Abstract
Despite the discovery of Epstein-Barr virus more than 35 years ago, a thorough understanding of gammaherpesvirus capsid composition and structure has remained elusive. We approached this problem by purifying capsids from Kaposi's sarcoma-associated herpesvirus (KSHV), the only other known human gammaherpesvirus. The results from our biochemical and imaging analyses demonstrate that KSHV capsids possess a typical herpesvirus icosahedral capsid shell composed of four structural proteins. The hexameric and pentameric capsomers are composed of the major capsid protein (MCP) encoded by open reading frame 25. The heterotrimeric complexes, forming the capsid floor between the hexons and pentons, are each composed of one molecule of ORF62 and two molecules of ORF26. Each of these proteins has significant amino acid sequence homology to capsid proteins in alpha- and betaherpesviruses. In contrast, the fourth protein, ORF65, lacks significant sequence homology to its structural counterparts from the other subfamilies. Nevertheless, this small, basic, and highly antigenic protein decorates the surface of the capsids, as does, for example, the even smaller basic capsid protein VP26 of herpes simplex virus type 1. We have also found that, as with the alpha- and betaherpesviruses, lytic replication of KSHV leads to the formation of at least three capsid species, A, B, and C, with masses of approximately 200, 230, and 300 MDa, respectively. A capsids are empty, B capsids contain an inner array of a fifth structural protein, ORF17.5, and C capsids contain the viral genome.
Kaposi's sarcoma (KS), a multicentric angiogenic tumor of mixed cellularity, is the leading neoplasm of patients with AIDS. Molecular and seroepidemiologic data demonstrate that a rhadinovirus, KS-associated herpesvirus (KSHV), also known as human herpesvirus 8, is the infectious cause of KS (17, 21, 26, 28, 35, 39). KSHV is also associated with primary effusion lymphoma (PEL), a clonal B-cell tumor, and multicentric Castleman's disease, a rare lymphoproliferative disorder (7, 36). Although the identity of the cell population initially infected with KSHV remains unclear, infected but asymptomatic individuals often demonstrate latent virus in their circulating B cells (47) and macrophages (1). In KS lesions, the virus is present in the hallmark spindle cells and in some of the endothelial cells lining vascular spaces (4, 37).
Since most otherwise healthy KSHV-infected individuals remain disease free, a healthy cellular immune response probably keeps active viral replication in check. In contrast, immunosuppression can lead to viral reactivation, replication, and widespread dissemination. It is in this setting that pathogenic progression can ensue. Tumor formation probably requires not only an initial infection of critical numbers of target cells but also the continual recruitment of new cells to replace those lysed from low levels of spontaneous lytic viral replication (11, 37). Similarly, human-to-human transmission, even in the absence of overt disease, presumably relies on horizontal spread of KSHV. Such processes clearly depend on viral replication, including successful formation of infectious particles. As with all herpesviruses, the first structures to appear following the initiation of KSHV replication are the capsids—the icosahedral particles that fill the nucleus and, when fully mature, harbor the linear viral genome.
Herpesvirus structure and assembly.
Studies of alpha- and betaherpesviruses indicate that mature herpesviruses comprise three distinct structural layers plus an inner DNA core. Most information on herpesvirus structure stems from work on these two branches of the herpesvirus family, with the largest portion reflecting data from herpes simplex virus type 1 (HSV-1). In alpha- and betaherpesviruses, the innermost layer is the capsid, consisting of a highly ordered icosahedral structure with a triangulation number (T) of 16 (reviewed in references 14 and 38). Only a portion of the synthesized capsids undergoes viral DNA packaging. When this occurs, the encapsidated DNA is present as a single linear copy of the viral genome and is free of nucleosomes or other DNA binding proteins (2). The KSHV genome also follows this paradigm, assuming a linear form in virions and a circular form during latency (9, 32). A fraction of these DNA-containing capsids, as well as some DNA-free capsids (empty or A capsids [see below]), then acquire first a spherical halo of proteins known as the tegument and second a surrounding envelope with a cadre of integrated proteins. Capsids probably acquire the two outer layers while budding through the nuclear membrane into the cytoplasm (5, 14). Groups of these enveloped particles then transcytose within vesicles toward the plasma membrane. Fusion of the vesicles with the plasma membrane releases the viral particles into the extracellular space.
Capsid architecture.
One of the first and essential steps in the cascade of events that leads to viral production is the synthesis of the highly ordered capsid structures. During lytic replication of HSV-1, for example, multiple capsid forms arise (12). These include (i) A capsids (empty icosahedral shells lacking DNA or any other discernible internal structure), (ii) B capsids (shells containing an inner array of scaffolding protein), and (iii) C capsids (shells with packaged DNA and no scaffolding protein). Although their interpretation remains controversial, early pulse-chase experiments suggest that B capsids may mature to C capsids that, in turn, serve as the infectious virus precursors (30). The A capsids probably represent dead-end products derived from either the inappropriate loss of DNA from a C capsid or the premature release of scaffolding protein from a B capsid (without concurrent DNA packaging) (18, 20).
All three capsid types, however, have a common shell structure that consists of 150 hexameric and 12 pentameric capsomers made up exclusively of the major capsid protein (MCP). MCP is the single largest contributor to the capsid's mass and is a protein that is well conserved throughout the herpesvirus family. The capsomeric pentons each have fivefold rotational symmetry, and one is located at each capsid vertex. The hexons have sixfold symmetry and compose the capsid edges and faces. The capsomers are connected in groups of three by the triplexes, asymmetric structures that lie on the capsid floor at the base of the capsomer protrusions (reviewed by Homa and Brown [14]). In HSV-1, each triplex is made up of one molecule of viral protein 19C (VP19C) and two molecules of VP23. Another small protein (VP26 in HSV-1) resides at the tips of each MCP subunit in the hexons (but not the pentons) (3, 50) and may interact with a component of the overlying tegument (44).
Alpha- and betaherpesvirus B capsids also contain a critical fifth protein, the scaffolding protein, which binds, through its C terminus, to a single MCP molecule (13, 27, 41). Elegant in vitro reconstitution experiments with cell extracts programmed with the essential HSV-1 capsid genes suggest that these scaffolding-MCP heteroduplexes probably self-aggregate in the nucleus, forming essentially three-dimensional arcs. These eventually grow either by adding more heterodimers to their edges or aggregating with other such arcs to form an intact spherical shell with an inner spoke-like array of scaffolding protein (24, 42).
In contrast to the investigations into the structure and assembly of the alpha- and betaherpesvirus capsids, similar attempts with gammaherpesviruses have only recently begun to make progress. The single examination of KSHV capsid structure published to date employed cryoelectron microscopy to generate a three-dimensional reconstruction of the capsid at 24-Å resolution. The work clearly demonstrated that the KSHV capsid possesses typical herpesvirus icosahedral geometry and compared its contours with that of HSV-1 (48). However, in the absence of complementary biochemical studies, reconstructions from cryoelectron micrographs are unable to support definitive conclusions regarding protein composition. We have purified structurally intact capsids in ways that allowed parallel biochemical, structural, and imaging analyses. We identified and purified three distinct capsid species that arise during lytic KSHV replication. We then defined the protein and nucleic acid composition of each species and ascertained the viral genes encoding their protein components. Together, the data provide a firm foundation from which to interpret KSHV capsid structure and assembly studies while also allowing comparisons with other herpesvirus subfamilies.
MATERIALS AND METHODS
Cell culture.
BCBL-1 is a B-cell line derived from a primary effusion lymphoma that is latently infected with KSHV; no Epstein-Barr virus (EBV) DNA is present in this line (33). The cells were maintained as described previously (33), except that the medium was additionally buffered with 20 mM HEPES (pH 7.3).
Isolation of KSHV capsids.
KSHV virions and released capsids were isolated essentially as we have described previously for whole virions (33). In brief, 1 to 2 liters of BCBL-1 grown to 2 × 105 to 3 × 105 cells/ml were treated with both 20 ng of 12-O-tetradecanoyl phorbol 13-acetate (TPA) per ml and 0.3 mM sodium butyrate for 12 to 18 h. The cells were then changed to their standard medium and incubated for another 6 to 7 days. The medium was centrifuged (600 × g for 5 min and then 2,000 × g for 30 min) to sediment the cells, nuclei, and large debris, and then the viral and subviral particles were pelleted from this cleared medium by ultracentrifugation (50,000 × g for 2 h). The pellet was resuspended in DNase buffer (10 mM MnCl2, 50 mM Tris HCl [pH 7.5]) with 0.03 U of DNase I (Roche Molecular Biochemicals) per ml and incubated for 30 min at 37°C. The reaction was stopped on ice with 20 mM EDTA (pH 8.0). Tris HCl (pH 8.0) and NaCl were then added to give final concentrations of 20 and 250 mM, respectively, and a protease inhibitor cocktail (PILL; Roche Molecular Biochemicals), diluted as specified by the manufacturer was added. Triton X-100 was then added to a final concentration of 2%, and the mixture was incubated overnight at 4°C. This mixture was sonicated in a bath for 15 s and then sedimented (75,000 × g for 30 min) through a 35% (wt/vol) sucrose cushion made up in 20 mM Tris HCl (pH 8)–250 mM NaCl–1 mM EDTA (MTNE). The resulting pellet was resuspended, sonicated as above, loaded (60 μl/gradient) onto a 600-μl 20 to 50% sucrose-MTNE gradient, and centrifuged at 75,000 × g for 40 min. We then collected 40-μl fractions from the top of the gradient for electron microscopy (EM) and protein analyses.
Detection of encapsidated KSHV DNA.
KSHV capsid DNA isolation was performed essentially as described previously (33). In brief, the capsid samples were incubated with DNase I as described above, followed by inactivation of the enzyme by the addition of 15 mM EGTA and then digestion with 20 μg of proteinase K (Gibco BRL) per ml in the presence of sodium dodecyl sulfate (SDS) for 1 h at 55°C. After phenol-chloroform-isoamyl alcohol extraction and ethanol precipitation of the encapsidated DNA, the samples were applied to a GeneScreen hybridization membrane (NEN) using a dot blot apparatus and probed for KSHV-specific DNA using a fluorescein-labeled single-stranded probe complementary to the 3′ end of KSHV open reading frame 73 (orf73) (random primer fluorescein labeling kit; NEN). The membrane was exposed to Hyperfilm (Amersham), the exposed film was digitally scanned, and the relative intensities of the signals were quantified using GelExpert Software (Nucleotech).
