Abstract
We have previously demonstrated that salmonellae, but not Escherichia coli or Yersinia enterocolitica, stimulates tumor necrosis factor alpha (TNFα) production in the human promonocytic cell line U38. Subsequent analysis revealed that the TNFα-inducing activity of salmonellae was associated with flagellin, a major component of flagella from gram-negative bacteria. In the present study, we have explored the basis for the apparent specificity of action of Salmonella flagella on TNFα expression in U38 cells and have extended this analysis to normal human peripheral blood mononuclear cells (PBMC). Flagella from the enteropathogenic E. coli strain E2348/69, Y. enterocolitica JB580, and Pseudomonas aeruginosa PAO1, which did not induce significant levels of TNFα production in U38 cells, were as potent as Salmonella flagella in terms of TNFα and interleukin 1β activation in PBMC. However, TNFα production in U38 cells was greatly enhanced when these cells were stimulated with flagella from E. coli, Y. enterocolitica, and P. aeruginosa in the presence of a costimulant, phorbol 13-myristate acetate. These findings are consistent with the hypothesis that the activation or differentiation state of a monocyte may have a substantial effect on the cell’s responsiveness to flagellum stimulation of cytokine synthesis. Furthermore, these results indicate that cytokine induction in monocytes may be a general property of flagella from gram-negative bacteria.
Tumor necrosis factor alpha (TNFα) is produced by activated macrophages and plays a central role in the defense against intracellular organisms such as Salmonella. Animal models have shown that host-derived inflammatory mediators such as TNFα promote the strong inflammatory response observed during Salmonella infection of the intestinal mucosa (2, 6, 38, 39). Despite its protective effect during infection (5, 13, 32, 37), elevated levels of serum TNFα in patients with endotoxic shock apparently correlate with the course and extent of pathology (31).
The most potent inducer of TNFα is the lipopolysaccharide (LPS) component of the outer membrane of gram-negative bacteria. In addition, Salmonella porins stimulate cytokine production in human monocytes (15). Using genetic and biochemical approaches, we demonstrated that Salmonella enteritidis and Salmonella typhimurium flagellin (the filament subunit protein of the flagellum) up regulates TNFα expression in the human promonocytic cell line U38 and in adherent peripheral blood mononuclear cells (PBMC) (9, 10). Because U38 cells are insensitive to LPS stimulation (4), and because PBMC were tolerized in vitro, this effect of Salmonella flagellin was independent of LPS in the cultures. More recently, Wyant et al. (43) reported that purified Salmonella typhi flagella stimulate TNFα production in PBMC.
Unlike Salmonella flagella, Escherichia coli and Yersinia enterocolitica flagella induce very low levels of TNFα production in U38 cells (10). Amino acid analysis of flagellin shows that the carboxy and amino termini are well conserved between Salmonella and other gram-negative bacteria. In contrast, the residues in the central portion of the protein are highly variable (22, 33). In order to explain the apparent specificity of Salmonella flagellin on TNFα production in U38 cells, the epitopes responsible for the activity would presumably have to be in the hypervariable domain. However, we were unable to identify any residues in the hypervariable region that were conserved in all of the Salmonella serotypes showing TNFα-inducing activity in U38 cells (10).
In the present study, we investigated the basis for the apparent specificity of action of Salmonella flagella in U38 cells. We compared the TNFα-inducing activity of flagella from Salmonella and from other gram-negative bacteria, including enteropathogenic E. coli, Y. enterocolitica, and Pseudomonas aeruginosa. In addition, we have extended the analysis of cell activation and induction of cytokine production by flagella to normal human blood monocytes. We found that Salmonella, E. coli, Y. enterocolitica, and P. aeruginosa flagella induce comparable levels of TNFα and interleukin 1β (IL-1β) in LPS-tolerized PBMC. However, neutralization of LPS with polymyxin B slightly decreased the effect of E. coli and P. aeruginosa flagella, but not the effect of Salmonella or Y. enterocolitica flagella, on TNFα expression in PBMC.
MATERIALS AND METHODS
Cells.
The human promonocytic cell line U38 is a stable derivative of U937 containing a human immunodeficiency virus long terminal repeat chloramphenicol acetyltransferase reporter construct (14). U38 was obtained from the National Institutes of Health AIDS Research and Reference Reagent Program (Rockville, Md.). U38 cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum and 50 μg of gentamicin per ml (complete RPMI medium) at 37°C in 5% CO2 atmosphere. U38 cells were subcultured daily and maintained at a density of approximately 5 × 105 cells/ml. PBMC were isolated from healthy donors. Heparinized whole blood was layered over isolymph (Gallard-Schlesinger Industries, Carle Place, N.Y.) at a 3:2 ratio and allowed to stand for 1 h at room temperature. The buffy coat was removed and layered again over isolymph at a 4:3 ratio. Following centrifugation at 1,200 × g for 30 min at room temperature, the top layer containing mononuclear cells was transferred and diluted at least 1:2 in phosphate-buffered saline (PBS) (pH 7.2). The cells were counted and collected by centrifugation, and the cell pellet was resuspended in serum-free RPMI medium containing gentamicin (50 μg/ml). Cells were seeded into 24-well plates at a density of approximately 5 × 106 cells/well and allowed to adhere for 2 h at 37°C in 5% CO2 atmosphere. Nonadherent cells were then removed, and the medium was replaced with complete RPMI medium. At this point, the monocyte-enriched monolayers were allowed to rest overnight at 37°C in 5% CO2 atmosphere. To induce LPS tolerance, PBMC were incubated overnight (16 to 20 h) in the presence of 1 μg of Salmonella LPS per ml (Sigma, St. Louis, Mo.).
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. S. enteritidis CDC5str (CD5), Y. enterocolitica JB580, and the bacteriophage P22 HT were the generous gifts of Virginia Miller. S. typhimurium SL1344 was kindly provided by Charles Dorman. The wild-type enteropathogenic E. coli strain E2348/69 was donated by James Kaper. P. aeruginosa PAO1 was a gift of Daniel Wozniak. S. typhimurium SJW86 and SJW1103 were provided by Robert Macnab. Frozen stocks of all strains were stored at −70°C in 25% (vol/vol) glycerol. Cultures of Salmonella, E. coli, and Pseudomonas strains were routinely grown in Luria-Bertani (LB) medium at 37°C with aeration. Yersinia cultures were grown at 25°C with aeration to promote flagellation.
TABLE 1.
Bacterial strains used in the study
| Strain | Parent (relevant genotype or phenotype) | References or source |
|---|---|---|
| S. enteritidis CDC5str | CDC5 (streptomycin resistance) | 36 |
| S. typhimurium | ||
| SL1344 | Wild type | 20 |
| SL1344H2 | SL1344 (fliC::Tn10) | This study |
| SJW1103 | H1-i− H2-I · 2straight vh2+ | 44 |
| SJW86 | SJW1103 (fliC::Tn10) | R. M. Macnab |
| P. aeruginosa PAO1 | Wild type | 19 |
| E. coli E2348/69 | Wild type | 12 |
| Y. enterocolitica JB580 | 8081v (yen IMR) | 24 |
Preparation of bacterial-conditioned medium and isolation of partially purified flagella.
