Abstract
Background
Kinase gene fusions are strong driver mutations in neoplasia; however, kinase intergenic-breakpoint rearrangements (IGRs) confound the detection of such fusions and of targeted treatments. We aim to provide an overview of kinase IGRs in a large lung cancer cohort and examine real-world survival outcomes of patients with such fusions.
Methods
Mutational profiles analyzed using targeted next-generation sequencing of 425 cancer-related genes between June 2016 and July 2019 were retrospectively reviewed. Patients’ demographic data, clinical characteristics, and survivals were analyzed. RNA sequencing or immunohistochemical assays were carried out to verify chimeric fusion products.
Results
We identified 3411 patients with kinase fusions from a cohort of 30 450 patients with lung cancer, and 624 kinase IGR events were identified in 538 of the 3411 patients. The most frequently identified kinase genes included anaplastic lymphoma kinase (ALK), RET proto-oncogene (RET), ROS proto-oncogene 1 (ROS1), Erb-B2 receptor tyrosine kinase 2/3 (ERBB2/3), and epidermal growth factor receptor (EGFR). Our data showed that most (67%) kinase IGRs occurred on the same chromosome and kinase domains remained intact at the 3′-end. Approximately 3% (19/624) of the kinase IGRs had one genomic breakpoint located in gene promoter regions, including nine fusion events involving ALK, RET, ROS1, EGFR, ERBB2, or fibroblast growth factor receptor 3 (FGFR3). Among the 538 patients with kinase IGRs, 167 (31%) lacked oncogenic driver mutations, among which 28 received targeted therapies in real-world practice. Notably, three ALK IGR patients who harbored no canonical oncogenic aberrations were confirmed with an EML4-ALK chimeric fusion product by RNA sequencing and/or ALK immunohistochemical assays. One patient demonstrated a favorable clinical outcome after 14 months on crizotinib. An additional two patients who had ROS1 IGRs demonstrated a clinical benefit after 13 and 19 months on crizotinib, respectively.
Conclusion
A large real-world lung cancer cohort with kinase IGRs was comprehensively analyzed for their molecular characteristics. The data indicated the potential oncogenic function of kinase IGRs and their outcomes following the administration of targeted therapies.
Key words: kinase fusion, intergenic breakpoints, targeted next-generation sequencing, real-world survival, lung cancer
Highlights
-
•
Kinase IGRs were recurrent in patients with lung cancer, with clinically-druggable kinase genes being the most frequent.
-
•
The molecular features of IGRs were studied, e.g. inter-/intra-chromosome, kinase domain integrity, and promoter involvement.
-
•
A subset of DNA-level IGR events was validated by RNA-seq and/or IHC assays which confirmed the chimeric fusion products.
-
•
The potential oncogenic function of kinase IGRs was illustrated and may facilitate the administration of targeted therapies.
Introduction
Since the first identification of an ELM4-anaplastic lymphoma kinase (ALK) fusion gene in a non-small-cell lung cancer (NSCLC) patient in 2007,1 increasing numbers of kinase fusions have been uncovered with multiple different partners. Such canonical gene fusions including, but not limited to, ELM4-ALK,1 CD74-ROS1,2 and CCDC6-RET3 were identified as oncogenic drivers in multiple cancer types. Encouragingly, existing targeted therapies, and tyrosine kinase inhibitors (TKIs)4 in particular, exhibit favorable responses, and have been approved by the Food and Drug Administration for treating fusion-positive patients. Until now, several companion diagnostic platforms for the clinical detection of actionable gene fusions have been used,5,6 including FISH, immunohistochemistry (IHC), RT-PCR, and next-generation sequencing (NGS). NGS can evaluate multiple targets simultaneously and its ability to detect fusion partners and breakpoints has led to the discovery of novel gene fusions7 that may be potential oncogenic drivers, especially in cases lacking canonical mutations or fusions.
Recently, InterGenic-breakpoint Rearrangements (IGRs) have drawn broad attention, in which at least one breakpoint occurs within an intergenic region. Such cases have reported IGR events in patients with lung cancer, some of which benefited from targeted therapies, such as crizotinib.8, 9, 10, 11, 12, 13, 14, 15, 16, 17 Whether the IGRs detected by DNA sequencing could result in the production of chimeric coding transcripts and translate into chimeric fusion proteins, however, is uncertain, and thus requires RNA and/or protein level validation.18 Nevertheless, IGRs may still be targetable typically in cases lacking canonical targets.
Herein, we sought to study the molecular characteristics of kinase IGR events in a large lung cancer cohort and their clinical outcomes.
Materials and methods
Patients
A large cohort of lung cancer cases (N = 30 450) was subjected to comprehensive genomic profiling using targeted NGS of 425 cancer-related genes in a Clinical Laboratory Improvement Amendments-certified, College of American Pathologists-accredited laboratory (Nanjing Geneseeq Technology, Jiangsu, China), as previously described.19 Patients who had kinase IGRs were identified in our Laboratory Information Management System using a natural language search program. Patients’ demographic and clinical data, including age, sex, histology type, pathological stage, and treatment history, were retrospectively evaluated. This study was approved by the ethics committee of the First Affiliated Hospital of Zhengzhou University Hospital, Henan, China. All participants provided written informed consent before sample collection.
DNA extraction and targeted enrichment
Circulating tumor DNA from plasma was purified using the Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. Genomic DNA from white blood cells was extracted using the DNeasy Blood & Tissue Kit (Qiagen), while formalin-fixed paraffin-embedded (FFPE) genomic DNA was purified using the QIAamp DNA FFPE Tissue Kit (Qiagen). All DNA was quantified using the dsDNA HS Assay Kit on a Qubit Fluorometer (Life Technologies, Bleiswijk, The Netherlands). Sequencing libraries were prepared using the KAPA Hyper Prep Kit (KAPA Biosystems, Wilmington, MA), as described previously.19 Indexed DNA libraries for all solid tumors were pooled for probe-based hybridization capture of the targeted 425 cancer-related genes.
