Abstract
We aimed to provide in vitro data on the neutralization capacity of different monoclonal antibody (mAb) preparations against the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) delta and omicron variant, respectively, and describe the in vivo RNA kinetics of coronavirus disease 2019 (COVID‐19) patients treated with the respective mAbs. Virus neutralization assays were performed to assess the neutralizing effect of the mAb formulations casirivimab/imdevimab and sotrovimab on the SARS‐CoV‐2 delta and omicron variant. Additionally, respiratory tract SARS‐CoV‐2 RNA kinetics are provided for 25 COVID‐19 patients infected with either delta variant (n = 18) or omicron variant (n = 7) treated with the respective mAb formulations during their hospital stay. In the virus neutralization assay, sotrovimab exhibits neutralizing capacity at therapeutically achievable concentrations against the SARS‐CoV‐2 delta and omicron variant. In contrast, casivirimab/imdevimab had neutralizing capacity against the delta variant but failed neutralization against the omicron variant except for a very high concentration above the currently recommended therapeutic dosage. In patients with delta variant infections treated with casivirimab/imdevimab, we observed a rapid decrease of respiratory viral RNA at day 3 after mAb therapy. In contrast, no such prompt decline was observed in patients with delta variant or omicron variant infections receiving sotrovimab.
Keywords: casirivimab/imdevimab, COVID‐19, delta variant, monoclonal antibodies, omicron variant, SARS‐CoV‐2, sotromivab, virus neutralization assay
1. INTRODUCTION
The newly emerged severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) omicron variant (B.1.1.529) was first reported to the WHO on November 24, 2021 and has since become the predominant variant in many countries. 1 It is characterized by a high number of mutations that may be associated with enhanced transmissibility and a potentially milder clinical course compared to the previous SARS‐CoV‐2 alpha (B.1.1.7) and delta variant (B.1.617). 2 High rates of reinfection by the omicron variant have been reported, and the omicron variant has been shown to evade infection‐ or vaccine‐induced immunity. Furthermore, recent cell‐culture‐based studies show that the omicron variant is resistant to neutralization of most currently available therapeutic monoclonal antibodies (mAbs), including casivirimab/imdivimab (US: REGEN‐COV, EU: Ronapreve, Roche) but remained susceptible to in vitro inhibition by sotrovimab (Xevudy, GlaxoSmithKline). 3 , 4 Previously published studies assessed the neutralization capacity of mAb at concentrations below those that can be theoretically achieved after recommended therapeutic dosages and do not provide clinical data on treatment or outcome. 3 , 4 , 5 To shed light on this aspect, we focused here on mAb concentrations in the range of the maximum theoretically achievable therapeutic concentration in a patient after mAb administration at recommended doses. These high concentrations were tested in a virus neutralization assay (VNT) against the delta and omicron variant. 6 Additionally, we provide viral RNA kinetics of patients infected with either SARS‐CoV‐2 delta or omicron variant treated with the respective mAb formulations.
2. METHODS
2.1. Study and ethics
Clinical data were collected from coronavirus disease 2019 (COVID‐19) patients hospitalized at either the University Medical Center Hamburg‐Eppendorf (UKE) (n = 17) or the Klinik Favoriten, Vienna, Austria (n = 8). Due to the retrospective nature of the study, the need for informed consent was waived by the Ethics Committee of the Medical Council of Hamburg (WF‐052/20) and the Ethics Committee of the City of Vienna.
2.2. SARS‐CoV‐2 molecular diagnostics
Nasopharyngeal swabs in UTM (MANTACC) or Amies Medium (E‐swab) were collected regularly during the hospital stay. SARS‐CoV‐2 RNA in specimens was detected and quantified as described previously 7 using commercially available assays Xpert Xpress SARS‐CoV‐2 (Cepheid), Cobas SARS‐CoV‐2 (Roche), or laboratory‐developed assays run on the Cobas6800 system (Roche), the NeuMoDx system (Qiagen) or the Light Cycler 480 II (Roche). 8 , 9 , 10 , 11 SARS‐CoV‐2 variants were discriminated via in‐house multiplex RT‐qPCR assays 12 and commercially available typing assays (VirSNiP, TiB Molbiol).