Conditioned media were obtained by growing bacteria overnight as described above. The optical density of each culture was determined spectrophotometrically at 510 nm. The values obtained were normalized using the optical density at 510 nm of strain CD5 as reference. Bacteria were pelleted, and the conditioned medium was sterilized by passage through a 0.2-μm-pore-size Millipore filter. Conditioned media were freshly prepared for each experiment.
Differential centrifugation was used to partially purify flagella from each strain (28). Briefly, bacteria were grown on LB agar plates for 24 h at 37°C (Yersinia strain JB580 was grown at 25°C). The cultures were then collected into 100 ml of PBS and centrifuged for 15 min at 5,000 × g and 4°C. The bacterial pellets were resuspended in 100 ml of PBS for each 6 g of dry weight and blended in a Waring blender for 5 min. The samples were then centrifuged at 16,000 × g for 15 min at 4°C to pellet whole cells, membranes, and other cell debris. The supernatants were centrifuged at 40,000 × g for 3 h at 4°C. The flagella thus obtained were resuspended in PBS and stored at 4°C. The protein concentration of each sample was determined spectrophotometrically by the method of Bradford (8) by using a commercial protein assay solution (Bio-Rad, Hercules, Calif.).
TNFα induction in U38 cells and primary human monocytes.
Just before stimulation, 5 × 106 U38 cells were seeded into each well of a 24-well plate in 0.5 ml of complete RPMI medium. The flagella were then added at various concentrations (see figure legends). When phorbol 13-myristate acetate (PMA) was used as a costimulant, it was added to a final concentration of 10 ng/ml at the same time as the flagella. U38 cells were incubated at 37°C in 5% CO2 for 4 h. Cells were then lysed in 0.5% Triton X-100 (TX-100). The lysates were briefly vortexed and incubated on ice for 5 min. The samples were then centrifuged at 16,000 × g for 5 min at 4°C, and the supernatants were stored at −20°C until they were assayed for TNFα content. We had previously determined that, on average, only 30% of the TNFα produced by U38 cells in response to Salmonella is released into the medium. The majority of induced TNFα was cell associated. Because TNFα is not stored intracellularly, the cell-associated TNFα is most likely the membrane form of the protein, which is biologically active (26) and converted into the soluble form by proteolysis (35). By adding TX-100 to the cell suspension, we were able to assess the total amount of TNFα induced by each stimulus. The synergism index (I) was calculated as follows: I = FP/(F + P), where FP is the amount of TNFα (in picograms per milliliter) produced in response to both flagella and PMA, F is the amount of TNFα (in picograms per milliliter) produced in response to flagella alone, and P is the amount of TNFα (in picograms per milliliter) produced in response to PMA alone.
Before stimulation, LPS-tolerized and nontolerized PBMC were washed three times in serum-free RPMI medium to remove free LPS and any remaining nonadherent cells. Conditioned media and flagella were diluted in serum-free RPMI medium to the final concentrations indicated in each figure legend. In the polymyxin B experiments, flagella or LPS (1.25 ng/ml) was incubated for 10 min at room temperature in serum-free RPMI medium either in the presence or absence of polymyxin B (10 μg/ml) before being added to the PBMC. Stimulated and unstimulated PBMC were then incubated for 4 h at 37°C in 5% CO2 and lysed as described for U38 cells. The concentration of U38 cells and PBMC in each experiment was held constant at 107 cells/ml, although the number of cells per sample varied slightly from experiment to experiment.
ELISA for TNFα.
The total amount of TNFα in each sample (soluble and cell associated) was determined with a commercial enzyme-linked immunosorbent assay (ELISA) kit (Cistron, Pine Brook, N.J.). Aliquots (100 μl) of each TX-100-treated cell lysate were used in the assay according to the manufacturer’s protocol. The addition of 0.5% TX-100 to the samples and to the standards had no effect on the sensitivity of the assay.
Generating an S. typhimurium mutant expressing only phase 2 flagellin (FljB).
Generalized transduction with bacteriophage P22 HT int was used to move the Tn10::fliC mutation from the S. typhimurium mutant SJW86 to the wild-type strain SL1344. P22 minilysates were prepared as described (27). To obtain P22 lysates from the donor strain SJW86, 0.5 ml of a saturated culture of SJW86 was incubated with 2 ml of P22 minilysates until the bacteria lysed (8 h). The culture was then transferred to 1.5-ml tubes and centrifuged at 23,000 × g for 2 min to pellet debris. The lysates were stored at 4°C in 1% (vol/vol) chloroform. For transduction, 10 μl of SJW86 lysates was mixed with 100 μl of an overnight culture of SL1344. The culture was grown for 10 min at 37°C without shaking, and 0.9 ml of LB medium supplemented with 10 mM EGTA was added to stop phage adsorption. The culture was incubated at 37°C for 15 min, and a 100-μl sample was plated on L agar containing tetracycline (15 μg/ml).
IL-1β induction and immunoprecipitation.
LPS-tolerized PBMC were washed three times in methionine-free RPMI medium (ICN, Costa Mesa, Calif.) and incubated for 1 h in methionine-free RPMI medium in the presence of flagella as described above. [35S]methionine (ICN) was then added to each well to a final concentration of 250 μCi/ml, and incubation was allowed to continue for an additional 90 min. At this point, the medium was discarded and the PBMC were lysed in 500 μl of immunoprecipitation buffer (IPB) (150 mM NaCl, 0.4% Nonidet P-40 [NP-40], 50 mM Tris [pH 8.0], 10 mM EDTA) containing 0.1% bovine serum albumin and 10 μg of anti-IL-1β antibody 3ZD per ml (Biological Response Modifiers Program, National Cancer Institute, Frederick, Md.). The samples were incubated overnight at 4°C with shaking. Protein G-agarose (20 μl) (Life Technologies, Gaithersburg, Md.) equilibrated in IPB was added to each sample, and the incubation was allowed to proceed for an additional 45 min at 4°C with shaking. The samples were washed once in IPB, once in wash buffer 2 (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% NP-40, 1 mM EDTA, 0.25% gelatin, 0.25% sodium azide, 0.1% sodium dodecyl sulfate [SDS]), and once in wash buffer 3 (10 mM Tris-HCl [pH 7.5], 0.1% NP-40). The pellets were resuspended in 20 μl of 2× treatment buffer (125 mM Tris-HCl [pH 6.8], 4% SDS, 20% glycerol, 10% 2-mercaptoethanol, trace bromophenol blue), boiled for 90 s, and diluted 1:2 with distilled H2O. Samples were electrophoresed in 12.5% SDS polyacrylamide gels. Bands were visualized by autoradiography with a Kodak BioMax TranScreen-LE intensifying screen (Eastman Kodak Co., Rochester, N.Y.) and were quantified using an Ambis radioimager (Ambis Systems, San Diego, Calif.).
RESULTS
Various bacterial flagella induce TNFα expression in U38 cells.
