ABSTRACT
Enterovirus 71 (EV71) is deemed a reemergent pathogen, with recent outbreaks worldwide. EV71 infection causes hand, foot, and mouth disease (HFMD) and has been associated with severe cardiac and central nervous system complications and even death. Viruses need host factors to complete their life cycle; therefore, the identification of the host factors for EV71 infection is pivotal to new antiviral research. Emerging evidence has highlighted the importance of protein acetylation during infection by various human viruses. The endoplasmic reticulum (ER), as the prominent organelle of EV71 replication, also has a unique acetylation regulation mechanism. However, the pathogenesis of EV71 and its relationship with the ER-based acetylation machinery are not fully understood. In this study, we demonstrated for the first time that the ER-resident acetyltransferase N-acetyltransferase 8 (NAT8) is a host factor for EV71 infection. Inhibiting NAT8 with CRISPR or a small compound significantly suppressed EV71 infection in SK-N-SH cells. NAT8 promoted EV71 replication in an acetyltransferase-activity-dependent manner. Additionally, we found that NAT8 facilitates EV71 infection by interacting with EV71 2B, 3AB, and 3C proteins and increasing the stability of these proteins. These results uncovered a novel function of NAT8 and elucidated a new mechanism underlying the regulation of EV71 replication.
IMPORTANCE EV71 is one of the most common pathogens causing HFMD in young children, and some patients experience severe or fatal neurological consequences. To ensure efficient replication, the virus must hijack multiple host factors for its own benefit. Here, we show that the ER-resident acetyltransferase NAT8 is a host factor for EV71 infection. EV71 fails to complete its infection in various cells in the absence of NAT8. We further show that NAT8 benefits EV71 replication in an acetyltransferase-activity-dependent manner. Finally, we show that NAT8 facilitates EV71 infection by interacting with EV71 2B, 3AB, and 3C proteins and increasing the stability of these proteins. These results uncovered a novel function of NAT8 in EV71 infection and elucidated a new mechanism underlying the regulation of EV71 replication.
KEYWORDS: acetylation, enterovirus, viral replication
INTRODUCTION
Enterovirus 71 (EV71) infection can cause hand, foot, and mouth disease (HFMD), which is usually self-limiting but is highly contagious in infants and children (1). In addition to HFMD, EV71 has been shown to infect the central nervous system (CNS) and cause various neurological complications, such as aseptic meningitis, brainstem encephalitis, acute flaccid paralysis, neurogenic pulmonary edema, delayed neurodevelopment, and reduced cognitive function (2, 3). EV71 is a nonenveloped positive single-stranded RNA virus that belongs to the family Picornaviridae, genus Enterovirus, species Enterovirus A (4). The EV71 genome is about 7.5 kb long and contains one open reading frame (ORF), which encodes 2,193 amino acids and is flanked by 5′ and 3′ untranslated regions (UTRs) (5). After translation, the polyprotein is processed into P1, P2, and P3 regions, which are subsequently cleaved into four structural proteins (VP1 to VP4) and seven nonstructural (NS) proteins (2A, 2B, 2C, 3A, 3B, 3C, and 3D) (4, 6).
It is widely accepted that viruses utilize host factors to complete their life cycles (7), and protein posttranslational modification (PTM) has also been shown to play important roles during virus infection (8–10). N-terminal acetylation is one of the most common protein modifications; it mainly occurs in the mitochondria, cytosol, and nucleus, but recent studies demonstrated that it can also happen in the endoplasmic reticulum (ER) (11, 12). The process of N-terminal acetylation in the lumen of the ER requires a membrane transporter, AT-1 (SLC33A1), which translocates acetyl-coenzyme A (CoA) from the cytoplasm to the ER lumen, and two specific ER-based acetyltransferases (N-acetyltransferase 8 [NAT8] and NAT8B), which transfer the acetyl group from the donor (acetyl-CoA) to the acceptor (the lysine residue) (13). NAT8 (ATase2) and NAT8B (ATase1) are single-pass type II membrane proteins whose C-terminal catalytic domain faces the lumen of the organelle, where the reaction of lysine acetylation occurs (14). The ER acetylation machinery has been shown to play important roles in maintaining protein homeostasis and regulating autophagy within the secretory pathway (12).
In this study, we identified NAT8 as a host factor for EV71 infection with genome-wide CRISPR/Cas9 screening and demonstrated that interfering with NAT8 in SK-N-SH cells significantly decreased EV71 infection. Further investigation revealed that NAT8 promotes EV71 propagation by benefiting viral replication, which is dependent on its acetyltransferase activity. In addition, we found that NAT8 interacted with 2B, 3AB, and 3C proteins of EV71, which eventually resulted in increased stability of those proteins. Moreover, the NAT8 inhibitor can suppress the infection of EV71 in various host cells, which indicates that NAT8 is a very important host factor for EV71 infection in various human organs. These results revealed a new mechanism underlying the regulation of EV71 replication mediated by NAT8, and they suggested that NAT8 might function as a potential target for the prevention and treatment of the diseases caused by EV71 infection.
RESULTS
NAT8 is a vital host factor for EV71 infection in SK-N-SH cells.
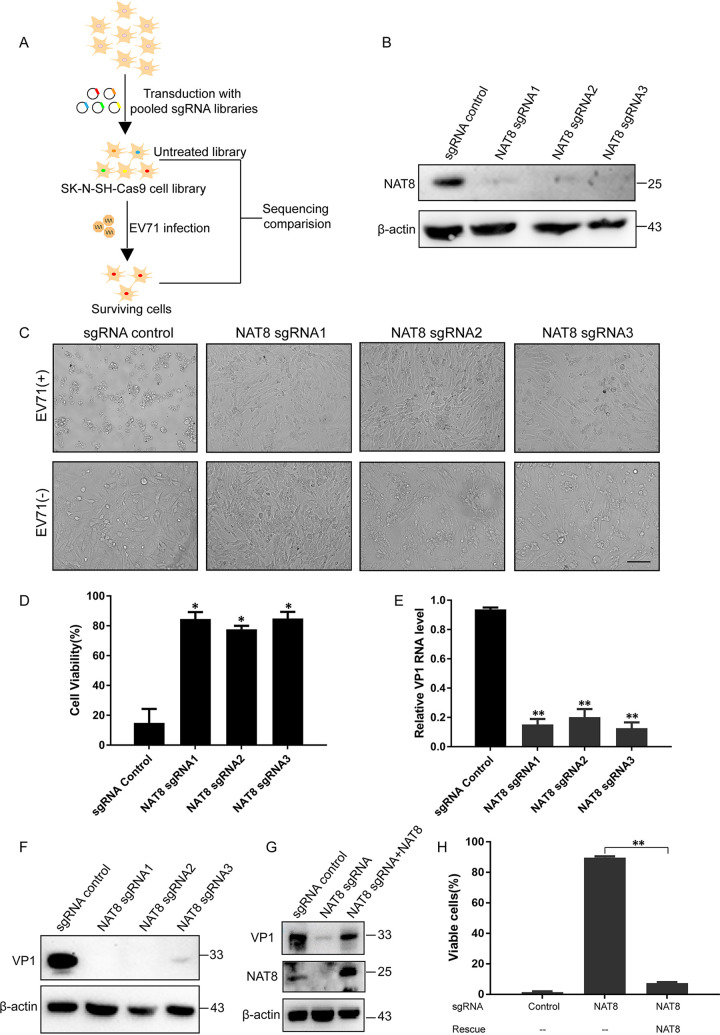
To identify the mechanisms by which EV71 hijacks the cellular machinery to promote replication and spread, we performed genome-wide CRISPR/Cas9 screening in SK-N-SH cells (Fig. 1A). Using the MAGeCK algorithm, we found strong enrichment of guide RNAs (gRNAs) targeting known host factors, including entry receptor SCARB2 and replication host factor ATG4B, suggesting that the screening was robust. Among the screened genes (see Table S1 in the supplemental material), the NAT8 gene attracted our interest for its special function and localization. NAT8 is an acetyltransferase located in the ER, where the enterovirus replication organelles form (12, 15). To verify the role of NAT8 in EV71 infection, we designed three single guide RNAs (sgRNAs) targeting NAT8. Immunoblotting results showed that all three sgRNAs could target NAT8 efficiently (Fig. 1B). When control and NAT8 knockout (KO) SK-N-SH cells were infected with EV71, cytopathic effect (CPE) was observed in control cells at 24 h postinfection (hpi), whereas it was undetectable in NAT8 KO cells (Fig. 1C). In consonance with this, cell viability of NAT8 KO cells was significantly higher than that of control cells after infection (Fig. 1D), indicating that NAT8 is required for EV71-induced cell death. Viral RNA and VP1 expression were also dramatically reduced in NAT8 KO cells (Fig. 1E and F), suggesting that NAT8 is essential for EV71 propagation.