Sequence data processing
Sequencing was carried out using the HiSeq 4000 platform (Illumina, Essex, UK), followed by data analysis, as previously described.20 In brief, sequencing data were analyzed by Trimmomatic21 to remove low-quality (quality <15) or N bases, and were then mapped to the human reference genome, hg19, using the Burrows-Wheeler Aligner (https://github.com/lh3/bwa/tree/master/bwakit). PCR duplicates were removed by Picard (available at https://broadinstitute.github.io/picard/). The Genome Analysis Toolkit (GATK) (https://software.broadinstitute.org/gatk/) was used to perform local realignments around indels and base quality reassurance. Gene fusions were identified by Factera.22 Somatic single-nucleotide polymorphisms (SNPs) and indels were analyzed by VarScan223 and Mutect2, with the mutant allele frequency cut-off at 2% for tissue samples, 0.2% for cell-free DNA samples, and a minimum of three unique mutant reads. Common SNPs were excluded if they were present in >1% population frequency in the 1000 Genomes Project or the Exome Aggregation Consortium 65 000 exomes database. The resulting mutation list was further filtered by an in-house list of recurrent artifacts based on a normal pool of whole blood samples. The gene rearrangements, with one breakpoint within a coding region of kinase genes and the other in an intergenic region, were identified as kinase IGRs, the allele frequency cut-off of which was 0.3%.
RNA sequencing
Total RNA from FFPE samples was extracted using the RNeasy FFPE kit (Qiagen). The total RNA was quantified using a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). Ribosomal RNA and residual genomic DNA were depleted using the KAPA Stranded RNA-Seq Kit (KAPA Biosystems) with RiboErase (HMR) and DNase digestion, followed by purification using the Agencourt RNA Clean XP Beads (Beckman Coulter, Beverly, MA) according to the manufacturer's protocol. The KAPA Stranded RNA-Seq Library Preparation Kit was used to construct Illumina-compatible sequencing libraries, including RNA fragmentation and priming, double-stranded cDNA synthesis, adaptor ligation, and PCR amplification.
Sequencing was carried out on Illumina HiSeq NGS platforms (Illumina). To generate sequence reads in the FASTQ format, base calling was carried out on bcl2fastq v2.16.0.10 (Illumina, Inc.) and quality control was carried out with Trimmomatic (version 0.33).21 RNA-Seq reads were mapped to the human genome (hg19, Genome Reference Consortium GRCh37) using STAR (version 2.5.3a)24 to identify individual exon, intron, and intergenic features. The average coverage of mapped reads across the base positions of the feature coordinates was calculated. Gene fusions were detected and visualized on the Integrative Genomics Viewer.25
IHC and assessment
IHC staining was carried out as previously described.26 In brief, paraffin embedded specimen (4 mm sections) slides were baked at 65°C for 30 min, then deparaffinized with xylene and rehydrated with ethanol. For antigen retrieval, slides were steamed in 1× EDTA (pH 9.0) at 99-100°C for 10 min. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide for 10 min. The slides were incubated with an anti-ALK antibody (1/250; D5F3; Cell Signaling Technology, Danvers, MA) overnight at 4°C in a humidified chamber. Using the OptView DAB IHC Detection Kit (Model: 6396500001; Roche, Basel, Switzerland), the tissue sections were treated with biotinylated anti-rabbit secondary antibody, followed by further incubation with streptavidin-horseradish peroxidase. The antigen-antibody complexes were visualized using 3,3′-diaminobenzidine (DAB) and counterstained with 10% Mayer’s hematoxylin, dehydrated, and mounted in Crystal Mount. The degree of immunostaining of the FFPE sections was reviewed and scored independently by two pathologists, based on both the proportion of positively stained tumor cells and the intensity of staining. Tumors with positive staining in >1% of tumor cells were defined as positive.
Results
The landscape of kinase IGRs in patients with lung cancer
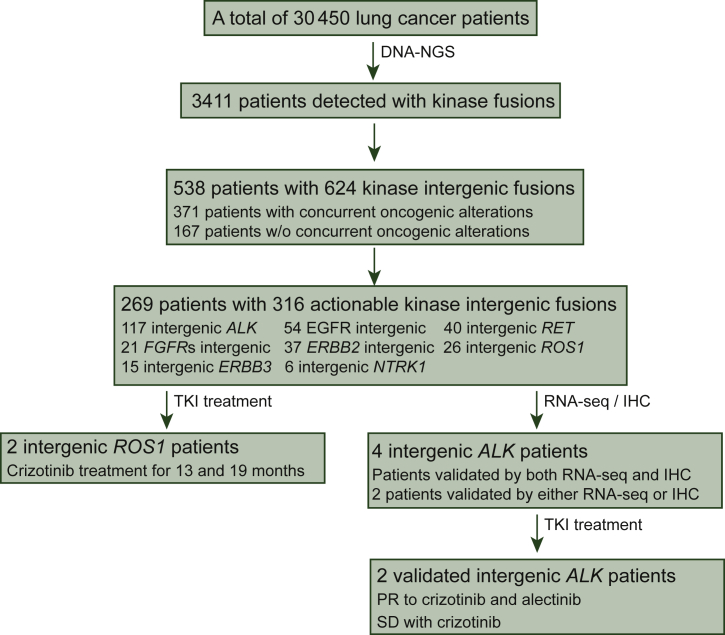
We retrospectively reviewed the mutational profiles of a large cohort of Chinese patients with lung cancer (N = 30 450) whose tumor tissue samples and/or liquid biopsies were subject to targeted NGS between June 2016 and July 2019. Approximately 11% of the cohort (3411/30 450) contained kinase gene rearrangements. In particular, a subset of 538 unique patients (1.8%) were identified to have kinase IGR events (Figure 1 and Table 1). Kinase IGRs were detected in diverse sample types, including tumor tissue (70%) and liquid biopsy specimens (30%). ALK was the most frequently rearranged kinase gene in the cohort, accounting for 117 out of the 624 kinase IGRs, followed by EGFR (54/624, 9%), RET (40/624, 6%), ERBB2 (37/624, 6%), and ROS1 (26/624, 4%) (Figure 2A). For each kinase IGR, the detection frequencies in tumor tissues and liquid biopsy samples were not significantly different (Supplementary Figure S1, available at https://doi.org/10.1016/j.esmoop.2022.100405). The other druggable kinase genes, including FGFRs and NTRK1, were also recurrent in the cohort with relatively low frequencies (3% and 1%, respectively).
