Abstract
Primary pulmonary hypertension (PPH) is a potentially lethal disorder, because the elevation of the pulmonary arterial pressure may result in right-heart failure. Histologically, the disorder is characterized by proliferation of pulmonary-artery smooth muscle and endothelial cells, by intimal hyperplasia, and by in situ thrombus formation. Heterozygous mutations within the bone morphogenetic protein type II receptor (BMPR-II) gene (BMPR2), of the transforming growth factor β (TGF-β) cell–signaling superfamily, have been identified in familial and sporadic cases of PPH. We report the molecular spectrum of BMPR2 mutations in 47 additional families with PPH and in three patients with sporadic PPH. Among the cohort of patients, we have identified 22 novel mutations, including 4 partial deletions, distributed throughout the BMPR2 gene. The majority (58%) of mutations are predicted to lead to a premature termination codon. We have also investigated the functional impact and genotype-phenotype relationships, to elucidate the mechanisms contributing to pathogenesis of this important vascular disease. In vitro expression analysis demonstrated loss of BMPR-II function for a number of the identified mutations. These data support the suggestion that haploinsufficiency represents the common molecular mechanism in PPH. Marked variability of the age at onset of disease was observed both within and between families. Taken together, these studies illustrate the considerable heterogeneity of BMPR2 mutations that cause PPH, and they strongly suggest that additional factors, genetic and/or environmental, may be required for the development of the clinical phenotype.
Introduction
Familial primary pulmonary hypertension (PPH [MIM 178600]) is an autosomal dominant disorder characterized by obliteration of the pulmonary arteries, which leads to sustained elevation of pulmonary vascular resistance (mean pressure >25 mmHg at rest, >30 mmHg during exercise) and progressive right-heart failure (Rubin 1997). The disease gene acts with markedly reduced penetrance, and the majority of patients (>90%) have no known family history of the disease. In both familial and sporadic PPH, the disease is twice as common in females as in males, and symptoms typically develop during the 3d decade of life, although the disease may occur at any age (Rich et al. 1987; Gaine and Rubin 1998). A number of potential environmental triggers for the development of the disease have been recognized in recent years, including prolonged use of appetite-suppressant drugs (e.g., dexfenfluramine and fenfluramine), infection with HIV, and consumption of adulterated rapeseed oil (Gomez-Sanchez et al. 1991; Abenhaim et al. 1996; Mesa et al. 1998; Thomson and Trembath, in press). Treatment strategies have relied on the use of oral anticoagulants and vasodilators. More recently, the use of prostacyclin analogues has improved life expectancy, with heart and/or lung transplantation as a further treatment option (Rich et al. 1992; McLaughlin et al. 1998).
Pulmonary hypertension may occur as a secondary phenomenon in patients with various diseases, including recurrent thromboembolism, chronic hypoxic lung disease, and left-heart disease. However, until recently the etiology of PPH has been poorly understood. Linkage analysis in affected families mapped the familial PPH locus to chromosome 2q33, with no evidence for locus heterogeneity (Morse et al. 1997; Nichols et al. 1997; Deng et al. 2000a; Machado et al. 2000). Using a positional candidate-gene strategy, we and others recently have demonstrated that mutations in the bone morphogenetic protein type II receptor (BMPR-II) gene (BMPR2) cause familial PPH (Deng et al. 2000b; The International PPH Consortium et al. 2000). We have also demonstrated that germline mutations of BMPR2 are found in ⩾26% of sporadic cases of PPH (Thomson et al. 2000).
BMPR2 encodes a type II receptor member (BMPR-II) of the transforming growth factor β (TGF-β) superfamily of cell-signaling molecules (Liu et al. 1995). After ligand binding, type II receptors, which have serine/threonine kinase activity, form heteromeric complexes with membrane-bound type I receptors, initiating phosphorylation of the type I receptor and downstream intracellular Smads (Massague and Chen 2000). This diverse pathway appears to be critical in both cell differentiation and cell growth, with specificity mediated through transcriptional regulation. Of considerable interest, defects of two additional components of the TGF-β pathway (endoglin (ENG) and the activin receptor–like kinase-1 gene [ACVRLK1]) have been demonstrated in some families with another vascular disorder, hereditary hemorrhagic telangiectasia (MIM 187300 and MIM 600376) (McAllister et al. 1994; Johnson et al. 1996; Marchuk 1998).
Our initial findings suggest that, although heterozygous germline mutations are required for inheritance of PPH, they may not be sufficient for the development of the disease phenotype. Furthermore, our findings raise important questions regarding the molecular mechanisms that lead to the pathogenesis of the vascular lesions seen in PPH. To obtain data to enhance the understanding of the functional significance of BMPR2 mutations in PPH, we have investigated both an additional 47 unrelated families with PPH and three patients with sporadic PPH. We report 23 novel mutations of the BMPR2 gene that, taken together with in vitro functional analyses, support a model of haploinsufficiency as the molecular mechanism for the dominant inheritance of this important vascular disorder.
Families and Methods
Families
Probands with familial PPH were identified in 47 unrelated families by specialist physicians at major organ-transplant centers. Individuals were defined as affected if cardiac catheterization revealed normal pulmonary-artery wedge pressure and pulmonary hypertension (mean pulmonary artery pressure >25 mmHg) or if autopsy results showed plexogenic pulmonary arteriopathy (Gaine and Rubin 1998). We excluded patients with secondary causes of pulmonary hypertension, including other diseases of the heart and lung, pulmonary embolism, connective-tissue diseases, and the use of appetite-suppressant drugs (Thomson and Trembath, in press). All families are of western European origin. For assessment of genotype/phenotype correlation, age at onset was defined as the first age at which relevant symptoms or clinical signs were recorded. To document the number of relatives affected with PPH, a structured interview was performed, including a detailed family history (by J.R.T., L.W., or the local PPH specialist team). Affected status was confirmed through autopsy reports, histological review by transplant center pathologists, and assessment of medical records. Unaffected status was assigned to related family members with either no symptoms or signs of disease and/or after indirect measurement of pulmonary artery pressures by Doppler echocardiography. All available data were assessed by a physician specializing in PPH (J.E.L., J.N., N.G., M.H., K.M., P.C., G.M., and L.T.), according to World Health Organization criteria (Thomson and Trembath, in press). DNA was extracted, by standard methods, from whole blood, lymphoblastoid cell lines, or pathological tissue specimens. All samples were collected after participants gave informed consent, and the study was approved by the Leicestershire Health Authority Ethics Committee, the Huntingdon Local Research Ethics Committee, the Royal Brompton and Harefield National Health Service Trust Ethics Committee, the South Manchester Local Research Ethics Committee, the West Glasgow Hospitals University National Health Service Trust, and the institutional review boards of the individual institutions.
