IE110(N) were described elsewhere (5, 56).
Immunoprecipitation.
U373-MG cells in 10-cm-diameter dishes were infected with HCMV(Towne) at an MOI of 5 for 72 h, harvested cell pellets were lysed by boiling with two volumes of 2% sodium dodecyl sulfate (SDS) in Tris-buffered saline (TBS; 10 mM Tris-Cl [pH 8.0] and 150 mM NaCl) supplemented with 5 mM N-ethylmaleimide (NEM; Sigma; E-1271) for 10 min. Eight volumes of 1% Triton X-100 in TBS with 5 mM NEM were added, and lysates were sonicated for 2 min. For immunoprecipitation, cell lysates were incubated with 5 μg of anti-SUMO-1 PAb FL101 at 4°C overnight before 30 μl of protein A- and G-Sepharose beads was added. The remaining steps were the same as those described by Buschmann et al. (10), and the immunoblot was first probed with MAb CH810 and then stripped and reprobed with MAb 6E1.
Immunoblot analysis.
Transfected 293T or infected U373 (U373-MG, U373-Neo, and U373-PML) cells were washed with phosphate-buffered saline and lysed with 200 μl of ice-cold radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with a protease inhibitor cocktail (Santa Cruz Biotechnology) and 5 mM NEM. Clarified cell extracts from the equivalent of 104 cells were separated on SDS–8% polyacrylamide gel, followed by standard immunoblot procedures with the enhanced chemiluminescence system (Amersham). Densitometry quantitation of protein band intensity was carried out with a Fluorchem 8000 digital imaging system (Alpha Innotech, San Leandro, Calif.) according to the user's manual.
In vitro SUMO-1 conjugation assay.
[35S]Met-labeled wild-type and mutant IE1 proteins were in vitro translated from pJHA303 and pYX118. The conjugation assay procedures were slightly modified from those described by Desterro et al. (15) and described in detail elsewhere (7). The GST-PML-L protein was prepared by standard procedures. In reaction mixtures containing GST-PML-L, its approximate final concentration was 1 μg/μl.
IFA.
Transfected or virus-infected cells were fixed in methanol or with 1% paraformaldehyde and permeabilized with 0.2% Triton X-100; all subsequent procedures were described previously (2).
CAT and luciferase reporter assays.
Transfected HeLa cells were lysed 48 h after transfection in six-well plates by three freeze-thaw cycles in 200 μl of 0.25 M Tris-HCl (pH 7.9) plus 1 mM dithiothreitol. Both the CAT assay (62) and luciferase assay (4) procedures were described previously.
RESULTS
The HCMV IE1 protein is covalently modified by SUMO-1 in DNA-transfected cells.
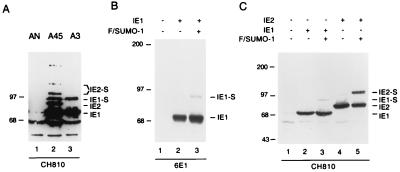
When we examined the relative amounts and levels of intactness of both the IE1 and IE2 proteins in two U373 cell lines stably expressing either IE1 (U373-A3) or IE1 plus IE2 (U373-A45), the cell lines were found to include aberrantly high-molecular-weight forms. For example, a 90-kDa IE1 isoform in both cell lines (Fig. 1A, lanes 3 and 2) and two novel IE2 isoforms in U373-A45 cells with apparent sizes of 105 and 120 kDa (Fig. 1A, lane 2) were detected with MAb CH810, in addition to the expected and more-abundant regular 72-kDa IE1 and 86-kDa IE2 protein bands. We have shown elsewhere that the two unusual IE2 isoforms represent conjugated mono- and disumoylated IE2 (7). Earlier studies by Muller and Dejean indicated that IE1 could also be a substrate for SUMO-1 modification in transiently transfected HeLa cells (58). Therefore, we were interested to examine whether the 90-kDa IE1 isoform detected in stable cell lines could be reproduced in transient cotransfections with SUMO-1. 293T cells were transfected with an IE1 expression plasmid in the presence or absence of Flag-tagged SUMO-1 plasmid. In this experiment, no 90-kDa IE1 isoform was detected by either MAb 6E1 or CH810 in cells transfected with IE1 alone (Fig. 1B and C, lanes 2), but when exogenous Flag-SUMO-1 was cotransfected, a small fraction of the 90-kDa isoform was detected by both antibodies (Fig. 1B and C, lanes 3), although the amount was not as great as the amount of the 105-kDa form of IE2 in parallel positive-control samples (Fig. 1C, lane 5).
FIG. 1.
Covalent modification of HCMV IE1 by SUMO-1 in DNA-transfected cells. (A) Detection of higher-molecular-weight IE1 and IE2 isoforms in U373 cell lines stably expressing IE1 (U373-A3) or IE1 plus IE2 (U373-A45). Lysate from a cell line carrying the NeoR selection marker (U373-AN) was used as a negative control. (B and C) Detection of SUMO-1-conjugated IE1 in transfected 293T cells. Immunoblot analysis was carried out with MAb CH810 (A and C) or 6E1 (B).
Because the cell lysates here were prepared with radioimmunoprecipitation assay buffer (without the supplementation of 5 mM NEM), these results are probably underestimates because of the known propensity for sumoylated proteins to be readily deconjugated by isopeptidases upon cell lysis in nondenaturing buffers (53). Overexpression of SUMO-1 increases the level of sumoylation and was necessary to detect the modification of IE1 in this experiment. However, when we used alternative experimental conditions that were expected to improve the identification of SUMO-1 modifications by lysing the cells under denaturing conditions or in the presence of 5 mM NEM to inhibit desumoylating isopeptidases (51, 58), both the 72- and the 90-kDa isoforms were detected in cells transfected with IE1 alone (see Fig. 3A, lane 2). Therefore, all subsequent immunoblot and immunoprecipitation experiments to study IE1 modification by endogenous SUMO were carried out in buffers supplemented with 5 mM NEM.
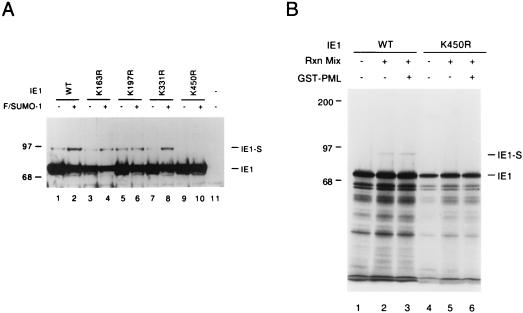
FIG. 3.
Identification of lysine residues in IE1 that are conjugated by SUMO-1. (A) Total extracts were prepared from 293T cells transfected with 2 μg of vector alone (lane 11) or 2 μg of pJHA303 encoding wild-type (WT) IE1 (lane 1), pYX108 encoding IE1(K163R) (lane 3), pYX109 encoding IE1(K197R) (lane 5), pYX102 encoding IE1(K331R) (lane 7), or pYX118 encoding IE1(K450R) (lane 9), or the same five plasmids plus 2 μg of pJHA312 encoding Flag-SUMO-1 (lanes 2, 4, 6, 8, and 10). Cell extracts were subjected to SDS–8% polyacrylamide gel electrophoresis (PAGE), and immunoblot analysis was carried out with MAb CH810. (B) In vitro-translated [35S]Met-labeled wild-type IE1 and K450R mutant IE1 were incubated at 37°C for 2 h in the absence or presence of reaction mixtures (Rxn Mix) containing ATP-regenerating buffer, GST-Ubc9, His-tagged SUMO-1, and a HeLa cell fraction with E1 activity for SUMO-1 conjugation. For reaction mixtures containing GST-PML-L, its approximate final concentration was 1 μg/μl. Reaction products were separated by SDS–8% PAGE and visualized by autoradiography.
