Abstract
Cell-free systems have enabled the development of genetically encoded biosensors to detect a range of environmental and biological targets. Encapsulation of these systems in synthetic membranes to form artificial cells can reintroduce features of the cellular membrane, including molecular containment and selective permeability, to modulate cell-free sensing capabilities. Here, we demonstrate robust and tunable performance of a transcriptionally regulated, cell-free riboswitch encapsulated in lipid membranes, allowing the detection of fluoride, an environmentally important molecule. Sensor response can be tuned by varying membrane composition, and encapsulation protects from sensor degradation, facilitating detection in real-world samples. These sensors can detect fluoride using two types of genetically encoded outputs, enabling detection of fluoride at the Environmental Protection Agency maximum contaminant level of 0.2 millimolars. This work demonstrates the capacity of bilayer membranes to confer tunable permeability to encapsulated, genetically encoded sensors and establishes the feasibility of artificial cell platforms to detect environmentally relevant small molecules.
Encapsulation of a cell-free expression-based riboswitch allows for detection of fluoride in real-world samples.
INTRODUCTION
Cell-free systems have emerged as a powerful technology to detect a wide variety of molecular signals, including chemical contaminants relevant to the environment and human health (1–9) and markers of disease and infection (10–17). By reconstituting purified cellular machinery in vitro, these systems enable the use of natural cellular biochemical sensing mechanisms in a low-cost, distributable, and easily tunable platform. Despite these key advantages, removal from the cell also eliminates certain features of the cell’s native membrane barrier—such as reaction containment, protection from reaction inhibitors, and selective gating—all of which can add important functionality to cell-free biosensors (18).
Efforts to deploy sensors highlight these limitations caused by the absence of cellular membranes. For example, without a barrier between the sensor and the sample, detecting targets in complex matrices such as polluted water or biological samples requires additional modifications to the reaction or preparation protocols (6, 19, 20). This is because cell-free reactions are easily inhibited by changes in salt concentrations, the presence of detergent-like molecules, and bioactive enzymes such as deoxyribonuclease/ribonuclease (RNase) that either degrade or compete with components of the reaction. Cell-free sensors are also sensitive to dilution and therefore require a controlled reaction environment (21). One strategy to mitigate these limitations is to recapitulate some of the lost features of the cell membrane by encapsulating cell-free sensors inside of synthetic membranes. These structures, often referred to as artificial cells, leverage powerful cellular protein expression mechanisms inside a cell-like compartment to carry out dedicated sensing behaviors. In these systems, encapsulation in a semipermeable compartment allows users to tune the reaction environment on a molecular scale, enabling control over molecular access to the sensor while maintaining many of the advantageous features of cell-free systems (18).
There are two major considerations in designing artificial cell-based sensors: determining the impacts of a confined reaction environment on sensor function and choosing an appropriate target molecule and application. In terms of reaction confinement, the small scale of the encapsulated environment can affect reactant loading, reaction time, and limit of detection (22–24). These effects have been shown to affect the basic processes of gene expression (25), which, in turn, affects cell-free biosensors that regulate reporter gene expression at the level of transcription or translation (26). Of the wide range of genetic regulatory networks used for biosensing, RNA-based biosensors that regulate transcription (mRNA synthesis) require the fewest components and operate on a faster time scale (3, 27), which may reduce the impacts of confinement on sensor function relative to biosensors that regulate translation (protein synthesis). Riboswitches—noncoding RNA elements upstream of protein coding genes that change their structure in response to specific ligands to regulate gene expression—could offer an opportunity to address these constraints due to reaction confinement.
Previous proof-of-concept studies have focused on encapsulation of two synthetic, translationally regulated riboswitches that respond to membrane-permeable signals: theophylline (21, 28, 29) and histamine (30). Both riboswitches have been successfully encapsulated in bilayer vesicles, generating either a fluorescent protein readout or a protein-mediated response upon analyte entry into the vesicle interior (21, 28–30). Encapsulation of transcriptionally regulated riboswitches has yet to be demonstrated. Transcriptional riboswitches require dynamic conformational changes during transcription to enact their mechanism—a process that could be affected by general features of confinement or electrostatic interactions with the lipid bilayers (31, 32). Despite these potential challenges, the mechanisms underlying transcriptionally regulated riboswitches are being further uncovered (33). These sensors have demonstrated the feasibility of detecting environmentally important analytes in cell-free systems and can function with RNA-level outputs (3)—a key feature that may mitigate resource constraints—motivating further efforts for their encapsulation and deployment.
A second major consideration in encapsulated sensor development is the selection of an appropriate target and application. Of the many potential uses of encapsulated biosensors, water quality monitoring is one of the most compelling from a global perspective. One in three people globally lack access to safe drinking water (34), and the ability to identify contaminated water sources is essential for their quarantine or remediation (35). Fluoride is among the most concerning of these contaminants; chronic exposure to fluoride binds it to the calcium in teeth and bones, weakening them and causing lifelong health consequences (36). From both environmental and anthropogenic sources, fluoride exposure is especially problematic in parts of China, Africa, South America, and India (36, 37), with high fluoride concentrations also found in groundwater across the United States (37). This diversity of sample sources comes with a corresponding increase in potential reaction inhibitors, presenting the need for a robust sensor that retains function in complex matrices. Encapsulated fluoride biosensing reactions would address this need, delivering far-reaching global health benefits and establishing a framework to address future water quality challenges.
In this study, we sought to develop artificial cell-based sensors for fluoride by encapsulating a transcriptionally regulated, fluoride-responsive riboswitch within bilayer membranes (Fig. 1). We first encapsulate the riboswitch, then demonstrate its ability to detect externally added fluoride, and show that membrane composition can be modified to tune sensitivity to exogenous ions. We also demonstrate that encapsulation protects cell-free reactions from sample degradation, particularly from extravesicular degradative enzymes. Last, we couple riboswitch output to both fluorescent and colorimetric reporters and show that vesicle-based sensors can detect fluoride at environmentally relevant concentrations and in real-world water samples. This work demonstrates the potential of encapsulated, riboswitch-based sensors for biosensing applications, complimenting existing cell-free sensor engineering strategies and enabling sensing in otherwise inhospitable environments.
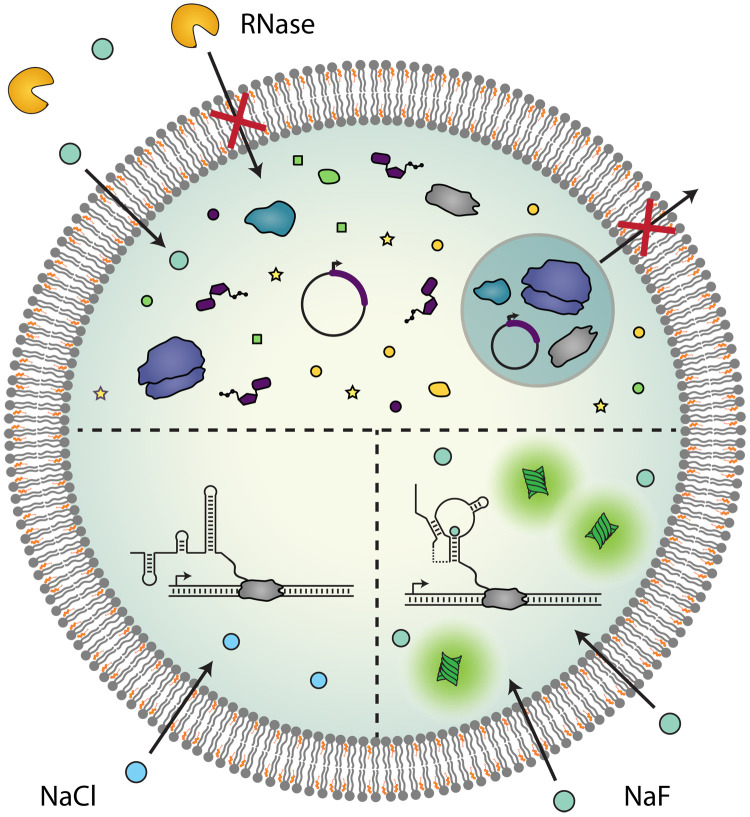
Fig. 1. Artificial cell sensors.
