Abstract
Specific and sensitive reverse transcription-PCR (RT-PCR) assays were developed for the detection of eastern, western, and Venezuelan equine encephalitis viruses (EEE, WEE, and VEE, respectively). Tests for specificity included all known alphavirus species. The EEE-specific RT-PCR amplified a 464-bp region of the E2 gene exclusively from 10 different EEE strains from South and North America with a sensitivity of about 3,000 RNA molecules. In a subsequent nested PCR, the specificity was confirmed by the amplification of a 262-bp fragment, increasing the sensitivity of this assay to approximately 30 RNA molecules. The RT-PCR for WEE amplified a fragment of 354 bp from as few as 2,000 RNA molecules. Babanki virus, as well as Mucambo and Pixuna viruses (VEE subtypes IIIA and IV), were also amplified. However, the latter viruses showed slightly smaller fragments of about 290 and 310 bp, respectively. A subsequent seminested PCR amplified a 195-bp fragment only from the 10 tested strains of WEE from North and South America, rendering this assay virus specific and increasing its sensitivity to approximately 20 RNA molecules. Because the 12 VEE subtypes showed too much divergence in their 26S RNA nucleotide sequences to detect all of them by the use of nondegenerate primers, this assay was confined to the medically important and closely related VEE subtypes IAB, IC, ID, IE, and II. The RT-PCR–seminested PCR combination specifically amplified 342- and 194-bp fragments of the region covering the 6K gene in VEE. The sensitivity was 20 RNA molecules for subtype IAB virus and 70 RNA molecules for subtype IE virus. In addition to the subtypes mentioned above, three of the enzootic VEE (subtypes IIIB, IIIC, and IV) showed the specific amplicon in the seminested PCR. The practicability of the latter assay was tested with human sera gathered as part of the febrile illness surveillance in the Amazon River Basin of Peru near the city of Iquitos. All of the nine tested VEE-positive sera showed the expected 194-bp amplicon of the VEE-specific RT-PCR–seminested PCR.
Venezuelan, eastern, and western equine encephalitis viruses (VEE, EEE, and WEE, respectively) are zoonotic viruses (genus Alphavirus, family Togaviridae) transmitted to equides and humans by mosquitos. They are maintained in enzootic transmission cycles by certain mosquito species and in either rodents or birds which serve as natural hosts. The ornithophilic mosquito Culiseta melanura transmits EEE along the eastern coast of the United States, while Culex species are involved in EEE transmission in Mexico, Central America, and southward to Argentina (46, 50). The less-host-restricted Aedes and Coquillettidia species transmit EEE to humans and horses. While there is little overlap between EEE and WEE in North America, both viruses are enzootic in Central and South America. WEE cycles between passeriform birds and Culex tarsalis, which also serves as the epizootic vector (28).
VEE consist of related, but differentiable viruses that are classified as a complex containing six antigenic subtypes (I to VI). Viruses of subtypes I and III are further differentiated into five (IAB to IF) and three (IIIA to IIIC) antigenic varieties, respectively (3, 4, 9, 15, 51). In contrast to both EEE and WEE, the distribution of VEE is limited primarily to Central and South America. The two exceptions are Everglades virus (EVE) found in Florida (6) and Bijou Bridge virus (20) found in Colorado. EVE is antigenically classified as a subtype II virus (51), but it is genetically more closely related to the epizootic subtype IAB and IC viruses than the VEE-IE virus strain Mena II (14, 48). Bijou Bridge virus is genetically and antigenically classified as subtype IIIB virus (20, 45). VEE epizootics, which were caused by subtype IAB and IC viruses, frequently occurred in South America, including a 1969 to 1972 pandemic which spread from Central America to Texas in the United States. After an absence of 20 years, epizootic VEE (subtype IC) reemerged in Venezuela in 1993 (29). Since then, VEE has caused concern because of (i) a large epizootic in Venezuela and Colombia in 1995 involving about 75,000 humans and 50,000 equids (30, 49), (ii) a small epidemic in Peru during 1994 caused by a VEE-ID virus (23, 44), and (iii) two epidemics affecting the southern Mexican horse population in 1993 and 1996 (22). Both outbreaks in Mexico were caused by VEE strains of subtype-variety IE, thus far considered enzootic and not pathogenic for horses (42).
The clinical signs of EEE-, WEE-, or VEE-infected humans and horses are nonspecific, and a panel of nonviral and noninfectious etiologies has to be considered (37). Although no specific treatment exists for infections by these viruses, a fast and specific diagnosis is needed to prevent the further spread of the disease by quarantine, trade restriction, vaccination, and vector control. Detection of the viral antigen or its nucleic acid in serum is only successful if the blood is collected during the viremic phase of the infection, which lasts for 3 to 5 days. Virus isolation by intracerebral inoculation of baby mice or cell cultures is the “gold standard” for virus detection (24), but it is very time-consuming. RT-PCR is a fast, sensitive, and specific alternative for the diagnosis of infections caused by RNA viruses (26). This technique has been described for the detection of EEE (1, 40), WEE (41), and VEE (2). In those studies, the specificity of the RT-PCRs was not thoroughly tested. Recent work has indicated that these assays failed to distinguish closely related heterologous alphaviruses (19). The intent of the present study was to provide specific diagnostic tests for each of the equine encephalitis viruses, as verified by evaluating amplimer reactivities with all known alphavirus species. Rapid diagnosis by sensitive, virus-specific RT-PCRs would permit the timely implementation of prevention and control measures. This is of particular importance in countries such as Mexico, where all three equine encephalitis viruses circulate simultaneously.
MATERIALS AND METHODS
Virus strains.
Each of the alphaviruses used in this study represented one of the currently classified alphavirus species (21). A description of the isolates, including the year and location of isolation, as well as their passage history, was published earlier (25). To evaluate each RT-PCR assay using a panel of virus strains, nine additional virus isolates each of EEE and WEE were chosen from the virus collection of the Division of Vector-Borne Infectious Diseases, National Center for Infectious Diseases, Centers for Disease Control, Fort Collins, Colo. The selection of isolates was intended to include those with maximal geographical distribution (South and North America) and various dates of virus isolation (Table 1). To further determine the specificity or cross-reactivity of the RT-PCR, representative strains of the 12 VEE subtypes were used as described earlier (14, 25). Recent VEE isolates from Mexico and Peru were used as the original sera or their first cell culture passage (Table 2).
TABLE 1.