Covalent modification of IE1 by SUMO-1 in virus-infected cells.
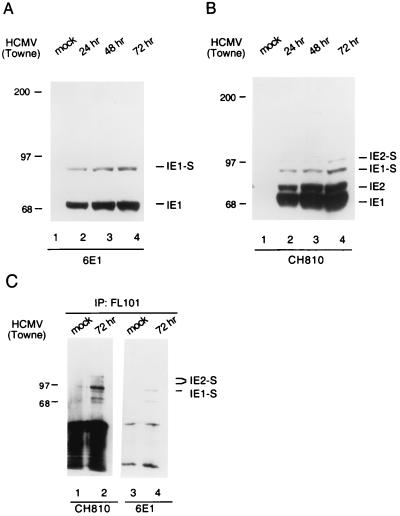
We then asked whether this posttranslational modification of IE1 also occurs during HCMV infection. We first examined U373-MG cell lysates infected with HCMV(Towne) for 24, 48, and 72 h by immunoblot analysis with MAb 6E1. An increasing amount of both the unmodified 72-kDa band and the 90-kDa SUMO-1-modified isoform accumulated throughout the time course of HCMV infection (Fig. 2A). The same set of lysates were also subjected to immunoblot analysis with MAb CH810, which is directed against epitopes shared by IE1 and IE2. The same 90-kDa band was also detected in increasing amounts during the time course of HCMV infection (Fig. 2B). In addition, MAb CH810 also detected the unmodified (86-kDa) and SUMO-1-modified (105-kDa) IE2 isoforms that we and others have already described (7, 26). A similar fraction of 90-kDa IE1 was also detected in HCMV-infected HF cells (not shown).
FIG. 2.
Covalent modification of IE1 by SUMO-1 in HCMV-infected cells. (A and B) Detection of the 90-kDa covalently modified isoform of IE1 in U373-MG cells infected with HCMV(Towne). Immunoblot analysis was carried out with mouse anti-IE1 MAb 6E1 (A) or mouse anti-IE1 and -IE2 MAb CH810 (B). (C) Mock-infected and HCMV-infected whole-cell lysates were immunoprecipitated (IP) with anti-SUMO-1 PAb FL101 and subjected to immunoblot analysis with MAb CH810 (lanes 1 and 2). The same blot was then stripped and reprobed with MAb 6E1 (lanes 3 and 4).
To obtain direct evidence that the 90-kDa IE1 isoform is indeed covalently conjugated by SUMO-1, we infected U373-MG cells with HCMV(Towne) and immunoprecipitated the whole-cell lysates with anti-SUMO-1 PAb FL101. Immunoblot analysis of the immunoprecipitates with MAb 6E1 detected the 90-kDa IE1 isoform (Fig. 2C, lane 4), while MAb CH810 was able to detect sumoylated isoforms of both IE1 and IE2 on the same immunoblot (Fig. 2C, lane 2). These results establish that IE1 is covalently modified by endogenous SUMO-1 during HCMV infection.
Mapping of SUMO-1 conjugation sites in the IE1 protein.
The consensus KXE motif (34) is a well-conserved pattern in almost all SUMO-1 substrates except when the target lysine residues are located within the RING finger domains of PML and MDM2 (10, 36). There are four KXE motifs at positions 163, 197, 331, and 450 in IE1 that represent potential SUMO-1 attachment sites. Each of these four candidate lysine residues was mutated to arginine, and the resulting mutant IE1 proteins were tested for SUMO-1 modification by transfecting them with or without Flag-SUMO-1 into 293T cells. Immunoblot analysis of the cell lysates showed that K163R, K197R, and K331R single point mutants were all still modified by SUMO-1, whereas no 90-kDa isoform was detected in cells transfected with the K450R mutant (Fig. 3A).
We confirmed these mapping data with an in vitro sumoylation assay system. In contrast to the high in vitro sumoylation efficiency of IE2 (7), wild-type IE1 was only modestly sumoylated in our assay system (Fig. 3B, lane 2). When we incubated in vitro-translated K450R mutant IE1 protein with the same reaction mixture, there was clearly no detectable SUMO-1 modification (Fig. 3B, lane 5). Taken together, these results mapped lysine residue 450 as the only major SUMO-1 conjugation site in the HCMV IE1 protein. Therefore, the known SUMO-1 conjugation target motifs in IE1 and IE2 are DTVSVKSEPVSEI and MLPLIKQEPIKPE, respectively (conjugation sites are in boldface).
IE1 does not interact with SUMO-1 or Ubc9 in yeast two-hybrid assays.
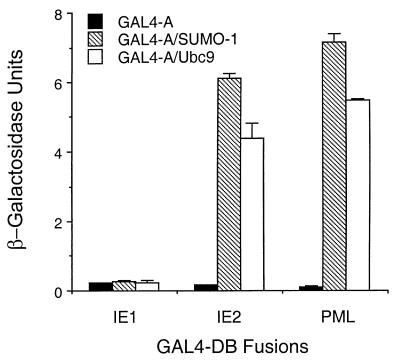
Because most SUMO-1 substrates, including HCMV IE2 and PML, interact with SUMO-1 and/or Ubc9 in yeast two-hybrid assays (7, 26, 53), we were interested to test whether IE1 also interacts with SUMO-1 or Ubc9. Surprisingly, we detected no IE1–SUMO-1 or IE1-Ubc9 interactions after transforming GAL4-DB/IE1 together with GAL4-A/SUMO-1 or GAL4-A/Ubc9 into Y190 yeast cells, whereas IE2 and PML both showed a high level of interaction with both SUMO-1 and Ubc9 (Fig. 4). In a parallel control experiment, this same GAL4-DB/IE1 expression plasmid was able to interact with GAL4-A/PML1 at a level similar to that described previously (2), confirming that the protein was stable and available for interaction in the yeast cells. In agreement with the results in yeast two-hybrid assays, GST-SUMO-1 and GST-Ubc9 both failed to bind to in vitro-translated IE1, whereas both bind to GST-IE2 with high affinity (7). These results suggest that the mechanism of IE1 sumoylation might be very different from that of most previously described SUMO-1 substrates, whose direct binding to Ubc9 and SUMO-1 presumably facilitates the transfer of the SUMO-1 moiety from the Ubc9 thioester intermediate.
FIG. 4.
HCMV IE1 does not interact with SUMO-1 or Ubc9 in yeast two-hybrid assays. Plasmids encoding GAL4-DB fusions and GAL4-A fusions were introduced together into Y190 cells. Transformants were selected on plates lacking Trp and Leu, and β-galactosidase (β-Gal) activities of the transformants were measured by the standard ONPG method as the interaction readout. The unit of β-Gal activity was defined as 1,000(A420 − 1.75A550)/(A600× t × v) (t, reaction time in minutes; v, reaction volume in milliliters). Values are means of duplicate assays; error ranges are indicated.
IE1 sumoylation is not required for its transient targeting to and subsequent disruption of PODs.
