Abstract
Objective
Immune function is multifaceted and characterizations based on single biomarkers may be uninformative or misleading, particularly when considered across ecological contexts. However, measuring the many facets of immunity in the field can be challenging, since many measures cannot be obtained on‐site, necessitating sample preservation and transport. Here we assess state‐of‐the‐art methods for measuring immunity, focusing on measures that require a minimal blood sample obtained from a finger prick, which can be: (1) dried on filter paper, (2) frozen in liquid nitrogen, or (3) stabilized with chemical reagents.
Results
We review immune measures that can be obtained from point‐of‐care devices or from immunoassays of dried blood spots (DBSs), field methods for flow cytometry, the use of RNA or DNA sequencing and quantification, and the application of immune activation assays under field conditions.
Conclusions
Stable protein products, such as immunoglobulins and C‐reactive protein are reliably measured in DBSs. Because less stable proteins, such as cytokines, may be problematic to measure even in fresh blood, mRNA from stabilized blood may provide a cleaner measure of cytokine and broader immune‐related gene expression. Gene methylation assays or mRNA sequencing also allow for the quantification of many other parameters, including the inference of leukocyte subsets, though with less accuracy than with flow cytometry. Combining these techniques provides an improvement over single‐marker studies, allowing for a more nuanced understanding of how social and ecological variables are linked to immune measures and disease risk in diverse populations and settings.
1. INTRODUCTION
The immune system is fundamental to understanding health. Immune function measures are useful for assessing a broad range of characteristics, at both the population‐level (i.e., infection prevalence (Blackwell et al., 2011); maintenance of immunity [Demonbreun et al., 2021]) and individual‐level (i.e., informing clinical care [Cals et al., 2010]). Immune characterization is also central to studies investigating life history allocations (Blackwell et al., 2010; McDade et al., 2008; Urlacher et al., 2018), behavioral responses (Cepon‐Robins et al., 2021), and interactive effects of infection on other disease risks (Schneider‐Crease et al., 2021). However, capturing the complexity of the immune system and its function outside of a clinical setting presents challenges, primarily arising from limitations on sample collection, preservation, and transport (Blackwell et al., 2021). Blood is the primary tissue used to analyze immune function, but blood collection from venous samples typically requires a trained phlebotomist and participants comfortable providing venous blood. As an alternative, finger prick blood collection is easily accomplished with limited training (McDade et al., 2007). With practice, a finger prick can typically provide ~250 to 500 μl of blood, especially with high‐flow lancets (we prefer the blue BD Microtainer Contact‐Activated Lancets #366594; Becton, Dickinson and Company). By using small samples and preservation methods such a drying, freezing, or mixing with stabilizers, sample transport from finger prick samples is simplified, relative to large venous draws or analyses with fresh blood. Finger prick blood can even be collected and mailed by participants themselves (e.g., Demonbreun et al., 2021).
The immune system is complex. It is composed of a wide array of cells, signaling molecules, and effector molecules that exist or act in coordination to prevent or limit infection. Consequently, there are a large number of parameters that studies might measure when seeking to quantify the immune system. For some studies, single measures may be sufficient. For example, C‐reactive protein (CRP) levels are useful for capturing a snapshot of systemic inflammation and infection (Black et al., 2004; McDade et al., 2012). However, single measures tell us little about interactions between the immune and other systems, or more abstract constructs like immunocompetence, immunosenescence, and immunological strategies or trade‐offs. Additionally, circulating levels may not always be reflective of immunological response to infection; measuring immune responses to stimuli—rather than levels at a single, arbitrary time point—may sometimes be more informative.
Here we evaluate current state‐of‐the‐art methods for measuring immune function in “non‐clinical” or field settings using blood collected from a minimally invasive finger prick. First, we provide a brief introduction to the components of immune function which might be measured, and discuss the theoretical and scientific contexts in which these measures might be useful. Next, we briefly discuss point‐of‐care devices which give immediate results in the field, and then focus the remaining bulk of this review on methods for collection and preservation of finger‐prick blood samples and their subsequent quantification for immune measures including immunoassays of dried blood spots (DBSs), flow cytometry, RNA sequencing, DNA quantification and sequencing, and direct, experimental measures of immune response. After reviewing these, we also briefly discuss prospects for measuring immune function in other biospecimens such as urine, saliva, and feces and discuss further considerations for sample collection and analysis.
2. APPROACHES TO IMMUNE FUNCTION
2.1. Aspects of immune function
The immune system is extraordinarily complex and a full accounting of its components and functioning is outside the scope of this review. (A number of excellent textbooks are available on this subject, such as [Parham, 2021]). However, we briefly summarize some major components, which are also listed in Table 1.
TABLE 1.
Measures of immunity and their applications
| Measure | Description | Proxy for | Short‐term variation a | Long‐term variation a |
|---|---|---|---|---|
| C‐reactive protein | Acute phase reactant | Infection; chronic inflammation | High | Medium |
| Cytokines | Immune signaling molecules | Infection; chronic inflammation | High | Medium |
| Leukocytes | All white blood cells | Infection; immune investment | High | Medium |
| Helper T cells | Recognize antigen and activate killer T cells and B cells | Cell mediated adaptive immunity; extracellular defense | Low | Medium |
| Cytotoxic T cells | Trigger apoptosis in cells infected by viruses | Cell mediated adaptive immunity; intracellular defense | Low | Medium |
| Natural Killer cells | Trigger apoptosis in cells infected by viruses; may defend against parasites | Innate cell mediated immunity | Low | Medium |
| B cells | Produce antibody | Cellular and humoral immunity | Low | Medium |
| Naïve T cells, sjTREC | Pool of cells with randomly generated receptors, from which those recognizing novel threats will be selected | Thymic output; investment into defense against future threats; immunosenescence | Low/medium | High |
| Eosinophils/ Basophils | Respond to macroparasite infections and contribute to allergy | Endo or ectoparasite infection and responses; allergic responses | Medium | Medium |
| Neutrophils | Phagocytic cells that migrate to infection sites to attack microbes | Infection; immune investment | High | Medium |
| Monocytes | Phagocytic cells that migrate to infection sites | Chronic inflammation; low levels in blood may indicate migration into tissues to fight infections | High | Medium |
| Regulatory T cells | Moderate other immune responses | Immunological tolerance; regulation of autoimmunity and allergy | Low | High |
| IgG | Long‐term antibodies to past infections | Humoral immunity; long term protection | Low | Low |
| IgE | Antibodies related to parasite defense and allergy | Humoral immunity; endoparasite exposure; allergy | Low | High |
| sIgA | Secreted antibody in saliva, milk, and other fluids | Secretory immunity | Medium | Medium |
| IgM | The first class of antibody produced after a new infection | Humoral immunity; recent infection | High | Medium |
| Gene expression | mRNA transcripts from particular genes; representative of production of the related gene product | Depends on the gene and analysis method. See Table 2. | Generally high but varies by gene | Generally medium but varies by gene |
| Gene methylation | Methylation of regulatory regions of genes, affecting mRNA expression levels | Depends on the gene and analysis method. | Generally low but varies by gene | Generally medium but varies by gene |
| Immune response assays | In vitro response to an antigen, mitogen, foreign erythrocyte, or microbe | Ability to respond to infection | Unknown | High |
Assessment of short‐term within individual lability (e.g., due to infection) versus long‐term differences between individuals or populations. Actual differences for any study will depend on context, for example, the degree of variation in exposures, genetic diversity in the study population, comparison groups, and so forth. Selected reference for variation in these measures: (Blackwell et al., 2011, 2016; Braun et al., 2018; Campisi et al., 2012; Garcia, Trumble, et al., 2020; Lorenzi et al., 2008; Mcdade et al., 2012; Ou et al., 2011; Petrovsky & Harrison, 1998; Sbiera et al., 2011; Urlacher et al., 2018).
The immune system consists primarily of specialized white blood cells (leukocytes) and their secreted products, as well as some components secreted by other tissues for the purposes of signaling or immune defense. These components are typically transported by and measured in the blood but exist throughout the tissues of the body. Note, however, components in the blood and tissues actually represent the last line of defense against parasites and pathogens, as there are many other barriers to infection that include behavioral avoidance and response (the behavioral immune system), physical barriers (such as skin), and secreted immune components at points of entry into the body (the mucosal immune system).
Leukocytes can be broken down into six main subsets: neutrophils, lymphocytes, basophils, eosinophils, monocytes, and macrophages. One subset, lymphocytes, can further be categorized into three main subtypes (T cells, B cells, and natural killer cells), with several distinct classes to each of these cell types. Humoral immunity consists of immunoglobulins (antibodies) secreted by B cells, the main classes of which are IgG (the predominate type), IgM (produced in response to new infections), IgE (a response to macroparasites), and IgA (most commonly investigated in its secreted form, sIgA, as the immunoglobulin in saliva, milk, and other secretions). A wide range of signaling molecules called cytokines (e.g., interleukins, chemokines) are secreted by leukocytes and other cells in the body. Finally, effector molecules (e.g., nitric oxide), members of the complement cascade, and inflammatory proteins such as CRP are secreted by various tissues and participate in immune defense and regulation of other responses.
2.2. Categorizing aspects of immune function
Given the complexity of the immune system, there are a number of ways to categorize its components for the purposes of simplifying reasoning and analysis. One common schema divide between innate and adaptive immunity (Blackwell et al., 2010; Garcia, Blackwell, et al., 2020; McDade, Georgiev, & Kuzawa, 2016). Innate immunity does not require experience with a pathogen, while adaptive immunity is acquired through exposure. Adaptive immunity primary involves T and B cells (as well as immunoglobulins), which undergo recombination of their receptors to generate novel variants capable of recognizing new pathogens. Some approaches further divide adaptive responses into cell‐mediated and humoral responses, reflecting the differing functions and costs of leukocytes versus antibodies (McDade, 2003, 2005a). However, the term cellular immunity is also often used to encompass all cellular responses, both innate and adaptive, and the term humoral immunity sometimes refers to all factors in the serum, not just antibody (Ruoss et al., 2019).
Other approaches group aspects differently, for example a pattern of gene expression dubbed the conserved transcriptional response to adversity (CTRA; Cole, 2019) has components representing antibody response, type 1 interferon responses (viral responses), and general proinflammatory responses. Approaches also sometimes differentiate between constitutive defenses and induced defenses (Heinrich et al., 2017; Schmid‐Hempel & Ebert, 2003). Still other approaches look at responses to specific classes of infection, for example, viral versus bacterial stimuli or indicators of responses specific to parasitic infections (Blackwell et al., 2010; Harrison et al., 2019; Trumble et al., 2016).
2.3. Tradeoffs and defense portfolios
Classifying aspects of immunity is important from both a theoretical and a practical perspective. Many studies in ecoimmunology and evolutionary anthropology use life history theory as an organizing framework. Life history theory examines the trade‐offs organisms make between competing demands on time, energy, and other resources. Most life history approaches decompose these demands into the categories of growth, reproduction, and maintenance, with the maintenance category encompassing tissue repair, immune response, and other factors.
Studies examining trade‐offs between immune function and other life history demands are often most interesting in quantifying broad measures of immunity, for example, proxies for immunocompetence or measures associated with the overall cost of immunity. However, most researchers recognize that immunocompetence is not unidimensional. Different individuals or populations may be more resistant to one kind of threat than to another. Thus, a more nuanced approaches examines trade‐offs between competence or investment in different categories of immune response, or in specific immune responses to specific threats. Other approaches think of the suite of investments and individual has as a defense portfolio (Schmid‐Hempel & Ebert, 2003), or as an immunophenotype, 1 which can be thought of as a snapshot of an individual's immune system allocations (McDade, Georgiev, & Kuzawa, 2016).
3. THE RIGHT MEASURES FOR THE QUESTION
An important consideration in designing a study investigating immune function is to measure the right aspect or aspects of immune function for the question under investigation. A well‐designed study cannot simply pick the most convenient measure and assume it represents “immune function,” since there are many facets to the immune system which may not be correlated or for which correlations may depend on context. We therefore review some general areas of inquiry and the markers researchers have used in those areas, before turning to practical concerns and field friendly methods.
3.1. Life history allocations, energetics, and individual variation
Central to the field of ecoimmunology is the study of how the energetic demands of immune defense and activation trade‐off against other demands such as growth and reproduction. Thus, the measures that are most appropriate for these studies are measures that (1) vary between individuals or populations and (2) are indicative of the energetic demands of immunity. In some contexts, indicators of acute infection may be most relevant, for example, CRP, while in others indicators of chronic infection or immune activation might be better, for example, concentration of immunoglobulin E (Blackwell et al., 2010; McDade et al., 2008). As we might expect from the lability of these measures, these indicate investments on different timescales, and thus trade‐offs on different timescales. Urlacher et al. (2018) found that CRP, which varies on short timescales, was associated with growth over a 1‐week period, while IgG was associated with growth over a 3‐month period, and IgE with growth over much longer periods.
Leukocyte counts are also likely to represent energy investments, since these cells are costly to manufacture and operate (Straub et al., 2010). However, some cell types may be costlier than others, particularly those involved in adaptive immunity, due to the need to maintain a wide repertoire of cells and antibodies with different receptors. For example, Garcia, Blackwell, et al. (2020) found that T cells and B cells were negatively associated with child height‐for‐age. Naïve subsets of adaptive cell types, or markers of thymic output like sjTREC may further be interpreted as investment into future immune defense, since these represent the production of new cell lines in preparation for new infections.
3.2. Immunosenescence and immunocompetence
Measures related to immunosenescence may also be useful for comparing individuals and making life history inferences. Naïve cell counts and sjTREC may be useful in this regard, as well as counts of senescent T cells (Franceschi et al., 2000). Methylation assays have been used to develop a so‐called “epigenetic clock” related to immune aging (Horvath et al., 2016).
Finally, many measures are intended to compare individuals in terms of immunocompetence—the hypothetical capacity of an individual to respond to a randomly encountered pathogen. The most direct assays of immunocompetence are functional assays such as antigen stimulation assays and bacterial killing assays. However, we caution that for the most part the results of these assays have not been directly validated against real‐life morbidity and mortality from illness. It is also probably impossible to create a single, global measure of immunocompetence since individuals will differ in response to different pathogens (Schmid‐Hempel & Ebert, 2003).
In animals, measures of immune response, such as the hemolysis‐hemagglutination assay (HHA), have been used as measures of immunocompetence and related to sex, reproductive status, and other factors (Gilot‐Fromont et al., 2012; Ruoss et al., 2019). These are usually used in concert with other measures such as cell counts and antibody levels in order to provide a multifaceted assessment.
3.3. The role of inflammation in disease
In the biomedical literature measures of inflammation have gained prominence due to their associations with what tend to be considered chronic diseases of modernity such as obesity, heart disease, and type‐2 diabetes (Gurven et al., 2009, 2016; Hersoug & Linneberg, 2007; West‐Eberhard, 2019). These are often straightforward to collect given their ease of measurement in DBSs. Measures usually include CRP and inflammatory cytokines such as IL‐6 and TNF‐α. However, these measures are quite labile and care must be taken to distinguish between baseline levels and acute elevations due to infection. Inflammatory markers may need to be interpreted differently in high‐ and low‐pathogen contexts: in low‐pathogen contexts baseline elevations of these markers are indicative of nonspecific inflammation and a variety of chronic diseases, whereas in high‐pathogen contexts these are more likely to indicate recent or ongoing acute infections (McDade et al., 2012).
