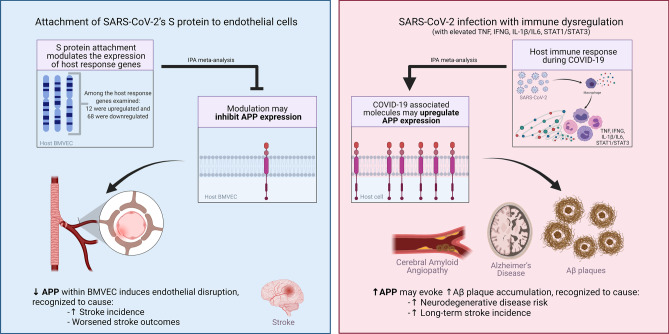
Abstract
SARS-CoV-2 infection begins with the attachment of its spike (S) protein to angiotensin-converting enzyme-2 (ACE2) followed by complex host immune responses with cardiovascular and neurological implications. Our meta-analyses used QIAGEN Ingenuity Pathway Analysis (IPA) and Knowledge Base (QKB) to investigate how the expression of amyloid precursor protein (APP) was modulated by attachment of SARS-CoV-2 S protein in the brain microvascular endothelial cells (BMVECs) and during COVID-19 in progress. Published 80 host response genes reported to be modulated in BMVECs following SARS-CoV-2 S protein binding were used to identify key canonical pathways and intermediate molecules mediating the regulation of APP production following the attachment of S protein to endothelial cells. This revealed that the attachment of SARS-CoV-2 S protein may inhibit APP expression in the BMVECs. Our results shed light on the molecular mechanisms by which SARS-CoV-2 infection may potentiate the incidence of stroke by inhibiting the production of APP in the BMVECs. We also analyzed molecules associated with COVID-19, which revealed six upstream regulators, TNF, IFNG, STAT1, IL1β, IL6, and STAT3. The upstream regulators mediate the increased production of APP via intermediators, with eleven regulated by all six upstream regulators. These COVID-19 upstream regulators increased APP expression with a statistically significant Z-score of 3.705 (p value = 0.000211). These findings have revealed molecular mechanisms by which COVID-19 disease may lead to long-term neurological manifestations resulting from the elevated APP expression in line with immune response in the host. Altogether, our study revealed two distinct scenarios which may have differential impact on APP expression.
Graphic Abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s11481-021-10012-9.
Keywords: COVID-19, Brain microvascular endothelial cells, Blood–brain barrier, Alzheimer’s disease, Neuroinflammation
Introduction
Infection with severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is the cause of coronavirus disease 2019 (COVID-19) in over 150 million people (Organization 2020). Common clinical symptoms of COVID-19 include respiratory issues, headaches, olfactory dysfunction, and delirium, and 2.1% of severe patients may experience acute ischemic strokes (Kennedy et al. 2020; Merkler et al. 2020; Taquet et al. 2021). A study on the post-infection outcomes of discharged patients reported that up to 76% of patients experience at least one of the symptoms six months after their release from the hospital, with 11% reporting loss of smell and 7% reporting taste disorder (Huang et al. 2021a). In addition, many studies have reported neurological pathologies associated with COVID-19, including ischemia-derived central nervous system (CNS) damage and cerebrovascular events, such as cerebral hemorrhagic or encephalitic incidences in hospitalized patients (Ellul et al. 2020; Wu et al. 2020). Furthermore, common COVID-19 pathophysiological processes of hypoxia, cerebral hypoperfusion, cerebrovascular coagulation, cerebral microvascular endothelia damage, dysregulation of the renin-angiotensin system, and encephalitis may also augment the risks for chronic neurological conditions (Miners et al. 2020).
A critical component of cerebrovascular damage is the disintegration of the blood–brain barrier (BBB). In particular, hypoxia/ischemia has been found to contribute to the degradation of BBB through the activation of various matrix metalloproteinases (MMPs), which break down the basal lamina and tight junctions of the endothelium, thereby disrupting the BBB (Yang and Rosenberg 2011). The compromised BBB integrity allows for transport of toxic amyloid-beta (Aβ), which may penetrate BBB and get deposited into the CNS (Deane et al. 2003; Kitazawa et al. 2005; Garwood et al. 2011; Walters et al. 2016). Aβ is a cleaved form of amyloid precursor protein (APP), which is an integral membrane receptor found in many tissues, including brain microvascular cells, and is believed to play a role in cell adhesion, neuronal development, and neural plasticity (d'Uscio et al. 2017; Montagna et al. 2017). Improper APP cleavage creates a toxic form of Aβ monomer that aggregates to form Aβ oligomers and plaques. Aβ plaques are seen in cerebral amyloid angiopathy (CAA), where they accumulate in brain arteries and increase stroke occurrence (Zipfel et al. 2009; Biffi and Greenberg, 2011). Loss of APP homeostasis within the cerebral arteries, through APP deficiency, has also been shown to trigger endothelial cell dysfunction both in vivo and in vitro (d’Uscio et al. 2018; Ristori et al. 2020). Aβ plaques are also found in the brain of Alzheimer’s disease (AD) patients, where they disrupt neural connections and initiate immune responses that further induce neuronal damage (Arai et al. 1991; Kinney et al. 2018). SARS-CoV-2 infection has been reported to alter BBB integrity (Reynolds and Mahajan 2021), and cerebral hypoperfusion may accelerate the accumulation of Aβ (Shang et al. 2019; Miners et al. 2020). Therefore, we hypothesized that APP may be involved in COVID-19-associated neurological manifestations, and this study was designed to investigate in silico the molecular relationships between COVID-19 and APP, which would lay a basis for further studies that investigate the expression of specific cleaved isoforms, such as Aβ.
The time from onset of COVID-19 symptoms to stroke diagnosis varies from 0 to 130 days (Merkler et al. 2020; Tu et al. 2021), which suggests that the development of COVID-19-associated stroke may involve multiple factors contributing to short and/or long term susceptibility. This highlights the importance of studies to evaluate and understand how different stages of COVID-19 infection may affect the activities of different key molecules contributing to symptom manifestations. Therefore, our study used the QIAGEN Ingenuity Pathway Analysis (IPA) bioinformatics tools to conduct network meta-analysis to identify potential connections between SARS-CoV-2 infection and APP expression. IPA utilizes QIAGEN Knowledge Base (QKB), a comprehensive database containing over 7 million manually and automated curated findings from published literature. The QKB encompasses relationships connecting biological diseases; genetic expression; proteins, metabolites, and chemicals; and processes including regulation, reaction, expression, and ubiquitination.