Mutation Analysis of the BMPR2 Gene in Families with PPH
The entire protein-coding region and intron/exon boundaries of the BMPR2 gene were amplified by PCR using DNA samples from affected individuals and primer pairs specific for all 13 exons, as described elsewhere (The International PPH Consortium et al. 2000). PCR products were separated by electrophoresis through 4% agarose (3% Nusieve [FMC Bioproducts] and 1% agarose [Gibco BRL]), to ensure the presence of sufficient quantities for sequence analysis, and were purified using the QIAquick 96 PCR purification kit (QIAGEN). PCR products were sequenced using the same primers as for the PCR reactions, on either an ABI 377 sequencer or an ABI 3700 DNA analyzer, with the Applied Biosystems DyeDeoxy or BigDye terminator kit. Using PCR amplification of the relevant exon, followed by either mutation-specific RFLP analysis or direct sequencing, we confirmed segregation of the mutations within families and excluded the presence of the mutations in a panel of 150 chromosomes from normal individuals.
Haplotype Analysis to Determine Origins of Recurrent Mutations
Genotyping of polymorphic markers around the BMPR2 gene was performed as described elsewhere for individuals from different families found to carry the same mutation (Machado et al. 2000). Alternatively, genotype analysis for a BMPR2 intron 1 tetranucleotide repeat, using primers 5′-TGGAAGGTTAGATGCAAATGG-3′ (forward) and 5′-GTGAAGGTTGCAGTGAGCAG-3′ (reverse), was performed by PCR amplification.
RNA Extraction and Reverse Transcriptase (RT) PCR
Total RNA was extracted from lymphoblasts or whole lung tissue, by use of the RNAzol B kit (AMS Biotechnology), and 2.5 μg of total RNA was reverse transcribed using 25 ng of primer R1 (5′-CACCCCGCCCGGTAAACATC-3′). After the RNA was heated for 10 min at 65°C, 1× RT buffer, 10 mM DTT, 500 μM of dNTP, 40 U of RNase inhibitor, and 200 U Moloney murine leukemia virus RT (Gibco BRL) were added, and the mix was incubated at 42°C for 1 h. The entire coding sequence was amplified in two overlapping fragments of 2,075 bp (exons 1–12) and 1,183 bp (exons 12 and 13). A primary PCR was performed for each fragment, as described above for genomic DNA amplification. A heminested PCR was then performed by addition of 1 μl of the primary PCR product to a secondary PCR mix, as described above. Secondary products were then separated by electrophoresis on a 1.5% agarose gel.
Fluorescent-Dosage PCR
A quantitative fluorescent multiplex PCR reaction was performed, as described elsewhere (Morgan et al. 1999), to amplify fragments of exons 1 and 12 of the BMPR2 gene, along with exon 4 of the SURF6 gene (primers 5′-AGTGGGTGCCTCTTTCC-3′ and 5′-TGAAGCTAACAAGGGGCAAC-3′), which served as an internal control. All forward primers were labeled with a fluorescent dye (Applied Biosystems). To keep the reactions within the exponential phase, only 19 cycles of PCR were performed in a 25-μl reaction with 125 ng of genomic DNA and 0.25 μM each of forward and reverse primer, as described above. A 1-μl aliquot of PCR product was added to 1.5 μl of formamide loading buffer (95% formamide in 1× Tris-borate EDTA and 5 mg dextran blue/ml) and 0.5 μl of internal lane size standard (Genescan-500 Rox; Applied Biosystems). Samples were denatured for 5 min at 95°C and were separated by electrophoresis on a 6% denaturing polyacrylamide gel for 3 h by an ABI 377 fluorescent DNA sequencer. Data were analyzed by GENESCAN and GENOTYPER software to obtain electropherograms for each sample. The copy numbers of BMPR2 exons 1 and 12 in the patient samples were determined by calculation of a dosage quotient, using the area below the peaks for the amplification products relative to those of the control sample. The dosage quotient for BMPR2 exon 1 is calculated as follows: [patient sample BMPR2 exon 1/patient sample SURF6 exon 4]/[control BMPR2 exon 1/control SURF6 exon 4].
In Vitro Expression of Recombinant BMPR-II
Plasmid constructs
Plasmids were constructed by digesting the BMPR2 cDNA from CL4-1 (kindly provided by H. Moses) with XhoI and SalI. This 3.3-kb fragment was ligated into XhoI/SalI-digested pCI-neo (Promega). Clones were confirmed by restriction digest and DNA sequencing. To generate the mutations used for the in vitro expression studies the initial construct was subjected to site-directed mutagenesis, according to the Stratagene QuickChange™ (200518) protocol. Independent transformants were screened by amplification of the appropriate fragment from recovered plasmid and by restriction digestion to confirm the altered site. Results of this assay were confirmed by restriction endonuclease digestion of the intact isolated plasmid.