Figure 1.
Patient enrollment and inclusion. This flow chart depicts the patients screened and the number of patients included in the analysis.
IHC, immunohistochemistry; NGS, next-generation sequencing; PR, partial response; RNA-seq, RNA sequencing; SD, stable disease; TKI, tyrosine kinase inhibitor.
Table 1.
Clinical characteristics of the study population
| Patient information | All (N = 538) |
|---|---|
| Median age, years (range) | 59 (11-96) |
| Sex, n (%) | |
| Male | 289 (53.7) |
| Female | 241 (44.7) |
| Unknown | 8 (1.5) |
| Lung cancer subtype, n (%) | |
| NSCLC | 424 (78.8) |
| SCLC | 7 (1.3) |
| Not determined | 107 (19.9) |
| Histology cancer type, n (%) | |
| Adenocarcinoma | 375 (69.7) |
| Squamous carcinoma | 32 (5.9) |
| Adenosquamous carcinoma | 4 (10.7) |
| Not determined | 127 (23.6) |
| Stage, n (%) | |
| I | 12 (2.2) |
| II | 9 (1.7) |
| III | 20 (3.7) |
| IV | 201 (37.4) |
| Not determined | 296 (55.0) |
NSCLC, non-small-cell lung cancer; SCLC, small-cell lung cancer.
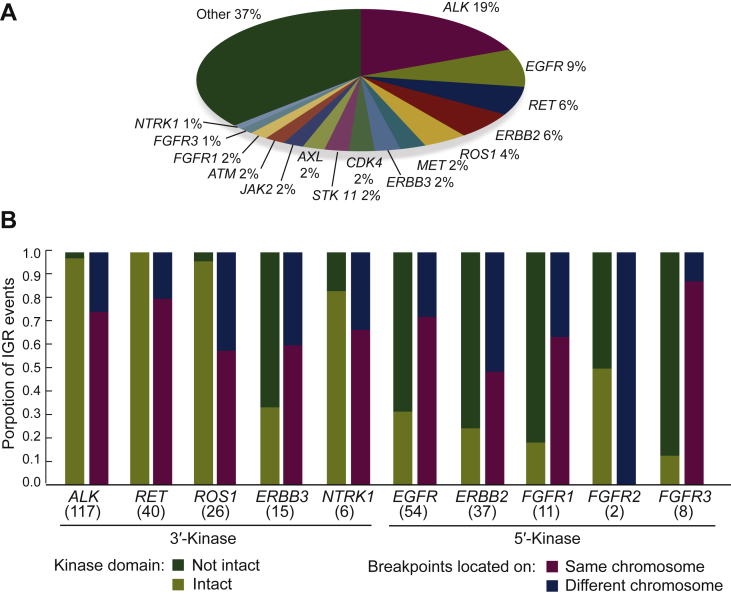
Figure 2.
Statistical analysis of the incidence of kinase IGRs and breakpoint locations.
(A) Pie chart shows the distribution of kinase genes in a total of 624 kinase IGR events. (B) Proportions of IGRs with intact and non-intact kinase domain are shown in the blue and orange bars on the y-axis. The two breakpoints of IGR located on the same chromosome are colored in green versus yellow for different chromosomes. The number of IGR events with the 10 actionable kinase genes are labeled below in parentheses.
IGR, intergenic-breakpoint rearrangement.
We next examined the genomic position of intergenic breakpoints. Kinase gene rearrangements often occurred with intergenic breakpoints on the same chromosome (67.3%, 420/624), and the majority (71.5%, 446/624) of the kinase IGRs were 3′-kinases (Figure 2B). Notably, clinically actionable kinase genes, including ALK, EGFR, RET, ERBB2/3, ROS1, FGFR1/2/3, and NTRK1 were detected in a total of 316 kinase IGRs in 269 patients (Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2022.100405). Fusions with an intact kinase domain were observed in 219 kinase IGRs (69.3%, 219/316). In most cases, the breakpoints spanned the entire coding regions of kinase genes, without significant hotspots. In contrast, ALK- and RET-involved IGR events tended to converge at the kinase domains. Specifically, 90.6% (106/117) of ALK breakpoints were located at exon 19/intron 19 and 94.9% (37/39) of that occurred between intron 10 and intron 11 of the RET gene. Intergenic breakpoints were rarely recurrent, however, except for one fusion that fused the intergenic region between NFYC and KCNQ4 to exon 48 of MTOR.
