Abstract
We describe a multiplex reverse transcription-PCR (m-RT-PCR) assay that is able to detect and differentiate all known human parainfluenza viruses (HPIVs). Serial dilution experiments with reference strains that compared cell culture isolation and m-RT-PCR showed sensitivities ranging from 0.0004 50% tissue culture infective dose (TCID50) for HPIV type 4B (HPIV-4B) to 32 TCID50s for HPIV-3. As few as 10 plasmids containing HPIV PCR products could be detected in all cases. When 201 nasopharyngeal aspirate specimens from pediatric patients hospitalized for lower respiratory illness were tested, m-RT-PCR assay detected 64 HPIVs (24 HPIV-3, 23 HPIV-1, 10 HPIV-4, and 7 HPIV-2), while only 42 of them (21 HPIV-1, 14 HPIV-3, 6 HPIV-2, and 1 HPIV-4 isolates) grew in cell culture. Our m-RT-PCR assay was more sensitive than either cell culture isolation or indirect immunofluorescence with monoclonal antibodies for the detection of HPIV infections. Also, HPIV-4 was more frequently detected than HPIV-2 in this study, suggesting that it may have been underestimated as a lower respiratory tract pathogen because of the insensitivity of cell culture.
Human parainfluenza viruses (HPIVs) are nonsegmented RNA viruses that belong to the Paramyxovirus (HPIV type 1 [HPIV-1] and HPIV-3) and Rubulavirus (HPIV-2 and HPIV-4) genera of the family Paramyxoviridae (23). HPIV-4 is further divided into two subtypes, subtypes A and B, on the basis of antigenic differences (1). HPIV-1, -2, and -3 are important respiratory pathogens and are major causes of croup, bronchiolitis, and pneumonia in infants and very young children (22, 31). They have been estimated to be the cause of 40% of acute respiratory tract illnesses in children from which a virus is recoverable and 20% of respiratory illnesses in hospitalized children (26). HPIV-4 has traditionally been associated with mild upper respiratory tract infections in children and adults (5).
The etiological diagnosis of HPIV infections cannot be based exclusively on clinical signs and symptoms because other pathogens cause similar syndromes. The use of classic diagnostic methods, such as viral isolation and serology, can result in delays of several weeks before test results are available (6). Rapid diagnosis is desirable both to assist the clinician in making therapeutic decisions and to prevent nosocomial infections (6, 21). Direct antigen detection with respiratory specimens provides rapid results, but different methods such as immunofluorescence (16, 25, 29, 32) or enzyme immunoassay (28) have been reported to have variable sensitivities depending on the virus. Molecular techniques based on reverse transcription (RT)-PCR constitute another approach to rapid diagnosis with expected high sensitivity. RT-PCR assays have been applied to the detection of HPIV-1 and HPIV-3 (10, 13, 18) in monospecific assays or the simultaneous amplification of HPIVs with other respiratory viruses (9, 11, 15, 24); multiplex RT-PCR (m-RT-PCR) assays permit the detection of several viruses simultaneously and consume less reagents, samples, and time than single RT-PCR assays, which can be an important consideration for high-volume diagnostic laboratories.
In a previous report (8) we described an m-RT-PCR for the detection of HPIV-1, -2, and -3. In the present work, this assay was evaluated with (i) a more complete panel of clinical samples, (ii) a simplified protocol that used a one-step RT and first PCR amplification, and (iii) an internal control for the detection of ineffective PCR amplification and primers for the detection of HPIV-4. The enlargement of the m-RT-PCR to detect HPIV-4 was motivated by previous reports that suggested that HPIV-4 can be underestimated as a cause of lower respiratory tract disease (20, 27).
MATERIALS AND METHODS
Virus.
Prototype strains of HPIV-1 (strain C35), HPIV-2 (strain Greer), HPIV-3 (strain C-243), HPIV-4A (strain M-25), and HPIV-4B (strain 19.153) were obtained from the Centers for Disease Control and Prevention collections. Wild-type HPIV isolates (two isolates each of HPIV-1, -2, and -3) from cell cultures inoculated with samples from multiple respiratory disease outbreak seasons were obtained from the Spanish National Center for Microbiology archives, as were three individual isolates of influenza A virus (two of subtype H3 and one of subtype H1), three individual isolates of influenza B virus, two individual isolates of adenovirus, two individual isolates of mumps virus, two individual isolates of measles virus, and three individual isolates of respiratory syncytial virus.