Functional studies
The expression system used the normal mouse mammary-gland epithelium (NMuMG) cell line that is constitutively competent for TGF-β signaling (Piek et al. 1999). NMuMG cells (American Type Culture Collection CRL-1636) were grown in Dulbecco's minimal essential medium (DMEM), supplemented with 4.5 g glucose/liter, 0.584 g l-glutamine/liter, 100 U/100 μg penicillin-streptomycin/ml, and 10% fetal bovine serum. Cells were plated at ∼3 × 105 cells per well, in six-well Falcon plates (Falcon). After 24 h, the media were removed, and the wells were washed once with Hanks' balanced salt solution (LifeTechnologies). Transfection was accomplished by addition, to each well, of 0.5 ml of OptiMEM (LifeTechnologies) containing 2 μg of plasmid DNA complexed with Lipofectamine™ (LifeTechnologies) as a transfection reagent. The transfection reagent was prepared by addition of 14 μg of plasmid DNA to 3.5 ml of OptiMEM in polystyrene tubes (Falcon), followed by addition of 70 μl of lipofectamine. The mixture was agitated lightly, was incubated at 37°C for 30 min, and then was applied to the washed wells. The plates were incubated at 37°C, under 5% CO2 for 8 h, and then 0.5 ml of supplemented DMEM was added to the wells, and the incubation continued for 16 h.
The transfection experiments consisted of cotransfection of three different plasmids. The reporter plasmid was pSBE, described by Jonk et al. (1998), which uses a Smad-binding element (SBE) and luciferase reporter. The plasmid for initiating the signal was either a BMPR2-containing plasmid, as described above, or a constitutively active activin type 1 receptor as a control for maximum signal amplitude. Finally, a plasmid expressing the heat-stable alkaline phosphatase (AP) gene under the control of the cytomegalovirus (CMV) promoter was added as a control for transfection efficiency. The ratio of the reporter:signal initiator:transfection control plasmids was 2:1:1. A CMV-driven chloramphenicol acetyl transferase reporter was added to adjust the plasmid quantity to 2 μg, where necessary.
The DNA liposome–containing medium was removed at the conclusion of the 16-h incubation and was replaced by DMEM, as described above, except that it contained 0.1% FBS and no antibiotic. After 8–16 h, 50 ng BMP4/ml was added to one-half of the wells. Incubation with ligand or a similar amount of diluent continued for 16–24 h, at which time the media were removed and assayed for heat-stable AP and the cells were lysed according to the manufacturer's instruction for the luciferase assay (Promega). The luciferase determination, measured as relative light units (RLU), was done according to the manufacturer's protocol and was normalized to the AP activity in the retained media.
Results
Mutation Analysis of the BMPR2 Gene
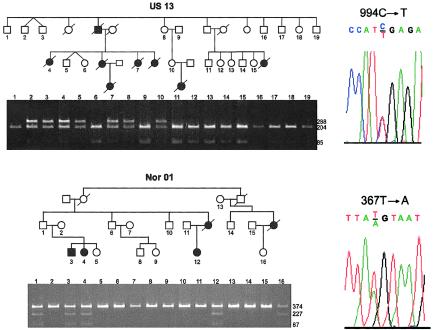
Results of the BMPR2 mutation analysis in families with PPH are summarized in table 1 and figure 1. A total of 26 mutations were identified, accounting for 23 of the 47 families with PPH and for the three patients with sporadic PPH. All these mutations were shown to segregate with the disease in families with additional members available, and none were detected in a panel of 150 chromosomes from normal individuals. Figure 2 demonstrates familial segregation for two of the newly identified mutations.
Table 1.
BMPR2 Mutations Identified in 23 Families with PPH and in Three Patients with Sporadic PPH
| Familya | Mutation Location | Nucleotide Changeb | Amino Acid Changec | Segregation | No. AffectedIndividuals/Generations | Age(s) at Onsetd(years) | Ages of Asymptomatic Obligate Gene Carriers(years) |
| UKSp14 | Exons 1–6 | 51–814 del | T17fs(+7 amino acids) | Direct sequencing | 1/1 | 26 | … |
| UKSp15 | Exon 1 | Undetermined 5′ deletion | … | Dosage | 1/1 | 31 | … |
| UK01 | Exon 1 | Undetermined 5′ deletion | … | Dosage | 8/3 | 1–42 | 57, 72 (living); 66 (deceasede) |
| US70 | Exon 1 | 44delC | P15fs(+30 amino acids) | NcoI | 2/2 | 36–38 | … |
| US42 | Exon 2 | 156–157delTC | S52fs(+0 amino acids) | Direct sequencing | 3/1 | 32–39 | … |
| UK09 | Exon 3 | 367T→C | C123R | MseI | 2/1 | 9–26 | … |
| NOR01 | Exon 3 | 367T→A | C123S | MseI | 5/2 | NR | … |
| US37 | Exon 4 | 439C→T | R147X | DdeI | 3/2 | 10–39 | … |
| UK21 | Exon 4 | 504insT | L168fs(+11 amino acids) | Direct sequencing | 2/2 | 13–19 | … |
| ITA01 | Exon 6 | 631C→T | R211X | Taq1 | 2/1 | 17–18 | … |
| US44 | Exon 6 | 689–690delAA | K230fs(+23 amino acids) | Direct sequencing | 3/2 | 4–46 | … |
| FRA04 | Exon 6 | 727G→T | E243X | Direct sequencing | 2/2 | 40 | … |
| UK04 | intron 7 | IVS7 958+3 delT | Inactivates exon 7 donor site | Direct sequencing | 8/4 | 14–60 | 39, 45, 52 (living); 86 (deceasedf) |
| US50 | Exon 8 | 994C→T | R332X | TaqI | 2/2 | 28–32 | … |
| US13 | Exon 8 | 994C→T | R332X | TaqI | 8/3 | 13–42 | 45, 71 |
| US94 | Exon 8 | 1076delC | T359fs(+14 amino acids) | BsrI, XcmI | 2/2 | 39–42 | … |
| US80 | intron 8 | IVS8 1129–3C→G | Inactivates exon 9 acceptor site | MnlI | 6/3 | 24–53 | … |
| US89 | Exon 9 | 1191/1192delTG | C397fs(+0 amino acids) | HinfI | 2/1 | 26 | … |
| GER01 | Exon 9 | 1258T→C | C420R | MaeI | 4/2 | 9–40 | … |
| GRE01 | Exon 11 | 1535A→C | K512T | Direct sequencing | 2/2 | 19–50 | … |
| SWE01 | Exon 12 | 1750C→T | R584X | Direct sequencing | 2/2 | 34 | … |
| UK22 | Exon 12 | 2292insA | N764fs(+47 amino acids) | Direct sequencing | 3/2 | 30–57 | … |
| US79 | Exon 12 | 2408insTG | T803fs(+3 amino acids) | HphI | 2/1 | 51–52 | … |
| UK11 | Exon 12 | 2579–2580delT | I860fs(+10 amino acids) | AseI | 3/2 | 2–22 | … |
| US91 | Exon 12 | 2695C→T | R899X | HaeIII | 2/1 | 27–32 | … |
| UKSp16 | Exon 12 | Undetermined 3′ deletion | … | Dosage | 1/1 | 22 | … |
FRA = France; GER = Germany; GRE = Greece; ITA = Italy; NOR = Norway; Sp = patient with sporadic PPH; SWE = Sweden; UK = United Kingdom; US = United States.
