Abstract
Antiretroviral therapy controls HIV replication but does not eliminate the virus from the infected host. The persistence of a small pool of cells harboring integrated and replication-competent HIV genomes impedes viral eradication efforts. The HIV reservoir was originally described as a relatively homogeneous pool of resting memory CD4+ T cells. Over the past 20 years, the identification of multiple cellular subsets of CD4+ T cells endowed with distinct biological properties shed new lights on the heterogeneity of HIV reservoirs. It is now clear that HIV persists in large variety of CD4+ T cells, which contribute to HIV persistence through different mechanisms. In this review, we summarize recent findings indicating that specific biological features of well-characterized subsets of CD4+ T cells individually contribute to the persistence of HIV. These include an increased sensitivity to HIV infection, specific tissue locations, enhanced survival and heightened capacity to proliferate. We also discuss the relative abilities of these cellular reservoirs to contribute to viral rebound upon ART interruption. Together, these findings reveal that the HIV reservoir is not homogeneous and should be viewed as a mosaic of multiple cell types that all contribute to HIV persistence through different mechanisms.
Keywords: HIV reservoir, CD4+ T cells, tissues, Tfh, central memory cells, latency
Graphical abstract
1. Introduction
More than 20 years ago, the discovery that HIV had the ability to persist in resting CD4+ T cells provided a likely explanation for the inability of ART alone at eradicating the virus [1–3]. It became rapidly clear that a viral reservoir from which HIV replication can reignite when the therapeutic pressure is withdrawn would represent a formidable challenge to HIV eradication efforts. The HIV reservoir was originally described as a small pool of resting CD4+ T cells harboring transcriptionally silent proviruses (“the latent HIV reservoir”) [4–6]. Since then, years of research revealed the complexity of cellular subsets and it is now clear that HIV persists in multiple types of cells that are endowed with distinct biological features and in which proviruses are expressed at different levels [7], further complicating the development of a safe and scalable cure for all people living with HIV (PLWH). Cellular reservoirs are numerous and different in nature. Myeloid reservoirs such as tissue macrophages remain understudied, primarily because they are difficult to access and because obtaining a sufficient number of pure myeloid cells for in vitro culture to demonstrate their potential as clinically relevant reservoirs remain technically challenging [8, 9]. Similarly, while circulating CD4+ T cells can be easily isolated from the blood, CD4+ T cells residing in tissues are difficult to study [10–13]. Recently, the use of nonhuman primate models of SIV infection [14–16] and the development of less invasive procedures to collect cells from tissues in PLWH [17] revealed the major contribution of cells residing in various tissues/lymphoid organs to viral persistence. The identification of these new viral reservoirs was largely made possible through the characterization of novel subsets of CD4+ T cells revealed by the field of human T cell immunology. These discoveries resulted in multiple classifications of CD4+ T cells, which are based on their functions, localization and memory differentiation status.
2. Flavors of CD4+ T cells
2.1. Functions of CD4+ T cells
Upon activation through TCR, naïve CD4+ T cells differentiate into lineage specific T helper (Th) subsets. Each subset produce distinct sets of cytokines that activates downstream signal transducer and activator of transcription (STAT) signaling proteins and dictates lineage commitment by expressing unique master transcription factors [18]. Th cells are typically classified by the cytokines they produce upon TCR engagement and were originally divided into two subsets named Th1 and Th2 [19]. Th1 cells, which are prominent during infections by viruses and intracellular bacteria, produce IFN-γ and IL-2, induce the differentiation and proliferation of cytotoxic T lymphocytes (CTL) and contribute to the activation of macrophages. In contrast, Th2 cells coordinate the immune response to large extracellular pathogens such as parasites and helminths and are characterized by IL-4 production, which contributes to the differentiation of B cells and the development of humoral immune responses. In 2005, Harrington [20] and Park [21] identified a third and distinct subset of Th effector cells named Th17 due to their capacity to produce IL-17. Under physiological conditions, Th17 cells reside mainly in the lamina propria of the small intestine and contribute to the integrity of the mucosal barrier [22]. During infection they are induced at other mucosal sites and contribute to the immune control of a variety of pathogens including Staphylococcus aureus, Citrobacter rodentium, and Salmonella [23], primarily through the recruitment of neutrophils. More recently, Th9 cells have been described a new lineage involved in the development of immune responses to helminthic infections through the production of IL-9 [24]. They also contribute to the development of allergic inflammatory diseases and play a role in anti-tumor immune responses [25]. The most recent members of the effector CD4+ T cells family are Th22 cells, which were first identified in skin tissues of patients with inflammatory skin diseases, in which they produce IL-22 [26]. Th22 cells resemble Th17 cells, but unlike Th17 cells, which produce IL-17 either alone or concomitantly with IL-22, the Th22 subset completely lacks expression of IL-17 [27]. Regulatory T cells (Tregs) represent another subset of CD4+ T cells that are induced during virtually all infections and contribute to tumor progression by suppressing anticancer immunity [28]. They control the magnitude of adaptive immune responses by producing the immunoregulatory cytokines IL-10 and TGF-β and contribute to the maintenance of self-tolerance to prevent auto-immune disease [29]. Finally, T follicular helper cells (Tfh), are located in the B cell follicles of secondary lymphoid organs and contribute to the maturation of B cells through the production of IL-21 and IL-4 [30]. Therefore, they are likely involved in humoral adaptive immune responses in all infectious diseases [31].
2.2. Anatomic locations of CD4+ T cells
The case of Tfh is the best illustration that the classification based on the function of CD4+ T cells somewhat overlaps with another classification that uses their specific anatomical locations. Although Tfh cells circulating in the periphery can be detected [32], they primarily exert their B cells helper function in the B cell follicles located in lymph nodes, the spleen and Peyer patches. Other examples are given by Th17 cells which primarily reside in the lamina propria of the gut during homeostasis, and Th22 cells which are essentially recruited to the skin. From the recent discovery that a subset of CD4+ T cells have the ability to persist in tissues without recirculating emerged the concept of tissue resident memory T cells (Trm) [33]. Recent studies suggest that a significant fraction of CD4+ Trm cells derive from effector Th17 cells [34], indicating that once again, these classifications may largely overlap.
2.3. Memory status of CD4+ T cells
The classifications described above are mainly based on the functions and locations of CD4+ T cells when they exert their effector functions. After the antigen is cleared, a fraction of these cells persist as memory cells which are maintained for decades in response to homeostatic signals such as IL-7. The memory compartment is heterogeneous, and two main subsets of memory cells named central (TCM) and effector (TEM) memory cells can be distinguished by multiple criteria: (i) the absence (TCM) or presence (TEM) of immediate effector functions [35]; (ii) the expression of the homing receptor CCR7 that allows cells to migrate to secondary lymphoid organs (TCM) versus nonlymphoid tissues (TEM) [36]; (iii) the capacity to produce IL-2 (TCM) or IFN-γ (TEM) upon antigen stimulation [37]; (iv) the prevalence of a pro-survival (TCM) or pro-apoptotic program (TEM) [38]. Upon antigenic stimulation, TCM differentiate into TEM cells, whereas TEM cannot revert back to a TCM phenotype [39]. Although the memory subsets are largely defined by their capacity to migrate to secondary lymphoid organs and to have immediate effector functions, it is important to keep in mind that when activated (i.e. upon secondary stimulation), they will exert specific effector functions and could be re-classified as Th1, Th2, Th9, Th17, Th22, Tfh or Treg cells according to the cytokines they produce.
Therefore the definition of CD4+ T cells based on their function, location and memory status are largely overlapping, which complicates the identification of a particular subset as a preferential cellular target or a preferred cellular reservoir for HIV. Theoretically, a cell that could serve as a long-lived viral reservoir should present at least two characteristics: 1) being susceptible to HIV infection up to the integration step and 2) having the ability to persist during ART. Both parameters greatly vary between subsets and CD4+ T cells and should be investigated independently.
3. Susceptibility of CD4+ T cells to HIV infection
Early studies on the susceptibility of different cell subsets to HIV infection revealed that infection of activated CD4+ T cells is much more efficient than resting cells [40–42]. Unlike that of activated CD4+ T cells, the viral genome is not completely reverse transcribed in quiescent cells [43], suggesting that a minimal state of activation is required to establish infection. Such cells with minimal levels of activation are found in tissues even in homeostatic conditions, which is in line with the high susceptibility of CD4+ T cells from the gut to HIV infection [44]. Among those, Th17 cells have been repeatedly shown to be preferentially infected by HIV [45–47]. Similarly, CD4+ T cells expressing the gut homing marker α4β7 and tissue resident memory CD4+ T cells all show enhanced susceptibility to HIV infection [48]. A high expression level of CCR5 is also a hallmark of enhanced susceptibility to HIV [49], particularly in tissues [50], which is in line with the observation that TEM cells, which express high levels of this HIV coreceptor, are preferentially infected during acute HIV infection [51]. This also partially explains the relative resistance of naïve cells to HIV infection by CCR5-using viral strains [52, 53], since this chemokine receptor is expressed at very low levels on these cells [54]. In addition to the activation status, Th17 lineage and CCR5 expression, the anatomic location of some CD4+ T cell subsets may contribute to their preferential infection. This is best exemplified by the case of Tfh cells, which are known to be major producers of HIV particles during untreated HIV infection [55], possibly because they are relatively protected from HIV-specific CTLs [56].
The antigen specificity of CD4+ T cells also influences their susceptibility to HIV infection. While CMV-specific CD4+ T cells may be relatively protected through the autocrine production of CCR5 ligands [57], CD4+ T cells specific to HIV [58], mycobacterium tuberculosis [59] as well as tetanus toxoid and Candida albicans [60] have been shown to be preferentially targeted by HIV. Whether the increased susceptibility of these cells to HIV infection depends on their function or anatomical location remains unclear.
4. Generation of latently infected cells
Unlike productively infected cells which are primarily found during untreated HIV infection, proviruses integrated in persistently infected CD4+ T cells identified in PLWH on ART display low to no transcriptional activity [61]. Whether these cells are derived from previously productively infected cells that reverted back to a resting state or whether they were directly infected as resting cells and immediately established latency is still a matter of debate. These two models of establishment of HIV latency are known as the post- and the pre-activation latency models, respectively.
4.1. Post-activation latency
Post-activation latency is based on the idea that the transition from an activated and productively infected CD4+ T cells to a resting memory state is accompanied by HIV transcriptional silencing. The transition from an activated state to quiescence may offer a narrow window of opportunity that permits HIV silencing and persistence of the infected cells [62]. During the contraction phase of the immune response, when the antigen load decreases and activated cells transition from an effector to a memory phenotype, a rare subset of cells expressing CCR5 are still permissive to HIV infection but also are transcriptionally programmed to become quiescent, a state that is favorable to HIV latency [62]. In addition, the strength of TCR stimulation is key to influence the generation of memory CD4+ T cells [63]. Analogously, intermediate and low TCR signals predispose cells towards latent infections that are refractory to reversal [64].