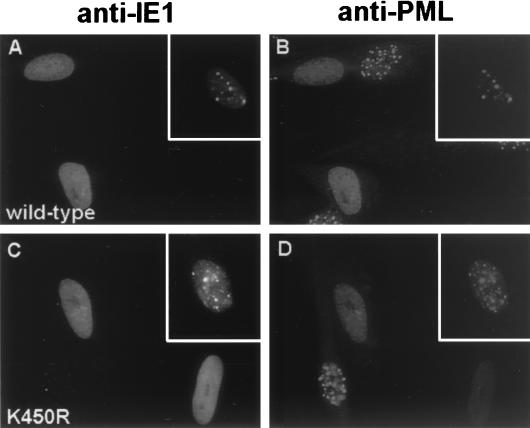
Our previous studies have shown that both the HCMV IE1 and IE2 proteins are targeted to PODs within the first a few hours after virus infection (5). However, IE1 only transiently colocalizes with PML in the PODs, and subsequently both IE1 and PML become redistributed in a nuclear diffuse pattern, whereas IE2 alone has no effect on the distribution of PML. It has been suggested that modification by SUMO-1 is required for the normal localization of PML and some other associated proteins such as hDaxx in the PODs (59, 83). Therefore, after generating the K450R IE1 mutant, which is not modified by SUMO-1, we asked whether the properties of transient targeting to and disruption of PODs might be affected in DNA-transfected HF cells. Double-label IFA analysis was carried out with the PML(C) PAb to detect endogenous punctate POD structures and with MAb 6E1 to detect the IE1 proteins. However, both wild-type IE1 and the mutant IE1(K450R) proteins were found to efficiently colocalize with PML in either nuclear punctate forms (Fig. 5A to D, insets) or as nuclear diffuse patterns (Fig. 5A to D). We confirmed these results by infecting HF cells with either Ad-IE1 or Ad-IE1(K450R) for 12 h in a similar double-label IFA experiment. Again both wild-type and K450R mutant IE1 displayed either nuclear punctate or nuclear diffuse colocalization with PML in the infected cells (data not shown). Therefore, the lack of SUMO-1 modification had no obvious effect on either the targeting to or the disruption of PODs by IE1.
FIG. 5.
Intracellular localization patterns of wild-type IE1 and the sumoylation-negative IE1(K450R) mutant. HF cells were transfected with expression plasmids encoding wild-type IE1 (A and B) or IE1(K450R) (C and D). At 24 h after transfection, the cells were fixed with methanol and double-label IFA was carried out with mouse MAb 6E1 for IE1 (A and C) and rabbit PAb PML(C) for endogenous PML (B and D). Fluorescein isothiocyanate-labeled anti-mouse immunoglobulin G (IgG) and rhodamine-coupled anti-rabbit IgG were used for visualization. Insets, transfected cells with transient punctate colocalization of IE1 and PML.
Stable expression of exogenous PML enhances the level of IE1 sumoylation during HCMV infection.
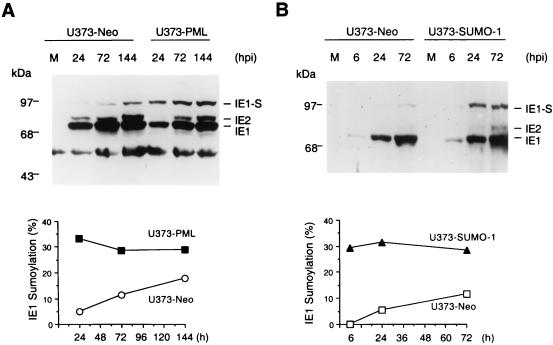
We previously generated a stable U373 cell line overexpressing PML1 (referred to as U373-PML cells) and showed that both the disruption of PODs and the formation of viral DNA replication compartments were delayed during HCMV infection of this cell line (4). Given that IE1 is a substrate for SUMO-1 modification and that the IE1-PML interaction is important for the disruption of PODs by IE1, we were interested to examine the extent of IE1 sumoylation in U373-PML cells. Both U373-PML and the control U373-Neo cells were infected with HCMV(Towne), and cell lysates harvested at various time points after infection were examined by immunoblot analysis with MAb CH810. Although the 90-kDa IE1 isoform accumulated in both cell lines during HCMV infection, the relative abundance of the sumoylated IE1 band was significantly higher in U373-PML cells especially at 24 h after infection (Fig. 6A). Interestingly, the fraction of sumoylated IE1 remained at close to 30% from 24 to 144 h after HCMV infection in U373-PML cells, whereas the sumoylated fraction only gradually increased from 5 to 18% in the control HCMV-infected U373-Neo cells over this same time course (Fig. 6A). These results suggest the existence of a dynamic equilibrium between IE1 sumoylation and desumoylation in HCMV-infected cells. The stable expression of exogenous PML evidently enhanced the steady-state level of IE1 sumoylation, either directly or indirectly, revealing a novel aspect of IE1-PML and IE1-POD interactions that may contribute to the slower virus growth in this cell line.
FIG. 6.
Overexpressed PML or SUMO-1 leads to up-regulated IE1 sumoylation during HCMV infection. U373-MG cells were stably transfected with pCMX-PML1 or pJHA312 encoding Flag-SUMO-1 or an empty vector carrying a Neo resistance marker to establish U373-PML, U373-SUMO-1, and U373-Neo cell lines. (A) Comparison of IE1 sumoylation in U373-PML and U373-Neo cells during HCMV infection. (B) Comparison of IE1 sumoylation in U373-SUMO-1 and U373-Neo cells during HCMV infection. Cells were either mock infected (M) or infected with HCMV(Towne) at an MOI of 5, and cell lysates were harvested at the indicated time points after infection (hours postinfection, hpi). Immunoblot analyses were performed with mouse MAb CH810 recognizing both HCMV IE1 and IE2. After enhanced chemiluminescence visualization of the immunoblots, densitometric quantitation of the 72-kDa unmodified IE1 and the 90-kDa sumoylated IE1 was acquired from X-ray films with a Fluorchem 8000 digital imaging system. The intensities were then used for the calculation of the abundance of the 90-kDa IE1 isoform relative to the total amount of IE1 protein at different time points after infection.
Because PML is a RING finger protein and because a growing number of RING finger domains have been shown to have E3 ubiquitin ligase activities (33, 82), we wondered whether PML might have a direct enzymatic effect on IE1 sumoylation given the fact that PML physically interacts with IE1, SUMO-1, and Ubc9. Therefore, to test whether the PML protein has any direct effect on IE1 sumoylation, we introduced purified GST-PML-L protein into the in vitro SUMO-1 conjugation assay. This form of GST-PML was able to interact with the in vitro-translated IE1 protein in GST binding assays (data not shown). However, no significant increase or decrease of the 90-kDa sumoylated IE1 was observed when the eluted GST-PML-L protein was added to the reaction (Fig. 3B; compare lanes 3 and 2). Since the PML protein is efficiently sumoylated in vitro, GST-PML might compete with IE1 for SUMO-1, thereby masking its potential catalytic effect on IE1 sumoylation. To exclude this possibility, we also fused an N-terminal RING finger fragment of PML with GST and introduced the resulting GST-PML(1-96) protein into the in vitro IE1 sumoylation assay. There is only one SUMO-1 target lysine residue (K65) present in this PML fragment, and Kamitani et al. have shown that sumoylation of K65 is dependent on the sumoylation of K160 and K490 in vivo (36). Therefore, GST-PML(1-96) is not expected to greatly reduce the amount of available SUMO-1 in the in vitro IE1 sumoylation assay. Again, no significant increase or decrease of IE1 sumoylation was observed upon addition of GST-PML(1-96) (data not shown).
Stable expression of exogenous SUMO-1 also enhances the level of IE1 sumoylation during HCMV infection.