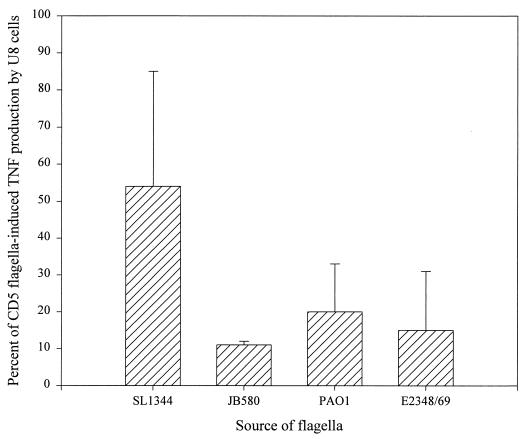
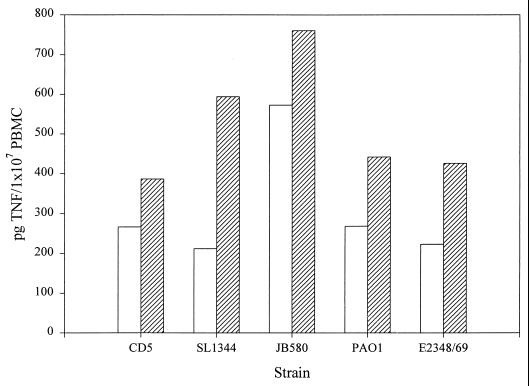
In a previous study, we reported that Salmonella-conditioned medium stimulates TNFα production in the U38 promonocytic cell line (10). In contrast, conditioned medium from E. coli or Y. enterocolitica induced very low levels of TNFα in these cells. The lack of strong TNFα-inducing activity by E. coli- or Y. enterocolitica-conditioned media might simply be due to differences in the amounts of flagella that are shed into the culture medium. To bypass this possibility and directly assess the ability of E. coli and Y. enterocolitica flagella to induce TNFα production, flagella were isolated from these strains, as well as from P. aeruginosa, and were tested for the ability to stimulate TNFα expression in U38 cells. Although it was not possible to determine the actual concentration of flagellin each sample due to the presence of other flagellar components, the design of these experiments controlled the total amount of protein used as stimulus. It should be noted, however, that approximately 20,000 flagellin monomers are assembled in each filament, making this protein by far the most abundant component of the flagellum. In addition to S. enteritidis CD5 and S. typhimurium SL1344 strains, flagella were obtained from enteropathogenic E. coli E2348/69, from Y. enterocolitica JB580, and from P. aeruginosa PAO1. Multiple titration experiments using CD5 and PAO1 flagella showed the optimal concentration of flagella to be 1.6 μg/ml (data not shown). All of the flagellum preparations were therefore tested on U38 cells at this concentration. The total amount of cell-associated and released TNFα produced was determined by ELISA following a 4-h incubation (Fig. 1). As previously observed with conditioned medium, Salmonella flagella induced significantly higher levels of TNFα in U38 cells than flagella from E. coli (E2348/69), Y. enterocolitica (JB580), or P. aeruginosa (PAO1). Interestingly, we found that, although Salmonella flagella were generally more potent inducers of TNFα production than E. coli, Y. enterocolitica, or P. aeruginosa flagella, S. enteritidis CD5 flagella consistently induced higher levels of TNFα in U38 cells than flagella from S. typhimurium SL1344.
FIG. 1.
TNFα induction by flagellar preparations from S. enteritidis (CD5), S. typhimurium (SL1344), enteropathogenic E. coli (E2348/69), Y. enterocolitica (JB580), and P. aeruginosa (PAO1). U38 cells (5 × 106) were incubated with each of the flagellum preparations at a final protein concentration of 1.6 μg/ml for 4 h. TX-100-treated cell lysates were then assayed for TNFα by ELISA. The results shown are the means and standard deviations from three independent experiments. Control values have been subtracted for each experiment. The amount of TNFα produced in response to the flagellum preparations from CD5 in each experiment was set as 100% induction and used to normalize the amount of TNFα in the other samples. The TNFα values obtained in the CD5 samples were 582, 108, and 389 pg/ml.
TNFα induction by phase 1 and phase 2 flagella.
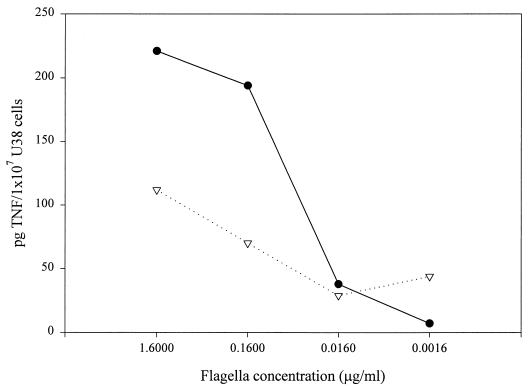
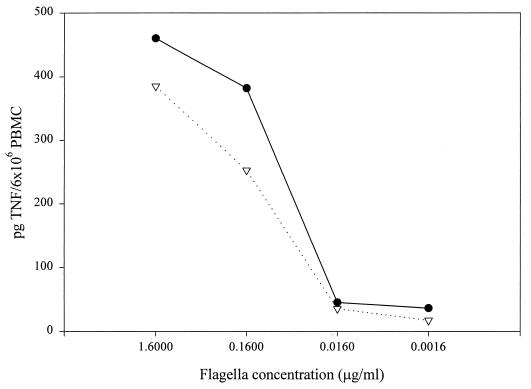
Unlike S. enteritidis strains, which possess only one form of flagellin (40), FliC, S. typhimurium strains alternate between the expression of two homologous forms of flagellin, FliC (also called phase 1 protein) and FljB (or phase 2 protein). Only one of the two proteins is expressed at a given time. This phase-variable system is controlled at the level of transcription by a site-specific recombinase, Hin. Because the frequency of recombination is relatively low (10−3 to 10−5 cells/generation) and there is no known selection, S. typhimurium cultures presumably contain a mixed population of organisms expressing both forms of flagellin. We had previously observed that conditioned medium from an E. coli strain expressing S. enteritidis FliC contained more TNFα-inducing activity than conditioned medium from the same strain expressing S. typhimurium FljB (9). If there were an intrinsic difference in the abilities of FliC and FljB to stimulate cytokine production (that is, if FljB were less efficient in terms of TNFα stimulation than FliC), the presence of FljB-containing flagella could explain the lower levels of TNFα-inducing activity found in conditioned medium and flagellum preparations from S. typhimurium as compared to preparations from S. enteritidis. We explored this possibility by testing the effect of flagella from S. typhimurium strains which carry only one functional flagellin gene, either fliC or fljB. One of these strains, SJW1103, expresses only the phase 1 protein due to the deletion of the fljB gene (44). The other strain, SL1344H2, was obtained by introducing a fliC::Tn10 mutation (from SJW86) into the wild-type strain SL1344 using P22 HT generalized transduction. Although fljB is functional in SL1344H2, this strain can still switch to expression of the now-inactivated fljC gene. As a consequence, SL1344H2 bacteria alternate between a flagellate (phase 2) and a nonflagellate state. The partially motile phenotype of SL1344H2 was confirmed on soft agar (data not shown). To determine whether there is an intrinsic difference in the abilities of FliC and FljB to induce TNFα production, U38 cells were incubated with flagella isolated from SJW1103 (FliC) or SL1344H2 (FljB) at various concentrations (Fig. 2). The results of these experiments show that TNFα production in U38 cells is lower in response to FljB-containing flagella than in response to FliC-containing flagella. This observation is consistent with the results obtained by using CD5 and SL1344 flagella, as well as with previously published data demonstrating that conditioned medium from E. coli expressing only S. enteritidis FliC induced higher levels of TNFα production than conditioned medium from E. coli expressing only S. typhimurium FljB (9). Taken together, these results indicate that FliC is a more effective stimulus for TNFα production in U38 cells than FljB.