Nucleotide numbers are according to published cDNA, with the A of the ATG designated as +1.
Frameshifts are denoted with the amino acid, position, and number of amino acids until the next stop codon.
NR = information not released.
Because of breast cancer.
Because of cerebrovascular accident.
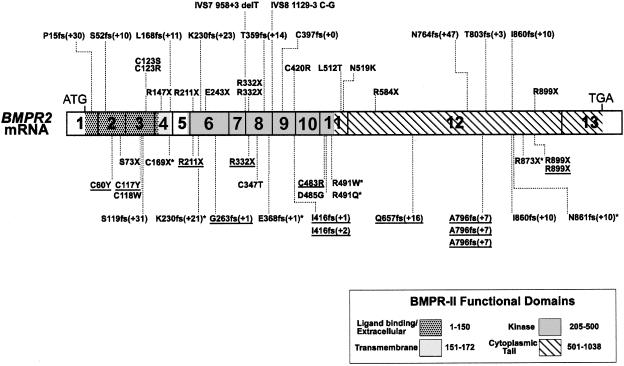
Figure 1.
BMPR2 cDNA structure and location of all reported PPH mutations. Exons are numbered 1–13, and their boundaries are indicated by the solid black lines segmenting the cDNA schematic. Extracellular/ligand binding, transmembrane, kinase, and cytoplasmic tail domains of BMPR-II are defined as indicated. Mutations shown above the BMPR2 cDNA are newly reported in this study, whereas those shown below have been reported elsewhere (Deng et al. 2000b;The International PPH Consortium et al. 2000; Thomson et al. 2000). Underlined mutations have been reported in patients with sporadic PPH (Thomson et al. 2000); asterisks (*) indicate mutations that were independently reported (Deng et al. 2000b). Frameshift mutations (fs) are denoted by the one-letter amino acid abbreviation and amino acid–position number at which the mutation occurs, followed by the number of extra amino acids before the next stop codon—for example, L168fs (+11).
Figure 2.
Cosegregation and sequence analysis of BMPR2 mutations in two kindreds with PPH. Black symbols denote affected individuals, and white symbols denote unaffected individuals. Nucleotides are numbered according to the cDNA sequence, with the adenosine of the initiation codon assigned +1. Family US13 (top) carries a C→T substitution at nucleotide 994, as shown in the chromatogram (top, right). Segregation of the mutations was tracked through the family by TaqI digestion of exon 8 PCR products shown on the agarose gel below the pedigree. Gene carriers are detected by presence of a 288-bp band due to loss of a TaqI site. Gel lanes correspond to the individuals numbered in the pedigree. Family Nor01 (bottom) carries a T→A substitution at nucleotide 367, as shown in the chromatogram (bottom, right). MseI digestion of exon 3 PCR products was used to track inheritance of the mutations through the family, as shown on the agarose gel below the pedigree. The mutation is evidenced by gain of an MseI site, resulting in the generation of 227- and 87-bp fragments. Individuals are identified by numbers corresponding to those on gels.
Fifteen of the 26 mutations are either frameshift (n=9) or nonsense (n=6) mutations that would be predicted to result in a truncated BMPR-II molecule. The nine frameshift mutations are dispersed throughout the gene, with the mutation closest to the 5′ end occurring in exon 1 and with the frameshift closest to the 3′ end found in exon 12. The exon 1 frameshift mutation would predict a protein containing only the 15 N-terminal amino acids of BMPR-II, with an additional 30 amino acids until the next stop codon.
Six nonsense mutations, accounting for seven of the families, are found in exons 4, 6, 8, and 12. The same exon 8 R332X mutation was detected in two unrelated families with two different nonsense mutations (R584X and R899X) in exon 12. An exon 6 nonsense mutation (E243X) results from a G→T substitution at nucleotide 727, changing a GAA glutamic acid codon to a TAA stop codon.
Four missense mutations are found in exons 3, 9, and 11. Two different exon 3 missense mutations were identified at codon 123. A T→C substitution at nucleotide 367 results in the substitution of cysteine 123 by arginine in family UK09, whereas a T→A change at the same nucleotide results in a substitution of cysteine 123 by serine in family Nor01. Shown in table 2 are six nucleotide changes believed to represent polymorphisms, because they were also observed in individuals not at risk for the disease. One of the polymorphisms, a silent mutation at codon 937 in exon 12, appears to be common, since it was detected in 15 unrelated families. The remaining five polymorphisms, each of which was found in three or fewer families, appear to be less common.
Table 2.
BMPR2 Polymorphisms
| Location of Polymorphism | Nucleotide Changea | Amino Acid Change | No. of Families |
| Exon 6 | 672G→T | E672D | 3 |
| Exon 8 | 1107A→G | E369E | 1 |
| Exon 12 | 2811G→A | R937R | 15 |
| Intron 3 | IVS3 420-43 delT | … | 2 |
| Intron 4 | IVS4 529+64 C→T | … | 1 |
Nucleotide numbers are according to published cDNA, with the A of the ATG designated as +1.