The above observations imply that the effect of overexpressed PML on IE1 sumoylation is indirect. One possibility is that overexpressed PML provides a larger intranuclear pool of conjugated forms of SUMO-1. Upon HCMV infection and subsequent IE1-dependent PML desumoylation (58), higher levels of free SUMO-1 may become available for IE1 sumoylation in U373-PML cells. To test this hypothesis, we established another stable U373 cell line overexpressing exogenous Flag-SUMO-1 (U373-SUMO-1 cells) and examined the level of IE1 sumoylation in HCMV-infected U373-SUMO-1 cells with MAb CH810. Similar to the situation in U373-PML cells, the fraction of sumoylated IE1 remained constant at between 28 and 33% from 6 to 72 h after HCMV infection in U373-SUMO-1 cells, whereas the sumoylated fraction again increased gradually from being undetectable at 6 h to up to 12% at 72 h after HCMV infection in the control U373-Neo cells (Fig. 6B). Therefore, stable expression of exogenous SUMO-1 enhances IE1 sumoylation to a level similar to that in U373-PML cells as early as 6 h after HCMV infection.
The displacement of PML from the PODs by IE1 is independent of proteasome activity.
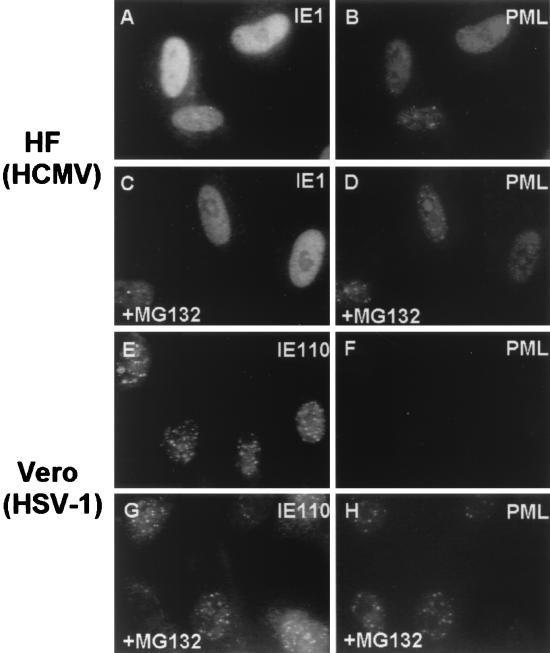
The disruption of PODs by the HSV-1 IE110 (ICP0) protein has been shown to correlate with the rapid proteasome-dependent degradation of PML, and Sp100 and proteasome inhibitors such as MG132 proved to stabilize PODs in HSV-1-infected cells (12, 18, 61). However, the PML signal is just displaced, not lost, in HCMV-infected cells (5), and therefore we were interested to test if proteasome inhibitors have a similar effect on the disruption of PODs during HCMV infection. HF cells were infected with HCMV(Towne) in the presence or absence of proteasome inhibitor MG132 and stained for IE1 and PML at 6 h after infection. Surprisingly, the infected cells showed similar efficiencies of POD disruption in both situations (Fig. 7A to D). In contrast, as a positive control for MG132 activity, Vero cells were infected with HSV-1(KOS) in the presence or absence of MG132 and stained for IE110 and PML at 4 h after infection. Without MG132, almost all of the infected cells had a barely detectable PML signal (Fig. 7E and F). However, in the presence of MG132, a majority of the infected cells retained punctate POD structures, with only a few cells still losing the PML signal (Fig. 7G and H). Therefore, we conclude that the displacement of PML from the PODs by IE1 during HCMV infection does not require proteasome activity and that IE1 utilizes a different mechanism for POD disruption than does HSV-1 IE110.
FIG. 7.
Proteasome-independent disruption of PODs during HCMV infection. HF cells were infected with HCMV(Towne) at an MOI of 5 in the absence (A and B) or presence (C and D) of 5 μM MG132 and stained for IE1 (A and C) and PML (B and D) at 6 h after infection. As a positive control for MG132 activity, Vero cells were infected with HSV-1(KOS) at an MOI of 20 in the absence (E and F) or presence (G and H) of MG132 (5 μM) and stained for IE110 (E and G) and PML (F and H) 4 h after infection.
PML1 represses basal transcription.
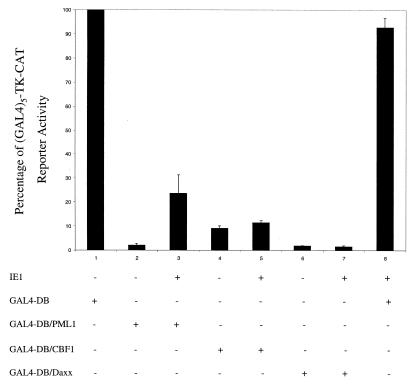
Several studies have documented that the 641-aa PML-L isoform can function as a transcriptional repressor when fused to the GAL4 DNA-binding domain (GAL4-DB) (75, 80). We were interested to test whether the 560-aa PML1 isoform used in all of our studies also has repressor activity in the mammalian GAL4 targeting assay. Therefore, we cotransfected GAL4-DB or GAL4-DB/PML1 with our standard target (GAL4)5-TK-CAT reporter used for repressor assays (27, 28). GAL4-DB/CBF1 (27) and GAL4-DB/Daxx (42) were also used as positive controls for transrepression. The basal activity of the (GAL4)5-TK-CAT target alone in HeLa cells gave 85% acetylation (Fig. 8, lane 1). Cotransfection with both PML1 and Daxx showed a 50-fold inhibition of CAT activity from the (GAL4)5-TK promoter, whereas CBF1 gave a 10-fold inhibition in the same experiment (Fig. 8, lanes 2, 4, and 6). Therefore, targeting PML1 to a promoter can also repress basal transcription despite the lack of the unique C-terminal domain of PML-L that is known to interact with HDAC. Control experiments confirmed that the GAL4-PML1 fusion protein still localizes normally in PODs and is displaced by IE1 cotransfection (not shown).
FIG. 8.
The HCMV IE1 protein specifically inhibits PML1-mediated transcriptional repression. HeLa cells were transiently cotransfected with the (GAL4)5-TK-CAT target gene and the indicated effector genes. Cell lysates were then harvested 48 h after transfection and assayed for CAT activity. Shown are mean values for the percentage of CAT activity in duplicate assays and error ranges. Basal CAT activity upon cotransfection with the control GAL4-DB gene is defined as 100%, which corresponds to 81.7% acetylation in one experiment and 75.2% acetylation in another.
HCMV IE1 specifically inhibits PML-mediated transcriptional repression.
PML1 has been reported recently to inhibit Daxx-mediated transcriptional repression via a physical interaction with Daxx and the consequent sequestration of Daxx to the PODs (42). Given that the proteasome-independent displacement of PML from PODs by IE1 may be mediated through a specific IE1-PML physical interaction (2), we also asked whether IE1 could regulate PML1-mediated transcriptional repression in a GAL4 targeting assay similar to that described above. GAL4-DB/PML1 was transfected with or without the wild-type IE1 expression plasmid into HeLa cells, and the activity of the target (GAL4)5-TK-CAT reporter gene was measured. Interestingly, IE1 coexpression reduced the 50-fold PML repression to less than 5-fold (Fig. 8, lanes 2 and 3). In contrast, IE1 coexpression had little effect on CBF1- or Daxx-mediated repression of the (GAL4)5-TK promoter (Fig. 8, lanes 4 to 7). These data suggest that expression of IE1 specifically overcomes PML-mediated transcriptional repression, presumably either through direct physical interaction with PML or through some associated effect connected to POD disruption.
Disruption of PODs by IE1 but not IE1 sumoylation is required for efficient inhibition of PML-mediated repression.