FIG. 2.
Effect of phase 1 and phase 2 flagellin on TNFα production in U38 cells. Flagella from SJW1103 (●) or SL1344H2 (▿) were used to stimulate U38 cells at the indicated concentrations. The amount of TNFα in each sample was determined by ELISA. This experiment was repeated three times with similar results.
Effect of bacterial-conditioned medium and flagella on TNFα expression in PBMC.
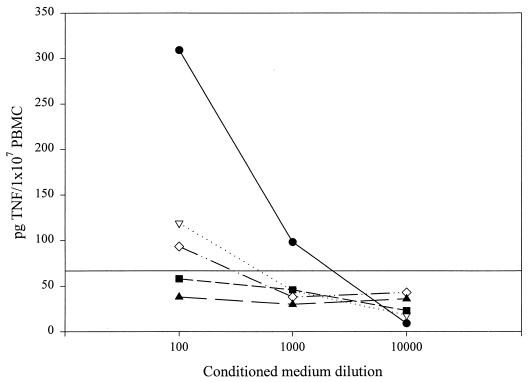
We have previously shown that conditioned medium and flagella from S. enteritidis CD5 stimulate TNFα production in LPS-tolerant PBMC (9). To determine whether flagella from E. coli, Y. enterocolitica, and P. aeruginosa can induce TNFα in PBMC, we tested conditioned medium from each strain on LPS-tolerized PBMC at various dilutions (Fig. 3). Blood monocytes, which are normally exquisitely sensitive to activation by LPS, become nonresponsive after prolonged exposure to LPS (18, 23). In order to differentiate between the effects of flagella and contaminating LPS on TNFα production, PBMC were LPS tolerized prior to testing. U38 cells, which do not express the CD14 receptor, are LPS tolerant and did not require any additional treatment prior to stimulation with bacteria-derived preparations. S. enteritidis CD5-conditioned medium induced the highest levels of TNFα in PBMC. This effect was concentration dependent and independent of LPS, since LPS itself induced a very low level of TNFα production. P. aeruginosa- and S. typhimurium-conditioned media were markedly less effective as TNFα inducers, whereas E. coli- and Y. enterocolitica-conditioned media induced little, if any, TNFα. Given these results, it is possible that flagella from Salmonella, especially S. enteritidis, are significantly more effective TNFα inducers than flagella from E. coli, Y. enterocolitica, or P. aeruginosa. Alternatively, the limited cytokine induction by conditioned media from non-S. enteritidis strains may simply reflect differences in the extent of flagella shed by each organism.
FIG. 3.
Conditioned media from S. enteritidis CD5 (●), S. typhimurium SL1344 (▿), Y. enterocolitica JB580 (■), P. aeruginosa PAO1 (◊), and enteropathogenic E. coli E2348/69 (▴) induce TNFα production in LPS-tolerized PBMC. Following overnight exposure to LPS (1 μg/ml), PBMC were stimulated with conditioned medium from each strain at the dilutions indicated or with LPS (1 μg/ml). The amount of TNFα in each sample (cell associated and released) was measured by ELISA after a 4-h incubation. The data are representative of three similar experiments. The horizontal line represents the amount of TNFα produced in response to LPS (1 μg/ml) in the experiment shown.
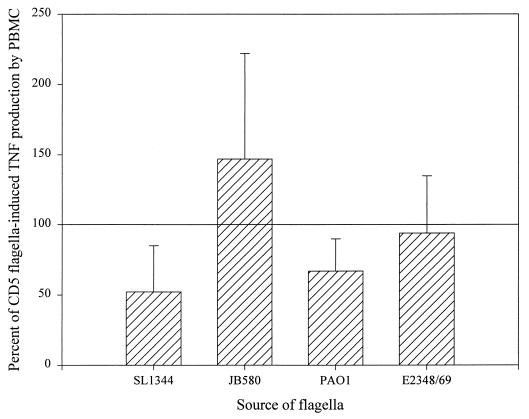
To test this second possibility, equal concentrations of flagella from the various bacterial species were tested on LPS-tolerant PBMC (Fig. 4). In contrast to U38 cells (Fig. 1) and the PBMC response to conditioned medium (Fig. 3), the levels of TNFα produced by PBMC were not significantly different in response to flagella from Salmonella, E. coli, Y. enterocolitica, or P. aeruginosa. The quantitative variability from experiment to experiment most likely reflects differences in the sensitivity of each individual blood donor to stimulation by the flagellum preparations and also to the extent of LPS tolerization. In general, however, E. coli, Y. enterocolitica, and P. aeruginosa flagella induced levels of TNFα comparable to those induced by S. enteritidis. Several conclusions can be drawn from these results. First, the differences in TNFα-inducing activity observed with conditioned media from Salmonella, E. coli, Y. enterocolitica, and P. aeruginosa most likely reflect differences in the amounts of flagella shed from different organisms. Second, the fact that Salmonella flagella are not unique in their ability to induce TNFα production in PBMC (as opposed to the situation in U38 cells) suggests that PBMC and U38 cells differ in their sensitivities to activation by gram-negative flagella. Finally, we observed that in PBMC, as in U38 cells, flagella from S. typhimurium SL1344 induced the production of approximately half as much TNFα as S. enteritidis CD5 (Fig. 1 and 4). To determine whether the response of PBMC to CD5 and SL1344 flagella was due to a difference in sensitivity of these cells to activation by FliC and FljB, we compared the TNFα-inducing activity of SJW1103 and SL1344H2 flagella on LPS-tolerized PBMC (Fig. 5). There was only a slight difference in the abilities of these two flagellum preparations to induce TNFα production in PBMC. This result contrasts with the data obtained in U38 cells (Fig. 2), as well as with our prior observation that conditioned medium from E. coli expressing S. typhimurium FljB was less effective as a TNFα inducer in U38 cells than conditioned medium from E. coli expressing S. enteritidis FliC (9).
FIG. 4.
Flagella from CD5, SL1344, JB580, PAO1, and E2348/69 up regulate TNFα expression in human blood monocytes. LPS-tolerized PBMC were incubated for 4 h in the presence of each flagellum preparation at a final protein concentration of 1.6 μg/ml. As described in Fig. 1, the amount of TNFα in each sample was measured by ELISA and then normalized against the value for the CD5 flagellum-treated cells. The means and standard deviations from five independent experiments are shown.
FIG. 5.
Induction of TNFα production in LPS-tolerized PBMC by FliC- and FljB-containing flagella. LPS-tolerized PBMC were incubated with flagella from SJW1103 (FliC) (●) or SL1344H2 (FljB) (▿) at the indicated concentrations for 4 h. The amount of TNFα in each sample was determined by ELISA. The graph shown is representative of three independent experiments.
Source of the TNFα-inducing activity in flagellum preparations.