EEE and WEE strains used in this study
| Virus and straina | Yr, Location of isolation | Source | Passage historyc | Reference |
|---|---|---|---|---|
| EEE | ||||
| NJ/60 (ref.) | 1960, United States (N.J.) | Mosquito | P?AM1SM4 | 46 |
| 663 | 1989, United States (Mass.) | Mosquito | P?V1 | |
| 17943 | 1989, United States (Del.) | Horse | P?V2 | |
| 89-42687 | 1989, United States (Tenn.) | Horse | P?V2 | 46 |
| 82V-2137 | 1982, United States (Fla.) | Mosquito | P?V2 | 50 |
| W24317 | 1966, United States (La.) | Bird | P?SM3 | |
| 76V-25343 | 1976, Brazil | Mosquito | P?SM3 | |
| BeAn 5122 | 1956, Brazil | Monkeyb | P?V1SM1 | 47 |
| 75V 1496 | 1974, Ecuador | Mosquito | P?V1SM3 | |
| 10365 | 1978, Dominican Republic | Horse | P?SM2 | 50 |
| WEE | ||||
| Fleming (ref.) | 1946, United States (Calif.) | Human | P?SM9 | 3 |
| McMillan | 1941, Canada (Ontario) | Human | P?M2SM2 | 3 |
| BFS 2005 | 1964, United States (Calif.) | Mosquito | P?DE1 | 12 |
| 73V 1492 | 1973, United States (Tex.) | Bird | P?WC1DE1SM1 | |
| R-43738 | 1983, United States (S.D.) | Human | P?V2SM3 | 3 |
| Montana 64 | 1967, United States (Mont.) | Horse | P?DE1 | |
| CBA 87 | 1958, Argentina | Horse | P?SM1 | |
| Ag80-646 | 1980, Argentina | Mosquito | P?V2SM3 | 3 |
| BeAn 112509 | 1966, Brazil | Mouseb | P?DE1 | |
| Rio-1257 | 1961, Brazil | Horse | P8WC1DE1SM1 |
The strain used as reference (ref.) for the respective alphavirus species is indicated.
Sentinel animals.
AM, adult mouse; SM, suckling mouse; V, Vero cells; DE, primary Pekin duck embryo; WC, 1.5-day-old chicken; ?, unknown.
TABLE 2.
VEE strains and VEE-positive serum samples used in this study
| Strain | Date (mo-day-yr), location of isolation | Source | Passage historya | Subtypeb | Reference |
|---|---|---|---|---|---|
| IQT 1724 | 5-2-95, Iquitos, Peru | Human | SM1 | VEE-ID | 43 |
| IQT 4191 | 8-6-97, Iquitos, Peru | Human | SM1 and V1 | ND | This study |
| IQT 5831 | 2-5-98, Iquitos, Peru | Human | C6/36-1 | ND | This study |
| IQT 6119 | 2-20-98, Iquitos, Peru | Human | C6/36-1 | ND | This study |
| IQT 6415 | 3-6-98, Iquitos, Peru | Human | V1 | ND | This study |
| IQT 6486 | 3-10-98, Iquitos, Peru | Human | V1 | ND | This study |
| IQT 7057 | 4-8-98, Iquitos, Peru | Human | C6/36-1 | ND | This study |
| IQT 7988 | 6-23-98, Iquitos, Peru | Human | V1 | ND | This study |
| IQT 8131 | 7-9-98, Iquitos, Peru | Human | C6/36 | ND | This study |
| 142-96 | 7-20-96, Tapanatepec, Mexico | Horse | SM1 | VEE-IE | 22 |
SM, suckling mouse; V, Vero cells; C6/36, C6/36 (Aedes albopticus) cells.
VEE subtype varieties were determined by nucleotide sequencing of parts of the PE2 genes as described in the references given; ND, not determined.
Extraction of viral RNA.
Viral RNA was extracted from a 200-μl aliquot of infectious Vero cell culture supernatant according to the method of Lewis et al. (18). The viral RNA was rehydrated in 50 μl of diethyl pyrocarbonate (DEPC)-treated water (25). The amount of RNA of each preparation was measured photometrically (GeneQuant; Pharmacia), and the number of RNA molecules was calculated on the basis of a genome length of 11,700 nucleotides and an average weight for each nucleotide of 336.3 g/mol, yielding a weight of 6.53 × 10−9 ng per RNA molecule.
Primer selection.
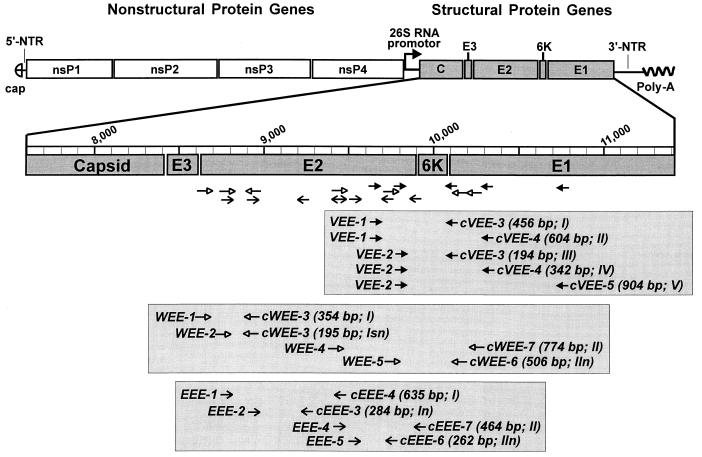
The oligonucleotide primers were chosen from the aligned nucleotide sequences of the structural polyprotein coding regions of Sindbis (38), Ockelbo (35), Aura (31), Ross River (8), O'Nyong Nyong (17), Chikungunya (M. Parker, GenBank accession no. L37661), Semliki Forest (10), EEE (7, 46, 47, 50), WEE (12), and representative strains of the VEE subtype viruses (14). For each virus-specific detection, two primer pairs each for primary RT-PCR and subsequent nested PCR were synthesized. Their sequences and genomic locations are presented in Table 3 and Fig. 1.
TABLE 3.
Oligonucleotide primers used for the specific amplification of WEE, EEE, or VEE by RT-PCR and nested PCR
| Virus species and primera | Positionb | Primer sequence (5′–3′) | Amplicon (bp) | Combinationc |
|---|---|---|---|---|
| EEE | ||||
| EEE-1 | 8763 (E2) | TCATGAACGGCAAAACGCAGAAATC | 635 | I |
| cEEE-4 | 9377 (E2) | TGCGGTGTTTGTGCTCAACTAG | ||
| EEE-2 | 8913 (E2) | CTAACCGCCATACGTGCACAGTTGCC | 284 | In |
| cEEE-3 | 9175 (E2) | CGCTGCTGGTAATTCCCTCTCGT | ||
| EEE-4 | 9377 (E2) | CTAGTTGAGCACAAACACCGCA | 464 | II |
| cEEE-7 | 9817 (E2) | CACTTGCAAGGTGTCGTCTGCCCTC | ||
| EEE-5 | 9457 (E2) | AAGTGATGCAAATCCAACTCGAC | 262 | IIn |
| cEEE-6 | 9697 (E2) | GGAGCCACACGGATGTGACACAA | ||
| WEE | ||||
| WEE-1 | 1157 (E2) | GTTCTGCCCGTATTGCAGACACTCA | 354 | I |
| cWEE-3 | 1490 (E2) | CCTCCTGATCTTTTTCTCCACG | ||
| WEE-2 | 1316 (E2) | GTCTTTCGACCACGACCATG | 195 | Isn |
| cWEE-3 | 1490 (E2) | CCTCCTGATCTTTTTCTCCACG | ||
| WEE-4 | 2016 (E2) | GGGCTGCGAGCAGACGCAACAG | 774 | II |
| cWEE-7 | 2767 (E1) | GCGGCATGTGTAATCCGCTTTTGA | ||
| WEE-5 | 2258 (E2) | CACTGCATCGTCAGCAGCTTGTATCG | 506 | IIn |
| cWEE-6 | 2741 (E1) | GAGGATGCCTTACACTCGAGGGAC | ||
| VEE | ||||
| VEE-1 | 9574 (E2) | GTTTTGGGCACAGGAAACAGC | 456 | I |
| cVEE-3 | 10009 (E1) | TTGGCTCGGCATCGTGTTCGCG | ||
| VEE-1 | 9574 (E2) | GTTTTGGGCACAGGAAACAGC | 604 | II |
| cVEE-4 | 10159 (E1) | TGGCTGGTGAATCCATTCCT | ||
| VEE-2 | 9836 (6K) | ACCACCTGGGAGTCCTTGGA | 342 | IV |
| cVEE-4 | 10159 (E1) | TGGCTGGTGAATCCATTCCT | ||
| VEE-2 | 9836 (6K) | ACCACCTGGGAGTCCTTGGA | 194 | IIIsn |
| cVEE-3 | 10009 (E1) | TTGGCTCGGCATCGTGTTCGCG | ||
| VEE-2 | 9836 (6K) | ACCACCTGGGAGTCCTTGGA | 904 | V |
| cVEE-5 | 10720 (E1) | TTATCTTTCTTCCATTGCTCA |
Primers for RT are indicated with a “c” before of the three-letter abbreviation of the equine encephalitis virus it was designed for.