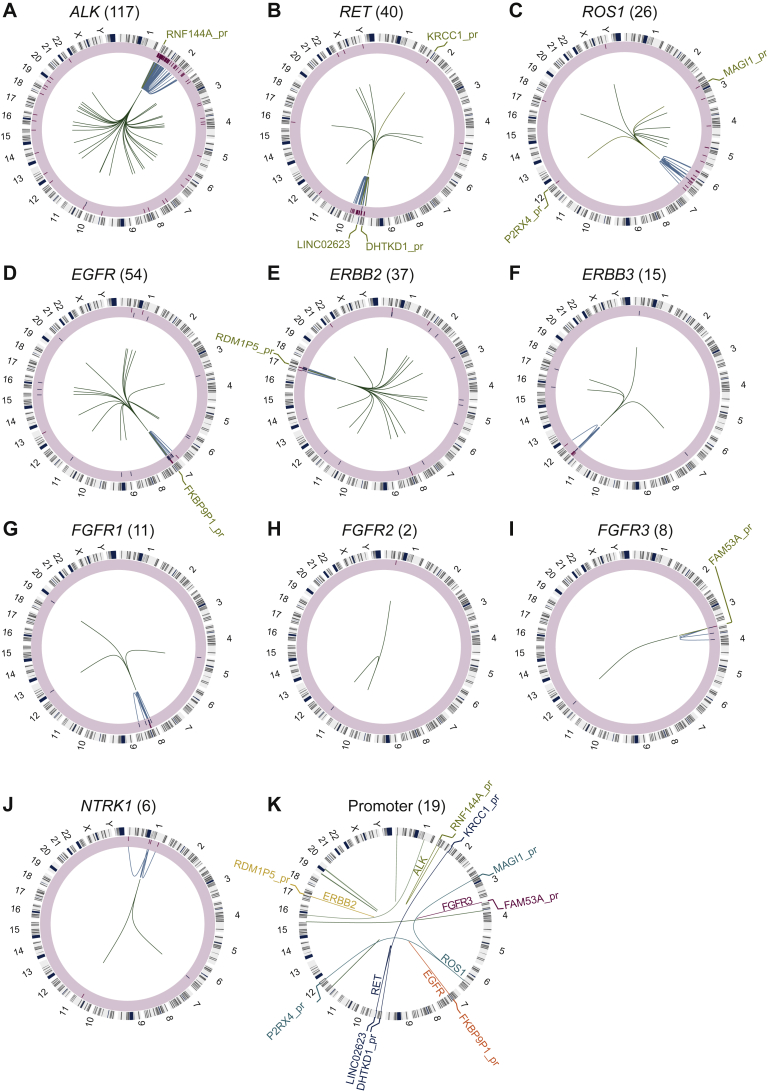
In addition, we identified 19 kinase IGR events in which a kinase gene was fused to a promoter region (Figure 3), including ALK (n = 1), RET (n = 3), ROS1 (n = 2), EGFR (n = 1), ERBB2 (n = 1), and FGFR3 (n = 1) (Supplementary Table S2, available at https://doi.org/10.1016/j.esmoop.2022.100405). ALK, RET, and ROS1 were observed to have an intact kinase domain in six cases, which may thereby result in the up-regulation of the oncogenic kinase caused by the promoter of the upstream fusion partner.
Figure 3.
Circos plots for the 10 actionable kinase gene IGRs.
(A-J) The circos plot for each kinase gene IGR. From outside to inside: track 1 is the chromosome annotation. Track 2 with a light-yellow shade represents the intactness of the kinase domain of the IGR (intact: red bars in the outer layer; non-intact: blue bars in the inner layer). Track 3 shows the intra-chromosomal rearrangements colored in light blue. The green lines in track 4 are the inter-chromosomal IGRs. The IGRs with promoter involvement are colored in orange with the gene promoter labeled. (K) All of the promoter-involved IGRs are shown. The actionable kinase genes are highlighted (ALK: olive green, ERBB2: yellow, EGFR: red, RET: light blue, ROS1: teal, and FGFR3: burgundy) and the rest are colored in green.
ALK, anaplastic lymphoma kinase; EGFR, epidermal growth factor receptor; ERBB2, Erb-B2 receptor tyrosine kinase 2; FGFR3, fibroblast growth factor receptor 3; RET, RET proto-oncogene; ROS1, ROS proto-oncogene 1.
Co-alterations
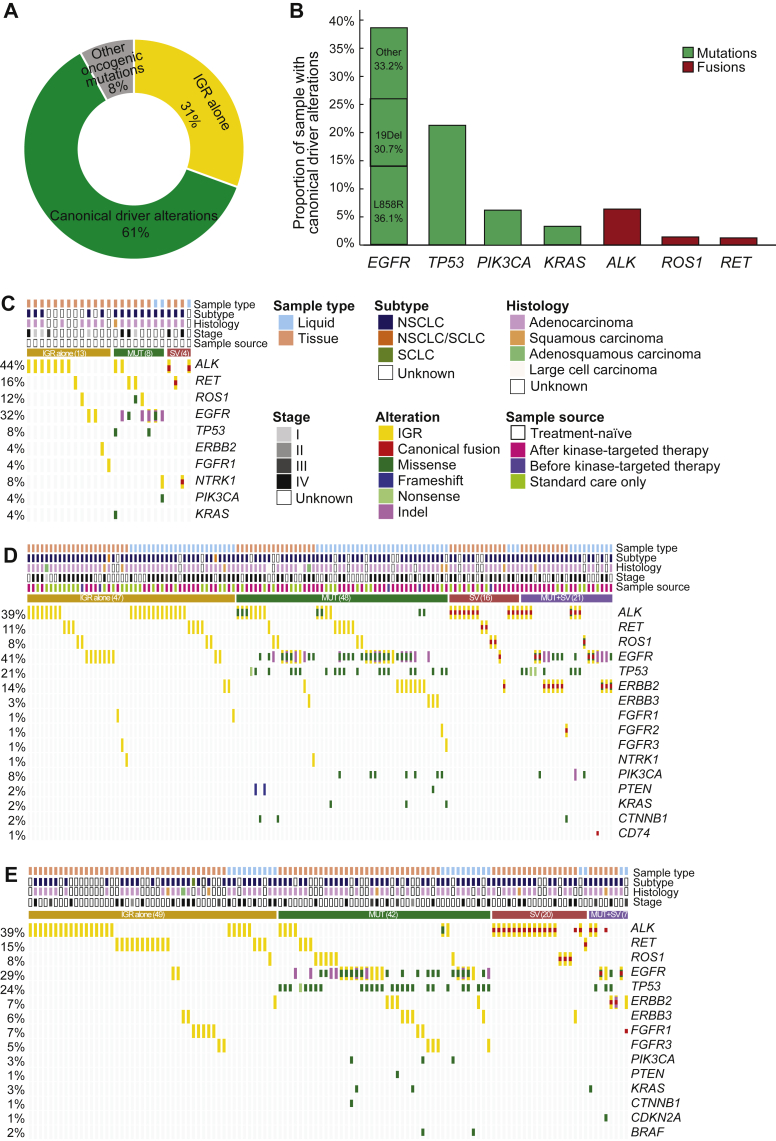
Analyses of concurrent alterations revealed that 31% of samples (167/546) lacked any oncogenic alterations (Figure 4A). EGFR mutations (38.6%) and ALK fusions (6.4%) were the most frequently detected mutations and structural variants in the entire cohort, followed by other oncogenic driver alterations, including TP53 (21.2%), PIK3CA (6.2%), KRAS (3.3%), ROS1 fusion (1.5%), and RET fusion (1.3%). EGFR L858R was the most abundant variant, accounting for 36.1% of all EGFR mutations (Figure 4B). To gain an understanding of the therapeutic implications of the druggable kinase IGRs, we focused on a total of 275 samples collected from 269 patients (Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2022.100405), which were classified into three categories: treatment-naïve (Figure 4C, N = 25), formally-treated (Figure 4D, N = 132), and treatment history unknown (Figure 4E, N = 118). In the treatment-naïve and treatment history unknown subgroups, the majority of kinase IGRs were detected in tissue samples (treatment-naïve: 22/25, 88%; unknown: 94/118, 80%). Relatively equal tissue and liquid samples were analyzed in the formally-treated subgroup (tissue: 65 versus liquid: 67); however, no significant differences in kinase IGR frequencies were found between tissue and liquid biopsies in all three categories. Consistent throughout the entire cohort, EGFR and TP53 were the most frequently mutated genes in all three categories.
