Abstract
Respiratory syncytial virus (RSV) is a major cause of viral lower respiratory tract infections among infants and young children in both developing and developed countries. There are two major antigenic groups of RSV, A and B, and additional antigenic variability occurs within the groups. The most extensive antigenic and genetic diversity is found in the attachment glycoprotein, G. During individual epidemic periods, viruses of both antigenic groups may cocirculate or viruses of one group may predominate. When there are consecutive annual epidemics in which the same group predominates, the dominant viruses are genetically different from year to year. The antigenic differences that occur among these viruses may contribute to the ability of RSV to establish reinfections throughout life. The differences between the two groups have led to vaccine development strategies that should provide protection against both antigenic groups. The ability to discern intergroup and intragroup differences has increased the power of epidemiologic investigations of RSV. Future studies should expand our understanding of the molecular evolution of RSV and continue to contribute to the process of vaccine development.
INTRODUCTION
Scope of the Problem
Respiratory syncytial virus (RSV) is a major cause of viral lower respiratory tract infections among infants and young children in both developing and developed countries (102). Severe RSV bronchiolitis and pneumonia requiring hospitalization typically occur in infants less than 9 months of age (27). RSV is the most common cause of bronchiolitis. The rates of hospitalization due to bronchiolitis increased between 1980 and 1995 and accounted for over one-third of the admissions due to lower respiratory tract disease among infants younger than 1 year in 1995 in the United States. (D. K. Shay, R. C. Holman, and L. J. Anderson, Abstract, Clin. Infect. Dis. 27:928, 1998). Children with underlying illnesses such as congenital heart disease and bronchopulmonary dysplasia are at increased risk for severe infections due to RSV (27). In addition, RSV is increasingly recognized as an important pathogen in other groups, including immunocompromised patients and the elderly (37, 39; A. R. Falsey, E. E. Walsh, and R. F. Betts, Letter, J. Infect. Dis. 162:568–569, 1990). RSV is also an important cause of community-acquired pneumonia among hospitalized adults of all ages (35).
The need for effective preventive and therapeutic approaches against RSV is clear. At present there is no licensed vaccine for routine use in active immunization (36). Passive immunization with polyclonal and monoclonal antibodies (MAbs) is being used for select groups of children at high risk for severe disease due to RSV (62, 96). The available therapeutic modalities are chiefly supportive, and the role of ribavirin therapy remains controversial (5).
Challenges to Vaccine Development
One of the features of RSV that poses a challenge to vaccine development is that infections may occur in the presence of preexisting immunity. Examples include the infections that occur in young infants in the presence of maternally derived antibodies and the reinfections that are the norm throughout life (51, 56). The specific aspects of the host-parasite relationship that allow these infections have yet to be defined. Reinfections may occur by repeated exposure to the same viral isolate, so that antigenic variation is not strictly required to allow reinfections (53). However, as described in detail below, several lines of evidence suggest that antigenic variation may play a role in the ability of RSV to escape the immune response and establish infections. Another obstacle to vaccine development is the need to immunize infants in the first months of life, at a time of immunologic immaturity and interference by maternal antibodies. In addition, the formalin-inactivated vaccine that was tested in the 1960s not only failed to protect but also resulted in enhanced disease among the recipients during naturally occurring RSV infections (27, 70).
Viral Genome and Proteins
RSV is a member of the genus Pneumovirus in the family Paramyxoviridae. It has a negative-sense, nonsegmented, single-stranded RNA genome. Thus, it does not have the capacity for reassortment of genome segments, the process by which influenza virus undergoes antigenic shifts leading to influenza virus pandemics (27). However, as with other RNA viruses, RSV has a quite mutable genome by virtue of its dependence on an RNA polymerase that lacks the capacity for RNA proofreading and editing. Populations of RNA viruses exist as quasispecies, with a distribution of related, nonidentical genomes that exist in equilibrium around a theoretical consensus sequence. This genetic heterogeneity is then shaped by the selective pressures of the environment, providing for great adaptability among these viruses (34).
The RSV genome encodes the synthesis of at least 10 viral proteins. There are three transmembrane glycoproteins, i.e., the attachment glycoprotein (G), the fusion protein (F), and the small hydrophobic protein (SH). There are two matrix proteins, M and M2 (or 22K); three proteins associated with the nucleocapsid, N, P, and L; and two nonstructural proteins, NS1 and NS2. The F and G proteins are important antigenically because they stimulate the production of protective immune responses. Responses to the F protein include humoral and cytotoxic T-lymphocyte responses (21). The fusion protein of RSV is similar to that of the other paramyxoviruses in structure and function. The F protein is a type I transmembrane glycoprotein with a cleaved N-terminal signal sequence and a transmembrane anchor near the C terminus. After synthesis and modification by the addition of N-linked sugars, it is cleaved into two subunits, F1 and F2, that are linked by disulfide bonds (27).
The G protein is recognized by neutralizing antibodies but does not stimulate significant cytotoxic T-lymphocyte responses (21, 27). The G protein is of particular interest because variability in this protein is greater than that in the other proteins both between and within the major antigenic groups of RSV (27, 67). Thus, much of the data reviewed here will emphasize the G protein. The G protein is presumed to be the attachment protein of RSV by analogy to the other paramyxoviruses and on the basis of experimental data (73). However, it is said to lack the hemagglutinin or neuraminidase activity of the other paramyxovirus attachment proteins. It also differs in size and biochemistry from these proteins (119). A candidate vaccine virus (cp52) that lacks both the G and SH proteins has been described. Thus, the G protein and its presumed attachment function are dispensable for replication in cell culture. The cp52 virus is, however, severely attenuated for replication in animals, so that the requirements for replication differ between in vitro and in vivo conditions (69).
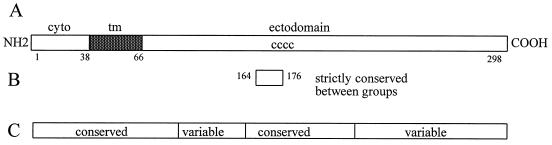
The A2 isolate of RSV, a prototype of the group A viruses, has a G mRNA of 918 nucleotides. The major open reading frame encodes a 298-amino-acid type II membrane protein with a predicted Mr of 32,600. The precursor form of the G protein is modified by the addition of N-linked sugars to Mr 45,000 and O-linked sugars to achieve the mature Mr 90,000 form. Ser or Thr, potential O-linked sugar acceptors, comprise 30% of the residues. The extensive O glycosylation, along with a proline content of 10%, gives the G protein features similar to those of the mucinous proteins of the respiratory tract. Four Cys residues in the ectodomain are conserved among all naturally occurring strains of RSV (Fig. 1) (119). Translation beginning at an internal AUG corresponding to Met48 on the full-length mRNA transcript produces a soluble form of the G protein (GS). Met48 is located in the membrane-spanning domain (residues 38 to 66). After synthesis and modification by glycosylation the protein undergoes proteolytic cleavage to produce the GS form with an N terminus corresponding to Asn66 (99). The biological functions of the GS form in the viral life cycle remain to be defined. The G and GS proteins may play an immunopathologic role in infections (68).
FIG. 1.
Variable and conserved domains of the G protein. (A) The primary amino acid structure of the G protein of the A2 isolate of RSV is shown. The cytoplasmic tail (cyto), transmembrane domain (tm, solid), and ectodomain are marked. Amino acid numbers are shown below the linear figure. The location of the four conserved cysteine residues in the ectodomain is shown (cccc). (B) The 13-amino-acid region that is strictly conserved among all naturally occurring human RSV G proteins is indicated (amino acids 164 to 176). (C) The linear regions of the G protein that are relatively conserved or variable within and between the antigenic groups are shown.
MAJOR ANTIGENIC GROUPS
Early Descriptions of Antigenic Diversity
The importance for vaccine design of developing a thorough understanding of the extent of antigenic variability of RSV was well recognized by earlier investigators of the virus. The Long strain (isolated in 1956) and the CH-18537 strain (isolated in 1962) were compared. Reciprocal antigenic differences were shown in cross-neutralization assays with postinfection ferret sera (23). These investigators raised the question, “Will it be necessary to include several different strains in order to prepare an effective RS virus vaccine?” The circulation of antigenically distinct viruses during a single epidemic period was also described. However, such antigenic variation was not thought to contribute to infections in humans (22).
Wulff et al. (136) compared the Long strain virus to viruses isolated in Kansas in 1962. Testing with animal sera revealed differences between the Long and Kansas viruses. Human sera failed to reveal such differences, although the investigators noted that children 3 to 6 months of age had higher antibody titers to the Kansas 87 virus than to the Long strain virus. Other investigators observed that the 8/60 virus (isolated in Sweden in 1960) differed immunologically from the other viruses tested. Based on antigenic differences between the 8/60 virus and same-year isolates from other countries, it was suggested that strains which vary antigenically probably circulate simultaneously in the world (32). The Long strain virus was compared to viruses from Japan isolated in 1963 and 1964. Immune sera prepared in guinea pigs revealed antigenic differences among the viruses in reciprocal neutralization tests. However, similar testing of a limited number of human sera failed to reveal antigenic differences (120).
A study of repeated infections with RSV examined 11 pairs of viruses from successive infections in children. Ferret antiserum revealed antigenic differences among six of nine pairs of viruses which were tested. However, testing with human sera from the infected patients did not reveal antigenic differences. It was concluded that antigenic variation did not explain the reinfections that occur with RSV (9).
An RSV isolate (strain 9320) from Massachusetts in 1977 was found to be antigenically distinct from the Long and CH-18537 viruses in cross-neutralization assays using animal antisera. This isolate was further examined. Virus-specific antisera to several isolates were raised in cotton rats. By using in vitro neutralization assays, only the CH-18537 virus was found to differ from the other viruses tested (this included the A2 and Long strains). The 9320 virus was different only from the CH-18537 virus (60). In vivo resistance to infection was compared after infection of cotton rats with selected viruses. Complete or almost complete resistance to challenge by both homologous and heterologous viruses was observed. It was concluded that antigenic instability of the virus was not going to be a concern for workers attempting to develop immunoprophylaxis against RSV (97).
Recognition of Two Distinct Antigenic Groups
The application of MAb technology provided new insights into RSV antigenic variability and led to the recognition that there are two major antigenic groups of these viruses. Anderson et al. (7) prepared MAbs against three strains of RSV, Long, CH-18537, and A2. These MAbs recognized the F, G, and N proteins. Three groups were identified, with the prototype virus and group as follows: Long, group 1; CH-18537, group 2; and A2, group 3. The group 1 and 3 viruses were found to be similar, while the group 2 viruses were antigenically more distinct (7). Mufson et al. (85) raised MAbs against the Long strain virus that recognized the F, G, N, M, and P proteins. Two different subtypes of the virus were identified. Subtype A included the Long and A2 viruses, while subtype B included four viruses isolated in West Virginia and the CH-18537 virus. The main differences among isolates were found on the attachment glycoprotein G. These investigators suggested that the two subtypes had evolved separately for a considerable period. They also pointed out that the existence of two subtypes had implications for understanding the ability of RSV to establish infections in the presence of a preexisting immune response and for efforts to produce a vaccine (85). These two publications were the first to clearly describe the existence of two major antigenic categories of RSV (7, 85).
Gimenez et al. (49) prepared MAbs against the RSN2 strain of RSV. Initial testing of a limited number of isolates with MAbs to the P and M proteins revealed that antigenic differences could be detected. A subsequent publication (50) reported that a greater number of isolates were tested with MAbs which recognized the F and P proteins. The reactivity with the P protein varied among the isolates tested, allowing the classification of the viruses into two antigenic types or groups. In addition, differences in P-protein mobility in sodium dodecyl sulfate-polyacrylamide gel electrophoresis were observed which corresponded to the antigenic classification of the isolates. Group 1 included the Long and A2 viruses, and group 2 included the RSN-2 virus that was classified as group 2 or subtype B in the studies described above. The authors commented that multiple antigenic variants might circulate in a geographic area over limited periods. Norrby et al. (90) also observed differences in the mobility of the P protein that correlated with the antigenic classification of the viruses. Akerlind and Norrby (1) used MAbs which recognized the G, F, N, P, and M2 proteins to analyze viruses isolated in Stockholm. The viruses were identified as subtype A or subtype B as described above (85). The major differences between the subtypes were on the G protein.