Clinical samples.
Two hundred thirty nasopharyngeal aspirate specimens were collected from pediatric patients who had a lower tract respiratory illness and who were recruited for a long-term prospective study of severe respiratory infections. These patients were referred to the emergency room or required hospitalization at the Severo Ochoa Hospital in Leganés (Madrid, Spain). These samples were collected during the periods of maximum HPIV activity detected by viral isolation and antigen detection. One hundred eighty-four specimens obtained from September 1997 to January 1998 were tested retrospectively and 46 specimens obtained from June 1998 to July 1998 were tested prospectively (see below for study design). Specimens were obtained with an aspirator device, placed in viral transport medium, and processed within 24 h of collection. When the specimens arrived in the laboratory, they were diluted to 5 ml with phosphate-buffered saline solution and homogenized before testing. Three 0.5-ml aliquots were stored at −70°C.
IF assay.
The indirect immunofluorescence (IF) assay was performed directly with cells from respiratory secretions that were concentrated by centrifugation and stained by standard methods. Commercial reagents (Chemicon, Temecula, Calif.) were used; however, monoclonal antibody (MAb) specific for HPIV-4 (MAb 531-3F) was obtained from the Centers for Disease Control and Prevention (17). For each MAb the results for two 7-mm-diameter wells were read before a sample was considered negative.
Virus isolation.
Human laryngeal epidermoid carcinoma (HEp-2) cells, human lung mucoepidermoid carcinoma (NCI-H292) cells, Madin-Darby canine kidney (MDCK) cells, and human embryonic lung fibroblast (Fp) cell cultures were used for primary viral isolation. Tubes with 80% confluent monolayers were inoculated with 0.3 ml of homogenized samples. The adsorption of HEp-2, Fp, and MDCK cell cultures was enhanced by centrifugation at 3,000 rpm (Labofuge GL, Heraeus Sepatech) for 45 min. HEp-2 and Fp cells were fed 2 ml of 2% fetal calf serum in Eagle basal medium containing antibiotics. For MDCK cells, Eagle minimal essential medium with antibiotics was supplemented with 3 μg of trypsin per ml. NCI-H292 cells were adsorbed for an hour without centrifugation and were fed Eagle minimal essential medium supplemented with 1.5 μg of trypsin per ml (4). Cell monolayers were observed for cytopathic effect every 48 h. When a cytopathic effect was observed or after 10 days, the monolayer was scraped and tested for respiratory viruses by IF assay, as described above. The monolayers with IF assay-negative culture results were subcultured and again submitted to blind IF assay after 10 days.
Primers.
Primers specific for HPIV-1, -2, and -3 (8) and internal control primers (2) were published previously. For design of the primer specific for HPIV-4, the sequences of the phosphoprotein P gene were obtained from GenBank and were aligned by using the Wisconsin Analysis Package, version 8 (Genetics Computer Group, Madison, Wis.). External generic primers PI4P+ (5′-CTGAACGGTTGCATTCAGGT-3′ [genome sense; bases 11 to 39]) and PI4P− (5′-AGGACTCATTCTTGATGCAA-3′ [genome antisense; bases 433 to 452]) were chosen from regions conserved between the HPIV-4A and HPIV-4B subtypes. A second pair of internal generic primers, PI4S+ (5′-AAAGAATTAGGTGCAACCAGTC-3′ [genome sense; bases 158 to 179]) and PI4S− (5′-GCTGCTTATGGGATCAGACAC-3′ [genome antisense; bases 382-402]) were selected for the nested reaction by using the same criteria described above. Subtype A- and B-specific primers PI4SA+ (5′-ATGATGGTGGAACCAAGATT-3′ [genome sense; bases 226 to 245]) and PIASB+ (5-AACCAGGGAAACAGAGCTC-3′ [genome sense; bases 320 to 339]), whose sequences were within the first amplification fragment, were also selected; PI4P+ annealed to the 5′ noncoding region, and the rest of the primers annealed to the P/V common region of phosphoprotein P (19).