Figure 4.
Concurrent mutational and clinical information analysis of IGRs.
(A) Proportion of samples from the entire cohort with only IGR (31%, 167/546), canonical oncogenic driver alterations listed in Figure 4B (61%, 335/546), or other oncogenic driver alterations (8%, 44/546). (B) The proportion of selected oncogenic driver alterations in the entire cohort. The detailed concurrent alterations of (C) treatment-naïve, (D) formally-treated, and (E) treatment information unavailable samples with actionable kinase intergenic fusions are shown by the oncoprint plots. For each category, samples were classified into four subgroups: IGR alone (golden), concurrent with MUT (canonical driver mutation) (green), SV (structure variant) (red), and both MUT and SV (purple). Different alterations are represented in different colors, as shown in the legend. Top annotations include the sample type, cancer subtype, tumor histology, disease stage, and treatment information (if available), as shown in the legend.
IGR, intergenic-breakpoint rearrangement; NSCLC, non-small-cell lung cancer; SCLC, small-cell lung cancer.
Notably, kinase IGRs were detected in 13 treatment-naïve samples (52%) that had no accompanying oncogenic driver mutations or canonical structural variants (IGR alone). Among them, one patient (patient 6: ALK IGR) received crizotinib treatment based on the NGS results, which led to a 14-month clinical benefit.
Clinical outcomes of targeted therapies in patients with ALK and ROS1 IGRs
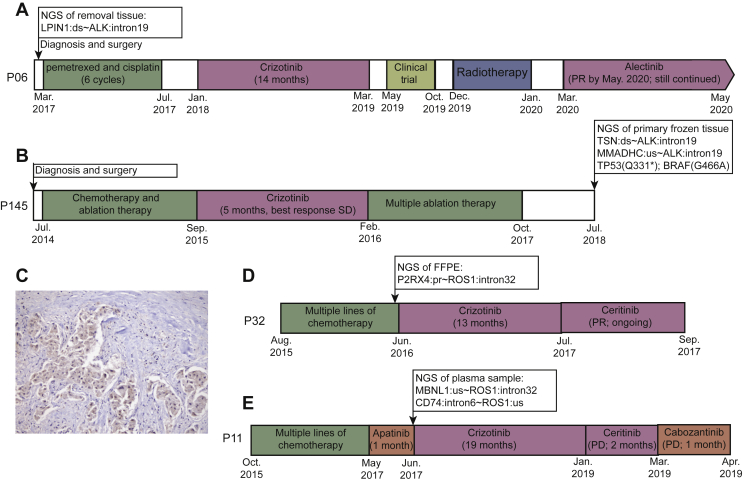
Among the 167 patients containing kinase IGRs without oncogenic alterations, 28 patients received targeted therapies in real-world practice. In particular, targeted treatments were administrated in six patients after kinase IGR detection, and three of which exhibited a favorable response from crizotinib (Figure 5A, D, and E). Therefore, the clinical significance of kinase intergenic fusions remained to be determined. Alternative detection methods such as RNA sequencing, IHC, or FISH are typically recommended to validate the presence of functional fusion products. As shown in Table 2, one patient, patient 6 (Figure 5A), who was diagnosed with NSCLC in March 2017, underwent surgical removal and received six cycles of adjuvant chemotherapy (pemetrexed and cisplatin). An ALK IGR, of which the 5′ intergenic breakpoint was located downstream of LPIN1 on chromosome 2, was identified in the patient’s tumor tissue, without any other concomitant oncogenic alterations. RNA sequencing further revealed messenger RNA transcripts of EML4-ALK in the same tumor sample, and the patient demonstrated clinical benefits from crizotinib (14 months) and alectinib. A similar observation was also made in a second patient (as shown in Table 2), patient 145 (Figure 5B), who harbored an ALK IGR, without other known oncogenic mutations. Patient 145 experienced a 5-month stable disease following crizotinib treatment, before the discovery and validation of an EML4-ALK fusion by RNA sequencing and IHC in the patient’s primary tumor tissue (Figure 5C).
Figure 5.
The treatment history of four patients benefiting from targeted TKI therapy.
The treatment history of patient 6 (A), patient 145 (B), patient 32 (D), and patient 11 (E) are shown. ALK-TKIs are indicated by the blue boxes. The arrows with boxes above the timeline indicate the timepoint of NGS with sequencing results (only fusions and oncogenic mutations are shown). (C) The ALK immunohistochemistry result from the primary tissue sample from patient 145 with ×20 magnification.
ALK, anaplastic lymphoma kinase; FFPE, formalin-fixed paraffin-embedded; NGS, next-generation sequencing; PR, partial response; SD, stable disease; TKI, tyrosine kinase inhibitor.
Table 2.
RNA sequencing and IHC validation of four ALK IGRs and their clinical outcomes
| Patient | DNA level ALK IGR | RNA sequencing | ALK IHC | ALK-TKI exposure | Best response |
|---|---|---|---|---|---|
| 6 | Downstream LPIN1∼ALK:intron19 | + | NA | Crizotinib alectinib | Partial response |
| 81 | Upstream TMEM178A∼ALK:intron19 | NA | + | NA | NA |
| 145 | Downstream TSN∼ALK:intron19; upstream MMADHC∼ALK:intron19 | + | + | Crizotinib | Stable disease |
| 198 | Upstream ABHD2∼ALK:intron19 | + | + | NA | NA |
ALK, anaplastic lymphoma kinase; IGR, intergenic-breakpoint rearrangement.
