Abstract
Human cytomegalovirus (HCMV) is a globally pervasive β-herpesvirus that causes lifelong infection. The lytic replication cycle of HCMV is characterized by global organelle remodeling and dynamic virus-host interactions, both of which are necessary for productive HCMV replication. With the advent of new technologies for investigating protein-protein and protein-nucleic acid interactions, numerous critical interfaces between HCMV and host cells have been identified. Here, we review temporal and spatial virus-host interactions that support different stages of the HCMV replication cycle. Understanding how HCMV interacts with host cells during entry, replication, and assembly, as well as how it interfaces with host cell metabolism and immune responses promises to illuminate processes that underlie the biology of infection and the resulting pathologies.
Introduction
Human cytomegalovirus (HCMV) is a ubiquitous pathogen, with a seroprevalence that exceeds 80% worldwide [1]. As a β-herpesvirus, HCMV establishes life-long latent infections in myeloid cells. Reactivation from these latent reservoirs can lead to disease in the immunocompromised or cause severe birth defects. There is currently no available vaccine and there are limited antiviral therapeutics. Knowledge of interfaces that mediate virus-host interactions is critical for understanding the biology and pathogenesis of infection, as well as for discovering targets for therapeutic intervention.
HCMV is an enveloped, double-stranded DNA (dsDNA) virus that replicates in the nucleus (Figure 1). As an obligate parasite, HCMV relies on the host to provide precursors and much of the machinery to produce its proteins, replicate its genome, and form its lipid envelope. HCMV accomplishes this through a remarkably long replication cycle of four to five days, underpinned by a global transformation of the host cell proteome, metabolism, and organelle morphology (reviewed in [2,3]). Meanwhile, viral factors must disable host intrinsic defense pathways and prevent cytokine production.
Figure 1. Overview of the HCMV replication cycle.
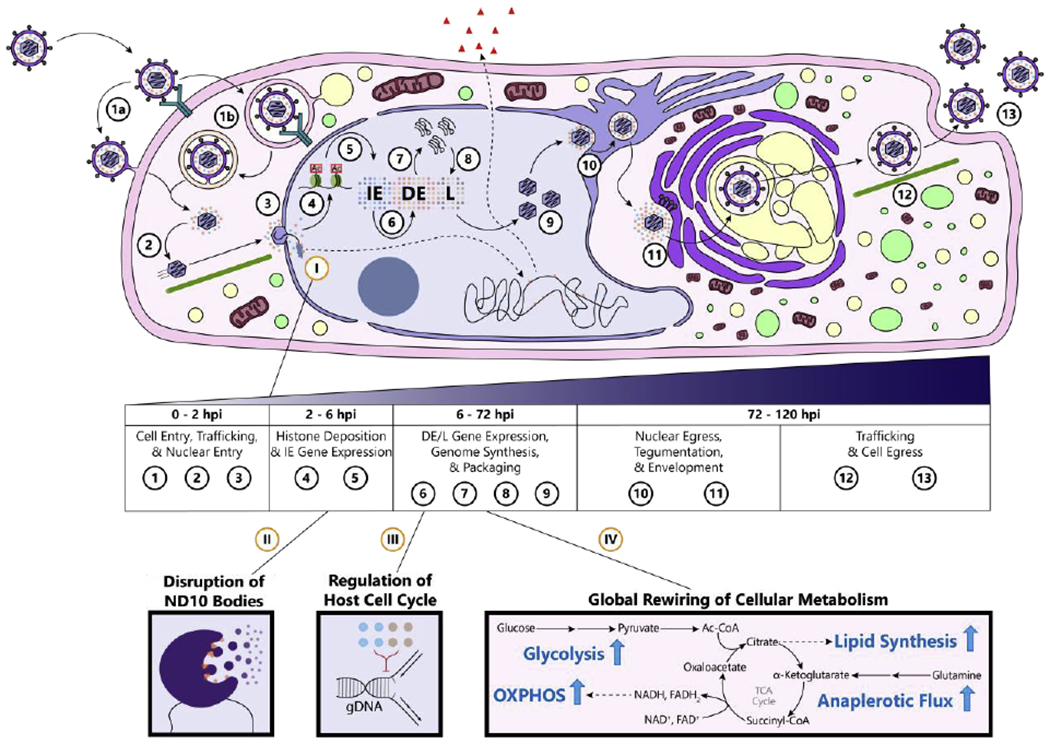
[1] The virion binds to host transmembrane proteins to promote internalization into the cell. The virion delivers the capsid and accompanying tegument proteins into the cytoplasm by either [a] direct fusion at the plasma membrane or [b] receptor-mediated endocytosis. [2] The capsid traffics along microtubules towards the nucleus. [3] The capsid docks on the nuclear pore and injects its stored genome. [4] The genome becomes histone-associated in a replication-independent manner. [5] Histone post-translational modifications (PTMs) are modified to remodel viral nucleosomes and promote immediate-early (IE) gene expression—the first group of genes expressed in the characteristic herpesvirus temporal cascade of gene expression. [6] IE proteins activate expression of delayed-early (DE) genes important for genome replication. [7] DE proteins replicate the HCMV genome. [8] Late (L) genes, which produce viral capsid proteins and glycoproteins, begin high-level gene expression. [9] Newly synthesized genomes are packaged into viral capsids. [10] The capsid acquires its first layer of tegument proteins and egresses from the nucleus by budding through both nuclear membranes. [11] The capsid acquires its second layer of tegument proteins and becomes enveloped at a new proviral organelle composed of secretory system components, the virion assembly complex. [12] Newly formed virions traffic to the plasma membrane. [13] Virions egress from the cytoplasm and spread to other cells.
Example host-virus interactions: (I) DNA sensors bind the incoming viral genome, both inducing cytokine expression from the host genome and repressing viral gene expression. These DNA sensors are targeted and inhibited by viral proteins. (II) The HCMV genome encounters nuclear domain 10 (ND10) bodies, which intrinsically repress expression from viral genomes. HCMV proteins deconstruct ND10 during the early stages of infection to relieve this repression. (III) Viral proteins stall the cell cycle at the G1/S boundary to prevent the competition over nucleotides. (IV) HCMV induces a global upregulation of cellular metabolic pathways to support different facets of its replication cycle (OXPHOS = oxidative phosphorylation).
Herpesviruses such as HCMV have coevolved with vertebrates over millions of years, leading to a myriad of virus-host interactions that occur through several core interfaces: protein-protein interactions (PPIs), post-translational modifications (PTMs) and non-coding RNA-based regulation. Here, we provide an overview of the current knowledge of critical virus-host interactions that mediate different stages of the HCMV lytic replication cycle, and highlight advanced technologies that can further our understanding of HCMV biology and pathogenesis.
Viral Entry and the Initiation of Host-Virus Interactions
Host-virus interactions during HCMV infection commence upon binding and entry of the virus into the host cell. Virus attachment and entry are achieved via protein interactions occurring between glycoproteins found on the viral envelope and cellular proteins embedded in the host plasma membrane. Initial virion-cell attachment is mediated by an interaction between the viral glycoproteins gM/gN or gB with heparin sulfate proteoglycans exposed on the cell surface [4–6]. After virion attachment to the host cell, the mechanism of entry and the complement of virus and host proteins involved is cell-type dependent.
HCMV contains trimeric and pentameric glycoprotein complexes within its viral envelope that are involved in cell-type dependent receptor binding and entry processes. Viral entry into fibroblast cells requires gB and the gH/gL/gO trimeric complex and proceeds via gB-mediated fusion with the plasma membrane [7–10]. In addition to direct fusion with the plasma membrane, macropinocytosis has been proposed as the entry mechanism for HCMV into fibroblasts [11,12]. PDGFRα has been shown to interact with gO and serve as an HCMV receptor for either mechanism of entry into fibroblasts [13–19]. In contrast, HCMV entry into endothelial and epithelial cells requires the trimeric complex, as well as the gH/gL/UL128-UL131 pentameric complex [18,20–23]. Virions enter epithelial and endothelial cells via endocytosis followed by endosomal acidification [24]. Neuropilin-2 (Nrp2) and OR14I1 have been identified as receptors for endothelial and epithelial cell entry [13,15]. In addition to PDGFRα, Nrp2, and OR14I1, several other cell surface proteins have been proposed to act as receptors or co-receptors for HCMV entry, including CD46 [25], CD90 [11,12,26], CD147 [27], integrins [28,29], EGFR [15,28,30,31], and annexin II [32,33]. The internalization of some of these co-receptors, such as the CD63-dependent internalization of integrin beta 1 [34], was observed to support virus production, suggesting a mechanism to prevent virion reattachment to the plasma membrane during egress from the cell.
Given the importance of viral entry for initiating the cascade of events resulting in the production of new virus progeny, several host proteins have been found to act in host defense by specifically interfering with the viral entry process. For example, galectin-9 (Gal-9) has been shown to become upregulated in the plasma of patients experiencing HCMV reactivation. Gal-9 binds to the HCMV virion, likely through interaction with the trimeric complex, and acts to inhibit viral fusion with both epithelial and fibroblast cells [35].
Initiation of Host Immune Responses and HCMV Immune Evasion Mechanisms
Following entry, virus capsids utilize the host cytoskeletal machinery for trafficking to the nucleus, where virus gene expression and genome replication occur [36,37]. The injection of the viral genome into the nucleus activates host intrinsic immune responses. The viral genome becomes exposed to nuclear DNA sensors, such as IFI16, which distinguish the HCMV dsDNA from host chromatin and induce antiviral responses upon binding to the viral DNA [38–42]. IFI16 was shown to both promote cytokine production and inhibit viral gene expression [38,39,42]. In response to host intrinsic immune responses, HCMV acquired a number of viral immune evasion mechanisms to suppress immune and inflammatory pathways. This is accomplished through the early actions of tegument proteins—a cargo of viral proteins packaged into virions along with the capsid—and the later actions of de novo synthesized viral proteins, in particular the immediate-early proteins. For example, the tegument protein pp65 (pUL83) binds to the pyrin domain of IFI16, inhibiting its oligomerization and intrinsic immune functions [39,43]. Similarly, several HCMV proteins—pUL31, pUL42, and pUL83—inhibit cytoplasmic DNA sensing and immune signaling by cyclic GMP-AMP synthase (cGAS) [44–46].
HCMV suppresses various host intrinsic and innate immune response pathways. HCMV gene products antagonize protein kinase R-dependent translation shutoff [47–49], MAVS and cGAS-STING immune signaling [44,45,50–53], and JAK-STAT immune signaling [54–59]. As another viral immune evasion strategy, HCMV encodes an IL-10 mimic to inhibit an inflammatory response [60]. One exception to this broad inhibition of immune signaling is the NF-κB pathway. NF-κB signaling and type I IFN is activated upon viral entry [61–65], but, perhaps in a counter-intuitive fashion, NF-κB transcription factors support virus replication by activating immediate-early genes [66–68]. As such, HCMV promotes expression of NF-κB subunits early in infection [69]. However, later in infection, NF-κB signaling is inhibited at several steps along the pathway. NF-κB signaling transduction and downstream expression of interferon-stimulated genes (ISGs) is inhibited by HCMV proteins IE2 [70–72], pUL26 [73–76], and pUL44 [77].