It is well-established that COVID-19 pathogenesis involves the initial attachment of SARS-CoV-2’s spike (S) protein to angiotensin-converting enzyme-2 (ACE2), a cell surface receptor and a key enzyme catalyzing the hydrolysis of angiotensin II into angiotensin (Gheblawi et al. 2020; Yang et al. 2020). ACE2 is not only found highly expressed in the upper and lower airway epithelial cells and the lung parenchyma, but also present in many other cell and tissue types such as the brain stem, vascular endothelium, kidney cells, and gastrointestinal tract (Yang et al. 2019, 2020; Gheblawi et al. 2020; Li et al. 2020; Rahman et al. 2021). Kaneko et al. recently examined the mRNA expression levels of a set of pre-specified 785 host response genes. These genes are involved in pathways of homeostasis, adaptive immune response, host susceptibility, and interferon response following the binding of the SARS-CoV-2 recombinant S-protein trimer to ACE2 receptor to human brain microvascular endothelial cells (BMVECs). They reported that the expression levels of 80 host response genes are significantly altered (Kaneko et al. 2021). These 80 genes were therefore used in our in silico analysis to examine how SARS-CoV-2 infection may induce cerebrovascular damage. IPA was used to characterize the 80 genes reportedly modulated in BMVEC in response to the binding of SARS-CoV-2 S protein. Among the 80 modulated genes, 12 were upregulated following S protein attachment (Additional file 1: Table 1), and 68 were downregulated after S protein attachment (Additional file 1: Table 2). The 12 upregulated genes include cytokines, chemokines, transmembrane receptors, and enzymes (Additional file 1: Table 1). The 68 downregulated genes include signaling kinases, enzymes, transcription regulators, cytokines, chemokines, transporters, and transmembrane receptors (Additional file 1: Table 2). With this already established and published data, we investigated whether the molecules affected by attachment of SARS-CoV-2 S protein may influence APP expression.
Considering that as the virus amplifies and COVID-19 symptoms progress, a cytokine storm may develop (Shi et al. 2020; Tian et al. 2020), we therefore also studied the impact of COVID-19 on expression of APP. By investigating how APP expression is affected by molecules differentially modulated in BMVECs following SARS-CoV-2 S-protein binding and as COVID-19 progresses, our study sheds light on the underlying molecular mechanisms by which COVID-19 modulates APP expression.
Methods and Materials
Ingenuity Pathway Analysis (IPA) Software
The IPA Analysis Match CL license utilized in this study was purchased from QIAGEN and has been renewed annually since 2018 (QIAGEN Inc., Germantown, MD, USA, https://www.qiagenbioinformatics.com/products). IPA is a bioinformatics tool that was used to analyze data and biological processes using the QKB repository composed of over seven million individually modeled relationships to produce and analyze networks through known metabolic and signaling pathways. It also allows for the visualization and exploration of omics data. The genes impacted by SARS-CoV-2 were inputted into IPA for analysis. The data used for this study were retrieved from February 4, 2021 to May 28, 2021.
Data Collection of IPA’s QIAGEN Knowledge Base (QKB)
IPA is a bioinformatics tool that utilizes the QKB repository to analyze, integrate, and provide an expansive and holistic interpretation of data from various sources, ranging from proteomics, metabolomics, RNA sequencing, miRNA, SNP microarrays, and differential gene expression. QKB spans across 40 public and proprietary databases curated and maintained by many well-trained data collectors. It includes an extensive network of published omics experiments and more than seven million individually modeled relationships that include diseases, drugs, genes, cells, tissue, and proteins applied in various processes such as expression, phosphorylation, and cleavage. The structured and detailed biological and chemical data in QKB are updated weekly with findings from new clinical trials, emerging therapeutics, and recently published primary literatures.
The Molecules Modulated Upon Binding of SARS-CoV-2 S Protein to BMVECs
SARS-CoV-2 S protein has been reported to bind to ACE2 receptor in BMVECs (Yang et al. 2020). Kaneko et al. utilized direct RNA hybridization assays to examine the expression of 785 host genes following 24-h exposure to 5 μg/mL of SARS-CoV-2 S protein. These host genes encompassed those involved in homeostasis, adaptive immune response, host susceptibility, and interferon response pathway, and reported that the expression of 80 genes was significantly altered in BMVECs following exposure to SARS-CoV-2 S protein (Kaneko et al. 2021). These 80 genes were listed in Additional file 1: Tables 1 and 2; and were uploaded to IPA as a dataset alongside their reported relative expression levels. Using IPA tools, the downstream effects of these 80 genes were analyzed to examine how SARS-CoV-2 infection may modulate the expression of APP.
Identification of the Overlapping Molecules Between the 80 Genes Affected by SARS-CoV-2 Infection, COVID-19, and APP
IPA’s “Grow” tool was utilized to generate a set of genes, chemicals, proteins, and data node points associated with the BMVEC modulated genes and APP, via QKB. This tool expanded the molecules affected by each gene of choice in the pathway. Because chemical drugs and toxicants are not naturally found in a biological system, IPA’s “Trim” tool was used to eliminate all chemicals and toxicants from the dataset; this ensured that a biological system was replicated as accurately as possible. IPA’s “Path Explorer” tool was used to discover the shortest path between the 80 molecules and APP, thereby identifying the overlapping molecules between the expanded sets of genes. This was also used to identify overlapping molecules between the molecules affected by COVID-19 and those associated with APP.
“Molecule Activity Predictor (MAP)” Simulation
IPA’s “MAP” tool was used on the molecules connected to APP to simulate how the activation or inhibition of those molecules would influence APP expression. The tool was used to simulate changes, such as transcription, activation, inhibition, or phosphorylation. This could identify how the impact of SARS-CoV-2 S protein binding on the 80 modulated genes would consequently influence APP expression, and how the activation of the upstream regulators of COVID-19 would influence APP expression.
Identification of COVID-19 Upstream Regulators and Canonical Pathways Through the “Core Analysis”
IPA’s “Grow” tool was utilized to generate a set of genes, chemicals, proteins, and data node points associated with COVID-19 and APP. This tool identified and added all the known intermediate molecules in QKB connecting COVID-19 to APP onto a pathway. The intermediate connections that were not naturally occurring, including chemical drugs and toxicants, were removed from the pathway to accurately represent the human biological system (Fig. 5). All remaining molecules connecting developed COVID-19 to APP in the synthesized pathway were compiled to form a list. “Core Analysis” was performed on this dataset which focused on “Expression Analysis”. The “Core Expression Analysis” revealed which canonical pathways within the molecular dataset were statistically predicted to be involved using a − log(p value) calculated by the Benjamini–Hochberg Corrected Fisher’s Exact test. The “Expression Analysis” also predicted which “Diseases and Biological Functions” the dataset would be involved in. Another feature was the “Upstream Analysis” which includes the top upstream regulators of a given dataset. The dataset of the 79 overlapping molecules between COVID-19 and APP revealed a series of upstream regulators. IPA’s identified upstream regulators are genes that have a downstream genetic cascade discovered to have a statistically significant overlap with a dataset. Thus, the upstream regulators found for this dataset can serve as genetic placeholders for COVID-19 for any subsequent analysis on this dataset; these regulators can help explain the biological actions occurring within cells. The top 6 upstream regulators were chosen as they had the most overlap with the molecules within the dataset; they were also found to be impacted by COVID-19 in various studies.
Fig. 5.