Identification of Deletion Variants by Fluorescent-Gene Dosage
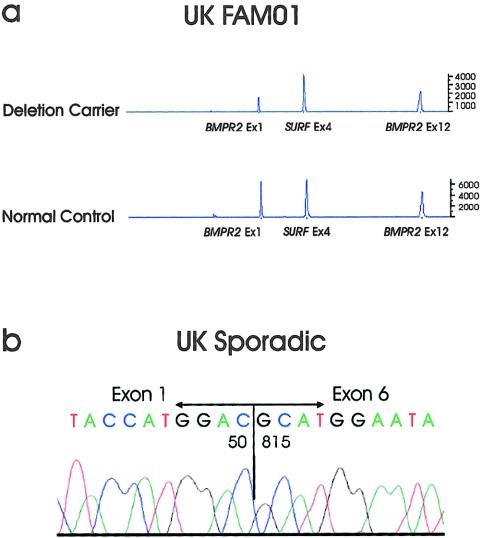
When fluorescent gene–dosage PCR was used, one affected individual from family UK01 was observed to have a dosage quotient of 0.46 for BMPR2 exon 1 PCR products, relative to an internal control PCR from exon 4 of the SURF6 gene (fig. 3). Normal levels of BMPR2 exon 12 PCR products were detected in this patient, indicating that this patient with PPH is heterozygous for at least a partial deletion of the BMPR2 gene, the extent of which remains to be determined. This result was confirmed in one obligate gene carrier and in four individuals bearing the disease haplotype in family UK01. Two additional partial gene deletions were detected by this technique in two patients with sporadic PPH. A dosage quotient of 0.41 for exon 1 was observed for patient UKSp15, indicating a heterozygous partial deletion in the 5′ portion of the gene. Similarly, UKSp16 yielded an exon 12 dosage quotient of 0.46, suggesting a heterozygous deletion in the 3′ portion of BMPR2. The extent of each deletion remains to be characterized.
Figure 3.
Detection of heterozygous deletions of the BMPR2 gene. A, Fluorescent-dosage PCR of an affected individual in family UK01. The patient demonstrates a reduction of 50% in peak intensity for exon 1 of the BMPR2 gene, compared with both exon 4 of the SURF6 gene and exon 12 of the BMPR2 gene. This indicates a heterozygous deletion encompassing at least exon 1 of the BMPR2 gene. Also shown is an electropherogram from a normal control subject, in whom peaks for all three PCR products are approximately equal. B, Sequence analysis of BMPR2 lung mRNA RT-PCR products of UK sporadic 14 subject. An RNA product with nucleotide 50 contiguous to nucleotide 815 was detected by sequence analysis, as seen in the chromatogram. The normal-sized RT-PCR product was also identified, indicating that this patient is heterozygous for a deletion of nucleotides 51–814.
Characterization of Deletion and Splice-Site Mutations by RT-PCR
RT-PCR using mRNA isolated from whole-lung tissue of a patient (UKSp14) with sporadic PPH yielded a PCR product deleted for nucleotides 51–814 of the BMPR2 cDNA, encompassing the last 26 nucleotides of exon 1, all of exons 2–5, and two-thirds of exon 6 (fig 3). The normal PCR product was also present, indicating heterozygosity for this event. This mRNA would be predicted to encode a protein consisting of the N-terminal 16 amino acids of BMPR-II plus 7 additional amino acids before the next stop codon.
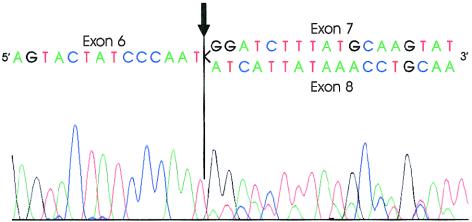
DNA sequence analysis in two families identified mutations in the consensus splice sites of two different introns, indicating potential splicing variants (fig. 2). The first of these is a heterozygous one-base deletion in the third nucleotide of intron 7 in family UK04 (table 1). RT-PCR products from lymphocyte mRNA from a patient yielded a heterozygous template consisting of exon 6 spliced either to exon 7 or directly to exon 8, with exon 7 skipped (fig. 4). This abnormally spliced mRNA would predict a protein consisting of the first 284 amino acids of BMPR-II, with 11 additional amino acids until the next stop codon. A C→G substitution in intron 8, three nucleotides 5′ of exon 9, in family US80 suggests an additional splice variant. The lack of BMPR2 mRNA from this patient prevents confirmation of this change as a splicing mutation.
Figure 4.
Sequence analysis of RT-PCR products for a heterozygous BMPR2 intron 7 splice-site mutant. The splice-site mutation leads to skipping of exon 7 in the mutant. Nucleotides are listed above the sequencing peaks and indicate the end of exon 6, joined with the beginning of exon 7 or exon 8 in the normal or mutant, respectively. The arrow indicates the splice junction between exon 6 and either exon 7 or 8. Exon 7 sequence can be seen as the higher peaks in the right half of the chromatogram, with exon 8 represented by the lower peaks.
In Vitro Expression Analysis of BMPR2 Kinase Domain Mutation
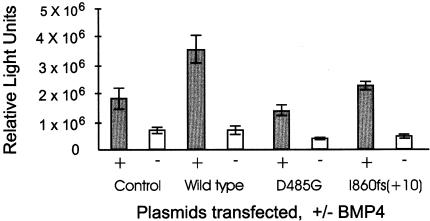
Using site-directed mutagenesis, we generated the previously reported kinase domain missense mutation D485G (The International PPH Consortium et al. 2000). Either full-length wild-type BMPR2 or full-length D485G recombinant BMPR2 was cotransfected with an SBE luciferase-reporter plasmid (pBSE) in NMuMG cells in the presence of the ligand BMP4. As seen in figure 5, cotransfection with the wild-type BMPR2 yielded a measurement of 4.3 × 106 RLU, whereas cotransfection with D485G BMPR2 resulted in the production of only 1.7 × 106 RLU. Results obtained with the recombinant molecule are similar to those obtained with transfection of only pBSE in the absence of any exogenous type II receptor (2.0 × 106 RLU), a result that is consistent with loss of function of the BMPR-II molecule containing the D485G substitution. Similar results were obtained on expression of a mutant plasmid containing a frameshift mutation in exon 12 (fig. 5).