Nucleotide position according to the genomic sequence data of EEE strain 82V-1237 (GenBank U01034), the 26S RNA sequence data of WEE strain BFS1703 (GenBank J03854), and the genomic sequence data of VEE-IAB vaccine strain TC-83 (L01443).
The primer combinations allowing the most sensitive detection of the respective equine encephalitis virus are in boldface. Each primer combination tested is numbered in consecutive order. The letters following the roman numbers indicate the type of reaction as follows: no letter, RT-PCR; n, nested PCR; and sn, seminested PCR.
FIG. 1.
Schematic drawing of the alphavirus genome organization (adapted from reference 39). The subgenomic 26S RNA coding for the viral structural proteins is enlarged and has been drawn to scale. The primers for the specific detection of each equine encephalitis virus by RT-PCR or nested PCR are shown in boxes. Their orientation is depicted by arrows and by their names (c, complementary to the viral RNA, priming the RT). Each primer combination tested is numbered in consecutive order. The letters following the roman numbers indicate the type of reaction as follows: no letter, RT-PCR; n, nested PCR; and sn, seminested PCR (see also Table 3). The size of each amplicon is given in brackets.
General PCR procedures.
All RT-PCRs and nested PCRs were carried out in 0.2 ml of thin-walled PCR tubes using a model 9600 or a model 2400 thermocycler (Perkin-Elmer Corp.). The final reaction volume was always 100 μl. During optimization of each of the specific RT-PCR and nested-PCR assays, the amount of the following reagents was varied: primers (0.1 to 1 μM, in 0.1 μM steps), MgCl2 (1 to 4 mM, in 0.5 mM steps), and deoxynucleoside triphosphate (dNTP) (100, 150, and 200 μM; MBI Fermentas). The remaining reagents were identical in all RT-PCRs (5 mM dithiothreitol, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 0.01% gelatin, 2.5 U of TaqExtender PCR additive (Stratagene, Heidelberg, Germany), 2 U of RAV-2 reverse transcriptase (Amersham, Braunschweig, Germany), 2 U of AmpliTaq DNA polymerase (Perkin-Elmer Corp., Weiterstadt, Germany), 5 μl of RNA template, and DEPC-treated water to give a final volume of 100-μl). Nested PCRs were performed in 100-μl reactions containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 0.01% gelatin, 5 mM dithiothreitol, 1.5 U of AmpliTaq DNA polymerase, and 2 μl of cDNA template from the previous RT-PCR. The annealing temperatures tested in both RT-PCRs and nested PCRs ranged from 55 to 72°C. For both RT-PCRs and nested-PCRs, the optimal concentrations of MgCl2 and dNTP were consistently 2 mM and 200 μM, respectively. Optimal results were defined as the virus-specific amplification obtained with the primer pairs, concentrations of reaction components, and cycling conditions that allowed the most sensitive detection of the respective viral RNA. When differing reactions resulted in the same sensitivity, criteria such as the lack of signs of nonspecific background bands were used as additional parameters to measure the success of the optimization.
To prevent carryover contaminations, pipetting of the reaction mixtures for the RT-PCRs was performed under a hood in a room separated from both the room where amplicons were analyzed by gel electrophoresis and the room where the pipetting of the nested PCRs was done. A 10-μl aliquot of each reaction was loaded onto 2% agarose gels (Eurogentec, Heidelberg, Germany) in TBE buffer (0.1 M Tris-HCl, pH 8.0; 0.091 M boric acid; 2 mM disodium EDTA). DNA fragments were visualized after staining with ethidium bromide on a UV transilluminator at 302 nm and recorded using a charge-coupled device CCD camera (Fröbel, Wasserburg, Germany) and the FotoTouch Color software (Logitech).
All RT-PCR and nested-PCR were tested with genomic RNA template of representative strains of all known alphavirus species to ensure virus specificity within the genus.
EEE-specific RT-PCR and nested PCR.
Cycling conditions of the uninterrupted RT-PCR program were 30 min at 50°C followed by one cycle with denaturing at 94°C for 90 s, annealing at 64°C for 90 s, and extension at 72°C for 90 s and 35 cycles of 94°C for 20 s, 64°C for 30 s, and 72°C for 20 s. The final extension step was prolonged to 5 min to ensure completion of the length of the amplicon. Optimal results were obtained with 0.1 μM concentrations each of primers EEE-4 and cEEE-7 (Table 3, combination II).
A 2-μl aliquot of the RT-PCR mixture was subjected to a nested PCR which, after an initial denaturation at 94°C for 90 s, consisted of 25 cycles of 94°C for 20 s, 65°C for 35 s, and 72°C for 17 s. The final elongation step was 4 min. EEE-5 and cEEE-6 primer concentrations of 0.3 μM each (combination IIn) yielded the best results.
WEE-specific RT-PCR and seminested PCR.
For the specific amplification of WEE, a set of three primers targeting the E2 gene and implemented as RT-PCR and seminested PCR (Table 3, combinations I and Isn) were found to be 300 times more sensitive than a RT-PCR–nested PCR primer set (combinations II and IIn) hybridizing to the E2 and E1 genes embracing the 6K gene (Fig. 1; see also below). The reaction conditions were as follows: 30 min at 50°C; one cycle consisting of 90 s at 94°C, 60 s at 68°C, and 90 s at 72°C; 35 cycles consisting of 20 s at 94°C, 30 s at 68°C, and 17 s at 72°C; and a final extension step of 5 min for the RT-PCR I (Table 3). Each primer (WEE-1 and cWEE-3) was at 0.2 μM, while in the subsequent seminested PCR (combination Isn) 0.3 μM concentrations each of primers WEE-2 and cWEE-3 gave the best results. The cycling conditions were as described for the EEE-specific nested PCR, except that the annealing temperature was set at 63°C and the elongation time was only 15 s.
VEE-specific RT-PCR and seminested PCR.
To develop VEE-specific primers, the aligned 26S RNA sequence data of representative strains of the 12 VEE subtype varieties were used (14) and compared with the available sequence data of other alphaviruses (see above). The level of nucleotide identity among VEE subtype varieties IAB, IC, and ID was >94%, while this level dropped to 70 to 88% when VEE-IAB was compared to the other subtype varieties (14). To avoid the use of degenerate primers, we focused on the genetically closely related and medically important VEE-IAB, -IC, -ID, -II, and -IE subtype varieties. Unfortunately, highly homologous regions among them also displayed a high level of nucleotide identity to EEE or WEE in these regions. Hence, two forward and three reverse primers were designed to function in RT-PCR–nested-PCR combinations (Table 3, Fig. 1).
