Valencia et al. show DSTYK, a dual serine/threonine and tyrosine kinase, altered in lung cancer and associated with poor clinical outcome. DSTYK controls cellular processes encompassing cytoprotective autophagy and mitochondrial fitness and its ablation sensitizes cancer cells to T cell–mediated killing.
Abstract
Lung cancer remains the leading cause of cancer-related death worldwide. We identify DSTYK, a dual serine/threonine and tyrosine non-receptor protein kinase, as a novel actionable target altered in non-small cell lung cancer (NSCLC). We also show DSTYK's association with a lower overall survival (OS) and poorer progression-free survival (PFS) in multiple patient cohorts. Abrogation of DSTYK in lung cancer experimental systems prevents mTOR-dependent cytoprotective autophagy, impairs lysosomal biogenesis and maturation, and induces accumulation of autophagosomes. Moreover, DSTYK inhibition severely affects mitochondrial fitness. We demonstrate in vivo that inhibition of DSTYK sensitizes lung cancer cells to TNF-α–mediated CD8+-killing and immune-resistant lung tumors to anti–PD-1 treatment. Finally, in a series of lung cancer patients, DSTYK copy number gain predicts lack of response to the immunotherapy. In summary, we have uncovered DSTYK as new therapeutic target in lung cancer. Prioritization of this novel target for drug development and clinical testing may expand the percentage of NSCLC patients benefiting from immune-based treatments.
Introduction
Lung cancer, as the leading cause of cancer-related death worldwide (Sung et al., 2021), is a major public health challenge in advanced societies (Siegel et al., 2020). It has been predicted that less-developed regions will face a 144% increase in new cancer cases (Pilleron et al., 2019). Lung cancer incidence ranges from 12 to 13%, and almost one in five deaths attributed to cancer are caused by lung cancer globally (Sung et al., 2021). Usually, two out of three lung cancer patients are diagnosed at advanced stages when the curative options are limited, and survival rates are poor. In stage IV cases, the 5-yr overall survival (OS) rate is <15%. Even among early-stage patients, the risk of mortality remains high, with relapse rates of 30–45% within 5 yr of diagnosis (Uramoto and Tanaka, 2014). Apart from primary prevention aimed at reducing smocking habits and low-dose computed tomography–based early detection, reducing the burden of lung cancer mortality requires optimization of personalized therapies targeting known or novel driver alterations and improved immunotherapy-based strategies. Although considerable improvement in survival has been achieved in the last decade, mainly due to the wide use of targeted therapies, a significant proportion of non-small cell lung cancer (NSCLC) patients are still not benefiting from these strategies (Jordan et al., 2017). In recent years, immune checkpoint inhibitors (ICIs) have emerged as a successful therapy for a subset of lung cancer patients. The use of monoclonal antibodies, such as the programmed cell death protein 1 (PD-1)/programmed death-ligand 1 (PD-L1) axis, has shown notable clinical results in NSCLC (Horn et al., 2018). These treatments boost the immune response against tumors by reactivating antigen-specific effector T cells. However, their efficiency in monotherapy is still limited to 10–20% of unselected patients (Hellmann et al., 2019; Herbst et al., 2020; Reck et al., 2016), and some initial responders eventually develop resistance. Therefore, combination therapies and optimization of sensitivity or resistance predictive biomarkers are emerging to further improve the efficacy of immunotherapy treatments (reviewed in Kinoshita et al. [2021] and Grant et al. [2021]).
Among the molecular hallmarks of cancer, the alteration of tyrosine protein kinases is one of the best known. The development of specific inhibitors for these proteins has revolutionized the management of cancer patients in the last 20 yr. Indeed, in lung cancer, the use of genotype-directed therapies has supplanted the use of systemic chemotherapy as gold standard for patients with advanced NSCLC. Eight different lung cancer genomic alterations have unique drug approvals to date. Although many molecular alterations are known in lung cancer, there is still a high percentage of patients without druggable targets (Cohen et al., 2021).
In recent years, modulating autophagy has been proposed as a promising treatment for lung cancer, as it plays a central role in tumorigenesis and tumor progression (Nam, 2021; Wang et al., 2022). Autophagy seems to be a double-edge sword, as it can play both cytoprotective and cytotoxic roles in cells, in a context-specific way (Colhado Rodrigues et al., 2020). In the early stages of cancer development, autophagy is believed to protect the cells against cancer initiation. Once a tumor is established, the neighboring microenvironment is often hypoxic and nutrient-deprived, and autophagy fuels and supports cancer cells growth, switching into a tumor promoter role (Wang et al., 2022). Autophagy also promotes epithelial–mesenchymal transition, migration, and invasion of tumoral cells (Li et al., 2013; Zhu et al., 2014; Luo et al., 2018). In addition, autophagy contributes to stemness and drug resistance (Hao et al., 2019). Strategies to inhibit autophagy are the object of intense research and a noticeable number of clinical trials in the search for new cancer therapies (Marinković et al., 2018; Xiao et al., 2021; Nam, 2021). In lung cancer, combinations of chloroquine or hydroxychloroquine with EGFR-TKI and chemotherapy (Malhotra et al., 2019) or MEK-1/2 inhibitors (NTC04735068) are being tested.
Recent reports in different types of cancers, including lung neoplasias, have also shown that autophagy plays a major role in tumor immune evasion. Autophagy-mediated tumor immune escape is achieved through different mechanisms such as alteration of HLA1-dependent antigen presentation or immune cells functional modifications (Yamamoto et al., 2020; Lawson et al., 2020; Wang et al., 2021a).
Following our recent finding that Dstyk was mutated in a cell line derived from a syngeneic lung squamous cell carcinoma (LUSC) mouse model (Valencia et al., 2021), we characterize the role of DSTYK as a driver of lung cancer initiation, progression and immune therapy resistance. We also show the prognostic value of its expression and gene copy number (CN) gain across the two main NSCLC subtypes, LUSC and lung adenocarcinoma (LUAD). Mechanistically, we demonstrate that DSTYK is a central regulator of autophagy and oxidative stress response, and its inhibition collapse tumor-sustaining autophagy and sensitize lung cancer cells to immune-based therapies. Our findings uncover DSTYK as a previously unrecognized dependency in lung cancer and support its relevance as a therapeutic target to advance personalized medicine efforts for NSCLC patients.
Results
Prioritization of tyrosine kinases identifies DSTYK as a prevalent alteration in NSCLC patients
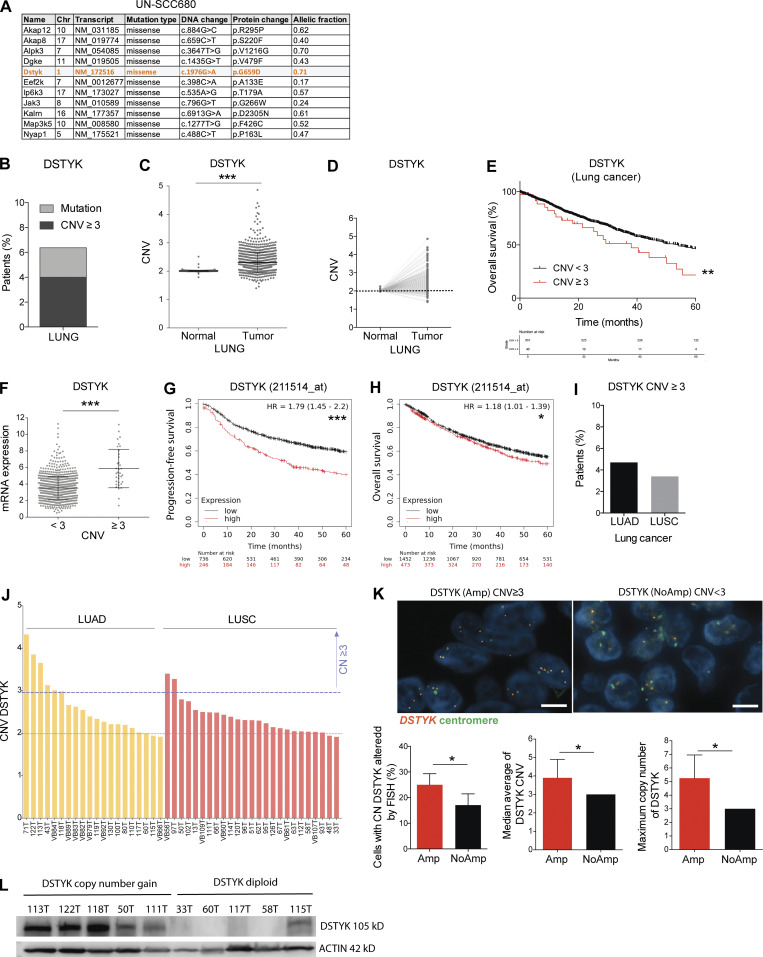
Aiming to search for novel kinases involved in lung cancer, exome analysis was performed in murine lung LUSC cell lines generated in our group (Valencia et al., 2021), leading to identify the tyrosine protein kinase Dstyk (dual-serine/threonine and tyrosine protein kinase) as a potentially interesting mutated gene for study in lung cancer (Fig. 1 A). In order to explore the clinical relevance of DSTYK alteration in lung cancer, we analyzed The Cancer Genome Atlas (TCGA) data and we found around 6.4% of lung cancer patients were reported to have alterations in DSTYK (Fig. 1 B). According to the TCGA dataset, CN gain affected a greater number of patients (4%) than mutations (2.4%) in the DSTYK gene. These percentages of alteration were comparable to those for most of the druggable targets in lung cancer, except for EGFR and K-RAS (Jordan et al., 2017; Campbell et al., 2016). Mutations were distributed homogenously throughout the gene sequence and showed no characteristic pattern or hot spot regions that would merit particular attention. Therefore, we decided to focus on the potential functional relevance of DSTYK genetic CN gain. In order to study the percentage of NSCLC patients with DSTYK CN gain, we compared the DSTYK CN in tumor samples and samples from healthy lungs from the TCGA cohort. Tumors showed a significant increase in DSTYK CN compared to normal lung (Fig. 1 C). More importantly, in the subgroup of patients where matched tumor and normal sample was available, increase in CN was found for 82% of cases (Fig. 1 D). The average DSTYK gene CN in tumors was 2.31 ± 0.011. Moreover, patients with DSTYK CN gain showed a poor survival rate (Fig. 1 E). Therefore, these data suggested that DSTYK was altered in NSCLC patients and that genomic gain/amplification might be relevant to the tumorigenic process in a percentage of NSCLC patients.
Figure 1.
DSTYK is amplified and high levels of the gene correlate with poor prognosis in NSCLC patients. (A) Mutated kinase coding genes of a syngeneic SCC cell line (UN-SCC680) derived from a N-nitroso-tris-chloroethylurea chemically induced mouse model in A/J mice. Orange highlights tyrosine kinase genes. (B) Percentage of patients with DSTYK alteration from lung cancer. From Pan-Cancer TCGA data. (C) DSTYK CNV in normal (n = 960) and tumoral (n = 904) samples from NSCLC patients from the TCGA project. A t test was performed to compared both groups; P value = 2.2 × 10−16. (D) DSTYK CNV in paired normal and tumoral samples (n = 832) of NSCLC patients from the TCGA project. 82% of tumor samples showed DSTYK CN gain. (E) Kaplan–Meier plot. OS significantly decreases in lung cancer patients from the TCGA project with DSTYK CN gain (CNV ≥ 3; n, CVN < 3 = 951; n, CNV ≥ 3 = 40). Log-rank test P = 0.01. (F) Dot plot shows the correlation between CNV and mRNA expression of DSTYK. Patients with CNV ≥3 present significantly higher expression of DSTYK mRNA (n, CVN < 3 = 954; n, CNV ≥ 3 = 54). A t test was performed to compared both groups; P value = 8.5 × 10−7. (G) Kaplan–Meier plot. PFS significantly decreases in lung cancer patients with high DSTYK mRNA expression (Q1). (n, low = 736; n, high = 246). Log-rank test P = 2.4 × 10−8. Data are from https://kmplot.com/. (H) Kaplan–Meier plot. OS significantly decreases in lung cancer patients with high DSTYK mRNA expression (Q1). n, low = 1452; n, high = 373. Log-rank test P = 0.036. Data are from https://kmplot.com/. (I) Percentage of LUAD and LUSC patients with CN gain of DSTYK (CN > 3). Data from the TCGA project. (J) DSTYK CNV evaluation in CIMA-CUN-cohort patients by qRT-PCR. (K) FISH analysis of DSTYK (Amp) and (NoAmp) patients. Scale bar: 5 µm. A t test was performed to compare cells with DSTYK CN (P value = 0.04); median average of DSTYK CN (P = 0.02) and maximum CN of DSTYK (P = 0.038). (L) Cropped images from the Western blot analysis of DSTYK in frozen tumor samples from CIMA-CUN cohort patients with DSTYK CN gain and diploid number of DSTYK. *, P < 0.05; ***, P < 0.001. Source data are available for this figure: SourceData F1.
As expected, the DSTYK CN was significantly correlated with DSTYK mRNA levels and patients with DSTYK CN gain (CN variation [CNV] ≥ 3) showed significantly higher mRNA expression (Fig. 1 F). Furthermore, in the Kaplan–Meier plotter dataset, patients with DSTYK expression in the first quartile (Q1) showed significantly worse progression-free survival (PFS) and OS rates (Fig. 1, G and H).
After obtaining a general picture of DSTYK alteration in NSCLC, we then studied CNV separately in LUAD and LUSC. We observed DSTYK CN gain (three or more copies) in 4.7% of LUAD and 3.4% of LUSC patients, respectively (Fig. 1 I).
To confirm publicly available data, we assessed DSTYK CN by qPCR in a series of 45 frozen samples from NSCLC patients who underwent surgical resection at the Clínica Universidad de Navarra (CIMA-CUN; Table S1). According to our analyses, 13.3% of patients showed a gain of DSTYK CN (Fig. 1 J). Fluorescence in situ hybridization (FISH) was performed to corroborate these results in formalin-fixed, paraffin-embedded samples from the same cohort. We observed that 25% of cancer cells showed three or more copies in those patients where DSTYK was found to be amplified by qPCR (Amp DSTYK) and 17% in patients with DSTYK CN < 3 by qPCR (NoAmp DSTYK). More interestingly, among cells that displayed DSTYK CNV by qPCR, Amp DSTYK patients presented 4.3 copies on average while 3 copies on average were counted in NoAmp DSTYK patients. Moreover, the maximum mean number of DSTYK copies found in altered cells in Amp and NoAmp DSTYK patients was 5.3 and 3, respectively (Fig. 1 K). The observed alterations in DSTYK CN were not due to aneuploidy as two centromeric copies from chromosome 1 were counted in all cases.
Finally, to confirm the correlation between CN and transcript levels of DSTYK, we performed a Western blot analysis of frozen tumor samples from CIMA-CUN cohort patients with DSTYK CN gain and diploid number. We observed higher levels of DSTYK protein in patients with CN gain comparing to those with diploid DSTYK, which showed low DSTYK protein levels (Fig. 1 L).