RNA extraction, RT, and primary amplification.
RNA was extracted from clinical samples and virus isolates as described previously (3). Briefly, 50 μl of each sample was treated with 200 μl of guanidinium thiocyanate extraction buffer including 100 molecules of a plasmid with an insert of the polymerase gene of the pseudorabies herpesvirus DNA as an internal control template (2), followed by alcohol precipitations. The pellet was resuspended in 10 μl of RNase-free water. A single-step RT-amplification reaction was performed by using the Promega Access RT-PCR system kit (Promega, Madison, Wis.), which consisted of a PCR mixture containing 3 mM MgSO4, 500 μM each dATP, dGTP, dCTP, and dTTP, 0.5 μM HPIV type 1- to 4-specific primers, 0.2 μM internal control-specific primary reaction primers, 10 μl of avian myeloblastosis virus-Tfl 5× reaction buffer, 5 U of avian myeloblastosis virus reverse transcriptase, and 5 U of Tfl DNA polymerase. The PCR mixtures were overlaid with mineral oil, and 5 μl of extracted RNA was added to a final volume of 50 μl. The tubes were centrifuged for a few seconds and were placed in an Autocycler Plus Termocycler (Linus, Cultek S. L., Spain). Cycling conditions were as published previously (8).
Nested amplification and product detection.
Analytical conditions were as published previously (8), but generic HPIV-4-specific and internal control-specific primers were added.
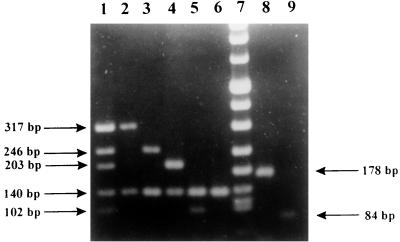
PCR products were sized by gel electrophoresis on 2% agarose. Expected band sizes were 317 bp for HPIV-1, 203 bp for HPIV-2, 102 bp for HPIV-3, 246 bp for HPIV-4, and 140 bp for the internal control (Fig. 1). For subtyping of HPIV-4, a nested reaction that included only primers PIS4A+, PIS4B+, and PIS4− was performed under the same conditions used for the nested PCR mentioned above with the first reaction products. Expected band sizes were 178 bp for HPIV-4A and 84 bp for HPIV-4B (Fig. 1). Positive samples showed the specific HPIV band and the internal control band. When only the internal control band appeared, samples were considered negative. Samples that showed no band were retested, and those that lacked any band after repeat testing were assumed to contain enzyme inhibitors.
FIG. 1.
Typing of HPIVs and subtyping of HPIV-4 by m-RT-PCR: mixture of HPIVs (lane 1), HPIV-1 (lane 2), HPIV-4 (lane 3), HPIV-2 (lane 4), HPIV-3 (lane 5), negative control (lane 6), marker (DNA molecular weight marker VIII; Boehringer Mannheim) (lane 7), HPIV-4A (lane 8), and HPIV-4B (lane 9).
Standard precautions were taken throughout the procedure to avoid cross-contamination. Negative and low-titer positive controls were included in every assay. All positive results were confirmed by retesting a different aliquot of the specimen. In case of disagreement of the results, a third m-RT-PCR was performed to resolve the results.
PCR product cloning.
Primary amplification products from prototype strains of HPIVs were purified by using the GeneClean II kit and were ligated into pGEM-T plasmid vectors with the pGEM-T plasmid vector system (Promega) by following the manufacturer's directions. Plasmids were transformed into high-efficiency competent cells (Epicurian coli XL1-Blue; Stratagene Cloning Systems, La Jolla, Calif.) by electroporation. Transformants were selected in Luria broth–ampicillin–isopropyl-β-d-thiogalactopyranoside–5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside plates, and the presence of the expected insert was confirmed by PCR. Plasmids were purified with the Wizard Plus SV Minipreps kit (Promega). The number of plasmid copies in the final suspensions was estimated by UV spectroscopy at an optical density of 260 nm.
