Abstract
A novel class of benzyl-free and benzyl-substituted carbamylated tryptamine derivatives (CDTs) was designed and synthesized to serve as effective building blocks for the development of novel multi-target directed ligands (MTDLs) for the treatment of neurological disorders linked to cholinesterase (ChE) activity. The majority of them endowed butyrylcholinesterase (BuChE) with more substantial inhibition potency than acetylcholinesterase (AChE), according to the full study of ChE inhibition. Particularly, hybrids with dibenzyl groups (2b-2f, 2j, 2o, and 2q) showed weak or no neuronal toxicity and hepatotoxicity and single-digit nanomolar inhibitory effects against BuChE. Through molecular docking and kinetic analyses, the potential mechanism of action on BuChE was first investigated. In vitro H2O2-induced HT-22 cells assay demonstrated the favorable neuroprotective potency of 2g, 2h, 2j, 2m, 2o, and 2p. Besides, 2g, 2h, 2j, 2m, 2o, and 2p endowed good antioxidant activities and COX-2 inhibitory effects. This study suggested that this series of hybrids can be applied to treat various ChE-associated neurodegenerative disorders such as Alzheimer’s disease (AD) and Parkinson’s disease (PD), as well as promising building blocks for further structure modification to develop efficient MTDLs.
Keywords: carbamylated tryptamine derivatives, benzylation, promising building blocks, MTDLs, cholinesterase inhibitors, neuroprotection, anti-neuroinflammation, neurodegenerative disorders
Introduction
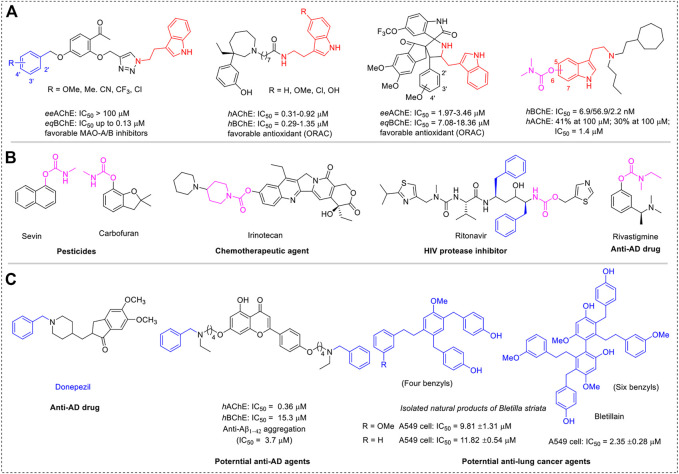
Because of the complex pathogenesis, neurodegenerative diseases (NDs), including Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease (HD), and amyotrophic lateral sclerosis (ALS), remain unclear and incurable, have seriously endangered human health, and brought huge economic burden to society (Armstrong, 2020). Numerous recent studies have shown that multi-target directed ligands (MTDLs) are the preferred method for developing new drugs for NDs (Cavalli et al., 2008; Geldenhuys and Van der Schyf, 2013; Wichur and Malawska, 2015). Thereinto, molecular hybridization is the main approach to achieving MTDLs. Tryptamine, a monoamine alkaloid that contains indole, can function in vivo as a neuromodulator or neurotransmitter (Mousseau, 1993; Wu et al., 2018). Due to their wide range of biological activities and widespread use in medicinal chemistry (Scheme 1A), tryptamine and its analogs have attracted intense interest in industry and academia. Evidence has shown that tryptamine derivatives obtained using tryptamine as a key synthetic block endow significant pharmaceutical activities (Cheng et al., 2015; Kumar et al., 2021; Liu et al., 2022; Meden et al., 2022; Oh et al., 2022). Serotonin, also known as 5-hydroxytryptamine, plays an important biological role as a neurotransmitter in the cerebral cortex and synapses, making it a prominent target for drug development in immunological and neurological illnesses (Zhang et al., 2022a; Fang et al., 2022; Kim et al., 2022; Meden et al., 2022; Shevchenko et al., 2022; Wang et al., 2022). The most common optimization strategies for serotonin alteration are focused on hydroxyl and amino groups because of their intrinsic structural features.
SCHEME 1.
(A) Structures of several represented tryptamine derivatives. (B) Structures of several represented carbamate-based drugs. (C) Structures of several represented benzyl-containing agents with pharmaceutical activities.
Many neurodegenerative illnesses, including AD, PD, and HD, have been linked to cholinergic system alterations, and the loss of cholinergic transmission, including acetylcholine (ACh), is crucial for cognitive function (Hampel et al., 2018; Petrova et al., 2020; van der Zee et al., 2021; Bohnen et al., 2022). As a preferred pharmacophore of cholinesterase (ChE) inhibitors, carbamate fragments have been extensively exploited to produce extremely potent ChE inhibitors for the treatment of a wide range of ailments, including AD, HIV inhibitors, pesticides, and chemotherapeutic agents against various cancers (Scheme 1B) (Matosevic and Bosak, 2020; Martins et al., 2021; Mishra et al., 2021; Zhang et al., 2022b; Mdeni et al., 2022). This is mostly because of the structural properties of carbamate fragments, which can form bidentate H-bonds to anchor more firmly in the active pocket and act as an H-bond acceptor and donor. Additionally, adding a carbamate fragment can make molecules more drug-like and improve their metabolic stability. Therefore, a promising method to produce multifunctional ChE inhibitors is a molecular hybridization approach incorporating the carbamate fragment and tryptamine skeleton.
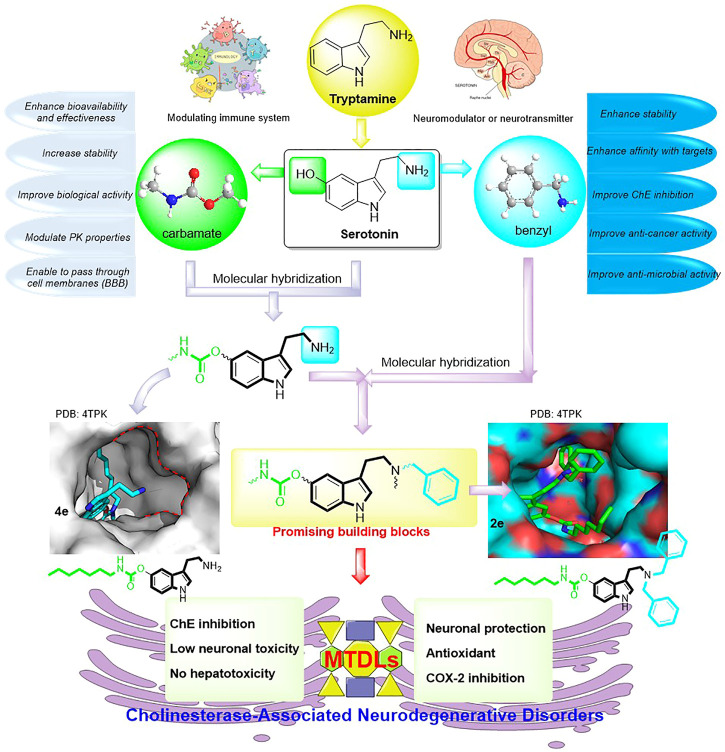
In this context and combined with the previous studies (Darras et al., 2012; Huang et al., 2014; Sawatzky et al., 2016; Scheiner et al., 2022), we first synthesized and evaluated the potential bioactivities of CDTs. Among them, 4e possessed highly selective BuChE inhibitory activity (Scheme 1C). Further molecular docking assay revealed that the amino group is oriented toward two unacted subpockets (Figure 1). To further improve its BuChE inhibition potency, we decide to further introduce an additional auxiliary fragment. Based on surveying extensive pieces of the literature, we find that the benzyl group is often involved in the design of pharmaceutical molecules to improve the stability and the biological activity of intrinsic compounds, which are also widely distributed in the structure of natural products (Scheme 1C) (Cacabelos, 2007; Guo et al., 2018; Jiang et al., 2019; Sang et al., 2020; Choubey et al., 2021; Saeedi et al., 2021; Zhu et al., 2021; Yang et al., 2022). Enlightened by this, benzyl-substituted CTDs were subsequently obtained, and the molecular docking assay confirmed that the benzyl group could interact with the two active subpockets (Figure 1). To develop potent benzyl-substituted CTDs for the discovery of multifunctional drugs against refractory diseases, herein we designed and synthesized a series of CTDs bearing benzyl groups and focused on their action modes of ChE inhibition and kinetic characteristics of interaction with BuChE (Figure 1). We also evaluated in vitro neuronal protection effects, antioxidant activities, and COX-2 inhibitory effects of the selected compounds with little neuronal toxicity and hepatotoxicity.
FIGURE 1.
Design strategy for developing promising building blocks for MTDLs against ChE-associated neurodegenerative disorders.
Experimental section
Chemistry
All reagents were used without further purification and bought from common commercial suppliers. Proton (1H) and carbon (13C) NMR spectra (400 or 300 MHz for 1HNMR; 101 or 75 MHz for 13CNMR) were recorded on a Bruker spectrometer (Bruker Company, Germany). NMR spectra used DMSO-d 6 , MeOD, or CDCl3 as a solvent. Proton chemical shifts are reported relative to a residual solvent peak (CDCl3 at 7.26 ppm, DMSO-d 6 at 2.50 ppm, MeOD at 3.31 ppm). Carbon chemical shifts are reported relative to a residual solvent peak (CDCl3 at 77.00 ppm, DMSO-d 6 at 39.60 ppm, MeOD at 49.00 ppm). The values of the chemical shifts are expressed in ppm and the coupling constants (J) in hertz. The following abbreviations were used to designate multiplicities: s = singlet, d = doublet, t = triplet, q = quartet, and m = multiplet. High-resolution mass spectrometry (HRMS) was obtained on an Agilent UPLC-IM-QTOF spectrometer (Agilent 6560, United States). Mass spectrometry (MS) analysis was performed using a liquid chromatograph-mass spectrometer (LC-MS). The purity was determined by high-performance liquid chromatography (HPLC). The purity of all final compounds was more than 95% (Agilent 1260 Infinity II; United States). The column was Eclipse Plus C18 (4.6 × 150 mm, 4 µm). Chromatographic conditions for all compounds: Mobile phase: 0–7 min, MeOH:H2O = 20:80; 7–16 min MeOH:H2O = 95:5, 16–25 min MeOH:H2O = 20:80; wavelength: 254 nm; column temperature: 25°C; flow rate of 0.5 ml/min. All synthesized compounds are >95% pure by HPLC analysis.
2a : 3-(2-(dibenzylamino)ethyl)-1H-indol-5-yl propylcarbamate: white solid (68% yield). 1H NMR (400 MHz, CDCl3-d) δ 7.88 (s, 1H, NH), 7.25 (d, J = 7.6 Hz, 4H, aromatic protons), 7.20 – 7.14 (m, 4H, aromatic protons), 7.09 (m, 3H, aromatic protons), 6.97 – 6.90 (m, 2H, aromatic protons), 6.72 (m, 1H, aromatic proton), 6.57 (s, 1H, CH), 3.53 (s, 4H, CH2), 3.11 (q, J = 6.7 Hz, 2H, CH2), 2.74 (m, 2H, CH2), 2.61 (m, 2H, CH2), 1.46 (m, 2H, CH2), 0.84 (t, J = 7.4 Hz, 3H, CH3). 13C NMR (75 MHz, CDCl3) δ 156.15, 144.36, 140.14, 134.15, 129.09, 128.45, 128.04, 127.05, 123.16, 116.37, 111.63, 111.28, 58.53, 54.01, 43.28, 23.43, 23.18, 11.57. MS (ESI+) m/z calcd for C28H32N3O2 + [M+H]+ 442.2, found 442.2. Purity = 96.4%.
2b : 3-(2-(dibenzylamino)ethyl)-1H-indol-5-yl butylcarbamate: orange solid (70% yield). 1H NMR (400 MHz, CDCl3-d) δ 7.89 (s, 1H, NH), 7.36 – 7.30 (m, 4H, aromatic protons), 7.27 – 7.21 (m, 4H, aromatic protons), 7.18 (m, 1H, aromatic proton), 7.16 (d, J = 2.3 Hz, 1H, aromatic proton), 7.15 – 7.10 (m, 1H, aromatic proton), 7.02 (d, J = 2.3 Hz, 1H, aromatic proton), 6.83 (m, 1H, aromatic proton), 6.75 (d, J = 2.3 Hz, 1H, CH), 3.61 (s, 4H, CH2), 3.23 (q, J = 6.7 Hz, 2H, CH2), 2.84 (m, 2H, CH2), 2.73 – 2.67 (m, 2H, CH2), 1.56 – 1.46 (m, 2H, CH2), 1.41 – 1.29 (m, 2H, CH2), 0.91 (t, J = 7.3 Hz, 3H, CH3). 13C NMR (101 MHz, CDCl3) δ 155.73, 144.26, 139.90, 133.89, 128.94, 128.83, 128.24, 128.19, 127.88, 126.79, 122.77, 116.30, 114.73, 111.27, 111.15, 58.31, 53.76, 41.04, 32.04, 22.97, 19.98, 13.80. MS (ESI+) m/z calcd for C29H34N3O2 + [M+H]+ 456.3, found 456.3. Purity = 96.2%.
2c : 3-(2-(dibenzylamino)ethyl)-1H-indol-5-yl pentylcarbamate: orange solid (63% yield). 1H NMR (400 MHz, CDCl3-d) δ 7.99 (s, 1H, NH), 7.36 (d, J = 7.1 Hz, 4H, aromatic protons), 7.28 (t, J = 7.4 Hz, 4H, aromatic protons), 7.22 (s, 1H, aromatic proton), 7.20 (d, J = 7.2 Hz, 2H, aromatic protons), 7.08 (d, J = 8.7 Hz, 1H, aromatic proton), 7.05 (d, J = 2.1 Hz, 1H, aromatic proton), 6.84 (m, 1H, aromatic proton), 6.69 (s, 1H, CH), 3.65 (s, 4H, CH2), 3.25 (q, J = 6.8 Hz, 2H, CH2), 2.89 – 2.82 (m, 2H, CH2), 2.72 (m, 2H, CH2), 1.57 (q, J = 7.0 Hz, 2H, CH2), 1.34 (m, 4H, CH2), 0.91 (t, J = 6.7 Hz, 3H, CH3). 13C NMR (101 MHz, CDCl3) δ 155.82, 144.13, 139.87, 133.88, 128.90, 128.81, 128.21, 128.17, 127.78, 126.76, 122.85, 116.13, 114.41, 111.33, 111.02, 58.26, 53.74, 41.30, 29.59, 28.93, 22.90, 22.36, 14.02. MS (ESI+) m/z calcd for C30H36N3O2 + [M+H]+ 470.3, found 470.3. Purity = 97.9%.
2d : 3-(2-(dibenzylamino)ethyl)-1H-indol-5-yl hexylcarbamate: brown solid (67% yield). 1H NMR (400 MHz, CDCl3-d) δ 7.93 (s, 1H, NH), 7.28 (s, 2H, aromatic protons), 7.19 (t, J = 7.4 Hz, 4H, aromatic protons), 7.11 (m, 3H, aromatic protons), 7.00 – 6.93 (m, 2H, aromatic protons), 6.74 (m, 1H, aromatic proton), 6.58 (s, 1H, CH), 3.55 (s, 4H, CH2), 3.16 (q, J = 6.8 Hz, 2H, CH2), 2.76 (m, 2H, CH2), 2.63 (m, 2H, CH2), 1.46 (t, J = 7.3 Hz, 2H, CH2), 1.29 – 1.17 (m, 6H, CH2), 0.83 – 0.77 (m, 3H, CH3). 13C NMR (101 MHz, CDCl3) δ 155.83, 144.11, 139.84, 133.88, 128.90, 128.81, 128.17, 127.77, 126.77, 122.87, 116.10, 114.35, 111.34, 111.00, 58.25, 53.73, 41.33, 31.47, 29.86, 26.45, 22.88, 22.57, 14.04. MS (ESI+) m/z calcd for C31H38N3O2 + [M+H]+ 484.3, found 484.3. Purity = 95.4%.
2e : 3-(2-(dibenzylamino)ethyl)-1H-indol-5-yl heptylcarbamate: white solid (65% yield). 1H NMR (300 MHz, CDCl3-d) δ 7.87 (s, 1H, NH), 7.29 (d, J = 2.1 Hz, 2H, aromatic protons), 7.19 (t, J = 7.3 Hz, 5H, aromatic protons), 7.12 (m, 2H, aromatic protons), 7.03 (d, J = 8.5 Hz, 1H, aromatic proton), 6.96 (d, J = 2.4 Hz, 1H, aromatic proton), 6.76 (m, 1H, aromatic proton), 6.65 (d, J = 2.3 Hz, 1H, CH), 3.56 (s, 4H, CH2), 3.17 (q, J = 6.7 Hz, 2H, CH2), 2.77 (m, 2H, CH2), 2.64 (m, 2H, CH2), 1.47 (d, J = 8.7 Hz, 2H, CH2), 1.34 – 1.11 (m, 8H, CH2), 0.87 – 0.72 (m, 3H, CH3). 13C NMR (101 MHz, CDCl3) δ 155.78, 144.19, 139.89, 133.89, 128.92, 128.82, 128.23, 128.18, 126.78, 122.82, 116.21, 114.56, 111.31, 111.08, 58.39, 58.29, 53.75, 41.35, 31.78, 29.93, 28.98, 26.77, 22.94, 22.62, 14.11. MS (ESI+) m/z calcd for C32H40N3O2 + [M+H]+ 498.3, found 498.3. Purity = 96.4%.
2f : 3-(2-(dibenzylamino)ethyl)-1H-indol-5-yl cyclopropylcarbamate: white solid (70% yield). 1H NMR (400 MHz, CDCl3-d) δ 7.97 (s, 1H, NH), 7.39 – 7.34 (m, 4H, aromatic protons), 7.32 – 7.25 (m, 4H, aromatic protons), 7.25 – 7.22 (m, 1H, aromatic proton), 7.22 – 7.17 (m, 2H, aromatic protons), 7.13 (d, J = 8.7 Hz, 1H, aromatic proton), 7.06 (d, J = 2.2 Hz, 1H, aromatic proton), 6.88 – 6.82 (m, 1H, aromatic proton), 6.75 (d, J = 2.3 Hz, 1H, CH), 3.66 (s, 4H, CH2), 2.87 (m, 2H, CH2), 2.79 – 2.66 (m, 3H, CH2, CH), 0.85 – 0.71 (m, 2H, CH2), 0.69 – 0.53 (m, 2H, CH2). 13C NMR (101 MHz, CDCl3) δ 156.38, 144.12, 139.93, 133.94, 128.84, 128.21, 127.85, 126.87, 126.80, 122.87, 116.17, 114.61, 111.33, 111.08, 58.32, 53.79, 23.37, 22.98, 6.93. MS (ESI+) m/z calcd for C28H30N3O2 + [M+H]+ 440.2, found 440.2. Purity = 97.4%.
