Abstract
The present study examined the regulatory and metabolic response of the aromatic degrader Pseudomonas putida F1 and its tod operon, controlling toluene degradation, to fluorinated aromatic and aliphatic compounds. The tod operon is upregulated by inducer binding to the TodS sensing domain of a two‐component regulator. The induced enzymes include toluene dioxygenase that initiates catabolic assimilation of benzenoid hydrocarbons. Toluene dioxygenase was shown to oxidize 6‐fluoroindole to a meta‐stable fluorescent product, 6‐fluoroindoxyl. The fluorescent output allowed monitoring relative levels of tod operon induction in whole cells using microtiter well plates. Mono‐ and polyfluorinated aromatic compounds were shown to induce toluene dioxygenase, in some cases to a greater extent than compounds serving as growth substrates. Compounds that are oxidized by toluene dioxygenase and undergoing defluorination were shown to induce their own metabolism. 1,2,4‐Trifluorobenzene caused significant induction and computational modelling indicated productive binding to the TodS sensor domain of the TodST regulator. Toluene dioxygenase also showed preferential binding of 1,2,4‐trifluorobenzene such that defluorination was favoured. Fluorinated aliphatic compounds were shown to induce toluene dioxygenase. An aliphatic ether with seven fluorine atoms, 1,1,1,2‐tetrafluoro‐2‐trifluoromethoxy‐4‐iodobutane (TTIB), was an excellent inducer of toluene dioxygenase activity and shown to undergo transformation in cultures of P. putida F1.
INTRODUCTION
There are no reported natural product polyfluorinated organic compounds (PFCs), but synthetic chemistry has prepared millions in the last century with more than 9000 fluorinated compounds used commercially (Caron, 2020; Cros et al., 2022; Han et al., 2021; Wackett, 2022). In that context, microbial exposure to PFCs has been extremely short on an evolutionary time scale. This suggests that dedicated regulatory and metabolic systems will be rare (Wackett, 2021); a notable exception being biodegradation of the natural product monofluoroacetate (Kurihara et al., 2003; Murphy et al., 2003). Given that more than 20% of new agrichemicals and pharmaceuticals contain fluorine and most contain benzene rings (Britton et al., 2021; Ogawa et al., 2020), microbes evolved for aromatic hydrocarbon metabolism may be good candidates to induce enzymes and metabolize PFCs.
Cleaving the C—F bond is difficult chemically, but it is necessary for the biodegradation and bioremediation of PFCs accumulating in the environment. Pseudomonas putida strains carry out defluorination of 3‐fluorotoluene (Renganathan, 1989) and more recently, were shown to defluorinate 23 mostly PFCs, including those with trifluoromethyl groups (Bygd et al., 2021, 2022). All of the demonstrated defluorination reactions were shown to require toluene dioxygenase, a highly promiscuous enzyme (Gibson & Parales, 2000), and, in some cases, further co‐induced enzymes. There are numerous examples of oxygenases catalyzing defluorination reactions at sp2 and sp3 carbons, using oxidative and, in some cases reductive, mechanisms (Huang et al., 2021; Wang & Liu, 2020; Xie et al., 2020). For bacterial strains such as P. putida F1 to biodegrade PFCs under environmental conditions, the required enzymes must be induced beyond their very low basal level (Finette & Gibson, 1988; Gibson et al., 1968), requiring recognition of unnatural fluorinated compounds by the regulatory system.
The regulation and expression of toluene metabolizing enzymes such as the toluene dioxygenase (TDO), and the entire tod operon, has been well‐studied (Busch et al., 2007, 2009). Expression of the TOD pathway is controlled via TodS/T, a two‐component regulatory system (Figure 1A). In the presence of glucose, expression of TodS/T is under catabolite repression resulting in low expression of the toluene dioxygenase (Busch et al., 2010; Finette & Gibson, 1988). In the absence of glucose, the sensor TodS binds toluene and similar small molecules to signal via the cognate response regulator, TodT, the induction of the TOD pathway that begins with the TDO reaction (Busch et al., 2007, 2010). Several studies have identified multiple aromatic hydrocarbon inducers of TDO expression (Busch et al., 2007; Cho et al., 2000; Lacal et al., 2006; Silva‐Jiménez et al., 2012). Using a β‐galactosidase reporter construct, the induction of TDO by a series of haloalkylbenzenes including bromo‐, chloro‐ and fluorotoluene isomers was determined. Fluorinated compounds previously known to induce TDO include fluorobenzene and all three isomers of fluorotoluene (Busch et al., 2010; Lacal et al., 2006).
FIGURE 1.
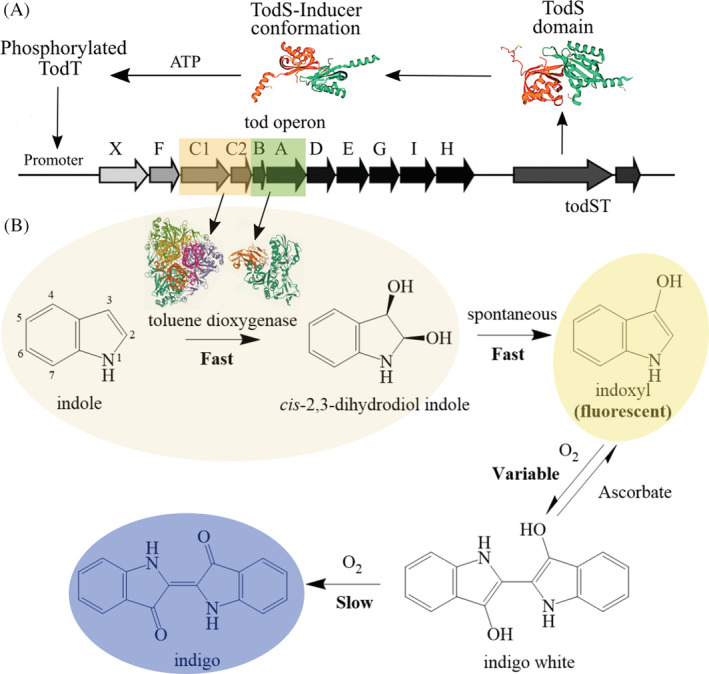
Schematic of toluene dioxygenase (TDO) regulation and oxidation of indole to indoxyl and indigo. (A) The tod operon is controlled by todST gene products in which ligands are bound by the TodS with inducers causing a conformational change and phosphorylation of TodT that can bind to the promoter. (B) The operon induction expresses toluene dioxygenase that is known to dioxygenate indole compounds, followed by spontaneous dehydration to produce fluorescent indoxyl and further spontaneous oxidation produces indigo white and then indigo. Indole dihydrodiol and indoxyl formation occur on the time scale of seconds, subsequent reactions on the time scale of minutes and hours. Ascorbate helps maintain indoxyl that is fluorescent.
The β‐galactosidase reporter previously used to assay TDO was informational but did not use the native strain directly as it required construction of a strain with the promoter (P todX ) located upstream of a reporter gene. Measuring oxygen consumption uses the native strain (Cho et al., 2000) but requires specialized equipment and has single sample throughput. Searching for a higher‐throughput assay that directly measures the activity of TDO in vivo, we were attracted to the possibility of a rapid assay for TDO activity via measuring fluorescence of an indoxyl intermediate, which forms spontaneously upon TDO oxidation of various indole compounds (Figure 1B) (Ensley et al., 1983; Woo et al., 2000). Further non‐enzymatic oxidation and dimerization reactions are known to form indigo white and indigo (Guilbault & Kramer, 1965).
To more efficiently measure induction by PFCs, we developed a high‐throughput assay for toluene dioxygenase activity based on the enzyme oxidation of an indole substrate to a fluorescent species, indoxyl (Figure 1B). Many oxygenases have been shown to oxidize indole to form insoluble indigo (Ameria et al., 2015; Fabara & Fraaije, 2020; Gillam et al., 2000). While the insoluble product is difficult to quantify, the formation of the bright blue indigo dye has been useful in numerous molecular genetic and enzymological studies (Jenkins & Dalton, 1985; Kim et al., 2003; O'Connor & Hartmans, 1998). Here, we made the assay quantitative by measuring a novel fluorescent indoxyl and increasing the linear range of measurement by adding ascorbate. Using the new method as a high‐throughput assay, we determined the varying levels of TDO induction in P. putida F1 for a series of mono‐ and poly‐fluorinated aromatics. Further, it was shown that several fluorinated aromatics were recognized by both TodS, to induce activity, and undergo defluorination by the induced enzymes. Surprisingly, a non‐aromatic, highly fluorinated alkyl compound that meets the organization for economic co‐operation and development (OECD) criteria of a per‐ and polyfluoroalkyl substances (PFAS) compound showed significant ability to induce the tod operon.