Encapsulation of cell-free systems in artificial cells creates a semipermeable barrier between sensor components and the environment, which modulates their molecular interactions. Reactants are contained within the vesicle interior, while proteins and other large molecules in the external sample are excluded from vesicle entry (top). Small, membrane-permeable molecules can diffuse into the vesicle interior, initiating a riboswitch-mediated response that is specific to an analyte of interest (bottom right). The riboswitch folds into a terminating conformation in the absence of sufficient concentrations of target analyte (bottom left).
RESULTS
A transcriptionally regulated fluoride riboswitch can function inside lipid vesicles
We first sought to confirm that a transcriptional riboswitch can function when encapsulated inside lipid vesicles. For the riboswitch, we chose the fluoride responsive riboswitch from Bacillus cereus, which we characterized previously (3). In earlier studies, we showed that this riboswitch can be used to control the expression of several different reporter proteins and fluorescent RNA aptamers in bulk Escherichia coli extract–based cell-free systems (3). In this system, the fluoride riboswitch is encoded within a single DNA template, downstream of a consensus E. coli promoter sequence, and upstream of a reporter coding sequence. In the absence of fluoride, E. coli polymerase transcribes the riboswitch sequence, causing it to fold into a conformation that exposes a transcriptional terminator hairpin and subsequently causes RNA polymerase to stop transcription (38). In the presence of fluoride, fluoride binding to the riboswitch aptamer domain prevents the terminator from folding, allowing transcriptional elongation of the reporter coding sequence.
In this study, we first chose to use a super folder green fluorescent protein (GFP) reporter, as it allows convenient measurement of riboswitch activity. For the cell-free system, we used an E. coli S30 lysate prepared with runoff and dialysis, which has been shown to allow the function of biosensors that require bacterial polymerases (39). Embedding the riboswitch DNA template into the extract system alongside varying concentrations of sodium fluoride (NaF) showed, as expected (3), an increase in GFP fluorescence as fluoride concentrations increased up to 3 mM, followed by a decrease in fluorescence at higher concentrations (Fig. 2A). This decrease is likely caused by fluoride inhibition of the gene expression machinery (40) and is consistent with our previous characterization of this construct (3). Accordingly, we used 3 mM NaF for the remainder of this study to obtain the expected maximum fluorescent output of the system.
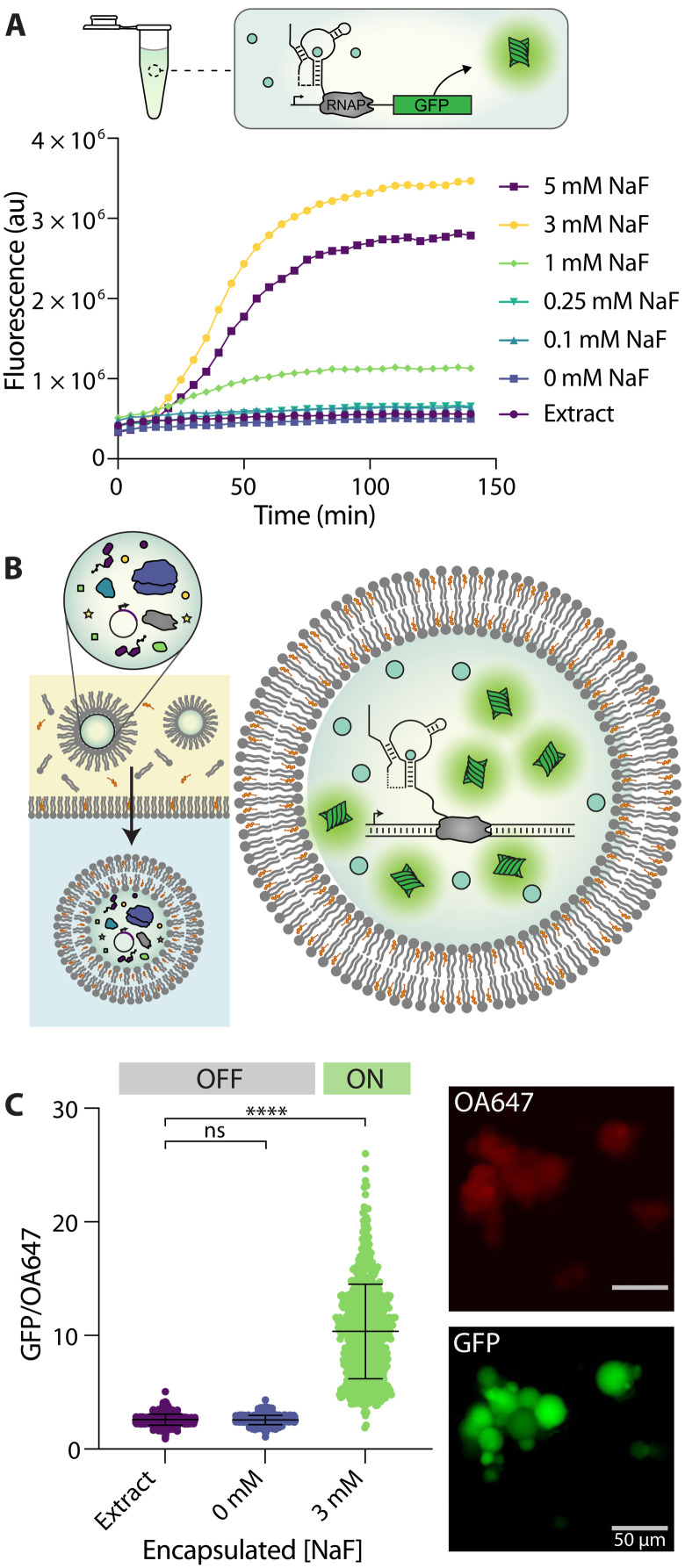
Fig. 2. A fluoride riboswitch can function within bilayer vesicles.
(A) Representative trace of riboswitch-regulated GFP expression under bulk conditions in response to increasing fluoride concentrations. In the presence of NaF, the riboswitch folds into an ON state, which allows the expression of a GFP reporter molecule. au, arbitrary units; RNAP, RNA polymerase. (B) Double emulsion assembly allows the encapsulation of functional cell-free reactions. Assembled reactions are vortexed into a lipid/oil mixture and then centrifuged into an aqueous solution (left). The resulting vesicles contain cell-free reactions, which can respond to coencapsulated fluoride by expressing GFP (right). (C) GFP/ovalbumin-conjugated Alexa Fluor 647 (OA647) fluorescence, which indicates GFP concentration relative to the OA647 volume marker inside each liposome. GFP/OA647 fluorescence increases inside of vesicles when 3 mM NaF is coencapsulated compared to no DNA (extract) or no fluoride (0 mM NaF) controls. Micrographs show variations in GFP fluorescence between vesicles from the same population, which results in a distribution of fluorescence values. Scale bars, 50 μm. Black lines indicate mean fluorescence and SD. ****P ≤ 0.0001; nonsignificant (ns), P > 0.05; P values are generated using a one-way analysis of variance (ANOVA) and Tukey’s multiple comparisons test.
We then set out to assess whether the fluoride riboswitch could retain functionality when encapsulated within lipid vesicles. Vesicles were synthesized using a water-in-oil emulsion transfer method (Fig. 2B) (41). In this method, various membrane amphiphiles (e.g., lipids, cholesterol, fatty acids, and diblock copolymers) are dissolved into an oil phase, and an emulsion is formed by vortexing the aqueous cell–free reaction into this mixture. The emulsion is then layered onto a second aqueous layer containing the small-molecule components of the cell-free reaction, and emulsified droplets are centrifuged through the oil-water interface to generate unilamellar vesicles. Previous characterization of vesicles formed using this method has yielded encapsulation efficiencies of ~98%, indicating high levels of entrapment of aqueous components (41). Vesicle synthesis using this approach yields a distribution of different vesicle sizes on the 5- to 50-μm scale, which could affect our quantification of fluorescence (42). To control for this, we also incorporated a protein-conjugated dye, ovalbumin-conjugated Alexa Fluor 647 (OA647), which served as a volume marker and allowed us to detect the vesicle interior regardless of GFP expression level (30, 43). After synthesis, vesicles were incubated under varying conditions at 37°C, and protein expression was assessed using epifluorescent microscopy after 6 hours of incubation (Fig. 2C and fig. S1) (30, 42). Vesicles were imaged using GFP and Cy5.5 channels, and images were analyzed using the NIS-Elements Advanced Research (AR) software program (44), which allowed us to automatically select vesicle interiors using the OA647 marker and report GFP fluorescence in those regions. This protocol allowed us to analyze hundreds of vesicles per sample, maintain the same selection parameters between samples, and minimize the impact of user selection bias in the analysis. In addition, the encapsulated volume marker allowed us to report GFP expression relative to OA647 fluorescence to control for possible variability in vesicle size or loading. Last, to control for variations in levels of extract autofluorescence between vesicle preparations, we analyzed all samples compared to an “extract-only” control population for each preparation. Using these methods, we were able to ensure that our measurements were isolated to intact (nonlysed) vesicles, which retained their protein cargo (fig. S2).