KSHV capsid antibodies.
Antibodies to the KSHV MCP were raised in rabbits injected with either internal peptide DQNYDNPQNR (antiserum was a gift from S.-J. Gao) or KAGVQTGSPGN (Animal Pharm Services, Inc., Healdsburg, Calif.). Rabbit polyclonal antisera specific for ORF65 were kind gifts from both S.-J. Gao (unpublished data) and G. Miller (15). Horseradish peroxidase (HRP)-conjugated anti-rabbit antibodies were from Jackson ImmunoResearch Laboratories, Inc. (West Grove, Pa.).
To raise antibodies to ORF17.5 (the scaffolding protein or mature assembly protein), the 3′ region of KSHV orf17 (encompassing the entire sequence encoding ORF17.5) was cloned into the bacterial expression vector pQE30 (Qiagen). PCR was used to amplify the DNA sequence between the ORF17 release (R) and maturation (M) sites (45) from plasmid pBSλ21-5.8 (49). The 5′ primer (GGG GGG GGA TCC AGC ATG AGC CAA TTC CCG GCC GGC ATC) used for amplification introduced a BamHI site upstream of the R site Ser (position 250) codon, and the 3′ primer (GGG GGG AAG CTT CTA GGC TTC AAG GCG GTT CGA TGT) introduced a stop codon and HindIII site directly downstream of the maturation site Ala (position 531) codon. Following digestion with these two restriction enzymes, the PCR fragment was ligated into pQE30, which had been similarly digested, to form pQE30-KS-orf17R-M. This ligation resulted in an in-frame His-tagged protein, ORF17R-M, that included the predicted coding region for the ORF17.5/scaffolding protein (below). The recombinant protein from this construct had an N-terminal leader sequence of MRGSHHHHHHGS, which allowed its purification by metal chelate affinity chromatography. The integrity of this construct was verified by DNA sequencing.
Expression of the fusion protein (H6-ORF17R-M) was carried out after transformation of Escherichia coli strain X-90 with the recombinant plasmid. A 1-liter Luria broth culture containing 100 μg of ampicillin per ml was inoculated with a 5-ml overnight culture of the transformed bacteria. His-tagged protein was extracted from inclusion bodies in 6 M urea before being run on the nickel column. The H6-ORF17.5 was then purified on a 5-ml Ni-nitrilotriacetic acid column (Qiagen) as specified by the manufacturer. Approximately 20 mg of H6-ORF17R-M was eluted in a 0 to 0.5 M imidazole gradient and dialyzed overnight against refolding buffer (50 mM Tris-HCl [pH 8.5], 100 mM NaCl, 1 mM β-mercaptoethanol) containing 6 M deionized urea, with stirring for 1 h at 4°C. To more fully renature the protein, it was then dialyzed against refolding buffer with 3 M urea and then against refolding buffer alone. To produce antiserum, partially purified H6-ORF17.5 was separated by polyacrylamide gel electrophoresis (PAGE) through a 10% polyacrylamide–SDS gel. Approximately 2 mg of protein, which migrated at 34 kDa on the gel, was prepared in this fashion and cut from the gel after Coomassie blue (Coomassie brilliant blue G; ICN, Aurora, Ohio) staining and destaining. This gel slice was sent to BAbCO/COVANCE (Berkeley, Calif.) for injection into rabbits for the production of polyclonal antiserum.
Protein electrophoresis and immunoblotting.
Samples containing capsid proteins were separated by SDS-PAGE. Proteins were detected either by staining with Coomassie blue or silver (Silver Stain Plus; Bio-Rad, Hercules, Calif.) or by electroblotting onto polyvinylidene difluoride membranes (Immobilon-P; Millipore) for Western analyses. Immunoblots were preincubated in Block (phosphate-buffered saline [PBS] containing 0.05% Tween 20 [PBST] and 5% nonfat dry milk) for 1 h at room temperature. Primary antibodies were added at the indicated dilutions, and the mixtures were incubated at room temperature for 1 to 2 h and washed three times for 5 min in PBST. The membranes were then incubated with the HRP-conjugated secondary antibody (Jackson Laboratories, Inc.) diluted (1:10,000) in PBST and washed as above. The membranes were then developed using the Renaissance detection kit (NEN) as recommended by the manufacturer, and reactive proteins were detected by autoradiography.
Protein band densitometry.
Coomassie blue-stained SDS-polyacrylamide gels underwent digital scanning using a Molecular Dynamics personal densitometer SI. The relative content of individual protein bands was determined using GelExpert Software (Nucleotech).
Mass spectrometry.
The details of the mass spectrometric (MS) determination of tryptic peptides are described elsewhere (16). In brief, the gel piece containing the protein band of interest was digested with trypsin and the peptides formed were extracted from the polyacrylamide in 50% acetonitrile–5% formic acid. These extracts then underwent liquid chromatography-MS analysis, and the peptides were eluted from the column by an acetonitrile–0.1 M acetic acid gradient. The digest was analyzed by acquiring full-scan mass spectra (LCQ ion trap mass spectrometer; Finnigan, San Jose, Calif.) to determine the peptide molecular weights and product ion spectra to determine the amino acid sequence in sequential scans. The data were analyzed by database searching using the Sequest search algorithm against the NR database (National Center for Biotechnology Information [NCBI]). Peptides that were not matched were searched against the EST database (NCBI) using Sequest and interpreted manually.
EM.
Pellets containing capsids were prepared for EM by fixation, embedding in Epon 812, and sectioning as described previously (19), except that fixation was carried out in 4.5% (wt/vol) glutaraldehyde–4% (wt/vol) paraformaldehyde in 0.1 M sodium phosphate (pH 7.2) overnight at room temperature. Negative staining was performed with 1% (wt/vol) uranyl acetate (40). All thin section and negative-stain electron micrographs were recorded on a Philips 400T transmission electron microscope operated at 80 keV.
Immuno-EM.
Samples containing capsids resuspended in MTNE were placed onto carbon-coated copper grids for 45 to 60 s, changed to TBS (100 mM Tris 7.5, 50 mM NaCl), and blocked for 1 h in BB (0.1% fish skin gelatin, 5% goat serum, and 5% bovine serum albumin in TBS). The grids were then incubated for 1 h in antiserum diluted 1:50 in BB and then washed six times for 15 s and then once for 15 min with BB. Colloidal (10-nm-diameter) gold-conjugated goat anti-rabbit immunoglobulin G (IgG) (Electron Microscopy Services) was then added and incubated for 0.5 to 1 h. The grid was washed three times for 15 s with BB and then four times for 15 s with TBS. The grid was then rinsed twice with PBS and fixed with 1% glutaraldhyde in PBS for 2 min. This solution was then removed, and the grid was rinsed three times with TBS. Samples were stained with uranyl acetate and viewed by EM as described above.
Capsid mass determinations.
Scanning transmission EM (STEM) determinations of the molecular mass of individual capsid particles were carried out at Brookhaven National Laboratories (Brookhaven, N.Y.). The STEM facility and its analysis capabilities have been reviewed (46). In brief, capsid samples were placed on a thin (2- to 3-nm) carbon film supported by a thick holey film over a titanium grid. Tobacco mosaic virus was also applied as an internal standard. The grid was washed five times with 300 mM ammonium acetate and then ten times with 20 mM ammonium acetate, blotted to a thin layer of liquid, plunged into liquid nitrogen slush, and freeze-dried under vacuum overnight before being transferred to the microscope. The microscope was operated in a dark-field mode in which annular detectors collect nearly all the scattered electrons. A digital image was obtained which consists of 512 by 512 pixels, showing the number of scattered electrons from each pixel. The number of scattered electrons in each pixel is directly proportional to the mass thickness in that pixel.
Mass measurements were made in areas with relatively clean backgrounds. The background was computed for clean areas and subtracted from the intensity summed over the particles. The microscope calibration factor was determined by measurements of tobacco mosaic virus, and the summed intensities (minus the background) multiplied by the calibration factor gave the mass values for the specimen. When particles were sparse (e.g., for the C capsids), they were selected manually and analyzed with the PCMass program 1.4 (J. S. Wall).
RESULTS
A source of stable KSHV capsids.
In a herpesvirus-infected cell, lytic replication often results in nuclei filled with viral capsids. Electron micrographs have documented similar phenomena within the nuclei of PEL cells supporting KSHV replication (29, 33). In studying HSV-1 and cytomegalovirus (CMV) capsid structures, investigators have used such nuclei as the source of capsids. However, as mentioned above, this approach has been unsuccessful with gammaherpesviruses such as EBV. The reason for this is unclear. One possibility is that for gammaherpesviruses many of the intranuclear capsids may be inherently unstable, acquiring additional structural integrity only as they mature and exit the nucleus. In fact, the only successful characterizations of the protein composition of EBV capsids used released virions as the starting material (10).
With this in mind, we reasoned that intact KSHV virions or maturing capsids that were undergoing, or just about to undergo, tegument and envelope addition might represent a more stable capsid population. We had previously demonstrated that a subset of the KSHV particles released into the medium from a TPA-induced PEL line represented enveloped virions containing KSHV genomic DNA (33). This work also revealed that KSHV replication led to the formation of intracellular subviral particles suggestive of genome-containing capsids. Lacking envelopes, these latter particles required no prior treatment with lipid-dissolving detergent to render them susceptible to proteolytic enzymes such as pronase.