3.4. Old friends, hygiene, and mismatches with ancestral environments
Complementary to the study of diseases of modernity is the study of how exposure to parasites and pathogens may regulate immune responses and limit disease. These studies are informed by the old friends and hygiene (i.e., biodiversity) hypotheses and often focus on the mismatch between modern and ancestral environments (Blackwell, 2022). These studies often involve measures of the disease state in question, often linked to either excess inflammation or to misdirected immunity, for example, autoimmunity or allergy. Measures of inflammation have been noted above, and for information on measures of autoimmunity, see Cepon‐Robins (2021). Additionally, measures related to the regulation of immunity are important for this area of study, including counts of regulatory T cells through flow cytometry and measurement of cytokines characteristic of Th1 or Th2 immune responses (e.g., IL‐10, IL‐4, IFN‐γ). Parasite exposure can be indirectly inferred through immune responses to parasites, including IgE levels, eosinophil counts, and the expression of genes related to both (Table 2).
TABLE 2.
Selected genes whose expression may be useful as immunological biomarkers
| Category | Gene name | Description | CTRA a |
|---|---|---|---|
| Cell type | CD3D | CD3 (T cell marker) | |
| CD4 | CD4 (helper T cell marker) | ||
| CD8A | CD8 (killer T cell marker) | ||
| FCGR3A | CD16 (neutrophil and natural killer cell marker) | ||
| CD14 | CD14 (monocyte marker) | ||
| CD19 | CD19 (B cell marker) | ||
| NCAM1 | CD56 (natural killer cells) | ||
| Humoral | IGHM | IgM heavy chain | |
| IGHG1/2/3/4 | IgG1 heavy chain 1/2/3/4 | ||
| IGHE | IgE heavy chain | ||
| CRP | C‐reactive protein | ||
| IGJ | Joining chain of Multimeric IgA and IgM | Antibody | |
| IGLL1/3 | B cell development | Antibody | |
| Cytokines | IFNG | IFN‐γ | |
| IFNB1 | Interferon Beta 1 | Type 1 IFN Antiviral | |
| TNF | TNF‐α | Pro‐inflammatory | |
| IL1A | Interleukin‐1A | Pro‐inflammatory | |
| IL1B | Interleukin‐1B | Pro‐inflammatory | |
| IL4 | Interleukin‐4 | ||
| IL6 | Interleukin‐6 | Pro‐inflammatory | |
| CXCL8 | Interleukin‐8 | Pro‐inflammatory | |
| IL10 | Interleukin‐10 | ||
| Eosinophil | EPX | Eosinophil peroxidase | |
| RNASE3 | Eosinophil cationic protein | ||
| CCL11 | Eotaxin (Eosinophil chemokine) | ||
| Pro‐inflammatory | PTGS1/PTGS2 | Prostaglandin‐endoperoxide synthase | Pro‐inflammatory |
| FOS/B, FOSL1/L2, JUN/B/D, NFKB1/2, REL/A/B | Transcription factors | Pro‐inflammatory | |
| Transcription factors | Pro‐inflammatory | ||
| Transcription factors | Pro‐inflammatory | ||
| Transcription factors | Pro‐inflammatory | ||
| Antiviral | GBP1, IFI16, IFI27, IFI27L1/2, IFI30, IFI35, IFI44, IFI44L, IFI6, IFIH1, IFIT1/2/3, IFIT5, IFIT1L, IFITM1‐3, IFITM4P, IFITM5, MX1/2, OAS1/2/3, OASL | Interferon induced proteins | Type 1 IFN antiviral |
| IRF2/7/8 | Interferon regulatory factors | Type 1 IFN antiviral |
Genes associated with one of the three conserved transcriptional response to adversity dimensions (Cole, 2019).
3.5. Stress, social inequality, status
Studies examining the regulation of immunity in relation to social environment have used several different approaches. Studies of chronic stress or inequality often focus on inflammatory markers, as above, as these relate to chronic diseases which are often associated with economic and social inequality. A number of studies (e.g., Dieckmann et al., 2020; Levine et al., 2017; Powell et al., 2013) have examined patterns of RNA expression associated with the “conserved transcriptional response to adversity” (Cole, 2019), which is characterized by upregulation of pro‐inflammatory responses and downregulation of type I interferon antiviral response genes and antibody synthesis genes (Table 1). Measures related to the reactivation of latent retroviruses such as Epstein–Barr virus and cytomegalovirus have also been used as proxy measures for stress related immunosuppression (Glaser et al., 1991; McDade et al., 2000). However, anti‐EBV antibody is an indirect measure and findings have been mixed (see below).
3.6. Choosing the right marker
In choosing biomarkers, often researchers will face a trade‐off between scientific and practical concerns. For example, the best measure is often not the most field friendly or cost‐effective. Thus, choosing the right measure is a matter of picking something that is a close proxy for what you really want to investigate, but which fits within logistical and financial constraints. Table 1 gives a general overview of the aspects of immunity we discuss in this review, as well as some general ideas for the theoretical concepts these might proxy. In the following sections we discuss more of the practical details for each of these measures when used in the field.
4. POINT OF CARE DEVICES
Point‐of‐care devices are typically small, portable devices that provide immediate results. When available, point‐of care devices have a number of advantages over other approaches since they eliminate the need for sample storage and transport and allow the researcher to provide immediate results to participants and local health professionals. To date, however, point‐of‐care devices are available for only a very narrow range of immune markers, and some of these devices have significant drawbacks.
For quantifying peripheral blood cells, there are two main point‐of‐care devices on the market that are compact enough to be convenient for field work. quantitative buffy coat (QBC) dry hematology analyzers (Autoread Plus and STAR; Drucker Diagnostic, Inc) provide a 2‐part differential with nine parameters: hematocrit, hemoglobin, MCHC, white blood cell count, granulocyte count, lymphocyte/monocyte count, and platelet count. The QBC analyzers requires 70 or 111 μl of capillary or venous blood (tube type dependent). Once blood is collected (or pipetted) into a capillary tube, a float is inserted, and the tube is then centrifuged in a system‐specific capillary centrifuge. The differing densities of blood components will cause them to separate into layers during centrifugation. This is optimized by the addition of the float—which physically expands the cell and platelet layers, and a metachromatic acridine orange stain that the capillary tubes are coated with which uniquely dyes the cell types. The resulting layers can then be read by the analyzer using a dual light source and optical detector (https://druckerdiagnostics.com/qbc-autoread-plus-dry-hematology-system/). The QBC is generally stable under hot and humid conditions and does not require refrigerated or liquid reagents. It does require AC electrical power but can be run off a battery with an AC adapter. The QBC provides high consistency across repeat measures taken by different individuals (Laney et al., 2019). Currently the QBC system (analyzer and centrifuge) runs around $10 000. Tests kits run about $8 per sample.
The HemoCue WBC System (Hemocue AB) provides only total white blood cell (WBC) counts. It offers some advantages over the QBC as it is smaller in size, has battery‐powered capacity, and requires less blood (10 μl vs. 70–111 μl). In controlled conditions the HemoCue WBC works well and provides high correlations with reference methods (Osei‐Bimpong et al., 2009). However, in our experience with this system in the field we found that it often fails when temperature and humidity are high. We learned a few hacks that did not affect the read reliability—like keeping the HemoCue WBC in a cooler with ice packs between tests—however these are less than ideal and data loss can still occur with higher than acceptable frequency. The HemoCue WBC system costs around $2000, while test cuvettes cost around $3.25 per sample. The same company also offers the HemoCue WBC Diff, which provides a 5‐part differential (neutrophils, lymphocytes, monocytes, eosinophils, and basophils). It is not currently available in the United States, and we have not had an opportunity to test it. Some anthropologists have successfully used this device in the field (Page et al., 2016). However, other reports in the literature suggest it may be somewhat inaccurate with finger prick samples (Karawajczyk et al., 2017).
Apart from white blood cell differentials, the only analyte directly relevant to immune function that can now be measured by a point‐of‐care device is CRP, using the NycoCard Reader II (Abbott). However, it has a measuring range of only 8 to 200 mg/L, making it functional for detecting acute infections (typical CRP >10 mg/L) but ineffective for detecting lower‐level variation in CRP, which is often of interest for assessing cardiometabolic risk, recent infection, or testing theories from eco‐immunology and developmental origins of health and disease. It is also not currently available in the United States. A number of other point‐of‐care devices are available for clinical settings, some of which measure high sensitivity CRP (hs‐CRP), but they are generally much larger. Given that CRP can be cheaply and easily measured in DBSs (see next section) we generally prefer this approach.
In addition to these general measures, there are a number of point‐of‐care options for diagnosing specific ailments. Most of you reading this (in 2022) are by now familiar with rapid tests for detecting SARS‐CoV‐2. Whether these tests will remain available or will be effective for future variants after the COVID‐19 pandemic remains to be seen. Rapid tests are also available for influenza A & B, for example with the BD Veritor Plus System (Becton, Dickinson and Company) which is around $300, with tests running around $12 each. Test kits are also available to use this system for respiratory syncytial virus (RSV), SARS‐CoV‐2, and Group A Streptococcus. The supplier Thermo Scientific makes rapid tests under the Xpect brand which can detect Flu A & B, RSV, C. difficile, rotavirus, giardia, cryptosporidium, and legionella, some of which work with nasal or throat swabs and other fecal samples. However, all are fairly expensive (around $40 per test). These kind of point‐of‐care rapid tests are likely to always be less sensitive than PCR analysis. If immediate results are not necessary, studies have shown that nasal swabs are stable for at least 21 days at room temperature and probably longer if refrigerated, at least for SARS‐CoV‐2 (Skalina et al., 2022). These can therfore be stored for later PCR analysis.
5. DRIED BLOOD SPOTS
DBSs are a favorite for minimally invasive measurements of immune function. Relative to other methods, they are easy to collect, easy to store, and easy to transport. The use of DBS has been reviewed extensively elsewhere (McDade et al., 2007) so we will highlight only a few key measures. DBS are best utilized for measuring the levels of stable and prevalent protein products, while more care must be used in measuring proteins that are less concentrated or less stable, for example many cytokines. Some examples of stable proteins for which enzyme‐linked immunosorbent assays (ELISAs) of DBS are particularly useful include:
5.1. Immunoglobulins
The major subclasses of immunoglobulins (IgG, IgA, IgE, and IgM) are robust against freeze–thaw cycles and are easily measured in DBSs with only slight modifications to commercially available kits (Tanner & McDade, 2007). In general, this means that DBS can be used to measure not only total immunoglobulin levels, but also specific antibodies against things such as Epstein–Barr virus capsid antigen (EBV), which is commonly measured as an index of stress‐related reactivation of latent virus and possibly T‐cell suppression (Glaser et al., 1991; McDade et al., 2000). However, anti‐EBV antibody may be a more complicated measure than is typically appreciated (e.g., Cacioppo et al., 2002) and many studies, particularly those including other biomarkers, do not find associations between EBV and other stress‐related measures (e.g., Panter‐Brick et al., 2020; Rudzik et al., 2014) raising the possibly of publication bias for positive findings. Specific antibodies against a number of pathogens have been measured in DBS, for example, Toxoplasma gondii (Lebech & Petersen, 1995), malaria (Corran et al., 2008), and measles (Colson et al., 2015). In principle antibodies should have similar properties in DBSs regardless of target, so adapting ELISAs devised for measuring immunoglobulins in serum for use with DBSs is often possible. DBSs can also be used to measures immunoglobin subtypes, that is, sub‐classes of IgG (Andersen et al., 2014).
5.2. C‐reactive protein
CRP is a favorite measure in human eco‐immunology given its stability and the availability of established ELISA protocols (i.e., Brindle et al., 2010; McDade et al., 2004). However, these protocols were published a number of years ago, and some of the antibodies used by them are no longer available commercially. The capture antibody used by Brindle et al., 2010 is currently available from Meridian (clone C5 anti‐CRP MAb, M86005M), however, the detection antibody is no longer listed. An alternate detection antibody (M01239M) is given as pairable with M86005M and will likely work similarly to the original antibody from the protocol. However, we would recommend validating this or any other replacement antibody pair before using them to analyze DBS since it is always possible for confirmational changes, elution buffer differences, or matrix effects to make a DBS assay behave differently from serum assays. DBS measures of CRP have been used extensively in a number of papers (e.g., Blackwell et al., 2010; Urlacher et al., 2018).
5.3. Cytokines
Cytokines are small molecules used for signaling between cells of the immune system. Because each cytokine has its own molecular structure, cytokines differ significantly in their stability at room temperature, loss due to freeze–thaw cycles, and recovery from DBS. One study found that interleukin (IL)‐6 and IL‐10 are stable in serum through freeze–thaws, but levels of IL‐4, IL‐13, IL‐15, IL‐17, tumor necrosis factor (TNF)α, interferon (IFN)γ and CXCL8 all changed with freeze–thaw cycles (De Jager et al., 2009). Skogstrand et al. (2005) used a Luminex xMAP assay to measure 25 cytokines and other biomarkers in DBS. The results show that in principle many cytokines can be measured in DBS and that cytokines in DBS are stable when kept frozen at −80°C, but the study did not compare values in plasma to DBS or assess the effect of freeze–thaw cycles. A second study (Skogstrand et al., 2008) assessed stability at room temperature, 35 and 4°C and found that a few cytokines, such as IL‐8, IL‐10, and TNF‐α appear to be relatively stable, even up to 30 days at room temperature, but the majority were not. Another concern is that the variability in cytokine levels due to differences in collection or storage is not consistently directional (i.e., values may go up or down). There may be a variety of reasons for this including differential degradation of cytokine molecules and release of cytokines from intracellular locations, binding molecules, or the DBS matrix. The study did find that one advantage of DBSs is that cytokine levels change rapidly in venous blood that is kept at room temperature or refrigerated, as the living cells in the blood become activated by the novel environment (Keustermans et al., 2013; Skogstrand et al., 2008). Since DBS are usually collected and dried immediately this is less of an issue.
More recently McDade et al. (2021) used a multiplex electrochemiluminescent immunoassay to simultaneously measure IL‐6, IL‐10, IL‐8, and TNF‐α in matched DBSs and plasma. They obtained good correspondence for IL‐6, IL‐10, and TNF‐α, but poor correspondence for IL‐8. They speculate this may be due to IL‐8 stored in red blood cells which is released on lysing, and which would not be present in plasma. An ELISA protocol for just IL‐6 has also been validated, with good correlation between DBS and serum (Miller & McDade, 2012).
In conclusion, DBSs seem to be reliable for the measurement of higher concentration, higher stability cytokines such as IL‐6, IL‐10, and TNF‐α. However, caution may be needed when measuring other cytokines in DBS, particularly those with lower concentrations or shorter half‐lives. An alternative may be to measure cytokine related gene (mRNA) expression (see below).
5.4. CD4 lymphocytes
For many assays where serum equivalents are desired the inclusion of red and white blood cells may be a nuisance. However, the fact that DBS contain whole blood presents an opportunity as well. Mwaba et al. (2003) tested an ELISA kit for measured CD4 protein in DBS and showed a reasonable correspondence between estimated CD4 counts from this method, and counts obtained by flow cytometry. Likely estimation of other cell types using ELISAs and DBS is possible, though we are not aware of publications validating these methods.
5.5. Single versus multiplex platforms
ELISAs can be used to measure many single analytes in DBS. ELISAs have the advantage of using relatively common equipment (an ELISA plate reader, at the minimum) and having a lower cost than multiplex assays. DBS kits typically cost around $300 to $700 for a 96‐well plate, good for running about 35 samples in duplicate, after accounting for standards and controls. Multiplex kits may be more than five times as expensive as ELISA kits. However, when measuring many analytes multiplex assays become more efficient, in terms of labor, sample used, and cost. However, multiplex assays also sometimes sacrifice sensitivity, particularly for lower concentration analytes, since they must compromise on assay dilutions that are effective for the many things being measured, which may differ in concentration by orders of magnitude. Common multiplex platforms include the MagPix xMAP instrument (Luminex Corp) which uses magnetic beads with bound antibodies, instruments from Meso Scale Diagnostics which use electrochemiluminescence, and flow cytometry‐based bead assays such as the Luminex 200 System and BD Cytometric Bead Array kits (Becton, Dickinson and Company) which can be used with any number of flow cytometers. However, only a handful of multiplex assays have been validated with DBS (McDade et al., 2021; Skogstrand et al., 2008), so other assays will require protocol development for use.