EXPERIMENTAL PROCEDURES
Chemicals
Most chemicals were purchased from Sigma‐Aldrich and were of the highest purity available, others are as indicated below. l‐Arginine was from Acros Organics. 6‐chloroindole was from Alfa Aesar. (Difluoromethoxy)benzene and 1,2,3 trifluorobenzene were from TCI. Toluene was from Fisher Scientific, and m‐xylene was obtained from J.T. Baker Chemicals. All difluorobenzenes, 1,2,4‐trifluorobenzene, 1,3,5‐trifluorobenzene, 4‐trifluoromethoxyfluorobenzene and 4‐fluorobenzotrifluoride were from Oakwood Chemical Company. 2,2‐Difluoro‐1,3‐benzodioxole, 97% purity, was obtained from Matrix Scientific. Deuterochloroform, 99% purity, was from Cambridge Isotope Laboratories. 1,1,1,2‐Tetrafluoro‐2‐trifluoromethoxy‐4‐iodobutane was procured from Apollo Scientific. N,
Bacterial strains and cell growth
Pseudomonas putida F1 (Finette & Gibson, 1988; Gibson et al., 1968) was grown on LB plates or in liquid on mineral salts media with Hutner's metals (MSB, also known as MM) (Turner et al., 2007) with 0.2% arginine or with toluene or other substrates supplied in the headspace of the culture through a vapour bulb (illustrated in Figure S6). Eschericia coli pDTG 602 was maintained on Luria Broth (LB) ampicillin (100 μg/ml) plates and grown on MSB with 0.2% glucose and ampicillin (100 μg/ml). Expression of the todABC 1 C 2 and todD genes was induced with isopropyl beta‐D‐1‐thiogalactopyranoside (IPTG) added to a final concentration of 1 mM.
Characterization of product from 6‐fluoroindole from P. putida F1
A 1 L MSB medium in a 2 L baffled flask with toluene supplied in vapour from a side arm flask was inoculated at 0D600 = 0.15 with an overnight culture of P. putida F1 grown on MSB + 0.2% l‐arginine. The culture was grown with shaking at 200 rpm at 30°C. At an OD600 of 0.8, toluene vapour was removed for 10 min with culture shaking prior to adding 80 mg of 6‐fluoroindole. The culture was further incubated with shaking overnight. Most of the coloured compound pelleted with the cells when harvested. The cells and compound were resuspended in 1/10 volume of the spent media and extracted with 1/5 volume dichloromethane in a separatory funnel. After removing the pink organic layer from the emulsion layer, the emulsion was extracted a second time and the organic layers were combined before drying with anhydrous MgSO4 and evaporating to dryness under rotary vacuum. An estimated 10–20 mg of purple residue remained. The UV–Visible spectrum of the extract resuspended at approximately 1 mg/ml in chloroform was collected on Cary 3500 double‐beam spectrophotometer with a xenon flash lamp (Agilent, Santa Clara, CA). The cell extract was dissolved in deuterochloroform at approximately 1 mg/ml for NMR acquisitions on a Varian INOVA 400‐MHz NMR spectrometer. 19F‐NMR (400 MHz, CDCl3) −121.87 ppm (dt, J = 5.2 Hz and 9.8 Hz). 1H‐NMR (400 MHz, CDCl3) 6.9 ppm (dt, J = 2.3 Hz and 9.2 Hz), 7.15 ppm (apparent dd, J = 9.5 Hz), 7.55 (dd, J = 8.5 Hz and 5.3 Hz).
Toluene dioxygenase activity and induction assay via fluorescence determination
Unless otherwise noted, 200 μl of P. putida F1 cells previously induced with toluene, or other compounds to be tested, were incubated in a polystyrene 96‐well plate with 10 μl of 300 mM ascorbic acid. The reaction was started by adding 2 μl 100 mM 6‐fluoroindole dissolved in ethanol. Fluorescence was measured in a SpectraMax Gemini EM plate reader (Molecular Devices, San Jose, CA) with an excitation wavelength of 365 nm and emission was measured at 470 nm. The increase in fluorescence was found to be linear for 10 min, the slope was determined, and the value expressed as a percentage of the rate with toluene induction that was set at 100%. A positive control was always run with toluene induction to correct for slightly different metabolic state of cell batches used for determining percentage of induction. Negative controls were always run to determine basal level of toluene dioxygenase expression which was <5% the level of toluene‐induced levels.
For induction, P. putida F1 cells were grown on MSB 0.2% arginine to an OD600 of 1. Additional arginine was added to the culture at 0.2% before distributing 12–15 ml of the culture into 125 ml baffled flasks. This was done because some tested inducers may not be metabolized and will not provide carbon and energy and it was necessary to have a universal carbon and energy source to support new transcription and translation. Each chemical tested for induction was provided to a separate flask in a vapour bulb. Duplicate biological replicates were each assayed in triplicate, and the six readings were averaged.
Fluoride determination
Fluoride release was determined with a fluoride specific electrode (ISE Ionplus Sure‐Flow Orion fluoride probe from Thermo Fisher and the Orion Star A214 meter) after 5 h incubation. The supernatant resulting from pelleting cells at 15,000 rpm for 1 min in a microcentrifuge was combined 1:1 with total ionic strength adjustment buffer (TISAB) prior to reading fluoride (Bygd et al., 2022).
Metabolism of fluorinated compounds determined by fluoride determination
After determining that many fluorinated compounds are able to induce the Tod pathway, we asked whether prior induction with toluene was necessary for metabolism of the compounds leading to fluoride release. Pseudomonas putida F1 was grown overnight on MSB 0.2% arginine. The culture was divided into six baffled flasks. Each chemical was added to a flask in a vapour bulb. Toluene was added to one flask as a positive control for induction. The flasks were shaken at 28C. At 2 h, aliquots were taken to test induction using the fluorescent 6‐F indole assay. After 5 h, cell supernatants were tested for fluoride release with the fluoride specific electrode.
Determination of metabolites from 1,2,4‐trifluorobenzene via GC–MS and NMR
Gas‐chromatography mass spectrometry experiments were done as by Bygd et al. (2022), except that the GC oven temperature started at 40°C instead of 50°C. Eschericia coli pDTG 602 was grown on MSB with 0.2% glucose and ampicillin (100 μg/ml) at 37°C to early exponential phase. The culture was then induced with 1 mM IPTG for 1 h at 30°C. After induction, 1,2,4‐trifluorobenzene was added to the culture in vapour form and left to incubate at 30°C for 3 h. One mL aliquots were taken and pelleted at 14,000 rpm for 1 min. The supernatant was extracted with an equal volume of methyl‐t‐butyl ether (MTBE). To one sample, 1 μl of N,O‐bis(trimethylsilyl) trifluoroacetamide was added as a derivatizing agent. Product ion spectra were identified in positive ion mode on an HP6890 gas chromatograph with an HP5973 MS detector.
For NMR analysis, E. coli pDTG 602 was grown as written above in a 200 ml culture volume to early exponential phase. The culture was induced for 1 h with 1 mM IPTG at 30°C and then provided 1,2,4‐trifluorobenzene in vapour form for a 2‐h incubation. At this time, the culture was centrifuged at 9000 rpm for 10 min. The supernatant was acidified with a few drops of 6 N HCl and stirred. The acidified medium was extracted using an equal volume of methyl‐t‐butyl ether (MTBE) and dried with anhydrous magnesium sulfate. The dried extract was decanted into a flask and then placed in a 500 ml round bottom flask to be dried via rotary evaporation. Remaining water was removed under vacuum. The residue was dissolved in deuterated chloroform and NMR analysis was done using a Varian INOVA 400‐MHz NMR spectrometer. Trimethylsilane (TMS) and trichlorofluoromethane (F11) served as a reference for 1H‐NMR and 19F‐NMR respectively. The products extracted from incubation of E. coli pDTG 602 with 1,2,4‐trifluorobenzene were analysed: 1H‐NMR (400 MHz, CDCl3) 6.58 ppm (t, J = 7.4 Hz and 7.0 Hz), 6.54 ppm (dt, J = 13.9 Hz, 3.5 Hz, and 6.8 Hz). 19F‐NMR (400 MHz, CDCl3) −165.33 (ddd, J = 22.5 Hz, 10.5 Hz, and 6.9 Hz), −146.85 ppm (ddd, J = 21.8 Hz, 10.3 Hz, and 2.9 Hz), −142.35 ppm (t, J = 7.5 Hz), −141.37 ppm (td, J = 10.0 Hz and 3.4 Hz). Fluorine NMR showed evidence for a third minor product in too low of abundance to identify.