Post-activation latency is likely to be an active rather than a passive phenomenon: During the resolution of immune responses, several pathways are known to dampen T cell activation and consequently could trigger HIV latency [65, 66]. T cell activation and proliferation can be modulated by anti-inflammatory cytokines such as TGF-β and IL-10. TGF-β acts on TCR-induced activation [67] but also on the proliferation induced by γ-c cytokines [68]. Although the role these immunomodulatory cytokines may exert on HIV latency has not been formally demonstrated in vivo, in vitro evidence are emerging: Combination of TGF-β, IL-10 and IL-8 induces T cell quiescence and HIV latency in differentiated effector CD4+ T cells [69], suggesting that HIV latency can be established in Th1, Th2, Th17 and Treg cells post-activation. Additionally, T cell activation can be dampened by the engagement of immune checkpoint molecules such as PD-1, CTLA-4, TIGIT, LAG-3 and TIM-3 [70]. For instance, PD-1 is actively promoting HIV transcriptional silencing in productively infected cells [71, 72]. Consequently, PD-1 expressing memory CD4+ T cells are more likely to become latently infected and persist during ART [72–74].
4.2. Pre-activation latency
An alternative way to generate latently infected cells is to increase susceptibility of resting CD4+ T cells to HIV infection. Resting CD4+ T cells are largely refractory to productive HIV infection due to blocks at the levels of entry, reverse transcription, nuclear import, and viral gene expression [43, 75, 76]. However, CCL19 and CCL20, two chemokines involved in the trafficking of cells to lymph node and the gut-associated lymphoid tissues (GALT) via CCR7 and CCR6 respectively, enhance HIV infection of resting CD4+ T cells by modifying the actin cytoskeleton, thereby increasing nuclear entry and integration of the viral DNA [77]. These findings from an in vitro model are in line with the important contribution of CCR7 expressing cells, such as TCM cells, to HIV reservoirs during ART [73]. An in vitro model of HIV latency that recapitulates the complex dynamics of the establishment and maintenance of the latent reservoir in different memory T cell subsets was recently developed [78]. Interestingly, the generation of latent cells in this LAtency and Reversion Assay (LARA) does not require polyclonal T cell activation before infection but only exposure of resting CD4+ T cells to TGF-β, IL-7 and conditioned medium containing TGF-β, IL-9 and IL-21 to promote the survival of infected cells in long-term culture. In this model, latently infected cells display various memory status and functions including TCM, TEM, Th1, Th2 and Th17 cells [78]. In addition, IL-7, a cytokine involved in T cell homeostasis, modulates the activity of the restriction factor SAMHD1 and increases the permissiveness of resting CD4+ T cells to HIV infection [79, 80]. It is important to note that CD127, the α chain of the IL-7 receptor, was recently identified as a marker of susceptibility to latent HIV infection of memory CD4+ T cell isolated from tissues [81]. This particular tonsillar memory subset (CD57-CD127+) is endowed with the transcriptional signature of quiescent T cells which prompts infected cells to HIV transcriptional silencing.
Altogether, these studies suggest that within the memory compartment, TCM CD4+ T cells displaying a CCR7+/CD127+/CCR5+ may represent a subset particularly favorable to the establishment of latent infection. Given the plasticity of CD4+ T cells, it is difficult to determine if the cells in which HIV latency is established retain their phenotype after prolonged ART. As discussed below, the number of cell types in which HIV persist may be even larger than the number of subsets in which latency can be efficiently established.
5. Persistence of HIV in CD4+ T cell subsets during ART
5.1. Dynamics of the HIV reservoir during ART
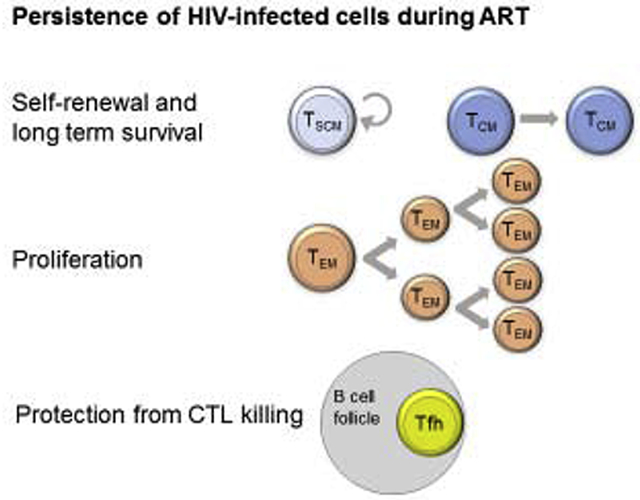
Following ART initiation, only a minute fraction of productively HIV-infected CD4+ T cells survive and are maintained as persistent and long-lived latently infected cells [51, 61]. Whereas some studies suggest that the bulk of the persistent reservoir is established at this time [82], others have reported the presence of archived sequences corresponding to transmitted founders in CD4+ T cells persisting on ART [83]. Therefore, the reservoir is likely made of a mix of cells infected at different times before ART initiation. Whether the reservoir is replenished through de novo infection of CD4+ T cells during ART remains controversial [84–87]. Independently of the possible generation of newly infected cells, several lines of evidence indicate that the reservoir is highly dynamic in virally suppressed individuals [88]. This dynamic is attributed to sustained as well as sequential clonal expansions of infected cells which are attributed to i) proviral integration in genes controlling cell growth [89, 90], ii) homeostatic proliferation [73, 91] and iii) clonal expansions of infected CD4+ T cell clones following antigenic stimulation [92]. Here, we describe these mechanisms contributing to HIV persistence during ART and elaborate on the cell subsets in which they are more likely to occur (Figure 1).
Figure 1. Contributions of CD4+ T cells subsets to HIV persistence.
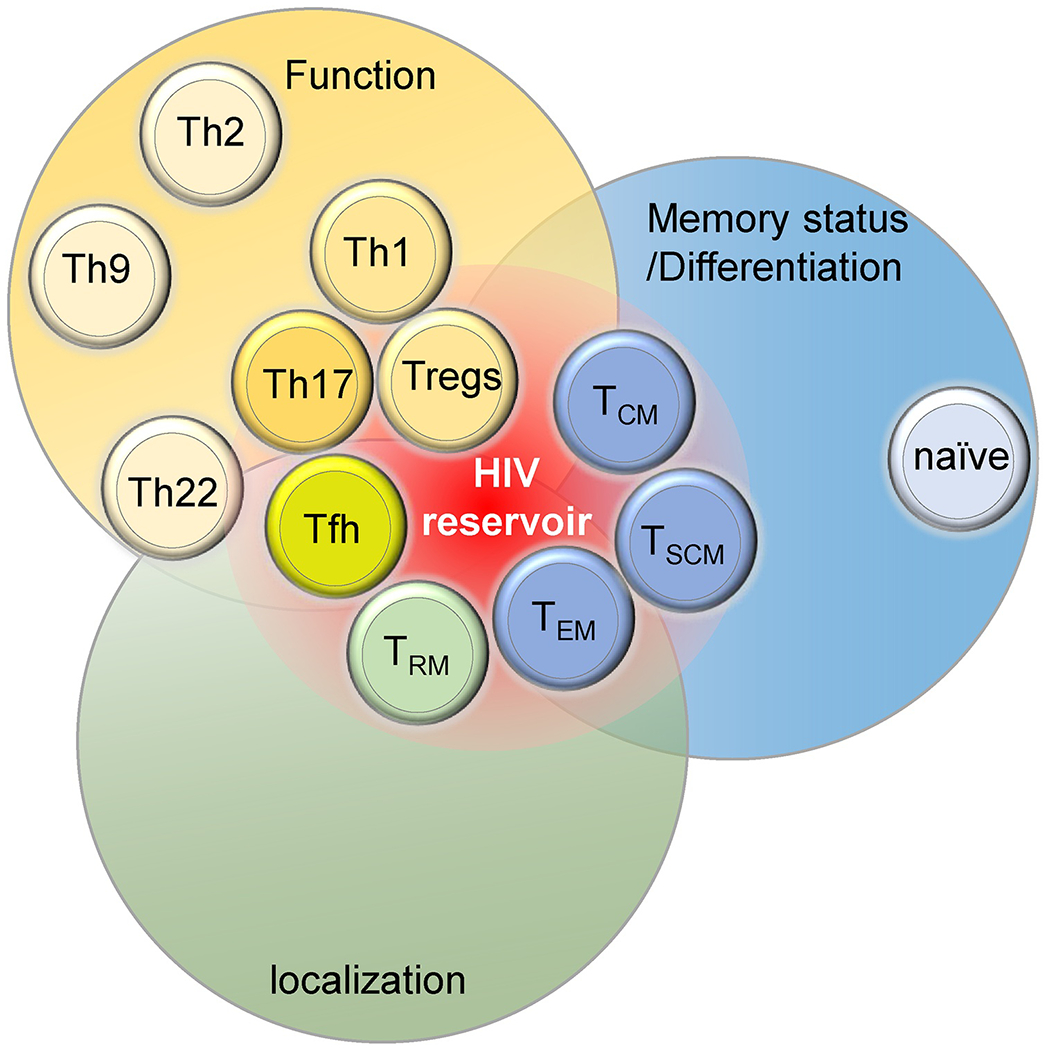
CD4+ T cell subsets can be classified according to their functions (yellow), memory status (blue) and localization (green). Some of these classifications largely overlap. The relative contributions to viral persistence in depicted by their proximity to the red zone representing the HIV reservoir.
5.2. HIV persistence in memory CD4+ T cells
HIV-infected cells need to survive for long periods of time to persist during ART. In the memory compartment, TCM CD4+ T cells, which are phenotypically defined as CD45RA−/CD27+/CCR7+, show exquisite survival and self-renew abilities [38, 93] and have a long half-life [94, 95]. Accordingly, TCM are a key player in HIV persistence, as they highly contribute to the pool of HIV-infected cells [73, 96]. In addition, TCM cells are the source of the more differentiated TEM cells (CD45RA−/CD27−/CCR7−) which are rapidly generated upon antigen stimulation [97]. Although TEM CD4+ T cells contribute less than TCM cells to the pool of cells harboring HIV DNA [73], they account for the majority of clonal expansions in the reservoir as a result of their elevated proliferative capacity [98, 99]. In addition, TEM cells may play a critical role in viral rebound since they harbor higher frequencies of intact and inducible genomes [78, 98, 100–102] (see section 6). Although memory CD4+ T cell subsets are the main reservoirs for HIV during ART, naïve cells may also contribute to HIV persistence [103, 104]. A limitation to these findings stems from the difficulty in defining the phenotype of truly naïve cells (i.e. non antigen-experienced cells). In the two studies mentioned above, the combination of three cell surface markers CD45RA+, CD27+, CCR7+ does not allow to distinguish stem-cell like memory CD4+ T cells (TSCM), which are known to contribute to HIV persistence [105, 106]. Zerbato et al. isolated rare naïve cells, from which TSCM were excluded by depleting CD95-expressing cells, and from which replication-competent HIV was detected [107]. These studies raise the question of the nature of the mechanisms by which naïve CD4+ T cells, which are resting CD4+ T cells expressing extremely low levels of CCR5, get initially infected. They also emphasize the importance of combining several cell-surface markers to precisely define antigen-naïve cells and of using flow cytometry cell sorting to obtain highly pure cellular populations.
