Abstract
S-acylation, also known as palmitoylation, is the most widely prevalent form of protein lipidation, whereby long-chain fatty acids get attached to cysteine residues facing the cytosol. In humans, 23 members of the zDHHC family of integral membrane enzymes catalyze this modification. S-acylation is critical for the life cycle of many enveloped viruses. The Spike protein of SARS-CoV-2, the causative agent of COVID-19, has the most cysteine-rich cytoplasmic tail among known human pathogens in the closely related family of β-coronaviruses; however, it is unclear which of the cytoplasmic cysteines are S-acylated, and what the impact of this modification is on viral infectivity. Here we identify specific cysteine clusters in the Spike protein of SARS-CoV-2 that are targets of S-acylation. Interestingly, when we investigated the effect of the cysteine clusters using pseudotyped virus, mutation of the same three clusters of cysteines severely compromised viral infectivity. We developed a library of expression constructs of human zDHHC enzymes and used them to identify zDHHC enzymes that can S-acylate SARS-CoV-2 Spike protein. Finally, we reconstituted S-acylation of SARS-CoV-2 Spike protein in vitro using purified zDHHC enzymes. We observe a striking heterogeneity in the S-acylation status of the different cysteines in our in cellulo experiments, which, remarkably, was recapitulated by the in vitro assay. Altogether, these results bolster our understanding of a poorly understood posttranslational modification integral to the SARS-CoV-2 Spike protein. This study opens up avenues for further mechanistic dissection and lays the groundwork toward developing future strategies that could aid in the identification of targeted small-molecule modulators.
Keywords: SARS-CoV-2, spike protein, posttranslational modification (PTM), membrane enzyme, in vitro reconstitution, membrane protein, infectious disease, viral protein, zDHHC enzyme, protein palmitoylation
Abbreviations: ACE2, angiotensin-converting enzyme 2; COVID-19, coronavirus disease 2019; DMEM, Dulbecco's modified Eagle's medium; ERGIC, ER-Golgi intermediate compartment; FBS, fetal bovine serum; PTM, posttranslational modification; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2
The Coronavirus disease 2019 (COVID-19) pandemic, the most recent epidemic caused by an outbreak of zoonotic coronaviruses in the past two decades, was preceded closely by MERS in 2012 and SARS in 2003 (1, 2). Since the onset of COVID-19, over 160 million cases have been recorded, resulting in more than 3.3 million deaths globally (3). These trends undoubtedly suggest that coronaviruses represent an ongoing threat to human health and economic stability for years to come (4). As the causative agent of COVID-19, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has rightly received an unprecedented amount of attention from the scientific community and been the focus of intense investigation (5). However, several aspects about this virus remain poorly understood. These gaps in knowledge underscore the need to investigate the chemistry and biology of SARS-CoV-2 in a manner that could lead to a greater comprehension of the unknown aspects of the disease.
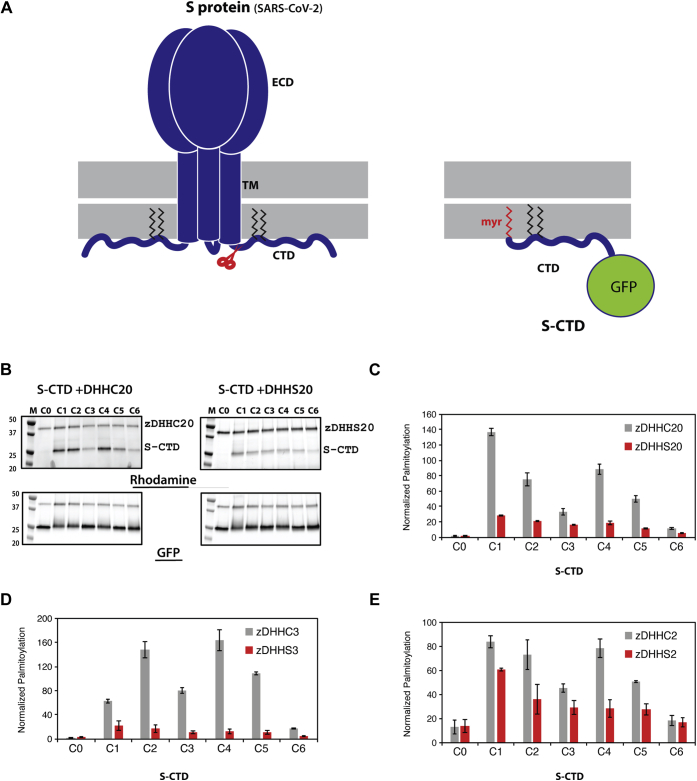
As a global initiative to eradicate COVID-19 and fortify a future defense against similar pandemics, not only is it essential to focus research on the discovery of antivirals, but it is also important to obtain insights into the cell biology of SARS-CoV-2 and identify pathways with the highest potential for the discovery of novel therapeutics. SARS-CoV-2 belongs to the β-Coronavirus family of enveloped, positive-strand RNA viruses(6). Upon infection, the Spike (S) protein (Fig. 1A), resident in the viral membrane, directly mediates the critical process of membrane fusion, which is initiated upon binding to the angiotensin-converting enzyme 2 (ACE2) receptor located on the host cell plasma membrane (7). Post infection, the viral genetic material encodes for several proteins along with the S protein, which traverse the ER-Golgi intermediate compartment (ERGIC) and Golgi where they get cleaved into the S1 and S2 proteins, that remain noncovalently attached to each other (8). Virions move through these organelles as membrane-bound carriers and eventually get trafficked out of the cell (9). Consequently, SARS-CoV-2 S protein has been the single most active target of various research studies in the last several months. Though most of the attention has been concentrated on the extracellular domain of S protein, resulting in a large body of structural and biochemical data, very little is known about the flexible C-terminal domain, located on the intracellular/intraviral side of the membrane.
Figure 1.
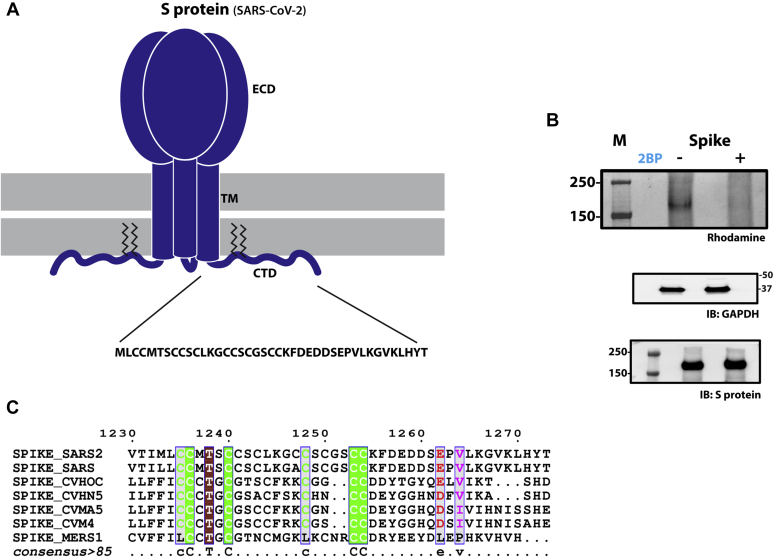
S-acylation of SARS-CoV-2 S protein.A, schematic showing the trimeric form of SARS-CoV-2 S protein with the ectodomain (ECD), Trans-membrane (TM) region, and C-terminal Domain (CTD). The CTD is further enhanced to show the cystine clusters, which are potential sites of S-acylation. B, S-acylation of S protein determined by click chemistry. Gel images detecting rhodamine signal at the position of S protein in the presence and absence of 2-Bromopalmitate (2BP), a global inhibitor of S-acylation. Western blots probed with antibodies against GAPDH and S protein respectively serve as controls for loading and total S protein expression. C, sequence alignment of the C-terminal domains of S protein from various coronaviruses.
It has been previously demonstrated that the C-terminal segment of the S protein from pathogenic viruses including closely related coronaviruses, such as SARS-CoV and MERS-CoV, are targets of posttranslational S-acylation (10, 11, 12), the most prevalent form of covalent lipid modification of proteins (13). Protein S-acylation, defined by the attachment of long-chain fatty acids to cysteine residues proximal to the cytosolic face of the membrane, is catalyzed by 23 members of the zDHHC family of integral membrane enzymes in humans (14, 15, 16). Both intracellular and transmembrane proteins are targets of S-acylation, which is important for a wide range of physiological processes; nearly 4000 proteins have been identified as substrates for zDHHC enzymes (13).