After we tested the specificity and sensitivity of each combination, we found that performing the RT-PCR with primers VEE-2 versus cVEE-4 (Table 3, combination IV) with a seminested PCR using primers VEE-2 versus cVEE-3 (combination IIIsn) yielded the best results (see below). The optimal reaction conditions were found to be an RT for 30 min at 50°C, followed by one cycle of 90 s at 94°C, 90 s at 61°C, and 90 s at 72°C and 35 cycles, each consisting of 20 s at 94°C, 40 s at 61°C, and 17 s at 72°C. The final extension step was 5 min. For this RT-PCR (IV), 0.2 μM concentrations of primers VEE-2 and cVEE-4 gave the best results. In the subsequent seminested PCR (IIIsn), the cycling conditions were as described for the WEE-specific seminested PCR (Isn), with VEE-2 and cVEE-3 primer concentrations of 0.3 μM each found to give the best results.
Investigation of serum samples.
The VEE-specific RT-PCR assay was applied to human sera gathered as part of the “febrile illness surveillance in the Amazon River Basin.” From all of the serum samples listed in Table 2 virus was isolated in either Vero cells, C6/36 cells, or newborn mice and subsequently identified as VEE in an indirect immunofluorescence assay as described previously (44). A Mayaro virus-positive serum sample served as a negative control. RNA extracted from this serum was amplified by using a genus-specific RT-PCR–seminested PCR as described earlier (25). Compared to the VEE-specific RT-PCR–seminested PCR method described above, the following slight modifications were made: the viral RNA was extracted with the QIAmp viral RNA kit according to the supplier's recommendation (Qiagen), no TaqExtender PCR additive was used, SuperScript reverse transcriptase (2 U; Gibco BRL) was used instead of RAV-2 reverse transcriptase, and the annealing temperature was set at 55°C in the model 9600 thermocycler for both RT-PCR and seminested PCR.
RESULTS
EEE-specific RT-PCR and nested PCR.
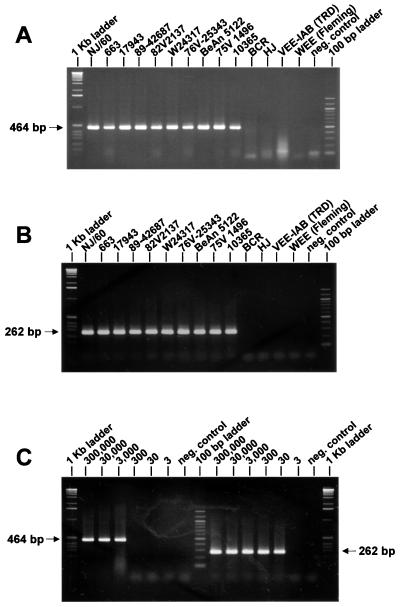
Based on the nucleotide and the deduced amino acid sequence alignments of the available alphaviral 26S RNA sequence data, a total of 10 regions with virus-specific nucleotide sequences sufficiently distinct from the remaining alphaviruses were identified. Primers for three regions showed a high degree of self-complementarity and were not used. Of the remaining seven regions, eight primers (one region was used for both a forward and a reverse primer) were made, resulting in two sets of RT-PCR–nested-PCR primer pairs. With the first RT-PCR (I in Table 3, and Fig. 1) Aura virus yielded an amplicon of about 600 bp, which was slightly smaller than the EEE-specific RT-PCR product of 635 bp (not shown). In the subsequent nested PCR (In), the 10 tested EEE strains exclusively yielded the expected 284-bp fragment rendering the I and In combinations EEE specific. The sensitivity of this combination was 3 × 105 RNA molecules in the RT-PCR and 3 × 103 RNA molecules in the nested PCR (not shown). The second set of primers for RT-PCR (II; EEE-4 versus cEEE-7) yielded the expected 464-bp product with all of the EEE strains tested (Fig. 2A) and did not amplify the RNA of any other alphavirus (not shown). Weak bands of 100 bp were visible for most alphaviruses and the negative control, suggesting them to be of primer dimer origin. In the respective nested PCR (IIn), the 262-bp fragment was also only amplified with the EEE strains (Fig. 2B). None of the other alphaviruses showed a visible amplicon (not shown). The second combinations (II plus IIn) were 100-fold more sensitive than the previous combination, detecting 3 × 103 (RT-PCR) and 30 RNA molecules (nested PCR), respectively (Fig. 2C).
FIG. 2.
Ethidium bromide-stained agarose gels showing the results of EEE-specific amplification. A 10-μl aliquot of each reaction was loaded onto 2% agarose gels. Amplicon sizes are depicted in the margin. (A) Amplification of the EEE-specific 464-bp product resulting from RT-PCR II of 10 different EEE strains (see Table 1) and the closely related alphaviruses Buggy Creek (BCR) and Highlands J (HJ), VEE-IAB strain Trinidad donkey (TRD), and WEE strain Fleming. The negative control contained water instead of RNA in the reaction. (B) The EEE-specific 262-bp amplification products resulting from nested PCR IIn of the template cDNA derived from RT-PCR II (combinations II plus IIn) shown in Fig. 2A. The negative control contained 2 μl of the RT-PCR II negative control reaction. (C) Serial dilutions containing 3 × 105 to 3 RNA molecules of EEE strain NJ/60 used as template for EEE-specific RT-PCR II (left) and subsequently performed nested-PCR IIn (right). The 262-bp amplification product was visible on an ethidium bromide-stained agarose gel down to the dilution containing 30 RNA molecules.
WEE-specific RT-PCR and seminested PCR.
Eleven regions with sufficient sequence divergence compared to the other alphaviruses were identified for WEE. Four possible primer sequences were excluded because of predicted primer dimer formations. The remaining seven sites were used to design primers for use in a RT-PCR–seminested PCR (Table 3, combinations I and Isn) and a RT-PCR–nested PCR (combinations II and IIn) (Fig. 1). Testing of the specificity of the RT-PCR primers revealed the expected 354-bp amplicon for all 10 WEE and slightly smaller amplicons for Mucambo, Pixuna, and Babanki viruses for RT-PCR I (Fig. 3A). The second primer pair (II) amplified only the 10 WEE by RT-PCR (not shown). In the subsequently performed seminested or nested round of amplification, either combination (Isn or IIn) amplified the expected band (195 or 506 bp, respectively) specifically for the 10 WEE strains tested (shown for Isn in Fig. 3B). Although the first RT-PCR (I) was only WEE specific in conjunction with the seminested PCR (Isn), it was found to be 300 times more sensitive than the WEE-specific combinations II plus IIn. The detection limits were 6 × 106 and 6 × 103 RNA molecules for combinations II plus IIn, respectively. The RT-PCR of combination I detected 2 × 103 RNA molecules. This sensitivity was increased by a factor of 100 in the seminested PCR (Isn), allowing the detection of as few as 20 RNA molecules (Fig. 4).
FIG. 3.