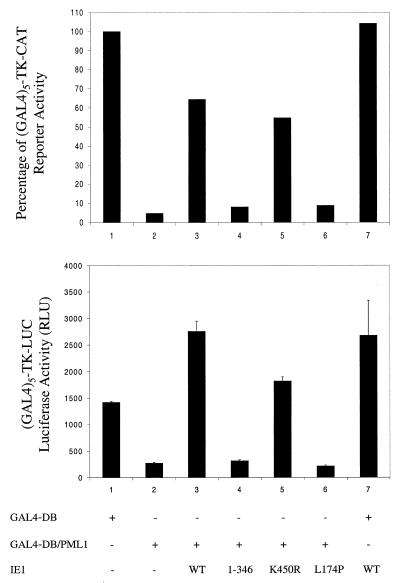
Our previous studies showed that a truncated IE1(1-346) mutant lacking the acidic C terminus was still targeted to and colocalized with PODs but that it nevertheless failed to displace PML from the PODs, thus uncoupling the requirements for targeting from subsequent disruption (2). Muller and Dejean found an IE1(L174P) point mutant that did not interact with PML in yeast and showed that this same mutant protein also failed to target or disrupt PODs (58). Therefore, we generated an L174P mutant IE1 expression plasmid by site-directed mutagenesis and confirmed that the mutant protein was localized exclusively in a nuclear diffuse pattern and failed to displace PML from PODs (data not shown). Given the observation that wild-type IE1 could inhibit PML-mediated transcriptional repression, we asked whether any of the three IE1 expression vector mutants, IE1(K450R), IE1(1-346), or IE1(L174P) (Table 1) retained this derepression activity. In this experiment, HeLa cells were cotransfected with either the (GAL4)5-TK-CAT or the (GAL4)5-TK-LUC reporter gene (67) together with GAL4-DB or GAL4-DB/PML1 in the presence or absence of wild-type or mutant IE1. The control samples (Fig. 9, lanes 3) showed that PML repression occurred equally in both the CAT and luciferase reporter assays, confirming that the effect is likely to be transcriptional and not a reporter gene-specific posttranscriptional-level event such as that mediated by the Epstein-Barr virus MTA protein. Furthermore, the sumoylation-negative IE1(K450R) mutant still overcame PML-mediated repression with an efficiency of greater than 60% of the level obtained with wild-type IE1 (Fig. 9, lanes 5), whereas both the IE1(1-346) and IE1(L174P) mutants failed to inhibit the PML-mediated repression to any measurable extent (Fig. 9, lanes 4 and 6). Importantly, although we cannot exclude the possibility that IE1 alone might have upregulated the (GAL4)5-TK-CAT target, the different kinetics of the luciferase assay show that IE1 alone has no significant effect on the (GAL4)5-TK-LUC target. Therefore, these data indicate that the inhibitory effect of IE1 on PML-mediated transcriptional repression may require its ability to target to and displace PML from the PODs but does not depend on its ability to be sumoylated.
TABLE 1.
Functional properties of wild-type and mutant IE1 proteins
| Protein | Interaction with PML | POD
|
Sumoylation | Reference or source | |
|---|---|---|---|---|---|
| Targeting | Disruption | ||||
| Wild-type IE1a | + | + | + | + | 5 |
| IE1(1-346) | + | + | − | − | 2 |
| IE1(L174P) | − | − | − | + | 58 |
| IE1(K450R) | + | + | + | − | This study |
491 amino acids.
FIG. 9.
The inhibitory effect of IE1 on PML-mediated repression requires its ability to displace PML from PODs. HeLa cells were transiently cotransfected with either the (GAL4)5-TK-CAT (top) or the (GAL4)5-TK-LUC (bottom) target gene and the indicated effector genes. Cell lysates were then harvested at 48 h after transfection and assayed for CAT activity or luciferase activity, respectively. (Top) Relative values of CAT activity. Basal CAT activity upon cotransfection with GAL4-DB is defined as 100%, which corresponds to 90.2% acetylation in one experiment. (Bottom) Mean values for relative light units (RLU) in duplicate assays, with error ranges indicated.
DISCUSSION
IE1 is the most abundant protein expressed from the strongly transcribed MIE locus of the HCMV genome, yet its functional role in HCMV infection and permissiveness remains elusive. In this study, we showed that IE1, like the other abundant MIE gene product, IE2 (7, 26), is an authentic SUMO-1 substrate both in vivo and in vitro and that its interaction with PML-containing PODs, a major site of SUMO-1 accumulation in the nucleus, has significant functional consequences. These two HCMV immediate-early gene products were the first viral proteins shown to be subject to this type of cellular posttranslational modification. More recently, the Zta protein of Epstein-Barr virus (1) and the E1 protein of bovine papillomavirus (64) were also found to be SUMO-1 substrates, implying the potentially broad significance of SUMO-1 modification for viral gene regulation. Here we provided evidence that IE1 can inhibit PML-mediated repression of basal transcription, revealing a novel functional aspect of HCMV IE1. We also suggest that this newly discovered derepression activity of IE1 is associated with its ability to displace PML from the PODs but not with its ability to undergo SUMO-1 modification.
Initially, to address the difficult question of the functional consequences of IE1 sumoylation, we had only two unambiguous assays for functional roles of IE1 to consider (1): the displacement of PML from PODs and (2) and the enhancement of plaque-forming efficiency and lytic-cycle progression upon low-MOI infection of permissive HF cells. Only the former seems to be a direct effect, whereas the latter potentially involves mechanisms to stimulate the activity of other viral regulatory proteins, most likely including IE2 (4). After mapping lysine residue 450 as the major SUMO-1 conjugation site in IE1, we found that the sumoylation-negative IE1(K450R) mutant protein still transiently colocalizes with and subsequently displaces PML. Therefore, SUMO-1 modification of IE1 is not an absolute requirement for its direct targeting to or disruptive effects on the POD structure. The IE1(K450R) mutant and wild-type IE1 proteins also showed no significant difference in efficiency of interaction with PML in yeast two-hybrid assays (data not shown). However, additional studies will be required to address whether IE1(K450R) has reduced affinity for PODs or displays an altered temporal pattern of displacement during virus infection. All of the known functional characteristics of different IE1 mutants are summarized in Table 1. Apparently, the transient targeting of PODs, displacement of PML from PODs, and covalent modification by SUMO-1 may be independent functional aspects of IE1, although the targeting to PODs may correlate with a physical interaction with PML. Whether or not IE1 requires PML to be sumoylated for POD targeting remains to be determined.
Surprisingly, unlike PML or IE2, IE1 did not interact significantly with either Ubc9 or SUMO-1 in yeast two-hybrid assays (Fig. 4). In fact, IE1 appears to be the first SUMO-1 target ever reported that shows no interaction with either Ubc9 or SUMO-1 in such assays. This observation immediately raises an intriguing question about the exact mechanism that leads to IE1 sumoylation. The observed low efficiency of IE1 sumoylation in vitro (in contrast to that of IE2 and PML) also might reflect a requirement for an unidentified E3 SUMO ligase that needs to bind to the substrate to promote the transfer of the SUMO moiety, either directly or indirectly, from the Ubc9-SUMO thioester intermediate to an amide linkage with the target lysine side chain. Alternatively, some intranuclear cofactor regulating the phosphorylation status of IE1 could perhaps contribute to the regulation of IE1 sumoylation.