Figure 5.
In vitro expression analysis of wild-type and mutant BMPR2 in NMuMG cells. The control transfection using only the SBE reporter plasmid in the presence (+) or absence (−) of the ligand BMP4 measures activation of endogenous type II receptor function. Cotransfection of the SBE reporter with either wild-type or mutant BMPR2 constructs determines functional activity of the exogenous BMPR-II. Cotransfection of wild-type BMPR2 and the SBE reporter yielded a twofold increase in reporter activity. In contrast, cotransfection of mutant BMPR2 yielded functional activity equivalent to that of the endogenous receptor, indicating loss of function for both mutants.
Haplotype Analysis for Recurrent BMPR2 Mutations
Three of the mutations in the present study have been reported elsewhere (The International PPH Consortium et al. 2000; Thomson et al. 2000). Affected individuals from two unrelated families, US13 and US50, were shown to carry the R332X nonsense mutation previously seen in a patient with sporadic PPH (Thomson et al. 2000). Another family (UK11) was shown to carry the previously reported I860fs(+10) familial mutation, and the R899X nonsense mutation, previously detected in both familial and sporadic PPH, was identified in 1 (US91) of the 47 families (The International PPH Consortium et al. 2000; Thomson et al. 2000). None of the individuals carrying the same mutations shared the same disease haplotype, indicating independent origins for all of the recurrent mutations.
Genotype and Disease Onset
The observed overall range for the age at onset of symptoms of PPH was 1–60 years (table 1). Marked variation in the age at onset was noted both within families and between families that had the same BMPR2 mutation. For example, in family UK04, the age at onset for the eight affected individuals was 14–60 years. In families US13 and US50, both carrying the R332X mutation, the mean age at onset was 28 and 30 years, respectively, with a range of 13–42 years in the eight affected individuals in US13, whereas the lowest age at onset in US50 was 28 years. These data show no correlation between type of BMPR2 mutation and penetrance of PPH, either in terms of number of affected individuals within a family or age at onset of disease. Furthermore, the range and nature of the presenting symptoms were similar between families with and without identified BMPR2 mutations.
Discussion
In this study, we have identified heterozygous mutations of the BMPR2 gene in 23 of 47 studied families as well as in three individuals with sporadic PPH. Twenty-two of the mutations are novel, and elsewhere we have reported four of the mutations in other patients with familial or sporadic PPH (The International PPH Consortium et al. 2000; Thomson et al. 2000). These data represent the first report of splice mutations as well as the first evidence of partial deletions of the BMPR2 gene in patients with PPH. To date, we have identified 28 novel familial PPH mutations, accounting for 32 of 55 families analyzed. We have also recently identified an additional 11 mutations in 13 of 50 individuals with sporadic PPH (Thomson et al. 2000). When a recent report of 7 additional mutations in 19 families (Deng et al. 2000b) is included, a total of 46 unique BMPR2 mutations have now been identified in patients with PPH (fig. 1). The mutations are widely dispersed throughout the gene and are found in all exons except 5, 10, and 13.
The first evidence that large deletions in the BMPR2 gene contribute to the development of PPH was detected, in the present study, in one patient with familial PPH and in three patients with sporadic PPH. In one of the patients with sporadic PPH deletions, the RT-PCR product encompassed the last 26 nucleotides of exon 1, all of exons 2–5, and two-thirds of exon 6. Interestingly, the sequence ATGGAC is found on the 5′ side of the deletion breakpoints in both exon 1 and exon 6, suggesting homologous recombination as the mechanism producing the deletion. The identified frameshift mutations are predicted to result from replication slippage, because all eight occur in regions of small mono- or dinucleotide repeats. Independent recurrence of the I860fs(+10) frameshift mutation, resulting from a single thymine deletion (ATTAATT→ATTAAT) at either nucleotide 2579 or nucleotide 2580 may be a consequence of the sequence surrounding the deleted nucleotide. Not surprisingly, five of the six nonsense mutations occur at CpG dinucleotides, changing a CGA arginine codon to a TGA stop codon.
Using direct sequence analysis, we and others have detected mutations in 40 (55%) of 73 families with PPH (Deng et al. 2000b; The International PPH Consortium et al. 2000). There are a number of possible explanations for the 45% of families with PPH in which BMPR2 gene mutations have not been identified. First, the direct-sequencing methods employed in the study may not detect all heterozygous mutations. In addition, mutations in portions of the BMPR2 gene that were not sequenced, such as upstream regulatory regions, 3′ UTR, or intronic sequences, could account for PPH in these patients, although mutations of this type are generally infrequent in other genetic disorders. Alternatively, a hotspot for recurrent mutation, similar to the one that results in the common FVIII gene inversion in hemophilia A, could account for this observation (Lakich et al. 1993). The extent to which gene rearrangements at the BMPR2 locus contribute to the development of PPH is still unknown. Finally, the possibility of genetic heterogeneity must be considered. Of the 23 families in which mutations have not been detected, 13 demonstrate either direct evidence for linkage to or allele sharing at the BMPR2 locus. Although we cannot exclude the possibility that one or more additional genes may contribute to PPH pathogenesis, ⩾72% of families studied either have an identified BMPR2 mutation or show evidence of linkage to the BMPR2 locus.
Through pedigree analysis of kindreds with PPH, including several families included in the present study, it has previously been shown that PPH is a disorder demonstrating genetic anticipation (Loyd et al. 1995). Examination of the BMPR2 coding sequence fails to reveal any triplet-repeat sequences, and PCR analysis of a GCC repeat ∼1 kb 5′ of the initiation codon failed to demonstrate expansion in a cohort of patients with PPH. Thus, molecular evidence for genetic anticipation has yet to be detected in any of the families in the present study that demonstrate anticipation. Although expansion of an undetected intronic triplet repeat in the BMPR2 gene may account for this anticipation, it is also possible that a novel mechanism of anticipation occurs in families with PPH.