Orthogonal to the aforementioned suppression of immediate immune responses, HCMV acquired means to hide the infection status of its host cell from immune effector cells. HCMV encodes protein and miRNA to inhibit several steps in the MHC-I antigen presentation pathway to prevent T cell- and natural killer (NK) cell-mediated lysis. Viral pUS3 and pUS6 prevent trafficking of mature MHC-I to the plasma membrane [78,79], while pUS2 and pUS11 promote outright degradation of MHC-I [80,81]. Degradation of MHC-like receptors MICA and MICB is induced upon infection to prevent NK-dependent lysis: MICA is targeted by viral proteins [82–86] and MICB by viral miRNAs [87,88]. HCMV further promotes surface exposure of NK inhibitory receptors by enabling HLA-E translocation [89] and encoding an MHC-I surrogate [90,91]. This suite of mechanisms allows HCMV to antagonize both cell-intrinsic and -extrinsic host defense pathways.
Virus-Host Interactions Affecting Viral Gene Expression and Genome Replication
The HCMV genome is naked while encapsidated, but becomes chromatinized via the addition of histones after its deposition into host nuclei [92–95]. In the same manner as host chromatin, viral chromatin must be remodeled to promote gene expression, which is dictated by the addition or removal of histone post-translational modifications (PTMs) [92,94,96–102]. Herpesvirus genes are expressed in a “temporal cascade,” whereby the first set of viral genes, the immediate-early (IE) genes, drive the subsequent expression of delayed-early (DE) and late (L) genes (Figure 1). The initial deposition of histones results in a “pre-immediate-early” repression of the major immediate-early promoter (MIEP) [94]. Viral and host proteins compete for this key regulatory node. Upstream of the MIEP are various transcription factor (TF) binding sites for viral proteins, as well as host proteins [66–68,103–107]. While NF-κB TFs activate the MIEP, some host restriction factors bind and repress the MIEP [96,98–100,102,108–111].
A prominent gene repression mechanism is driven by nuclear domain 10 (ND10), also known as PML bodies. These macromolecular structures activate both intrinsic and innate immune responses against herpesviruses. Components of ND10 are interferon-induced and can themselves promote induction of interferons [57,58,112,113]. ND10 recruits histone deacetylases and other repressive proteins to the HCMV genome to prevent gene expression [99,100,109,114]. The HCMV protein IE1 dissolves these structures to relieve transcriptional repression [115–117], which is accomplished by antagonizing SUMOylation of ND10 components PML and Sp100 [115,118–120]. In parallel with these deSUMOylation events, IE2 becomes SUMOylated to bolster its transactivation of viral promoters [121–123]. Regulation of this PTM is key to galvanizing early gene expression events.
As gene expression proceeds through the temporal cascade and genome synthesis begins, HCMV is regulating core host pathways to maximize replication. HCMV maintains a robust pool of dNTPs by antagonizing the dNTPase SAMHD1 [124–126]. HCMV prevents degradation of certain viral mRNAs through stem-loop secondary structures that recruit 3’ tail extension machinery [127]. A critical requirement for effective replication is the manipulation of the cell cycle. HCMV encodes a cell cycle sensor, pp150 (pUL32), that blocks IE gene expression in S/G2 until the cell returns to G1 [128,129]. Once in a permissive state, the serine/threonine protein kinase pUL97 and pUL21a inhibit progression into S-phase via degradation and inactivation of the anaphase-promoting complex (APC/C) [130–133]. HCMV proteins further inactivate tumor-suppressor proteins p53 [134–137] and Rb [138–142]. This stops cells at the G1/S boundary, which is believed to establish an S-phase proteome conducive to viral genome replication without allowing host DNA replication to compete for replication machinery and dNTPs. With favorable conditions present, the interaction between pUL44 and the host nucleolin protein establishes nuclear replication compartments for the synthesis of new genomes [143,144]. These genomes are then packaged into viral capsids and must egress from the nucleus.
Host-Virus Interactions in Metabolic Regulation
To promote productive infection, HCMV interfaces with host cell metabolic pathways to induce energetic and biosynthetic output necessary for the production of progeny virions. The specific metabolic program activated during HCMV infection consists of upregulated glycolysis, glutaminolysis, lipid biosynthesis, and oxidative phosphorylation [145–149]. These metabolic changes begin around 48 hours post infection and are necessary for productive virus replication [146,149–152].
The importance of cellular metabolism for HCMV replication is reflected by the diverse mechanisms acquired by the virus to alter metabolic processes during infection. For instance, glycolytic induction is achieved via a finely orchestrated modulation of signaling pathways, enzyme activity, and protein localization. The increases in glucose uptake and glycolysis during infection are linked to HCMV induction of the calmodulin-dependent kinase kinase (CaMKK) and AMP-activated protein kinase (AMPK) signaling pathways [152–155]. The induction of AMPK is achieved by HCMV protein pUL37x1-mediated targeting of the host protein viperin to the mitochondria [155,156]. At the mitochondria, viperin interacts with the mitochondrial trifunctional protein to decrease fatty acid oxidation, resulting in decreased ATP production, AMPK activation, and increased GLUT4 expression [155,156]. Mitochondrial localized viperin also results in induction and nuclear localization of ChREBP, which further increases GLUT4 expression [157]. GLUT4, a high-capacity glucose transporter, replaces the lower-capacity glucose transporter GLUT1 at the plasma membrane, resulting in increased glucose uptake [158,159].
It has been recently demonstrated that, in spite of the activation of glycolysis during infection, HCMV also induces and requires mitochondrial respiration for energy production [148,149,160,161]. Upregulated oxidative phosphorylation during infection is partly achieved via increased transcription and translation of mitochondrial DNA genes, resulting in elevated electron transport chain protein abundances [160]. An HCMV non-coding RNA (ncRNAβ2.7) has been shown to contribute to increased ATP production by interacting with complex I of the electron transport chain to stabilize mitochondrial membrane potential [162]. Furthermore, the HCMV protein pUL13 has been demonstrated to target the mitochondria during infection, where it interacts with the mitochondrial contact site and cristae organizing system (MICOS) complex and modulates cristae ultrastructure to increase electron transport chain efficiency and ATP production [161].
A major source of biosynthetic output during HCMV infection relates to lipid biosynthesis, which is important for viral envelopment and cellular membrane remodeling. The increase in lipid biosynthesis is due to the activation and nuclear translocation of the SREBP and ChREBP transcription factors during infection, resulting in increased lipogenic gene expression [151,155,157,163]. This is achieved through the actions of the HCMV proteins pUL38 and pUL37x1. pUL38 contributes to SREBP activation by binding to and antagonizing the function of tuberous sclerosis protein complex (TSC1/2), resulting in mTOR activation and induction of SREBP processing [163,164], pUL37x1-mediated translocation of viperin to the mitochondria also induces nuclear localization of ChREBP, resulting in lipogenic gene transcription [155,157]. In addition to inducing ChREBP, pUL37x1 also localizes to the peroxisomes during infection [165,166]. There, it activates the peroxisome fission factor PEX11β resulting in increased peroxisome abundance [167]. This serves to increase plasmalogen synthesis, which is important for virion envelopment [166]. In addition to phospholipids, cholesterol is required during HCMV replication for inclusion in the viral envelope [168]. Virions depleted of cholesterol are unable to fuse with the plasma membrane of successive host cells [168]. HCMV primarily drives increased cellular cholesterol levels by regulating the abundance of cholesterol receptors and transporters at the plasma membrane. The levels of low-density lipoprotein scavenger receptors at the plasma membrane are increased by HCMV [169]. As this occurs, HCMV activates calpain-mediated cleavage of the cholesterol efflux transporter ABCA1 [170] and metalloproteinase-mediated cleavage of the low-density lipoprotein receptor protein LRP1 [168].
Remodeling of Subcellular Organization during Virion Assembly and Trafficking
The remainder of the HCMV replication cycle is characterized by a striking remodeling of organelle structure and location for supporting virus assembly processes. Hallmarks of HCMV-infected cells are the formation of a kidney bean-shaped nucleus with infoldings present at the nuclear periphery and the dynamic reorganization of host secretory membranes into the perinuclear viral assembly complex (vAC) [171] (Figure 1).
The nuclear egress of newly formed virus capsids requires the remodeling of the nuclear periphery via the formation of infoldings of the lamina. To accomplish this remodeling, capsids first associate with myosin 5A to move from replication compartments to the nuclear periphery via nuclear actin, which is both induced and bundled into thick filaments connecting replication compartments to the nuclear membrane [172,173]. Once at the nuclear periphery, capsids dock to the multimeric nuclear egress complex (NEC). The primary function of the NEC is to orchestrate phosphorylation of nuclear lamin proteins, resulting in disruption of the nuclear lamina and capsid egress to the cytoplasm [174]. The core components of the NEC are the viral proteins pUL50, pUL53, and pUL97 [174,175]. However, it has been shown that host proteins, including the structural proteins p32 [176,177], emerin [177], and WDR5 [178] and cellular kinases CDK1 [179] and PKC [179,180] are also recruited to the NEC and serve critical roles in NEC formation and lamina disruption. It was demonstrated that capsid nuclear egress can be inhibited by the host cell via acetylation of lamin B1, which protects the integrity of the nuclear periphery [181].
Once capsids egress from the nucleus, virion envelopment and egress from the cell commence. The processes of virion envelopment at the vAC and cellular egress are not fully understood; however, there is accumulating evidence that the virus utilizes the host exosome machinery for these processes [182,183]. Additionally, it has been demonstrated that virion tegument and envelope proteins possess diverse means for trafficking to the vAC. The envelope glycoprotein gB has been shown to interact with the host secretory protein PACS-1 for retrograde transport from endosomes to the Golgi [184,185]. In order for this to occur, viral protein pUL35 binds and inactivates sorting nexin SNX5 to regulate retrograde shuttling [186]. In contrast, gM localization to the vAC requires host RAB11-GTPase effector protein FIP4 [187]. Viral tegument protein pUL99 traffics to the vAC using the ESCRT pathway, while pUL32 traffics via an interaction with BicD1, a protein involved in dynein-mediated microtubule transport [188–190].