FIG 1.
CRISPR screening identified NAT8 as a vital host factor for EV71 infection in SK-N-SH cells. (A) Schematic diagram of the genome-wide CRISPR/Cas9 screening. A library of CRISPR/Cas9 KO SK-N-SH cells were infected with EV71, and cells resistant to viral infection were selected. Genes enriched were then compared with those from uninfected cells. (B) SK-N-SH cells stably transduced with lentivirus encoding Cas9 and a sgRNA targeting the indicated gene were immunoblotted for the indicated proteins. (C) SK-N-SH cell pools as described in panel B were infected with EV71 at an MOI of 5, and CPE was examined by light microscopy at 24 hpi. Bar, 100 μm. (D) SK-N-SH cell pools as described in panel B were infected with EV71, and cell viability was determined with the CellTiter-Glo assay at 24 hpi. *, P < 0.05, compared to control sgRNA. (E) SK-N-SH cell pools as described in panel B were infected with EV71, and viral RNA was determined with RT-qPCR at 24 hpi. **, P < 0.01, compared to control sgRNA. (F) SK-N-SH cell pools as described in panel B were infected with EV71, and cells were then immunoblotted for VP1 and β-actin. (G) Control cells, NAT8 KO cells, and NAT8 KO cells transduced with sgRNA-resistant NAT8 were infected with EV71 and immunoblotted with the indicated antibodies. (H) Stably transduced cells as described in panel G were infected with EV71, and cell viability was determined 24 h later. **, P < 0.01 for NAT8 rescue, compared to no rescue.
In order to confirm the specificity of the KO cell lines, sgRNA-resistant NAT8 was introduced back into KO cell lines to restore NAT8 expression (Fig. 1G). Exogenous expression of NAT8 also restored VP1 expression (Fig. 1G) and reduced cell viability significantly when cells were infected with EV71, as expected (Fig. 1H). All of these results suggested that NAT8 is an essential host factor required for EV71 infection.
NAT8 is required for EV71 replication.
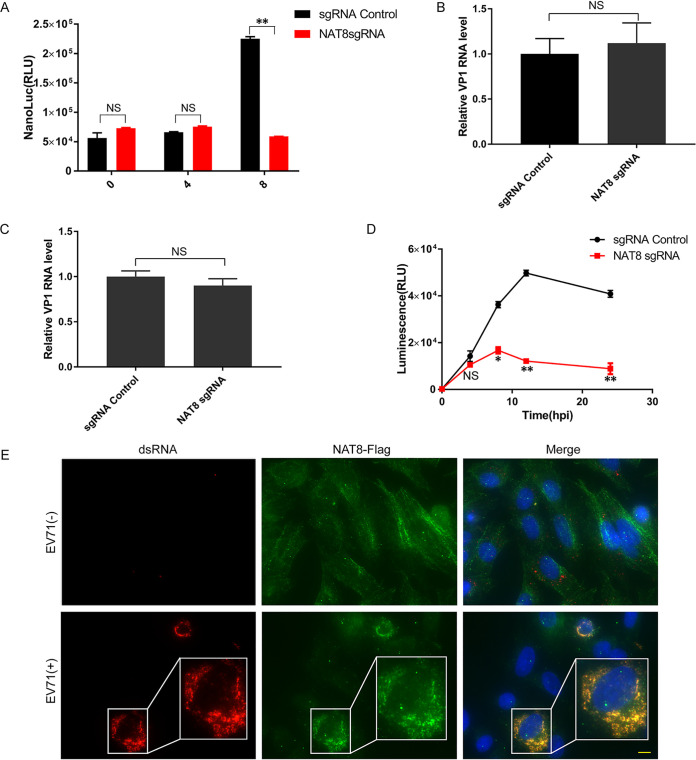
To gain more mechanistic insight into NAT8, we sought to determine at which stage of the viral life cycle NAT8 functions. For this purpose, we infected wild-type and NAT8 KO cells with a full-length virus with a NanoLuc (NL) luciferase reporter (NL-EV71) (16), and luciferase activity was measured at various time points postinfection. Comparable luciferase activity was observed at 4 hpi, while the luciferase activity was significantly lower in NAT8 KO cells at 8 hpi (Fig. 2A), suggesting that NAT8 functions at a later stage of viral infection (viral replication, virus assembly, or virus release). To confirm this, we tested whether NAT8 could influence virus attachment and internalization. Figure 2B shows that comparable amounts of virus were attached to control and NAT8 KO cells, and similar results were obtained in virus internalization assays (Fig. 2C), suggesting that NAT8 is not required for virus entry. To test the role of NAT8 in virus replication, we employed a subgenomic replicon system (SGR) containing a Renilla luciferase reporter that bypassed the virus entry process (16). When cells were transfected with this SGR RNA, NAT8 KO cells generated luciferase amounts comparable to those of control cells at 4 h posttransfection. However, when luciferase levels were determined at 8 h posttransfection or beyond, dramatic decreases were observed for NAT8 KO cells, suggesting that NAT8 is important for viral genome replication (Fig. 2D).
FIG 2.
NAT8 is required for EV71 replication. (A) Control or NAT8 KO cells were infected with NL-EV71 reporter virus, and luciferase activity was measured at the indicated time points postinfection. **, P < 0.01, compared to control sgRNA; NS, not significant. (B) NAT8 does not affect EV71 binding to the host cells. Viral RNA from the indicated SK-N-SH cells incubated with EV71 (MOI of 5) at 4°C for 1 h was determined by RT-qPCR. (C) NAT8 does not affect EV71 internalization. Viral RNA from the indicated SK-N-SH cells incubated with EV71 (MOI of 5) at 4°C for 1 h followed by 37°C for another 1 h was determined by RT-qPCR. (D) Control or NAT8 KO cells were transfected with EV71 SGR RNA, and luciferase activity was measured at the indicated time points postinfection. *, P < 0.05; **, P < 0.01, compared to control. (E) SK-N-SH cells infected with EV71 for 24 h were immunostained for dsRNA (red) and NAT8 (green). Nuclei were counterstained with DAPI (blue). Bar, 10 μm.