Interestingly, S-acylation was first discovered in viral S proteins (17) and is a well-conserved feature of the S proteins in many enveloped RNA viruses (18). S-acylation has been conclusively shown to be a crucial determinant for multiple facets of the viral replication cycle across a wide range of virus families (reviewed in (19)). Notably, mutation of S-acylation sites in the S protein of SARS-CoV severely compromises viral fusion. A sequence alignment between the cytoplasmic domains of SARS-CoV and SARS-CoV-2 shows that they are virtually identical with the exception of an additional cysteine residue in the SARS-CoV-2 sequence (Fig. 1C). Given the prevalence and importance of S-acylation in viruses, it is surprising that there is currently no biochemical dissection of the S-acylation of any viral protein that can offer a framework for the mechanistic understanding of this important PTM. Here, we demonstrate that the S protein of SARS-CoV-2 (hereafter referred to as S protein) is S-acylated, identify the sites of S-acylation, and unravel their role in viral infectivity. To accomplish this, we have developed a library of human zDHHC enzymes in mammalian expression vectors in order to identify putative members of the zDHHC family that are involved in S protein S-acylation. Finally, we reconstitute in vitro the S-acylation of an S protein fragment with purified zDHHC enzymes, which provides valuable insight into the mechanisms of substrate specificity, allows for further biochemical dissection, and ultimately lays the groundwork for the development of novel assays for the identification of small-molecule inhibitors (20, 21).
Results
SARS-CoV-2 spike protein is S-acylated at multiple sites within the cytoplasmic domain
Protein S-acylation involves cysteine residues that face the cytosol. The C-terminal cytoplasmic tail of SARS-CoV-2 S protein contains a series of membrane-proximal cysteines that lie adjacent to the transmembrane domain (Fig. 1A). To determine the S-acylation status of the cytoplasmic tail, we first utilized an in cellulo click chemistry assay. This assay was carried out by metabolic labeling of wild-type S protein expressed in HEK293T cells with 17-octadecynoic acid (17-ODYA), a fatty acid alkyne, followed by the conjugation and subsequent detection of a rhodamine fluorophore (22), which revealed robust S-acylation (Fig. 1B). A Δcys construct, in which all the cysteines in the intracellular C-terminus were mutated, showed no detectable S-acylation. The use of 2-bromopalmitate (2BP), a global inhibitor of protein S-acylation, completely abolished palmitoylation of S protein indicating the role of zDHHCs in this posttranslational modification (PTM) (Fig. 1B). Together, these experiments confirmed that the S protein is S-acylated by zDHHC enzymes at one or more cysteine residues within its cytoplasmic tail.
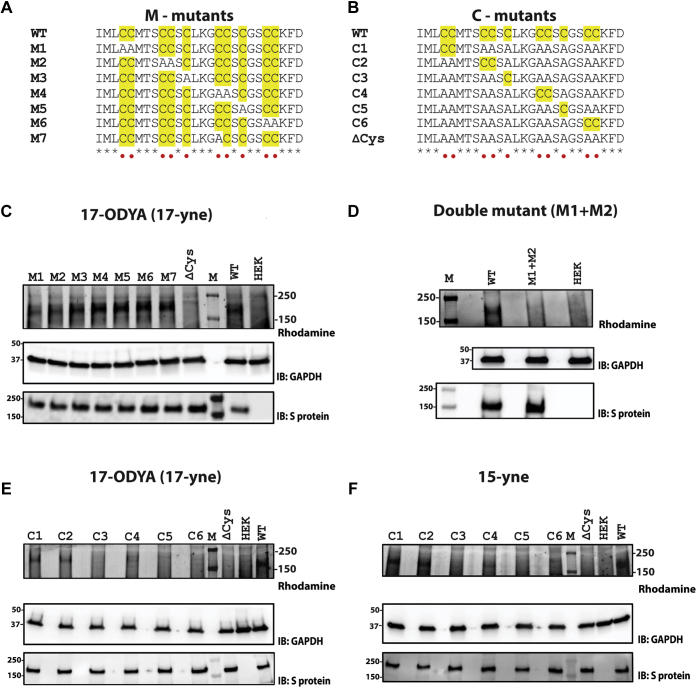
Sequence alignment of the cytoplasmic tail of the S proteins of SARS-CoV-2 together with those of SARS-CoV, MERS-CoV, and other closely related coronaviruses revealed that, in addition to the highly conserved cysteines in the C-terminus, SARS-CoV-2 S protein contains an additional cysteine (Fig. 1C). Consequently, the C-terminus of SARS-CoV-2 S protein appears to be the most cysteine-rich in sequence among the known human coronaviruses. In SARS-CoV-2, these cysteines can be broadly divided into six groups, with four groups each containing two cysteine residues and two groups with a single cysteine (Fig. 2A). To interrogate which of these sites are targets for S-acylation, we used the previously described click chemistry experiments with S protein constructs (M1 through M6) that have had each of the putative S-acylation sites removed by mutagenesis of the cysteine residue to alanine (Fig. 2A). An additional mutant, M7, represents a construct where the cysteine unique to SARS-CoV-2 S protein was mutated to mimic the sequence of SARS-CoV S protein (Figs. 1C and 2A). Expression of the S protein mutants in HEK293T cells, followed by click chemistry, revealed diminished S-acylation of the M1 and M2 mutants compared with wild-type, implicating the involvement of those cysteines in S-acylation (Fig. 2C). Comparison of the double mutant, M1+M2, showed a complete lack of detectable palmitoylation compared with the wild-type S protein (Fig. 2D).
Figure 2.
S-acylation of SARS-CoV-2 S protein mutants. Sequence alignment of the two classes of S protein mutants. Cysteine residues are highlighted in yellow. A, built on the backbone of wild-type (WT) S protein, M1 through M6 indicate either an individual cysteine or di-cysteine motif mutated to alanine. M7 mutant represents a construct where the additional cysteine in SARS-CoV-2 with respect to SARS-Co-V has been mutated out. B, built on the backbone of a cys-less (ΔCys) construct of SARS-CoV-2 S protein, C1 through C6 indicate either an individual cysteine or a di-cysteine motif reintroduced. S-acylation determined by click chemistry, using 17-ODYA for M-mutants (C), for M1+M2 double-mutant (D), for C-mutants (E); and using 15-yne for C-mutants (F). Lanes HEK, ΔCys, and WT serve as controls. Western blots probed with antibodies against GAPDH and S protein serve as controls for loading and total S protein expression respectively.
To gain further understanding, we performed the reciprocal experiment, in which we started from a construct in which all the cysteines on the intracellular C-terminus were mutated to alanine (Δcys) and, on this background, each of the six groups of cysteine was reintroduced individually (C1 through C6) (Fig. 2B). Thus, in each of the constructs from C1 through C6, only one of the six groups of cysteines was retained. Consistent with our previous experiments, C1 and C2 showed the most prominent levels of S-acylation, thus reinforcing findings from the aforementioned experiments with the corresponding M constructs (Fig. 2E). Additionally, modest level of palmitoylation was also detected at position C4. We also conducted experiments for C1 through C6 mutants with 15-yne, a fatty acid alkyne that more closely resembles palmitic acid (23) and is gaining traction in the field as a replacement for 17-ODYA. Similar results to those obtained with 17-ODYA were observed with C1 and C2 being the major site of S-acylation together with C4 being a third minor site (Fig. 2F). These experiments altogether revealed that the S protein is S-acylated and that the S-acylation status of the individual groups of cysteines within the cytosolic tail is markedly heterogeneous.