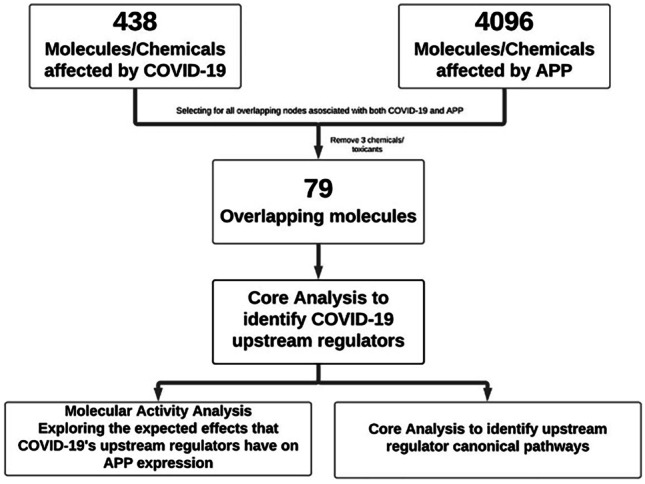
Flow chart for the analysis of COVID-19’s impact on APP
Quantitative Analysis of the Impact of COVID-19 Upstream Regulators on APP Expression
This study utilized the “Downstream Effect Analysis” algorithm, as described by Krämer et al. (2014), to provide a quantitative perspective on the extent of how much APP expression was impacted by the six upregulated COVID-19 upstream regulators. The individual contribution weight of each connection between the 6 upstream regulators and the 11 intermediates on APP expression was computed. The algorithm used QKB references as data points to confirm a confidence for the change in APP expression upon the upregulation of the 6 COVID-19 upstream regulators.
| 1 |
| 2 |
And we define the activation Z-score as
| 3 |
The formulas identified by Krämer were used to compute a Z-score for the individual involvement of each intermediate downstream of the upstream regulators on APP expression. The range of Z-scores that reflects the individual expression change caused by a molecule is between − 2 and 2; the low end, − 2, indicates a strong inhibitory relationship and the high end, 2, indicates a strong activation relationship. Each intermediate is considered to be an edge, e, in any given relationship with an upstream and downstream molecule. The of each edge reflects data points stored in QKB upstream and downstream of it. s(e) identifies the overall sign of a particular direction from the edge, ranging from {− 1, 0, 1}; w(e) identifies the weight of the edge, ranging from {0, 1} (Alabed et al. 2021). The data points plugged into the formulas use the findings, f, in QKB, with either a 1 for a positive relationship or a -1 for a negative relationship. SD is the sign of the upstream molecule in one particular relationship {− 1, 1}. z(r) reflects the overall change in expression of the target molecule in a relationship, which is computed by combining the values obtained upstream of an edge, e, and downstream of that same edge, e.
The same z(r) formula was used to compute an overall Z-score of the impact of each upstream regulator on APP expression by aggregating the individual Z-scores of the intermediates with a slight difference. WR, which corresponds to w(e) in the individual Z-score computation, now corresponded to s(e), which can only be {− 1, 1}. The same calculations were then performed (Alabed et al. 2021).
Furthermore, another formula was used to compute an overall Z-score of the entire network by combing all 6 overall Z-scores of each upstream regulator. The formula is outlined by Stouffer and computes a Z-score by aggregating independently found Z-statistics into a two tailed standard normal distribution (Stouffer et al. 1949; WHITLOCK 2005).
| 4 |
ZS is the overall Z-score of the two-tailed standard normal distribution; Zi is the individual one tailed Z-score; and k is the total number of individual Z-scores in the standard normal distribution (Alabed et al. 2021).
Results
Molecules Affected by Attachment of SARS-CoV-2 S Protein that Influence APP Expression
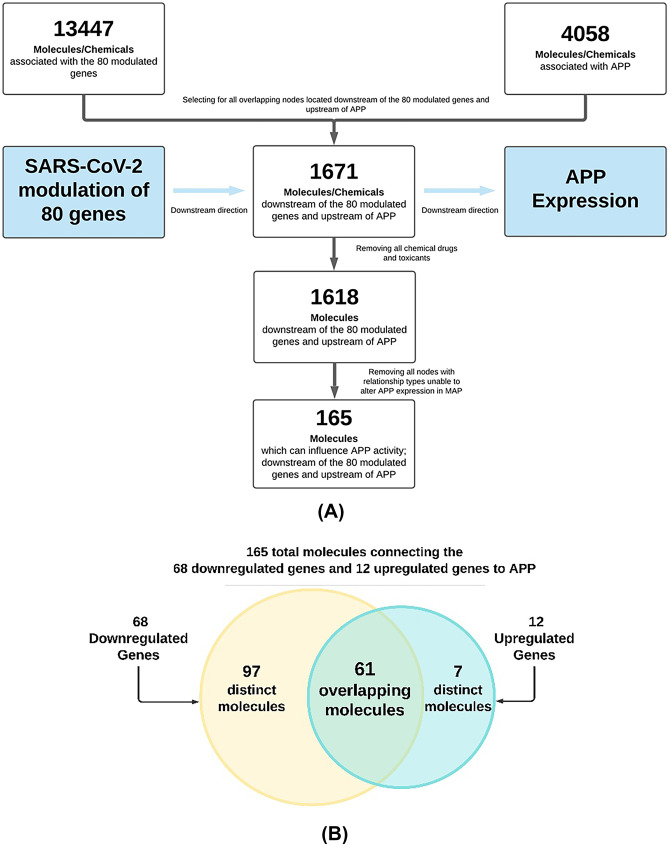
The 80 genes modulated in BMVECs following the attachment of SARS-CoV-2 S protein identified by Kaneko et al. were uploaded into IPA. 13,447 molecules and chemicals were found to be associated with these 80 genes (Fig. 1A). Similarly, 4058 molecules and chemicals were found to be associated with APP in QKB. Among these two datasets, 1671 molecules and chemicals were found to be downstream of the 80 modulated genes and upstream of APP. Because chemicals and drugs are not found naturally in a biological system, the fifty-three chemicals and drugs were then removed from this set of 1671 molecules to replicate the biological system as accurately as possible, which produced a dataset of 1618 biological molecules that connect early modulation of BMVECs by S protein to downstream APP production. Lastly, the molecules that did not connect SARS-CoV-2 to APP via a relationship that could alter APP expression were trimmed, producing a dataset of 165 molecules (Fig. 1A). This final dataset of 165 molecules consisted of molecules discovered to be affected by binding of SARS-CoV-2 S protein to BMVECs and to also influence APP expression within BMVECs (Additional file 1: Table 3). Figure 1B shows how the final dataset of 165 molecules is composed of distinct and shared molecules from the downregulated and upregulated genes.
Fig. 1.
The identification process and composition of the 165 molecules connecting the 80 genes modulated by the binding of SARS-CoV-2 S protein and APP. A Flow chart of the stepwise process to identify the 165 molecules associated with the 80 genes modulated by the binding of SARS-CoV-2 S protein to BMVEC, and also influence APP expression; B Venn diagram displaying the makeup of the 165 genes with respect to the 12 upregulated genes and 68 downregulated genes
The Top 15 Canonical Pathways Implicated by the 165 Overlapping Intermediates Between Molecules Associated with SARS-CoV-2 S Protein Binding and APP
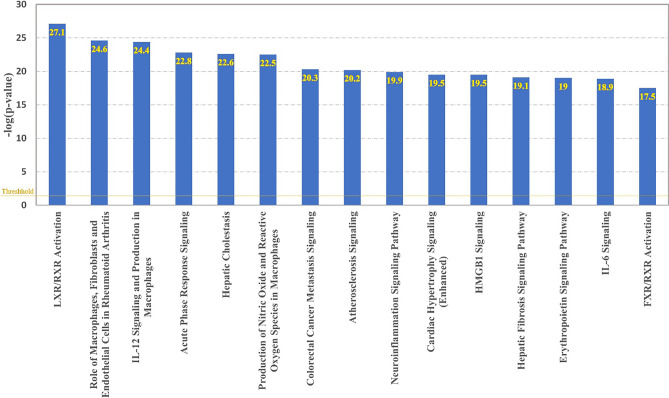
The biological mechanisms and pathways affected by the attachment of SARS-CoV-2 S protein to BMVECs and APP were investigated by running a “Core Expression Analysis” on the 165 overlapping molecules connecting them. Figure 2 shows the top 15 canonical pathways found to be involved with the dataset of the 165 overlapping molecules connecting the 80 SARS-CoV-2 S protein-modulated BMVEC genes to APP. These pathways included LXR/RXR Activation, Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid Arthritis, Acute Phase Response Signaling, Atherosclerosis Signaling, and Neuroinflammation Signaling Pathway.