The final processes of virion assembly and cellular egress require global organelle remodeling and repurposing of host machinery that together serve as a platform for supporting virus production. The vAC contains early endosomes at the center, surrounded by fragmented Golgi networks that form concentric rings where viral capsids obtain their tegument layer and envelope [171,191–194]. Although the full panel of host proteins involved in vAC formation are still being identified, crucial roles for the ER chaperone BiP/GRP78 [195,196], SNARE proteins [182,197–199], GTPases [200,201], and the v-ATPase proton pump [202] have been demonstrated, supporting that host endosome and secretory pathway machinery is repurposed to function at the vAC during HCMV infection. Furthermore, viral miRNAs target secretory pathway proteins, which is necessary for vAC formation [203].
Beyond its translocation to the vAC, fragmentation of the Golgi is necessary for proper vAC formation. This is achieved via virus-mediated phosphorylation of the cellular Grasp65 protein, which functions in regulating Golgi structural integrity [204]. Phosphorylation of Grasp65 typically occurs during cellular mitosis and results in Golgi fragmentation. Therefore, this represents an instance of HCMV supporting replication by inducing a process that typically occurs in uninfected cells. At the vAC, Golgi fragments function in microtubule nucleation [205,206]. The vAC was previously shown to form around a microtubule organizing center (MTOC) via a microtubule-dependent process [171,189]. It has now been demonstrated that the microtubule end binding protein EB3 is phosphorylated by the host kinase CDK and virus kinase pUL97, resulting in its stabilization. It is then recruited to the vAC where it nucleates Golgi-derived microtubules that are rapidly acetylated and stabilized, resulting in nuclear rotation and vAC formation [205,206].
Perspective: Methods for Investigating Virus-Host Interactions and Future Directions
As highlighted above, HCMV infection dramatically alters the composition and function of the cellular proteome in a temporal manner. A range of biochemical assays have been used to gain insights into dynamic virus-host protein interactions, viewed either through the perspective of single or multiple proteins of interest (Figure 2). Immunoaffinity purification coupled with mass spectrometry (IP-MS) [161,167,207] and yeast-two hybrid screens (Y2H) [117,121,208] have proved valuable for investigations focused on specific virus or host proteins of interest. Global omic techniques have allowed researchers to acquire a holistic understanding of host cell remodeling by determining: temporal changes in the transcriptome and proteome of an infected cell [209,210]; changes in organellar proteomes through spatial proteomics [211,212]; regulation of protein functions via PTM states through PTM-enrichment methods [181,213,214]; and global protein-protein interaction dynamics through thermal-proximity coaggregation assays (TPCA) [34]. This has been augmented by the integration and computational mining of these datasets [167,215,216]. Despite the depth of information that can be garnered from these approaches, only a handful of PTM types have been investigated during infection and targeted or global omic investigations have been performed in limited cell types and upon infection with few viral strains. Another consideration is that many of these studies obtain information that is averaged across cell populations, thereby being impacted by cell-to-cell heterogeneity. Single-cell technologies [217] are poised to fill this niche, providing a complementary toolset for virus-host interaction studies in HCMV and beyond.
Figure 2. Methods used for investigating protein-protein and protein-nucleic acid interactions during HCMV infection.
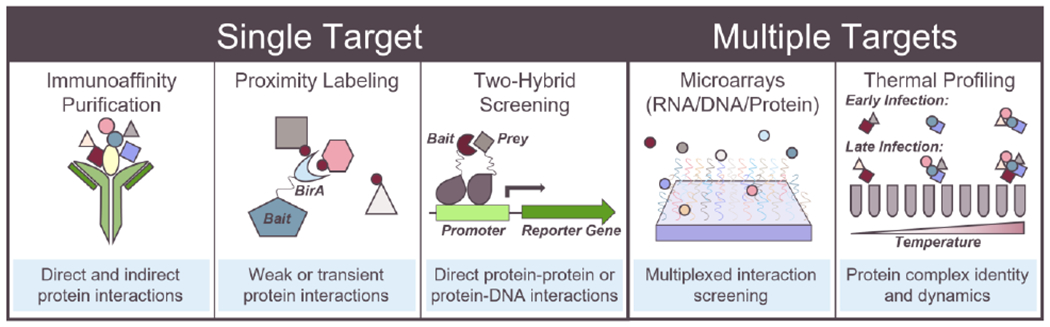
Immunoaffinity purification coupled with mass spectrometry or western blotting, proximity labeling, and yeast or bacterial two-hybrid screening are powerful techniques for investigating interactions of single viral or host proteins of interest. Conversely, microarrays and thermal proximity coaggregation profiling provide global insight into virus-host interactions.
Acknowledgments:
We are grateful for funding from the NIH NIGMS (GM114141) and the Stand-Up-to-Cancer to I. M.C., NIH NIAID Ruth L. Kirschstein NRSA fellowship F31AI154796 to C.N.B., and NIGMS T32GM007388 to M.D.T. The funders had no role in the decision to submit the work for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interest:
The authors declare that there is no conflict of interest.
Declarations of interest: none
References:
- 1.Zuhair M, Smit GSA, Wallis G, Jabbar F, Smith C, Devleesschauwer B, Griffiths P: Estimation of the worldwide seroprevalence of cytomegalovirus: A systematic review and meta-analysis. Rev Med Virol 2019, 29:1–6. [DOI] [PubMed] [Google Scholar]
- 2.Shenk T, Alwine JC: Human Cytomegalovirus: Coordinating Cellular Stress, Signaling, and Metabolic Pathways. Annu Rev ofVirology 2014, doi: 10.1146/annurev-virology-031413-085425. [DOI] [PubMed] [Google Scholar]
- 3.Beltran PMJ, Cristea IM: The life cycle and pathogenesis of human cytomegalovirus infection: lessons from proteomics. Expert Rev Proteomics 2015, 11:697–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyle KA, Compton T: Receptor-binding properties of a soluble form of human cytomegalovirus glycoprotein B. J Virol 1998, 72:1826–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Compton T, Nowlin DM, Cooper NR: Initiation of HCMV infection requires initial interaction with cell surface heparan sulfate. Virology 1993, 193:834–841. [DOI] [PubMed] [Google Scholar]
- 6.Kari B, Gehrz R: A human cytomegalovirus glycoprotein complex designated gC-II is a major heparin-binding component of the envelope. J Virol 1992, 66:1761–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wille PT, Wisner TW, Ryckman B, Johnson DC: Human cytomegalovirus (HCMV) glycoprotein gB promotes virus entry in Trans acting as the viral fusion protein rather than as a receptor-binding protein. MBio 2013, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Isaacson MK, Compton T: Human Cytomegalovirus Glycoprotein B Is Required for Virus Entry and Cell-to-Cell Spread but Not for Virion Attachment, Assembly, or Egress. J Virol 2009, 83:3891–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Navarro D, Paz P, Tugizov S, Topp K, La Vail J, Pereira L: Glycoprotein B of human cytomegalovirus promotes virion penetration into cells, transmission of infection from cell to cell, and fusion of infected cells. Virology 1993, 197:143–158. [DOI] [PubMed] [Google Scholar]
- 10.Compton T, Nepomuceno RR, Nowlin DM: Human cytomegalovirus penetrates host cells by pH-independent fusion at the cell surface. Virology 1992, 191:387–395. [DOI] [PubMed] [Google Scholar]; * Demonstrates that HCMV enters fibroblast cells via fusion rather than endocytosis.
- 11.Li Q, Fischer E, Cohen JI: Cell Surface THY-1 Contributes to Human Cytomegalovirus Entry via a Macropinocytosis-Like Process. J Virol 2016, 90:9766–9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hetzenecker S, Helenius A, Krzyzaniak MA: HCMV Induces Macropinocytosis for Host Cell Entry in Fibroblasts. Traffic 2016, 17:351–368. [DOI] [PubMed] [Google Scholar]
- 13.Xiaofei E, Meraner P, Lu P, Perreira JM, Aker AM, McDougall WM, Zhuge R, Chan GC, Gerstein RM, Caposio P, et al. : OR14I1 is a receptor for the human cytomegalovirus pentameric complex and defines viral epithelial cell tropism. Proc Natl Acad Sci U S A 2019, 116:7043–7052. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Performed a genome-wide CRISPR/Cas9 screen to investigate host factors required for HCMV infection and identified OR14I1 as a pentamer-dependent receptor for HCMV infection of epithelial cells.
- 14.Wu K, Oberstein A, Wang W, Shenk T: Role of PDGF receptor-α during human cytomegalovirus entry into fibroblasts. Proc Natl Acad Sci U S A 2018, 115:E9889–E9898. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Mapped the essential domains of PDGFRα that are required for HCMV entry into fibroblast cells.
- 15.Martinez-Martin N, Marcandalli J, Huang CS, Arthur CP, Perotti M, Foglierini M, Ho H, Dosey AM, Shriver S, Payandeh J, et al. : An Unbiased Screen for Human Cytomegalovirus Identifies Neuropilin-2 as a Central Viral Receptor. Cell 2018, 174:1158–1171.e19. [DOI] [PubMed] [Google Scholar]; ** Constructed a library of human receptors for investigating receptor-ligand interactions in an unbiased fashion, leading to the identification of neuropilin-2 as a receptor for the HCMV pentamer and validating PDGFRα as the trimer receptor.
- 16.Stegmann C, Hochdorfer D, Lieber D, Subramanian N, Stöhr D, Laib Sampaio K, Sinzger C: A derivative of platelet-derived growth factor receptor alpha binds to the trimer of human cytomegalovirus and inhibits entry into fibroblasts and endothelial cells. PLoS Pathog 2017, 13:1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y, Prager A, Boos S, Resch M, Brizic I, Mach M, Wildner S, Scrivano L, Adler B: Human cytomegalovirus glycoprotein complex gH/gL/gO uses PDGFR-α as a key for entry. PLoS Pathog 2017, 13:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kabanova A, Marcandalli J, Zhou T, Bianchi S, Baxa U, Tsybovsky Y, Lilleri D, Silacci-Fregni C, Foglierini M, Fernandez-Rodriguez BM, et al. : Platelet-derived growth factor-α receptor is the cellular receptor for human cytomegalovirus gHgLgO trimer. Nat Microbiol 2016, 1:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soroceanu L, Akhavan A, Cobbs CS: Platelet-derived growth factor-α receptor activation is required for human cytomegalovirus infection. Nature 2008, 455:391–395. [DOI] [PubMed] [Google Scholar]
- 20.Zhou M, Lanchy J-M, Ryckman BJ: Human Cytomegalovirus gH/gL/gO Promotes the Fusion Step of Entry into All Cell Types, whereas gH/gL/UL128-131 Broadens Virus Tropism through a Distinct Mechanism. J Virol 2015, 89:8999–9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wille PT, Knoche AJ, Nelson JA, Jarvis MA, Johnson DC: A Human Cytomegalovirus gO-Null Mutant Fails To Incorporate gH/gL into the Virion Envelope and Is Unable To Enter Fibroblasts and Epithelial and Endothelial Cells. J Virol 2010, 84:2585–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryckman BJ, Chase MC, Johnson DC: HCMV gH/gL/UL128-131 interferes with virus entry into epithelial cells: Evidence for cell type-specific receptors. Proc Natl Acad Sci USA 2008, 105:14118–14123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang D, Shenk T: Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc Natl Acad Sci USA 2005, 102:18153–18158. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Define the pentameric complex as necessary for HCMV infection of endothelial and epithelial cells.