Effect of the S protein mutations on particle infectivity, incorporation into virus particles, subcellular localization, and cell-surface expression
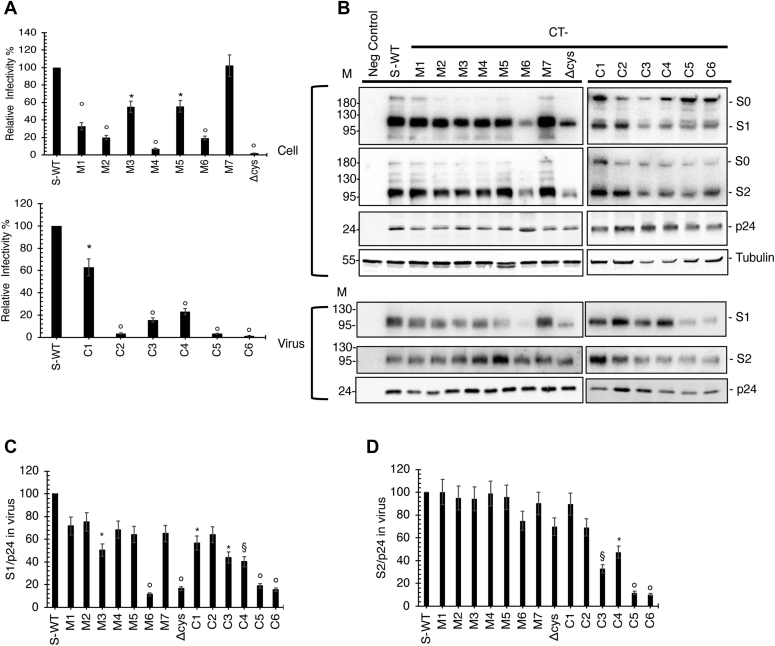
To determine the effects of the S protein mutations on particle infectivity, HIV-1 pseudotypes bearing WT and mutant S protein were generated and their infectivities measured in HEK293T cells expressing human TMPRESS2 and ACE2 (see Experimental procedures). The mutations were observed to elicit a range of effects on particle infectivity in this system. The M7 mutant showed WT levels of infectivity. M3, M5, and C1 were reduced by around 50% relative to WT; M1 was reduced by about 30% while M2, M6, C3, and C4 exhibited ∼20% WT levels of infectivity. The M4, ΔCys, C2, C5, and C6 showed a nearly complete loss of infectivity (Fig. 3A). Likewise, the M1+M2 mutation largely abrogated pseudotype particle infectivity (Fig. S1A).
Figure 3.
Effect of SARS-CoV-2 S protein mutations on pseudotype particle infectivity and S protein incorporation.A, infectivity of HIV-1 particles pseudotyped with S protein. HEK293T cells in 6-well dishes were cotransfected with pNL4-3.Luc.R-E- (3 mg) and vectors expressing WT or mutant SARS-CoV-2 S (300 ng). Virus supernatants were collected 2 days posttransfection, normalized for RT activity, and infectivity was measured by luciferase assay using TMPRESS2-transfected HEK293T cells stably expressing hACE2. Infectivity of HIV-1 particles pseudotyped with WT SARS-CoV-2 S protein was set to 100%. Data are derived from four independent experiments. B, Western blotting of cell- and particle-associated S protein. HEK293T cells were cotransfected with pNL4-3.Luc.R-E- (3 mg) and vectors expressing WT or mutant SARS-CoV-2 S (300 ng). Cell and virus lysates were prepared and subjected to Western blot analysis with anti-SARS-CoV-2 S1 or S2 Ab to detect S protein expression. HIV-Ig was used to detect HIV-1 Gag proteins. Mobility of molecular mass standards is shown on the left side of the blots. The levels of S1 (C) and S2 (D) in virus were quantified and normalized to p24 (CA) and set to 100% for WT S protein. p values using two-tailed unpaired t test: ∗p < 0.01, §p< 0.001, °p < 0.0001. Values that are not statistically significantly different from WT are not labeled.
To investigate the effects of the mutations on S protein expression, processing, and particle incorporation, we performed western blotting analysis of cell and virus lysates obtained from cells cotransfected with the HIV-1 reporter vector and the S protein expression vectors. A range of phenotypes was observed. The ΔCys mutant showed a marked reduction in levels of S1 in virions (approximately 20% of WT levels) (Fig. 3, B and C); however, levels of particle-associated S2 were close to those of WT (Fig. 3, B and D). The presence of WT levels of particle-associated S2 while having severely reduced incorporation of S1 suggests the possibility of increased S1 shedding from the surface of cells and/or particles. The M6 mutant showed a similar phenotype. Several of the mutants (e.g., C5 and C6) displayed severe reductions in the levels of both S1 and S2 in particles. The M2 mutant showed a pattern of S protein expression and incorporation that was similar to that of the WT, despite severely compromised infectivity (Fig. 3, A–D). Thus, mutation of Cys residues in the cytoplasmic tail of the S protein imposed a range of defects; in some cases, reduced infectivity was associated with reductions in the levels of S1 and/or S2 in virions, whereas for other mutants, reduced infectivity was not linked to defects in expression or incorporation. The M1+M2 mutant exhibited essentially WT levels of particle-associated S1 and S2 protein (Fig. S1, B–D).
We next examined the effect of a set of S protein mutations—ΔCys, M2, M4, and C2—on the subcellular localization of the S protein by confocal microscopy. Cells transfected with S protein expression vectors were stained with antibodies specific for the S protein and the lysosomal marker LAMP-1. The S protein showed a high level of colocalization with LAMP-1 (Pearson correlation coefficient r ∼0.8). For the mutants tested, the degree of colocalization between the S protein and LAMP-1 was similar to that of the WT (Fig. S2). We also investigated the cell-surface expression of WT and the ΔCys, M2, and M4 mutants by flow cytometry using antibodies specific for the S1 and S2 subunits of the S protein complex. The results indicated that these mutations did not significantly affect S protein expression at the cell surface (Fig. S3).
Identification of DHHC enzymes that catalyze spike S-acylation
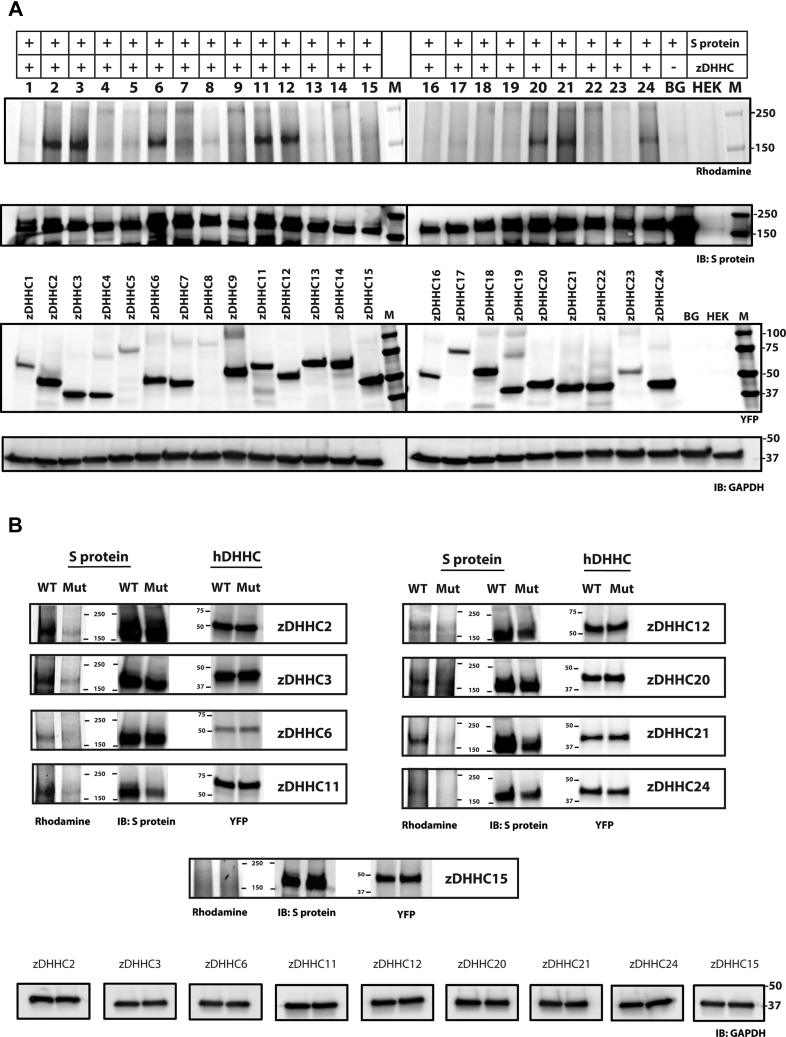
Bacterial and viral pathogens do not encode for enzymes that catalyze S-acylation. Instead, they rely on the S-acylation machinery of the host. In humans, protein S-acylation is carried out by 23 members of the zDHHC family of integral membrane enzymes that reside in various organellar membranes as well as the plasma membrane. zDHHC family of enzymes are promiscuous in their substrates with no defined consensus motif for S-acylation. Consequently, the zDHHC enzymes that S-acylate a target substrate need to be determined empirically. The most prevalent method, by far, has been to individually coexpress a target with each zDHHC member (24). The zDHHC members that increase the levels of S-acylation of a target can be reliably interpreted as the zDHHC enzymes which canonically act on that target, or at the very least, are capable of acting on that target. Historically, this has been achieved with a library of murine zDHHCs originally constructed by the Fukata lab (24). Presumably, this was because of the generous availability of the “Fukata library” as a reagent. However, in the context of SARS-CoV-2, we deemed it imperative to develop a generic screen (which we call the HeaTil screen named after two of the coauthors of this manuscript) with human zDHHCs that, at the outset, could provide more pertinent information to human disease. All 23 ZDHHC genes (Table S1) were inserted into the Bacmam vector (25) and optimized for overexpression with a terminal YFP fusion. Click-chemistry-based analyses of coexpression of S protein with each human zDHHC clearly identified multiple enzymes that palmitoylate S protein (Fig. 4A). Remarkably, S protein expression varied over a wide range when coexpressed with different zDHHC members (Fig. 4A). Although such variation is uncommon in the vast majority of cases where zDHHC enzymes for a specific target have been identified by this method, S-acylation has indeed been shown, in certain cases, to modulate protein levels by affecting protein turnover rates (26).