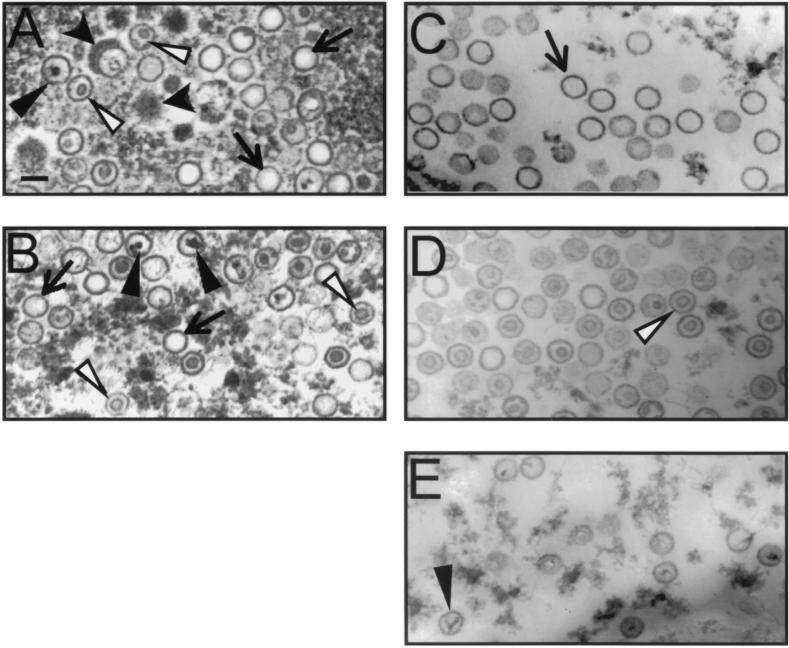
To obtain KSHV capsids sufficiently stable for biochemical and imaging analyses, we induced viral lytic replication by treating BCBL-1 cells with a combination of the phorbol ester TPA and sodium butyrate and then pelleted all viral and subviral particles from the medium by ultracentrifugation (see Materials and Methods). We hypothesized that cell lysis, a consequence of lytic replication, would lead to the premature release of particles in all stages of viral assembly, including capsids prior to their acquisition of tegument and envelope. Moreover, virion production in herpesvirus replication is often an inefficient process, leading to formation of a significant number of “dead-end” or abortive capsids that resemble either empty A capsids or scaffolding protein-filled B capsids. A priori, there is no reason to assume that KSHV assembly is any more efficient than that of other herpesviruses. TEM of the pelleted particles demonstrated a mixture of three basic types of KSHV capsids (Fig. 1A). The most obvious distinguishing characteristic among these species lies in the morphology of their centers. The first type appears empty, the second contains an inner ring-like structure, and the third, a more rare species, demonstrates a dense irregularly shaped inner core, often with fine strands that extend to the capsid's perimeter. The above three phenotypes are reminiscent of the A, B, and C capsids that arise during the replication of other herpesviruses such as HSV-1 (see above). Prior to detergent treatment, the collection of capsids includes, in addition, a small subset of KSHV capsids that appear to be wrapped, or partially wrapped, in a thick radial layer of darkly staining material (Fig. 1A, wide black arrowheads). This dense material is suggestive of a tegument layer and, in our preparations, could surround either A, B, or C capsid morphologies.
FIG. 1.
Transmission electron micrographs of KSHV capsid species at sequential stages of purification. (A and B) Capsid mixtures pelleted from BCBL-1 medium before (A) and after (B) Triton X-100 extraction. Capsids surrounded with material suggestive of a tegument layer are indicated by wide black arrowheads, empty capsids are indicated by arrows, ring-filled capsids are indicated by white arrowheads, and core-containing capsids are indicated by narrow black arrowheads. (C to E) Gradient fractions enriched for each of these last three capsid morphologies (fractions 7, 9, and 12 from the gradient depicted in Fig. 4). Bar, 0.1 μm.
To maximize the yield of KSHV capsids for biochemical and morphologic studies, we treated the pelleted particles with nonionic detergent (2% Triton X-100), thereby adding virion-derived capsids to the unenveloped capsids already present in the medium. Nonionic detergents in low concentrations have little effect on HSV-1 and CMV capsids but strip the envelope from virions. The result of this addition is shown in Fig. 1B. Again, three species of capsids remain but, interestingly, the putative tegument layer no longer seems to associate with capsids (compare Fig. 1A and B). At this stage in the preparation, the mixture typically comprises approximately 40 to 50% A capsids, 35 to 45% B capsids, and 10 to 15% C capsids (Fig. 1B).
Isolation and purification of distinct KSHV capsid species.
To characterize more carefully the different KSHV capsid species we observed by EM, we next isolated each subpopulation. If the mixture of particles present in the pelleted medium represented the KSHV homologs of the A, B, and C capsids of alpha- and betaherpesviruses, it follows that they would probably have masses sufficiently different to allow their separation by velocity sedimentation. In fact, after sedimentation of the KSHV capsid mixture through a linear sucrose gradient, two closely spaced, light-scattering bands were present toward the center of the gradient and a third, quite faint band was present toward the bottom. Since the A, B, and C capsids of HSV-1 display a similar sedimentation profile under identical conditions (data not shown), we reasoned that the light-scattering bands in our KSHV capsid preparations probably represented the KSHV homologs of these three capsid species.
EM supported this notion, showing that each of the visible bands in the gradient contained a moderately pure collection of a single type of capsid. The predominant capsid population within the upper, lower, and middle gradient bands was either empty, ring filled, or core filled, respectively (Fig. 1C to E). Examination by EM demonstrated that the capsid particle density dropped off dramatically in the fractions from regions of the gradient outside the visible bands (data not shown).
Protein characterization of KSHV capsids.
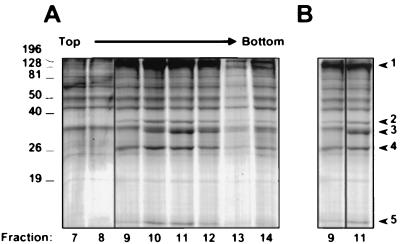
To determine the protein composition of the capsids, we analyzed the gradient fractions from a series of KSHV capsid preparations by PAGE, concentrating our initial efforts on the two distinct and more abundant capsid species that sedimented toward the middle of the sucrose gradients. Coomassie blue staining of the gels revealed a unique set of protein bands that arose in the same fractions that also contained EM evidence of either empty or ring-filled capsids (Fig. 2A). This set comprised four protein bands (Fig. 2B, bands 1, 2, 4, and 5) that had apparent molecular masses of approximately 140 to 160, 36, 28, and 16 kDa. In addition, a fifth species (labeled band 3 in Fig. 2B), migrating with an apparent molecular mass of approximately 34 kDa, was disproportionately represented in the fraction that contained predominantly capsids with the inner ring structure (Fig. 1D and 2, fraction 11). Importantly, neither Coomassie blue nor silver staining detected this 34-kDa protein in fractions containing only empty capsids (Fig. 1C and 2, fraction 9) despite the presence of the other four capsid-associated protein bands discussed above. The consistently low yield of the capsids with a dense core (the most rapidly sedimenting species) prevented their detection on Coomassie blue-stained gels. However, silver staining of gradient fractions subjected to SDS-PAGE revealed that this underrepresented population of capsids likewise contained the same four proteins shared by the other two capsid species but, as with the empty capsids, lacked the 34-kDa protein (data not shown). The presence of this third population of capsids and its sucrose gradient sedimentation profile relative to those of the other two species was best discerned in Western and silver-stained blots probed for the largest capsid-associated band (see below). We have yet to determine the origin of the other proteins present throughout the sucrose fractions, but their appearance even in regions free of capsids argues that they most probably represent proteins from or adherent to cellular debris of different sizes.
FIG. 2.
Velocity sedimentation of KSHV capsids through a 20 to 50% sucrose gradient. (A) Coomassie blue-stained SDS–10% polyacrylamide gel of eight sequential fractions (numbered at the bottom) containing KSHV capsids sedimented through the gradient (note that fraction 13 was underloaded). (B) Fractions 9 and 11 (from panel A) juxtaposed for comparison and with a slightly lighter exposure. The five protein bands that rose above background and sedimented as components of a particle through the gradient are indicated to the right. Note that the capsids in fraction 9 lack band 3. Molecular mass markers (in Kilodaltons) are indicated to the left.
KSHV major capsid protein.
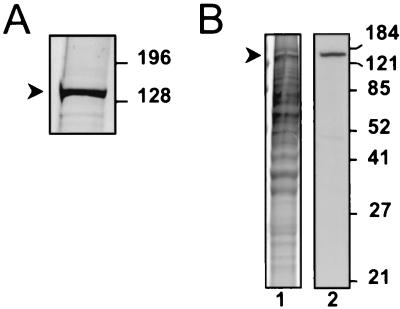
Since the single largest contributor to the overall mass of the capsid shell of herpesvirus capsids is MCP, it seemed reasonable to predict that the most prominent capsid-associated protein in our preparations (Fig. 2B, band 1) might represent the KSHV MCP. Sequence homology suggests that orf25 in KSHV encodes the MCP homolog and that the protein, ORF25, would have a molecular mass of 153 kDa (34). We subjected the fraction with the largest amount of band 1 from another capsid preparation to electrophoresis through an SDS–4 to 20% gradient polyacrylamide gel. Better separation of the larger proteins in this type of gel demonstrated more clearly that the band 1 protein migrated with an approximate molecular mass of 150 kDa (Fig. 3A), close to that predicted for ORF25. We confirmed that this band was the KSHV ORF25/MCP by using rabbit polyclonal antiserum that recognizes a unique peptide (KAGVQTGSPGN) encoded by orf25. Figure 3B shows that the MCP peptide-directed serum specifically detected the protein even in the relatively crude pregradient capsid mixture. The antiserum did not react significantly with the other bands in the gel that were detected by Coomassie blue. A second antiserum raised against another MCP peptide (DQNYDNPQNR) likewise specifically reacted with capsid band 1 (data not shown).
FIG. 3.
Capsid band 1 is ORF25/MCP. (A) Magnified view of band 1 (arrowhead) stained with Coomassie blue after separation on a 4 to 20% polyacrylamide gel. The band migrates at approximately 150 kDa. (B) Coomassie blue-stained 12% polyacrylamide gel (lane 1) and immunoblot (lane 2) of the capsid mixture prior to gradient purification. The immunoblot was probed with rabbit anti-MCP peptide polyclonal antisera. Molecular mass markers (in kilodaltons) are indicated to the right of each panel.
