Abstract
We analyzed the performance characteristics of the qualitative AMPLICOR CMV Test (Roche Molecular Systems, Pleasanton, Calif.) and quantitative COBAS AMPLICOR CMV MONITOR Test (Roche Molecular Systems) assays and compared the performance of the AMPLICOR quantitative assay with an in-house-developed cytomegalovirus (CMV) DNA PCR assay. The quantitative AMPLICOR assay was found to be more sensitive than the qualitative AMPLICOR assay. The quantitative AMPLICOR assay has a lower limit of sensitivity of 400 CMV DNA copies/ml of plasma and is linear to 50,000 CMV DNA copies/ml of plasma. Compared to the in-house PCR assay, the AMPLICOR quantitative assay gave lower viral load values at all concentrations tested, but the difference between the two assays was not consistent across the entire dynamic range of the AMPLICOR quantitative assay. At the lower end of the assay, the viral load values obtained with the in-house PCR assay were three- to fivefold (0.5 to 0.7 log units) higher than those measured with the AMPLICOR assay. At higher input concentrations, the differences between the two assays approached 10-fold. This direct comparison of the in-house assay and the quantitative AMPLICOR assay provides the ability to compare previously published in-house data with an assay widely available for future research and clinical monitoring of patients with CMV infections.
There is increasing evidence that the risk of developing cytomegalovirus (CMV) disease in AIDS patients is directly related to the quantity of CMV DNA in plasma (8, 9). A recent study has shown that, in AIDS patients with advanced disease, the CMV DNA load is an independent marker of CMV disease and survival and is more predictive of these events than human immunodeficiency virus type 1 (HIV-1) viral load (8). The detection and quantification of CMV DNA are often done using PCR-based assays, and the majority of the assays are developed in-house. For in-house-developed assays, the individual laboratory determines the performance, verification, and validation of the assays. As a result, these assays may vary with regard to specimen type, nucleic acid extraction method, target DNA, or detection method. Moreover, the limit of sensitivity and the linear range vary, making it difficult to compare results from different studies. A standardized assay for the detection and quantification of CMV DNA is needed, as this will allow clinically significant cutoffs to be determined and broadly applied in clinical practice. To address this issue, we analyzed the lower limit of detection, inter- and intra-assay variability, linear range, and upper limit of quantification of commercial qualitative (AMPLICOR CMV Test; Roche Molecular Systems, Pleasanton, Calif.) and quantitative (COBAS AMPLICOR CMV MONITOR Test; Roche Molecular Systems) CMV DNA PCR assays. In addition, the correspondence between the commercial quantitative PCR assay and the in-house assay used in the studies mentioned above (8, 9) was analyzed.
MATERIALS AND METHODS
CMV DNA quantitation panel: input CMV DNA concentration.
The stock virus used as the quantitation panel was a cell-free purified preparation of CMV strain AD169 from culture supernatants. Purification of viral particles was done by high-speed centrifugation (23,000 × g) to remove cellular debris. The concentration of the stock was determined by counting viral particles using electron microscopy. A twofold dilution series was created with a single batch of plasma that was DNA-free and seronegative for CMV. The concentration of the quantitation standard used in these studies ranged from 8 to 1,000,000 CMV DNA copies/ml. An aliquot of the plasma used for the dilutions was included as a negative control. The concentration of the quantitation standard added to the amplification reaction mixture is referred to as the input CMV DNA concentration.
Assay methods.
The AMPLICOR CMV Test (Roche Molecular Systems), a qualitative PCR assay, amplifies a 365-bp fragment of the CMV polymerase gene (4). The assay includes an internal control, which uses the same primer binding sites as the CMV target sequence, and the amplified product is the same size as the CMV-specific target sequence. The internal-control DNA is coamplified with the CMV target DNA but does not cross-react with the CMV-specific detection probe and is detected separately with a specific probe. The amplified products are detected by a colorimetric method using probes complementary to a sequence internal to the primers. The internal control allows monitoring for the presence of inhibitors of amplification. Results are expressed as negative or positive for CMV DNA.