TSCM cells own unique sternness properties which contribute to their ability to serve as a stable reservoir for HIV [108–110]. Since TSCM cells (and to a lower extent, TCM cells) have the ability to self-renew and to generate a progeny of more differentiated cells, they could represent an infinite source of infected cells. Interfering with the Wnt/β-catenin signaling pathways to induce the differentiation of TSCM and TCM cells has recently been proposed as a possible eradication strategy [111].
5.3. HIV persistence in CD4+ T cells expressing immune checkpoint molecules
As mentioned above (section 4.1), immune checkpoint molecules, and particularly PD-1, actively promote HIV latency [71, 72]. These receptors may also favor the persistence of HIV-infected cells over time by preventing reactivation of the latent provirus. Indeed, PD-1, LAG-3, TIGIT and CTLA-4 have been identified as markers enriching in HIV/SIV infected cells during ART, both in peripheral blood and tissues [74, 101, 112]. Of note, PD-1 is also a marker of T-cell activation and infected cells expressing PD-1 cells may also represent a labile pool of activated infected cells, particularly during the first months of ART [113]. After prolonged ART, HIV genomes found in the less differentiated memory subsets (TCM and TTM) expressing PD-1 may have a selective advantage to persist over time compared to cells that do not express this molecule [74]. Importantly, CD4+ T cells co-expressing multiple immune checkpoint molecules (PD-1, LAG-3 and TIGIT) are further enriched for integrated viral genomes, suggesting an enhanced capacity to persist during ART. Since the co-expression of these molecules is a hallmark of profound immune exhaustion, it is possible that infected cells expressing multiple immune checkpoint molecules harbor deeply latent proviruses.
Besides their role in T cell exhaustion, some immune checkpoint molecules are constitutively expressed by subsets of cells in which HIV persists, independently of the functional role played by these receptors. For instance, PD-1 and TIGIT are markers of Tfh cells [32, 114], which are major cellular reservoirs for HIV during ART [115]. In addition to their high susceptibility to HIV infection [55], productively infected Tfh may escape CD8+ T cell killing by being localized in the germinal centers within the lymph node B-cell follicles, from which CTL are largely excluded [56, 116]. Additional factors may contribute to HIV persistence in Tfh cells, since their circulating counterparts (CXCR5+/PD-1+/CXCR3−) are also enriched in HIV [117]. Another example is CTLA-4, which identifies CD4+ T cell with regulatory properties [118, 119]. Using a model of virally suppressed SIV-infected rhesus macaques, McGary et al. recently characterized the contribution of CTLA-4 expressing T cells to viral persistence [112]. CTLA-4+/PD-1− memory CD4+ T cells residing outside of the lymph node follicle were enriched in replication-competent virus. Their ability to support viral persistence was not related to spatial escape from CD8+ T cell killing but more likely to increased potential of survival (high Bcl-2 expression) and homeostatic proliferation (high levels of phosphorylated STAT5). Whether these cells expressing CTLA-4 exert regulatory functions remains to be determined. Indeed, the contribution of Tregs in HIV persistence remains unclear: Initial studies of the latent HIV reservoir were performed using “resting CD4+ T cells” from which CD25+ cells were depleted, which obviously excluded Tregs from these analyses. More recently, several studies highlighted that Treg cells (typically identified as CD25hi/CD127lo) are enriched in HIV DNA and have the ability to produce infectious virus [120–122]. Since Tregs are hyporesponsive to stimulation and relatively resistant to killing, they may represent a particularly challenging reservoir to eliminate [123]
5.4. HIV persistence in functional CD4+ T cell subsets
As discussed above (section 2.1), CD4+ T cells can be defined by their functional properties. Although the spectrum of cytokines they produced remains the gold standard way to characterize these subsets, expression of chemokine receptors are commonly used as surrogate markers to identify functionally polarized CD4+ T cell subsets such as Th1 (CXCR3+/CCR4−/CCR6−), Th2 (CXCR3−/CCR4+/CCR6−), Th17 (CXCR3−/CCR4+/CCR6+) and Th1/Th17 cells (CXCR3+/CCR4−/CCR6+) [124]. Extensive work using CD4+ T cells isolated from the blood of ART-treated PLWH allowed the identification of CCR6 as a marker of HIV susceptibility and persistence during ART [125, 126]. In addition, CCR6+ cells are imprinted with gut homing properties, which is reflected by preferential persistence of HIV in this subset in the gut [11, 47]. Th17, and by extension Th1/Th17, are relatively heterogenous and plastic in their fate. Thus, a fraction of Treg and Th1 cells could be the progeny of subsets of Th17 cells [127, 128]. In addition, Th17 cells are endowed with sternness properties supporting their long-lived capacity [128, 129]. Such properties support the ability of Th17 cells to serve as long-lived viral reservoir for HIV. Of note, CD161, a marker of Th17 and Th17 precursor cells [130], identifies HIV-infected cells which have the ability to persist through proliferation during ART [131]. Remarkably, a recent study characterized HIV persistence in polarized CD4+ T cell subsets defined by their cytokines expression [132]. Th9 cells, specialized in antitumor immune responses [25], were enriched for HIV genomes, but these were mostly defective, while Th1 cells harbored clonally expanded intact HIV genomes. Interestingly, despite their relatively short half-life, Th1 cells may significantly contribute to HIV persistence through antigen-induced proliferation. This is well supported by a recent study in which antigen induced clonal expansion of HIV proviruses was observed in HIV- and CMV-specific CD4+ T cells [92]. A broader assessment of the contribution of different antigens to HIV persistence will be key to the development of targeted HIV cure strategies.
5.5. Additional cellular markers associated with HIV persistence
In addition to the subsets described above and that are usually defined by combinations of cellular markers, individual markers highly expressed at the surface of HIV-infected CD4+ T cells persisting during ART have been identified. Some of these receptors can be targeted by antibodies to specifically deplete the infected cells, which make them particularly attractive for HIV eradication strategies: This is the case of CD20-expressing cells which can be depleted by rituximab [133] and CD30-expressing cells which are targeted by brentuximab vedotin [134]. Interestingly, the latter is a marker of transcriptionally active HIV-infected cells persisting during ART, highlighting the potential and controversial contribution of leaky latency to HIV persistence [135, 136]. Finally, CD32a, also known as Fc gamma receptor IIa (FcγIIa), is expressed on rare CD4+ T cells which are enriched for HIV DNA at a unprecedently reported high level (up to a 1,000-fold enrichment when compared to their negative counterparts) [137]. Although the role of CD32-expressing CD4+ T cell in the persistence of the latent and replication-competent HIV reservoir remains controversial [138–142], several reports indicate that CD32 may be preferentially expressed by HIV-infected and transcriptionally active CD4+ T cells, particularly in tissues [143, 144].
6. Contributions of CD4+ T cell subsets to HIV rebound
Although persistently infected macrophages [8] and viral particles retained in follicular dendritic cells [145] can contribute to rebound, HIV-infected CD4+ T cells persisting represent a likely source of viral recrudescence upon ART cessation. The identification of the CD4+ T cells subsets from which infection reignites will be key to develop eradication strategies based on the prevention of burst of viral replication. Obviously, only cells harboring intact HIV genomes can contribute to viral rebound. Recent studies indicate that these cells have a shorter half-life than those carrying defective proviruses [146, 147]. Whether this difference is due to intrinsic properties of the cells harboring intact genomes (shorter half-life) or to a greater immune pressure that negatively select for defective viruses over time remains to be determined. In any case, viral rebound may originate both from latently infected cells or from cells harboring transcriptionally active proviruses, as long as they carry intact genomes.
6.1. Transcriptionally active cells as a source of HIV rebound
Transcriptionally active HIV-infected cells persist during ART and may represent the first cells to fuel viral rebound. Indeed, the size of the active reservoir, as measured by cell-associated viral RNA, predicts time to viral rebound [148, 149]. Phylogenetic studies identified these transcriptionally active cells present before analytical treatment interruption as the source of plasma viral rebound [150, 151]. Interestingly these cells tend to harbor clonally expanded proviruses suggesting that proliferating cells are more likely to be the source of rebounding viruses. In addition, a single-genome sequencing approach combined with quantification of cell-associated HIV RNA revealed the transcriptional activity of expanded proviruses [152]. Although, the phenotype of HIV-infected cells was not determined in this study, this active reservoir maybe less stable [153]. TEM cells own these characteristics (active viral transcriptional and proliferation), suggesting their potential prominent role in viral rebound, although this remains to be formally demonstrated (Figure 2). Interestingly, CD32a and CD30 identify actively transcribing cells persisting in blood and tissues during ART [134, 143], but whether the viral genomes persisting in these cells are intact and can produce replication-competent HIV is unknown. Circulating CD4+ T cells expressing CD32a display a TEM phenotype and co-express multiple markers of T cell-activation such as CD69, CD25, HLA-DR, CD38 and Ki67 [143]. Remarkably, the frequency of CD30+ CD4+ T cells increases before viral rebound, suggesting that CD30 may represent a surrogate marker of early replication or transcriptional activity during analytical treatment interruption (ATI) [154]. To characterize the source of viral rebound phenotypically and virologically, an SIV barcoded virus, which allows infection of rhesus macaques with more than a thousand different viral variants, has recently been developed [155]. This novel tool will certainly be used in the near future to molecularly track viral rebound after ART cessation and to characterize the phenotype of the cells responsible for the initial burst of replication A way to assess the potential ability of a viral genome to generate replication-competent HIV particles is to evaluate the intactness of the provirus using near full length genome sequencing [156]. During the past few years, several groups characterized the phenotype of CD4+ T cell subsets harboring intact proviruses [98, 99, 132]. Collectively, the results from these studies indicate that TEM cells (CD45RA−/CD27−/CCR7−), Th1 cells (IFN-γ+) and activated CD4+ T cells (HLA-DR+) are enriched in intact genomes. Of note, the markers used in these studies are not mutually exclusive and their combination may identify a subset of proliferating cells enriched in intact genomes [157] and from which infection may reignite. The possibility that viral recrudescence may originate from multiple tissues [158] and that recombinant viruses may contribute to viral rebound [159] complicate the efforts to identify the cellular sources of HIV rebound.
Figure 2. Cellular features involved in the establishment and maintenance of HIV reservoirs and in viral rebound.