5.6. Complications for DBS samples
Although DBSs are convenient, recent studies have shown that there are a number of ways to introduce errors into blood spot analyses. In addition to the aforementioned effects of freeze–thaws and storage temperatures, humidity may also impact certain analytes measured in DBS. DBSs differ from serum measures due to the inclusion of white and red blood cells, which lyse, and the filter paper matrix itself which may create assay nonlinearities. At a minimum, the analysis of whole blood, rather than serum, will dilute concentrations by a factor of about 2 to 2.5, equivalent to the proportional volume of red blood cells in blood (hematocrit). Because of this and other factors, many protocols will include conversion equations for estimated serum equivalent values from DBS results. However, in the absence of data on serum equivalence it is problematic to compare DBS and serum values, so caution is needed. Most commercial kits are developed for analysis of serum and thus it is also inadvisable to perform DBS assays without validating the protocol against serum, given the issues noted above. Considerations and requirements for protocol validation has been described in a number of papers (e.g., McDade et al., 2007; Sultana et al., 2022). Additionally, the use of small blood spots, overlapping drops, smeared drops or the edges of blood spots can create issues, as these all affect the concentration per unit of filter paper and may also affect matrix effects and elution (Crimmins et al., 2020). Finally, because most commercial assay kits are not designed for DBS, maintaining consistency in the kit manufacturer used across a study sample is necessary. This means that if a researcher shifts to a different antibody or assay manufacturer (e.g., if one brand goes off the market) the burden is on them to do a new validation. To ensure continuity and comparability of biomarker data across a study sample, researchers should purchase enough antibody or kits for their entire study, while also needing to be aware of expiration dates. Planning out the materials needed and timeline to conduct laboratory analyses are as important as planning the data collection itself.
6. FLOW CYTOMETRY
Flow cytometry is used to analyze the relative proportions of different white blood cells by tagging cells with fluorescently marked antibodies for specific cell‐surface (or sometimes interior) proteins. Cells are suspended in a salt‐based buffer and single cells are then rapidly analyzed as they flow past a single (or set of) laser(s). Cells are analyzed based on the presence or absence of each tag, and the visible light scatter, which roughly corresponds to size and shape of the cell.
Blackwell et al. (2016) used flow cytometry to characterize leukocyte subtypes in participants as part of the Tsimane Health and Life history Project (THLHP). For that project, a small Accuri C6 (Becton, Dickinson and Company) flow cytometer was taken to Bolivia for use in the THLHP field clinic (Figure 1D). In this study, venous blood was collected from participants and analyzed fresh using flow cytometry. While analyzing fresh samples is ideal and it is certainly more convenient to avoid transporting samples, this setup presents a number of challenges. First, a flow cytometer must be available for transport, which precludes the use of core facilities. Second, reagents require refrigeration, and may be unavailable in country, and thus require temperature‐controlled transport and storage. Third, if the instrument requires maintenance, it may be difficult to obtain service in country (team members took ours from Bolivia to Chile for service). Finally, a clean area with regular power is needed for analyses.
FIGURE 1.

Methods for measuring immune function and collecting samples. (A) Dried blood spots covered by a net to prevent mosquitos from landing (photo: Lawrence Sugiyama). (B) Field antigen stimulation, in which a candle is lit and then sealed in a plastic container to deplete oxygen and increase carbon dioxide. (the jaguar skull looks cool but has not been shown to improve results). (C) a liquid nitrogen shipping tank for sample transport. (D) THLHP field setup for flow cytometry and antigen stimulation
Given the difficulty of this procedure, we recently validated a field protocol for freezing finger prick blood samples in the field for later flow cytometry (Blackwell et al., 2021). The procedure utilizes a liquid nitrogen (LN2) storage or shipping tank, sometimes called a dewar (Figure 1C), which can keep samples frozen without electricity for a month or more. Such tanks can be bought from scientific suppliers like Thermo Scientific, but may also be sold in some countries for the purpose of livestock breeding (e.g., sementanks.com). The key is to freeze samples in the vapor phase, which freezes at a slower rate and causes less lysing than freezing in the liquid. In short, the steps are as follows:
Prepare aliquots of 20% DMSO/80% RPMI 1640 medium. These may be prepared early and stored frozen in the LN2 tank.
Collect blood from a finger prick in in a heparin coated tube, such as the BD Microtainer Capillary Blood Collector (Becton, Dickinson and Company).
In a cryotube, mix 100 ul blood +100 ul RPMI/DMSO. Do not invert to keep blood from getting stuck in the cap. Instead, gently tap, vortex, or pipette to mix.
Do not allow to sit for more than 5 min before freezing
Freeze in the vapor phase of a LN2 tank with ~1 inch LN2 in the bottom, using only the top two cane positions in the tank, or in a LN2 shipper with the liquid absorbed.
After ~6 to 24 h, move to liquid phase in second tank
Frozen blood can be transported in the liquid nitrogen shipping tank or on dry ice and then stored in a freezer at −80°C. When ready to analyze, cells are thawed in a water bath and quickly washed with fresh medium (full protocol details are available in Blackwell et al., 2021).
Overall, analyses with previously frozen cells produce equivalent results to those with fresh cells (Figure 2). However, there are some notable exceptions. First, freezing lyses granulocytes more than lymphocytes, and may alter the lymphocyte percentage in samples. Thus, measuring white blood cell differentials at the time of collection with a point of care device (see above) can be very useful if absolute differential counts are desired. Major lymphocyte subsets (i.e., CD4 T cells, CD8 T cells, natural killer cells, and B cells) can be measured accurately in frozen cells. However, some minor subsets, such as the percent naïve and senescent were altered with freezing. Additionally, some antibody‐fluorochrome combinations performed poorly with previously frozen cells. We recommend testing these on frozen control samples before analyzing precious field samples.
FIGURE 2.
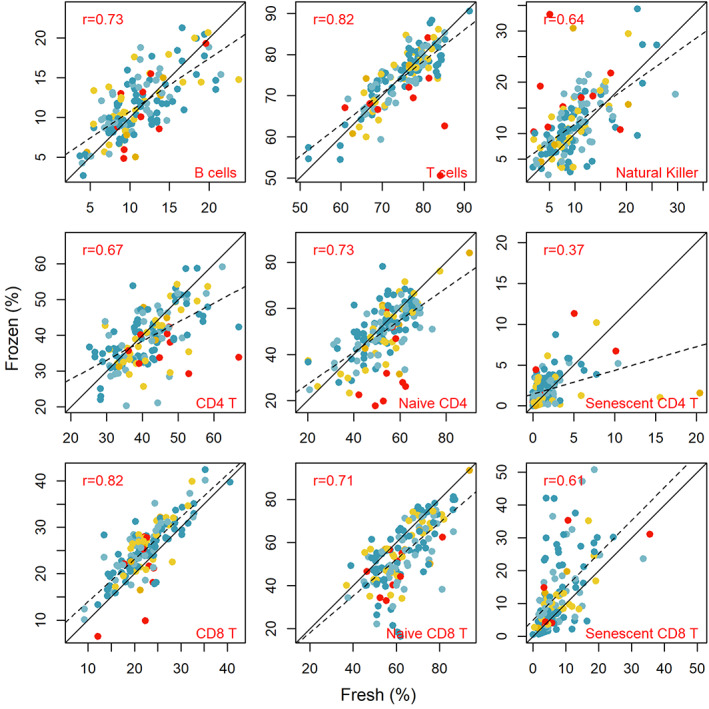
Percent of CD45+ cells or percent of respective T cell subset (CD4 or CD8 for naive and senescent cells) in matched fresh and frozen samples analyzed by flow cytometry. Black line is the 1:1 slope. Dashed line is the best linear fit line. Reprinted from (Blackwell et al., 2021). See original reference for further details
Apart from the cost of the flow cytometer, expenses for flow cytometry can be minimized by using small samples and no more reagents than are necessary. Typical flow cytometry protocols call for 100 μl of blood and 5 μl of each labeled antibody. However, we have had good results with as little as 10 μl of blood and 0.5 μl of each antibody per sample, cutting the sample required and expense by a factor of 10 (Blackwell et al., 2021).
7. RNA SEQUENCING
RNA sequencing (RNA‐seq) allows for the quantification of mRNA transcripts and is used to compare gene expression between individuals or experimental conditions. RNA‐seq relies on next generation sequencing (NGS), a technology that sequences multiple DNA fragments (reverse transcribed from mRNA) in parallel. Because most immunological processes of interest require mRNA transcription for protein synthesis, gene expression levels are likely to be correlated with many other biomarkers and are informative of a wide range of processes. As such, RNA sequencing may prove a more cost effective and informative method than direct measurement of isolated biomarkers.
RNA‐seq data can be analyzed in several ways, for example, to infer broad patterns of immune investment, to infer the representation of different leukocyte proportions, or to infer cytokine or antibody production (Table 1). Cell proportions can be inferred through RNA transcripts for canonical cell type markers, for example, CD14 for monocytes, CD3D and CD3E for T‐cells, CD4 for helper t‐cells, CD8A for cytotoxic t‐cells, CD19 for B cells, CD16 (FCGR3A) for neutrophils and monocytes, and CD56 (NCAM1) for natural killer cells. They can also be inferred through transcript origin analysis which uses full profiles of gene expression in multiple tissues (e.g., Cole et al., 2011; Palmer et al., 2006; Su et al., 2004).
Functional genomics also uses RNA‐seq data to study gene and protein expression and function on a global (genome‐ or system‐wide) scale. Research by Cole (2019), focusing on individuals exposed to chronic psychosocial threats has led to the identification of a pattern of gene expression that associates with such experiences. This pattern, referred to as the CTRA, involves upregulation of pro‐inflammatory responses and downregulation of type I interferon antiviral response genes and antibody synthesis genes (Table 1). Differences in patterns of gene expression across these markers has been increasingly used to investigate the biological effects of psychosocial stress (Dieckmann et al., 2020; Levine et al., 2017; Powell et al., 2013).
Finally, with network‐based approaches RNA‐seq data can be used to assess the effects of different environmental exposures on the function of biological pathways. For example, PathOlogist, an analytical tool developed by Greenblum et al. (2011), utilizes pathway structure to integrate biological information, and assesses differences in activity and consistency states of networks of genes within pathways of known biological processes and functions. While such network‐based genomic approaches tend to be used to assess differences by disease state (e.g., gene expression profiles in healthy versus tumorous tissue), applying them to questions relevant to eco‐immunology opens opportunities for in‐depth investigation of how the environment influences biologic pathways and processes related to life history processes (e.g., cellular energy metabolism or immune investment).
7.1. Isolating RNA from finger‐prick blood samples
Typical RNA‐seq blood collection protocols, such as the PAXgene Blood RNA extraction system (Qiagen, Inc) call for the collection of 2.5 ml venous blood into PAXgene vacutainers. PAXgene documents report that 2.5 ml blood yields ≥3 μg RNA for ≥95% of samples (mean ~ 8 μg) (PreAnalytiX., 2020). However, RNA sequencing requires only 50 to 100 ng RNA for evaluation of transcripts, and only 500 to 1000 ng for de novo transcriptome assembly, suggesting that 2.5 ml produces roughly 60× the minimum RNA yield needed for 3′ sequencing and that less blood could be collected.
Krawiec et al. (2009) developed a protocol for isolation of RNA from small samples of blood, to be used with mice who cannot spare 2.5 ml. Although not yet validated with human blood, for mice 50 μl of blood yielded 2300 ng of RNA, more than sufficient for sequencing. ADB also successfully collected and extracted RNA from great‐tailed grackles, with yields from 50 μl blood averaging 3152 ng of RNA. While birds have nucleated red blood cells, mice and humans have roughly similar white blood cell counts, suggesting results for humans may be similar to mice. While reported RNA yields from PAXgene documents suggest possible yields as low as 60 ng from 50 μl blood, other references suggest typical RNA levels for humans are actually higher than this, with a mean of 15 μg/ml (range 7–23 μg/ml) (Chomczynski et al., 2016), which would yield 750 ng from 50 μl. Lower yields in some PAXgene protocols may in fact be due to a ceiling effect from the RNA spin columns used, with higher recovery possible for lower volumes of blood (Krawiec et al., 2009).
Together, these results suggest that blood collected via finger‐prick should produce usable yields for RNAseq in humans. We plan to validate this procedure in the near future, but prior to that we recommend verifying RNA yields before using this protocol. An example finger‐prick collection protocol, modified from the protocols used for mice and birds, might read as follows: (1) Aliquot PAXgene reagent into smaller tubes at a ratio of 2.76 reagent/blood (e.g., 138 μl PAXgene/50 μl blood); (2) Pierce the fingertip with a sterile lancet and collect blood into a finger‐prick blood collection tube; (3) pipette 50 μl of blood from the collection tube into the PAXgene aliquot; (4) store at room temperature for at least 2 h (no longer than 72 hrs), then freeze. PAXgene aliquots may be frozen at −20C, −80C, or placed in liquid nitrogen. PAXgene‐preserved samples are also stable across at least three freeze–thaws, making them ideal for easy transport. Extraction and analysis should follow standard protocols.
7.2. Isolated RNA from DBS
Multiple authors have successfully sequenced RNA from DBS (McDade, Ross, et al., 2016; Reust et al., 2018). In these studies, relative transcript reads derived from DBS were highly correlated with PAXgene derived values and yielded largely similar associations with predictors, though with some discrepancies. Most of these discrepancies may be due to the small amount of RNA recovered from DBS. For example, McDade, Ross, et al. (2016) did not quantify RNA yields since these were expected to be too low for accurate quantification. The authors note that due to the increased noise from a small RNA sample, a sample size ~5% greater than in PAXgene studies might be required to maintain statistical power. These studies require a large number of blood spots, in the range of two to four complete blood spots (McDade, Ross, et al., 2016) or 8 × 3 mm2 punches (Reust et al., 2018). Overall, DBS are likely to be simpler to store and transport, but small liquid aliquots with PAXgene are likely to yield better results.
7.3. Sample preparation and sequencing for RNA‐seq
Sequencing is typically done either by reading from the 3′ end of each cDNA transcript, which only partially reads the transcript, or by fragmenting the mRNA into smaller pieces for whole transcript sequencing before reverse transcribing into cDNA. Each approach has its own advantages and disadvantages, which we will briefly detail in the next sections (for a more thorough comparison see Ma et al., 2019). For both methods, after collecting the sample, RNA is isolated and purified, usually with a kit such as the RNeasy kit (Qiagen).
Due to the high volume of RNA in red blood cells, globin‐based reads can consequently comprise a large proportion (>80%) of sequencing reads. Thus, when preparing whole blood for 3′ RNA sequencing, it may be advisable to include a globin depletion step in the protocol. However, globin reads can also be removed from analyses after sequencing, and depletion may introduce biases to other reads (Harrington et al., 2020). Consequently, there are costs and benefits to globin depletion with the primary benefit being a reduction in sequencing depth and expense, and the cost being an increase in protocol complexity and read bias. Globin reads may also sometimes be of interest.
For whole transcript sequencing an enrichment step is commonly added after RNA isolation and purification to remove ribosomal RNA repeats which would otherwise swamp the sequencing and be uninformative for many purposes. Some protocols also enrich for RNAs with poly‐adenine tails, which helps to eliminate immature RNAs which may contain introns that have not yet been spliced out. These steps are not necessary for 3′ sequencing, as 3′ sequencing only targets the poly‐adenine tails at the 3′ ends of mature mRNAs. After enrichment the RNA is converted to complementary double‐stranded DNA (cDNA) via a reverse transcriptase enzyme and prepared for sequencing.
