Abstract
Since the 20th century, humans have lived through five pandemics caused by influenza A viruses (IAVs) (H1N1/1918, H2N2/1957, H3N2/1968, and H1N1/2009) and the coronavirus (CoV) severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). IAVs and CoVs both have broad host ranges and share multiple hosts. Virus co-circulation and even co-infections facilitate genetic reassortment among IAVs and recombination among CoVs, further altering virus evolution dynamics and generating novel variants with increased cross-species transmission risk. Moreover, SARS-CoV-2 may maintain long-term circulation in humans as seasonal IAVs. Co-existence and co-infection of both viruses in humans could alter disease transmission patterns and aggravate disease burden. Herein, we demonstrate how virus-host ecology correlates with the co-existence and co-infection of IAVs and/or CoVs, further affecting virus evolution and disease dynamics and burden, calling for active virus surveillance and countermeasures for future public health challenges.
Keywords: SARS-CoV-2, co-infection, influenza A virus, coronavirus, co-existence, public health challenges
Graphical abstract
Public summary
-
•
Influenza A viruses (IAVs) and coronaviruses (CoVs) have broad host ranges and share multiple hosts
-
•
Co-existence and co-infection of IAVs and/or CoVs are inevitable based on virus-host ecology
-
•
Co-circulation and co-infection could alter virus evolution and drive novel variant emergence
-
•
Co-circulation and co-infection could affect disease transmission and burden in humans
-
•
Active surveillance and countermeasures are necessary for the public health challenges
Introduction
Five pandemics have been documented in history since the 20th century. The first four pandemics were caused by influenza A virus (IAV) H1N1 (in 1918 and 2009), H2N2 (1957), and H3N2 (1968) subtypes, which led to substantial economic losses and severe social panic.1,2 After the pandemics, H1N1 and H3N2 subtype IAVs have become seasonal influenza viruses and currently cause annual epidemics in humans. Notably, avian influenza viruses (AIVs) contributed gene segments to the genesis of causative pathogens for each influenza pandemic. In addition, sporadic human cases infected by AIVs, including H3, H5, H7, H9, and H10 subtypes, were also reported.3,4 The seasonal influenza epidemics and occasional zoonotic influenza infections in humans have become a significant disease burden of great concern globally.
Especially, the current coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has led to unprecedented challenges to public health and devastating economic losses. As of July 21, 2022, WHO reported 564.12 million confirmed cases of COVID-19, including about 6.37 million deaths in over 200 countries and regions (https://covid19.who.int). Although a series of stringent non-pharmaceutical and pharmaceutical interventions have been implemented to contain the spread of the disease,5, 6, 7 the pandemic has not been brought under control. Moreover, the emerging novel variant, Omicron, has caused a rapid resurgence of cases worldwide since late 2021 (https://covid19.who.int).8 Hence, SARS-CoV-2 may become a long-term problem for humans, similar to seasonal influenza. Furthermore, another six human coronaviruses (HCoVs) are known to infect humans. The HCoVs HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU1 are circulating around the world and typically result in mild diseases, with symptoms resembling the common cold.9 In addition, SARS-CoV and Middle East respiratory syndrome CoV (MERS-CoV) can cause severe respiratory disease in humans. SARS-CoV has vanished in the human population after its sudden emergence in 2002, but MERS-CoV still circulates between dromedary camels and humans in the Middle East region.10, 11, 12
Etiological pathogens responsible for past pandemics could continue to plague humans even after the declared end of the pandemic. Given the virus-host ecology of IAVs and CoVs, multiple influenza subtypes and CoVs could simultaneously circulate in animals and humans and even co-infect one host.13, 14, 15, 16 This review will describe the virus-host ecology and host range for IAVs and CoVs and highlight the inevitable co-existence and co-infection of multiple IAVs or CoVs or IAVs and CoVs in one host. Secondly, the co-infection of different IAV subtypes or multiple CoV lineages or two types of viruses could alter the genetic evolution dynamics, molecular characteristics, and related phenotypes for both viruses. Lastly, co-existence and co-infection of IAVs and SARS-CoV-2 in humans have been reported, which could alter disease transmission dynamics and increase disease burden.17, 18, 19 The cell tropism20, 21, 22 for viruses and respective receptors could be a mechanism for co-infection. Hence, global cooperation on preparedness and response to the current and next pandemics or disease outbreaks comes into focus.
Ecology of IAVs and CoVs
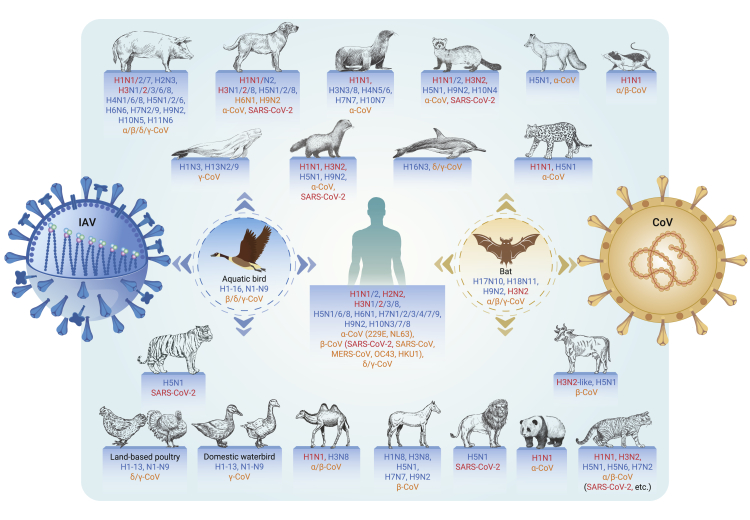
Typically, wild waterfowls of the order Anseriformes (especially the family Anatidae, eg, ducks, geese, and swans) and Charadriiformes (mainly gulls and shorebirds) are considered the natural reservoirs for IAVs.16 The H1-H16 and N1-N9 influenza subtypes have all been isolated in waterfowls (Figure 1; Table S1), which are usually asymptomatic.16,23,24 In addition, the H17N10 and H18N11 subtype influenza viruses have been identified from bats.25,26 Of note, the recent increasing outbreaks in aquatic birds with significant morbidity and mortality caused by highly pathogenic AIVs (HPAIVs) H5N1, H5N8, and H5N6 have been reported27, 28, 29, 30, 31, 32, 33, 34 after the first identification of H5N1 outbreak among migratory birds at Qinghai Lake in 2005.35,36
Figure 1.
The shared host species of influenza A viruses and coronaviruses
The reservoir hosts for influenza A viruses (IAVs; aquatic birds) and multiple coronaviruses (CoVs; bats) are highlighted by a dashed circle in blue and orange color, respectively. Dominant IAVs and CoVs isolated in each host species are listed in the text next to the stick figure. Names of IAVs and CoVs are colored in blue and orange, respectively. The same subtypes of IAVs and CoVs related to the pandemics are colored in red.