2.3. SARS‐CoV‐2 serology
Serum samples were collected as part of the clinical routine. Elecsys Anti‐SARS‐CoV‐2‐S (Roche) assay and Elecsys anti‐SARS‐CoV‐2 (Roche) assay were performed as recommended by the manufacturer on the appropriate automated device (Cobas e411, Roche Diagnostics).
2.4. Virus naturalization assays
For the in vitro neutralization assays with the delta and the omicron SARS‐CoV‐2 variants, mAb formulations were diluted to reflect potential plasma levels in treated patients. The calculation was performed based on current recommendations: For patients older than 12 years and weighing at least 40 kg, the recommended therapeutic dose is 600/600 mg for casirivimab/imdevimab 13 and 500 mg for sotrovimab. 14 Given an estimated blood volume of about 5 l in an average adult individual, maximum plasma levels will be 120/120 µg/ml for casirivimab/imdevimab and 100 µg/ml for sotrovimab. Based on this calculation, we selected a maximum mAb concentration exceeding ≥2x the conceivable plasma levels in patients for our assay. Triplicates of the mAb dilutions were mixed with an equal volume of SARS‐CoV‐2 clinical isolates, equivalent to 60 times the Median Tissue Culture Infectious Dose (TCID50) of the SARS‐CoV‐2 delta variant isolate and 55 times the TCID50 of the omicron variant isolate per sample, respectively. These concentrations represent the lowest virus concentration at which a cytopathic effect (CPE) is certain to occur after infection with the respective isolate and allows for comparability of virus concentrations used. Briefly, neutralization tests were performed as described previously. 6 After incubation at 37°C for one hour, the serum/virus mixtures were transferred to 96‐well plates containing 5.0 × 106 cells/plate of Vero cells (ATCC CRL‐1008) seeded the previous day. Following incubation for 96 h at 37°C, supernatants were discarded. The plates were fixed in 4% formaldehyde and stained with crystal violet. The highest dilution protecting two of three wells from CPE was taken as the neutralizing antibody titer.
2.5. Statistical analysis
Analysis of viral RNA kinetics was done by comparing nasopharyngeal RNA concentration on the day of mAb treatment, day 3 (±1 day), and day 7 (±1 day) within the three different treatment groups (Kruskall–Wallis test) and were performed using GraphPad Prism software version 9.0.0 (GraphPad Software).
3. RESULTS
3.1. Patients and clinical data
All 25 COVID‐19 patients were symptomatic for less than 7 days and were at high risk for progressing to severe disease due to underlying medical comorbidities. In addition, all patients were either shown to be seronegative for anti‐SARS‐CoV‐2 Spikeprotein antibodies at baseline, were unvaccinated or were at high risk of poor antibody responses following SARS‐CoV‐2 vaccination (e.g., due to immunosuppression or immunocompromise). Of those 25 patients, 72% (n = 18) had SARS‐CoV‐2 delta variant and 28% (n = 7) were confirmed for omicron variant infection. According to the SARS‐CoV‐2 variant and mAb treatment, patients were assigned to the following subgroups:
-
(i)
delta variant infection, t/w (treatment with) casirivimab/imdevimab (n = 10),
-
(ii)
delta variant infection, t/w sotrovimab (n = 8),
-
(iii)
omicron variant infection, t/w sotrovimab (n = 7).
The patients showed rather similar baseline characteristics with respect to age, sex, previous COVID‐19 vaccinations, and immunosuppression (Table 1).
Table 1.
Baseline patient characteristics for the patients with delta or omicron variant infections treated with monoclonal antibodies
| Delta, t/w casirivimab/imdevimab | Delta, t/w sotrovimab | Omicron, t/w sotrovimab | |
|---|---|---|---|
| n | 10 | 8 | 7 |
| Age, median (IQR) | 63.5 (36;67) | 71 (57;75) | 42 (33;63.5) |
| Female, n (%) | 6 (60) | 4 (50) | 3 (43) |
| Vaccination status | |||
| Unknown, n (%) | 1 (10) | 0 | 1 (14) |
| Unvaccinated, n (%) | 5 (50) | 6 (75) | 1 (14) |
| 1 Dose, n (%) | 3 (30) | 1 (13) | 0 |
| 2 Doses, n (%) | 1 (10) | 1 (13) | 4 (57) |
| 3 Doses, n (%) | 0 | 0 | 1 (14) |
| Anti‐SARS‐CoV‐2 Spike (S1 RBD) a | |||
| N/A | 1 (10) | 0 | 3 (43) |
| <0.8AU/ml | 7 (70) | 8 (100) | 2 (29) |
| >0.8AU/ml | 2 (20) | 0 | 2 (29) |
| Immunocompromise, n (%) b | 6 (60) | 3 (38) | 5 (71) |
Abbreviations: IQR, interquartile range; N/A, not available; RBD, receptor binding domain; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2; t/w, treatment with.