Because NAT8 mainly localizes to the ER, where EV71 replication occurs, we then evaluated the subcellular localization of NAT8 during EV71 infection. SK-N-SH cells overexpressing NAT8-Flag were immunolabeled with anti-double-stranded RNA (dsRNA) antibody for viral RNA and anti-Flag antibody for NAT8. Figure 2E shows that NAT8 protein colocalized with EV71 dsRNA in the cytoplasm, suggesting that NAT8 functions at the places of viral replication. All of these results revealed that NAT8 mainly participated in viral replication during EV71 infection.
Acetyltransferase activity is required for NAT8 to support EV71 infection.
NAT8 is a specific ER-based acetyltransferase, which transfers the acetyl group from the donor to the acceptor. To test whether the acetyltransferase activity of NAT8 is required for viral infection, we made a catalytically dead mutant of NAT8. A previous study reported that a mutation affecting a conserved residue (R149K) abolished the enzymatic activity of NAT8 while barely interfering with the expression of the recombinant protein (17). We then tested whether the impaired infection caused by NAT8 deficiency could be rescued by complementation with NAT8/R149K. Our results showed that, while wild-type NAT8 could rescue EV71 infection and cause cell death, NAT8/R149K failed to support EV71 infection (Fig. 3A and B). These results suggest that the catalytic activity of NAT8 is required to support EV71 infection.
FIG 3.
Acetyltransferase activity is required for NAT8 to support EV71 infection. (A) Control or NAT8 KO cells transduced with lentiviral vectors expressing wild-type or R149K mutant NAT8 were infected with EV71, and cell viability was determined at 24 hpi. **, P < 0.01 for NAT8 rescue, compared to no rescue; NS, not significant for R149K mutant rescue, compared to no rescue. (B) Cells as described in panel A were infected with EV71, and then they were immunoblotted with the indicated antibodies. (C) SK-N-SH cells infected with NL-EV71 were treated with the indicated concentration of compound 9 (C9), and luciferase activity was determined at 24 hpi. Cell viability was determined with the CellTiter-Glo assay. *, P < 0.05, compared to control; NS, not significant. (D) SK-N-SH cells infected with EV71 were treated with indicated concentrations of compound 9, and NAT8 and viral RNA were determined by RT-qPCR. *, P < 0.05, compared to control; NS, not significant. (E) Cells were treated as in panel D, and VP1 and NAT8 protein were determined by immunoblotting.
To further prove that the enzymatic activity of NAT8 is required for EV71 infection, we treated NL-EV71-infected SK-N-SH cells with a specific NAT8 inhibitor, i.e., compound 9, which has been reported to inhibit NAT8 acetyltransferase activity in vitro and in vivo (18). The results showed that the luciferase activity decreased in a dose-dependent manner, while cell viability was largely not affected (Fig. 3C). Consistent with this, viral RNA was decreased in a dose-dependent manner while NAT8 mRNA was unaffected when virus-infected cells were treated with compound 9 (Fig. 3D). However, when infected cells were tested for VP1 expression, we found both VP1 protein and NAT8 protein were inhibited after compound 9 treatment (Fig. 3E). The discrepancy between NAT8 mRNA and protein levels after compound 9 treatment was suggested by a previous report (18). Overall, these results suggest that compound 9 could inhibit EV71 infection by both inhibiting NAT8 acetyltransferase activity and downregulating its expression.
NAT8 is required for EV71 infection in different cell lines.
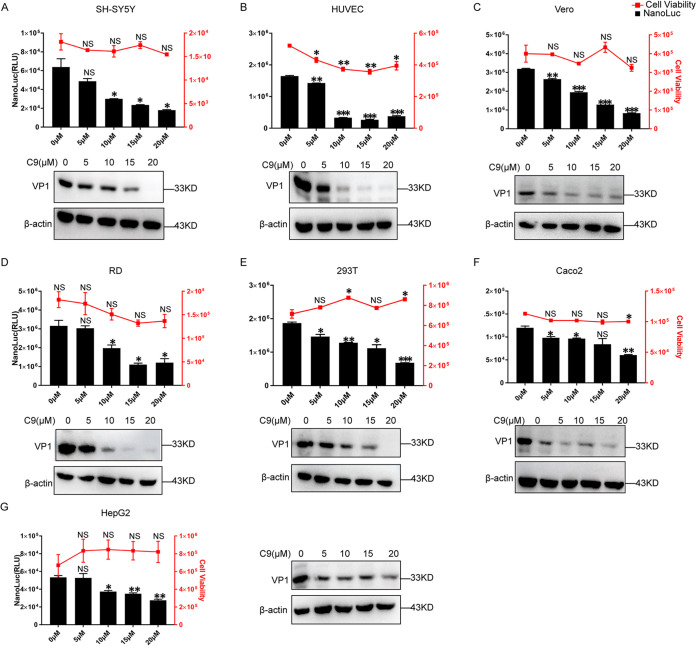
EV71 has been reported to infect different cell lines from various tissue origins (19), and we next tested whether NAT8 is required for EV71 infection in different cell lines. When SH-SY5Y, HUVEC, Vero, RD, 293T, Caco2, or HepG2 cells were infected with NL-EV71 and treated with compound 9, dose-dependent inhibition was observed for all cell lines tested (Fig. 4A to G, upper). Similarly, when these cells were infected with EV71 and VP1 expression was examined, compound 9 could inhibit VP1 expression in a dose-dependent manner (Fig. 4A to G, lower), suggesting NAT8 might play important roles in different cell lines with EV71 infection.
FIG 4.
Compound 9 (C9) inhibits EV71 infection in different cell lines. (A to G) Upper, SH-SY5Y, HUVEC, Vero, RD, 293T, Caco2, and HepG2 cells were infected with NL-EV71 at an MOI of 1, 1, 1, 0.05, 0.1, 5, and 5, respectively; 4 h later, cells were treated with the indicated amount of compound 9. NL luciferase was determined at 24 hpi. Cell viability was measured with uninfected cells to indicate the cytotoxicity of compound 9. Lower, SH-SY5Y, HUVEC, Vero, RD, 293T, Caco2, and HepG2 cells were infected with EV71 at an MOI of 1, 1, 1, 0.05, 0.1, 5, and 5, respectively; 4 h later, cells were treated with the indicated amount of compound 9. VP1 expression was determined by immunoblotting at 24 hpi. *, P < 0.05; **, P < 0.01; ***, P < 0.001, compared to control; NS, not significant.
NAT8B and AT-1 are also required for EV71 infection.