Morgan et al. (80) tested isolates from Newcastle with MAbs to the G and M2 (22K) proteins. Subgroup A viruses resembled the Long and A2 strains, while subgroup B viruses were similar in reactivity to the subgroup B prototype 8/60 virus. Differences in P-protein mobility were observed. The cocirculation of the two subgroups was noted, although the subgroup B strains were less frequently isolated. The authors commented that the subgroup B viruses replicate slowly in cell culture. Orvell et al. (92) raised MAbs to a subgroup B strain of RSV, strain WV4843; these MAbs recognized the F, M, N, P, and G proteins. The MAbs to the G protein were subgroup B specific. Most of the other MAbs were reactive with viruses of both antigenic subgroups, although two F-protein MAbs were specific for the subgroup B viruses. Storch and Park (109) produced MAbs to the Long, A2, CH-18537, and 9320 strains of RSV. Strain-specific and cross-reactive MAbs were identified, and the MAbs chosen for use recognized the G protein. Isolates from St. Louis were analyzed and found to have patterns of reactivity with the MAbs that resembled both the A2 and Long strains or the 9320 strain. Thus, two types of MAb reactivity were observed, as was also observed in the above studies.
The above studies demonstrated that there were two major antigenic categories of RSV that were discernible with MAbs. The categories revealed that a larger number of antigenic differences occurred on the attachment glycoprotein G than on the other viral proteins. The antigenic categories were variously described as groups, subgroups, types, and subtypes and were differentiated by using either number or letter designations. The group appellation has seen increasing (although not universal) usage (12, 36, 77, 87, 94), with the two groups being designated A and B; this nomenclature is followed here. Some authors have used a nomenclature for individual isolates that indicates the place of isolation, a number, and the year of isolation (46). The Long and A2 viruses have served as the prototype group A viruses, and the CH-18537 and 8/60 viruses have been the standard group B viruses with which comparisons have been made.
GENETIC CONFIRMATION OF TWO GROUPS
Nucleotide Sequence Analysis
At the same time that newer immunological tools were being used to study RSV, the techniques of molecular biology were also being applied. The cloning of cDNAs and the determination of nucleotide sequences for the genes provided new insights into the antigenic and genetic variability of the virus. Comparisons were made among two of the group A viruses, A2 and Long, and a group B virus, CH-18537 (67). The G proteins of the two group A viruses were very similar (6% amino acid differences), whereas the group A and group B viruses had extensive differences (47% amino acid differences between the A2 and CH-18537 G proteins). Although the ectodomain of the G protein was variable between the groups, a strictly conserved 13-amino-acid region in the central ectodomain was noted (amino acids 164 to 176). The other RSV proteins are more similar between the antigenic groups, although SH protein comparisons reveal variability (8, 27, 75) (Table 1).
TABLE 1.
Sequence differences among group A (A2) and group B (CH-18537) virus genes and proteinsa
| Protein | Nucleotide differences (%) | Amino acid differences (%) |
|---|---|---|
| G | 33 | 47 |
| SH | 28 | 24 |
| NS1 | 22 | 13 |
| F | 21 | 11 |
| M2 | 22 | 8 |
| NS2 | 22 | 8 |
| N | 14 | 4 |
These observations were extended by the analysis of another group B RSV (8/60) G protein, which also allowed comparisons among the group B viruses. Again, genetic similarity within the group was noted (2% amino acid differences between 8/60 and CH-18537), whereas the group A and B G-protein gene sequences were quite divergent (44% amino acid differences between 8/60 and A2) (112).
Other Genetic Techniques
The recognition that there were extensive glycoprotein G gene differences between the groups and relative conservation within the groups allowed the development of new techniques for the differentiation of the two groups. A nucleic acid hybridization assay used G-protein gene cDNAs from the A2 and 8/60 viruses as probes (113). Viral samples were tested in dot blot format and could be classified into group A or B. This study provided additional genetic confirmation of the validity of the classification of RSV into two groups by using MAbs. Synthetic oligonucleotide probes were also shown to discriminate among the groups in a hybridization assay (118).
PCR technology was used for the discrimination of RSV antigenic groups. All of the PCR techniques used with RSV require an initial first-strand cDNA synthesis from RNA of viral origin. Cane and Pringle (19) amplified the SH gene by PCR; in this study, group A and group B viruses were discriminated by a combination of group-specific and group-cross-reactive primers. The nucleotide sequences of the SH genes were determined, revealing a relative conservation of the SH gene sequences within the groups, particularly compared to the differences between the two groups. The N gene was also amplified and analyzed by restriction endonuclease digestion.
A PCR-based approach was developed which used group A- or B-specific 5′-end primers which annealed within the G gene and a 3′-end primer which annealed within the F gene of both groups of viruses. The placement of the 5′-end primers was such that the sizes of the PCR products varied by group and thus analysis of a single PCR product by simple agarose gel electrophoresis provided a group assignment. In addition, restriction fragment analysis of the amplified cDNAs permitted the detection of genetic variability within the groups (117). Another assay relied on amplification from a smaller region within the G gene. In this assay, the size of the products again allowed group discrimination (52). A strain-specific PCR assay was developed to discriminate among wild-type and vaccine strains of RSV. The assay designed for group B viruses was found to amplify both group A and B viruses, with the two groups being differentiated by PCR product size (137).
PCR can also be used, in combination with classification into viral groups, for the direct detection of RSV in clinical specimens. Primers which spanned from the 1B into the N genes were selected for the detection of both viral groups. Group assignments were made by hybridization with group-specific oligonucleotide probes (127). PCR amplification directly from nasal aspirates followed by detection with group-specific probes in a DNA-enzyme immunoassay has also been reported (44). PCR-based assays which detect group A and B RSV and other respiratory viruses from a single sample have been developed (38, 41, 93).
Mobility differences are found by polyacrylamide gel electrophoresis for the P proteins of different RSV isolates. These differences may be used to classify individual isolates into groups (128).
VARIATION WITHIN THE TWO GROUPS
The studies cited above, which identified the existence of the two major groups of RSV, also revealed that there was additional variability within an individual group. These studies and additional studies that added to the information about the two groups and variability within the groups are discussed in this section.
Antigenic Variation
Antigenic variation in the N protein was described based on the reactivity of N-protein MAbs in competitive radioimmunoassays (132). MAbs to the P and M proteins also revealed antigenic differences among isolates (49). As part of the initial description of antigenic groups, variability of MAb reactivity within the group A and B viruses was noted (7, 80, 85).
In another investigation, isolates were tested with a panel of G-protein-specific MAbs that had been raised against group A and B viruses (109). Reactivity varied among viruses within individual groups. Differences in G protein mobility by sodium dodecyl sulfate-polyacrylamide gel electrophoresis were also observed, with the group B virus G protein migrating more rapidly than the group A virus G protein. Similarly, intragroup variability in reactivity with a G-protein MAb was noted when MAbs raised against a group B virus were used (92). Subsequent analysis of additional isolates with MAbs against group A and B viruses revealed variability among the group B isolates. Two subsets of the group B viruses were identified based on differences in G-protein reactivity. It was suggested that the group B viruses may be a more heterogeneous group and the group A viruses may be a more homogeneous group (2). However, testing of other isolates with expanded panels of MAbs demonstrated that there is also heterogeneity among the group A viruses (58, 124).
Garcia-Barreno et al. (47) tested isolates with MAbs raised against the group A Long strain virus. In addition to confirming the delineation into two antigenic groups, extensive antigenic variability among the group A viruses was noted. This variability was particularly evident in testing with G-protein MAbs, in which some MAbs showed a strain-specific reactivity. In this investigation, variant viruses were selected by growth in the presence of F- or G-protein MAbs. The variant viruses selected with a G-protein MAb not only lost reactivity with most of the G-protein MAbs but also were no longer recognized by polyclonal antivirus antiserum. This suggested that the antigenic structure of the G protein had been significantly altered. Subsequent work revealed that these G proteins had changed dramatically due to frameshift mutations (48).
The studies cited above demonstrate that although two antigenic groups of RSV can be delineated, there is additional antigenic variability within the individual groups. This antigenic variability was demonstrated by testing with MAbs, which revealed that the greatest variability occurred on the attachment protein G. In the next section, genetic evidence of intragroup variability is presented. Subsequently, additional studies that demonstrated intragroup variability are reviewed in the context of RSV epidemiology.
Genetic Variation
RNase protection was used to study genomic variation among group A RSV isolates (110). Viral RNAs from virus infected cells were hybridized with radiolabeled RNA probes synthesized from a G-protein cDNA. After incubation with RNase A, RNA cleavage patterns were discriminated. Substantial genetic heterogeneity was observed among circulating isolates of group A RSV. RNase protection analysis revealed genetic differences among viruses that were indistinguishable by MAb reactivity. The authors concluded that there might be well-defined genome variants among the group A viruses and that RNase protection analysis could be used for the study of RSV molecular epidemiology.
RNase protection analysis was applied to both group A and B viruses in a study that used probes corresponding to the P, N, M2, G, and F genes (29). Because the group A and B viruses were found to have different cleavage patterns, this assay could be used to differentiate the two viral groups. Within each group, additional genetic variability was evident. Viruses isolated over a short time span could have similar cleavage patterns, but some viruses from a single outbreak had substantial genetic differences (29). The G and P genes of group A RSV isolated in a variety of international locales between 1961 and 1989 were analyzed by RNase A mismatch cleavage. Heterogeneity was seen for both the G and P genes. This heterogeneity was greatest among strains isolated during different epidemics and more restricted in strains isolated during individual epidemics (30).
As noted above, genetic variability within and between the groups was also assessed by nucleotide sequence analysis (67, 112). These initial studies made comparisons among remotely isolated prototype strains. Subsequent investigations would evaluate samples collected over broader expanses of time and place.
Nucleotide sequence determination and restriction mapping were used to analyze PCR-amplified SH and N gene products (19). SH gene sequence analysis showed limited intragroup differences but did demonstrate that such differences occur among both A and B group viruses. Restriction mapping of N gene products also revealed intragroup genetic variability. Discrete genetic lineages of the viruses were observed in a single epidemic period. Subsequently, the same investigators analyzed the G-protein gene of group A viruses by nucleotide sequence determination (15) and found differences of up to 20% among the group A RSV G proteins. The central region of the ectodomain of the protein was found to be highly conserved and was flanked by variable domains (Fig. 1). The high proportion of nucleotide changes that resulted in amino acid coding changes suggested that there might be immunologic pressure for change in the G protein. The techniques for discrimination between the groups were further modified so that PCR amplification of the N gene followed by restriction mapping would delineate the two groups and provide information about variability within the groups. For group A viruses, intragroup variability was assessed by restriction mapping of the G-protein gene PCR products (18).
Genetic diversity among group B viruses was assessed for viruses isolated between 1977 and 1989 (114). Amino acid sequence differences of as great as 12% occurred in the ectodomain of the G protein. In addition to single amino acid changes due to substitution mutations, changes in termination codons resulted in proteins of different lengths. A frameshift mutation also was observed, similar to that described for G-protein escape mutants (48). As described above for the group A viruses, the central ectodomain was conserved and was flanked by more variable regions (Fig. 1). Overall, 51% of the nucleotide changes resulted in amino acid changes, suggesting that there may be a selective pressure for change in the G protein. The authors suggested that this selective pressure was likely to be the host antibody response. Restriction fragment analysis of G-protein gene PCR products also revealed intragroup genetic heterogeneity. Such heterogeneity occurred among viruses of both groups and among viruses which had revealed no differences by testing with MAbs (117).
RNase protection assays, analysis of restriction patterns of PCR-amplified cDNAs, and nucleotide sequence analysis demonstrated that there is genetic variability within the A and B groups of RSV. These genetic assays confirmed the variability that had been shown by testing with MAbs.
EPIDEMIOLOGY AND VIRAL VARIABILITY
The availability of tools for the discrimination of antigenic and genetic variability among RSV isolates provided an opportunity to extend the understanding of the epidemiology of these viruses. In the following sections, insights gained from antigenic and molecular studies of viral epidemiology are reviewed.
Epidemiology
Early studies of antigenic variability performed by using cross-neutralization assays demonstrated variability among isolates. It was suggested that strains that vary antigenically circulate simultaneously in the world (32). The circulation of distinct viruses during a single epidemic period was also described (22).