Study design. (i) Prospective panel.
Viral isolation and antigen detection by IF assay with all HPIV-specific MAbs and m-RT-PCR were performed with all fresh specimens (n = 46) that arrived at the laboratory during the study period; the results for m-RT-PCR-positive specimens were confirmed with frozen aliquots.
(ii) Retrospective panel.
Because an HPIV-4-specific MAb was not used for cell culture screening prior to the study, frozen aliquots from all archived specimens (n = 184) were recultured in NCI-H292 cells, and the cultures were rescreened for all HPIVs by IF assay. The results were the same as those obtained with the fresh samples except for the results for one specimen from which HPIV-1 was previously isolated but for which the result was negative on rescreening. Only rescreening results are considered for further analysis. The IF assay was performed only with fresh samples. Consequently, no IF assay results are available for HPIV-4. The m-RT-PCR was performed with frozen aliquots, and the viruses in samples positive for HPIV-4 were subtyped by using the primary amplification products as templates.
RESULTS
Evaluation of m-RT-PCR sensitivity and specificity.
DNA bands of the expected size were obtained by m-RT-PCR with all reference HPIV strains and all wild-type HPIV isolates. No amplification was observed with the other respiratory viruses tested. A comparison of the results of m-RT-PCR with those of virus culture in NCI-H292 cells by using serial 10-fold dilutions of the reference HPIV strains obtained sensitivities of 0.01 50% tissue culture infective dose (TCID50), calculated by the Reed-Muench method, for HPIV-1, 0.02 TCID50 for HPIV-2, 32 TCID50s for HPIV-3, 0.001 TCID50 for HPIV-4A, and 0.0004 TCID50 for HPIV-4B. In dilution experiments with plasmids containing cloned HPIV cDNA, m-RT-PCR was able to detect as few as 10 molecules for all HPIVs.
Evaluation of m-RT-PCR with clinical specimens.
Of the 230 clinical specimens, 22 from the retrospective panel and 7 from the prospective panel were not suitable for use in comparisons of the techniques. Sixteen (6.9%) exhibited microbial contamination in the cell culture, 8 (3.4%) had scarce respiratory tract epithelial cells by IF assay, 1 (0.4%) had both problems, and 4 (1.7%) were suspected of having enzymatic inhibition when tested by m-RT-PCR. Among the samples with contamination in the cell cultures, m-RT-PCR and IF assay detected two HPIVs (one HPIV-1 and one HPIV-3), and m-RT-PCR alone detected five HPIVs (two HPIV-1, one HPIV-2, and two HPIV-3). Both culture and m-RT-PCR detected one HPIV-1 and one HPIV-3 in samples for which results were not available by IF assay. One HPIV-3 in the specimen with both problems was amplified by m-RT-PCR. No HPIVs were detected by any other technique in samples for which the m-RT-PCR was inhibited.
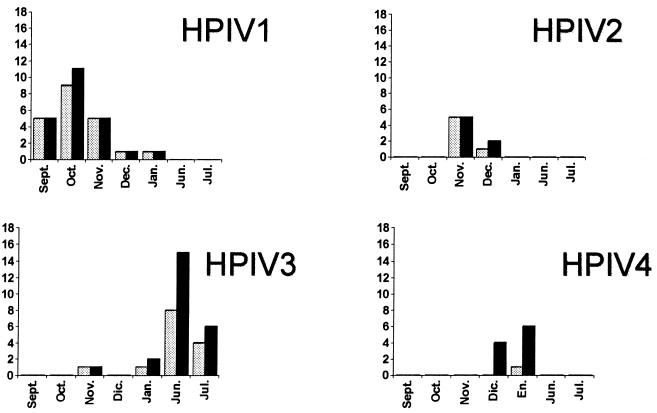
The remaining 201 samples for which results were available by all techniques were analyzed further. For all samples, the same virus was detected by m-RT-PCR and culture. Other respiratory viruses were identified in 50 specimens (Table 1). Similar monthly distributions of positive results for the different HPIVs were observed by cell culture isolation and m-RT-PCR (Fig. 2).
TABLE 1.