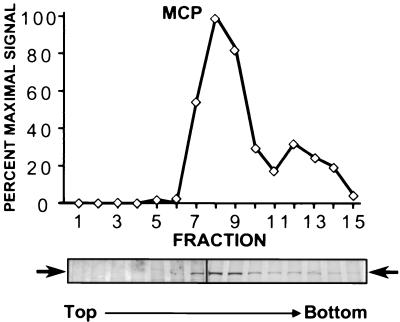
In a subsequent capsid preparation, in which the gradient was centrifuged for 30 min, silver staining revealed that the MCP band when plotted against fraction number gave rise to two distinct peaks (Fig. 4). (Note that the shorter centrifugation time better retained the faster-sedimenting core-containing capsids on the gradient.) EM examination of each fraction from this profile confirmed that the first peak reflected the convergence of the two closely sedimenting and more abundant KSHV capsid populations, one with empty centers and the other with inner ring-like structures. The second, smaller peak, in contrast, reflected the faster-sedimenting and less abundant core-containing capsids. In the particular experiment depicted in Fig. 4, moderately pure populations of each of the three capsid species were present in fractions 7, 9, and 12, respectively (EM analyses of a similar set of fractions are shown in Fig. 1C to E). By counting the number and types of capsids within multiple fields on the EM grid from each of the pelleted fractions, we found that fraction 7 contained nearly 100% A capsids, fraction 9 contained approximately 85% B capsids and 15% A capsids, and fraction 12 contained a mixture of approximately 74% C capsids, 16% B capsids, and 10% A capsids. The fraction containing the absolute maximum concentration of KSHV MCP (Fig. 4, fraction 8), in contrast, comprised a mixture of approximately equal numbers of empty and ring-structure-containing capsids.
FIG. 4.
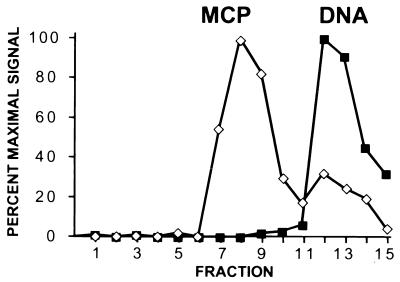
MCP profile across fractions from a sucrose gradient reveals distinct sedimentation velocities of the different KSHV capsid species. The intensity of silver staining of MCP (band 1) is plotted against fraction number (fractions were collected from the top of the gradient as indicated). The first peak (fractions 7 to 9) contains empty and ring-filled capsids, and the second, smaller peak (fractions 12 and 13) contains mainly the less abundant core-filled capsids (see the text). Maximal Coomassie blue staining of MCP (fraction 8) was designated 100%. The silver-stained gels containing fractions 1 to 7 and 8 to 16, respectively, are aligned below the graph, and the position of MCP is indicated by horizontal arrows (a vertical black line marks the break between the two gels).
Identification of the 34-kDa capsid protein as the KSHV scaffolding homolog.
B capsids of HSV-1 and CMV capsids have a protein scaffolding within their shells. This structure is absent in the other (A and C) capsid species that arise during lytic replication. The presence of the scaffolding endows B capsids with a larger mass than A capsids, so that the former sediment at slightly higher rates through a sucrose gradient. Our EM and sedimentation velocity data support the notion that KSHV lytic replication similarly leads to the accumulation of A and B capsid species. We observed that the two closely spaced bands present in our gradients corresponded to two distinct capsid morphologies present in the fractions from these bands (Fig. 1C and D). The only discernible morphologic difference between the two capsid populations was the inner ring structure. As a result, these capsids would probably have similar protein profiles, differing only by the one or two proteins that would explain the discrepancies in their morphology and sedimentation behavior. In fact, as shown above, both had the common set of four capsid-associated proteins, but the slightly faster-sedimenting capsids demonstrated a unique protein migrating on SDS-PAGE with a mass of 34 kDa. This protein was absent from the lane containing the uppermost gradient band of capsids (compare fractions 9 and 11 in Fig. 2B).
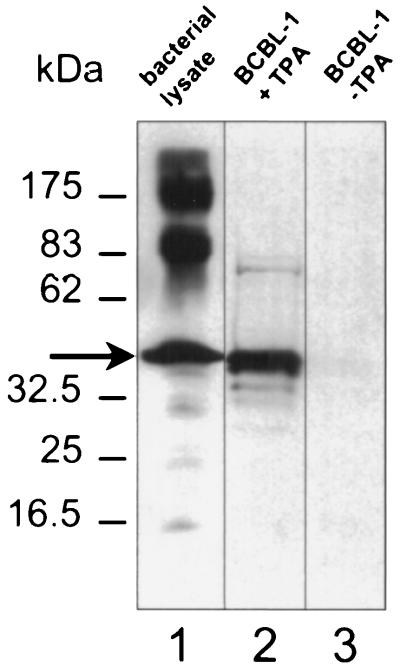
In light of these results, the 34-kDa protein was an obvious candidate to be the KSHV scaffolding protein homolog (also known as assembly protein). We ascertained its identity by using polyclonal antibodies specific for a recombinant polypeptide that included the entire predicted amino acid sequence encoded by the 3′ end of orf17, the region that includes the scaffolding protein (see Materials and Methods) (8, 45). The rabbit antiserum (anti-ORF17.5) recognized both the recombinant protein and a comigrating protein from TPA-induced BCBL-1 cells supporting active KSHV replication and capsid formation (Fig. 5). This latter protein was noticeably absent from uninduced BCBL-1 cells in which nearly all the virus is in a latent state.
FIG. 5.
Rabbit antisera raised against a recombinant protein encoded by the 3′ half of orf17 recognizes a KSHV lytic protein in TPA-induced BCBL-1 cells. Overproduced recombinant protein (arrow) from bacterial lysates (lane 1) comigrates with its endogenous counterpart from TPA-induced BCBL-1 cells (lane 2) in Western analyses probed with the rabbit antiserum (anti-ORF17.5). The protein is essentially absent in extracts from uninduced BCBL-1 cells (lane 3). The secondary antibody was HRP-conjugated goat anti-rabbit IgG. Molecular mass markers are shown to the left. Vertical lines indicate where separate lanes from the single gel were juxtaposed for optimal comparison.
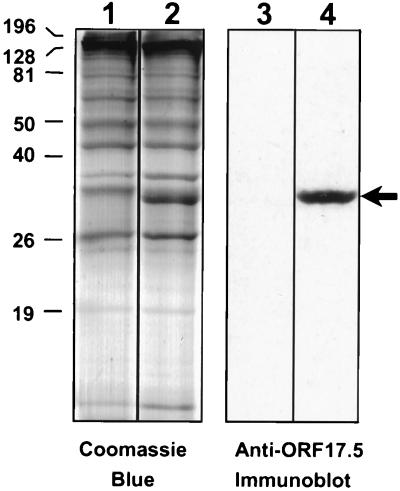
When used in immunoblot analyses of gradient-purified KSHV capsids, this antiserum recognized the 34-kDa protein present in the species that sedimented with intermediate velocity (Fig. 6, lanes 2 and 4). In fractions containing mainly empty capsids, little to no reactivity was present (lanes 1 and 3). Coupled with the corresponding electron micrographs and gradient-banding profiles, these immunoblot results (we have repeated such analyses on multiple capsid preparations) suggest that the 34-kDa protein (band 3 in Fig. 2) is the KSHV scaffolding protein. It follows that the capsids containing this protein (Fig. 1D) are the KSHV homologs of HSV-1 and CMV B capsids. Similarly, the lack of scaffolding protein in the empty (and slowest-sedimenting) species of KSHV capsids provides further evidence for their identification as the A capsids (Fig. 1C).
FIG. 6.
The 34-kDa capsid band 3 reacts with anti-ORF17.5 antiserum. KSHV capsids from gradient fractions 9 and 11 (Fig. 2B) were subjected to SDS-PAGE and then either stained with Coomassie blue (lanes 1 and 2) or immunoblotted and probed with anti-ORF17.5 antiserum (lanes 3 and 4), as in Fig. 5. The 34-kDa ORF17.5 capsid protein (arrow) was present in the gradient fraction containing ring-filled capsids (lanes 2 and 4) but absent in the fraction containing empty capsids (lanes 1 and 3). Molecular mass markers (in kilodaltons) are indicated to the left. Vertical lines between lanes indicate where separate lanes from the gel and Western blot were juxtaposed for direct comparison.
MS identification of the remaining three protein components common to all the KSHV capsid populations.
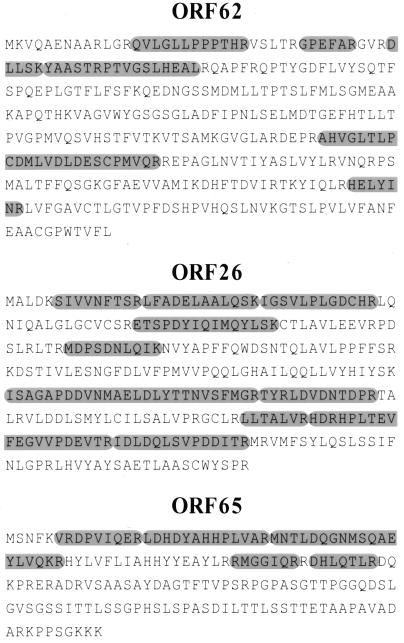
Although immunoblots helped identify the KSHV capsid proteins MCP (ORF25) and scaffolding protein (ORF17.5) (Fig. 2, bands 1 and 3, respectively), we used MS to help assign the appropriate KSHV orfs to the three remaining candidate capsid proteins (Fig. 2B, bands 2, 4, and 5). MS determines both the overall mass and amino acid sequence of tryptic peptide fragments from protein bands removed from Coomassie blue-stained gels (see Materials and Methods). The technique usually provides a sufficient portion of a protein's sequence to allow it to be identified unambiguously (see below). Since the KSHV genome is essentially completed sequenced, identification of the proteins that give rise to MS-derived peptide sequences should, in theory, be possible by searching the protein and DNA databases (22, 34). To rule out the formal possibility that the proteins we identified as capsid associated were instead cellular contaminants, we searched the MS-derived peptide sequences against entire protein databases rather than focusing exclusively on those predicted from the sequence of the KSHV genome.