2g : 3-(2-(dibenzylamino)ethyl)-1H-indol-5-yl cyclopentylcarbamate: white solid (69% yield). 1H NMR (300 MHz, CDCl3-d) δ 7.93 (s, 1H, NH), 7.34 (d, J = 7.1 Hz, 4H, aromatic protons), 7.26 (t, J = 7.3 Hz, 4H, aromatic protons), 7.19 (d, J = 6.7 Hz, 2H, aromatic protons), 7.10 (d, J = 8.5 Hz, 1H, aromatic proton), 7.03 (d, J = 2.2 Hz, 1H, aromatic proton), 6.87 – 6.79 (m, 1H, aromatic proton), 6.73 (s, 1H, CH), 4.94 (d, J = 7.3 Hz, 1H, CH), 3.63 (s, 4H, CH2), 2.91 – 2.79 (m, 2H, CH2), 2.71 (m, 2H, CH2), 2.07 – 1.93 (m, 2H, CH2), 1.76 – 1.55 (m, 4H, CH2), 1.47 (m, 2H). 13C NMR (75 MHz, CDCl3) δ 144.39, 140.11, 134.10, 129.05, 128.41, 128.06, 127.01, 123.01, 116.49, 114.80, 111.51, 111.34, 58.51, 53.97, 53.24, 33.51, 23.82, 23.17. MS (ESI+) m/z calcd for C30H34N3O2 + [M+H]+ 468.3, found 468.3. Purity = 96.4%.
2h : 3-(2-(dibenzylamino)ethyl)-1H-indol-5-yl cyclohexylcarbamate: bright solid (61% yield). 1H NMR (300 MHz, CDCl3-d) δ 8.05 (s, 1H, NH), 7.43 (d, J = 6.5 Hz, 4H, aromatic protons), 7.35 (t, J = 7.4 Hz, 4H, aromatic protons), 7.27 (m, 2H, aromatic protons), 7.15 – 7.08 (m, 2H, aromatic protons), 6.90 (m, 1H, aromatic proton), 6.75 (m, 1H, CH), 5.00 (d, J = 8.1 Hz, 1H, CH), 3.71 (s, 4H, CH2), 2.91 (m, 2H, CH2), 2.78 (m, 2H, CH2), 2.07 (d, J = 10.8 Hz, 2H, CH2), 1.72 (m, 3H, CH2, CH), 1.50 – 1.35 (m, 2H, CH2), 1.26 (q, J = 11.6 Hz, 3H, CH3). 13C NMR (75 MHz, CDCl3) δ 154.95, 144.05, 139.85, 133.83, 128.88, 128.79, 128.16, 127.73, 126.75, 122.82, 116.13, 114.32, 111.31, 111.01, 58.31, 58.22, 53.70, 50.08, 33.32, 25.46, 24.78, 22.87. MS (ESI+) m/z calcd for C31H36N3O2 + [M+H]+ 482.3, found 482.3. Purity = 96.1%.
2i : 3-(2-(dibenzylamino)ethyl)-1H-indol-5-yl phenylcarbamate: orange solid (66% yield). 1H NMR (400 MHz, CDCl3-d) δ 7.95 (s, 1H, NH), 7.44 (d, J = 8.0 Hz, 2H, aromatic protons), 7.35 (d, J = 7.5 Hz, 4H, aromatic protons), 7.30 (s, 1H, aromatic proton), 7.25 (d, J = 7.4 Hz, 3H, aromatic protons), 7.18 (q, J = 8.3, 6.7 Hz, 3H, aromatic protons), 7.13 – 7.08 (m, 1H, aromatic proton), 7.07 – 7.04 (m, 2H, aromatic protons), 7.03 – 7.00 (m, 1H, aromatic proton), 6.89 (m, 1H, aromatic proton), 6.76 (s, 1H, CH), 3.64 (s, 4H, CH2), 2.86 (m, 2H, CH2), 2.73 (m, 2H, CH2). 13C NMR (101 MHz, CDCl3) δ 143.60, 139.79, 137.66, 134.01, 129.10, 128.76, 128.14, 127.81, 126.75, 123.67, 122.93, 118.63, 115.99, 114.60, 111.36, 111.15, 58.25, 53.62, 22.90. MS (ESI+) m/z calcd for C31H30N3O2 + [M+H]+ 476.2, found 476.2. Purity = 95.2%.
2j : 3-(2-(dibenzylamino)ethyl)-1H-indol-5-yl dimethylcarbamate: white solid (64% yield). 1H NMR (400 MHz, CDCl3-d) δ 7.91 (s, 1H, NH), 7.42 – 7.36 (m, 4H, aromatic protons), 7.30 (t, J = 7.5 Hz, 4H, aromatic protons), 7.25 – 7.19 (m, 3H, aromatic protons), 7.06 (d, J = 2.2 Hz, 1H, aromatic proton), 6.89 (m, 1H, aromatic proton), 6.85 (d, J = 2.3 Hz, 1H, CH), 3.68 (s, 4H, CH2), 3.15 (s, 3H, CH3), 3.05 (s, 3H, CH3), 2.91 (m, 2H, CH2), 2.78 (m, 2H, CH2). 13C NMR (75 MHz, CDCl3) δ 156.44, 144.81, 140.20, 134.13, 129.08, 128.44, 128.03, 127.01, 123.09, 116.49, 114.57, 111.57, 111.32, 58.60, 58.51, 54.02, 37.02, 36.74, 23.20. MS (ESI+) m/z calcd for C27H30N3O2 + [M+H]+ 428.2, found 428.2. Purity = 95.8%.
2k : 3-(2-(dibenzylamino)ethyl)-1H-indol-5-yl diethylcarbamate: orange solid (66% yield). 1H NMR (300 MHz, CDCl3-d) δ 8.03 (s, 1H, NH), 7.40 (d, J = 7.1 Hz, 4H, aromatic protons), 7.32 (t, J = 7.3 Hz, 4H, aromatic protons), 7.27 – 7.21 (m, 2H, aromatic protons), 7.14 (d, J = 8.7 Hz, 1H, aromatic proton), 7.08 (d, J = 2.2 Hz, 1H, aromatic proton), 6.87 (m, 1H, aromatic proton), 6.75 (d, J = 2.2 Hz, 1H, CH), 3.69 (s, 4H, CH2), 3.48 (q, J = 8.0 Hz, 4H, CH2), 2.91 (m, 2H, CH2), 2.77 (m, 2H, CH2), 1.35 – 1.22 (m, 6H, CH3). 13C NMR (75 MHz, CDCl3) δ 155.34, 144.54, 139.83, 133.75, 128.76, 128.11, 127.75, 126.71, 122.64, 116.30, 114.36, 111.16, 111.01, 58.18, 53.66, 41.80, 22.84, 14.28, 13.42. MS (ESI+) m/z calcd for C29H34N3O2 + [M+H]+ 456.3, found 456.3. Purity = 95.5%.
2l : 3-(2-(dibenzylamino)ethyl)-1H-indol-5-yl dipropylcarbamate: orange solid (61% yield). 1H NMR (300 MHz, CDCl3-d) δ 9.39 (s, 1H, NH), 8.74 (t, J = 5.0 Hz, 4H, aromatic protons), 8.66 (m, 4H, aromatic protons), 8.58 (m, 2H, aromatic protons), 8.44 (m, 2H, aromatic protons), 8.19 (q, J = 3.6 Hz, 1H, aromatic proton), 8.06 (s, 1H, CH), 5.03 (d, J = 3.7 Hz, 4H, CH2), 4.70 (m, 4H, CH2), 4.32 – 4.18 (m, 2H, CH2), 4.11 (m, 2H, CH2), 3.06 (s, 4H, CH2), 2.35 (m, 6H, CH3). 13C NMR (101 MHz, CDCl3) δ 155.94, 144.67, 139.97, 133.85, 128.95, 128.85, 128.21, 127.83, 126.79, 122.75, 116.32, 114.42, 111.27, 111.02, 58.29, 53.80, 49.58, 49.27, 22.93, 22.13, 21.39, 11.40. MS (ESI+) m/z calcd for C31H38N3O2 + [M+H]+ 484.3, found 484.3. Purity = 96.5%.
2m : 3-(2-(dibenzylamino)ethyl)-1H-indol-5-yl dibutylcarbamate: orange solid (60% yield). 1H NMR (300 MHz, CDCl3-d) δ 9.27 (s, 1H, NH), 8.67 (d, J = 7.3 Hz, 4H, aromatic protons), 8.58 (t, J = 7.3 Hz, 4H, aromatic protons), 8.51 (d, J = 7.5 Hz, 2H, aromatic protons), 8.43 (d, J = 8.7 Hz, 1H, aromatic proton), 8.35 (d, J = 2.3 Hz, 1H, aromatic proton), 8.13 (d, J = 8.6 Hz, 1H, aromatic proton), 8.03 (s, 1H, CH), 4.96 (m, 4H, CH2), 4.67 (m, 4H, CH2), 4.17 (d, J = 7.9 Hz, 2H, CH2), 4.10 – 3.98 (m, 2H, CH2), 2.94 (m, 4H, CH2), 2.69 (q, J = 9.8, 8.3 Hz, 4H, CH2), 2.26 (q, J = 7.3 Hz, 6H, CH3). 13C NMR (101 MHz, CDCl3) δ 155.80, 144.73, 139.94, 133.81, 128.83, 128.19, 127.86, 126.77, 122.66, 116.38, 114.55, 111.20, 111.07, 58.28, 53.78, 47.57, 47.23, 31.04, 30.28, 22.93, 20.14, 13.96. MS (ESI+) m/z calcd for C33H42N3O2 + [M+H]+ 512.3, found 512.3. Purity = 98.1%.
2n: 3-(2-(dibenzylamino)ethyl)-1H-indol-5-yl diphenylcarbamate: orange solid (67% yield). 1H NMR (300 MHz, CDCl3-d) δ 7.90 (s, 1H, NH), 7.41 – 7.32 (m, 12H, aromatic protons), 7.29 (d, J = 1.3 Hz, 1H, aromatic proton), 7.28 – 7.23 (m, 4H, aromatic protons), 7.23 – 7.15 (m, 3H, aromatic protons), 7.12 (d, J = 8.6 Hz, 1H, aromatic proton), 7.09 (d, J = 2.3 Hz, 1H, aromatic proton), 6.88 (m, 1H, aromatic proton), 6.73 (d, J = 2.3 Hz, 1H, CH), 3.65 (s, 4H, CH2), 2.87 (m, 2H, CH2), 2.73 (m, 2H, CH2). 13C NMR (75 MHz, CDCl3) δ 154.09, 144.23, 142.53, 139.82, 133.84, 128.95, 128.74, 128.12, 127.71, 126.91, 126.73, 126.23, 122.74, 115.87, 114.58, 111.16, 110.87, 58.21, 53.63, 22.88. MS (ESI+) m/z calcd for C37H34N3O2 + [M+H]+ 552.3, found 552.3. Purity = 98.9%.
2o : 3-(2-(dibenzylamino)ethyl)-1H-indol-5-yl methoxy(methyl)carbamate: brown solid (65% yield). 1H NMR (300 MHz, CDCl3-d) δ 7.96 (s, 1H, NH), 7.38 (d, J = 7.4 Hz, 4H, aromatic protons), 7.29 (m, 4H, aromatic protons), 7.25 – 7.16 (m, 3H, aromatic protons), 7.09 (s, 1H, aromatic proton), 6.90 (d, J = 8.3 Hz, 1H, aromatic proton), 6.82 (s, 1H, CH), 3.84 (d, J = 2.4 Hz, 3H, CH3), 3.68 (s, 4H, CH2), 3.33 (d, J = 2.4 Hz, 3H, CH3), 2.95 – 2.85 (m, 2H, CH2), 2.77 (d, J = 7.9 Hz, 2H, CH2). 13C NMR (101 MHz, CDCl3) δ 156.32, 144.08, 139.83, 133.98, 128.79, 128.15, 127.82, 126.76, 122.86, 115.99, 114.69, 111.28, 111.03, 61.78, 58.26, 53.68, 35.77, 22.93. HRMS (ESI+) m/z calcd for C27H30N3O3 + [M+H]+ 444.2282, found 444.2307. Purity = 96.7%.
2p : 3-(2-(dibenzylamino)ethyl)-1H-indol-5-yl methyl(phenyl)carbamate: orange solid (70% yield). 1H NMR (400 MHz, CDCl3-d) δ 7.91 (s, 1H, NH), 7.42 (d, J = 4.3 Hz, 4H, aromatic protons), 7.38 (d, J = 7.5 Hz, 4H, aromatic protons), 7.29 (t, J = 7.4 Hz, 5H, aromatic protons), 7.21 (m, 3H, aromatic protons), 7.07 (s, 1H, aromatic proton), 6.89 (d, J = 8.7 Hz, 1H, aromatic proton), 6.84 (s, 1H, CH), 3.68 (s, 4H, CH2), 3.48 (m, 3H, CH3), 2.91 (m, 2H, CH2), 2.77 (m, 2H, CH2). 13C NMR (101 MHz, CDCl3) δ 154.97, 144.54, 143.25, 133.85, 128.94, 128.82, 128.16, 127.80, 126.79, 125.78, 122.69, 116.22, 111.13, 111.06, 58.23, 53.63, 38.13, 22.89. HRMS (ESI+) m/z calcd for C32H32N3O2 + [M+H]+ 490.2489, found 490.2508. Purity = 96.9%.
2q : 3-(2-(dibenzylamino)ethyl)-1H-indol-5-yl azetidine-1-carboxylate: brown solid (50% yield). 1H NMR (400 MHz, CDCl3-d) δ 7.96 (s, 1H, NH), 7.37 (d, J = 7.0 Hz, 4H, aromatic protons), 7.29 (t, J = 7.5 Hz, 4H, aromatic protons), 7.24 – 7.19 (m, 2H, aromatic protons), 7.14 (m, 1H, aromatic proton), 7.06 (d, J = 2.3 Hz, 1H, aromatic proton), 6.86 (m, 1H, aromatic proton), 6.77 (d, J = 2.7 Hz, 1H, CH), 4.35 – 4.03 (m, 4H, CH2), 3.66 (s, 4H, CH2), 2.88 (m, 2H, CH2), 2.79 – 2.71 (m, 2H, CH2), 2.40 – 2.27 (m, 2H, CH2). 13C NMR (101 MHz, CDCl3) δ 155.53, 144.15, 139.86, 133.83, 128.78, 128.13, 127.80, 126.71, 122.70, 116.19, 114.52, 111.19, 111.02, 58.22, 53.74, 22.92, 15.79. HRMS (ESI+) m/z calcd for C28H30N3O2 + [M+H]+ 440.2333, found 440.2332. Purity = 95.3%.
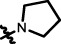
2r : 3-(2-(dibenzylamino)ethyl)-1H-indol-5-yl pyrrolidine-1-carboxylate: brown solid (53% yield). 1H NMR (300 MHz, CDCl3-d) δ 7.96 (s, 1H, NH), 7.42 – 7.34 (m, 4H, aromatic protons), 7.33 – 7.26 (m, 4H, aromatic protons), 7.23 (m, 1H, aromatic proton), 7.20 (d, J = 2.7 Hz, 1H, aromatic proton), 7.18 – 7.12 (m, 1H, aromatic proton), 7.07 (d, J = 2.2 Hz, 1H, aromatic proton), 6.88 (m, 1H, aromatic proton), 6.77 (d, J = 2.3 Hz, 1H, CH), 3.66 (s, 4H, CH2), 3.60 (t, J = 6.5 Hz, 2H, CH2), 3.51 (t, J = 6.5 Hz, 2H, CH2), 2.89 (dd, J = 9.4, 5.8 Hz, 2H, CH2), 2.81 – 2.70 (m, 2H, CH2), 2.05 – 1.87 (m, 4H, CH2). 13C NMR (75 MHz, CDCl3) δ 154.32, 144.49, 139.86, 133.77, 128.76, 128.11, 127.78, 126.68, 122.61, 116.39, 114.50, 111.13, 58.19, 53.71, 46.41, 46.30, 25.87, 25.03, 22.91. MS (ESI+) m/z calcd for C29H32N3O2 + [M+H]+ 454.2, found 454.2. Purity = 96.0%.
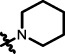
2s : 3-(2-(dibenzylamino)ethyl)-1H-indol-5-yl piperidine-1-carboxylate: brown solid (57% yield). 1H NMR (300 MHz, CDCl3-d) δ 8.02 (s, 1H, NH), 7.37 (d, J = 7.1 Hz, 4H, aromatic protons), 7.33 – 7.25 (m, 4H, aromatic protons), 7.20 (m, J = 6.1, 1.7 Hz, 2H, aromatic protons), 7.08 (d, J = 8.8 Hz, 1H, aromatic proton), 7.03 (d, J = 2.3 Hz, 1H, aromatic proton), 6.83 (dd, J = 8.6, 2.3 Hz, 1H, aromatic proton), 6.68 (d, J = 2.3 Hz, 1H, CH), 3.65 (s, 6H, CH2), 3.55 (s, 2H, CH2), 2.86 (dd, J = 9.6, 5.7 Hz, 2H, CH2), 2.72 (dd, J = 10.2, 5.9 Hz, 2H, CH2), 1.65 (s, 6H, CH2). 13C NMR (75 MHz, CDCl3) δ 154.91, 144.50, 139.79, 133.75, 128.75, 128.11, 127.70, 126.70, 122.69, 116.21, 114.22, 111.20, 110.97, 58.14, 53.65, 45.34, 45.02, 25.86, 25.58, 24.34, 22.82. MS (ESI+) m/z calcd for C30H34N3O2 + [M+H]+ 468.3, found 468.3. Purity = 96.1%.