Furthermore, we also observed favorable clinical outcomes in two other patients who harbored ROS1 IGRs. As shown in Figure 5D and E, after the failure of multiple lines of chemotherapy, patients 32 and 11 underwent targeted NGS which uncovered a ROS1 IGR, but without any other canonical oncogenic mutations. ROS1 was fused to the promoter region of P2RX4, which may have up-regulated the expression of the ROS1 kinase domain. Both patient 32 and patient 11 achieved favorable clinical benefits from crizotinib treatment for 13 and 19 months, respectively. Neither case, however, was validated by alternative approaches due to sample shortages.
Discussion
The largest lung cancer IGR cohort was reported in this study. We sought to investigate the molecular characteristics and the clinical applications of these rarely detected gene rearrangements in a real-world setting. Consistent with previous studies, ALK IGR was detected with the highest incidence in our cohort. Interestingly, all of the 10 actionable kinases were detected with only one type of rearrangement, either 5′- or 3′-kinase. Moreover, the 3′-kinase rearrangements tended to retain the intact kinase domain. It is possible that the kinase domain in these proteins was localized to the C-terminus, which was more likely to be retained when the gene was rearranged to the 3′-end of the IGR.
We also paid particular attention to breakpoints within promoter regions. The rearrangements causing the translocation of promoter regions were reported to up-regulate gene expression in specific diseases such as medulloblastoma27 and aromatase excess syndrome.28 In the entire cohort, 3.0% (19/624) of IGRs were promoter-involved rearrangements and a similar proportion (2.8%, 9/316) was observed in the subgroup of the 10 actionable kinase-involved IGRs. Furthermore, six of the nine promoters were rearranged with the intact kinase domain of ALK, RET, and ROS1, and thus, we focused on these gene promoters. For example, RNF144A, whose promoter was fused with ALK kinase domain in our study, is a tumor suppressor gene that is epigenetically silenced by promoter hypermethylation in patients with breast cancer.29 Whereas in ovarian cancer, KRCC1 was found to be highly expressed and associated with poor prognosis.30 We identified one IGR in which the KRCC1 promoter and the RET kinase domain were rearranged together. The third example was a ROS1 IGR where the intact kinase domain of ROS1 was fused to the promoter of MAGI1. MAGI1 acts as a tumor suppressor in multiple cancer types, such as colorectal cancer,31 gastric cancer,32 and estrogen receptor-positive breast cancer.33 Thus, it would be of great value to quantify the expression of these IGRs to further investigate the outcome of promoter-involved IGRs at the RNA and/or protein level. To comprehensively study the discovery of IGR events in lung cancer, we searched PubMed publications and conference abstracts for reports of intergenic fusion cases. By June 2021, a total of 74 IGR cases were identified by DNA-based NGS in 19 single-case reports or cohort studies8, 9, 10, 11, 12, 13, 14, 15, 16, 17,34, 35, 36, 37, 38, 39, 40, 41, 42 (Supplementary Table S3, available at https://doi.org/10.1016/j.esmoop.2022.100405). Fifty-three of those cases were ALK fusions. Additionally, IHC, which is the most frequently used validation method for chimeric fusion products, was applied in 12 of the publications. Notably, 17 patients with ALK IGRs responded to ALK-TKIs (14 were treated with crizotinib, 1 with alectinib, 1 with ensartinib, and 1 with crizotinib and alectinib) with partial responses, 11 of which were validated by IHC. The remaining six cases that responded to ALK-TKIs were not validated by any RNA or protein level methods. Surprisingly, three cases with positive FISH or IHC results showed no response to crizotinib. The remaining 54 patients were not exposed to crizotinib or their treatment information was not available. Taken together, the IGR cases in patients with lung cancer were rarely reported and it is not possible to predict prognosis following targeted therapies.
In a retrospective study with IHC/FISH-confirmed ALK-positive NSCLC patients, ALK IGR was detected by DNA NGS in 15 patients and the median progression-free survival of those who received first-line crizotinib did not significantly differ from others with known partners of ALK gene rearrangements, suggesting the potential role of ALK IGR as a therapeutic target for ALK inhibitors.40 The majority of samples in our database (84.1%, 132/157), however, were collected after disease progression with the physician’s treatment choice. Therefore, we only identified three IGRs without any canonical driver mutations and structural variants before targeted therapies. With the available sample from one of the three cases, the EML4-ALK fusion was identified by RNA sequencing resulting from the ALK IGR. This case indicated the existence of a complicated splicing mechanism that could transcribe IGRs into functional chimeric RNAs. As expected, the patient achieved a favorable response to crizotinib for 14 months. Due to the unavailability of sufficient samples for further RNA- or protein level tests, however, whether the majority of kinase IGR events produced chimeric fusions is unknown and is one of the limitations of this retrospective study. Thus, it was not possible to determine the presence or absence of fused transcripts and/or proteins in this study. Furthermore, we identified ∼0.5% (167/30 450) of patients who carried kinase IGRs without oncogenic mutations. With the broader application of TKIs for treating fusion-positive patients, we believe that the DNA level detection of kinase IGRs should be considered when administrating targeted therapies, especially when no canonical biomarkers are detected. Of note, other RNA/protein level examinations to confirm the production of gene fusions are also strongly recommended.
Another limitation of this study was the lack of paired tissue and liquid biopsy samples collected simultaneously from the same patient to evaluate the consistency of kinase IGR detection. The frequencies of kinase IGRs detected in tissue samples and liquid biopsies, however, were not significantly different in the entire cohort. ALK IGR was the most frequent in tissue samples and liquid biopsies, followed by EGFR and RET IGRs.
Conclusion
IGR is rarely detected and reported in patients with lung cancer, with very limited clinical information available. Our study substantially expanded the dataset of NGS-detected IGRs in a real-world setting. Using comprehensive molecular analyses and clinical information, we reported on the potential of IGRs to serve as the druggable targets, however, further validation at the RNA and/or protein levels is highly recommended.