Using the above approach, we encapsulated cell-free reactions with and without fluoride present in the bulk reaction mixture. We chose to use a 2:1 ratio of cholesterol and 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC) phospholipid as membrane amphiphiles due to their previous use in similar encapsulated expression studies (21, 28–30, 43). Upon coencapsulation of the riboswitch with 3 mM NaF, we observed GFP expression inside vesicles, indicating the riboswitch was in the “ON” state (Fig. 2C). In contrast, in the absence of DNA (extract only) or in the absence of fluoride (0 mM NaF), we observed minimal GFP expression, indicating an “OFF” state (Fig. 2C). This high level of GFP induction inside vesicles by fluoride indicates that membrane encapsulation does not eliminate the ability of the riboswitch to fold properly and does not cause significant nonspecific expression.
We observed that populations of vesicles exhibited variations in GFP fluorescence between individual liposomes after 6 hours of incubation (Fig. 2C), a phenomenon that has been observed in similar studies across multiple encapsulation protocols (21, 24, 30, 42, 45–47). It has been hypothesized that these variations in gene expression may be caused by variability in vesicle loading and/or varied levels of molecular exchange with the surrounding buffer for vesicles of different sizes (24, 45, 46, 48). To report this variability across vesicle populations, we have included metrics of skew for each population result (tables S1 to S3). Even after taking this variability into account, however, induction of GFP expression is clearly observable across the vesicle population, indicating proper riboswitch sensor activity and a robust response to fluoride in encapsulated sensors.
External fluoride can be detected by an encapsulated riboswitch
We next sought to determine whether the encapsulated riboswitch could detect fluoride added to the external solution of preassembled sensor vesicles. To assess this, we prepared vesicles containing cell-free reactions without NaF present in the reaction mixture. We then titrated in NaF into the solution surrounding the vesicles (Fig. 3A) and imaged them following incubation for 6 hours at 37°C. We observed increasing GFP expression with increasing concentrations of NaF up to 3 mM and a slight decrease in average fluorescence at 5 mM, consistent with bulk studies (Fig. 3, B to D). Vesicle populations exhibited increases in both mean GFP/OA647 fluorescence and population skew in response to increasing fluoride, either of which could serve as a metric of fluoride detection (table S1). All fluoride-containing conditions exhibited a significant increase in fluorescence compared to no DNA and no fluoride controls (Fig. 3, B and C, and table S1). When incubated with chloride, a similarly monovalent anion, a slight response to increasing ion concentration was observed; however, these responses were significantly lower than any response to fluoride and did not exhibit any of the highly active vesicles that were observed under all fluoride-containing conditions (fig. S3). These responses were easily distinguishable between fluoride and chloride, indicating sufficient specificity to fluoride, as has been observed previously (3). Together, these results indicate that increasing concentrations of fluoride added to the extravesicular environment can be detected by the encapsulated riboswitch.
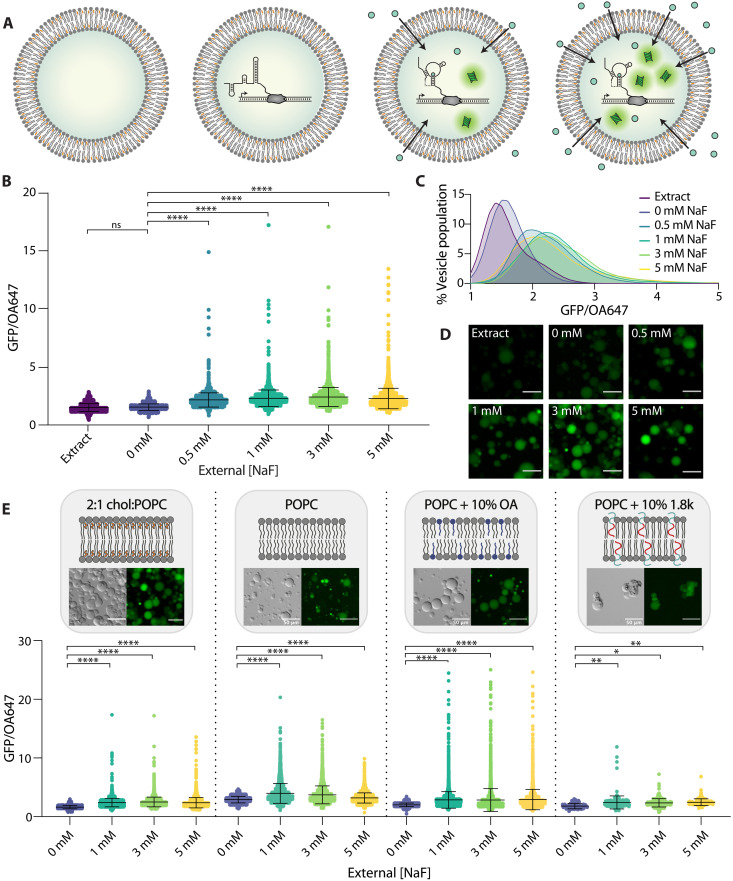
Fig. 3. Encapsulated sensors detect external fluoride.
(A) Schematic of conditions. Vesicles were prepared encapsulating extract only (left) or fully assembled reactions without NaF. Upon addition of increasing fluoride in the external solution, expression of GFP inside vesicles increases (right). (B) GFP/OA647 fluorescence as a result of riboswitch activity in 2:1 cholesterol:POPC vesicles in response to increasing NaF added externally. Black lines indicate mean fluorescence ratio and SD. (C) Histogram of vesicle populations shown in (B). Data are plotted with lowless curve fitting. (D) GFP fluorescence in micrographs of vesicles with increasing external concentrations of NaF. Scale bars, 50 μm. (E) GFP/OA647 fluorescence in response to increasing fluoride shown from left to right: 2:1 cholesterol:POPC membranes, pure POPC lipid membranes, POPC + 10% oleic acid (OA) membranes, and POPC + 10% 1.8k (molecular weight = 1.8 kDa) PEO-b-PBD membranes. Composition and morphology of each membrane composition are indicated by schematics and micrographs, respectively. ****P ≤ 0.0001; **P ≤ 0.01; *P ≤ 0.05; ns, P > 0.05; P values are generated using one-way ANOVA and Tukey’s multiple comparisons test.
This result was somewhat unexpected, as we anticipated that the membrane would be relatively impermeable to charged fluoride ions. The observed magnitude of fluoride permeability may be explained in part by the transient formation of hydrofluoric acid (HF). HF has been shown to exhibit a permeability coefficient that is seven orders of magnitude greater than fluoride anions through lipid/cholesterol bilayers, indicating that HF travels through the membrane much more readily than its anionic F− counterpart (40, 49). We confirmed this effect by encapsulating a pH sensitive dye, 8-hydroxypyrene-1,3,6-trisulfonic acid trisodium salt (HPTS), which reported a slight decrease in pH in the vesicle interior upon the addition of fluoride to the external buffer (fig. S4). This result indicates an increase in proton concentration inside the vesicle as fluoride concentration increases, consistent with cross-membrane transport of HF.
Because we observed that fluoride could pass through the membrane to interact with the encapsulated riboswitch, we wondered whether we could alter the composition of vesicle membranes to modulate membrane permeability and thereby modulate sensitivity of these sensors to external fluoride. Membrane permeability to small molecules depends significantly on membrane composition, as various lipid chain chemistries and contributions from other amphiphilic components can impart an effect on membrane physical properties. Cholesterol, a major component of our original 2:1 cholesterol:POPC lipid composition, is known to decrease membrane permeability by increasing lipid packing and altering membrane fluidity and rigidity (50). Poly(ethylene oxide)-b-poly(butadiene) (PEO-b-PBD) diblock copolymers are similarly known to reduce membrane permeability by increasing membrane viscosity, introducing steric barriers from the polyethylene glycol groups that assemble at the membrane interface, and increasing thickness and chain entanglements within the hydrophobic portions of the membrane (51, 52). In contrast, fatty acids such as oleic acid (OA) have been shown to increase membrane permeability to ionic solutes by incorporating single hydrocarbon chains that have a different shape and amphipathicity than diacyl chains, reducing lipid chain packing and enhancing fluidity of the bilayer (53, 54). Using this series of amphiphilic molecules, we set out to assess the capacity of membrane amphiphiles and the resulting membrane permeability to modulate the performance of an encapsulated cell-free sensor.