Transformation of 1,1,1,2‐tetrafluoro‐2‐trifluoromethoxy‐4‐iodobutane
Pseudomonas putida F1 was grown on MSB and 0.2% arginine to mid‐exponential phase. A small glass tube containing 1,1,1,2‐tetrafluoro‐2‐trifluoromethoxy‐4‐iodobutane (TTIB) and a cotton plug was placed into the vial suspended by a thin wire. The tube was sealed with a rubber septum and crimp‐sealed. The normal atmosphere was replaced with oxygen via an inserted needle and a vent needle. The exit needle was removed first to build up a slight positive pressure from excess oxygen. The vial was placed at 28C and incubated overnight. Trichlorofluoromethane (F11) was used to extract the contents of the sealed vial, injecting 6 ml into the 7 ml culture. Before removing the seal, the vial was gently mixed without tipping the tube and then the cap, seal, wire and glass tube containing TTIB were removed. The vial was then re‐capped and vortexed for thorough mixing. After settling, the aqueous layer was removed and placed in a separate tube. A disposable glass pipette was used to remove the organic layer, which was filtered through cotton into a clean glass tube. The sample was sealed and stored at 4°C prior to analysis.
At time of NMR analysis, the trichlorofluoromethane extract was placed in an NMR tube with ~20% (v/v) deuterated benzene for the purpose of locking on a deuterated solvent. 1H‐NMR and 19F‐NMR analyses were done using a Varian INOVA 400‐MHz NMR spectrometer. The products extracted from incubation of P. putida F1 with TTIB were analysed: 19F‐NMR (400 MHz, F11 and benzene D6) −68.0 ppm (d, J = 9.0 Hz), −72.1 ppm (m, J = 9.0 Hz), −78.5 ppm (m, J = 4.6 Hz and 3.5 Hz). The fluorine NMR identified one major product, which is described. There is evidence for additional minor products in too low of abundance to identify.
Molecular modelling methods
The modelling of the ligand 1,2,4 trifluorobenzene in TodS (PDB 5HWV) and TDO (PDB 3EN1) was done using Coot, v0.8.9.2 (Emsley et al., 2010). The ligand was positioned into the existing electron density of toluene in both TodS and TDO and the structures were refined with default restraints using the Refmac5 tool within the software, CCP4, version 7.0 (Winn et al., 2011). The refinement minimized the energy of the bonded and non‐bonded interactions of the modelled ligand and the side chains of the TodS and TDO proteins which resulted in the final models shown in Figures 5A and 7A. Space‐filling models and manipulations were derived using the Galaxy Visualizer tool from MolInspiration (Hadda et al., 2021).
FIGURE 5.
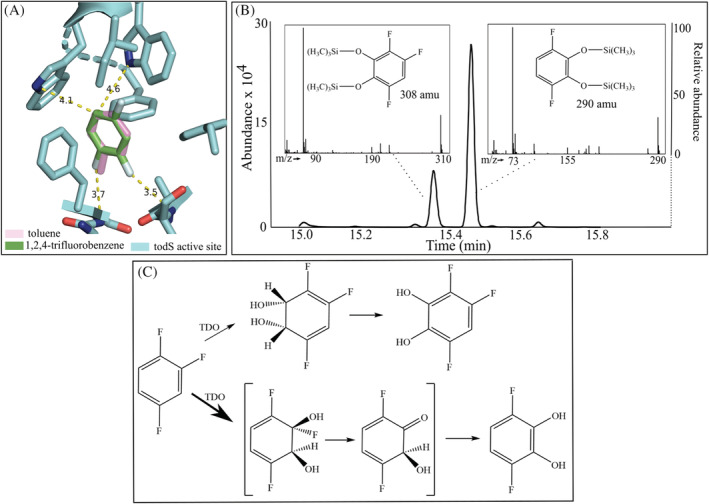
1,2,4‐Trifluorobenzene induction by binding to TodS and metabolism by toluene dioxygenase and diol dehydrogenase with formation of 3,4,6 trifluorobenzene diol and 3,6‐difluorobenzene diol. (A) Docking and energy minimization with 1,2,4‐benzene, shown overlapped with toluene, bound in TodS as described in Experimental Procedures. (B) GC separation of catechols doubly derivatized with trimethylsilane and mass spectra of the respective peaks. (C) Pathways leading to a trifluorocatechol (top) and difluorocatechol and hydrogen fluoride (bottom). The latter derives from gem‐elimination of a dihydrodiol.
FIGURE 7.

Substrates and position of dioxygenation by toluene dioxygenase with known reactions and as proposed for 3,4,4,4‐tetrafluoro‐3‐trifluoromethoxy‐butene, the elimination product of 1,1,1,2‐tetrafluoro‐2‐trifluoromethoxy‐4‐iodobutane (TTIB). (A) Modelling of toluene (purple) and 1,2,4‐trifluorobenzene (green) in the active site of toluene dioxygenase showing the direction of dioxygen attack consistent with known absolute stereochemistry of the toluene dihydrodiol and of the major fluorinated product as demonstrated here. The red sphere represents the active site iron atom that binds and activates dioxygen for attack from the top face as shown by the blue arrow. (B) Space‐filling model for toluene showing attack from the top face of the ring with the methyl group oriented down. (C) Space‐filling model showing proposed direction of dioxygen attack catalyzed by TDO with 3,4,4,4‐tetrafluoro‐3‐trifluoromethoxy‐butene. (D) Space‐filling model showing known position of attack on styrene by TDO. (E) Space‐filling model showing known position of attack on indene by toluene dioxygenase.
RESULTS
Development of a new in vivo assay for toluene dioxygenase
We initially tested activity for P. putida F1 TDO using indole in a microtiter plate assay. Indole (500 μM) was added to toluene‐grown P. putida F1 cells (200 μl) in MSB media and fluorescence was determined with an excitation wavelength of 365 nm and an emission wavelength of 470 nm. We observed an increase in fluorescence with a linear response for ~6 min. The fluorescent indoxyl intermediate is relatively short‐lived because it is oxidized and dimerized to indigo white (Figure 1B). However, the addition of ascorbic acid has been demonstrated to slow a decrease in the fluorescence signal, presumably by simultaneously keeping indoxyl in the reduced state and by lowering pH (Cotson & Holt, 1958; Guilbault & Kramer, 1965). We found the addition of 15 mM ascorbic acid increased the linear period and overall signal intensity (Figure S1).
Knowing that several indoles substituted on the six‐membered ring are substrates of TDO (Kim et al., 2003), we tested a series of 5‐, 6‐ and 7‐ substituted indoles to determine whether substituted indoles resulted in higher fluorescence signal compared to unsubstituted indole (Figure 2A). Substitution at the 6‐position with fluorine or chlorine both improved the assay signal. Using 6‐fluoroindole as the substrate for TDO instead of indole resulted in a three times higher fluorescence signal intensity (Figure 2A). TDO was shown to be responsible for the signal by observing no fluorescence with P. putida F106, a mutant that is specifically defective in TDO (Finette et al., 1984).
FIGURE 2.
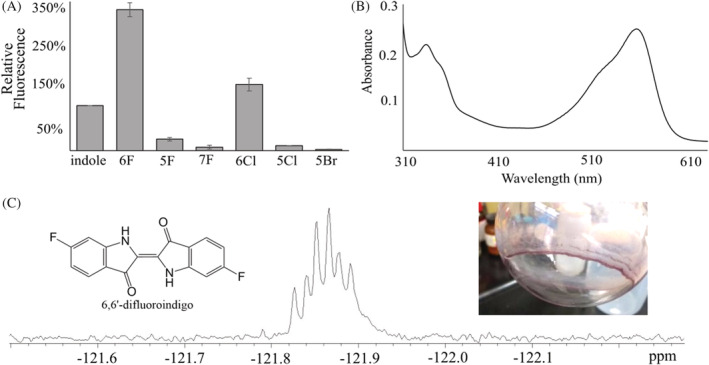
Fluorescence and stable structural products from the oxidation of 6‐fluoroindole by toluene dioxygenase. (A) Relative fluorescence for each substituted indole substrate of toluene dioxygenase compared to indole. (B) Characterization of the extracted product of toluene‐grown Pseudomonas putida F1 incubated with 6‐fluoroindole by UV–vis spectroscopy. (C) 19F‐NMR spectrum (at 400 MHz 1H) of extract dissolved in deuterochloroform (CDCl3) (dt, J 5 Hz, 10 Hz, F‐11 reference). The inset shows the material analysed by NMR and UV–vis after extraction from culture media.
There are several possible explanations for the increased signal with 6‐fluoroindole. TDO may oxidize 6‐fluoroindole faster than indole, 6‐fluoroindoxyl may be inherently more fluorescent than indoxyl, and/or the indoxyl may be more stable. However, indoxyl has never been purified and characterized due to its instability. Nonetheless, we found here the use of 1 mM 6‐fluoroindole in an aqueous solution containing 1% (v/v) ethanol to be a sensitive and reliable indicator for toluene dioxygenase activity (Figures S2 and S3).