Fig. 2.
Top 15 canonical pathways in the network of all known connections between the 80 genes modulated by binding of SARS-CoV-2 to BMVEC and APP
To confirm that the canonical pathways identified above were specific to SARS-CoV-2 S protein binding and APP expression, the top 6th canonical pathway (p value < 4.96E10-21), Colorectal Cancer Metastasis Signaling, was selected as a negative control since it is the most significant canonical pathway with no a-priori associations. Connecting this Colorectal Cancer Metastasis Signaling pathway and APP identified 14 overlapping molecules. Canonical pathway analysis of these 14 overlapping molecules found that none of their top 15 canonical pathways overlap with those shown in Fig. 2, except for the Colorectal Cancer Metastasis Signaling pathway. This negative control analysis further validated that the top 15 canonical pathways shown in Fig. 2 were specifically related to SARS-CoV-2 S protein binding and APP expression rather than random associations.
The Impact of the 165 Overlapping Molecules Associated with SARS-CoV-2 S Protein-Modulated Genes on APP Expression
The “MAP” tool was used to simulate the activation of the 80 modulated genes according to their reported relative expression levels following SARS-CoV-2 S protein binding (Kaneko et al. 2021). Among the 12 genes upregulated in response to SARS-CoV-2 S protein binding, 8 molecules, CD163, CXCL8, EBI3, IL9, IL15, ITGAL, OSM, and TNFRSF17, were found to increase APP expression, while C3 was found to inhibit APP expression. The remaining 3 molecules, APOBEC3G, CCL8, and CCL24, had no direct or intermediate relationship that influenced APP expression and, therefore, had no lines connecting them with APP. Figure 3A shows the direct relationships (1) and relationships with a single intermediate molecule (119) between the 12 genes and APP identified through QKB. 68 molecules were found to mediate the effects of the 12 genes upregulated by SARS-CoV-2 S protein binding on APP expression in BMVECs.
Fig. 3.
Potential pathways connecting the 12 upregulated and 68 downregulated genes in BMVEC following SARS-CoV-2 S protein attachment and APP. A “MAP” simulated activation of the 12 genes according to their relative expression fold changes, leading to increased APP production directly or indirectly via 68 intermediary molecules; B “MAP” simulated decrease of the 68 genes according to their relative expression fold changes, leading to decreased APP production directly or indirectly via 158 intermediary molecules
The impact of the 68 downregulated genes following SARS-CoV-2 S protein binding in BMVECs on APP expression was also investigated. Figure 3B shows that the 68 downregulated genes may influence APP through 158 intermediate molecules. Figure 3B shows the direct relationships (7) and relationships with a single intermediate molecule (740) between the 68 genes and APP identified through QKB. The “MAP” tool was then used to simulate the downregulation of those genes as reported in the study (Kaneko et al. 2021), which resulted in an overall decrease in APP expression. Of the 68 genes downregulated in response to SARS-CoV-2 S protein binding, 52 were shown to alter APP expression and 16 had no predicted effects on APP (Fig. 3B).
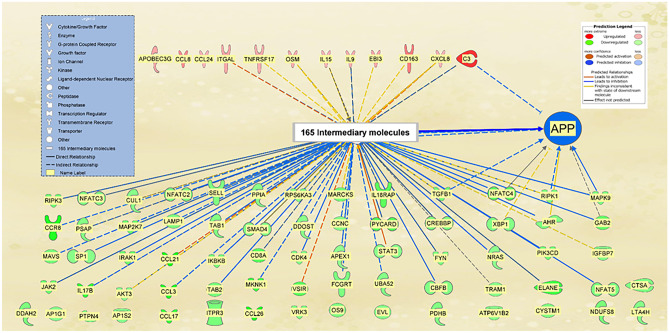
The overall impact of the 12 upregulated genes and the 68 downregulated genes by SARS-CoV-2 binding was then combined to provide a holistic view of the change in APP expression following SARS-CoV-2 infection and modulation of those genes. Figure 4 shows that the 80 modulated genes may influence APP through 165 intermediate molecules. These 165 intermediate molecules are the nodes identified in Fig. 1A. The 165 intermediate molecules consist of 7 intermediates unique to the 12 upregulated genes; 97 intermediates unique to the 68 downregulated genes; 61 intermediates that are shared by both the 12 upregulated and 68 downregulated genes (Fig. 1B). Figure 4 shows the collective effects from 80 genes modulated by SARS-CoV-2 binding in BMVECs. The “MAP” tool was used to simulate the relative expression fold changes of these 80 modulated genes in BMVECs. APP expression was found to be strongly inhibited by these 80 modulated genes in BMVECs.
Fig. 4.
Influence of the 80 SARS-CoV-2 S-protein-modulated genes in BMVEC on APP expression. “MAP” simulated modulation of the 80 genes according to their relative expression fold changes, resulted in the inhibition of APP expression. This inhibition occurred through 165 intermediary molecules, represented in the figure through the intermediary molecule nodes
Identification of COVID-19 and APP Overlapping Molecules
IPA’s “Grow” tool was used to expand the molecules affected by COVID-19 and APP, respectively, based on the data stored in QKB. A total of 438 molecules were identified to be affected by COVID-19 and 4096 molecules were found to be affected by APP. Of those, 79 molecules were found to overlap between both COVID-19 and APP (Fig. 5). A “Core Analysis” was run on the 79-molecule dataset, revealing a series of upstream regulators listed in order of highest overlap with the molecules in the dataset (Table 1). The top six molecules were tumor necrosis factor (TNF), interferon gamma (IFNG), signal transducer and activator of transcription 1 (STAT1), interleukin 1 beta (IL1β), interleukin 6 (IL6), and signal transducer and activator of transcription 3 (STAT3).
Molecules Affected by the COVID-19 Upstream Regulators and APP
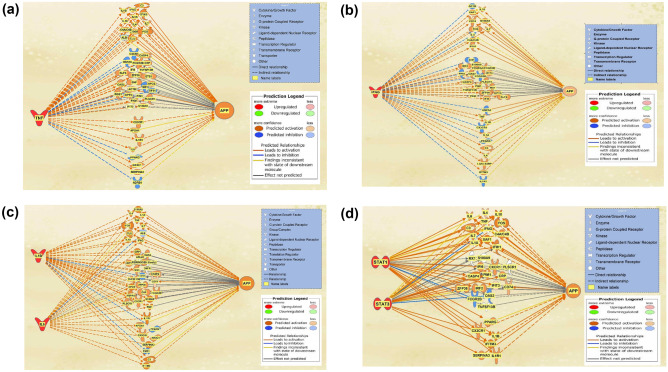
The molecules affected by each of the upstream regulators were added into separate pathways and connected to APP using the “Path Explorer” tool. TNF was an upstream regulator for a total of 44 molecules of the 80-molecule dataset; IFNG overlapped with 39 molecules; IL1β and IL6 overlapped with 39 molecules; STAT1 and STAT3 overlapped with 33 molecules. The “MAP” tool was used to simulate the upregulation of those upstream regulators as COVID-19 infection causes an increase in the expression of those regulators. Figures 6A–D show the categorization of those molecules in the following order: the top cluster includes molecules that are downstream from TNF, IFNG, IL-1β/IL6, STAT1/STAT3 (the upstream regulators), respectively, but also downstream from APP; the middle cluster includes molecules with no determined expression change either by TNF, IFNG, IL-1β/IL6, STAT1/STAT3 (upstream regulators) or on APP expression; the bottom cluster includes the intermediates that exhibited change in expression upon the upregulation of the TNF, IFNG, IL-1β/IL6, STAT1/STAT3 (upstream regulators), and also had a known influence on APP expression.