Assessment of the functional impact of the heterogeneous mutations in the BMPR2 gene is likely to provide insight into the molecular mechanisms associated with the initiation and progression of the vascular pathological lesions characteristic of PPH. The majority of nonsense and frameshift mutations, of which the mutation closest to the 5′ end occurs in exon 1, lead to premature truncation of the BMPR2 transcript. These mutations predict nonsense-mediated mRNA decay and absence of the production of the mutated BMPR-II polypeptide (Maquat 1995).
The 10 different missense mutations identified in this and other studies, each altering highly conserved amino acids, require further investigation of functional impact. As evidence of the pathogenicity of these mutations, none were detected among a panel of 150 normal chromosomes. Five missense mutations have been identified in the region coding for the extracellular/ligand-binding domain, four in the kinase domain–encoding region, and one immediately distal to the kinase domain–encoding region (fig. 1). Each of the 10 missense mutations introduces a nonconservative change that alters the charge, polarity, hydrophobicity, and/or size of the substituted amino acid residue. The five missense mutations in the ligand-binding domain involve 4 of the 10 highly conserved cysteine residues. If expressed and transported to the cell membrane, such mutant BMPR-II polypeptides are predicted to alter the three-dimensional structure of the ligand-binding domain. Such alterations may affect either ligand binding, type I–receptor binding, or propagation of signal across the membrane to the kinase domain. In vitro expression of recombinant BMPR-II containing either the D485G mutation or the I860fs(+10) frameshift mutation, predicted to encode a protein containing 83% of the normal BMPR-II, demonstrated complete loss of function.
Taken together, these data strongly support the hypothesis that the predominant molecular mechanism in the dominant inheritance of PPH is one of haploinsufficiency. This suggests that the target cell(s) within the pulmonary arterial wall are sensitive to BMPR2 gene dosage and that the TGF-β pathway, mediated through BMPR-II, is critical for maintenance and/or normal response to injury of the pulmonary vasculature. However, the age at onset of disease is variable, both within families and between subjects carrying identical but recurrent mutations (table 1). These findings, together with the previously reported large number of individuals in whom BMPR2 lacked penetrance (Deng et al. 2000b; The International PPH Consortium et al. 2000), point to the possibility that additional factors, either environmental or genetic, are required for the pathogenesis of the disease. The marked sex bias (female:male ratio 2:1) for presentation of the disease suggests a role for either a hormonal factor or an X-linked locus in disease predisposition (Loyd et al. 1995).
The identification of germline mutations of the BMPR2 gene as the inherited basis of PPH has important implications for assessment and management of patients with this dramatic vascular disease. However, the detailed molecular mechanisms associated with the pathogenesis of the disorder will require further analysis, including identification of target genes whose transcription is regulated by BMPR-II–mediated cell signaling. The demonstration that gene dosage and receptor activity are critical will aid the development of functional experiments designed to delineate the detailed molecular pathology of PPH.
Acknowledgments
We wish to express our thanks to the many patients and their parents who provided samples and details of their medical and family histories and to the Pulmonary Hypertension Association, for its encouragement and support. We acknowledge the many clinicians and colleagues who have provided information for the patients described, in particular, E. W. Benbow, M. K. Bennet, M. Burke, C. Chapman, J. Egan, M. Goldberg, G. B. M. Lindop, B. Meyrick, A. Peacock, D. Smith, and S. Stuart. This work has financial support from a Medical Research Council of the United Kingdom Clinical Training Fellowship (to J.R.T.), British Heart Foundation project grant 97054 (to R.C.T.), and National Institutes of Health grants HL61997 (to W.C.N. and J.E.L.) and HL48164 (to J.E.L.).
Electronic-Database Information
Accession numbers and the URL for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for familial PPH [MIM 178600] and hereditary hemorrhagic telangiectasia [MIM 187300 and 600376])
References
- Abenhaim L, Moride Y, Brenot F, Rich S, Benichou J, Kurz X, Higenbottam T, Oakley C, Wouters E, Aubier M, Simonneau G, Begaud B (1996) Appetite-suppressant drugs and the risk of primary pulmonary hypertension. International Primary Pulmonary Hypertension Study Group. N Engl J Med 335:609–616 [DOI] [PubMed] [Google Scholar]
- Deng Z, Haghighi F, Helleby L, Vanterpool K, Horn EM, Barst RJ, Hodge SE, Morse JH, Knowles JA (2000a) Fine mapping of PPH1, a gene for familial primary pulmonary hypertension, to a 3-cM region on chromosome 2q33. Am J Respir Crit Care Med 161:1055–1059 [DOI] [PubMed] [Google Scholar]
- Deng Z, Morse JH, Slager SL, Cuervo N, Moore KJ, Venetos G, Kalachikov S, Cayanis E, Fischer SG, Barst RJ, Hodge SE, Knowles JA (2000b) Familial primary pulmonary hypertension (gene PPH1) is caused by mutations in the bone morphogenetic protein receptor-II gene. Am J Hum Genet 67:737–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaine SP, Rubin LJ (1998) Primary pulmonary hypertension. Lancet 352:719–725 [DOI] [PubMed] [Google Scholar]