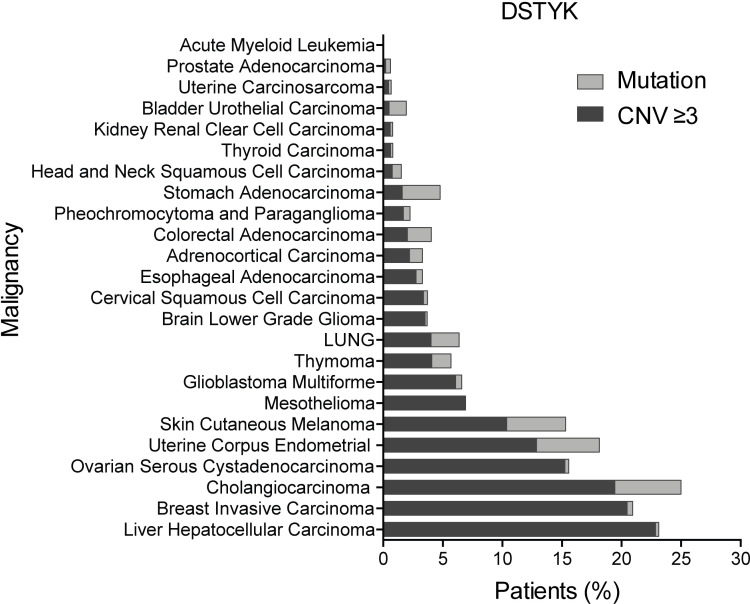
In order to explore the clinical relevance of DSTYK alteration in other human cancers, we analyzed TCGA data, and we found widespread incidence of CN gain and/or mutation in DSTYK across cancer types (Fig. S1 A). Alterations were highly prevalent in many cancer types and were found in >20% of patients with hepatocarcinoma, breast cancer, or cholangiocarcinoma and around 15% of patients with melanoma, ovarian, or uterine carcinoma, which suggests that DSTYK alteration was important in a very relevant number of human cancer types.
Figure S1.
DSTYK alterations in cancer. Percentage of patients with DSTYK alteration from 24 different cancer types. From Pan-Cancer TCGA data.
Taken together, these findings suggested that amplification of DSTYK was commonly altered in NSCLC and showed prognostic relevance in lung cancer patients.
DSTYK is located in autophagosomes where it performs its molecular functions
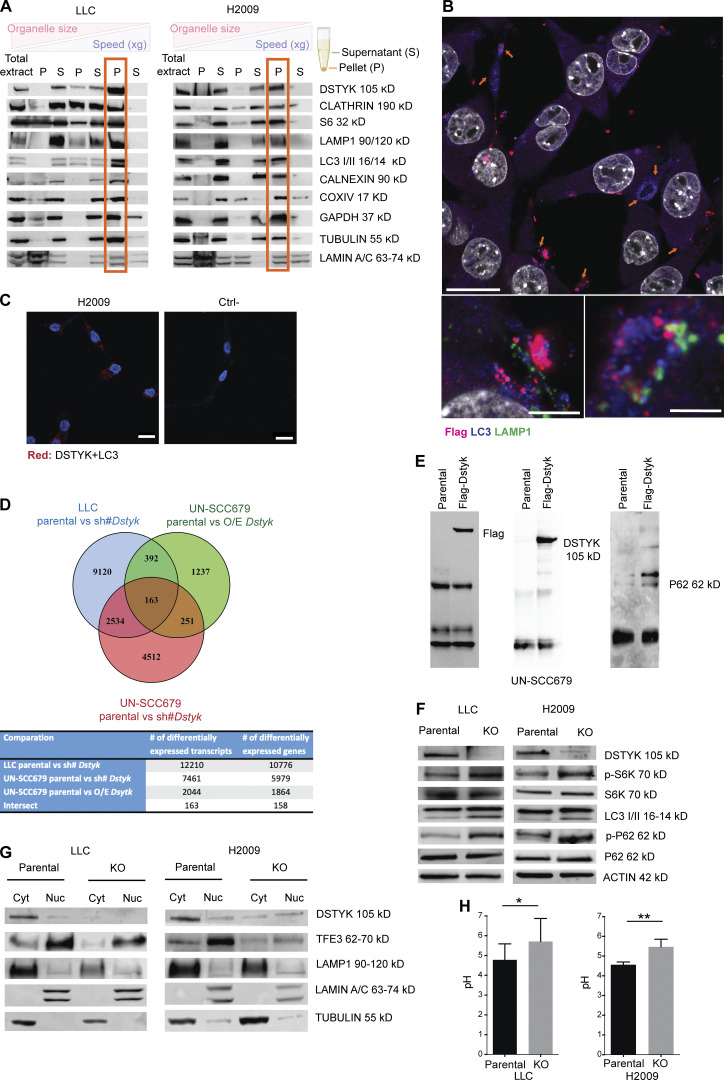
The function and subcellular location of DSTYK within the cell remain largely unknown. Through cellular fractionation experiments in murine and human lung cancer cell lines, we observed DSTYK was enriched in the microsomal fraction (Fig. 2 A). To delve into the nature of vesicles that were associated with DSTYK, we studied markers from the main vesicles by immunofluorescence (IF) in a Flag-DSTYK cell line. We observed that DSTYK, together with the autophagy marker LC3, co-localized in large, rounded cytoplasmic structures mainly located at the periphery of cells. The shape and LC3 protein location in these structures pointed to the fact that DSTYK was mainly located in autophagosomes (Fig. 2 B and Fig. S2 A). We assessed a proximity ligation assay (PLA) to corroborate that DSTYK and LC3 were in close proximity, verifying both markers were part of a single structure, located at least in contiguous spots (40 nm of maximum distance) and configured autophagosomes in a cooperative way (Fig. 2 C). Flag (DSTYK) signal did not co-localize in any other of the evaluated vesicle types, for which IF markers were used (Fig. S2 B).
Figure 2.
DSTYK is located in autophagosomes and regulates processes related to those vesicles. (A) Subcellular fractionation of LLC and H2009 cells. The following fractions were shown in the Western blot from left to right: total extract; nuclear extract (P3000); cytosolic extract (S3000); large organelle extract (P10000); cytosolic extract without large organelles (S10000); microsomal fraction (P100000); and soluble cytosolic extract (S100000). Different organelle markers were tested to control proper fractionation. (B) Top: Double IF of Flag-Dstyk LLC cells. Flag-DSTYK (magenta) showing co-localization with LC3 (blue). Scale bar: 20 µm. Bottom: Triple IF of Flag-Dstyk LLC cells. Detailed image of autophagosomes: Flag-DSTYK (magenta), LC3 (blue), and LAMP1 (green). Scale bars: 5 and 2 µm. Observed by super-resolution/Airyscan based/confocal microscopy. (C) PLA images of H2009 cells. Left: Red fluorescent dots result from DSTYK+LC3 proteins proximity (maximum distance: 40 nm). Right: Technical negative control. Scale bar: 20 µm. (D) Venn diagram picturing RNAseq analysis. Three RNAseq analyses were carried out to identify the differentially expressed transcripts: LLC comparing parental line vs. silenced (sh#) Dstyk (blue circle), UN-SCC679 line comparing parental vs. overexpression (O/E) Dstyk (green circle), and UN-SCC679 comparing parental vs. silenced (sh#) Dstyk (red circle). Once differentially expressed transcripts were selected (adjusted P value <0.05) in each contrast, all experiments were merged to verify the intersection of the transcripts based on name and the direction. 163 transcripts were finally selected. (E) Immunoprecipitation of Flag-DSTYK in the UN-SCC-679 cell line validating Western blots showing DSTYK binding to P62 protein. (F) Cropped images from the Western blot analysis of autophagy pathway proteins in LLC and H2009 parental and DSTYK-KO cells. (G) Cropped images from the Western blot analysis of lysosomal biogenesis pathway proteins in LLC and H2009 parental and DSTYK-KO cells in cytoplasmic/nuclear fractions. All Western blots show representative results from three independent experiments. (H) Lysosomal pH measurement in LLC and H2009 parental and DSTYK-KO cells. A t test was performed to compared both groups: LLC P value = 0.05; H2009 P value = 0.01. Images shown in A, B, F, G, and H show a representative experiment out of three similarly performed. *, P < 0.05; **, P < 0.01. Source data are available for this figure: SourceData F2.
Figure S2.
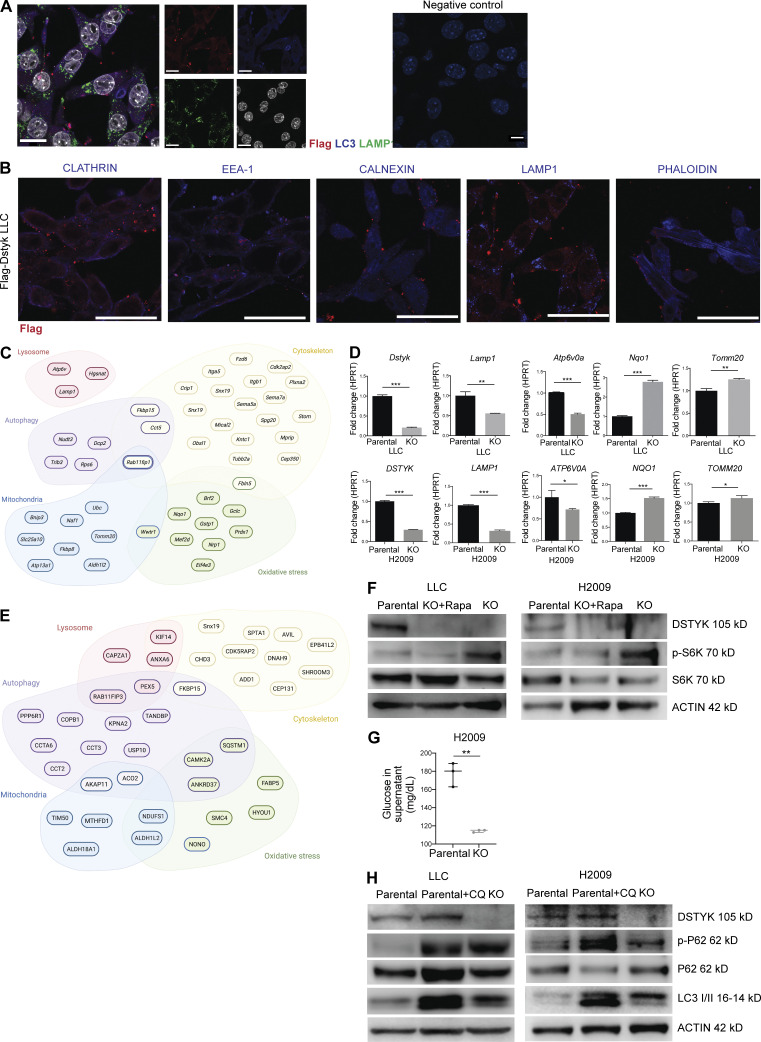
DSTYK is involved in autophagic molecular pathways. (A) Left: Double IF of Flag-Dstyk LLC cells. DSTYK (magenta) showing co-localization with LC3 (blue). Scale bar: 20 µm. Middle: Single channel images correspondent to triple IF of Flag-Dstyk LLC cells. DSTYK (red), LC3 (blue), LAMP1 (green), and nuclei (white). Scale bars: 20 µm. Right: Technical negative control. Scale bars: 10 µm. Observed by super-resolution/Airyscan based/confocal microscopy. (B) Double IF of Flag-Dstyk LLC cells. DSTYK (red) do not co-localize with cellular vesicles markers (blue). Scale bar: 20 µm. (C) Scheme of differentially expressed genes from the RNAseq analysis enriched in the following categories: lysosome, autophagy, mitochondria, oxidative stress, and cytoskeleton. (D) Validation of mRNA expression of candidate genes from the RNAseq analysis in LLC and H2009 parental and CRISPRed cells by quantitative PCR. Analysis was assessed by t test. P values for parental vs. KO comparation were the following: Dstyk in LLC, P < 0.0001 and DSTYK in H2009, P < 0.0001; Lamp1 in LLC, P = 0.001 and LAMP1 in H2009, P < 0.0001; Atp6v0a in LLC, P < 0.0001 and ATP6V0A in H2009, P < 0.03; Nqo1 in LLC, P < 0.0001 and NQO1 in H2009, P < 0.0001; Tomm20 in LLC, P < 0.001 and TOMM20 in H2009, P < 0.03. Three experimental replicates were performed for each experimental condition. (E) Scheme of DSTYK interactor proteins from immunoprecipitation analysis enriched in the following categories: lysosome, autophagy, mitochondria, oxidative stress, and cytoskeleton. (F) Cropped images from Western blot analysis of autophagy pathway proteins in LLC and H2009 parental, DSTYK-KO, and KO treated with rapamycin cells. A representative experiment out of two similarly performed is shown. (G) Glucose levels (mg/dl) in cell culture supernatants of parental H2009 and KO cells. Analysis by t test rendered P = 0.001. Three experimental replicates were performed for each experimental condition. (H) Cropped images from Western blot analysis of autophagy pathway proteins in LLC and H2009 parental, parental treated with chloroquine, and DSTYK-KO cells. *, P < 0.05; **, P < 0.01; ***, P < 0.001. A representative experiment out of two similarly performed is shown. Source data are available for this figure: SourceData FS2.
Autophagy has been associated with carcinogenesis in several tumor types including lung cancer (Nam, 2021). To confirm whether DSTYK role was related to autophagy in lung cancer and to ascertain the potential molecular carcinogenesis processes in which DSTYK was involved, we performed an RNA sequencing (RNAseq) analysis of two different lung cancer cell lines with either overexpressed or inhibited DSTYK. Inhibition was achieved through shRNA technology (Fig. 2 D). After data filtering, we observed a significant enrichment of genes related to lysosomes, autophagy, mitochondria, oxidative stress, and cytoskeleton. These genes were differentially expressed in an altered DSTYK context (Fig. S2 C and Table S2). We confirmed these shRNA-based results in two independent DSTYK-CRISPRed partially KO cell lines, both murine and human (Lewis lung carcinoma [LLC] and H2009, respectively; Fig. S2 D). The altered RNA expression data from both technologies (shRNA and CRISPR) supported the previous data obtained through protein analysis of fractionated cell compartments and the morphological IF results. The integrated interpretation of these experiments strongly suggested that DSTYK was involved in autophagy processes presumably related to mitochondria (mitophagy).
In order to further dissect the potential molecular role of DSTYK in autophagy in a lung cancer context, we evaluated the DSTYK protein interactions in lung cancer cell lines by immunoprecipitation followed by mass spectrometry. These experiments yielded several interesting proteins (Table S3 and Fig. S2 E). Among these proteins, consistent with our transcriptomic and proteomic results suggesting a role for DSTYK in autophagy, we validated a protein–protein interaction of DSTYK with P62 (SQSTM1; Fig. 2 E). P62 is upregulated in many cancer types. It is a key participant in the formation of the autophagosome, first identified as an autophagy adaptor, residing in the late endosome lysosome (Moscat and Diaz-Meco, 2009).
To further elucidate the specific molecular role of DSTYK in autophagy in lung cancer, we analyzed the expression or activation of key members of the autophagy cell protein pathway in our CRISPRed cellular models. We found an increase in p-S6K, LC3, and p-P62 in DSTYK KO cells compared to parental cells, which suggested that DSTYK inhibited mTORC1 and, consequently, fueled autophagy. To verify that the phenotypes observed were mediated by mTOR, we treated KO cells with rapamycin (mTOR inhibitor), recovering the autophagy values of parental cell lines (Fig. S2 G). Moreover, other mTOR-related functions such as glucose uptake were impacted by DSTYK inhibition (Fig. S2 G). These data were consistent with previous work reporting a role for DSTYK in regulating mTORC1 in zebrafish models (Sun et al., 2020). Nevertheless, the accumulation of p-P62 (and stable total P62) protein that we found in DSTYK inhibited lung cancer cell lines suggested that the autophagosome content was not being processed; this indicated that the autophagic process was not fully functional in these cell lines (Fig. 2 F). To verify that DSTYK inhibition interrupted autophagy, we treated parental cells with chloroquine, which showed a similar pattern of autophagic markers to DSTYK KO cells (Fig. S2 H). In summary, these data suggested that DSTYK induces autophagy, and its deficiency caused autophagy interruption, which led to a progressive accumulation of autophagosomes.
mTORC1 is known to phosphorylate TFEB/3, avoiding its nuclear translocation and preventing the expression of its target genes, which trigger lysosome biogenesis (Roczniak-Ferguson et al., 2012). Having found Lamp1 to be downregulated in DSTYK KO cells, we hypothesized that DSTYK could also regulate lysosome biogenesis in lung cancer. For this purpose, we studied the presence of TFE3 and LAMP1 proteins in the nucleus and in the cytoplasm, and we observed that DSTYK KO cells showed reduced levels of nuclear TFE3 and subsequently reduced levels of LAMP1 (Fig. 2 G). Therefore, the absence of DSTYK negatively affected lysosomal biogenesis.