- 24.Ryckman BJ, Jarvis MA, Drummond DD, Nelson JA, Johnson DC: Human Cytomegalovirus Entry into Epithelial and Endothelial Cells Depends on Genes UL128to UL150 and Occurs by Endocytosis and Low-pH Fusion. J Virol 2006, 80:710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Demonstrates that HCMV enters epithelial and endothelial cells via endocytosis followed by low-pH-dependent fusion.
- 25.Stein KR, Gardner TJ, Hernandez RE, Kraus TA, Duty JA, Ubarretxena-Belandia I, Moran TM, Tortorella D: CD46 facilitates entry and dissemination of human cytomegalovirus. Nat Commun 2019, 10:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q, Wilkie AR, Weller M, Liu X, Cohen Jl: THY-1 Cell Surface Antigen (CD90) Has an Important Role in the Initial Stage of Human Cytomegalovirus Infection. PLoS Pathog 2015, 11:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vanarsdall AL, Pritchard SR, Wisner TW, Liu J, Jardetzky TS, Johnson DC: CD147 Promotes Entry of Pentamer-Expressing Human Cytomegalovirus into Epithelial and Endothelial Cells. MBio 2018, 9:e00781–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Huang DY, Huong SM, Huang ES: Integrin αvβ3 is a coreceptor for human cytomegalovirus. Nat Med 2005, 11:515–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feire AL, Koss H, Compton T: Cellular integrins function as entry receptors for human cytomegalovirus via a highly conserved disintegrin-like domain. Proc Natl Acad Sci U S A 2004, 101:15470–15475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Isaacson MK, Feire AL, Compton T: Epidermal Growth Factor Receptor Is Not Required for Human Cytomegalovirus Entry or Signaling. J Virol 2007, 81:6241–6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Huong SM, Chiu ML, Raab-Traub N, Huang ES: Epidermal growth factor receptor is a cellular receptor for human cytomegalovirus. Nature 2003, 424:456–461. [DOI] [PubMed] [Google Scholar]
- 32.Pietropaolo R, Compton T: Interference with annexin II has no effect on entry of human cytomegalovirus into fibroblast cells. J Gen Virol 1999, 80:1807–1816. [DOI] [PubMed] [Google Scholar]
- 33.Pietropaolo RL, Compton T: Direct interaction between human cytomegalovirus glycoprotein B and cellular annexin II. J Virol 1997, 71:9803–9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hashimoto Y, Sheng X, Murray-Nerger LA, Cristea IM: Temporal dynamics of protein complex formation and dissociation during human cytomegalovirus infection. Nat Commun 2020, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** First application of thermal coaggregation profiling to defining global alterations in protein interactions during a viral infection.
- 35.Machala EA, Avdic S, Stern L, Zajonc DM, Benedict CA, Blyth E, Gottlieb DJ, Abendroth A, McSharry BP, Slobedman B: Restriction of Human Cytomegalovirus Infection by Galectin-9. J Virol 2019, 93:e01746–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller MS, Hertel L: Onset of Human Cytomegalovirus Replication in Fibroblasts Requires the Presence of an Intact Vimentin Cytoskeleton. J Virol 2009, 83:7015–7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogawa-Goto K, Tanaka K, Gibson W, Moriishi E, Miura Y, Kurata T, Irie S, Sata T: Microtubule Network Facilitates Nuclear Targeting of Human Cytomegalovirus Capsid. J Virol 2003, 77:8541–8547. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Found that capsid trafficking to the nucleus is dependent on the microtubule network.
- 38.Gariano GR, Dell’Oste V, Bronzini M, Gatti D, Luganini A, de Andrea M, Gribaudo G, Gariglio M, Landolfo S: The intracellular DNA sensor IFI16 gene acts as restriction factor for human Cytomegalovirus replication. PLoS Pathog 2012, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li T, Chen J, Cristea IM: Human cytomegalovirus tegument protein pUL83 inhibits IFI16-mediated DNA sensing for immune evasion. Cell Host Microbe 2013, 14:591–599. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Discovery that the HCMV protein pUL83 prevents IFI16-mediated DNA sensing by interfering with IFI16 pyrin domain-based oligomerization.
- 40.Dell’Oste V, Gatti D, Gugliesi F, De Andrea M, Bawadekar M, Lo Cigno I, Biolatti M, Vallino M, Marschall M, Gariglio M, et al. : Innate Nuclear Sensor IFI16 Translocates into the Cytoplasm during the Early Stage of In Vitro Human Cytomegalovirus Infection and Is Entrapped in the Egressing Virions during the Late Stage . J Virol IMA, 88:6970–6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diner BA, Lum KK, Toettcher JE, Cristea IM: Viral DNA sensors IFI16 and cyclic GMP-AMP synthase possess distinct functions in regulating viral gene expression, immune defenses, and apoptotic responses during herpesvirus infection. MBio 2016, 7:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Demonstration that the DNA sensor IFI16 binds to incoming HCMV and HSV-1 genomes within puncta at the nuclear periphery
- 42.Biolatti M, Dell’Oste V, Pautasso S, von Einem J, Marschall M, Plachter B, Gariglio M, De Andrea M, Landolfo S: Regulatory Interaction between the Cellular Restriction Factor IFI16 and Viral pp65 (pUL83) Modulates Viral Gene Expression and IFI16 Protein Stability. J Virol 2016, 90:8238–8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cristea IM, Moorman NJ, Terhune SS, Cuevas CD, O’Keefe ES, Rout MP, Chait BT, Shenk T: Human Cytomegalovirus pUL83 Stimulates Activity of the Viral Immediate-Early Promoter through Its Interaction with the Cellular IFI16 Protein. J Virol 2010, 84:7803–7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang ZF, Zou HM, Liao BW, Zhang HY, Yang Y, Fu YZ, Wang SY, Luo MH, Wang YY: Human Cytomegalovirus Protein UL31 Inhibits DNA Sensing of cGAS to Mediate Immune Evasion. Cell Host Microbe 2018, 24:69–80.e4. [DOI] [PubMed] [Google Scholar]; * Discovery that pUL31 inhibits DNA sensing by cGAS to prevent antiviral immune signaling.
- 45.Biolatti M, Oste D, Pautasso S, Gugliesi F, Einem J Von, Krapp C, Jakobsen MR, Borgogna C, Gariglio M, Andrea M De: Human Cytomegalovirus Tegument Protein pp65 (pUL83) Dampens Type I Interferon Production by Inactivating the DNA Sensor cGAS without Affecting STING. J Virol 2018, 92:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu Y, Guo Y, Zou H, Su S, Wang S, Yang Q, Id HL, Id YW: Human cytomegalovirus protein UL42 antagonizes cGAS / MITA-mediated innate antiviral response. PLoS Pathog 2019, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hakki M, Marshall EE, De Niro KL, Geballe AP: Binding and Nuclear Relocalization of Protein Kinase R by Human Cytomegalovirus TRS1. J Virol 2006, 80:11817–11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ziehr B, Vincent HA, Moorman NJ: Human Cytomegalovirus pTRS1 and plRS1 Antagonize Protein Kinase R To Facilitate Virus Replication. J Virol 2016, 90:3839–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vincent HA, Ziehr B, Moorman NJ: Mechanism of Protein Kinase R Inhibition by Human Cytomegalovirus pTRS1. J Virol 2017, 91:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi HJ, Park A, Kang S, Lee E, Lee TA, Ra EA, Lee J, Lee S, Park B: Human cytomegalovirus-encoded US9 targets MAVS and STING signaling to evade type i interferon immune responses. Nat Commun 2018, 9:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lio C-WJ, McDonald B, Takahashi M, Dhanwani R, Sharma N, Huang J, Pham E, Benedict CA, Sharma S: cGAS-STING Signaling Regulates Initial Innate Control of Cytomegalovirus Infection. J Virol 2016, 90:7789–7797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fu YZ, Su S, Gao YQ, Wang PP, Huang ZF, Hu MM, Luo WW, Li S, Luo MH, Wang YY, et al. : Human Cytomegalovirus Tegument Protein UL82 Inhibits STING-Mediated Signaling to Evade Antiviral Immunity. Cell Host Microbe 2017, 21:231–243. [DOI] [PubMed] [Google Scholar]
- 53.Zou H, Huang Z, Yang Y, Luo W, Wang S: Human Cytomegalovirus Protein UL94 Targets MITAto Evade the Antiviral Immune Response. J Virol 2020, 94:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paulus C, Krauss S, Nevels M: A human cytomegalovirus antagonist of type I IFN-dependent signal transducer and activator of transcription signaling. Proc Natl Acad Sci U S A 2006, 103:3840–3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huh YH, Kim YE, Kim ET, Park JJ, Song MJ, Zhu H, Hayward GS, Ahn J-H: Binding STAT2 by the Acidic Domain of Human Cytomegalovirus IE1 Promotes Viral Growth and Is Negatively Regulated by SUMO. J Virol 2008, 82:10444–10454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krauss S, Kaps J, Czech N, Paulus C, Nevels M: Physical Requirements and Functional Consequences of Complex Formation between the Cytomegalovirus IE1 Protein and Human STAT2. J Virol 2009, 83:12854–12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim YE, Ahn JH: Positive Role of Promyelocytic Leukemia Protein in Type I Interferon Response and Its Regulation by Human Cytomegalovirus. PLoS Pathog 2015, 11:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scherer M, Otto V, Stump JD, Klingl S, Muller R, Reuter N, Muller YA, Sticht H, Stamminger T: Characterization of Recombinant Human Cytomegaloviruses Encoding IE1 Mutants L174P and 1-382 Reveals that Viral Targeting of PML Bodies Perturbs both Intrinsic and Innate Immune Responses. J Virol 2016, 90:1190–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Le-Trilling VTK, Becker T, Nachshon A, Stern-Ginossar N, Scholer L, Voigt S, Hengel H, Trilling M: The Human Cytomegalovirus pUL145 Isoforms Act as Viral DDB1-Cullin-Associated Factors to Instruct Host Protein Degradation to Impede Innate Immunity. Cell Rep 2020, 30:2248–2260.e5. [DOI] [PubMed] [Google Scholar]
- 60.Kotenko SV, Saccani S, Izotova LS, Mirochnitchenko OV, Pestka S: Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10). Proc Natl Acad Sci U S A 2000, 97:1695–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yurochko AD, Hwang ES, Rasmussen L, Keay S, Pereira L, Huang ES: The human cytomegalovirus UL55 (gB) and UL75 (gH) glycoprotein ligands initiate the rapid activation of Sp1 and NF-kappaB during infection. J Virol 1997, 71:5051–5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boyle KA, Pietropaolo RL, Compton T: Engagement of the Cellular Receptor for Glycoprotein B of Human Cytomegalovirus Activates the Interferon-Responsive Pathway. Mol Cell Biol 1999, 19:3607–3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Browne EP, Wing B, Coleman D, Shenk T: Altered Cellular mRNA Levels in Human Cytomegalovirus-Infected Fibroblasts: Viral Block to the Accumulation of Antiviral mRNAs. J Virol 2001, 75:12319–12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Simmen KA, Singh J, Luukkonen BGM, Lopper M, Bittner A, Miller NE, Jackson MR, Compton T, Früh K: Global modulation of cellular transcription by human cytomegalovirus is initiated by viral glycoprotein B. Proc Natl Acad Sci U S A 2001, 98:7140–7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boehme KW, Guerrero M, Compton T: Human Cytomegalovirus Envelope Glycoproteins B and H Are Necessary for TLR2 Activation in Permissive Cells. J Immunol 2006, 177:7094–7102. [DOI] [PubMed] [Google Scholar]