Fig. 6.
Molecules affected by COVID-19 Upstream Regulators and APP. A TNF was revealed as an upstream regulator with the most significant p-value of 1.85E−28. There was a total of 44 intermediates, 13 of which were downstream from APP, 23 had no determined impact by TNF or on APP, and 8 were impacted by TNF and influenced APP expression; B IFNG was revealed as an upstream regulator with a p-value of 3.6E−27. There was a total of 39 intermediates, 10 of which were downstream from APP, 19 had no determined impact by IFNG or on APP, and 11 were impacted by IFNG and influenced APP expression. C IL1B and IL6 were revealed as upstream regulators with a p-value of 3.23E−24 and 8.35E−22, respectively. There was a total of 39 intermediates between both, 10 of which were downstream from APP, 20 had no determined impact by IL1B and IL6, or on APP, and 9 were impacted by IL1B or IL6 and influenced APP expression. D STAT1 and STAT3 were revealed as upstream regulators with a p-value of 9.35E−27 and 1.15E−20, respectively. There was a total of 33 intermediates between both, 11 of which were downstream from APP, 16 had no determined impact by STAT1 or STAT3, or on APP, and 6 were impacted by STAT1 and STAT3 and influenced APP expression
To investigate how these 6 upstream regulators would influence APP expression when acting together, the overlapping molecules with a defined relationship from the upstream regulators and a known influence on APP from each pathway (Fig. 6) were taken and put together. Figure 7A shows the holistic impact of the 11 overlapping intermediates between TNF, IFNG, IL1β, IL6, STAT1, STAT3 on APP expression. The 11 overlapping intermediates include genes that exhibited a change in expression upon the upregulation of TNF, IFNG, IL-1β/IL6, STAT1/STAT3 (upstream regulators), and also had a known influence on APP expression. The “MAP” tool was used to simulate the upregulation of the 6 upstream regulators, which resulted in an overall increase in APP expression.
Fig. 7.
The holistic impact of the 6 COVID-19 upstream regulators on APP expression and top 15 canonical pathways of the 11 intermediates. A After combining the intermediates of each of the upstream regulators into one pathway, the “MAP” tool was used to simulate the activation of TNF, IFNG, IL1B, IL6, STAT1, and STAT3. The resulting impact on the 11 intermediates led to an increase in the expression of APP; B The “Core Analysis” of the 11-molecule dataset, obtained from the overlapping molecules of COVID-19 upstream regulators and APP, revealed a list of canonical pathways in which they overlapped. The top canonical pathway was the Acute Phase Response Signaling, following by the Neuroinflammation Signaling pathway
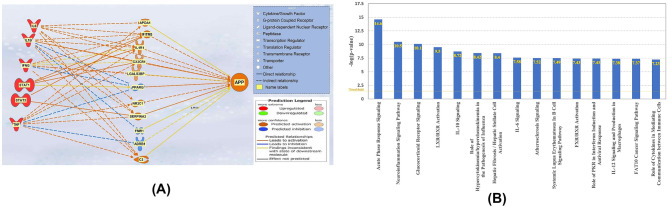
“Core Analysis” of the 11 Intermediates Between the COVID-19 Upstream Regulators and APP
A dataset of the 11 intermediates from Fig. 7 was created and “Core Analysis” was run. The analysis identified the canonical pathways in which the 11 intermediates overlap. Figure 7B lists the canonical pathways in order of decreasing − log(p value). The most significant signaling pathway was the Acute Phase Response Signaling pathway, with a p value of 2.81E−15; the second most significant canonical pathway was the Neuroinflammation Signaling Pathway, which had a p value of 3.34 E−11.
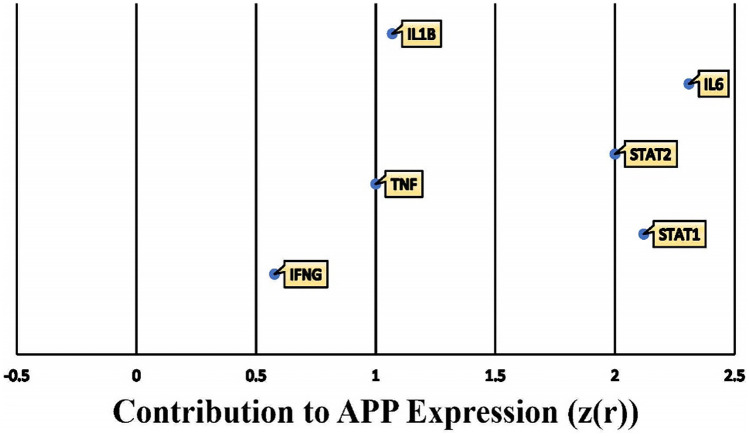
In order to assign weights to the impact of the 6 identified COVID-19 upstream regulators on APP following “MAP” activation, Z-scores were calculated using the “Downstream Effect Analysis” algorithm described by Krämer et al. (2014). The algorithm computes a Z-statistic based on the consistency of the expression and directional changes of each relationship with the scientific literature stored in QKB as data points. A total of 322 references of prior known scientific literature were used as data points to calculate how the upregulation of the 6 COVID-19 upstream regulators contribute to the change in APP expression. Figure 8 shows the overall Z-score for each upstream regulator; the overall Z-score of each upstream regulator was calculated by aggregating the individual Z-scores of each individual intermediate downstream of each upstream regulator, which can be referred to in Additional file 1: Table 4. Among the 6 COVID-19 upstream regulators, IL6 and STAT1 were found to affect APP expression with a Z-score of higher than 2, while TNF, IL1β, and STAT2 had a Z-score between 1 and 2; lastly, IFNG had the lowest, but still positive Z-score of 0.577.
Fig. 8.
Individual contribution of the 6 COVID-19 upstream regulators on APP expression. The overall change in APP expression by the upregulation of the 6 COVID-19 upstream regulators was measured by the individual involvement, z(r), of each of the 6 upstream regulators
The results indicated an increase in APP expression with an overall calculation that gave a Z-score of 3.705 (Stouffer et al. 1949; Kramer et al. 2014), which corresponds to a p-value of 0.000211 in a two tailed hypothesis, suggesting that the likelihood of finding equally strong consistency of the results occurs at a less than 1% chance.