Interestingly, our results showed that stable overexpression of either exogenous SUMO-1 or PML1 protein in U373 cells enhanced the relative level of in vivo IE1 sumoylation in mammalian cells up to sixfold early during HCMV infection (Fig. 6). However, addition of GST-PML-L to the reaction mixture gave no increase in in vitro IE1 sumoylation (Fig. 3B). These observations suggest that the effect of PML on IE1 sumoylation levels is indirect. IE1 sumoylation reached a peak level of about 30% in U373-PML cells as early as 24 h after HCMV infection, whereas the relative abundance of sumoylated IE1 built up slowly from a much lower level in the control U373-Neo cells from 24 to 144 h after infection (Fig. 6A). Our previous studies showed that PML displacement during HCMV infection was highly delayed in U373-PML cells, with only 20% of the cells showing nuclear diffuse PML at 9 h, whereas at the same time point PML was redistributed in at least 95% of HCMV-infected U373-Neo cells (4). Nevertheless, PML was eventually displaced together with IE1 at later time points, with only 5% of U373-PML cells still showing punctate PML by 22 h after HCMV infection (4). Although Muller and Dejean showed that IE1(L174P) could still be sumoylated (58), it remains to be determined whether the disruption of PODs contributes to the regulation of IE1 and IE2 sumoylation during HCMV infection.
How can the indirect effect of PML on IE1 sumoylation be explained? Given the observation that the steady-state amount of unconjugated free SUMO-1 is very limited in normal cells (15, 51), it is conceivable that SUMO-1 needs to be deconjugated from cellular substrates for newly synthesized viral proteins to be sumoylated. The various PML isoforms are major intranuclear SUMO-1 substrates which contain three SUMO-1 conjugation sites per molecule. Muller and Dejean showed that high levels of wild-type IE1 either blocked or removed conjugated SUMO from cotransfected exogenous PML and Sp100, whereas the L174P mutant IE1 did not have this activity (58). We propose that overexpression of exogenous PML results in more SUMO-1 molecules being conjugated to POD component proteins such as PML and Sp100. Upon HCMV infection of U373-PML cells and the eventual disruption of the larger granular PODs, a larger pool of free SUMO-1 is released; this tips the dynamic conjugation-deconjugation equilibrium toward IE1 sumoylation and accelerates the establishment of a steady-state level of sumoylated IE1. This hypothesis was supported by our observation that stable expression of exogenous Flag-SUMO-1 in U373 cells enhanced the level of IE1 sumoylation during HCMV infection, with a kinetic profile similar to that for U373-PML cells (Fig. 6B). However, whether HCMV infection is also slowed down in the latter cell line remains to be determined. An anti-Flag immunoblot analysis of U373-SUMO-1 cell lysates revealed only a small amount of the 20-kDa free Flag-SUMO-1, suggesting that even overexpressed exogenous SUMO-1 exists mainly as conjugated forms (J.-H. Ahn, Y. Xu, and G. S. Hayward, unpublished data). Therefore, the deconjugation of SUMO-1 from cellular proteins such as PML and Sp100 might provide an important reservoir for HCMV IE1 and IE2 proteins to be efficiently modified by SUMO-1.
Recent studies demonstrate that the longer PML-L isoform can repress basal transcription by functionally and physically interacting with HDAC (80). This physical interaction requires the unique C-terminal domain of PML-L, which is missing in the 560-aa PML1 isoform. As a result, PML1 was found not to interact with any of the three HDAC isoforms directly (42, 80). Nevertheless, we found that the shorter PML1 isoform still represses basal transcription with high efficiency (Fig. 8). This finding suggests either of two possible scenarios: (i) PML1 may repress basal transcriptional via an HDAC-independent mechanism or (ii) PML1 might still recruit HDAC for transcriptional repression via an adapter protein, such as Rb or hDaxx, which interacts with both PML and HDAC (8, 9, 30, 42, 46, 47). Future investigations are necessary to distinguish between these two possibilities.
We found that IE1-mediated displacement of PML from PODs is a proteasome-independent process distinctly different from that for HSV IE110, which tends to lead to degradation of PML. However, inhibition of PML repression by IE1 and degradation of PML by IE110 could produce similar overall results in the two viruses. Furthermore, the implication that the ability of IE1 to inhibit PML-mediated transcriptional repression depends on POD disruption strengthens our earlier concept that disruption of PODs by IE1 is important for efficient viral gene expression during HCMV infection (4). When the PML1-overexpressing U373-PML cells were infected with recombinant HCMV expressing an extragenic luciferase reporter gene under the control of a viral early (Pol) or late (pp28) promoter, their transcriptional activation was reduced up to fivefold at both low and high MOI (4). These observations suggest that the transcriptional-repression activity of PML1 might be utilized by the host cell to delay viral gene expression. However, it is important to point out that other POD component proteins probably also play a role in regulating gene transcription (84), and it is unknown at present whether IE1 also affects the transcriptional roles of any other POD proteins in addition to PML.
ACKNOWLEDGMENTS
Y. Xu and J.-H. Ahn contributed equally to this work.
This study was funded by Public Health Service research grant RO1 AI24576 to G.S.H. from the National Institute of Allergy and Infectious Diseases.
We thank Dolores Ciufo for generating the PML(C) and IE110(N) polyclonal antibodies and Robert LaFemina for establishing the U373-A3 cell line. We are grateful to Stephen Hardy for the pAdlox vector, Ψ5 virus, and CRE8 cells, Kun-Sang Chang for the GST-PML-L plasmid, Susan Michelson for the U373-AN and U373-A45 cell lines, and Stephen Baylin for the (GAL4)5-TK-LUC plasmid.
REFERENCES
- 1.Adamson A L, Kenney S. Epstein-Barr virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J Virol. 2001;75:2388–2399. doi: 10.1128/JVI.75.5.2388-2399.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn J H, Brignole III E J, Hayward G S. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol Cell Biol. 1998;18:4899–4913. doi: 10.1128/mcb.18.8.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn J H, Chiou C J, Hayward G S. Evaluation and mapping of the DNA binding and oligomerization domains of the IE2 regulatory protein of human cytomegalovirus using yeast one and two hybrid interaction assays. Gene. 1998;210:25–36. doi: 10.1016/s0378-1119(98)00056-0. [DOI] [PubMed] [Google Scholar]
- 4.Ahn J H, Hayward G S. Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection. Virology. 2000;274:39–55. doi: 10.1006/viro.2000.0448. [DOI] [PubMed] [Google Scholar]
- 5.Ahn J H, Hayward G S. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol. 1997;71:4599–4613. doi: 10.1128/jvi.71.6.4599-4613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahn J H, Jang W J, Hayward G S. The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10) J Virol. 1999;73:10458–10471. doi: 10.1128/jvi.73.12.10458-10471.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahn J H, Xu Y, Jang W J, Matunis M J, Hayward G S. Evaluation of interactions of the human cytomegalovirus immediate-early IE2 regulatory protein with small ubiquitin-like modifiers and their conjugation enzyme Ubc9. J Virol. 2001;75:3859–3872. doi: 10.1128/JVI.75.8.3859-3872.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alcalay M, Tomassoni L, Colombo E, Stoldt S, Grignani F, Fagioli M, Szekely L, Helin K, Pelicci P G. The promyelocytic leukemia gene product (PML) forms stable complexes with the retinoblastoma protein. Mol Cell Biol. 1998;18:1084–1093. doi: 10.1128/mcb.18.2.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 10.Buschmann T, Fuchs S Y, Lee C G, Pan Z Q, Ronai Z. SUMO-1 modification of Mdm2 prevents its self-ubiquitination and increases Mdm2 ability to ubiquitinate p53. Cell. 2000;101:753–762. doi: 10.1016/s0092-8674(00)80887-9. [DOI] [PubMed] [Google Scholar]