MS of the 36-, 28-, and 16-kDa proteins (Fig. 2B, bands 2, 4, and 5) gave rise to 6, 10, and 5 discrete peptides that together spanned 70, 137, and 55 amino acids, respectively. When we used these sequences to search the SWISS-PROT protein database, the resultant matches led to unambiguous assignment of the 36-, 28-, and 16-kDa capsid proteins to KSHV orf62, orf26, and orf65, respectively (Fig. 7). A BLASTP search (NCBI) against four nonredundant protein databases (including SWIS-PROT) gave identical results. Using the ORF nomenclature for clarity, we have designated the proteins ORF62, ORF26, and ORF65, respectively. With the exception of ORF26, which migrates faster than expected, the predicted molecular masses of these three proteins (36.3, 34.3, and 18.6 kDa, respectively) fit well with those we observed.
FIG. 7.
Results of MS of KSHV capsid bands 2, 4, and 5. Coomassie blue-stained capsid bands 2, 4, and 5 (Fig. 2B) were subjected to MS (see Materials and Methods). The resultant peptides from each band and their respective sequences (oval shaded regions) are shown superimposed on the deduced amino acid sequences of ORF62, ORF26, and ORF65, respectively.
ORF62 and ORF26 are the KSHV capsid triplex homologs.
As discussed above, the capsids of alpha- and betaherpesviruses studied to date contain, in between each adjacent set of MCP capsomers, a heterotrimeric complex referred to as the triplex. Triplexes in these other herpesviruses are composed of two distinct proteins. In HSV-1, for example, each triplex consists of one molecule of VP19C and two molecules of VP23. Although the predicted sequence of ORF62 is only loosely homologous to VP19C, it is 31% identical and 51% similar to the predicted EBV triplex counterpart, the BORF1 gene product. (Amino acid comparison of these EBV and HSV-1 triplex homologs, in turn, reveals 30% identity and 49% similarity.) Likewise, the KSHV ORF26 is 21% identical and 41% similar to the HSV-1 triplex component VP23, whereas it is 49% identical and 69% similar to the predicted amino acid sequence of the protein encoded by the EBV homolog, BDLF1. To test further the notion that ORF62 and ORF26 represent the two components of the putative KSHV triplex, we calculated whether they were present in purified capsids with the expected 1:2 stoichiometry. We estimated the relative amounts of these proteins in both silver-stained and Coomassie blue-stained gels of our capsid preparations (compare, as an example, Fig. 2B, bands 2 and 4, in fractions 9 and 11), quantifying the staining intensity of each (see Materials and Methods) and taking into account the predicted molecular masses of the two proteins. Although these measurements are only rough estimates, given the potential variability in staining between these two proteins, in five capsid preparations we found that the molar ratio of ORF62 to ORF26, in both A- and B-type capsids, was approximately 1:2 (range 1:1.7 to 1:2.2) regardless of which of the two stains was used. These data, coupled with the sequence homologies we describe above, argue for the identification of ORF62 and ORF26 as the components of the KSHV triplex.
The smallest capsid-associated structural protein, ORF65, decorates the outside of KSHV capsids.
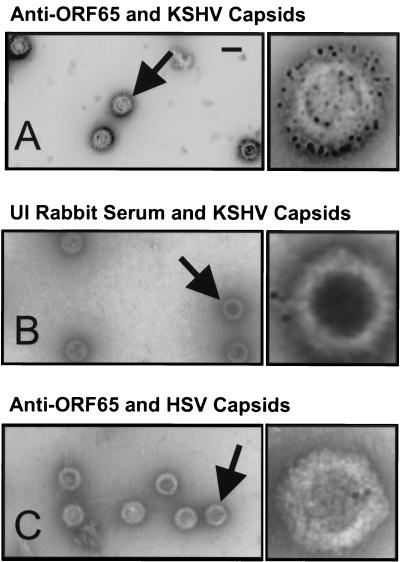
In the hexomeric capsomers of alpha- and betaherpesviruses, each MCP component is associated with a single small capsid protein in a 1:1 ratio (3). Across the herpesvirus family, these MCP-interacting proteins often show little sequence homology to one another; however, each is the smallest structural component of their respective capsids and each has a highly basic isoelectric point. The predicted EBV counterpart, the 20-kDa small viral capsid antigen (sVCA, encoded by BFRF3), has a pI of 10.8. In the case of HSV-1, the likely structural homolog, VP26, has a mass of 12.1 kDa and a pI of 11.1. Since ORF65, with a predicted molecular mass of 18.6 kDa and a predicted pI of 9.6, is the smallest structural capsid protein present in stoichiometric amounts in our purified KSHV capsid preparations (Fig. 2B, band 5), it seemed reasonable to propose that it is the likely KSHV homolog of sVCA and VP26 (even though the latter has no significant amino acid homology to ORF65) (3).
Although the size, stoichiometry and pI of ORF65 combine to argue for a role as the structural homolog of the well-studied HSV-1 VP26, more direct proof depends on demonstrating that it occupies at least a similar relative position on the capsid. We reasoned that antibodies directed against ORF65 should be able to bind to the surface of purified intact capsids. To test this hypothesis, we incubated KSHV A capsids with rabbit polyclonal anti-ORF65 antibodies followed by donkey anti-rabbit secondary antibodies conjugated to colloidal gold (see Materials and Methods). EM of these particles revealed an extensive gold signal surrounding the capsid, whereas capsids similarly treated but with serum from unimmunized rabbits showed minimal to no reactivity (Fig. 8B). Likewise, the anti-ORF65 antibodies did not react with purified HSV-1 capsids (Fig. 8C). Identical experiments with B capsids gave similar results (data not shown).
FIG. 8.
Immuno-EM reveals the presence of ORF65 on the surface of purified KSHV B capsids. (A and B) Purified KSHV B capsids were incubated with either anti-ORF65 rabbit antiserum (A) or with unimmunized rabbit serum (B) followed by colloidal gold-conjugated goat anti-rabbit IgG antibodies and then subjected to EM (see Materials and Methods). (C) Purified HSV-1 B capsids were also incubated with the anti-ORF65 rabbit antiserum and then secondary antibody as above. Arrows indicate the single capsids magnified approximately fivefold to the right of each panel in the electron micrographs. Colloidal gold appears as dark, uniformly sized circles. Bar, 0.1 μm.
Only the rapidly sedimenting capsids contain KSHV DNA.
Although nonlinear with absolute protein concentration, the relative signal intensities from HRP-conjugated secondary antibodies on immunoblots probed for ORF25/MCP provided a sensitive and rapid means of identifying the fractions containing each of the capsid populations and, particularly, the more elusive core-filled species that sedimented toward the bottom of the gradient (Fig. 1E). Armed with the ability to detect the different capsid populations, we tested the three capsid species for the presence of encapsidated KSHV DNA. Our prediction was that the third gradient band, composed of core-filled capsids, represented KSHV C capsids, each containing a single copy of the viral genome. We isolated potentially encapsidated (DNase-resistant) DNA from each gradient fraction and analyzed it by dot blot Southern analysis using a chemiluminescent probe complementary to KSHV orf73 (see Materials and Methods). The digitized profile of the resultant autoradiograph, superimposed on the profile of MCP shown above (Fig. 4), revealed that the majority of KSHV DNA cosediments with fractions containing the low-abundance, rapidly sedimenting capsid population (Fig. 9). In contrast, the middle gradient fractions containing the more abundant A and B capsid species showed, as expected, an absence of KSHV DNA. These results indicate, therefore, that the capsids shown in Fig. 1E and sedimenting in the second, smaller peak within sucrose gradients are KSHV C capsids.
FIG. 9.
Profile of encapsidated DNA from KSHV capsids separated by velocity sedimentation through a sucrose gradient. Relative amounts of KSHV-specific DNA (solid squares) in each fraction from the same gradient shown in Fig. 4 were measured by Southern dot blot analysis (see Materials and Methods). For direct comparison, the MCP profile (open diamonds) from Fig. 4 is also shown. The KSHV DNA peaks in fractions 12 and 13, correlating with the second MCP peak, which contains the core-filled capsids (Fig. 1E).
Capsid mass determinations reflect those predicted by the stoichiometry and molecular masses of the individual capsid components.
The relative sedimentation velocities of the three KSHV capsid species suggested that the mass of a B capsid is slightly greater than that of an A capsid but that both are lower than that of the genome containing C capsid. Knowing the orf for each of the five major capsid protein components and therefore the predicted molecular mass of each, we were able to estimate the overall molecular mass of each capsid species. Such calculations were based on the assumption that KSHV capsid geometry parallels that of the alpha- and betaherpesviruses, namely, that each shell contains 150 hexameric and 12 pentameric capsomers, giving a total of 960 molecules of MCP. Likewise, each would also have 320 triplexes (since each triplex joins three MCP molecules, each on a different capsomer). Comparisons of electron micrographs of KSHV and HSV-1 capsids (data not shown) helped support these compositional inferences. Furthermore, our three-dimensional reconstructions of KSHV capsid cryoelectron micrographs demonstrated that KSHV capsids share, with other herpesviruses, the same basic capsomer and triplex architecture (see the accompanying paper [44a]).
In contrast to the deduced mass contributions from the capsomers, the triplexes, and, for the C capsids, the genome, those from ORF65 and scaffolding are less clear. For example, the likely homolog of ORF65 in HSV-1, VP26, interacts with the HSV-1 MCP in a 1:1 molar ratio but only when the latter is in a hexameric (and not a pentameric) capsomer. As a result, there are 900 (150 × 6) copies of VP26 in HSV-1 capsids. Although the immunoelectron microscopic evidence argued that ORF65 is present on the capsid surface (Fig. 8), the molar ratio between ORF25/MCP and ORF65 in multiple capsid preparations (see, for example, Fig. 2, bands 1 and 5) ranged from 0.8:1 to 1.1:1. Such measurements lacked the necessary precision to deduce whether the stoichiometry of ORF65 parallels exactly that of its VP26 counterpart, interacting only with hexameric rather than all capsomeric MCP molecules. (This issue is addressed further in the analysis of KSHV capsid reconstructions in the accompanying paper [44a].) Nevertheless, the present data are consistent with each capsid containing between 900 and 960 copies of ORF65. Similarly, the contribution of scaffolding (ORF17.5) to the overall mass of B capsids was not immediately obvious, although stoichiometric determinations of the scaffolding homolog in HSV-1 capsids have suggested that this protein exists in either a 1:1 or 2:1 molar ratio with MCP (14). Extrapolation from such data would predict, therefore, that a KSHV B capsid would have between 960 and 1,920 copies of the scaffolding protein.