Discussion
In this study, we sought to explore and comprehend the molecular mechanisms by which SARS-CoV-2 S protein attachment modulates APP expression in BMVEC, and COVID-19 modulates the expression of APP. Our network meta-analysis revealed that SARS-CoV-2 infection may cause differential expression of APP at different stages of viral infection. The initial attachment of SARS-CoV-2 S protein modulates the expression of host response genes including the 80 reported by Kaneko et al. (2021). Our results show that the altered expression of these 80 genes may result in an overall inhibition on APP expression in BMVEC. This decrease in APP expression inside BMVECs may lead to the disruption of brain vascular endothelial integrity. Such endothelial disruption has been known to cause worsened stroke outcomes and has recently been implicated as an underlying factor involved in COVID-19’s increased cerebrovascular events (d’Uscio et al. 2018; Ristori et al. 2020). Because COVID-19’s hallmark symptoms can manifest days after initial exposure, we analyzed how APP would be affected during this later viral infection stage. Using molecules found to be statistically significant upstream regulators modulated by COVID-19, we found that the activated pathways found in a SARS-CoV-2 infection strongly activated APP expression. Depending on where SARS-CoV-2-induced increase of APP expression occurs, patients can be at higher risk for neurogenerative diseases and CAA.
To investigate SARS-CoV-2’s effects during the initial stage of infection, we used the reported unique genetic profile of the reported 80 host response genes modulated in BMVEC following SARS-CoV-2 S protein attachment (Kaneko et al. 2021). The implications on APP from SARS-CoV-2 S protein attachment through these modulated genes were analyzed through a stepwise categorization, which revealed that 165 overlapping molecules, associated with the 80 modulated genes, influenced APP expression. Running this group of 165 overlapping molecules in the “Core Expression Analysis” tool allowed us to identify which canonical pathways were involved in the effects following the initial binding of SARS-CoV-2 S protein on APP (Table 1). Key pathways included the “Acute Phase Response Pathway” involved in the early cellular responses following viral or bacterial detection. Both the “Neuroinflammation Signaling” and “HMGB1 Signaling” pathways involve BBB homeostasis. As explained previously, the experienced decrease in brain endothelium APP following SARS-CoV-2’s attachment can consequently induce BBB disruption (d’Uscio et al. 2018; Yang et al. 2019). In addition, the “Production of Nitric Oxide and Reactive Oxygen Species in Macrophages” pathway was also found among the top 15 canonical pathways. Although the study by Kaneko et al. was conducted in BMVECs, APP protection of brain endothelial stability involves maintaining eNOS expression (d’Uscio et al. 2018; Ristori et al. 2020), which explains why the 165 overlapping molecules identified in our analysis also involved Nitric Oxide pathways.
Table 1.
List of top 10 upstream regulators identified by the “Core Analysis” of the 79 overlapping molecules between COVID-19 and APP; the regulators are listed in the order of most significant p-value of overlap
| Upstream regulator | Molecule type | p value of overlap |
|---|---|---|
| TNF | Cytokine | 1.85E−28 |
| IFNG | Cytokine | 3.6E−27 |
| STAT1 | Transcription regulator | 9.35E−27 |
| IL1β | Cytokine | 3.23E−24 |
| IL6 | Cytokine | 8.35E−22 |
| STAT3 | Transcription regulator | 1.15E−20 |
| IL4 | Cytokine | 1.26E−19 |
| TLR3 | Transmembrane receptor | 2.33E−18 |
| IRF1 | Transcription regulator | 7.12E−18 |
| Interferon alpha | Group | 9.16E−18 |
Though individually not all of the 80 genes modulated by SARS-CoV-2 binding to ACE2 in BMVEC affected APP expression, the overall effects of these 80 modulated genes were predicted to inhibit APP expression (Fig. 4). This holds direct biological implications to BBB homeostasis and subsequent pathological manifestations. Disruption of APP homeostasis has been known to cause endothelial cell dysfunction within cerebral arteries (d’Uscio et al. 2018; Ristori et al. 2020), which may lead to neuroinflammation, neurodegeneration (Obermeier et al. 2013), and worsened stroke outcomes as reported in COVID-19 patients (Yang et al. 2019). While the increased incidence of stroke in COVID-19 patients may be multifaceted and may take place early on and months after SARS-CoV-2 infection, the decrease in APP within BMVECs and subsequent endothelial dysfunction may help explain the increased stroke at earlier stages of SARS-CoV-2 infection.
It should also be noted that although these findings are based on the unique genetic profile in BMVEC following the binding of SARS-CoV-2 S protein to ACE2 receptor, ACE2 receptor is expressed in many tissues throughout the body including the gastrointestinal tract, kidney cells, lung tissue, some neural tissue, and microvascular endothelial cells. Thus, it is likely that the ACE2 expressing tissues where SARS-CoV-2 can attach may have a similarly modulated genetic profile and therefore exhibit inhibition of APP expression. Areas that APP expression has been reported includes basal ephemeral cells, human adipocytes, gastrointestinal tract enterocytes, neurons, and the adrenal glands (Arai et al. 1991; Puig and Combs 2013) The effect of this inhibition in respect to the manifestation of COVID-19 symptoms ultimately depends on the role of APP in cellular homeostasis.
As SARS-CoV-2 infection progresses, the host continues to mount on the immune response. This development may lead to host immune responses with hyperactive cytokine release causing inflammatory driven damage. Analysis of COVID-19’s associated markers later in the course of SARS-CoV-2 infection revealed upstream regulators that caused an increase in APP expression. Our analysis revealed two distinct scenarios which may have differential impact on APP expression. During the initial viral transmission phase, the attachment and transmission of SARS-CoV-2 take place without significant pre-existing inflammatory factors, and APP expression may be decreased (Shi et al. 2020; Tian et al. 2020). The ensuing SARS-CoV-2 infection stage with excessive production of pro-inflammatory cytokines and inflammatory damage is expected to cause an increase in APP expression.
Figure 6 shows that COVID-19 increased the activities of TNF, IL1β, IL6, and IFNG. Clinical data show that elevated levels of TNF are associated with a higher COVID-19 mortality, Anti-TNF therapy has been used to reduce the severity of COVID-19 outcomes and deaths (Robinson et al. 2020). Elevated levels of TNF and other cytokines, such as IFNG and IL1β, can stimulate γ-secretase directly, which in turn increases the production of Aβ in the brain (Liao et al. 2004). It would be worthwhile to develop further studies on how the modulation of TNF, IL1β, IL6, and IFNG during COVID-19 may affect the activities of different secretases responsible for cleaving APP into various isoforms of Aβ (Nunan and Small 2000; Zhang et al. 2011), which may help to understand how the dysregulated cytokine production during COVID-19 might manifest in different neurological pathologies.
The buildup of APP and subsequent Aβ plaque levels in the later stages of COVID-19 patients has various short-and long-term implications. Patients who have COVID-19-induced APP buildup may exhibit increased artery blockage, thrombosis events, ischemic strokes, and neurodegenerative disease prevalence. Multiple APP-related diseases such as CAA directly rely on the buildup of Aβ, which is the cleaved product of APP (Biffi and Greenberg 2011; Ghiso et al. 2014). Furthermore, Aβ plaque buildup could occur naturally over time, and CAA symptoms have been reported to naturally bolster with age (Biffi and Greenberg 2011). Aging has been identified as a neuroinflammatory process; neuroinflammation is known to lead to the onset and progression of neurodegenerative diseases, including AD (McManus and Heneka 2017; Bossù et al. 2020; Huang et al. 2021b). Moreover, it has been reported that the number of naïve T-cells decreases with age, which is thought to be one of the factors contributing to a decreased capacity to induce an immune response following new infections (McManus and Heneka 2017); moreover, AD patients have also been reported to have significantly low levels of naïve T-cells (McManus and Heneka 2017). These findings may help to explain why older COVID-19 patients may likely experience worsened Aβ plaque related symptoms and outcomes, and experience higher COVID-19 mortality rate. On the other hand, it is likely that the observed increase of ischemic incidence immediately following SARS-CoV-2 infection within younger populations may be due in part to a greater degree to the APP-triggered endothelial disruption in BMVEC instead of Aβ plaque deposits. This is because while the buildup and subsequent effects of Aβ plaque deposits can take time to develop, if not already present, endothelial dysfunction requires no prerequisites and thus consequences can be more easily seen in younger populations.