In addition, IAVs circulating in waterfowls often jump to infect domestic poultry and occasionally mammalian species.16 Seasonal influenza H1N1 and H3N2 viruses have been well adapted to and maintained in humans, resulting in annual recurrence of seasonal epidemics in the temperate zones and year-round circulation in the tropical zone.37 Moreover, multiple IAVs (eg, H9N2, H5N1, and H5N6 AIVs) have persisted in poultry for a long time and have evolved into different genetic lineages.38,39 Notably, at least 15 AIV subtypes (H1N2, H3N8, H5N1, H5N6, H5N8, H6N1, H7N2, H7N3, H7N4, H7N7, H7N9, H9N2, H10N3, H10N7, and H10N8) could sporadically overcome species barriers to cause human infections directly (https://www.who.int/teams/global-influenza-programme/avian-influenza).3,4 Of grave concern are H5 and H7 AIVs due to their persistence in poultry and occasional human infections. Several IAV subtypes have also been identified in other animals,40, 41, 42 such as H5 in captive tigers and lions40 and domestic pigs and H4, H13, and H16 in sea animals.41 Moreover, some IAVs persist in several animals, such as the Eurasian avian-like H1N1 virus in pigs, H3N8 in horses, and H3N2 in dogs (Figure 1).41,43,44
The host range for CoVs is also as broad as influenza, ranging from mammals and reptiles to rodents and birds (Figure 1). Bats may be the major natural reservoirs for alpha- and beta-CoVs.45,46 Regarding HCoVs, bats are likely the natural hosts for SARS-CoV-2, SARS-CoV, MERS-CoV, HCoV-NL63, and HCoV-229E; rodents are speculated to be the natural hosts for HCoV-OC43 and HCoV-HKU1.45 During the genetic origin tracing for SARS-CoV-2, multiple SARS-CoV-2-related CoVs have been identified from horseshoe bats (Rhinolophus) in Southeast Asian regions13,47,48 and in Malayan pangolins (Manis javanica) smuggled into southern China.14,49 The genetically closest SARS-CoV-2-related CoV was identified in Rhinolophus bats from Laos with up to 96.8% sequence identity compared with the whole-genome sequence of SARS-CoV-2.50 Live SARS-CoV-2 virus has not been isolated from these bats to date. However, animal infections with SARS-CoV-2 have been reported in 14 species, including cats, dogs, minks, otters, pet ferrets, tigers, lions, snow leopards, pumas, gorillas, white-tailed deer, fishing cats, binturongs, and South American coatis (https://www.oie.int/en/what-we-offer/emergency-and-resilience/covid-19/). Of note, SARS-CoV-2 caused mink infections in farms, and the mink-derived mutant was found to be transmitted back to humans, highlighting the animal-to-human transmission risk for SARS-CoV-2.51,52 Experimentally, SARS-CoV-2 can infect hamsters, ferrets, dogs, rhesus macaques (Macaca mulatta), cynomolgus monkeys (Macaca fascicularis), and African green monkeys (Chlorocebus sabaeus).15,53, 54, 55, 56 The potential host range of SARS-CoV-2 has also been estimated according to the binding ability between SARS-CoV-2 and angiotensin-converting enzyme 2 (ACE2) receptors of various species.57
Given the overlapping ecology of IAVs and CoVs, multiple influenza subtypes and CoV variants co-circulate in wild and domestic animals and humans,9,13, 14, 15, 16 which undoubtedly increases the probabilities of co-infections among different IAVs, different CoVs, or both IAVs and CoVs in one host. The co-infections may alter the genetic evolution trajectory of viruses and facilitate mutations related to cross-species transmission and adaption to humans. Moreover, the potentially long-term co-existence and co-infections of IAVs and SARS-CoV-2 in humans could aggravate the disease burden compared with independent infections.
Genetic evolution and molecular characteristics
Phylogenetic dynamics for IAVs and SARS-CoV-2
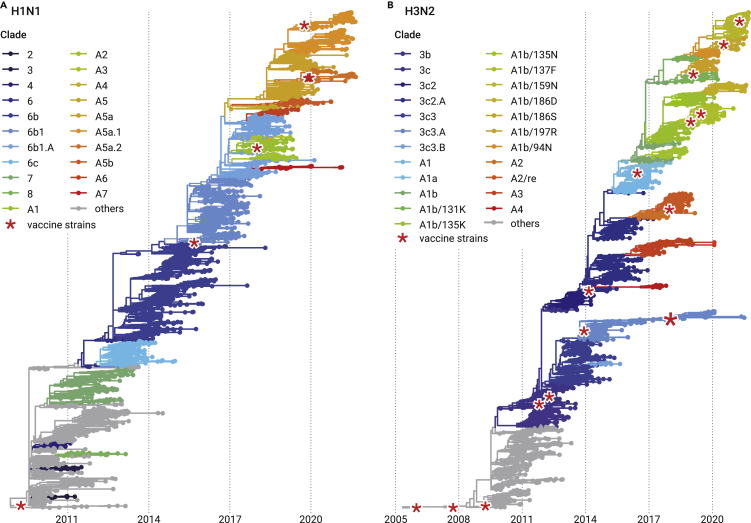
IAVs (eg, H1, H3, H5, and H7) have experienced rapid evolution and established divergent lineages and genotypes. Phylogenies of hemagglutinin (HA) genes of seasonal influenza H1N1 and H3N2 viruses exhibit ladder-like tree topologies and constant genetic diversity within lineages across time, and some lineages persist from one influenza season through to the next years.58, 59, 60 Given the genetic divergence and phylogeny relationship of HA genes, global H1N1 and H3N2 viruses can be divided into different clades (Figure 2).61,62 Of multiple co-existent clades, H1N1 clade A5a and H3N2 clade A1b are the currently dominant clades circulating the world (Figure 2; https://nextstrain.org/flu/seasonal/).62,63 Regarding H5, H7, and H9 AIVs of public health concern, H5 influenza was designated into multiple clades by the WHO/OIE/FAO H5N1 Evolution Working Group according to the phylogenetic topology and genetic divergence of HA genes.64 Two main lineages of novel H7N9 have been established based on the HA phylogeny relationship, the Yangtze River Delta lineage and the Pearl River Delta lineage, since its emergence in 2013.65,66 At least three HA clades of H9 AIVs are co-circulating among poultry.39 Moreover, new subclades or lineages are gradually emerging as the evolution of H5, H7, and H9 AIVs.38,39
Figure 2.
Time-scaled phylogenies of global H1N1 and H3N2 seasonal IAVs
(A) Time-scaled phylogeny of H1N1 seasonal IAVs around the globe. Current seasonal influenza H1N1 viruses derivate from the A(H1N1)2009 pandemic strains and completely replaced the seasonal H1N1 circulating before 2009 with different genetic and antigenic characteristics.
(B) Time-scaled phylogeny of H3N2 seasonal IAVs around the globe. The tips in the tree are colored by the clade classification. The red asterisks represent the vaccine strains. The ladder-like evolution dynamics and seasonal epidemics of H1N1 and H3N2 are, to a large extent, attributed to their partial cross-immunity and the acute and short infectious period. Figures are reannotated from Nextstrain (https://nextstrain.org/influenza/, adapted from and courtesy of Creative Commons Attribution Licensing).
A dynamic nomenclature system has been proposed for the expanding phylogenetically divergent SARS-CoV-2 viruses (https://cov-lineages.org/lineage_list.html).67 The phylogeny of global SARS-CoV-2 can be grouped into two main lineages: lineage A and lineage B. Lineage A is a minor group and shares two nucleotides, at 8782 nt of ORF1ab and 28144 nt of ORF8, with the genetically close SARS-CoV-2-related bat CoVs RaTG13 and RmYN02.13,47 Lineage B includes the currently persistent and dominant SARS-CoV-2 variants. Based on the increased risk of SARS-CoV-2 variants to public health and corresponding biological and clinical features, WHO designated the variants of concern (VOC) and variants of interest (VOI) using the Greek alphabet (https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/). On July 14, 2022, the VOC group includes Omicron (B.1.1.529) SARS-CoV-2 variants, and previously circulating VOCs, Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), and Delta (B.1.617.2), have been removed from this group. The VOCs and VOIs can be reclassified given the circulation, epidemiological situation, and biological properties of corresponding variants. In addition, the within host diversity of SARS-CoV-2 is relatively low, with narrow transmission bottlenecks at high viral loads (at least at the early infection), but the transmitted SARS-CoV-2 variant could spread rapidly.68
The IAV and SARS-CoV-2 underwent genetic diversity and relatively rapid evolution dynamics. The partial cross-immunity and the acute and short infectious period, to a large extent, facilitate the evolutionary dynamics and seasonal epidemics of human H1N1 and H3N2 IAVs.69 Of note, the vaccine breakthrough infection and reinfection with SARS-CoV-270,71 mean that vaccine-induced and natural immunity are not enough to protect humans from virus infection, especially for the current variants, suggesting probably long-term co-circulation of SARS-CoV-2 with seasonal influenza viruses. In addition, the rapid evolution with antigenic changes and global persistence of seasonal influenza require the intensive surveillance of influenza activity and formulation of well-matched vaccines before each annual influenza season.72 These valuable lessons and experiences should be learned to control the current pandemic and possibly annual SARS-CoV-2 epidemics in the future.
Genetic reassortment and recombination in IAVs and HCoVs
Virus ecology affects the genetic evolution of IAVs and CoVs. When co-infection of multiple IAVs or CoVs happens in the same host, genetic reassortment among IAVs and recombination among CoVs potentially occur and further facilitate virus evolution and novel variant emergence. To our knowledge, the genetic interactions between IAVs and HCoVs have not yet been documented, while the potential recombination between the two types of viruses might also occur during their co-infections in one host.