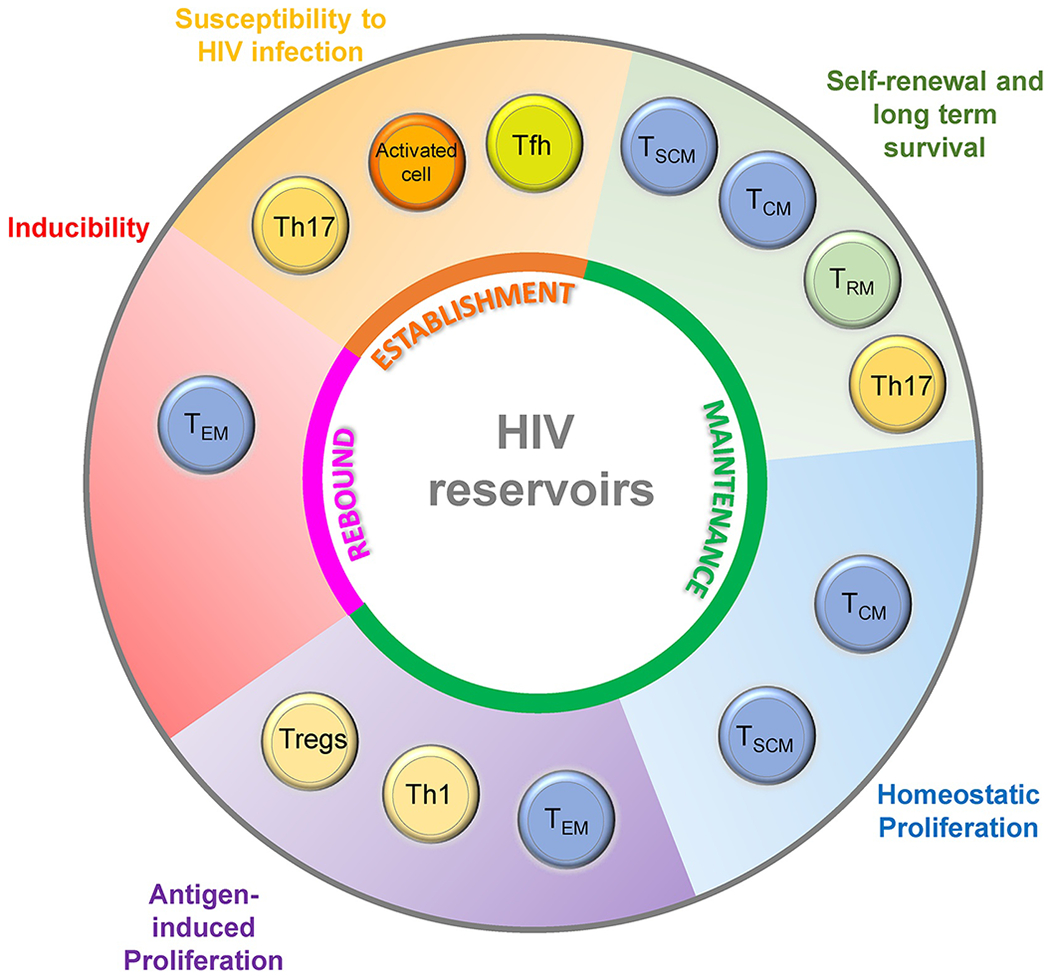
Common features of HIV-infected cells (outer cyle) are required for the establishment, maintenance and rebound of the HIV reservoirs (inner cycle). Multiple CD4+ T cell subsets likely contribute to each phenomenon.
6.2. Latently infected cells as a source of HIV rebound
A prerequisite to viral rebound from latently infected cells is an efficient viral reactivation of the latent provirus to generate infectious viral particles. Several studies identified TEM cells as the memory subset harboring the highest frequency of inducible proviruses [78, 100, 101]. These findings have recently been challenged by a study suggesting that all subsets have an equal ability to generate infectious HIV particles upon activation [160]. However, the exclusion of CD69+/CD25+/HLA-DR+ CD4+ T cells, which are enriched in intact genomes [99] and from which latent HIV may preferentially reactivate [102], provides a possible explanation for the discrepancy with the aforementioned studies.
An additional layer of complexity emerged from a recent study that combined integration sites and near full length proviral sequencing. This approach revealed that intact HIV genomes are characterized by a particular integration landscape and are more frequently found in non-genic chromosomal positions, in opposite orientation relative to host genes and distant from accessible chromatin regions [161]. These observations suggest that intact proviruses integrated in more silent regions of the host genome may be selected over time, resulting in a viral reservoir characterized by a deeper degree of viral latency after prolonged ART.
Altogether, these studies suggest that in addition to the intactness of the HIV genomes, their inducibility (i.e their capacity to produce viral particles upon stimulation) should be assessed to better identify potential sources of rebound upon ATI.
7. Perspective: Single cell approaches to study HIV cellular reservoirs
Most studies describing the phenotypic heterogeneity of HIV-infected cells during the course of HIV infection mainly used well-characterized cell subsets to identify distinct cellular reservoirs. This approach may not suffice to grasp the complex heterogeneity of HIV reservoirs during ART. During the past five years, single-cell approaches opened new avenues to analyze the HIV reservoir dynamics with an unprecedented depth.
Single-cell transcriptomic studies identified a new type of cell (CD25+CD298+CD63+BST-2+) highly permissive to HIV infection in vitro [162]. This cell subset expressing activation markers is imprinted with a downregulated interferon-mediated response and low expression levels of several known restriction factors. Single-cell RNA sequencing analysis using in vitro models of HIV latency highlighted the heterogeneity of the cell types in which HIV latency is established and from which HIV can be reactivated by latency-reversing agents (LRAs) [163–165]. Ex vivo studies identifying HIV-infected cells through the detection of viral proteins or transcripts confirmed the downregulation of cellular antiviral immunity pathways and the presence of pro-survival factors as common features of persistently HIV-infected cells [166, 167]. Of note, a limitation to these studies is the need for a stimulation step to reveal latently infected cells, which likely induces transcriptomic and phenotypic changes.
Single-cell flow cytometry based analysis of the phenotype of HIV-infected cells is currently going through a revolution supported by the usage of multiparametric flow cytometry and mass cytometry (CyTOF) associated with high-dimensional analysis. They confirmed CD127 as a marker of cells permissive to latent HIV infection [81, 168] and TIGIT as a cellular marker of persistently infected cells [169]. The heterogeneity of different cellular reservoirs in response to a variety of LRAs was formally demonstrated by single-cell flow cytometry based studies [170–172]. These studies revealed that CD4+ T cells displaying a TEM phenotype are generally more responsive to current LRAs when compared to TCM cells. Single-cell epigenetic studies using ATAC-seq should help to further understand the molecular mechanisms responsible for these differential responses to LRAs.
In situ hybridization methods to visualize single infected cells have also been developed and present the advantage of visualizing HIV-infected cells in the context of a preserved tissue architecture [14, 173]. Remarkably, the development of new whole body imaging positron emission tomography coupled with magnetic resonance imaging allows the visualization of foci of HIV-infected cells which co-localize with activated T cells in lymph nodes of PLWH [174, 175]. Although this novel technology will require an increased resolution to detect HIV at the single-cell level, it will certainly contribute to a better understanding of the biology of persistent HIV reservoirs in the near future.
More than 20 years of research on HIV reservoirs have revealed the heterogeneity of the cells in which HIV persists during ART. Although these discoveries may be seen as an increased level of complexity and an additional obstacle to the development of a cure for HIV infection, they also reveal common features of HIV-infected cells shared by cells displaying distinct phenotypes. Therefore, rather than adding more cellular reservoirs to the list, it is now time to identify shared cellular markers, metabolic pathways and functions that are hallmarks of persistently infected cells. The development of single cell approaches that can identify reservoir cells with an unprecedented level of specificity and which allow the combination of multiple parameters will certainly help in this endeavor.
Highlights.
HIV persists in a large variety of CD4+ T cells
Cellular reservoirs contribute to HIV persistence through different mechanisms
Tissue locations, survival and capacity to proliferate promote HIV persistence
Cellular reservoirs differ in their ability to cause viral rebound
Novel single-cell approaches revolutionized this area of research
Acknowledgements
The authors are grateful to the individuals who volunteered to participate in the studies reviewed in this article. We also thank Nancie Archin, Marlene Bras, Mathias Lichterfeld, Viviana Simon and Lydie Trautmann for their contribution to the title of this manuscript.
Funding
This work was supported by the Canadian Institutes for Health Research (CIHR; operating grant #364408 and the Canadian HIV Cure Enterprise (CanCURE) Team Grant HB2 - 164064), the National Institute of Allergy and Infectious Diseases (UM1AI126611: Delaney AIDS Research Enterprise (DARE) to Find a Cure), and the Réseau SIDA et maladies infectieuses du Fonds de Recherche du Québec - Santé (FRQ-S). N.C. is supported by Research Scholar Career Awards of the FRQ-S (#253292).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
The authors declare no competing interests.