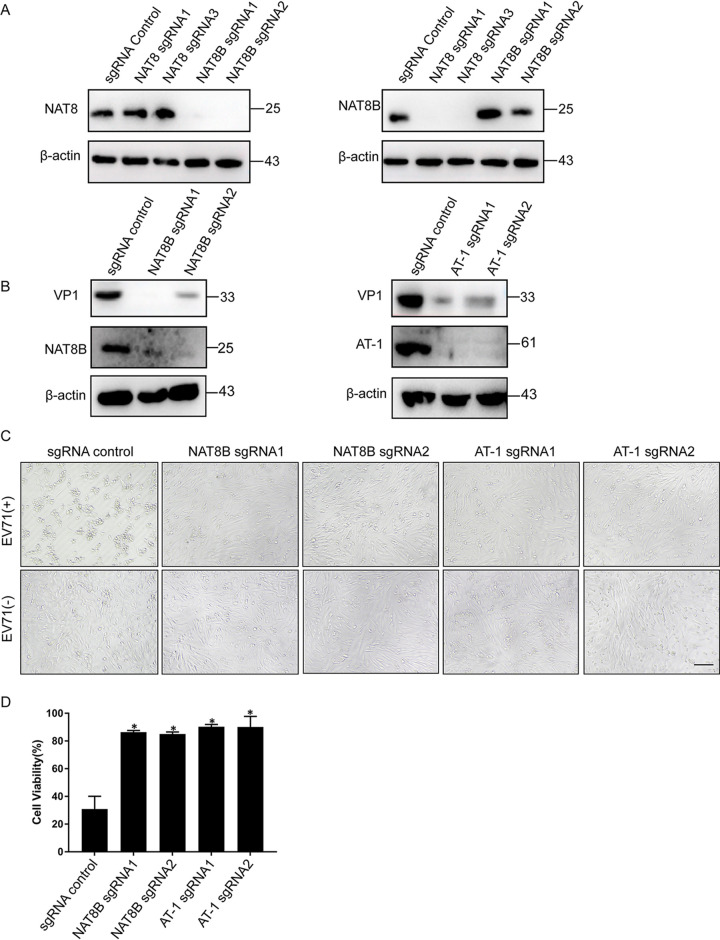
NAT8 forms homodimers or heterodimers with another acetyltransferase, NAT8B, and also requires the acetyl-CoA transporter AT-1 to exert this N-lysine acetylation activity (12, 20). We next wanted to test whether these two proteins are also required for EV71 infection. Since NAT8 and NAT8B are paralogs and have a high level of sequence similarity, we first tested whether the sgRNAs we used to generate KO cells had off-target effects. Figure 5A shows that NAT8 sgRNAs inhibit only NAT8 expression, without affecting NAT8B expression. Similarly, NAT8B sgRNAs KO only NAT8B expression, suggesting that the specificity of the sgRNAs we use is robust. Next, when NAT8B KO or AT-1 KO SK-N-SH cells were infected with EV71, viral VP1 protein was dramatically reduced (Fig. 5B). In addition, compared to control cells, CPE was undetectable in NAT8B or AT-1 KO cells (Fig. 5C), and cell viability was significantly increased when these cells were infected with EV71 (Fig. 5D). These results suggested that NAT8B and AT-1 are also important for EV71 infection.
FIG 5.
NAT8B and AT-1 are required for EV71 infection. (A) SK-N-SH cells stably transduced with lentivirus encoding Cas9 and a sgRNA targeting the indicated gene were immunoblotted for the indicated proteins. (B) SK-N-SH cells stably transduced with lentivirus encoding Cas9 and a sgRNA targeting the indicated gene were infected with EV71 and then immunoblotted for the indicated proteins. (C) SK-N-SH cell pools as described in panel B were infected with EV71 at an MOI of 5, and CPE was examined by light microscopy at 24 hpi. Bar, 100 μm. (D) SK-N-SH cell pools as described in panel B were infected with EV71, and cell viability was examined at 24 hpi. *, P < 0.05, compared to control sgRNA.
NAT8 interacts with viral NS proteins.
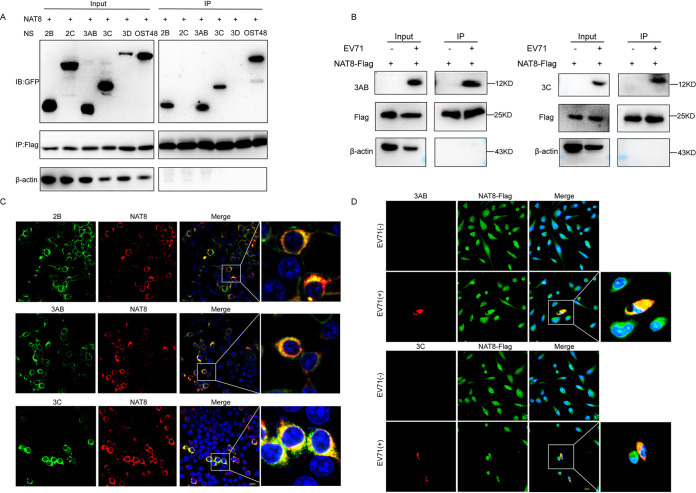
Because NAT8 resides mainly on ER and is colocalized with viral dsRNA, we speculated that NAT8 might interact with NS proteins of EV71 to promote viral replication. To test this hypothesis, we first cotransfected 293T cells with the plasmid expressing NAT8-Flag and plasmids expressing green fluorescent protein (GFP)-tagged EV71 NS proteins 2B, 2C, 3AB, 3C, or 3D. OST48, which has been reported to interact with NAT8 (20), was used as a positive control. Figure 6A shows that NS proteins 2B, 3AB, and 3C and OST48 coimmunoprecipitated with NAT8-Flag, while 2C and 3D did not, indicating that NAT8 could interact with EV71 2B, 3AB, and 3C proteins. To test whether NAT8 interacts with NS proteins during EV71 infection, SK-N-SH cells expressing NAT8-Flag were infected with EV71. Figure 6B shows that NS protein 3AB or 3C coimmunoprecipitated with NAT8. NS protein 2B was not tested due to the lack of suitable antibody. These results suggest that NAT8 interacts with viral NS proteins during EV71 infection.
FIG 6.
NAT8 interacts with the NS proteins. (A) 293T cells cotransfected with NAT8-Flag and plasmids encoding GFP-tagged 2B, 2C, 3AB, 3C, 3D, or OST48 were immunoprecipitated (IP) with anti-Flag antibody, and interactions between NAT8 and viral proteins were detected by Western blotting (immunoblot [IB]) analyses using anti-GFP antibody. (B) SK-N-SH cells overexpressing NAT8-Flag were infected with EV71 and immunoprecipitated with anti-Flag antibody, and interactions between NAT8 and 3AB or 3C protein were analyzed. (C) Colocalization of NAT8 and 2B, 3AB, and 3C in 293T cells. 293T cells cotransfected with NAT8-Flag and GFP-tagged 2B, 3AB, or 3C were immunostained with anti-Flag (red) and anti-GFP (green). Nuclei were counterstained with DAPI (blue). Bar, 10 μm. (D) Colocalization of NAT8 and 3AB or 3C in EV71-infected SK-N-SH cells. SK-N-SH cells overexpressing NAT8-Flag were infected with EV71 and immunostained with anti-Flag (green) and anti-3AB (upper, red) or anti-3C (lower, red). Nuclei were counterstained with DAPI (blue). Bar, 10 μm.