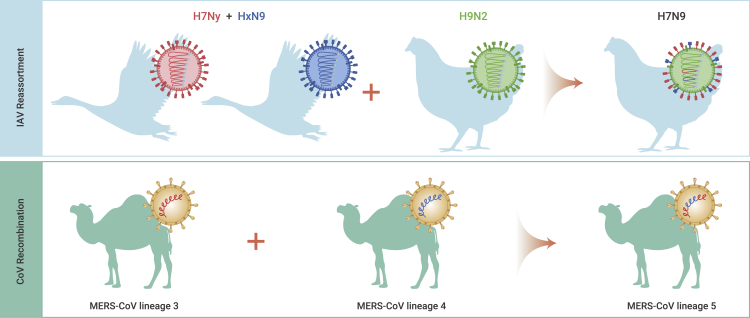
IAV is an enveloped virus with a negative-sense, RNA-segmented genome that can encode for more than 17 proteins.73 The segmented genome drives the exchange of gene segments between IAVs (genetic reassortment) when they simultaneously infect the same host or cell.74 Reassortment facilitates the formation of novel influenza variants with new genomic constellations. Moreover, the reassortant virus could obtain fitness advantage, cross-species transmission, evasion from host immune responses, and even cause pandemics/epidemics in humans.16 Since the 20th century, at least three of four influenza pandemics were caused by reassortants: the 1957/H2N2 virus (HA, NA, and PB1 from AIV, the other five genes from human IAV), the 1968/H3N2 virus (HA and PB1 from AIV, the other six genes from human IAV), and the 2009/H1N1 virus (PB2 and PA from AIV, PB1 from human IAV, and others from swine IAV). Notably, the novel H7N9 AIVs also emerged by reassortment in 2013 and have caused five infection waves in humans.65,75 The internal genes of H7N9 originate from H9N2 AIVs that adapted well in chickens and H7 and N9 genes from viruses found in aquatic birds (Figure 3). Later, the novel H7N9 strain evolved into more genotypes by further reassortments with diverse H9N2 variants and other AIV subtypes.66,76
Figure 3.
Reassortment and recombination facilitate the emergence of novel variants for IAVs and CoVs
The novel H7N9 avian influenza virus (AIV) was found to have emerged by reassortment, obtaining the HA and NA genes from viruses circulating in waterfowls and the internal genes from H9N2 AIV circulating in chickens. MERS-CoV lineage 5 was generated by the genetic recombination between lineage 3 and 4 strains.
CoV is also an enveloped virus but carries a large, positive-sense, single-stranded RNA genome.77 The common mutations and recombination in the positive-strand RNA viruses with the largest genome contribute to genetic divergence and novel CoV variant emergence.9,78,79 Following co-infection with more than one CoVs, recombination may occur during virus replication when multiple subgenomic RNAs are generated, and genetically related genes are readily recombinant among different CoVs by template switching.80 Genetic recombination has been reported in human and animal CoVs, but the recombination breakpoints are commonly random.9,12,81,82 In addition, the novel recombinant CoVs could lead to cross-species transmission and outbreaks in humans and/or animals.12,81,82 The MERS-CoV lineage 5 is a recombinant between lineage 3 and 4 and caused human outbreaks in Saudi Arabia in 2015 (Figure 3).12 The SARS-CoV TOR2 strain might originate from recombination among SARS-CoV, alpha CoV (HCoV-229E), and gamma CoV (avian infectious bronchitis virus).81 Further, genetic evidence13,14,47, 48, 49 suggests that SARS-CoV-2 is of natural origin by genetic recombination and is related to bat CoVs.
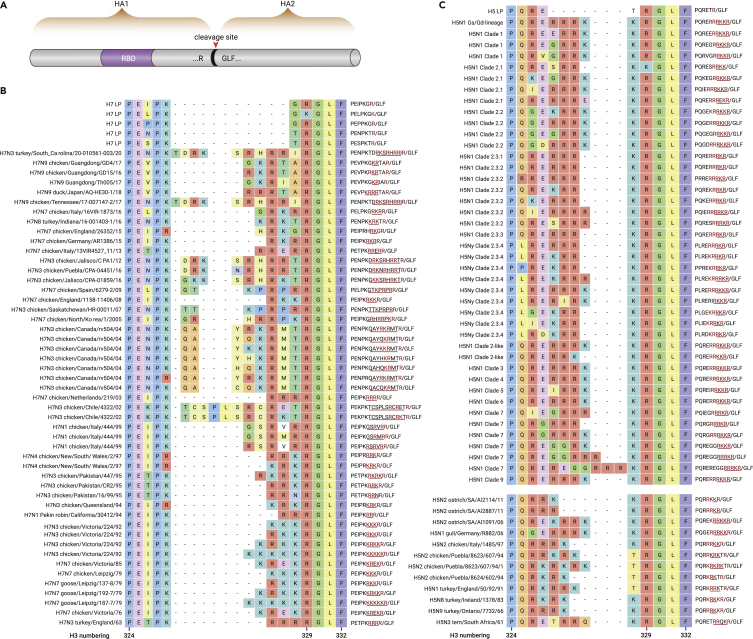
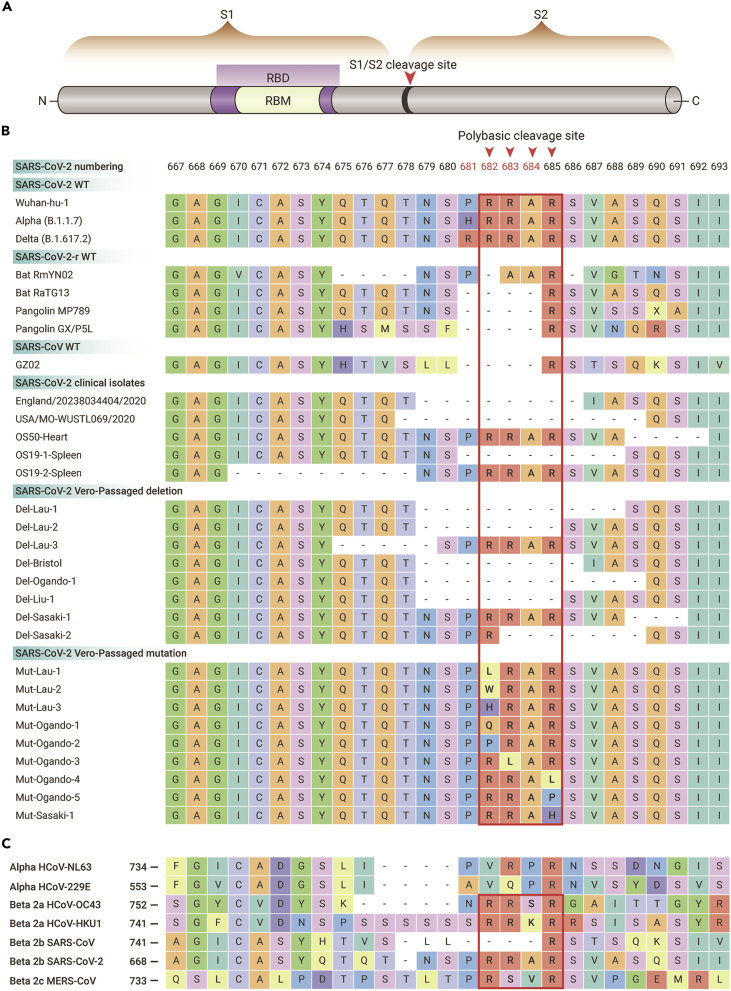
Cleavage sites with polybasic amino residues in the surface proteins of AIVs and HCoVs
The HA protein of IAVs and spike (S) protein of HCoVs can be cleaved into two subunits at their respective cleavage sites (CS) by host cell proteases. The seasonal IAVs H1N1 and H3N2 do not contain multibasic amino acids at their CSs. In the case of AIVs, HPAIVs always possess a motif with polybasic amino acids at the HA cleavage site (HA1/HA2 junction) (Figure 4), which is a genetic marker for H5 and H7 HPAIVs.83,84 Also, a polybasic motif was identified at the S cleavage site (S1/S2 junction) in four HCoVs, including SARS-CoV-2, MERS-CoV, HCoV-OC43, and HCoV-HKU1 (Figure 5). Furthermore, the analogous polybasic CS on HA of HPAIV and S of SARS-CoV-2 could be correlated with increased virus virulence in poultry and/or mammals.13,85, 86, 87
Figure 4.
Cleavage site motif on HA protein of H5 and H7 AIVs
(A) Schematic pattern for the HA protein of IAV. The HA protein can be cleaved into HA1 and HA2 subunits at the cleavage site. The receptor-binding domain (RBD) is located in HA1.
(B) The amino acid sequences at the cleavage site for H7 AIVs. H7 LP means low pathogenic AIVs, and other H7 strains are highly pathogenic AIVs.
(C) The amino acid sequences at cleavage site for H5. The H5 LP is a low pathogenic AIV, and other H5 strains are highly pathogenic AIVs. In the rightmost column, the red colors represent the key basic residues; “/” represents the cleavage position; the residues of insertion mutation are underlined.
Figure 5.
Cleavage site motif of the spike protein of SARS-CoV-2, SARS-CoV-2-related viruses, and other human CoVs
(A) Schematic pattern for SARS-CoV-2 spike (S) protein. The S protein can be cleaved into S1 and S2 proteins at the S1/S2 junction site. The receptor-binding domain (RBD) is located in S1.