Whole transcript sequencing generates more cDNA fragments for longer genes, and consequentially more reads for these genes, relative to shorter genes. In contrast 3′ sequencing sequences from the 3′ end of the mRNA. Since mRNA is not fragmented, gene length does not influence sequencing reads and the number of reads is proportional to the original mRNA transcripts. Ma et al. (2019) compared the two methods, and as expected found that 3′ sequenced performed better for shorter transcripts (particularly <1000 bp) because they receive equal reads. Many genes of interest for quantifying immune function are small genes, for example, cytokines (e.g., the mRNA for IL‐10 is approximately 500 bases). 3′ can also be substantially cheaper than whole transcript sequencing (Sholder et al., 2020). There are, however, advantages of whole transcript sequencing. First among these is that whole transcript sequencing allows for the discovery and counting of alternative splice variants and noncoding RNAs. Depending on the study aims this may or may not be relevant.
Another important consideration—which depends on the sequencing technique and the question—is sequencing depth. For RNA‐seq, sequencing depth refers to the total number of reads obtained in a sequencing run. For example, if a gene of interest is lowly expressed, it may appear in only one out of a million transcripts. If a sequencing depth of 5 million is used, on average there will be 5 reads, but with a 95% confidence interval ranging from 1 to 10 reads, leading to an estimated relative expression of between 2 and 20 per 107 reads. Increasing sequencing depth to 20 million narrows this range to 6 to 14.5 per 107 reads. A main disadvantage of higher sequencing depth is increased cost.
The required read depth also depends on whether one is doing whole transcriptome sequencing or expression profiling sequencing (i.e., 3′ sequencing). Because only a single fragment is generated for a transcript with 3′ sequencing, this method attains a similar coverage as whole transcriptome sequencing at a fraction of the depth, and there are diminishing returns for increasing depth past ~10 million reads for 3′ sequencing (Moll et al., 2014). Thus, depending on the study design and budget a researcher might have to choose between sequencing many samples at low depth or few at high depth. One analysis of this question found that sequencing more replicates at lower depth provided more power than sequencing fewer replicates at higher depth (Liu et al., 2014). Thus, if the object of a study is to compare two groups of individuals, a lower sequencing depth and a greater sample might be preferred. If, however, it is essential to quantify the gene expression for a particular individual (rather than a group of individuals) a greater sequencing depth might be more important.
7.4. Post‐sequencing processing and analysis workflow
After samples are sequenced (i.e., “read”) raw read data go through multiple processing steps before they are converted to raw count data, and ready for downstream analysis. Post‐sequencing processing steps include quality assessment, transcript assembly, alignment, analysis of raw counts, and annotation. For the trainee interested in integrating RNA‐seq into their project, an important consideration is that each of these steps relies on bioinformatic tools and pipelines, which require a level of fluency and comfort with programming languages (e.g., R, Python) and statistical software and packages. As an example, here is an abbreviated description of an RNA‐seq pipeline for samples sequenced on an Illumina sequencing platform (full pipeline available at https://github.com/heinin):
Raw RNA‐seq data exist as FASTQ files, which contain the short sequence reads and quality scores. The quality of the FASTQ files is assessed with a tool called FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/)
The raw sequence reads often contain adapter sequences that were used to create the Illumina sequencing libraries. These adapters and low‐quality bases at the ends of each read are trimmed using Trimmomatic (https://anaconda.org/bioconda/trimmomatic). Trimming improves read mapping and affects all downstream analyses.
After trimming, the FASTQ quality is assessed again, until quality checks are passed, after which we continue with read mapping. The trimmed reads are mapped to the human reference genome using HISAT2 (https://ccb.jhu.edu/software/hisat2/index.shtml) (this is one of many tools for DNA and RNAseq alignment). The result is one SAM file (Sequence Alignment Map) for each sample. The SAM file is converted to a binary version using SAMtools (http://samtools.sourceforge.net/; https://samtools.github.io/hts-specs/SAMv1.pdf), and these BAM files are sorted based on genomic coordinates (also with SAMtools). Most tools that take BAMs as input files require them to be coordinate sorted.
Next, the expression levels of genes are obtained by quantifying the read counts mapping to each gene. One commonly used tool is called featureCounts (http://bioinf.wehi.edu.au/featureCounts/). For each sample, the output is a text file containing the read counts in each gene. These files can then be combined to create a count table with samples in columns and genes in rows, and that count table is used in downstream analyses (e.g., differential expression).
For researchers new to RNAseq, the learning curve for bioinformatic processing can be steep but may be worthwhile depending on your approach to a research question. There are a number of resources that describe workflows, pipelines, and workflow managers, including https://bioinformatics.uconn.edu/resources-and-events/tutorials-2/rna-seq-tutorial-with-reference-genome/ and https://rnaseq.uoregon.edu/. The Biostar Handbook is also an invaluable guide to bioinformatics that is updated multiple times a year and has topic‐specific (https://www.biostarhandbook.com/). The learning curve can also be buffered by teaming up with other researchers that are experts in the use of a particular method(s), as they can guide you through parts of the process and direct you to relevant resources. It is also possible to have a majority of these steps done “behind the scenes” (e.g., among the services offered by a Core Lab), though we suggest familiarizing yourself with the overall process in order to better understand the data you are working with, including its limitations.
7.5. Statistical analysis
The statistical approach for downstream analysis of count data will depend on the specific goals and research question being asked and is thus outside the scope of this review. Here we briefly discuss the process of count normalization and its importance for the interpretation of RNA‐seq data in field studies. In both whole transcript sequencing and 3′ RNA‐seq reads must be normalized for between sample variation in library size, that is, the total of all reads for a sample. For whole transcript sequencing counts are also normalized for gene length because gene length affects the number of cDNA fragments produced and thus the number of reads mapped to a particular gene; this is not usually necessary for 3′ sequencing because only one fragment is generated per transcript (Moll et al., 2014). Because library size is more likely to reflect variation in sequencing depth due to methodological stochasticity, rather than real‐world variation in total mRNA expression, counts can be normalized in a number of ways which reflect different assumptions (Evans et al., 2018).
One approach is to normalize to total library size, which converts values into counts per million reads (CPM). When using whole blood rather than isolated PBMCs normalized values will reflect the total proportion of all transcripts in all blood cells including red blood cells which contain RNA from a large number of genes (Kabanova et al., 2009). If red blood cell globin transcripts are depleted (either before sequencing or by removing these reads after sequencing) the interpretation of CPMs will change to represent the proportion of nonglobin reads. However, in some circumstances, normalization by read count may lead to faulty conclusions if the total mRNA differs between samples for biological reasons; for example, if some prevalent genes are upregulated in one sample this could instead be interpreted as a downregulation of the remaining genes, since these transcript proportions would decrease. To avoid this issue, a number of other normalization procedures are possible, including normalizing or controlling for sample RNA concentration while also normalizing for library size. Other approaches include normalization for housekeeping genes (genes thought to be expressed at relatively constant levels across cells), normalization based on distribution (quantile normalization), normalization based on cell‐type specific genes, and normalization based on cell counts measured independently in the sample (Dillies et al., 2013; Eisenberg & Levanon, 2013; Evans et al., 2018).
The choice of normalization procedure should depend on: (1) the initial sample (e.g., whole blood), (2) whether the RNA was enriched before sequencing, (3) whether the desired end product is mRNA/cell, mRNA/leukocyte, mRNA/total transcript, mRNA per volume of blood or some other value. These values will have different interpretations, for example, mRNA/leukocyte may be most appropriate for inferring leukocyte differentials, while mRNA/volume of blood may speak more to cell counts or systemic expression levels for the whole organism. Like most statistical analyses, there are numerous ways to approach normalization and we strongly advice investigating the advantages and constraints of each method before deciding which method is most appropriate for answering a specific question. There are a number of studies that describe the details of each method and provide useful insights on appropriate choice of RNA‐seq normalization and analysis methods (Abrams et al., 2019; Evans et al., 2018; Zyprych‐Walczak et al., 2015).
In sum, the decision to incorporate RNA‐seq into one's research should be driven by a triangulation between the question, the cost, and the learning curve. All specific methods and analyses will follow from the question.
8. DNA‐BASED ASSAYS
8.1. Thymic output
Like RNA, it is also possible to extract DNA from DBS for analysis. In addition to genotyping, there are several assays of interest when measuring immune function. When T cells mature, they undergo a process wherein the DNA coding for the T‐cell receptor is randomly rearranged to create a unique receptor for each T cell lineage. Leftover DNA from this process is excised from the T cell receptor arrangement and circularized into DNA molecules called signal joint TCR excision circles (sjTRECs). New T cells leave the thymus with one or two sjTREC molecules, and as they mature and divide these become distributed among the daughter cells (Lorenzi et al., 2008). As such, sjTREC levels in the blood directly correspond to thymic output, that is, production of new naïve T cell lines by the thymus. sjTREC levels correlate with naïve cell counts measured in the blood, but imperfectly so, as naïve cells may also replicate in the periphery (though divergences are themselves informative) (den Braber et al., 2012). Thymic output declines with age in a consistent enough fashion that sjTREC has been used to estimate age from bloodstains (Ou et al., 2011; Ou et al., 2012). However, thymic output can also be temporarily upregulated during infections (Permar et al., 2003) and is influenced by various immunological (Cho et al., 2017) and physiological (Tarcic et al., 1998) conditions. These divergences from general age patterns are probably of the most interest in eco‐immunological studies, as they are useful measures of variation in immune investment. However, this means that analyses using sjTREC need to consider how to parse out infection or exposure‐related variation from effects due to initial thymic size and typical age‐related decline.
Quantitative PCR (qPCR) is used to quantify sjTREC DNA, as well as levels of a housekeeping gene for normalization. To quantify sjTREC in DBS, DNA is first extracted with a kit such as the QIAamp 96 DNA Blood Kit or Gentra Puregene Tissue Kit (Qiagen); Qiagen also provides an adapted protocol specifically for DBS. Extracted DNA samples are analyzed for quantity and quality with a spectrometer (e.g., NanoDrop, Thermo Scientific.). qPCR is then performed on an instrument such as the Bio‐Rad CFX96 (Bio‐Rad Laboratories) using appropriate dyes and primers for sjTREC and any normalization genes (see Lang et al., 2011 for primer sequences).
Lang et al. (2011) used ALB2 for initial normalization, to obtain a ratio of sjTREC per total nucleated cells. They also quantified the unrearranged configuration of the VD junction region (VD‐J) of the TCR‐β, which is the version present in non‐T cells. By subtracting the estimate of non‐T cells from total cells they were able to obtain an estimate of T cells, and ultimately calculate an sjTREC/T cell ratio. As with the RNA‐seq assays above, the choice of normalization alters interpretation. If peripheral cells replicate, for example in response to infection, then the sjTREC/T cell ratio will go down, even though thymic output might actually increase. Thus, some advocate for measuring sjTREC/ml (Lorenzi et al., 2008). An alternative approach when blood volume is not precise is to measure sjTREC/cell by PCR and use a point‐of‐care device to measure total leukocyte count. Multiplying sjTREC/cell times cell count will yield sjTREC/ml.
8.2. Lymphocyte cell subsets
As should be apparent from the sjTREC protocol above, qPCR can also be used to estimate the proportion of non‐T cells (and by extension T cells) in a sample. Though we know of no validations, a similar approach is likely possible for estimating B cells using primers for the non‐rearranged heavy and light immunoglobulin chains. Assessment of rearrangements is currently described for monitoring of B cell disorders (Cho et al., 2017). Measurement of non‐rearranged sequenced should be considerably simpler.
8.3. Methylation
DNA methylation can be measured in minimally invasive blood samples, like DBS, but can also be measured in buccal swabs or saliva (Non & Thayer, 2015). However, because buccal swab and saliva samples contain epithelial cells as well as white blood cells, blood may be the best sample for measuring immune function directly. Methylation is stable in DBS for considerable lengths of time, even at room temperature (Sasaki et al., 2020), and protocols for measuring methylation in DBS are similar to those for fresh blood (Walker et al., 2019). DNA is first extracted with a DNA purification kit before analysis. Methylation analyses may look at global methylation, may compare groups or individuals for differentially methylated genes, or may look at methylation of specific genes. The choice of assay method will differ depending on which of these approaches a study takes; methylation assays may be done through sequencing, mass spectrometry, the reading of microarrays, or through qPCR (Bock et al., 2016; Kurdyukov & Bullock, 2016; Non & Thayer, 2015). We refer the reader to the citations referenced here for more information.
For the purposes of eco‐immunology, methylation may be useful for examining particular immune related genes, particularly with regard to chronic effects due to lifestyle (Ligthart et al., 2016; McDade et al., 2019; Nakajima et al., 2010). Methylation is part of the regulatory pathway for gene expression, and may be important, for example, in TH1/TH2 balance (Brand et al., 2012). Patterns of methylation have also been found to associate with levels of CRP (Sun et al., 2013). However, methylation is not the only factor in gene expression as transcription factors and other regulators make many short‐term adjustments (Makar & Wilson, 2004). Thus, for measuring expression at a particular timepoint the measurement of RNA expression or protein product level is probably complementary. Similar to RNA‐seq data, methylation results can also be used to infer underlying cell subset proportions and thus may be another useful alternative to flow cytometry (Titus et al., 2017).
9. IMMUNE RESPONSE
The methods described above measure the state of the immune system at the time a blood sample is taken. However, interpretation of such measures is sometimes difficult. Does a high value indicate an immune system more prepared to face an infection? Or does it represent the consequence of an infection that evaded initial immune defenses and triggered a more widespread response? In some cases, the difference between baseline immune levels and activated responses can be identified, but in others they cannot. For this reason, it is sometimes informative to measure immune responses to experimental stimuli. In humans, in vivo tests are occasionally possible, usually in the case of vaccination campaigns in which response to a vaccine is measured. However, generally in vitro stimulations provide more flexibility and are more feasible.
9.1. Antigen stimulation
Antigen stimulation assays involve mixing blood with an antigen or mitogen and incubating in medium, followed by measurement of the response to the treatment. Common antigens include lipopolysaccharide (LPS), a cell wall component of gram‐negative bacteria that initiates B‐cell division and activation of antigen presenting cells, and phytohemagglutinin (PHA) a mitogen that activates T cells; other studies have also used antigens ranging from fragments of helminths (van Riet et al., 2009) to H1N1 flu vaccine (Schneider‐Crease et al., 2021). Typical protocols call for incubation in a CO2 enriched incubator at 37°C. However, May, van Bodegom, et al. (2009) developed a field‐friendly method for whole blood stimulation that does not require the CO2 tank. They created a CO2 enriched environment by sealing the samples in a plastic container containing water and a burning candle. The candle depletes the oxygen and enriches the CO2 in the contained before going out (Figure 1B). This procedure has been successfully carried out for a number of studies (May, van Den Biggelaar, et al., 2009; Schneider‐Crease et al., 2021; Trumble et al., 2016), however, because it still requires the ability to maintain samples at 37°C—usually in an incubator—it is not the most field‐friendly protocol. The incubation time ranges from 24 to 72 h, depending on whether the assay is designed to capture initial, innate responses, or slower adaptive immune responses.
Antigen stimulations are usually done with 100 μl of whole blood per sample, so it is feasible to conduct them with finger‐prick collections. However, the blood must be fresh (ideally stimulation is done within an hour of collection) and cannot be frozen. After stimulation, the supernatant is collected and frozen (which necessitates a freezer or liquid nitrogen tank). The supernatant is typically transported frozen and assayed for cytokines using ELISA or multiplex assays. Cytokine levels in stimulated supernatants are usually much higher than they are in circulating blood samples. In principle, therefore, assays could be developed which avoid freezing, for example, by spotting stimulated samples on filter paper, or by preserving stimulated cells with PAXgene reagent and performing RNA‐seq instead of cytokine measurement (e.g., Meitern et al., 2014; Pulido‐Salgado et al., 2018).