- 66.Yurochko AD, Mayo MW, Poma EE, Baldwin AS, Huang ES: Induction of the transcription factor Sp1 during human cytomegalovirus infection mediates upregulation of the p65 and p105/p50 NF-kappaB promoters. J Virol 1997, 71:4638–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Caposio P, Dreano M, Garotta G, Gribaudo G, Landolfo S: Human Cytomegalovirus Stimulates Cellular IKK2 Activity and Requires the Enzyme for Productive Replication. J Virol 2004, 78:3190–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.DeMeritt IB, Milford LE, Yurochko AD: Activation of the NF-κB Pathway in Human Cytomegalovirus-Infected Cells Is Necessary for Efficient Transactivation of the Major Immediate-Early Promoter. J Virol 2004, 78:4498–4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yurochko AD, Kowalik TF, Huong SM, Huang ES: Human cytomegalovirus upregulates NF-kappa B activity by transactivating the NF-kappa B p105/p50 and p65 promoters. J Virol 1995, 69:5391–5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Taylor RT, Bresnahan WA: Human Cytomegalovirus Immediate-Early 2 Gene Expression Blocks Virus-Induced Beta Interferon Production. J Virol 2005, 79:3873–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Taylor RT, Bresnahan WA: Human Cytomegalovirus Immediate-Early 2 Protein IE86 Blocks Virus-Induced Chemokine Expression. J Virol 2006, 80:920–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Taylor RT, Bresnahan WA: Human Cytomegalovirus IE86 Attenuates Virus- and Tumor Necrosis Factor Alpha-Induced NFκB-Dependent Gene Expression. J Virol 2006, 80:10763–10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mathers C, Schafer X, Martínez-Sobrido L, Munger J: The Human Cytomegalovirus UL26 Protein Antagonizes NF-κB Activation. J Virol 2014, 88:14289–14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hancock MH, Hook LM, Mitchell J, Nelson JA: Human cytomegalovirus microRNAs miR-US5-1 and miR-UL112-3p block proinflammatory cytokine production in response to NF-κB-activating factors through direct downregulation of IKKα and IKKβ. MBio 2017, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Goodwin CM, Munger J: The IκB Kinases Restrict Human Cytomegalovirus Infection. J Virol 2019, 93:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goodwin CM, Schafer X, Munger J: UL26 Attenuates IKKB-Mediated Induction of Interferon- Stimulated Gene (ISG) Expression and Enhanced Protein ISGylation during Human Cytomegalovirus Infection Christopher. J Virol 2019, 93:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fu Y-Z, Su S, Zou H, Guo Y, Wang S-Y, Li S, Luo M-H, Wang Y-Y: Human Cytomegalovirus DNA Polymerase Subunit UL44 Antagonizes Antiviral Immune Responses by Suppressing IRF3- and NF-κB-Mediated Transcription. J Virol 2019, 93:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ahn K, Angulo A, Ghazal P, Peterson PA, Yang Y, Früh K: Human cytomegalovirus inhibits antigen presentation by a sequential multistep process. Proc Natl Acad Sci U S A 1996, 93:10990–10995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lehner PJ, Karttunen JT, Wilkinson GWG, Cresswell P: The human cytomegalovirus US6 glycoprotein inhibits transporter associated with antigen processing-dependent peptide translocation. Proc Natl Acad Sci U S A 1997, 94:6904–6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wiertz EJHJ, Tortorella D, Bogyo M, Yu J, Mothes W, Jonest TR, Rapoport TA, Ploegh HL: Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Science (80- ) 1996, 384:636–638. [DOI] [PubMed] [Google Scholar]
- 81.Wiertz EJHJ, Jones TR, Sun L, Bogyo M, Geuze HJ, Ploegh HL: The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 1996, 84:769–779. [DOI] [PubMed] [Google Scholar]
- 82.Wills MR, Ashiru O, Reeves MB, Okecha G, Trowsdale J, Tomasec P, Wilkinson GWG, Sinclair J, Sissons JGP: Human Cytomegalovirus Encodes an MHC Class I-Like Molecule (UL142) That Functions to Inhibit NK Cell Lysis. J Immunol 2005, 175:7457–7465. [DOI] [PubMed] [Google Scholar]
- 83.Chalupny NJ, Rein-Weston A, Dosch S, Cosman D: Down-regulation of the NKG2D ligand MICA by the human cytomegalovirus glycoprotein UL142. Biochem Biophys Res Commun 2006, 346:175–181. [DOI] [PubMed] [Google Scholar]
- 84.Ashiru O, Bennett NJ, Boyle LH, Thomas M, Trowsdale J, Wills MR: NKG2D Ligand MICA Is Retained in the cis -Golgi Apparatus by Human Cytomegalovirus Protein UL142. J Virol 2009, 83:12345–12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fielding CA, Aicheler R, Stanton RJ, Wang ECY, Han S, Seirafian S, Davies J, McSharry BP, Weekes MP, Antrobus PR, et al. : Two Novel Human Cytomegalovirus NK Cell Evasion Functions Target MICA for Lysosomal Degradation. PLoS Pathog 2014, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dassa L, Seidel E, Oiknine-Djian E, Yamin R, Wolf DG, Le-Trilling VTK, Mandelboim O: The Human Cytomegalovirus Protein UL148A Downregulates the NK Cell-Activating Ligand MICA To Avoid NK Cell Attack. J Virol 2018, 92:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, Horwitz E, Prokocimer Z, Prichard M, Hahn G, et al. : Host Immune System Gene Targeting by a Viral miRNA. Science (80- ) 2007, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nachmani D, Lankry D, Wolf DG, Mandelboim O: The human cytomegalovirus microRNA miR-UL112 acts synergistically with a cellular microRNA to escape immune elimination. Nat Immunol 2010, 11:806–813. [DOI] [PubMed] [Google Scholar]; * Work investigating the interplay of cellular and host miRNAs in regulating MICB expression.
- 89.Tomasec P, Braud VM, Rickards C, Powell MB, McSharry BP, Gadola S, Cerundolo V, Borysiewicz LK, McMichae AJ, Wilkinson GWG: Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science (80- ) 2000, 287:1031–1033. [DOI] [PubMed] [Google Scholar]; ** Discovery that the HCMV glycoprotein UL40 contains a leader peptide mimicking that of MHC-I proteins, promoting surface expression of HLA-E to prevent natural killer cell-mediated lysis while MHC-I proteins themselves are being degraded.
- 90.Cosman D, Fanger N, Borges L, Kubin M, Chin W, Peterson L, Hsu ML: A novel immunoglobulin superfamily receptor for cellular and viral MHC class I molecules. Immunity 1997, 7:273–282. [DOI] [PubMed] [Google Scholar]
- 91.Prod’homme V, Griffin C, Aicheler RJ, Wang ECY, McSharry BP, Rickards CR, Stanton RJ, Borysiewicz LK, López-Botet M, Wilkinson GWG, et al. : The Human Cytomegalovirus MHC Class I Homolog UL18 Inhibits LIR-1 + but Activates LIR-1 – NK Cells. J Immunol 2007, 178:4473–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Murphy JC, Fischle W, Verdin E, Sinclair JH: Control of cytomegalovirus lytic gene expression by histone acetylation. EMBO J 2002, 21:1112–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nitzsche A, Paulus C, Nevels M: Temporal Dynamics of Cytomegalovirus Chromatin Assembly in Productively Infected Human Cells. J Virol 2008, 82:11167–11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Groves IJ, Reeves MB, Sinclair JH: Lytic infection of permissive cells with human cytomegalovirus is regulated by an intrinsic “pre-immediate-early” repression of viral gene expression mediated by histone post-translational modification. J Gen Virol 2009, 90:2364–2374. [DOI] [PubMed] [Google Scholar]
- 95.Albright ER, Kalejta RF: Canonical and Variant Forms of Histone H3 Are Deposited onto the Human Cytomegalovirus Genome during Lytic and Latent Infections. J Virol 2016, 90:10309–10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nevels M, Paulus C, Shenk T: Human cytomegalovirus immediate-early 1 protein facilitates viral replication by antagonizing histone deacetylation. Proc Natl Acad Sci U S A 2004, 101:17234–17239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zalckvar E, Paulus C, Tillo D, Asbach-Nitzsche A, Lubling Y, Winterling C, Strieder N, Mucke K, Goodrum F, Segal E, et al. : Nucleosome maps of the human cytomegalovirus genome reveal a temporal switch in chromatin organization linked to a major IE protein. Proc Natl Acad Sci U S A 2013, 110:13126–13131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wright E, Bain M, Teague L, Murphy J, Sinclair J: Ets-2 repressor factor recruits histone deacetylase to silence human cytomegalovirus immediate-early gene expression in non-permissive cells. J Gen Virol 2005, 86:535–544. [DOI] [PubMed] [Google Scholar]
- 99.Woodhall DL, Groves IJ, Reeves MB, Wilkinson G, Sinclair JH: Human Daxx-mediated repression of human cytomegalovirus gene expression correlates with a repressive chromatin structure around the major immediate early promoter. J Biol Chem 2006, 281:37652–37660. [DOI] [PubMed] [Google Scholar]
- 100.Saffert RT, Kalejta RF: Inactivating a Cellular Intrinsic Immune Defense Mediated by Daxx Is the Mechanism through Which the Human Cytomegalovirus pp71 Protein Stimulates Viral Immediate-Early Gene Expression. J Virol 2006, 80:3863–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Park JJ, Kim YE, Pham HT, Kim ET, Chung YH, Ahn JH: Functional interaction of the human cytomegalovirus IE2 protein with histone deacetylase 2 in infected human fibroblasts. J Gen Virol 2007, 88:3214–3223. [DOI] [PubMed] [Google Scholar]
- 102.Bigley TM, Reitsma JM, Mirza SP, Terhune SS: Human Cytomegalovirus pUL97 Regulates the Viral Major Immediate Early Promoter by Phosphorylation-Mediated Disruption of Histone Deacetylase 1 Binding. J Virol 2013, 87:7393–7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Niller HH, Hennighausen L: Formation of several specific nucleoprotein complexes on the human cytomegalovirus immediate early enhancer. Nucleic Acids Res 1991, 19:3715–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wade EJ, Klucher KM, Spector DH: An AP-1 binding site is the predominant cis-acting regulatory element in the 1.2-kilobase early RNA promoter of human cytomegalovirus. J Virol 1992, 66:2407–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rideg K, Hirka G, Prakash K, Bushar LM, Nothias JY, Weinmann R, Andrews PW, Gönczöl E: DNA-binding proteins that interact with the 19-base pair (CRE-like) element from the HCMV major immediate early promoter in differentiating human embryonal carcinoma cells. Differentiation 1994, 56:119–129. [DOI] [PubMed] [Google Scholar]