Figure 4.
S-acylation of SARS-CoV-2 S protein using the human zDHHC screen. S-acylation of SARS-CoV-2 S protein detected by azido rhodamine using click chemistry, when coexpressed with individual zDHHCs (1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24) (A); and with zDHHC and zDHHS of the selected candidates (B). Lane BG indicates transfection with S protein without any zDHHC while HEK serves as a negative control. Detection of YFP fluorescence indicates expression levels of zDHHC or zDHHS when coexpressed with S protein. Western blots probed with antibodies against GAPDH and S protein serve as controls for loading and total S protein expression respectively.
To further validate our findings, we tested the coexpression of the S protein together with each candidate zDHHC enzyme and its catalytically inactive zDHHS mutant (Fig. 4B). This experiment clearly demonstrated the ability of the selected zDHHC members to S-acylate the S protein and helped further substantiate our findings from the initial screen. For example, coexpression of S protein with zDHHC2 increased S protein S-acylation levels, whereas coexpression with the catalytically inactive zDHHS2 mutant did not (Fig. 4B). On the contrary, coexpression of either zDHHC15 or the catalytically inactive zDHHS15 did not change the S-acylation levels of S protein (Fig. 4B). Altogether, these experiments identified zDHHCs 2, 3, 6, 11, 20, 21, and 24 as putative S-acylation enzymes for the SARS-CoV-2 S protein.
In vitro reconstitution of spike S-acylation
An essential step towards the mechanistic dissection of S-acylation of any substrate protein is the development of in vitro assays using purified proteins, that together with cell-based assays, can provide discrete biochemical insights. However, remarkably few reports of in vitro reconstitution of substrate S-acylation by zDHHC enzymes exist in the literature (27, 28), most of them being carried out with very small peptide fragments of substrates (29). S protein, on the other hand, is a complex protein with a very large N-terminal ectodomain leading into a single transmembrane helix, followed by the relatively small C-terminal cytoplasmic domain, which contains the sites for S-acylation. To simplify the design of our in vitro assay, we focused on the C-terminal cytosolic domain, which harbors the sites for S-acylation (Fig. 5A, left panel). To facilitate detection, we appended a C-terminal GFP and replaced the transmembrane and the extracellular domain with an N-terminal myristoylation sequence (Fig. 5A, right panel). A cysteineless construct was generated on this backbone, S-CTD-ΔCys. Subsequently, individual clusters of cysteines were introduced onto this background to generate the S protein substrates, S-CTD-C1 through CTD-C6, as designed previously for the C-mutants (Fig. 2B). These substrates were purified from Escherichia coli and tested in a click-chemistry-based in vitro assay (see Experimental procedures) with palmitoyl alkyne-coenzyme A and purified zDHHCs to append an alkyne analog of palmitic acid onto the substrate followed by conjugation of a rhodamine fluorophore to the alkyne moiety for detection. Thus, the rhodamine channel revealed the extent of S-acylation while the GFP channel enabled independent assessment of total substrate protein levels in the assay (Fig. 5B).
Figure 5.
In vitro reconstitution of S protein CTD (S-CTD) S-acylation using purified proteins.A, development of S-CTD for in vitro biochemical studies. Schematic of full-length trimeric spike protein from which the S-CTD construct was derived (left panel). Schematic of the S-CTD protein with N-terminal myristoyl group (red), palmitoyl group (black) appended to a GFP at its C-terminal end (right panel). B, representative gel images displaying in vitro click chemistry results for S-CTD (C0 through C6) using zDHHC 20 and zDHHS 20. Top gels detect S-acylation through rhodamine signal while bottom gels display protein expression through GFP fluorescence. C–E, relative quantification of S-acylation of S-CTD (C0–C6) mutants by 2, 3, and 20 zDHHC and zDHHS enzymes. The intensity of the S-CTD band in the rhodamine detection gel was divided by the intensity of the S-CTD band in the GFP detection gel. Histograms represent average ± SD (n = 3).
Based on the zDHHCs identified from our in cellulo experiments (Fig. 4) and the availability of an established protocol for the purification of human zDHHC20 (28), we first focused on zDHHC20 for the in vitro assay. zDHHC20 showed robust S-acylation of the S-CTD-C1 through S-CTD-C6 constructs with a clear difference from the level of S-acylations detected with the catalytically inactive DHHS20 (Fig. 5B). There is a striking heterogeneity between the levels of acylation in S-CTD-C1 through S-CTD-C6 with the highest levels of S-acylation in S-CTD-C1, C2, and C4 (Fig. 5C). Intriguingly, these are the same cysteines that showed the highest levels of S-acylation in our in cellulo experiments with full-length S protein (Fig. 2), indicating that we are recapitulating salient aspects of the in cellulo S-acylation in our in vitro assay. We next tested both human zDHHC2 and human zDHHC3 using the same in vitro S-acylation assay. Again, we observed robust S-acylation of the S protein substrates in comparison with the respective catalytically inactive mutants and a similar level of heterogeneity among the levels of S-acylation in the different substrates (Fig. 5, D and E). However, the different zDHHC members showed a distinct spectrum of activities among the individual cysteines. Notably, S-CTD-C1, C2, and C4 showed the highest activity with zDHHC2, while S-CTD-C2 and C4 showed the highest activity with zDHHC3. Taken together, these experiments show that between zDHHC2, zDHHC3, and zDHHC20, S-CTD-C1, C2 and C4 showed the highest level of S-acylation activity, in agreement with our in cellulo experiments.
Discussion
S-acylation of viral proteins has been known since the discovery of this PTM (17). SARS-CoV-2 S protein harbors the most cysteine-rich C-terminal cytosolic tail compared with other related coronaviruses such as SARS-CoV and MERS-CoV (Fig. 1). However, it has been unclear whether some or all of these cysteines are S-acylated and how that might impact viral infectivity. Herein, we demonstrate conclusively that the S protein of SARS-CoV-2 is S-acylated and identify the sites of S-acylation. We also identify specific cysteines that are targets for this modification. Although the clusters of cysteines are all positioned in the membrane-proximal segment of the C-terminus within a stretch of 20 residues, yet our in cellulo assay demonstrated a remarkable heterogeneity in the ability of different cysteines to become S-acylated. Our initial analysis identified cysteines corresponding to positions C1 and C2 as the major site of palmitoylation along with C4, which demonstrated modest S-acylation. Remarkably, mutating cysteines at these positions selectively reduced viral infectivity. Even the mutation of a single cluster of cysteines, such as the M2 construct, led to an 80% reduction in comparison to the wild-type S protein. This reduction was not due to aberrant S protein expression, processing, or maturation. Notably, C6 is a fourth cysteine cluster whose mutation (construct M6) showed a similar effect on viral infectivity. We did not identify C6 as a target site for S-acylation. However, mutating C6 (construct M6) results in aberrant S protein maturation and incorporation in the virus (Fig. 3, B–D), and this is likely the cause for the reduced infectivity of this mutant. Interestingly, the M7 mutant, which mimics the arrangement of cysteines in SARS-CoV, differs from the M4 mutant by the position of one additional cysteine found in SARS-CoV-2 (Fig. 3A). The presence of this additional cysteine led to a marked difference in infectivity, underscoring its likely importance in SARS-CoV-2.