In sum, the mass of a C capsid would be identical to that of an A capsid but for the additional genome mass of approximately 109 MDa (165 kbp) (32). By multiplying the molecular mass of each protein component by its predicted copy number, we determined that the A, B, and C capsids would possess masses of approximately 198, 227 to 254, and 309 MDa, respectively.
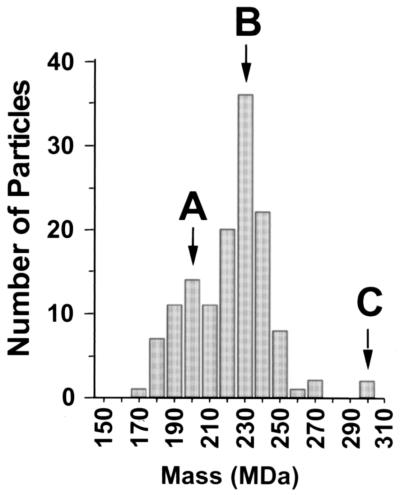
To verify these estimates, we employed STEM, using tobacco mosaic virus as an internal standard (see Materials and Methods). STEM is unique in its ability to visualize individual biological molecules directly without staining, fixing, or shadowing. We employed this technique to measure the mass of individual particles in a mixed preparation of A, B, and C species (Fig. 10). We interpreted the results to indicate that the mass of the capsids with A, B, and C morphologies (see above) had modes of 200, 230, and 300 MDa, respectively. (The error in these mass determinations in this range was approximately 10 MDa.) A comparison of the estimated and empirical STEM mass measurement for each capsid type is shown in Table 1. In these calculations, we used the molecular masses predicted for the capsid proteins rather than those observed during PAGE, since empirical measurement is often somewhat variable, depending on the characteristics of both the specific protein and the gel system used. Interestingly, the 30-MDa difference between the STEM-determined masses of KSHV A capsids and B capsids (divided by the predicted scaffolding mass of 28.5 kDa) is most consistent with a 1:1 scaffolding-to-MCP ratio.
FIG. 10.
STEM analysis of a mixed population of KSHV capsids. KSHV capsids pelleted from the medium of TPA-induced BCBL-1 cells were subjected to Triton X-100 extraction (but no gradient fractionation) and then analyzed by STEM to determine their masses. Each bar in the graph indicates the frequency with which individually identified particles had a specific mass. Downward arrows indicate the modes and corresponding A, B, and C capsid morphologies.
TABLE 1.
Predicted and observed masses of A, B, and C KSHV capsids and their components
| Component | Mass (kDa)
|
Predicted copies/capsid for capsid speciesa:
|
|||
|---|---|---|---|---|---|
| Predicted | Observed | A | B | C | |
| ORF25 | 153 | 150 | 960 | 960 | 960 |
| ORF62 | 36 | 36 | 320 | 320 | 320 |
| ORF26 | 34 | 28 | 640 | 640 | 640 |
| ORF17.5 | 29 | 34 | 960–1,920 | ||
| ORF65 | 19 | 16 | 900–960 | 900–960 | 900–960 |
| KSHV DNA (165 kbp) | 109 | 1 | |||
| Total predicted mass (MDa) | 198–199 | 226–255 | 307–308 | ||
| Total observed mass (MDa) | 200 | 230 | 300 | ||
The sequence-predicted mass for each of the major capsid components was multiplied by its predicted copy number per capsid. The sum of the appropriate combination of these components then gave a predicted overall mass for each capsid species.
Also apparent from the height of each mode in the STEM measurements was that (at least in this KSHV capsid preparation) B capsids were the dominant species accumulating in the medium, followed closely by A capsids and then distantly by the more rare C capsids. These results were consistent with out earlier characterization of the relative frequency of each capsid species in the BCBL-1 medium (see, for example, the distribution of capsid species in Fig. 1A and the relative intensities of Coomassie blue-stained capsid proteins from fractions containing A, B, and C capsid species, respectively [Fig. 2]).
DISCUSSION
Beginning with infected-cell supernatant, we have isolated intact KSHV capsids in quantities sufficient for structural and biochemical analyses. To date, this has not been possible for gammaherpesvirus capsids derived from the nuclei of lytically infected cells. Although the reasons for the apparent inability of these intranuclear capsids to withstand the stresses inherent in standard herpesvirus purification procedures remain unclear, recent data suggest that very early forms of HSV-1 capsids (procapsids) possess a more spherical and less angular shell and that this species has less structural integrity compared with the angularized and presumably more mature form that arises subsequently (24, 43). If parallel processes occur in KSHV formation but such maturation is delayed relative to that in the alpha- or betaherpesviruses, then selecting a pool of KSHV capsids at a later stage of development might permit their successful isolation. KSHV capsids that accumulate in the medium of induced PEL cells represent such a population. Supporting this notion, Wu et al. have recently isolated KSHV capsids from the medium of infected cells (48). Our work has shown, in thin-section electron micrographs, that the capsid mixture pelleted from the medium prior to detergent treatment contains a variety of capsid species that are released from TPA-induced BCBL-1 cells (Fig. 1A). This release of previrion forms probably arises from the lysis of cells supporting productive virus replication. Since capsid formation is probably a continuous process during productive infection, it follows that cell death and the accompanying cellular disruption lead to the release of partially formed particles still in the early stages of formation. In addition to mature virions, this would include, therefore, partially and fully formed capsids with and without overlying tegument and envelope layers.
The ability to purify A, B, and C KSHV capsids has allowed us to determine the proteins common to each as well as to delineate the compositional differences among them. Western analyses and MS on preparations of the capsids demonstrated that the basic shell of the KSHV capsid consists of four major protein components, ORF25, ORF62, ORF26, and ORF65. Integrating the present data and those from capsid reconstructions (44a) with the known capsid composition of other herpesviruses, we propose tentative names for the capsid proteins that reflect more accurately their structural roles in the virion (Table 2). When not in conflict with this goal, these names also preserve consistency with the nomenclature of homologous proteins from the other herpesvirus subfamilies. Therefore, we suggest continuing to use MCP (the major capsid protein) for ORF25 but TRI-1 and TRI-2 (the two putative triplex components) for ORF62 and ORF26, respectively, and SCIP (the small capsomer-interacting protein) for ORF65. In particular, we suggest dropping the somewhat misleading designation “minor capsid protein” for ORF26 (and replacing it with TRI-2) since the protein is the second most abundant and third largest component of the capsid shell. In turn, we propose SCIP over sVCA, the name adopted from the EBV nomenclature (15), for two reasons: (i) the EBV nomenclature implies that only this capsid protein is antigenic, an untested hypothesis in KSHV, and (ii) SCIP refers to the likely structural position of the protein in KSHV (as well as that of its homologs in other herpesviruses).
TABLE 2.
Comparison of the capsid components of KSHV with those of HSV-1 and with those predicted for EBV based on sequence homologya
| KSHV capsid protein
|
HSV-1 homolog
|
EBV homolog
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Systematic | Structural | Other | Mass (kDa) | Systematic | Mass (kDa) | Identity (%) | Systematic | Mass (kDa) | Identity (%) |
| ORF25 | MCP | MCP | 153 | VP5 | 150 | 28 | BcLF | 154 | 56 |
| ORF62 | TRI-1 | 36 | VP19C | 50 | NSb | BORF1 | 39 | 31 | |
| ORF26 | TRI-2 | Minor capsid protein | 34 | VP23 | 34 | 21 | BDLF1 | 34 | 49 |
| ORF17.5 | Scaffolding | Assembly protein | 29 | VP22a | 31 | 23 | BdRF1 | 32 | 22 |
| ORF65 | SCIP | sVCA | 19 | VP26c | 12 | NS | BFRF3 | 20 | 22 |
For KSHV capsid components, the first and second columns in the table indicate the systematic and suggested structurally oriented ORF-based nomenclatures, respectively. The third column gives a partial list of previously employed KSHV protein designations. For comparison purposes, we have included only the molecular masses of the proteins predicted by their amino acid sequence rather than those observed by PAGE. The percent identity compares the sequences of KSHV to their counterparts from HSV-1 and EBV.
NS. not found to be significant by NCBI search program BLASTP 2.1.2 (13 Nov 2000).
No direct sequence identity exists between the VP26 and either ORF65 or BFRF3, but the three are likely to be structural homologs (see text).
Our analyses have also identified a fifth capsid structural protein, the scaffolding protein, present in stoichiometric amounts only in B capsids and demonstrating an apparent molecular mass of 34 kDa. (We have chosen to use the name “scaffolding protein” rather than “assembly protein” since the former designation is not only descriptive but also the most widely used throughout the alpha- and betaherpesvirus structural and assembly literature.) Interestingly, as with other herpesviruses, the KSHV protease mRNA (encoded by orf17) encodes a long precursor protein with downstream sequence that is colinear and in frame with that of the scaffolding proteins (34, 45). The scaffolding protein, in alpha- and betaherpesviruses, however, derives not from proteolytic processing of the protease precursor but, rather, from the translation of a distinct coterminal mRNA that initiates downstream from the protease termination codon (14). Northern analyses have demonstrated that replicating KSHV gives rise to an approximately 950-base (in addition to the longer pre-protease) mRNA that hybridizes with sequences complementary to the downstream half of KSHV orf17 (45). Recently, Chang and Ganem have mapped the 5′ end of this KSHV transcript and have named the corresponding viral gene orf17.5 (8). These data, along with our own (M. Cruise, unpublished results), suggest that KSHV scaffolding expression probably follows the same pattern as seen with the other herpesvirus subfamilies.