We then immunostained NAT8 with 2B, 3AB, or 3C proteins in cotransfected 293T cells and found that they were perfectly colocalized (Pearson correlation coefficients of 0.89, 0.94, and 0.92, respectively) (Fig. 6C). Furthermore, colocalization of NAT8 with 3AB or 3C was also observed in NAT8-Flag-expressing SK-N-SH cells infected with EV71 (Pearson correlation coefficients of 0.83 and 0.72, respectively) (Fig. 6D), further suggesting that NAT8 could interact with EV71 NS proteins.
NAT8 enhances the stability of NS proteins.
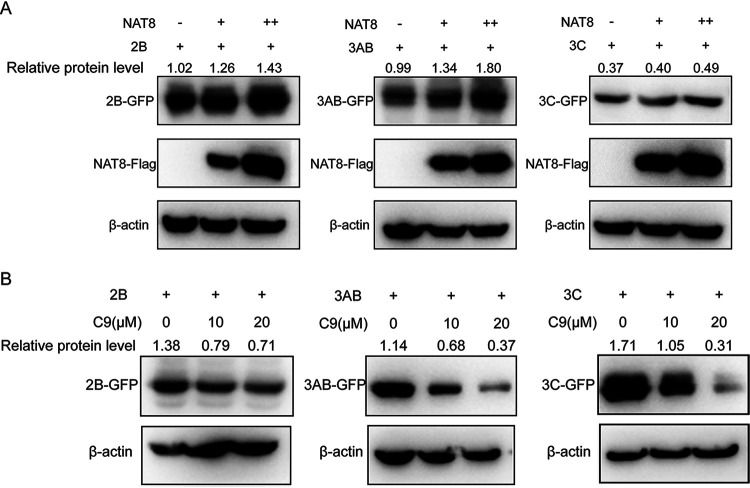
To test the effect of NAT8 on NS protein expression, 293T cells were transfected with expression vectors for GFP-NS proteins with or without NAT8 coexpression, and then the GFP-NS protein expression level was examined. Figure 7A shows that the expression of 2B, 3AB, and 3C proteins was upregulated in the presence of NAT8 proteins. On the other hand, when 293T cells transfected with expression vectors for GFP-NS proteins were treated with compound 9, which inhibited NAT8 function, the GFP-NS protein expression level was decreased in a dose-dependent manner (Fig. 7B), suggesting that NAT8 could upregulate NS protein expression.
FIG 7.
NAT8 regulates viral NS protein expression. (A) 293T cells were transfected with GFP-tagged 2B, 3AB, or 3C with increasing amounts of NAT8 expression vectors (0, 0.5, or 1 μg), and then the protein mount of NS proteins was examined by immunoblotting using an anti-GFP antibody. (B) 293T cells transfected with expression plasmids for GFP-tagged 2B, 3AB, or 3C were incubated in the presence of the NAT8 inhibitor compound 9 (C9) (0, 10, or 20 μM) for 24 h. The cell lysates were then immunoblotted with anti-GFP antibody.
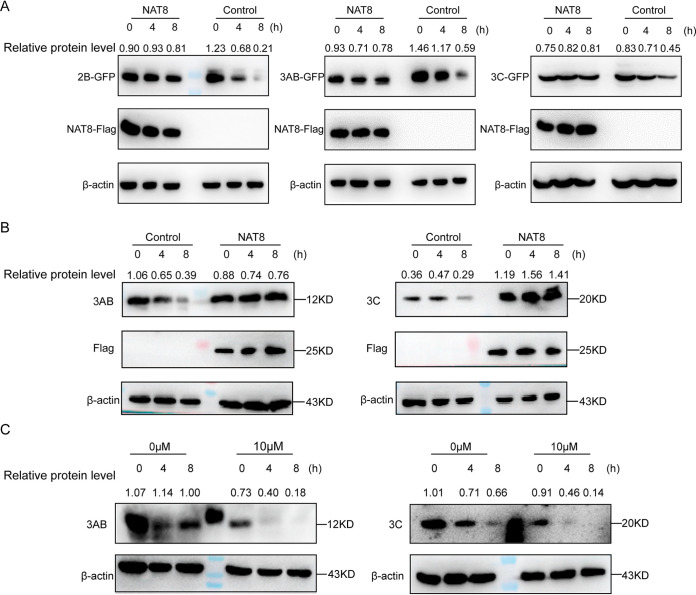
Previous studies reported that CD133 protein expression is positively regulated by NAT8 via increased stability (21), and we then wanted to test whether NAT8 increased the expression of 2B, 3AB, and 3C by enhancing the protein stability. To test this possibility, the amounts of 2B, 3AB, and 3C were determined in the presence of the protein synthesis inhibitor cycloheximide with or without NAT8 coexpression in 293T cells. In control cells, the amounts of 2B, 3AB, and 3C decreased significantly over 8 h of cycloheximide treatment (Fig. 8A); in contrast, proteins maintained similar amounts in the presence of NAT8 expression (Fig. 8A), suggesting that NAT8 could enhance 2B, 3AB, or 3C stability. Similar results were obtained when EV71-infected SK-N-SH cells were treated with cycloheximide; overexpression of NAT8 significantly increased viral 3AB and 3C stability, compared to control cells (Fig. 8B). On the other hand, when EV71-infected SK-N-SH cells were treated with compound 9 (10 μM) in the presence of cycloheximide, accelerated degradation of 3AB or 3C was observed, compared to control cells (Fig. 8C), Taken together, these results indicated that NAT8 increased the protein stability of viral NS proteins.
FIG 8.
NAT8 enhances the stability of NS proteins. (A) 293T cells transfected with a GFP-tagged 2B, 3AB, or 3C construct with or without NAT8-Flag coexpression were treated with cycloheximide (125 μM), and the amount of NS protein expression was determined with anti-GFP antibody at the indicated time points. (B) Control or NAT8-Flag-overexpressing SK-N-SH cells were infected with EV71; 24 h later, cells were treated with 125 μM cycloheximide, and the amounts of 3AB (left) or 3C (right) protein expression were determined at the indicated time points. (C) SK-N-SH cells were infected with EV71 for 4 h before they were treated with 0 or 10 μM compound 9; 24 h later, cells were treated with 125 μM cycloheximide, and the amounts of 3AB (left) or 3C (right) protein expression were determined at the indicated time points.
NAT8 stabilizes NS proteins by inhibiting autophagy-lysosome degradation.