To explore the effect of these amphiphiles on membrane permeability to fluoride, we prepared vesicles with (i) pure POPC lipid, (ii) POPC lipid + 10% OA, or (iii) POPC lipid + 10% PEO14-b-PBD22 diblock copolymer [molecular weight = 1.8 kDa; hereafter referred to as 1.8k] components in the lipid/oil mixture, encapsulating cell-free reactions as normal (Fig. 3E). We observed an increase in overall GFP expression in both pure POPC lipid and POPC + 10% OA compositions compared to our original 2:1 cholesterol:POPC lipid composition (for 3 mM external fluoride; 2:1 cholesterol:POPC versus POPC, P < 0.0001; 2:1 cholesterol:POPC versus POPC + 10% OA, P < 0.0001). These results are consistent with the removal of cholesterol and the addition of OA, respectively, both of which are expected to increase membrane permeability. In addition, the inclusion of OA in vesicle membranes led to a reduction of sensor dynamic range, measured via a reduced concentration dependence of GFP expression on NaF concentration, consistent with our hypothesis of increased permeability to any amount of external fluoride (10% OA + 1 mM NaF versus 10% OA + 3 mM NaF, P > 0.9999; 10% OA + 1 mM NaF versus 10% OA + 5 mM NaF, P = 0.8149). In contrast, vesicles containing 10% 1.8k diblock copolymer exhibited reduced responsiveness to increasing external NaF, indicating reduced membrane permeability. Mean POPC vesicle fluorescence peaked at 1 mM NaF, while 10% 1.8k diblock copolymer responses were maximum at 5 mM NaF (table S2). All fluoride-containing conditions for samples containing 10% OA were statistically indistinguishable. Together, these results indicate that exchanging membrane components to control membrane permeability provides a handle to tune the sensitivity of an encapsulated riboswitch to an analyte of interest. Furthermore, the selection of highly permeable amphiphiles does not necessarily improve sensor performance and may instead increase overall signal but limit sensor resolution. A balance between analyte access and desired sensing behavior is likely an important consideration for engineering encapsulated biosensing systems depending on the desired application.
Encapsulation protects sensor components from degradation
Having established that these vesicle sensors can detect external fluoride, we next wanted to explore how they might function in complex samples. One major benefit of membrane encapsulation is the ability to leverage the semipermeable barrier formed by the membrane to contain and protect encapsulated components. Cell-free reactions, particularly those using riboswitches, are highly sensitive to the presence of nucleases and proteases, which can degrade sensor components before a target analyte is encountered (19). Because of their large size, however, enzymes are unable to pass through the vesicle membrane to access encapsulated reactants.
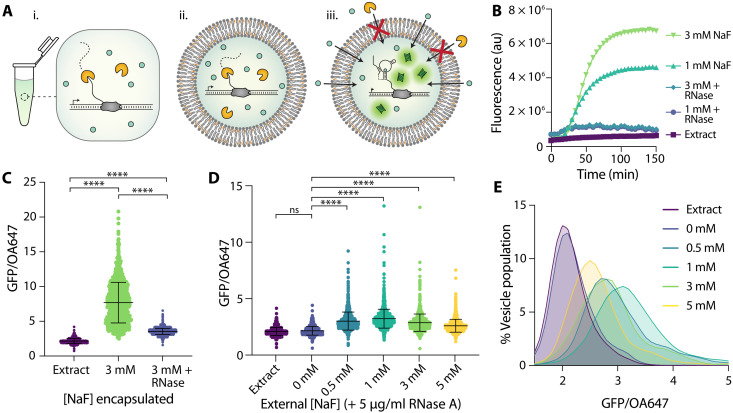
To determine whether the vesicle membrane can sufficiently protect encapsulated reactions from external degradation, we tested various vesicle assemblies in the presence of RNase A (Fig. 4A). We observed that RNase completely eliminated the riboswitch response to NaF both under bulk conditions and when RNase was coencapsulated with the cell-free reaction inside vesicles (Fig. 4, B and C). In contrast, encapsulated sensors maintained the ability to respond to externally added NaF when RNase was present in the external sample (Fig. 4, D and E, and table S3). We noticed a decrease in mean GFP fluorescence at higher external NaF concentrations—a trend that we did not observe in non–RNase-exposed samples. We hypothesized that this difference was due to slightly higher degrees of membrane permeability from the addition of small amounts of glycerol in the RNase buffer (fig. S5), which could cause increased reaction poisoning with high fluoride concentrations. In addition, glycerol has been demonstrated to inhibit cell-free protein synthesis at high concentrations and may be causing slight effects even at the low concentrations used here (55). Nevertheless, all vesicle populations exhibited increased GFP expression in the presence of fluoride, demonstrating simultaneous permeation of fluoride into the vesicle interior and exclusion of RNase A from the cell-free reaction. These results indicate that encapsulation in bilayer membranes can sufficiently exclude RNase from the reaction environment, thereby protecting the cell-free sensor within a semipermeable compartment. The external addition of RNase A, demonstrated here, serves as a proof-of-concept step toward cell-free detection in complex environments, such as biological samples or highly contaminated environmental samples.
Fig. 4. Encapsulation protects from degradation by RNase A.
(A) Schematic of RNase-containing conditions. RNase A degrades the riboswitch (i) under bulk conditions and (ii) when coencapsulated with reactants but is unable to reach reactants contained within vesicles (iii). (B) Riboswitch response to NaF under bulk conditions with and without RNase A added to reaction. (C) Riboswitch activity as indicated by GFP/OA647 fluorescence when encapsulated with 3 mM NaF compared to the coencapsulation of both 3 mM NaF and RNase A. (D) Response of encapsulated riboswitch to externally added NaF with RNase A present in external solution. Black lines indicate mean and SD. (E) Histogram of data in (D). Data are plotted with lowless curve fitting. ****P ≤ 0.0001; ns, P > 0.05; P values are generated using a one-way ANOVA and Tukey’s multiple comparisons test.
Vesicle-based sensors can incorporate a colorimetric readout and detect fluoride in real-world samples
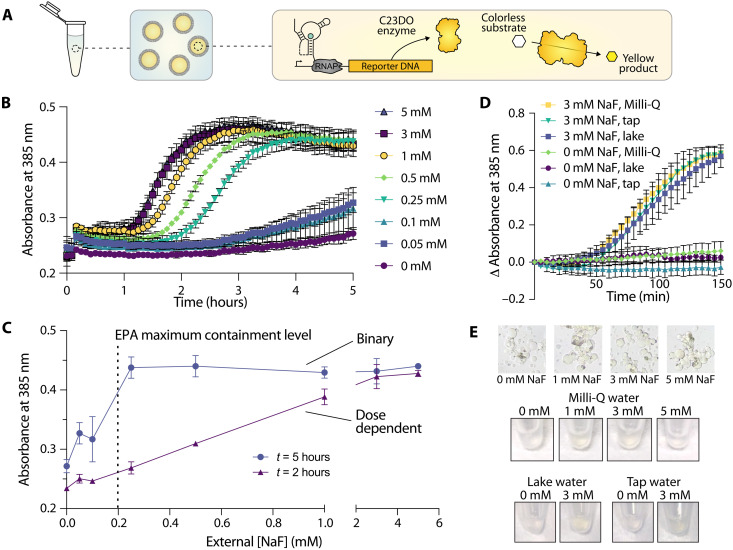
Last, we wondered whether we could extend these results to conditions that would be more relevant for real-world environmental sensing. Although fluorescence is a common readout for many biological assays, GFP fluorescence inside vesicles is difficult to monitor with common equipment, particularly in nonlaboratory settings. In addition, with our vesicle-based construct, GFP fluorescence was too low to measure in vesicle populations in bulk, necessitating more sensitive microscopy analysis methods. To address this limitation, we coupled fluoride detection to an alternative reporter enzyme, catechol (2,3)-dioxygenase (C23DO) (3). In this system, the riboswitch ON state leads to the expression of C23DO, which catalyzes the conversion of its colorless substrate, catechol, to the yellow-colored 2-hydroxymuconate semialdehyde to generate a colorimetric response (Fig. 5A). Under bulk conditions, this construct exhibits a fast and robust response to fluoride, and the colorimetric output generated is clearly distinguishable by eye for both laboratory- and field-collected water samples (3).