To further assess the effects of DSTYK on lysosome function, we measured their pH in parental and KO human and murine cells. Surprisingly, lung cancer cells lacking DSTYK expression showed less acidic pH (5.6 ± 0.08) in lysosomes than parental cells (4.6 ± 0.2), suggesting an impaired lysosomal functionality (Fig. 2 H). This fact was in accordance with the lower expression of V-Atpases shown in DSTYK knockdown cells in the RNAseq analysis (Fig. S2, C and D), which might partially explain a dysregulation in proton uptake in KO cells lysosomes among other possible mechanisms.
Our data, thus, strongly supported an involvement of DSTYK in molecular pathways related to autophagy through its interaction with the key autophagy regulating protein P62.
DSTYK regulates the NRF2 pathway
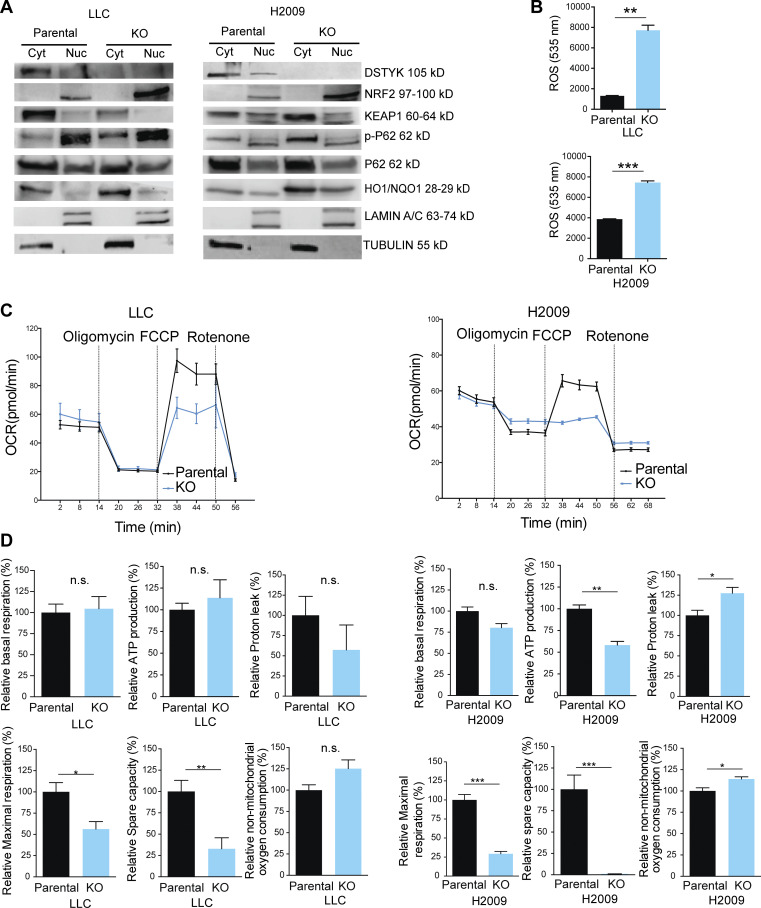
Since P62 is also a key regulatory protein of the KEAP1-NRF2 pathway and we found oxidative stress genes expression to be increased upon DSTYK inhibition (Table S2 and Fig. S2 B), we assessed the implication of DSTYK KO in this scenario. Through Western blot experiments, we showed that the increase in P62 consequently to DSTYK knockdown led to a greater interaction P62-KEAP1, which allowed NRF2 to translocate to the nucleus and expressed its target genes such as HO1 or NQO1, involved in the oxidative stress response (Fig. 3 A).
Figure 3.
DSTYK role in oxidative stress in lung cancer cells. (A) Cropped images from the Western blot analysis of KEAP1/NRF2 pathway proteins in LLC and H2009 parental and DSTYK-KO cells in cytoplasmic/nuclear fractions. Western blots show representative results from three independent experiments. (B) ROS measurements in LLC and H2009 parental and DSTYK-KO cells in basal conditions. Three experimental replicates were performed for each experimental condition. Experimental groups were compared by t test: LLC, P = 0.0031; and H2009, P = 0.0011. (C) Seahorse extracellular flux analysis shows dynamic representation of oxygen consumption rate (OCR) in LLC and H2009 cells under stress conditions. Three experimental replicates were performed for each experimental condition. (D) Comparison of different phases of mitochondrial respiration. Parental or KO LCC and H2009 cells share alteration of maximal respiratory and spare respiratory capacity in stress conditions. Experimental groups were compered by t test. LLC relative maximal respiration, P = 0.03; relative spare capacity, P = 0.01. H2009 relative ATP production, P = 0.002; relative proton leak, P = 0.04; relative maximal respiration, P = 0.0008; relative spare capacity, P < 0.0001; relative non-mitochondrial oxygen consumption, P = 0.03. *, P < 0.05; **, P < 0.01; ***, P < 0.001. All images are representative results out of three experimental replicates. Source data are available for this figure: SourceData F3.
Taken together, these results suggested that DSTYK could modulate oxidative stress response.
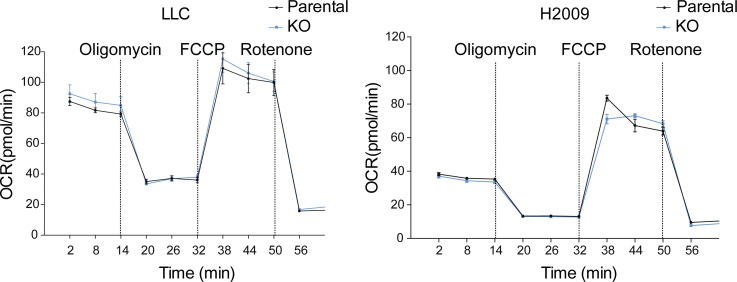
Oxidative stress can be triggered by ROS as a consequence of mitochondrial respiration. Following our RNAseq and immunoprecipitation data pointing to a functional role of DSTYK in mitochondrial biology, we studied mitochondrial functionality in our models. As compared to parental cells, ROS levels in KO cells under normal conditions were dramatically increased (Fig. 3 B). ROS production may be due to dysfunctional mitochondrial respiration. To further investigate the potential functional alteration in mitochondria, we performed a mitostress test (Seahorse). Under basal conditions, the two types of cells did not exhibit respiratory differences (Fig. S3 A) but under stress conditions, DSTYK KO cells did not respond to the FCCP treatment, which is a potent mitochondrial oxidative phosphorylation uncoupler that disrupts ATP synthesis by transporting protons across mitochondrial inner membranes and depolarizes mitochondrial membrane potential, displaying a reduced maximal respiratory and spare respiratory capacities (Fig. 3, C and D).
Figure S3.
Mitochondrial respiration under baseline conditions in an altered DSTYK context. Seahorse extracellular flux analysis shows dynamic representation of oyxgen consumption rate (OCR) in LLC and H2009 cells under basal conditions. Three experimental replicates were performed for each experimental condition.
These data suggested that DSTYK sustains mitochondrial fitness, and its depletion triggered mitochondrial dysfunction and consequently raises oxidative stress conditions.
DSTYK maintains mitochondrial integrity
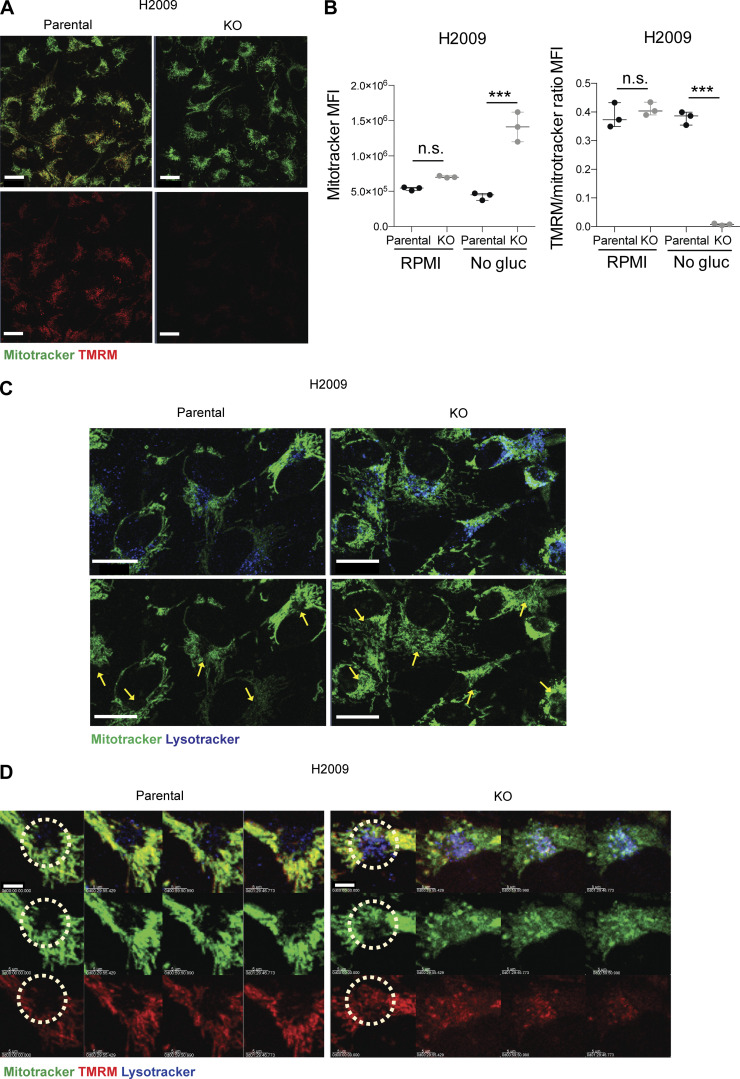
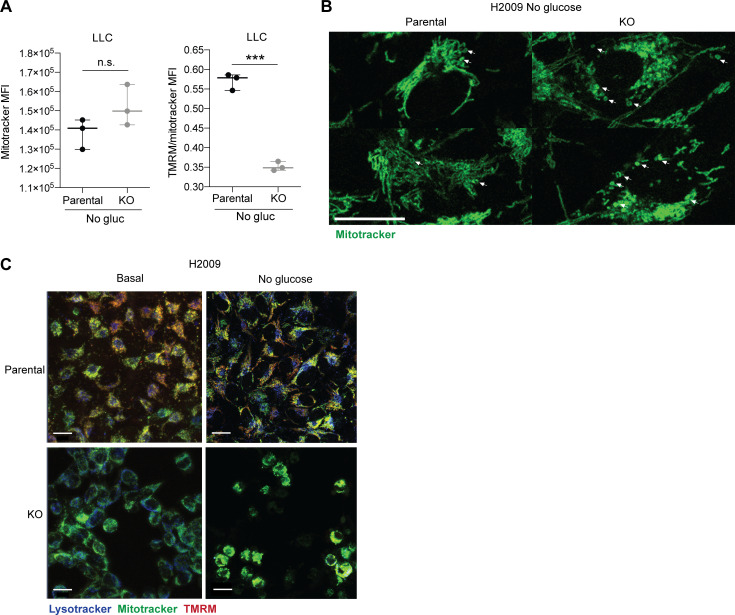
An increase in ROS production may be the consequence of a loss of mitochondrial membrane (Zorova et al., 2018). To further investigate the integrity or potential functional alteration in mitochondria, we performed an in vivo IF, monitoring mitochondria within parental and KO cells with Mitotracker Green and tetramethylrhodamine methyl ester perchlorate (TMRM) fluorescent dye, to assess its membrane potential (Momcilovic et al., 2019; Fig. 4 A). In parallel, in an independent experiment, we evaluated and quantified labeled cells by flow cytometry. In normal conditions, cells showed no differences neither in mitochondrial mass nor in membrane potential. However, under stress conditions, DSTYK KO cells showed a dramatic TMRM reduction (loss of 98.5%) and a significant increase in mitochondrial mass (gain of 327%; Fig. 4, A and B; and Fig. S4 A). These results were in agreement with our previous data, suggesting that in DSTYK KO cells mitochondria were damaged, and cells were incapable of eliminating them through selective autophagy (mitophagy). This is likely the consequence of a deficient autophagy, and subsequently, damaged mitochondria were accumulated. Notably, ring-shaped mitochondria were observed in DSTYK KO cells under stress conditions, which is a sign of their damage (Ahmad et al., 2013; Fig. S4 B). To better understand mitophagy alterations in the absence of DSTYK, we tracked in vivo mitochondria and lysosomes by multiphoton confocal microscopy. In parental cells, we found that most foci of active mitophagy, surrounded by lysosome accumulation, exhibited a complete lack of mitochondrial labeling, whereas, in DSTYK KO cells, mitochondria were not able to be eliminated in those spots and lose TMRM labeling along time (Video 1 and Fig. 4, C and D). More importantly, under stress conditions, lung cancer DSTYK KO cells succumbed to ROS levels and accumulation of undigested mitochondria and died. In contrast, parental cells conserved the cytoprotective autophagy and normal mitochondrial respiration as mechanisms of cell survival and tumor progression (Fig. S4 C).
Figure 4.
DSTYK is involved in mitochondrial fitness. (A) Confocal microscopy images. Parental or DSTYK KO H2009 tumor cells were plated, and mitochondria were stained using Mitotracker Deep Red and TMRM and treated with RPMI glucose-free serum to induce stress. Scale bar: 20 µm. (B) Flow cytometry was performed for parental or DSTYK KO H2009 cells in basal and stress conditions and stained with Mitotracker Green and TMRM to assess mitochondrial mass and membrane potential. The results shown are triplicates from a representative experiment out of two performed similarly. Experimental groups were compered by t test. H2009 Mitotracker mean fluorescence intensity (MFI) under stress condition, P = 0.0014; H2009 TMRM/Mitotracker ratio MFI under no glucose condition, P < 0.0001. (C) Confocal microscopy images of lysosomes and mitochondria in parental or DSTYK KO H2009 cells were stained using Mitotracker Deep Red (red) and Lysotracker (green) and treated with RPMI glucose-free serum to induce stress for 4 h. Yellow arrows point to lysosome-enriched areas. Scale bar: 20 µm. (D) Confocal microscopy snapshots zoomed in on mitophagy spots in the time-lapse videos taken of parental or DSTYK KO H2009 cells stained using Mitotracker (green), TMRM (red), and Lysotracker (blue) and treated with glucose-free medium for 4 h to induce stress. Light blue staining is the sum of Lysotracker and Mitotracker. ***, P < 0.001. Representative timeframes corresponding to Video 1. Scale bar: 5 µm.
Figure S4.
DSTYK inhibition affects mitochondrial morphology and functionality. (A) FACs analysis of mitochondrial mass (Mitotracker) and membrane potential (TMRM) of parental or KO LLC and H2009 cells in stress conditions. Three experimental replicates were performed for each experimental condition. Analysis by t test rendered TMRM/Mitotracker ratio MFI, P < 0.0001. (B) Detailed confocal microscopy images of mitochondrial ring-shape in parental or KO H2009 cells under stress conditions. Scale bar: 20 µm. (C) Confocal microscopy images of parental or KO H2009 cells under basal or stress conditions 16 h after treatment. Scale bar: 20 µm. ***, P < 0.001. Panels show representative results from at least two independent experiments performed.
Video 1.