Acknowledgements
We thank the patients and family members who provided their consent to present the data in this study, as well as the investigators and research staff involved in this study.
Funding
None declared.
Disclosure
YM, QO, and XW are employees of Nanjing Geneseeq Technology Inc., Jiangsu, China. All other authors have declared no conflicts of interest.
Contributor Information
D. Lu, Email: deganlu@126.com.
X. Li, Email: fcclixy1@zzu.edu.cn.
Supplementary data
The detection frequencies of kinase IGRs in tumor tissue or liquid biopsy samples
The absolute number (label in each column) and the percentage of patients (y-axis) who were detected of each kinase-IGR in tumor tissue (blue) and liquid biopsies (orange) are shown. Note: n.s., not significant.
References
- 1.Soda M., Choi Y.L., Enomoto M., et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 2.Jun H.J., Johnson H., Bronson R.T., de Feraudy S., White F., Charest A. The oncogenic lung cancer fusion kinase CD74-ROS activates a novel invasiveness pathway through E-Syt1 phosphorylation. Cancer Res. 2012;72(15):3764–3774. doi: 10.1158/0008-5472.CAN-11-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang R., Hu H., Pan Y., et al. RET fusions define a unique molecular and clinicopathologic subtype of non-small-cell lung cancer. J Clin Oncol. 2012;30(35):4352–4359. doi: 10.1200/JCO.2012.44.1477. [DOI] [PubMed] [Google Scholar]
- 4.Pottier C., Fresnais M., Gilon M., Jerusalem G., Longuespee R., Sounni N.E. Tyrosine kinase inhibitors in cancer: breakthrough and challenges of targeted therapy. Cancers (Basel) 2020;12(3):731. doi: 10.3390/cancers12030731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong D., Yip S., Sorensen P.H. Methods for identifying patients with tropomyosin receptor kinase (TRK) fusion cancer. Pathol Oncol Res. 2020;26(3):1385–1399. doi: 10.1007/s12253-019-00685-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heyer E.E., Blackburn J. Sequencing strategies for fusion gene detection. Bioessays. 2020;42 doi: 10.1002/bies.202000016. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J., White N.M., Schmidt H.K., et al. INTEGRATE: gene fusion discovery using whole genome and transcriptome data. Genome Res. 2016;26(1):108–118. doi: 10.1101/gr.186114.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fei X., Zhu L., Zhou H., Qi C., Wang C. A novel intergenic region between CENPA and DPYSL5-ALK exon 20 fusion variant responding to crizotinib treatment in a patient with lung adenocarcinoma. J Thorac Oncol. 2019;14(9):e191–e193. doi: 10.1016/j.jtho.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 9.Li W., Liu Y., Li W., Chen L., Ying J. Intergenic breakpoints identified by DNA sequencing confound targetable kinase fusion detection in NSCLC. J Thorac Oncol. 2020;15:1223–1231. doi: 10.1016/j.jtho.2020.02.023. [DOI] [PubMed] [Google Scholar]
- 10.Chen X., Zhao G., Zhong P., Zhang M., Chen R., Zhang D. Chr2 30297612-ALK, a novel intergenic fusion with exon18 of ALK, responds to crizotinib. Clin Lung Cancer. 2020;21:e524–e566. doi: 10.1016/j.cllc.2020.04.014. [DOI] [PubMed] [Google Scholar]
- 11.Zhao R., Zhang J., Han Y., et al. Clinicopathological features of ALK expression in 9889 cases of non-small-cell lung cancer and genomic rearrangements identified by capture-based next-generation sequencing: a Chinese retrospective analysis. Mol Diagn Ther. 2019;23(3):395–405. doi: 10.1007/s40291-019-00389-y. [DOI] [PubMed] [Google Scholar]
- 12.Tian P., Liu Y., Zeng H., et al. Unique molecular features and clinical outcomes in young patients with non-small cell lung cancer harboring ALK fusion genes. J Cancer Res Clin Oncol. 2020;146(4):935–944. doi: 10.1007/s00432-019-03116-6. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y., Zeng L., Zhou C., et al. Detection of nonreciprocal/reciprocal ALK translocation as poor predictive marker in patients with first-line crizotinib-treated ALK-rearranged NSCLC. J Thorac Oncol. 2020;15(6):1027–1036. doi: 10.1016/j.jtho.2020.02.007. [DOI] [PubMed] [Google Scholar]
- 14.Qiu Y., Li B., Zhang Y., et al. ALK-rearranged lung adenocarcinoma patient with development of severe sinus bradycardia after treatment with crizotinib: a case report. Medicine (Baltimore) 2019;98(11) doi: 10.1097/MD.0000000000014826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J., Zou C., Zhou C., et al. A novel linc00308/D21S2088E intergenic region ALK fusion and its enduring clinical responses to crizotinib. J Thorac Oncol. 2020;15(6):1073–1077. doi: 10.1016/j.jtho.2020.03.009. [DOI] [PubMed] [Google Scholar]
- 16.Xia H., Xue X., Ding H., et al. Evidence of NTRK1 fusion as resistance mechanism to EGFR TKI in EGFR+ NSCLC: results from a large-scale survey of NTRK1 fusions in Chinese patients with lung cancer. Clin Lung Cancer. 2020;21(3):247–254. doi: 10.1016/j.cllc.2019.09.004. [DOI] [PubMed] [Google Scholar]
- 17.Ou S.I., Lee T.K., Young L., et al. Dual occurrence of ALK G1202R solvent front mutation and small cell lung cancer transformation as resistance mechanisms to second generation ALK inhibitors without prior exposure to crizotinib. pitfall of solely relying on liquid re-biopsy? Lung Cancer. 2017;106:110–114. doi: 10.1016/j.lungcan.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Davies K.D., Le A.T., Sheren J., et al. Comparison of molecular testing modalities for detection of ROS1 rearrangements in a cohort of positive patient samples. J Thorac Oncol. 2018;13(10):1474–1482. doi: 10.1016/j.jtho.2018.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shu Y., Wu X., Tong X., et al. Circulating tumor DNA mutation profiling by targeted next generation sequencing provides guidance for personalized treatments in multiple cancer types. Sci Rep. 2017;7(1):583. doi: 10.1038/s41598-017-00520-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Z., Yang N., Ou Q., et al. Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor osimertinib in non-small cell lung cancer patients. Clin Cancer Res. 2018;24(13):3097–3107. doi: 10.1158/1078-0432.CCR-17-2310. [DOI] [PubMed] [Google Scholar]