(B) The amino acid diversity at cleavage site for prototype type, clinical isolates (isolated by Vero cells), and Vero cell-passaged viruses of SARS-CoV-2 as well as SARS-CoV-2-related viruses.
(C) Diversity at cleavage sites for seven human CoVs that have been identified to date. Polybasic amino acid insertions are observed in SARS-CoV-2, MERS-CoV, HCoV-OC43, and HCoV-HKU1.
Regarding AIVs, the CS is flanked by PQ/L for H5 and PE for H7 at the N terminus and by GLF for both H5 and H7 AIVs at the C terminus (Figure 4). H5 and H7 low pathogenic AIVs (LPAIVs) can evolve into HPAIVs by the insertion of a polybasic motif at the HA CS (Figure 4).88 In addition, varied lengths and polymorphisms of the CS were observed in naturally occurring viruses.89 There are several potential mechanisms for the insertion of a polybasic CS: (1) substitution of non-basic amino acids with the basic R or K residues, (2) insertion related to duplication of purine triplets due to the polymerase slippage, and (3) recombination with other gene segments of IAVs or host 28S ribosomal RNA.83,88,89
The polybasic CS is a crucial determinant for the infectivity and virulence of AIVs.89 Generally, multibasic CSs of HPAIV HA protein can be recognized and cleaved by the ubiquitous expressed cellular proteases such as furin and PC6, causing systemic infection and even a fatal outcome in poultry.88 However, the monobasic CS of LPAIV HA can merely be cleaved by trypsin and trypsin-like proteases.88,89 The distribution of proteases for LPAIV HA cleavage restricts virus replication in respiratory and/or intestinal tracts and causes asymptomatic or mild symptoms in birds.88,89 Further, the multibasic motif at CS also affects AIV virulence in mammals.90
The SARS-CoV-2 also has a polybasic CS (PRRAR) and forms a furin site at the S1/S2 junction of the S protein; this insertion has not been previously observed in other beta-CoVs of clade 2b (lineage B) (Figure 5).47,85,91 The mutations at the CS have been found in the Alpha (HRRAR), Delta (RRRAR), and Omicron (HRRAR) SARS-CoV-2 variants. In addition, SARS-CoV-2 variants with deletions and mutations at the CS were readily found during virus passage in Vero-E6 cells (Figure 5).92,93 Furthermore, a three-residue (PAA) insertion at the S1/S2 junction was identified in a bat SARS-CoV-2-like virus (RmYN02).47 Currently, how the insertion of polybasic residues into the SARS-CoV-2 CS occurred is still elusive. However, these results support the natural diversity of the polybasic CS in SARS-CoV-2 or SARS-CoV-2-related viruses.
The correlation between the furin CS and the infectivity and pathogenicity of SARS-CoV-2 was explored in in vivo and in vitro experiments. Loss of the furin CS (PRRA) decreased SARS-CoV-2 replication in human respiratory cells and attenuated virus pathogenesis in both hamster and mouse models compared with its parental virus.85,86,93 Further, the cell-passaged SARS-CoV-2 variant with eight-residue deletions at CS was deficient in transmission among co-housed ferrets.94 These results about AIVs and SARS-CoV-2 suggest that the polybasic CS is a crucial determinant for infection and replication ability, virulence, and transmissibility of HPAIVs and SARS-CoV-2 in poultry and/or mammals.
Receptors, target cells, and mechanisms of IAV and HCoV co-infections
Sialic acid receptor and cell and organ tropisms for IAVs
IAVs utilize sialic acid receptors linked to glycoproteins and gangliosides for cell entry, especially N-acetyl-neuraminic acids, which are crucial in influenza viruses infecting hosts.21 The tissue distribution and expression of the sialic acid receptors are different in humans and avian hosts.95 In humans, α-2,6 sialic acid (α2-6-SA) is predominantly expressed in the upper respiratory tract, while α2-3-SA is mainly expressed in the lower respiratory tract.96 However, α2-6-SA and α2-3-SA were also occasionally detected in the lower respiratory tract and nasal mucosa, respectively.97,98 However, in avian species, α2-3-SA predominates in both upper and lower respiratory tracts, and it is also extensively expressed in the intestinal epithelial cells of birds.99 Typically, human IAVs (eg, H3N2) preferentially bind to human-type receptors (α2-6-SA), while AIVs preferentially bind to avian-type receptors (α2-3-SA). Hence, receptor specificity is a major determinant for the host range of influenza viruses. The preference of AIVs for human-type receptors was considered a crucial warning for cross-species infection of AIVs to humans.
The tissue distribution of sialic acid receptors corresponds to cell and organ tropisms for IAVs. The respiratory system is the primary target for IAV infections. Human IAVs have been documented to infect nasal mucosa, the epithelial cells of trachea, bronchi, and bronchioles, and type I pneumocytes (AT1) in the alveoli (Figure 6).99,100 In contrast, the antigens of AIVs were primarily detected in the lower respiratory tract of humans;99 AIVs tend to target cells in bronchioles and also infect epithelial cells, AT2, and macrophages in the alveoli (Figure 6).99,100
Figure 6.
Cell tropism for SARS-CoV-2 and IAVs and their receptors in the human respiratory system
(A) Cell tropism for SARS-CoV-2 and its receptor hACE2 in human lung and trachea.
(B) Cell tropism for avian and human influenza viruses and distribution of sialic acid receptors in human lung and trachea. α2-3-SA and α2-6-SA represent the α-2,3 and α-2,6 sialic acid receptors, respectively, with a preference for binding to avian and human influenza viruses. The α2-6-SA and α2-3-SA receptors are mainly located in the upper and lower respiratory tracts of humans, respectively. α2-3-SA and α2-6-SA are also occasionally detected in human nasal mucosa and the lower respiratory tract, respectively. AT1 and AT2 represent type I and type II pneumocytes in the alveoli, respectively.
Sialic acids also act as receptors for some alpha-, beta-, and gamma-CoVs.21 Specifically, O-acetylated sialic acids serve as receptors for beta-CoVs, eg, HCoV-OC43 and bovine CoV.21 N-acetyl- and N-glycolylneuraminic acids were recognized as receptor determinants of cell infection for transmissible gastroenteritis virus (alpha-CoV) and infectious bronchitis virus (gamma-CoV).21
ACE2 receptor and cell and organ tropisms for SARS-CoV-2
ACE2 serves as a receptor for SARS-CoV-2, SARS-CoV, and HCoV-NL63 to enter cells.20,101 SARS-CoV-2 can efficiently use ACE2 as a receptor for cell entry, with up to 10- to 20-fold higher affinity than for SARS-CoV.102 ACE2 is expressed in the lungs, hearts, kidneys, livers, testes, and intestines of humans.103 The ratio of ACE2-expressing cells of human lungs is relatively low.104,105 About 0.64% cells that can express ACE2 in normal human lungs were profiled by single-cell RNA sequencing.104 Specifically, in human lungs, ACE2 expresses in AT2, AT1, airway epithelial cells (ciliated and club cells), fibroblasts, endothelial cells, and immune cells (macrophages and T cells).104,105 In the trachea, ACE2-expressing cells include club, goblet, ciliated, and proliferative cells.106 The expression patterns of ACE2 are variable in current reports, so more samples and studies on ACE2 expression and distribution are needed to obtain solid results and understanding.