- 11.Chang C P, Malone C L, Stinski M F. A human cytomegalovirus early gene has three inducible promoters that are regulated differentially at various times after infection. J Virol. 1989;63:281–290. doi: 10.1128/jvi.63.1.281-290.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chelbi-Alix M K, de The H. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene. 1999;18:935–941. doi: 10.1038/sj.onc.1202366. [DOI] [PubMed] [Google Scholar]
- 13.Cherrington J M, Khoury E L, Mocarski E S. Human cytomegalovirus IE2 negatively regulates alpha gene expression via a short target sequence near the transcription start site. J Virol. 1991;65:887–896. doi: 10.1128/jvi.65.2.887-896.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cherrington J M, Mocarski E S. Human cytomegalovirus IE1 transactivates the alpha promoter-enhancer via an 18-base-pair repeat element. J Virol. 1989;63:1435–1440. doi: 10.1128/jvi.63.3.1435-1440.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desterro J M, Rodriguez M S, Hay R T. SUMO-1 modification of IkappaBalpha inhibits NF-κB activation. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 16.de The H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- 17.Doucas V, Evans R M. Human T-cell leukemia retrovirus-Tax protein is a repressor of nuclear receptor signaling. Proc Natl Acad Sci USA. 1999;96:2633–2638. doi: 10.1073/pnas.96.6.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett R D, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J Virol. 1998;72:6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fagioli M, Alcalay M, Pandolfi P P, Venturini L, Mencarelli A, Simeone A, Acampora D, Grignani F, Pelicci P G. Alternative splicing of PML transcripts predicts coexpression of several carboxy-terminally different protein isoforms. Oncogene. 1992;7:1083–1091. [PubMed] [Google Scholar]
- 20.Gostissa M, Hengstermann A, Fogal V, Sandy P, Schwarz S E, Scheffner M, Del Sal G. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 1999;18:6462–6471. doi: 10.1093/emboj/18.22.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greaves R F, Mocarski E S. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J Virol. 1998;72:366–379. doi: 10.1128/jvi.72.1.366-379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guiochon-Mantel A, Savouret J F, Quignon F, Delabre K, Milgrom E, De The H. Effect of PML and PML-RAR on the transactivation properties and subcellular distribution of steroid hormone receptors. Mol Endocrinol. 1995;9:1791–1803. doi: 10.1210/mend.9.12.8614415. [DOI] [PubMed] [Google Scholar]
- 23.Hardy S, Kitamura M, Harris-Stansil T, Dai Y, Phipps M L. Construction of adenovirus vectors through Cre-lox recombination. J Virol. 1997;71:1842–1849. doi: 10.1128/jvi.71.3.1842-1849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayhurst G P, Bryant L A, Caswell R C, Walker S M, Sinclair J H. CCAAT box-dependent activation of the TATA-less human DNA polymerase alpha promoter by the human cytomegalovirus 72-kilodalton major immediate-early protein. J Virol. 1995;69:182–188. doi: 10.1128/jvi.69.1.182-188.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hochstrasser M. Evolution and function of ubiquitin-like protein-conjugation systems. Nat Cell Biol. 2000;2:E153–E157. doi: 10.1038/35019643. [DOI] [PubMed] [Google Scholar]
- 26.Hofmann H, Floss S, Stamminger T. Covalent modification of the transactivator protein IE2–p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J Virol. 2000;74:2510–2524. doi: 10.1128/jvi.74.6.2510-2524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsieh J J, Hayward S D. Masking of the CBF1/RBPJ kappa transcriptional repression domain by Epstein-Barr virus EBNA2. Science. 1995;268:560–563. doi: 10.1126/science.7725102. [DOI] [PubMed] [Google Scholar]
- 28.Hsieh J J, Zhou S, Chen L, Young D B, Hayward S D. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc Natl Acad Sci USA. 1999;96:23–28. doi: 10.1073/pnas.96.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishov A M, Maul G G. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishov A M, Sotnikov A G, Negorev D, Vladimirova O V, Neff N, Kamitani T, Yeh E T, Strauss III J F, Maul G G. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J Cell Biol. 1999;147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishov A M, Stenberg R M, Maul G G. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J Cell Biol. 1997;138:5–16. doi: 10.1083/jcb.138.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeang K T, Gibson W. A cycloheximide-enhanced protein in cytomegalovirus-infected cells. Virology. 1980;107:362–374. doi: 10.1016/0042-6822(80)90304-9. [DOI] [PubMed] [Google Scholar]
- 33.Joazeiro C A, Weissman A M. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 34.Johnson E S, Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol. 1999;147:981–994. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kakizuka A, Miller W H, Umesono K, Warrell R P, Frankel S R, Murty V V, Dmitrovsky E, Evans R M. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 36.Kamitani T, Kito K, Nguyen H P, Wada H, Fukuda-Kamitani T, Yeh E T. Identification of three major sentrinization sites in PML. J Biol Chem. 1998;273:26675–26682. doi: 10.1074/jbc.273.41.26675. [DOI] [PubMed] [Google Scholar]
- 37.Klucher K M, Sommer M, Kadonaga J T, Spector D H. In vivo and in vitro analysis of transcriptional activation mediated by the human cytomegalovirus major immediate-early proteins. Mol Cell Biol. 1993;13:1238–1250. doi: 10.1128/mcb.13.2.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korioth F, Maul G G, Plachter B, Stamminger T, Frey J. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp Cell Res. 1996;229:155–158. doi: 10.1006/excr.1996.0353. [DOI] [PubMed] [Google Scholar]
- 39.LaFemina R L, Hayward G S. Differences in cell-type-specific blocks to immediate early gene expression and DNA replication of human, simian and murine cytomegalovirus. J Gen Virol. 1988;69:355–374. doi: 10.1099/0022-1317-69-2-355. [DOI] [PubMed] [Google Scholar]
- 40.LaFemina R L, Pizzorno M C, Mosca J D, Hayward G S. Expression of the acidic nuclear immediate-early protein (IE1) of human cytomegalovirus in stable cell lines and its preferential association with metaphase chromosomes. Virology. 1989;172:584–600. doi: 10.1016/0042-6822(89)90201-8. [DOI] [PubMed] [Google Scholar]
- 41.Lang D, Stamminger T. The 86-kilodalton IE-2 protein of human cytomegalovirus is a sequence-specific DNA-binding protein that interacts directly with the negative autoregulatory response element located near the cap site of the IE-1/2 enhancer-promoter. J Virol. 1993;67:323–331. doi: 10.1128/jvi.67.1.323-331.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Leo C, Zhu J, Wu X, O'Neil J, Park E J, Chen J D. Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol Cell Biol. 2000;20:1784–1796. doi: 10.1128/mcb.20.5.1784-1796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu B, Hermiston T W, Stinski M F. A cis-acting element in the major immediate-early (IE) promoter of human cytomegalovirus is required for negative regulation by IE2. J Virol. 1991;65:897–903. doi: 10.1128/jvi.65.2.897-903.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lukac D M, Harel N Y, Tanese N, Alwine J C. TAF-like functions of human cytomegalovirus immediate-early proteins. J Virol. 1997;71:7227–7239. doi: 10.1128/jvi.71.10.7227-7239.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lukac D M, Manuppello J R, Alwine J C. Transcriptional activation by the human cytomegalovirus immediate-early proteins: requirements for simple promoter structures and interactions with multiple components of the transcription complex. J Virol. 1994;68:5184–5193. doi: 10.1128/jvi.68.8.5184-5193.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 47.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain J P, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 48.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 49.Malone C L, Vesole D H, Stinski M F. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J Virol. 1990;64:1498–1506. doi: 10.1128/jvi.64.4.1498-1506.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Margolis M J, Pajovic S, Wong E L, Wade M, Jupp R, Nelson J A, Azizkhan J C. Interaction of the 72-kilodalton human cytomegalovirus IE1 gene product with E2F1 coincides with E2F-dependent activation of dihydrofolate reductase transcription. J Virol. 1995;69:7759–7767. doi: 10.1128/jvi.69.12.7759-7767.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matunis M J, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matunis M J, Wu J, Blobel G. SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1 to the nuclear pore complex. J Cell Biol. 1998;140:499–509. doi: 10.1083/jcb.140.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Melchior F. SUMO—nonclassical ubiquitin. Annu Rev Cell Dev Biol. 2000;16:591–626. doi: 10.1146/annurev.cellbio.16.1.591. [DOI] [PubMed] [Google Scholar]