Anti‐SARS‐CoV‐2 Spike (S1 RBD) titer of patients before administration of the respective monoclonal antibody preparation (cut‐off: ≥0.8 AU/ml).
Number of patients with underlying immunocompromising conditions or immunosuppressive medication at the time of diagnosis of SARS‐CoV‐2 infection.
3.2. Virus neutralzation assays
The antibody combination casivirimab/imdivimab had a neutralizing effect on the SARS‐CoV‐2 delta variant at all concentrations analyzed (range: 0.006/0.006–600/600 µg/ml). In contrast, for SARS‐CoV‐2 omicron variant, only a partial neutralization was observed for the two highest concentrations of casivirimab/imdivimab analyzed (17% neutralization at 240/240 µg/ml, 67% neutralization at 600/600 µg/ml) (Figure 1A). Sotrovimab resulted in complete neutralization of the SARS‐CoV‐2 delta variant at all concentrations analyzed (range; 0.01–200 µg/ml). Similarly, sotrovimab resulted in neutralization of the omicron variant with effective neutralization observed at 0.1 µg/ml sotrovimab in our assay (Figure 1B). Notably, the lowest sotrovimab concentration used here (0.01 µg/ml) failed omicron variant neutralization. To investigate a possible effect of mutual enhancement or inhibition of sotrovimab and casivirimab/imdivimab, we additionally performed VNTs with combinations of the 2 mAB formulations. Sotrovimab was added to a final concentration of 100 µg/ml ≥10 min after initial incubation of the virus variants with imdivimab/casivirimab at varying concentrations (range: 0.006/0.006–600/600 µg/ml) resulted in complete neutralization for both SARS‐CoV‐2 variants (Figure 1C). Similarly, the addition of a fixed concentration of casirivimab/imdevimab (240/240 µg/ml) to sotrovimab in varying concentrations (range: 0.01–200 µg/ml) led to complete neutralization of both variants (Figure 1D). A combination of serial dilutions of both casirivimab/imdevimab and sotrovimab at the same concentrations used in individual assays again led to complete neutralization of the delta variant. Notably, the omicron variant was effectively neutralized at 0.1 µg/ml sotrovimab combined with 3/3 µg/ml casivirimab/imdivimab. A partial neutralization (50%) was observed at 0.01 µg/ml sotrovimab combined with 0.06/0.06 µg/ml casivirimab/imdivimab (Figure 1E).
Figure 1.