Given difficulties in isolating the fluorescent intermediate 6‐fluoroindoxyl formed upon dehydration of the TDO dioxygenation product of 6‐fluoroindole, we sought to determine stable products, since indigo derivatives are stable and known to derive from indoxyl intermediates (Figure 2B,C). Whole cells of P. putida F1 grown on MSB with toluene vapour were incubated with 6‐fluoroindole overnight with shaking. The next‐day culture was purple, consistent with the formation of a substituted indigo. The purple compound was extracted and characterization was consistent with the structure being 6,6′‐difluoroindigo. The absorbance spectrum of the extracted material dissolved in chloroform was similar to the spectrum of indigo in dimethylformamide (Sousa et al., 2008) (Figure 2B), and the λmax was 570 nm, consistent with the λmax published by P. Sadler for 6,6′‐difluoroindigo dissolved in tetrachloroethane (Sadler, 1960). The 19F‐NMR of the extract in CDCl3 shows a single signal centered at −121.87 ppm. This indicated the presence of a fluorinated compound in which there is only one nuclear environment for fluorine (Figure 2C) consistent with fluorine in the symmetrical 6 and 6′‐positions of an indigo backbone. The 19F‐NMR spectrum of 6,6′‐difluoroindigo is not available for comparison; however, the chemical shift observed for the extracted material is similar to −121.83 ppm reported for 6‐fluoroindole in CDCl3 (Božilović et al., 2007). The 1H‐NMR spectrum of the extract is also consistent with the formation of 6,6′‐difluoroindigo (Tanoue et al., 2004) (Figure S4).
We also considered that a contribution to the fluorescence signal could also derive from an isomer of indoxyl. After cis‐2,3‐dihydroxyindoline formation, spontaneous dehydration could result in the formation of either indoxyl or oxindole. It is possible that 6‐fluoro substitution favours the formation of 6‐fluorooxindole over 6‐fluoroindoxyl. To determine whether there could be a contribution to fluorescence from the oxindole, we measured the fluorescence of the stable and commercially available compound, 6‐fluorooxindole. Only weak fluorescence (82 RFU) was detected for 10 mg/ml 6‐fluorooxindole in ethanol with an excitation of 350 nm and emission of 470 nm. Taken together, both the low fluorescence for 6‐fluorooxindole and the formation of 6,6‐difluoroindigo upon overnight incubation with TDO‐expressing cells is most consistent with the formation of a transient, highly fluorescent 6‐fluoroindoxyl species.
Application of the new assay to study induction in comparison to previous methods
With a robust TDO activity assay, we could sensitively determine induction levels of the native TDO in P. putida F1. First, reasoning that incubation of the cells with a weak inducer for a long time will eventually result in full induction of TDO activity, we tested the induction of TDO in P. putida F1 grown to medium and high optical densities for 1 and 2 h to determine a short incubation time with toluene that results in strong fluorescent signal (Figure S5). A 2 h incubation of cells grown to 0.9 OD600 with toluene vapour resulted in a strong and linear response in the TDO in vivo activity assay. Growth of cells to late exponential phase continuously exposed to toluene gave an overall higher level of induction. However, the shorter 2 h induction period gave a much better discrimination between known weak and strong inducers of toluene dioxygenase activity and was largely consistent with previous induction assays, as described below.
Next, we compared results of the induction of TDO using our fluorescent TDO activity to previously reported results. These experiments were conducted with the non‐fluorinated chemicals m‐ xylene, p‐xylene and styrene, and for fluorobenzene and fluorotoluenes (Figure 3). These chemicals have been reported to be moderate inducers of P todx using a β‐galactosidase reporter assay with 1 mM of each chemical added directly to the media (Busch et al., 2007; Lacal et al., 2006). We found general agreement between previous reports and the current method, the latter showing somewhat higher levels, perhaps due to measuring the native enzyme in the native strain (Figure 3).
FIGURE 3.

Comparison of toluene dioxygenase (TDO) induction using the 6‐fluoroindole activity assay from this article (light bars) to previously published results using a β‐galactosidase reporter (dark bars). The percent induction for each chemical was determined by normalizing to TDO activity with toluene induction that was set at 100%.
Testing of polyfluorinated benzene compounds
Monofluorinated benzene and toluenes were shown to induce toluene dioxygenase substantially, here and in previous studies. To follow up, it was of interest to determine the response of the TodS‐mediated induction system to polyfluorinated aromatic compounds. The compounds ranged from 2 fluorine substituents to 4 and with fluorine atoms substituted on methyl or methylene carbon atoms or with different isomeric displacement on the benzene ring (Figure 4). In all experiments, the extent of induction was compared to toluene. Controls were always run with no inducer to rule out induction by a contaminant and this negative control invariably showed minimal TDO activity.
FIGURE 4.
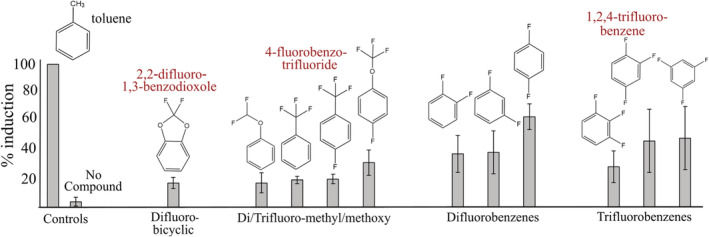
Induction of toluene dioxygenase activity with four different classes of polyfluorinated benzene compounds as a percent of induction observed with toluene using a protocol described in the Experimental Procedures section. The four compounds named have been subjected to metabolic studies, including toluene, the archetypal substrate for the tod operon. The three fluorinated compounds highlighted in red have been shown to undergo toluene dioxygenase‐dependent defluorination: 2,2‐difluoro‐1,3‐benzodioxole (Bygd et al., 2021); 4‐fluorobenzotrifluoride (Bygd et al., 2022); and 1,2,4‐trifluorobenzene (this study, in a subsequent section).
A previous study showed defluorination of the difluoro‐bicyclic compound, 2,2‐difluoro‐1,3‐benzodioxole (DFBD) (Bygd et al., 2021) after inducing P. putida F1 with toluene, because previous biotransformation experiments with bicyclic ring compounds had required pre‐induction (Kwit et al., 2008; Wackett et al., 1988). Indeed, previous direct induction experiments with multicyclic compounds 1‐naphthol, 1,2‐dihydronapthalene and anthracene had shown no induction (Busch et al., 2007). Here, we showed modest induction with DFBD at a level of 20% that observed with toluene (Figure 4). Testing of compounds with polyfluorinated methyl groups appended directly to benzene rings, or bridged by an oxygen in ether linkage, also gave modest induction, in the range of 20%–30%. Better induction was observed when fluorine substituents were directly bonded to the benzene ring, in the range of 35%–50% (Figure 4). This was comparable to induction by known toluene dioxygenase substrates such as styrene, m‐xylene and p‐xylene.
Fluorinated compounds mediating their induction and defluorination
In previous studies on toluene dioxygenase‐mediated defluorination, P. putida F1 cells were pre‐incubated with toluene before transformation of the fluorinated substrate (Bygd et al., 2021, 2022). Here, we investigated whether specific fluorinated compounds would induce the tod operon enzymes, undergo metabolic transformation and defluorination as indicated by fluoride release into the medium. The results here demonstrated that toluene pre‐induction is not required for fluoride release, induction does not guarantee fluoride release, and there are different mechanisms of defluorination (Table 1). 1,3,5‐Trifluorobenzene induces moderate TDO activity, but it does not release fluoride indicating that the dioxygenase pathway is not able to release fluoride from this compound. DFBD is known to undergo defluorination via a hydroxide‐mediated ring‐opening reaction (Bygd et al., 2021) and here, relatively low levels of TDO activity resulted in measurable fluoride release from DFBD. 3‐Fluorotoluene, which shows high levels of induction, has been previously shown to undergo 2,3‐dioxygenation and gem‐elimination of fluoride from a resultant fluorinated alcohol (Renganathan, 1989). A similar mechanism of defluorination is proposed here with 1,2,4‐trifluorobenzene and that was investigated in experiments described in the next section.
TABLE 1.
Fluorinated compounds with P. putida F1 inducing toluene dioxygenase activity, measured fluoride release, and mechanism of defluorination
| Compound | % Induction | F − (μM) | Defluorination mechanism | References |
|---|---|---|---|---|
| None (control) | <5 | 0 | None | ‐ |
| 1,3,5‐Trifluorobenzene | 40 | 0 | None | ‐ |
| 1,2,4‐Trifluorobenzene | 45 | 200 | 2,3‐dioxygenation and gem‐HF elimination | This paper |
| 2,2‐Difluoro‐1,3‐benzo‐dioxole | 20 | 200 | −OH ring opening | Bygd et al. (2021) |
| 3‐Fluorotoluene | 85 | 500 | 2,3‐Dioxygenation and gem‐HF elimination | Renganathan (1989) |
Note: Growth, induction and fluoride measurements were carried out as described in the Experimental Procedures section.