References
- [1].Bukrinsky MI, Stanwick TL, Dempsey MP, Stevenson M, Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection, Science 254(5030) (1991) 423–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF, In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency, Nat Med 1(12) (1995) 1284–90. [DOI] [PubMed] [Google Scholar]
- [3].Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo YH, Brookmeyer R, Zeiger MA, Barditch-Crovo P, Siliciano RF, Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection, Nature 387(6629) (1997) 183–8. [DOI] [PubMed] [Google Scholar]
- [4].Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF, Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy, Science 278(5341) (1997) 1295–300. [DOI] [PubMed] [Google Scholar]
- [5].Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, Lloyd AL, Nowak MA, Fauci AS, Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy, Proc Natl Acad Sci U S A 94(24) (1997) 13193–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, Richman DD, Recovery of replication-competent HIV despite prolonged suppression of plasma viremia, Science 278(5341) (1997)1291–5. [DOI] [PubMed] [Google Scholar]
- [7].Darcis G, Berkhout B, Pasternak AO, The Quest for Cellular Markers of HIV Reservoirs: Any Color You Like, Front Immunol 10 (2019) 2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Andrade VM, Mavian C, Babic D, Cordeiro T, Sharkey M, Barrios L, Brander C, Martinez-Picado J, Dalmau J, Llano A, Li JZ, Jacobson J, Lavine CL, Seaman MS, Salemi M, Stevenson M, A minor population of macrophage-tropic HIV-1 variants is identified in recrudescing viremia following analytic treatment interruption, Proc Natl Acad Sci U S A 117(18) (2020) 9981–9990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Clayton KL, Garcia JV, Clements JE, Walker BD, HIV Infection of Macrophages: Implications for Pathogenesis and Cure, Pathog Immun 2(2) (2017) 179–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Costiniuk CT, Salahuddin S, Farnos O, Olivenstein R, Pagliuzza A, Orlova M, Schurr E, De Castro C, Bourbeau J, Routy JP, Ancuta P, Chomont N, Jenabian MA, HIV persistence in mucosal CD4+ T cells within the lungs of adults receiving long-term suppressive antiretroviral therapy, AIDS 32(16) (2018) 2279–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gosselin A, Wiche Salinas TR, Planas D, Wacleche VS, Zhang Y, Fromentin R, Chomont N, Cohen EA, Shacklett B, Mehraj V, Ghali MP, Routy JP, Ancuta P, HIV persists in CCR6+CD4+ T cells from colon and blood during antiretroviral therapy, AIDS 31(1) (2017) 35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jenabian MA, Costiniuk CT, Mehraj V, Ghazawi FM, Fromentin R, Brousseau J, Brassard P, Belanger M, Ancuta P, Bendayan R, Chomont N, Routy JP, g. Orchid study, Immune tolerance properties of the testicular tissue as a viral sanctuary site in ART-treated HIV-infected adults, AIDS 30(18) (2016) 2777–2786. [DOI] [PubMed] [Google Scholar]
- [13].Nolan DJ, Rose R, Rodriguez PH, Salemi M, Singer EJ, Lamers SL, McGrath MS, The Spleen Is an HIV-1 Sanctuary During Combined Antiretroviral Therapy, AIDS Res Hum Retroviruses 34(1) (2018) 123–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Estes JD, Kityo C, Ssali F, Swainson L, Makamdop KN, Del Prete GQ, Deeks SG, Luciw PA, Chipman JG, Beilman GJ, Hoskuldsson T, Khoruts A, Anderson J, Deleage C, Jasurda J, Schmidt TE, Hafertepe M, Callisto SP, Pearson H, Reimann T, Schuster J, Schoephoerster J, Southern P, Perkey K, Shang L, Wietgrefe SW, Fletcher CV, Lifson JD, Douek DC, McCune JM, Haase AT, Schacker TW, Defining total-body AIDS-virus burden with implications for curative strategies, Nat Med 23(11) (2017) 1271–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rabezanahary H, Moukambi F, Palesch D, Clain J, Racine G, Andreani G, Benmadid-Laktout G, Zghidi-Abouzid O, Soundaramourty C, Tremblay C, Silvestri G, Estaquier J, Despite early antiretroviral therapy effector memory and follicular helper CD4 T cells are major reservoirs in visceral lymphoid tissues of SIV-infected macaques, Mucosal immunology 13(1) (2020) 149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Abreu C, Shirk EN, Queen SE, Beck SE, Mangus LM, Pate KAM, Mankowski JL, Gama L, Clements JE, Brain macrophages harbor latent, infectious simian immunodeficiency virus, AIDS 33Suppl 2 (2019) S181–S188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hey-Nguyen WJ, Xu Y, Pearson CF, Bailey M, Suzuki K, Tantau R, Obeid S, Milner B, Field A, Carr A, Bloch M, Cooper DA, Kelleher AD, Zaunders JJ, Koelsch KK, Quantification of Residual Germinal Center Activity and HIV-1 DNA and RNA Levels Using Fine Needle Biopsies of Lymph Nodes During Antiretroviral Therapy, AIDS Res Hum Retroviruses 33(7) (2017) 648–657. [DOI] [PubMed] [Google Scholar]
- [18].Mirlekar B, Co-expression of master transcription factors determines CD4(+) T cell plasticity and functions in auto-inflammatory diseases, Immunol Lett 222 (2020) 58–66. [DOI] [PubMed] [Google Scholar]
- [19].Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL, Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins, J Immunol 136(7) (1986) 2348–57. [PubMed] [Google Scholar]
- [20].Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT, Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages, Nat Immunol 6(11) (2005) 1123–32. [DOI] [PubMed] [Google Scholar]
- [21].Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q, Dong C, A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17, Nat Immunol 6(11) (2005) 1133–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Omenetti S, Bussi C, Metidji A, Iseppon A, Lee S, Tolaini M, Li Y, Kelly G, Chakravarty P, Shoaie S, Gutierrez MG, Stockinger B, The Intestine Harbors Functionally Distinct Homeostatic Tissue-Resident and Inflammatory Th17 Cells, Immunity 51(1) (2019) 77–89 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Muranski P, Restifo NP, Essentials of Th17 cell commitment and plasticity, Blood 121(13) (2013) 2402–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Veldhoen M, Uyttenhove C, van Snick J, Helmby H, Westendorf A, Buer J, Martin B, Wilhelm C, Stockinger B, Transforming growth factor-beta ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset, Nat Immunol 9(12) (2008) 1341–6. [DOI] [PubMed] [Google Scholar]
- [25].Lu Y, Hong S, Li H, Park J, Hong B, Wang L, Zheng Y, Liu Z, Xu J, He J, Yang J, Qian J, Yi Q, Th9 cells promote antitumor immune responses in vivo, J Clin Invest 122(11) (2012) 4160–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Eyerich S, Eyerich K, Pennino D, Carbone T, Nasorri F, Pallotta S, Cianfarani F, Odorisio T, Traidl-Hoffmann C, Behrendt H, Durham SR, Schmidt-Weber CB, Cavani A, Th22 cells represent a distinct human T cell subset involved in epidermal immunity and remodeling, J Clin Invest 119(12) (2009) 3573–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Plank MW, Kaiko GE, Maltby S, Weaver J, Tay HL, Shen W, Wilson MS, Durum SK, Foster PS, Th22 Cells Form a Distinct Th Lineage from Th17 Cells In Vitro with Unique Transcriptional Properties and Tbet-Dependent Th1 Plasticity, J Immunol 198(5) (2017) 2182–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Togashi Y, Shitara K, Nishikawa H, Regulatory T cells in cancer immunosuppression - implications for anticancer therapy, Nat Rev Clin Oncol 16(6) (2019) 356–371. [DOI] [PubMed] [Google Scholar]
- [29].Wing K, Sakaguchi S, Regulatory T cells exert checks and balances on self tolerance and autoimmunity, Nat Immunol 11(1) (2010) 7–13. [DOI] [PubMed] [Google Scholar]
- [30].Crotty S, T Follicular Helper Cell Biology: A Decade of Discovery and Diseases, Immunity 50(5) (2019) 1132–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Greczmiel U, Krautler NJ, Pedrioli A, Bartsch I, Agnellini P, Bedenikovic G, Harker J, Richter K, Oxenius A, Sustained T follicular helper cell response is essential for control of chronic viral infection, Sci Immunol 2(18) (2017). [DOI] [PubMed] [Google Scholar]
- [32].Locci M, Havenar-Daughton C, Landais E, Wu J, Kroenke MA, Arlehamn CL, Su LF, Cubas R, Davis MM, Sette A, Haddad EK, A.V.I.P.C.P.I. International, Poignard P, Crotty S, Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses, Immunity 39(4) (2013) 758–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sathaliyawala T, Kubota M, Yudanin N, Turner D, Camp P, Thome JJ, Bickham KL, Lerner H, Goldstein M, Sykes M, Kato T, Farber DL, Distribution and compartmentalization of human circulating and tissue-resident memory T cell subsets, Immunity 38(1) (2013) 187–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Amezcua Vesely MC, Pallis P, Bielecki P, Low JS, Zhao J, Harman CCD, Kroehling L, Jackson R, Bailis W, Licona-Limon P, Xu H, Iijima N, Pillai PS, Kaplan DH, Weaver CT, Kluger Y, Kowalczyk MS, Iwasaki A, Pereira JP, Esplugues E, Gagliani N, Flavell RA, Effector TH17 Cells Give Rise to Long-Lived TRM Cells that Are Essential for an Immediate Response against Bacterial Infection, Cell 178(5) (2019) 1176–1188 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Geginat J, Sallusto F, Lanzavecchia A, Cytokine-driven proliferation and differentiation of human naive, central memory and effector memory CD4+ T cells, Pathol Biol (Paris) 51(2) (2003) 64–6. [DOI] [PubMed] [Google Scholar]
- [36].Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A, Two subsets of memory T lymphocytes with distinct homing potentials and effector functions, Nature 401(6754) (1999) 708–12. [DOI] [PubMed] [Google Scholar]
- [37].Sallusto F, Geginat J, Lanzavecchia A, Central memory and effector memory T cell subsets: function, generation, and maintenance, Annu Rev Immunol 22 (2004) 745–63. [DOI] [PubMed] [Google Scholar]
- [38].Riou C, Yassine-Diab B, Van grevenynghe J, Somogyi R, Greller LD, Gagnon D, Gimmig S, Wilkinson P, Shi Y, Cameron MJ, Campos-Gonzalez R, Balderas RS, Kelvin D, Sekaly RP, Haddad EK, Convergence of TCR and cytokine signaling leads to FOXO3a phosphorylation and drives the survival of CD4+ central memory T cells, J Exp Med 204(1) (2007) 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Jameson SC, Masopust D, Understanding Subset Diversity in T Cell Memory, Immunity 48(2) (2018) 214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Folks T, Kelly J, Benn S, Kinter A, Justement J, Gold J, Redfield R, Sell KW, Fauci AS, Susceptibility of normal human lymphocytes to infection with HTLV-III/LAV, J Immunol 136(11) (1986) 4049–53. [PubMed] [Google Scholar]
- [41].Zack JA, Cann AJ, Lugo JP, Chen IS, HIV-1 production from infected peripheral blood T cells after HTLV-I induced mitogenic stimulation, Science 240(4855) (1988) 1026–9. [DOI] [PubMed] [Google Scholar]