The tissue expression and distribution of ACE2 receptors correlate with the cell and organ tropisms for SARS-CoV-2 infection. SARS-CoV-2 can infect multiple organs, including the lung, trachea, pharynx, intestine, kidneys, pancreas, brain, and heart of humans.22,107 The respiratory system is also the primary target for SARS-CoV-2 infection. The co-location of viral antigens with cell markers by immunofluorescence staining was used to uncover cell tropism details for SARS-CoV-2.22 In lungs of postmortem specimens, viral antigens were found in basal cells, ciliated cells, club cells, AT2, AT1, proliferative cells, and vascular endothelial cells (Figure 6).22,108 SARS-CoV-2 infections in alveolar cells and airway cells have also been reported in human distal lung organoids109 and ex vivo cultures of human bronchus and lung.110 In AT2-AT1 cell cultures, AT2 cells are preferentially infected by SARS-CoV-2 over AT1 cells.108 In the trachea of autopsied humans, viral infection was reported in ciliated and goblet cells of the mucosa and epithelial cells of conduits and glands.22
Target cells and receptors responsible for co-infection of IAV and SARS-CoV-2
The IAV and SARS-CoV-2 are airborne pathogens, and they primarily target the respiratory system of humans, including the nasal mucosa, trachea, bronchi, and alveoli.111 The co-location and expression of sialic acid and ACE2 receptors may contribute to the co-infection of IAVs and SARS-CoV-2 in the same organs and cells. For example, both IAV and SARS-CoV-2 could target and infect AT2 cells in alveoli.108,112 Further, co-infections in the same organs and cells could facilitate pathogenic and immunological interactions between the two types of viruses. Moreover, IAVs could promote the infectivity of SARS-CoV-2, probably due to IAV pre-infection elevating ACE2 expression in a human cell line.111
Disease burden and dynamics for co-infection of IAVs and SARS-CoV-2
Disease burden of co-infection with seasonal IAVs and SARS-CoV-2
During the COVID-19 pandemic, co-infections of SARS-CoV-2 with other respiratory pathogens have been reported, and IAVs were one of the most common co-infections.113 Patients co-infected with IAV and SARS-CoV-2 have been sporadically identified in multiple countries, such as China, Japan, Iran, the United States, Turkey, and Germany.17,18 However, patients co-infected with IAV and SARS-CoV-2 or only infected with either virus presented similar clinical respiratory symptoms and radiological images (Table S2). The most common symptoms of co-infections are fever, cough, dyspnea, and myalgia, and the typical changes in chest radiology images include ground-glass opacities.18,19,114,115 However, patients with IAV and SARS-CoV-2 co-infection usually presented more severe clinical outcomes compared with those with SARS-CoV-2 infection alone.18 The concentrations of serum cytokines/chemokines in co-infections should be further studied to understand the potential immunological mechanism for the observed clinical manifestations.116 Some contradictory disease severity of co-infection could result from the small sample size or insufficient consideration of the confounding factors, including the order of infection, virus subtypes, and underlying comorbidity.117,118
The clinical outcomes of co-infection with IAV and SARS-CoV-2 have been evaluated in laboratory-based studies.111,113 Co-infection of SARS-CoV-2 with IAVs caused more severe disease in vivo than mono-infection with either virus. Further, the IAV pre-infection could interfere with the SARS-CoV-2 replication but increase disease severity.113 However, another study indicated that IAV pre-infection promoted the infectivity and viral load of SARS-CoV-2 and caused more severe lung damage in mice, while several other respiratory viruses did not enhance SARS-CoV-2 infectivity in cells.111
During the COVID-19 pandemic, the prevalence of patients co-infected with IAV and SARS-CoV-2 varied substantially across regions and studies.18,19,119,120 Some studies documented a co-infection rate of influenza and COVID-19 as high as 57.3% in Wuhan,119,120 while a relatively low ratio of influenza infections, 0.8%, was also reported in patients with COVID-19 based on a systematic review and meta-analysis.19 Non-pharmaceutical interventions for the pandemic, including mask wearing, social distancing, and travel restrictions, could reduce the transmission of IAVs, resulting in a low co-infection rate.121
Disease dynamics caused by the co-existence of seasonal IAVs and SARS-CoV-2
The COVID-19 pandemic suppressed the number of influenza infections in the 2020–2021 season.122,123 In the Southern and Northern Hemispheres, seasonal influenza cases reduced drastically in the 2020 and 2020–2021 influenza seasons, respectively.122 In the United States and Europe, the positive influenza rate of samples in the 2020–2021 influenza season is about 0.15%–0.2%, which is estimated to be 18%–23% in the 2019–2020 influenza season.122 The decreased influenza burdens may have resulted from several factors. One explanation could be the interference and competition between distinct viruses in shared niches.122 At the onset of the A(H1N1)2009 pandemic, rhinovirus infections delayed the circulation of the A(H1N1)2009 in France.124 Also, the negative transmission dynamics between the seasonal IAV and rhinovirus were documented in the UK.125 Influenza infections could destroy cells and/or the surface receptors of the cells and thus decrease the number of susceptible cells for subsequent infection by other viruses.125 Additionally, non-specific innate immune responses, such as interferon secretion, could be activated by prior virus infection and then suppress other virus infections.124,125 Third, the non-pharmaceutical mitigation measures for the current pandemic could limit the influenza transmission routes analogous to SARS-CoV-2.126 Also, the changed physician visiting behaviors and limited surveillance capacity could affect the documented transmission patterns of influenza during the pandemic.123 Fourth, many countries (eg, the United States, Australia, and France) increased and accelerated the influenza vaccine uptakes to reduce the burden of overwhelming health systems in the COVID-19 pandemic.122 Given the above factors, the ecological and immunological relationship between SARS-CoV-2 and IAVs is a key determinant for their future infection dynamics,125, 126, 127 which should be evaluated in a unified model incorporating the interaction of multiple pathogens.
The atypically low influenza circulation in the seasons overlapping with the COVID-19 pandemic indicated a reduction in natural population-level immunity for influenza, which could shift the immune landscape, build up a susceptible pool, and further alter the timing, trajectories, and severity of influenza in future seasonal epidemics.122,123 Moreover, the low influenza circulation could challenge the choice of vaccine strains and the vaccine efficacy and therefore increase the influenza disease burden, including overwhelmed healthcare capacities and severe morbidity and mortality, in the next season. Further, rapid virus evolution and immunity dynamics in humans could complicate the forecast modeling of influenza.123 Anticipating the potentially atypical transmission patterns of influenza epidemics, especially after the COVID-19 pandemic, alongside well-prepared vaccines and antivirals, could guide targeted interventions and reduce the burden on healthcare systems.
Mitigation strategies of co-infection with seasonal IAVs and SARS-CoV-2
As the pandemic wanes and the countermeasures against COVID-19 become relaxed worldwide, circulation of IAVs and HCoVs may resume at pre-pandemic levels, together with SARS-CoV-2 continuing to circulate in humans. Co-infections with IAVs and SARS-CoV-2 may increase, complicating the diagnosis, treatment, and prognosis for patients and aggravating the disease burden for the medicine and healthcare system.121 Given this, the early detection of co-infection by multiplex molecular diagnostics should be implemented to timely initiate antiviral therapy and improve the prognosis of patients.113,121 Practical preventive actions (eg, wearing masks, washing hands, and social distancing) could protect against infection with these airborne pathogens.121 Further, vaccinations could reduce co-infections, clinical severity, and disease burden, especially in high-risk individuals and the elderly.128 Moreover, the live attenuated influenza virus-vectored COVID-19 vaccine has been designed to induce co-immunity against influenza virus and SARS-CoV-2,129 and vaccine deployments will simplify the vaccination strategy against these two types of viruses.
Conclusions
Given that the current pandemic is not yet under control, the long-term co-existence and co-infection of seasonal influenza and SARS-CoV-2 in humans may be unpreventable. Hence, targeted development and distribution of antiviral drugs and therapeutics should be underscored for long-standing disease control. Moreover, non-pharmaceutical interventions containing hand washing, mask wearing, and social distancing should be highlighted, especially during regional outbreaks of these viruses.
As etiological agents for pandemics documented, the co-existence and co-infection of IAVs and CoVs could be potential candidates for the next pandemic and come into focus. Given the virus-host ecology circles for IAVs and CoVs, proactive surveillance and evaluations for emerging virus variants, along with inter-species transmission, outbreak, and even pandemic risks, should be strengthened in animals (eg, birds and bats). Sporadically, novel AIV variants could directly infect humans from birds; the spillover of SARS-CoV-2 from humans to minks could transmit back to humans, and the mink SARS-CoV-2 mutant could further transmit between humans.52 Early discovery and identification of VOIs/VOCs can provide sufficient time for technological preparedness about diagnosis and antiviral drugs and vaccines prior to the virus spilling over from animals to humans. In addition, real-time surveillance of human infections with novel variants or pathogens is crucial to accomplish early diagnosis, intervention, and quarantine of confirmed cases, especially super-spreaders, and hereafter timely containing further disease outbreaks and even pandemics in humans. At least, lessons and experiences from past pandemics should be learned in order to be better prepared against the next one, with enhanced global cooperation.
Acknowledgments
This work is supported by the National Key R&D Program of China (2021YFC2300903, 2021YFE0109100, and 2021YFC2301300); Strategic Priority Research Program of Chinese Academy of Sciences (CAS) (XDB29010102); National Natural Science Foundation of China (NSFC) (32161123001, 82161148010, and 32041010); Beijing Natural Science Foundation (M22029 and L192007); CAS Southeast Asia Biodiversity Research Institute (151C53KYSB20210023); Shenzhen Science and Technology Research and Development Project (JCYJ20180504165549581); and the National Science and Technology Infrastructure of China (National Pathogen Resource Center-NPRC-32). J.Y. is supported by the Special Program of China Postdoctoral Science Foundation (2020T130123ZX). Y.B. is supported by National Natural Science Foundation of China (NSFC) Outstanding Young Scholars (31822055), Youth Innovation Promotion Association of CAS (Y2021034), and the Innovation Team and Talents Cultivation Program of National Administration of Traditional Chinese Medicine (ZYYCXTD-D-202208).