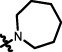
2t : 3-(2-(dibenzylamino)ethyl)-1H-indol-5-yl azepane-1-carboxylate: orange solid (58% yield). 1H NMR (400 MHz, CDCl3-d) δ 7.97 – 7.90 (m, 1H, NH), 7.32 (d, J = 7.1 Hz, 4H, aromatic protons), 7.24 (m, 4H, aromatic protons), 7.18 (t, J = 1.8 Hz, 1H, aromatic protons), 7.16 – 7.13 (m, 1H, aromatic protons), 7.07 (d, J = 8.7 Hz, 1H, aromatic proton), 7.01 (d, J = 2.2 Hz, 1H, aromatic protons), 6.80 (dd, J = 8.7, 2.3 Hz, 1H, aromatic protons), 6.68 (d, J = 2.3 Hz, 1H, CH), 3.61 (s, 4H, CH2), 3.59 – 3.54 (m, 2H, CH2), 3.50 (t, J = 6.1 Hz, 2H, CH2), 2.83 (m, 2H, CH2), 2.70 (m, 2H, CH2), 1.84 – 1.68 (m, 4H, CH2), 1.60 (m, 4H, CH2). 13C NMR (101 MHz, CDCl3) δ 155.82, 144.72, 139.94, 133.84, 128.84, 128.19, 127.86, 126.78, 122.70, 116.42, 114.51, 111.24, 111.09, 58.28, 53.79, 47.39, 47.15, 28.82, 28.24, 27.59, 27.03, 22.95. MS (ESI+) m/z calcd for C31H36N3O2 + [M+H]+ 482.3, found 482.3. Purity = 96.3%.
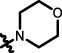
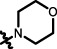
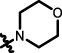
2u : 3-(2-(dibenzylamino)ethyl)-1H-indol-5-yl morpholine-4-carboxylate: white solid (59% yield). 1H NMR (400 MHz, CDCl3-d) δ 7.98 (s, 1H, NH), 7.36 (d, J = 7.0 Hz, 4H, aromatic protons), 7.28 (t, J = 7.5 Hz, 4H, aromatic protons), 7.22 (m, 1H, aromatic proton), 7.21 – 7.18 (m, 1H, aromatic proton), 7.16 – 7.11 (m, 2H, aromatic proton), 7.05 (d, J = 2.3 Hz, 1H, aromatic proton), 6.85 (dd, J = 8.7, 2.3 Hz, 1H, aromatic proton), 6.73 (d, J = 2.2 Hz, 1H, CH), 3.76 (m, 4H, CH2), 3.72 – 3.54 (m, 8H, CH2), 2.87 (m, 2H, CH2), 2.74 (m, 2H, CH2). 13C NMR (101 MHz, CDCl3) δ 154.95, 144.37, 139.91, 133.97, 128.85, 128.57, 128.22, 127.90, 126.80, 122.94, 116.14, 114.57, 111.37, 111.08, 66.73, 58.29, 53.79, 44.88, 44.23, 22.98. MS (ESI+) m/z calcd for C29H30N3O3 + [M+H]+ 470.2, found 470.2. Purity = 95.3%.
3a: 3-(2-(benzylamino)ethyl)-1H-indol-5-yl propylcarbamate: white solid (85% yield). 1H NMR (400 MHz, MeOD) δ 7.42 – 7.34 (m, 5H, aromatic protons), 7.32 (d, J = 8.7 Hz, 1H, NH), 7.25 (d, J = 2.2 Hz, 1H, aromatic proton), 7.14 (s, 1H, aromatic proton), 6.86 (dd, J = 8.7, 2.3 Hz, 1H, CH), 4.00 (s, 2H, CH2), 3.15 (t, J = 7.1 Hz, 2H, CH2), 3.11 (m, 2H, CH2), 3.07 – 3.00 (m, 2H, CH2), 1.59 (m, 2H, CH2), 0.97 (t, J = 7.4 Hz, 3H, CH3). 13C NMR (101 MHz, MeOD) δ 158.31, 145.49, 135.63, 135.02, 130.27, 129.78, 129.59, 128.24, 125.29, 117.05, 112.55, 111.37, 52.67, 49.06, 43.60, 23.90, 23.85, 11.42. MS (ESI+) m/z calcd for C21H26N3O2 + [M+H]+ 352.2, found 352.2. Purity = 95.1%.
3b : 3-(2-(benzylamino)ethyl)-1H-indol-5-yl butylcarbamate: white solid (88% yield). 1H NMR (400 MHz, MeOD) δ 7.46 (m, J = 10.9, 3.9, 2.1 Hz, 5H, aromatic protons), 7.34 (d, J = 8.7 Hz, 1H, NH), 7.27 (d, J = 2.2 Hz, 1H, aromatic proton), 7.21 (s, 1H, aromatic proton), 6.87 (dd, J = 8.7, 2.2 Hz, 1H, CH), 4.16 (s, 2H, CH2), 3.27 (t, J = 7.7 Hz, 2H, CH2), 3.20 (t, J = 7.0 Hz, 2H, CH2), 3.15 – 3.08 (m, 2H, CH2), 1.61 – 1.50 (m, 2H, CH2), 1.42 (m, 2H, CH2), 0.97 (t, J = 7.3 Hz, 3H, CH3). 13C NMR (101 MHz, MeOD) δ 158.28, 145.59, 135.66, 132.47, 130.79, 130.41, 130.05, 128.08, 125.58, 117.20, 112.67, 111.32, 110.30, 52.04, 48.62, 41.54, 32.81, 22.99, 20.79, 13.93. MS (ESI+) m/z calcd for C22H28N3O2 + [M+H]+ 366.2, found 366.2. Purity = 95.7%.
3c : 3-(2-(benzylamino)ethyl)-1H-indol-5-yl pentylcarbamate: white solid (90% yield). 1H NMR (400 MHz, MeOD) δ 7.50 (m, J = 7.2, 3.6 Hz, 5H, aromatic protons), 7.38 (d, J = 8.6 Hz, 1H, NH), 7.30 (d, J = 2.3 Hz, 1H, aromatic proton), 7.24 (s, 1H, aromatic proton), 6.91 (dd, J = 8.7, 2.3 Hz, 1H, CH), 4.21 (s, 2H, CH2), 3.34 (s, 2H, CH2), 3.22 (t, J = 7.1 Hz, 2H, CH2), 3.16 (t, J = 7.7 Hz, 2H, CH2), 1.61 (t, J = 7.1 Hz, 2H, CH 2), 1.52 – 1.32 (m, 4H, CH2), 0.98 (t, J = 6.5 Hz, 3H, CH3). 13C NMR (101 MHz, MeOD) δ 158.27, 145.61, 135.67, 132.38, 130.79, 130.45, 130.07, 128.08, 125.59, 117.21, 112.67, 111.31, 110.27, 52.03, 49.23, 41.84, 30.37, 29.92, 23.24, 22.97, 14.17. MS (ESI+) m/z calcd for C23H30N3O2 + [M+H]+ 380.2, found 380.2. Purity = 95.6%.
3d : 3-(2-(benzylamino)ethyl)-1H-indol-5-yl hexylcarbamate: white solid (93% yield). 1H NMR (300 MHz, MeOD) δ 7.35 – 7.19 (m, 7H, aromatic protons), 7.06 (s, 1H, NH), 6.84 (dd, J = 8.8, 2.2 Hz, 1H, CH), 3.77 (s, 2H, CH2), 3.18 (t, J = 7.0 Hz, 2H, CH2), 2.91 (q, J = 4.4 Hz, 4H, CH2), 1.63 – 1.49 (m, 2H, CH2), 1.37 (m, 6H, CH2), 0.92 (t, J = 6.5 Hz, 3H, CH3). 13C NMR (75 MHz, CD3OD) δ 157.30, 144.36, 138.41, 134.62, 128.49, 128.38, 127.57, 127.27, 123.84, 115.87, 112.21, 111.37, 110.52, 52.84, 48.89, 40.89, 31.55, 29.70, 26.44, 24.52, 22.52, 13.27. MS (ESI+) m/z calcd for C24H32N3O2 + [M+H]+ 394.2, found 394.2. Purity = 98.8%.
3e : 3-(2-(benzylamino)ethyl)-1H-indol-5-yl heptylcarbamate: white solid (95% yield). 1H NMR (400 MHz, MeOD) δ 7.47 (m, J = 6.9, 2.8 Hz, 6H, aromatic protons), 7.35 (d, J = 8.7 Hz, 1H, NH), 7.27 (d, J = 2.2 Hz, 1H, aromatic proton), 7.22 (s, 1H, aromatic proton), 6.87 (dd, J = 8.7, 2.3 Hz, 1H, CH), 4.19 (d, J = 3.4 Hz, 2H, CH2), 3.28 (d, J = 7.8 Hz, 2H, CH2), 3.19 (t, J = 7.1 Hz, 2H, CH2), 3.13 (t, J = 8.0 Hz, 2H, CH2), 1.57 (t, J = 7.1 Hz, 2H, CH2), 1.46 – 1.28 (m, 8H, CH2), 0.96 – 0.88 (m, 3H, CH3). 13C NMR (101 MHz, MeOD) δ 158.27, 145.61, 135.68, 132.34, 130.80, 130.47, 130.08, 128.07, 125.59, 117.21, 112.67, 111.31, 110.24, 52.03, 49.23, 41.87, 32.77, 30.69, 29.94, 27.69, 23.48, 22.97, 14.22. HRMS (ESI+) m/z calcd for C25H34N3O2 + [M+H]+ 408.2646, found 408.2642. Purity = 98.0%.
3f : 3-(2-(benzylamino)ethyl)-1H-indol-5-yl cyclopropylcarbamate: white solid (88% yield). 1H NMR (400 MHz, MeOD) δ 7.38 (d, J = 3.3 Hz, 6H, aromatic protons), 7.32 (d, J = 8.7 Hz, 1H, NH), 7.25 (d, J = 2.2 Hz, 1H, aromatic proton), 7.13 (s, 1H, aromatic proton), 6.85 (dd, J = 8.7, 2.2 Hz, 1H, CH), 3.98 (s, 2H, CH2), 3.09 (m, 2H, CH2), 3.01 (m, 2H, CH2), 2.62 (t, J = 3.7 Hz, 1H, CH), 0.71 (m, 2H, CH2), 0.59 (q, J = 3.7 Hz, 2H, CH2). 13C NMR (101 MHz, MeOD) δ 159.08, 145.37, 135.62, 134.85, 130.29, 129.77, 129.60, 128.22, 125.34, 117.01, 112.58, 111.38, 111.33, 52.60, 49.44, 23.82, 6.50. MS (ESI+) m/z calcd for C21H24N3O2 + [M+H]+ 350.2, found 350.2. Purity = 97.0%.
3g : 3-(2-(benzylamino)ethyl)-1H-indol-5-yl cyclopentylcarbamate: white solid (89% yield). 1H NMR (400 MHz, MeOD) δ 7.50 – 7.42 (m, 5H, aromatic protons), 7.34 (d, J = 8.7 Hz, 1H, NH), 7.27 (d, J = 2.2 Hz, 1H, aromatic proton), 7.21 (s, 1H, aromatic proton), 6.87 (dd, J = 8.7, 2.2 Hz, 1H, CH), 4.18 (s, 2H, CH2), 4.04 – 3.91 (m, 1H, CH), 3.30 – 3.25 (m, 2H, CH2), 3.17 – 3.08 (m, 2H, CH2), 1.95 (m, 2H, CH2), 1.76 (m, 2H, CH2), 1.59 (m, 4H, CH2). 13C NMR (101 MHz, MeOD) δ 156.58, 144.40, 134.48, 131.18, 129.61, 129.27, 128.89, 126.88, 124.38, 116.05, 111.47, 110.15, 109.07, 52.76, 50.84, 32.28, 23.24, 21.79. HRMS (ESI+) m/z calcd for C23H28N3O2 + [M + H]+ 378.2176, found 378.2189. Purity = 97.6%.
3h : 3-(2-(benzylamino)ethyl)-1H-indol-5-yl cyclohexylcarbamate: white solid (86% yield). 1H NMR (400 MHz, MeOD) δ 7.50 – 7.40 (m, 5H, aromatic protons), 7.34 (d, J = 8.7 Hz, 1H, NH), 7.27 (d, J = 2.2 Hz, 1H, aromatic proton), 7.19 (s, 1H, aromatic proton), 6.87 (dd, J = 8.7, 2.3 Hz, 1H, CH), 4.14 (s, 2H, CH2), 3.45 (m, 1H, CH), 3.24 (m, 2H, CH2), 3.15 – 3.05 (m, 2H, CH2), 2.01 – 1.89 (m, 2H, CH2), 1.78 (m, 2H, CH2), 1.70 – 1.58 (m, 1H, CH2), 1.42 – 1.16 (m, 5H, CH2). 13C NMR (101 MHz, MeOD) δ 157.44, 145.56, 135.62, 132.50, 130.77, 130.36, 130.02, 128.08, 125.56, 117.22, 112.65, 111.36, 110.33, 52.03, 51.43, 33.87, 26.40, 25.98, 22.99. MS (ESI+) m/z calcd for C24H30N3O2 + [M+H]+ 392.2, found 392.2. Purity = 96.0%.
3i : 3-(2-(benzylamino)ethyl)-1H-indol-5-yl phenylcarbamate: white solid (93% yield). 1H NMR (400 MHz, MeOD) δ 7.55 – 7.49 (m, 2H, aromatic protons), 7.47 – 7.36 (m, 6H, aromatic protons), 7.35 (d, J = 2.2 Hz, 1H, NH), 7.33 – 7.27 (m, 2H, aromatic protons), 7.21 (s, 1H, aromatic proton), 7.09 – 7.03 (m, 1H, aromatic proton), 6.96 (dd, J = 8.7, 2.2 Hz, 1H, CH), 4.11 (d, J = 3.5 Hz, 2H, CH2), 3.22 (t, J = 7.2 Hz, 2H, CH2), 3.15 – 3.05 (m, 2H, CH2). 13C NMR (101 MHz, MeOD) δ 155.30, 145.15, 139.77, 135.80, 133.45, 130.58, 130.08, 129.94, 129.71, 128.19, 125.59, 124.21, 119.78, 117.08, 112.73, 111.46, 110.82, 52.26, 40.22, 23.36. MS (ESI+) m/z calcd for C24H24N3O2 + [M+H]+ 386.2, found 386.2. Purity = 96.3%.
3j : 3-(2-(benzylamino)ethyl)-1H-indol-5-yl dimethylcarbamate: white solid (92% yield). 1H NMR (400 MHz, CDCl3) δ 8.35 (s, 1H, NH), 7.30 (d, J = 4.4 Hz, 4H, aromatic protons), 7.25 – 7.23 (m, 2H, aromatic protons), 7.22 (s, 1H, aromatic proton), 6.92 – 6.86 (m, 2H, aromatic protons), 3.82 (s, 2H, CH2), 3.20 (s, 1H, CH), 3.14 (s, 3H, CH3), 3.03 (s, 3H, CH3), 2.91 (s, 4H, CH2). 13C NMR (101 MHz, MeOD) δ 155.75, 145.91, 136.03, 133.12, 130.66, 130.19, 129.98, 128.17, 125.97, 116.10, 112.91, 110.92, 110.79, 54.59, 52.24, 30.52, 23.24. MS (ESI+) m/z calcd for C20H24N3O2 + [M+H]+ 338.2, found 338.2. Purity = 95.3%.
3k : 3-(2-(benzylamino)ethyl)-1H-indol-5-yl diethylcarbamate: white solid (90% yield). 1H NMR (400 MHz, CDCl3) δ 8.50 (s, 1H, NH), 7.31 – 7.19 (m, 6H, aromatic protons), 7.15 (d, J = 8.7 Hz, 1H, aromatic proton), 6.87 (dd, J = 8.7, 2.3 Hz, 1H, aromatic proton), 6.79 (d, J = 1.8 Hz, 1H, CH), 3.78 (s, 2H, CH2), 3.44 (m 4H, CH2), 2.88 (s, 4H, CH2), 1.26 (m, 6H, CH3). 13C NMR (101 MHz, CDCl3) δ 155.40, 144.63, 139.58, 133.99, 128.35, 128.24, 127.56, 126.96, 123.37, 116.38, 113.35, 111.42, 110.94, 53.52, 49.01, 43.15, 42.17, 25.32, 14.22, 13.44. MS (ESI+) m/z calcd for C22H28N3O2 + [M+H]+ 366.2, found 366.2. Purity = 98.2%.
3l : 3-(2-(benzylamino)ethyl)-1H-indol-5-yl dipropylcarbamate: white solid (87% yield). 1H NMR (400 MHz, CDCl3) δ 8.42 (s, 1H, NH), 7.27 (d, J = 5.7 Hz, 4H, aromatic protons), 7.22 (m, J = 6.1, 1.4 Hz, 2H, aromatic protons), 7.14 (d, J = 8.7 Hz, 1H, aromatic proton), 6.85 (dd, J = 8.7, 2.3 Hz, 1H, aromatic proton), 6.79 (d, J = 1.9 Hz, 1H, CH), 3.77 (s, 2H, CH2), 3.36 (t, J = 7.6 Hz, 2H, CH2), 3.29 (t, J = 7.6 Hz, 2H, CH2), 2.87 (s, 4H, CH2), 1.67 (m, 4H, CH2), 0.94 (m, 6H, CH3). 13C NMR (101 MHz, CDCl3) δ 155.85, 144.71, 139.63, 133.97, 128.36, 128.23, 127.57, 126.96, 123.32, 116.40, 113.44, 111.39, 110.93, 53.55, 49.48, 49.16, 49.04, 25.35, 22.02, 21.27, 11.29. MS (ESI+) m/z calcd for C24H32N3O2 + [M+H]+ 394.2, found 394.2. Purity = 97.7%.
3m : 3-(2-(benzylamino)ethyl)-1H-indol-5-yl dibutylcarbamate: white solid (91% yield). 1H NMR (300 MHz, CDCl3) δ 9.44 (s, 1H, NH), 7.52 – 7.39 (m, 2H, aromatic protons), 7.37 – 7.18 (m, 5H, aromatic protons), 7.08 (d, J = 2.2 Hz, 1H, aromatic proton), 6.76 (dd, J = 8.7, 2.2 Hz, 1H, aromatic proton), 6.49 (s, 1H, CH), 3.89 (s, 2H, CH2), 3.39 (t, J = 7.5 Hz, 2H, CH2), 3.28 (t, J = 7.5 Hz, 2H, CH2), 2.95 – 2.80 (m, 2H, CH2), 2.71 (t, J = 7.8 Hz, 2H, CH2), 1.74 – 1.52 (m, 4H, CH2), 1.36 (m, 4H, CH2), 0.96 (m, 6H, CH3). 13C NMR (75 MHz, CDCl3) δ 156.05, 144.35, 133.86, 132.75, 129.68, 128.75, 128.61, 126.98, 124.26, 115.98, 111.92, 110.51, 110.11, 51.27, 47.46, 47.17, 46.92, 30.84, 30.12, 22.40, 20.01, 13.85. MS (ESI+) m/z calcd for C26H36N3O2 + [M+H]+ 422.3, found 422.3. Purity = 96.7%.