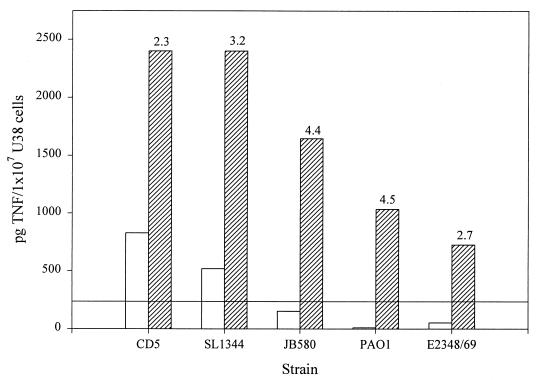
Our results demonstrate that, unlike U38 cells, LPS-tolerant PBMC respond equally to flagellum preparations from Salmonella, E. coli, Y. enterocolitica, and P. aeruginosa. To rule out the possibility that the non-Salmonella flagellum preparations contained a soluble component other than flagella that may be responsible for TNFα induction, each preparation was centrifuged at 100,000 × g to sediment the flagella. The level of TNFα-inducing activity in the sedimented flagella was then compared to the activity of the nonfractionated preparations in cultures of LPS-tolerized PBMC (Fig. 6). The results of these experiments demonstrate that all of the TNFα-inducing activity in flagellum preparations from Salmonella, E. coli, Y. enterocolitica, and P. aeruginosa is found in the fractions pelleted at 100,000 × g. Surprisingly, the centrifuged samples showed increased levels of activity relative to the original preparations, possibly because of the presence of an inhibitor in the uncentrifuged preparations. Taken together, these data support the conclusion that the flagella from Salmonella, E. coli, Y. enterocolitica, and P. aeruginosa are responsible for TNFα induction in U38 cells and PBMC.
FIG. 6.
Association of the TNFα-inducing activity in flagellum preparations from Salmonella, E. coli, Yersinia, and Pseudomonas with intact flagella. Active flagellum preparations from each bacterial strain were centrifuged at 100,000 × g for 30 min. The pelleted fractions were resuspended in PBS and used to stimulate LPS-tolerized PBMC at a final protein concentration of 1.6 μg/ml (hatched bars). The original preparations (open bars) were also tested at the same concentration (1.6 μg/ml). The level of TNFα in each sample was measured by ELISA.
Flagella from Salmonella, E. coli, Y. enterocolitica, and P. aeruginosa induce IL-1β in PBMC.
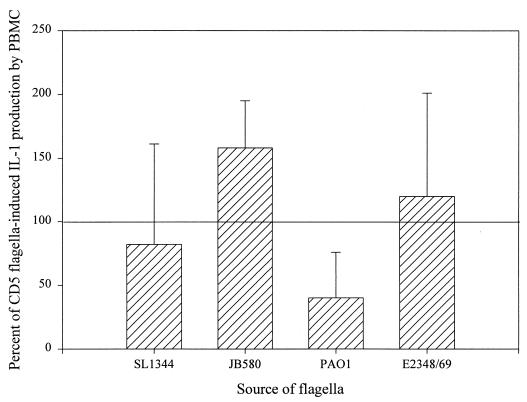
In addition to TNFα, components of the cell wall of Salmonella such as LPS and porins trigger the release of another potent cytokine, IL-1, from human monocytes (15). In view of the shared role of TNFα and IL-1β in host defense and inflammation, we determined whether flagella are able to stimulate IL-1β production. Since the process of tolerizing PBMC (overnight exposure to 1 μg of LPS per ml) induces high levels of IL-1β, immunoprecipitation, rather than an ELISA, was used to measure IL-1β production in flagellum-treated cells. By labeling newly synthesized protein with 35S, IL-1β produced in response to the flagella could be distinguished from that produced in response to the LPS used to tolerize cells. LPS-tolerant PBMC were incubated with or without flagella from Salmonella, E. coli, Y. enterocolitica, and P. aeruginosa for 1 h, then [35S]methionine was added and the incubation was allowed to proceed for an additional 1.5 to 3 h (Fig. 7). Because using different blood donors results in a certain degree of variability, the data for each experiment were normalized with the levels of IL-1β induced by CD5 flagella. The results obtained closely resemble those for TNFα induction in PBMC (Fig. 4); flagella from each of the five strains tested induced IL-1β production in LPS-tolerant PBMC (Fig. 7). Even accounting for some variability between experiments, flagella from Y. enterocolitica and E. coli were, on average, stronger cytokine inducers than flagella from P. aeruginosa. The effect of flagella is, therefore, not limited to TNFα up regulation but includes at least one other important proinflammatory cytokine, IL-1β. Previous reports showed that IL-8 is induced in epithelial cells, monocytes, and neutrophils by various E. coli and P. aeruginosa strains (1, 11, 25). Using reverse transcription-PCR, we were unable to detect an increase in IL-8 or -12 mRNA levels in response to Salmonella, E. coli, Y. enterocolitica, or P. aeruginosa flagella in either U38 cells or PBMC (data not shown).
FIG. 7.
Flagella from gram-negative bacteria induce IL-1β synthesis in LPS-tolerized PBMC. PBMC were incubated with 1.6 μg of each flagella per ml for 1 h in methionine-free RPMI medium. [35S]methionine (250 μCi/ml) was added, and the samples were incubated for an additional 1.5 to 3 h. The amount of cell-associated IL-1β in each sample was determined by immunoprecipitation with an anti-IL-1β monoclonal antibody as described in Materials and Methods. This figure summarizes the results obtained in four separate immunoprecipitation experiments.
Effect of costimulation on flagellum-induced TNFα production in U38 cells.
The differential responsiveness of U38 cells and PBMC to FliC- versus FljB-containing flagella (Fig. 2 and 5) and to flagella from non-Salmonella species (Fig. 1 and 4) may be due to differences in the activation state or state of differentiation of these cells, or to differences in the ability of U38 cells and PBMC to recognize and bind flagellin. Given these possibilities, we explored whether U38 cells might be converted (i.e., primed) to a heightened state of flagellum responsiveness by a costimulant such as PMA. PMA is a protein kinase C agonist and a strong TNFα inducer in monocytes and macrophages, including U38 cells. In addition, PMA treatment promotes terminal differentiation of U937 cells, the parent line from which U38 cells were obtained (3, 7, 34, 42). Therefore, we reasoned that exposure to PMA would cause U38 cells to become more monocyte-macrophage-like, and consequently increase their sensitivity to flagellin. Preliminary titration experiments showed that induction of TNFα expression by PMA was concentration dependent, with approximately 20 to 30% of maximal TNFα levels being produced at a PMA concentration of 10 ng/ml (data not shown). U38 cells were incubated with flagella from S. enteritidis, S. typhimurium, E. coli, Y. enterocolitica, and P. aeruginosa in the presence or absence of 10 ng of PMA per ml. For each bacterial strain, the effect of PMA costimulation was determined using a synergism index (see Materials and Methods). The value of I is defined as the ratio of TNFα produced when the two stimuli (flagella and PMA) are used together to the sum of the amounts of TNFα produced when each stimulus is used independently. Therefore, a value of 1 for I indicates no synergism between flagella and PMA, whereas values of I above 1 suggest a synergistic effect of these two stimuli. As shown in Fig. 8, the levels of TNFα induced in U38 cells by the combination of flagella and PMA are two- to fivefold greater than would be predicted if the two stimuli were acting independently of each other. The synergism between PMA and flagella was strong when E. coli, Y. enterocolitica, and P. aeruginosa flagella were used. However, the level of TNFα production in the presence of PMA and any of the non-Salmonella flagella was less than that achieved with PMA and S. enteritidis or S. typhimurium flagella. Nonetheless, these results clearly demonstrate that U38 cells, like PBMC, possess the intrinsic potential to respond to flagella from a spectrum of gram-negative organisms. When the state of cellular differentiation of U38 cells is changed (i.e., the cells are primed) by PMA, the effect of flagella on TNFα expression is greatly enhanced. Similarly, it is possible that TNFα production in response to flagella from E. coli, Y. enterocolitica, and P. aeruginosa is higher in PBMC than in U38 cells because LPS tolerization, or residual LPS in the flagellum preparations, acts like PMA to prime cells to respond to flagella.