Molecular basis of induction and defluorination with 1,2,4‐trifluorobenzene
Here, we investigated how 1,2,4‐trifluorobenzene induced TDO activity, underwent defluorination and formed organic products. Tod operon induction is mediated through productive binding to TodS (Koh et al., 2016). In that context, computational binding of 1,2,4‐trifluorobenzene was compared to toluene, as described in the Experimental Procedures section (Figure 5A). Modelling suggested a very close superpositioning of the benzene rings of toluene and 1,2,4‐trifluorobenzene. Moreover, the 1‐fluorine substituent of 1,2,4‐trifluorobenzene positioned similarly to the methyl group of toluene. There is significant open volume to accommodate the 4‐fluorine substituent and this is consistent with experimental data that benzene rings with para‐substituents (i.e. para‐xylene) tend to be good inducers. The active site in the vicinity of a ring ortho‐position that accommodates the 2‐fluoro‐substituent packs closer to substrates. However, in the model (Figure 5A), the 2‐fluorine‐substituent, being not much larger than a hydrogen atom, can fit and may be stabilized by a weak polar interaction with a backbone amide group that is within 3.5 Å of the fluorine.
Metabolic studies were conducted and products extracted as described in the Experimental Procedures section. Initial studies were conducted with P. putida F1 and showed evidence for two distinct catechols with two and three fluorine atoms, respectively. To obtain more products, E. coli (pDTG602) was used. That strain expresses toluene dioxygenase and dihydrodiol dehydrogenase, recombinantly. As with P. putida F1, two catechols were observed and mass spectra were consistent with the structures assigned in the insets in Figure 5B. The proposed pathways leading to each catechol are shown (Figure 5C) and this is consistent with the appearance of fluoride in the medium following induction by, and metabolism of, 1,2,4‐trifluorobenzene. Direct 19F‐NMR of materials extracted from cell cultures was conducted and allowed us to discern the relative proportions of the dilfuorocatechol and trifluorocatechol shown in Figure 5C. Similar to the results observed by derivatization and GC separation (Figure 5B), the difluorocatechol was predominant. NMR coupling patterns were consistent with the specific catechols shown. Additionally, docking studies with toluene dioxygenase are consistent with the catechols shown, as discussed later in Results section. These data suggested that toluene dioxygenase preferentially oxidizes the face of the aromatic ring containing the 2‐fluoro‐substituent and is consistent with the observation of significant fluoride release (Table 1 and Figure 5C).
Non‐aromatic fluorinated inducers
Given the hint that fluorine substituents might show favourable binding interactions with TodS, we tested a series of non‐aromatic fluorinated compounds (Figure 6). Previously, no non‐aromatic compounds tested were shown to induce (Busch et al., 2007; Cho et al., 2000; Lacal et al., 2006; Silva‐Jiménez et al., 2012), although toluene dioxygenase has been shown to oxidize numerous non‐aromatic compounds (Lange & Wackett, 1997; Li & Wackett, 1992). Most polyfluorinated aliphatic compounds failed to cause significant induction, but two fluorinated olefins and a perfluoroether, that resembles important commercial compounds such as GenX, hexafluoropropylene oxide dimer acid, induced toluene dioxygenase activity. Induction by 1,2‐dichloro‐3,3‐difluoro‐1‐propene is relatively poor but trichlorofluoroethylene gave significant levels of toluene dioxygenase induction. Interestingly, trichloroethylene had been previously reported to induce toluene metabolic enzymes in a different Pseudomonas strain (Shingleton et al., 1998). Most surprisingly, 1,1,1,2‐tetrafluoro‐2‐trifluoromethoxy‐4‐iodobutane, a compound on the OECD PFAS registry list (Wang et al., 2021), showed higher levels of induction than benzene compounds considered to be good inducers of toluene dioxygenase activity, such as p‐xylene (Figures 3 and 6). This was surprising given that 1,1,1,2‐tetrafluoro‐2‐trifluoro‐methoxy‐4‐iodobutane has only sp3 carbon centers and contains seven fluorine substituents. Previously, only benzenoid aromatic compounds had been shown to induce the tod operon, and 24 aromatic compounds were shown to be non‐inducers. It is presently unclear what role water solubility, vapour pressure, and cell penetration have on the level of induction observed. The types of fluorinated compounds examined here are similar to toluene in having low water solubility and a high vapour pressure.
FIGURE 6.

Induction of toluene dioxygenase activity by fluorinated aliphatic compounds. Induction controls were toluene, set at 100%, and no inducer that gave basal level of toluene dioxygenase. Compounds giving no significant induction were as follows: (I) heptafluoro‐2‐iodopropane; (II) 1,2‐dichloro‐3,3,3‐trifluoropropene; (III) hexafluoroacetylacetone; (IV) perfluoropropyl iodide. The compounds highlighted in red showed significant levels of induction.
Metabolic studies with 1,1,1,2‐tetrafluoro‐2‐trifluoromethoxy‐4‐iodobutane
Subsequently, metabolic studies were conducted with 1,1,1,2‐tetrafluoro‐2‐trifluoromethoxy‐4‐iodobutane as described in the Methods and 1H‐NMR and 19F‐NMR revealed transformation of the substrate. We propose that the transformation proceeds via an HI elimination to generate the fluorinated olefin, 3,4,4,4‐tetrafluoro‐3‐trifluoromethoxybutene (TTMB) and that serves as both the inducer of the tod operon and the substrate for toluene dioxygenase. While the molecule is not intuitively perceived to be a likely toluene dioxygenase substrate in a traditional two‐dimensional stick representation, three‐dimensional space‐filling models of TTMB and known toluene dioxygenase (TDO) substrates better represent what the regulatory and metabolic proteins recognize (Figure 7). Figure 7A shows a model of the TDO active site with bound toluene and with 1,2,4‐trifluorobenzene, in which the methyl group of toluene and fluorine‐1 of the trifluorinated benzene superimpose. Moreover, the computational model is consistent with the known stereochemistry of TDO oxidation of toluene, styrene and indene in which the enzyme introduces dioxygen into the ring in a cis‐(1R,2S) configuration relative to a substituent in the 3‐position (Friemann et al., 2009; Wackett et al., 1988; Ziffer et al., 1977). Space‐filling models and known positions of TDO dioxygenation are shown for aromatic substrates and as proposed for TTMB based on experiments and substrate shape. Surprisingly, TTMB occupies molecular space not unlike the aromatic substrates shown in Figure 7. There is precedent for toluene dioxygenase reacting with aliphatic olefins. For example, dichlorinated butene compounds have previously been shown to undergo oxygenation by TDO (Lange & Wackett, 1997). In that study, TDO was also shown to dioxygenate non‐aromatic C5–C8 olefins. The much smaller surface area of fluorinated aliphatic compounds compared to chlorine and methyl substituents (Gavezzotti, 1985) is suggestive that other fluorinated aliphatic compounds might both induce and react with toluene dioxygenase.
DISCUSSION
PFCs are often highly stable, have unique properties, can form a fluorous phase distinct from organic solvents and water, and microbes have not been exposed to them until very recently in evolutionary time (Dobbs & Kimberley, 2002; O'Hagan & Rzepa, 1997; Wackett, 2021). As such, cellular macromolecules may bind and react with PFCs differently than they would with structurally analogous polychlorinated compounds or hydrocarbons, despite most commercial PFCs having significant hydrophobicity as defined by their octanol:water partitional coefficients (Han et al., 2021; Johnson et al., 2020; Olsen et al., 2003). The current study was conducted to examine microbial recognition of fluorinated compounds by using well‐studied proteins involved in aromatic hydrocarbon enzyme induction and metabolism by P. putida F1. In that system, enzyme induction is dependent on recognition by the regulatory sensor domain TodS and initial metabolic recognition is mediated by TodC1C2 (toluene dioxygenase). Both systems have been well‐studied previously by X‐ray crystallography and site‐directed mutagenesis to define mechanisms of ligand recognition and molecular function (Busch et al., 2007; Jiang et al., 1996; Koh et al., 2016; Lee et al., 2005; Wissner et al., 2021).