Fig. 5. Colorimetric sensors detect fluoride at environmentally relevant concentrations and in real-world samples.
(A) Schematic of encapsulated enzymatic readout. Riboswitch activation inside vesicles leads to the expression of C23DO, resulting in the production of a yellow product that is localized to the vesicle interior. (B) Absorbance over time in populations of vesicles encapsulating a catecholase-based readout in response to external NaF. n = 2 technical replicates. (C) Absorbance as a function of external NaF concentration at two time points; t = 2 and 5 hours since initiating vesicle exposure to fluoride. n = 2 technical replicates. (D) Changes in absorbance over time inside of populations of vesicles incubated in samples derived from Milli-Q water, tap water, and lake water supplemented with either 0 or 3 mM NaF. n = 2 independent vesicle preparations. (E) Colorimetric changes in vesicles as viewed through a microscope eyepiece and by eye in Eppendorf tubes.
To investigate whether this enzymatic reporter could function within our sensor vesicles, we encapsulated cell-free reactions with DNA coding for the riboswitch-C23DO construct and supplemented them with 1 mM catechol (Fig. 5A). We then titrated NaF into the outer solution and monitored color changes in each population of vesicles via changes in absorbance at 385 nm. In contrast to our GFP-based readout, signal amplification from the enzyme-regulated construct allowed us to assess absorbance changes in an entire population of vesicles rather than on a vesicle-by-vesicle basis. RNase A was added to the outer vesicle solution to control for any unencapsulated reactions caused by vesicle lysis and to assess the membrane’s ability to protect encapsulated sensor components. Sensor responses were significantly lower than those observed in bulk without RNase (fig. S6); however, despite this reduction in signal magnitude, sensors exhibited significantly increased absorbance in response to increasing fluoride concentrations (Fig. 5B). Vesicle sensors exhibited sensitivity to fluoride at a range of concentrations; here, readout time is an important consideration, as enzymatic signal amplification can lower the resolution of a dose response curve and even reduce overall signal at long reaction times (fig. S6) (3). Sensor responses to fluoride reach statistical significance at varying times depending on the external concentration of fluoride encountered (fig. S7). This variability in response as a function of both time-to-detection and fluoride concentration can enable these sensors to serve as dose-responsive or binary (yes/no) sensors depending on the time at which the response is assessed (Fig. 5C and fig. S7). Sensors could detect fluoride concentrations at levels consistent with the Environmental Protection Agency (EPA)’s maximum contaminant level for fluoride (~0.21 mM fluoride) and secondary contaminant standard (~0.1 mM) within 3 and 5 hours, respectively (Fig. 5C and fig. S7) (56). Responses were significant but noisy at 0.05 and 0.1 mM fluoride, however, indicating that these sensors would be most useful for detection of fluoride concentrations above the maximum contaminantt limit of 0.2 mM (Fig. 5C). By accounting for both the detection time and the final absorbance of these colorimetric vesicle sensors, health-relevant concentrations of fluoride can ultimately be detected in either a binary or a dose-dependent manner.
Last, we set out to test whether these sensors could be used to monitor fluoride concentrations in real-world samples. We collected water samples from Lake Michigan and the Evanston, IL municipal tap water supply and used each sample to prepare the vesicle outer solution supplemented with the small molecule components of the reaction. We supplemented these outer solutions with either 0 or 3 mM NaF and RNase A, which removes sensor activity from any ruptured vesicles, and added vesicles to the sample. We first assessed the performance of encapsulated GFP sensors and observed increased GFP expression in the presence of 3 mM NaF, with slightly higher levels of GFP expression in both lake and tap water samples compared to those incubated with laboratory-grade Milli-Q water (fig. S8). We next assessed the performance of encapsulated C23DO enzyme sensors and similarly observed increased colorimetric outputs, measured via absorption at 385 nm, in all populations of enzyme-expressing vesicles incubated with 3 mM NaF (Fig. 5D). We did not observe increases in absorbance in the absence of supplemented fluoride, with the exception of municipal tap water samples (fig. S6). Color changes were visible by eye in tubes containing vesicles, and changes in color inside of individual vesicles could be observed on the microscope when imaged through the eyepiece (Fig. 5E). By incorporating a colorimetric detection method as demonstrated here, these sensors could ultimately be used to detect fluoride in samples containing inhospitable components and in settings that prohibit the use of laboratory-based equipment. The results observed here highlight the feasibility of these vesicle-based sensors to detect environmentally relevant small molecules in complex, real-world samples, a step toward encapsulation to generate deployable cell-free sensors.
DISCUSSION
To our knowledge, this work represents the first demonstrated function of a transcriptionally regulated riboswitch encapsulated in bilayer vesicles. We have demonstrated that this encapsulated riboswitch can detect exogenous fluoride through permeability-based sensing, generating both fluorescent and colorimetric outputs. In addition, we have shown that membrane composition can serve as an additional handle by which cell-free sensing reactions can be modulated by altering permeability and therefore the sensor’s access to an analyte of interest. Furthermore, we demonstrated that encapsulation allows a cell-free sensor to outperform those in bulk solution when the solution contains degradative enzymes that inhibit sensor performance. Looking ahead toward sensor deployment, this work establishes that encapsulation within artificial cells can not only protect cell-free sensors from degradative sample components but can also allow analyte detection in real-world samples. Here, the encapsulation of a colorimetric reporter system allowed detection of fluoride in both a dose-responsive and binary manner at two EPA containment levels, even while in the presence of degradative sample components. While cell-free sensors have been previously used for the detection of environmental molecules of interest (1–9), encapsulation of these systems in artificial cells may ultimately diversify the contexts within which cell-free sensors can operate.
Although encapsulation can provide powerful advantages to cell-free sensing, it also brings some limitations. The variability observed in the responses of individual GFP-expressing liposomes within a vesicle population would likely serve as a hurdle for technological use of fluorescence-based sensors in future applications, which may necessitate alternative vesicle assembly techniques, such as microfluidics (42), and a better understanding of the underlying biophysics of cell-free reactions inside membranes. In addition, encapsulation reduces the overall signal magnitude compared to bulk reactions—a limitation that we show can be offset by the protection encapsulation confers to the sensor—which could be improved by loading higher amounts of sensor components. Finally, the reintroduction of a barrier between sample and sensor requires strategies to transport specific analytes into the vesicle. To address these limitations, future sensor development may require incorporation of alternative expression systems and membrane components. While we explored protein-based outputs here, riboswitch expression could also be coupled to transcription-based reporting, such as aptamer-dye outputs (3), to build a transcription-only sensor that would require encapsulation of fewer reaction components relative to sensors that encode translation-mediated outputs and which should operate on quicker time scales. Membrane compositional changes can enable permeability-based import for certain small analytes, such as fluoride, with many natural and synthetic amphiphiles to choose from. As we observed with the inclusion of OA into vesicle membranes, enhanced permeability can also have negative effects on sensor performance by reducing the sensor dynamic range. For this condition with OA, we hypothesize that this result may be due not only to enhanced permeability of fluoride into vesicles at lower concentrations but also to the subsequent retention of the ion in the vesicle upon interaction with the riboswitch and cell-free components, ultimately leading to a similar GFP signal for all the fluoride concentrations tested. Thus, not all amphiphiles that improve membrane permeability to the analyte of interest will necessarily improve sensor performance. Moving forward, we can gain even finer control of analyte transport into and out of the vesicle by incorporating transmembrane proteins to enhance sensing capabilities and introduce more advanced sensing or responsive functions. These strategies could ultimately allow new functions for these types of sensors, including conjugation-based capture methods, deployment and transport of cell-free reactions, controlled sensor degradation, or enhanced sensor biocompatibility (18).