The recognition of the two major antigenic groups of RSV and the variability that occurs within these groups provided an opportunity to better define the epidemiology of these viruses. The work of Anderson et al. (7) showed that the two groups have existed for over 20 years and have a worldwide distribution and that both groups may occur during a single season. Group A viruses were identified more often than the group B viruses. Mufson et al. (85) suggested that the two groups had evolved separately for a considerable period.
Hendry et al. (59) analyzed isolates collected in Boston in 1981 to 1982 and in 1983 to 1984. The two antigenic groups were found to circulate concurrently. Temporal and geographic clustering of the groups was observed. The two groups were isolated with similar frequencies in one period, and the group A viruses predominated in the other period. In a later study, the prevalence of the two antigenic groups in six RSV outbreaks in Boston from 1981 to 1987 was assessed (58). Of 981 viruses, 60% were group A and 39% were group B. In two of the six periods the groups were found in equal proportions, in three periods the group A viruses were dominant, and in one period the group B viruses were dominant. Heterogeneity among the group A viruses was noted.
Akerlind and Norrby (1) found that both antigenic groups also circulate in Sweden. Group B viruses isolated in West Virginia were classified as B1 or B2 based on differences in G-protein MAb reactivity. The B2 viruses predominated in a single epidemic year and were not detected subsequently (2, 82). Similarly, a variant group A RSV with altered reactivity to F MAbs was identified in only one epidemic period (86). The distributions of group A and B viruses during five epidemic periods from 1981 to 1986 in West Virginia were analyzed (83). Of 211 viruses tested, 76% were group A and 24% were group B. Both groups were isolated each year, but the group A viruses were found more often in most epidemic periods. The studies in West Virginia were later extended to include the epidemic periods from 1978 to 1988. Of 405 viruses tested, 79% were group A and 21% were group B. The group B viruses were dominant in one epidemic (82).
Storch and Park (109) analyzed 114 viruses isolated from 1981 to 1986 in St. Louis, Mo., and found that the group A viruses were predominant (80% of the isolates), although both groups were found. Analysis of RSV isolates collected in Newcastle upon Tyne, United Kingdom, between 1965 and 1986 showed that the two antigenic groups cocirculated; group B viruses were less frequently isolated (80). Viruses isolated during five epidemic periods in Sapporo, Japan, were analyzed (125). The first and last periods showed a group A virus predominance, but during the three intervening periods the group B viruses were isolated more frequently. RSV isolates collected during three epidemic periods in Uruguay from 1987 to 1987 were analyzed; group A isolates were dominant in two epidemic periods, and group B isolates were dominant in one (100).
RSV isolates collected in Tecumseh, Mich., between 1965 and 1971 and between 1976 and 1981 were classified into groups (79). Group A and B viruses were present as remotely as 1965; overall, the group A viruses predominated. In Hawaii, RSV was isolated each month of the year, although the peak incidence was in the winter wet season (98). Group A and B viruses cocirculated except during the months of lowest incidence.
The prevalence of the group A and B viruses in Caen, France, during eight epidemic periods from 1982 to 1990 was determined. Both subgroups occurred each year, and there was a gradual change in the predominant group over a period of about 5 years (45). In Finland, a regular alteration of group prevalence was observed, suggesting that antigenic variation of RSV influenced the epidemiology of the virus (133).
RSV isolates from Rochester, N.Y., spanning a 15-year collection period were classified into groups, with variants within the group being designated subgroups. Three patterns of group distributions were seen, group A predominance (9 years), equal proportions of group A and B (4 years), and group B predominance (2 years). Overall, the pattern was that 1 or 2 years of group A predominance was followed by a year in which group B viruses would make up at least 40% of the isolates. Within each of the groups, subgroups were identified. Among the group A viruses, subgroups A1 and A2 predominated; among the group B viruses, three subgroups were found in similar numbers (54).
A total of 483 RSV isolates collected in 14 laboratories in the United States and Canada during two epidemic periods were analyzed. Group A viruses accounted for 63% of isolates, group B accounted for 24%, and 14% could not be grouped. Within the group A and B viruses, there were six and three subclassifications, respectively. Isolates varied among laboratories during the same year. The authors concluded that RSV outbreaks may be community or regional phenomena but were not national ones (6). Antigenic and genomic diversity among group A viruses was assessed for 47 isolates collected during four RSV seasons in St. Louis. Based on reactivity with MAbs to the G protein, four subgroups within the group A viruses were identified. RNase protection patterns were similar with two of the antigenic subgroups and more diverse with one of the subgroups. Thus, this study also demonstrated that there is viral heterogeneity within a single epidemic period (107).
The group classification was determined for 87 viruses isolated in Rio de Janeiro, Brazil, from 1982 to 1988. Both group A and B viruses were identified in each epidemic period, although their relative predominance varied. Four antigenic variants among the group A viruses and three variants among the group B viruses were recognized (105). Among viruses isolated in Cleveland, Ohio, over three epidemic periods between 1985 and 1988, 176 were group A and 21 were group B (106). A total of 53 isolates of group B RSV obtained in Sapporo, Japan, from 1980 to 1989 were tested for intragroup antigenic variability. Three different patterns of antigenic reactivity were reported (88). Analysis of 613 viruses isolated in Vancouver, Canada, revealed that both group A and B viruses were circulating from 1987 to 1992. The group B viruses predominated during only one season (121). RSV isolates from a single epidemic period in Texas were found to be almost equally distributed between A and B group viruses, and intragroup variability was observed (78).
RSV strains from Australia and Papua New Guinea were characterized. In Australia the RSV season lasts from April through September (61). In Papua New Guinea the RSV season is year-round. Both group A and B RSV isolates were identified. Group A viruses were found to replicate better than the group B viruses in cell culture. In Mexico City, RSV of both antigenic groups was identified, with a preponderance of group A viruses (122).
Cane and Pringle analyzed viral isolates from a single epidemic period in Birmingham, United Kingdom (19). PCR products were used for the partial sequencing of the SH-protein gene and restriction fragment analysis of a region of the N-protein gene. From this outbreak, four group A and two group B lineages were identified. Genetic variability among group A viruses isolated worldwide was also assessed. The viruses had been isolated in the United Kingdom, Germany, Malaysia, Finland, and Uruguay. This study showed that multiple RSV lineages can cocirculate and that viruses isolated on separate continents can be very similar (13).
Five successive epidemics in Birmingham, United Kingdom, were assessed for variability among 187 isolates. PCR products from parts of the N-, SH-, and G-protein genes were analyzed by restriction digestion or nucleotide sequencing. Six group A and two group B lineages were identified; different lineages predominated in each epidemic. Interestingly, the isolation of some lineages increased for a few years and then declined. This result suggested the possibility that immunity interferes with the spread of particular viruses in the community (14).
A total of 13 clinical isolates of group A RSV obtained at different times and places in the United States were analyzed by sequencing a variable region of the glycoprotein G gene. These results were compared to the sequences from three reference strains of RSV, and extensive genetic diversity was noted among the isolates. The genetic relationships had little correlation with the date or place of isolation or with the antigenic subgroupings based on MAb reactivity. These results showed that multiple RSV genotypes might circulate in individual communities. In addition, very similar genotypes may be found in geographically distant sites. The observation that half the nucleotide changes encoded amino acid changes suggested that the G protein may be subject to immune selection (101).
The genetic and antigenic variability of the G protein among 76 group A viruses collected in Montevideo, Uruguay, and Madrid, Spain, during six epidemic periods was assessed (46). RNase A mismatch cleavage, nucleotide sequencing, and MAb reactivity were used. A phylogenetic tree that included the sequences determined in this investigation and the sequences of isolates from Birmingham, United Kingdom, was constructed. Two main branches and several subbranches were identified. During a single epidemic, viruses from different branches were found. Very similar viruses could also be found in geographically distant locations and during different epidemic periods, indicating the ability of these viruses to spread globally. When G-protein MAbs with strain-specific patterns of reactivity were used, a close relationship was observed between genetic and antigenic relatedness. As was seen in earlier studies, changes accumulated in the two variable regions of the G protein, and the pattern of the changes suggested that there may be a selective pressure for change. The pattern of evolution for RSV was suggested to be similar to that for the influenza B viruses. However, the degree of divergence for the G protein was found to be the highest reported for any RNA virus gene product (46).
Nucleotide sequence analysis and MAb reactivity for 48 group A viruses isolated during a 38-year period measured the variability of the attachment protein G. Nucleotide sequences were determined for the two variable regions of the G-protein gene. Phylogenetic analysis revealed that close clustering tended to be related to the date of isolation. However, in some years, two or more clusters might be distant from one another. Recent viral isolates were found to lie at the ends of the branches of the phylogenetic tree. There appeared to be an accumulation of amino acid changes with time; for the variable regions examined in this study, the rate of change was 0.5 to 1% per year. The reactivity of the viruses with G-protein MAbs in general mirrored the results of the genetic groupings. The pattern of evolution for RSV was said to be similar to that for influenza B virus, with cocirculation of multiple lineages and evidence for sequential evolution. The authors concluded that the current genotypes of RSV are widespread and that the genotypes are changing with time. Thus, vaccines modeled on historical prototype viruses may become antigenically less and less similar to currently circulating viruses (16).
RSV isolates collected during three epidemics in Denmark were analyzed as to group (63). The group B viruses were dominant during the first and last years, and the group A viruses were dominant during the intervening year. Restriction pattern analysis revealed diversity within the groups. Comparison of these results with previous work suggested that viruses similar in restriction patterns to the viruses found in Denmark had been found much earlier in Australia and the United States (117). The authors commented that this apparent identity among isolates collected more than 20 years earlier in other parts of the world suggested that the globally circulating virus types might be relatively stable. They speculated that temporal fluctuations in the dominant viruses might be due to immune status favoring certain viruses from the circulating pool rather than the process of antigenic drift (63).
Circulating RSV isolates collected in Liverpool, United Kingdom, during two epidemic periods were analyzed (43). The viruses were categorized by restriction fragment analysis of N and G gene PCR products. Most (91%) of the isolates were members of group A. At least 10 different genotypes were found to cocirculate.
The analysis of a collection of RSV isolates from Cuba yielded fascinating results (126). The G-protein genes of 23 viruses isolated in 1994 and 1995 had only five nucleotide differences from that of the Long strain virus isolated in 1956 in the United States. In addition, comparisons to viruses from later epidemic periods revealed that viruses with G proteins identical to those from 1994 to 1995 were present. No identical viruses had been reported from different epidemics previously. Thus, the Cuban isolates had unique features. The authors noted that previous studies had suggested that viruses from different lineages cocirculate and that over time there is an accumulation of changes among viruses in the same lineage. These studies were performed predominantly on viruses isolated in developed countries in temperate climates. The authors speculated that differences in the seasonality of RSV infections or restrictions on travel might have favored viral stability. The Cuban viruses demonstrate the uncertainties surrounding RSV epidemiology and provide an example of a lack of accumulation of change over time.
Most of the studies described above which assessed intragroup genetic variability to the level of nucleotide sequence determination analyzed only group A RSV isolates. Thus, until recently there was less information about the extent of variation among the group B viruses. Peret et al. (94) characterized the molecular epidemiology of both antigenic groups of RSV in Rochester, N.Y., during five epidemic periods. The group A or B viruses predominated for 2 years each and were found in similar proportions in 1 year. The group B viruses were less variable than the group A viruses. Based on phylogenetic analyses of the nucleotide sequences of the gene encoding a region of the G protein, gene clades or genotypes were identified and were further classified into subtypes within the genotypes. Among the group A viruses, five genotypes and 22 subtypes and among the group B viruses four genotypes and 6 subtypes were described. Each year, one or two genotypes or subtypes accounted for at least half of the isolates. The predominant genotype or subtype was different each year. The authors hypothesized that strain differences affect protective immunity such that a novel virus may be transmitted more efficiently or may be more pathogenic. Thus, globally circulating strains could be introduced into a community and factors such as the levels of strain specific immunity would determine which strains might become epidemic strains (94).