Ethidium bromide-stained agarose gels showing the results of WEE-specific amplification. A 10-μl aliquot of each reaction was loaded onto 2% agarose gels. Amplicon sizes are depicted in the margins. (A) Amplification of the WEE-specific 354-bp product resulting from RT-PCR I of 10 different WEE strains (see Table 1) and the closely related alphaviruses VEE-IAB strain Trinidad donkey (TRD), VEE-IIIA strain Mucambo (MUC), VEE-IV strain Pixuna (PIX), EEE strain NJ/60, Aura, Babanki (BBK), Buggy Creek (BCK), Highlands J (HJ), and Fort Morgan (FM). The negative control contained water instead of RNA in the reaction. While all 10 WEE strains showed the amplicon with the expected size, MUC, PIX, and BBK showed a band smaller than 354 bp. (B) The WEE-specific 195-bp amplification products resulting from seminested PCR Isn of the amplicons derived from RT-PCR I shown in Fig. 3A (combinations I and Isn). The negative control contained 2 μl of the RT-PCR I negative control reaction. In this subsequent reaction the 195-bp amplicon was visible only for the WEE strains, rendering the I plus Isn combinations WEE specific.
FIG. 4.
Serial dilutions containing 2 × 109 to 2 RNA molecules of WEE strain Fleming used as a template for WEE-specific RT-PCR I (A) and subsequently performed seminested PCR Isn (B). After RT-PCR, the expected 354-bp amplification product was visible down to the reaction containing 2,000 RNA molecules (A). After the subsequently performed seminested PCR Isn, the 195-bp amplicon was visible on an ethidium bromide-stained agarose gel down to the dilution containing 20 RNA molecules, enhancing the detection limit by a factor of 100 (B).
VEE-specific RT-PCR and seminested PCR.
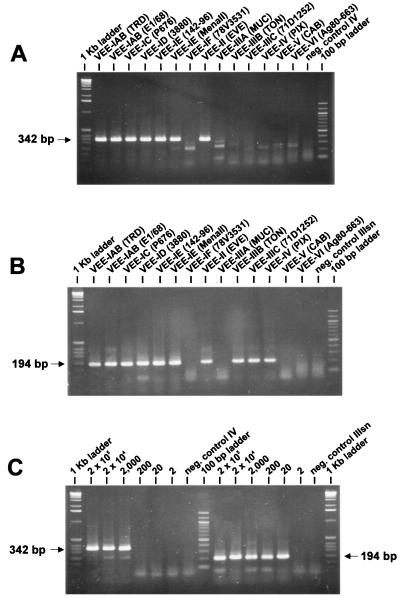
All five primer combinations (Fig. 1) exclusively amplified VEE and no other alphavirus when used in RT-PCRs (not shown). However, their reactivity with the various VEE subtypes differed considerably. While all five RT-PCRs amplified VEE-IAB, -IC, -ID, and -II equally well, RNA of VEE-IE strain Mena II was amplified by RT-PCRs I and II to a much lower extent. None of the five RT-PCRs amplified the RNA of VEE-IF strain 78V-3531. RT-PCR I was the only RT-PCR that amplified the remaining VEE subtypes. RT-PCR II additionally amplified the three varieties of VEE subtype III (IIIA, IIIB, and IIIC), while RT-PCR III additionally amplified RNA of VEE-IIIB, -IIIC, and -IV. Only RT-PCRs IV and V reacted specifically with the subtype viruses (VEE-IAB, -IC, -ID, -IE, and -II) that the primers had been designed for (shown for RT-PCR IV in Fig. 5A). Smaller bands of lighter intensity of about 100, 250, and 300 bp were produced with the other VEE subtypes tested (Fig. 5A). Because detection of VEE-IE is important in light of the recent outbreaks in Mexico, RT-PCRs I and II were not chosen for further testing. For a subsequent round of amplification, three seminested PCRs using the templates from RT-PCR IV or V were possible, i.e., IIIsn (194 bp) out of IV or V and IVsn (342 bp) out of V. Cross-reactivity among the VEE subtypes existed with the first combinations (IV and IIIsn) for VEE-IIIB, -IIIC, and -IV (Fig. 5B), which resembled the reaction pattern of RT-PCR III. In the combinations V plus IIIsn, the 194-bp band was additionally produced for VEE subtypes IIIB and IV, while the second combinations (V plus IVsn) did not amplify RNA of VEE subtypes other than VEE-IAB to IE, and II (data not shown). We determined the sensitivity of combinations IV plus IIIsn and V plus IIIsn for both VEE-IAB strain Trinidad donkey (TRD) and for VEE-IE strain Mena II. The sensitivity for VEE-IAB was the same for both RT-PCR and seminested-PCR combinations, allowing the amplification of 20 RNA molecules of strain TRD (shown for IV plus IIIsn in Fig. 5C). For the amplification of VEE-IE strain Mena II, combinations IV plus IIIsn detected 70 RNA molecules of VEE subtype variety IE. This was 10 times more sensitive than the V and IIIsn combinations with a detection limit of 700 RNA molecules.
FIG. 5.
Ethidium bromide-stained agarose gels showing the results of VEE-specific amplification. A 10-μl aliquot of each reaction was loaded onto 2% agarose gels. Amplicon sizes are depicted in the margins. (A) Amplification of the VEE-specific 342-bp product resulting from RT-PCR IV (see Table 3) of 12 VEE subtype varieties. The negative control contained water instead of RNA in the reaction. Only the VEE-IAB to -IE and the VEE-II strains showed the specific amplification product. The remaining VEE subtypes displayed a weak band ca. 100 bp shorter than that of the VEE subtypes above. (B) VEE-specific amplification products of 194 bp resulting from seminested PCR IIIsn of the amplicons derived from RT-PCR IV shown in Fig. 5A. The negative control contained 2 μl of the RT-PCR IV negative control reaction. Besides the VEE subtypes demonstrating a specific amplification during RT-PCR IV, three VEE subtypes (IIIB, IIIC, and IV) showed the specific 194-bp amplicon. (C) Serial dilutions containing 2 × 105 to 2 RNA molecules of VEE-IAB strain Trinidad donkey (TRD) used as a template for VEE-specific RT-PCR IV (left) and subsequently performed seminested PCR IIIsn (right). The 194-bp amplification product was visible on an ethidium bromide-stained agarose gel down to the dilution containing 20 RNA molecules of VEE-IAB strain TRD.
VEE-specific combinations IV plus IIIsn were successfully applied to serum samples known to be VEE positive by virus isolation. By using RT-PCR IV, one serum sample (IQT 8131) showed the specific 342-bp amplicon. All samples that were once passaged in cell cultures or suckling mouse brain displayed the specific 342-bp band as well (Fig. 6A). After the subsequent seminested PCR (IIIsn), all samples, including the serum samples that had been negative after RT-PCR, showed the VEE-specific 194-bp amplicon (Fig. 6B).
FIG. 6.
Ethidium bromide-stained agarose gels showing the results of VEE-specific amplification as applied to the VEE strains and serum samples listed in Table 2. A 10-μl aliquot of each reaction was loaded onto the 2% agarose gels. Amplicon sizes are depicted in the margin. (A) Amplification of the VEE-specific 342-bp product resulting from RT-PCR IV (see Table 3) was successful with RNA of all VEE isolates from Peru and Mexico and with RNA of serum sample IQT 8131. The negative control contained RNA of a Mayaro virus-positive human serum sample in the reaction. (B) In addition to the samples found to be positive by RT-PCR alone, all samples showed the VEE-specific 194-bp amplification products resulting from seminested PCR IIIsn of the amplicons derived from RT-PCR IV shown in Fig. 6A. The negative control contained 2 μl of the RT-PCR IV-negative control reaction.