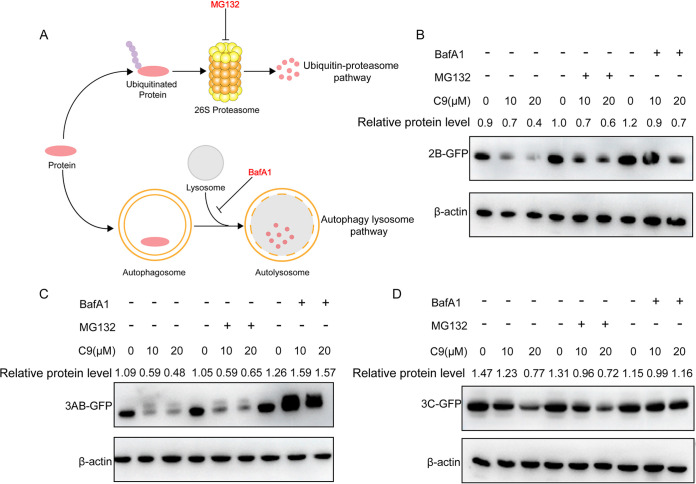
The ubiquitin-proteasome system and autophagy-lysosome degradation are the two major intracellular protein degradation pathways in eukaryotes (22, 23). Proteins degraded by proteasomes are modified by ubiquitin chains, and this pathway could be blocked by MG132 treatment, whereas autophagy-mediated protein degradation relies on fusion with lysosomes and thus can be blocked by bafilomycin A1 (BafA1) treatment (Fig. 9A). To test which pathway was involved in NS protein degradation during NAT8 inhibition, 293T cells were transfected with 2B-, 3AB-, or 3C-expressing construct and then treated with compound 9 in the presence of MG132 or BafA1. We found that BafA1antagonized the protein degradation by compound 9 treatment, while MG132 did not (Fig. 9B to D), suggesting that NAT8 increased the stability of 2B, 3AB, and 3C proteins by inhibiting autophagy-lysosome pathway-dependent degradation.
FIG 9.
NAT8 stabilizes NS proteins by inhibiting autophagy-lysosome degradation. (A) Diagram of proteasome-mediated (upper) and autophagy-mediated (lower) protein degradation. The specific inhibitor (MG132 or BafA1) targeting each pathway is indicated. (B to D) 293T cells were transfected with expression plasmids for GFP-tagged 2B (B), 3AB (C), or 3C (D) and were incubated in the presence of the NAT8 inhibitor compound 9 (C9) (0, 10, or 20 μM) with MG132 (10 μM) or BafA1 (10 nM) treatment for 24 h. The cell lysates were then immunoblotted with anti-GFP antibody.
DISCUSSION
Since EV71 was first described during an outbreak in California, its infection has emerged as a serious hazard that threatens the health of children and causes tremendous damage to both families and society (24). It is important to understand the mechanisms underlying viral infection and replication in order to prevent and control this disease. In this study, we investigated the mechanisms involved in the regulation of EV71 replication. We identify the ER-based acetyltransferase NAT8 as a pivotal host factor for EV71infection. We initially demonstrated that NAT8 KO SK-N-SH cells displayed a severe defect in EV71 infection. Further, we confirmed that NAT8 mainly targets viral RNA replication, whereas viral entry and initial translation are undisturbed.
As an important NAT on ER, NAT8 acts as an integral component of ER quality control and regulates the proteostasis of the secretory pathway (12). It is known that EV71 replication occurs in the cytoplasm, and its membranous vesicles are closely associated with the ER (15). We hypothesized that NAT8 interacted with viral NS proteins and regulated their function. Consistent with this, we showed that NAT8 could interact with 2B, 3AB, and 3C proteins and regulated their stability in transfected 293T cells. The 2A expression plasmid we constructed fail to express, possibly due to its concomitant restriction of its own expression, as reported previously (25). Furthermore, we showed that, in EV71-infected cells, NAT8 also interacted with NS proteins 3AB and 3C and enhanced their stability. These results suggest that NAT8 helps to protect viral NS proteins from degradation during viral replication and thus enhances EV71 infection.
NAT8 and NAT8B are acetyltransferases that are responsible for N-terminal acetylation in the lumen of the ER. Pehar et al. reported that a total of 143 ER-resident or ER-transiting proteins were acetylated in the ER lumen in human neuroglioma cells (26), suggesting that N-terminal acetylation in the ER lumen could display diverse biochemical functions. Therefore, NAT8 inhibition with Crispr/Cas9 or compound 9 would have a broad effect on the function of ER, and we cannot rule out the possibility that acetylation of cell host proteins could also have an effect on EV71 infection. Further research is needed to prove this.
Another important function of NAT8 is regulating the induction of autophagy (12). It was shown that ATG9A was a substrate of NAT8 and increased acetylation of this protein acted as an inhibitory signal to induce autophagy (27, 28). On the other hand, several studies showed that EV71 infection induced autophagy, which would be beneficial to viral replication (29–31). Therefore, how the virus manipulates NAT8 to regulate autophagy for its own replication during EV71 infection requires further elucidation.
In conclusion, the finding that an acetyltransferase acts as a proviral host factor for the pathogenesis of EV71 provides interesting insight into how EV71 benefits from acetyltransferase for viral replication. In addition, NAT8 may serve as an attractive host-directed antiviral target to combat EV71 infection, and the specific inhibitor compound 9 could be a prospective antiviral drug.
MATERIALS AND METHODS
Cells and reagents.
SK-N-SH and SH-SY5Y cells were maintained at 37°C in modified Eagle’s medium (MEM) (Gibco) containing 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 1 μg/mL streptomycin, in the presence of 5% CO2. 293T, RD, Caco2, HepG2, and Vero cells were maintained at 37°C in Dulbecco’s MEM (DMEM) (Gibco) supplemented with 10% FBS, 100 U/mL penicillin, and 1 μg/mL streptomycin, in the presence of 5% CO2. HUVEC cells were maintained at 37°C in endothelial cell medium (ECM) (ScienCell) containing 10% FBS, 100 U/mL penicillin, and 1 μg/mL streptomycin, in the presence of 5% CO2. The following antibodies were used in this study: NAT8 (ABclonal), NAT8B (Bioss), SLC33A1 (Bioss), β-actin (ABclonal, Woburn, MA), GFP (Thermo Fisher scientific), and Flag (Origene) antibodies. EV71 VP1, 3AB, and 3C antibodies were purchased from GeneTex. dsRNA antibody (J2) was from English and Scientific Consulting (Hungary). 4′,6-Diamidino-2-phenylindole (DAPI)- or Alexa Fluor-conjugated secondary antibodies were purchased from Thermo Fisher Scientific. Compound 9 was kindly provided by Luigi Puglielli (University of Wisconsin-Madison).
Viruses, reporter viruses, and infectious clones.
The EV71 strain and the full-length EV71 with a luciferase reporter (NL-EV71) and subgenomic replicon SGR-EV71 were described previously (16). They were propagated and titrated on RD cells.
Constructs.
The cDNA encoding human NAT8 was obtained by reverse transcription (RT) of total RNA from SK-N-SH cells, followed by PCR using the specific primers listed in Table S2 in the supplemental material. The cDNAs were subcloned into a lentiviral expression vector (pHW200) with a C-terminal Flag tag with MluI and BsiwI sites. NAT8sg3Mut and NAT8R149K mutant were obtained with overlap extension PCR amplification using the primers listed in Table S2 and then cloned were into the same vector between the MluI and BsiwI sites. To construct plasmids expressing 2B, 2C, 3AB, 3C, and 3D, the fragments were amplified from EV71 cDNA and then cloned into the lentiviral expression vector with a C-terminal GFP tag with MluI and BsiwI sites; the primers are listed in Table S2. All constructs were confirmed by sequencing.
Lentiviral packaging and transduction.
Lentivirus stock was prepared as described previously (32). Briefly, 293T cells were cotransfected with psPAX2, pMD2.G, and pLentiCrispr (for KO) or lentiviral expression vector (for overexpression). The sgRNA sequences used are listed in Table S3. Forty-eight hours posttransfection, lentivirus was harvested from the supernatant, filtered through a 0.45-μm filter, and used for transduction. Lentiviral transduction was performed on designated cell lines, and stable cell lines were generated with puromycin or blastcidin S (InVivoGen) selection.