The COBAS AMPLICOR CMV MONITOR Test (Roche Molecular Systems), a quantitative PCR assay, utilizes the same primer pair as the AMPLICOR CMV Test assay. CMV viral DNA is quantified by coamplifying a region of the CMV DNA polymerase gene in the presence of a known amount of quantitation standard. The quantitation standard is the same as the internal standard described for the qualitative assay. The amplified target and quantitation standard are detected separately with specific probes. The amplification and detection reactions are performed on the semiautomated COBAS system. Results are expressed as CMV DNA copies of CMV DNA per milliliter of plasma. According to the manufacturer the quantitative assay has a lower limit of sensitivity at 400 CMV DNA copies/ml.
The in-house quantitative CMV DNA PCR assays were performed as previously described (7, 12). The primer pair was constructed from the EcoRI fragment D region of CMV strain AD169. The quantitation standard was the same as the target except for a 20-nucleotide insertion in the middle of the amplicon. The detection method utilized a 32P-labeled probe. Results are expressed as CMV DNA copies per milliter of plasma. The assay limit of detection is 500 CMV DNA copies/ml, and the limit of quantification is 2,500 CMV DNA copies/ml of plasma.
Testing laboratories and study design.
The PCR testing was performed in four laboratories. All of the laboratories were experienced with PCR testing. Laboratories 1 and 2 performed the Roche qualitative and quantitative DNA PCR assays using kit lots 1 through 6. Laboratory 3 performed the Roche qualitative and quantitative DNA PCR assays using kit lots 7 through 10, and laboratory 4 performed the in-house quantitative CMV DNA PCR assay.
Initially small numbers of samples were tested in the qualitative and quantitative assays with the laboratories unblinded to the CMV DNA concentrations of the specimens; for the remainder of the study the laboratories were blinded to the sample concentration. Table 1 outlines the concentrations tested for the blinded and unblinded testing that was performed by laboratories 1 to 3.
TABLE 1.
Blinded and unblinded testing performed by laboratories 1 to 3 using the AMPLICOR qualitative and quantitative CMV DNA PCR assays
| Quantitation panel input concn (copies of CMV DNA/ml) | No. of replicates/dilution/laboratory by:
|
|||
|---|---|---|---|---|
| Qualitative PCR
|
Quantitative PCR
|
|||
| Unblinded | Blinded | Unblinded | Blinded | |
| 0 | 4 | 12 | 2 | 6 |
| 8 | 6 | 6 | ||
| 16 | 2 | 6 | 2 | 6 |
| 31 | 6 | 6 | ||
| 63 | 2 | 6 | 2 | 6 |
| 125 | 6 | |||
| 250 | 2 | 6 | 2 | 6 |
| 500 | ||||
| 1,000 | 2 | 6 | 2 | 6 |
| 2,000 | ||||
| 4,000 | 2 | 6 | 2 | 6 |
| 8,000 | 6 | |||
| 40,000 | 6 | 1 | 4 | |
| 200,000 | 4 | |||
| 1,000,000 | 6 | 1 | 4 | |
Statistical analysis.
Results of the quantitative assays were transformed to a log10 scale for analysis. The linear range of the AMPLICOR CMV MONITOR Test was evaluated with a “lowess line” to illustrate concordance with a nominal 45° line. The standard deviations presented are based on crude within-laboratory variances uncorrected for lot and unequal sample size. Variance components for lot, laboratory, and error variances were evaluated in the two laboratories that used lots 1 to 6 (laboratories 1 and 2). Estimations were performed in PROC VARCOMP (SAS) using all four algorithms offered. All produced nearly identical results and REML estimates are presented.
RESULTS
Performance of the AMPLICOR qualitative and quantitative CMV DNA PCR assays.