Mitochondrial and lysosomal dynamics in DSTYK KO cells under metabolic stress. H2009 tumor cells were stained for mitochondria (Mitotracker Deep Red, Green), mitochondrial transmembrane potential (TMRM, red), and lysosomes (Lysotracker Green FM, Blue) and deprived of glucose for 4 h before starting live imaging. Time-lapse videos are 3D reconstructions of two imaging fields in parental and DSTYK KO cells recorded simultaneously. The video is representative of two experiments performed similarly.
In conclusion, these data showed the involvement of DSTYK in the maintenance of an operative mitophagy and its depletion affected mitochondrial morphology, physiology and interrupted mitophagy.
DSTYK inhibition impairs tumorigenesis in vivo by sensitizing lung cancer cells to TNF-α–mediated CD8+ killing
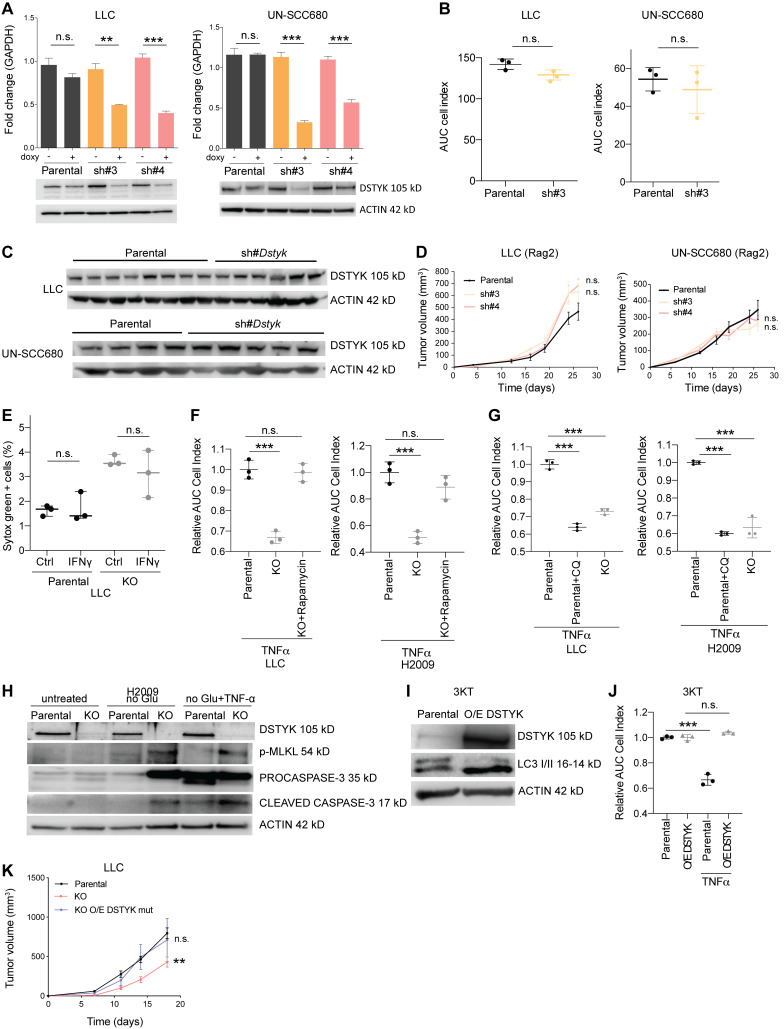
In the organism, cancer cells face different stresses such as hypoxia, nutrient deprivation, or immunosurveillance. To assess the functional effects of DSTYK decreased levels in vivo, we performed experiments in syngeneic mouse preclinical models. We used cell lines from the main histological subtypes of NSCLC: LUAD and LUSC (LLC and UN-SCC680, respectively) and inhibited DSTYK using an inducible vector (Tet-pLKO) that activated an shRNA against DSTYK upon doxycycline administration (Fig. S5 A). The generated cell lines with inducible DSTYK-knockdown showed no cell proliferation differences in vitro (Fig. S5 B). sh#DSTYK was activated after tumor establishment in vivo (75–100 mm3 in size) by doxycycline administration. Both models showed a dramatic decrease (P < 0.001) in tumor size under DSTYK inhibition, which was directly proportional to the inhibition degree achieved by the sh#DSTYK, achieving tumor regression in both syngeneic models 2–3 wk after tumor cell injections (Fig. 5 A).
Figure S5.
DSTYK inhibition promotes immunogenic death of lung cancer cells. (A) Top: DSTYK mRNA expression by quantitative PCR in LLC and UN-SCC680 parental and sh#DSTYK cells (n = 3). Bottom: DSTYK protein expression by Western blot in LLC and UN-SCC680 parental and sh#DSTYK cells. Analysis was done by t test. Comparison LLC sh#3 −/+ doxy, P = 0.003; sh#4 −/+ doxy, P < 0.001. Comparison UN-SCC680 sh#3 −/+ doxy and sh#4 −/+ doxy, P < 0.001. (B) Proliferation assay measured by xCELLigence Technology for 96 h in LLC and UN-SCC680 parental and sh#DSTYK cells (n = 3). Analysis was done by t test. (C) DSTYK protein expression by Western blot in parental and sh#DSTYK tumors from LLC and UN-SCC680 preclinical models at the end point of in vivo experiment. (D) Tumor volume of LLC and UN-SCC680 parental or sh#DSTYK cells in immunocompromised mice. Analysis was done by ANOVA. Number of Rag−/− IL2Rg−/− mice per group = 5. (E) T cell killing of LLC parental and KO cell lines was performed in the presence or absence of IFN-γ (n = 3). (F) TNF-α–mediated killing of LLC or H2009 parental, KO, and KO treated with rapamycin cell lines (n = 3). Analysis was performed by t test. Parental vs. KO cells treated with TNF-α, P < 0.001 both in LLC and H2009 cell lines. (G) TNF-α–mediated killing of LLC or H2009 parental, parental treated with chloroquine, and KO cell lines (n = 3). Analysis was performed by t test. Parental vs. parental + chloroquine and parental vs. KO cells treated with TNF-α showed P < 0.001 both in LLC and H2009 cell lines. (H) Cropped images from Western blot analysis of apoptosis and necroptosis-related proteins in H2009 parental and DSTYK-KO cells under normal and stress conditions. (I) Cropped images from Western blot analysis of autophagic protein (LC3) in immortalized 3KT parental and overexpressing DSTYK. (J) T cell killing of 3KT parental and overexpressing DSTYK cell lines was performed in the presence or absence of TNF-α (n = 3). Analysis was performed by t test. P < 0.001 for parental vs. parental treated with TNF-α 3KT cell lines. (K) Tumor volume of parental, KO, and KO overexpressing mutated DSTYK LLC tumor–bearing mice. Analysis was performed by ANOVA. P = 0.002 (parental vs. KO tumor-bearing mice). Number of C57BL/6J mice per group = 7. **, P < 0.01; ***, P < 0.001. Source data are available for this figure: SourceData FS5.
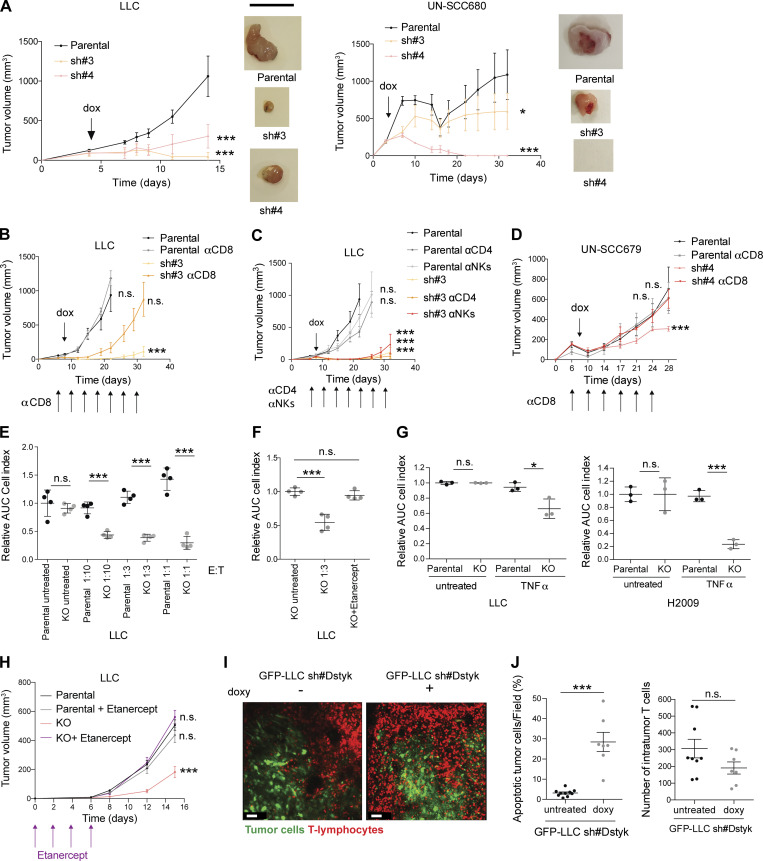
Figure 5.
DSTYK depletion sensitizes lung cancer cells to TNF-α CD8+ killing. (A) Syngeneic mouse models for sh#DSTYK LUAD (left) and squamous carcinoma cell lines (right) respectively. sh#DSTYK was activated when tumors reached 50–100 mm3. Analysis by ANOVA. LLC parental vs. sh#3 and parental vs. sh#4, P < 0.001; UN-SCC680 parental vs. sh#3, P = 0.04; and parental vs. sh#4, P < 0.001. Number of C57BL/6J or A/J mice per group = 7. Scale bar: 1 cm. (B) LLC tumor–bearing mice were treated with doxycycline to induce sh#DSTYK in the presence or absence of depleting antibodies (anti-CD8). Statistical analysis was performed by ANOVA comparing tumor volumes of experimental groups at the end point day. LLC parental vs. sh#3, P < 0.001, while no differences were found in other comparations. Number of C57BL/6J mice per group = 7. (C) LLC tumor–bearing mice were treated with doxycycline to induce sh#DSTYK in the presence or absence of depleting antibodies (anti-CD4, anti-NK). Statistical analysis was performed by ANOVA comparing tumor volumes of experimental groups at the end point day. LLC parental vs. sh#3, sh#3+anti-CD4, and sh#3+anti-NKs, P < 0.001. Number of C57BL/6J mice per group = 7. (D) UN-SCC679 tumor–bearing mice were treated with doxycycline to induce sh#DSTYK in the presence or absence of depleting antibodies (anti-CD8). Statistical analysis was performed by ANOVA. Parental vs. sh#4, P < 0.001, while no differences were found in other comparations. Number of A/J mice per group = 7. (E) Control or DSTYK KO LLC cells were pulsed with Ova peptide and incubated with OT-1 T cells at the indicated E:T ratios for 96 h. The dot plot graph shows the relative area under the curve (n = 3) compared with cells incubated in the absence of T cells. t test was used to compared experimental groups and rendered P < 0.001 in every treated condition. (F) LLC T cell killing was performed in the presence of the TNF-α–blocking molecule etanercept. Analysis by t test. KO untreated vs. KO 1:3, P < 0.001. (G) T cell killing of LLC or H2009 parental and KO cell lines was performed in the presence or absence of TNF-α. Analysis by t test. LLC parental vs. KO cells treated with TNF-α, P = 0.02; H2009 parental vs. KO cells treated with TNF-α, P < 0.001. (H) Parental and DSTYK inhibited LLC tumor–bearing mice were treated with etanercept. Statistical analysis was performed by ANOVA. Parental vs. KO, P < 0.0001, while no differences were found in other comparations. Number of C57BL/6J mice per group = 7. (I) Intravital microscopy of T cell responses in LLC DSTYK wildtype and deficient tumors. GFP-LLC#sh tumor cells were injected in the dorsal part of the ear of hCD2dsRED mice in which T cells are red fluorescent. 7 d later, animals were given or not given doxycycline in the drinking water for 24 h and then imaged in a confocal microscope. Images of a representative experiment and corresponding to Video 2. Scale bar: 50 µm. (J) Dot plot quantification of percentage of apoptotic tumor cells per field and number of intratumor T cells in LLC DSTYK wildtype and deficient tumors from F. n = 4 mice per group; each dot represents a single quantified HPF. Statistical analysis of apoptotic tumor cells per field was performed by t test; P < 0.001 and no differences were found in the number of intratumor T cells. *, P < 0.05; ***, P < 0.001.
To understand the mechanism by which DSTYK depletion affected tumor size without modifying cell proliferation, we evaluated DSTYK expression levels in tumors at the end point of in vivo experiments by Western blot (Fig. S5 C). Surprisingly, we observed similar DSTYK expression levels in parental and DSTYK inhibited tumors, indicating that a very potent selective pressure was being produced against DSTYK KO cells in vivo, and only the clones that maintained DSTYK expression were able to survive and grow. Therefore, DSTYK was a dependency of lung cancer cells required to cope with stress in vivo.
Because DSTYK inhibition sensitized cells to stress and immune cells may threaten tumor stability, we hypothesized that the immune system could be mediating these potent rejections shown in our syngeneic models upon DSTYK ablation. First, to distinguish the direct growth effects of DSTYK depletion on cells from the effects derived from its interaction with the immune system, we performed a subcutaneous injection of control and sh#DSTYK murine cells in immunocompromised mice (Fig. S5 D). DSTYK inhibition did not affect cell proliferation in vivo suggesting that the effect observed in the syngeneic models might be non-autonomous and related to the immune system. To identify the effector cells responsible for these rejections, key immune populations were selectively depleted in vivo in parallel experiments. Elimination of CD8+ T cells completely abrogated the antitumoral effects of DSTYK knockdown in LLC tumors. However, neither CD4+ T nor natural killer depletion had any impact on LLC tumor rejection (Fig. 5, B and C). These effects were verified in an independent LUSC model: UN-SCC679 (Fig. 5 D). These results suggested that DSTYK depletion sensitized lung tumor cells to the action of cytotoxic CD8 cells.
To validate the role of DSTYK in limiting tumor cell killing by T cells, we assessed the behavior of CRISPRed KO cells in co-culture in vitro. DSTYK depletion sensitized LLC cells to T cell killing in vitro, dependent on T cell/tumor cell ratio excluding off target effects (Fig. 5 E). We next explored the mechanism by which CD8+ T cells were killing. A recent work showed how autophagy inhibited TNF-α–induced apoptosis (Young et al., 2020). In our cells, CD8+ T cell killing was reversed by capturing and blocking TNF-α with etanercept (Fig. 5 F). However, no effect was observed upon INF-γ treatment (Fig. S5 E). Consistently, tumoral killing by T cells was mimicked upon TNF-α treatment in KO LLC cells (Fig. 5 G). Similar results were observed in human cell lines (Fig. 5 G). These results were consistent with above-mentioned publication.
Interestingly, when we inhibited mTOR with rapamycin in KO cells, we rescued a parental phenotype, resistant to TNF-α–mediated killing (Fig. S5 F). Besides, parental cells treated with chloroquine, which prevent autophagy, became sensitive to TNF-α–mediated killing similarly to DSTYK KO cells (Fig. S5 G).
To verify whether these effects were maintained in vivo, parental and DSTK KO LLC tumor–bearing mice were treated with etanercept every 2 d after cell inoculation to block TNF-α. DSTYK-KO tumors treated with etanercept rescued the parental cell phenotype, results were consistent with those previously observed in vitro (Fig. 5 H).
In order to clarify the underlying cell death mechanism related to DSTYK depletion and TNF-α–induced apoptosis, we exposed H2009 parental and KO cells to stress conditions and we observed an increase of both activated caspase and p-MLKL in KO cells (Fig. S5 H). Thus, cell death related to DSTYK inhibition seemed to involve both caspase-dependent and independent mechanisms.