One important caveat about antigen stimulation assays is to remember that the assays measure the response to stimulation, but not how effective that response is in fighting the infection. Cytokines are signaling molecules and measuring them is equivalent to measuring how loud the immune system yells when it feels threatened. Indeed, in some cases strong inflammatory responses may be detrimental or represent a failure of specific responses, so care in interpretation is needed.
9.2. Bacterial killing assays
Bacterial killing assays involve in vitro introduction of microbes to blood, plasma, or saliva in a dish or well. Microbes such as Candida albicans, Escherichia coli, and Staphylococcus aureus can be obtained as lyophilized pellets and reconstituted as needed. Liebl and Martin II (2009) report on a procedure designed for small birds that uses only 1.5 μl of plasma or blood. The sample is diluted in medium and mixed with the microbe solution, then incubated for 30 min to 1 h at a temperature optimal for the particular microbe used. The microbes are then diluted in nutrient medium and incubated for an additional 12 h. The bacterial concentration is then measured with a small Nanodrop Spectrophotometer (Thermo Scientific). As an alternate measurement, bacteria can also be spread on plates and the colonies counted (Millet et al., 2007). Since bacterial killing largely depends on the action of whole cells which are likely to be lysed or stressed by freezing it is unlikely to produce useful results from frozen blood, though we are not aware of protocols that have directly evaluated this.
9.3. Hemolysis‐hemagglutination assay
The HHA is an assay utilized by field biologists to measure complement and natural antibody activity via the ability of plasma to lyse or agglutinate foreign red blood cells. Natural antibodies are antibodies produced without previous exposure to an antigen and which are genetically encoded rather than requiring gene rearrangement to produce, while the complement system consists of a number of small molecules that bind to invaders and trigger further responses to clear infections. The HHA assay is similar to the hemolytic complement assay (HCA) developed in the 1960s to measure complement activity in humans and aid in diagnosing complement related disorders (Kirschfink & Mollnes, 2003; Mayer, 1961; Morgan, 2000; Rapp & Borsos, 1970). However, it is simpler to perform in the field with small volumes and results that are read by eye or with a scanner or camera, rather than a spectrophotometer. The HCA assay uses antibody‐sensitized sheep erythrocytes to evaluate the classical complement pathway and rabbit erythrocytes to assesses the alternative complement pathway; the HHA does not distinguish lysis by pathway, however it does assess agglutination which requires natural antibodies.
The HHA was first developed in birds, in which bird plasma was treated with a sample of rabbit blood (Matson et al., 2005), but has since been used for mammals, with chicken blood replacing rabbit blood for the assay (Gilot‐Fromont et al., 2012; Heinrich et al., 2017; Ruoss et al., 2019). To the best of our knowledge the HHA has not yet been used to assess human immune response, but it is use in other mammals suggests it should produce results. As little as 10 μl of plasma is serially diluted in PBS into 11 wells in a 96‐well plate. A 1% suspension of chicken red blood cells is added. Plates are incubated at 37°C for 110 min and then photographed or scanned to measure agglutination due to natural antibodies and then again after 70 more minutes to measure lysis, which is caused by complement. The scans are scored manually based on appearance. As with the other response assays, this assay requires an incubator or other method of maintaining a controlled temperature. Most assays use commercially available red blood cells, which may be difficult to obtain and maintain in the field. However, it is likely possible to use fresh erythrocytes which have been washed and resuspended in an appropriate buffer. Given the relatively complexity of performing the assay in the field, it is possible to use frozen plasma for the assay. Most protocols caution against freeze–thaw cycles, however evaluations of freeze‐thaw cycles on complement activity show that these have little effect (Hegemann et al., 2017; O'Shaughnessy et al., 2012; Yang et al., 2015).
10. FURTHER CONSIDERATIONS
10.1. Technical variation (batch effects)
For a number of assays, samples are analyzed in a series of “batches.” For example, ELISA and microarray assays are typically run on 96‐well plates that hold anywhere between ~36 and 96 samples, depending on protocols; qPCR assays are run on a 384‐well plate, and other genomic assays that use NGS technologies (e.g., RNA‐seq) are run on flowcells that have between one and eight lanes, and can analyze up to 400 samples. For ELISAs, metrics like the coefficient of variation (CV) are typically used describe variation within and between plates. The CV is the ratio of the standard deviation to the mean (typically multiplied by 100 to report as a percentage). Within each plate the CV is assessed for each sample, run in duplicate, to identify idiosyncrasies, many of which are due to pipetting errors or samples that are on the high or low end of the assay range. Samples with high CVs (>10%) are usually run again, sometimes with a different dilution. “High” and “low” controls, which have a known quantity of the analyte being measured, are included in most assay kits and should be run on each plate. CVs for the control duplicates are used to report variation across plates, due to minor differences in handling and processing. This noise is known as “technical variation.” It can be minimized through careful laboratory technique, but some variation in usually unavoidable. In some cases, it may be useful to statistically control for batch in the data analysis step, however this is only feasible if batch is not correlated with another variable of interest. For example, if using two plates to compare samples from two villages, it would be better to run each plate with a mix of samples from both villages, rather than one plate for each village, in order to minimize confounding of village and batch.
Unfortunately, the practice of including something like a “control” in next‐gen sequencing assays has yet to be adopted, and it remains unclear how best to accomplish this given the way NGS operates. There can be a significant amount of variation between batches due to technical factors in RNA‐seq and other NGS data. There are some statistical packages (e.g., Combat‐seq, variancePartition [Hoffman & Schadt, 2016; Zhang et al., 2020]) that quantify sources of variation that may be used to adjust the substantive NGS data for better comparability. Still, it is important to try to sequence all samples for a specific project together to limit such batch effects.
10.2. Age, sex, and reproductive status
Many immunological values vary with age, sex, and reproductive status. Age‐related patterns are apparent in many measures due to development and late‐life senescence (e.g., Blackwell et al., 2011, 2016; McDade, 2005b). Studies should evaluate whether it is appropriate to use age‐standardized z‐scores or to otherwise control for age. Note, however, that many age‐related changes are not strictly linear, so linear terms in regression models may not be sufficient. Differences by sex and reproductive status (e.g., menstrual cycle, pregnancy) are also apparent (Aghaeepour et al., 2017; Hové et al., 2020; Klein & Flanagan, 2016; Natri et al., 2019; Wander et al., 2008) and so studies should take these into account as part of study design and analysis.
10.3. Diurnal pattens
Many papers studying immune function do not report or control for time of blood collection. However, many components of the immune system vary diurnally, with many being lower in the morning and higher in the afternoon, opposite the diurnal rhythm of cortisol (Ackermann et al., 2012; Labrecque & Cermakian, 2015; Petrovsky & Harrison, 1998). Diurnal regulation of immunity may be relevant for many research questions (e.g., Garcia, Blackwell, et al., 2020), but even when it is not the focus, time of sample collection is an important variable that should be captured and accounted for in study designs.
10.4. Chronic versus acute elevations
Some immunological measures are likely to be much more variable within an individual. McDade et al. (2012) analyzed repeat samples of CRP in an Amazonian population. Even though 35% had elevated CRP at one timepoint, no participant had more than one elevated measure, indicating that these were acute elevations rather than chronic elevations. Blackwell et al. (2016) calculated intra‐class correlations for 19 immune measures based on repeat samples. Some measures, such as IgE concentration, CD4 counts, and CD8 counts were relatively stable across repeat measures, while others such as neutrophil count and natural killer cell count appeared more labile. The stability of a measure should also be considered in study design (repeat vs. single measures) and interpretation (see Table 1).
10.5. Capillary versus venous blood
For the most part, measures from capillary and venous blood correlate strongly. However, some differences have been noted (Sitoe et al., 2011; Srisala et al., 2019). Study designs should consider whether this is relevant to a particular study design and may wish to validate against serum measures.
10.6. Measures in urine or feces
Blood is the optimal tissue in which to measures systemic immunity, since blood distributes white blood cells, cytokines, and other factors throughout the body. However, if blood is unavailable, what, if any, immunological measures are available? The short answer is: it is difficult to measure systemic immunity in other fluids. The long answer is that while many things can be measured, care must be taken in interpretation. This is because markers in urine or feces are much more indicative of localized bladder or renal (in the case of urine) or gut (in the case of feces) infections than systemic immune allocations. Higham et al. (2015) compared serum measures with several fecal and urinary measures. They found that while serum and urinary neopterin were correlated (r = 0.664), fecal concentrations were not correlated with serum. Serum CRP was not correlated with urinary or fecal CRP levels, but there was a short‐term rise in fecal and urinary CRP following a surgery. Serum and urinary haptoglobin were also not correlated, and haptoglobin could not be measured in feces.
10.7. Measures in saliva and breastmilk
Many studies have measured secretory IgA in saliva, breastmilk, and other secreted substances (e.g., Hodges‐Simeon et al., 2017; Miller & McConnell, 2011; Van Anders, 2010). However, this is probably best interpreted as a marker of mucosal immunity, a complex system in its own right. Caution should be used when interpreting whether secretary IgA is indicative of overall immune status since mucosal immunity is not necessarily correlated with systemic immunity, and can be indicative of better health or worse health depending on the circumstances (Drummond & Hewson‐Bower, 1997; Gornowicz et al., 2014; Koss et al., 2016; Nakamura et al., 2006; Reid et al., 2001; Thaweboon et al., 2008). Cytokines can also be measured in saliva, but salivary cytokine levels are generally uncorrelated with serum levels (Riis et al., 2014) and may be indicative of periodontal disease (Riis et al., 2014). Saliva is also a source of white blood cells, which might be used for RNA‐seq, methylation, or other assays. The concentration of leukocytes in saliva is ~1.6 × 105 cells/ml (Vidović et al., 2012), meaning ~2 ml of saliva would be equivalent to 50 ul blood. However, cells in saliva are only ~60% leukocytes with the remaining 40% being buccal cells (Theda et al., 2018), which may change the interpretation of these assays. Salivary leukocytes are also affected by gingivitis and other oral conditions, and in general proportions differ from blood, with monocytes overrepresented. Because of these differences, further validation is needed to understand how best to interpret measures in saliva, and how they correlate (or perhaps trade‐off) with measures in blood.
11. CONCLUSIONS
Many studies have relied on single measures of immunity to draw broad conclusions about the regulation of immune function and how immune function trades‐off with other life history demands. However, immunity is multifaceted and single measures are unlikely to accurately capture the full range and plasticity of human immune function. Luckily, the repertoire of methods for the minimally invasive measurement of immune function is rapidly expanding. These measures differ in the aspects of immunity they measure, as well as in the inferences that can be made based on them (Figure 3). They also differ in cost and complexity, which are summarized in Table 3. Traditional measures, such as ELISAs for DBSs remain the simplest and cheapest to implement. These measures are accurate and easy to implement but are limited in scope since they typically measure only one analyte at a time. Multiplex immunoassays overcome this limitation, but usually with a cost to sensitivity, which may be particularly detrimental to measures of low‐concentration analytes such as cytokines. Point‐of‐care devices are often the most convenient because they require no sample transport. However, sensitive point‐of‐care devices are only available for a few measures, though many more will likely become available in the relatively near future.
FIGURE 3.
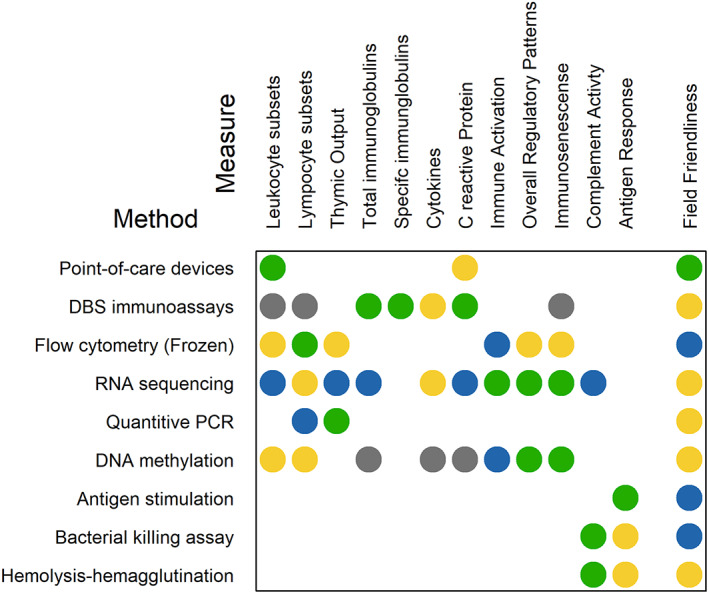
Summary of ecoimmunology methods. Colors indicate our subjective assessment of the suitability of a method for a particular measure. Green = optimal measure. Yellow = good measure but may be error prone, difficult, or indirect. Blue = indirect measure that can still be used to infer values. Gray = untested or unvalidated, but perhaps possible. For field friendliness: Green = immediate results, no sample transport; yellow = sample transport or equipment needed; blue = equipment needed and or difficult sample handling
TABLE 3.
Key equipment, expenses, and references for each method
| Assay type | Major equipment | Cost of equipment a | Cost per sample b | Expertise | References |
|---|---|---|---|---|---|
| Point of care | |||||
| White blood cell differential | QBC hematology analyzer; microcapillary tube centrifuge | $9–$14 k | $6–$8 | Minimal | Laney et al., 2019 |
| HemoCue WBC system | $2.5 k | $3–$4 | Minimal | Osei‐Bimpong et al., 2009; Karawajczyk et al., 2017 | |
| Influenza A & B | BD veritor system plus | $300 | $20 | Minimal | Hassan et al., 2014 |
| Immunoassays | |||||
| Immunoglobulins | Microplate reader; software | $6–$56 k | $14–$18 | Intermediate | Tanner & McDade, 2007; Andersen et al., 2014 |
| C‐reactive protein | Microplate reader | $6–56 k | $1–$15 | Intermediate | McDade et al., 2004; Brindle et al., 2010 |
| Cytokines (single) | Microplate reader | $6–56 k | $9–$15 | Intermediate | McDade et al., 2007, 2021 |
| Cytokines (multiplex) | Multiplex reader | $40–$80 | Intermediate | Skogstrand et al., 2005; McDade et al., 2021 | |
| CD4 Lymphocytes | Microplate reader; software | $6–56 k | $12–20 | Intermediate | Mwaba et al., 2003 |
| Flow cytometry | |||||
| Sample collection | Liquid nitrogen tank/dry shipper | $300–$1200 | $2 | Intermediate | |
| Analysis | Flow cytometer | $40–$60 k | $3–$10 | Intermediate | Blackwell et al., 2021 |
| Nucleic acid assays | |||||
| Collection, extraction | Qubit fluorometer | $3200 | $20–40 | Intermediate | Krawiec et al., 2009 |
| 3′ QuantSeq | Next generation sequencer | $100–$500 k | $40–$100 | Advanced | Liu et al., 2014; Moll et al., 2014; Zyprych‐Walczak et al., 2015; McDade, Ross, et al., 2016; Evans et al., 2018; Reust et al., 2018; Abrams et al., 2019; Cole, 2019; Ma et al., 2019; Harrington et al., 2020; Sholder et al., 2020; https://rnaseq.uoregon.edu/ |
| Whole transcript RNA‐Seq | Next generation sequencer | $100–$500 k | $200–$400 | Advanced | |
| sjTREC, lymphocyte subsets | qPCR machine | $15–90 k | $2–$10 | Intermediate | Lorenzi et al., 2008; Lang et al., 2011; den Braber et al., 2012; Cho et al., 2017 |
| DNA methylation | Pyrosequencer; qPCR; mass spectrometer; microarray scanner c | $15–$500 k | $50–$1000 | Advanced | Non & Thayer, 2015; Bock et al., 2016; Kurdyukov & Bullock, 2016; Sasaki et al., 2020 |
| Immune response | |||||
| Antigen stimulation | Incubator | $500–$2000 | $2–$5 d | Intermediate | May, van Den Biggelaar, et al., 2009; van Riet et al., 2009; Trumble et al., 2016; Schneider‐Crease et al., 2021 |
| Bacterial killing | Nanodrop spectrophotometer; incubator | $12–$17 k | $5–$10 | Intermediate | Liebl & Martin Ii, 2009 |
| HHA | Flatbed scanner; incubator | $500–2000 | $5–$10 | Intermediate | Matson et al., 2005; Gilot‐Fromont et al., 2012; Heinrich et al., 2017; Ruoss et al., 2019 |
Note: Cost per sample is an estimate as of time of writing and excludes cost of major equipment. Expertise needed is coded as follows: minimal = able to be performed by a novice, with some practice on elements of protocol; intermediate = bench‐based laboratory training; statistical analysis; advanced = some bench‐based laboratory training, fluency in software and programming, statistical packages, computational analysis.