- 21.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newman A.M., Bratman S.V., Stehr H., et al. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics. 2014;30(23):3390–3393. doi: 10.1093/bioinformatics/btu549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koboldt D.C., Zhang Q., Larson D.E., et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dobin A., Davis C.A., Schlesinger F., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2012;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thorvaldsdottir H., Robinson J.T., Mesirov J.P. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mino-Kenudson M., Chirieac L.R., Law K., et al. A novel, highly sensitive antibody allows for the routine detection of ALK-rearranged lung adenocarcinomas by standard immunohistochemistry. Clin Cancer Res. 2010;16(5):1561–1571. doi: 10.1158/1078-0432.CCR-09-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Northcott P.A., Lee C., Zichner T., et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511(7510):428–434. doi: 10.1038/nature13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Demura M., Martin R.M., Shozu M., et al. Regional rearrangements in chromosome 15q21 cause formation of cryptic promoters for the CYP19 (aromatase) gene. Hum Mol Genet. 2007;16(21):2529–2541. doi: 10.1093/hmg/ddm145. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y., Yang Y.L., Zhang F.L., Liao X.H., Shao Z.M., Li D.Q. Epigenetic silencing of RNF144A expression in breast cancer cells through promoter hypermethylation and MBD4. Cancer Med. 2018;7(4):1317–1325. doi: 10.1002/cam4.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dwivedi S.K.D., Shameer K., Dey A., et al. KRCC1: A potential therapeutic target in ovarian cancer. FASEB J. 2020;34(2):2287–2300. doi: 10.1096/fj.201902259R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaric J., Joseph J.M., Tercier S., et al. Identification of MAGI1 as a tumor-suppressor protein induced by cyclooxygenase-2 inhibitors in colorectal cancer cells. Oncogene. 2012;31(1):48–59. doi: 10.1038/onc.2011.218. [DOI] [PubMed] [Google Scholar]
- 32.Jia S., Lu J., Qu T., et al. MAGI1 inhibits migration and invasion via blocking MAPK/ERK signaling pathway in gastric cancer. Chin J Cancer Res. 2017;29(1):25–35. doi: 10.21147/j.issn.1000-9604.2017.01.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alday-Parejo B., Richard F., Worthmuller J., et al. MAGI1, a new potential tumor suppressor gene in estrogen receptor positive breast cancer. Cancers (Basel) 2020;12(1):223. doi: 10.3390/cancers12010223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng W., Li S., Li L., Xiao M., Zhong J. A novel LINC00478/LINC01549 intergenic region-ALK fusion responded well to alectinib in a patient with lung adenocarcinoma. JTO Clin Res Rep. 2020;2:100112. doi: 10.1016/j.jtocrr.2020.100112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang M., Zhou H., Liu D., Yu R., Chen J. A case of lung adenocarcinoma harboring a rare LOC285000-ALK-NCK2 gene fusion identified by next-generation sequencing with long-term response to crizotinib. JTO Clin Res Rep. 2021;2(2):100106. doi: 10.1016/j.jtocrr.2020.100106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dou Y., Duan Q., Qi C., Hou L., Wang H. An intergenic region ALK fusion identified by DNA sequencing and validated by IHC in an early-stage lung adenocarcinoma. J Cancer Res Clin Oncol. 2021;147:1865–1867. doi: 10.1007/s00432-021-03526-5. [DOI] [PubMed] [Google Scholar]
- 37.Zhou H., Xu B., Xu J., Zhu G., Guo Y. Novel MRPS9-ALK fusion mutation in a lung adenocarcinoma patient: a case report. case report. Front Oncol. 2021;11:670907. doi: 10.3389/fonc.2021.670907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao G., Chen L., Xiao M., Yang S. Rare coexistence of three novel CDCA7-ALK, FSIP2-ALK, ALK-ERLEC1 fusions in a lung adenocarcinoma patient who responded to Crizotinib. Lung Cancer. 2021;152:189–192. doi: 10.1016/j.lungcan.2020.12.013. [DOI] [PubMed] [Google Scholar]
- 39.Zhao R., Yao F., Xiang C., et al. Identification of NTRK gene fusions in lung adenocarcinomas in the Chinese population. J Pathol Clin Res. 2021;7(4):375–384. doi: 10.1002/cjp2.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai C., Tang Y., Li Y., et al. Distribution and therapeutic outcomes of intergenic sequence-ALK fusion and coexisting ALK fusions in lung adenocarcinoma patients. Lung Cancer. 2021;152:104–108. doi: 10.1016/j.lungcan.2020.12.018. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y.L., Wu Z.Z., Zhang H.R., Chen D.S., Zhao X. Coexistence of a novel RGS18 downstream intergenic region ALK fusion and a THUMPD2-ALK fusion in a lung adenocarcinoma patient and response to crizotinib. Lung Cancer. 2021;154:216–218. doi: 10.1016/j.lungcan.2021.02.008. [DOI] [PubMed] [Google Scholar]
- 42.Qiu H., Li Q., Xiao Y., Wu D., Meng R. A novel intergenic region between KLHL31 and LRRC1-ALK exon 20 fusion variant in advanced lung adenocarcinoma and its remarkable response to ALK Inhibitor. J Thorac Oncol. 2021;16(4):e21–e23. doi: 10.1016/j.jtho.2020.12.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The detection frequencies of kinase IGRs in tumor tissue or liquid biopsy samples
The absolute number (label in each column) and the percentage of patients (y-axis) who were detected of each kinase-IGR in tumor tissue (blue) and liquid biopsies (orange) are shown. Note: n.s., not significant.