FIG. 8.
Flagella and PMA act synergistically to stimulate TNFα expression in U38 cells. U38 cells (5 × 106) were incubated in PMA alone (10 ng/ml) (indicated by the horizontal line), flagella alone (1.6 μg/ml) (open bars), or PMA and flagella together (10 ng/ml and 1.6 μg/ml, respectively) (hatched bars) for 4 h. The synergism index, which is indicated for each strain, was calculated as the ratio of the amount of TNFα produced in response to both stimuli and the sum of the levels of TNFα in the samples independently incubated with each stimulus. This experiment was repeated three times with similar results.
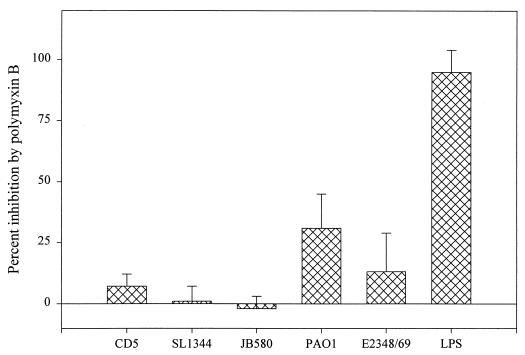
Effect of polymyxin B on the ability of gram-negative flagella to induce TNFα production in nontolerized PBMC.
To explore the possibility that exposure to LPS either during the tolerization process or during the 4-h stimulation period may contribute to PBMC activation by non-Salmonella flagella, nontolerized PBMC were incubated with flagella treated with polymyxin B. Polymyxin B binds to lipid A with high affinity and inhibits the biological activity of LPS (29). Flagella from Salmonella, E. coli, Y. enterocolitica, and P. aeruginosa were treated with polymyxin B and then used to stimulate nontolerized PBMC (Fig. 9). Polymyxin B treatment completely abolished the stimulatory effect of added LPS. However, the drug had no effect on the TNFα-inducing activity of Salmonella or Y. enterocolitica flagella, confirming that flagellin, and not contaminating LPS, is responsible for the biological activity found in these flagellum preparations. Furthermore, neutralization of LPS by polymyxin B did cause a small but significant decrease (approximately 30%) in TNFα induction by P. aeruginosa and an intermediate (approximately 12%) decrease in activity in response to E. coli flagella. Taken together, these results suggest that priming is not required for the stimulatory effect of non-Salmonella-derived flagella on PBMC cytokine production. Although LPS contamination is not responsible for TNFα production in response to Salmonella or Y. enterocolitica flagella, very low levels of LPS in E. coli and P. aeruginosa flagellum preparations may be required for optimal TNFα induction.
FIG. 9.
Effect of polymyxin B on the ability of flagella to induce TNFα expression in nontolerized PBMC. Flagella from each bacterial strain was incubated in the presence or absence of 10 μg of polymyxin B per ml at room temperature for 10 min and then used to stimulate nontolerized PBMC. Polymyxin B-treated and untreated LPS (1.25 ng/ml) was used as a control for polymyxin B activity. Following a 4-h incubation of the cells in the presence or absence of stimulation, the amount of TNFα produced was measured by ELISA. The means and standard deviations from three independent experiments are shown.
DISCUSSION
As with many organisms, the pathogenic potential of salmonellae is determined to a large extent by their effect on the release of inflammatory mediators by host cells. In experimental models of Salmonella infection, for example, low levels of TNFα are protective (5, 32, 37), whereas increased production of this cytokine by macrophages is associated with damage to the epithelial layer of the small intestine (2, 6, 38, 39). Similarly, the pathophysiology of septic shock is primarily due to TNFα, IL-1, and IL-6 that are released into the bloodstream in response to circulating LPS (16, 30, 31). Host cell activation and TNFα expression are therefore essential aspects of Salmonella infection. In addition to LPS, a very potent TNFα inducer both in vivo and in vitro (17), outer membrane porins from Salmonella increase TNFα synthesis in human blood monocytes (15). As we (9) and others (43) have shown, Salmonella flagella also causes TNFα and IL-1β production in monocytes. This effect of flagella on cytokine synthesis is independent of LPS, since U38 cells are insensitive to LPS stimulation (they lack the CD14 receptor), and LPS-tolerized PBMC retain their responsiveness to flagella, even when the flagella are treated with polymyxin B. Although Salmonella flagella are equally effective on U38 cells and primary human monocytes, E. coli, Y. enterocolitica, and P. aeruginosa showed remarkable differences in their abilities to induce TNFα in U38 cells and PBMC. Flagella from E. coli, Y. enterocolitica, and P. aeruginosa were minimally effective as TNFα-inducers in U38 cells but were as effective as Salmonella in cultures of PBMC. Taken together, these results show a fundamental difference in the way U38 cells and PBMC respond to non-Salmonella flagella. Genetic evidence from a previous study (9) demonstrated that flagellin is responsible for the induction of cytokine synthesis in PBMC. In lieu of genetic evidence, the results of the ultracentrifugation experiments support the assumption that the TNFα-inducing activity in E. coli, Y. enterocolitica, and P. aeruginosa flagella preparations is due to flagella (as is the case for Salmonella) and not due to a soluble contaminant.
To reconcile the discrepancy between the responses of U38 cells and PBMC to E. coli, Y. enterocolitica, and P. aeruginosa flagella, we hypothesized that LPS contamination of the flagellum preparations may prime PBMC to respond more strongly to E. coli and P. aeruginosa flagella. Consistent with this hypothesis, we observed that when U38 cells were costimulated with PMA, their response to E. coli, Y. enterocolitica, and P. aeruginosa flagella was greatly enhanced. Although these results demonstrate a synergism between flagella and PMA, the extent of the synergistic effect of flagella and LPS does not fully account for the differences between Salmonella and the other gram-negative bacteria in terms of the induction of cytokine synthesis. Particularly evident is the fact that LPS neutralization by polymyxin B had no effect on the TNFα-inducing activity of Y. enterocolitica flagella, suggesting that LPS did not contribute to this activity with PBMC. However, when tested on U38 cells, Y. enterocolitica flagella did not induce significant levels of TNFα. Therefore, priming may play a role in TNFα stimulation by P. aeruginosa and perhaps, to a lesser extent, E. coli, but it does not appear to be a factor in activation by Salmonella or Y. enterocolitica. A second possible explanation for the different responses of U38 cells and PBMC is that PBMC may be intrinsically more sensitive than U38 cells to the effects of flagella, including non-Salmonella flagella. Several factors may increase the sensitivity of PBMC. Some of these are apparently linked to the differentiation state of the cells, since PMA treatment of U38 cells heightens the response of these cells to non-Salmonella flagella. It is possible that PBMC express higher levels of a putative surface receptor for flagellin or a form of receptor that has higher affinity for flagellin. In addition, coupling of this receptor to cellular second messengers may be more efficient in PBMC than in U38 cells.