Results obtained for 201 nasopharyngeal aspirates in the prospective and retrospective panels with suitable results by tissue culture, IF assay, and m-RT-PCR
| No. of
specimens
|
Combinations of results
founda
|
Distribution of HPIVs (no.
of specimens)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Retrospective panel (n = 162) | Prospective panel (n = 39) | Culture | IF assayb | m-RT-PCR | Retrospective
panel
|
Prospective panel, HPIV-3 | |||
| HPIV-1 | HPIV-2 | HPIV-3 | HPIV-4 | ||||||
| 19 | 7 | + | + | + | 16 | 2 | 1 | 0 | 7 |
| 12 | 5 | + | − | + | 5 | 4 | 1 | 1 | 5 |
| 13 | 9 | − | − | +c | 2 | 1 | 1 | 9 | 9 |
| 0 | 1 | − | + | − | 0 | 0 | 0 | 0 | NTd |
| 118e | 16f | − | − | − | 0 | 0 | 0 | 0 | 0 |
| Total | 42 | 27b | 56 | 23 | 7 | 3 | 10 | 22 | |
+, positive result; −, negative result.
An MAb to HPIV-4 only was not included in the retrospective panel (see text).
In three samples positive for HPIV-3 by m-RT-PCR and one sample positive for HPIV-4 by m-RT-PCR, three adenoviruses and 1 respiratory syncytial virus were isolated, respectively.
NT, not tested.
The panel included 45 samples positive for other respiratory viruses (36 for respiratory syncytial viruses, 5 for adenoviruses, and 4 for influenza A viruses).
The panel included five samples positive for other respiratory viruses (four for adenovirus, and one for influenza B virus).
FIG. 2.
Comparison of the temporal distribution between culture-positive ( ) and m-RT-PCR-positive (■) results. Dic., December; En., January.
HPIVs were recovered from 32 of the 162 suitable samples belonging to the retrospective panel; HPIV-1 from 21 (12.9%), HPIV-2 from 6 (3.7%), HPIV-3 from 2 (1.2%), and HPIV-4 from 1 (0.6%). The IF assay detected 16 (9.8%) HPIV-1, 2 (1.2%) HPIV-2, and 1 (0.6%) HPIV-3. m-RT-PCR yielded 23 (14.2%) HPIV-1, 6 (3.7%) HPIV-2, 3 (1.8%) HPIV-3, and 10 (6.1%) HPIV-4 (Table 1). All 10 HPIV-4 were subtyped as HPIV-4A with the subtype-specific primers. One sample previously positive for respiratory syncytial virus was found to be HPIV-4 positive by m-RT-PCR. Dual infections were not observed by either culture or IF assay.
Only HPIV-3 was detected within the prospective panel. Twelve (30.7%) of the 39 suitable clinical samples yielded viral isolates, with 7 (17.9%) of these being virus positive by IF assays. Twenty-one (53.8%) of the total had a positive result by m-RT-PCR (Table 1). One sample had a positive result by IF assay but negative results by both cell culture isolation and m-RT-PCR. HPIV-3 could be amplified from three samples known to contain adenoviruses. As for the retrospective panel, no coinfection was observed by IF assay or cell culture.
DISCUSSION
Our data demonstrate that all four HPIVs could be detected by m-RT-PCR with clinical specimens. Previously reported m-RT-PCR assays for detection of HPIVs were not designed to detect HPIV-4 (8, 11, 15, 24) or HPIV-2 (15) and did not include an internal control. Although the sensitivity of m-RT-PCR was only 32 TCID50s for HPIV-3, it was able to detect as few as 10 plasmids containing cloned DNAs of all HPIVs. More importantly, the m-RT-PCR assay was able to identify a greater number of positive clinical specimens than IF assay or cell culture. Of the 22 specimens that were positive only by m-RT-PCR, 4 were compromised by coinfection with other faster-growing viruses that could mask HPIVs. Detection of HPIVs in the remaining 18 specimens was likely due to the higher sensitivity of the m-RT-PCR assay; false-positive results caused by cross-contamination could account for this difference, but it seems unlikely, since the temporal distributions of m-RT-PCR-positive and cell culture-positive results matched (Fig. 2), and the results for all m-RT-PCR-positive specimens were confirmed by retesting of a separate aliquot of the specimen. RT-PCR seemed to be a better method for detecting coinfections, which were missed by culture and IF assay, as observed before by others (7, 13).