RSV strains isolated during three epidemic periods in Alabama were characterized by Coggins et al. (25) as to group and intragroup differences by restriction pattern analysis and nucleotide sequencing of a variable region of the G-protein gene. Restriction fragment analysis revealed that the dominant pattern was different each year. Thus, even though the group A viruses were dominant for 2 years, the prevalent circulating virus was different each year. The deduced amino acid sequences for the variable region were compared, and it was found that among the group A viruses differences as great as 38% were found over the three epidemic periods. However, the viruses in one of the group A restriction patterns were noted to have no nucleotide differences observed over the three epidemic periods. The group B viruses displayed less variability than the group A viruses over this same period, with amino acid differences for the variable region being as great as 14%. The authors speculated that the limited variability among the group B viruses might contribute to a more restricted spread of these viruses, leading to the predominance of group A over group B viruses in many studies of RSV epidemiology. The group B viruses have the potential for greater variation than was seen among the Alabama isolates, as indicated by differences of up to 27% from a prototype group B virus. The percent nucleotide changes resulting in amino acid changes was calculated for the variable region analyzed in the study, and it was found that over 60% of the nucleotide changes resulted in amino acid changes among both the group A and B viruses. Thus, for both viral groups it is possible that there is a selective advantage, such as avoidance of the host immune response, associated with the G-protein changes.
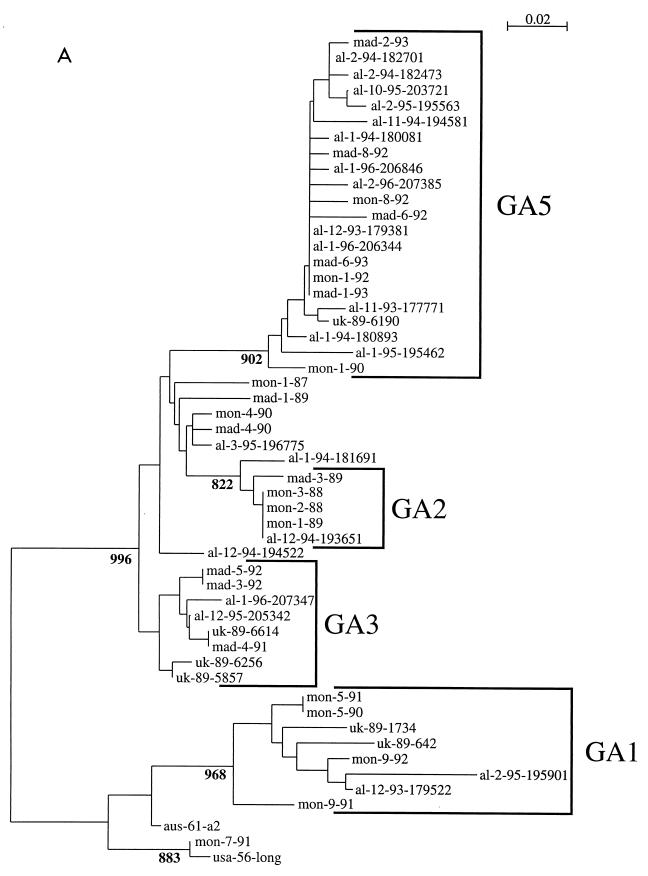
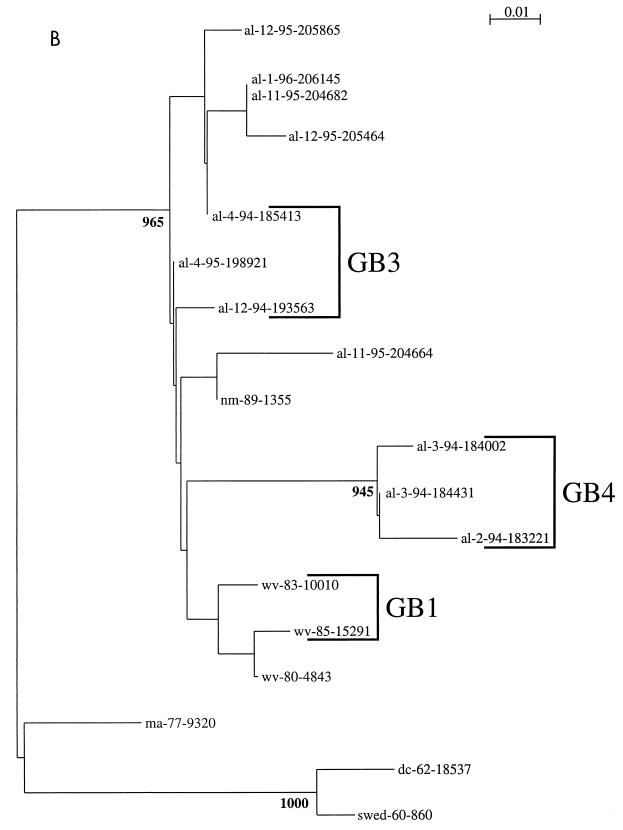
Phylogenetic comparisons were performed for the Alabama isolates and compared to RSV G-protein gene sequences available through GenBank. As described for the studies discussed above, multiple lineages were evident among both group A and B viruses (25). The dendrograms from this investigation were modified to show the corresponding genotype classifications described by Peret et al. (94) (Fig. 2). Not all of the isolates were classified, because they, or similar viruses, were not included in the earlier classification scheme. Such designations may be expected to undergo modification as more data become available and as formal phylogenetic comparisons are made among the growing number of available RSV gene sequences. The dendrograms show that viruses isolated on different continents several years apart may be phylogenetically very similar. Viruses from a single location and epidemic period may be placed on distant phylogenetic branches or may be phylogenetically very similar. These dendrograms may be compared to earlier work by other investigators (16, 17, 46, 77). The general groupings of the isolates are very similar among these different publications, suggesting that the broad outlines of these trees are likely to be maintained even as the specifics are refined. Among the group A viruses, the genotype designations of Peret et al. (GA1, GA2, etc.) correspond to the classifications of Cane et al. as follows, with branch designations for the G gene-based groupings and SHL for the classifications from the SH gene data: GA1 corresponds to branch 4 and SHL5; GA3 corresponds to branch 1 and SHL1, SHL3, and SHL4; and GA5 corresponds to branch 5 and SHL2 (16, 94).
FIG. 2.
Group A (A) and B (B) RSV G-protein phylogenetic relationships. Partial G-protein sequences from RSV strains isolated in the Children's Hospital of Alabama were compared to published G-protein sequences available through GenBank (25). Viruses are identified by the geographic location (from the United States, al, Alabama; wv, West Virginia [114]; dc, District of Columbia [67]; ma, Massachusetts [114]; md, Maryland [67]; nm, New Mexico [114]; from the United Kingdom, uk [15]; from Spain, mad, Madrid; from Uruguay, mon, Montevideo [46]; from Sweden, swed [112]; and from Australia, aus [134]), year of isolation (or number and year for isolates from Montevideo and Madrid), and, for isolates from the United States and the United Kingdom, a number designation. Nucleotide sequence alignments of either group A or group B sequences were used to create the neighbor-joining trees in the figure. The scales represent either 0.02 (group A) or 0.01 (group B) substitutions per base per indicated horizontal distance. The numbers present at some of the internal nodes of the trees represent the number of bootstrap replicates of a total of 1,000 that display the indicated sequence groupings. Only significant bootstrap replicate numbers with values greater than 800 are shown. Tentative assignments to the genotypes described by Peret et al. (94) are indicated by the brackets and the designations GA1, GA2, GA3, GA5, GB1, GB3, and GB4.
Nosocomial Infections
RSV isolates obtained during investigations of nosocomial outbreaks of infection have also been assessed for antigenic and genetic variability. Analysis of RSV isolates obtained from institutionalized adults revealed that among the group A viruses in an outbreak there were antigenic differences discernible by testing with G-protein MAbs (42). Isolates from nine immunocompromised adult patients, including three who died of respiratory failure, were collected during two winter periods. During one season, four patients were infected by four antigenically distinct viruses (one group A virus and three variants of group B virus). In the next epidemic period, three isolates were group A and two were group B among isolates collected from five patients. No intragroup variability was noted among these viruses. Thus, in the first winter, the patients appeared to have acquired their viruses from different sources, whereas in the second winter, there was a possibility that some of the patients had acquired their infection, directly or indirectly, from the same source (37).
An outbreak of RSV infections among bone marrow transplant patients was investigated. Viral isolates were available from 13 patients and 1 employee. Eleven isolates were antigenic group A and had two distinct RNase protection assay patterns. The three group B viruses also had two distinct RNase protection assay patterns. Epidemiologic correlation with these results suggested that there might have been one cluster of infections due to a single virus whereas the other patients were infected with different strains from other sources (55). In a study of therapy for RSV infections in bone marrow transplant patients, six hospital-acquired infections were due to group B viruses whereas community-acquired infections were due to group A (two patients) and group B (two patients) viruses. These infections occurred during a community outbreak of RSV among children, in which 76% of the isolates examined were group A RSV (135).
Nosocomial isolates of group A RSV collected during two epidemic periods were assessed for antigenic and genomic diversity (108). Three distinct subgroups and 12 discrete viral variants were identified. Although there was diversity among the isolates, an outbreak that included indistinguishable isolates was identified. Compared to isolates from community-acquired infections, the nosocomial isolates had a more limited diversity.
Virulence Differences
It is possible that there are virulence differences between the two groups of RSV. Studies which address this issue were recently summarized (131) and are not reviewed in detail here. Walsh et al. (131) found that among infants without underlying medical conditions, those infected with group A RSVs were more likely to require ventilatory support and had higher indices of disease severity than those infected with group B RSV. These observations are in agreement with some previous studies, whereas other investigators have reported no differences in clinical severity by antigenic group (131). Differences in viral replication between the two groups have been described. The group A viruses replicate to higher titers than the group B viruses in both cell culture and animals (31, 61, 80, 111, 112). The pathogenic outcome may be due in part to the level of viral replication. Thus, these in vitro and in vivo observations are compatible with the hypothesis that the group A viruses are more virulent than the group B viruses in humans. At present, there are no reports of the application of this information to decision making in a clinical setting. It may be that certain viruses within an individual group are more virulent than others, further complicating the comparisons of clinical severity (43, 54, 131).
IMPLICATIONS FOR VACCINE DEVELOPMENT
Studies with Animals
The implications that antigenic variation may have for vaccine development have been fundamental to the study of RSV variability (23). The current status of RSV vaccine development has been recently summarized (24, 36, 87). In this section the significance of RSV antigenic variation for vaccine development as revealed by studies with animals and with humans is reviewed.
Coincident with the recognition of the two antigenic groups, individual RSV protein genes were being cloned and expressed from recombinant vectors. The F and G proteins were compared for their ability to protect against homologous and heterologous group challenge (66, 111). The F protein of a group A virus expressed from recombinant vaccinia viruses was used to immunize cotton rats. The F protein provided nearly complete protection against viral replication in the lungs after group A or group B virus challenge. Immunization with a group A virus G protein provided much better protection against group A than group B viral challenge. These results were confirmed by the observation that immunization with a group B virus G protein protected against group B and not against group A viral challenge (112). Thus, a subunit vaccine based on the G protein might need G proteins of both antigenic groups to provide broad protection. An F protein vaccine might protect equally well against viruses of both antigenic groups. A chimeric FG protein used to immunize cotton rats reduced viral titers after challenge with either group A or group B viruses (10).
These results have been extended by studies with synthetic peptides and expressed fragments of the G protein. Synthetic peptides which provide protection against viral replication in rodent models have been described (123). These peptides, derived from the central conserved region of the G protein, are group specific in their protective capacity (104).
Attenuated live virus vaccines are also being developed. Candidate vaccines derived from both group A and group B viruses have been described (31, 36). Each of the vaccine approaches will have to meet stringent standards for both safety and efficacy. This is particularly an issue for the subunit vaccines due to the previous experience with the formalin-inactivated vaccine. In addition, G-protein immunization of mice leads to pulmonary eosinophilia upon viral challenge (91).