To dynamically understand the immune mechanisms that mediated tumor rejection in the absence of DSTYK, we performed intravital microscopy studies. Inducible sh#DSTYK LLC lung cancer lines were transfected with fluorescent proteins and injected subcutaneously into fluorescent T lymphocytes reporter mice. We dynamically observed that fluorescent T cells successfully attacked and eliminated knockdown-DSTYK tumors in 72 h, whereas parental tumors were insensible to T cell killing (Video 2 and Fig. 5 I). Our results showed a significant increase (P < 0.001) in apoptotic tumor cells per field in DSTYK-inhibited tumors but no differences in the number of intratumoral T cells (Fig. 5 J), suggesting that tumor elimination was a consequence of tumoral T cell killing sensitizing rather than tumor immune infiltrate changes.
Video 2.
Intravital microscopy of T cell responses in LLC DSTYK–deficient tumors. GFP-LLC#sh tumor cells were injected into the dorsal part of the ear of hCD2dsRED mice in which T cells are fluorescent red. 7 d later, animals were given or not given doxycycline in drinking water for 24 h and then imaged in a confocal microscope. Time-lapse videos of T cells in the tumors showing T cell interactions with tumor cells of both conditions. The video is representative of intravital microscopy videos performed in at least three mice per group.
To determine whether there was a therapeutic index for targeting DSTYK, we overexpressed the protein in immortalized human lung epithelial cells (3KT), and we demonstrated that amplification resulted in a gain of function both, raising autophagy markers and acquiring resistance to TNF-α treatment (Fig. S5, I and J). Interestingly, when we overexpressed Dstyk mutation found in UN-SCC680 in an independent murine cell line LLC, tumors of KO overexpressing mutated-DSTYK bearing mice rescued parental tumor growth (Fig. S5 K).
DSTYK inhibition sensitizes lung cancer cells to immunotherapy
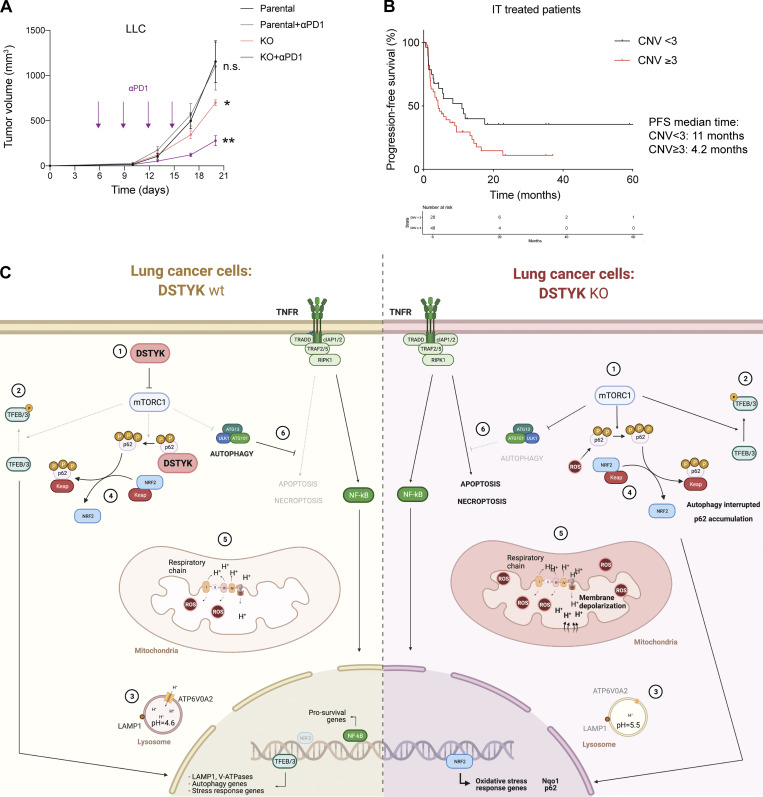
Despite great advances in oncological therapies, still a high percentage of patients remain refractory to current treatments (Grant et al., 2021). More personalized treatments are required in NSCLC, not only to expand the targeted therapy but also to design immunotherapy regimens according to the individual NSCLC patient immune and genetic landscape (Wang et al., 2021b). We postulated that DSTYK downregulated could sensitize lung cancer tumors to immunotherapy. To test this, we treated our syngeneic mouse models harboring wildtype or DSTYK-deficient tumors with checkpoint inhibitors. For that, we selected our LLC CRISPRed model. Established LLC tumors, normally refractory to checkpoint inhibitor treatments, partially responded to anti–PD-1 administration under DSTYK inhibition conditions, showing a synergistic effect (Fig. 6 A).
Figure 6.
DSTYK inhibition sensitizes lung cancer cells to immunotherapy. (A) Parental and KO LLC tumor–bearing mice were treated with or without anti–PD-1. Statistical analysis was performed by ANOVA. Parental vs. KO, P = 0.05; parental vs. KO + anti–PD-1, P = 0.008. No differences between parental and parental + anti–PD-1 treated mice. Number of C57BL/6J mice per group = 7. (B) Kaplan–Meier plot showing PFS of lung cancer patients treated with immunotherapy and classified by the presence of absence of DSTYK amplification (n = 76). Log-rank test, P = 0.06. (C) Proposed model of DSTYK molecular mechanism in lung cancer cells. (1) DSTYK inhibits mTOR activity; (2) mTOR mediates TFEB/3 phosphorylation preventing its nuclear translocation; (3) TFEB/3 transcribe genes related to lysosomal biogenesis and maturation; (4) DSTYK binds P62, which is involved in the NRF2/KEAP1 pathway; (5) DSTYK protects mitochondrial integrity; (6) autophagy prevents TNF-α mediated apoptosis. *, P < 0.05; **, P < 0.01.
More importantly, to confirm the translational value of our findings, we carried out a study on a cohort of 76 patients treated with ICI, for which both outcome and genomic traits were known (Frigola et al., 2022). According to our analysis, lung cancer patients who underwent immunotherapy and presented DSTYK amplification showed poorer PFS rates (P = 0.06). On average, the median PFS time for patients with DSTYK amplification was 11 mo, whereas 4.2 mo was the median PFS for patients with only two DSTYK genomic copies (Fig. 6 B). These results suggested DSTYK amplification as a predictive biomarker for immunotherapy resistance. More interesting, a combination of DSTYK inhibition and immunotherapy may be a potential strategy for lung cancer treatment where DSTYK blockage could sensitize tumor cells to ICIs.
Discussion
This work uncovers a new role of DSTYK in lung cancer and characterizes it as an important oncogene in tumor progression, and immunotherapy resistance. Mechanistically, DSTYK is demonstrated to be a master regulator of autophagy and oxidative stress response, whose inhibition collapses tumor-sustaining autophagy and sensitizes lung cancer cells to immune-based therapies.
The dual serine/threonine and tyrosine kinase (DSTYK), also known as CAKUT1, DustyPK, SPG23 has been classically considered as RIPK5. The RIPK family includes a group of kinase proteins which are key mediators of the cellular response to stress caused by infections, inflammation, or cell damage (Colhado Rodrigues et al., 2020). They are related to membrane receptors such as IRAK, FGFR, or TLRs and mediate the intracellular response through various pathways such as the activation of NF-κB or AP-1 (Meylan and Tschopp, 2005). These proteins display significant homology in the kinase domains but otherwise share limited structural similarities (Cuny and Degterev, 2020). Nowadays, there is some controversy regarding which kinase represents RIPK5 as there are two kinases (SgK288/ANKK1 and SgK496/RIPK5) assigned to this name (Peng et al., 2006; Meylan and Tschopp, 2005). DSTYK is highly conserved across species and shares high kinase homology to the RIPK family (Cuny and Degterev, 2020). It is a cytoplasmic protein kinase expressed in multiple human tissues including the brain, heart, kidney, lung, colon, and muscle (Peng et al., 2006). The functions of DSTYK are poorly understood, although recent reports have described a role in development and homeostasis.
Based on our previous data showing Dstyk mutated in a cell line derived from a syngeneic LUSC mouse model (Valencia et al., 2021), we study the impact of DSTYK in the field. Among the few DSTYK mutations reported in literature, most occur in the non-catalytic region 1 (NCR1) rather than in the catalytic domain (kinase domain), suggesting that the NCR domains might be crucial in regulating the catalytic activity of the kinase domain. DSTYK loss-of-function germline mutations have been related to autosomal dominant congenital anomalies of the kidney and urinary tract (Sanna-Cherchi et al., 2013) and with autosomal-recessive hereditary spastic paraplegia type 23 (Lee et al., 2017; Vidic et al., 2021). Furthermore, DSTYK kinase domain KO mice had impaired learning and memory capabilities (Li et al., 2014). Interestingly, no total DSTYK KO have been reported, suggesting that this protein is essential for development. In accordance, we were not able to generate total but partial CRISPRed DSTYK KO cell lines. On the other hand, gain-of-function mutations or high DSTYK expression has been linked to tumorigenicity and tumor progression (Tang et al., 2019; Zhang et al., 2020; Ogbu et al., 2021). Contrary to a recent work (Zhong et al., 2021), here we report DSTYK to be altered in a very significant proportion of lung cancer patients, showing gain of function as a consequence of a genetic gain or gene amplification.
An important detail that could partially explain the contradictory results between Zhong et al. (2021) and our data is the probe chosen to perform survival analyses based on DSTYK expression. We choose 211514_at, as it includes genomic locations corresponding to a central part of the gene, mainly the scaffold domain, contrary to other probes (229017_s_at and 214663_at) that mainly encompass exons relative to the kinase domain of the gene. Taking these probe design differences into account is very important when analyzing subsequent results. First, there are two known DSTYK isoforms and another predicted one that differ in the kinase domain, which is missing in two of the three isoforms. Besides, the kinase domain is known to >90% of homology with other kinases such as RIPK1, constituting a poor specific region to detect. Based on these data, we think that the choice of a probe that recognizes the scaffold domain would render more robust and specific results. Biological and phenotypic data obtained subsequently validate this choice.
Other mechanisms sustaining DSTYK expression dysregulation such as epigenetic factors are also possible, and we do not discard them although they are not considered in this work.
In lung cancer, several genes altered in <5% patients, such as MET or HER2, are widely studied during the diagnostic process to be considered for FDA-approved targeted therapy. Targeting infrequent oncogenic drivers in NSCLC has expanded the spectrum of targeted therapy (Guo et al., 2019; Nokin et al., 2021), contributing to advance personalized medicine efforts for NSCLC patients. Although around 4% of patients present DSTYK amplification, up to 40% show moderate alteration of DSTYK CN (>2.3 copies), which constitutes a very frequent alteration for this malignancy with unknown functional implications. Our data show that alteration of CN could potentially be used as a prognostic marker for lung cancer. According to our FISH data, the number of copies in altered tumoral cells is not very high. In fact, the average CN of DSTYK in lung tumors is 2.3. Interestingly, those patients with poor prognosis (Amp DSTYK) show three or more copies of DSTYK, whereas altered cells in NoAmp DSTYK patients always show three copies. In addition, we observe a different proportion of patients with DSTYK CN alterations among clinical cohorts used, showing VH series, composed of patients in advanced stages (Fig. 6 B), a higher percentage. Although we cannot discard that these discrepancies are due entirely to differences in the patient’s profile that compose them, and taking into account technical differences to measure DSTYK CN in each case, these data are in line to our hypothesis that DSTYK amplification may be a prognostic factor.
Nothing was known about DSTYK location within the cell except that it is a cytoplasmic protein and seems to colocalizes with FGFR1/2 in 293T cells (Sanna-Cherchi et al., 2013). We find DSTYK in vesicles compartment. More importantly, for the first time, DSTYK is shown to colocalize with LC3, which is consistent with the DSTYK immunoprecipitation experiment which showed P62 interaction. Hence, our data demonstrate a link between DSTYK and autophagy in different ways. All three methods used—IF, RNAseq analysis, and immunoprecipitation—show a direct role for DSTYK in autophagy. A very interesting recent study has reported that DSTYK regulated mTOR in monkey renal fibroblasts injected in a zebrafish-based developmental model, contributing to congenital scoliosis-like vertebral malformations in the zebrafish model (Sun et al., 2020).
We demonstrate that DSTYK depletion prevents autophagy through the confluence of several mechanisms. First, DSTYK inhibits mTOR, verified by the activation of p-S6K. Additionally, autophagy is interrupted by the accumulation of p-P62 and LC3 in KO cells. Furthermore, the absence of DSTYK and subsequent activation of mTOR prevents TFEB/3 to translocate to the nucleus and blocks the transcription of genes related to lysosomal biogenesis and maturation, causing a decrease in the number of lysosomes (Roczniak-Ferguson et al., 2012) and the formation of less acid, dysfunctional lysosomes, which are not able to digest the autophagosome content. Our results agree with previous work where colorectal cancer cells induced with TGF-β (which also triggers autophagy) underwent epithelial-mesenchymal-transition and became more malignant and less sensitive to apoptosis (Zhang et al., 2020). In accordance with this observation, we show how DSTYK KO lung cancer cells are sensitized to stress and, therefore, they are more prone to apoptosis. In fact, DSTYK KO cells are severely damaged under stressful conditions.
A noticeable number of reports have shown autophagy as a regulator of drug resistance in cancer, including lung tumors. Specifically, the potential drug resistance role of autophagy has been described for treatments very relevant to lung cancer such as Osimertinib (Chen et al., 2021), the ALK inhibitor Ceritinib (Schläfli et al., 2021), B-RAF inhibitors (Foth and McMahon, 2021) and others. The recently started LIMIT trial (NCT04735068) will evaluate the use of hydroxychloroquine along with the MEK1- and MEK2-inhibitor Binimetinib, as an effective method for treating K-RAS–dependent metastatic cancers, focusing mainly in LUAD.
Despite the above-mentioned dual role, most of the clinical trials with drugs related to autophagy have been focused on the tumor promoter and therapy resistance mechanisms of autophagy in cancer.
A recent report shows that mTORC1-mediated suppression of autophagy causes a metabolic vulnerability leading to energy crisis and apoptosis, which could be used as a potential druggable strategy against lung and other tumors (Gremke et al., 2020). Moreover, P62 cooperates with hyperactive mTORC1 to maintain mitochondrial integrity and promote tumorigenesis (Lam et al., 2017).
It is widely accepted that tumors develop under the influence of mitochondrial metabolism on all steps of oncogenesis (Porporato et al., 2018) and that the mitochondrial metabolism remains vital for both the production of ATP and the supply of biosynthetic intermediates (Bhandari et al., 2019). In fact, both, the human and mouse lung cancer cell lines used in the study have many mitochondria.
The levels of ROS in oxidative cancer cells are usually high, facilitating, in many cancers, cell survival, and cancer progression. However, excessive ROS causes cytochrome C release into the cytoplasm and triggers apoptosis (Zhang et al., 2016). High levels of ROS constitute a serious risk to mitochondrial integrity and, therefore, cell viability. Consequently, cancer cells need to strike a balance between mitochondrial biogenesis and mitophagy, to maintain mitochondrial fitness (Kenny et al., 2019). We observe a significant increase in ROS levels in the absence of DSTYK that cause DNA damage (data not shown) and activate the Nrf2/Keap1 pathway in an attempt to deal with oxidative stress. All these mechanisms are a consequence of the severe mitochondrial damage induced by DSTYK ablation. To our knowledge, the role of DSTYK in regulating the mitochondrial integrity has not been previously described. In a baseline situation, without cellular stress, the cell is able to deal with the DSTYK ablation but in the presence of stress (i.e., starvation), damaged mitochondria cannot respond to these cellular metabolic needs. We have also shown by fluorescent confocal microscopy some features indicative of mitochondrial dysfunction in DSTYK KO cells, such as TMRM decrease or the development of a ring-shape (Ahmad et al., 2013; Zorova et al., 2018). Damaged mitochondria normally undergo mitophagy to allow their elimination and minimize the damage induced to the cell. Yet, DSTYK KO cells undergo interrupted autophagy and therefore, damage mitochondria are accumulated in autophagosomes.