To best investigate the coupled induction by, and metabolism of, fluorinated compounds, we developed a new, sensitive in vivo assay for toluene dioxygenase. The assay is based on the fluorescence of unstable 3‐hydroxyindole (indoxyl) derivatives and 6‐fluoroindole was shown here to greatly enhance the assay signal, presumably by stabilizing the indoxyl intermediate. Indole oxidation to indoxyl has been used previously, but the signal decayed within 1–2 min, limiting the assay (Jenkins & Dalton, 1985). The greater fluorescent detection lifetime we achieved both increased sensitivity and permitted assays to be conducted with cell suspensions in microtiter well plates. We expect that this assay could be used for measuring enzyme activity in vitro, although that was beyond the scope of the present study. Many broad‐specificity oxygenases are reported to produce indoxyls from substituted indoles (Ameria et al., 2015; Ensley et al., 1983; Fabara & Fraaije, 2020; Gillam et al., 2000; O'Connor & Hartmans, 1998). Thus, the use of 6‐fluoroindole, that generates a strong and stable signal, may be broadly useful in that context.
Most studies on fluoro‐compound binding to proteins have been carried out with mammalian proteins; for example, in investigating the mode of action for new pharmaceuticals, many of which are fluorinated aromatic compounds (Olsen et al., 2003; Swallow, 2015; Xing et al., 2017). This study represents a much rarer investigation into molecular recognition of fluorinated compounds by bacterial proteins. For the regulatory element TodS, tightness of binding is not the only determinant, proper orientation also is important for the system to initiate induction of the Tod operon (Koh et al., 2016). Naturally occurring polyfluorinated compounds are unknown to science and so their recognition by the TodS sensor is likely not an intrinsically evolved property of the TodS binding site. Hence, the response of TodS and similar regulatory proteins to anthropogenic fluorinated compounds has remained largely undefined. Previous studies on tod operon induction by fluorinated compounds were confined to the monofluorinated fluorobenzene and fluorotoluenes (Busch et al., 2007, 2010; Lacal et al., 2006) (Table S1). The study here showed that the level of enzyme expression induced by polyfluorinated compounds such 1,2,4‐trifluorobenzene was comparable to or greater than petroleum hydrocarbons such as benzene, ethylbenzene, o‐xylene, m‐xylene, and p‐xylene, that comprise the characteristic BTEX compounds in liquid fossil fuels.
Since 1,2,4‐trifluorobenzene induced toluene dioxygenase and subsequently underwent defluorination, it was modelled in the ligand binding site of TodS, that mediates priming of the two‐component regulatory system. 1,2,4‐Trifluorobenzene was observed in the model to be co‐planar with toluene, with fluorine‐1 modelled in the position occupied by the methyl group of toluene. Fluorine‐4 was in a position between two tryptophans and a phenylalanine that has sufficient space to accommodate a methyl group or a larger halogen. It is known that para‐substituted toluenes with modest‐sized substituents are reasonable inducers of the tod operon (Busch et al., 2007, 2010; Lacal et al., 2006). However, ortho substituents are generally not tolerated; o‐xylene, o‐chlorotoluene and o‐bromotoluene are reported not to induce the operon (Table S1). Moreover, 1,2,4‐trimethylbenzene is not an inducer of the tod operon (Table S1) nor is it a substrate for toluene dioxygenase (Leahy et al., 2003). The tolerance to fluorine is likely linked to its low molecular free surface area relative to the other substituents, reported to be 12.1 squared Angstrom compared to 29.4, 33.4, and 37.1 for a chlorine, methyl group, and bromide, respectively (Gavezzotti, 1985). Moreover, the fluorine is modelled to be 3.5 Å distant to a carbon and nitrogen comprising a backbone amide. There are many examples of fluorobenzene drugs in tight‐binding interactions with proteins showing comparable interactions with protein backbone amide functionalities. In fluorine‐protein studies, this has been dubbed ‘multipolar interactions’, observed in many x‐ray structures of mammalian proteins with a ligand C—F dipole within 3.6 Å to backbone carbonyl carbons, amide NHs, α‐carbons hydrogens and guanidinium groups (Han et al., 2021; Swallow, 2015; Xing et al., 2017). The stabilization energies imparted are relatively small, 0.3–2.0 kcal/mol, but nonetheless are sufficient to increase binding affinity of drugs to targeted proteins. While binding to bacterial regulatory elements has been much less studied, the data presented here showed how a system evolved for aromatic hydrocarbons might be triggered to biodegrade anthropogenic PFCs.
The studies conducted here have clearly demonstrated that fluorinated compounds can mediate induction of, and metabolism by, enzymes evolved to handle natural petroleum hydrocarbons and this has relevance to the microbial world beginning to respond to new fluorinated drugs and agricultural chemicals. In the period of 1998–2020, greater than 50% of newly introduced agrichemicals are fluorinated, and most of those are aromatic compounds (Ogawa et al., 2020). Major blockbuster drugs are ingested by millions of people and much of the fluorinated benzene moieties are passed and enter wastewater treatment plants (Bhat et al., 2022; Britton et al., 2021; Murphy, 2016). For example, sitagliptin, a diabetes drug currently enjoying greater than $5.5 billion in sales, contains a 1,2,4‐trifluorobenzene moiety (Han et al., 2021). In the study presented here, 1,2,4‐trifluorobenzene was shown to induce TodC1C2 activity and undergo metabolic transformation with release of fluoride anion. Entering sitagliptin into the Eawag‐PPS system (Gao et al., 2011) to predict metabolites based on similar known reactions produced a trifluorobenzene catechol, one of the metabolites demonstrated in the present study. However, defluorination was not predicted. The knowledge generated here, and in other studies (Renganathan, 1989) on oxidation and gem‐elimination pathways begins to develop predicted frameworks for the environment fate of hundreds of fluorinated commercial chemicals.
Perhaps the most surprising element of the current study was the observation that fluorinated aliphatic compounds can induce enzymes that principally act on aromatic hydrocarbons. Trichlorofluoroethylene, which induced low toluene dioxygenase activity, resembles trichloroethylene, a compound that is reported to strongly and partially induce the toluene metabolizing enzymes in P. putida TAV8 (Shingleton et al., 1998) and P. putida F1, respectively. Interestingly, trichloroethylene is a substrate for toluene dioxygenase (Li & Wackett, 1992). Most surprisingly, 1,1,1,2‐tetrafluoro‐2‐trifluoromethoxy‐4‐iodobutane (TTIB) was observed to induce higher levels of toluene dioxygenase than most aromatic compounds. TTIB is listed in the OECD PFAS classification, indicated on PubChem (Kim et al., 2021). The structure of TTIB is reminiscent of the perfluorinated ether moiety of the widespread industrial chemical known as GenX. Moreover, evidence of metabolic transformation of TTIB observed here demonstrated that broad‐specificity recognition and metabolic systems provide recognition frameworks that can evolve naturally, or by laboratory engineering, into systems capable of handling polyfluorinated environmental contaminants.
AUTHOR CONTRIBUTIONS
Kelly G. Aukema and Madison D. Bygd carried out most microbiological experiments, conceived of key components of the research. Lambros J. Tassoulas contributed modelling and enzyme structure analyses. Jack E. Richman contributed to chemical analyses, analytical methods and data analysis. Lawrence P. Wackett conceived of key experiments, coordinated the research and obtained funding. All authors contributed to data analysis and writing the manuscript.
CONFLICT OF INTEREST
The authors declare that there are no competing interests associated with this manuscript.
Supporting information
Appendix S1 Supplementary Information
ACKNOWLEDGEMENTS
The authors thank Kristin Boardman and Wen Cai for assistance with initial experiments, and Tom Niehaus for the use of the Cary spectrophotometer. The authors thank Rebecca Parales for providing the recombinant E. coli pDTG602a strain. The authors acknowledge the support of MnDRIVE Industry and the Environment (to Madison D. Bygd and Jack E. Richman). The authors acknowledge support of National Institutes of Health Biotechnology training grant (5T32GM008347–27) and the Informatics Institute of the University of Minnesota for support (to Lambros J. Tassoulas).
Aukema, K.G. , Bygd, M.D. , Tassoulas, L.J. , Richman, J.E. & Wackett, L.P. (2022) Fluoro‐recognition: New in vivo fluorescent assay for toluene dioxygenase probing induction by and metabolism of polyfluorinated compounds. Environmental Microbiology, 24(11), 5202–5216. Available from: 10.1111/1462-2920.16187
Funding information National Institute of General Medical Sciences, Grant/Award Number: 5T32GM008347‐27; University of Minnesota
DATA AVAILABILITY STATEMENT
Data will be made available upon request.