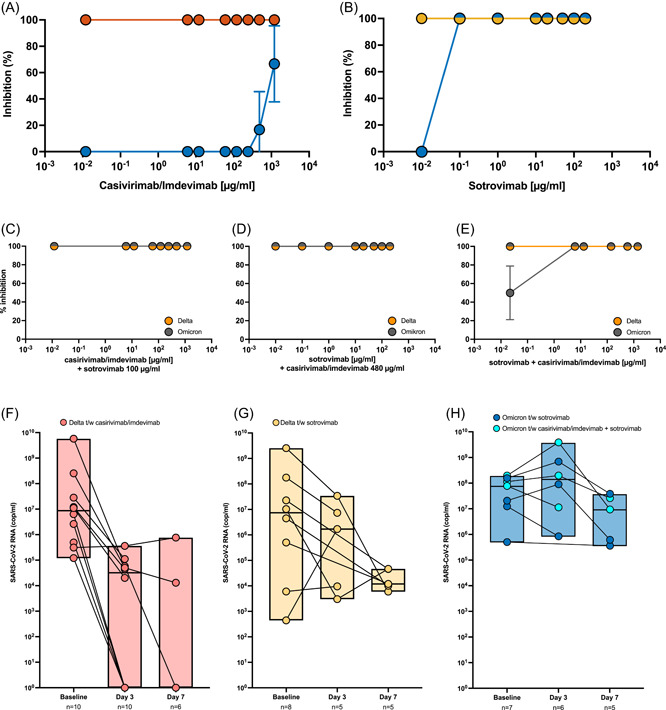
Virus neutralization and clinical efficacy by monoclonal antibody formulations. The percentage of neutralization of the severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) delta variant (A: red; B: yellow) or omicron variant (blue) by casivirimab/imdivimab (A) or sotrovimab (B) or combined monoclonal antibody (mAB) formulations (C–E) at the indicated concentrations is illustrated. (C) Sotrovimab was added to a final concentration of 100 µg/ml >10 min after initial incubation of the virus variants with casivirimab/imdevimab at the indicated concentrations. (D) A fixed concentration of casirivimab/imdevimab (240/240 µg/ml) was added, sotrovimab was assessed at indicated concentrations (E) Serial dilutions of both casivirimab/imdevimab and sotrovimab were combined. For each mAB concentration tested, triplicate dilutions were incubated with the respective variant isolate in virus neutralization assay. Kinetics of nasopharyngeal SARS‐CoV‐2 RNA loads of patients with delta variant infections treated with casirivimab/imdevimab (F) and sotrovimab (G), respectively and of patients with omicron variant infections treated with sotrovimab (dark blue) or sequential treatment with casirivimab/imdevimab and sotrovimab (light blue).
3.3. Kinetics of RNA copies in patients receiving mAB formulations
Baseline (day 0) SARS‐CoV‐2 RNA copies were comparable throughout groups (median [IQR]; delta t/w casirivimab/imdevimab: 8.65 × 106 SARS‐CoV‐2 RNA copies/ml [4.45 × 105; 8.35 × 107], delta t/w sotrovimab: 7.37 × 106 SARS‐CoV‐2 RNA copies/ml [1.30 × 105; 1.38 × 108], omicron t/w sotrovimab: 7.5 × 107 SARS‐CoV‐2 RNA copies/ml [1.2 × 107; 1.5 × 108]; p = 0.40) (Figure 1F–H).
In the group delta t/w casirivimab/imdevimab a significant decline in RNA levels was observed at Day 3 (median [IQR]; 3.2 × 104 SARS‐CoV‐2 RNA copies/ml [<level of detection (LOD); 7.1 × 104]; p = 0.004) and RNA levels remained at low to undetectable levels at Day 7 (median [IQR]; < LOD [<LOD; 2.0 × 105] in that group (Figure 1F). In the group delta t/w sotrovimab no significant decrease of viral RNA was observed at Day 3 (median [IQR]; 1.7 × 106 SARS‐CoV‐2 RNA copies/ml [6.27 × 103; 2.07 × 107], p = 0.32) and RNA levels remained at detectable levels at Day 7 (median [IQR]; 1.19 × 104 SARS‐CoV‐2 RNA copies/ml [8.01 × 103; 4.62 × 104]) in that group (Figure 1G). Similarly, no decrease in viral RNA levels was observed in omicron infected patients who received sotrovimab on Day 3 (median [IQR] 8.46 × 106 SARS‐CoV‐2 RNA copies/ml [1.39 × 108; 1.42 × 109], p = 0.16) and persistently high RNA levels were detected in this group on Day 7 (median [IQR] 4.75 × 105 SARS‐CoV‐2 RNA copies/ml [9.1 × 106; 3.1 × 107]) (Figure 1H). Among patients with omicron infections treated with sotrovimab, three patients initially received casvirimab/imdevimab at the time of diagnosis of SARS‐CoV‐2 infection followed by administration of sotrovimab the next day as virus typing, and confirmation of omicron infection was available. No difference in SARS‐CoV‐2 RNA kinetics was observed for those patients (Figure 1H, light blue dots).