Our data show how the interruption in autophagy caused by DSTYK inhibition sensitizes tumor cell killing by a TNF-α–dependent mechanism. The protective effect of autophagy in the context of T cell killing is mediated primarily through inhibition of TNF-α–dependent apoptosis (rather than of perforin/granzyme-mediated killing). Very recent reports indicate that autophagy can limit antitumor immunity (Yamamoto et al., 2020; Lawson et al., 2020). In agreement with our data, recent work reports how autophagy inhibits TNF-α–induced apoptosis by limiting the formation and/or activity of the FADD/caspase-8 complex (Young et al., 2020). However, the exact mechanism through which autophagy limits TNF-α–dependent apoptosis remains unknown.
Notably, we demonstrate a sensitivity switch of tumoral cells to T cell–mediated killing rather than a change in tumoral immune infiltrate. Consistent with the sensitization of cancer cells to T cell killing, lack of DSTYK boosts immunotherapy effects. Some reported targeted therapies against tumors affect pathways that are crucial for immune function or sensitize tumor cells to immune-mediated killing. Therefore, targeted therapies and immunotherapy offer a number of potential possible synergies in treatment when used together (Vanneman and Dranoff, 2012). Some recent examples are dasatinib plus anti–PD-1 for NSCLC patients with YES1 alteration (Redin et al., 2021) or the pro-inflammatory tumor microenvironment caused by AMG 510 in RAS-dependent lung cancers, which trigger response to immune-checkpoint inhibitors (Canon et al., 2019).
Taken together, our results support that DSTYK is a new target and that its inhibition could form the basis of new therapies, which would benefit a subgroup of NSCLC patients both at early (adjuvant) or late (single or combined treatment) stages of the disease. The combination of targeted therapies as modulators of immunotherapy response is an exciting new field that can change the paradigm of current therapies. In addition, we hypothesize that DSTYK inhibition can sensitize tumor cells to other standard therapies such as chemotherapy or radiation, as all of them cause cellular stress, with a potential benefit for a large subset of patients.
Thus, we believe that DSTYK inhibition might constitute a step forward in personalized medicine in NSCLC.
Materials and methods
Public data analysis
TCGA related data were downloaded from the TCGA bioportal using the TCGA biolinks R package (Colaprico et al., 2016). CNV and transcriptome profile data were downloaded for 24 cancer types. CN segment data type was used to measure the CNV in the segment where the DSTYK gene was identified. Log transformation was applied to define logCNV values. For transcriptome data, Gene Expression Quantification data were downloaded, quantified by HTSeq, and transformed in FPKM.
To assess the prognostic value of DSTYK expression, the online tool kmplot.com (http://kmplot.com/analysis/; Györffy et al., 2010) was used. The corresponding Affymetrix ID for DSTYK was 211514_at.
Cell lines and reagents
The human cell lines H2009 and H226 were purchased from ATCC. The human cancer cell lines were authenticated by the Genomics Unit at CIMA and grown according to ATCC specifications. The mouse LLC cell line was from the ATCC. UN-SCC679 and UN-SCC680 lung squamous carcinoma cell lines were previously established by our group from an N-nitroso-tris-chloroethylurea (NTCU) chemically induced mouse model (Valencia et al., 2021). 393P LUAD cells, derived from KrasLA1/+; p53R172 HΔG mice, were a gift from J.M. Kurie (The University of Texas MD Anderson Cancer Center, Houston, TX). All cells were cultured in RPMI 1640 medium (Invitrogen), 10% FetalClone (Thermo Fisher Scientific), 100 U ml−1 penicillin, and 100 U ml−1 streptomycin (Invitrogen). HBEC-3KT cells (Kerafast Inc.) were grown in keratinocyte medium (GIBCO). Only mycoplasma negative cells tested using the MycoAlert Mycoplasma Detection Kit (LONZA) were used.
Inducible knockdown constructs are as follows: Lentiviral transduction of shRNAs against DSTK (TRCN0000275831 [#1] and TRCN0000195393 [#2] for human; TRCN0000295440 [#3] and TRCN0000088496 [#4] for mouse; Sigma-Aldrich) was performed as previously described (Vallejo et al., 2017). Transduction of the empty vector (TetPLKO.1-puro; Sigma-Aldrich) was used as a control. CRISPR Cas9 strategy for DSTYK KO cell clones generation: single-guide RNAs complementary to the DSTYK sequence were designed using a GPP Web Portal tool from the Broad Institute (https://portals.broadinstitute.org/gpp/public/) and cloned into lentiCRISPRv2 plasmid from Addgene (Vallejo et al., 2017). Puromycin-resistant single cells were sorted in a p96 plate using MoFlo Astrios EQ (Beckman Coulter), characterized by quantitative RT-PCR (qRT-PCR) and Western blot analysis and expanded for experiments.
The single-guide RNA sequences used in this study are listed in Table S5.
Flag-DSTYK constructs are as follows: Flag sequence (5′-GATTACAAGGATGACGACGATAAG-3′) was added to 5′-end DSTYK cDNA sequence and cloned in pBABE puro plasmid. Mouse LLC and UN-SCC679 cell lines were then infected to overexpress the tagged protein.
DSTYK overexpressing constructs are as follows: Wildtype mouse and human DSTYK cDNA sequence or mouse containing punctual mutation c.1976G>A were cloned in pLenti6/V5-DEST plasmid. Cell lines were then infected to overexpress the wildtype or mutated protein.
To induce stress, cells were cultured in RPMI 1640 no glucose (#11879-020; Gibco) or standard RPMI without the FetalClone supplement up to 24 h.
To impact the autophagy pathway, cells were treated with 25 µM of chloroquine (#C6628; Sigma-Aldrich) or 300 nM of rapamycin (#R8781; Sigma-Aldrich) for 24 h.
Glucose was measured in supernatant in Cobas c-311 analyzer using a glucose (HK) assay kit (#04404483; Roche) following the manufacturer’s instructions.
Whole-exome, shallow whole-genome, and RNAseq analysis
Shallow whole-genome sequencing data (cohort VH) were obtained from Frigola et al. (2022). For RNAseq, samples were prepared with the Illumina TruSeq Stranded mRNA kit following the manufacturer’s indications and sequenced as reverse paired-end (75 bp) runs on the Nextseq sequencer. For the RNAseq analysis, raw FASTQ files were trimmed with Trimmomatic/0.36. Pseudoalignment was carried out to the mm10 reference genome and gene level counts were determined with Kallisto (Bray et al., 2016). Quantification was carried out with 100 boostraps. Differential gene expression analysis was conducted with R using Sleuth R package (Pimentel et al., 2017). Significant genes were selected based on the false discovery rate–corrected P value <0.05. The following comparisons were carried out in the study: (A) LLC parental vs. (B) LLC sh#Dstyk; (A) UN-SCC679 parental vs. (B) UN-SCC679 O/E Dstyk wt; (A) UN-SCC679 parental vs. (B) UN-SCC679 sh#Dstyk.
Once differentially expressed genes were selected in each A vs. B contrast, all experiments were merged to verify the intersection of the genes based on name and the direction.
Immunoprecipitation
Flag-DSTYK and control cells were seeded in 150 mm2 plates and incubated at 37°C until 80% confluency was reached. Cells were lysed (20 mM HEPES, 150 mM NaCl, 1% NP40, 10 mM MgCl2 + phosphatase and protease inhibitors [#04906845001 and #11836170001; Roche]) on ice for 10 min and centrifuged at 13,000 g for 15 min at 4°C. Protein was quantified using the BCA assay kit (#23227; Pierce). 1 mg/ml of cell lysates was incubated with the indicated antibody (Table S5) overnight at 4°C. The day after, Dynabeads Protein G (#10003D; Thermo Fisher Scientific), were added and mixed for 2–4 h in rotation at 4°C. Immunoprecipitates were washed with lysis buffer five times and lysates were denatured in lysis buffer with LDS Sample buffer (NuPage #2201446) and β-mercaptoethanol (Sigma-Aldrich). Mass spectrometry was performed at the Harvard Medical School (https://taplin.med.harvard.edu/home). Eluted products were analyzed by Western blotting.
Western blotting and antibodies
After treatments were administered to plated tumor cells, these were washed with PBS and lysed in radioimmunoprecipitation assay buffer and with Nuclear and Cytoplasmic Extraction Reagents (NE-PER; cat #78835) plus phosphatase and protease inhibitors (#04906845001 and #11836170001; Roche). Lysates were centrifuged at 13,200 rpm at 4°C and quantified using a BCA protein assay kit (#23227; Pierce).
Samples were prepared using LDS Sample buffer (NuPage #2201446) and β-mercaptoethanol (Sigma-Aldrich). Protein samples were incubated for 15 min at 95°C before being loaded 10–50 μg on 10–15% acrylamide gels. Electrophoresis was performed in a running 10× Tris-Glycine SDS buffer, and nitrocellulose membranes were used for proteins transfer (#1620112; Bio-Rad). Membranes were incubated overnight with the indicated antibodies, washed with TBS-Tween, and incubated for 1 h with anti-rabbit (#GENA934; Sigma-Aldrich), anti-mouse (#NA931; GE Healthcare), or anti-goat (#P0449; Agilent) secondary antibodies.
Nuclear and cytoplasmic fraction extraction were performed using the NE-PER Nuclear and Cytoplasmic Extraction Reagents kit (#78835; Thermo Fisher Scientific) according to the manufacturer’s protocol: 100 μl of CERI and 5.5 μl of the CERII reagents for the cytoplasmic extraction and 50 μl of the NER reagent for the nuclear extractions, adding protease, and phosphatase inhibitors to CERI and NER to minimize reagent dilution.
For subcellular fractionation experiments, different cell fractions were obtained by serial centrifugation steps at increasing speeds.
Briefly, a pellet of cells was washed in cold PBS and then lysed in extraction buffer (Tris HCl pH8.5, 10 mM; NaCl, 150 mM; MgCl2, 1.5 mM; IGEPAL, 0.5%; protease inhibitors). Cells were disrupted for 10 min on ice and then centrifuged at increasing speeds. Pellets and supernatants of serial centrifugations were recovered and then loaded onto an acrylamide gel with equivalent amounts of the obtained fractions. The following conditions were followed to obtain subcellular fractions: Total extract, cells pelleted and resuspended directly in Laemmli buffer 1×; nuclear extract (P3000), pellet of extraction buffer lysed cells centrifuged 2 min, 3,000 g, 4°C; cytosolic extract (S3000), supernatant resulting from the previous centrifugation step; Large organelle extract (P10000), pellet resulting from S3000 centrifugation 10 min, 10,000 g, 4°C; cytosolic extract without large organelles (S10000), supernatant resulting from previous centrifugation; microsomal extract (P100000), pellet resulting from S10000 centrifugation 2 h, 100,000 g, 4°C; coluble cytosolic extract (S100000), supernatant resulting from previous centrifugation.
Western blot band detection was performed using Image Studio software. Densitometry quantification was done using ImageJ (National Institutes of Health). All measurements can be found in Table S4.
The antibodies used are listed in Table S5.
qRT-PCR
Genomic DNA was extracted from frozen samples from the CIMA-CUN series and CN was measured by qPCR using the TaqMan Copy Number assay (#Hs01236436-cn for DSTYK and #4401631 for RNAse P; Applied Biosystems) according to the manufacturer’s protocol.
Total RNA from cells was isolated with the NucleoSpin RNA isolation kit (#740955; Macherey-Nagel). 1 µg RNA was used to synthesize complementary DNA using a High-Capacity cDNA Reverse Transcription kit with RNase inhibitor (#4374966; Applied Biosystems). Quantitative PCR was performed with the PowerUp SYBR Green PCR Master Mix (#100029284; Thermo Fisher Scientific). The primers designed for quantitative PCR are listed in Table S5.
FISH
DSTYK CNV was evaluated by FISH. Samples were dewaxed, hydrated, and heated in citrate buffer pH 6 for 30 min. Specimens were then treated with 4 mg/ml of pepsin in 0.01 N HCl for 5 min at 37°C and dehydrated. Next, a dual DSTYK1q32.1/CEN1p11.1 probe (Empire Genomics; #DSTYK-CHR01-20-ORGR) was added, denatured for 7 min at 75°C, and incubated 24 h at 37°C to allow hybridization. Samples were immersed in a post-hybridization saline-sodium citrate buffer (SCC; 0.3 M NaCl; 0.03 M sodium citrate; pH 7) for 2 min and then in SCC with 0.3% of NP-40 (Sigma-Aldrich) for 1 min at 70°C. Nuclei were counterstained with DAPI 1,500 ng/ml (#U0031; Abnovas). CNV variation was evaluated with a Nikon E800 microscope provided with a Nikon DS-Qi1-Mc camera and NIS-Elements software. Images were processed with ZEISS ZEN software.
IF
Cells were seeded in an 8-well culture slide (354108; Falcon) and cultured for 48 h. Then, cells were fixed with 4% formaldehyde methanol-free (28908; Thermo Fisher Scientific) for 10 min at 37°C and IF was performed in a BSA 1.5% saponin 0.05% buffer. All primary antibody incubations were carried out overnight at 4°C. Secondary antibodies were incubated for 1 h at room temperature. Nuclei were dyed with Hoechst (1:1,000). For negative control, cells were incubated directly with secondary antibody (1:100) 1 h at room temperature. Images were taken at 63× using a confocal microscope LSM 800 with Airyscan (Zeiss). IF antibodies are listed in Table S5.
For confocal microscopy experiments of mitochondrial and lysosome dynamics, parental, or DSTYK KO H2009 tumor cells (30,000 cells) were plated in 8-well LabTek microscopy plates (Nunc). 24 h later cells were stained using Mitotracker Deep Red (0.1 µM, Thermo Fisher Scientific), TMRM (125 ng/ml; Sigma-Aldrich) and Lysotracker Green DND-26 (0.1 µM; Thermo Fisher Scientific) for 20 min at 37°C in RPMI 10% FBS. The cells were then washed twice with warmed media and treated with RPMI glucose-free serum.
Time-lapse videos were recorded on an LSM880-inverted confocal microscope (Zeiss) equipped with an incubator to maintain 5% CO2, humidity, and a temperature of 37°C throughout the experiment. Images were taken using a 40× water plan apochromat LD objective (NA, 1.2). An Argon 488 laser line and two He/Ne lasers (543 and 633 nm) were used to simultaneously excite the three dyes. 3D Z-stacks of 15–20 μm were obtained by acquiring images every 0.5–0.7 µm.
Intravital microscopy experiments
For intravital imaging, sh#DSTYK LLC cells were transfected with pMAX EGFP and isolated by FACS. 1 × 106 LLC sh#DSTYK-EGFP cells were injected into the ears of hCD2RFP mice. 7 d after ear injection, sh#DSTYK was activated by adding doxycycline to drinking water for 24 h. Other tumors remained inactivated to maintain DSTYK expression. Then, intravital imaging in the mouse ears was performed. Anesthetized and temperature-controlled mice were placed on a custom-built stage and imaged with a LSM880-inverted microscope equipped with a 25× water immersion objective (NA, 0.8). Imaging sessions took from 2 to 4 h and time-lapse acquisitions in several regions of the tumor lasted 90 min with frames taken every 60 s. At least three mice were imaged per condition. Z-stacks were blindly selected according only to the presence of tumor cells. Videos were analyzed using IMARIS software, performing semi-automated surface-based segmentation to identify individual tumor cells, apoptotic tumor cells, and T cells. Time-lapse videos were generated with IMARIS software and edited with Final Cut Pro.
PLA
30,000 H2009 cells were plated in Lab-Tek Chambered coverglass (#155411; Thermo Fisher Scientific). After 24 h, cells were fixed and permeabilized 10 min with Triton X-100 0.5%. Primary antibodies were diluted to suitable concentration (1:500) and incubated overnight (Table S5).