The observation that flagella can stimulate TNFα and IL-1β production in both LPS-tolerant and nontolerant PBMC indicates that monocyte activation by bacterial components other than LPS may contribute to the development of inflammatory responses during gram-negative infections. Similar to the in vitro situation, blood monocytes, which are highly sensitive to LPS stimulation, become refractory to further activation in septic patients. This often results in the inability to mount an effective immune response to a secondary infection (41). Likewise, there is evidence that macrophages of the lamina propria, which are constantly exposed to the LPS shed by the normal intestinal flora, are tolerant to LPS in vivo (21). The ability of macrophages to become LPS tolerant is a protective mechanism to prevent the host from developing a chronic state of inflammation. Although these macrophages are no longer responsive to LPS stimulation, our results suggest that the ability to respond to flagella is not compromised. Secondary stimuli such as flagella may reverse the nonresponsive state induced by LPS and trigger cytokine production in these cells. On the other hand, the presence of flagella during LPS stimulation of monocytes (as occurs in the blood of patients infected with a gram-negative organism), may enhance the cytokine response and possibly lead to more severe symptoms in these individuals. Ultimately, new therapies may seek to neutralize the effects of flagellin on monocyte and macrophage activation during sepsis.
ACKNOWLEDGMENTS
We thank Virginia Miller for strains CDC5str and JB580 and for bacteriophage P22 HT int. We also thank Charles Dorman for strain SL1344, Robert Macnab for strains SJW1103 and SJW86, James Kaper for strain E2348/69, and Daniel Wozniak for strain PAO1.
This study was supported by NIH Grant R01-AI38670.
REFERENCES
- 1.Agace W W, Hedges S R, Ceska M, Svanborg C. Interleukin-8 and the neutrophil response to mucosal gram-negative infections. J Clin Investig. 1993;92:780–785. doi: 10.1172/JCI116650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold J W, Niesel D W, Annable C R, Hess C B, Asuncion M, Ja Cho Y, Peterson J W, Klimpel G R. Tumor necrosis factor-α mediates the early pathology in Salmonella infection of the gastrointestinal tract. Microb Pathog. 1993;14:217–227. doi: 10.1006/mpat.1993.1021. [DOI] [PubMed] [Google Scholar]
- 3.Asiedu C, Biggs J, Kraft A S. Complex regulation of CDK2 during phorbol ester-induced hematopoietic differentiation. Blood. 1997;90:3430–3437. [PubMed] [Google Scholar]
- 4.Bagasra O, Wright S D, Seshamma T, Oakes J W, Pomerantz R J. CD14 is involved in control of human immunodeficiency virus type 1 expression in latently infected cells by lipopolysaccharide. Proc Natl Acad Sci USA. 1992;89:6285–6289. doi: 10.1073/pnas.89.14.6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beutler B, Grau G E. Tumor necrosis factor in the pathogenesis of infectious diseases. Crit Care Med. 1993;21:S423–S435. [PubMed] [Google Scholar]
- 6.Beutler B, Milsark I W, Cerami A C. Passive immunization against cachectin/tumor necrosis factor protects mice from lethal effects of endotoxin. Science. 1985;229:869–871. doi: 10.1126/science.3895437. [DOI] [PubMed] [Google Scholar]
- 7.Biggs J R, Ahn N G, Kraft A S. Activation of the mitogen-activated protein kinase pathway in U937 leukemic cells induces phosphorylation of the amino terminus of the TATA-binding protein. Cell Growth Differ. 1998;9:667–676. [PubMed] [Google Scholar]
- 8.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 9.Ciacci-Woolwine F, Blomfield I C, Richardson S H. Salmonella flagellin induces tumor necrosis factor alpha in a human promonocytic cell line. Infect Immun. 1998;66:1127–1134. doi: 10.1128/iai.66.3.1127-1134.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciacci-Woolwine F, Kucera L S, Richardson S H, Iyer N P, Mizel S B. Salmonellae activate tumor necrosis factor alpha production in a human promonocytic cell line via a released polypeptide. Infect Immun. 1997;65:4624–4633. doi: 10.1128/iai.65.11.4624-4633.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiMango E, Zar H J, Bryan R, Prince A. Diverse Pseudomonas aeruginosa gene products stimulate respiratory epithelial cells to produce interleukin-8. J Clin Investig. 1995;96:2204–2210. doi: 10.1172/JCI118275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donnenberg M S, Yu J, Kaper J B. A second chromosomal gene necessary for intimate attachment of enteropathogenic Escherichia coli to epithelial cells. J Bacteriol. 1993;175:4670–4680. doi: 10.1128/jb.175.15.4670-4680.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everest P, Roberts M, Dougan G. Susceptibility to Salmonella typhimurium infection and effectiveness of vaccination in mice deficient in the tumor necrosis factor alpha p55 receptor. Infect Immun. 1998;66:3355–3364. doi: 10.1128/iai.66.7.3355-3364.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felber B K, Pavlakis G N. A quantitative bioassay for HIV-1 based on transactivation. Science. 1988;239:184–186. doi: 10.1126/science.3422113. [DOI] [PubMed] [Google Scholar]
- 15.Galdiero F, Cipollaro de L’Ero G, Benedetto N, Galdiero M, Tufano M A. Release of cytokines induced by Salmonella typhimurium porins. Infect Immun. 1993;61:155–161. doi: 10.1128/iai.61.1.155-161.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glauser M P. The inflammatory cytokines: new developments in the pathophysiology and treatment of septic shock. Drugs. 1996;52(Suppl. 2):9–17. doi: 10.2165/00003495-199600522-00004. [DOI] [PubMed] [Google Scholar]
- 17.Goldfeld A E, Doyle C, Maniatis T. Human tumor necrosis factor α gene regulation by virus and lipopolysaccharide. Proc Natl Acad Sci USA. 1990;87:9769–9773. doi: 10.1073/pnas.87.24.9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haas J G, Baeuerle P A, Riethmüller G, Ziegler-Heitbrock H W L. Molecular mechanisms in down-regulation of tumor necrosis factor expression. Proc Natl Acad Sci USA. 1990;87:9563–9567. doi: 10.1073/pnas.87.24.9563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holloway B W. Genetic recombination in Pseudomonas aeruginosa. J Gen Microbiol. 1955;13:572–581. doi: 10.1099/00221287-13-3-572. [DOI] [PubMed] [Google Scholar]