The diversity of existing cell-free sensors could ultimately lead to a new generation of artificial cell biosensors for a wide array of analytes. With the modular nature and signal processing capabilities of components in these systems, various vesicle-based sensors could be engineered, which use membrane components, genetic circuits, and triggered responses to detect small molecules of interest (30, 57, 58). As focus shifts toward sensor application, these platforms could offer additional handles with which to tune sensor kinetics, stability, and sensitivity to advance the types of contexts in which cell-free sensing can operate, allowing for detection in environments such as soil, groundwater, or biological samples. Finally, the incorporation of additional transcription-based cell-free systems, particularly those using riboswitch-based sensing, may ultimately allow the development of a family of encapsulated sensors that are fast, specific, and deployable.
MATERIALS AND METHODS
Chemicals
POPC and cholesterol were purchased from Avanti Polar Lipids Inc. OA, glycerol, sucrose, glucose, HPTS, BioUltra Mineral Oil, phosphate-buffered saline (PBS), bovine serum albumin (BSA), and NaF were purchased from MilliporeSigma. 1.8k PEO-b-PBD polymer was purchased from Polymer Source. OA647, calcein dye, and Hepes buffer were purchased from Thermo Fisher Scientific. RNase A was purchased from New England Biolabs.
Plasmid construction
Plasmids were assembled using Gibson assembly (New England Biolabs, catalog no. E2611S) and purified using a QIAGEN QIAfilter Midiprep Kit (QIAGEN, catalog no. 12143). Coding sequences of the plasmids consist of the crcB fluoride riboswitch from B. cereus regulating either superfolder GFP (pJBL3752) or C23DO (pJBL7025), all expressed under constitutive Anderson promoter J23119. Plasmid sequences are available on Addgene with accession numbers 128809 (pJBL3752) and 128810 (pJBL7025).
Cell-free reaction assembly
Cell-free extract and reactions were prepared according to established protocols (3, 39). Briefly, cell-free reactions were assembled by mixing cell extract, a reaction buffer containing the small molecules required for transcription and translation (nucleoside triphosphates, amino acids, buffering salts, crowding agents, and an energy source), and DNA templates and inducers at a ratio of approximately 30:30:40 (table S4). Sucrose was added to a final concentration of 200 mM to allow vesicle formation through the density-dependent double emulsion technique. Each reaction was prepared on ice to 16.5 μl of final volume in batches of seven. Reactions were prepared with 10 nM pJBL3752 (riboswitch-GFP plasmid) or pJBL7025 (riboswitch-enzyme plasmid) + 1 mM catechol. Reaction master mix was assembled and then added to DNA, inducers, sucrose, and water to a final volume of 16.5 μl per reaction aliquot. For reactions containing volume marker, reaction mix was supplemented with 1.4 μl of OA647. Preparation conditions were kept consistent between reactions, only varying NaF concentration or omitting DNA for extract-only controls.
Encapsulation of cell-free reactions
Encapsulated sensors were prepared via water-in-oil double emulsion methods. Lipid films were prepared by mixing amphiphiles (lipid, cholesterol, fatty acid, or polymers) in chloroform to a final amphiphile concentration of 25 mM at a volume of 200 μl. Films were dried onto the side of a glass vial under nitrogen gas and then placed in a vacuum oven overnight. A total of 200 μl of BioUltra mineral oil was added to lipid films and heated at 80°C for 30 min, followed by 10 s of vortexing to incorporate amphiphiles into the oil. Lipid/oil mixtures were cooled to room temperature and then placed on ice during cell-free reaction assembly. Cell-free reactions were prepared on ice as described above. Reactions were layered on top of lipid/oil mixture and then vortexed for 30 s to form an emulsion. Emulsions were incubated at 4°C for 5 min and then layered onto outer solution containing all small molecules required for transcription and translation, 100 mM Hepes buffer (pH 8), and 200 mM glucose, which is used to create a density gradient with the encapsulated sucrose. Samples were again incubated at 4°C for 5 min and then centrifuged for 15 min at 18,000 relative centrifugal force at 4°C to spin the denser, sucrose-containing vesicles into a pellet. Vesicle pellets were collected by pipette and placed into fresh Eppendorf tubes. Prepared vesicles were then added in 10 μl of aliquots to 20 μl of fresh outer solution supplemented with NaF, certain water samples, and/or RNase A (final concentration, 5 μg/ml). Osmolarity of NaF stock solution was adjusted to match that of the outer solution by adding glucose to minimize osmotic effects on vesicles.
Cell-free protein expression
For bulk assays, unencapsulated reactions were prepared as described above and added to 384-well plates. Protein expression was monitored at 37°C in a SpectraMax i3x plate reader (Molecular Devices). GFP was monitored at an excitation of 485 nm and an emission of 510 nm. Catechol absorbance was monitored at 385 nm.
Encapsulated sensors with a colorimetric readout were monitored at an absorbance of 385 nm using a SpectraMax i3x plate reader at 37°C until expression reached a plateau, about 2.5 hours, after which samples were removed from plates and placed into Eppendorf tubes or microscopy chambers for imaging. Images of tubes and through the microscope eyepiece were taken using an iPhone 8. Absorbance measurements in the plate reader are reported relative to initial absorbance to control for slight differences in vesicle concentration between vesicle preparations.
Encapsulated sensors expressing GFP were incubated in outer solution for 6 hours at 37°C and then imaged on a Nikon Ti2 inverted microscope. Imaging chambers were blocked with BSA for 20 min and then triple rinsed with 766 mosM of PBS. Vesicles were added to equiosmolar PBS and allowed to settle for 5 min before imaging. Images were taken using differential interference contrast, GFP (excitation, 470 nm; emission, 525 nm) and Cy 5.5 (excitation, 650 nm; emission, 720 nm) filters under ×10 magnification, 20% laser intensity, and 1-s exposure. Images were analyzed using Nikon NIS-Elements AR software Advanced Analysis tool (44): Vesicles were selected using the OA647 channel. General analysis protocol was set with the following settings: preprocessing: local contrast; size, 105; power, 50%; threshold minimum, 393; smooth, 1×; clean, 1×; size minimum, 2 μm; return mean, GFP; mean, OA647; max, GFP.
Encapsulated dye assays
HPTS assay
Vesicles were prepared via thin film hydration with 33% cholesterol and 66% POPC. Lipid and cholesterol in chloroform were dried onto the side of a glass vial under nitrogen gas to form a lipid film. Vesicle films were hydrated with Hepes + 0.5 mM HPTS dye overnight at 60°C. Vesicles were extruded to 400 nm, purified via size exclusion chromatography (SEC), and added to a 384-well plate with equiosmolar Hepes buffer + varying concentrations of NaCl and NaF. HPTS fluorescence was monitored with excitation at 405 and 450 nm and emission at 510 nm, as characterized by Hilburger et al. (59). HPTS fluorescence is reported as the ratio of emission intensities when excited at 450 nm/405 nm.
Calcein assay
Vesicles were prepared via thin film hydration with 33% cholesterol and 66% POPC. Lipid and cholesterol in chloroform were dried onto the side of a glass vial under nitrogen gas to form a lipid film. Vesicle films were hydrated with Hepes + 20 mM calcein dye overnight at 60°C. Vesicles were extruded to 400 nm, purified via SEC, and added to a 384-well plate with equiosmolar Hepes buffer and increasing volumes of 0.02% glycerol solution or RNase prepared in buffer to the same final glycerol concentration (1.25 μl corresponds to the concentration used for manuscript studies). Vesicles were incubated for 4 hours at 37°C, and calcein fluorescence was measured (excitation, 495 nm; emission, 515 nm). Vesicles were lysed with 1 μl of 10% Triton X-100, and total calcein fluorescence was measured to determine fraction release.
Statistical analysis
All graphing and statistical analysis were conducted in GraphPad Prism (60). Populations of vesicles were analyzed using one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons test or two-way ANOVA and Dunnett’s multiple comparisons test and descriptive statistics. Significance is reported as follows: nonsignificant (ns), P > 0.05; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001. Symbols and error bars on line plots represent mean and SD, respectively.
Acknowledgments
We thank Z. Jurado and the Murray Lab as well as the Build-a-Cell community for providing a water-in-oil vesicle formation protocol, which we adapted for these studies. We also thank A. Silverman and D. Brown for preparing cell extract and reaction materials.
Funding: This study was funded by Northwestern University Chemistry of Life Processes Institute Cornew Innovation Award (to N.P.K. and J.B.L.), National Science Foundation grant 1844219 (to N.P.K.), National Science Foundation grant 1844336 (to N.P.K. and J.B.L.), National Science Foundation grant 2145050 (to N.P.K.), and U.S. Department of Defense National Science and Engineering Graduate Fellowship (to M.A.B.).