Genome-scale CRISPR/Cas9 KO screens.
The pooled CRISPR screening was conducted as described previously (33). Briefly, the human GeCKOv2 plasmid library from Addgene was first prepared as lentiviral pools, as described above. SK-N-SH cells transduced with a lentiviral pool at a multiplicity of infection (MOI) of 0.3 were then selected with 1 μg/mL puromycin for stable cell lines. Cells were then infected with EV71 at an MOI of 0.1 for 2 weeks. Genomic DNA from surviving cells was prepared with the Quick-gDNA midikit (Zymo Research, Irvine, CA), and the integrated sgRNAs were amplified using PCR. Next-generation sequencing was performed at Oebiotech (Shanghai, China) on a MiSeq instrument (Illumina, CA), and data were analyzed with the MAGeCK algorithm (34).
Western blotting.
Cells with proper treatment were harvested with LDS sample buffer (Thermo Fisher Scientific), followed by separation on a 10% SDS-PAGE gel. Proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane before they were incubated with primary antibodies. Following a Tris-buffered saline with Tween20 (20mM Tris-Cl [pH 7.5], 150mM NaCl, 0.05% Tween20) (TBST) wash, corresponding secondary antibodies were added and bands were visualized with SuperSignal West Femto maximum sensitivity substrate (Thermo Fisher Scientific).
RNA extraction and RT-qPCR.
Different cells with or without infection were harvested, and total cellular RNA was isolated with the GeneJET RNA purification kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. cDNA was synthesized using a high-capacity cDNA RT kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. cDNA was diluted 1:20 and used directly for quantitative PCR (qPCR) analysis using specific primers and the iTaq universal SYBR green master mix (Bio-Rad). Primer sequences are listed in Table S4.
Luciferase assay.
Cells infected with NL-EV71 were harvested at 24 hpi, and luciferase signals were determined with the Nano-Glo luciferase assay system (Promega Corp.) according to the manufacturer’s instructions. For cells transfected with SGR-EV71, viral RNA was in vitro transcribed as described previously (16) and was transfected into SK-N-SH cells or NAT8 KO SK-N-SH cells using the TransIT-mRNA transfection reagent (Mirus Bio) according to the manufacturer’s instructions. Renilla luciferase was measured using a Renilla luciferase assay system (Promega Corp.) at the indicated time points. All luciferase was measured with a BioTek Neo2 microplate reader.
Viral binding and internalization assays.
Control or NAT8 KO SK-N-SH cells were incubated with EV71 (MOI of 5) at 4°C for 1 h (binding assay) or incubated at 4°C for 1 h followed by 1 h of incubation at 37°C (internalization assay). Cell media were then removed, and cells were washed five times with cold phosphate-buffered saline (PBS). Total RNA was extracted, and viral RNA was determined by qPCR as described above.
Compound 9 treatment assay.
Cells were seeded on a 24-well plate 1 day before they were infected with EV71 or NL-EV71. Four hours later, the cells were treated with compound 9 at the indicated concentrations. Cells were harvested for immunoblotting, RT-qPCR, or the luciferase assay as described above.
Protein degradation assay.
293T cells were transfected with EV71 NS-GFP proteins, and compound 9, MG132 (10 μM), or BafA1 (10 nM) was added at 12 h posttransfection and maintained for 24 h. Cells were harvested for immunoblotting as described above. For EV71-infected cells, SK-N-SH cells were either treated with 10 μM compound 9 or overexpressed NAT8-Flag. Twenty-four hours after EV71 infection, cells were treated with 125 μM cycloheximide and harvested at 0, 4, or 8 h for immunoblotting.
Coimmunoprecipitation.
For 293T cells, cells were first cotransfected with NAT8-Flag- and EV71 NS-GFP-encoding plasmids. For SK-N-SH/NAT8-Flag cells, cells were infected with EV71 at an MOI of 5. Twenty-four hours later, the cells were harvested and washed twice with cold PBS, followed by disruption with immunoprecipitation buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1% Nonidet P-40, 0.1% SDS) at 4°C for 15 min. Cell lysates were clarified by centrifugation at 1,000 × g for 5 min at 4°C. Anti-Flag antibody was mixed with the supernatant and incubated at 4°C for 1 h; the precleared beads with protein A/G were then added to the mixture and incubated at 4°C for 1 h on a roller. The reaction mixtures were then washed four times with cold wash buffer (0.05% Triton X-100) and subsequently analyzed by immunoblotting with the indicated antibodies.
Confocal microscopy.
Immunofluorescence staining was carried out as described previously (16). Briefly, cells were seeded on coverslips coated with poly-d-lysine 1 day before they were infected with EV71 (MOI of 5) and incubated for another 24 h. Cells were then fixed with ice-cold methanol for 10 min, followed by blocking with 2% FBS for 1 h. Cells were then stained with the corresponding antibodies. The images were acquired with a Nikon C2 confocal microscope.
Statistics.
Unless otherwise indicated, all values are means ± standard deviations and represent the results of a minimum of three independent experiments. The two-tailed Student's t test was used to compare the means of control and experimental groups.
ACKNOWLEDGMENTS
We thank Luigi Puglielli (University of Wisconsin-Madison) for the generous gift of compound 9.
This work was supported by the National Natural Science Foundation of China (grant 81871662) and Fundamental Research Funds for the Central Universities (grants xzy012019066 and xzy032020037).
Footnotes
Supplemental material is available online only.
Contributor Information
Hongliang Wang, Email: hongliangwang@xjtu.edu.cn.
Susana López, Instituto de Biotecnologia/UNAM.