- Gomez-Sanchez MA, de la Saenz C, Gomez-Pajuelo C, Martinez-Tello FJ, Mestre de Juan MJ, James TN (1991) Clinical and pathologic manifestations of pulmonary vascular disease in the toxic oil syndrome. J Am Coll Cardiol 18:1539–1545 [DOI] [PubMed] [Google Scholar]
- Johnson DW, Berg JN, Baldwin MA, Gallione CJ, Marondel I, Yoon SJ, Stenzel TT, Speer M, Pericak-Vance MA, Diamond A, Guttmacher AE, Jackson CE, Attisano L, Kucherlapati R, Porteous ME, Marchuk DA (1996) Mutations in the activin receptor-like kinase 1 gene in hereditary haemorrhagic telangiectasia type 2. Nat Genet 13:189–195 [DOI] [PubMed] [Google Scholar]
- Jonk LJ, Itoh S, Heldin CH, ten Dijke P, Kruijer W (1998) Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-β, activin, and bone morphogenetic protein-inducible enhancer. J Biol Chem 273:21145–21152 [DOI] [PubMed] [Google Scholar]
- Lakich D, Kazazian HH Jr, Antonarakis SE, Gitschier J (1993) Inversions disrupting the factor VIII gene are a common cause of severe haemophilia A. Nat Genet 5:236–241 [DOI] [PubMed] [Google Scholar]
- Liu F, Ventura F, Doody J, Massague J (1995) Human type II receptor for bone morphogenic proteins (BMPs): extension of the two–kinase receptor model to the BMPs. Mol Cell Biol 15:3479–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyd JE, Butler MG, Foroud TM, Conneally PM, Phillips JA III, Newman JH (1995) Genetic anticipation and abnormal gender ratio at birth in familial primary pulmonary hypertension. Am J Respir Crit Care Med 152:93–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado RD, Pauciulo MW, Fretwell N, Veal C, Thomson JR, Vilarino GC, Aldred M, Brannon CA, Trembath RC, Nichols WC (2000) A physical and transcript map based upon refinement of the critical interval for PPH1, a gene for familial primary pulmonary hypertension. Genomics 68:220–228 [DOI] [PubMed] [Google Scholar]
- Maquat LE (1995) When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA 1:453–465 [PMC free article] [PubMed] [Google Scholar]
- Marchuk DA (1998) Genetic abnormalities in hereditary hemorrhagic telangiectasia. Curr Opin Hematol 5:332–338 [DOI] [PubMed] [Google Scholar]
- Massague J, Chen YG (2000) Controlling TGF-β signaling. Genes Dev 14:627–644 [PubMed] [Google Scholar]
- McAllister KA, Grogg KM, Johnson DW, Gallione CJ, Baldwin MA, Jackson CE, Helmbold EA, Markel DS, McKinnon WC, Murrell J, McCormick MK, Pericak-Vance MA, Heutnik P, Oostra BA, Haitjema T, Westerman CJJ, Porteous ME, Guttmacher AE, Letarte M, Marchuk DA (1994) Endoglin, a TGF-β binding protein of endothelial cells, is the gene for hereditary haemorrhagic telangiectasia type 1. Nat Genet 8:345–351 [DOI] [PubMed] [Google Scholar]
- McLaughlin VV, Genthner DE, Panella MM, Rich S (1998) Reduction in pulmonary vascular resistance with long-term epoprostenol (prostacyclin) therapy in primary pulmonary hypertension. N Engl J Med 338:273–277 [DOI] [PubMed] [Google Scholar]
- Mesa RA, Edell ES, Dunn WF, Edwards WD (1998) Human immunodeficiency virus infection and pulmonary hypertension: two new cases and a review of 86 reported cases. Mayo Clin Proc 73:37–45 [DOI] [PubMed] [Google Scholar]
- Morgan NV, Tipping AJ, Joenje H, Mathew CG (1999) High frequency of large intragenic deletions in the Fanconi anemia group A gene. Am J Hum Genet 65:1330–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse JH, Jones AC, Barst RJ, Hodge SE, Wilhelmsen KC, Nygaard TG (1997) Mapping of familial primary pulmonary hypertension locus (PPH1) to chromosome 2q31-q32. Circulation 95:2603–2606 [DOI] [PubMed] [Google Scholar]
- Nichols WC, Koller DL, Slovis B, Foroud T, Terry VH, Arnold ND, Siemieniak DR, Wheeler L, Phillips JA III, Newman JH, Conneally PM, Ginsburg D, Loyd JE (1997) Localization of the gene for familial primary pulmonary hypertension to chromosome 2q31-32. Nat Genet 15:277–280 [DOI] [PubMed] [Google Scholar]
- Piek E, Moustakas A, Kurisaki A, Heldin CH, ten Dijke P (1999) TGF-β type I receptor/ALK-5 and Smad proteins mediate epithelial to mesenchymal transdifferentiation in NMuMG breast epithelial cells. J Cell Sci 112:4557–4568 [DOI] [PubMed] [Google Scholar]
- Rich S, Dantzker DR, Ayres SM, Bergofsky EH, Brundage BH, Detre KM, Fishman AP, Goldring RM, Groves BM, Koerner SK (1987) Primary pulmonary hypertension: a national prospective study. Ann Intern Med 107:216–223 [DOI] [PubMed] [Google Scholar]
- Rich S, Kaufmann E, Levy PS (1992) The effect of high doses of calcium-channel blockers on survival in primary pulmonary hypertension. N Engl J Med 327:76–81 [DOI] [PubMed] [Google Scholar]
- Rubin LJ (1997) Primary pulmonary hypertension. N Engl J Med 336:111–117 [DOI] [PubMed] [Google Scholar]
- The International PPH Consortium, Lane KB, Machado RD, Pauciulo MW, Thomson JR, Phillips JA III, Loyd JE, Nichols WC, Trembath RC (2000) Heterozygous germline mutations in BMPR2, encoding a TGF-β receptor, cause familial primary pulmonary hypertension. Nat Genet 26:81–84 [DOI] [PubMed] [Google Scholar]
- Thomson JR, Machado RD, Pauciulo MW, Morgan NV, Humbert M, Elliott GC, Ward K, Yacoub M, Mikhail G, Rogers P, Newman JH, Wheeler L, Higenbottam T, Gibbs JSR, Egan J, Crozier A, Peacock A, Allcock R, Corris P, Loyd JE, Trembath RC, Nichols WC (2000) Sporadic primary pulmonary hypertension is associated with germline mutations of the gene encoding BMPR-II, a receptor member of the TGF-β family. J Med Genet 37:741–745 [DOI] [PMC free article] [PubMed]
- Thomson JR, Trembath RC (2000) Primary pulmonary hypertension: the pressure rises for a gene. J Clin Pathol 53:899–903 [DOI] [PMC free article] [PubMed] [Google Scholar]