- [42].Zagury D, Bernard J, Leonard R, Cheynier R, Feldman M, Sarin PS, Gallo RC, Long-term cultures of HTLV-III--infected T cells: a model of cytopathology of T-cell depletion in AIDS, Science 231(4740) (1986) 850–3. [DOI] [PubMed] [Google Scholar]
- [43].Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, Chen IS, HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure, Cell 61(2) (1990)213–22. [DOI] [PubMed] [Google Scholar]
- [44].Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, Nguyen PL, Khoruts A, Larson M, Haase AT, Douek DC, CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract, J Exp Med 200(6) (2004) 749–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Monteiro P, Gosselin A, Wacleche VS, El-Far M, Said EA, Kared H, Grandvaux N, Boulassel MR, Routy JP, Ancuta P, Memory CCR6+CD4+ T cells are preferential targets for productive HIV type 1 infection regardless of their expression of integrin beta7, J Immunol 186(8) (2011) 4618–30. [DOI] [PubMed] [Google Scholar]
- [46].Planas D, Zhang Y, Monteiro P, Goulet JP, Gosselin A, Grandvaux N, Hope TJ, Fassati A, Routy JP, Ancuta P, HIV-1 selectively targets gut-homing CCR6+CD4+ T cells via mTOR-dependent mechanisms, JCI Insight 2(15) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Anderson JL, Khoury G, Fromentin R, Solomon A, Chomont N, Sinclair E, Milush JM, Hartogensis W, Bacchetti P, Roche M, Tumpach C, Gartner M, Pitman MC, Epling CL, Hoh R, Hecht FM, Somsouk M, Cameron PU, Deeks SG, Lewin SR, Human Immunodeficiency Virus (HIV)-Infected CCR6+ Rectal CD4+ T Cells and HIV Persistence On Antiretroviral Therapy, J Infect Dis 221(5) (2020) 744–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Tokarev A, McKinnon LR, Pagliuzza A, Sivro A, Omole TE, Kroon E, Chomchey N, Phanuphak N, Schuetz A, Robb ML, Eller MA, Ananworanich J, Chomont N, Bolton DL, Preferential infection of alpha4beta7+ memory CD4+ T cells during early acute HIV-1 infection, Clin Infect Dis (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Blaak H, Ran LJ, Rientsma R, Schuitemaker H, Susceptibility of in vitro stimulated PBMC to infection with NSI HIV-1 is associated with levels of CCR5 expression and beta-chemokine production, Virology 267(2) (2000) 237–46. [DOI] [PubMed] [Google Scholar]
- [50].Joag VR, McKinnon LR, Liu J, Kidane ST, Yudin MH, Nyanga B, Kimwaki S, Besel KE, Obila JO, Huibner S, Oyugi JO, Arthos J, Anzala O, Kimani J, Ostrowski MA, Toronto HIVRG, Kaul R, Identification of preferential CD4+ T-cell targets for HIV infection in the cervix, Mucosal immunology 9(1) (2016) 1–12. [DOI] [PubMed] [Google Scholar]
- [51].Leyre L, Kroon E, Vandergeeten C, Sacdalan C, Colby DJ, Buranapraditkun S, Schuetz A, Chomchey N, de Souza M, Bakeman W, Fromentin R, Pinyakorn S, Akapirat S, Trichavaroj R, Chottanapund S, Manasnayakorn S, Rerknimitr R, Wattanaboonyoungcharoen P, Kim JH, Tovanabutra S, Schacker TW, O’Connell R, Valcour VG, Phanuphak P, Robb ML, Michael N, Trautmann L, Phanuphak N, Ananworanich J, Chomont N, R.V.S.S.s.g. Rv254/Search, Abundant HIV-infected cells in blood and tissues are rapidly cleared upon ART initiation during acute HIV infection, Sci Transl Med 12(533) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Spina CA, Prince HE, Richman DD, Preferential replication of HIV-1 in the CD45RO memory cell subset of primary CD4 lymphocytes in vitro, J Clin Invest 99(7) (1997) 1774–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Helbert MR, Walter J, L’Age J, Beverley PC, HIV infection of CD45RA+ and CD45RO+ CD4+ T cells, Clin Exp Immunol 107(2) (1997) 300–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Blaak H, van’t Wout AB, Brouwer M, Hooibrink B, Hovenkamp E, Schuitemaker H, In vivo HIV-1 infection of CD45RA(+)CD4(+) T cells is established primarily by syncytium-inducing variants and correlates with the rate of CD4(+) T cell decline, Proc Natl Acad Sci U S A 97(3) (2000) 1269–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, De Leval L, Graziosi C, Pantaleo G, Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production, J Exp Med 210(1) (2013) 143–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Connick E, Mattila T, Folkvord JM, Schlichtemeier R, Meditz AL, Ray MG, McCarter MD, Mawhinney S, Hage A, White C, Skinner PJ, CTL fail to accumulate at sites of HIV-1 replication in lymphoid tissue, J Immunol 178(11) (2007) 6975–83. [DOI] [PubMed] [Google Scholar]
- [57].Casazza JP, Brenchley JM, Hill BJ, Ayana R, Ambrozak D, Roederer M, Douek DC, Betts MR, Koup RA, Autocrine production of beta-chemokines protects CMV-Specific CD4 T cells from HIV infection, PLoS Pathog 5(10) (2009) e1000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, Grossman Z, Dybul M, Oxenius A, Price DA, Connors M, Koup RA, HIV preferentially infects HIV-specific CD4+ T cells, Nature 417(6884) (2002) 95–8. [DOI] [PubMed] [Google Scholar]
- [59].Geldmacher C, Ngwenyama N, Schuetz A, Petrovas C, Reither K, Heeregrave EJ, Casazza JP, Ambrozak DR, Louder M, Ampofo W, Pollakis G, Hill B, Sanga E, Saathoff E, Maboko L, Roederer M, Paxton WA, Hoelscher M, Koup RA, Preferential infection and depletion of Mycobacterium tuberculosis-specific CD4 T cells after HIV-1 infection, J Exp Med 207(13) (2010) 2869–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hu H, Nau M, Ehrenberg P, Chenine AL, Macedo C, Zhou Y, Daye ZJ, Wei Z, Vahey M, Michael NL, Kim JH, Marovich M, Ratto-Kim S, Distinct gene-expression profiles associated with the susceptibility of pathogen-specific CD4 T cells to HIV-1 infection, Blood 121(7) (2013) 1136–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano RF, Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy, Nat Med 5(5) (1999) 512–7. [DOI] [PubMed] [Google Scholar]
- [62].Shan L, Deng K, Gao H, Xing S, Capoferri AA, Durand CM, Rabi SA, Laird GM, Kim M, Hosmane NN, Yang HC, Zhang H, Margolick JB, Li L, Cai W, Ke R, Flavell RA, Siliciano JD, Siliciano RF, Transcriptional Reprogramming during Effector-to-Memory Transition Renders CD4(+) T Cells Permissive for Latent HIV-1 Infection, Immunity 47(4) (2017) 766–775 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Snook JP, Kim C, Williams MA, TCR signal strength controls the differentiation of CD4(+) effector and memory T cells, Sci Immunol 3(25) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Gagne M, Michaels D, Schiralli Lester GM, Gummuluru S, Wong WW, Henderson AJ, Strength of T cell signaling regulates HIV-1 replication and establishment of latency, PLoS Pathog 15(5) (2019) e1007802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Vandergeeten C, Fromentin R, Chomont N, The role of cytokines in the establishment, persistence and eradication of the HIV reservoir, Cytokine Growth Factor Rev 23(4-5) (2012) 143–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Wykes MN, Lewin SR, Immune checkpoint blockade in infectious diseases, Nat Rev Immunol 18(2) (2018) 91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Tu E, Chia CPZ, Chen W, Zhang D, Park SA, Jin W, Wang D, Alegre ML, Zhang YE, Sun L, Chen W, T Cell Receptor-Regulated TGF-beta Type I Receptor Expression Determines T Cell Quiescence and Activation, Immunity 48(4) (2018) 745–759 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Nguyen TP, Sieg SF, TGF-beta inhibits IL-7-induced proliferation in memory but not naive human CD4(+) T cells, J Leukoc Biol 102(2) (2017) 499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Dobrowolski C, Valadkhan S, Graham AC, Shukla M, Ciuffi A, Telenti A, Karn J, Entry of Polarized Effector Cells into Quiescence Forces HIV Latency, mBio 10(2) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Attanasio J, Wherry EJ, Costimulatory and Coinhibitory Receptor Pathways in Infectious Disease, Immunity 44(5) (2016) 1052–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Evans VA, van der Sluis RM, Solomon A, Dantanarayana A, McNeil C, Garsia R, Palmer S, Fromentin R, Chomont N, Sekaly RP, Cameron PU, Lewin SR, Programmed cell death-1 contributes to the establishment and maintenance of HIV-1 latency, AIDS 32(11) (2018) 1491–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Fromentin R, DaFonseca S, Costiniuk CT, El-Far M, Procopio FA, Hecht FM, Hoh R, Deeks SG, Hazuda DJ, Lewin SR, Routy JP, Sekaly RP, Chomont N, PD-1 blockade potentiates HIV latency reversal ex vivo in CD4(+) T cells from ART-suppressed individuals, Nature communications 10(1) (2019) 814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy JP, Haddad EK, Sekaly RP, HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation, Nat Med 15(8) (2009) 893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Fromentin R, Bakeman W, Lawani MB, Khoury G, Hartogensis W, DaFonseca S, Killian M, Epling L, Hoh R, Sinclair E, Hecht FM, Bacchetti P, Deeks SG, Lewin SR, Sekaly RP, Chomont N, CD4+ T Cells Expressing PD-1, TIGIT and LAG-3 Contribute to HIV Persistence during ART, PLoS Pathog 12(7) (2016) e1005761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, Schenkova K, Ambiel I, Wabnitz G, Gramberg T, Panitz S, Flory E, Landau NR, Sertel S, Rutsch F, Lasitschka F, Kim B, Konig R, Fackler OT, Keppler OT, SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells, Nat Med 18(11) (2012) 1682–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Descours B, Cribier A, Chable-Bessia C, Ayinde D, Rice G, Crow Y, Yatim A, Schwartz O, Laguette N, Benkirane M, SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4(+) T-cells, Retrovirology 9 (2012) 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Cameron PU, Saleh S, Sallmann G, Solomon A, Wightman F, Evans VA, Boucher G, Haddad EK, Sekaly RP, Harman AN, Anderson JL, Jones KL, Mak J, Cunningham AL, Jaworowski A, Lewin SR, Establishment of HIV-1 latency in resting CD4+ T cells depends on chemokine-induced changes in the actin cytoskeleton, Proc Natl Acad Sci U S A 107(39) (2010) 16934–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Kulpa DA, Talla A, Brehm JH, Ribeiro SP, Yuan S, Bebin-Blackwell AG, Miller M, Barnard R, Deeks SG, Hazuda D, Chomont N, Sekaly RP, Differentiation into an Effector Memory Phenotype Potentiates HIV-1 Latency Reversal in CD4(+) T Cells, J Virol 93(24) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Pace MJ, Graf EH, Agosto LM, Mexas AM, Male F, Brady T, Bushman FD, O’Doherty U, Directly infected resting CD4+T cells can produce HIV Gag without spreading infection in a model of HIV latency, PLoS Pathog 8(7) (2012) e1002818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Coiras M, Bermejo M, Descours B, Mateos E, Garcia-Perez J, Lopez-Huertas MR, Lederman MM, Benkirane M, Alcami J, IL-7 Induces SAMHD1 Phosphorylation in CD4+ T Lymphocytes, Improving