Author contributions: Conceptualization: M.A.B., W.T., J.B.L., and N.P.K. Methodology: M.A.B., W.T., J.B.L., and N.P.K. Investigation: M.A.B. and W.T. Data analysis: M.A.B., W.T., J.B.L., and N.P.K. Visualization: M.A.B. Writing (original draft): M.A.B. and N.P.K. Writing (review and editing): M.A.B., W.T., N.P.K., and J.B.L.
Competing interests: The authors declare the following competing financial interest(s): W.T. and J.B.L. are inventors on a patent related to this work filed by Northwestern University (serial no: 17/593,026, filed 3 September 2021; no. PCT/US2020/020800, filed 3 March 2020; and no. 62/813,368, filed 4 March 2019). J.B.L. is a cofounder of Stemloop Inc. J.B.L.’s interests are reviewed and managed by Northwestern University in accordance with their conflict-of-interest policies. The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials via Open Science Framework at DOI 10.17605/OSF.IO/CNWR5. Plasmid sequences are available on Addgene with accession numbers 128809 (pJBL3752) and 128810 (pJBL7025). The riboswitch and extracts can be provided by Northwestern University pending scientific review and a completed material transfer agreement. Requests for these materials should be submitted to J.B.L.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Tables S1 to S4
REFERENCES AND NOTES
- 1.P. Zhang, H. Feng, J. Yang, H. Jiang, H. Zhou, Y. Lu, Detection of inorganic ions and organic molecules with cell-free biosensing systems. J. Biotechnol. 300, 78–86 (2019). [DOI] [PubMed] [Google Scholar]
- 2.A. Gräwe, A. Dreyer, T. Vornholt, U. Barteczko, L. Buchholz, G. Drews, U. L. Ho, M. E. Jackowski, M. Kracht, J. Lüders, T. Bleckwehl, L. Rositzka, M. Ruwe, M. Wittchen, P. Lutter, K. Müller, J. Kalinowski, A paper-based, cell-free biosensor system for the detection of heavy metals and date rape drugs. PLOS ONE 14, e0210940 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.W. Thavarajah, A. D. Silverman, M. S. Verosloff, N. Kelley-Loughnane, M. C. Jewett, J. B. Lucks, Point-of-use detection of environmental fluoride via a cell-free riboswitch-based biosensor. ACS Synth. Biol. 9, 10–18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.X. Liu, A. D. Silverman, K. K. Alam, E. Iverson, J. B. Lucks, M. C. Jewett, S. Raman, Design of a transcriptional biosensor for the portable, on-demand detection of cyanuric acid. ACS Synth. Biol. 9, 84–94 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.A. D. Silverman, U. Akova, K. K. Alam, M. C. Jewett, J. B. Lucks, Design and optimization of a cell-free atrazine biosensor. ACS Synth. Biol. 9, 671–677 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.P. L. Voyvodic, A. Pandi, M. Koch, I. Conejero, E. Valjent, P. Courtet, E. Renard, J.-L. Faulon, J. Bonnet, Plug-and-play metabolic transducers expand the chemical detection space of cell-free biosensors. Nat. Commun. 10, 1697 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.T. T. M. Duyen, H. Matsuura, K. Ujiie, M. Muraoka, K. Harada, K. Hirata, Paper-based colorimetric biosensor for antibiotics inhibiting bacterial protein synthesis. J. Biosci. Bioeng. 123, 96–100 (2017). [DOI] [PubMed] [Google Scholar]
- 8.T. Pellinen, T. Huovinen, M. Karp, A cell-free biosensor for the detection of transcriptional inducers using firefly luciferase as a reporter. Anal. Biochem. 330, 52–57 (2004). [DOI] [PubMed] [Google Scholar]
- 9.S. Gupta, S. Sarkar, A. Katranidis, J. Bhattacharya, Development of a cell-free optical biosensor for detection of a broad range of mercury contaminants in water: A plasmid DNA-based approach. ACS Omega 4, 9480–9487 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.K. Pardee, A. A. Green, M. K. Takahashi, D. Braff, G. Lambert, J. W. Lee, T. Ferrante, D. Ma, N. Donghia, M. Fan, N. M. Daringer, I. Bosch, D. M. Dudley, D. H. O’Connor, L. Gehrke, J. J. Collins, Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell 165, 1255–1266 (2016). [DOI] [PubMed] [Google Scholar]
- 11.D. Ma, L. Shen, K. Wu, C. W. Diehnelt, A. A. Green, Low-cost detection of norovirus using paper-based cell-free systems and synbody-based viral enrichment. Synth. Biol. 3, ysy018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.M. Verosloff, J. Chappell, K. L. Perry, J. R. Thompson, J. B. Lucks, PLANT-Dx: A molecular diagnostic for point-of-use detection of plant pathogens. ACS Synth. Biol. 8, 902–905 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.M. K. Takahashi, X. Tan, A. J. Dy, D. Braff, R. T. Akana, Y. Furuta, N. Donghia, A. Ananthakrishnan, J. J. Collins, A low-cost paper-based synthetic biology platform for analyzing gut microbiota and host biomarkers. Nat. Commun. 9, 3347 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.K. Y. Wen, L. Cameron, J. Chappell, K. Jensen, D. J. Bell, R. Kelwick, M. Kopniczky, J. C. Davies, A. Filloux, P. S. Freemont, A cell-free biosensor for detecting quorum sensing molecules in P. aeruginosa-infected respiratory samples. ACS Synth. Biol. 6, 2293–2301 (2017). [DOI] [PubMed] [Google Scholar]
- 15.J. P. Hunt, R. J. Barnett, H. Robinson, M. Soltani, J. A. D. Nelson, B. C. Bundy, Rapid sensing of clinically relevant glutamine concentrations in human serum with metabolically engineered E. coli-based cell-free protein synthesis. J. Biotechnol. 325, 389–394 (2021). [DOI] [PubMed] [Google Scholar]
- 16.J. P. Hunt, E. L. Zhao, T. J. Free, M. Soltani, C. A. Warr, A. B. Benedict, M. K. Takahashi, J. S. Griffitts, W. G. Pitt, B. C. Bundy, Towards detection of SARS-CoV-2 RNA in human saliva: A paper-based cell-free toehold switch biosensor with a visual bioluminescent output. N. Biotechnol. 66, 53–60 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.A. S. M. Salehi, M. J. Shakalli Tang, M. T. Smith, J. M. Hunt, R. A. Law, D. W. Wood, B. C. Bundy, Cell-free protein synthesis approach to biosensing hTRβ-specific endocrine disruptors. Anal. Chem. 89, 3395–3401 (2017). [DOI] [PubMed] [Google Scholar]
- 18.M. A. Boyd, N. P. Kamat, Designing artificial cells towards a new generation of biosensors. Trends Biotechnol. 39, 927–939 (2021). [DOI] [PubMed] [Google Scholar]
- 19.M. Soltani, J. P. Hunt, B. C. Bundy, Rapid RNase inhibitor production to enable low-cost, on-demand cell-free protein synthesis biosensor use in human body fluids. Biotechnol. Bioeng. 118, 3973–3983 (2021). [DOI] [PubMed] [Google Scholar]
- 20.M. P. McNerney, Y. Zhang, P. Steppe, A. D. Silverman, M. C. Jewett, M. P. Styczynski, Point-of-care biomarker quantification enabled by sample-specific calibration. Sci. Adv. 5, eaax4473 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.K. P. Adamala, D. A. Martin-Alarcon, K. R. Guthrie-Honea, E. S. Boyden, Engineering genetic circuit interactions within and between synthetic minimal cells. Nat. Chem. 9, 431–439 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.C. Tan, S. Saurabh, M. P. Bruchez, R. Schwartz, P. LeDuc, Molecular crowding shapes gene expression in synthetic cellular nanosystems. Nat. Nanotechnol. 8, 602–608 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Z. Nourian, C. Danelon, Linking genotype and phenotype in protein synthesizing liposomes with external supply of resources. ACS Synth. Biol. 2, 186–193 (2013). [DOI] [PubMed] [Google Scholar]
- 24.T. P. de Souza, P. Stano, P. L. Luisi, The minimal size of liposome-based model cells brings about a remarkably enhanced entrapment and protein synthesis. Chembiochem. 10, 1056–1063 (2009). [DOI] [PubMed] [Google Scholar]
- 25.R. Sakamoto, V. Noireaux, Y. T. Maeda, Anomalous scaling of gene expression in confined cell-free reactions. Sci. Rep. 8, 7364 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.A. D. Silverman, A. S. Karim, M. C. Jewett, Cell-free gene expression: An expanded repertoire of applications. Nat. Rev. Genet. 21, 151–170 (2020). [DOI] [PubMed] [Google Scholar]
- 27.S. W. Schaffter, R. Schulman, Building in vitro transcriptional regulatory networks by successively integrating multiple functional circuit modules. Nat. Chem. 11, 829–838 (2019). [DOI] [PubMed] [Google Scholar]
- 28.R. Lentini, S. P. Santero, F. Chizzolini, D. Cecchi, J. Fontana, M. Marchioretto, C. del Bianco, J. L. Terrell, A. C. Spencer, L. Martini, M. Forlin, M. Assfalg, M. D. Serra, W. E. Bentley, S. S. Mansy, Integrating artificial with natural cells to translate chemical messages that direct E. coli behaviour. Nat. Commun. 5, 4012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.L. Martini, S. S. Mansy, Cell-like systems with riboswitch controlled gene expression. Chem. Commun. 47, 10734–10736 (2011). [DOI] [PubMed] [Google Scholar]
- 30.M. Dwidar, Y. Seike, S. Kobori, C. Whitaker, T. Matsuura, Y. Yokobayashi, Programmable artificial cells using histamine-responsive synthetic riboswitch. J. Am. Chem. Soc. 141, 11103–11114 (2019). [DOI] [PubMed] [Google Scholar]
- 31.T. Janas, T. Janas, M. Yarus, Specific RNA binding to ordered phospholipid bilayers. Nucleic Acids Res. 34, 2128–2136 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.T. Czerniak, J. P. Saenz, Lipid membranes modulate the activity of RNA through sequence-dependent interactions. Proc. Natl. Acad. Sci. U.S.A. 119, e2119235119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.E. J. Strobel, L. Cheng, K. E. Berman, P. D. Carlson, J. B. Lucks, A ligand gated strand displacement mechanism for ZTP riboswitch transcription control. Nat. Chem. Biol. 15, 1067–1076 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.UNICEF, WHO, “Progress on household drinking water, sanitation and hygiene 2000–2017: Special focus on inequalities” (Publication 978-92-415-1623-5, UNICEF, 2019); www.unicef.org/reports/progress-on-drinking-water-sanitation-and-hygiene-2019.