Inclusion of an internal positive control template in all clinical specimens prevented reporting of false-negative results for 13 specimens (data not shown), illustrating the importance of establishing assay controls specific for each specimen. Four specimens repeatedly inhibited the internal control, probably because of the presence of enzyme inhibitors. The remaining nine specimens were positive on repeat testing, suggesting that handling error was the cause of the first inhibition. Removal of supernatant during the RNA precipitation steps of the sample extraction procedure could be a critical point, since the RNA pellets can be lost. The inclusion of the internal template in the extraction buffer can control for this possibility as well (3). Consequently, enzyme inhibitors or mishandling could account for m-RT-PCR negativity among cell culture-positive samples observed in studies that did not use internal controls (8, 12). Alternatively, unexpected primer mismatches with different virus strains could also account for a lack of reactivity. To address this possibility, our primers specific for HPIV-1, -2, and -3 were designed by use of multiple sequences of each virus and were tested with temporally and geographically diverse isolates (8). However, only one reference strain of each HPIV-4 subtype and only HPIV-4A strains from a single outbreak in Spain were detected. Additional isolates of both HPIV-4 subtypes will be required to complete the evaluation of this method.
Of particular interest was the large number of HPIV-4 isolates identified in the present study by m-RT-PCR. HPIV-4 appears to be the most difficult HPIV to grow in cell culture and is rarely isolated, despite serologic studies showing that it is relatively ubiquitous (5). MAbs to HPIV-4 have only recently become available commercially, which has also hindered identification of this virus (27). A recent study of hospitalized patients identified several patients with severe respiratory illnesses caused by HPIV-4 (20), suggesting that HPIV-4 is not the mild respiratory pathogen once thought. Even though HPIV-1 and HPIV-3 were the most prevalent HPIVs identified in this study, as expected (14, 30), HPIV-4 infections were more frequent than HPIV-2 infections and were associated with severe clinical disease.
In conclusion, our m-RT-PCR assay provides both a sensitive and a specific means of identification of HPIVs in clinical specimens and constitutes a more sensitive alternative to the IF assay as a rapid diagnostic method. It is especially convenient for the detection of HPIV-4 isolates, whose clinical impact may have been underestimated because of the insensitivity of cell culture.
ACKNOWLEDGMENTS
We are very grateful to Francisco Pozo for help with the cloning experiments and Angel del Pozo for photographic work.
This work was supported in part by “Fondo de Investigaciones Sanitarias” grant 98/0310 from the Spanish Ministry of Health.
REFERENCES
- 1.Canchola J, Vargosko A J, Kim H W, Parrot R H, Chritsmas F, Jeffries B, Chanock R M. Antigenic variation among new isolated strains of parainfluenza type 4 virus. Am J Hyg. 1964;79:357–364. doi: 10.1093/oxfordjournals.aje.a120390. [DOI] [PubMed] [Google Scholar]
- 2.Casas I, Tenorio A, Echevarría J M, Klapper P E, Cleator G M. Detection of enteroviral RNA and specific DNA of herpesviruses by multiplex genome amplification. J Virol Methods. 1997;66:39–50. doi: 10.1016/s0166-0934(97)00035-9. [DOI] [PubMed] [Google Scholar]
- 3.Casas I, Powel L, Klapper P E, Cleator G M. New method for the extraction of viral RNA and DNA from cerebrospinal fluid for use in the polymerase chain reaction assay. J Virol Methods. 1995;53:25–36. doi: 10.1016/0166-0934(94)00173-e. [DOI] [PubMed] [Google Scholar]