- 20.Hosieth S K, Stocker B A D. Aromatic-dependent S. typhimurium are nonvirulent and are effective as live vaccines. Nature (London) 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 21.Introna M, Bast R C, Jr, Johnston P A, Adams D O, Hamilton T A. Homologous and heterologous desensitization of proto-oncogene cfos expression in murine peritoneal macrophages. J Cell Physiol. 1987;131:36–42. doi: 10.1002/jcp.1041310107. [DOI] [PubMed] [Google Scholar]
- 22.Kanto S, Okino H, Aizawa S-I, Yamaguchi S. Amino acids responsible for flagellar shape are distributed in terminal regions of flagellin. J Mol Biol. 1991;219:471–480. doi: 10.1016/0022-2836(91)90187-b. [DOI] [PubMed] [Google Scholar]
- 23.Kastenbauer S, Löms Ziegler-Heitbrock H W. NF-κB1 (p50) is upregulated in lipopolysaccharide tolerance and can block tumor necrosis factor gene expression. Infect Immun. 1999;67:1553–1559. doi: 10.1128/iai.67.4.1553-1559.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinder S A, Badger J L, Bryant G O, Pepe J C, Miller V L. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O8 and construction of a transformable R−M+ mutant. Gene. 1993;136:271–275. doi: 10.1016/0378-1119(93)90478-l. [DOI] [PubMed] [Google Scholar]
- 25.König B, Ceska M, König W. Effect of Pseudomonas aeruginosa on interleukin-8 release from human phagocytes. Int Arch Allergy Appl Immunol. 1995;106:357–365. doi: 10.1159/000236867. [DOI] [PubMed] [Google Scholar]
- 26.Kriegler M, Perez C, DeFay K, Albert I, Lu S D. A novel form of TNF/cachectin is a cell surface cytotoxic transmembrane protein: ramifications for the complex physiology of TNF. Cell. 1988;53:45–53. doi: 10.1016/0092-8674(88)90486-2. [DOI] [PubMed] [Google Scholar]
- 27.Maloy S R, Stewart V J, Taylor R K. Genetic analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 28.Montie T C, Craven R C, Holder I A. Flagellar preparations from Pseudomonas aeruginosa: isolation and characterization. Infect Immun. 1982;35:281–288. doi: 10.1128/iai.35.1.281-288.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrison D C, Jacobs D M. Binding of polymyxin B to the lipid A portion of bacterial lipopolysaccharides. Immunochemistry. 1976;13:813–818. doi: 10.1016/0019-2791(76)90181-6. [DOI] [PubMed] [Google Scholar]
- 30.Morrison D C, Ryan J L. Bacterial endotoxins and host immune responses. Adv Immunol. 1979;28:293–450. doi: 10.1016/s0065-2776(08)60802-0. [DOI] [PubMed] [Google Scholar]
- 31.Munoz C, Carlet J, Fitting C, Misset B, Blériot J-P, Cavaillon J-M. Dysregulation of in vitro cytokine production by monocytes during sepsis. J Clin Investig. 1991;88:1747–1754. doi: 10.1172/JCI115493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakano Y, Onozuka K, Terada Y, Shinomiya H, Nakano M. Protective effect of recombinant tumor necrosis factor-α in murine Salmonellosis. J Immunol. 1990;144:1935–1941. [PubMed] [Google Scholar]
- 33.Newton S M C, Wasley R D, Wilson A, Rosenberg L T, Miller J F, Stocker B A D. Segment IV of a Salmonella flagellin gene specifies flagellar antigen epitopes. Mol Microbiol. 1991;5:419–425. doi: 10.1111/j.1365-2958.1991.tb02124.x. [DOI] [PubMed] [Google Scholar]
- 34.Raggs S J, Kaga S, Berg K A, Ochi A. The mitogen-activated protein kinase pathway inhibits ceramide-induced terminal differentiation of a human monoblastic leukemia cell line, U937. J Immunol. 1998;161:1390–1398. [PubMed] [Google Scholar]
- 35.Robache-Gallea S, Bruneau J M, Robbe H, Morand V, Capdevila C, Bhatnagar N, Chouaib S, Roman-Roman S. Partial purification and characterization of a tumor necrosis factor-alpha converting activity. Eur J Immunol. 1997;27:1275–1282. doi: 10.1002/eji.1830270532. [DOI] [PubMed] [Google Scholar]
- 36.Stone B J, Garcia C M, Badger J L, Hassett T, Smith R I F, Miller V L. Identification of novel loci affecting entry of Salmonella enteritidis into eukaryotic cells. J Bacteriol. 1992;174:3945–3952. doi: 10.1128/jb.174.12.3945-3952.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tite J P, Dougan G, Chatfield S N. The involvement of tumor necrosis factor in immunity to Salmonella infection. J Immunol. 1991;147:3161–3164. [PubMed] [Google Scholar]
- 38.Tracey K J, Beutler B, Lowry S F, Merryweather J, Wolpe S, Milsark I W, Hariri R J, Fahey III T J, Zentella A, Albert J D, Shires G T, Cerami A. Shock and tissue injury induced by recombinant human cachectin. Science. 1986;234:470–474. doi: 10.1126/science.3764421. [DOI] [PubMed] [Google Scholar]
- 39.Tracey K J, Fong Y, Hesse D G, Manogue K R, Lee A T, Kuo G C, Lowry S F, Cerami A. Anti-cachectin/TNF monoclonal antibodies prevent septic shock during lethal bacteremia. Nature. 1987;330:662–666. doi: 10.1038/330662a0. [DOI] [PubMed] [Google Scholar]
- 40.van Asten A J A M, Zwaagstra K A, Baay M F, Kusters J G, Huis in’t Veld J H J, van der Zeijst B A M. Identification of the domain which determines the g,m serotype of the flagellin of Salmonella enteritidis. J Bacteriol. 1995;177:1610–1613. doi: 10.1128/jb.177.6.1610-1613.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Volk H D, Reinke P, Krausch D, Zuckermann H, Asadullah K, Muller J M, Docke W D, Kox W J. Monocyte deactivation-rationale for a new therapeutic strategy in sepsis. Intensive Care Med. 1996;22(Suppl. 4):S474–S481. doi: 10.1007/BF01743727. [DOI] [PubMed] [Google Scholar]
- 42.Vrana J A, Saunders A M, Chellappan S P, Grant S. Divergent effects of bryostatin 1 and phorbol myristate acetate on cell cycle arrest and maturation in human myelomonocytic leukemia cells (U937) Differentiation. 1998;63:33–42. doi: 10.1046/j.1432-0436.1998.6310033.x. [DOI] [PubMed] [Google Scholar]
- 43.Wyant T L, Tanner M K, Sztein M B. Potent immunoregulatory effects of Salmonella typhi flagella on antigenic stimulation of human peripheral blood mononuclear cells. Infect Immun. 1999;67:1338–1346. doi: 10.1128/iai.67.3.1338-1346.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamaguchi S, Fujita H, Sugata K, Taira T, Iino T. Genetic analysis of H2, the structural gene for phase-2 flagellin in Salmonella. J Gen Microbiol. 1984;130:255–265. doi: 10.1099/00221287-130-2-255. [DOI] [PubMed] [Google Scholar]