3n : 3-(2-(benzylamino)ethyl)-1H-indol-5-yl diphenylcarbamate: white solid (93% yield). 1H NMR (400 MHz, CDCl3) δ 8.29 (s, 1H, NH), 7.33 – 7.25 (m, 8H, aromatic protons), 7.22 (d, J = 2.3 Hz, 1H, aromatic proton), 7.20 – 7.10 (m, 7H, aromatic protons), 7.06 (d, J = 8.7 Hz, 1H, aromatic proton), 6.83 (dd, J = 8.7, 2.3 Hz, 1H, aromatic proton), 6.73 (d, J = 1.9 Hz, 1H, CH), 3.69 (s, 2H, CH2), 2.81 (s, 4H, CH2), 2.41 – 2.32 (m, 1H, NH). 13C NMR (101 MHz, CDCl3) δ 154.22, 144.33, 142.46, 139.44, 134.07, 128.98, 128.37, 128.23, 127.48, 127.01, 126.30, 123.49, 115.99, 113.49, 111.43, 110.80, 53.50, 48.96, 25.31. MS (ESI+) m/z calcd for C30H28N3O2 + [M+H]+ 462.2, found 462.2. Purity = 95.6%.
3o : 3-(2-(benzylamino)ethyl)-1H-indol-5-yl methoxy(methyl)carbamate: brown solid (85% yield). 1H NMR (300 MHz, CDCl3) δ 9.09 (s, 1H, NH), 7.44 – 7.37 (m, 2H, aromatic protons), 7.32 – 7.21 (m, 4H, aromatic protons), 7.22 – 7.16 (m, 2H, aromatic protons), 6.84 (dd, J = 8.6, 2.2 Hz, 1H, aromatic proton), 6.69 (s, 1H, CH), 5.86 (s, 1H, NH), 3.88 (s, 2H, CH2), 3.80 (s, 3H, CH3), 3.28 (s, 3H, CH3), 2.94 (t, J = 7.5 Hz, 2H, CH2), 2.85 (t, J = 6.8 Hz, 2H, CH2). 13C NMR (75 MHz, CDCl3) δ 156.40, 143.97, 134.12, 133.74, 129.47, 128.74, 128.42, 127.11, 124.36, 115.87, 111.95, 110.94, 110.59, 61.72, 51.76, 47.32, 35.61, 23.04. MS (ESI+) m/z calcd for C20H24N3O2 + [M+H]+ 462.2, found 462.2. Purity = 95.1%.
3p : 3-(2-(benzylamino)ethyl)-1H-indol-5-yl methyl(phenyl)carbamate: white solid (90% yield). 1H NMR (400 MHz, MeOD) δ 7.42 (d, J = 4.2 Hz, 3H, aromatic protons), 7.40 – 7.35 (m, 5H, aromatic protons), 7.33 (d, J = 8.7 Hz, 1H, aromatic proton), 7.28 (m, 2H, aromatic protons), 7.14 (s, 1H, aromatic proton), 6.88 (d, J = 8.7 Hz, 1H, CH), 3.98 (s, 2H, CH2), 3.42 (s, 3H, CH3), 3.10 (m, 2H, CH2), 3.03 (t, J = 7.4 Hz, 2H, CH2). 13C NMR (101 MHz, MeOD) δ 156.79, 145.57, 144.26, 135.73, 134.52, 130.35, 129.98, 129.80, 129.71, 128.20, 127.62, 127.02, 125.47, 116.85, 112.66, 111.28, 111.24, 52.50, 48.91, 38.46, 23.71. MS (ESI+) m/z calcd for C25H26N3O2 + [M+H]+ 400.2, found 400.2. Purity = 96.9%.
3q : 3-(2-(benzylamino)ethyl)-1H-indol-5-yl azetidine-1-carboxylate: brown solid (85% yield). 1H NMR (300 MHz, CDCl3) δ 9.23 (d, J = 11.6 Hz, 1H, NH), 7.62 (d, J = 23.6 Hz, 3H, aromatic protons), 7.34 (q, J = 3.1 Hz, 2H, aromatic protons), 7.31 – 7.28 (m, 1H, aromatic proton), 7.19 – 7.10 (m, 2H, aromatic protons), 6.82 (m, 1H, aromatic proton), 6.60 (d, J = 3.1 Hz, 1H, CH), 4.15 (d, J = 32.9 Hz, 4H, CH2), 3.85 (d, J = 2.9 Hz, 2H, CH2), 2.95 – 2.75 (m, 4H, CH2), 2.31 (m, 2H, CH2). 13C NMR (75 MHz, CDCl3) δ 155.63, 144.04, 134.05, 129.40, 128.69, 128.32, 127.08, 124.13, 116.11, 111.78, 110.86, 110.60, 51.45, 50.10, 49.19, 47.11, 22.92, 15.72. HRMS (ESI+) m/z calcd for C21H24N3O2 + [M+H]+ 350.1863, found 350.1860. Purity = 97.6%.
3r : 3-(2-(dibenzylamino)ethyl)-1H-indol-5-yl pyrrolidine-1-carboxylate: brown solid (88% yield). 1H NMR (400 MHz, MeOD) δ 7.32 (d, J = 3.2 Hz, 4H, aromatic protons), 7.31 – 7.25 (m, 2H, aromatic protons), 7.23 (d, J = 2.2 Hz, 1H, aromatic proton), 7.12 (s, 1H, CH), 6.86 (dd, J = 8.7, 2.3 Hz, 1H, aromatic proton), 3.87 (s, 2H, CH2), 3.62 (t, J = 6.6 Hz, 2H, CH2), 3.45 (t, J = 6.6 Hz, 2H, CH2), 2.99 (s, 4H, CH2), 1.99 (m, 4H, CH2). 13C NMR (101 MHz, MeOD) δ 154.94, 144.25, 137.20, 134.52, 128.47, 128.27, 127.37, 127.28, 123.78, 115.70, 111.59, 111.18, 110.29, 52.37, 48.47, 46.13, 46.07, 25.38, 24.56, 23.94. HRMS (ESI+) m/z calcd for C22H26N3O2 + [M+H]+ 364.2020, found 364.2025. Purity = 96.7%.
3s : 3-(2-(benzylamino)ethyl)-1H-indol-5-yl piperidine-1-carboxylate: brown solid (87% yield). 1H NMR (300 MHz, CDCl3) δ 9.14 (s, 1H, NH), 7.41 (d, J = 7.2 Hz, 2H, aromatic protons), 7.29 (m, 3H, aromatic protons), 7.21 (dd, J = 8.8, 1.4 Hz, 1H, aromatic proton), 7.11 (d, J = 1.8 Hz, 1H, aromatic proton), 5.96 (s, 1H, CH), 3.87 (s, 2H, CH2), 3.62 (s, 2H, CH2), 3.51 – 3.39 (m, 2H, CH2), 2.85 (t, J = 7.4 Hz, 2H, CH2), 2.75 (d, J = 7.2 Hz, 2H, CH2), 1.64 (s, 6H, CH2). 13C NMR (75 MHz, CDCl3) δ 155.12, 144.46, 133.92, 129.46, 128.72, 128.34, 127.13, 124.07, 116.16, 111.85, 110.81, 110.69, 51.78, 47.42, 45.15, 25.53, 24.27, 22.94. MS (ESI+) m/z calcd for C23H28N3O2 + [M+H]+ 378.2, found 378.2. Purity = 95.2%.
3t : 3-(2-(benzylamino)ethyl)-1H-indol-5-yl azepane-1-carboxylate: white solid (93% yield). 1H NMR (300 MHz, CDCl3) δ 8.96 (s, 1H, NH), 7.41 – 7.34 (m, 2H, aromatic protons), 7.34 – 7.25 (m, 3H, aromatic protons), 7.21 (d, J = 8.7 Hz, 1H, aromatic proton), 7.15 (d, J = 2.2 Hz, 1H, aromatic proton), 6.82 (dd, J = 8.6, 2.2 Hz, 1H, aromatic proton), 6.65 (s, 1H, CH), 4.86 (s, 1H, NH), 3.84 (s, 2H, CH2), 3.65 – 3.56 (m, 2H, CH2), 3.49 (t, J = 6.0 Hz, 2H, CH2), 2.82 (m, 4H, CH2), 1.87 – 1.72 (m, 4H, CH2), 1.72 – 1.53 (m, 4H, CH2). 13C NMR (75 MHz, CDCl3) δ 155.97, 144.54, 135.58, 133.94, 129.12, 128.62, 127.97, 127.26, 123.89, 116.26, 111.72, 111.53, 110.77, 47.85, 47.35, 47.10, 28.65, 28.10, 27.46, 26.94, 23.59. MS (ESI+) m/z calcd for C24H30N3O2 + [M+H]+ 392.2, found 392.2. Purity = 97.4%.
3u: 3-(2-(benzylamino)ethyl)-1H-indol-5-yl morpholine-4-carboxylate: white solid (92% yield). 1H NMR (400 MHz, CDCl3) δ 8.97 (s, 1H, NH), 7.44 – 7.36 (m, 2H, aromatic protons), 7.29 (t, J = 7.0 Hz, 2H, aromatic protons), 7.26 – 7.19 (m, 2H, aromatic protons), 7.17 (d, J = 2.3 Hz, 1H, aromatic proton), 6.82 (dd, J = 8.7, 2.2 Hz, 1H, aromatic proton), 6.66 (s, 1H, CH), 5.96 (s, 1H, NH), 3.86 (s, 2H, CH2), 3.79 – 3.71 (m, 4H, CH2), 3.68 (s, 2H, CH2), 3.52 (s, 2H, CH2), 2.91 (t, J = 6.9 Hz, 2H, CH2), 2.83 (t, J = 6.9 Hz, 2H, CH2). 13C NMR (101 MHz, CDCl3) δ 155.00, 144.30, 134.62, 134.04, 129.30, 128.70, 128.23, 127.22, 124.09, 116.09, 111.82, 111.31, 110.72, 66.55, 52.02, 47.63, 44.80, 44.11, 23.32. MS (ESI+) m/z calcd for C22H26N3O3 + [M+H]+ 380.2, found 380.2. Purity = 97.4%.
4c: 3-(2-aminoethyl)-1H-indol-5-yl pentylcarbamate: white solid (90% yield). 1H NMR (400 MHz, CDCl3) δ 8.82 (s, 1H, NH), 7.28 – 7.21 (m, 1H, aromatic proton), 7.12 (d, J = 8.7 Hz, 1H, aromatic proton), 6.88 – 6.78 (m, 2H, aromatic protons), 5.27 (t, J = 5.8 Hz, 1H, CH), 3.22 (q, J = 6.7 Hz, 2H, CH2), 2.89 (t, J = 6.6 Hz, 2H, CH2), 2.75 (t, J = 6.6 Hz, 2H, CH2), 1.63 – 1.46 (m, 4H, CH2), 1.36 – 1.24 (m, 4H, CH2), 0.92 – 0.82 (m, 3H, CH3). 13C NMR (101 MHz, CDCl3) δ 155.95, 144.25, 134.21, 127.68, 123.65, 116.21, 113.34, 111.56, 110.96, 42.12, 41.31, 29.57, 29.18, 28.95, 22.36, 14.01. MS (ESI+) m/z calcd for C16H24N3O2 + [M+H]+ 290.2, found 290.2. Purity = 95.0%.
4d : 3-(2-aminoethyl)-1H-indol-5-yl hexylcarbamate: white solid (93% yield). 1H NMR (400 MHz, CDCl3) δ 8.80 (s, 1H, NH), 7.27 – 7.20 (m, 1H, aromatic proton), 7.12 (d, J = 8.7 Hz, 1H, aromatic proton), 6.88 – 6.77 (m, 2H, aromatic protons), 5.25 (t, J = 5.9 Hz, 1H, CH), 3.21 (q, J = 6.7 Hz, 2H, CH2), 2.89 (t, J = 6.6 Hz, 2H, CH2), 2.75 (t, J = 6.6 Hz, 2H, CH2), 1.53 (m, 8.4 Hz, 4H, CH2), 1.34 – 1.19 (m, 6H, CH2), 0.86 (t, J = 6.7 Hz, 3H, CH3). 13C NMR (101 MHz, CDCl3) δ 155.94, 144.26, 134.21, 127.69, 123.64, 116.22, 113.37, 111.56, 110.96, 42.12, 41.34, 31.48, 29.85, 29.20, 26.47, 22.58, 14.03. MS (ESI+) m/z calcd for C17H26N3O2 + [M+H]+ 304.2, found 304.2. Purity = 95.8%.
4e : 3-(2-aminoethyl)-1H-indol-5-yl heptylcarbamate: white solid (93% yield). 1H NMR (400 MHz, MeOD) δ 7.33 (d, J = 8.7 Hz, 1H, aromatic proton), 7.27 (d, J = 2.3 Hz, 1H, aromatic proton), 7.16 (s, 1H, CH), 6.85 (dd, J = 8.7, 2.2 Hz, 1H, aromatic proton), 3.18 (t, J = 7.1 Hz, 2H, CH2), 3.06 (t, J = 6.9 Hz, 2H, CH2), 2.97 (t, J = 7.0 Hz, 2H, CH2), 1.56 (q, J = 7.0 Hz, 2H, CH2), 1.39 – 1.30 (m, 8H, CH2), 0.95 – 0.89 (m, 3H, CH3). 13C NMR (101 MHz, MeOD) δ 157.13, 144.28, 134.52, 127.19, 124.14, 115.78, 111.30, 110.53, 110.17, 40.66, 40.62, 31.58, 29.48, 28.74, 26.48, 25.18, 22.28, 13.03. MS (ESI+) m/z calcd for C18H28N3O2 + [M+H]+ 318.2, found 318.2. Purity = 95.5%.
Inhibition assay on AChE and BuChE
The ChE inhibition activity of the synthesized CTDs was performed according to modified Ellman’s method. Donepezil and rivastigmine were used as reference standards. Butyrylcholinesterase (BuChE, E.C. 3.1.1.8, from equine serum), acetylcholinesterase (AChE, E.C. 3.1.1.7, Type V-S, lyophilized powder, from electric eel, 1000 unit), butyrylthiocholine iodide (BTCI), acetylthiocholine iodide (ATCI), and 5,5-dithiobis-(2-nitrobenzoic acid) (DTNB) were purchased from Sigma-Aldrich. The tested compounds were dissolved in DMSO (1%, analytical grade) and then diluted in phosphate buffer (75 mM, pH 7.4) to obtain the terminal concentrations required. At least ten concentration gradients were set for each compound in triplicate to detect the inhibition rate for BuChE, and five concentration gradients were set for each compound in triplicate to detect the inhibition rate for AChE. Specific experiment processes were conducted as in previous works (Zhang et al., 2022a). Specifically, 10 µl prepared AChE or BuChE solution (1.0 U/ml), 25 µl prepared tested compounds solution, and 65 µl phosphate buffer (pH 8.0, 0.1 mol/L) were mixed in each well of 96-well plates. Then, the 96-well plates were preincubated for 20 min at 37°C in a constant temperature incubator. The wells in the control group were operated under the same conditions without inhibitors, and the wells in the blank group were operated under the same conditions without enzyme solutions and inhibitors. Subsequently, 100 µl prepared DTNB solution (0.35 mM) and 50 µl prepared BTCI or ATCI solution (1.0 mM) were quickly added to each well, and the mixture reacted 5 min at room temperature. The OD values were tested at 412 nm using a microplate reader (Bio-Rad Laboratories, CA, United States). The IC50 values were calculated using IBM SPSS Statistics 25.0 software. All experiments were performed in triplicate, and the results were shown as the mean ± SD.
Kinetic study
In the kinetic study, the preincubated time of the mixture (BuChE, inhibitors solution, and phosphate buffer) were 2, 5, 10, 15, 20, 30, 40, 50, and 60 min, respectively. Other than that, the rest operation was the same as mentioned above. The obtained activities of BuChE were plotted against time and fitted to equation (A = A0 • e-kobst + A∞) to determine rate constant kobs by GraphPad Prism 8.0 software. Thereinto, A stands for the activity of BuChE at each time t, A0 stands for the activity of BuChE at time t = 0, and A∞ stands for the activity of BuChE at infinite time. Then, the created reciprocal kobs −1 values needed to be plotted against the reciprocal concentration C−1. According to the plot, k3 is calculated from the Y-intercept, and kc was calculated from the slope of the resulting linearization according to equation (kobs −1 = kc • k3 −1 • C−1 + k3 −1) by GraphPad Prism 8.0 software.
Molecular docking study
Schrodinger software (Release 2019-2, Schrodinger, LLC, New York, NY, 2019) is used to conduct a molecular docking study. The structures of the hBuChE (PDB code: 4TPK) and hAChE (PDB code: 4M0F) are retrieved from the RCSB Protein Data Bank (http://www.rcsb.org/pdb/). According to the previous method (Liu et al., 2022), the crystal structure profiles of BuChE and AChE are prepared by the Maestro Protein Preparation Wizard model by adding hydrogen atoms and straining minimization using the OPLS3 force field. And the ionization state is set at pH 7.0 ± 2.0. Then, the structures of the tested compounds are added to the hydrogen atoms at a neutralized environment, minimized by MMFFs forcefield, generating 3D coordinates. The binding sites of N-((1-(2,3-dihydro-1H-inden-2-yl) piperidin-3-yl) methyl)-N-(2-methoxyethyl)-2-naphthamide and territrem B in BuChE and AChE, respectively, are selected as the active sites for docking. The receptor grid is created at the selected residues using the grid box at the size of 20Å. Ultimately, molecular docking is run using the standard precision (SP). Other than above mentioned, the other docking parameters are set as default.