Early Steps of HIV-1 Life Cycle, Cell reports 14(9) (2016) 2100–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Hsiao F, Frouard J, Gramatica A, Xie G, Telwatte S, Lee GQ, Roychoudhury P, Schwarzer R, Luo X, Yukl SA, Lee S, Hoh R, Deeks SG, Jones RB, Cavrois M, Greene WC, Roan NR, Tissue memory CD4+ T cells expressing IL-7 receptor-alpha (CD127) preferentially support latent HIV-1 infection, PLoS Pathog 16(4) (2020) el008450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Abrahams M-R, Joseph SB, Garrett N, Tyers L, Moeser M, Archin N, Council OD, Matten D, Zhou S, D’oolabh D, Anthony C, Goonetilleke N, Abdool Karim S, Margolis D, Kosavovsky Ponds S, Williamson C, Swanstrom R, The Replication-Competent HIV-1 Latent Reservoir is Primarily Established Near the Time of Therapy Initiation, bioRxiv (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Brooks K, Jones BR, Dilernia DA, Wilkins DJ, Claiborne DT, Mclnally S, Gilmour J, Kilembe W, Joy JB, Allen SA, Brumme ZL, Hunter E, HIV-1 variants are archived throughout infection and persist in the reservoir, PLoS Pathog 16(6) (2020) el008378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Fletcher CV, Staskus K, Wietgrefe SW, Rothenberger M, Reilly C, Chipman JG, Beilman GJ, Khoruts A, Thorkelson A, Schmidt TE, Anderson J, Perkey K, Stevenson M, Perelson AS, Douek DC, Haase AT, Schacker TW, Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues, Proc Natl Acad Sci U S A (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Lorenzo-Redondo R, Fryer HR, Bedford T, Kim EY, Archer J, Pond SLK, Chung YS, Penugonda S, Chipman J, Fletcher CV, Schacker TW, Malim MH, Rambaut A, Haase AT, McLean AR, Wolinsky SM, Persistent HIV-1 replication maintains the tissue reservoir during therapy, Nature 530(7588) (2016) 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Kearney M, Wiegand A, Shao W, McManus WR, Bale MJ, Luke B, Maldarelli F, Mellors JW, Coffin JM, Ongoing HIV Replication During ART Reconsidered, Open Forum Infectious Diseases (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Van Zyl GU, Katusiime MG, Wiegand A, McManus WR, Bale MJ, Halvas EK, Luke B, Boltz VF, Spindler J, Laughton B, Engelbrecht S, Coffin JM, Cotton MF, Shao W, Mellors JW, Kearney MF, No evidence of HIV replication in children on antiretroviral therapy, J Clin Invest 127(10) (2017) 3827–3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Wang Z, Gurule EE, Brennan TP, Gerold JM, Kwon KJ, Hosmane NN, Kumar MR, Beg SA, Capoferri AA, Ray SC, Ho YC, Hill AL, Siliciano JD, Siliciano RF, Expanded cellular clones carrying replication-competent HIV-1 persist, wax, and wane, Proc Natl Acad Sci US A 115(11) (2018) E2575–E2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, Spindler J, Ferris AL, Mellors JW, Kearney MF, Coffin JM, Hughes SH, HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells, Science 345(6193) (2014) 179–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Wagner TA, McLaughlin S, Garg K, Cheung CY, Larsen BB, Styrchak S, Huang HC, Edlefsen PT, Mullins JI, Frenkel LM, HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection, Science 345(6196) (2014) 570–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Vandergeeten C, Fromentin R, DaFonseca S, Lawani MB, Sereti I, Lederman MM, Ramgopal M, Routy JP, Sekaly RP, Chomont N, Interleukin-7 promotes HIV persistence during antiretroviral therapy, Blood 121(21) (2013) 4321–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Mendoza P, Jackson JR, Oliveira TY, Gaebler C, Ramos V, Caskey M, Jankovic M, Nussenzweig MC, Cohn LB, Antigen-responsive CD4+ T cell clones contribute to the HIV-1 latent reservoir, J Exp Med 217(7) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Michie CA, McLean A, Alcock C, Beverley PC, Lifespan of human lymphocyte subsets defined by CD45 isoforms, Nature 360(6401) (1992) 264–5. [DOI] [PubMed] [Google Scholar]
- [94].Hellerstein MK, Hoh RA, Hanley MB, Cesar D, Lee D, Neese RA, McCune JM, Subpopulations of long-lived and short-lived T cells in advanced HIV-1 infection, J Clin Invest 112(6) (2003)956–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].De Boer RJ, Perelson AS, Quantifying T lymphocyte turnover, J Theor Biol 327 (2013) 45–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Soriano-Sarabia N, Bateson RE, Dahl NP, Crooks AM, Kuruc JD, Margolis DM, Archin NM, Quantitation of replication-competent HIV-1 in populations of resting CD4+ T cells, J Virol 88(24) (2014) 14070–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Macallan DC, Wallace D, Zhang Y, De Lara C, Worth AT, Ghattas H, Griffin GE, Beverley PC, Tough DF, Rapid turnover of effector-memory CD4(+) T cells in healthy humans, J Exp Med 200(2) (2004) 255–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Hiener B, Horsburgh BA, Eden JS, Barton K, Schlub TE, Lee E, von Stockenstrom S, Odevall L, Milush JM, Liegler T, Sinclair E, Hoh R, Boritz EA, Douek D, Fromentin R, Chomont N, Deeks SG, Hecht FM, Palmer S, Identification of Genetically Intact HIV-1 Proviruses in Specific CD4(+) T Cells from Effectively Treated Participants, Cell reports 21(3) (2017)813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Horsburgh BA, Lee E, Hiener B, Eden JS, Schlub TE, von Stockenstrom S, Odevall L, Milush JM, Liegler T, Sinclair E, Hoh R, Boritz EA, Douek DC, Fromentin R, Chomont N, Deeks SG, Hecht FM, Palmer S, High levels of genetically-intact HIV in HLA-DR+ memory T-cells indicates their value for reservoir studies, AIDS (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Grau-Exposito J, Serra-Peinado C, Miguel L, Navarro J, Curran A, Burgos J, Ocana I, Ribera E, Torrella A, Planas B, Badia R, Castellvi J, Falco V, Crespo M, Buzon MJ, A Novel Single-Cell FISH-Flow Assay Identifies Effector Memory CD4(+) T cells as a Major Niche for HIV-1 Transcription in HIV-Infected Patients, MBio 8(4) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Pardons M, Baxter AE, Massanella M, Pagliuzza A, Fromentin R, Dufour C, Leyre L, Routy JP, Kaufmann DE, Chomont N, Single-cell characterization and quantification of translation-competent viral reservoirs in treated and untreated HIV infection, PLoS Pathog 15(2) (2019) e1007619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Wonderlich ER, Subramanian K, Cox B, Wiegand A, Lackman-Smith C, Bale MJ, Stone M, Hoh R, Kearney MF, Maldarelli F, Deeks SG, Busch MP, Ptak RG, Kulpa DA, Effector memory differentiation increases detection of replication-competent HIV-l in resting CD4+ T cells from virally suppressed individuals, PLoS Pathog 15(10) (2019) e1008074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Venanzi Rullo E, Cannon L, Pinzone MR, Ceccarelli M, Nunnari G, O’Doherty U, Genetic Evidence That Naive T Cells Can Contribute Significantly to the Human Immunodeficiency Virus Intact Reservoir: Time to Re-evaluate Their Role, Clin Infect Dis 69(12) (2019) 2236–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Roche M, Tumpach C, Symons J, Gartner M, Anderson JL, Khoury G, Cashin K, Cameron PU, Churchill MJ, Deeks SG, Gorry PR, Lewin SR, CXCR4-Using HIV Strains Predominate in Naive and Central Memory CD4(+) T Cells in People Living with HIV on Antiretroviral Therapy: Implications for How Latency Is Established and Maintained, J Virol 94(6) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Buzon MJ, Sun H, Li C, Shaw A, Seiss K, Ouyang Z, Martin-Gayo E, Leng J, Henrich TJ, Li JZ, Pereyra F, Zurakowski R, Walker BD, Rosenberg ES, Yu XG, Lichterfeld M, HIV-1 persistence in CD4(+) T cells with stem cell-like properties, Nat Med 20(2) (2014) 139–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Jaafoura S, de Goer de Herve MG, Hernandez-Vargas EA, Hendel-Chavez H, Abdoh M, Mateo MC, Krzysiek R, Merad M, Seng R, Tardieu M, Delfraissy JF, Goujard C, Taoufik Y, Progressive contraction of the latent HIV reservoir around a core of less-differentiated CD4(+) memory T Cells, Nature communications 5 (2014) 5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Zerbato JM, McMahon DK, Sobolewski MD, Mellors JW, Sluis-Cremer N, Naive CD4+ T Cells Harbor a Large Inducible Reservoir of Latent, Replication-competent Human Immunodeficiency Virus Type 1, Clin Infect Dis 69(11) (2019) 1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Gattinoni L, Lugli E, Ji Y, Pos Z, Paulos CM, Quigley MF, Almeida JR, Gostick E, Yu Z, Carpenito C, Wang E, Douek DC, Price DA, June CH, Marincola FM, Roederer M, Restifo NP, A human memory T cell subset with stem cell-like properties, Nat Med 17(10) (2011) 1290–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Gattinoni L, Zhong XS, Palmer DC, Ji Y, Hinrichs CS, Yu Z, Wrzesinski C, Boni A, Cassard L, Garvin LM, Paulos CM, Muranski P, Restifo NP, Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells, Nat Med 15(7) (2009) 808–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Lugli E, Dominguez MH, Gattinoni L, Chattopadhyay PK, Bolton DL, Song K, Klatt NR, Brenchley JM, Vaccari M, Gostick E, Price DA, Waldmann TA, Restifo NP, Franchini G, Roederer M, Superior T memory stem cell persistence supports long-lived T cell memory, J Clin Invest (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Mavigner M, Zanoni M, Tharp GK, Habib J, Mattingly CR, Lichterfeld M, Nega MT, Vanderford TH, Bosinger SE, Chahroudi A, Pharmacological Modulation of the Wnt/beta-Catenin Pathway Inhibits Proliferation and Promotes Differentiation of Long-Lived Memory CD4(+) T Cells in Antiretroviral Therapy-Suppressed Simian Immunodeficiency Virus-Infected Macaques, J Virol 94(1) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].McGary CS, Deleage C, Harper J, Micci L, Ribeiro SP, Paganini S, Kuri-Cervantes L, Benne C, Ryan ES, Balderas R, Jean S, Easley K, Marconi V, Silvestri G, Estes JD, Sekaly RP, Paiardini M, CTLA-4(+)PD-1(−) Memory CD4(+) T Cells Critically Contribute to Viral Persistence in Antiretroviral Therapy-Suppressed, SIV-Infected Rhesus Macaques, Immunity 47(4) (2017) 776–788 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Eller MA, Hong T, Creegan M, Nau ME, Sanders-Buell E, Slike BM, Krebs SJ, Ratto-Kim S, McElrath MJ, Katabira ET, Bolton DL, Michael NL, Robb ML, Tovanabutra S, Baeten JM, Sandberg JK, Activated PD-1+ CD4+ T cells represent a short-lived part of the viral reservoir and predict poor immunologic recovery upon initiation of ART, AIDS 34(2) (2020) 197–202. [DOI] [PubMed] [Google Scholar]
- [114].Crotty S, Follicular helper CD4 T cells (TFH), Annu Rev Immunol 29 (2011) 621–63. [DOI] [PubMed] [Google Scholar]
- [115].Banga R, Procopio FA, Noto A, Pollakis G, Cavassini M, Ohmiti K, Corpataux JM, de Leval L, Pantaleo G, Perreau M, PD-1(+) and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals, Nat Med 22(7) (2016) 754–61. [DOI] [PubMed] [Google Scholar]