Results of the AMPLICOR qualitative and quantitative CMV DNA PCR assays are shown in Table 2. The viral load obtained with the AMPLICOR quantitative test is included and is expressed as 10 raised to the mean of the log10 of the CMV DNA copies per milliliter from the three laboratories. When the input concentration was 63 CMV DNA copies/ml, the sensitivity of the qualitative assay was 50% (12 of 24 replicates positive) and the sensitivity was 100% (24 of 24 replicates positive) at an input concentration of 1,000 CMV DNA copies/ml. The specificity of the AMPLICOR qualitative assay was 100% (i.e., none of the 48 negative specimens tested had CMV DNA detected by this assay).
TABLE 2.
Combined blinded- and unblinded-testing results from laboratories 1 to 3 using the AMPLICOR qualitative and quantitative CMV DNA PCR assays
| Quantitation panel input concn (CMV DNA copies/ml) | Quantitative AMPLICOR result (CMV DNA copies/ml)a | Qualitative PCR results
|
No. of replicates at indicated level of CMV DNA copies/ml by quantitative PCR
|
|||
|---|---|---|---|---|---|---|
| No. of replicates pos/negc | % Pos | >400 | <400 | BLDb | ||
| 0 | 0/48 | 0 | 24 | |||
| 8 | 0/18 | 0 | 2 | 0 | 16 | |
| 16 | 2/22 | 9 | 3 | 2 | 19 | |
| 31 | 9/9 | 50 | 6 | 5 | 7 | |
| 63 | 760 | 12/12 | 50 | 20 | 3 | 1 |
| 125 | 1,300 | 18 | 0 | 0 | ||
| 250 | 2,400 | 21/3 | 88 | 24 | 0 | 0 |
| 1,000 | 12,000 | 24/0 | 100 | 24 | 0 | 0 |
| 4,000 | 36,000 | 24/0 | 100 | 24 | 0 | 0 |
| 8,000 | 50,000 | 17 | 0 | 0 | ||
| 40,000 | 127,000 | 18/0 | 100 | 15 | 0 | 0 |
| 200,000 | 180,000 | 12 | 0 | 0 | ||
| 1,000,000 | 428,000 | 18/0 | 100 | 15 | 0 | 0 |
AMPLICOR quantitative assay results were expressed as 10 raised to the mean of the log10 of the CMV DNA copies per milliliter from the three laboratories.
BLD, below the limit of detection.
Pos, positive; neg, negative.
Overall, the viral load as measured by the quantitative AMPLICOR assay was approximately 10-fold greater than the input concentration (Table 2). For example, with an input concentration of 250 CMV DNA copies/ml, the mean viral load obtained from the quantitative AMPLICOR assay was 2,400 CMV DNA copies/ml. When the input concentration was 4,000 CMV DNA copies/ml, the mean viral load from the quantitative AMPLICOR assay was 36,000 CMV DNA copies/ml.
The quantitative AMPLICOR assay was more sensitive than the qualitative AMPLICOR assay. At an input concentration of 125 CMV DNA copies/ml of plasma, all 18 replicates had a viral load of >400 CMV DNA copies/ml, with a mean viral load of 1,300 CMV DNA copies/ml. The sensitivity of the AMPLICOR qualitative assay reached 100% at an input concentration of 1,000 CMV DNA copies/ml (mean viral load, 12,000 CMV DNA copies/ml).
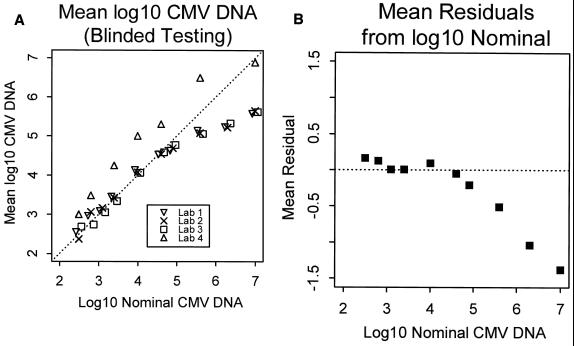
The AMPLICOR quantitative CMV DNA assay is linear to 50,000 CMV DNA copies/ml (Fig. 1A). There was excellent reproducibility of the viral load between the three laboratories using the AMPLICOR quantitative assay. Figure 1B presents the mean residuals (i.e., observed value minus expected value) for all three laboratories performing the quantitative AMPLICOR assay as a guide for evaluating where there is sizeable deviation from identity. This indicates that the assay is no longer linear beyond 50,000 CMV DNA copies/ml.