REFERENCES
- Ameria, S.P.L. , Jung, H.S. , Kim, H.S. , Han, S.S. , Kim, H.S. & Lee, J.H. (2015) Characterization of a flavin‐containing monooxygenase from Corynebacterium glutamicum and its application to production of indigo and indirubin. Biotechnology Letters, 37, 1637–1644. [DOI] [PubMed] [Google Scholar]
- Bhat, A.P. , Mundhenke, T.F. , Whiting, Q.T. , Peterson, A.A. , Pomerantz, W.C. & Arnold, W.A. (2022) Tracking fluorine during aqueous photolysis and advanced UV treatment of fluorinated phenols and pharmaceuticals using a combined 19F‐NMR, chromatography, and mass spectrometry approach. ACS Environmental Au, 2, 242–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Božilović, J. , Bats, J.W. & Engels, J.W. (2007) Synthesis and structure of fluoroindole nucleosides. Canadian Journal of Chemistry, 85, 283–292. [Google Scholar]
- Britton, R. , Gouverneur, V. , Lin, J.H. , Meanwell, M. , Ni, C. , Pupo, G. et al. (2021) Contemporary synthetic strategies in organofluorine chemistry. Nature Reviews Methods Primers, 1, 1–22. [Google Scholar]
- Busch, A. , Guazzaroni, M.E. , Lacal, J. , Ramos, J.L. & Krell, T. (2009) The sensor kinase TodS operates by a multiple step phosphorelay mechanism involving two autokinase domains. The Journal of Biological Chemistry, 284, 10353–10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch, A. , Lacal, J. , Martos, A. , Ramos, J.L. & Krell, T. (2007) Bacterial sensor kinase TodS interacts with agonistic and antagonistic signals. Proceedings of the National Academy of Sciences of the United States of America, 104, 13774–13779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch, A. , Lacal, J. , Silva‐Jímenez, H. , Krell, T. & Ramos, J.L. (2010) Catabolite repression of the TodS/TodT two‐component system and effector‐dependent transphosphorylation of TodT as the basis for toluene dioxygenase catabolic pathway control. Journal of Bacteriology, 192, 4246–4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bygd, M.D. , Aukema, K.G. , Richman, J.E. & Wackett, L.P. (2021) Unexpected mechanism of 2,2‐difluoro‐1,3‐benzodioxole defluorination by Pseudomonas putida F1. mBio, 12, e3001–e3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bygd, M.D. , Aukema, K.G. , Richman, J.E. & Wackett, L.P. (2022) Microwell fluoride screen for chemical, enzymatic and cellular reactions reveals latent microbial defluorination capacity for –CF3 groups. Applied and Environmental Microbiology, 88, e0028822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron, S. (2020) Where does the fluorine come from? A review on the challenges associated with the synthesis of organofluorine compounds. Organic Process Research & Development, 24, 470–480. [Google Scholar]
- Cho, M.C. , Kang, D.O. , Yoon, B.D. & Lee, K. (2000) Toluene degradation pathway from Pseudomonas putida F1: substrate specificity and gene induction by 1‐substituted benzenes. Journal of Industrial Microbiology & Biotechnology, 25, 163–170. [Google Scholar]
- Cotson, S. & Holt, S.J. (1958) Studies in enzyme cytochemistry. IV. Kinetics of aerial oxidation of indoxyl and some of its halogen derivatives. Proceedings of the Royal Society of London. Series B, 148, 506–519. [DOI] [PubMed] [Google Scholar]
- Cros, A. , Alfaro‐Espinoza, G. , De Maria, A. , Wirth, N.T. & Nikel, P.I. (2022) Synthetic metabolism for biohalogenation. Current Opinion in Biotechnology, 74, 180–193. [DOI] [PubMed] [Google Scholar]
- Dobbs, A.P. & Kimberley, M.R. (2002) Fluorous phase chemistry: a new industrial technology. Journal of Fluorine Chemistry, 118, 3–17. [Google Scholar]
- Emsley, P. , Lohkamp, B. , Scott, W.G. & Cowtan, K. (2010) Features and development of coot. Acta Crystallographica. Section D, Biological Crystallography, 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensley, B.D. , Ratzkin, B.J. , Osslund, T.D. , Simon, M.J. , Wackett, L.P. & Gibson, D.T. (1983) Expression of naphthalene oxidation genes in Escherichia coli results in the biosynthesis of indigo. Science, 222, 167–169. [DOI] [PubMed] [Google Scholar]
- Fabara, A.N. & Fraaije, M.W. (2020) An overview of microbial indigo‐forming enzymes. Applied Microbiology and Biotechnology, 104, 925–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finette, B.A. & Gibson, D.T. (1988) Initial studies on the regulation of toluene degradation by Pseudomonas putida F1. Biocatalysis, 2, 29–37. [Google Scholar]
- Finette, B.A. , Subramanian, V. & Gibson, D.T. (1984) Isolation and characterization of Pseudomonas putida PpF1 mutants defective in the toluene dioxygenase enzyme system. Journal of Bacteriology, 160, 1003–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friemann, R. , Lee, K. , Brown, E.N. , Gibson, D.T. , Eklund, H. & Ramaswamy, S. (2009) Structures of the multicomponent Rieske non‐heme iron toluene 2,3‐dioxygenase enzyme system. Acta Crystallographica Section D: Biol Crystallography, 65, 24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, J. , Ellis, L.B.M. & Wackett, L.P. (2011) The University of Minnesota Pathway prediction system: multi‐level prediction and visualization. Nucleic Acids Research, W406–W411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavezzotti, A. (1985) Molecular free surface: a novel method of calculations and its uses in conformational studies and in organic crystal chemistry. Journal of the American Chemical Society, 107, 962–967. [Google Scholar]
- Gibson, D.T. , Koch, J.R. & Kallio, R.E. (1968) Oxidative degradation of aromatic hydrocarbons by microorganisms. I. Enzymic formation of catechol from benzene. Biochemistry, 7, 2653–2662. [DOI] [PubMed] [Google Scholar]
- Gibson, D.T. & Parales, R.E. (2000) Aromatic hydrocarbon dioxygenases in environmental biotechnology. Current Opinion in Biotechnology, 11, 236–243. [DOI] [PubMed] [Google Scholar]
- Gillam, E.M. , Notley, L.M. , Cai, H. , De Voss, J.J. & Guengerich, F.P. (2000) Oxidation of indole by cytochrome P450 enzymes. Biochemistry, 39, 13817–13824. [DOI] [PubMed] [Google Scholar]
- Guilbault, G.G. & Kramer, D.N. (1965) Resorufin butyrate and indoxyl acetate as fluorogenic substrates for cholinesterase. Analytical Chemistry, 37, 120–123. [DOI] [PubMed] [Google Scholar]
- Hadda, T.B. , Rastija, V. , AlMalki, F. , Titi, A. , Touzani, R. , Mabkhot, Y.N. et al. (2021) Petra/Osiris/Molinspiration and molecular docking analyses of 3‐hydroxy‐indolin‐2‐one derivatives as potential antiviral agents. Current Computer‐Aided Drug Design, 17, 123–133. [DOI] [PubMed] [Google Scholar]
- Han, J. , Kiss, L. , Mei, H. , Remete, A.M. , Ponikvar‐Svet, M. , Sedgwick, D.M. et al. (2021) Chemical aspects of human and environmental overload with fluorine. Chemical Reviews, 121, 4678–4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Q. , Zhang, X. , Chen, Q. , Tian, S. , Tong, W. , Zhang, W. et al. (2021) Discovery of a P450‐catalyzed oxidative defluorination mechanism toward chiral organofluorines: uncovering a hidden pathway. ACS Catalysis, 12, 265–272. [Google Scholar]
- Jenkins, R.O. & Dalton, H. (1985) The use of indole as a spectrophotometric assay substrate for toluene dioxygenase. FEMS Microbiology Letters, 30, 227–231. [Google Scholar]
- Jiang, H. , Parales, R.E. , Lynch, N.A. & Gibson, D.T. (1996) Site‐directed mutagenesis of conserved amino acids in the alpha subunit of toluene dioxygenase: potential mononuclear non‐heme iron coordination sites. Journal of Bacteriology, 178, 3133–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, B.M. , Shu, Y.Z. , Zhuo, X. & Meanwell, N.A. (2020) Metabolic and pharmaceutical aspects of fluorinated compounds. Journal of Medicinal Chemistry, 63, 6315–6386. [DOI] [PubMed] [Google Scholar]
- Kim, J.Y. , Lee, K. , Kim, Y. & Kim, C.K. (2003) Production of dyestuffs from indole derivatives by naphthalene dioxygenase and toluene dioxygenase. Letters in Applied Microbiology, 36, 343–348. [DOI] [PubMed] [Google Scholar]