- 4.Castells E, George V G, Hierholzer J C. NCI-H292 as an alternative cell line for the isolation and propagation of the human paramyxoviruses. Arch Virol. 1990;115:277–288. doi: 10.1007/BF01310536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins P L, Chanock R M, McIntosh K. Parainfluenza viruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publications; 1996. pp. 1205–1241. [Google Scholar]
- 6.Downham M A P S, McQuillin J, Gardner P S. Diagnosis and clinical significance of parainfluenza virus infections in children. Arch Dis Child. 1974;49:8–15. doi: 10.1136/adc.49.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drews A L, Atmar R L, Glezen W P, Baxter B D, Piedra P A, Greenberg S B. Dual respiratory virus infections. Clin Infect Dis. 1997;25:1421–1429. doi: 10.1086/516137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Echevarría J E, Erdman D D, Swierkosz E M, Holloway B P, Anderson L J. Simultaneous detection and identification of human parainfluenza viruses 1, 2, and 3 from clinical samples by multiplex PCR. J Clin Microbiol. 1998;36:1388–1391. doi: 10.1128/jcm.36.5.1388-1391.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eugene-Ruellan G, Freymuth F, Bahloul C, Badrane H, Vabret A, Tordo N. Detection of respiratory syncytial virus A and B and parainfluenzavirus 3 sequences in respiratory tracts of infants by a single PCR with primers targeted to the l-polymerase gene and differential hybridization. J Clin Microbiol. 1998;36:796–801. doi: 10.1128/jcm.36.3.796-801.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan J, Henrickson K J. Rapid diagnosis of human parainfluenza type 1 infection by quantitative reverse transcription-PCR-enzyme hybridization assay. J Clin Microbiol. 1996;34:1914–1917. doi: 10.1128/jcm.34.8.1914-1917.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan J, Henrickson K J, Savatsky L L. Rapid simultaneous diagnosis of infections with respiratory syncytial viruses A and B, influenza viruses A and B, and human parainfluenza virus type 1, 2, and 3 by multiplex reverse transcription-polymerase chain reaction hybridization assay (Hexaplex) Clin Infect Dis. 1998;26:1397–1402. doi: 10.1086/516357. [DOI] [PubMed] [Google Scholar]
- 12.Freymuth F, Vabret A, Galateau-Salle F, Ferey J, Eugene G, Petitjean J, Gennetay E, Brouard J, Jokik M, Duhamel J-F, Guillois B. Detection of respiratory syncytial virus, parainfluenzavirus 3, adenovirus and rhinovirus sequences in respiratory tract of infants by polymerase chain reaction and hybridization. Clin Diagn Virol. 1997;8:31–40. doi: 10.1016/s0928-0197(97)00060-3. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert L L, Dakhama A, Bone B M, Thomas E E, Hegele R G. Diagnosis of viral respiratory tract infections in children by using a reverse transcription-PCR panel. J Clin Microbiol. 1996;34:140–143. doi: 10.1128/jcm.34.1.140-143.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glezen W P, Frank A L, Taber L H, Kasel J A. Parainfluenza virus type 3: seasonality and risk of infection and reinfection in young children. J Infect Dis. 1984;150:851–857. doi: 10.1093/infdis/150.6.851. [DOI] [PubMed] [Google Scholar]
- 15.Gröndahl B, Puppe W, Hoppe A, Kuhne I, Weigl J A, Schmitt H J. Rapid identification of nine microorganisms causing acute respiratory tract infections by single-tube multiplex reverse transcription-PCR: feasibility study. J Clin Microbiol. 1999;37:1–7. doi: 10.1128/jcm.37.1.1-7.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henrickson K J, Kuhn S M, Savatski L L, Sedmak J. Recovery of human parainfluenza virus types one and two. J Virol Methods. 1994;46:189–206. doi: 10.1016/0166-0934(94)90103-1. [DOI] [PubMed] [Google Scholar]
- 17.Hierholzer J C, Bingham G P, Castells E, Coombs R A. Time-resolved fluoroimmunoassay with monoclonal antibodies for rapid identification of parainfluenza type 4 and mumps viruses. Arch Virol. 1993;130:335–352. doi: 10.1007/BF01309665. [DOI] [PubMed] [Google Scholar]