3.4. Clinical outcome in patients receiving mAb formulations
None of the patients in our study had disease progression to severe COVID‐19 with systemic inflammation, pulmonary infiltrates, and respiratory compromise. However, some patients were in critical clinical condition at the time of diagnosis of SARS‐CoV‐2 infection due to underlying comorbidities, and five patients (one with delta variant infection and treatment with casirivimab/imdevimab, one with delta variant infection and treatment with sotrovimab, three with omicron variant infection and treatment with sotrovimab) died during the hospitalization. All other patients were discharged from the hospital without developing symptomatic COVID‐19.
4. DISCUSSION
Therapeutical intervention in the early phase of SARS‐CoV‐2 infection remains challenging. While both casivirimab/imdevimab and sotrovimab did show high clinical potency in preventing severe disease in clinical trials, 15 , 16 these studies were performed at a time when the omicron variant was not yet circulating. After first in vitro reports showed that the mAb preparation casivirimab/imdevimab, which was previously used with high clinical success, does not have potency against the newly emerged omicron variant, sotrovimab is recommended for patients with omicron infections qualifying for treatment with mAbs. We extended previous in vitro studies 3 , 4 by testing higher concentrations of both mAb formulations. We could show that the omicron variant was only neutralized by the highest concentrations of casivirimab/imdevimab used, whereas sotrovimab had a neutralizing effect on the omicron variant in vitro at concentrations far below the theoretically achieved therapeutic concentrations. We further demonstrate that treatment with sotrovimab in patients infected with either the delta or the omicron variant did not lead to the same rapid viral clearance in the upper respiratory tract that was observed in patients with delta variant infections treated with casivirimab/imdevimab. Notably, no difference in nasopharyngeal SARS‐CoV‐2 RNA kinetics was observed in three patients with omicron variant infections who received sequential administration of casivirimab/imdevimab and sotrovimab compared to those receiving sotrovimab alone. Our retrospective study is subject to important limitations. First, patient subgroups are relatively small and heterologous with regard to comorbidities and vaccination status. Second, clinical patient outcomes do not necessarily correlate with nasopharyngeal viral RNA concentration. Our study solely shows the in vivo effect on nasopharyngeal viral RNA concentration and is therefore not able to draw reliable and generalizable conclusions on the impact of the mAb preparations on clinical patient outcomes. Third, results of nasopharyngeal viral RNA concentration were only available for the first week after treatment for most patients so we are not able to provide viral kinetics for a longer time period. Third, since we did not treat any patients with omicron infections with casirivimab/imdevimab alone, we are not able to assess the impact of this preparation on nasopharyngeal RNA concentration or clinical outcomes in COVID‐19 patients.
5. CONCLUSION
In summary, we confirm that sotrovimab exhibits good neutralizing capacity in vitro both against both the SARS‐CoV‐2 delta and the omicron variant, while casivirimab/imdevimab has no significant efficacy against the omicron variant even at high dosages exceeding plasma levels conceivable at the current recommendations for therapy. In addition, our clinical data show that SARS‐CoV‐2 RNA concentration in nasopharyngeal swabs declines more rapidly in patients infected with delta variant receiving mAb treatment with casivirimab/imdevimab compared to patients with delta or omicron variant infections treated with sotrovimab. These data suggest that the in vitro potency of sotrovimab may not be easily transferred to a clinical effect in patients with SARS‐CoV‐2 infections and that further studies are needed to evaluate the benefits of mAB therapy for patients with omicron variant infections based on clinical outcomes.
AUTHOR CONTRIBUTIONS
Conceptualization: Thomas Theo Brehm, Susanne Pfefferle, Stefan Schmiedel, Marc Lütgehetmann; formal analysis: Thomas Theo Brehm, Susanne Pfefferle, Ronald von Possel, Marc Lütgehetmann; project administration: Thomas Theo Brehm, Susanne Pfefferle, Ronald von Possel, Stefan Schmiedel, Marc Lütgehetmann; resources: Thomas Theo Brehm, Susanne Pfefferle, Ronald von Possel, Mario Karolyi, Alexander Zoufaly, Dominic Wichmann, Robin Kobbe, Petra Emmerich, Dominik Nörz, Martin Aepfelbacher, Julian Schulze zur Wiesch, Marylyn M. Addo, Stefan Schmiedel, ML; supervision: Stefan Schmiedel, Marc Lütgehetmann; writing—original draft: Thomas Theo Brehm, Susanne Pfefferle, ML; writing—review and editing: all authors.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
Informed consent was waived by the Ethics Committee of the Medical Council of Hamburg (WF‐052/20) and the Ethics Committee of the City of Vienna in view of the retrospective nature of the study and all the procedures being performed were part of the routine care. Only aggregate data, not personal data, is published to ensure the anonymity of patients.