FIG. 1.
(A) Mean log10 CMV DNA for each laboratory versus the input concentration of the quantitation standard (nominal CMV DNA scaled by +1 log10 unit). The diagonal line indicates identity. Laboratories 1 to 3 used the quantitative AMPLICOR assay; laboratory 4 performed an in-house PCR assay. (b) Mean residuals (i.e., observed value minus expected value) for all three laboratories performing the quantitative AMPLICOR assay.
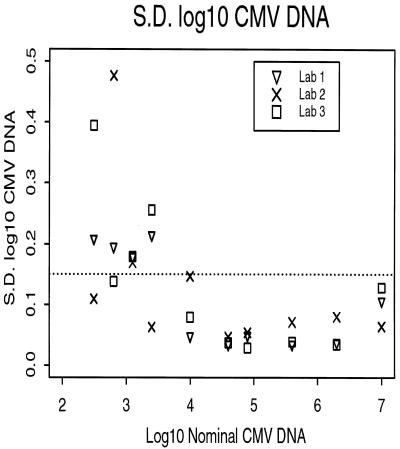
For the quantitative AMPLICOR assay, the variation is greater at the lower copy number. When the viral load obtained was less than 1,000 CMV DNA copies/ml (input concentrations of 31 and 64 CMV DNA copies/ml), the standard deviation ranged from 0.11 to 0.48 log10 unit (Fig. 2). When the viral load reached or exceeded 10,000 CMV DNA copies/ml (input concentration, 1,000 CMV DNA copies/ml and greater) the standard deviation was consistently at or below 0.15 log10 unit. A standard deviation of 0.15 log10 unit has been used by the Virology Quality Assurance program as part of the HIV-1 RNA quality assurance program to ensure that a laboratory could maintain the precision required to have 90% power to detect a fivefold difference in RNA copy number between two samples in the same batch (13).
FIG. 2.
The standard deviation of the log10 CMV DNA copy number versus the input concentration of the quantitation standard (nominal CMV DNA scaled by +1 log10 unit) for the three laboratories performing the quantitative AMPLICOR assay. The horizontal dotted line is at 0.15 log10 unit.
Table 3 presents the results of the analysis of the variance due to laboratory, kit lot, and replication (i.e., random error). Only the assays performed in laboratories 1 and 2 are included because they used the same kit lots for the same viral DNA concentrations. Results include samples analyzed both blinded and unblinded. Variance estimates are almost identical whether the samples analyzed unblinded were included or not. The stock with 31 CMV DNA copies/ml is not presented because so many of the results were <400 CMV DNA copies/ml, and stocks with over 8,000 CMV DNA copies/ml are not presented because they are out of the linear range of the assay. Table 3 shows that the error variance was greater than laboratory or lot variance.
TABLE 3.
Estimated variance components for lots 1 to 6 of the quantitative AMPLICOR assay when tested in laboratories 1 and 2
| Quantitation panel input concn (CMV DNA copies/ml) | Variance of:
|
||
|---|---|---|---|
| Laboratory | Lot | Error | |
| 63 | 0 | 0 | 0.1230 |
| 125 | 0 | 0.0026 | 0.0270 |
| 250 | 0.0032 | 0 | 0.0315 |
| 1,000 | 0 | 0 | 0.0143 |
| 4,000 | 0 | 0 | 0.0014 |
| 8,000 | 0.0015 | 0 | 0.0026 |
Comparison between the quantitative AMPLICOR and an in-house-developed CMV DNA PCR assay.