- 106.Lang D, Gebert S, Arlt H, Stamminger T: Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB. J Virol 1995, 69:6030–6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schwartz R, Helmich B, Spector DH: CREB and CREB-binding proteins play an important role in the IE2 86-kilodalton protein-mediated transactivation of the human cytomegalovirus 2.2-kilobase RNA promoter. J Virol 1996, 70:6955–6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Prösch S, Heine AK, Volk HD, Krüger DH: CCAAT/Enhancer-binding Proteins α and/3 β Negatively Influence the Capacity of Tumor Necrosis Factor α to Up-regulate the Human Cytomegalovirus IE1/2 Enhancer/Promoter by Nuclear Factor κB during Monocyte Differentiation. J Biol Chem 2001, 276:40712–40720. [DOI] [PubMed] [Google Scholar]
- 109.Kim Y-E, Lee J-H, Kim ET, Shin HJ, Gu SY, Seol HS, Ling PD, Lee CH, Ahn J-H: Human Cytomegalovirus Infection Causes Degradation of Sp100 Proteins That Suppress Viral Gene Expression. J Virol 2011, 85:11928–11937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Martinez FP, Cruz R, Lu F, Plasschaert R, Deng Z, Rivera-Molina YA, Bartolomei MS, Lieberman PM, Tang Q: CTCF Binding to the First Intron of the Major Immediate Early (MIE) Gene of Human Cytomegalovirus (HCMV) Negatively Regulates MIE Gene Expression and HCMV Replication. J Virol 2014, 88:7389–7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Reichel A, Stilp A-C, Scherer M, Reuter N, Lukassen S, Kasmapour B, Schreiner S, Cicin-Sain L, Winterpacht A, Stamminger T: Chromatin-Remodeling Factor SPOC1 Acts as a Cellular Restriction Factor against Human Cytomegalovirus by Repressing the Major Immediate Early Promoter. J Virol 2018, 92:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Grötzinger T, Sternsdorf T, Jensen K, Will H: Interferon-modulated expression of genes encoding the nuclear-dot-associated proteins Sp100 and promyelocytic leukemia protein (PML). Eur J Biochem 1996, 238:554–560. [DOI] [PubMed] [Google Scholar]
- 113.Gongora R, Stephan RP, Zhang Z, Cooper MD: An essential role for Daxx in the inhibition of B lymphopoiesis by type I interferons. Immunity 2001, 14:727–737. [DOI] [PubMed] [Google Scholar]
- 114.Lukashchuk V, McFarlane S, Everett RD, Preston CM: Human Cytomegalovirus Protein pp71 Displaces the Chromatin-Associated Factor ATRX from Nuclear Domain 10 at Early Stages of Infection. J Virol 2008, 82:12543–12554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lee H-R, Kim D-J, Lee J-M, Choi CY, Ahn B-Y, Hayward GS, Ahn J-H: Ability of the Human Cytomegalovirus IE1 Protein To Modulate Sumoylation of PML Correlates with Its Functional Activities in Transcriptional Regulation and Infectivity in Cultured Fibroblast Cells. J Virol 2004, 78:6527–6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Korioth F, Maul GG, Plachter B, Stamminger T, Frey J: The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp Cell Res 1996, 229:155–158. [DOI] [PubMed] [Google Scholar]
- 117.Ahn J-H, Brignole EJ, Hayward GS: Disruption of PML Subnuclear Domains by the Acidic IE1 Protein of Human Cytomegalovirus Is Mediated through Interaction with PML and May Modulate a RING Finger-Dependent Cryptic Transactivator Function of PML. Mol Cell Biol 1998, 18:4899–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tavalai N, Adler M, Scherer M, Riedl Y, Stamminger T: Evidence for a Dual Antiviral Role of the Major Nuclear Domain 10 Component Sp100 during the Immediate-Early and Late Phases of the Human Cytomegalovirus Replication Cycle. J Virol 2011, 85:9447–9458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Scherer M, Klingl S, Sevvana M, Otto V, Schilling EM, Stump JD, Müller R, Reuter N, Sticht H, Muller YA, et al. : Crystal Structure of Cytomegalovirus IE1 Protein Reveals Targeting of TRIM Family Member PML via Coiled-Coil Interactions. PLoS Pathog 2014, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schilling E-M, Scherer M, Reuter N, Schweininger J, Muller YA, Stamminger T: The Human Cytomegalovirus IE1 Protein Antagonizes PML Nuclear Body-Mediated Intrinsic Immunity via the Inhibition of PML De Novo SUMOylation. J Virol 2017, 91:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hofmann H, Flöss S, Stamminger T: Covalent Modification of the Transactivator Protein IE2-p86 of Human Cytomegalovirus by Conjugation to the Ubiquitin-Homologous Proteins SUMO-1 and hSMT3b. J Virol 2000, 74:2510–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ahn J-H, Xu Y, Jang W-J, Matunis MJ, Hayward GS: Evaluation of Interactions of Human Cytomegalovirus Immediate-Early IE2 Regulatory Protein with Small Ubiquitin-Like Modifiers and Their Conjugation Enzyme Ubc9. J Virol 2001, 75:3859–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kim ET, Kim Y-E, Huh YH, Ahn J-H: Role of Noncovalent SUMO Binding by the Human Cytomegalovirus IE2 Transactivator in Lytic Growth. J Virol 2010, 84:8111–8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Deutschmann J, Schneider A, Gruska I, Vetter B, Thomas D, Kießling M, Wittmann S, Herrmann A, Schindler M, Milbradt J, et al. : A viral kinase counteracts in vivo restriction of murine cytomegalovirus by SAMHD1. Nat Microbiol 2019, 4:2273–2284. [DOI] [PubMed] [Google Scholar]
- 125.Businger R, Deutschmann J, Gruska I, Milbradt J, Wiebusch L, Gramberg T, Schindler M: Human cytomegalovirus overcomes SAMHD1 restriction in macrophages via pUL97. Nat Microbiol 2019, 4:2260–2272. [DOI] [PubMed] [Google Scholar]
- 126.Zhang K, Lv DW, Li R: Conserved Herpesvirus Protein Kinases Target SAMHD1 to Facilitate Virus Replication. Cell Rep 2019, 28:449–459.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kim D, Lee Y suk, Jung SJ, Yeo J, Seo JJ, Lee YY, Lim J, Chang H, Song J, Yang J, et al. : Viral hijacking of the TENT4-ZCCHC14 complex protects viral RNAs via mixed tailing. Nat Struct Mol Biol 2020, 27:581–588. [DOI] [PubMed] [Google Scholar]
- 128.Fortunato EA, Sanchez V, Yen JY, Spector DH: Infection of Cells with Human Cytomegalovirus during S Phase Results in a Blockade to Immediate-Early Gene Expression That Can Be Overcome by Inhibition of the Proteasome. J Virol 2002, 76:5369–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bogdanow B, Weisbach H, Von Einem J, Straschewski S, Voigt S, Winkler M, Hagemeier C, Wiebusch L: Human cytomegalovirus tegument protein pp150 acts as a cyclin A2-CDK-dependent sensor of the host cell cycle and differentiation state. Proc Natl Acad Sci U S A 2013, 110:17510–17515. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Discovery that HCMV encodes a cell cycle sensor, pp150, which blocks immediate-early gene expression until the host cell returns to G1.
- 130.Wiebusch L, Bach M, Uecker R, Hagemeier C: Human cytomegalovirus inactivates the G0/G1-APC/C ubiquitin ligase by Cdh1 dissociation. Cell Cycle 2005, 4:1435–1439. [DOI] [PubMed] [Google Scholar]
- 131.Tran K, Kamil JP, Coen DM, Spector DH: Inactivation and Disassembly of the Anaphase-Promoting Complex during Human Cytomegalovirus Infection Is Associated with Degradation of the APC5 and APC4 Subunits and Does Not Require UL97-Mediated Phosphorylation of Cdh1. J Virol 2010, 84:10832–10843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fehr AR, Gualberto NC, Savaryn JP, Terhune SS, Yu D: Proteasome-dependent disruption of the E3 ubiquitin ligase anaphase-promoting complex by HCMV protein pUL21a. PLoS Pathog 2012, 8:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Clark E, Spector DH: Studies on the Contribution of Human Cytomegalovirus UL21a and UL97 to Viral Growth and Inactivation of the Anaphase-Promoting Complex/Cyclosome (APC/C) E3 Ubiquitin Ligase Reveal a Unique Cellular Mechanism for Downmodulation of the APC/C Subunits APC1,. J Virol 2015, 89:6928–6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Speir E, Huang ES, Modali R, Leon MB, Shawl F, Finkel T, Epstein SE: Potential Role of Human Cytomegalovirus and p53 Interaction in Coronary Restenosis. Science (80- ) 1996, 265:78–81. [DOI] [PubMed] [Google Scholar]
- 135.Tsai HL, Kou GH, Chen SC, Wu CW, Lin YS: Human cytomegalovirus immediate-early protein IE2 tethers a transcriptional repression domain to p53. J Biol Chem 1996, 271:3534–3540. [PubMed] [Google Scholar]
- 136.Hwang E-S, Zhang Z, Cai H, Huang DY, Huong S-M, Cha C-Y, Huang E-S: Human Cytomegalovirus IE1-72 Protein Interacts with p53 and Inhibits p53-Dependent Transactivation by a Mechanism Different from That of IE2-86 Protein. J Virol 2009, 83:12388–12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Savaryn JP, Reitsma JM, Bigley TM, Halligan BD, Qian Z, Yu D, Terhune SS: Human Cytomegalovirus pUL29/28 and pUL38 Repression of p53-Regulated p21CIP1 and Caspase 1 Promoters during Infection. J Virol 2013, 87:2463–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kalejta RF, Bechtel JT, Shenk T: Human Cytomegalovirus pp71 Stimulates Cell Cycle Progression by Inducing the Proteasome-Dependent Degradation of the Retinoblastoma Family of Tumor Suppressors. Mol Cell Biol 2003, 23:1885–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kalejta RF, Shenk T: Proteasome-dependent, ubiquitin-independent degradation of the Rb family of tumor suppressors by the human cytomegalovirus pp71 protein. Proc Natl Acad Sci U S A 2003, 100:3263–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hume AJ, Finkel JS, Kamil JP, Coen DM, Culbertson MR, Kalejta RF: Phosphorylation of retinoblastoma protein by viral protein with cyclin-dependent kinase function. Science (80- ) 2008, 320:797–799. [DOI] [PubMed] [Google Scholar]
- 141.Prichard MN, Sztul E, Daily SL, Perry AL, Frederick SL, Gill RB, Hartline CB, Streblow DN, Varnum SM, Smith RD, et al. : Human Cytomegalovirus UL97 Kinase Activity Is Required for the Hyperphosphorylation of Retinoblastoma Protein and Inhibits the Formation of Nuclear Aggresomes. J Virol 2008, 82:5054–5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Iwahori S, Hakki M, Chou S, Kalejta RF: Molecular determinants for the inactivation of the retinoblastoma tumor suppressor by the viral cyclin-dependent kinase UL97. J Biol Chem 2015, 290:19666–19680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Strang BL, Boulant S, Coen DM: Nucleolin Associates with the Human Cytomegalovirus DNA Polymerase Accessory Subunit UL44 and Is Necessary for Efficient Viral Replication. J Virol 2010, 84:1771–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Strang BL, Boulant S, Kirchhausen T, Coen DM: Host cell nucleolin is required to maintain the architecture of human cytomegalovirus replication compartments. MBio 2012, 3:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Munger J, Bajad SU, Coller HA, Shenk T, Rabinowitz JD: Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathog 2006, 2:1165–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chambers JW, Maguire TG, Alwine JC: Glutamine Metabolism Is Essential for Human Cytomegalovirus Infection. J Virol 2010, 84:1867–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Demonstrates that HCMV infected cells possess a Warburg-like metabolic phenotype in that glutamine uptake and utilization for ATP production are increased during infection.