Duolink In Situ PLA probes, MINUS mouse (#DUO92004) and PLUS rabbit (#DUO92002), and Duolink In situ Detection Reagents Red (#DUO92008) were obtained from Sigma-Aldrich. PLA assay was performed according to the manufacturer’s instructions.
Flow cytometry experiments
Mitochondrial mass and transmembrane potential were assessed by FACS staining. Parental or DSTYK KO H2009 tumor cells were cultured in complete media or glucose-free media overnight. Cells were then stained with Mitotracker Green FM 0.1 µM (#M7514; Thermo Fisher Scientific) and TMRM 125 ng/ml (#T668; Thermo Fisher Scientific) for 20 min at 37°C in RPMI 10% FBS. Cells were washed once with FACS buffer and analyzed in a Cytoflex S cytometer. Live cells were gated based on forward scatter (FSC) and side scatter (SSC) parameters and singlets were selected according to FSC-H, FSC-A parameters. Then, TMRM and Mitotracker Green fluorescence was assessed in the corresponding FITC and PE channels.
Mitochondrial respiration experiments
Mitochondrial respiration was measured using a Seahorse XFp Cell Mito Stress test kit (#103010-100; Agilent). Briefly, 15,000 cells of parental and KO LLC and 10,000 cells of the corresponding H2009 were seeded in triplicates in 8-well plates (#103022-100; Agilent). 24 h after seeding, cells were subjected to metabolic stress conditions through glucose or serum deprivation from 16 to 24 h. Plates were measured in a Seahorse XFp Analyzer (Agilent).
ROS measurement
120,000 cells were seeded in a 6-well plate, and after 24 h, a 50 µM DCFDA probe (#D6883; Sigma-Aldrich) was added for 30 min. Then, cells were trypsinized, pelleted, and resuspended in PBS. Fluorescence was measured in a Microplate Infinite Pro 200 reader (Tecan) in a black 96-well plate (#Rev. C/0815; PerkinElmer) at 485–530 nm (excitation-emission). Data were normalized by cell number.
Lysosomal pH measurement
20,000 cells were plated in black p96 plate with transparent bottom (#Rev. C/0815; PerkinElmer). A 2 mM LysoSensor Yellow/Blue dextran probe (#22460; Thermo Fisher Scientific) was added, and fluorescence was measured after 24 h in a Microplate Infinite Pro 200 reader (Tecan).
Proliferation assays
1,000 cells of parental and KO cells were seeded in XCelLigence plates in triplicates (#300600890; Agilent). Cells were treated with the corresponding treatment (i.e., either no glucose or no FBS) 24 h after seeding and measurements were taken every 6 min for 96 h. The area under the curve was calculated for data plotting.
T cell cytotoxicity experiments
Mouse CD8+ cells were isolated and purified from OTI mice spleens. OTI mice harbor a transgenic TCR recognizing the SIINFEKL peptide from ovalbumin on CD8+ T cells (InvivoGen; #vac-sin). Briefly, spleens were disaggregated in PBS and the cell suspension was filtered through a 40-µm cell strainer, erythrocytes were lysed with ACK lysis buffer (NH4Cl, KHCO3, EDTA Na.2H2O, pH 7.4) and after washing, cells were resuspended in autoMACS buffer (25 mM EDTA, 1% FBS, 1% P/E) and isolated from the spleens using the MACS CD8+ T Cell Isolation Kit (#130-095-236; Miltenyi Biotec) following the manufacturer’s instructions. CD8+ OTI cells were activated using CD3/CD28 activator beads (#11452D; Thermo Fisher Scientific) one bead/two cells and 30 U/ml of hIL-2 (#CN70389; Novartis).
15,000 DSTYK KO or control LLC cells were seeded in XCeLLigence E-Plates 16 PET (#300600890; Agilent). Tumor cells were then stimulated with 50 U/ml mouse IFN-γ overnight and then pulsed with SIINFEKL peptide (JPT; 1 ng/ml) for an additional 4 h. LLC cells were washed and activated CD8+ OTI T cells were added on top at different tumor cell/T cell ratios. Tumor cell growth and cytotoxicity were measured overtime in an xCELLigence Real-Time Cell Analysis MP instrument (Agilent) for 96 h.
To study the toxic effect of TNF-α on our cells, a similar procedure was performed. 15,000 LLC or 10,000 H2009 cells were seeded. 24 h after cell seeding, 20 ng/ml of mouse TNF-α (#259411; Abcam) or human TNF-α (#9642; Abcam) were added to LCC or H2009, respectively, and the toxicity effects were measured for 96 h. Etanercept (Enbrel; Pfizer) was used to block TNF-α.
To modulate the autophagy pathway, plated cells were treated with 25 μM of chloroquine (#C6628; Sigma-Aldrich) or 300 nM of rapamycin (#R8781; Sigma-Aldrich) and 4 h later, 20 ng/ml of mouse TNF-α (#259411; Abcam), or human TNF-α (#9642; Abcam) were added to corresponding wells.
In vivo studies
All animal experiments were conducted in accordance with the protocols approved by the institutional animal care committee (references 049-18, 035c-20, and 037-19).
1 × 106 LLC or 2 × 106 UN-SCC680 were subcutaneously inoculated in one flank of 8-wk-old immunocompetent female C57BL/6J or A/J mice, respectively (Harlan-Winkelmann).
shRNAs were activated by adding doxycycline to mice drinking water when tumors reached a volume of 50–100 mm3. Tumor volume (weight2 × length × 0.52) was measured with a digital caliper twice a week until sacrifice.
For tumor growth rate studies of mouse-derived cell lines, 3 × 106 LLC, UN-SCC679 or UN-SCC680 cells were subcutaneously inoculated in both flanks of 8-wk-old female Rag−/− IL2Rg−/− mice (Harlan-Winkelmann).
Depletion of CD8, CD4 or natural killer cells was performed by intraperitoneal injection of 100 μg anti-mouse CD8a (clone 2.43; Bio X Cell), CD4 (clone GK1.5; BioXCell) or NK1.1 (clone PK136; Bio X Cell), respectively, at days 6, 10, 14, 18, 22, 26, and 30 after cell inoculation.
For sh#Dstyk + anti–PD-1 combination treatment, LLC tumor–bearing mice were treated when tumors reached 75–100 mm3 with 200 μg anti–PD-1 blocking antibody (#RMP1-14; Bio X Cell) intraperitoneally at days 6, 9, 12, and 15.
LLC tumor–bearing mice were intraperitoneally treated with 40 mg of etanercept (Enbrel; Pfizer) to block TNF-α at days 0, 2, 4, and 6 after inoculation.
hCD2RFP mice, used for intravital microscopy experiments were a kind gift from Mark Coles from the University of York, York, UK, and OTI mice were purchased from Jackson. Ages of mice included in experiments ranged from 8 to 12 wk.
Patient samples
Two series of patients were used. A series of frozen material available from 45 patients with NSCLC whose tumors underwent surgical resection at CIMA-CUN from 2000 to 2012 was evaluated. The inclusion criteria were NSCLC histology, stage I to III, no neoadjuvant or adjuvant chemotherapy or radiotherapy, and absence of cancer within the 5 yr prior to lung cancer surgery. A series of 76 patients from Vall d’Hebron hospital (VH), previously reported (Frigola et al., 2022), diagnosed with advanced NSCLC and treated with ICIs as monotherapy. 54 (70.1%) had adenocarcinoma histology, 70 (90.1%) were current or former smokers, and 48 (62.3%) were males. 62 (80.5%) were treated with ICIs as monotherapy. 30 (39%) and 34 (44.2%) patients were treated with ICIs in their first or second line of therapy, respectively, whereas 13 (16.9%) patients were treated in ≥3 line.
Statistics
Sample size was chosen based on similar experiments previously published by the authors. When possible, all experiments were reproduced a minimum of three times (independent biological replicates). For comparing two groups, samples normality (Shapiro–Wilk test) and variance (Levene test) were explored. Groups with a normal distribution were compared using a two-tailed t test. Non-normal samples were analyzed using a Mann–Whitney test (equal variances) or a median test (unequal variances).
Online supplemental material
Fig. S1 shows the percentage of patients with DSTYK alteration from 24 different cancer types. Fig. S2 shows validation of IF in microvesicles as well as confirmation of different molecular pathways resulted from RNAseq and immunoprecipitation studies. Fig. S3 shows no alteration in mitochondrial respiration under baseline conditions in an altered DSTYK context. Fig. S4 shows mitochondrial morphological and functional features in an altered DSTYK context. Fig. S5 describes DSTYK KO cell lines behavior in vitro and in vivo and shows their response to different agents. Finally, Fig. S5 pictures the role of DSTYK amplification in a non-tumoral context and the role of DSTYK mutation in a tumoral context. Table S1 summarizes CIMA-CUN cohort clinical data. Table S2 shows differentially expressed genes in mouse cells in an altered DSTYK context. Table S3 shows a list of genes co-precipitated with Flag-DSTYK in an immunoprecipitation experiment using LLC cells. Table S4 shows Western blot densitometry analysis. Table S5 shows antibody references and dilutions as well as primers sequences used along the work. Video 1 shows mitochondrial and lysosomal dynamics in DSTYK KO cells under metabolic stress. Video 2 shows intravital microscopy of T cell responses in LLC DSTYK deficient tumors.
Supplementary Material
shows CIMA-CUN cohort clinical data summary.
shows differentially expressed genes in mouse cells in an altered DSTYK context.
lists genes co-precipitated with Flag-DSTYK in an immunoprecipitation experiment using LLC cells.
shows Western blot densitometry analysis.
shows antibody references and dilutions as well as primers sequences used along the work.
contains original blots for Fig. 1.
contains original blots for Fig. 2.
contains original blots for Fig. 3.
contains original blots for Fig. S2.
contains original blots for Fig. S5.
Acknowledgments
We thank Ana M. Aransay and Ana Rosa Cortázar for their bioinformatics analyses and support; and the Morphology and Imaging and animal care facilities at the Center for Applied Medical Research (CIMA) of the University of Navarra. We also acknowledge Diego Alignani, Tomás Aragón, Roberto Ferrero, Maria Eugenia Echarri, and Aroa Irigoyen (CIMALabs Diagnostics) for technical help. Illustrations were created with Biorender.com.
This work was supported by Fundación para la investigación médica aplicada (FIMA), Centro de Investigación Biomédica en Red de Cáncer (CIBERONC; CB16/12/00443), Spanish Association Against Cancer Scientific Foundation (AECC; GCB14-2170), Fundación Ramón Areces, Instituto de Salud Carlos III, and co-funded by the European Union (European Regional Development Fund, “A way to make Europe”; PI19/00098; PI19/00230; PI20/00419), Fundación Roberto Arnal Planelles, and International Association for the Study of Lung Cancer (IASLC) Fellowship funding (K. Valencia). M. Echepare was supported by Contratos Predoctorales de Formación en Investigación en Salud (PFIS), Instituto de Salud Carlos III, and co-funded by the European Union (European Social Fund, "Investing in your future"; FI20/00295).
Author contribution: K. Valencia and L.M. Montuenga conceived the experiments and supervised the work. C. Sainz, C. Bértolo, and K. Valencia performed the in vivo experiments. C. Bértolo, A. Pasquier, M. Echepare, and B. Picabea performed the in vitro experiments. A. Teijeira carried out the microscopy experiments. G. Bosco, R. Thomas, J. Frigola, and R. Amat performed the NGS analysis. I. Tamayo carried out the bioinformatic analysis. K. Valencia and A. Teijeira designed the experiments and analyzed their results. K. Valencia performed the statistical analysis. All authors critically reviewed the manuscript.
Data availability
RNAseq data are available on the Gene Expression Omnibus website under accession no. GSE212858.