Testing results from the laboratory performing the in-house quantitative assay are shown in Fig. 1A along with the results from the three laboratories using the quantitative AMPLICOR assay. The difference between the two assays varied depending on the concentration of input DNA. At lower concentrations, the AMPLICOR assay was one-fifth to one-third (0.2 to 0.5 log10 unit, respectively) lower than the in-house PCR assay. At higher input DNA concentrations, the difference between the two assays approached 1 log10 unit, with the in-house PCR assay giving higher concentrations. The in-house assay has a broader linear range than the AMPLICOR assay. The AMPLICOR assay remains linear down to a lower input concentration than the in-house assay, as the lowest concentration on Fig. 1A is no longer linear for the in-house assay. However, the in-house assay remains linear to a viral load of 1,000,000 CMV DNA copies/ml compared to 50,000 CMV DNA copies/ml for the AMPLICOR assay.
DISCUSSION
There is a need for standardized assays that detect and quantify CMV DNA in clinical specimens. Such assays would reduce the variation in viral load results among different in-house-developed CMV PCR assays. Though in-house assays within a given laboratory are reproducible, viral load results from different assays (and thus from different laboratories) often do not correlate (1, 2, 6, 11). This is due to differences in assay design, including specimen volume, specimen type, quantitation standard, and detection methods. We present data on the evaluation of standardized qualitative and quantitative AMPLICOR CMV DNA PCR assays and have compared the quantitative assay to the in-house-developed assay that was used to determine the clinical significance of CMV viral load in patients with AIDS in previous studies (8, 9).
The comparison between the qualitative and quantitative AMPLICOR assays showed that the quantitative assay is more sensitive than the qualitative assay. This difference in analytical sensitivity might be due to the different volumes of plasma used in the amplification step of both assays. For the qualitative assay, the equivalent of 5 μl of plasma is used for amplification, while the quantitative assay amplifies CMV DNA from the equivalent of 50 μl of plasma. The difference in assay design was due to concerns about the lack of clinical specificity of qualitative CMV DNA assays. Though these assays are sensitive, in transplant recipients they often detect CMV DNA in the absence of active CMV disease (3, 5, 10). Whether the difference in analytical sensitivity between the qualitative and quantitative versions of the AMPLICOR assays has clinical significance remains to be determined. Both assays showed good specificity. All of the samples that did not contain CMV DNA tested negative in both assays.
The viral load as measured by the AMPLICOR quantitative assay was approximately 10-fold greater than the input concentration. The input concentration of CMV DNA was based on a viral particle count by electron microscopy. This method would not account for free DNA that may be present in the solution, which would be detected by PCR but not by a particle count by electron microscopy. In addition, variability due to pipetting could account for the differences between input copy and detected viral load. As a result of this consistent 10-fold increase, the input concentration has been increased by 1 log unit in the plots of input versus observed CMV DNA copies per milliliter.
The AMPLICOR quantitative assay was linear to 50,000 CMV DNA copies/ml of plasma, and there was excellent agreement in the viral load values obtained from the three laboratories using this assay. An evaluation of the results from the two laboratories using the same lots of kits showed that the error variance (or interassay variance) was greater than the laboratory or lot variance. The low variance among laboratories and kit lots is encouraging and would facilitate using the AMPLICOR assays in multicenter clinical trials.
The standard deviation of the AMPLICOR quantitative assay varies throughout its dynamic range. When the viral load is greater than or equal to 12,000 CMV DNA copies/ml of plasma, the standard deviation of the log transformed data for the three testing laboratories was consistently below 0.15 log10 unit. However, at a viral load less than 1,000 CMV DNA copies/ml of plasma the standard deviation was quite variable, ranging from 0.11 to 0.48 log10 unit. This suggests that at the lower end of the assay, it may not be possible to reliably distinguish samples that have a fivefold difference in viral load.