To further identify zDHHCs that palmitoylate S protein, we developed a library of human zDHHC enzymes. Our screen identified zDHHCs 2, 3, 6, 11, 12, 20, 21, and 24 as candidate enzymes that potentially palmitoylate the S protein. These zDHHCs intriguingly overlap with zDHHCs identified in the S-acylation of other viral proteins such as hemagglutinin of Influenza A virus (30), GP5/M protein from Porcine reproductive and respiratory syndrome virus (PRRSV) (31), and BEnv from the bovine foamy virus (32). Interestingly, the level of S protein expression varied considerably with coexpression of the identified zDHHC, indicating a connection between S-acylation and biogenesis/stability of S protein, as observed in a recent study (33). To further validate the results of our in cellulo screen, we demonstrate the first in vitro reconstitution of S-acylation of a viral protein using the SARS-CoV-2 S protein. There are no reports of an in vitro reconstitution of S-acylation for any viral protein. Yet, this is an essential first step toward the mechanistic dissection of S-acylation any protein substrate, especially the S protein. zDHHC20 was one of the prominent enzymes that palmitoylates S protein as identified both in the present and another recent study (33). A ready purification protocol for zDHHC20 was available to us since we solved high-resolution structures of human zDHHC20 (28). With these considerations, we chose to first focus on developing an in vitro assay for S protein S-acylation with the entire cytosolic fragment and purified zDHHC20. Remarkably, the pattern of S-acylation activity on the individual clusters of cysteines was distinctly heterogeneous. Moreover, the highest activity was seen with C2 and C4, two of the three sites identified by our in cellulo S-acylation assays. These are also the same two sites that, when mutated, severely compromised viral infectivity. In sharp contrast to these results, when we reconstituted the S-acylation of S protein with zDHHC2 or zDHHC3, the pattern of reactivity, although similarly heterogeneous, was notably different. Most striking was the fact that zDHHC3 activity on the C1 site was greatly diminished compared with that of zDHHC2 and zDHHC20. Interestingly, previous studies analyzing the acyl chain length selectivities of zDHHC 2, 3, and 20 demonstrated that, while zDHHC2 and zDHHC20 are capable of utilizing acyl-CoAs consisting of chain lengths greater than 16 carbons, zDHHC3 discriminates against acyl chains of such length (28, 34). This observation provides insight into our results when considering studies that demonstrated that the viral spike protein for influenza A (hemagglutinin) is primarily modified with a 16-carbon fatty acid at most sites on its cytoplasmic tail accept for the site closest to the transmembrane domain, which is predominantly modified with an 18-carbon fatty acid (30). Our data suggest a mechanism for such heterogeneity by providing evidence that certain zDHHCs, which cannot utilize 18-carbon fatty acids (like zDHHC3), are incapable of acylating sites adjacent to the transmembrane region. Nevertheless, for all three zDHHC enzymes, 2, 3, and 20, C1, C2, and C4 cysteine clusters displayed the highest levels of S-acylation. These experiments not only provide a blueprint for an in vitro reconstitution of viral protein S-acylation with purified zDHHC enzymes but also set the stage for further probing of the mechanistic underpinnings of S-acylation of SARS-CoV-2 S protein.
What could be the possible role of S-acylation of S protein in the life cycle of SARS-CoV-2? S-acylation is known to target proteins to cholesterol-rich membrane microdomains as has been shown for SARS-CoV (10). Moreover, it has been shown for other enveloped viruses that cholesterol is an important determinant of the modulation of local membrane curvature by surface glycoproteins (35), (similar to S protein in SARS-CoV-2) as well as a determinant of viral fusion with the target membrane (36, 37). A recent study showed that cholesterol and S protein association plays a crucial role in SARS-CoV-2 infection (38). Furthermore, we also find that the cys-less (ΔCys) form of S protein was compromised in its ability to incorporate into virions and could be attributed to the improper translocation to cholesterol-rich domains. Future studies delineating the precise structural and chemical underpinnings of this process will shed more light on the interplay between S-acylation and the SARS-CoV-2 life cycle. In this context, it is intriguing that in comparison with other related coronaviruses, SARS-CoV-2 has the most cysteine-rich cytoplasmic tail. Finally, these findings also highlight S-acylation as a possible new therapeutic target against viral diseases. Although targeting zDHHC enzymes with small molecules had been proposed as a therapeutic strategy for several other diseases (21), the successful development of a substrate-selective small-molecule inhibitor of S-acylation has not yet been realized. The recent high-resolution structures of zDHHC20 and zDHHC15 together with the in vitro assays described here are the first steps toward that goal.
Experimental procedures
Plasmids
HeaTil screen (human zDHHC screen)
Plasmids pLH1-24, expressing Homo sapiens zDHHC enzymes, were cloned into modified pEG BacMam vector (Addgene, # 160451) containing N or C-Terminal YFP-tag. ZDHHC 1–24 sequences (Table S1) were derived from DNASU and Genomics Online. Amplification of individual ZDHHC sequences was performed using specific primers. Each ZDHHC PCR product was gel purified using NucleoSpin Gel and PCR Clean-up kit (Takara Bio). N or C terminal YFP- tagged pEG BacMam vectors were cut with restriction enzymes EcoRI/XhoI (New England Biolabs). Each PCR fragment contained 20–40 bp of overlap with digested pEG BacMam vector and integrated into the modified pEG BacMam vector using Gibson Assembly Master Mix (New England Biolabs) (39).
Human zDHHS and spike
pLHs1–24 containing zDHHS enzymes in modified pEG BacMam vectors, and M1–M7, Δcys, and C1-C6 were generated using and QuikChange II Site-Directed Mutagenesis Kit (Agilent). M1-M7, Δcys, and C1-C6 DNA were cloned from pcDNA3.1 (Addgene) containing Human Codon-Optimized 2019-nCov-S Protein. All plasmids were transformed into XL1-Blue super-competent Cells (Agilent), subsequently miniprepped with NucleoSpin Plasmid kit (Takara Bio) and sequence verified.
S-C-terminal domain (S-CTD)
pET25b vector (Sigma) was modified to contain the superfold GFP (sfGFP) (Addgene, #85492). Δcys construct, developed above, of SARS-CoV-2 was used as a backbone. The portion of the C-terminal Domain, which contains the putative S-acylation sites (1235–1260) of Δcys, was appended with sfGFP (Addgene, #85492) at its C-terminus and a consensus sequence of NMT protein substrates (MGXXXT/S) at its N-terminus denoted as the C0 construct. C1–C6 constructs were built on the C0 DNA using and QuikChange II Site-Directed Mutagenesis Kit. NMT (pBB131) was originally developed by the Gordon lab (40)
Mammalian cell culture
Human embryonic kidney cells 293 (HEK-293) ATCC CRL-1573 were cultured in Dulbecco's modified Eagle's medium without glutamine (DMEM; Corning) supplemented with 10% fetal bovine serum (FBS; Corning), L-glutamine (Corning), and penicillin/streptomycin (Corning). Cell were propagated at 37 °C with 5% CO2 and humidity. Transfection was carried out at 80% confluency of HEK293T cells in a 6-well tissue culture plate (Costar; Corning). For transfection, DNA, purified in elution buffer (AE; MACHERY-NAGEL), was mixed with Polyethylenimine MAX, (PEI; Polyscience) dissolved in PBS at 1 mg/ml and readjusted to pH 7.0, at a ratio of 1:3:: DNA:PEI in unsupplemented DMEM media. After 15–20 min at room temperature, the DNA-PEI mixture was added dropwise to the cells and left for 36 h.
HeaTil screen (human ZDHHC 1–24)
pLH 1–24 (human ZDHHC 1–24) with either N or C terminal YFP were expressed by transfecting HEK293T cells with 2 μg DNA. Cells were harvested at room temperature with two washes in 1× PBS (10XPBS; Corning) and resuspended in 150 μl of 1× PBS supplemented with 1.5 mM PMSF (GoldBio). Protein extraction was initiated by lysing with a short sonication using microtip followed by extraction in 1.5% n-dodecyl-β-D-maltoside- cholesteryl hemisuccinate (DDM-CHS) prepared in 40 mM HEPES, pH 8.0 for 90 min at 4 °C. The lysates were clarified by centrifugation at 21,000g, 4 °C for 12 min. The clarified supernatant was mixed with 5× SDS-PAGE loading dye and run on a precast 4–20% gel (Mini-PROTEAN TGX; Bio-Rad). Gels were with screened for fluorescence from YFP. For each individual zDHHC, a selection was made based on the expression levels of either the N or C terminal YFP tagged construct. A pool of zDHHC (1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24) was thus selected to constitute the HeaTil screen. pLHs 1–24 (human ZHHDS 1–24) were designed on the pool of ZDHHC constructs that formed the HeaTil screen.