- Kim, S. , Chen, J. , Cheng, T. , Gindulyte, A. , He, J. , He, S. et al. (2021) PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Research, 49, D1388–D1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh, S. , Hwang, J. , Guchhait, K. , Lee, E.G. , Kim, S.Y. , Kim, S. et al. (2016) Molecular insights into toluene sensing in the TodS/TodT signal transduction system. The Journal of Biological Chemistry, 291, 8575–8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara, T. , Yamauchi, T. , Ichiyama, S. , Takahata, H. & Esaki, N. (2003) Purification, characterization, and gene cloning of a novel fluoroacetate dehalogenase from Burkholderia sp. FA1. Journal of Molecular Catalysis B: Enzymatic, 23, 347–355. [Google Scholar]
- Kwit, M. , Gawronski, J. , Boyd, D.R. , Sharma, N.D. , Kaik, M. , More O'Ferrall, R.A. et al. (2008) Toluene dioxygenase‐catalyzed synthesis of cis‐dihydrodiol metabolites from 2‐substituted naphthalene substrates: assignments of absolute configurations and conformations from circular dichroism and optical rotation measurements. Chemistry–A European Journal, 14, 11500–11511. [DOI] [PubMed] [Google Scholar]
- Lacal, J. , Busch, A. , Guazzaroni, M.E. , Krell, T. & Ramos, J.L. (2006) The TodS–TodT two‐component regulatory system recognizes a wide range of effectors and works with DNA‐bending proteins. Proceedings of the National Academy of Sciences of the United States of America, 103, 8191–8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange, C.C. & Wackett, L.P. (1997) Oxidation of aliphatic olefins by toluene dioxygenase: enzyme rates and product identification. Journal of Bacteriology, 179, 3858–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leahy, J.G. , Tracy, K.D. & Eley, M.H. (2003) Degradation of mixtures of aromatic and chloroaliphatic hydrocarbons by aromatic hydrocarbon‐degrading bacteria. FEMS Microbiology Ecology, 43, 271–276. [DOI] [PubMed] [Google Scholar]
- Lee, K. , Friemann, R. , Parales, J.V. , Gibson, D.T. & Ramaswamy, S. (2005) Purification, crystallization and preliminary X‐ray diffraction studies of the three components of the toluene 2, 3‐dioxygenase enzyme system. Acta Crystallographica Section F, Structural Biology and Crystallization Communications, 61, 669–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. & Wackett, L.P. (1992) Trichloroethylene oxidation by toluene dioxygenase. Biochemical and Biophysical Research Communications, 58, 2820–2826. [DOI] [PubMed] [Google Scholar]
- Murphy, C.D. (2016) Microbial degradation of fluorinated drugs: biochemical pathways, impacts on the environment and potential applications. Applied Microbiology and Biotechnology, 100, 2617–2627. [DOI] [PubMed] [Google Scholar]
- Murphy, C.D. , Schaffrath, C. & O'Hagan, D. (2003) Fluorinated natural products: the biosynthesis of fluoroacetate and 4‐fluorothreonine in Streptomyces cattleya . Chemosphere, 52, 455–461. [DOI] [PubMed] [Google Scholar]
- O'Connor, K.E. & Hartmans, S. (1998) Indigo formation by aromatic hydrocarbon‐degrading bacteria. Biotechnology Letters, 20, 219–223. [Google Scholar]
- Ogawa, Y. , Tokunaga, E. , Kobayashi, O. , Hirai, K. & Shibata, N. (2020) Current contributions of organofluorine compounds to the agrochemical industry. iScience, 23, 101467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan, D. & Rzepa, H.S. (1997) Some influences of fluorine in bioorganic chemistry. Chemical Communications, 7, 645–652. [Google Scholar]
- Olsen, J.A. , Banner, D.W. , Seiler, P. , Obst Sander, U. , D'Arcy, A. , Stihle, M. et al. (2003) A fluorine scan of thrombin inhibitors to map the fluorophilicity/fluorophobicity of an enzyme active site: evidence for C‐F⋯C=O interactions. Angewandte Chemie (International Ed. in English), 42, 2507–2511. [DOI] [PubMed] [Google Scholar]
- Renganathan, V. (1989) Possible involvement of toluene‐2,3‐dioxygenase in defluorination of 3‐fluoro‐substituted benzenes by toluene‐degrading pseudomonas sp. strain T‐12. Applied and Environmental Microbiology, 55, 330–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler, P.W. (1960) Ultra‐violet and infra‐red spectra of substituted isoindigos and indirubins. Spectrochimica Acta, 16, 1094–1099. [Google Scholar]
- Shingleton, J.T. , Applegate, B.M. , Nagel, A.C. , Bienkowski, P.R. & Sayler, G.S. (1998) Induction of the tod operon by trichloroethylene in Pseudomonas putida TVA8. Applied and Environmental Microbiology, 64, 5049–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva‐Jiménez, H. , García‐Fontana, C. , Cadirci, B.H. , Ramos‐González, M.I. , Ramos, J.L. & Krell, T. (2012) Study of the TmoS/TmoT two‐component system: towards the functional characterization of the family of TodS/TodT like systems. Microbial Biotechnology, 5, 489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa, M.M. , Miguel, C. , Rodrigues, I. , Parola, A.J. , Pina, F. , Seixas de Melo, J.S. et al. (2008) A photochemical study on the blue dye indigo: from solution to ancient Andean textiles. Photochemical & Photobiological Sciences, 7, 1353–1359. [DOI] [PubMed] [Google Scholar]
- Swallow, S. (2015) Fluorine in medicinal chemistry. In: Progress in medicinal chemistry, Vol. 54. Amsterdam: Elsevier BV. [DOI] [PubMed] [Google Scholar]
- Tanoue, Y. , Sakata, K. , Hashimoto, M. , Hamada, M. , Kai, N. & Nagai, T. (2004) A facile synthesis of 6,6′‐ and 5,5′‐dihalogenoindigos. Dyes and Pigments, 62, 101–105. [Google Scholar]
- Turner, K. , Xu, S. , Pasini, P. , Deo, S. , Bachas, L. & Daunert, S. (2007) Hydroxylated polychlorinated biphenyl detection based on a genetically engineered bioluminescent whole‐cell sensing system. Analytical Chemistry, 79, 5740–5745. [DOI] [PubMed] [Google Scholar]
- Wackett, L.P. (2021) Why is the biodegradation of polyfluorinated compounds so rare? mSphere, 6, e0072121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wackett, L.P. (2022) Nothing lasts forever: understanding microbial biodegradation of polyfluorinated compounds, including PFAS. Microbial Biotechnology, 15, 773–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wackett, L.P. , Kwart, L.D. & Gibson, D.T. (1988) Benzylic monooxygenation catalyzed by toluene dioxygenase from Pseudomonas putida . Biochemistry, 27, 1360–1367. [DOI] [PubMed] [Google Scholar]
- Wang, Y. & Liu, A. (2020) Carbon–fluorine bond cleavage mediated by metalloenzymes. Chemical Society Reviews, 49, 4906–4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Buser, A.M. , Cousins, I.T. , Demattio, S. , Drost, W. , Johansson, O. et al. (2021) A new OECD definition for per‐and polyfluoroalkyl substances. Environmental Science & Technology, 55, 15575–15578. [DOI] [PubMed] [Google Scholar]
- Winn, M.D. , Ballard, C.C. , Cowtan, K.D. , Dodson, E.J. , Emsley, P. , Evans, P.R. et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallographica. Section D, Biological Crystallography, 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissner, J.L. , Schelle, J.T. , Escobedo‐Hinojosa, W. , Vogel, A. & Hauer, B. (2021) Semi‐rational engineering of toluene dioxygenase from Pseudomonas putida F1 towards oxyfunctionalization of bicyclic aromatics. Advanced Synthesis and Catalysis, 363, 4905–4914. [Google Scholar]
- Woo, H.‐J. , Sanseverino, J. , Cox, C.D. , Robinson, K.G. & Sayler, G.S. (2000) The measurement of toluene dioxygenase activity in biofilm culture of Pseudomonas putida F1. Journal of Microbiological Methods, 40, 181–191. [DOI] [PubMed] [Google Scholar]
- Xie, Y. , Chen, G. , May, A.L. , Yan, J. , Brown, L.P. , Powers, J.B. et al. (2020) Pseudomonas sp. strain 273 degrades fluorinated alkanes. Environmental Science & Technology, 54, 14994–15003. [DOI] [PubMed] [Google Scholar]
- Xing, L. , Keefer, C. & Brown, M.F. (2017) Fluorine multipolar interaction: toward elucidating its energetics in binding recognition. Journal of Fluorine Chemistry, 198, 47–53. [Google Scholar]
- Ziffer, H. , Kabuto, K. , Gibson, D.T. , Kobal, V.M. & Jerina, D.M. (1977) The absolute stereochemistry of several cis‐dihydrodiols microbially produced from substituted benzenes. Tetrahedron, 33, 2491–2496. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Supplementary Information
Data Availability Statement
Data will be made available upon request.