Evaluation of cell viability
The cell lines present in this study were purchased from the China Center for Type Collection (CCTCC, China). For cytotoxicity assay, HT-22, BV2, SH-SY5Y, HepG2, and LO2 cells were incubated in 96-well plates at the density of 5 × 10³ per well. After incubation for 12 h at 37°C, the tested compounds with the required concentrations were added to the corresponding wells and continuously incubated for 24 h at 37°C. Then, the prepared MTT (3-[4,5-dimethyl-2-thiazolyl]-2,5-diphenyl-2H-tetrazolium bromide) solution was added to each well and co-incubated for another 4 h at 37°C. Then, the supernatant was removed, and 100 μl DMSO was added to each well. The OD value of each well was detected at 570 nm using a microplate reader (Bio-Rad Laboratories, CA, United States). The IC50 values were calculated using IBM SPSS Statistics 25.0 software. All experiments were performed in triplicate, and the results were shown as the mean ± SD.
For the neuroprotection assay, HT-22 cells were incubated in 96-well plates at the density of 5 × 10³ per well. After incubation for 12 h at 37°C, the tested compounds with required concentrations and H2O2 (500 μM) were added to the corresponding wells and continuously incubated for 24 h at 37°C. Then, the prepared MTT (3-[4,5-dimethyl-2-thiazolyl]-2,5-diphenyl-2H-tetrazolium bromide) solution was added to each well and co-incubated for another 4 h at 37°C. Then, the supernatant was removed, and 100 μl DMSO was added to each well. The OD value of each well was detected at 570 nm using a microplate reader (Bio-Rad Laboratories, CA, United States). The cell viability values were calculated following the following equation: (OD570 (tested compounds) –OD570 (blank))/(OD570 (control) – OD570 (blank)) × 100%. All experiments were performed in triplicate, and the results were depicted as the mean ± SD.
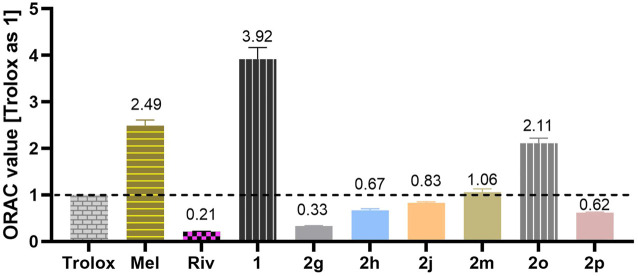
ORAC assay
Trolox was considered as standard, and its ORAC value was set as 1. Tested compounds in required concentrations (20 μl) and fluorescein solution (120 μl) were first co-incubated in a 96-well plate for 15 min at 37°C. Next, AAPH solution (60 µl) was added to each well. Then, the plate was immediately placed into a microplate reader, and the fluorescence of each well was detected every 2 min for 90 min with excitation at 485 nm and emission at 535 nm. After that, the fluorescence intensity was plotted on the vertical axis, and time was plotted on the horizontal axis. The antioxidant curves were normalized to the curve of the blank in the same assay. The AUC (area under the fluorescence decay curve) was obtained using GraphPad Prism 8.0 software. Net AUC = AUCantioxidant – AUCblank. Subsequently, the net AUC value was plotted on the vertical axis, and antioxidant concentration was plotted on the horizontal axis. ORAC value = slopeantioxidant/slopeTrolox. All experiments were conducted in triplicate, and the terminal ORAC values were depicted as the mean ± SD.
Inhibition assay on COX-2
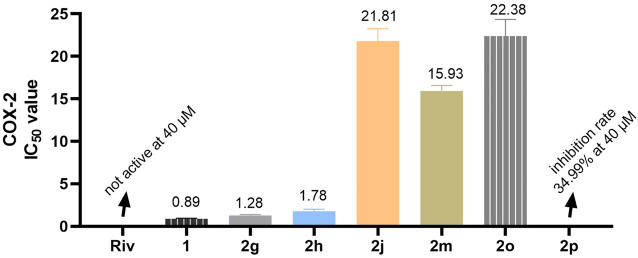
Inhibition assay on COX-2 was performed using a commercially available COX-2 screening assay kit (Beyotime, China, lot: S0168). At the initial screening, the tested concentrations of the selected compounds were 40, 20, 10, 5, and 2.5 μM. The reference concentrations (celecoxib) were 1.28, 0.64, 0.32, 0.16, and 0.08 μM. The specific experimental operations were conducted following the instruction of the assay kit. All experiments were conducted in triplicate, and the terminal inhibitory results were depicted as the mean ± SD.
Statistical analysis
All results were determined by ANOVA (analysis of variance) following Fisher’s PLSD procedure for post hoc comparison to verify the significance between the two means. p values less than 0.05 were considered statistically significant. The data were handled by SPSS 25.0 software.
Results and Discussion
Synthesis of target compounds
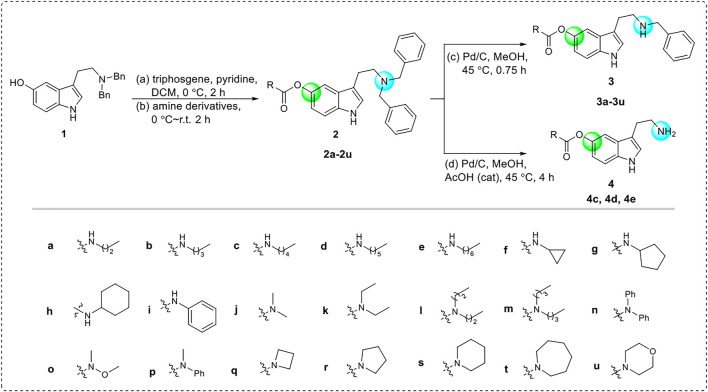
The starting material 1 was synthesized from commercially available 5-methoxytryptamine via the process of amino protection and demethylation (Zhang et al., 2022a). Subsequently, the benzyl-protected tryptamine analog 1 was acylated under the condition of triphosgene and then reacted with diverse amines to give the CTDs 2a-2u. Then, selectively removing one or two benzyl groups by controlling the reaction conditions, the CTDs 3a-3u or 4c-4e were obtained, respectively (Scheme 2). The structures of these CTDs were characterized by HRMS and nuclear magnetic resonance (NMR). All of them were produced in mild-to-good yields (53%–95%). The purity of these products was detected by high-performance liquid chromatography (HPLC) analysis and reached more than 95%.
SCHEME 2.
Synthetic method for the targeted carbamylated tryptamine derivatives.
Inhibition assay on AChE and BuChE
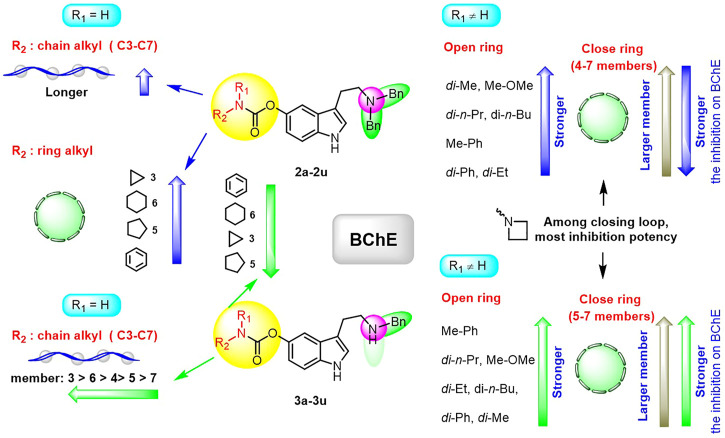
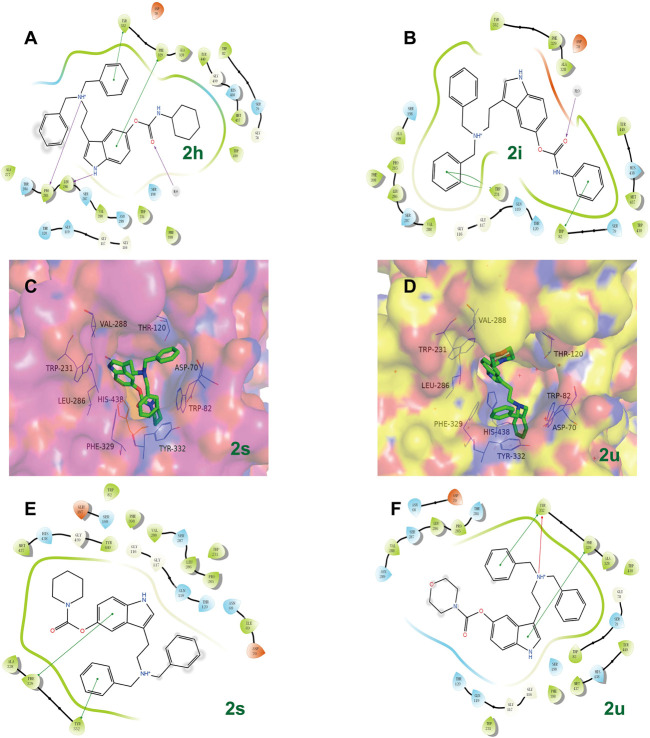
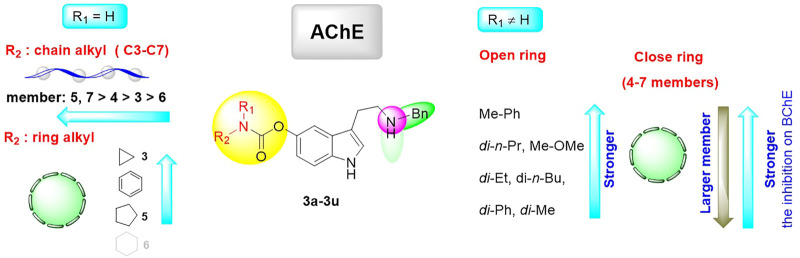
Ellman’s colorimetric assay was performed to evaluate the inhibitory activity of synthesized compounds on acetylcholinesterase (AChE) and BuChE, and donepezil and rivastigmine were selected as the reference ChE inhibitors (Hoffmann et al., 2019). The dibenzyl-substituted carbamylated compounds (2a-2u) were first selected to study the structure-activity relationships (SARs) of different carbamate moieties for ChE inhibition (Table 1; Figure 3). The results showed that most of these compounds had a poor inhibitory effect on AChE, except for N-methoxymethylamine (2o) and azetidine (2p) substituted compounds. However, all these compounds possessed good-to-excellent inhibition potency on BuChE. In detail, for the compounds bearing terminal secondary amine carbamate group were chain alkyl amine, compounds 2a-2e bearing three to seven carbon chains showed great inhibitory activity. Among them, the inhibition efficacy of 2a with three-carbon chain was slightly weaker than that of 2c-2e with longer carbon chains. When the terminal residues were ring carbon chains, the activity of ternary ring (2f) was significantly stronger than that of the six-membered ring (2h) and five-membered ring (2g). When the alkyl ring was replaced with a benzene ring, the activity of 2i was obviously decreased, indicating that the existence of π electrons for this scaffold probably had a negative influence on the inhibition potency of BuChE. In order to preliminarily verify this hypothesis, the molecular docking study was performed. The result showed that 2h could form a hydrogen bond interaction with Pro285 and Leu286, which was the key residue in the acyl-binding pocket of BuChE (PDB: 4TPK). Meanwhile, it could also form π–π stacking interactions with Tyr332 and Phe329, which were the residues in the peripheral anion site (PAS) (Figures 2A,B). Differently, 2i could only form π–π stacking interactions with Trp231 and Trp82, which were the key residues in the acyl-binding pocket and choline-binding pocket of BuChE, respectively. This observation indicated that the subtle difference might be the reason for the lower activity of 2i than 2h. Moreover, when the terminal residue was replaced with tertiary amine carbamate moieties, the different sizes and steric hindrance of substituents had different effects on the activity. For the ring-opening amino segment, dimethylamine (2j) and methoxymethylamine (2o) substituted compounds possessed great inhibition potency. Compounds with dipropylamine (2l), dibutylamine (2m), and N-methylaniline (2p) also exhibited favorable inhibition potency. However, the inhibition potency of compounds with diethylamine (2k) and diphenylamine (2n) was relatively weak. All these results showed that the inhibition potency on BuChE was not only regulated by the steric hindrance, but also adjusted by the size of the carbamate residue. When the carbamate residues were relatively rigid alkylamine fragments (2q-2t), the inhibition efficacy was decreased with the expanding ring. Among them, 2p with azetidine residue exhibited quite inhibitory activity against BuChE. Besides, when the cyclohexane was replaced with morpholine (2u), the inhibition efficacy was obviously decreased. According to the phenotypic result of the molecular docking study, the introduction of morpholine changed the lowest energy conformation of the compound, and the subtle variation weakened the affinity of the indole ring to the acyl-binding pocket and the whole compound to the active sites of BuChE (Figures 2C–F). As for the subtle changes in the study of SAR, further in-depth research is ongoing to clarify the observation.
TABLE 1.
AChE and BuChE inhibitory activity (IC50) of compounds 2a-2u.
| Comp. | R | IC50 ± SD | SI b (BuChE) | Comp. | R | IC50 ± SD | SI b (BuChE) | ||
|---|---|---|---|---|---|---|---|---|---|
| BuChE a (nM) | AChE a (nM) | BuChE a (nM) | AChE a (nM) | ||||||
| Don. | 3702.48 + 28.43 | 2450.14 ± 80.29 | 0.66 | 2k |
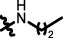
|
320.88 ± 2.06 | >100 μM | >312 | |
| Riv. | 231.84 ± 20.09 | 371.23 ± 10.46 | 1.60 | 2l |
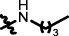
|
15.90 ± 0.55 | >100 μM | >6,289 | |
| 2a |
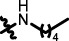
|
33.18 ± 4.99 | >100 μM | >3,013 | 2m |
|
13.82 ± 0.67 | >100 μM | >7,236 |
| 2b |
|
3.00 ± 0.21 | >100 μM | >33,333 | 2n |
|
262.27 ± 2.48 | >100 μM | >381 |
| 2c |
|
2.10 ± 0.14 | >100 μM | >47,619 | 2o |
|
8.65 ± 0.34 | 2,931.26 ± 17.98 | 339 |
| 2d |
|
1.47 ± 0.15 | >100 μM | >68,027 | 2p |
|
18.85 ± 1.34 | >100 μM | >5,305 |
| 2e |
|
1.42 ± 0.004 | >100 μM | >70,422 | 2q |
|
1.65 ± 0.03 | 742.16 ± 9.87 | 450 |
| 2f |
|
8.75 ± 0.23 | >100 μM | >11,428 | 2r |
|
20.47 ± 1.70 | >100 μM | >4,885 |
| 2g |
|
33.69 ± 1.62 | >100 μM | >2,968 | 2s |
|
33.35 ± 5.21 | >100 μM | >2,998 |
| 2h |

|
15.98 ± 0.32 | >100 μM | >6,258 | 2t |
|
67.97 ± 8.42 | >100 μM | >1,471 |
| 2i |
|
55.78 ± 3.14 | >100 μM | >1,793 | 2u |
|
114.49 ± 1.45 | >100 μM | >873 |
| 2j |
|
6.77 ± 0.27 | >100 μM | >14,771 | |||||
AChE from electric eel and BuChE from equine serum.
SI (BuChE) = IC50(AChE)/IC50(BuChE). SI: selection index. Enzyme activity was examined by Ellman’s colorimetric assay. IC50 values were calculated as the mean ± SD of triplicate in three independent experiments. Don, donepezil. Riv, rivastigmine.
FIGURE 3.
Outline of SARs of compounds 2a-2u and 3a-3u on BuChE.
FIGURE 2.
The possible binding mode for the compounds 2h, 2i, 2s, 2u in the BuChE (PDB code: 4TPK) binding sites. (A,B,E,F) The 2D images of 2h, 2i, 2s, 2u, respectively, binding to BuChE predicted by Schrodinger software (Release 2019-2, Schrodinger, LLC, New York, NY, 2019). The purple arrow indicates hydrogen bond, the red arrow indicates π–cation interaction, and the green line indicates π–π interaction. (C,D) The potential distribution surface diagrams of 2s and 2u, respectively, created by Pymol (http://www.pymol.org).
By considering the influence of the benzyl group on the affinity of compounds to the enzyme, further SAR studies on 3a-3u with monobenzyl substituent and 4c-4e without benzyl group were evaluated, and the results are depicted in Table 2 and Figure 3. Overall, the reduction or elimination of benzyl groups decreased the BuChE inhibitory activity of the compounds. Noteworthily, most monobenzyl-substituted CTDs possessed mild-to-good inhibition efficacy on AChE, significantly different from dibenzyl-substituted carbamylated tryptamine derivatives.
TABLE 2.
AChE and BuChE inhibitory activity (IC50) of compounds 3a-3u, 4c-4e.
| Comp. | R | IC50 ± SD a | SI b (BuChE) | Comp. | R | IC50 ± SD a | SI b (BuChE) | ||
|---|---|---|---|---|---|---|---|---|---|
| BuChE a (nM) | AChE a (nM) | BuChE a (nM) | AChE a (nM) | ||||||
| Don. | 3,702.48 + 28.43 | 2,450.14 ± 80.29 | 0.66 | 3l |
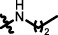
|
43.97 ± 1.84 | >100 μM | >2,274 | |
| Riv. | 231.84 ± 20.09 | 371.23 ± 10.46 | 1.60 | 3m |

|
97.43 ± 8.53 | 19,782.51 ± 32.69 | 203 | |
| 3a |
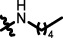
|
18.17 ± 0.68 | 1,952.31 ± 14.96 | 107 | 3n |
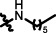
|
155.57 ± 3.58 | >100 μM | >643 |
| 3b |
|
23.69 ± 1.18 | 1,692.45 ± 12.69 | 71 | 3o |
|
58.83 ± 2.92 | 2,613.24 ± 7.69 | 44 |
| 3c |
|
23.89 ± 2.90 | 1,133.26 ± 11.54 | 47 | 3p |
|
25.43 ± 0.27 | 7,053.68 ± 41.62 | 277 |
| 3d |
|
19.01 ± 0.29 | 2,443.63 ± 16.95 | 128 | 3q |
|
2.58 ± 0.13 | 482.16 ± 5.99 | 187 |
| 3e |
|
33.57 ± 1.64 | 1,132.45 ± 9.89 | 34 | 3r |
|
40.49 ± 1.79 | 1,815.47 ± 14.11 | 45 |
| 3f |
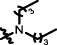
|
15.74 ± 1.30 | 3,174.86 ± 14.57 | 202 | 3s |
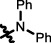
|
33.86 ± 2.14 | 9,076.38 ± 62.31 | 268 |
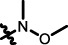
| 3g |
|
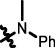
13.59 ± 0.70 | 27,183.67 ± 18.74 | 2,000 | 3t |
|
23.24 ± 0.94 | >100 μM | >4,303 |
| 3h |
|
43.55 ± 3.35 | >100 μM | >2,296 | 3u |
|
112.15 ± 7.37 | >100 μM | >892 |
| 3i |
|
110.44 ± 5.22 | 23,291.64 ± 43.12 | 211 | 4c |
|
120.91 ± 8.38 | >100 μM | >827 |
| 3j |
|
131.52 ± 3.59 | 1,913.07 ± 15.74 | 15 | 4d |
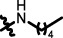
|
27.69 ± 2.04 | >100 μM | >3,611 |
| 3k |
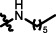
|
72.46 ± 6.55 | >100 μM | >1,380 | 4e |
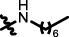
|
10.05 ± 0.83 | >100 μM | >9,950 |
AChE from electric eel and BuChE from equine serum.