In cellulo assay
Spike mutants
Expressions of Spike and its various mutants were carried out by transfecting HEK293T cells with 3 μg DNA and PEI. After 36 h the media was replaced with 1 ml of DMEM conditioned with 1%, fatty acid free, Bovine Serum Albumin (BSA; Sigma). The same media contained 50 μM 2-bromopalmitate (2BP; Sigma) solubilized in DMSO for 2BP experiments. Post 2 h, cells were fed ∼0.1 mM saponified (41) fatty acid alkynes 17-Octadecynoic Acid (17-ODYA; Cayman Chemical) or 15-hexadecynoic acid (15-yne; Avanti Polar) dissolved in DMEM condition with 20%, fatty acid free, BSA. After 6 hours cells were harvested into microfuge tubes with two washes of 1× PBS.
Spike and HeaTil screen
A stock of wild-type Spike DNA was prepared in unconditioned DMEM media at ∼0.8 μg/100 μl. This DNA was further aliquoted to multiple 1.5 ml microfuge tubes. Human ZDHHC constructs from the HeaTil screen (∼1–1.5 μg) were individually added to each tube and mixed gently. PEI solubilized in unconditioned DMEM was added in three times excess to the concentration of total DNA and incubated for 20 min. This DNA-PEI mixture was then added to each well of a 6-well plate. Additionally, spike DNA, devoid of ZDHHC, was also transfected as a control to check for background S-acylation. 17-ODYA was fed to the cells and harvested as described above.
Cells were resuspended in 1× PBS supplemented with 1.5 mM PMSF and extracted in 1.5% DDM-CHS for 90 min at 4 °C as detailed before. The clarified lysate was then subjected to copper-based click chemistry analysis (22) with tetramethyl-rhodamine azide (TAMRA; Lumiprobe) as a reporter group. Briefly, clarified lysate was mixed with 100 μM TAMRA (dissolved in DMSO) and 100 μM Tris[(1-benzyl-1H-1,2,3-triazol-4-yl) methyl] amine (TBTA) (Sigma), followed by the addition of 1 mM CuSO4 and 1 mM TCEP (GodlBio), both dissolved in water. This mixture was allowed to react for 1 h with intermittent mixing. 5× SDS sample loading buffer was added to the reaction mixture and separated on precast 4–20% gel (Mini-PROTEAN TGX; Bio-Rad). S-acylated proteins were visualized by detecting fluorescence from the rhodamine channel using a ChemiDoc MP System (Bio-Rad). Expression of YFP tagged zDHHC and Spike proteins were respectively detected using YFP fluorescence and Western blot using anti-Spike antibodies (SARS-CoV-2 Spike RBD Antibody; Sino-Biological, # 40592-T62). Loading controls were determined through blots developed with anti-GADPH (Invitrogen, #AM4300) antibodies.
Expression and purification of spike-CTD-GFP mutants (C0-C6)
E. coli BL21(DE3)-RIL competent cells were cotransformed with NMT and S-CTD mutants (C0-C6). Large 1 L cultures were grown at 37 °C from a 5 ml overnight starter culture in LB containing 100 μg/ml of ampicillin (GoldBio) and 50 μg/ml kanamycin (Amresco). When the cell density reached an OD600 of 0.2, the media was supplemented with 60 μM myristic acid (Acros Organic) from a 1 M stock in 100% ethanol. 1 mM IPTG (GoldBio) was added to the cells when they reached an OD600 of 0.8. After 4 h, cells were harvested by centrifugation at 4000g for 10 min at 4 °C. Cell pellets were flash frozen in liquid nitrogen and stored at –80 °C.
For purification, ∼ 2 g of cell pellet was resuspended in 80 ml of buffer A (50 mM NaPO4 (pH 7.5), 300 mM NaCl, 5% glycerol, 5 mM βME, 30 mM imidazole) supplemented with 1 mM benzamidine HCl (GoldBio) and 0.1 mM PMSF. Cells were lysed by sonication and clarified by centrifugation at 38,000g, 4 °C for 30 min. The supernatant was bound to 5 ml of Talon (Clontech) resin pre-equilibrated with buffer A. The resin was washed with five column volumes (CV) of buffer A. Protein was eluted with 3–4 CV of buffer B (50 mM NaPO4 [pH 7.5], 300 mM NaCl, 5% glycerol, 5 mM βME, 300 mM imidazole). The eluate was dialyzed overnight into buffer C (20 mM NaPO4 [pH 7.0], 1 M (NH4)2SO4, 5 mM DTT). This dialyzed sample was loaded onto a HiTrap 5 ml Butyl FF (Cytiva) column equilibrated in buffer C. Protein was eluted in a 60 ml gradient of buffer D (20 mM NaPO4 [pH 7.0], 5 mM DTT). Fraction containing myristoylated S-CTD-GFP was pooled and dialyzed in storage buffer (20 mM HEPES [pH 7.4], 150 mM NaCl, 1 mM TCEP) and later concentrated to ∼12 μM and stored at –80 °C.
Expression and purification of zDHHC/zDHHS enzymes
YFP-zDHHC and YFP-zDHHS-2, 3, and 20 were expressed in HEK293T cells grown in 15 cm tissue culture dish (Corning) as described above in “mammalian cell culture.” Cells were transfected with 20 μg of DNA using PEI at a ratio of 1:3:: DNA:PEI. After 48 h, cells were harvested and washed with 1× PBS before being flash frozen in liquid nitrogen and stored at –80 °C.
Frozen cells were thawed on ice and resuspended in 1.35 ml of extraction buffer (50 mM HEPES, pH 7.5), 250 mM NaCl, 5 mM βME, Protease inhibitors (Benzamidine HCl, PMSF, AEBSF (GoldBio), Aprotinin (Sigma), Pepstatin (RPI), Leupeptin (RPI), and DNase (Worthington). Extraction was initiated with addition of 150 ul of 20% DDM and left on a rotator for 2 h at 4 °C. Post extraction, lysate was clarified at 16,000g, 4 °C for 20 min. The supernatant was applied to 200 μl of pre-equilibrated TALON resin and mixed on a rotator for ∼1 h at 4 °C. The resin was first washed twice with 700 μl of 50 mM HEPES (pH 7.4) 250 mM NaCl, 5 mM βME, and 2 mM DDM, and then twice with 700 μl of 20 mM HEPES (pH 7.4) 250 mM NaCl, 5 mM βME, 2 mM DDM, and 30 mM imidazole. Protein was eluted in 700 μl of 20 mM HEPES (pH 7.4) 250 mM NaCl, 5 mM βME, 2 mM DDM, and 300 mM imidazole. Protein was exchanged into a buffer containing 20 mM HEPES (pH 7.4), 150 mM NaCl, and 2 mM DDM using a 50 kDa molecular weight cutoff (MWCO) centrifugal filter (Amicon-15, EMD-Millipore). The protein was concentrated down to 1 μM and used immediately for in vitro assay.
In vitro S-acylation assay
Frozen aliquots of the S-CTD mutants were thawed before being diluted to 2 μM in buffer containing 20 mM HEPES, pH 7.4, 150 mM NaCl. Ten microliter of 1 μM zDHHC enzyme was added to 13 μl of S-CTD mutant. 0.5 μl of 4 mM Palmitoyl Alkyne Coenzyme A-trifluoroacetate salt (Cayman chemicals) was then added and incubated at room temperature for an hour. Copper-assisted click chemistry reaction was initiated by the addition of the following reagents: 0.5 μl of 10 mM tetramethylrhodamine azide (TAMRA-Lumiprobe), 0.5 μl of 2 mM Tris[(1-benzyl-1H-1,2,3-triazol-4-yl) methyl] amine (TBTA-Sigma), 0.3 μl of 50 mM TCEP, and 0.5 μl of 50 mM CuSO4. The reaction proceeded for an hour at room temperature before being quenched with 5× SDS sample buffer. Twenty-five microliter of each sample was loaded onto a 12% SDS-PAGE gel (Mini-PROTEAN TGX; Bio-Rad). Gels were visualized using a ChemiDoc MP System (Bio-Rad) where S-acylation was observed in the rhodamine channel while protein expression was detected using fluorescence from GFP/YFP.