More expensive equipment is often available at core facilities for a recharge fee, which is usually much more cost effective than purchasing for an individual lab.
Excludes the cost of equipment.
There are several different methods, each using different equipment.
Does not include the cost of immunoassays to measure cytokines.
Apart from these practical concerns, we have given a general overview of the theoretical contexts in which different biomarkers may be useful. However, biomarkers are never just markers but are part of the biological system in which they are embedded. As such these should not be used as proxies without considering how their meaning might vary with context and what the implications of an interpretation are for the underlying biological process.
At present we are most excited by measures that allow for broad characterizations of immunity. RNA‐seq and DNA methylation, in particular, can be used in an exploratory fashion, but also with targeted analyses that can be used to infer cell compositions, cytokine expression levels, and many other aspects of immunity. The cost of sequencing continues to decline precipitously, while sensitivity and analysis tools increase. That said, many analyses require further validation, for example, against gold standard methods such as flow cytometry for immunophenotyping of leukocytes. RNA‐seq and methylation analysis are also limited in their ability to differentiate expression by cell type, which can be done with flow cytometry. Single‐cell RNA‐seq is becoming commonplace (Luecken & Theis, 2019), yet for field studies single cell analysis runs into the same limitation that flow cytometry has, namely the need to preserve and transport blood without cellular lysing.
The measures we catalog here are primarily intended to measure human immune function. However, these methods have much in common with those used in other species (e.g., Boughton et al., 2011; Prall & Muehlenbein, 2014). Methods designed for small animals like mice and birds are easily translatable to finger prick samples of capillary blood from humans. Point‐of‐care devices designed for medical professions are also likely to become increasingly useful in the future. The suite of tools available for eco‐immunology in the field continues to rapidly expand meaning that future studies will increasingly be able to integrate multiple measures of immunity with less effort.
ACKNOWLEDGMENTS
Thanks to Josh Snodgrass and Geeta Eick for organizing this special issue and the AAPA session on which it is based. We also thank two anonymous reviewers for their helpful comments.
Blackwell, A. D. , & Garcia, A. R. (2022). Ecoimmunology in the field: Measuring multiple dimensions of immune function with minimally invasive, field‐adapted techniques. American Journal of Human Biology, 34(11), e23784. 10.1002/ajhb.23784
Endnote
To avoid confusion, we also note that the term immunophenotype also often refers to the phenotype of individual immune cells, as determined by flow cytometry or other methods.
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- Abrams, Z. B. , Johnson, T. S. , Huang, K. , Payne, P. R. O. , & Coombes, K. (2019). A protocol to evaluate RNA sequencing normalization methods. BMC Bioinformatics, 20, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann, K. , Revell, V. L. , Lao, O. , Rombouts, E. J. , Skene, D. J. , & Kayser, M. (2012). Diurnal rhythms in blood cell populations and the effect of acute sleep deprivation in healthy young men. Sleep, 35, 933–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghaeepour, N. , Ganio, E. A. , Mcilwain, D. , Tsai, A. S. , Tingle, M. , Van, G. S. , Gaudilliere, D. K. , Baca, Q. , Mcneil, L. , Okada, R. , Ghaemi, M. S. , Furman, D. , Wong, R. J. , Winn, V. D. , Druzin, M. L. , El‐Sayed, Y. Y. , Quaintance, C. , Gibbs, R. , Darmstadt, G. L. , … Gaudilliere, B. (2017). An immune clock of human pregnancy. Science Immunology, 2946, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, N. J. , Mondal, T. K. , Preissler, M. T. , Freed, B. M. , Stockinger, S. , Bell, E. , Druschel, C. , Buck Louis, G. M. , & Lawrence, D. A. (2014). Detection of immunoglobulin isotypes from dried blood spots. Journal of Immunological Methods, 404, 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, S. , Kushner, I. , & Samols, D. (2004). C‐reactive protein. The Journal of Biological Chemistry, 279, 48487–48490. [DOI] [PubMed] [Google Scholar]
- Blackwell, A. D. (2022). The Ecoimmunology of health and disease: The hygiene hypothesis and plasticity in human immune function. Annual Review of Anthropology, 51. [Google Scholar]
- Blackwell, A. D. , Garcia, A. R. , Keivanfar, R. L. , & Bay, S. (2021). A field method for cryopreservation of whole blood from a finger prick for later analysis with flow cytometry. American Journal of Physical Anthropology, 174, 670–685. [DOI] [PubMed] [Google Scholar]
- Blackwell, A. D. , Gurven, M. , Sugiyama, L. S. , Madimenos, F. C. , Liebert, M. A. , Martin, M. A. , Kaplan, H. S. , & Snodgrass, J. J. (2011). Evidence for a peak shift in a humoral response to helminths: Age profiles of IgE in the Shuar of Ecuador, the Tsimane of Bolivia, and the U.S. NHANES. PLoS Neglected Tropical Diseases, 5, e1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell, A. D. , Snodgrass, J. J. , Madimenos, F. C. , & Sugiyama, L. S. (2010). Life history, immune function, and intestinal helminths: Trade‐offs among immunoglobulin E, C‐reactive protein, and growth in an Amazonian population. American Journal of Human Biology, 22, 836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell, A. D. , Trumble, B. C. , Maldonado Suarez, I. , Stieglitz, J. , Beheim, B. , Snodgrass, J. J. , Kaplan, H. , & Gurven, M. (2016). Immune function in Amazonian horticulturalists. Annals of Human Biology, 43, 382–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock, C. , Halbritter, F. , Carmona, F. J. , Tierling, S. , Datlinger, P. , Assenov, Y. , Berdasco, M. , Bergmann, A. K. , Booher, K. , Busato, F. , Campan, M. , Dahl, C. , Dahmcke, C. M. , Diep, D. , Fernández, A. F. , Gerhauser, C. , Haake, A. , Heilmann, K. , Holcomb, T. , … Zhang, K. (2016). Quantitative comparison of DNA methylation assays for biomarker development and clinical applications. Nature Biotechnology, 34, 726–737. [DOI] [PubMed] [Google Scholar]
- Boughton, R. K. , Joop, G. , & Armitage, S. A. O. (2011). Outdoor immunology: Methodological considerations for ecologists. Functional Ecology, 25, 81–100. [Google Scholar]
- Brand, S. , Kesper, D. A. , Teich, R. , Kilic‐Niebergall, E. , Pinkenburg, O. , Bothur, E. , Lohoff, M. , Garn, H. , Pfefferle, P. I. , & Renz, H. (2012). DNA methylation of TH1/TH2 cytokine genes affects sensitization and progress of experimental asthma. The Journal of Allergy and Clinical Immunology, 129, 1602–1610.e6. [DOI] [PubMed] [Google Scholar]
- Braun, R. , Kath, W. L. , Iwanaszko, M. , Kula‐Eversole, E. , Abbott, S. M. , Reid, K. J. , Zee, P. C. , & Allada, R. (2018). Universal method for robust detection of circadian state from gene expression. Proceeding of the National Academy of Sciences of the United States of America, 115, E9247–E9256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindle, E. , Fujita, M. , Shofer, J. , & O'Connor, K. A. (2010). Serum, plasma, and dried blood spot high‐sensitivity C‐reactive protein enzyme immunoassay for population research. Journal of Immunological Methods, 362, 112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacioppo, J. T. , Kiecolt‐Glaser, J. K. , Malarkey, W. B. , Laskowski, B. F. , Rozlog, L. A. , Poehlmann, K. M. , Burleson, M. H. , & Glaser, R. (2002). Autonomic and glucocorticoid associations with the steady‐state expression of latent Epstein‐Barr virus. Hormones and Behavior, 42, 32–41. [DOI] [PubMed] [Google Scholar]
- Cals, J. W. L. , Schot, M. J. C. , de Jong, S. A. M. , Dinant, G. J. , & Hopstaken, R. M. (2010). Point‐of‐care C‐reactive protein testing and antibiotic prescribing for respiratory tract infections: A randomized controlled trial. Annals of Family Medicine, 8, 124–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi, J. , Bravo, Y. , Cole, J. , & Gobeil, K. (2012). Acute psychosocial stress differentially influences salivary endocrine and immune measures in undergraduate students. Physiology & Behavior, 107, 317–321. [DOI] [PubMed] [Google Scholar]
- Cepon‐Robins, T. J. (2021). Measuring attack on self: The need for field‐friendly methods development and research on autoimmunity in human biology. American Journal of Human Biology, 33, 1–23. [DOI] [PubMed] [Google Scholar]
- Cepon‐Robins, T. J. , Blackwell, A. D. , Gildner, T. E. , Liebert, M. A. , Urlacher, S. S. , Madimenos, F. C. , Eick, G. N. , Snodgrass, J. J. , & Sugiyama, L. S. (2021). Pathogen disgust sensitivity protects against infection in a high pathogen environment. Proceedings of the National Academy of Sciences, 118, e2018552118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, S. , Seo, H. J. , Lee, J. H. , Kim, M. Y. , & Lee, S. D. (2017). Influence of immunologic status on age prediction using signal joint T cell receptor excision circles. International Journal of Legal Medicine, 131, 1061–1067. [DOI] [PubMed] [Google Scholar]
- Chomczynski, P. , Wilfinger, W. W. , Eghbalnia, H. R. , Kennedy, A. , Rymaszewski, M. , & Mackey, K. (2016). Inter‐individual differences in RNA levels in human peripheral blood. PLoS One, 11, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, S. W. (2019). The conserved transcriptional response to adversity. Current Opinion in Behavioral Sciences, 28, 31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, S. W. , Hawkley, L. C. , Arevalo, J. M. G. , & Cacioppo, J. T. (2011). Transcript origin analysis identifies antigen‐presenting cells as primary targets of socially regulated gene expression in leukocytes. Proceedings of the National Academy of Sciences, 108, 3080–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson, K. E. , Potter, A. , Conde‐Glez, C. , Hernandez, B. , Ríos‐Zertuche, D. , Zúñiga‐Brenes, P. , Collaborators, S. , Iriarte, E. , & Mokdad, A. H. (2015). Use of a commercial ELISA for the detection of measles‐specific immunoglobulin G (IgG) in dried blood spots collected from children living in low‐resource settings. Journal of Medical Virology, 87, 1491–1499. [DOI] [PubMed] [Google Scholar]
- Corran, P. H. , Cook, J. , Lynch, C. , Leendertse, H. , Manjurano, A. , Griffin, J. , Cox, J. , Abeku, T. , Bousema, T. , Ghani, A. C. , Drakeley, C. , & Riley, E. (2008). Dried blood spots as a source of anti‐malarial antibodies for epidemiological studies. Malaria Journal, 7, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crimmins, E. M. , Zhang, Y. S. , Kim, J. K. , Frochen, S. , Kang, H. , Shim, H. , Ailshire, J. , Potter, A. , Cofferen, J. , & Faul, J. (2020). Dried blood spots: Effects of less than optimal collection, shipping time, heat, and humidity. American Journal of Human Biology, 32, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jager, W. , Bourcier, K. , Rijkers, G. T. , Prakken, B. J. , & Seyfert‐Margolis, V. (2009). Prerequisites for cytokine measurements in clinical trials with multiplex immunoassays. BMC Immunology, 10, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demonbreun, A. R. , Sancilio, A. , Velez, M. P. , Ryan, D. T. , Saber, R. , Vaught, L. A. , Reiser, N. L. , Hsieh, R. R. , D'Aquila, R. T. , Mustanski, B. , McNally, E. M. , & McDade, T. W. (2021). Comparison of IgG and neutralizing antibody responses after one or two doses of COVID‐19 mRNA vaccine in previously infected and uninfected individuals. EClinicalMedicine, 38, 101018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Braber, I. , Mugwagwa, T. , Vrisekoop, N. , Westera, L. , Mögling, R. , Bregje de Boer, A. , Willems, N. , Schrijver, E. H. R. , Spierenburg, G. , Gaiser, K. , Mul, E. , Otto, S. A. , Ruiter, A. F. C. , Ackermans, M. T. , Miedema, F. , Borghans, J. A. M. , de Boer, R. J. , & Tesselaar, K. (2012). Maintenance of peripheral naive T cells is sustained by thymus output in mice but not humans. Immunity, 36, 288–297. [DOI] [PubMed] [Google Scholar]
- Dieckmann, L. , Cole, S. , & Kumsta, R. (2020). Stress genomics revisited: Gene co‐expression analysis identifies molecular signatures associated with childhood adversity. Transl. Psychiatry, 10, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillies, M. A. , Rau, A. , Aubert, J. , Hennequet‐Antier, C. , Jeanmougin, M. , Servant, N. , Keime, C. , Marot, N. S. , Castel, D. , Estelle, J. , Guernec, G. , Jagla, B. , Jouneau, L. , Laloë, D. , Le Gall, C. , Schaëffer, B. , Le Crom, S. , Guedj, M. , & Jaffrézic, F. (2013). A comprehensive evaluation of normalization methods for illumina high‐throughput RNA sequencing data analysis. Briefings in Bioinformatics, 14, 671–683. [DOI] [PubMed] [Google Scholar]
- Drummond, P. D. , & Hewson‐Bower, B. (1997). Increased psychosocial stress and decreased mucosal immunity in children with recurrent upper respiratory tract infections. Journal of Psychosomatic Research, 43, 271–278. [DOI] [PubMed] [Google Scholar]
- Eisenberg, E. , & Levanon, E. Y. (2013). Human housekeeping genes, revisited. Trends in Genetics, 29, 569–574. [DOI] [PubMed] [Google Scholar]
- Evans, C. , Hardin, J. , & Stoebel, D. M. (2018). Selecting between‐sample RNA‐Seq normalization methods from the perspective of their assumptions. Briefings in Bioinformatics, 19, 776–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi, C. , Bonafè, M. , & Valensin, S. (2000). Human immunosenescence: The prevailing of innate immunity, the failing of clonotypic immunity, and the filling of immunological space. Vaccine, 18, 1717–1720. [DOI] [PubMed] [Google Scholar]
- Garcia, A. R. , Blackwell, A. D. , Trumble, B. C. , Stieglitz, J. , Kaplan, H. , & Gurven, M. D. (2020). Evidence for height and immune function trade‐offs among preadolescents in a high pathogen population. Evolution Medicine and Public Health, 2020, 86–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, A. R. , Trumble, B. , Kraft, T. S. , Murillo, S. , Marquez, M. , Gurven, M. , & Blackwell, A. D. (2020). Does exposure to parasites modify relationships between diurnal cortisol and leukocytes among Honduran women? American Journal of Physical Anthropology, 173, 463–479. [DOI] [PubMed] [Google Scholar]
- Gilot‐Fromont, E. , Jégo, M. , Bonenfant, C. , Gibert, P. , Rannou, B. , Klein, F. , & Gaillard, J. M. (2012). Immune phenotype and body condition in roe deer: Individuals with high body condition have different, not stronger immunity. PLoS One, 7(9), e45576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser, R. , Pearson, G. , Jones, J. , Hillhouse, J. , Kennedy, S. , Mao, H. , & Kiecolt‐Glaser, J. (1991). Stress‐related activation of Epstein‐Barr virus. Brain, Behavior, and Immunity, 5, 219–232. [DOI] [PubMed] [Google Scholar]