SI (BuChE) = IC50(AChE)/IC50(BuChE). SI: selection index. Enzyme activity was examined by Ellman’s colorimetric assay. IC50 values were calculated as the mean ± SD of triplicate in three independent experiments. Don, donepezil. Riv, rivastigmine.
For more details, the BuChE inhibitory activity of these compounds exhibited different SAR trends from the dibenzyl-substituted compounds, which might be due to the variation of the carrier scaffold. When the carbamate residue was the terminal secondary alkyl amine, the length of carbon chains or the size of ring carbon chains exhibited a slight influence on the inhibitory activity (3a-3h). Similar to the dibenzyl-substituted tryptamine derivatives, the replacement of cyclohexane with the benzene ring, the inhibitory activity of 3i obviously decreased. When the terminal residue was the ring-opening tertiary amine carbamate moiety, the trend of the inhibitory activity of 3j-3p against BuChE was significantly different from the results of 2j-2p. Combining the SARs results of dibenzyl-substituted tryptamine derivatives, the variation further suggested that the inhibition potency was regulated by both the carbamate moiety and the size of the carrier scaffold. When the terminal residue was replaced with the relatively rigid alkylamine fragments, the compound with azetidine residue (3q) exhibited significant inhibition potency on BuChE, and the morpholine substituted compound 3u exhibited weak inhibition potency, similar to the observed phenomenon in 2q and 2u. However, the inhibitory activity was increased with the expanding ring with five-to-seven members (3r-3t), inconsistent with the observed trend of disubstituted compounds.
Besides, according to the IC50 values of the AChE inhibition assay, the length of the carbon chain of secondary amine in the carbamate group had a weak influence on AChE inhibition (3a-3e). When the unbranched alkyl group of 3a-3e was replaced with cycloalkyl (3f-3h), the resulting compounds suffered a 1.3–88-fold decrease in the AChE inhibitory activity. When aniline replaced cyclohexylamine, the inhibitory activity on AChE of 3i was markedly enhanced. The AChE inhibitory activity of different tertiary amine derivatives (3j-3n and 3p) was also investigated. Among them, compound 3i with a small bulky tertiary amine had the optimal potency on AChE. The methoxamine analog 3o displayed comparable activity against AChE to that of 3i. In addition, we explored the effect of different nitrogen-containing heterocycles (3q-3u) on the inhibitory activity against AChE. Interestingly, compound 3q bearing an azetidinyl group showed comparable AChE inhibitory activity to that of rivastigmine, providing valuable guidance for further research. The summary of the SARs study is depicted in Figure 4.
FIGURE 4.
Outline of SARs of compounds 3a-3u on AChE.
In addition, the CTDs 4c-4e without benzyl groups also possessed good inhibitory activity on BuChE (Table 2). The longer length of carbon chains had positive effects on the BuChE inhibition efficacy. However, the overall activity of the non-benzyl group was weaker than that of benzyl-substituted compounds. At the same time, considering the stability of this series of compounds, the study only evaluated the inhibition potency of 4c-4e, and the results further confirmed the importance of the benzyl group for the affinity of tryptamine derivatives to the enzyme.
Molecular docking
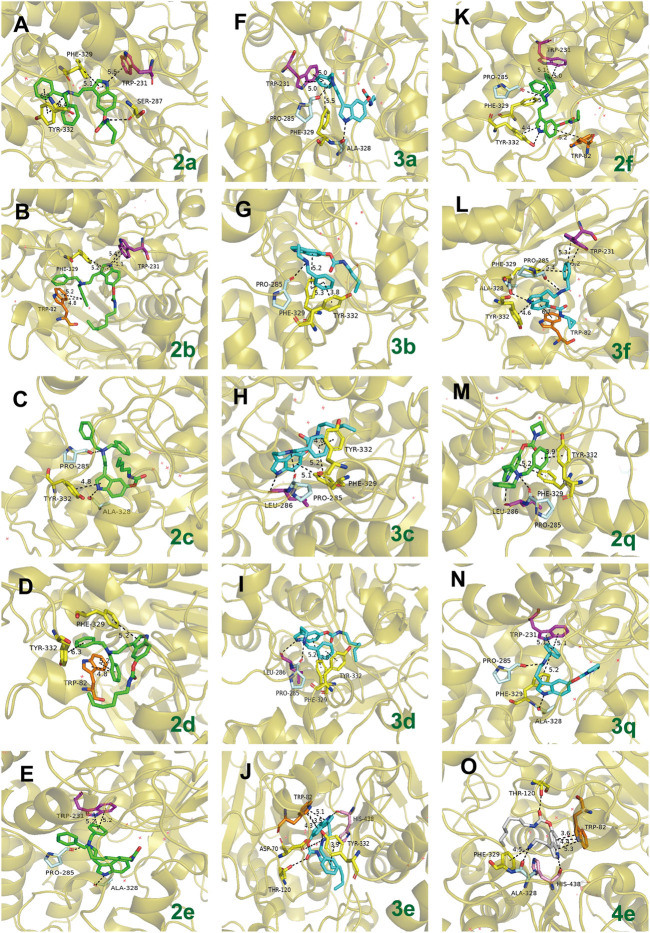
A molecular docking study was performed to predict the binding mode of the synthesized CTDs to hBuChE (PDB code: 4TPK) and hAChE (PDB code: 4M0F) (Brus et al., 2014; Liu et al., 2022). Given that the majority of synthesized tryptamine derivatives have significant inhibition efficacy on BuChE, we conducted a preliminary investigation into the action mode of various series of compounds (Figure 5). Docking poses indole fragments in compounds with 3- and 4-carbon chains (2a and 2b, respectively) can form π–π stacking interactions and/or π–cation interactions with residues in the acyl-binding pocket (Trp231) and PAS (Phe329). Only stacking interactions with PAS (Tyr332, Phe329) or H-bond interactions with Ala328 can form the indole ring as increasing the alky carbon chain lengthens (2c-2e). The 2e-bearing 7-carbon chain can form two stacking interactions with benzyl benzene and Trp231, but the indole fragment cannot form any interactions with the acyl-binding pocket or PAS in its structure. Furthermore, one of the benzyls in 2a and 2d can interact with PAS via an π–π stacking interaction (Tyr332). The slight difference is that 2b and 3d can form π–π stacking interaction with the choline-binding site (Trp82). However, the benzyl group in 2c cannot form any interaction with active residues. Moreover, the tertiary amine nitrogen atom tends to form ions in the body and form H-bond interaction with part of the site cavity (Pro285), which can be observed in 2c, 2e, 2f, and 2q. By the way, due to the initial characteristic of the nitrogen atom, a similar phenomenon can also be observed in 3a-3d, 3f, and 3q. All these results indicate that the variation of chain length has a certain effect on the action mode of compounds with BuChE. Notably, when one benzyl group is eliminated, the action mode is also changed. In contrast to 2a, 3a’s benzyl benzene can form stacking interactions with an acyl-binding pocket (Trp231) and a PAS (Phe329) rather than an indole fragment. Differently, 3c bearing 5-carbon chain and 3d bearing 6-carbon chain can form H-bond with acyl-binding pocket (Leu286) and π–π stacking interaction with PAS (Phe329) in their indole fragments, and their benzyl group can also form π–π stacking interactions with PAS (Tyr332 and Phe329). In addition, 3b bearing 4-carbon chain and 3e bearing 7-carbon chain can only form interactions with PAS. Thereinto, 3e can also form H-bond with the key residue (His438) of the catalytic active site (CAS) and π–π stacking interactions with the choline-binding site (Trp82). When the terminal residue is a 3-carbon ring chain, the 2f bearing dibenzyl and 3f bearing monobenzyl groups all form π–π stacking interactions between the benzyl group and the acyl-binding pocket (Trp231) and PAS (Phe329), and the indole fragment with PAS (Tyr332). Furthermore, their indole fragment also can form π–π stacking interactions with the choline-binding site (Trp82). When the amino residue is relatively rigid alkyl amine, compounds bearing azetidine-based carbamate residue showed different modes of action. Among them, 2q with dibenzyl groups can form H-bond interactions with acyl-binding pockets (Leu286) and π–π stacking interactions with PAS (Phe329), and benzyl groups can also form π–π stacking interactions with PAS (Tyr332). 3q with a monobenzyl group, on the contrary, can only form π–π stacking interactions with acyl-binding pockets (Trp231) and PAS (Phe329) in its benzyl group. When the benzyl group is completely removed, 4e in its indole fragment can form stacking interactions with the choline-binding site (Trp82) and H-bond interactions with CAS (His328). At the same time, it can form π–cation interaction and H-bond interaction with PAS (Phe329 and Thr120, respectively). All these results of the predicted action mode further indicate that the inhibition efficacy of this kind of compounds on BuChE is modulated by both the carbamate residues and carrier scaffolds. The subtle difference may be the key reason for their different inhibition potency because their different conformations have been varied.
FIGURE 5.
The possible binding mode for the selected compounds in the BuChE (PDB code: 4TPK) binding sites (A–O). The 3D images of selected compounds bind to BuChE polished by Pymol (http://www.pymol.org). Residues of the acyl-binding pocket (Leu286, Trp231) are shown in magenta, the CAS (His438) is shown in pink, the choline-binding site (Trp82) is shown in orange, the residues of PAS (Asp70, Thr120, Tyr332, and Phe329) are shown in yellow, and parts of the side cavity (Pro285, Ala328) are in pale cyan. The dashed lines indicate hydrogen bond interaction, π–π stacking interactions, or π–cation interactions.
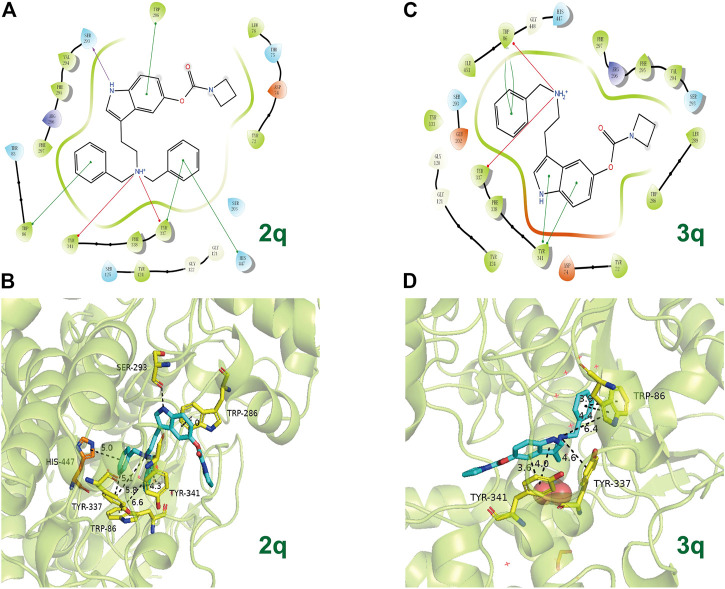
In view of 2q and 3q bearing azetidine-based carbamate residue possessing better inhibition efficacy on AChE than other synthesized compounds, the action modes of 2q and 3q with AChE are preliminarily predicted by molecular docking (Figure 6). Docking poses of 2q and 3q with hAChE (PDB code: 4M0F) reveal that the compounds can form π–π stacking interactions with PAS (Trp86, Tyr341) in their benzyl group and indole fragment, as well as cation interactions with PAS (Tyr337) in their tertiary or secondary amine. Because the number of the containing benzyl group is different, one of the benzyl groups in 2q can also form π–π stacking interaction with CAS (His447). However, the obtained IC50 value of 2q is larger than that of 3q. Given that the distances of π–π stacking interactions between compounds and PAS differ, we assume that more benzyl groups reduce the affinity of 2q to AChE, and research into the specific action mode of compounds on AChE is ongoing.
FIGURE 6.
The possible binding mode for the compounds 2q and 3q in the AChE (PDB code: 4M0F) binding sites. (A,C) The 2D images of 2q and 3q binding to AChE predicted by Schrodinger software (Release 2019-2, Schrodinger, LLC, New York, NY, 2019). The purple arrow indicates hydrogen bond, the red arrow indicates π–cation interaction, and the green line indicates π–π stacking interaction. (B,D) The 3D images of 2q and 3q binding to AChE polished by Pymol (http://www.pymol.org). Residues of the CAS (His447) are shown in orange, and the residues of PAS (Ser293, Trp286, Tyr337, Trp86, and Tyr341) are shown in yellow. The dashed lines indicate hydrogen bond interaction, π–π stacking interactions, or π cation interactions.
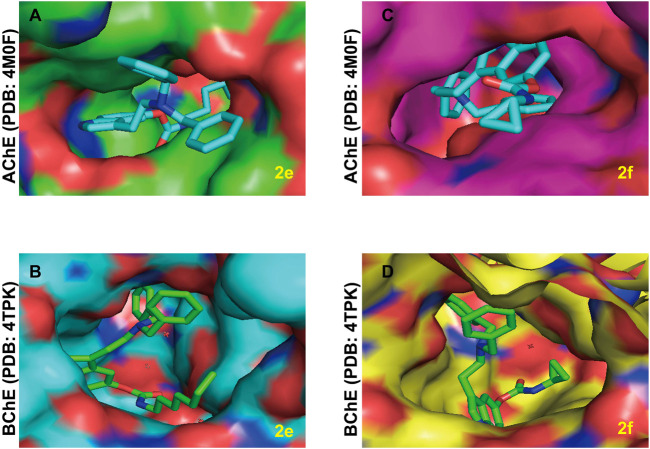
According to previous studies, compounds with larger molecular shapes are preferred to bind to BuChE because of the larger size of the BuChE binding pocket compared to AChE (Sawatzky et al., 2016). In view of the curiosity about the selective BuChE inhibitory activity of the series of bisbenzyl-substituted CTDs, we explored the possible mode of action of 2e and 2f for AChE and BuChE (Figure 7). As expected, the bisbenzyl-substituted CTDs 2e and 2f cannot completely bind to the AChE binding pocket. Thereinto, the benzyl groups of 2e and the carbamate residue of 2f are distributed outside the active pocket due to the relatively small binding pocket of AChE and the relatively large molecular scaffold. Differently, due to the relatively large binding pocket of BuChE, the whole skeletons of 2e and 2f exhibit favorable affinity with BuChE active sites, which explains the possible mechanism of the selectivity obtained in the enzyme inhibition assay.
FIGURE 7.
The possible binding mode for the compounds 2e and 2f in the AChE (PDB code: 4M0F) and BuChE (PDB code: 4TPK) binding sites. (A,B) The potential distribution surface diagrams of 2e in AChE and BuChE binding sites, respectively, created by Pymol (http://www.pymol.org). (C,D) The potential distribution surface diagrams of 2f in AChE and BuChE binding sites, respectively, created by Pymol (http://www.pymol.org).
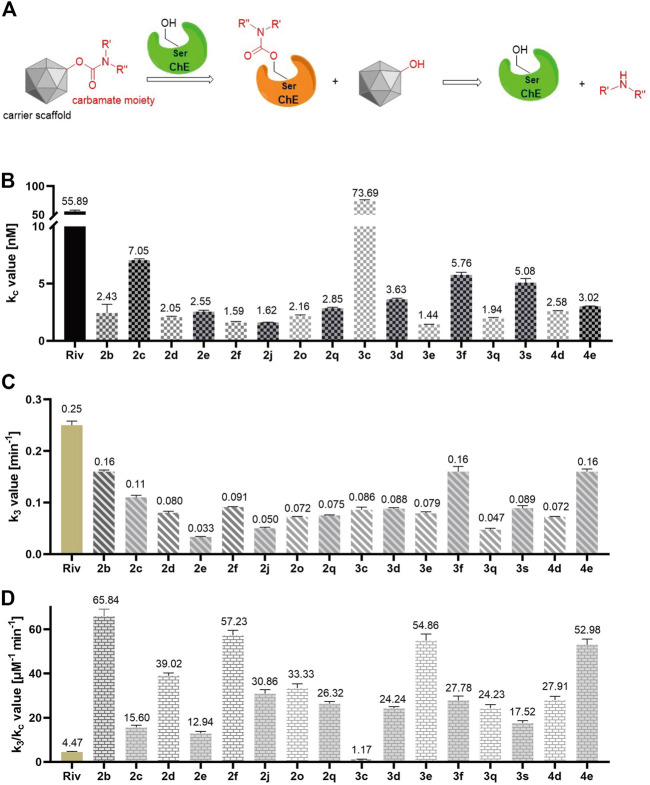
Kinetic characteristic of BuChE inhibition
Convincing evidence has demonstrated that carbamylated derivatives with ChE inhibition efficacy exhibit a pseudo-irreversible inhibition mode (Figure 8A) (Hoffmann et al., 2019; Zhang et al., 2022b). Enlightened by this, we further conducted a kinetic study to explore the inhibition type of the synthesized CTDs, which possessed excellent inhibitory activity on BuChE. According to the results of the equilibrium constant (kc) for reversible combinations (Figure 8B), the carrier skeleton has a pronounced influence on the affinity between compounds and BuChE. Overall, compounds with monobenzyl substituents exhibit larger kc values than those with dibenzyl groups, suggesting that the expansion of the carrier skeleton structure can decrease the affinity of compounds for BuChE. Combining the predicted action mode of these compounds with BuChE by molecular docking, the tryptamine skeleton is a key fragment that binds effectively to the enzyme. Besides, among the compounds with dibenzyl groups and compounds without benzyl group, the alkyl carbon chains in carbamate residues exhibit subtle influence for the affinity of compounds with enzyme, except for 2c bearing a 5-carbon chain, which possesses stronger affinity than others in this series of compounds. Particularly, 3c with the monobenzyl group shows a stronger affinity than that of rivastigmine. Furthermore, 3f bearing 3-ring alkyl carbon chain and 3s bearing piperidine-based carbamate residue have high affinity for BuChE. In addition, the results of the carbamylation rate constant (k3, Figure 8C) showed that the increasing carbon chain has a negative effect on carbamate transfer from the carrier scaffold to the enzyme in dibenzyl-substituted compounds 2b-2e. However, there is no obvious effect of the carbamate residues in monobenzyl-substituted compounds 3c-3e on k3. Compared with 2e and 3e, the benzyl-free compound 4e bearing a 7-carbon side chain shows a significantly larger carbamate transfer rate, suggesting that the carrier scaffold and side residue of carbamate moiety synergistically determined the carbamoylation rate. Besides, the cyclopropanamine residues of carbamate moiety have a positive effect on the carbamoylation rate (2f and 3f). Noteworthily, most of the tested compounds performed better affinity to BuChE and better carbamoylation efficacy because the k3/kc value of compounds was greater than that of rivastigmine (Figure 8D). Therefore, this series of compounds is worthy of further study to perform structure optimization and excavate novel tryptamine-based MTDLs.