Transfection and infectivity assays
All cell lines were cultured in DMEM containing 10% FBS (HyClone) and 1% penicillin-streptomycin (Lonza) at 37 °C with 5% CO2. HEK293T cells were purchased from ATCC. HIV-1 particles pseudotyped with SARS-CoV-2 S protein were generated by transfecting HEK293T cells with the HIV-1 luciferase-encoding reporter vector pNL4-3.Luc.R-E- (obtained from the NIH AIDS Reagent Program) and a pcDNA-SARS-CoV-2 S protein expression vector (a gift from Thomas Gallagher, Loyola University) using Lipofectamine 2000 (Invitrogen) according to manufacturer's instructions. Virus-containing supernatants were then filtered through 0.45-μm membrane 48 h posttransfection and virus was quantified by measuring RT activity. The HEK293T-hACE2 cell line (BEI; NR-52511) was transfected with pCAGGS-TMPRESS2 (a gift from Dr Stefan Pöhlman, German Primate Center) and was inoculated with the RT-normalized S protein-pseudotyped virus for 48 h in the presence of 10 μg/ml DEAE-dextran. Cells were then lysed with BriteLite Luciferase reagent (PerkinElmer) and luciferase was measured in a GloMax Navigator Microplate Luminometer (Promega) at 48 h postinfection. Relative infectivities were normalized to that of pseudotyped particles bearing WT S protein.
Western blotting
Cells were lysed in lysis buffer (10 mM iodoacetamide [Sigma-Aldrich], Complete protease inhibitor tablets [Roche], 300 mM sodium chloride, 50 mM Tris-HCl [pH 7.5], and 0.5% Triton X-100 [Sigma-Aldrich]). Virus was pelleted through 20% sucrose and resuspended in lysis buffer. Cell and virus lysates were heated to 95 °C for 5 min. Samples were analyzed on 10% 1.5 mm Tris-glycine gels, followed by transfer to PVDF membrane (BioRad) using a BioRad Trans-Blot Turbo Transfer system according to manufacturer's instructions. Blots were probed with the following antibodies: rabbit anti-SARS-CoV-2 S (Sino Biological; #40589), rabbit anti-SARS-CoV-2 S2 (Sino Biological; #40590), human anti-HIV immune globulin (HIV-Ig; NIH AIDS Reagent Program), and mouse anti-tubulin (Invitrogen; #62204). Secondary antibodies conjugated with horseradish peroxidase were used for chemiluminescent detection for SARS-CoV-2 S and other proteins were detected using near-IR fluorescently labeled secondary antibodies (Azure Biosystems 650 and 800; AC2165 and AC2135). Protein bands were visualized using a Sapphire Biomolecular Imager (Azure Biosystems) and analyzed with AzureSpot (Azure Biosystems).
Confocal microscopy
HEK293T cells transfected with vectors expressing WT or mutant SARS-CoV-2 S protein were seeded onto 18 mm coverslips (Electron Microscopy Sciences; 72291-06) pretreated with fibronectin (Sigma-Aldrich; GC010) at 1:50 dilution in PBS for 30 min at room temperature. One day posttransfection cells were fixed with 4% paraformaldehyde (Electron Microscopy Sciences, BM-155) for 1 h and quenched with 0.1 M glycine in PBS for 10 min. Cells were then permeabilized with 0.1% Triton X-100 (Sigma-Aldrich; T-8787) in PBS for 3 min, blocked with 10% BSA/PBS (Sigma Aldrich) for 30 min, stained with primary antibodies (BEI Resources, NIAID, NIH): monoclonal anti-SARS coronavirus recombinant human IgG1, Clone CR3022 (NR 52392) and Invitrogen Anti-LAMP1 monoclonal antibody, Ly1C6 (MA1-164) at a 1:400 dilution with 0.1% Triton and 1% BSA in PBS for 1 h. Cells were then washed 3× in PBS and incubated with secondary antibodies (Invitrogen, Alexa Fluor555, and Alexa Fluor488) and DAPI stain and mounted with Fluoromount-G (Electron Microscopy Sciences; 0100-01). Imaging was performed with a Leica TCS SP8 microscope (Leica Microsystems Inc) using a 63× oil-immersion objective. 3-D images were generated using ImageJ (NIH) from z-stack images. Background was subtracted using ImageJ's built-in “rolling ball” background subtraction process. Colocalization analyses were performed using the colocalization test within ImageJ. Untransfected cells were excluded from analyses.
Flow cytometry
HEK293T cells transfected with vectors expressing WT or mutant SARS-CoV-2 S protein were seeded in 6-well plates and collected. Cells were pelleted and stained with antibodies targeting the S1 subunit (Sinobiological, #40589-T62) or S2 subunit (Invitrogen, #MA5-35946) at 1 μg/μl, Alexa Fluor 647 at 2 μg/μl was used for secondary antibody. Cells were fixed with 4% paraformaldehyde prior to flow cytometry analysis with a FACSCalibur.
Data availability
All data are available upon request from the authors and are included in the article.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The following reagent was obtained through BEI Resources, NIAID, National Institutes of Health: Monoclonal Anti-SARS Coronavirus Recombinant Human IgG1, Clone CR3022 (produced in Nicotiana benthamiana), NR-52392. We are grateful for support through a Strategic Planning Award from the Office of the Director, NICHD, National Institutes of Health.
Author contributions
A. B. conceptualization; R. P., C. M. L., R. E. M., L. B. H., G. V., and E. T. C. formal analysis; E. O. F. and A. B. funding acquisition; R. P., C. M. L., R. E. M., L. B. H., G. V., and E. T. C. investigation; A. B. project administration; E. O. F. and A. B. supervision; E. O. F. and A. B. writing–original draft; R. P., R. E. M., L. B. H., E. O. F., and A. B. writing–review and editing
Funding and additional information
This research is supported by the Intramural Research Program of the Center for Cancer Research, NCI, National Institutes of Health (to E. O. F.) and the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), National Institutes of Health (A. B.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Craig Cameron
Supporting information
References
- 1.Sharif-Yakan A., Kanj S.S. Emergence of MERS-CoV in the Middle East: Origins, transmission, treatment, and perspectives. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cherry J.D. The chronology of the 2002-2003 SARS mini pandemic. Paediatr. Respir. Rev. 2004;5:262–269. doi: 10.1016/j.prrv.2004.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicola M., Alsafi Z., Sohrabi C., Kerwan A., Al-Jabir A., Iosifidis C., Agha M., Agha R. The socio-economic implications of the coronavirus pandemic (COVID-19): A review. Int. J. Surg. 2020;78:185–193. doi: 10.1016/j.ijsu.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.COVID research: A year of scientific milestones. Nature. 2021 doi: 10.1038/d41586-020-00502-w. [DOI] [PubMed] [Google Scholar]
- 6.Machhi J., Herskovitz J., Senan A.M., Dutta D., Nath B., Oleynikov M.D., Blomberg W.R., Meigs D.D., Hasan M., Patel M., Kline P., Chang R.C., Chang L., Gendelman H.E., Kevadiya B.D. The natural history, pathobiology, and clinical manifestations of SARS-CoV-2 infections. J. Neuroimmune Pharmacol. 2020;15:359–386. doi: 10.1007/s11481-020-09944-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffmann M., Kleine-Weber H., Pohlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784.e775. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nal B., Chan C., Kien F., Siu L., Tse J., Chu K., Kam J., Staropoli I., Crescenzo-Chaigne B., Escriou N., van der Werf S., Yuen K.Y., Altmeyer R. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J. Gen. Virol. 2005;86:1423–1434. doi: 10.1099/vir.0.80671-0. [DOI] [PubMed] [Google Scholar]
- 9.Tooze J., Tooze S.A., Fuller S.D. Sorting of progeny coronavirus from condensed secretory proteins at the exit from the trans-Golgi network of AtT20 cells. J. Cell Biol. 1987;105:1215–1226. doi: 10.1083/jcb.105.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McBride C.E., Machamer C.E. Palmitoylation of SARS-CoV S protein is necessary for partitioning into detergent-resistant membranes and cell-cell fusion but not interaction with M protein. Virology. 2010;405:139–148. doi: 10.1016/j.virol.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petit C.M., Chouljenko V.N., Iyer A., Colgrove R., Farzan M., Knipe D.M., Kousoulas K.G. Palmitoylation of the cysteine-rich endodomain of the SARS-coronavirus spike glycoprotein is important for spike-mediated cell fusion. Virology. 2007;360:264–274. doi: 10.1016/j.virol.2006.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shulla A., Gallagher T. Role of spike protein endodomains in regulating coronavirus entry. J. Biol. Chem. 2009;284:32725–32734. doi: 10.1074/jbc.M109.043547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blanc M., David F.P.A., van der Goot F.G. SwissPalm 2: Protein S-palmitoylation database. Methods Mol. Biol. 2019;2009:203–214. doi: 10.1007/978-1-4939-9532-5_16. [DOI] [PubMed] [Google Scholar]