As the extent of variability within the individual groups was recognized, questions arose about the implications that this might have for vaccine design. As discussed above, the epidemiologic patterns and genetic features are compatible with there being a selective advantage for G-protein changes. The postulated advantage is avoidance of the host immune response. To address these issues, the G proteins of a pair of group A viruses which had caused an infection and later reinfection in a child were assessed (116). The two G proteins were found to have 14% amino acid differences and differences in reactivity with G-protein-specific MAbs. Vaccinia viruses that expressed the two G proteins were constructed. Cotton rats immunized with the vaccinia virus recombinants were equally protected against challenge by either of the two RSV isolates. Thus, this study did not reveal differences in protection after immunization with the G proteins from two different viruses from the same antigenic group. These data differ from those after cross-group (A versus B) G-protein immunization and challenge, which results in better homologous than heterologous protection (described above). The lack of intragroup differences in protection may be encouraging news for vaccine design, because it suggests that intragroup variability in the G protein may not be an obstacle to developing a broadly protective vaccine. However, it would be premature to make this conclusion on the basis of a single set of experiments in a rodent model (116). This is particularly true since the results of molecular epidemiologic studies in humans suggest that intragroup variability may play a role in the establishment of infections in humans.
The approaches taken to understanding the significance of viral variability in humans have included serologic assays and epidemiologic investigations. The scope and content of the broad-based epidemiologic studies are reviewed above. Information about reinfections in humans and the antibody responses to infections is presented below.
Reinfections in Humans
Mufson et al. (84) assessed the group characteristics of the viruses which caused reinfections in 13 children. Of the 10 children with initial group A virus infections, 6 had group B viruses and 4 had group A viruses upon reinfection. This represented more group B virus infections than would have been expected by chance, since the ratio in the community of group A to group B viruses was 3:1. The authors concluded that an initial infection with a group A virus provides some degree of protection against reinfection by group A viruses. They speculated that a broadly protective vaccine would require incorporation of both antigenic groups. Waris (133) observed that children older than 6 months during their first RSV infection were more resistant to homologous than heterologous group reinfections. Two children who experienced reinfections in a single epidemic period were infected by RSV isolates that varied in reactivity with MAbs (P. Pothier, S. Ghim, T. B. Bour, J. B. Guyon, and M. Dauvergne, Letter, Eur. J. Clin. Microbiol. 6:212, 1987).
Antigenic Variability and Antibody Responses
The antibody responses to group A and group B viruses have been assessed (129). Immunoaffinity-purified F and G proteins from group A and B viruses were used to raise antisera in rabbits. Antisera to either group A or group B F proteins reacted similarly by enzyme-linked immunosorbent assay (ELISA) or neutralization to group A and B viruses. However, anti-G antisera reacted preferentially with the homologous protein and neutralized the homologous virus to higher titers than it neutralized the heterologous group virus.
Hendry et al. (57) measured acute- and convalescent-phase antibody responses after primary infections shown to be due to group A or group B viruses in children. Antibody titers to the F and G proteins were determined by ELISA with F and G proteins purified from infected cells. Antibody responses to the F protein were found to be cross-reactive between the two antigenic groups, whereas responses to the G protein were largely group specific. Neutralizing-antibody titers to the homologous RSV group were also higher than to the heterologous RSV group. The authors noted that efficient neutralization of the virus might require antibodies to both the F and G proteins. In addition, candidate RSV vaccines should probably include G proteins from both antigenic groups as well as the F protein. Muelenaer et al. (81) found that antibody responses after infections with group A viruses were more cross-reactive than were the responses which followed primary infection by group B viruses. They suggested that this one-way pattern of cross-reactivity might contribute to the predominance of group A over group B RSV in epidemiologic studies.
Synthetic peptides have also been used to measure antibody responses to RSV (89). Peptides synthesized to represent the central region of the G protein, corresponding to amino acids 174 to 188 of the group A or group B G proteins, were compared for the detection of antibodies against RSV among 42 children with documented RSV infections. Preliminary work with a limited number of sera had suggested that these peptides might be useful for the detection of antibodies (3). However, the peptides were found to lack sufficient sensitivity and specificity to allow the group-specific detection of RSV antibodies in children (4). Peptides were also synthesized for bovine, ovine, and human RSV G proteins (amino acids 158 to 189) and used to detect antibodies against RSV (72). Testing of paired human sera with the group A and B human RSV peptides revealed that they could be used as antigens in immunoassays and that the peptides reacted in a group-preferential manner (71).
Group A and group B RSV G proteins have also been expressed from recombinant baculoviruses and used to detect antibodies to RSV (11, 115). Group specificity was not tested for human sera, but comparisons of sera from animals suggested that the expressed G proteins should display a group-specific reactivity as described for immunoaffinity-purified G proteins.
Human antibody responses to RSV have also been measured with portions of the G protein. Six group A RSV isolates representing six different genotypes were compared. The carboxy-terminal 84 to 85 amino acids of the G proteins were expressed from Escherichia coli. Children were shown to develop an antibody response to these protein fragments after RSV infections. In addition, the pattern of the antibody responses was related to the infecting genotype (20). Similar results were obtained with synthetic peptides corresponding to the same carboxy-terminal region of the protein (12). These results show that the carboxy-terminal region of the G protein, which is known to be highly variable, is recognized by the human immune response. These responses occur in a genotype-specific manner.
Thus, studies in animals and in humans have demonstrated that the differences between the two antigenic groups of RSV should be considered as vaccine approaches are planned. The differences between the two groups are discernible in serologic studies in humans by both neutralizing-antibody and G-protein ELISA determinations. The antigenic group of the previous infection may influence the pattern of reinfections in humans. Immunization with individual F or G proteins in animals has shown that the F protein is broadly cross-protective whereas the G protein provides a more group-specific protection. The importance of intragroup variability for vaccine design remains to be determined. Studies with variable regions of the G protein have shown that human antibody responses may have genotypic or viral isolate-specific components. However, a comparison of two group A virus G proteins as immunogens showed equal protection against the same and different viral isolate challenge.
CONCLUSIONS
The antigenic variability among isolates of RSV that was first recognized by evaluation with polyclonal antibodies has seen further definition by testing with MAbs (23, 85). The existence of two major antigenic groups of RSV, groups A and B, has been clearly established. The demarcation into two groups has been confirmed by molecular analyses of individual viral protein genes (67). The most extensive antigenic and nucleotide sequence differences between the two groups are found on the attachment glycoprotein, G (27). There is also antigenic and genetic variability within the individual groups of RSV (109).
Epidemiologic investigations have shown that the two antigenic groups may cocirculate in a single epidemic period. Although the antigenic groups cocirculate, temporal and geographic clustering may occur (59). The antigenic differences between the two groups are revealed serologically in young children after RSV infections, with differences being demonstrated by neutralizing-antibody and G-protein antibody determinations (57). In animals, the G protein provides a largely group-specific protection against challenge (111). Thus, it seems likely that antigenic differences among RSV isolates contribute to the ability of these viruses to establish infections in the presence of preexisting immunity (84).
The additional variability that occurs within the major antigenic groups has provided opportunities to gain further insights into the epidemiology of RSV. For instance, although a single antigenic group may be dominant for more than one epidemic year, a genotypically different virus of that group is dominant each year (14). This observation leaves clinical-outcome studies that rely on historical controls suspect, since the viruses that are compared are unlikely to be identical. The viruses that are found in a single community in an epidemic period will include viruses that are very similar, but other cocirculating viruses will be quite different from the predominant local viruses. Viruses from one locale may be quite similar to isolates from geographically or temporally remote sites (16, 46). There is evidence that there is a sequential accumulation of change in the G protein of RSV, perhaps in response to selective immune pressure (16). However, there are also instances in which viruses isolated almost 40 years apart are very similar to one another (126).
The epidemiology of RSV has been postulated to resemble that of influenza B virus, with multiple viral lineages undergoing change in response to selective pressure (15, 16, 46, 114). Future work should help to define whether the epidemiology of RSV should be modeled on that of the influenza viruses or whether different models and mechanisms of escape from immune surveillance should be considered. Foot-and-mouth disease virus serotype C undergoes significant antigenic variation. This variation occurs due to fluctuation among a few amino acid residues without the accumulation of amino acid changes over time (76). A MAb-resistant cytomegalovirus varies the amount of the target gH protein expressed. This variation in protein production occurs only when the selecting antibody is present (74). While these mechanisms may not apply to RSV, they provide examples of the diverse means by which viruses may vary antigenically. Antigenic variation may also occur among RNA viruses in the absence of immune selection (33). Studies of MAb escape mutants of the G protein of RSV have revealed novel mechanisms of change, including frameshift mutations and reduced G-protein exposure on virions (48, 130). Future studies which define the molecular evolution of RSV and the mechanisms by which these viruses evade the immune response should facilitate the rational design of vaccines against RSV.
ACKNOWLEDGMENTS
Support was received from Public Health service grants AI33425 and AI37197.
I thank Dana Pinson for secretarial support and Elliot Lefkowitz for assistance with the phylogenetic figures.
REFERENCES
- 1.Akerlind B, Norrby E. Occurrence of respiratory syncytial virus subtypes A and B strains in Sweden. J Med Virol. 1986;19:241–274. doi: 10.1002/jmv.1890190306. [DOI] [PubMed] [Google Scholar]
- 2.Akerlind B, Norrby E, Orvell C, Mufson M A. Respiratory syncytial virus: heterogeneity of subgroup B strains. J Gen Virol. 1988;69:2145–2154. doi: 10.1099/0022-1317-69-9-2145. [DOI] [PubMed] [Google Scholar]