DISCUSSION
The recent emergence and reemergence of equine encephalitis viruses, in particular of VEE subtype varieties IC, ID, and IE in Central and South America, is an important health concern (22, 23, 29, 30, 44, 49). Because of the high epidemic potential of these viruses, rapid confirmatory diagnosis is critical in order to quickly launch countermeasures such as quarantine, vaccination, and mosquito control (37). High-titer viremias in equine encephalitis virus-infected individuals are rather short-lived, lasting only 3 to 5 days (36). Because these alphaviruses are able to infect a variety of different hosts, there is little to no growth restriction in the usual cell culture systems or suckling mice, making virus isolation the “gold standard” for antigen detection (24). However, virus isolation is time-consuming and requires sterile handling of tissue cultures under biosafety level 3 conditions. Further, crude mosquito extracts are known to hamper virus isolation (33, 40, 41). In contrast, molecular techniques such as the RT-PCR can yield results within 1 to 2 working days. RT-PCR has successfully amplified viral RNA from EEE-infected mosquitos and bird tissues (1, 40) and WEE-infected mosquitos (41). Recently performed evaluations of the specificity of the latter two assays have revealed that they also efficiently amplify other alphaviruses, i.e., Buggy Creek virus and several subtypes of VEE (19). This is not surprising given the 50% or greater amino acid identities among the structural proteins of the three equine encephalitis viruses and the current lack of sequence data for all known alphavirus species (39). Hence, the specificities of the assays presented here were tested with representative strains of all 27 alphavirus species (21). To further ascertain the specificity of an RT-PCR, subsequent reactions with the amplicon, such as restriction enzyme digestion, hybridization with specific DNA probes, or nested PCR should be performed (1, 25, 34, 41). We chose the latter option because a single nucleotide substitution within the amplicon may affect the cleavage site of a restriction enzyme but may not adversely affect hybridization or nested-PCR priming. By using RT-PCR–nested-PCR combinations, we rescued the specificities for EEE using combinations I plus In and for WEE using combinations I plus Isn. Armstrong and coworkers (1) achieved EEE specificity after dot hybridization with an internal probe, whereas the corresponding amplicon of WEE did not react. The EEE-specific assay described here was able to amplify the RNA of two EEE isolates from Brazil. This is in contrast to the EEE-specific assay described by Armstrong and coworkers, which failed to detect the RNA of EEE strains from Panama and Brazil (16). Because there was no heterologous amplification in the subsequent nested PCRs, we did not investigate the origin of the nonspecific Aura virus-derived amplicon in the EEE-specific RT-PCR I or the reaction of the MUC, PIX, and BBK strains with WEE RT-PCR I. Amplicons that differ in size from the specific PCR product have been observed with various bacterial DNAs or mosquito-borne flaviviral RNAs in a recently described panel of VEE-specific RT-PCR assays (2). Such circumstances require additional tests to confirm the amplicon's origin. By using EEE combinations I plus In and WEE combinations I plus Isn, we not only demonstrated virus specificity but also enhanced the sensitivity 100-fold. This increase of the detection limit corresponds to the findings of Sellner et al. (34) and Hörling et al. (13), who published specific RT-PCR–nested-PCR combinations for Ross River and Ockelbo viruses. Applying the calculation that 3,000 RNA molecules are equal to 1.3 PFU of Ross River virus (34) to the three equine encephalitis viruses, the methods described here are likely to exceed the sensitivity of enzyme immunoassay-based antigen detection procedures (11, 32). The differences in the sensitivity of the VEE-specific combinations IV plus IIIsn to detect IAB and IE subtype varieties might be due to a mismatch of up to three nucleotides at the 5′ end of each primer to the sequence of VEE-IE strain Mena II (14). Interestingly, these differences are reflected by the VEE subtype-specific reactivity of the primer pairs described by Brightwell et al. (2). Based on their results, VEE-IF strain 78V-3531 reacted like VEE-VI strain Ag80-663. This further supports recent studies suggesting that the antigenetically classified VEE-IF strain is genetically closer related to VEE-VI (14, 27, 45). Since our assay focused on VEE-IAB, -IC, -ID, and -II, the VEE-specific combinations IV and IIIsn are most probably not suitable for the screening of mosquito pools in enzootic foci because the enzootic VEE subtypes are either not detected or are amplified with poor sensitivity. However, the specific assays described here may be useful in rapidly detecting and elucidating the occurrence, spread, and epidemiology of the medically important equine encephalitis viruses. Although it was only applied to a limited number of serum samples, the VEE-specific combinations IV plus IIIsn proved to be as sensitive as virus isolation but were completed after 2 working days.
ACKNOWLEDGMENTS
We thank Grit Kermes for expert technical assistance. We are indebted to Moises Fraire, Comision Mexico-Estados Unidos Para Aftosa, and Rebeca Rico-Hesse, Southwest Foundation for Biomedical Research, San Antonio, Tex., for providing the RNA of VEE-IE strain 142-96.
This work was supported by the Fraunhofer Gesellschaft (no. 2087-V-4390), Munich, Germany, and by the U.S. Naval Medical Research and Development Command NNMC, Bethesda, Md., work unit no. 62787A8701612.