REFERENCES
- 1.Yi EJ, Shin YJ, Kim JH, Kim TG, Chang SY. 2017. Enterovirus 71 infection and vaccines. Clin Exp Vaccine Res 6:4–14. 10.7774/cevr.2017.6.1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mao LX, Wu B, Bao WX, Han FA, Xu L, Ge QJ, Yang J, Yuan ZH, Miao CH, Huang XX, Zhang C, Xu H. 2010. Epidemiology of hand, foot, and mouth disease and genotype characterization of enterovirus 71 in Jiangsu, China. J Clin Virol 49:100–104. 10.1016/j.jcv.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 3.Chang LY, Lin HY, Gau SS, Lu CY, Hsia SH, Huang YC, Huang LM, Lin TY. 2019. Enterovirus A71 neurologic complications and long-term sequelae. J Biomed Sci 26:57. 10.1186/s12929-019-0552-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solomon T, Lewthwaite P, Perera D, Cardosa MJ, McMinn P, Ooi MH. 2010. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect Dis 10:778–790. 10.1016/S1473-3099(10)70194-8. [DOI] [PubMed] [Google Scholar]
- 5.Chen L, He YQ, Meng J, Xiong LH, Wang C, Yao XJ, Zhang HL, Zhang RL, Yang H. 2016. Full-Genome Sequences of Seven Fatal Enterovirus 71 Strains Isolated in Shenzhen, China, in 2014. Genome Announc 4:e00316-16. 10.1128/genomeA.00316-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H, Li Y. 2019. Recent progress on functional genomics research of enterovirus 71. Virol Sin 34:9–21. 10.1007/s12250-018-0071-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loboda AP, Soond SM, Piacentini M, Barlev NA. 2019. Lysine-specific post-translational modifications of proteins in the life cycle of viruses. Cell Cycle 18:1995–2005. 10.1080/15384101.2019.1639305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu B, Gao C. 2018. Regulation of MAVS activation through post-translational modifications. Curr Opin Immunol 50:75–81. 10.1016/j.coi.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 9.Zhou Y, He C, Wang L, Ge B. 2017. Post-translational regulation of antiviral innate signaling. Eur J Immunol 47:1414–1426. 10.1002/eji.201746959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Proulx J, Borgmann K, Park IW. 2020. Post-translational modifications inducing proteasomal degradation to counter HIV-1 infection. Virus Res 289:198142. 10.1016/j.virusres.2020.198142. [DOI] [PubMed] [Google Scholar]
- 11.Aksnes H, Ree R, Arnesen T. 2019. Co-translational, post-translational, and non-catalytic roles of N-terminal acetyltransferases. Mol Cell 73:1097–1114. 10.1016/j.molcel.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farrugia MA, Puglielli L. 2018. Nε-lysine acetylation in the endoplasmic reticulum: a novel cellular mechanism that regulates proteostasis and autophagy. J Cell Sci 131:jcs221747. 10.1242/jcs.221747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pehar M, Puglielli L. 2013. Lysine acetylation in the lumen of the ER: a novel and essential function under the control of the UPR. Biochim Biophys Acta 1833:686–697. 10.1016/j.bbamcr.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ko MH, Puglielli L. 2009. Two endoplasmic reticulum (ER)/ER Golgi intermediate compartment-based lysine acetyltransferases post-translationally regulate BACE1 levels. J Biol Chem 284:2482–2492. 10.1074/jbc.M804901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melia CE, Peddie CJ, de Jong AWM, Snijder EJ, Collinson LM, Koster AJ, van der Schaar HM, van Kuppeveld FJM, Barcena M. 2019. Origins of enterovirus replication organelles established by whole-cell electron microscopy. mBio 10:e00951-19. 10.1128/mBio.00951-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang H, Zhao X, Xun M, Ma C, Wang H. 2021. Reverse genetic approaches for the generation of full length and subgenomic replicon of EV71 virus. Front Microbiol 12:665879. 10.3389/fmicb.2021.665879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Veiga-da-Cunha M, Tyteca D, Stroobant V, Courtoy PJ, Opperdoes FR, Van Schaftingen E. 2010. Molecular identification of NAT8 as the enzyme that acetylates cysteine S-conjugates to mercapturic acids. J Biol Chem 285:18888–18898. 10.1074/jbc.M110.110924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding Y, Ko MH, Pehar M, Kotch F, Peters NR, Luo Y, Salamat SM, Puglielli L. 2012. Biochemical inhibition of the acetyltransferases ATase1 and ATase2 reduces β-secretase (BACE1) levels and Aβ generation. J Biol Chem 287:8424–8433. 10.1074/jbc.M111.310136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shih C, Liao CC, Chang YS, Wu SY, Chang CS, Liou AT. 2018. Immunocompetent and immunodeficient mouse models for enterovirus 71 pathogenesis and therapy. Viruses 10:674. 10.3390/v10120674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding Y, Dellisanti CD, Ko MH, Czajkowski C, Puglielli L. 2014. The endoplasmic reticulum-based acetyltransferases, ATase1 and ATase2, associate with the oligosaccharyltransferase to acetylate correctly folded polypeptides. J Biol Chem 289:32044–32055. 10.1074/jbc.M114.585547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mak AB, Pehar M, Nixon AM, Williams RA, Uetrecht AC, Puglielli L, Moffat J. 2014. Post-translational regulation of CD133 by ATase1/ATase2-mediated lysine acetylation. J Mol Biol 426:2175–2182. 10.1016/j.jmb.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun-Wang JL, Ivanova S, Zorzano A. 2020. The dialogue between the ubiquitin-proteasome system and autophagy: implications in ageing. Ageing Res Rev 64:101203. 10.1016/j.arr.2020.101203. [DOI] [PubMed] [Google Scholar]
- 23.Cohen-Kaplan V, Livneh I, Avni N, Cohen-Rosenzweig C, Ciechanover A. 2016. The ubiquitin-proteasome system and autophagy: coordinated and independent activities. Int J Biochem Cell Biol 79:403–418. 10.1016/j.biocel.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt NJ, Lennette EH, Ho HH. 1974. An apparently new enterovirus isolated from patients with disease of the central nervous system. J Infect Dis 129:304–309. 10.1093/infdis/129.3.304. [DOI] [PubMed] [Google Scholar]
- 25.Wang B, Xi X, Lei X, Zhang X, Cui S, Wang J, Jin Q, Zhao Z. 2013. Enterovirus 71 protease 2Apro targets MAVS to inhibit anti-viral type I interferon responses. PLoS Pathog 9:e1003231. 10.1371/journal.ppat.1003231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pehar M, Lehnus M, Karst A, Puglielli L. 2012. Proteomic assessment shows that many endoplasmic reticulum (ER)-resident proteins are targeted by Nε-lysine acetylation in the lumen of the organelle and predicts broad biological impact. J Biol Chem 287:22436–22440. 10.1074/jbc.C112.362871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee IH, Finkel T. 2009. Regulation of autophagy by the p300 acetyltransferase. J Biol Chem 284:6322–6328. 10.1074/jbc.M807135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. 2008. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci USA 105:3374–3379. 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu ZW, Zhuang ZC, Chen R, Wang XR, Zhang HL, Li SH, Wang ZY, Wen HL. 2019. Enterovirus 71 VP1 protein regulates viral replication in SH-SY5Y cells via the mTOR autophagy signaling pathway. Viruses 12:11. 10.3390/v12010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu Y, Xu W, Chen D, Feng C, Zhang L, Wang X, Lv X, Zheng N, Jin Y, Wu Z. 2015. Enterovirus 71 induces autophagy by regulating has-miR-30a expression to promote viral replication. Antiviral Res 124:43–53. 10.1016/j.antiviral.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 31.Huang SC, Chang CL, Wang PS, Tsai Y, Liu HS. 2009. Enterovirus 71-induced autophagy detected in vitro and in vivo promotes viral replication. J Med Virol 81:1241–1252. 10.1002/jmv.21502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Perry JW, Lauring AS, Neddermann P, De Francesco R, Tai AW. 2014. Oxysterol-binding protein is a phosphatidylinositol 4-kinase effector required for HCV replication membrane integrity and cholesterol trafficking. Gastroenterology 146:1373–1385.e1-11. 10.1053/j.gastro.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin DL, Cherepanova NA, Bozzacco L, MacDonald MR, Gilmore R, Tai AW. 2017. Dengue virus hijacks a noncanonical oxidoreductase function of a cellular oligosaccharyltransferase complex. mBio 8:e00939-17. 10.1128/mBio.00939-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, Liu XS. 2014. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 15:554. 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S4. Download jvi.00119-22-s0001.xlsx, XLSX file, 1.5 MB (1.5MB, xlsx)