- 3.Akerlind-Stopner B, Utter G, Mufson M A, Orvell C, Lerner R A, Norrby E. A subgroup-specific antigenic site in the G protein of respiratory syncytial virus forms a disulfide-bonded loop. J Virol. 1990;64:5143–5148. doi: 10.1128/jvi.64.10.5143-5148.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akerlind-Stopner B, Utter G, Norrby E, Mufson M A. Evaluation of subgroup-specific peptides of the G protein of respiratory syncytial virus for characterization of the immune response. J Med Virol. 1995;47:120–125. doi: 10.1002/jmv.1890470203. [DOI] [PubMed] [Google Scholar]
- 5.American Academy of Pediatrics Committee on Infectious Diseases. 1997 Redbook. Report of the Committee on Infectious Diseases. 24th ed. Elk Grove Village: AAP; 1997. [Google Scholar]
- 6.Anderson L J, Hendry R M, Pierik L T, Tsou C, McIntosh K. Multicenter study of strains of respiratory syncytial virus. J Infect Dis. 1991;163:687–692. doi: 10.1093/infdis/163.4.687. [DOI] [PubMed] [Google Scholar]
- 7.Anderson L J, Hierholzer J C, Tsou C, Hendry R M, Fernie B F, Stone Y, McIntosh K. Antigenic characterization of respiratory syncytial virus strains with monoclonal antibodies. J Infect Dis. 1985;151:626–633. doi: 10.1093/infdis/151.4.626. [DOI] [PubMed] [Google Scholar]
- 8.Baybutt H N, Pringle C R. Molecular cloning and sequencing of the F and 22K membrane protein genes of the RSS-2 strain of respiratory syncytial virus. J Gen Virol. 1987;68:2789–2796. doi: 10.1099/0022-1317-68-11-2789. [DOI] [PubMed] [Google Scholar]
- 9.Beem M. Repeated infections with respiratory syncytial virus. J Immunol. 1967;98:1115–1122. [PubMed] [Google Scholar]
- 10.Brideau R J, Walters R R, Stier M A, Wathen M W. Protection of cotton rats against human respiratory syncytial virus by vaccination with a novel chimeric FG glycoprotein. J Gen Virol. 1989;70:2637–2644. doi: 10.1099/0022-1317-70-10-2637. [DOI] [PubMed] [Google Scholar]
- 11.Buraphacheep W, Britt W J, Sullender W. Detection of antibodies to respiratory syncytial virus attachment and nucleocapsid proteins with recombinant baculovirus-expressed antigens. J Clin Microbiol. 1997;35:354–357. doi: 10.1128/jcm.35.2.354-357.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cane P A. Analysis of linear epitopes recognised by the primary human antibody response to a variable region of the attachment (G) protein of respiratory syncytial virus. J Med Virol. 1997;51:297–304. doi: 10.1002/(sici)1096-9071(199704)51:4<297::aid-jmv7>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 13.Cane P A, Matthews D A, Pringle C R. Analysis of relatedness of subgroup A respiratory syncytial viruses isolated worldwide. Virus Res. 1992;25:15–22. doi: 10.1016/0168-1702(92)90096-r. [DOI] [PubMed] [Google Scholar]
- 14.Cane P A, Matthews D A, Pringle C R. Analysis of respiratory syncytial virus strain variation in successive epidemics in one city. J Clin Microbiol. 1994;32:1–4. doi: 10.1128/jcm.32.1.1-4.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cane P A, Matthews D A, Pringle C R. Identification of variable domains of the attachment (G) protein of subgroup A respiratory syncytial viruses. J Gen Virol. 1991;72:2091–2096. doi: 10.1099/0022-1317-72-9-2091. [DOI] [PubMed] [Google Scholar]
- 16.Cane P A, Pringle C R. Evolution of subgroup A respiratory syncytial virus: evidence for progressive accumulation of amino acid changes in the attachment protein. J Virol. 1995;69:2918–2925. doi: 10.1128/jvi.69.5.2918-2925.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cane P A, Pringle C R. Molecular epidemiology of human respiratory syncytial virus. Semin Virol. 1995;6:371–378. [Google Scholar]
- 18.Cane P A, Pringle C R. Molecular epidemiology of respiratory syncytial virus: rapid identification of subgroup A lineages. J Virol Methods. 1992;40:297–306. doi: 10.1016/0166-0934(92)90088-u. [DOI] [PubMed] [Google Scholar]
- 19.Cane P A, Pringle C R. Respiratory syncytial virus heterogeneity during an epidemic: analysis by limited nucleotide sequencing (SH gene) and restriction mapping (N gene) J Gen Virol. 1991;72:349–357. doi: 10.1099/0022-1317-72-2-349. [DOI] [PubMed] [Google Scholar]
- 20.Cane P A, Thomas H M, Simpson A F, Evans J E, Hart C A, Pringle C R. Analysis of the human serological immune response to a variable region of the attachment (G) protein of respiratory syncytial virus during primary infection. J Med Virol. 1996;48:253–261. doi: 10.1002/(SICI)1096-9071(199603)48:3<253::AID-JMV7>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 21.Cherrie A H, Anderson K, Wertz G W, Openshaw P J M. Human cytotoxic T cells stimulated by antigen on dendritic cells recognize the N, SH, F, M, 22K, and 1b proteins of respiratory syncytial virus. J Virol. 1992;66:2102–2110. doi: 10.1128/jvi.66.4.2102-2110.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coates H V, Alling D W, Chanock R M. An antigenic analysis of respiratory syncytial virus isolates by a plaque reduction neutralization test. Am J Epidemiol. 1966;83:299–313. doi: 10.1093/oxfordjournals.aje.a120586. [DOI] [PubMed] [Google Scholar]
- 23.Coates H V, Kendrick L, Chanock R M. Antigenic differences between two strains of respiratory syncytial virus. Proc Soc Exp Biol Med. 1963;12:958–964. doi: 10.3181/00379727-112-28221. [DOI] [PubMed] [Google Scholar]
- 24.Coffin S E, Offit P A. New vaccines against mucosal pathogens: rotavirus and respiratory syncytial virus. Adv Pediatr Infect Dis. 1997;13:333–348. [PubMed] [Google Scholar]
- 25.Coggins W B, Lefkowitz E J, Sullender W M. Genetic variability among group A and group B respiratory syncytial viruses in a children's hospital. J Clin Microbiol. 1998;36:3552–3557. doi: 10.1128/jcm.36.12.3552-3557.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collins P L, Hill M G, Johnson P R. The two open reading frames of the 22k mRNA of human respiratory syncytial virus: sequence comparison of antigenic subgroups A and B and expression in vitro. J Gen Virol. 1990;71:3015–3020. doi: 10.1099/0022-1317-71-12-3015. [DOI] [PubMed] [Google Scholar]
- 27.Collins P L, McIntosh K, Chanock R M. Respiratory syncytial virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1313–1351. [Google Scholar]
- 28.Collins P L, Olmsted R A, Johnson P R. The small hydrophobic protein of human respiratory syncytial virus: comparison between antigenic subgroups A and B. J Gen Virol. 1990;71:1571–1576. doi: 10.1099/0022-1317-71-7-1571. [DOI] [PubMed] [Google Scholar]
- 29.Cristina J, Lopez J A, Albo C, Garcia-Barreno B, Garcia J, Melero J A, Portela A. Analysis of genetic variability in human respiratory syncytial virus by the RNase A mismatch cleavage method: subtype divergence and heterogeneity. Virology. 1990;174:126–134. doi: 10.1016/0042-6822(90)90061-u. [DOI] [PubMed] [Google Scholar]
- 30.Cristina J, Moya A, Arbiza J, Russi J, Hortal M, Albo C, Garcia-Barreno B, Garcia O, Melero J A, Portela A. Evolution of the G and P genes of human respiratory syncytial virus (subgroup A) studied by the RNase A mismatch cleavage method. Virology. 1991;184:210–218. doi: 10.1016/0042-6822(91)90837-2. [DOI] [PubMed] [Google Scholar]
- 31.Crowe J E, Bui P T, Firestone C Y, Connors M, Elkins W R, Chanock R M, Murphy B R. Live subgroup B respiratory syncytial virus vaccines that are attenuated, genetically stable, and immunogenic in rodents and nonhuman primates. J Infect Dis. 1996;173:829–839. doi: 10.1093/infdis/173.4.829. [DOI] [PubMed] [Google Scholar]
- 32.Doggett J E, Taylor-Robinson D. Serological studies with respiratory syncytial virus. Arch Virusforsch. 1965;15:601–608. doi: 10.1007/BF01245207. [DOI] [PubMed] [Google Scholar]
- 33.Domingo E, Diez J, Martinez M A, Hernandez J, Holguin A, Borrego B, Mateu M G. New observations on antigenic diversification of RNA viruses. Antigenic variation is not dependent on immune selection. J Gen Virol. 1993;74:2039–2045. doi: 10.1099/0022-1317-74-10-2039. [DOI] [PubMed] [Google Scholar]
- 34.Domingo E, Holland J J. Mutation rates and rapid evolution of RNA viruses. In: Morse S S, editor. The evolutionary biology of viruses. New York, N.Y: Raven Press; 1994. pp. 161–184. [Google Scholar]
- 35.Dowell S F, Anderson L J, Gary H E, Erdman D D, Plouffe J F, File T M, Marston B J, Breiman R F. Respiratory syncytial virus is an important cause of community-acquired lower respiratory infection among hospitalized adults. J Infect Dis. 1996;174:456–462. doi: 10.1093/infdis/174.3.456. [DOI] [PubMed] [Google Scholar]
- 36.Dudas R A, Karron R A. Respiratory syncytial virus vaccines. Clin Microbiol Rev. 1998;11:430–439. doi: 10.1128/cmr.11.3.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Englund J A, Anderson L J, Rhame F S. Nosocomial transmission of respiratory syncytial virus in immunocompromised adults. J Clin Microbiol. 1991;29:115–119. doi: 10.1128/jcm.29.1.115-119.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eugene-Ruellan G, Freymuth F, Bahloul C, Badrane H, Vabret A, Tordo N. Detection of respiratory syncytial virus A and B and parainfluenza virus 3 sequences in respiratory tracts of infants by a single PCR with primers targeted to the L-polymerase gene and differential hybridization. J Clin Microbiol. 1998;36:796–801. doi: 10.1128/jcm.36.3.796-801.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Falsey A R, Cunningham C K, Barker W H, Kouides R W, Yuen J B, Menegus M, Weiner L B, Bonville C A, Betts R F. Respiratory syncytial virus and influenza A infections in the hospitalized elderly. J Infect Dis. 1995;172:389–394. doi: 10.1093/infdis/172.2.389. [DOI] [PubMed] [Google Scholar]
- 40.Reference deleted.
- 41.Fan J, Henrickson K J, Savatski L L. Rapid simultaneous diagnosis of infections with respiratory syncytial viruses A and B, influenza viruses A and B, and human parainfluenza virus types 1, 2, and 3 by multiple quantitative reverse transcription—polymerase chain reaction—enzyme hybridization assay (Hexaplex) Clin Infect Dis. 1998;26:1397–1402. doi: 10.1086/516357. [DOI] [PubMed] [Google Scholar]
- 42.Finger F, Anderson L J, Dicker R C, Harrison B, Doan R, Downing A, Corey L. Epidemic infections caused by respiratory syncytial virus in institutionalized young adults. J Infect Dis. 1987;155:1335–1339. doi: 10.1093/infdis/155.6.1335. [DOI] [PubMed] [Google Scholar]
- 43.Fletcher J N, Smyth R L, Thomas H M, Ashby D, Hart C A. Respiratory syncytial virus genotypes and disease severity among children in hospital. Arch Dis Child. 1997;77:508–511. doi: 10.1136/adc.77.6.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freymuth F, Eugene G, Vabret A, Petitjean J, Gennetay E, Brouard J, Duhamel J F, Guillois B. Detection of respiratory syncytial virus by reverse transcription-PCR and hybridization with a DNA enzyme immunoassay. J Clin Microbiol. 1995;33:3352–3355. doi: 10.1128/jcm.33.12.3352-3355.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freymuth F, Petitjean J, Pothier P, Brouard J, Norrby E. Prevalence of respiratory syncytial virus subgroups A and B in France from 1982 to 1990. J Clin Microbiol. 1991;29:653–655. doi: 10.1128/jcm.29.3.653-655.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garcia O, Martin M, Dopazo J, Arbiza J, Frabasile S, Russi J, Hortal M, Perez-Brena P, Martinez I, Garcia-Barreno B, Melero J A. Evolutionary pattern of human respiratory syncytial virus (subgroup A): cocirculating lineages and correlation of genetic and antigenic changes in the G glycoprotein. J Virol. 1994;68:5448–5459. doi: 10.1128/jvi.68.9.5448-5459.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garcia-Barreno B, Palomo C, Penas C, Delgado T, Perez-Brena P, Melero J A. Marked differences in the antigenic structure of human respiratory syncytial virus F and G glycoproteins. J Virol. 1989;63:925–932. doi: 10.1128/jvi.63.2.925-932.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garcia-Barreno B, Portela A, Delgado T, Lopez J A, Melero J A. Frame shift mutations as a novel mechanism for the generation of neutralization resistant mutants of human respiratory syncytial virus. EMBO J. 1990;9:4181–4187. doi: 10.1002/j.1460-2075.1990.tb07642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gimenez H B, Cash P, Melson W T. Monoclonal antibodies to human respiratory syncytial virus and their use in comparison of different virus isolates. J Gen Virol. 1984;65:963–971. doi: 10.1099/0022-1317-65-5-963. [DOI] [PubMed] [Google Scholar]