- Gornowicz, A. , Tokajuk, G. , Bielawska, A. , Maciorkowska, E. , Jabłoński, R. , Wójcicka, A. , & Bielawski, K. (2014). The assessment of sIgA, histatin‐5, and lactoperoxidase levels in saliva of adolescents with dental caries. Medical Science Monitor, 20, 1095–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblum, S. I. , Efroni, S. , Schaefer, C. F. , & Buetow, K. H. (2011). The PathOlogist: An automated tool for pathway‐centric analysis. BMC Bioinformatics, 12, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurven, M. D. , Kaplan, H. , Winking, J. , Eid Rodriguez, D. , Vasunilashorn, S. , Kim, J. K. , Finch, C. , & Crimmins, E. (2009). Inflammation and infection do not promote arterial aging and cardiovascular disease risk factors among lean horticulturalists. PLoS One, 4, e6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurven, M. D. , Trumble, B. C. , Stieglitz, J. , Blackwell, A. D. , Michalik, D. E. , Finch, C. E. , & Kaplan, H. S. (2016). Cardiovascular disease and type 2 diabetes in evolutionary perspective: A critical role for helminths? Evolution Medicine and Public Health, 2016, 338–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington, C. A. , Fei, S. S. , Minnier, J. , Carbone, L. , Searles, R. , Davis, B. A. , Ogle, K. , Planck, S. R. , Rosenbaum, J. T. , & Choi, D. (2020). RNA‐Seq of human whole blood: Evaluation of globin RNA depletion on Ribo‐zero library method. Scientific Reports, 10, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, G. F. , Sanz, J. , Boulais, J. , Mina, M. J. , Grenier, J.‐C. , Leng, Y. , Dumaine, A. , Yotova, V. , Bergey, C. M. , Nsobya, S. L. , Elledge, S. J. , Schurr, E. , Quintana‐Murci, L. , Perry, G. H. , & Barreiro, L. B. (2019). Natural selection contributed to immunological differences between hunter‐gatherers and agriculturalists. Nature Ecology and Evolution, 3, 1253–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan, F. , Nguyen, A. , Formanek, A. , Bell, J. J. , & Selvarangan, R. (2014). Comparison of the BD Veritor system for flu A + B with the Alere BinaxNOW influenza A&B card for detection of influenza A and B viruses in respiratory specimens from pediatric patients. Journal of Clinical Microbiology, 52, 906–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegemann, A. , Pardal, S. , & Matson, K. D. (2017). Indices of immune function used by ecologists are mostly unaffected by repeated freeze‐thaw cycles and methodological deviations. Frontiers in Zoology, 14, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich, S. K. , Hofer, H. , Courtiol, A. , Melzheimer, J. , Dehnhard, M. , Czirják, G. , & Wachter, B. (2017). Cheetahs have a stronger constitutive innate immunity than leopards. Scientific Reports, 7, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersoug, L. G. , & Linneberg, A. (2007). The link between the epidemics of obesity and allergic diseases: Does obesity induce decreased immune tolerance? Allergy, 62, 1205–1213. [DOI] [PubMed] [Google Scholar]
- Higham, J. P. , Kraus, C. , Stahl‐Hennig, C. , Engelhardt, A. , Fuchs, D. , & Heistermann, M. (2015). Evaluating noninvasive markers of nonhuman primate immune activation and inflammation. American Journal of Physical Anthropology, 158, 673–684. [DOI] [PubMed] [Google Scholar]
- Hodges‐Simeon, C. R. , Prall, S. P. , Blackwell, A. D. , Gurven, M. , & Gaulin, S. J. C. (2017). Adrenal maturation, nutritional status, and mucosal immunity in Bolivian youth. American Journal of Human Biology, 29, e23025. [DOI] [PubMed] [Google Scholar]
- Hoffman, G. E. , & Schadt, E. E. (2016). variancePartition: Interpreting drivers of variation in complex gene expression studies. BMC Bioinformatics, 17, 17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath, S. , Gurven, M. , Levine, M. E. , Trumble, B. C. , Kaplan, H. , Allayee, H. , Ritz, B. R. , Chen, B. , Lu, A. T. , Rickabaugh, T. M. , Jamieson, B. D. , Sun, D. , Li, S. , Chen, W. , Quintana‐Murci, L. , Fagny, M. , Kobor, M. S. , Tsao, P. S. , Reiner, A. P. , … Assimes, T. L. (2016). An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biology, 17, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hové, C. , Trumble, B. C. , Anderson, A. S. , Stieglitz, J. , Kaplan, H. , Gurven, M. D. , & Blackwell, A. D. (2020). Immune function during pregnancy varies between ecologically distinct populations. Evolution Medicine amd Public Health, 2020, 114–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabanova, S. , Kleinbongard, P. , Volkmer, J. , Andrée, B. , Kelm, M. , & Jax, T. W. (2009). Gene expression analysis of human red blood cells. International Journal of Medical Sciences, 6, 156–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karawajczyk, M. , Haile, S. , Grabski, M. , & Larsson, A. (2017). The HemoCue WBC DIFF system could be used for leucocyte and neutrophil counts but not for full differential counts. Acta Paediatrica International Journal of Paediatrica, 106, 974–978. [DOI] [PubMed] [Google Scholar]
- Keustermans, G. C. E. , Hoeks, S. B. E. , Meerding, J. M. , Prakken, B. J. , & de Jager, W. (2013). Cytokine assays: An assessment of the preparation and treatment of blood and tissue samples. Methods, 61, 10–17. [DOI] [PubMed] [Google Scholar]
- Kirschfink, M. , & Mollnes, T. E. (2003). Modern complement analysis. Clinical and Diagnostic Laboratory Immunology, 10, 982–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, S. L. , & Flanagan, K. L. (2016). Sex differences in immune responses. Nature Reviews. Immunology, 16, 626–638. [DOI] [PubMed] [Google Scholar]
- Koss, M. A. , Castro, C. E. , Gramajo, A. M. , & López, M. E. (2016). sIgA, peroxidase and collagenase in saliva of smokers aggressive periodontal patients. Journal of Oral Biology Craniofacial Research, 6, S24–S28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawiec, J. A. , Chen, H. , Alom‐Ruiz, S. , & Jaye, M. (2009). Modified PAXgene method allows for isolation of high‐integrity total RNA from microlitre volumes of mouse whole blood. Laboratory Animals, 43, 394–398. [DOI] [PubMed] [Google Scholar]
- Kurdyukov, S. , & Bullock, M. (2016). DNA methylation analysis: Choosing the right method. Biology (Basel), 5, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrecque, N. , & Cermakian, N. (2015). Circadian clocks in the immune system. Journal of Biological Rhythms, 30, 277–290. [DOI] [PubMed] [Google Scholar]
- Laney, W. , Mawelele, R. , & Omar, S. (2019). The accuracy of a point‐of‐care test among different operators using the QBC autoread plus analyzer for the measurement of a basic full blood count. South African Medical Journal, 109, 952–956. [DOI] [PubMed] [Google Scholar]
- Lang, P. O. , Mitchell, W. A. , Govind, S. , & Aspinall, R. (2011). Real time‐PCR assay estimating the naive T‐cell pool in whole blood and dried blood spot samples: Pilot study in young adults. Journal of Immunological Methods, 369, 133–140. [DOI] [PubMed] [Google Scholar]
- Lebech, M. , & Petersen, E. (1995). Detection by enzyme immunosorbent assay of toxoplasma gondii IgG antibodies in dried blood spots on PKU‐filter paper from newborns. Scandinavian Journal of Infectious Diseases, 27, 259–263. [DOI] [PubMed] [Google Scholar]
- Levine, M. E. , Crimmins, E. M. , Weir, D. R. , & Cole, S. W. (2017). Contemporaneous social environment and the architecture of late‐life gene expression profiles. American Journal of Epidemiology, 186, 503–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebl, A. L. , & Martin II, L. B. (2009). Simple quantification of blood and plasma antimicrobial capacity using spectrophotometry. Functional Ecology, 23, 1091–1096. [Google Scholar]
- Ligthart, S. , Marzi, C. , Aslibekyan, S. , Mendelson, M. M. , Conneely, K. N. , Tanaka, T. , Colicino, E. , Waite, L. L. , Joehanes, R. , Guan, W. , Brody, J. A. , Elks, C. , Marioni, R. , Jhun, M. A. , Agha, G. , Bressler, J. , Ward‐Caviness, C. K. , Chen, B. H. , Huan, T. , … Dehghan, A. (2016). DNA methylation signatures of chronic low‐grade inflammation are associated with complex diseases. Genome Biology, 17, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Zhou, J. , & White, K. P. (2014). RNA‐seq differential expression studies: More sequence or more replication? Bioinformatics, 30, 301–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzi, A. R. , Patterson, A. M. , Pratt, A. , Jefferson, M. , Chapman, C. E. , Ponchel, F. , & Isaacs, J. D. (2008). Determination of thymic function directly from peripheral blood: A validated modification to an established method. Journal of Immunological Methods, 339, 185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luecken, M. D. , & Theis, F. J. (2019). Current best practices in single‐cell RNA‐seq analysis: A tutorial. Molecular Systems Biology, 15, e8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, F. , Fuqua, B. K. , Hasin, Y. , Yukhtman, C. , Vulpe, C. D. , Lusis, A. J. , & Pellegrini, M. (2019). A comparison between whole transcript and 3’ RNA sequencing methods using Kapa and Lexogen library preparation methods. BMC Genomics, 20, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makar, K. W. , & Wilson, C. B. (2004). DNA methylation is a nonredundant repressor of the Th2 effector program. Journal of Immunology, 173, 4402–4406. [DOI] [PubMed] [Google Scholar]
- Matson, K. D. , Ricklefs, R. E. , & Klasing, K. C. (2005). A hemolysis‐hemagglutination assay for characterizing constitutive innate humoral immunity in wild and domestic birds. Developmental and Comparative Immunology, 29, 275–286. [DOI] [PubMed] [Google Scholar]
- May, L. , van Bodegom, D. , Kuningas, M. , Meij, J. J. , de Craen, A. J. M. , Frölich, M. , & Westendorp, R. G. J. (2009). Performance of the whole‐blood stimulation assay for assessing innate immune activation under field conditions. Cytokine, 45, 184–189. [DOI] [PubMed] [Google Scholar]
- May, L. , van Den Biggelaar, A. H. , van Bodegom, D. , Meij, H. J. , de Craen, A. J. , Amankwa, J. , Frölich, M. , Kuningas, M. , & Westendorp, R. G. (2009). Adverse environmental conditions influence age‐related innate immune responsiveness. Immunity & Ageing, 6, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, M. M. (1961). Complement and complement fixation. In Kabat E. & Mayer M. M. (Eds.), Experimental immunochemistry (pp. 133–240). Springfield, Ill: C. C. Thomas. [Google Scholar]
- McDade, T. W. (2003). Life history theory and the immune system: Steps toward a human ecological immunology. American Journal of Physical Anthropology, 122, 100–125. [DOI] [PubMed] [Google Scholar]
- McDade, T. W. (2005a). The ecologies of human immune function. Annual Review of Anthropology, 34, 495–521. [Google Scholar]
- McDade, T. W. (2005b). Life history, maintenance, and the early origins of immune function. American Journal of Human Biology, 17, 81–94. [DOI] [PubMed] [Google Scholar]
- McDade, T. W. , Burhop, J. , & Dohnal, J. (2004). High‐sensitivity enzyme immunoassay for C‐reactive protein in dried blood spots. Clinical Chemistry, 50, 652–654. [DOI] [PubMed] [Google Scholar]
- McDade, T. W. , Georgiev, A. V. , & Kuzawa, C. W. (2016). Trade‐offs between acquired and innate immune defenses in humans. Evolution Medicine and Public Health, 2016, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDade, T. W. , Miller, A. , Tran, T. T. , Borders, A. E. B. , & Miller, G. (2021). A highly sensitive multiplex immunoassay for inflammatory cytokines in dried blood spots. American Journal of Human Biology, 33, 1–7. [DOI] [PubMed] [Google Scholar]
- McDade, T. W. , Reyes‐Garcia, V. , Tanner, S. , Huanca, T. , & Leonard, W. R. (2008). Maintenance versus growth: Investigating the costs of immune activation among children in lowland Bolivia. American Journal of Physical Anthropology, 136, 478–484. [DOI] [PubMed] [Google Scholar]
- McDade, T. W. , Ross, K. M. , Fried, R. L. , JMG, A. , Ma, J. , Miller, G. E. , & Cole, S. W. (2016). Genome‐wide profiling of RNA from dried blood spots: Convergence with Bioinformatic results derived from whole venous blood and peripheral blood mononuclear cells. Biodemography and Social Biology, 62, 182–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDade, T. W. , Ryan, C. P. , Jones, M. J. , Hoke, M. K. , Borja, J. , Miller, G. E. , Kuzawa, C. W. , & Kobor, M. S. (2019). Genome‐wide analysis of DNA methylation in relation to socioeconomic status during development and early adulthood. American Journal of Physical Anthropology, 169, 3–11. [DOI] [PubMed] [Google Scholar]
- McDade, T. W. , Stallings, J. F. , Angold, A. , Costello, E. J. , Burleson, M. , Cacioppo, J. T. , Glaser, R. , & Worthman, C. M. (2000). Epstein‐Barr virus antibodies in whole blood spots: A minimally invasive method for assessing an aspect of cell‐mediated immunity. Psychosomatic Medicine, 62, 560–568. [DOI] [PubMed] [Google Scholar]
- McDade, T. W. , Tallman, P. S. , Madimenos, F. C. , Liebert, M. A. , Cepon, T. J. , Sugiyama, L. S. , & Snodgrass, J. J. (2012). Analysis of variability of high sensitivity C‐reactive protein in lowland Ecuador reveals no evidence of chronic low‐grade inflammation. American Journal of Human Biology, 24, 675–681. [DOI] [PubMed] [Google Scholar]
- McDade, T. W. , Williams, S. , & Snodgrass, J. J. (2007). What a drop can do: Dried blood spots as a minimally invasive method for integrating biomarkers into population‐based research. Demography, 44, 899–925. [DOI] [PubMed] [Google Scholar]
- Meitern, R. , Andreson, R. , & Hõrak, P. (2014). Profile of whole blood gene expression following immune stimulation in a wild passerine. BMC Genomics, 15, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, E. M. , & McConnell, D. S. (2011). The stability of immunoglobulin a in human milk and saliva stored on filter paper at ambient temperature. American Journal of Human Biology, 825, 823–825. [DOI] [PubMed] [Google Scholar]
- Miller, E. M. , & McDade, T. W. (2012). A highly sensitive immunoassay for interleukin‐6 in dried blood spots. American Journal of Human Biology, 24, 863–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet, S. , Bennett, J. , Lee, K. A. , Hau, M. , & Klasing, K. C. (2007). Quantifying and comparing constitutive immunity across avian species. Developmental and Comparative Immunology, 31, 188–201. [DOI] [PubMed] [Google Scholar]
- Moll, P. , Ante, M. , Seitz, A. , & Reda, T. (2014). QuantSeq 3′ mRNA sequencing for RNA quantification. Nature Methods, 11, i–iii. [Google Scholar]
- Morgan, B. P. (2000). Measurement of complement hemolytic activity, generation of complement‐depleted. In Morgan B. P. (Ed.), Complement methods and protocols. Methods in molecular biology (Vol. 150, pp. 61–71). Human Press. [DOI] [PubMed] [Google Scholar]