Author contributions
This study was conceived and designed by Y.B. J.Y. drafted the manuscript, and Y.B., J.Y., Y.G., C.Z., J.S., G.W., W.S., W.L., and G.F.G. discussed and edited the manuscript.
Declaration of interests
The authors declare no competing interests.
Published Online: August 17, 2022
Footnotes
Supplemental information is available at https://doi.org/10.1016/j.xinn.2022.100306.
Lead contact website
https://www.im.cas.cn/jgsz2018/yjtx/zgkxybywswymyxzdsys/201911/t20191125_5442572.html.
Supplemental information
References
- 1.Taubenberger J.K., Reid A.H., Krafft A.E., et al. Initial genetic characterization of the 1918 "Spanish" influenza virus. Science. 1997;275:1793–1796. doi: 10.1126/science.275.5307.1793. [DOI] [PubMed] [Google Scholar]
- 2.Kilbourne E.D. Influenza pandemics of the 20th century. Emerg. Infect. Dis. 2006;12:9–14. doi: 10.3201/eid1201.051254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao G.F. From "A"IV to "Z"IKV: attacks from emerging and re-emerging pathogens. Cell. 2018;172:1157–1159. doi: 10.1016/j.cell.2018.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization Disease outbreak news; avian influenza A (H3N8) – China. 2022. https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON378
- 5.Yang J., Li J., Lai S., et al. Uncovering two phases of early intercontinental COVID-19 transmission dynamics. J. Travel. Med. 2020;27:taaa200. doi: 10.1093/jtm/taaa200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai L., Zheng T., Xu K., et al. A universal design of betacoronavirus vaccines against COVID-19, MERS, and SARS. Cell. 2020;182:722–733. doi: 10.1016/j.cell.2020.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor P.C., Adams A.C., Hufford M.M., et al. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021;21:382–393. doi: 10.1038/s41577-021-00542-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma K., Chen J. Omicron XE emerges as SARS-CoV-2 keeps evolving. Innovation. 2022;3:100248. doi: 10.1016/j.xinn.2022.100248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su S., Wong G., Shi W., et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peiris J.S., Lai S.T., Poon L.L., et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raj V.S., Osterhaus A.D., Fouchier R.A., Haagmans B.L. MERS: emergence of a novel human coronavirus. Curr. Opin. Virol. 2014;5:58–62. doi: 10.1016/j.coviro.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabir J.S., Lam T.T., Ahmed M.M., et al. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science. 2016;351:81–84. doi: 10.1126/science.aac8608. [DOI] [PubMed] [Google Scholar]
- 13.Zhou P., Yang X.L., Wang X.G., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lam T.T., Jia N., Zhang Y.W., et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 15.Shi J., Wen Z., Zhong G., et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wille M., Holmes E.C. The ecology and evolution of influenza viruses. Cold. Spring. Harb. Perspect. Med. 2020;10:a038489. doi: 10.1101/cshperspect.a038489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ozaras R., Cirpin R., Duran A., et al. Influenza and COVID-19 coinfection: report of six cases and review of the literature. J. Med. Virol. 2020;92:2657–2665. doi: 10.1002/jmv.26125. [DOI] [PubMed] [Google Scholar]
- 18.Xiang X., Wang Z.H., Ye L.L., et al. Co-infection of SARS-CoV-2 and influenza A virus: a case series and fast review. Curr. Med. Sci. 2021;41:51–57. doi: 10.1007/s11596-021-2317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dadashi M., Khaleghnejad S., Abedi Elkhichi P., et al. COVID-19 and influenza co-infection: a systematic review and meta-analysis. Front. Med. 2021;8:681469. doi: 10.3389/fmed.2021.681469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Q., Zhang Y., Wu L., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matrosovich M., Herrler G., Klenk H.D. Sialic acid receptors of viruses. Top. Curr. Chem. 2015;367:1–28. doi: 10.1007/128_2013_466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J., Li Y., Liu Q., et al. SARS-CoV-2 cell tropism and multiorgan infection. Cell Discov. 2021;7:17. doi: 10.1038/s41421-021-00249-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krauss S., Webster R.G. Avian influenza virus surveillance and wild birds: past and present. Avian Dis. 2010;54:394–398. doi: 10.1637/8703-031609-Review.1. [DOI] [PubMed] [Google Scholar]
- 24.Liu W.J., Wu Y., Bi Y., et al. Emerging HxNy influenza A viruses. Cold. Spring. Harb. Perspect. Med. 2022;12:a038406. doi: 10.1101/cshperspect.a038406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tong S., Li Y., Rivailler P., et al. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. USA. 2012;109:4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tong S., Zhu X., Li Y., et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J., Zhang C., Cao J., et al. Re-emergence of H5N8 highly pathogenic avian influenza virus in wild birds. China. Emerg. Microbes. Infect. 2021;10:1819–1823. doi: 10.1080/22221751.2021.1968317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwon H.I., Kim E.H., Kim Y.I., et al. Comparison of the pathogenic potential of highly pathogenic avian influenza (HPAI) H5N6, and H5N8 viruses isolated in South Korea during the 2016-2017 winter season. Emerg. Microbes Infect. 2018;7:29. doi: 10.1038/s41426-018-0029-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim H.R., Kwon Y.K., Jang I., et al. Pathologic changes in wild birds infected with highly pathogenic avian influenza A(H5N8) viruses, South Korea, 2014. Emerg. Infect. Dis. 2015;21:775–780. doi: 10.3201/eid2105.141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwon J.H., Bahl J., Swayne D.E., et al. Domestic ducks play a major role in the maintenance and spread of H5N8 highly pathogenic avian influenza viruses in South Korea. Transboundary Emerg. Dis. 2020;67:844–851. doi: 10.1111/tbed.13406. [DOI] [PubMed] [Google Scholar]
- 31.Mine J., Uchida Y., Nakayama M., et al. Genetics and pathogenicity of H5N6 highly pathogenic avian influenza viruses isolated from wild birds and a chicken in Japan during winter 2017-2018. Virology. 2019;533:1–11. doi: 10.1016/j.virol.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 32.Bi Y., Chen J., Zhang Z., et al. Highly pathogenic avian influenza H5N1 clade 2.3.2.1c virus in migratory birds. Virol. Sin. 2016;31:300–305. doi: 10.1007/s12250-016-3750-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bi Y., Liu H., Xiong C., et al. Novel avian influenza A (H5N6) viruses isolated in migratory waterfowl before the first human case reported in China, 2014. Sci. Rep. 2016;6:29888. doi: 10.1038/srep29888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi W., Gao G.F. Emerging H5N8 avian influenza viruses. Science. 2021;372:784–786. doi: 10.1126/science.abg6302. [DOI] [PubMed] [Google Scholar]
- 35.Liu J., Xiao H., Lei F., et al. Highly pathogenic H5N1 influenza virus infection in migratory birds. Science. 2005;309:1206. doi: 10.1126/science.1115273. [DOI] [PubMed] [Google Scholar]
- 36.Chen H., Smith G.J., Zhang S.Y., et al. Avian flu: H5N1 virus outbreak in migratory waterfowl. Nature. 2005;436:191–192. doi: 10.1038/nature03974. [DOI] [PubMed] [Google Scholar]
- 37.Petrova V.N., Russell C.A. The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 2018;16:47–60. doi: 10.1038/nrmicro.2017.118. [DOI] [PubMed] [Google Scholar]
- 38.Bi Y., Chen Q., Wang Q., et al. Genesis, evolution and prevalence of H5N6 avian influenza viruses in China. Cell Host Microbe. 2016;20:810–821. doi: 10.1016/j.chom.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 39.Bi Y., Li J., Li S., et al. Dominant subtype switch in avian influenza viruses during 2016-2019 in China. Nat. Commun. 2020;11:5909. doi: 10.1038/s41467-020-19671-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Q., Wang H., Zhao L., et al. First documented case of avian influenza (H5N1) virus infection in a lion. Emerg. Microbes. Infect. 2016;5:e125. doi: 10.1038/emi.2016.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu S., Ji K., Chen J., et al. Panorama phylogenetic diversity and distribution of type A influenza virus. PLoS One. 2009;4:e5022. doi: 10.1371/journal.pone.0005022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.L'vov D.K., Easterday B., Hinshow W., et al. Isolation of strains of the Hong Kong complex (H3N2) influenza virus from Nyctalus noctula bats in Kazakhstan. Vopr. Virusol. 1979;4:338–341. [PubMed] [Google Scholar]