Assuming that the scaffolding protein initiates at the first methionine downstream of the protease R site (as in other herpesviruses), the resultant mature scaffolding protein would have a predicted molecular mass of approximately 28.5 kDa rather than the 34 kDa we observed. This discrepancy, however, is not unexpected since the scaffolding proteins from other herpesviruses also demonstrate aberrantly low electrophoretic mobilities. In HSV-1, for example, the scaffolding protein homolog, VP22a, in its mature state has a molecular mass of 31 kDa but an electrophoretic mobility close to 40 kDa (23). The apparent conservation of expression in these regions among the different herpesviruses argues that the systematic designation of ORF17.5 is appropriate for this KSHV protein. However, in light of the present data confirming its structural role in KSHV capsids and in keeping with the nomenclature goals stated above, we suggest the use of the more descriptive name of scaffolding.
The least abundant capsid species that we detected were the C capsids. Although these capsids demonstrated the same four shell proteins as A and B capsids, they were the only capsids that contained KSHV DNA (Fig. 9 and Table 1). Their low abundance in medium from induced BCBL-1 cells may explain, at least in part, the low infectivity found in experiments using this cell line as a source of infectious particles (31). We are presently evaluating other PEL lines to determine if infectivity rates correlate with C capsid and/or virion production.
By comparing the total signal from the encapsidated KSHV DNA on such blots to that from cloned orf73 DNA standards (data not shown), we were able to estimate that the final yield of purified C capsids (i.e., genome equivalents) was approximately 2 × 105 per ml of medium. (Note that there were 2 × 105 BCBL-1 cells/ml at the time of TPA induction.) Since our EM analyses of pelleted medium indicated that approximately 10 to 15% of the total capsids accumulated during the 7 days after TPA induction are C capsids (see above and Fig. 1B), it follows that the final yield of all capsid particles was approximately 1.3 × 106 to 2 × 106/ml. Finally, if 10 to 20% of the BCBL-1 cells (passaged for 2 to 4 months) supported lytic KSHV replication after TPA induction, each such cell gave rise to 30 to 100 total capsid particles.
It is also important to note that such calculations of the absolute yield of capsids from the BCBL-1 cells are probably minimal estimates since a portion of the capsids probably degrade during the prolonged period between TPA induction and capsid harvest. Further, even the relative contributions of A, B, and C capsids to this total may be somewhat distorted. Our work with HSV-1 capsid preparations, for example, indicates that HSV-1 B and C capsids slowly lose their contents over time even at 4°C (W. W. Newcomb, unpublished observations). This phenomenon, if it occurred during our KSHV capsid preparations, would lead to an underestimation of B and C capsid production and an overestimation of A (empty) capsid production.
KSHV capsids as a source of more detailed structural analyses.
The identification, stochiometry, and localization of the major structural proteins of alpha- and betaherpesvirus capsids are well known from a combination of biochemical and cryoEM studies (14, 38). The present work provides the first such insights into a gammaherpesvirus, combining KSHV capsid purification with compositional and stoichiometric determinations. Consistent with the structure of capsids from the other two herpesvirus subfamilies, KSHV capsids demonstrate the expected arrangements of capsomers and triplexes as well as the other capsid proteins (namely, SCIP/ORF65 and the scaffolding protein). Further, the results have allowed us to interpret accurately three-dimensional reconstructions from cryoEM analysis in a separate study (44a), which would otherwise rely solely on extrapolation from work on other herpesviruses. Wu et al. recently presented a three-dimensional reconstruction of the KSHV capsid, suggesting that ORF65 was absent in their preparations (48). This conclusion was based on the absence of density near the tip of the KSHV MCP, where, in HSV-1 capsids, the supposed structural homolog (VP26) lies. In contrast, our data demonstrate that ORF65 is in fact present in KSHV capsids. This is clear from four sets of experiments employing (i) PAGE, (ii) Western analyses, (iii) MS, and (iv) immuno-EM. Since the isolation procedure used by Wu et al. is not radically different from ours, loss of ORF65 in their samples as they suggest, although formally possible, seems improbable. A more likely explanation is that ORF65 is present in their capsid samples as well but that it lies in a region distinct from that occupied by VP26 in HSV-1 capsids. Only a through biochemical analysis of their capsids would bear this out. We address this discrepancy more thoroughly in the accompanying structrual study (44a).
Investigators studying alphaherpesvirus capsid assembly have speculated that A capsids may represent aborted forms of B or C capsids that have lost their inner contents. Others have suggested that B capsids may be the precursors to C capsids (30) and that the latter, in turn, acquire the tegument and envelope layers during their egress. Such precursor-product relationships, however, remain controversial. Recent work with HSV-1, in fact, has demonstrated the presence of a rounded procapsid form within infected cell nuclei (24, 25, 43). These early capsids contain immature scaffolding protein, lack well-defined angularity, and are reminiscent of similar bacteriophage precursors (6). Procapsids may therefore represent the true precursors of C type capsids or even virions themselves, but these notions also remain speculative.
Despite this incomplete understanding of herpesvirus capsid assembly, the present work on KSHV demonstrates that A-, B-, and C-type capsids also arise in a gammaherpesvirus. This observation, coupled with the parallels in the composition of each of the capsid types across all three herpesvirus subfamilies, also argues that capsid assembly for each probably follows a similar pathway. In contrast, the regulation and kinetics of capsid assembly (issues not addressed directly in the present study) probably do vary markedly among the different herpesvirus subfamilies. Such differences may explain, for example, the disparate lag phases that each herpesvirus seems to display (at least in culture) prior to the appearance of detectable mature virions. Studies to address this latter issue are under way in our laboratory.
ACKNOWLEDGMENTS
We thank N. Sherman and J. Shannon at the W. M. Keck Biomedical Mass Spectrometry Laboratory, University of Virginia Biomolecular Research Facility, which is supported by a grant from the University of Virginia Pratt Committee. In addition, we thank George Miller and S.-J. Gao for generous gifts of capsid-specific antibodies and for helpful discussions. We thank Martha Simon at The Brookhaven National Laboratories for help in the analysis of capsids by STEM.
The STEM is an NIH-supported resource center (NIH P41-RR01777), with additional support provided by Department of Energy and Office of Biological and Environmental Research. This work was supported by P30-CA44579 (D.H.K.), NIH R-01 5-23924 (D.H.K.), The Pew Memorial Trust P0320SC (D.H.K.), The Doris Duke Charitable Foundation 20000355 (D.H.K.), NIH R-01 AI41644-04 (J.C.B.), NSF MCB-9904879 (J.C.B.), and NIH GM 56531 (C.S.C.)
REFERENCES
- 1.Blasig C, Zietz C, Haar B, Neipel F, Esser S, Brockmeyer N H, Tschachler E, Colombini S, Ensoli B, Sturzl M. Monocytes in Kaposi's sarcoma lesions are productively infected by human herpesvirus 8. J Virol. 1997;71:7963–7968. doi: 10.1128/jvi.71.10.7963-7968.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Booy F P, Newcomb W W, Trus B L, Brown J C, Baker T S, Steven A C. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell. 1991;64:1007–1015. doi: 10.1016/0092-8674(91)90324-r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Booy F P, Trus B L, Newcomb W W, Brown J C, Conway J F, Steven A C. Finding a needle in a haystack: detection of a small protein (the 12-kDa VP26) in a large complex (the 200-MDa capsid of herpes simplex virus) Proc Natl Acad Sci USA. 1994;91:5652–5656. doi: 10.1073/pnas.91.12.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boshoff C, Schulz T F, Kennedy M M, Graham A K, Fisher C, Thomas A, McGee J O, Weiss R A, O'Leary J J. Kaposi's sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat Med. 1995;1:1274–1278. doi: 10.1038/nm1295-1274. [DOI] [PubMed] [Google Scholar]
- 5.Campadelli-Fiume G, Farabegoli F, Di Gaeta S, Roizman B. Origin of unenveloped capsids in the cytoplasm of cells infected with herpes simplex virus 1. J Virol. 1991;65:1589–1595. doi: 10.1128/jvi.65.3.1589-1595.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casjens S, Hendrix R. Control mechanisms in dsDNA bacteriophage assembly. In: Calendar R, editor. The bacteriophages. Vol. 1. New York, N.Y: Plenum Press; 1988. pp. 15–91. [Google Scholar]
- 7.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 8.Chang J, Ganem D. On the control of late gene expression in Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) J Gen Virol. 2000;81:2039–2047. doi: 10.1099/0022-1317-81-8-2039. [DOI] [PubMed] [Google Scholar]
- 9.Decker L L, Shankar P, Khan G, Freeman R B, Dezube B J, Lieberman J, Thorley-Lawson D A. The Kaposi sarcoma-associated herpesvirus (KSHV) is present as an intact latent genome in KS tissue but replicates in the peripheral blood mononuclear cells of KS patients. J Exp Med. 1996;184:283–288. doi: 10.1084/jem.184.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolyniuk M, Wolff E, Kieff E. Proteins of Epstein-Barr virus. II. Electrophoretic analysis of the polypeptides of the nucleocapsid and the glucosamine- and polysaccharide-containing components of enveloped virus. J Virol. 1976;18:289–297. doi: 10.1128/jvi.18.1.289-297.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganem D. KSHV and Kaposi's sarcoma: the end of the beginning? Cell. 1997;91:157–160. doi: 10.1016/s0092-8674(00)80398-0. [DOI] [PubMed] [Google Scholar]