- 147.Koyuncu E, Purdy JG, Rabinowitz JD, Shenk T: Saturated Very Long Chain Fatty Acids Are Required for the Production of Infectious Human Cytomegalovirus Progeny. PLoS Pathog 2013, 9:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kaarbø M, Ager-Wick E, Osenbroch PØ, Kilander A, Skinnes R, Müller F, Eide L: Human cytomegalovirus infection increases mitochondrial biogenesis. Mitochondrion 2011, 11:935–945. [DOI] [PubMed] [Google Scholar]
- 149.Combs JA, Norton EB, Saifudeen ZR, Bentrup KHZ, Katakam PV, Morris CA, Myers L, Kaur A, Sullivan DE, Zwezdaryk KJ: Human Cytomegalovirus Alters Host Cell Mitochondrial Function during Acute Infection. J Virol 2019, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Munger J, Bennett BD, Parikh A, Feng X-J, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD: Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol 2008, 26:1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Used liquid chromatography-tandem mass spectrometry based metabolite profiling to monitor cellular metabolic flux throughout HCMV infection and demonstrated upregulated glycolysis, TCA cycle, and fatty acid biosynthesis during infection.
- 151.Spencer CM, Schafer XL, Moorman NJ, Munger J: Human Cytomegalovirus Induces the Activity and Expression of Acetyl-Coenzyme A Carboxylase, a Fatty Acid Biosynthetic Enzyme Whose Inhibition Attenuates Viral Replication. J Virol 2011, 85:5814–5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.McArdle J, Schafer XL, Munger J: Inhibition of Calmodulin-Dependent Kinase Kinase Blocks Human Cytomegalovirus-Induced Glycolytic Activation and Severely Attenuates Production of Viral Progeny. J Virol 2011, 85:705–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.McArdle J, Moorman NJ, Munger J: HCMV targets the metabolic stress response through activation of AMPK whose activity is important for viral replication. PLoS Pathog 2012, 8:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Terry LJ, Vastag L, Rabinowitz JD, Shenk T: Human kinome profiling identifies a requirement for AMP-activated protein kinase during human cytomegalovirus infection. Proc Natl Acad Sci U S A 2012, 109:3071–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Seo JY, Cresswell P: Viperin Regulates Cellular Lipid Metabolism during Human Cytomegalovirus Infection. PLoS Pathog 2013, 9:1003497. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Discovered that viperin is targeted to the mitochondria by viral protein pUL37x1 during infection, where it interacts with the mitochondrial trifunctional protein to decrease fatty acid oxidation, resulting in a cascade of signaling events that ultimately increases glucose import and lipogenesis.
- 156.Seo J, Yaneva R, Hinson ER, Cresswell P: Human cytomegalovirus directly induces the antiviral protein viperin to enhance infectivity. Science (80- ) 2011, 332:1093–1097. [DOI] [PubMed] [Google Scholar]
- 157.Yu Y, Maguire TG, Alwine JC: ChREBP, a glucose-responsive transcriptional factor, enhances glucose metabolism to support biosynthesis in human cytomegalovirus-infected cells. Proc Natl Acad Sci U S A 2014, 111:1951–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Revealed that the high-capacity glucose transporter GLUT4 is induced during infection and replaces the lower-capacity glucose transporter, GLUT1, at the plasma membrane of fibroblasts, resulting in increased glucose uptake.
- 158.Landini MP: Early enhanced glucose uptake in human cytomegalovirus-infected cells. J Gen Virol 1984, 65:1229–1232. [DOI] [PubMed] [Google Scholar]
- 159.Yu Y, Maguire TG, Alwine JC: Human Cytomegalovirus Activates Glucose Transporter 4 Expression To Increase Glucose Uptake during Infection. J Virol 2011, 85:1573–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Karniely S, Weekes MP, Antrobus R, Rorbach J, Van Haute L, Umrania Y, Smith DL, Stanton RJ, Minczuk M, Lehner PJ, et al. : Human cytomegalovirus infection upregulates the mitochondrial transcription and translation machineries. MBio 2016, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Betsinger CN, Jankowski CSR, Hofstadter WA, Federspiel JD, Otter CJ, Jean Beltran PM, Cristea IM: The human cytomegalovirus protein pUL13 targets mitochondrial cristae architecture to increase cellular respiration during infection. Proc Natl Acad Sci U S A 2021, 118:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Reeves MB, Davies AA, McSharry BP, Wilkinson GW, Sinclair JH: Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science (80- ) 2007, 316:1345–1348. [DOI] [PubMed] [Google Scholar]
- 163.Purdy JG, Shenk T, Rabinowitz JD: Fatty acid elongase 7 catalyzes lipidome remodeling essential for human cytomegalovirus replication. Cell Rep 2015, 10:1375–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Moorman NJ, Cristea IM, Terhune SS, Rout MP, Chait BT, Shenk T: Human Cytomegalovirus Protein UL38 Inhibits Host Cell Stress Responses by Antagonizing the Tuberous Sclerosis Protein Complex. Cell Host Microbe 2008, 3:253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Magalhães AC, Ferreira AR, Gomes S, Vieira M, Gouveia A, Valença I, Islinger M, Nascimento R, Schrader M, Kagan JC, et al. : Peroxisomes are platforms for cytomegalovirus’ evasion from the cellular immune response. Sci Rep 2016, 6:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Discovery that pUL37x1 localizes to peroxisomes to inhibit MAVS-dependent antiviral signaling early in infection.
- 166.Jean Beltran PM, Cook KC, Hashimoto Y, Galitzine C, Murray LA, Vitek O, Cristea IM: Infection-Induced Peroxisome Biogenesis Is a Metabolic Strategy for Herpesvirus Replication. Cell Host Microbe 2018, 24:526–541.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Discovery that HCMV infection drives peroxisome structural remodeling to induce plasmalogen synthesis for secondary envelopment during virion assembly.
- 167.Federspiel Joel D.; Cook Katelyn C.; Kennedy Michelle A.; Venkatesh Samvida S.; Otter Clayton J.; Hofstadter William A.; Jean Beltran Pierre M.; Cristea IM: Mitochondria and peroxisome remodeling across cytomegalovirus infection time viewed through the lens of Inter-ViSTA. Cell Reports 2020, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gudleski-O’Regan N, Greco TM, Cristea IM, Shenk T: Increased expression of LDL receptor-related protein 1 during human cytomegalovirus infection reduces virion cholesterol and infectivity. Cell Host Microbe 2012, 12:86–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhou YF, Guetta E, Yu ZX, Finkel T, Epstein SE: Human cytomegalovirus increases modified low density lipoprotein uptake and scavenger receptor mRNA expression in vascular smooth muscle cells. J Clin Invest 1996, 98:2129–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sanchez V, Dong JJ: Alteration of lipid metabolism in cells infected with human cytomegalovirus. Virology 2010, 404:71–77. [DOI] [PubMed] [Google Scholar]
- 171.Sanchez V, Greis KD, Sztul E, Britt WJ: Accumulation of Virion Tegument and Envelope Proteins in a Stable Cytoplasmic Compartment during Human Cytomegalovirus Replication: Characterization of a Potential Site of Virus Assembly. J Virol 2000, 74:975–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wilkie AR, Sharma M, Pesola JM, Ericsson M, Fernandez R, Coen DM: A Role for Myosin Va in Human Cytomegalovirus Nuclear Egress. J Virol 2018, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wilkie AR, Lawler JL, Coen DM: A role for nuclear F-actin induction in human cytomegalovirus nuclear egress. MBio 2016, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hamirally S, Kamil JP, Ndassa-Colday YM, Lin AJ, Jahng WJ, Baek MC, Noton S, Silva LA, Simpson-Holley M, Knipe DM, et al. : Viral mimicry of Cdc2/cyclin-dependent kinase 1 mediates disruption of nuclear lamina during human cytomegalovirus nuclear egress. PLoS Pathog 2009, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Camozzi D, Pignatelli S, Valvo C, Lattanzi G, Capanni C, Dal Monte P, Landini MP: Remodelling of the nuclear lamina during human cytomegalovirus infection: Role of the viral proteins pUL50 and pUL53. J Gen Virol 2008, 89:731–740. [DOI] [PubMed] [Google Scholar]
- 176.Marschall M, Marzi A, Aus Dem Siepen P, Jochmann R, Kalmer M, Auerochs S, Lischka P, Leis M, Stamminger T: Cellular p32 recruits cytomegalovirus kinase pUL97 to redistribute the nuclear lamina. J Biol Chem 2005, 280:33357–33367. [DOI] [PubMed] [Google Scholar]
- 177.Milbradt J, Kraut A, Hutterer C, Sonntag E, Schmeiser C, Ferro M, Wagner S, Lenac T, Claus C, Pinkert S, et al. : Proteomic analysis of the multimeric nuclear egress complex of human cytomegalovirus. Mol Cell Proteomics 2014, 13:2132–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yang B, Liu X-J, Yao Y, Jiang X, Wang X-Z, Yang H, Sun J-Y, Miao Y, Wang W, Huang Z-L, et al. : WDR5 Facilitates Human Cytomegalovirus Replication by Promoting Capsid Nuclear Egress. J Virol 2018, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sonntag E, Milbradt J, Svrlanska A, Strojan H, Häge S, Kraut A, Hesse AM, Amin B, Sonnewald U, Couté Y, et al. : Protein kinases responsible for the phosphorylation of the nuclear egress core complex of human cytomegalovirus. J Gen Virol 2017, 98:2569–2581. [DOI] [PubMed] [Google Scholar]
- 180.Milbradt J, Auerochs S, Marschall M: Cytomegaloviral proteins pUL50 and pUL53 are associated with the nuclear lamina and interact with cellular protein kinase C. J Gen Virol 2007, 88:2642–2650. [DOI] [PubMed] [Google Scholar]
- 181.Murray LA, Sheng X, Cristea IM: Orchestration of protein acetylation as a toggle for cellular defense and virus replication. Nat Commun 2018, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** First acetylome investigation during HCMV infection and the finding that lamin B1 acetylation protects the integrity of the nuclear periphery.