- 43.Liu J., Bi Y., Qin K., et al. Emergence of European avian influenza virus-like H1N1 swine influenza A viruses in China. J. Clin. Microbiol. 2009;47:2643–2646. doi: 10.1128/JCM.00262-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun H., Xiao Y., Liu J., et al. Prevalent Eurasian avian-like H1N1 swine influenza virus with 2009 pandemic viral genes facilitating human infection. Proc. Natl. Acad. Sci. USA. 2020;117:17204–17210. doi: 10.1073/pnas.1921186117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woo P.C., Lau S.K., Lam C.S., et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou H., Chen X., Hu T., et al. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr. Biol. 2020;30:2196–2203. doi: 10.1016/j.cub.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou H., Ji J., Chen X., et al. Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses. Cell. 2021;184:4380–4391. doi: 10.1016/j.cell.2021.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiao K., Zhai J., Feng Y., et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020;583:286–289. doi: 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- 50.Mallapaty S. Closest known relatives of virus behind COVID-19 found in Laos. Nature. 2021;597:603. doi: 10.1038/d41586-021-02596-2. [DOI] [PubMed] [Google Scholar]
- 51.Oude Munnink B.B., Sikkema R.S., Nieuwenhuijse D.F., et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371:172–177. doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang L., Didelot X., Bi Y., Gao G.F. Assessing the extent of community spread caused by mink-derived SARS-CoV-2 variants. Innovation. 2021;2:100128. doi: 10.1016/j.xinn.2021.100128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim Y.I., Kim S.G., Kim S.M., et al. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe. 2020;27:704–709. doi: 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Munster V.J., Feldmann F., Williamson B.N., et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585:268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rockx B., Kuiken T., Herfst S., et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368:1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cross R.W., Agans K.N., Prasad A.N., et al. Intranasal exposure of African green monkeys to SARS-CoV-2 results in acute phase pneumonia with shedding and lung injury still present in the early convalescence phase. Virol. J. 2020;17:125. doi: 10.1186/s12985-020-01396-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu L., Chen Q., Liu K., et al. Broad host range of SARS-CoV-2 and the molecular basis for SARS-CoV-2 binding to cat ACE2. Cell Discov. 2020;6:68. doi: 10.1038/s41421-020-00210-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lemey P., Rambaut A., Bedford T., et al. Unifying viral genetics and human transportation data to predict the global transmission dynamics of human influenza H3N2. PLoS Pathog. 2014;10:e1003932. doi: 10.1371/journal.ppat.1003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chan M., Wang M.H., Chen Z., et al. Frequent genetic mismatch between vaccine strains and circulating seasonal influenza viruses, Hong Kong, China, 1996-2012. Emerg. Infect. Dis. 2018;24:1825–1834. doi: 10.3201/eid2410.180652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ferguson N.M., Galvani A.P., Bush R.M. Ecological and immunological determinants of influenza evolution. Nature. 2003;422:428–433. doi: 10.1038/nature01509. [DOI] [PubMed] [Google Scholar]
- 61.Nelson M., Spiro D., Wentworth D., et al. The early diversification of influenza A/H1N1pdm. PLoS Curr. 2009;1:RRN1126. doi: 10.1371/currents.RRN1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Potter B.I., Kondor R., Hadfield J., et al. Evolution and rapid spread of a reassortant A(H3N2) virus that predominated the 2017-2018 influenza season. Virus. Evol. 2019;5:vez046. doi: 10.1093/ve/vez046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Al-Qahtani A.A., Mubin M., Dela Cruz D.M., et al. Phylogenetic and nucleotide sequence analysis of influenza A (H1N1) HA and NA genes of strains isolated from Saudi Arabia. J. Infect. Dev. Ctries. 2017;11:81–88. doi: 10.3855/jidc.9259. [DOI] [PubMed] [Google Scholar]
- 64.Smith G.J., Donis R.O. World health organization/world organisation for animal health/food and agriculture organization (WHO/OIE/FAO) H5 evolution working group. Influenza. Other. Respir. Viruses. 2015;9:271–276. nomenclature updates resulting from the evolution of avian influenza A(H5) virus clades 2.1.3.2a, 2.2.1, and 2.3.4 during 2013-2014. [Google Scholar]
- 65.Yang L., Zhu W., Li X., et al. Genesis and spread of newly emerged highly pathogenic H7N9 avian viruses in mainland China. J. Virol. 2017;91 doi: 10.1128/JVI.01277-17. e01277–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quan C., Shi W., Yang Y., et al. New threats from H7N9 influenza virus: spread and evolution of high- and low-pathogenicity variants with high genomic diversity in wave five. J. Virol. 2018;92:e00301–e00318. doi: 10.1128/JVI.00301-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rambaut A., Holmes E.C., O'Toole Á., et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lythgoe K.A., Hall M., Ferretti L., et al. SARS-CoV-2 within-host diversity and transmission. Science. 2021;372:eabg0821. doi: 10.1126/science.abg0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grenfell B.T., Pybus O.G., Gog J.R., et al. Unifying the epidemiological and evolutionary dynamics of pathogens. Science. 2004;303:327–332. doi: 10.1126/science.1090727. [DOI] [PubMed] [Google Scholar]
- 70.Emary K., Golubchik T., Aley P.K., et al. Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 variant of concern 202012/01 (B.1.1.7): an exploratory analysis of a randomised controlled trial. Lancet. 2021;397:1351–1362. doi: 10.1016/S0140-6736(21)00628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garcia-Beltran W.F., Lam E.C., St Denis K., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372–2383. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.World Health Organization (WHO) 2020. Recommended composition of influenza virus vaccines for use in the 2020-2021 northern hemisphere influenza season.https://www.who.int/influenza/vaccines/virus/recommendations/202002_recommendation.pdf?ua=1 [Google Scholar]
- 73.Vasin A.V., Temkina O.A., Egorov V.V., et al. Molecular mechanisms enhancing the proteome of influenza A viruses: an overview of recently discovered proteins. Virus Res. 2014;185:53–63. doi: 10.1016/j.virusres.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 74.McDonald S.M., Nelson M.I., Turner P.E., Patton J.T. Reassortment in segmented RNA viruses: mechanisms and outcomes. Nat. Rev. Microbiol. 2016;14:448–460. doi: 10.1038/nrmicro.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu D., Shi W., Shi Y., et al. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet. 2013;381:1926–1932. doi: 10.1016/S0140-6736(13)60938-1. [DOI] [PubMed] [Google Scholar]
- 76.Lam T.T.Y., Zhou B., Wang J., et al. Dissemination, divergence and establishment of H7N9 influenza viruses in China. Nature. 2015;522:102–105. doi: 10.1038/nature14348. [DOI] [PubMed] [Google Scholar]
- 77.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Woo P.C., Lau S.K., Huang Y., Yuen K.Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp Biol Med (Maywood). 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- 79.Bentley K., Evans D.J. Mechanisms and consequences of positive-strand RNA virus recombination. J. Gen. Virol. 2018;99:1345–1356. doi: 10.1099/jgv.0.001142. [DOI] [PubMed] [Google Scholar]
- 80.Lau S.K., Woo P.C., Yip C.C., et al. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J. Clin. Microbiol. 2006;44:2063–2071. doi: 10.1128/JCM.02614-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stanhope M.J., Brown J.R., Amrine-Madsen H. Evidence from the evolutionary analysis of nucleotide sequences for a recombinant history of SARS-CoV. Infect. Genet. Evo. 2004;4:15–19. doi: 10.1016/j.meegid.2003.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang X.W., Yap Y.L., Danchin A. Testing the hypothesis of a recombinant origin of the SARS-associated coronavirus. Arch. Virol. 2005;150:1–20. doi: 10.1007/s00705-004-0413-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.OIE OIE Terrestrial Manual 2021. https://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/3.03.04_AI.pdf
- 84.OIE. Influenza A cleavage site. 2021. http://www.offlu.org/wp-content/uploads/2021/01/Influenza_A_Cleavage_Sites.pdf
- 85.Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Johnson B.A., Xie X., Bailey A.L., et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature. 2021;591:293–299. doi: 10.1038/s41586-021-03237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kawaoka Y., Webster R.G. Sequence requirements for cleavage activation of influenza virus hemagglutinin expressed in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 1988;85:324–328. doi: 10.1073/pnas.85.2.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alexander D.J., Brown I.H. History of highly pathogenic avian influenza. Rev. Sci. Tech. 2009;28:19–38. doi: 10.20506/rst.28.1.1856. [DOI] [PubMed] [Google Scholar]