- 12.Gibson W, Roizman B. Proteins specified by herpes simplex virus. 8. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J Virol. 1972;10:1044–1052. doi: 10.1128/jvi.10.5.1044-1052.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haanes E, Thomsen D, Martin S, Homa F, Lowery D. The bovine herpesvirus 1 maturational proteinase and scaffold proteins can substitute for the homologous herpes simplex virus type 1 proteins in the formation of hybrid type B capsids. J Virol. 1995;69:7375–7379. doi: 10.1128/jvi.69.11.7375-7379.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Homa F, Brown J. Capsid assembly and DNA packaging in herpes simplex virus. Rev Med Virol. 1997;7:107–122. doi: 10.1002/(sici)1099-1654(199707)7:2<107::aid-rmv191>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 15.Lin S F, Sun R, Heston L, Gradoville L, Shedd D, Haglund K, Rigsby M, Miller G. Identification, expression, and immunogenicity of Kaposi's sarcoma-associated herpesvirus-encoded small viral capsid antigen. J Virol. 1997;71:3069–3076. doi: 10.1128/jvi.71.4.3069-3076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mandal A, Naaby-Hansen S, Wolkowicz M J, Klotz K, Shetty J, Retief J D, Coonrod S A, Kinter M, Sherman N, Cesar F, Flickinger C J, Herr J C. FSP95, a testis-specific 95-kilodalton fibrous sheath antigen that undergoes tyrosine phosphorylation in capacitated human spermatozoa. Biol Reprod. 1999;61:1184–1197. doi: 10.1095/biolreprod61.5.1184. [DOI] [PubMed] [Google Scholar]
- 17.Martin J N, Ganem D E, Osmond D H, Page-Shafer K A, Macrae D, Kedes D H. Sexual transmission and the natural history of human herpesvirus 8 infection. N Eng J Med. 1998;338:948–954. doi: 10.1056/NEJM199804023381403. [DOI] [PubMed] [Google Scholar]
- 18.Martinez R, Sarisky R T, Weber P C, Weller S K. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J Virol. 1996;70:2075–2085. doi: 10.1128/jvi.70.4.2075-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matusick-Kumar L, Hurlburt W, Weinheimer S P, Newcomb W W, Brown J C, Gao M. Phenotype of the herpes simplex virus type 1 protease substrate ICP35 mutant virus. J Virol. 1994;68:5384–5394. doi: 10.1128/jvi.68.9.5384-5394.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McNab A R, Desai P, Person S, Roof L L, Thomsen D R, Newcomb W W, Brown J C, Homa F L. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J Virol. 1998;72:1060–1070. doi: 10.1128/jvi.72.2.1060-1070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melbye M, Cook P M, Hjalgrim H, Begtrup K, Simpson G R, Biggar R J, Ebbesen P, Schulz T F. Risk factors for Kaposi's-sarcoma-associated herpesvirus (KSHV/HHV-8) seropositivity in a cohort of homosexual men, 1981–1996. Int J Cancer. 1998;77:543–548. doi: 10.1002/(sici)1097-0215(19980812)77:4<543::aid-ijc12>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 22.Neipel F, Albrecht J C, Fleckenstein B. Human herpesvirus 8—the first human rhadinovirus. J Natl Cancer Inst Monogr. 1998;23:73–77. doi: 10.1093/oxfordjournals.jncimonographs.a024178. [DOI] [PubMed] [Google Scholar]
- 23.Newcomb W W, Brown J C. Structure of the herpes simplex virus capsid: effects of extraction with guanidine hydrochloride and partial reconstitution of extracted capsids. J Virol. 1991;65:613–620. doi: 10.1128/jvi.65.2.613-620.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newcomb W W, Homa F L, Thomsen D R, Booy F P, Trus B L, Steven A C, Spencer J V, Brown J C. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J Mol Biol. 1996;263:432–446. doi: 10.1006/jmbi.1996.0587. [DOI] [PubMed] [Google Scholar]
- 25.Newcomb W W, Trus B L, Cheng N, Steven A C, Sheaffer A K, Tenney D J, Weller S K, Brown J C. Isolation of herpes simplex virus procapsids from cells infected with a protease-deficient mutant virus. J Virol. 2000;74:1663–1673. doi: 10.1128/jvi.74.4.1663-1673.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Brien T R, Kedes D, Ganem D, Macrae D R, Rosenberg P S, Molden J, Goedert J J. Evidence for concurrent epidemics of human herpesvirus 8 and human immunodeficiency virus type 1 in US homosexual men: rates, risk factors, and relationship to Kaposi's sarcoma. J Infect Dis. 1999;180:1010–1017. doi: 10.1086/315039. [DOI] [PubMed] [Google Scholar]
- 27.Oien N L, Thomsen D R, Wathen M W, Newcomb W W, Brown J C, Homa F L. Assembly of herpes simplex virus capsids using the human cytomegalovirus scaffold protein: critical role of the C terminus. J Virol. 1997;71:1281–1291. doi: 10.1128/jvi.71.2.1281-1291.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olsen S J, Chang Y, Moore P S, Biggar R J, Melbye M. Increasing Kaposi's sarcoma-associated herpesvirus seroprevalence with age in a highly Kaposi's sarcoma endemic region, Zambia in 1985. AIDS. 1998;12:1921–1925. doi: 10.1097/00002030-199814000-00024. [DOI] [PubMed] [Google Scholar]
- 29.Orenstein J M, Alkan S, Blauvelt A, Jeang K T, Weinstein M D, Ganem D, Herndier B. Visualization of human herpesvirus type 8 in Kaposi's sarcoma by light and transmission electron microscopy. AIDS. 1997;11:F35–F45. doi: 10.1097/00002030-199705000-00001. [DOI] [PubMed] [Google Scholar]
- 30.Perdue M L, Cohen J C, Randall C C, O'Callaghan D J. Biochemical studies of the maturation of herpesvirus nucleocapsid species. Virology. 1976;74:194–208. [PubMed] [Google Scholar]
- 31.Renne R, Blackbourn D, Whitby D, Levy J, Ganem D. Limited transmission of Kaposi's sarcoma-associated herpesvirus in cultured cells. J Virol. 1998;72:5182–5188. doi: 10.1128/jvi.72.6.5182-5188.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Renne R, Lagunoff M, Zhong W, Ganem D. The size and conformation of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) DNA in infected cells and virions. J Virol. 1996;70:8151–8154. doi: 10.1128/jvi.70.11.8151-8154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 34.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarid R, Olsen S J, Moore P S. Kaposi's sarcoma-associated herpesvirus: epidemiology, virology, and molecular biology. Adv Virus Res. 1999;52:139–232. doi: 10.1016/s0065-3527(08)60299-7. [DOI] [PubMed] [Google Scholar]
- 36.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay M F, Clauvel J P, Raphael M, Degos L, et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 37.Staskus K A, Zhong W, Gebhard K, Herndier B, Wang H, Renne R, Beneke J, Pudney J, Anderson D J, Ganem D, Haase A T. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol. 1997;71:715–719. doi: 10.1128/jvi.71.1.715-719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steven A C, Trus B L, Booy F P, Cheng N, Zlotnick A, Caston J R, Conway J F. The making and breaking of symmetry in virus capsid assembly: glimpses of capsid biology from cryoelectron microscopy. FASEB J. 1997;11:733–742. doi: 10.1096/fasebj.11.10.9271358. [DOI] [PubMed] [Google Scholar]
- 39.Sturzl M, Ensoli B. Big but weak: how many pathogenic genes does human herpesvirus-8 need to cause Kaposi's sarcoma? Int J Oncol. 1999;14:287–289. doi: 10.3892/ijo.14.2.287. [DOI] [PubMed] [Google Scholar]
- 40.Thomas D, Newcomb W W, Brown J C, Wall J S, Hainfeld J F, Trus B L, Steven A C. Mass and molecular composition of vesicular stomatitis virus: a scanning transmission electron microscopy analysis. J Virol. 1985;54:598–607. doi: 10.1128/jvi.54.2.598-607.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomsen D R, Newcomb W W, Brown J C, Homa F L. Assembly of the herpes simplex virus capsid: requirement for the carboxyl-terminal twenty-five amino acids of the proteins encoded by the UL26 and UL26.5 genes. J Virol. 1995;69:3690–3703. doi: 10.1128/jvi.69.6.3690-3703.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomsen D R, Roof L L, Homa F L. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. J Virol. 1994;68:2442–2457. doi: 10.1128/jvi.68.4.2442-2457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trus B L, Booy F P, Newcomb W W, Brown J C, Homa F L, Thomsen D R, Steven A C. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J Mol Biol. 1996;263:447–462. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- 44.Trus B L, Gibson W, Cheng N, Steven A C. Capsid structure of simian cytomegalovirus from cryoelectron microscopy: evidence for tegument attachment sites. J Virol. 1999;73:2181–2192. doi: 10.1128/jvi.73.3.2181-2192.1999. . (Erratum, 73:4530.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44a.Trus B L, Heymann J B, Nealon K, Cheng N, Newcomb W W, Brown J C, Kedes D H, Steven A C. Capsid structure of Kaposi's sarcoma-associated herpesvirus, a gammaherpesvirus, compared to those of an alphaherpesvirus, herpes simplex virus type 1, and a betaherpesvirus, cytomegalovirus. J Virol. 2001;75:2879–2890. doi: 10.1128/JVI.75.6.2879-2890.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Unal A, Pray T R, Lagunoff M, Pennington M W, Ganem D, Craik C S. The protease and the assembly protein of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) J Virol. 1997;71:7030–7038. doi: 10.1128/jvi.71.9.7030-7038.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wall J S, Hainfeld J F, Simon M N. Scanning transmission electron microscopy (STEM) of nuclear structures. In: Berrios M, editor. Methods in cell biology. New York, N.Y: Academic Press, Inc.; 1998. pp. 139–166. [DOI] [PubMed] [Google Scholar]
- 47.Whitby D, Howard M R, Tenant-Flowers M, Brink N S, Copas A, Boshoff C, Hatzioannou T, Suggett F E, Aldam D M, Denton A S, et al. Detection of Kaposi sarcoma associated herpesvirus in peripheral blood of HIV-infected individuals and progression to Kaposi's sarcoma. Lancet. 1995;346:799–802. doi: 10.1016/s0140-6736(95)91619-9. [DOI] [PubMed] [Google Scholar]
- 48.Wu L, Lo P, Yu X, Stoops J K, Forghani B, Zhou Z H. Three-dimensional structure of the human herpesvirus 8 capsid. J Virol. 2000;74:9646–9654. doi: 10.1128/jvi.74.20.9646-9654.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci USA. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou Z H, He J, Jakana J, Tatman J D, Rixon F J, Chiu W. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat Struct Biol. 1995;2:1026–1030. doi: 10.1038/nsb1195-1026. [DOI] [PubMed] [Google Scholar]