- 35.R. Damania, S. Desbureaux, A.-S. Rodella, J. Russ, E. Zaveri, “Quality unknown: The invisible water crisis” (World Bank, 2019); hdl.handle.net/10986/32245.
- 36.R. Fuge, Fluorine in the environment, a review of its sources and geochemistry. Appl. Geochem. 100, 393–406 (2019). [Google Scholar]
- 37.P. B. McMahon, C. J. Brown, T. D. Johnson, K. Belitz, B. D. Lindsey, Fluoride occurrence in United States groundwater. Sci. Total Environ. 732, 139217 (2020). [DOI] [PubMed] [Google Scholar]
- 38.K. E. Watters, E. J. Strobel, A. M. Yu, J. T. Lis, J. B. Lucks, Cotranscriptional folding of a riboswitch at nucleotide resolution. Nat. Struct. Mol. Biol. 23, 1124–1131 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.A. D. Silverman, N. Kelley-Loughnane, J. B. Lucks, M. C. Jewett, Deconstructing cell-free extract preparation for in vitro activation of transcriptional genetic circuitry. ACS Synth. Biol. 8, 403–414 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O. Barbier, L. Arreola-Mendoza, L. M. del Razo, Molecular mechanisms of fluoride toxicity. Chem. Biol. Interact. 188, 319–333 (2010). [DOI] [PubMed] [Google Scholar]
- 41.S. Pautot, B. J. Frisken, D. A. Weitz, Production of unilamellar vesicles using an inverted emulsion. Langmuir 19, 2870–2879 (2003). [Google Scholar]
- 42.D. T. Gonzales, N. Yandrapalli, T. Robinson, C. Zechner, T. Y. D. Tang, Cell-free gene expression dynamics in synthetic cell populations. ACS Synth. Biol. 11, 205–215 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.K. Nishimura, T. Matsuura, K. Nishimura, T. Sunami, H. Suzuki, T. Yomo, Cell-free protein synthesis inside giant unilamellar vesicles analyzed by flow cytometry. Langmuir 28, 8426–8432 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Nikon Instruments Inc., NIS-Elements Advanced Research; www.microscope.healthcare.nikon.com/products/software/nis-elements/nis-elements-advanced-research.
- 45.K. Nishimura, S. Tsuru, H. Suzuki, T. Yomo, Stochasticity in gene expression in a cell-sized compartment. ACS Synth. Biol. 4, 566–576 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Z. Nourian, W. Roelofsen, C. Danelon, Triggered gene expression in fed-vesicle microreactors with a multifunctional membrane. Angew. Chem. Int. Ed. 51, 3114–3118 (2012). [DOI] [PubMed] [Google Scholar]
- 47.H. Saito, Y. Kato, M. le Berre, A. Yamada, T. Inoue, K. Yosikawa, D. Baigl, Time-resolved tracking of a minimum gene expression system reconstituted in giant liposomes. Chembiochem. 10, 1640–1643 (2009). [DOI] [PubMed] [Google Scholar]
- 48.D. Garenne, V. Noireaux, Analysis of cytoplasmic and membrane molecular crowding in genetically programmed synthetic cells. Biomacromolecules 21, 2808–2817 (2020). [DOI] [PubMed] [Google Scholar]
- 49.J. Gutknecht, A. Walter, Hydrofluoric and nitric acid transport through lipid bilayer membranes. Biochim. Biophys. Acta. 644, 153–156 (1981). [DOI] [PubMed] [Google Scholar]
- 50.D. Papahadjopoulos, S. Nir, S. Ohki, Permeability properties of phospholipid membranes: Effect of cholesterol and temperature. Biochim. Biophys. Acta. 266, 561–583 (1972). [DOI] [PubMed] [Google Scholar]
- 51.B. M. Discher, Y.-Y. Won, D. S. Ege, J. C.-M. Lee, F. S. Bates, D. E. Discher, D. A. Hammer, Polymersomes: Tough vesicles made from diblock copolymers. Science 284, 1143–1146 (1999). [DOI] [PubMed] [Google Scholar]
- 52.M. L. Jacobs, M. A. Boyd, N. P. Kamat, Diblock copolymers enhance folding of a mechanosensitive membrane protein during cell-free expression. Proc. Natl. Acad. Sci. U.S.A. 116, 4031–4036 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.H. Jespersen, J. H. Andersen, H. J. Ditzel, O. G. Mouritsen, Lipids, curvature stress, and the action of lipid prodrugs: Free fatty acids and lysolipid enhancement of drug transport across liposomal membranes. Biochimie. 94, 2–10 (2012). [DOI] [PubMed] [Google Scholar]
- 54.P.-A. Monnard, D. W. Deamer, Membrane self-assembly processes: Steps toward the first cellular life. Anat. Rec. 268, 196–207 (2002). [DOI] [PubMed] [Google Scholar]
- 55.N. Jiang, X. Ding, Y. Lu, Development of a robust Escherichia coli-based cell-free protein synthesis application platform. Biochem. Eng. J. 165, 107830 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Environmental Protection Agency (EPA), “National primary drinking water regulations” (Code of Federal Regulations Title 40 Part 141, EPA, 2021); www.ecfr.gov/current/title-40/chapter-I/subchapter-D/part-141.
- 57.N. S. Kruyer, W. Sugianto, B. I. Tickman, D. Alba Burbano, V. Noireaux, J. M. Carothers, P. Peralta-Yahya, Membrane augmented cell-free systems: A new frontier in biotechnology. ACS Synth. Biol. 10, 670–681 (2021). [DOI] [PubMed] [Google Scholar]
- 58.J. T. Lazar, J. J. Tabor, Bacterial two-component systems as sensors for synthetic biology applications. Curr. Opin. Syst. Biol. 28, 100398 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.C. E. Hilburger, M. L. Jacobs, K. R. Lewis, J. A. Peruzzi, N. P. Kamat, Controlling secretion in artificial cells with a membrane AND gate. ACS Synth. Biol. 8, 1224–1230 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.GraphPad, Prism; www.graphpad.com.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Tables S1 to S4