References
- Ahmad, T., Aggarwal K., Pattnaik B., Mukherjee S., Sethi T., Tiwari B.K., Kumar M., Micheal A., Mabalirajan U., Ghosh B., et al. 2013. Computational classification of mitochondrial shapes reflects stress and redox state. Cell Death Dis. 4:e461. 10.1038/cddis.2012.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari, V., Hoey C., Liu L.Y., Lalonde E., Ray J., Livingstone J., Lesurf R., Shiah Y.J., Vujcic T., Huang X., et al. 2019. Molecular landmarks of tumor hypoxia across cancer types. Nat. Genet. 51:308–318. 10.1038/s41588-018-0318-2 [DOI] [PubMed] [Google Scholar]
- Bray, N.L., Pimentel H., Melsted P., and Pachter L.. 2016. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34:525–527. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Campbell, J.D., Alexandrov A., Kim J., Wala J., Berger A.H., Pedamallu C.S., Shukla S.A., Guo G., Brooks A.N., Murray B.A., et al. 2016. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat. Genet. 48:607–616. 10.1038/ng.3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canon, J., Rex K., Saiki A.Y., Mohr C., Cooke K., Bagal D., Gaida K., Holt T., Knutson C.G., Koppada N., et al. 2019. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 575:217–223. 10.1038/s41586-019-1694-1 [DOI] [PubMed] [Google Scholar]
- Chen, H., Lu C., Lin C., Li L., Wang Y., Han R., Hu C., and He Y.. 2021. VPS34 suppression reverses osimertinib resistance via simultaneously inhibiting glycolysis and autophagy. Carcinogenesis. 42:880–890. 10.1093/carcin/bgab030 [DOI] [PubMed] [Google Scholar]
- Cohen, P., Cross D., and Jänne P.A.. 2021. Kinase drug discovery 20 years after imatinib: Progress and future directions. Nat. Rev. Drug Discov. 20:551–569. 10.1038/s41573-021-00195-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaprico, A., Silva T.C., Olsen C., Garofano L., Cava C., Garolini D., Sabedot T.S., Malta T.M., Pagnotta S.M., Castiglioni I., et al. 2016. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 44:e71. 10.1093/nar/gkv1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuny, G.D., and Degterev A.. 2020. RIPK protein kinase family: Atypical lives of typical kinases. Semin. Cell Dev. Biol. 109:96–105. 10.1016/j.semcdb.2020.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foth, M., and McMahon M.. 2021. Autophagy inhibition in braf-driven cancers. Cancers. 13:3498. 10.3390/cancers13143498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigola, J., Carbonell C., Irazno P., Pardo N., Callejo A., Cedres S., Martinez-Marti A., Navarro A., Soleda M., Jimenez J., et al. 2022. High levels of chromosomal aberrations negatively associate with benefit to checkpoint inhibition in NSCLC. J. Immunother. Cancer. 10:e004197. 10.1136/jitc-2021-004197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, M.J., Herbst R.S., and Goldberg S.B.. 2021. Selecting the optimal immunotherapy regimen in driver-negative metastatic NSCLC. Nat. Rev. Clin. Oncol. 18:625–644. 10.1038/s41571-021-00520-1 [DOI] [PubMed] [Google Scholar]
- Gremke, N., Polo P., Dort A., Schneikert J., Elmshäuser S., Brehm C., Klingmüller U., Schmitt A., Reinhardt H.C., Timofeev O., et al. 2020. mTOR-mediated cancer drug resistance suppresses autophagy and generates a druggable metabolic vulnerability. Nat. Commun. 11:4684. 10.1038/s41467-020-18504-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y., Cao R., Zhang X., Huang L., Sun L., Zhao J., Ma J., and Han C.. 2019. Recent progress in rare oncogenic drivers and targeted therapy for non-small cell lung cancer. Onco Targets Ther. 12:10343–10360. 10.2147/OTT.S230309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Györffy, B., Lanczky A., Eklund A.C., Denkert C., Budczies J., Li Q., and Szallasi Z.. 2010. An online survival analysis tool to rapidly assess the effect of 22, 277 genes on breast cancer prognosis using microarray data of 1, 809 patients. Breast Cancer Res. Treat. 123:725–731. 10.1007/s10549-009-0674-9 [DOI] [PubMed] [Google Scholar]
- Hao, C., Liu G., and Tian G.. 2019. Autophagy inhibition of cancer stem cells promotes the efficacy of cisplatin against non-small cell lung carcinoma. Ther. Adv. Respir. Dis. 13:1753466619866097. 10.1177/1753466619866097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann, M.D., Paz-Ares L., Bernabe Caro R., Zurawski B., Kim S.-W., Carcereny Costa E., Park K., Alexandru A., Lupinacci L., de la Mora Jimenez E., et al. 2019. Nivolumab plus ipilimumab in advanced non–small-cell lung cancer. N. Engl. J. Med. 381:2020–2031. 10.1056/nejmoa1910231 [DOI] [PubMed] [Google Scholar]
- Herbst, R.S., Giaccone G., de Marinis F., Reinmuth N., Vergnenegre A., Barrios C.H., Morise M., Felip E., Andric Z., Geater S., et al. 2020. Atezolizumab for first-line treatment of PD-L1–selected patients with NSCLC. N. Engl. J. Med. 383:1328–1339. 10.1056/nejmoa1917346 [DOI] [PubMed] [Google Scholar]
- Horn, L., Mansfield A.S., Szczęsna A., Havel L., Krzakowski M., Hochmair M.J., Huemer F., Losonczy G., Johnson M.L., Nishio M., et al. 2018. First-line atezolizumab plus chemotherapy in extensive-stage small-cell lung cancer. N. Engl. J. Med. 379:2220–2229. 10.1056/nejmoa1809064 [DOI] [PubMed] [Google Scholar]
- Jordan, E.J., Kim H.R., Arcila M.E., Barron D., Chakravarty D., Gao J., Chang M.T., Ni A., Kundra R., Jonsson P., et al. 2017. Prospective comprehensive molecular characterization of lung adenocarcinomas for efficient patient matching to approved and emerging therapies. Cancer Discov. 7:596–609. 10.1158/2159-8290.CD-16-1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny, T.C., Craig A.J., Villanueva A., and Germain D.. 2019. Mitohormesis primes tumor invasion and metastasis. Cell Rep. 27:2292–2303.e6. 10.1016/j.celrep.2019.04.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, T., Terai H., and Yaguchi T.. 2021. Clinical efficacy and future prospects of immunotherapy in lung cancer. Life. 11:1029. 10.3390/life11101029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, H.C., Baglini C.V., Lope A.L., Parkhitko A.A., Liu H.J., Alesi N., Malinowska I.A., Ebrahimi-Fakhari D., Saffari A., Yu J.J., et al. 2017. P62/SQSTM1 cooperates with hyperactive mTORC1 to regulate glutathione production, maintain mitochondrial integrity, and promote tumorigenesis. Cancer Res. 77:3255–3267. 10.1158/0008-5472.CAN-16-2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson, K.A., Sousa C.M., Zhang X., Kim E., Akthar R., Caumanns J.J., Yao Y., Mikolajewicz N., Ross C., Brown K.R., et al. 2020. Functional genomic landscape of cancer-intrinsic evasion of killing by T cells. Nature. 586:120–126. 10.1038/s41586-020-2746-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.Y.W., Hsu C.K., Michael M., Nanda A., Liu L., McMillan J.R., Pourreyron C., Takeichi T., Tolar J., Reid E., et al. 2017. Large intragenic deletion in DSTYK underlies autosomal-recessive complicated spastic paraparesis, SPG23. Am. J. Hum. Genet. 100:364–370. 10.1016/j.ajhg.2017.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Yang B., Zhou Q., Wu Y., Shang D., Guo Y., Song Z., Zheng Q., and Xiong J.. 2013. Autophagy promotes hepatocellular carcinoma cell invasion through activation of epithelial-mesenchymal transition. Carcinogenesis. 34:1343–1351. 10.1093/carcin/bgt063 [DOI] [PubMed] [Google Scholar]
- Li, K., Liu J.W., Zhu Z.C., Wang H.T., Zu Y., Liu Y.J., Yang Y.H., Xiong Z.Q., Shen X., Chen R., et al. 2014. DSTYK kinase domain ablation impaired the mice capabilities of learning and memory in water maze test. Int. J. Clin. Exp. Pathol. 7:6486–6492 [PMC free article] [PubMed] [Google Scholar]
- Luo, D., Hu S., Tang C., and Liu G.. 2018. Mesenchymal stem cells promote cell invasion and migration and autophagy-induced epithelial-mesenchymal transition in A549 lung adenocarcinoma cells. Cell Biochem. Funct. 36:88–94. 10.1002/cbf.3320 [DOI] [PubMed] [Google Scholar]
- Malhotra, J., Jabbour S., Orlick M., Riedlinger G., Guo Y., White E., and Aisner J.. 2019. Phase Ib/II study of hydroxychloroquine in combination with chemotherapy in patients with metastatic non-small cell lung cancer (NSCLC). Cancer Treat. Res. Commun. 21:100158. 10.1016/j.ctarc.2019.100158 [DOI] [PubMed] [Google Scholar]
- Marinković, M., Šprung M., Buljubašić M., and Novak I.. 2018. Autophagy modulation in cancer: Current knowledge on action and therapy. Oxid. Med. Cell. Longev. 2018:8023821. 10.1155/2018/8023821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan, E., and Tschopp J.. 2005. The RIP kinases: Crucial integrators of cellular stress. Trends Biochem. Sci. 30:151–159. 10.1016/j.tibs.2005.01.003 [DOI] [PubMed] [Google Scholar]
- Momcilovic, M., Jones A., Bailey S.T., Waldmann C.M., Li R., Lee J.T., Abdelhady G., Gomez A., Holloway T., Schmid E., et al. 2019. In vivo imaging of mitochondrial membrane potential in non-small-cell lung cancer. Nature. 575:380–384. 10.1038/s41586-019-1715-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscat, J., and Diaz-Meco M.T.. 2009. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 137:1001–1004. 10.1016/j.cell.2009.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam, H.J. 2021. Autophagy modulators in cancer: Focus on cancer treatment. Life. 11:839. 10.3390/life11080839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nokin, M.J., Ambrogio C., Nadal E., and Santamaria D.. 2021. Targeting infrequent driver alterations in non-small cell lung cancer. Trends Cancer. 7:410–429. 10.1016/j.trecan.2020.11.005 [DOI] [PubMed] [Google Scholar]
- Ogbu, S.C., Rojas S., Weaver J., Musich P.R., Zhang J., Yao Z.Q., and Jiang Y.. 2021. DSTYK Enhances chemoresistance in triple-Negative breast cancer cells. Cells. 11:97. 10.3390/cells11010097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., Dong W., Chen Y., Mo R., Cheng J.F., Hui C.C., Mohandas N., and Huang C.H.. 2006. Dusty protein kinases: Primary structure, gene evolution, tissue specific expression and unique features of the catalytic domain. Biochim. Biophys. Acta. 1759:562–572. 10.1016/j.bbaexp.2006.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilleron, S., Sarfati D., Janssen-Heijnen M., Vignat J., Ferlay J., Bray F., and Soerjomataram I.. 2019. Global cancer incidence in older adults, 2012 and 2035: A population-based study. Int. J. Cancer. 144:49–58. 10.1002/ijc.31664 [DOI] [PubMed] [Google Scholar]
- Pimentel, H., Bray N.L., Puente S., Melsted P., and Pachter L.. 2017. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods. 14:687–690. 10.1038/nmeth.4324 [DOI] [PubMed] [Google Scholar]
- Porporato, P.E., Filigheddu N., Pedro J.M.B., Kroemer G., and Galluzzi L.. 2018. Mitochondrial metabolism and cancer. Cell Res. 28:265–280. 10.1038/cr.2017.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck, M., Rodríguez-Abreu D., Robinson A.G., Hui R., Csőszi T., Fülöp A., Gottfried M., Peled N., Tafreshi A., Cuffe S., et al. 2016. Pembrolizumab versus chemotherapy for PD-L1–positive non–small-cell lung cancer. N. Engl. J. Med. 375:1823–1833. 10.1056/nejmoa1606774 [DOI] [PubMed] [Google Scholar]
- Redin, E., Garmendia I., Lozano T., Serrano D., Senent Y., Redrado M., Villalba M., De Andrea C.E., Exposito F., Ajona D., et al. 2021. SRC family kinase (SFK) inhibitor dasatinib improves the antitumor activity of anti-PD-1 in NSCLC models by inhibiting Treg cell conversion and proliferation. J. Immunother. Cancer. 9:e001496. 10.1136/jitc-2020-001496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roczniak-Ferguson, A., Petit C.S., Froehlich F., Qian S., Ky J., Angarola B., Walther T.C., and Ferguson S.M.. 2012. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 5:ra42. 10.1126/scisignal.2002790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colhado Rodrigues, B.L., Lallo M.A., and Perez E.C.. 2020. The controversial role of autophagy in tumor development: A systematic review. Immunol. Invest. 49:386–396. 10.1080/08820139.2019.1682600 [DOI] [PubMed] [Google Scholar]
- Sanna-Cherchi, S., Sampogna R.V., Papeta N., Burgess K.E., Nees S.N., Perry B.J., Choi M., Bodria M., Liu Y., Weng P.L., et al. 2013. Mutations in DSTYK and dominant urinary tract malformations. N. Engl. J. Med. 369:621–629. 10.1056/NEJMoa1214479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schläfli, A.M., Tokarchuk I., Parejo S., Jutzi S., Berezowska S., Engedal N., and Tschan M.P.. 2021. ALK inhibition activates LC3B-independent, protective autophagy in EML4-ALK positive lung cancer cells. Sci. Rep. 11:9011. 10.1038/s41598-021-87966-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel, R.L., Miller K.D., and Jemal A.. 2020. Cancer statistics, 2020. CA A Cancer J. Clin. 70:7–30. 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- Sun, X., Zhou Y., Zhang R., Wang Z., Xu M., Zhang D., Huang J., Luo F., Li F., Ni Z., et al. 2020. Dstyk mutation leads to congenital scoliosis-like vertebral malformations in zebrafish via dysregulated mTORC1/TFEB pathway. Nat. Commun. 11:479. 10.1038/s41467-019-14169-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., and Bray F.. 2021. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 71:209–249. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- Tang, G., Yang Y., Shang L., Jun F., and Liu Q.. 2019. A DSTYK mutation activates ERK1/2 signaling to promote intraspinal dissemination in a case of solitary fibrous tumor/hemangiopericytoma. Lab. Invest. 99:1501–1514. 10.1038/s41374-019-0274-1 [DOI] [PubMed] [Google Scholar]
- Uramoto, H., and Tanaka F.. 2014. Recurrence after surgery in patients with NSCLC. Transl. Lung Cancer Res. 3:242–249. 10.3978/j.issn.2218-6751.2013.12.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia, K., Sainz C., Bértolo C., de Biurrun G., Agorreta J., Azpilikueta A., Larrayoz M.J., Bosco G., Zandueta C., Redrado M., et al. 2021. Two alternative cell line models for the study of multiorganic metastasis and immunotherapy in Lung Squamous Cell Carcinoma. Dis. Model. Mech. 15:dmm049137. 10.1242/dmm.049137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo, A., Perurena N., Guruceaga E., Mazur P.K., Martinez-Canarias S., Zandueta C., Valencia K., Arricibita A., Gwinn D., Sayles L.C., et al. 2017. An integrative approach unveils FOSL1 as an oncogene vulnerability in KRAS-driven lung and pancreatic cancer. Nat. Commun. 8:14294. 10.1038/ncomms14294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneman, M., and Dranoff G.. 2012. Combining immunotherapy and targeted therapies in cancer treatment. Nat. Rev. Cancer. 12:237–251. 10.1038/nrc3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidic, C., Zaniew M., Jurga S., Thiele H., Reutter H., and Hilger A.C.. 2021. Exome sequencing implicates a novel heterozygous missense variant in DSTYK in autosomal dominant lower urinary tract dysfunction and mild hereditary spastic paraparesis. Mol. Cell. Pediatr. 8:13. 10.1186/s40348-021-00122-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Gong M., Fan X., Huang D., Zhang J., and Huang C.. 2022. Autophagy-related signaling pathways in non-small cell lung cancer. Mol. Cell. Biochem. 477:385–393. 10.1007/s11010-021-04280-5 [DOI] [PubMed] [Google Scholar]
- Wang, M., Herbst R.S., and Boshoff C.. 2021b. Toward personalized treatment approaches for non-small-cell lung cancer. Nat. Med. 27:1345–1356. 10.1038/s41591-021-01450-2 [DOI] [PubMed] [Google Scholar]
- Wang, Z., Zhou C., and Yang S.. 2022. The roles, controversies, and combination therapies of autophagy in lung cancer. Cell Biol. Int. 46:3–11. 10.1002/cbin.11704 [DOI] [PubMed] [Google Scholar]
- Xiao, M., Benoit A., Hasmim M., Duhem C., Vogin G., Berchem G., Noman M.Z., and Janji B.. 2021. Targeting cytoprotective autophagy to enhance anticancer therapies. Front. Oncol. 11:626309. 10.3389/fonc.2021.626309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, K., Venida A., Yano J., Biancur D.E., Kakiuchi M., Gupta S., Sohn A.S.W., Mukhopadhyay S., Lin E.Y., Parker S.J., et al. 2020. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature. 581:100–105. 10.1038/s41586-020-2229-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, T.M., Reyes C., Pasnikowski E., Castanaro C., Wong C., Decker C.E., Chiu J., Song H., Wei Y., Bai Y., et al. 2020. Autophagy protects tumors from T cell-mediated cytotoxicity via inhibition of TNFα-induced apoptosis. Sci. Immunol. 5:eabb9561. 10.1126/sciimmunol.abb9561 [DOI] [PubMed] [Google Scholar]
- Zhang, J., Wang X., Vikash V., Ye Q., Wu D., Liu Y., and Dong W.. 2016. ROS and ROS-mediated cellular signaling. Oxid. Med. Cell. Longev. 2016:4350965. 10.1155/2016/4350965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Miller Z., Musich P.R., Thomas A.E., Yao Z.Q., Xie Q., Howe P.H., and Jiang Y.. 2020. DSTYK promotes metastasis and chemoresistance via EMT in colorectal cancer. Front. Pharmacol. 11:1250. 10.3389/fphar.2020.01250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, C., Chen M., Chen Y., Yao F., and Fang W.. 2021. Loss of DSTYK activates Wnt/β-catenin signaling and glycolysis in lung adenocarcinoma. Cell Death Dis. 12:1122. 10.1038/s41419-021-04385-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, H., Wang D., Zhang L., Xie X., Wu Y., Liu Y., Shao G., and Su Z.. 2014. Upregulation of autophagy by hypoxia-inducible factor-1α promotes EMT and metastatic ability of CD133+ pancreatic cancer stem-like cells during intermittent hypoxia. Oncol. Rep. 32:935–942. 10.3892/or.2014.3298 [DOI] [PubMed] [Google Scholar]
- Zorova, L.D., Popkov V.A., Plotnikov E.Y., Silachev D.N., Pevzner I.B., Jankauskas S.S., Babenko V.A., Zorov S.D., Balakireva A.V., Juhaszova M., et al. 2018. Mitochondrial membrane potential. Anal. Biochem. 552:50–59. 10.1016/j.ab.2017.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows CIMA-CUN cohort clinical data summary.
shows differentially expressed genes in mouse cells in an altered DSTYK context.
lists genes co-precipitated with Flag-DSTYK in an immunoprecipitation experiment using LLC cells.
shows Western blot densitometry analysis.
shows antibody references and dilutions as well as primers sequences used along the work.
contains original blots for Fig. 1.
contains original blots for Fig. 2.
contains original blots for Fig. 3.
contains original blots for Fig. S2.
contains original blots for Fig. S5.
Data Availability Statement
RNAseq data are available on the Gene Expression Omnibus website under accession no. GSE212858.