- 54.Michelson S, Alcami J, Kim S J, Danielpour D, Bachelerie F, Picard L, Bessia C, Paya C, Virelizier J L. Human cytomegalovirus infection induces transcription and secretion of transforming growth factor β1. J Virol. 1994;68:5730–5737. doi: 10.1128/jvi.68.9.5730-5737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mocarski E S. Cytomegalovirus and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2447–2492. [Google Scholar]
- 56.Mullen M A, Gerstberger S, Ciufo D M, Mosca J D, Hayward G S. Evaluation of colocalization interactions between the IE110, IE175, and IE63 transactivator proteins of herpes simplex virus within subcellular punctate structures. J Virol. 1995;69:476–491. doi: 10.1128/jvi.69.1.476-491.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muller S, Berger M, Lehembre F, Seeler J S, Haupt Y, Dejean A. c-Jun and p53 activity is modulated by SUMO-1 modification. J Biol Chem. 2000;275:13321–13329. doi: 10.1074/jbc.275.18.13321. [DOI] [PubMed] [Google Scholar]
- 58.Muller S, Dejean A. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J Virol. 1999;73:5137–5143. doi: 10.1128/jvi.73.6.5137-5143.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muller S, Matunis M J, Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 61.Parkinson J, Everett R D. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J Virol. 2000;74:10006–10017. doi: 10.1128/jvi.74.21.10006-10017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pizzorno M C, Hayward G S. The IE2 gene products of human cytomegalovirus specifically down-regulate expression from the major immediate-early promoter through a target sequence located near the cap site. J Virol. 1990;64:6154–6165. doi: 10.1128/jvi.64.12.6154-6165.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pizzorno M C, O'Hare P, Sha L, LaFemina R L, Hayward G S. trans-Activation and autoregulation of gene expression by the immediate-early region 2 gene products of human cytomegalovirus. J Virol. 1988;62:1167–1179. doi: 10.1128/jvi.62.4.1167-1179.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rangasamy D, Woytek K, Khan S A, Wilson V G. SUMO-1 modification of bovine papillomavirus E1 protein is required for intranuclear accumulation. J Biol Chem. 2000;275:37999–38004. doi: 10.1074/jbc.M007777200. [DOI] [PubMed] [Google Scholar]
- 65.Rodriguez M S, Desterro J M, Lain S, Midgley C A, Lane D P, Hay R T. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 1999;18:6455–6461. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 67.Rountree M R, Bachman K E, Baylin S B. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 68.Sarisky R T, Hayward G S. Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays. J Virol. 1996;70:7398–7413. doi: 10.1128/jvi.70.11.7398-7413.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Satijn D P, Gunster M J, van der Vlag J, Hamer K M, Schul W, Alkema M J, Saurin A J, Freemont P S, van Driel R, Otte A P. RING1 is associated with the polycomb group protein complex and acts as a transcriptional repressor. Mol Cell Biol. 1997;17:4105–4113. doi: 10.1128/mcb.17.7.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shi Y, Seto E, Chang L S, Shenk T. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 71.Stenberg R M, Fortney J, Barlow S W, Magrane B P, Nelson J A, Ghazal P. Promoter-specific trans-activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J Virol. 1990;64:1556–1565. doi: 10.1128/jvi.64.4.1556-1565.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stenberg R M, Witte P R, Stinski M F. Multiple spliced and unspliced transcripts from human cytomegalovirus immediate-early region 2 and evidence for a common initiation site within immediate-early region 1. J Virol. 1985;56:665–675. doi: 10.1128/jvi.56.3.665-675.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vallian S, Chin K V, Chang K S. The promyelocytic leukemia protein interacts with Sp1 and inhibits its transactivation of the epidermal growth factor receptor promoter. Mol Cell Biol. 1998;18:7147–7156. doi: 10.1128/mcb.18.12.7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vallian S, Gaken J A, Gingold E B, Kouzarides T, Chang K S, Farzaneh F. Modulation of Fos-mediated AP-1 transcription by the promyelocytic leukemia protein. Oncogene. 1998;16:2843–2853. doi: 10.1038/sj.onc.1201837. [DOI] [PubMed] [Google Scholar]
- 75.Vallian S, Gaken J A, Trayner I D, Gingold E B, Kouzarides T, Chang K S, Farzaneh F. Transcriptional repression by the promyelocytic leukemia protein, PML. Exp Cell Res. 1997;237:371–382. doi: 10.1006/excr.1997.3801. [DOI] [PubMed] [Google Scholar]
- 76.Vo N, Goodman R H. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- 77.Waheed I, Chiou C J, Ahn J H, Hayward G S. Binding of the human cytomegalovirus 80-kDa immediate-early protein (IE2) to minor groove A/T-rich sequences bounded by CG dinucleotides is regulated by protein oligomerization and phosphorylation. Virology. 1998;252:235–257. doi: 10.1006/viro.1998.9448. [DOI] [PubMed] [Google Scholar]
- 78.Wathen M W, Stinski M F. Temporal patterns of human cytomegalovirus transcription: mapping the viral RNAs synthesized at immediate early, early, and late times after infection. J Virol. 1982;41:462–477. doi: 10.1128/jvi.41.2.462-477.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wilkinson G W, Kelly C, Sinclair J H, Rickards C. Disruption of PML-associated nuclear bodies mediated by the human cytomegalovirus major immediate early gene product. J Gen Virol. 1998;79:1233–1245. doi: 10.1099/0022-1317-79-5-1233. [DOI] [PubMed] [Google Scholar]
- 80.Wu W S, Vallian S, Seto E, Yang W M, Edmondson D, Roth S, Chang K S. The growth suppressor PML represses transcription by functionally and physically interacting with histone deacetylases. Mol Cell Biol. 2001;21:2259–2268. doi: 10.1128/MCB.21.7.2259-2268.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yoo Y D, Chiou C J, Choi K S, Yi Y, Michelson S, Kim S, Hayward G S, Kim S J. The IE2 regulatory protein of human cytomegalovirus induces expression of the human transforming growth factor β1 gene through an Egr-1 binding site. J Virol. 1996;70:7062–7070. doi: 10.1128/jvi.70.10.7062-7070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zheng N, Wang P, Jeffrey P D, Pavletich N P. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 83.Zhong S, Muller S, Ronchetti S, Freemont P S, Dejean A, Pandolfi P P. Role of SUMO-1-modified PML in nuclear body formation. Blood. 2000;95:2748–2752. [PubMed] [Google Scholar]
- 84.Zhong S, Salomoni P, Pandolfi P P. The transcriptional role of PML and the nuclear body. Nat Cell Biol. 2000;2:E85–E90. doi: 10.1038/35010583. [DOI] [PubMed] [Google Scholar]
- 85.Zhu H, Shen Y, Shenk T. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J Virol. 1995;69:7960–7970. doi: 10.1128/jvi.69.12.7960-7970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]