REFERENCES
- 1.Armstrong P, Borovsky D, Shope R E, Morris C D, Mitchell C J, Karabatsos N, Komar N, Spielman A. Sensitive and specific colorimetric dot assay to detect eastern equine encephalomyelitis viral RNA in mosquitos (Diptera: Culicidae) after polymerase chain reaction amplification. J Med Entomol. 1995;32:42–52. doi: 10.1093/jmedent/32.1.42. [DOI] [PubMed] [Google Scholar]
- 2.Brightwell G, Brown J M, Coates D M. Genetic targets for the detection and identification of Venezuelan equine encephalitis viruses. Arch Virol. 1998;143:731–742. doi: 10.1007/s007050050326. [DOI] [PubMed] [Google Scholar]
- 3.Calisher C H, Karabatsos N, Lazuick J S, Monath T P, Wolff K L. Reevaluation of the western equine encephalitis antigenic complex of alphaviruses (family Togaviridae) as determined by neutralization tests. Am J Trop Med Hyg. 1988;38:447–452. doi: 10.4269/ajtmh.1988.38.447. [DOI] [PubMed] [Google Scholar]
- 4.Calisher C H, Kinney R M, de Souza Lopes O, Trent D W, Monath T P, France D B. Identification of a new Venezuelan equine encephalitis virus from Brazil. Am J Trop Med Hyg. 1982;31:1260–1272. doi: 10.4269/ajtmh.1982.31.1260. [DOI] [PubMed] [Google Scholar]
- 5.Calisher C H, Monath T P, Sabattini M S, Cropp C B, Kerschner J, Hunt A R, Lazuick J S. Arbovirus investigations in Argentinia, 1977–1980. III. Identification and characterization of viruses isolated, including new subtypes of western and Venezuelan equine encephalitis viruses and four new bunyaviruses (Las Malayos, Resistencia, Barranqueras, and Antequera) Am J Trop Med Hyg. 1985;34:956–965. [PubMed] [Google Scholar]
- 6.Chamberlain R W, Sudia W D, Coleman P H, Work T H. Venezuelan equine encephalitis virus from south Florida. Science. 1964;145:272–274. doi: 10.1126/science.145.3629.272. [DOI] [PubMed] [Google Scholar]
- 7.Chang G-J J, Trent D W. Nucleotide sequence of the genome region encoding the 26S mRNA of eastern equine encephalomyelitis virus and the deduced amino acid sequence of the viral structural proteins. J Gen Virol. 1987;68:2129–2142. doi: 10.1099/0022-1317-68-8-2129. [DOI] [PubMed] [Google Scholar]
- 8.Faragher S G, Meek A D J, Rice C M, Dalgarno L. Genome sequences of a mouse-avirulent and a mouse-virulent strain of Ross River virus. Virology. 1988;163:509–526. doi: 10.1016/0042-6822(88)90292-9. [DOI] [PubMed] [Google Scholar]
- 9.France J K, Wyrick B C, Trent D W. Biochemical and antigenic comparison of the envelope glycoproteins of Venezuelan equine encephalomyelitis strains. J Gen Virol. 1979;44:725–740. doi: 10.1099/0022-1317-44-3-725. [DOI] [PubMed] [Google Scholar]
- 10.Garoff H, Frischauf A-M, Simons K, Lehrbach H, Delius H. Nucleotide sequence of cDNA coding for Semliki Forest virus membrane glycoproteins. Nature (London) 1980;288:236–241. doi: 10.1038/288236a0. [DOI] [PubMed] [Google Scholar]
- 11.Greiser-Wilke I M, Moennig V, Kaaden O-R, Shope R E. Detection of alphaviruses in a genus-specific antigen capture enzyme immunoassay using monoclonal antibodies. J Clin Microbiol. 1991;29:131–137. doi: 10.1128/jcm.29.1.131-137.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hahn C S, Lustig S, Strauss E G, Strauss J H. Western equine encephalitis virus is a recombinant virus. Proc Natl Acad Sci USA. 1988;85:5997–6001. doi: 10.1073/pnas.85.16.5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hörling J, Vene S, Franzén C, Niklasson B. Detection of Ockelbo virus RNA in skin biopsies by polymerase chain reaction. J Clin Microbiol. 1993;31:2004–2009. doi: 10.1128/jcm.31.8.2004-2009.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kinney R M, Pfeffer M, Tsuchiya K R, Chang G-J J, Roehrig J T. Nucleotide sequences of the 26S mRNAs of the viruses defining the Venezuelan equine encephalitis antigenic complex. Am J Trop Med Hyg. 1998;59:952–964. doi: 10.4269/ajtmh.1998.59.952. [DOI] [PubMed] [Google Scholar]
- 15.Kinney R M, Trent D W, France J K. Comparative immunological and biochemical analyses of viruses in the Venezuelan equine encephalitis complex. J Gen Virol. 1983;64:135–147. doi: 10.1099/0022-1317-64-1-135. [DOI] [PubMed] [Google Scholar]
- 16.Kuno G. Universal diagnostic RT-PCR protocol for arboviruses. J Virol Methods. 1998;72:27–41. doi: 10.1016/s0166-0934(98)00003-2. [DOI] [PubMed] [Google Scholar]
- 17.Levinson R S, Strauss J H, Strauss E G. Complete sequence of the genomic RNA of O'Nyong-Nyong virus and its use in the construction of alphavirus phylogenetic trees. Virology. 1990;175:110–123. doi: 10.1016/0042-6822(90)90191-s. [DOI] [PubMed] [Google Scholar]
- 18.Lewis J G, Chang G J, Lanciotti R S, Trent D W. Direct sequencing of large flavivirus PCR products for analysis of genome variation and molecular epidemiological investigations. J Virol Methods. 1992;38:11–24. doi: 10.1016/0166-0934(92)90165-a. [DOI] [PubMed] [Google Scholar]
- 19.Linssen B, Kinney R M, Kaaden O-R, Pfeffer M. In: Specific detection of equine encephalitis viruses by RT-PCR. Wernery U, Wade J F, Mumford J A, Kaaden O-R, editors. Newmarket, United Kingdom: Equine infectious diseases VIII. R & W Publications, Ltd.; 1999. pp. 280–285. [Google Scholar]
- 20.Monath T P, Lazuick J S, Cropp C B, Rush W A, Calisher C H, Kinney R M, Trent D W, Kemp G E, Bowen G S, France D B. Recovery of Tonate virus (“Bijou Bridge” strain), a member of the Venezuelan equine encephalomyelitis virus complex, from Cliff Swallow nest bugs (Oeciacus vicarium) and nestling birds in North America. Am J Trop Med Hyg. 1980;29:969–983. doi: 10.4269/ajtmh.1980.29.969. [DOI] [PubMed] [Google Scholar]
- 21.Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D. Arch. Virol. Suppl. 10:428–433. 1995. Virus taxonomy: classification and nomenclature of viruses. Sixth report of the International Committee on Taxonomy of Viruses. [Google Scholar]
- 22.Oberste M S, Fraire M, Navarro R, Zepeda C, Zarate M L, Ludwig G V, Kondig J F, Weaver S C, Smith J F, Rico-Hesse R. Association of Venezuelan equine encephalitis virus subtype IE with two equine epizootics in Mexico. Am J Trop Med Hyg. 1998;59:100–107. doi: 10.4269/ajtmh.1998.59.100. [DOI] [PubMed] [Google Scholar]
- 23.Oberste M S, Weaver S C, Watts D M, Smith J F. Identification and genetic analysis of Panama-genotype Venezuelan equine encephalitis virus subtype ID in Peru. Am J Trop Med Hyg. 1998;58:41–46. doi: 10.4269/ajtmh.1998.58.41. [DOI] [PubMed] [Google Scholar]
- 24.Office International des Epizooties. Manual of standards for diagnostic tests and vaccines. OIE Standards Commission (ed.) 3rd ed. Paris, France: OIE; 1996. Venezuelan equine encephalomyelitis; pp. 452–456. [Google Scholar]
- 25.Pfeffer M, Proebster B, Kinney R M, Kaaden O-R. Genus-specific detection of alphaviruses by a semi-nested reverse transcription-polymerase chain reaction. Am J Trop Med Hyg. 1997;57:709–718. doi: 10.4269/ajtmh.1997.57.709. [DOI] [PubMed] [Google Scholar]