- 50.Gimenez H B, Hardman N, Keir H M, Cash P. Antigenic variation between human respiratory syncytial virus isolates. J Gen Virol. 1986;67:863–870. doi: 10.1099/0022-1317-67-5-863. [DOI] [PubMed] [Google Scholar]
- 51.Glezen W P, Paredes A, Allison J E, Taber L H, Frank A L. Risk of respiratory syncytial virus infection for infants from low-income families in relationship to age, sex, ethnic group, and maternal antibody level. J Pediatr. 1981;98:708–715. doi: 10.1016/s0022-3476(81)80829-3. [DOI] [PubMed] [Google Scholar]
- 52.Gottschalk J, Zbinden R, Kaempf L, Heinzer I. Discrimination of respiratory syncytial virus subgroups A and B by reverse transcription-PCR. J Clin Microbiol. 1996;34:41–43. doi: 10.1128/jcm.34.1.41-43.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hall C B, Walsh E E, Long C E, Schnabel K C. Immunity to and frequency of reinfection with respiratory syncytial virus. J Infect Dis. 1991;163:693–698. doi: 10.1093/infdis/163.4.693. [DOI] [PubMed] [Google Scholar]
- 54.Hall C B, Walsh E E, Schnabel K C, Long C E, McConnochie K M, Hildreth S W, Anderson L J. Occurrence of groups A and B of respiratory syncytial virus over 15 years: associated epidemiologic and clinical characteristics in hospitalized and ambulatory children. J Infect Dis. 1990;162:1283–1290. doi: 10.1093/infdis/162.6.1283. [DOI] [PubMed] [Google Scholar]
- 55.Harrington R D, Hooton T M, Hackman R C, Storch G A, Osborne B, Gleaves C A, Benson A, Meyers J D. An outbreak of respiratory syncytial virus in a bone marrow transplant center. J Infect Dis. 1992;165:987–993. doi: 10.1093/infdis/165.6.987. [DOI] [PubMed] [Google Scholar]
- 56.Henderson F W, Collier A M, Clyde W A, Denny F W. Respiratory syncytial virus infections, reinfections and immunity. N Engl J Med. 1979;300:530–534. doi: 10.1056/NEJM197903083001004. [DOI] [PubMed] [Google Scholar]
- 57.Hendry R M, Burns J C, Walsh E E, Graham B S, Wright P F, Hemming V G, Rodriguez W J, Kim H W, Gregory A P, McIntosh K, Chanock R M, Murphy B R. Strain-specific serum antibody responses in infants undergoing primary infection with respiratory syncytial virus. J Infect Dis. 1988;157:640–647. doi: 10.1093/infdis/157.4.640. [DOI] [PubMed] [Google Scholar]
- 58.Hendry R M, Pierik L T, McIntosh K. Prevalence of respiratory syncytial virus subgroups over six consecutive outbreaks: 1981–1987. J Infect Dis. 1989;160:185–190. doi: 10.1093/infdis/160.2.185. [DOI] [PubMed] [Google Scholar]
- 59.Hendry R M, Talis A L, Godfrey E, Anderson L J, Fernie B F, McIntosh K. Concurrent circulation of antigenically distinct strains of respiratory syncytial virus during community outbreaks. J Infect Dis. 1986;153:291–297. doi: 10.1093/infdis/153.2.291. [DOI] [PubMed] [Google Scholar]
- 60.Hierholzer J C, Hirsch M S. Croup and pneumonia in human infants associated with a new strain of respiratory syncytial virus. J Infect Dis. 1979;140:826–828. doi: 10.1093/infdis/140.5.826. [DOI] [PubMed] [Google Scholar]
- 61.Hierholzer J C, Tannock G A, Hierholzer C M, Coombs R A, Kennett M L, Phillips P A, Gust I D. Subgrouping of respiratory syncytial virus strains from Australia and Papua New Guinea by biological and antigenic characteristics. Arch Virol. 1994;136:133–147. doi: 10.1007/BF01538823. [DOI] [PubMed] [Google Scholar]
- 62.Impact-RSV Study Group. Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. Pediatrics. 1998;102:531–537. [PubMed] [Google Scholar]
- 63.Johansen J, Christensen L S, Hornsleth A, Klug B, Hansen K S, Nir M. Restriction pattern variability of respiratory syncytial virus during three consecutive epidemics in Denmark. APMIS. 1997;105:303–308. doi: 10.1111/j.1699-0463.1997.tb00573.x. [DOI] [PubMed] [Google Scholar]
- 64.Johnson P R, Collins P L. The 1B (NS2), 1C (NS1) and N proteins of human respiratory syncytial virus (RSV) of antigenic subgroups A and B: sequence conservation and divergence within RSV genomic RNA. J Gen Virol. 1989;70:1539–1547. doi: 10.1099/0022-1317-70-6-1539. [DOI] [PubMed] [Google Scholar]
- 65.Johnson P R, Collins P L. The fusion glycoproteins of human respiratory syncytial virus of subgroups A and B: sequence conservation provides a structural basis for antigenic relatedness. J Gen Virol. 1988;69:2623–2628. doi: 10.1099/0022-1317-69-10-2623. [DOI] [PubMed] [Google Scholar]
- 66.Johnson P R, Olmsted R A, Prince G A, Murphy B R, Alling D W, Walsh E E, Collins P L. Antigenic relatedness between glycoproteins of human respiratory syncytial virus subgroups A and B: evaluation of the contributions of F and G glycoproteins to immunity. J Virol. 1987;61:3163–3166. doi: 10.1128/jvi.61.10.3163-3166.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Johnson P R, Spriggs M K, Olmsted R A, Collins P L. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc Natl Acad Sci USA. 1987;84:5625–5629. doi: 10.1073/pnas.84.16.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Johnson T R, Johnson J E, Roberts S R, Wertz G W, Parker R A, Graham B S. Priming with secreted glycoprotein G of respiratory syncytial virus (RSV) augments interleukin-5 production and tissue eosinophilia after RSV challenge. J Virol. 1998;72:2871–2880. doi: 10.1128/jvi.72.4.2871-2880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karron R A, Bounagurio D A, Georgius A F, Whitehead S S, Adamus J E, Clements-Mann M L, Harris D O, Randolph V B, Udem S A, Murphy B R, Sidhu M S. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc Natl Acad Sci USA. 1997;94:13961–13966. doi: 10.1073/pnas.94.25.13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim H W, Canchola J G, Brandt C D, Pyles G, Chanock R M, Jensen K, Parrott R H. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am J Epidemiol. 1969;89:422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 71.Langedijk J P M, Brandenburg A H, Middel W G J, Osterhaus A, Meloen R H, van Oirschot J T. A subtype-specific peptide-based enzyme immunoassay for detection of antibodies to the G protein of human respiratory syncytial virus is more sensitive than routine serological tests. J Clin Microbiol. 1997;35:1656–1660. doi: 10.1128/jcm.35.7.1656-1660.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Langedijk J P M, Middel W G J, Schaaper W M M, Meloen R H, Kramps J A, Brandenburg A H, van Oirschot J T. Type-specific serologic diagnosis of respiratory syncytial virus infection, based on a synthetic peptide of the attachment protein G. J Immunol Methods. 1996;193:157–166. doi: 10.1016/0022-1759(96)00039-7. [DOI] [PubMed] [Google Scholar]
- 73.Levine S, Klaiber-Franco R, Paradiso P R. Demonstration that glycoprotein G is the attachment protein of respiratory syncytial virus. J Gen Virol. 1987;68:2521–2524. doi: 10.1099/0022-1317-68-9-2521. [DOI] [PubMed] [Google Scholar]
- 74.Li L, Coelingh K L, Britt W J. Human cytomegalovirus neutralizing antibody-resistant phenotype is associated with reduced expression of glycoprotein H. J Virol. 1995;69:6047–6053. doi: 10.1128/jvi.69.10.6047-6053.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lopez J A, Villanueva N, Melero J A, Portela A. Nucleotide sequence of the fusion and phosphoprotein genes of human respiratory syncytial (RS) virus Long strain: evidence of subtype genetic heterogeneity. Virus Res. 1988;10:249–261. doi: 10.1016/0168-1702(88)90020-2. [DOI] [PubMed] [Google Scholar]
- 76.Martinez M A, Dopazo J, Hernandez J, Mateu M G, Sobrino F, Domingo E, Knowles N J. Evolution of the capsid protein genes of foot-and-mouth disease virus: antigenic variation without accumulation of amino acid substitutions over six decades. J Virol. 1992;66:3557–3565. doi: 10.1128/jvi.66.6.3557-3565.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Melero J A, Garcia-Barreno B, Martinez I, Pringle C R, Cane P A. Antigenic structure, evolution and immunobiology of human respiratory syncytial virus attachment (G) protein. J Gen Virol. 1997;78:2411–2418. doi: 10.1099/0022-1317-78-10-2411. [DOI] [PubMed] [Google Scholar]
- 78.Mlinaric-Galinovic G, Chonmaitree T, Cane P A, Pringle C R, Ogra P L. Antigenic diversity of respiratory syncytial virus subgroup B strains circulating during a community outbreak of infection. J Med Virol. 1994;42:380–384. doi: 10.1002/jmv.1890420410. [DOI] [PubMed] [Google Scholar]
- 79.Monto A S, Ohmit S. Respiratory syncytial virus in a community population: circulation of subgroups A and B since 1965. J Infect Dis. 1990;161:781–783. doi: 10.1093/infdis/161.4.781. [DOI] [PubMed] [Google Scholar]
- 80.Morgan L A, Routledge E G, Wilcocks M M, Samson A C R, Scott R, Toms G L. Strain variation of respiratory syncytial virus. J Gen Virol. 1987;68:2781–2788. doi: 10.1099/0022-1317-68-11-2781. [DOI] [PubMed] [Google Scholar]
- 81.Muelenaer P M, Henderson F W, Hemming V G, Walsh E E, Anderson L J, Prince G A, Murphy B R. Group-specific serum antibody responses in children with primary and recurrent respiratory syncytial virus infections. J Infect Dis. 1991;164:15–21. doi: 10.1093/infdis/164.1.15. [DOI] [PubMed] [Google Scholar]
- 82.Mufson M A, Akerlind-Stopner B, Orvell C, Belshe R B, Norrby E. A single-season epidemic with respiratory syncytial virus subgroup B2 during 10 epidemic years, 1978 to 1988. J Clin Microbiol. 1991;29:162–165. doi: 10.1128/jcm.29.1.162-165.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mufson M A, Belshe R B, Orvell C, Norrby E. Respiratory syncytial virus epidemics: variable dominance of subgroups A and B strains among children, 1981–1986. J Infect Dis. 1988;157:143–148. doi: 10.1093/infdis/157.1.143. [DOI] [PubMed] [Google Scholar]
- 84.Mufson M A, Belshe R B, Orvell C, Norrby E. Subgroup characteristics of respiratory syncytial virus strains recovered from children with two consecutive infections. J Clin Microbiol. 1987;25:1535–1539. doi: 10.1128/jcm.25.8.1535-1539.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mufson M A, Orvell C, Rafnar B, Norrby E. Two distinct subtypes of human respiratory syncytial virus. J Gen Virol. 1985;66:2111–2124. doi: 10.1099/0022-1317-66-10-2111. [DOI] [PubMed] [Google Scholar]
- 86.Mufson M A, Stanek R J. Identification of a variant subgroup A strain of respiratory syncytial virus. J Clin Microbiol. 1996;34:2493–2496. doi: 10.1128/jcm.34.10.2493-2496.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Murphy B R, Collins P L. Current status of respiratory syncytial virus (RSV) and parainfluenza virus type 3 (PIV3) vaccine development: memorandum from a joint WHO/NIAID meeting. Bull W H O. 1997;75:307–313. [PMC free article] [PubMed] [Google Scholar]
- 88.Nagai K, Tsutsumi H, Yamazaki H, Pattamadilok S, Chiba S. Three antigenic variant groups in human respiratory syncytial virus subgroup B isolated in Japan. Arch Virol. 1993;128:55–63. doi: 10.1007/BF01309788. [DOI] [PubMed] [Google Scholar]
- 89.Norrby E, Mufson M A, Alexander H, Houghten R A, Lerner R A. Site-directed serology with synthetic peptides representing the large glycoprotein G of respiratory syncytial virus. Proc Natl Acad Sci USA. 1987;84:6572–6576. doi: 10.1073/pnas.84.18.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Norrby E, Mufson M A, Sheshberadaran H. Structural differences between subtype A and B strains of respiratory syncytial virus. J Gen Virol. 1986;67:2721–2729. doi: 10.1099/0022-1317-67-12-2721. [DOI] [PubMed] [Google Scholar]
- 91.Openshaw P J M, Clarke S L, Record F M. Pulmonary eosinophilic response to respiratory syncytial virus infection in mice sensitized to the major surface glycoprotein G. Int Immunol. 1992;4:493–500. doi: 10.1093/intimm/4.4.493. [DOI] [PubMed] [Google Scholar]
- 92.Orvell C, Norrby E, Mufson M A. Preparation and characterization of monoclonal antibodies directed against five structural components of human respiratory syncytial virus subgroup B. J Gen Virol. 1987;68:3125–3135. doi: 10.1099/0022-1317-68-12-3125. [DOI] [PubMed] [Google Scholar]
- 93.Osiowy C. Direct detection of respiratory syncytial virus, parainfluenza virus, and adenovirus in clinical respiratory specimens by a multiplex reverse transcription-PCR assay. J Clin Microbiol. 1998;36:3149–3154. doi: 10.1128/jcm.36.11.3149-3154.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Peret T C T, Hall C B, Schnabel K C, Golub J A, Andeson L J. Circulation patterns of genetically distinct group A and B strains of human respiratory syncytial virus in a community. J Gen Virol. 1998;79:2221–2229. doi: 10.1099/0022-1317-79-9-2221. [DOI] [PubMed] [Google Scholar]
- 95.Reference deleted.