Clinical studies using the in-house PCR assay we used in this study have shown that the risk of developing CMV disease in patients with AIDS is greater with increasing CMV viral load and that CMV DNA viral load is an independent marker of CMV disease and survival. The correspondence between this in-house-developed quantitative CMV DNA assay and the quantitative AMPLICOR assay was established. Though the dynamic ranges of the two assays were not the same, a comparison of values between the two assays was possible. The quantitative in-house assay was linear to approximately 1,000,000 CMV DNA copies/ml, while the AMPLICOR assay was linear to 50,000 CMV DNA copies/ml. The in-house assay was not linear at the lower concentrations (input CMV DNA copy numbers, 31 and 63 CMV DNA copies/ml). Though the AMPLICOR assay gave lower viral load values at all concentrations tested, the difference between the two assays was not consistent across the entire dynamic range of the AMPLICOR assay. At the lower end of the assay, the viral load values obtained with the in-house assay were three- to fivefold (0.5 to 0.7 log unit) higher than those seen with the AMPLICOR assay. At higher input concentrations, the differences between the two assays approached 10-fold. This direct comparison of the in-house and AMPLICOR assays is very important, as it provides the data necessary to compare viral load values obtained with either assay. Laboratories doing CMV PCR testing with the AMPLICOR assay can now establish clinically relevant viral load cut-off values based on the clinical trials done by Spector et al. (8, 9) using the in-house PCR assay evaluated in this study.
The AMPLICOR assays we evaluated here offer several advantages for the clinical laboratory, one of which is that a small volume of plasma is required for the test. The starting plasma volumes for the qualitative and quantitative assays are 50 and 200 μl, respectively. Since the assays are plasma based, sensitivity will not be influenced by leukopenia, which can be a problem in HIV-infected patients at risk for CMV infection. The quantitative assay test is performed using the semiautomated COBAS AMPLICOR format, so the technical time required to perform the PCR test is much less than that required for many in-house assays. Currently, the quantitative AMPLICOR assay has not been approved by the Food and Drug Administration; however, it is available as a “research use only” product.
Results of this study show that the AMPLICOR quantitative assay is more sensitive than the AMPLICOR qualitative assay. In addition, the correspondence between the in-house-developed quantitative CMV DNA assay used in this study and the quantitative AMPLICOR assay has been established. The availability of standardized assays is an important step for CMV diagnostics, as it eliminates the current problem of trying to compare viral load levels obtained with different in-house-developed assays. A standardized assay will allow results of studies at different laboratories to be compared and may allow for the determination of viral load cutoffs for identifying AIDS patients at high risk of developing CMV disease.
ACKNOWLEDGMENTS
A. M. Caliendo and R. Schuurman contributed equally to this work.
We thank Colleen Starkey of the Cleveland Clinic and Jessica Allega of Emory University for technical assistance and Roche Molecular Systems for generously donating the reagents for the AMPLICOR testing.
This work was supported by the Complications of HIV Disease RAC, AIDS Clinical Trials Group and Virology Quality Assurance contract no. NO1 AI85354.