- 14.Stix R., Lee C.J., Faraldo-Gomez J.D., Banerjee A. Structure and mechanism of DHHC protein acyltransferases. J. Mol. Biol. 2020;432:4983–4998. doi: 10.1016/j.jmb.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linder M.E., Deschenes R.J. Palmitoylation: Policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- 16.Jiang H., Zhang X., Chen X., Aramsangtienchai P., Tong Z., Lin H. Protein lipidation: Occurrence, mechanisms, biological functions, and enabling technologies. Chem. Rev. 2018;118:919–988. doi: 10.1021/acs.chemrev.6b00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt M.F., Schlesinger M.J. Fatty acid binding to vesicular stomatitis virus glycoprotein: A new type of post-translational modification of the viral glycoprotein. Cell. 1979;17:813–819. doi: 10.1016/0092-8674(79)90321-0. [DOI] [PubMed] [Google Scholar]
- 18.Kordyukova L.V., Serebryakova M.V., Khrustalev V.V., Veit M. Differential S-acylation of enveloped viruses. Protein Pept. Lett. 2019;26:588–600. doi: 10.2174/0929866526666190603082521. [DOI] [PubMed] [Google Scholar]
- 19.Gadalla M.R., Veit M. Toward the identification of ZDHHC enzymes required for palmitoylation of viral protein as potential drug targets. Expert Opin. Drug Discov. 2020;15:159–177. doi: 10.1080/17460441.2020.1696306. [DOI] [PubMed] [Google Scholar]
- 20.Lanyon-Hogg T., Ritzefeld M., Sefer L., Bickel J.K., Rudolf A.F., Panyain N., Bineva-Todd G., Ocasio C.A., O'Reilly N., Siebold C., Magee A.I., Tate E.W. Acylation-coupled lipophilic induction of polarisation (Acyl-cLIP): A universal assay for lipid transferase and hydrolase enzymes. Chem. Sci. 2019;10:8995–9000. doi: 10.1039/c9sc01785b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fraser N.J., Howie J., Wypijewski K.J., Fuller W. Therapeutic targeting of protein S-acylation for the treatment of disease. Biochem. Soc. Trans. 2020;48:281–290. doi: 10.1042/BST20190707. [DOI] [PubMed] [Google Scholar]
- 22.Martin B.R., Cravatt B.F. Large-scale profiling of protein palmitoylation in mammalian cells. Nat. Methods. 2009;6:135–138. doi: 10.1038/nmeth.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yap M.C., Kostiuk M.A., Martin D.D., Perinpanayagam M.A., Hak P.G., Siddam A., Majjigapu J.R., Rajaiah G., Keller B.O., Prescher J.A., Wu P., Bertozzi C.R., Falck J.R., Berthiaume L.G. Rapid and selective detection of fatty acylated proteins using omega-alkynyl-fatty acids and click chemistry. J. Lipid Res. 2010;51:1566–1580. doi: 10.1194/jlr.D002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fukata M., Fukata Y., Adesnik H., Nicoll R.A., Bredt D.S. Identification of PSD-95 palmitoylating enzymes. Neuron. 2004;44:987–996. doi: 10.1016/j.neuron.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 25.Goehring A., Lee C.H., Wang K.H., Michel J.C., Claxton D.P., Baconguis I., Althoff T., Fischer S., Garcia K.C., Gouaux E. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protoc. 2014;9:2574–2585. doi: 10.1038/nprot.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaballa M.E., van der Goot F.G. The molecular era of protein S-acylation: Spotlight on structure, mechanisms, and dynamics. Crit. Rev. Biochem. Mol. Biol. 2018;53:420–451. doi: 10.1080/10409238.2018.1488804. [DOI] [PubMed] [Google Scholar]
- 27.Dong X., Mitchell D.A., Lobo S., Zhao L., Bartels D.J., Deschenes R.J. Palmitoylation and plasma membrane localization of Ras2p by a nonclassical trafficking pathway in Saccharomyces cerevisiae. Mol. Cell Biol. 2003;23:6574–6584. doi: 10.1128/MCB.23.18.6574-6584.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rana M.S., Kumar P., Lee C.J., Verardi R., Rajashankar K.R., Banerjee A. Fatty acyl recognition and transfer by an integral membrane S-acyltransferase. Science. 2018;359 doi: 10.1126/science.aao6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gottlieb C.D., Zhang S., Linder M.E. The cysteine-rich domain of the DHHC3 palmitoyltransferase is palmitoylated and contains tightly bound zinc. J. Biol. Chem. 2015;290:29259–29269. doi: 10.1074/jbc.M115.691147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gadalla M.R., Abrami L., van der Goot F.G., Veit M. Hemagglutinin of Influenza A, but not of Influenza B and C viruses is acylated by ZDHHC2, 8, 15 and 20. Biochem. J. 2020;477:285–303. doi: 10.1042/BCJ20190752. [DOI] [PubMed] [Google Scholar]
- 31.Zhang M., Han X., Osterrieder K., Veit M. Palmitoylation of the envelope membrane proteins GP5 and M of porcine reproductive and respiratory syndrome virus is essential for virus growth. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chai K., Wang Z., Xu Y., Zhang J., Tan J., Qiao W. Palmitoylation of the bovine foamy virus envelope glycoprotein is required for viral replication. Viruses. 2020;13:31. doi: 10.3390/v13010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mesquita F.S., Abrami L., Sergeeva O., Turelli P., Kunz B., Raclot C., Montoya J.P., Abriata L.A., Peraro M.D., Trono D., D'Angelo G., van der Goot F.G. S-acylation controls SARS-Cov-2 membrane lipid organization and enhances infectivity. bioRxiv. 2021 doi: 10.1101/2021.03.14.435299. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jennings B.C., Linder M.E. DHHC protein S-acyltransferases use similar ping-pong kinetic mechanisms but display different acyl-CoA specificities. J. Biol. Chem. 2012;287:7236–7245. doi: 10.1074/jbc.M111.337246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chlanda P., Mekhedov E., Waters H., Sodt A., Schwartz C., Nair V., Blank P.S., Zimmerberg J. Palmitoylation contributes to membrane curvature in influenza A virus assembly and hemagglutinin-mediated membrane fusion. J. Virol. 2017;91 doi: 10.1128/JVI.00947-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang S.T., Kiessling V., Simmons J.A., White J.M., Tamm L.K. HIV gp41-mediated membrane fusion occurs at edges of cholesterol-rich lipid domains. Nat. Chem. Biol. 2015;11:424–431. doi: 10.1038/nchembio.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee J., Kreutzberger A.J.B., Odongo L., Nelson E.A., Nyenhuis D.A., Kiessling V., Liang B., Cafiso D.S., White J.M., Tamm L.K. Ebola virus glycoprotein interacts with cholesterol to enhance membrane fusion and cell entry. Nat. Struct. Mol. Biol. 2021;28:181–189. doi: 10.1038/s41594-020-00548-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanders D.W., Jumper C.C., Ackerman P.J., Bracha D., Donlic A., Kim H., Kenney D., Castello-Serrano I., Suzuki S., Tamura T., Tavares A.H., Saeed M., Holehouse A.S., Ploss A., Levental I. SARS-CoV-2 requires cholesterol for viral entry and pathological syncytia formation. ELife. 2021 doi: 10.7554/eLife.65962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., 3rd, Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 40.Duronio R.J., Jackson-Machelski E., Heuckeroth R.O., Olins P.O., Devine C.S., Yonemoto W., Slice L.W., Taylor S.S., Gordon J.I. Protein N-myristoylation in Escherichia coli: Reconstitution of a eukaryotic protein modification in bacteria. Proc. Natl. Acad. Sci. U. S. A. 1990;87:1506–1510. doi: 10.1073/pnas.87.4.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao L.M.Q., Gray R.A.V., Martin D.D.O. Optimized incorporation of alkynyl fatty acid analogs for the detection of fatty acylated proteins using click chemistry. J. Vis. Exp. 2021 doi: 10.3791/62107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available upon request from the authors and are included in the article.