FIGURE 8.
(A) The reported pseudo-irreversible inhibition mode of carbamylated derivatives on ChE. (B) The equilibrium constant (kc) values of tested compounds. (C) The carbamylation rate constant (k3) values of tested compounds. (D) The k3/kc values of the tested compounds. All these data are the mean ± SD of triplicate in three independent experiments.
Neuronal cytotoxicity assay
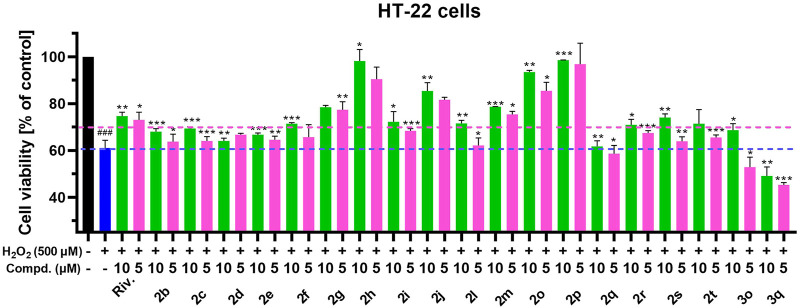
In order to explore the applicability of the synthesized CTDs in neurological diseases, the neuronal cytotoxicity of the synthesized compounds was evaluated on mouse hippocampal neuronal cell line HT-22. Table 3 shows that the neuronal cytotoxicity of compounds (2a-2u) bearing dibenzyl groups on HT-22 is lower than that of compounds (3a-3u) bearing monobenzyl group in general. The cytotoxicity of benzyl-free compounds (4c-4e) exhibited a large difference that the 4c bearing 5-carbon alky chain residue showed little toxicity, but 4d and 4e bearing increasing carbon alkyl chain residues showed certain toxicity. In view of these CTDs endowed strong inhibition efficacy on BuChE, we preliminarily evaluated their safety range by comparing their IC50 values on cytotoxicity and BuChE inhibitory activity. The results showed that most of the tested compounds possessed considerable safety range, and only a few compounds (3m and 3n) showed obvious toxicity. Therefore, the series of compounds with dibenzyl groups, especially compounds bearing ring carbon alkyl chains (2f-2h) and tertiary amine residues (2j-2u) with little cytotoxicity on HT-22, can be considered as potential lead building blocks for further structure optimization to develop novel effective agents against neurological diseases.
TABLE 3.
Cytotoxicity of the synthesized compounds on HT-22 cell line.
| Comp. | R | IC50 ± SD | Safety vs. BuChE b | Comp. | R | IC50 ± SD | Safety vs. BuChE b |
|---|---|---|---|---|---|---|---|
| HT-22 a (μM) | HT-22 a (μM) | ||||||
| Don. | 56.42 ± 0.25 | 15 | 3a |
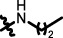
|
31.29 ± 2.32 | 1722 | |
| Riv. | >100 | >431 | 3b |
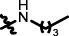
|
18.52 ± 0.81 | 782 | |
| 2a |
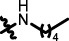
|
51.58 ± 0.05 | 1,554 | 3c |
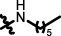
|
10.64 ± 0.24 | 445 |
| 2b |
|
40.59 ± 1.12 | 13,530 | 3d |
|
16.28 ± 1.19 | 856 |
| 2c |
|
37.47 ± 0.23 | 17,842 | 3e |
|
6.49 ± 0.34 | 193 |
| 2d |
|
37.63 ± 1.92 | 25,598 | 3f |
|
15.00 ± 1.17 | 953 |
| 2e |
|
35.65 ± 2.45 | 25,105 | 3g |
|
18.21 ± 0.95 | 1,340 |
| 2f |
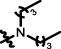
|
60.25 ± 3.85 | 6,886 | 3h |
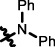
|
11.61 ± 0.34 | 266 |
| 2g |
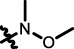
|
57.27 ± 4.30 | 1670 | 3i |
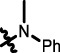
|
18.35 ± 0.43 | 166 |
| 2h |

|
74.18 ± 1.50 | 4,642 | 3j |
|
21.67 ± 0.72 | 165 |
| 2i |
|
41.78 ± 2.31 | 749 | 3k |
|
15.95 ± 0.39 | 220 |
| 2j |
|
>100 | >14,771 | 3l |
|
6.31 ± 0.39 | 144 |
| 2k |
|
54.68 ± 0.42 | 170 | 3m |
|
3.87 ± 0.35 | 40 |
| 2l |
|
38.58 ± 0.90 | 2,426 | 3n |
|
0.54 ± 0.03 | 3.5 |
| 2m |
|
65.28 ± 0.09 | 4,723 | 3o |
|
50.42 ± 1.07 | 857 |
| 2n |
|
>100 | 381 | 3p |
|
11.02 ± 1.12 | 433 |
| 2o |
|
72.08 ± 2.23 | 8,333 | 3q |
|
57.79 ± 2.88 | 22,399 |
| 2p |
|
>100 | 5,305 | 3r |
|
31.72 ± 1.64 | 783 |
| 2q |
|
86.54 ± 1.82 | 52,448 | 3s |
|
13.83 ± 0.91 | 408 |
| 2r |
|
79.77 ± 5.36 | 3,897 | 3t |

|
11.90 ± 0.39 | 512 |
| 2s |
|
42.73 ± 2.09 | 1,281 | 3u |
|
51.29 ± 2.55 | 457 |
| 2t |
|
>100 | 1471 | 4c |
|
>100 | >827 |
| 2u |
|
78.72 ± 3.79 | 688 | 4d |
|
25.25 ± 1.37 | 912 |
| 4e |
|
26.06 ± 2.29 | 2,593 |
Cell viability was examined by MTT assay.
IC50 values were calculated as the mean ± SD of triplicate in three independent experiments.
Safety vs. BuChE: IC50 (HT-22)/IC50 (BuChE). Don, donepezil. Riv, rivastigmine.
In order to further evaluate the applicability of the compounds with dibenzyl groups for neurological diseases, we also tested the cytotoxicity of the selected compounds (2f, 2h, 2j, 2m, 2o-2r, 2t, and 2u) bearing little toxicity on HT-22 cells, microglia cell line BV2, and human neuroblastoma cell line SH-SY5Y, which are widely used in the study of neurological diseases. Table 4 shows that all these tested compounds exhibited little or no toxicity on BV2 and SH-SY5Y cells, except for 2f and 2q, which exhibited certain toxicity. In view of some carbamate-based ChE inhibitors reported to be hepatotoxic, we further performed the preliminary evaluation of the hepatotoxicity of the selected compounds on human normal liver cell line LO2 and human hepatocellular carcinoma cell line HepG2. The results showed that all the tested compounds exhibited no hepatoxicity, suggesting that these CTDs can be safely used in drug discovery for further study.
TABLE 4.
Cytotoxicity of the tested compounds on BV2, SH-SY5Y, LO2, and HepG2 cell lines.
| Comp. | R | a IC50 ± SD (μM) | |||
|---|---|---|---|---|---|
| BV2 | SH-SY5Y | LO2 | HepG2 | ||
| Riv. | >100 | >100 | >100 | >100 | |
| 2f |
|
60.25 + 3.85 | 42.86 + 3.27 | 91.27 + 9.01 | 53.781 + 2.40 |
| 2h |
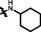
|
74.18 + 1.50 | >100 | >100 | 89.37 + 5.77 |
| 2j |
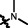
|
>100 | >100 | >100 | >100 |
| 2m |
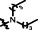
|
65.28 + 0.09 | >100 | >100 | >100 |
| 2o |
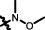
|
72.08 + 2.23 | >100 | 74.44 + 4.26 | 95.23 + 0.22 |
| 2p |
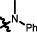
|
>100 | >100 | >100 | >100 |
| 2q |
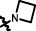
|
86.54 + 1.82 | 42.51 + 3.11 | 72.13 + 0.84 | >100 |
| 2r |
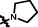
|
79.77 + 5.36 | >100 | >100 | >100 |
| 2t |
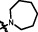
|
>100 | 98.68 + 4.82 | >100 | >100 |
| 2u |
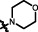
|
78.72 + 3.79 | >100 | >100 | >100 |
Cell viability was examined by MTT assay.
IC50 values were calculated as the mean ± SD of triplicate in three independent experiments. Riv, rivastigmine.
Neuroprotective effects of the selected compounds
Neuronal damage caused by oxidative stress has been widely recognized in various neurological diseases. It is of great significance to protect neurons damaged by oxidative stress for the treatment of neurological diseases (Bai et al., 2022; Wakhloo et al., 2022). To date, many tryptamine derivatives have been reported to endow favorable neuronal protective functions (Hanikoglu et al., 2020; Forman and Zhang, 2021). Given this, we performed an elementary evaluation of neuronal protection against neuronal death elicited by H2O2 of the selected CTDs, which possessed little neuronal toxicity. Figure 9 shows that most of the tested compounds exhibited certain neuronal protection efficacy compared with the model group, in which the cell viability of the model group significantly decreased to 61.07% after treatment with H2O2 (500 μM). Among them, 2g, 2h, 2j, 2m, 2o, and 2p showed relatively favorable neuroprotective effects (the cell viability was more than 70% at 5 and 10 μM), so further structure optimization based on these compounds is of great value for the development of neuronal protective agents. By the way, the compounds bearing azetidine-based carbamate residues (2q and 3q) exhibited no protective effects on H2O2-induced neuronal death. However, considering their extraordinary inhibitory activity on AChE and BuChE, this kind of compounds could be considered for drug development in peripheral cholinergic-related disease, which is ongoing in our current study.
FIGURE 9.
Cell viability was examined by MTT assay. Neuroprotective effects of tested compounds against the toxicity elicited by 500 μM H2O2. HT-22 cells were co-incubated with H2O2 and the tested compounds (5 and 10 μM) for 24 h. The values were expressed as mean ± SD of three independent experiments. ### p < 0.001 compared to the control group. ***p < 0.001, **p < 0.01, or *p < 0.05 compared to the H2O2 induced model group. Riv, rivastigmine.
Evaluation of ORAC values of the selected compounds
Antioxidative effects are crucial in the treatment of many diseases (Ansari et al., 2020; Dumanovic et al., 2021). The Oxygen Radical Absorbance Capacity-Fluorescein (ORAC-FL) assay was used to preliminarily evaluate the antioxidant activities of the selected compounds (2g, 2h, 2j, 2m, 2o, and 2p), which endowed favorable BuChE inhibition efficacy, little neuronal toxicity, and neuroprotective effects. The vitamin E analog Trolox was used as a standard, and its ORAC value was set as 1. Melatonin was used as a positive control with an ORAC value equal to 2.551. At the same time, the non-carbamoylated compound 1 of the tested CTDs was also evaluated as a reference. As shown in Figure 10, non-carbamoylated compound 1 was endowed with favorable oxygen radical scavenging property with a Trolox equivalent value equal to 3.92. However, the carbamylated derivatives showed decreasing antioxidant activities, suggesting that the hydroxyl group played an important role in radical scavenging. Among them, 2o bearing N-methoxymethylamine-based carbamate moiety exhibited a good antioxidant activity with Trolox equivalents value equal to 2.11, indicating that the special characteristic of methyl-methoxylamine fragment played certain modulation for the antioxidant activity. Besides, we also detected the inhibition efficacy of these compounds on NO production using the Griess reagent method. However, the results showed that most of this series of compounds had a weak inhibitory activity or even no effect, indicating that this series of compounds were insensitive to nitrogen radical.
FIGURE 10.
Results are expressed in the Trolox equivalents (TE) unit. The values were expressed as mean ± SD of three independent experiments.
Inhibition assay on COX-2 of the selected compounds
Glia cells play a significant role in neurological diseases (Zhang et al., 2021; Cai et al., 2022; Rauf et al., 2022). Microglia can release large amounts of COX-2 after being stimulated by inflammatory substances. Similarly, the expression of COX-1 in astrocytes was largely unchanged after stimulation, but the expression of COX-2 was significantly increased (Fan et al., 2021; Ghazanfari et al., 2021; Iwata et al., 2021; Nagano et al., 2021; Kuo et al., 2022). Therefore, inhibition of COX-2 expression in glia cells is of great value in the treatment of neurological diseases. The COX-2 inhibitory activity of the selected compounds was preliminarily evaluated using a commercial assay kit. As shown in Figure 11, 2g and 2h bearing cyclopentanamine-based carbamate residue and cyclohexanamine-based carbamate residue, respectively, had good COX-2 inhibition efficacy compared with the non-carbamylated compound 1. However, other compounds bearing tertiary amine-based carbamate residues (2j, 2m, 2o, and 2p) showed mild-to-weak inhibition efficacy on COX-2. By analyzing the structures of the tested compounds, we assumed that hydrogen bond interaction plays a certain role in the inhibitory activity of COX-2.
FIGURE 11.
IC50 is the concentration (μМ) that causes 50% inhibition of COX-2 enzymatic activity. The values were expressed as mean ± SD of three independent experiments.
Drug-like prediction of synthesized compounds
According to the drug discovery, “the rule of 5” predicts that the poor druggability of the compound is more likely to appear when the molecular weight (MWT) is greater than 500, calculated Log P (MLog P) is greater than 4.15, H-bond acceptors are greater than 10, H-bond donors are greater than 5, and the number of rotatable bonds is greater than 10 (Lipinski et al., 2022). Given this, the predicted druggability of synthesized compounds was calculated by the SwissADME. Table 5 shows that the MWT values of all the synthesized compounds were less than 500, except for 2m and 2n. The number of H-bond acceptors and H-bond donors of all these compounds was in accordance with Lipinski’s rule. Most compounds possessed favorable MLog P and BBB permeation properties. All these predicted results provide guidance for further study and suggest the potential value of the tryptamine derivatives.
TABLE 5.
The predicted druggability of synthesized compounds.
| Comp. | MWT | MLog P | HA | HD | BBB | Comp. | MWT | MLog P | HA | HD | BBB |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2a | 441.56 | 3.89 | 3 | 2 | No | 3c | 379.50 | 3.08 | 3 | 3 | Yes |
| 2b | 455.59 | 4.08 | 3 | 2 | No | 3d | 393.52 | 3.30 | 3 | 3 | Yes |
| 2c | 469.62 | 4.28 | 3 | 2 | No | 3e | 407.55 | 3.50 | 3 | 3 | No |
| 2d | 483.64 | 4.47 | 3 | 2 | No | 3f | 349.43 | 2.65 | 3 | 3 | Yes |
| 2e | 497.67 | 4.66 | 3 | 2 | No | 3g | 377.48 | 3.08 | 3 | 3 | Yes |
| 2f | 439.55 | 3.89 | 3 | 2 | Yes | 3h | 391.51 | 3.30 | 3 | 3 | Yes |
| 2g | 467.60 | 4.28 | 3 | 2 | No | 3i | 385.46 | 3.35 | 3 | 3 | Yes |
| 2h | 481.63 | 4.47 | 3 | 2 | No | 3j | 337.42 | 2.43 | 3 | 2 | Yes |
| 2i | 475.58 | 4.53 | 3 | 2 | No | 3k | 365.47 | 2.87 | 3 | 2 | Yes |
| 2j | 427.54 | 3.69 | 3 | 1 | Yes | 3l | 393.52 | 3.30 | 3 | 2 | Yes |
| 2k | 455.59 | 4.08 | 3 | 1 | No | 3m | 421.58 | 3.71 | 3 | 2 | No |
| 2l | 483.64 | 4.47 | 3 | 1 | No | 3n | 461.55 | 4.61 | 3 | 2 | No |
| 2m | 511.70 | 4.84 | 3 | 1 | No | 3o | 353.41 | 2.43 | 4 | 2 | Yes |
| 2n | 551.68 | 5.69 | 3 | 1 | No | 3p | 399.48 | 3.55 | 3 | 2 | Yes |
| 2o | 443.54 | 3.69 | 4 | 1 | Yes | 3q | 349.43 | 2.65 | 3 | 2 | Yes |
| 2p | 489.61 | 4.72 | 3 | 1 | No | 3r | 363.45 | 2.87 | 3 | 2 | Yes |
| 2q | 439.55 | 3.89 | 3 | 1 | Yes | 3s | 377.48 | 3.08 | 3 | 2 | Yes |
| 2r | 453.58 | 4.08 | 3 | 1 | Yes | 3t | 391.51 | 3.30 | 3 | 2 | Yes |
| 2s | 467.60 | 4.28 | 3 | 1 | Yes | 3u | 379.45 | 2.06 | 4 | 2 | Yes |
| 2t | 481.63 | 4.47 | 3 | 1 | No | 4c | 289.37 | 1.72 | 3 | 2 | No |
| 2u | 469.57 | 3.27 | 4 | 1 | Yes | 4d | 303.40 | 1.96 | 3 | 3 | No |
| 3a | 351.44 | 2.65 | 3 | 3 | Yes | 4e | 317.43 | 2.20 | 3 | 3 | No |
| 3b | 365.47 | 2.87 | 3 | 3 | Yes |
Lipinski’s rules: MWT < 500; MW: MLog p < 4.15; HA < 10; HD < 5. MWT, molecular weight; MLog P, Log P o/w ; HA, H-bond acceptors; HD, H-bond donors; BBB, BBB permeant. All these parameters were predicted by http://www.swissadme.ch/.