SARS-CoV-2 pathophysiology may be marked by two distinct stages: an initial period of heightened virus propagation and infection via ACE2 receptors; then, a second phase of uncontrolled cytokine and inflammatory damage (Trougakos et al. 2021). Considering the broader consequences of COVID-19 infection, the predicted buildup of APP later in COVID-19 can lead to an increase in neurodegenerative disease prevalence. APP proteins can be broken down to form Aβ peptides which form plaque deposits (Roberts et al. 1994; Verdile et al. 2004; de Paula et al. 2009). If Aβ is deposited as plaques within the brain, the signaling process can be interrupted and neuronal damage may occur. Similar to how overactive C3 causes neural damage in an attempt to remove TAU tangles near neurons, the attempt of immune activation to remove these Aβ plaques could lead to neuronal damage and thus increase neurodegenerative disease risk. This suggests that SARS-CoV-2 may lead to increased neurodegenerative disease risk, such as AD, stemming from the buildup of APP during COVID-19 with enhanced production of upstream regulators, such as TNF, IFNG, STAT1, IL1β, IL6 and STAT3.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank Tanvi Patel for her assistance in seeking clarifications from QIAGEN on molecules associated with COVID-19, and Dr. Eric Seiser for initial use of QIAGEN Knowledge Base and QIAGEN Ingenuity Pathway Analysis tools. This study is partially supported by National Institutes of Health grants AA026071, and DA046258 to SLC. Graphical abstract was created with BioRender.com.
Declarations
Human and Animal Rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict Interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ryan C. Camacho and Sedra Alabed contributed equally to this work
Contributor Information
Heping Zhou, Email: heping.zhou@shu.edu.
Sulie L. Chang, Email: sulie.chang@shu.edu
References
- Alabed S, Zhou H, Sariyer IK, Chang SL. Meta-analysis of methamphetamine modulation on amyloid precursor protein through HMGB1 in Alzheimer’s disease. Int J Mol Sci. 2021;22:4781. doi: 10.3390/ijms22094781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai H, Lee VM, Messinger ML, Greenberg BD, Lowery DE, Trojanowski JQ. Expression patterns of beta-amyloid precursor protein (beta-APP) in neural and nonneural human tissues from Alzheimer's disease and control subjects. Ann Neurol. 1991;30:686–693. doi: 10.1002/ana.410300509. [DOI] [PubMed] [Google Scholar]
- Biffi A, Greenberg SM. Cerebral amyloid angiopathy: a systematic review. J Clin Neurol. 2011;7:1. doi: 10.3988/jcn.2011.7.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossù P, Toppi E, Sterbini V, Spalletta G. Implication of aging related chronic neuroinflammation on COVID-19 pandemic. J Pers Med. 2020;10:102. doi: 10.3390/jpm10030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Uscio LV, He T, Santhanam AV, Katusic ZS. Endothelium-specific amyloid precursor protein deficiency causes endothelial dysfunction in cerebral arteries. J Cereb Blood Flow Metab. 2018;38:1715–1726. doi: 10.1177/0271678X17735418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Paula VJR, Guimaraes FM, Diniz BS, Forlenza OV. Neurobiological pathways to Alzheimer's disease: amyloid-beta, TAU protein or both? Dement Neuropsychol. 2009;3:188–194. doi: 10.1590/S1980-57642009DN30300003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane R, et al. RAGE mediates amyloid-beta peptide transport across the blood-brain barrier and accumulation in brain. Nat Med. 2003;9:907–913. doi: 10.1038/nm890. [DOI] [PubMed] [Google Scholar]
- d'Uscio LV, He T, Katusic ZS. Expression and processing of amyloid precursor protein in vascular endothelium. Physiology. 2017;32:20–32. doi: 10.1152/physiol.00021.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellul MA, Benjamin L, Singh B, Lant S, Michael BD, Easton A, Kneen R, Defres S, Sejvar J, Solomon T. Neurological associations of COVID-19. Lancet Neurol. 2020;19:767–783. doi: 10.1016/S1474-4422(20)30221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garwood CJ, Pooler AM, Atherton J, Hanger DP, Noble W. Astrocytes are important mediators of Abeta-induced neurotoxicity and tau phosphorylation in primary culture. Cell Death Dis. 2011;2:e167. doi: 10.1038/cddis.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheblawi M, Wang K, Viveiros A, Nguyen Q, Zhong J-C, Turner AJ, Raizada MK, Grant MB, Oudit GY. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the Renin-Angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ Res. 2020;126:1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiso J, Fossati S, Rostagno A. Amyloidosis associated with cerebral amyloid angiopathy: cell signaling pathways elicited in cerebral endothelial cells. JAD. 2014;42:S167–S176. doi: 10.3233/JAD-140027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, et al. 6-month consequences of COVID-19 in patients discharged from hospital: a cohort study. The Lancet. 2021;397:220–232. doi: 10.1016/S0140-6736(20)32656-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Zhou H, Hodgkinson C, Montero A, Goldman D, Chang SL. Network meta-analysis on the mechanisms underlying alcohol augmentation of COVID-19 pathologies. Alcohol Clin Exp Res. 2021;45:675–688. doi: 10.1111/acer.14573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko N, Satta S, Komuro Y, Muthukrishnan SD, Kakarla V, Guo L, An J, Elahi F, Kornblum HI, Liebeskind DS, Hsiai T, Hinman JD. Flow-mediated susceptibility and molecular response of cerebral endothelia to SARS-CoV-2 infection. Stroke. 2021;52:260–270. doi: 10.1161/STROKEAHA.120.032764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M, et al. Delirium in older patients with COVID-19 presenting to the emergency department. JAMA Netw Open. 2020;3:e2029540. doi: 10.1001/jamanetworkopen.2020.29540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney JW, Bemiller SM, Murtishaw AS, Leisgang AM, Salazar AM, Lamb BT. Inflammation as a central mechanism in Alzheimer's disease. Alzheimers Dement (n y) 2018;4:575–590. doi: 10.1016/j.trci.2018.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa M, Oddo S, Yamasaki TR, Green KN, LaFerla FM. Lipopolysaccharide-induced inflammation exacerbates tau pathology by a cyclin-dependent kinase 5-mediated pathway in a transgenic model of Alzheimer's disease. J Neurosci. 2005;25:8843–8853. doi: 10.1523/JNEUROSCI.2868-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YC, Bai WZ, Hashikawa T. The neuroinvasive potential of SARS-CoV2 may play a role in the respiratory failure of COVID-19 patients. J Med Virol. 2020;92:552–555. doi: 10.1002/jmv.25728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao YF, Wang BJ, Cheng HT, Kuo LH, Wolfe MS. Tumor necrosis factor-alpha, interleukin-1beta, and interferon-gamma stimulate gamma-secretase-mediated cleavage of amyloid precursor protein through a JNK-dependent MAPK pathway. J Biol Chem. 2004;279:49523–49532. doi: 10.1074/jbc.M402034200. [DOI] [PubMed] [Google Scholar]
- McManus RM, Heneka MT. Role of neuroinflammation in neurodegeneration: new insights. Alzheimers Res Ther. 2017;9:14–14. doi: 10.1186/s13195-017-0241-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkler AE, et al. Risk of Ischemic stroke in patients with Coronavirus disease 2019 (COVID-19) vs patients with influenza. JAMA Neurol. 2020;77:1366. doi: 10.1001/jamaneurol.2020.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miners S, Kehoe PG, Love S. Cognitive impact of COVID-19: looking beyond the short term. Alz Res Therapy. 2020;12:170. doi: 10.1186/s13195-020-00744-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagna E, Dorostkar MM, Herms J. The role of APP in structural spine plasticity. Front Mol Neurosci. 2017;10:136. doi: 10.3389/fnmol.2017.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunan J, Small DH. Regulation of APP cleavage by α-, β- and γ-secretases. FEBS Lett. 2000;483:6–10. doi: 10.1016/S0014-5793(00)02076-7. [DOI] [PubMed] [Google Scholar]
- Obermeier B, Daneman R, Ransohoff RM. Development, maintenance and disruption of the blood-brain barrier. Nat Med. 2013;19:1584–1596. doi: 10.1038/nm.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organization GWH (2020) WHO COVID-19 Dashboard. In.