- Mwaba, P. , Cassol, S. , Pilon, R. , Chintu, C. , Janes, M. , Nunn, A. , & Zumla, A. (2003). Use of dried whole blood spots to measure CD4+ lymphocyte counts in HIV‐1‐infected patients. Lancet, 362, 1459–1460. [DOI] [PubMed] [Google Scholar]
- Nakajima, K. , Takeoka, M. , Mori, M. , Hashimoto, S. , Sakurai, A. , Nose, H. , Higuchi, K. , Itano, N. , Shiohara, M. , Oh, T. , & Taniguchi, S. (2010). Exercise effects on methylation of ASC gene. International Journal of Sports Medicine, 31, 671–675. [DOI] [PubMed] [Google Scholar]
- Nakamura, D. , Akimoto, T. , Suzuki, S. , & Kono, I. (2006). Daily changes of salivary secretory immunoglobulin a and appearance of upper respiratory symptoms during physical training. The Journal of Sports Medicine and Physical Fitness, 46, 152–157. [PubMed] [Google Scholar]
- Natri, H. , Garcia, A. R. , Buetow, K. H. , Trumble, B. C. , & Wilson, M. A. (2019). The pregnancy pickle: Evolved immune compensation due to pregnancy underlies sex differences in human diseases. Trends in Genetics, 35, 478–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Non, A. , & Thayer, Z. (2015). Epigenetics for anthropologists: An introduction to methods. American Journal of Human Biology, 27, 295–303. [DOI] [PubMed] [Google Scholar]
- Osei‐Bimpong, A. , Jury, C. , Mclean, R. , & Lewis, S. M. (2009). Point‐of‐care method for total white cell count: An evaluation of the HemoCue WBC device. International Journal of Laboratory Hematology, 31, 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shaughnessy, C. M. , Cunningham, A. F. , & MacLennan, C. A. (2012). The stability of complement‐mediated bactericidal activity in human serum against salmonella. PLoS One, 7, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou, X. , Gao, J. , Wang, H. , Sheng, W. H. , Ling, L. H. , & Yu, S. H. (2012). Predicting human age with bloodstains by sjTREC quantification. PLoS One, 7, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou, X. , Zhao, H. , Sun, H. , Yang, Z. , Xie, B. , Shi, Y. , & Wu, X. (2011). Detection and quantification of the age‐related sjTREC decline in human peripheral blood. International Journal of Legal Medicine, 125, 603–608. [DOI] [PubMed] [Google Scholar]
- Page, A. E. , Viguier, S. , Dyble, M. , Smith, D. , Chaudhary, N. , Salali, G. D. , Thompson, J. , Vinicius, L. , Mace, R. , & Migliano, A. B. (2016). Reproductive trade‐offs in extant hunter‐gatherers suggest adaptive mechanism for the Neolithic expansion. Proceedings of National Academy and Science, 113, 4694–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, C. , Diehn, M. , Alizadeh, A. A. , & Brown, P. O. (2006). Cell‐type specific gene expression profiles of leukocytes in human peripheral blood. BMC Genomics, 7, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panter‐Brick, C. , Wiley, K. , Sancilio, A. , Dajani, R. , & Hadfield, K. (2020). C‐reactive protein, Epstein‐Barr virus, and cortisol trajectories in refugee and non‐refugee youth: Links with stress, mental health, and cognitive function during a randomized controlled trial. Brain, Behavior, and Immunity, 87, 207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham, P. (2021). The immune system (5th ed.). W.W. Norton & Company. [Google Scholar]
- Permar, S. R. , Moss, W. J. , Ryon, J. J. , Douek, D. C. , Monze, M. , & Griffin, D. E. (2003). Increased thymic output during acute measles virus infection. Journal of Virology, 77, 7872–7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovsky, N. , & Harrison, L. C. (1998). The chronobiology of human cytokine production. International Reviews of Immunology, 16, 635–649. [DOI] [PubMed] [Google Scholar]
- Powell, N. D. , Sloan, E. K. , Bailey, M. T. , Arevalo, J. M. G. , Miller, G. E. , Chen, E. , Kobor, M. S. , Reader, B. F. , Sheridan, J. F. , & Cole, S. W. (2013). Social stress up‐regulates inflammatory gene expression in the leukocyte transcriptome via β‐adrenergic induction of myelopoiesis. Proceedings of the National Academy of Sciences of the United States of America, 110, 16574–16579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prall, S. P. , & Muehlenbein, M. P. (2014). Testosterone and immune function in primates: A brief summary with methodological considerations. International Journal of Primatology, 35, 805–824. [Google Scholar]
- PreAnalytiX . (2020). PAXgene Blood RNA Kit Handbook. Version 2. PreAnalytix GmbH. [Google Scholar]
- Pulido‐Salgado, M. , Vidal‐Taboada, J. M. , Barriga, G. G. D. , Solà, C. , & Saura, J. (2018). RNA‐Seq transcriptomic profiling of primary murine microglia treated with LPS or LPS + IFNγ. Scientific Reports, 8, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp, H. J. , & Borsos, T. (1970). Molecular basis of complement action. Appleton Century Crofts. [Google Scholar]
- Reid, M. R. , Mackinnon, L. T. , & Drummond, P. D. (2001). The effects of stress management on symptoms of upper respiratory tract infection, secretory immunoglobulin a, and mood in young adults. Journal of Psychosomatic Research, 51, 721–728. [DOI] [PubMed] [Google Scholar]
- Reust, M. J. , Lee, M. H. , Xiang, J. , Zhang, W. , Xu, D. , Batson, T. , Zhang, T. , Downs, J. A. , & Dupnik, K. M. (2018). Dried blood spot RNA transcriptomes correlate with transcriptomes derived from whole blood RNA. The American Journal of Tropical Medicine and Hygiene, 98, 1541–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riis, J. L. , Out, D. , Dorn, L. D. , Beal, S. J. , Denson, L. A. , Pabst, S. , Jaedicke, K. , & Granger, D. A. (2014). Salivary cytokines in healthy adolescent girls: Intercorrelations, stability, and associations with serum cytokines, age, and pubertal stage. Developmental Psychobiology, 56, 797–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudzik, A. E. F. , Breakey, A. , & Bribiescas, R. G. (2014). Oxytocin and Epstein‐Barr virus: Stress biomarkers in the postpartum period among first‐time mothers from São Paulo, Brazil. American Journal of Human Biology, 26, 43–50. [DOI] [PubMed] [Google Scholar]
- Ruoss, S. , Becker, N. I. , Otto, M. S. , Czirják, G. , & Encarnação, J. A. (2019). Effect of sex and reproductive status on the immunity of the temperate bat Myotis daubentonii. Mammalian Biology, 94, 120–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, A. , Kim, B. , Murphy, K. E. , & Matthews, S. G. (2020). Impact of ex vivo sample handling on DNA methylation profiles in human cord blood and neonatal dried blood spots. Frontiers in Genetics, 11, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbiera, S. , Dexneit, T. , Reichardt, S. D. , Michel, K. D. , van den Brandt, J. , Schmull, S. , Kraus, L. , Beyer, M. , Mlynski, R. , Wortmann, S. , Allolio, B. , Reichardt, H. M. , & Fassnacht, M. (2011). Influence of short‐term glucocorticoid therapy on regulatory T cells in vivo. PLoS One, 6, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid‐Hempel, P. , & Ebert, D. (2003). On the evolutionary ecology of specific immune defence. Trends in Ecology & Evolution, 18, 27–32. [Google Scholar]
- Schneider‐Crease, I. A. , Blackwell, A. D. , Kraft, T. S. , Emery Thompson, M. , Maldonado Suarez, I. , Cummings, D. K. , Stieglitz, J. , Snyder‐Mackler, N. , Gurven, M. , Kaplan, H. , & Trumble, B. C. (2021). Helminth infection is associated with dampened cytokine responses to viral and bacterial stimulations in Tsimane forager‐horticulturalists. Evolution Medicine and Public Health, 9, 349–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sholder, G. , Lanz, T. A. , Moccia, R. , Quan, J. , Aparicio‐Prat, E. , Stanton, R. , & Xi, H. S. (2020). 3′Pool‐seq: An optimized cost‐efficient and scalable method of whole‐transcriptome gene expression profiling. BMC Genomics, 21, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitoe, N. , Luecke, E. , Tembe, N. , Matavele, R. , Cumbane, V. , Macassa, E. , Vaz, P. , Sheppard, H. , & Jani, I. V. (2011). Absolute and percent CD4+ T‐cell enumeration by flow cytometry using capillary blood. Journal of Immunological Methods, 372, 1–6. [DOI] [PubMed] [Google Scholar]
- Skalina, K. A. , Goldstein, D. Y. , Sulail, J. , Hahm, E. , Narlieva, M. , Szymczak, W. , & Fox, A. S. (2022). Extended storage of SARS‐CoV‐2 nasopharyngeal swabs does not negatively impact results of molecular‐based testing across three clinical platforms. Journal of Clinical Pathology, 75, 61–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skogstrand, K. , Ekelund, C. K. , Thorsen, P. , Vogel, I. , Jacobsson, B. , Nørgaard‐Pedersen, B. , & Hougaard, D. M. (2008). Effects of blood sample handling procedures on measurable inflammatory markers in plasma, serum and dried blood spot samples. Journal of Immunological Methods, 336, 78–84. [DOI] [PubMed] [Google Scholar]
- Skogstrand, K. , Thorsen, P. , Norgaard‐Pedersen, B. , Schendel, D. E. , Sorensen, L. C. , & Hougaard, D. M. (2005). Simultaneous measurement of 25 inflammatory markers and neurotrophins in neonatal dried blood spots by immunoassay with xMAP technology. Clinical Chemistry, 51, 1854–1866. [DOI] [PubMed] [Google Scholar]
- Srisala, S. , Pongsakul, N. , Sahakijpicharn, T. , Hongeng, S. , Chutipongtanate, S. , & Apiwattanakul, N. (2019). Capillary blood as an alternative specimen for enumeration of percentages of lymphocyte subsets. BMC Research Notes, 12, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub, R. H. , Cutolo, M. , Buttgereit, F. , & Pongratz, G. (2010). Energy regulation and neuroendocrine‐immune control in chronic inflammatory diseases. Journal of Internal Medicine, 267, 543–560. [DOI] [PubMed] [Google Scholar]
- Su, A. I. , Wiltshire, T. , Batalov, S. , Lapp, H. , Ching, K. A. , Block, D. , Zhang, J. , Soden, R. , Hayakawa, M. , Kreiman, G. , Cooke, M. P. , Walker, J. R. , & Hogenesch, J. B. (2004). A gene atlas of the mouse and human protein‐encoding transcriptomes. Proceedings of the National Academy of Sciences, 101, 6062–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana, R. , Bhuiyan, T. R. , Sathi, A. S. , Sharmin, S. , Yeasmin, S. , Uddin, M. I. , Bhuiyan, M. S. , Mannoor, K. , Karim, M. M. , Zaman, K. , & Qadri, F. (2022). Developing and validating a modified enzyme linked immunosorbent assay method for detecting HEV IgG antibody from dried blood spot (DBS) samples in endemic settings. Microbes and Infection, 24, 104890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y. V. , Lazarus, A. , Smith, J. A. , Chuang, Y. H. , Zhao, W. , Turner, S. T. , & Kardia, S. L. R. (2013). Gene‐specific DNA methylation association with serum levels of C‐reactive protein in African Americans. PLoS One, 8, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner, S. , & McDade, T. W. (2007). Enzyme immunoassay for total immunoglobulin E in dried blood spots. American Journal of Human Biology, 19, 440–442. [DOI] [PubMed] [Google Scholar]
- Tarcic, N. , Ovadia, H. , Weiss, D. W. , & Weidenfeld, J. (1998). Restraint stress‐induced thymic involution and cell apoptosis are dependent on endogenous glucocorticoids. Journal of Neuroimmunology, 82, 40–46. [DOI] [PubMed] [Google Scholar]
- Thaweboon, S. , Thaweboon, B. , Nakornchai, S. , & Jitmaitree, S. (2008). Salivary secretory IgA, pH, flow rates, mutans streptococci and Candida in children with rampant caries. The Southeast Asian Journal of Tropical Medicine and Public Health, 39, 900–905. [PubMed] [Google Scholar]
- Theda, C. , Hwang, S. H. , Czajko, A. , Loke, Y. J. , Leong, P. , & Craig, J. M. (2018). Quantitation of the cellular content of saliva and buccal swab samples. Scientific Reports, 8, 4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titus, A. J. , Gallimore, R. M. , Salas, L. A. , & Christensen, B. C. (2017). Cell‐type deconvolution from DNA methylation: A review of recent applications. Human Molecular Genetics, 26, R216–R224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumble, B. C. , Blackwell, A. D. , Stieglitz, J. , Thompson, M. E. , Suarez, I. M. , Kaplan, H. , & Gurven, M. (2016). Associations between male testosterone and immune function in a pathogenically stressed forager‐horticultural population. American Journal of Physical Anthropology, 161, 494–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlacher, S. S. , Ellison, P. T. , Sugiyama, L. S. , Pontzer, H. , Eick, G. , Liebert, M. A. , Cepon‐Robins, T. J. , Gildner, T. E. , & Snodgrass, J. J. (2018). Tradeoffs between immune function and childhood growth among Amazonian forager‐horticulturalists. Proceedings of National Academy and Science, 115, E3914–E3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Anders, S. M. (2010). Gonadal steroids and salivary IgA in healthy young women and men. American Journal of Human Biology, 22, 348–352. [DOI] [PubMed] [Google Scholar]
- van Riet, E. , Everts, B. , Retra, K. , Phylipsen, M. , van Hellemond, J. J. , Tielend, A. G. M. , van Der Kleij, D. , Hartgers, F. C. , & Yazdanbakhsh, M. (2009). Combined TLR2 and TLR4 ligation in the context of bacterial or helminth extracts in human monocyte derived dendritic cells: Molecular correlates for Th1/Th2 polarization. BMC, 10, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidović, A. , Vidović Juras, D. , Vučićević Boras, V. , Lukač, J. , Grubišić‐Ilić, M. , Rak, D. , & Sabioncello, A. (2012). Determination of leucocyte subsets in human saliva by flow cytometry. Archives of Oral Biology, 57, 577–583. [DOI] [PubMed] [Google Scholar]
- Walker, R. M. , MacGillivray, L. , McCafferty, S. , Wrobel, N. , Murphy, L. , Kerr, S. M. , Morris, S. W. , Campbell, A. , McIntosh, A. M. , Porteous, D. J. , & Evans, K. L. (2019). Assessment of dried blood spots for DNA methylation profiling. Wellcome Open Research, 4, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wander, K. , Brindle, E. , & O'Connor, K. A. (2008). C‐reactive protein across the menstrual cycle. American Journal of Physical Anthropology, 136, 138–146. [DOI] [PubMed] [Google Scholar]
- West‐Eberhard, M. J. (2019). Nutrition, the visceral immune system, and the evolutionary origins of pathogenic obesity. Proceedings of the National Academy of Sciences of the United States of America, 116, 723–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S. , McGookey, M. , Wang, Y. , Cataland, S. R. , & Wu, H. M. (2015). Effect of blood sampling, processing, and storage on the measurement of complement activation biomarkers. American Journal of Clinical Pathology, 143, 558–565. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Parmigiani, G. , & Johnson, W. E. (2020). ComBat‐seq: Batch effect adjustment for RNA‐seq count data. NAR Genomics and Bioinformatics, 2, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zyprych‐Walczak, J. , Szabelska, A. , Handschuh, L. , Górczak, K. , Klamecka, K. , Figlerowicz, M. , & Siatkowski, I. (2015). The Impact of Normalization Methods on RNA‐Seq Data Analysis. BioMed Research International, 2015, 621690. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