ACKNOWLEDGMENTS
Open Access funding enabled and organized by Projekt DEAL.
Brehm TT, Pfefferle S, von Possel R, et al. Clinical efficacy and in vitro neutralization capacity of monoclonal antibodies for severe acute respiratory syndrome coronavirus‐2 delta and omicron variants. J Med Virol. 2022;94:5038‐5043. 10.1002/jmv.27916
Thomas Theo Brehm and Susanne Pfefferle contributed equally to this study.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Karim SSA, Karim QA. Omicron SARS‐CoV‐2 variant: a new chapter in the COVID‐19 pandemic. Lancet. 2021;398(10317):2126‐2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nealon J, Cowling BJ. Omicron severity: milder but not mild. Lancet. 2022;399(10323):412‐413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. VanBlargan LA, Errico JM, Halfmann PJ, et al. An infectious SARS‐CoV‐2 B.1.1.529 omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat Med. 2022;28:1‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hoffmann M, Krüger N, Schulz S, et al. The omicron variant is highly resistant against antibody‐mediated neutralization: implications for control of the COVID‐19 pandemic. Cell. 2021;S0092‐8674(0021):01495‐01491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Takashita E, Kinoshita N, Yamayoshi S, et al. Efficacy of antibodies and antiviral drugs against Covid‐19 omicron variant. N Engl J Med. 2022;386:995‐998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brehm TT, Pfefferle S, von Possel R, et al. SARS‐CoV‐2 reinfection in a healthcare worker despite the presence of detectable neutralizing antibodies. Viruses. 2021;13(4):661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Heinrich F, Nentwich MF, Bibiza‐Freiwald E, et al. SARS‐CoV‐2 blood RNA load predicts outcome in critically ill COVID‐19 patients. Open Forum Infect Dis. 2021;8(11):ofab509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nörz D, Fischer N, Schultze A, et al. Clinical evaluation of a SARS‐CoV‐2 RT‐PCR assay on a fully automated system for rapid on‐demand testing in the hospital setting. J Clin Virol. 2020;128:104390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pfefferle S, Reucher S, Norz D, Lutgehetmann M. Evaluation of a quantitative RT‐PCR assay for the detection of the emerging coronavirus SARS‐CoV‐2 using a high throughput system. Euro Surveill. 2020;25(9):2000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Norz D, Hoffmann A, Aepfelbacher M, Pfefferle S, Lutgehetmann M. Clinical evaluation of a fully automated, laboratory‐developed multiplex RT‐PCR assay integrating dual‐target SARS‐CoV‐2 and influenza A/B detection on a high‐throughput platform. J Med Microbiol . 2021;70:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nörz D, Frontzek A, Eigner U, et al. Pushing beyond specifications: evaluation of linearity and clinical performance of the cobas 6800/8800 SARS‐CoV‐2 RT‐PCR assay for reliable quantification in blood and other materials outside recommendations. J Clin Virol. 2020;132:104650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nörz D, Grunwald M, Tang HT, et al. Rapid automated screening for SARS‐CoV‐2 B.1.617 lineage variants (Delta/Kappa) through a versatile toolset of qPCR‐Based SNP detection. Diagnostics. 2021;11(10):1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. https://www.ema.europa.eu/en/documents/product-information/ronapreve-epar-product-information_en.pdf
- 14. https://www.ema.europa.eu/en/documents/product-information/xevudy-epar-product-information_en.pdf
- 15. Weinreich DM, Sivapalasingam S, Norton T, et al. REGN‐COV2, a neutralizing antibody cocktail, in outpatients with Covid‐19. N Engl J Med. 2021;384(3):238‐251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gupta A, Gonzalez‐Rojas Y, Juarez E, et al. Early treatment for Covid‐19 with SARS‐CoV‐2 neutralizing antibody sotrovimab. N Engl J Med. 2021;385(21):1941‐1950. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