- Puig KL, Combs CK. Expression and function of APP and its metabolites outside the central nervous system. Exp Gerontol. 2013;48:608–611. doi: 10.1016/j.exger.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MA, Islam K, Rahman S, Alamin M. Neurobiochemical cross-talk between COVID-19 and Alzheimer’s disease. Mol Neurobiol. 2021;58:1017–1023. doi: 10.1007/s12035-020-02177-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds JL, Mahajan SD. SARS-COV2 alters blood brain barrier integrity contributing to neuro-inflammation. J Neuroimmune Pharmacol. 2021;16:4–6. doi: 10.1007/s11481-020-09975-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristori E, Donnini S, Ziche M. New insights into blood–brain barrier maintenance: the homeostatic role of β-amyloid precursor protein in cerebral vasculature. Front Physiol. 2020;11:1056. doi: 10.3389/fphys.2020.01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts GW, Gentleman SM, Lynch A, Murray L, Landon M, Graham DI. Beta amyloid protein deposition in the brain after severe head injury: implications for the pathogenesis of Alzheimer's disease. J Neurol Neurosurg Psychiatry. 1994;57:419–425. doi: 10.1136/jnnp.57.4.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PC, Liew DFL, Liew JW, Monaco C, Richards D, Shivakumar S, Tanner HL, Feldmann M. The potential for repurposing anti-TNF as a therapy for the treatment of COVID-19. Med. 2020;1:90–102. doi: 10.1016/j.medj.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J, Yamashita T, Tian F, Li X, Liu X, Shi X, Nakano Y, Tsunoda K, Nomura E, Sasaki R, Tadokoro K, Sato K, Takemoto M, Hishikawa N, Ohta Y, Abe K. Chronic cerebral hypoperfusion alters amyloid-beta transport related proteins in the cortical blood vessels of Alzheimer's disease model mouse. Brain Res. 2019;1723:146379. doi: 10.1016/j.brainres.2019.146379. [DOI] [PubMed] [Google Scholar]
- Shi Y, Wang Y, Shao C, Huang J, Gan J, Huang X, Bucci E, Piacentini M, Ippolito G, Melino G. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27:1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouffer SA, Suchman EA, Devinney LC, Star SA, Williams RM., Jr . The American soldier: Adjustment during army life (Studies in social psychology in World War II) Oxford: Princeton Univ. Press; 1949. [Google Scholar]
- Taquet M, Geddes JR, Husain M, Luciano S, Harrison PJ. 6-month neurological and psychiatric outcomes in 236 379 survivors of COVID-19: a retrospective cohort study using electronic health records. Lancet Psychiatry. 2021;8:416–427. doi: 10.1016/S2215-0366(21)00084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W, Zhang N, Jin R, Feng Y, Wang S, Gao S, Gao R, Wu G, Tian D, Tan W, Chen Y, Gao GF, Wong CCL. Immune suppression in the early stage of COVID-19 disease. Nat Commun. 2020;11:5859. doi: 10.1038/s41467-020-19706-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trougakos IP, Stamatelopoulos K, Terpos E, Tsitsilonis OE, Aivalioti E, Paraskevis D, Kastritis E, Pavlakis GN, Dimopoulos MA. Insights to SARS-CoV-2 life cycle, pathophysiology, and rationalized treatments that target COVID-19 clinical complications. J Biomed Sci. 2021;28:9. doi: 10.1186/s12929-020-00703-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu TM, Seet CYH, Koh JS, Tham CH, Chiew HJ, De Leon JA, Chua CYK, Hui AC-F, Tan SSY, Vasoo SS, Tan BY-Q, Umapathi NT, Tambyah PA, Yeo LLL. Acute Ischemic stroke during the convalescent phase of asymptomatic COVID-2019 infection in men. JAMA Netw Open. 2021;4:e217498. doi: 10.1001/jamanetworkopen.2021.7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdile G, Fuller S, Atwood CS, Laws SM, Gandy SE, Martins RN. The role of beta amyloid in Alzheimer's disease: still a cause of everything or the only one who got caught? Pharmacol Res. 2004;50:397–409. doi: 10.1016/j.phrs.2003.12.028. [DOI] [PubMed] [Google Scholar]
- Walters A, Phillips E, Zheng R, Biju M, Kuruvilla T. Evidence for neuroinflammation in Alzheimer's disease. Prog Neurol Psychiatry. 2016;20:25–31. doi: 10.1002/pnp.444. [DOI] [Google Scholar]
- WHITLOCK MC Combining probability from independent tests: the weighted Z-method is superior to Fisher's approach. J Evol Biol. 2005;18:1368–1373. doi: 10.1111/j.1420-9101.2005.00917.x. [DOI] [PubMed] [Google Scholar]
- Wu Y, Xu X, Chen Z, Duan J, Hashimoto K, Yang L, Liu C, Yang C. Nervous system involvement after infection with COVID-19 and other coronaviruses. Brain Behav Immun. 2020;87:18–22. doi: 10.1016/j.bbi.2020.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Rosenberg GA. Blood-brain barrier breakdown in acute and chronic cerebrovascular disease. Stroke. 2011;42:3323–3328. doi: 10.1161/STROKEAHA.110.608257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Hawkins KE, Doré S, Candelario-Jalil E. Neuroinflammatory mechanisms of blood–brain barrier damage in ischemic stroke. Am J Physiol Cell Physiol. 2019;316:C135–C153. doi: 10.1152/ajpcell.00136.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Petitjean SJL, Koehler M, Zhang Q, Dumitru AC, Chen W, Derclaye S, Vincent SP, Soumillion P, Alsteens D. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat Commun. 2020;11:4541. doi: 10.1038/s41467-020-18319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y-w, Thompson R, Zhang H, Xu H. APP processing in Alzheimer's disease. Mol Brain. 2011;4:3. doi: 10.1186/1756-6606-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel GJ, Han H, Ford AL, Lee JM. Cerebral amyloid angiopathy: progressive disruption of the neurovascular unit. Stroke. 2009;40:S16–S19. doi: 10.1161/STROKEAHA.108.533174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.