- 182.Turner DL, Korneev DV., Purdy JG, de Marco A, Mathias RA: The host exosome pathway underpins biogenesis of the human cytomegalovirus virion. Elife 2020, 9:1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Proteomic investigation of virions discovered that the host exosome machinery is utilized for envelopment.
- 183.Tandon R, AuCoin DP, Mocarski ES: Human Cytomegalovirus Exploits ESCRT Machinery in the Process of Virion Maturation. J Virol 2009, 83:10797–10807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Fish KN, Soderberg-Naucler C, Nelson JA: Steady-State Plasma Membrane Expression of Human Cytomegalovirus gB Is Determined by the Phosphorylation State of Ser 900 . J Virol 1998, 72:6657–6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Crump CM, Hung C-H, Thomas L, Wan L, Thomas G: Role of PACS-1 in Trafficking of Human Cytomegalovirus Glycoprotein B and Virus Production. J Virol 2003, 77:11105–11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Maschkowitz G, Gärtner S, Hofmann-Winkler H, Fickenscher H, Winkler M: Interaction of Human Cytomegalovirus Tegument Proteins ppUL35 and ppUL35A with Sorting Nexin 5 Regulates Glycoprotein B (gpUL55) Localization. J Virol 2018, 92:e00013–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Krzyzaniak MA, Mach M, Britt WJ: HCMV-encoded glycoprotein M (UL100) interacts with rab11 effector protein FIP4. Traffic 2009, 10:1439–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Indran SV, Britt WJ: A Role for the Small GTPase Rab6 in Assembly of Human Cytomegalovirus. J Virol 2011, 85:5213–5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Indran SV, Ballestas ME, Britt WJ: Bicaudal D1-Dependent Trafficking of Human Cytomegalovirus Tegument Protein pp150 in Virus-Infected Cells. J Virol 2010, 84:3162–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Moorman NJ, Sharon-Friling R, Shenk T, Cristea IM: A targeted spatial-temporal proteomics approach implicates multiple cellular trafficking pathways in human cytomegalovirus virion maturation. Mol Cell Proteomics 2010, 9:851–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Taisne C, Lussignol M, Hernandez E, Moris A, Mouna L, Esclatine A: Human cytomegalovirus hijacks the autophagic machinery and LC3 homologs in order to optimize cytoplasmic envelopment of mature infectious particles. Sci Rep 2019, 9:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Cepeda V, Esteban M, Fraile-Ramos A: Human cytomegalovirus final envelopment on membranes containing both trans-Golgi network and endosomal markers. Cell Microbiol 2010, 12:386–404. [DOI] [PubMed] [Google Scholar]
- 193.Das S, Pellett PE: Spatial Relationships between Markers for Secretory and Endosomal Machinery in Human Cytomegalovirus-Infected Cells versus Those in Uninfected Cells. J Virol 2011, 85:5864–5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Homman-Loudiyi M, Hultenby K, Britt W, Söderberg-Nauclér C: Envelopment of Human Cytomegalovirus Occurs by Budding into Golgi-Derived Vacuole Compartments Positive for gB, Rab 3, Trans-Golgi Network 46, and Mannosidase II. J Virol 2003, 77:3191–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Buchkovich NJ, Maguire TG, Paton AW, Paton JC, Alwine JC: The Endoplasmic Reticulum Chaperone BiP/GRP78 Is Important in the Structure and Function of the Human Cytomegalovirus Assembly Compartment. J Virol 2009, 83:11421–11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Buchkovich NJ, Maguire TG, Yu Y, Paton AW, Paton JC, Alwine JC: Human Cytomegalovirus Specifically Controls the Levels of the Endoplasmic Reticulum Chaperone BiP/GRP78, Which Is Required for Virion Assembly. J Virol 2008, 82:31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Cruz L, Streck NT, Ferguson K, Desai T, Desai DH, Amin SG, Buchkovich NJ: Potent Inhibition of Human Cytomegalovirus by Modulation of Cellular SNARE Syntaxin 5. J Virol 2017, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Liu STH, Sharon-Friling R, Ivanova P, Milne SB, Myers DS, Rabinowitz JD, Brown HA, Shenk T: Synaptic vesicle-like lipidome of human cytomegalovirus virions reveals a role for SNARE machinery in virion egress. Proc Natl Acad Sci U S A 2011,108:12869–12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Cepeda V, Fraile-Ramos A: A role for the SNARE protein syntaxin 3 in human cytomegalovirus morphogenesis. Cell Microbiol 2011, 13:846–858. [DOI] [PubMed] [Google Scholar]
- 200.Goulidaki N, Alarifi S, Alkahtani SH, Al-Qahtani A, Spandidos DA, Stournaras C, Sourvinos G: RhoB is a component of the human cytomegalovirus assembly complex and is required for efficient viral production. Cell Cycle 2015, 14:2748–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Fraile-Ramos A, Cepeda V, Elstak E, van der Sluijs P: Rab27a is required for human cytomegalovirus assembly. PLoS One 2010, 5:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Pavelin J, McCormick D, Chiweshe S, Ramachandran S, Lin YT, Grey F: Cellular v-ATPase is required for virion assembly compartment formation in human cytomegalovirus infection. Open Biol 2017, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Hook LM, Grey F, Grabski R, Tirabassi R, Doyle T, Hancock M, Landais I, Jeng S, McWeeney S, Britt W, et al. : Cytomegalovirus miRNAs target secretory pathway genes to facilitate formation of the virion assembly compartment and reduce cytokine secretion. Cell Host Microbe 2014, 15:363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Rebmann GM, Grabski R, Sanchez V, Britt WJ: Phosphorylation of Golgi peripheral membrane protein Grasp65 is an integral step in the formation of the human cytomegalovirus cytoplasmic assembly compartment. MBio 2016, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Procter DJ, Furey C, Garza-Gongora AG, Kosak ST, Walsh D: Cytoplasmic control of intranuclear polarity by human cytomegalovirus. Nature 2020, 587:109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Procter DJ, Banerjee A, Nukui M, Kruse K, Gaponenko V, Murphy EA, Komarova Y, Walsh D: The HCMV Assembly Compartment Is a Dynamic Golgi-Derived MTOC that Controls Nuclear Rotation and Virus Spread. Dev Cell 2018, 45:83-100.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Show that microtubule end binding protein EB3 is recruited to the viral assembly complex during infection, where it nucleates Golgi-derived microtubules that are rapidly acetylated, driving nuclear rotation and assembly complex formation.
- 207.Nobre L, Nightingale K, Ravenhill BJ, Antrobus R, Soday L, Nichols J, Davies J, Seirafian S, Wang ECY, Davison AJ, et al. : Human cytomegalovirus interactome analysis identifies degradation hubs, domain associations and viral protein functions. Elite 2019, 8:1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Hofmann H, Sindre H, Stamminger T: Functional Interaction between the pp71 Protein of Human Cytomegalovirus and the PML-Interacting Protein Human Daxx. J Virol 2002, 76:5769–5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Weekes MP, Tomasec P, Huttlin EL, Fielding CA, Nusinow D, Stanton RJ, Wang ECY, Aicheler R, Murrell I, Wilkinson GWG, et al. : Quantitative temporal viromics: An approach to investigate host-pathogen interaction. Cell 2014, 157:1460–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]; * In-depth quantitative characterization of global temporal proteome changes during HCMV infection.
- 210.Nightingale K, Lin KM, Ravenhill BJ, Davies C, Nobre L, Fielding CA, Ruckova E, Fletcher-Etherington A, Soday L, Nichols H, et al. : High-Definition Analysis of Host Protein Stability during Human Cytomegalovirus Infection Reveals Antiviral Factors and Viral Evasion Mechanisms. Cell Host Microbe 2018, 24:447–460.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Jean Beltran PMM, Mathias RAA, Cristea IMM: A Portrait of the Human Organelle Proteome In Space and Time during Cytomegalovirus Infection. Cell Syst 2016, 3:361–373.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** First application of spatial proteomics and machine learning to characterize temporal organelle organization during HCMV infection.
- 212.Christopher JA, Stadler C, Martin CE, Morgenstern M, Pan Y, Betsinger CN, Rattray DG, Mahdessian D, Gingras A-C, Warscheid B, et al. : Subcellular proteomics. Nat Rev Methods Prim 2021. 11 2021, 1:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Oberstein A, Perlman DH, Shenk T, Terry LJ: Human cytomegalovirus pUL97 kinase induces global changes in the infected cell phosphoproteome. Proteomics 2015, 15:2006–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Griffante G, Gugliesi F, Pasquero S, Dell’Oste V, Biolatti M, Salinger AJ, Mondal S, Thompson PR, Weerapana E, Lebbink RJ, et al. : Human cytomegalovirus-induced host protein citrullination is crucial for viral replication. Nat Commun 2021, 12:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Tirosh O, Cohen Y, Shitrit A, Shani O, Le-Trilling VTK, Trilling M, Friedlander G, Tanenbaum M, Stern-Ginossar N: The Transcription and Translation Landscapes during Human Cytomegalovirus Infection Reveal Novel Host-Pathogen Interactions. PLoS Pathog 2015, 11:1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Kennedy MA, Hofstadter WA, Cristea IM: TRANSPIRE: A Computational Pipeline to Elucidate Intracellular Protein Movements from Spatial Proteomics Data Sets. J Am Soc Mass Spectrom 2020, 31:1422–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Hein MY, Weissman JS: Functional single-cell genomics of human cytomegalovirus infection. Nat Biotechnol 2021, doi: 10.1038/s41587-021-01059-3. [DOI] [PubMed] [Google Scholar]