- 96.PREVENT Study Group. Reduction of respiratory syncytial virus hospitalization among premature infants and infants with bronchopulmonary dysplasia using respiratory syncytial virus immune globulin prophylaxis. Pediatrics. 1997;99:93–99. doi: 10.1542/peds.99.1.93. [DOI] [PubMed] [Google Scholar]
- 97.Prince G A, Horswood R L, Koenig D W, Chanock R M. Antigenic analysis of a putative new strain of respiratory syncytial virus. J Infect Dis. 1985;151:634–637. doi: 10.1093/infdis/151.4.634. [DOI] [PubMed] [Google Scholar]
- 98.Reese P E, Marchette N J. Respiratory syncytial virus infection and prevalence of subgroups A and B in Hawaii. J Clin Microbiol. 1991;29:2614–2615. doi: 10.1128/jcm.29.11.2614-2615.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Roberts S R, Lichtenstein D, Ball A, Wertz G W. The membrane-associated and secreted forms of the respiratory syncytial virus attachment glycoprotein G are synthesized from alternative initiation codons. J Virol. 1994;68:4538–4546. doi: 10.1128/jvi.68.7.4538-4546.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Russi J C, Delfraro A, Arbiza J R, Chiparelli H, Orvell C, Grandien M, Hortal M. Antigenic characterization of respiratory syncytial virus associated with acute respiratory infections in Uraguayan children from 1985 to 1987. J Clin Microbiol. 1989;27:1464–1466. doi: 10.1128/jcm.27.7.1464-1466.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sanz M C, Kew O M, Anderson L J. Genetic heterogeneity of the attachment glycoprotein G among group A respiratory syncytial viruses. Virus Res. 1994;33:203–217. doi: 10.1016/0168-1702(94)90103-1. [DOI] [PubMed] [Google Scholar]
- 102.Selwyn B J. The epidemiology of acute respiratory tract infection in young children: comparison of findings from several developing countries. Rev Infect Dis. 1990;12:S870–S888. doi: 10.1093/clinids/12.supplement_s870. [DOI] [PubMed] [Google Scholar]
- 103.Reference deleted.
- 104.Simard C, Nadon F, Seguin C, Thien N N, Binz H, Basso J, Laliberte J F, Trudel M. Subgroup specific protection of mice from respiratory syncytial virus infection with peptides encompassing the amino acid region 174–187 from the G glycoprotein: the role of cysteinyl residues in protection. Vaccine. 1997;15:423–432. doi: 10.1016/s0264-410x(97)00189-8. [DOI] [PubMed] [Google Scholar]
- 105.Siqueira M M, Nascimento J P, Anderson L J. Antigenic characterization of respiratory syncytial virus group A and B isolates in Rio de Janeiro, Brazil. J Clin Microbiol. 1991;29:557–559. doi: 10.1128/jcm.29.3.557-559.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stark J M, Fatemi S H, Amini S B, Huang Y T. Occurrence of respiratory syncytial virus subtypes in hospitalized children in Cleveland, Ohio from 1985 to 1988. Pediatr Pulmonol. 1991;11:98–102. doi: 10.1002/ppul.1950110204. [DOI] [PubMed] [Google Scholar]
- 107.Storch G A, Anderson L A, Park C S, Tsou C, Dohner D E. Antigenic and genomic diversity within group A respiratory syncytial virus. J Infect Dis. 1991;163:858–861. doi: 10.1093/infdis/163.4.858. [DOI] [PubMed] [Google Scholar]
- 108.Storch G A, Hall C B, Anderson L J, Park C S, Dohner D E. Antigenic and nucleic acid analysis of nosocomial isolates of respiratory syncytial virus. J Infect Dis. 1993;167:562–566. doi: 10.1093/infdis/167.3.562. [DOI] [PubMed] [Google Scholar]
- 109.Storch G A, Park C S. Monoclonal antibodies demonstrate heterogeneity in the G glycoprotein of prototype strains and clinical isolates of respiratory syncytial virus. J Med Virol. 1987;22:345–356. doi: 10.1002/jmv.1890220407. [DOI] [PubMed] [Google Scholar]
- 110.Storch G A, Park C S, Dohner D E. RNA fingerprinting of respiratory syncytial virus using ribonuclease protection. Application to molecular epidemiology. J Clin Investig. 1989;83:1894–1902. doi: 10.1172/JCI114096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Stott E J, Taylor G, Ball L A, Anderson K, Young K Y, King A M Q, Wertz G W. Immune and histopathological responses in animals vaccinated with recombinant vaccinia viruses that express individual genes of human respiratory syncytial virus. J Virol. 1987;61:3855–3861. doi: 10.1128/jvi.61.12.3855-3861.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sullender W M, Anderson K, Wertz G W. The respiratory syncytial virus subgroup B attachment glycoprotein: analysis of sequence, expression from a recombinant vector, and evaluation as an immunogen against homologous and heterologous subgroup virus challenge. Virology. 1990;178:195–203. doi: 10.1016/0042-6822(90)90394-7. [DOI] [PubMed] [Google Scholar]
- 113.Sullender W M, Anderson L J, Anderson K, Wertz G W. Differentiation of respiratory syncytial virus subgroups with cDNA probes in a nucleic acid hybridization assay. J Clin Microbiol. 1990;28:1683–1687. doi: 10.1128/jcm.28.8.1683-1687.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sullender W M, Anderson L J, Mufson M A, Wertz G W. Genetic diversity of the attachment protein of subgroup B respiratory syncytial viruses. J Virol. 1991;65:5425–5434. doi: 10.1128/jvi.65.10.5425-5434.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sullender W M, Britt W J. Antigenic and immunogenic analysis of group A and group B respiratory syncytial virus G proteins expressed from recombinant baculoviruses. J Gen Virol. 1996;77:641–648. doi: 10.1099/0022-1317-77-4-641. [DOI] [PubMed] [Google Scholar]
- 116.Sullender W M, Mufson M A, Prince G A, Anderson L J, Wertz G W. Antigenic and genetic diversity among the attachment proteins of group A respiratory syncytial viruses that have caused repeat infections in children. J Infect Dis. 1998;178:925–932. doi: 10.1086/515697. [DOI] [PubMed] [Google Scholar]
- 117.Sullender W M, Sun L, Anderson L J. Analysis of respiratory syncytial virus genetic variability with amplified cDNAs. J Clin Microbiol. 1993;31:1224–1231. doi: 10.1128/jcm.31.5.1224-1231.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sullender W M, Wertz G W. Synthetic oligonucleotide probes differentiate respiratory syncytial virus subgroups in a nucleic acid hybridization assay. J Clin Microbiol. 1991;29:1255–1257. doi: 10.1128/jcm.29.6.1255-1257.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sullender W M, Wertz G W. The unusual attachment glycoprotein of the respiratory syncytial viruses: structure, maturation, and role in immunity. In: Kingsbury D, editor. The Paramyxoviruses. New York, N.Y: Plenum Press; 1991. pp. 383–406. [Google Scholar]
- 120.Suto T, Yano N, Ideda M, Miyamoto M, Takai S, Shigeta S, Hinuma Y, Ishida N. Respiratory syncytial virus infection and its serologic epidemiology. Am J Epidemiol. 1965;82:211–224. doi: 10.1093/oxfordjournals.aje.a120545. [DOI] [PubMed] [Google Scholar]
- 121.Thomas E, Margach M J, Orvell C, Morrison B, Wilson E. Respiratory syncytial virus subgroup B dominance during one winter season between 1987 and 1992 in Vancouver, Canada. J Clin Microbiol. 1994;32:238–242. doi: 10.1128/jcm.32.1.238-242.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tirado R, Sarmiento R E, Bustos J, Thompson O, Gomez B. Occurrence of respiratory syncytial virus subtypes in Mexican infants with acute lower respiratory tract disease. Arch Med Res. 1995;26:121–126. [PubMed] [Google Scholar]
- 123.Trudel M, Seguin N C, Binz H. Protection of Balb/c mice from respiratory syncytial virus infection by immunization with a synthetic peptide derived from the G glycoprotein. Virology. 1991;185:749–757. doi: 10.1016/0042-6822(91)90546-n. [DOI] [PubMed] [Google Scholar]
- 124.Tsutsumi H, Nagai K, Suga K, Chiba Y, Chiba S, Tsugawa S, Ogra P L. Antigenic variation of human RSV strains isolated in Japan. J Med Virol. 1989;27:124–130. doi: 10.1002/jmv.1890270211. [DOI] [PubMed] [Google Scholar]
- 125.Tsutsumi H, Onuma M, Suga K. Occurrence of respiratory syncytial virus subgroups A and B strains in Japan, 1980 to 1987. J Clin Microbiol. 1988;26:1171–1174. doi: 10.1128/jcm.26.6.1171-1174.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Valdes O, Martinez I, Valdivia A, Cancio R, Savon C, Goyenechea A, Melero J A. Unusual antigenic and genetic characteristics of human respiratory syncytial viruses isolated in Cuba. J Virol. 1998;72:7589–7592. doi: 10.1128/jvi.72.9.7589-7592.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.van Milaan A J, Sprenger M J, Rothbarth P H, Brandenburg A H, Masurel N, Claas E C. Detection of respiratory syncytial virus by RNA-polymerase chain reaction and differentiation of subgroups with oligonucleotide probes. J Med Virol. 1994;44:80–87. doi: 10.1002/jmv.1890440115. [DOI] [PubMed] [Google Scholar]
- 128.Walpita P, Mufson M A, Stanek R J, Pfeifer D, Connor J D. Distinguishing between respiratory syncytial virus subgroups by protein profile analysis. J Clin Microbiol. 1992;30:1030–1032. doi: 10.1128/jcm.30.4.1030-1032.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Walsh E E, Brandriss M W, Schlesinger J J. Immunological differences between the envelope glycoproteins of two strains of human respiratory syncytial virus. J Gen Virol. 1987;68:2169–2176. doi: 10.1099/0022-1317-68-8-2169. [DOI] [PubMed] [Google Scholar]
- 130.Walsh E E, Falsey A R, Sullender W M. Monoclonal antibody neutralization escape mutants of respiratory syncytial virus with unique alterations in the attachment (G) protein. J Gen Virol. 1998;79:479–487. doi: 10.1099/0022-1317-79-3-479. [DOI] [PubMed] [Google Scholar]
- 131.Walsh E E, McConnochie K M, Long C E, Hall C B. Severity of respiratory syncytial virus infection is related to virus strain. J Infect Dis. 1997;175:814–820. doi: 10.1086/513976. [DOI] [PubMed] [Google Scholar]
- 132.Ward K A, Everson J S, Lambden P R, Watt P J. Antigenic and structural variation in the major nucleocapsid protein of respiratory syncytial virus. J Gen Virol. 1984;65:1749–1757. doi: 10.1099/0022-1317-65-10-1749. [DOI] [PubMed] [Google Scholar]
- 133.Waris M. Pattern of respiratory syncytial virus epidemics in Finland: two-year cycles with alternating prevalence of groups A and B. J Infect Dis. 1991;163:464–469. doi: 10.1093/infdis/163.3.464. [DOI] [PubMed] [Google Scholar]
- 134.Wertz G W, Collins P L, Huang Y, Gruber C, Levine S, Ball L A. Nucleotide sequence of the G protein gene of human respiratory syncytial virus reveals an unusual type of viral membrane protein. Proc Natl Acad Sci USA. 1985;82:4075–4079. doi: 10.1073/pnas.82.12.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Whimbey E, Champlin R E, Englund J A, Mirza N Q, Piedra P A, Goodrich J M, Przepiorka D, Luna M A, Morice R C, Neumann J L, Elting L S, Bodey G P. Combination therapy with aerosolized ribavirin and intravenous immunoglobulin for respiratory syncytial virus disease in adult bone marrow transplant recipients. Bone Marrow Transplant. 1995;16:393–399. [PubMed] [Google Scholar]
- 136.Wulff H, Kidd P, Wenner H A. Respiratory syncytial virus: observations on antigenic heterogeneity. Proc Soc Exp Biol Med. 1964;115:240–243. doi: 10.3181/00379727-115-28880. [DOI] [PubMed] [Google Scholar]
- 137.Zheng H, Peret T C T, Randolph V B, Crowley J C, Anderson L J. Strain-specific reverse transcriptase PCR assay: means to distinguish candidate vaccine from wild-type strains of respiratory syncytial virus. J Clin Microbiol. 1996;34:334–337. doi: 10.1128/jcm.34.2.334-337.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]