REFERENCES
- 1.Cope A V, Sabin C, Burroughs A, Rolles K, Griffiths P D, Emery V C. Interrelationships among quantity of human cytomegalovirus (HCMV) DNA in blood, donor-recipient serostatus, and administration of methylprednisolone as risk factors for HCMV disease following liver transplantation. J Infect Dis. 1997;176:1484–1490. doi: 10.1086/514145. [DOI] [PubMed] [Google Scholar]
- 2.Ferreira-Gonzalez A, Fisher R A, Weymouth L A, Langley M R, Wolfe L, Wilkinson D S, Garrett C T. Clinical utility of a quantitative polymerase chain reaction for diagnosis of cytomegalovirus disease in solid organ transplant patients. Transplantation. 1999;68:991–996. doi: 10.1097/00007890-199910150-00016. [DOI] [PubMed] [Google Scholar]
- 3.Gerna G, Zipeto D, Parea M, Revello M G, Silini E, Percivalle E, Zavattoni M, Grossi P, Milanesi G. Monitoring of human cytomegalovirus infections and ganciclovir treatment in heart transplant recipients by determination of viremia, antigenemia and DNAemia. J Infect Dis. 1991;164:488–498. doi: 10.1093/infdis/164.3.488. [DOI] [PubMed] [Google Scholar]
- 4.Kouzarides T, Bankier A T, Satchwell S C, Weston K, Tomlinson P, Barrell B G. Sequence and transcription analysis of the human cytomegalovirus DNA polymerase gene. J Virol. 1987;61:125–133. doi: 10.1128/jvi.61.1.125-133.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nolte F S, Emmens R K, Thurmond C, Mitchell P S, Pascuzzi C, Devine S M, Saral R, Wingard J R. Early detection of human cytomegalovirus viremia in bone marrow transplant recipients with DNA amplification. J Clin Microbiol. 1995;33:1263–1266. doi: 10.1128/jcm.33.5.1263-1266.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts T C, Brennan D C, Buller R S, Gaudreault-Keener M, Schnitzler M A, Sternhell K E, Garlock K A, Singer G G, Storch G A. Quantitative polymerase chain reaction to predict occurrence of symptomatic cytomegalovirus infection and assess response to ganciclovir therapy in renal transplant recipients. J Infect Dis. 1998;178:626–635. doi: 10.1086/515383. [DOI] [PubMed] [Google Scholar]
- 7.Shinkai M, Bozzette S A, Powderly W, Frame P, Spector S A. Utility of urine and leukocyte cultures and plasma DNA polymerase chain reaction for identification of AIDS patients at risk for developing human cytomegalovirus disease. J Infect Dis. 1997;175:302–308. doi: 10.1093/infdis/175.2.302. [DOI] [PubMed] [Google Scholar]
- 8.Spector S A, Hsia K, Crager M, Pilcher M, Cabral S, Stempien M J. Cytomegalovirus (CMV) DNA load is an independent predictor of CMV disease and survival in advanced AIDS. J Virol. 1999;73:7027–7030. doi: 10.1128/jvi.73.8.7027-7030.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spector S A, Wong R, Hsia K, Pilcher M, Stempien M J. Plasma cytomegalovirus (CMV) DNA load predicits CMV disease and survival in AIDS patients. J Clin Investig. 1998;101:497–502. doi: 10.1172/JCI1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanabe K, Tokumoto T, Ishikawa N, Koyama I, Takahashi K, Fuchinoue S, Kawai T, Koga S, Yagisawa T, Toma H, Ota K, Nakajima H. Comparative study of CMV antigenemia assay, polymerase chain reaction, serology, and shell vial assay in the early diagnosis and monitoring of CMV infection after renal transplantation. Transplantation. 1997;64:1721–1725. doi: 10.1097/00007890-199712270-00016. [DOI] [PubMed] [Google Scholar]
- 11.Weinberg A, Hodges T N, Li S, Cai G, Zamora M R. Comparison of PCR, antigenemia assay, and rapid blood culture for detection and prevention of cytomegalovirus disease after lung transplantation. J Clin Microbiol. 2000;38:768–772. doi: 10.1128/jcm.38.2.768-772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolf D G, Spector S A. Early diagnosis of human cytomegalovirus disease in transplant recipients by DNA amplification in plasma. Transplantation. 1993;56:330–334. doi: 10.1097/00007890-199308000-00014. [DOI] [PubMed] [Google Scholar]
- 13.Yen-Lieberman B, Brambilla D, Jackson B, Bremer J, Coombs R, Cronin M, Herman S, Katzenstein D, Leung S, Lin H J, Palumbo P, Rasheed S, Todd J, Vahey M, Reichelderfer P. Evaluation of a quality assurance program for quantitation of human immunodeficiency virus type 1 RNA in plasma by the AIDS Clinical Trials Group virology laboratories. J Clin Microbiol. 1996;34:2695–2701. doi: 10.1128/jcm.34.11.2695-2701.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]