- 26.Pfeffer M, Wiedmann M, Batt C A. Applications of DNA amplification methods in veterinary diagnostics. Vet Res Commun. 1995;19:375–407. doi: 10.1007/BF01839319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Powers A M, Oberste M S, Brault A C, Rico-Hesse R, Schmura S M, Smith J F F, Kang W, Sweeney W P, Weaver S C. Repeated emergence of epidemic/epizootic Venezuelan equine encephalitis from a single genotype of enzootic subtype ID virus. J Virol. 1997;71:6697–6705. doi: 10.1128/jvi.71.9.6697-6705.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reisen W K, Monath T P. Western equine encephalomyelitis. In: Monath T P, editor. The arboviruses: epidemiology and ecology. V. Boca Raton, Fla: CRC Press; 1988. pp. 89–137. [Google Scholar]
- 29.Rico-Hesse R, Weaver S C, De Singer J, Medina G, Salas R A. Emergence of a new epidemic/epizootic Venezuelan equine encephalitis virus in South America. Proc Natl Acad Sci USA. 1995;92:5278–5281. doi: 10.1073/pnas.92.12.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rivas F, Diaz L A, Cardenas V M, Daza E, Bruzon L, Alcala A, de la Hoz O, Caceres F M, Aristizabal G, Martinez J W, Revelo D, de la Hoz F, Boshell J, Camacho T, Calderon L, Olano V A, Villarreal L I, Roselli D, Alvarez G, Ludwig G, Tsai T. Epidemic Venezuelan equine encephalitis in La Guarjira, Colombia, 1995. J Infect Dis. 1997;174:828–832. doi: 10.1086/513978. [DOI] [PubMed] [Google Scholar]
- 31.Rümenapf T, Strauss E G, Strauss J H. Aura virus is a New World representative of Sindbis-like viruses. Virology. 1995;208:621–633. doi: 10.1006/viro.1995.1193. [DOI] [PubMed] [Google Scholar]
- 32.Scott T W, Olson J G. Detection of eastern equine encephalomyelitis viral antigen in avian blood by enzyme immunoassay: a laboratory study. Am J Trop Med Hyg. 1986;35:611–618. doi: 10.4269/ajtmh.1986.35.611. [DOI] [PubMed] [Google Scholar]
- 33.Sellner L N, Coelen R J, Mackenzie J S. A one-tube, one manipulation RT-PCR reaction for detection of Ross River virus. J Virol Methods. 1992;40:255–264. doi: 10.1016/0166-0934(92)90084-q. [DOI] [PubMed] [Google Scholar]
- 34.Sellner L N, Coelen R J, Mackenzie J S. Sensitive detection of Ross River virus—a one tube nested RT-PCR. J Virol Methods. 1994;49:47–58. doi: 10.1016/0166-0934(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 35.Shirako Y, Niklasson B, Dalrymple J M, Strauss E G, Strauss J H. Structure of the Ockelbo virus genome and its relationship to other Sindbis viruses. Virology. 1991;182:753–764. doi: 10.1016/0042-6822(91)90616-j. [DOI] [PubMed] [Google Scholar]
- 36.Shope R E. Medical significance of togaviruses: an overview of the diseases caused by togaviruses in man and in domestic and wild vertebrate animals. In: Schlesinger R W, editor. The togaviruses. New York, N. Y: Academic Press; 1980. pp. 47–83. [Google Scholar]
- 37.Smith J F, Davis K, Hart M K, Ludwig G V, McClain D J, Parker M D, Pratt W D. Viral encephalitides. In: Zajtchuk R, editor. Textbook of military medicine part I: warfare, weaponry, and the casualty. Medical aspects of chemical and biological warfare. Washington, D. C.: Office of the Surgeon General, Department of the Army; 1997. pp. 561–589. [Google Scholar]
- 38.Strauss E G, Rice C M, Strauss J H. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology. 1984;133:92–110. doi: 10.1016/0042-6822(84)90428-8. [DOI] [PubMed] [Google Scholar]
- 39.Strauss J H, Strauss E G. Alphaviruses. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vodkin M H, McLaughlin G L, Day J F, Shope R E, Novak R J. A rapid diagnostic assay for eastern equine encephalomyelitis viral RNA. Am J Trop Med Hyg. 1993;49:772–776. doi: 10.4269/ajtmh.1993.49.772. [DOI] [PubMed] [Google Scholar]
- 41.Vodkin M H, Streit T, Mitchell C J, McLaughlin G L, Novak R J. PCR-based detection of arboviral RNA from mosquitos homogenized in detergent. BioTechniques. 1994;17:114–116. [PubMed] [Google Scholar]
- 42.Walton T E, Alvarez O, Jr, Buckwalter R M, Johnson K M. Experimental infection of horses with enzootic and epizootic strains of Venezuelan equine encephalomyelitis virus. J Infect Dis. 1973;128:272–282. doi: 10.1093/infdis/128.3.271. [DOI] [PubMed] [Google Scholar]
- 43.Watts D M, Callahan J, Rossi C, Oberste M S, Roehrig J T, Wooster M T, Smith J F, Cropp C B, Gentrau E M, Karabatsos N, Gubler D, Hayes C G. Venezuelan equine encephalitis febrile cases among humans in the Peruvian Amazon River region. Am J Trop Med Hyg. 1998;58:35–40. doi: 10.4269/ajtmh.1998.58.35. [DOI] [PubMed] [Google Scholar]
- 44.Watts D M, Lavera V, Callahan J, Rossi C, Oberste M S, Roehrig J T, Cropp C B, Karabatsos N, Smith J F, Gubler D J, Wooster M T, Nelson W M, Hayes C G. Venezuelan equine encephalitis and Oropouche virus infections among Peruvian army troops in the Amazon region of Peru. Am J Trop Med Hyg. 1997;56:661–667. doi: 10.4269/ajtmh.1997.56.661. [DOI] [PubMed] [Google Scholar]
- 45.Weaver S C, Bellew L A, Rico-Hesse R. Phylogenetic analysis of alphaviruses in the Venezuelan equine encephalitis complex and identification of epizootic viruses. Virology. 1992;191:282–290. doi: 10.1016/0042-6822(92)90190-z. [DOI] [PubMed] [Google Scholar]
- 46.Weaver S C, Hagenbaugh A, Bellew L A, Gousset L, Mallampalli V, Holland J J, Scott T W. Evolution of alphaviruses in the eastern equine encephalomyelitis complex. J Virol. 1994;68:158–169. doi: 10.1128/jvi.68.1.158-169.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weaver S C, Hagenbaugh A, Bellew L A, Netesov S V, Volchkov V E, Chang G-J J, Clarke D K, Gousset L, Scott T W, Trent D W, Holland J J. A comparison of the nucleotide sequence of eastern and western equine encephalomyelitis viruses with those of other alphaviruses and related RNA viruses. Virology. 1993;197:375–390. doi: 10.1006/viro.1993.1599. [DOI] [PubMed] [Google Scholar]
- 48.Weaver S C, Kang W, Shirako Y, Rümenapf T, Strauss E G, Strauss J H. Recombinational history and molecular evolution of western equine encephalomyelitis complex alphaviruses. J Virol. 1997;71:613–623. doi: 10.1128/jvi.71.1.613-623.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weaver S C, Salas R, Rico-Hesse R, Ludwig G V, Oberste M S, Boshell J, Tesh R B for the VEE Study Group. Re-emergence of epidemic Venezuelan equine encephalomyelitis in South America. Lancet. 1996;348:436–440. doi: 10.1016/s0140-6736(96)02275-1. [DOI] [PubMed] [Google Scholar]
- 50.Weaver S C, Scott T W, Rico-Hesse R. Molecular evolution of eastern equine encephalomyelitis virus in North America. Virology. 1991;182:774–784. doi: 10.1016/0042-6822(91)90618-l. [DOI] [PubMed] [Google Scholar]
- 51.Young N A, Johnson K M. Antigenic variants of Venezuelan equine encephalitis virus: their geographic distribution and epidemiologic significance. Am J Epidemiol. 1969;89:286–307. doi: 10.1093/oxfordjournals.aje.a120942. [DOI] [PubMed] [Google Scholar]