Conclusion
In this study, we designed and synthesized a novel series of benzyl-free, monobenzyl-, and bisbenzyl-substituted tryptamine derivatives bearing functional carbamate groups to explore promising building blocks for the discovery of efficient MTDLs against ChE-associated neurological disorders. The ChE inhibition assay revealed that the majority of these hybrids had good-to-excellent BuChE inhibitory activities. In particular, several dibenzyl-substituted CTDs (2b-2f, 2j, 2o, and 2q) possessed excellent BuChE inhibition efficacy with IC50 values in a single-digit nanomolar level. A molecular docking study on BuChE showed that the bisbenzyl-substituted CTDs could interact with the key residues of active binding sites, and the kinetic study indicated that the tested compounds performed pseudo-irreversible inhibition models. The in vitro neuronal cytotoxicity assay suggested that 2g, 2h, 2j, 2m, 2o, and 2p showed favorable neuroprotective potency on H2O2-induced HT-22 cells. The ORAC assay and COX-2 inhibition screening assay indicated that 2g, 2h, 2j, 2m, 2o, and 2p were also endowed with good antioxidant activities and COX-2 inhibitory effects. In view of the overall promising findings mentioned above, this kind of CTDs is used as the lead scaffold for further structural modification to develop efficient MTDLs agents against ChE-associated neurodegenerative disorders, such as AD and PD.
Associated content
The spectral data and additional experimental information, except for those stated above, are presented in the Supplementary Material.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
HZ and JW are responsible for the whole project, including project design, experimental operation and analysis, and article writing. YW, GY, and QL helped analyze the data and revise the article. LZ, HC, and ZW designed and supervised the study. All authors contributed to the article and approved the submitted version.
Funding
Recruitment Program of Global Experts (1000 Talents Plan) Gansu Education Department: “Star of Innovation” Project for Outstanding Graduate Students (2021CXZX-149) General Project of Natural Science Foundation of Qinghai Province (2019-ZJ-907).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.1036030/full#supplementary-material
References
- Ansari M. Y., Ahmad N., Haqqi T. M. (2020). Oxidative stress and inflammation in osteoarthritis pathogenesis: Role of polyphenols. Biomed. Pharmacother. 129, 110452. 10.1016/j.biopha.2020.110452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong R. (2020). What causes neurodegenerative diseaseδ Folia Neuropathol. 58 (2), 93–112. 10.5114/fn.2020.96707 [DOI] [PubMed] [Google Scholar]
- Bai R., Guo J., Ye X.-Y., Xie Y., Xie T. (2022). Oxidative stress: The core pathogenesis and mechanism of Alzheimer's disease. Ageing Res. Rev. 77, 101619. 10.1016/j.arr.2022.101619 [DOI] [PubMed] [Google Scholar]
- Bohnen N. I., Yarnall A. J., Weil R. S., Moro E., Moehle M. S., Borghammer P., et al. (2022). Cholinergic system changes in Parkinson's disease: Emerging therapeutic approaches. Lancet Neurology 21 (4), 381–392. 10.1016/s1474-4422(21)00377-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brus B., Kosak U., Turk S., Pislar A., Coquelle N., Kos J., et al. (2014). Discovery, biological evaluation, and crystal structure of a novel nanomolar selective butyrylcholinesterase inhibitor. J. Med. Chem. 57 (19), 8167–8179. 10.1021/jm501195e [DOI] [PubMed] [Google Scholar]
- Cacabelos R. (2007). Donepezil in Alzheimer's disease: From conventional trials to pharmacogenetics. Neuropsychiatr. Dis. Treat. 3 (3), 303–333. [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Liu J., Wang B., Sun M., Yang H. (2022). Microglia in the neuroinflammatory pathogenesis of Alzheimer's disease and related therapeutic targets. Front. Immunol. 13, 856376. 10.3389/fimmu.2022.856376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli A., Bolognesi M. L., Minarini A., Rosini M., Tumiatti V., Recanatini M., et al. (2008). Multi-target-directed ligands to combat neurodegenerative diseases. J. Med. Chem. 51 (3), 347–372. 10.1021/jm7009364 [DOI] [PubMed] [Google Scholar]
- Cheng S., Zheng W., Gong P., Zhou Q., Xie Q., Yu L., et al. (2015). (-)-meptazinol-melatonin hybrids as novel dual inhibitors of cholinesterases and amyloid-beta aggregation with high antioxidant potency for Alzheimer's therapy. Bioorg. Med. Chem. 23 (13), 3110–3118. 10.1016/j.bmc.2015.04.084 [DOI] [PubMed] [Google Scholar]
- Choubey P. K., Tripathi A., Tripathi M. K., Seth A., Shrivastava S. K. (2021). Design, synthesis, and evaluation of n-benzylpyrrolidine and 1, 3, 4-oxa-diazole as multitargeted hybrids for the treatment of Alzheimer's disease. Bioorg. Chem. 111, 104922. 10.1016/j.bioorg.2021.104922 [DOI] [PubMed] [Google Scholar]
- Darras F. H., Kling B., Heilmann J., Decker M. (2012). Neuroprotective tri- and tetracyclic BChE inhibitors releasing reversible inhibitors upon carbamate transfer. ACS Med. Chem. Lett. 3 (11), 914–919. 10.1021/ml3001825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumanovic J., Nepovimova E., Natic M., Kuca K., Jacevic V. (2021). The significance of reactive oxygen species and antioxidant defense system in plants: A concise overview. Front. Plant Sci. 11, 552969. 10.3389/fpls.2020.552969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Li J., Long L., Shi T., Liu D., Tan W., et al. (2021). Design, synthesis and biological evaluation of n-anthraniloyl tryptamine derivatives as pleiotropic molecules for the therapy of malignant glioma. Eur. J. Med. Chem. 222, 113564. 10.1016/j.ejmech.2021.113564 [DOI] [PubMed] [Google Scholar]
- Fang L., He S., Yin P., Wang N., Zhang B., Jin H. (2022). Design, synthesis, and structure-activity relationship studies of novel tryptamine derivatives as 5-ht 1b receptor agonists. J. Mol. Struct. 1265, 133320. 10.1016/j.molstruc.2022.133320 [DOI] [Google Scholar]
- Forman H. J., Zhang H. (2021). Targeting oxidative stress in disease: Promise and limitations of antioxidant therapy. Nat. Rev. Drug Discov. 20 (9), 689–709. 10.1038/s41573-021-00233-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldenhuys W. J., Van der Schyf C. J. (2013). Rationally designed multi-targeted agents against neurodegenerative diseases. Curr. Med. Chem. 20 (13), 1662–1672. 10.2174/09298673113209990112 [DOI] [PubMed] [Google Scholar]
- Ghazanfari N., van Waarde A., Dierckx R. A. J. O., Doorduin J., de Vries E. F. J. (2021). Is cyclooxygenase-1 involved in neuroinflammationδ J. Neurosci. Res. 99 (11), 2976–2998. 10.1002/jnr.24934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Zhai L., Long H., Chen N., Gao C., Ding Z., et al. (2018). Genetic diversity of bletilla striata assessed by scot and irap markers. Hereditas 155, 35. 10.1186/s41065-018-0074-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampel H., Mesulam M. M., Cuello A. C., Farlow M. R., Giacobini E., Grossberg G. T., et al. (2018). The cholinergic system in the pathophysiology and treatment of Alzheimer's disease. Brain 141, 1917–1933. 10.1093/brain/awy132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanikoglu A., Ozben H., Hanikoglu F., Ozben T. (2020). Hybrid compounds & oxidative stress induced apoptosis in cancer therapy. Curr. Med. Chem. 27 (13), 2118–2132. 10.2174/0929867325666180719145819 [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Stiller C., Endres E., Scheiner M., Gunesch S., Sotriffer C., et al. (2019). Highly selective butyrylcholinesterase inhibitors with tunable duration of action by chemical modification of transferable carbamate units exhibit pronounced neuroprotective effect in an Alzheimer's disease mouse model. J. Med. Chem. 62 (20), 9116–9140. 10.1021/acs.jmedchem.9b01012 [DOI] [PubMed] [Google Scholar]
- Huang G., Kling B., Darras F. H., Hellmann J., Decker M. (2014). Identification of a neuroprotective and selective butyrylcholinesterase inhibitor derived from the natural alkaloid evodiamine. Eur. J. Med. Chem. 81, 15–21. 10.1016/j.ejmech.2014.05.002 [DOI] [PubMed] [Google Scholar]
- Iwata Y., Miyao M., Hirotsu A., Tatsumi K., Matsuyama T., Uetsuki N., et al. (2021). The inhibitory effects of orengedokuto on inducible pge2 production in bv-2 microglial cells. Heliyon 7 (8), e07759. 10.1016/j.heliyon.2021.e07759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F., Li M., Wang H., Ding B., Zhang C., Ding Z., et al. (2019). Coelonin, an anti-inflammation active component of bletilla striata and its potential mechanism. Int. J. Mol. Sci. 20 (18), 4422. 10.3390/ijms20184422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Choi W., Yoon J., Jeong B.-k., Pagire S. H., Pagire H. S., et al. (2022). Synthesis and biological evaluation of tyrosine derivatives as peripheral 5ht(2a) receptor antagonists for nonalcoholic fatty liver disease. Eur. J. Med. Chem. 239, 114517. 10.1016/j.ejmech.2022.114517 [DOI] [PubMed] [Google Scholar]
- Kumar R. S., Almansour A. I., Arumugam N., Kotresha D., Manohar T. S., Venketesh S. (2021). Cholinesterase inhibitory activity of highly functionalized fluorinated spiropyrrolidine heterocyclic hybrids. Saudi J. Biol. Sci. 28 (1), 754–761. 10.1016/j.sjbs.2020.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo H.-C., Lee K.-F., Chen S.-L., Chiu S.-C., Lee L.-Y., Chen W.-P., et al. (2022). Neuron-microglia contacts govern the pge(2) tolerance through tlr4-mediated de novo protein synthesis. Biomedicines 10 (2), 419. 10.3390/biomedicines10020419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski C. A., Lombardo F., Dominy B. W., Feeney P. J. (2022). Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46 (1-3), 3–26. 10.1016/s0169-409x(00)00129-0 [DOI] [PubMed] [Google Scholar]
- Liu D., Zhang H., Wang Y., Liu W., Yin G., Wang D., et al. (2022). Design, synthesis, and biological evaluation of carbamate derivatives of n-salicyloyl tryptamine as multifunctional agents for the treatment of Alzheimer's disease. Eur. J. Med. Chem. 229, 114044. 10.1016/j.ejmech.2021.114044 [DOI] [PubMed] [Google Scholar]
- Martins F. C. O. L., Batista A. D., Melchert W. R. (2021). Current overview and perspectives in environmentally friendly microextractions of carbamates and dithiocarbamates. Compr. Rev. Food Sci. Food Saf. 20 (6), 6116–6145. 10.1111/1541-4337.12821 [DOI] [PubMed] [Google Scholar]
- Matosevic A., Bosak A. (2020). Carbamate group as structural motif in drugs: A review of carbamate derivatives used as therapeutic agents. Arh. Hig. Rada Toksikol. 71 (4), 285–299. 10.2478/aiht-2020-71-3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mdeni N. L., Adeniji A. O., Okoh A. I., Okoh O. O. (2022). Analytical evaluation of carbamate and organophosphate pesticides in human and environmental matrices: A review. Molecules 27 (3), 618. 10.3390/molecules27030618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meden A., Knez D., Brazzolotto X., Nachon F., Dias J., Svete J., et al. (2022). From tryptophan-based amides to tertiary amines: Optimization of a butyrylcholinesterase inhibitor series. Eur. J. Med. Chem. 234, 114248. 10.1016/j.ejmech.2022.114248 [DOI] [PubMed] [Google Scholar]
- Mishra S., Pang S., Zhang W., Lin Z., Bhatt P., Chen S., et al. (2021). Carbofuran toxicity and its microbial degradation in contaminated environments. Chemosphere 279, 127419. 10.1016/j.chemosphere.2020.127419 [DOI] [PubMed] [Google Scholar]
- Mousseau D. D. (1993). Tryptamine: A metabolite of tryptophan implicated in various neuropsychiatric disorders. Metab. Brain Dis. 8 (1), 1–44. 10.1007/BF01000528 [DOI] [PubMed] [Google Scholar]
- Nagano T., Tsuda N., Fujimura K., Ikezawa Y., Higashi Y., Kimura S. H. (2021). Prostaglandin e-2 increases the expression of cyclooxygenase-2 in cultured rat microglia. J. Neuroimmunol. 361, 577724. 10.1016/j.jneuroim.2021.577724 [DOI] [PubMed] [Google Scholar]
- Oh J. M., Kang Y., Hwang J. H., Park J.-H., Shin W.-H., Mun S.-K., et al. (2022). Synthesis of 4-substituted benzyl-2-triazole-linked-tryptamine-paeonol derivatives and evaluation of their selective inhibitions against butyrylcholinesterase and monoamine oxidase-b. Int. J. Biol. Macromol. 217, 910–921. 10.1016/j.ijbiomac.2022.07.178 [DOI] [PubMed] [Google Scholar]
- Petrova T., Orellana C., Jelic V., Oeksengaard A.-R., Snaedal J., Hogh P., et al. (2020). Cholinergic dysfunction, neurodegeneration, and amyloid-beta pathology in neurodegenerative diseases. Psychiatry Res. Neuroimaging 302, 111099. 10.1016/j.pscychresns.2020.111099 [DOI] [PubMed] [Google Scholar]
- Rauf A., Badoni H., Abu-Izneid T., Olatunde A., Rahman M. M., Painuli S., et al. (2022). Neuroinflammatory markers: Key indicators in the pathology of neurodegenerative diseases. Molecules 27 (10), 3194. 10.3390/molecules27103194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeedi M., Felegari P., Iraji A., Hariri R., Rastegari A., Mirfazli S. S., et al. (2021). Novel n-benzylpiperidine derivatives of 5-arylisoxazole-3-carboxamides as anti-Alzheimer's agents. Arch. Pharm. 354 (3), e2000258. 10.1002/ardp.202000258 [DOI] [PubMed] [Google Scholar]
- Sang Z., Wang K., Shi J., Liu W., Cheng X., Zhu G., et al. (2020). The development of advanced structural framework as multi-target-directed ligands for the treatment of Alzheimer's disease. Eur. J. Med. Chem. 192, 112180. 10.1016/j.ejmech.2020.112180 [DOI] [PubMed] [Google Scholar]
- Sawatzky E., Wehle S., Kling B., Wendrich J., Bringmann G., Sotriffer C. A., et al. (2016). Discovery of highly selective and nanomolar carbamate-based butyrylcholinesterase inhibitors by rational investigation into their inhibition mode. J. Med. Chem. 59 (5), 2067–2082. 10.1021/acs.jmedchem.5b01674 [DOI] [PubMed] [Google Scholar]
- Scheiner M., Sink A., Hoffmann M., Vrigneau C., Endres E., Carles A., et al. (2022). Photoswitchable pseudoirreversible butyrylcholinesterase inhibitors allow optical control of inhibition in vitro and enable restoration of cognition in an Alzheimer's disease mouse model upon irradiation. J. Am. Chem. Soc. 144 (7), 3279–3284. 10.1021/jacs.1c13492 [DOI] [PubMed] [Google Scholar]
- Shevchenko V. P., Andreeva L. A., Shevchenko K. V., Nagaev I. Y., Myasoedov N. F. (2022). Investigation of the membranotropic properties of acylated peptide derivatives of dopamine and serotonin using the pampa method. Pharm. Chem. J. 56, 602–606. 10.1007/s11094-022-02683-2 [DOI] [Google Scholar]
- van der Zee S., Muller M. L. T. M., Kanel P., van Laar T., Bohnen N. I. (2021). Cholinergic denervation patterns across cognitive domains in Parkinson's disease. Mov. Disord. 36 (3), 642–650. 10.1002/mds.28360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakhloo D., Oberhauser J., Madira A., Mahajani S. (2022). From cradle to grave: Neurogenesis, neuroregeneration and neurodegeneration in Alzheimer's and Parkinson's diseases. Neural Regen. Res. 17 (12), 2606–2614. 10.4103/1673-5374.336138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang H., Liu D., Li X., Long L., Peng Y., et al. (2022). Discovery of carbamate-based n-salicyloyl tryptamine derivatives as novel pleiotropic agents for the treatment of Alzheimer's disease. Bioorg. Chem. 127, 105993. 10.1016/j.bioorg.2022.105993 [DOI] [PubMed] [Google Scholar]
- Wichur T., Malawska B. (2015). Multifunctional ligands - a new approach in the search for drugs against multi-factorial diseases. Postepy Hig. Med. Dosw. 69, 1423–1434. [PubMed] [Google Scholar]
- Wu H., Denna T. H., Storkersen J. N., Gerriets V. A. (2018). Beyond a neurotransmitter: The role of serotonin in inflammation and immunity. Pharmacol. Res. 140, 100–114. 10.1016/j.phrs.2018.06.015 [DOI] [PubMed] [Google Scholar]
- Yang J., Zhou Y., Ban Y., Mi J., He Y., Li X., et al. (2022). Development of naringenin-o-alkylamine derivatives as multifunctional agents for the treatment of Alzheimer's disease. J. Enzyme Inhibition Med. Chem. 37 (1), 792–816. 10.1080/14756366.2022.2041627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Wang Z., Hu H., Zhao M., Sun L. (2021). Microglia in alzheimer's disease: A target for therapeutic intervention. Front. Cell. Neurosci. 15, 749587. 10.3389/fncel.2021.749587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Wang Y., Liu D., Li J., Feng Y., Lu Y., et al. (2022). Carbamate-based n-substituted tryptamine derivatives as novel pleiotropic molecules for alzheimerδS disease. Bioorg. Chem. 125, 105844. 10.1016/j.bioorg.2022.105844 [DOI] [PubMed] [Google Scholar]
- Zhang H., Wang Y., Wang Y., Li X., Wang S., Wang Z. (2022). Recent advance on carbamate-based cholinesterase inhibitors as potential multifunctional agents against Alzheimer's disease. Eur. J. Med. Chem. 240, 114606. 10.1016/j.ejmech.2022.114606 [DOI] [PubMed] [Google Scholar]
- Zhu H., Dai O., Zhou F., Yang L., Liu F., Liu Y., et al. (2021). Discovery of bletillain, an unusual benzyl polymer with significant autophagy-inducing effects in A549 lung cancer cells through the Akt/GSK-3β/β-catenin signaling pathway. Bioorg. Chem. 117, 105449. 10.1016/j.bioorg.2021.105449 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.