- 89.Luczo J.M., Stambas J., Durr P.A., et al. Molecular pathogenesis of H5 highly pathogenic avian influenza: the role of the haemagglutinin cleavage site motif. Rev. Med. Virol. 2015;25:406–430. doi: 10.1002/rmv.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang Y., Sun Y., Sun H., et al. A single amino acid at the hemagglutinin cleavage site contributes to the pathogenicity and neurovirulence of H5N1 influenza virus in mice. J. Virol. 2012;86:6924–6931. doi: 10.1128/JVI.07142-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Andersen K.G., Rambaut A., Lipkin W.I., et al. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang P., Lau S.Y., Deng S., et al. Characterization of an attenuated SARS-CoV-2 variant with a deletion at the S1/S2 junction of the spike protein. Nat. Commun. 2021;12:2790. doi: 10.1038/s41467-021-23166-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lau S.Y., Wang P., Mok B.W., et al. Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerg. Microbes Infect. 2020;9:837–842. doi: 10.1080/22221751.2020.1756700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Peacock T.P., Goldhill D.H., Zhou J., et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021;6:899–909. doi: 10.1038/s41564-021-00908-w. [DOI] [PubMed] [Google Scholar]
- 95.Sorrell E.M., Schrauwen E.J., Linster M., et al. Predicting 'airborne' influenza viruses: (trans-) mission impossible? Curr. Opin. Virol. 2011;1:635–642. doi: 10.1016/j.coviro.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shinya K., Ebina M., Yamada S., et al. Avian flu: influenza virus receptors in the human airway. Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 97.Rajao D.S., Vincent A.L., Perez D.R. Adaptation of human influenza viruses to swine. Front. Vet. Sci. 2019;5:347. doi: 10.3389/fvets.2018.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shinya K., Ebina M., Yamada S., et al. Influenza virus receptors in the human airway. Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 99.Sriwilaijaroen N., Nakakita S.I., Kondo S., et al. N-glycan structures of human alveoli provide insight into influenza A virus infection and pathogenesis. FEBS J. 2018;285:1611–1634. doi: 10.1111/febs.14431. [DOI] [PubMed] [Google Scholar]
- 100.van Riel D., Munster V.J., de Wit E., et al. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am. J. Pathol. 2007;171:1215–1223. doi: 10.2353/ajpath.2007.070248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li W., Sui J., Huang I.C., et al. The S proteins of human coronavirus NL63 and severe acute respiratory syndrome coronavirus bind overlapping regions of ACE2. Virology. 2007;367:367–374. doi: 10.1016/j.virol.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wrapp D., Wang N., Corbett K.S., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tipnis S.R., Hooper N.M., Hyde R., et al. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 2000;275:33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 104.Zhao Y., Zhao Z., Wang Y., et al. Single-cell RNA expression profiling of ACE2, the receptor of SARS-CoV-2. Am. J. Respir. Crit. Care Med. 2020;202:756–759. doi: 10.1164/rccm.202001-0179LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lukassen S., Chua R.L., Trefzer T., et al. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39:e105114. doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Barker H., Parkkila S. Bioinformatic characterization of angiotensin-converting enzyme 2, the entry receptor for SARS-CoV-2. PLoS One. 2020;15:e0240647. doi: 10.1371/journal.pone.0240647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Puelles V.G., Lütgehetmann M., Lindenmeyer M.T., et al. Multiorgan and renal tropism of SARS-CoV-2. N. Engl. J. Med. 2020;383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hou Y.J., Okuda K., Edwards C.E., et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182:429–446. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang T., Zhang N., Fan S., et al. Establishment of human distal lung organoids for SARS-CoV-2 infection. Cell Discov. 2021;7:108. doi: 10.1038/s41421-021-00346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hui K., Cheung M.C., Perera R., et al. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: an analysis in ex-vivo and in-vitro cultures. Lancet Respir. Med. 2020;8:687–695. doi: 10.1016/S2213-2600(20)30193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bai L., Zhao Y., Dong J., et al. Coinfection with influenza A virus enhances SARS-CoV-2 infectivity. Cell Res. 2021;31:395–403. doi: 10.1038/s41422-021-00473-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Traylor Z.P., Aeffner F., Davis I.C. Influenza A H1N1 induces declines in alveolar gas exchange in mice consistent with rapid post-infection progression from acute lung injury to ARDS. Influenza. Other. Respir. Viruses. 2013;7:472–479. doi: 10.1111/j.1750-2659.2012.00414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang A.J., Lee A.C., Chan J.F., et al. Coinfection by severe acute respiratory syndrome coronavirus 2 and influenza A(H1N1)pdm09 virus enhances the severity of pneumonia in golden syrian hamsters. Clin. Infect. Dis. 2021;72:e978–e992. doi: 10.1093/cid/ciaa1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang D., Hu B., Hu C., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Huang C., Wang Y., Li X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bi Y., Tan S., Yang Y., et al. Clinical and immunological characteristics of human infections with H5N6 avian influenza virus. Clin. Infect. Dis. 2019;68:1100–1109. doi: 10.1093/cid/ciy681. [DOI] [PubMed] [Google Scholar]
- 117.Guan Z., Chen C., Li Y., et al. Impact of coinfection with SARS-CoV-2 and Influenza on disease severity: a systematic review and meta-analysis. Front. Public Health. 2021;9:773130. doi: 10.3389/fpubh.2021.773130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Akhtar Z., Islam M.A., Aleem M.A., et al. SARS-CoV-2 and influenza virus coinfection among patients with severe acute respiratory infection during the first wave of COVID-19 pandemic in Bangladesh: a hospital-based descriptive study. BMJ Open. 2021;11:e053768. doi: 10.1136/bmjopen-2021-053768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yue H., Zhang M., Xing L., et al. The epidemiology and clinical characteristics of co-infection of SARS-CoV-2 and influenza viruses in patients during COVID-19 outbreak. J. Med. Virol. 2020;92:2870–2873. doi: 10.1002/jmv.26163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Swets M.C., Russell C.D., Harrison E.M., et al. SARS-CoV-2 co-infection with influenza viruses, respiratory syncytial virus, or adenoviruses. Lancet. 2022;399:1463–1464. doi: 10.1016/S0140-6736(22)00383-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Olsen S.J., Winn A.K., Budd A.P., et al. Changes in influenza and other respiratory virus activity during the COVID-19 pandemic - United States, 2020-2021. MMWR Morb. Mortal. Wkly. Rep. 2021;70:1013–1019. doi: 10.15585/mmwr.mm7029a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zipfel C.M., Colizza V., Bansal S. The missing season: the impacts of the COVID-19 pandemic on influenza. Vaccine. 2021;39:3645–3648. doi: 10.1016/j.vaccine.2021.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gomez G.B., Mahé C., Chaves S.S. Uncertain effects of the pandemic on respiratory viruses. Science. 2021;372:1043–1044. doi: 10.1126/science.abh3986. [DOI] [PubMed] [Google Scholar]
- 124.Casalegno J.S., Ottmann M., Duchamp M.B., et al. Rhinoviruses delayed the circulation of the pandemic influenza A (H1N1) 2009 virus in France. Clin. Microbiol. Infect. 2010;16:326–329. doi: 10.1111/j.1469-0691.2010.03167.x. [DOI] [PubMed] [Google Scholar]
- 125.Nickbakhsh S., Mair C., Matthews L., et al. Virus-virus interactions impact the population dynamics of influenza and the common cold. Proc. Natl. Acad. Sci. USA. 2019;116:27142–27150. doi: 10.1073/pnas.1911083116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Baker R.E., Park S.W., Yang W., et al. The impact of COVID-19 nonpharmaceutical interventions on the future dynamics of endemic infections. Proc. Natl. Acad. Sci. USA. 2020;117:30547–30553. doi: 10.1073/pnas.2013182117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rohani P., Green C.J., Mantilla-Beniers N.B., Grenfell B.T. Ecological interference between fatal diseases. Nature. 2003;422:885–888. doi: 10.1038/nature01542. [DOI] [PubMed] [Google Scholar]
- 128.Rossman H., Shilo S., Meir T., et al. COVID-19 dynamics after a national immunization program in Israel. Nat. Med. 2021;27:1055–1061. doi: 10.1038/s41591-021-01337-2. [DOI] [PubMed] [Google Scholar]
- 129.Chen J.Y., Wang P., Yuan L., et al. A live attenuated virus-based intranasal COVID-19 vaccine provides rapid, prolonged, and broad protection against SARS-CoV-2. Sci. Bull. 2022;67:1372–1387. doi: 10.1016/j.scib.2022.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.