- 18.Karron R A, Frohelich J L, Bobo L, Belshe R B, Yolken R. Rapid detection of parainfluenza virus type 3 RNA in respiratory specimens: use of reverse transcription-PCR-enzyme immunoassay. J Clin Microbiol. 1994;32:484–488. doi: 10.1128/jcm.32.2.484-488.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kondo K, Bando H, Tsurudome M, Kawano M, Nishio M, Ito Y. Sequence analysis of the phosphoprotein (P) genes of human parainfluenza type 4A and 4B viruses and RNA editing at transcript of the P genes: the number of G residues added is imprecise. Virology. 1990;178:321–326. doi: 10.1016/0042-6822(90)90413-l. [DOI] [PubMed] [Google Scholar]
- 20.Lindsquist S W, Darnule A, Istas A, Demmler G J. Parainfluenza virus type 4 infections in pediatric patients. Pediatr Infect Dis J. 1997;16:34–38. doi: 10.1097/00006454-199701000-00008. [DOI] [PubMed] [Google Scholar]
- 21.Moisiuk S E, Robson D, Klass L, Kliewer G, Wasyliuk W, Davi M, Plourde P. Outbreak of parainfluenza virus type 3 in an intermediate care neonatal nursery. Pediatr Infect Dis J. 1998;17:49–53. doi: 10.1097/00006454-199801000-00011. [DOI] [PubMed] [Google Scholar]
- 22.Monto A S. The Tecumseh study of respiratory illness. V. Patterns of infection with the parainfluenzaviruses. Am J Epidemiol. 1973;97:338–348. doi: 10.1093/oxfordjournals.aje.a121514. [DOI] [PubMed] [Google Scholar]
- 23.Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jawis A W, Martelli G P, Mayo M A, Summers M D, editors. Virus taxonomy. Sixth report of the International Committee on Taxonomy of Viruses. Arch Virol Suppl. 1995;10:1–586. [PubMed] [Google Scholar]
- 24.Osiowy C. Direct detection of respiratory syncytial virus, parainfluenza virus, and adenovirus in clinical respiratory specimens by a multiplex reverse transcription-PCR assay. J Clin Microbiol. 1998;36:3149–3154. doi: 10.1128/jcm.36.11.3149-3154.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ray C G, Minnich L L. Efficiency of immunofluorescence for rapid detection of common respiratory viruses. J Clin Microbiol. 1987;25:355–357. doi: 10.1128/jcm.25.2.355-357.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reed G, Jewett P H, Thompson J, Tollefson S, Wright P F. Epidemiology and clinical impact of parainfluenza virus infections in otherwise healthy infants and young children <5 years old. J Infect Dis. 1997;175:807–813. doi: 10.1086/513975. [DOI] [PubMed] [Google Scholar]
- 27.Rubin E E, Quennec P, McDonald J C. Infections due to parainfluenza type 4 in children. Clin Infect Dis. 1993;17:998–1002. doi: 10.1093/clinids/17.6.998. [DOI] [PubMed] [Google Scholar]
- 28.Sarkkinen H K, Halonen P E, Salmi A A. Type specific detection of parainfluenza viruses by enzyme-immunoassay and radioimmunoassay in nasopharyngeal specimens of patients with acute respiratory disease. J Gen Virol. 1981;56:49–57. doi: 10.1099/0022-1317-56-1-49. [DOI] [PubMed] [Google Scholar]
- 29.Stout C, Murphy M D, Lawrence S, Julian S. Evaluation of a monoclonal antibody pool for rapid diagnosis of respiratory viral infections. J Clin Microbiol. 1989;27:448–452. doi: 10.1128/jcm.27.3.448-452.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tellez A, Perez-Breña P, Fernández-Patiño M V, León P, Anda P, Nájera R. Acute respiratory disease in Spain: seven years of experience. Rev Infect Dis. 1990;12:745–753. doi: 10.1093/clinids/12.5.745. [DOI] [PubMed] [Google Scholar]
- 31.Vainionpää R, Hyypiä T. Biology of parainfluenza viruses. Clin Microbiol Rev. 1994;7:265–275. doi: 10.1128/cmr.7.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong D T, Welliver R C, Riddlesberger K R, Sun M S, Ogra P L. Rapid diagnosis of parainfluenzavirus infection in children. J Clin Microbiol. 1982;16:164–167. doi: 10.1128/jcm.16.1.164-167.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]