- [116].Fukazawa Y, Lum R, Okoye AA, Park H, Matsuda K, Bae JY, Hagen SI, Shoemaker R, Deleage C, Lucero C, Morcock D, Swanson T, Legasse AW, Axthelm MK, Hesselgesser J, Geleziunas R, Hirsch VM, Edlefsen PT, Piatak M Jr., Estes JD, Lifson JD, Picker LJ, B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers, Nat Med 21(2) (2015) 132–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Banga R, Procopio FA, Ruggiero A, Noto A, Ohmiti K, Cavassini M, Corpataux JM, Paxton WA, Pollakis G, Perreau M, Blood CXCR3(+) CD4 T Cells Are Enriched in Inducible Replication Competent HIV in Aviremic Antiretroviral Therapy-Treated Individuals, Front Immunol 9 (2018) 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Read S, Malmstrom V, Powrie F, Cytotoxic T lymphocyte-associated antigen 4 plays an essential role in the function of CD25(+)CD4(+) regulatory cells that control intestinal inflammation, J Exp Med 192(2) (2000) 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T, Miyara M, Fehervari Z, Nomura T, Sakaguchi S, CTLA-4 control over Foxp3+ regulatory T cell function, Science 322(5899) (2008) 271–5. [DOI] [PubMed] [Google Scholar]
- [120].Tran TA, de Goer de Herve MG, Hendel-Chavez H, Dembele B, Le Nevot E, Abbed K, Pallier C, Goujard C, Gasnault J, Delfraissy JF, Balazuc AM, Taoufik Y, Resting regulatory CD4 T cells: a site of HIV persistence in patients on long-term effective antiretroviral therapy, PLoS One 3(10) (2008) e3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Jiao YM, Liu CE, Luo LJ, Zhu WJ, Zhang T, Zhang LG, Su LS, Li HJ, Wu H, CD4+CD25+CD127 regulatory cells play multiple roles in maintaining HIV-1 p24 production in patients on long-term treatment: HIV-1 p24-producing cells and suppression of anti-HIV immunity, Int J Infect Dis 37 (2015) 42–9. [DOI] [PubMed] [Google Scholar]
- [122].Dunay GA, Solomatina A, Kummer S, Hufner A, Bialek JK, Eberhard JM, Tolosa E, Hauber J, Schulze Zur Wiesch J, Assessment of the HIV-1 reservoir in CD4+ regulatory T cells by a Droplet Digital PCR based approach, Virus Res 240 (2017) 107–111. [DOI] [PubMed] [Google Scholar]
- [123].Rocco J, Mellors JW, Macatangay BJ, Regulatory T cells: the ultimate HIV reservoir?, J Virus Erad 4(4) (2018) 209–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Sallusto F, Heterogeneity of Human CD4(+) T Cells Against Microbes, Annu Rev Immunol 34 (2016) 317–34. [DOI] [PubMed] [Google Scholar]
- [125].Gosselin A, Monteiro P, Chomont N, Diaz-Griffero F, Said EA, Fonseca S, Wacleche V, El-Far M, Boulassel MR, Routy JP, Sekaly RP, Ancuta P, Peripheral blood CCR4+CCR6+ and CXCR3+CCR6+CD4+ T cells are highly permissive to HIV-1 infection, J Immunol 184(3) (2010) 1604–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Khoury G, Anderson JL, Fromentin R, Hartogenesis W, Smith MZ, Bacchetti P, Hecht FM, Chomont N, Cameron PU, Deeks SG, Lewin SR, Persistence of integrated HIV DNA in CXCR3 + CCR6 + memory CD4+ T cells in HIV-infected individuals on antiretroviral therapy, AIDS 30(10) (2016) 1511–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Gagliani N, Amezcua Vesely MC, Iseppon A, Brockmann L, Xu H, Palm NW, de Zoete MR, Licona-Limon P, Paiva RS, Ching T, Weaver C, Zi X, Pan X, Fan R, Garmire LX, Cotton MJ, Drier Y, Bernstein B, Geginat J, Stockinger B, Esplugues E, Huber S, Flavell RA, Th17 cells transdifferentiate into regulatory T cells during resolution of inflammation, Nature 523(7559) (2015) 221–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Karmaus PWF, Chen X, Lim SA, Herrada AA, Nguyen TM, Xu B, Dhungana Y, Rankin S, Chen W, Rosencrance C, Yang K, Fan Y, Cheng Y, Easton J, Neale G, Vogel P, Chi H, Metabolic heterogeneity underlies reciprocal fates of TH17 cell stemness and plasticity, Nature 565(7737) (2019) 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Muranski P, Borman ZA, Kerkar SP, Klebanoff CA, Ji Y, Sanchez-Perez L, Sukumar M, Reger RN, Yu Z, Kern SJ, Roychoudhuri R, Ferreyra GA, Shen W, Durum SK, Feigenbaum L, Palmer DC, Antony PA, Chan CC, Laurence A, Danner RL, Gattinoni L, Restifo NP, Th17 cells are long lived and retain a stem cell-like molecular signature, Immunity 35(6) (2011) 972–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Cosmi L, De Palma R, Santarlasci V, Maggi L, Capone M, Frosali F, Rodolico G, Querci V, Abbate G, Angeli R, Berrino L, Fambrini M, Caproni M, Tonelli F, Lazzeri E, Parronchi P, Liotta F, Maggi E, Romagnani S, Annunziato F, Human interleukin 17-producing cells originate from a CD161+CD4+ T cell precursor, J Exp Med 205(8) (2008) 1903–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Li X, Liu Z, Li Q, Hu R, Zhao L, Yang Y, Zhao J, Huang Z, Gao H, Li L, Cai W, Deng K, CD161(+) CD4(+) T Cells Harbor Clonally Expanded Replication-Competent HIV-1 in Antiretroviral Therapy-Suppressed Individuals, mBio 10(5) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Lee GQ, Orlova-Fink N, Einkauf K, Chowdhury FZ, Sun X, Harrington S, Kuo HH, Hua S, Chen HR, Ouyang Z, Reddy K, Dong K, Ndung’u T, Walker BD, Rosenberg ES, Yu XG, Lichterfeld M, Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells, J Clin Invest 127(7) (2017) 2689–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Serra-Peinado C, Grau-Exposito J, Luque-Ballesteros L, Astorga-Gamaza A, Navarro J, Gallego-Rodriguez J, Martin M, Curran A, Burgos J, Ribera E, Raventos B, Willekens R, Torrella A, Planas B, Badia R, Garcia F, Castellvi J, Genesca M, Falco V, Buzon MJ, Expression of CD20 after viral reactivation renders HIV-reservoir cells susceptible to Rituximab, Nature communications 10(1) (2019) 3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Hogan LE, Vasquez J, Hobbs KS, Hanhauser E, Aguilar-Rodriguez B, Hussien R, Thanh C, Gibson EA, Carvidi AB, Smith LCB, Khan S, Trapecar M, Sanjabi S, Somsouk M, Stoddart CA, Kuritzkes DR, Deeks SG, Henrich TJ, Increased HIV-1 transcriptional activity and infectious burden in peripheral blood and gut-associated CD4+ T cells expressing CD30, PLoS Pathog 14(2) (2018) e1006856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Yukl SA, Kaiser P, Kim P, Telwatte S, Joshi SK, Vu M, Lampiris H, Wong JK, HIV latency in isolated patient CD4(+) T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing, Sci Transl Med 10(430) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Telwatte S, Lee S, Somsouk M, Hatano H, Baker C, Kaiser P, Kim P, Chen TH, Milush J, Hunt PW, Deeks SG, Wong JK, Yukl SA, Gut and blood differ in constitutive blocks to HIV transcription, suggesting tissue-specific differences in the mechanisms that govern HIV latency, PLoS Pathog 14(11) (2018) e1007357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Descours B, Petitjean G, Lopez-Zaragoza JL, Bruel T, Raffel R, Psomas C, Reynes J, Lacabaratz C, Levy Y, Schwartz O, Lelievre JD, Benkirane M, CD32a is a marker of a CD4 T-cell HIV reservoir harbouring replication-competent proviruses, Nature 543(7646) (2017) 564–567. [DOI] [PubMed] [Google Scholar]
- [138].Badia R, Ballana E, Castellvi M, Garcia-Vidal E, Pujantell M, Clotet B, Prado JG, Puig J, Martinez MA, Riveira-Munoz E, Este JA, CD32 expression is associated to T-cell activation and is not a marker of the HIV-1 reservoir, Nature communications 9(1) (2018) 2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Bertagnolli LN, White JA, Simonetti FR, Beg SA, Lai J, Tomescu C, Murray AJ, Antar AAR, Zhang H, Margolick JB, Hoh R, Deeks SG, Tebas P, Montaner LJ, Siliciano RF, Laird GM, Siliciano JD, The role of CD32 during HIV-1 infection, Nature 561(7723) (2018) E17–E19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Osuna CE, Lim SY, Kublin JL, Apps R, Chen E, Mota TM, Huang SH, Ren Y, Bachtel ND, Tsibris AM, Ackerman ME, Jones RB, Nixon DF, Whitney JB, Evidence that CD32a does not mark the HIV-1 latent reservoir, Nature 561(7723) (2018) E20–E28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Perez L, Anderson J, Chipman J, Thorkelson A, Chun TW, Moir S, Haase AT, Douek DC, Schacker TW, Boritz EA, Conflicting evidence for HIV enrichment in CD32(+) CD4 T cells, Nature 561(7723) (2018) E9–E16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Darcis G, Kootstra NA, Hooibrink B, van Montfort T, Maurer I, Groen K, Jurriaans S, Bakker M, van Lint C, Berkhout B, Pasternak AO, CD32(+)CD4(+) T Cells Are Highly Enriched for HIV DNA and Can Support Transcriptional Latency, Cell reports 30(7) (2020) 2284–2296 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Abdel-Mohsen M, Kuri-Cervantes L, Grau-Exposito J, Spivak AM, Nell RA, Tomescu C, Vadrevu SK, Giron LB, Serra-Peinado C, Genesca M, Castellvi J, Wu G, Del Rio Estrada PM, Gonzalez-Navarro M, Lynn K, King CT, Vemula S, Cox K, Wan Y, Li Q, Mounzer K, Kostman J, Frank I, Paiardini M, Hazuda D, Reyes-Teran G, Richman D, Howell B, Tebas P, Martinez-Picado J, Planelles V, Buzon MJ, Betts MR, Montaner LJ, CD32 is expressed on cells with transcriptionally active HIV but does not enrich for HIV DNA in resting T cells, Sci Transl Med 10(437) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Noto A, Procopio FA, Banga R, Suffiotti M, Corpataux JM, Cavassini M, Riva A, Fenwick C, Gottardo R, Perreau M, Pantaleo G, CD32(+) and PD-1(+) Lymph Node CD4 T Cells Support Persistent HIV-1 Transcription in Treated Aviremic Individuals, J Virol 92(20) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Heesters BA, Lindqvist M, Vagefi PA, Scully EP, Schildberg FA, Altfeld M, Walker BD, Kaufmann DE, Carroll MC, Follicular Dendritic Cells Retain Infectious HIV in Cycling Endosomes, PLoS Pathog 11(12) (2015) e1005285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Peluso MJ, Bacchetti P, Ritter KD, Beg S, Lai J, Martin JN, Hunt PW, Henrich TJ, Siliciano JD, Siliciano RF, Laird GM, Deeks SG, Differential decay of intact and defective proviral DNA in HIV-1-infected individuals on suppressive antiretroviral therapy, JCI Insight 5(4) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Antar AA, Jenike KM, Jang S, Rigau DN, Reeves DB, Hoh R, Krone MR, Keruly JC, Moore RD, Schiffer JT, Nonyane BA, Hecht FM, Deeks SG, Siliciano JD, Ho YC, Siliciano RF, Longitudinal study reveals HIV-1-infected CD4+ T cell dynamics during long-term antiretroviral therapy, J Clin Invest 130(7) (2020) 3543–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Li JZ, Etemad B, Ahmed H, Aga E, Bosch RJ, Mellors JW, Kuritzkes DR, Lederman MM, Para M, Gandhi RT, The size of the expressed HIV reservoir predicts timing of viral rebound after treatment interruption, AIDS 30(3) (2016) 343–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Pasternak AO, Grijsen ML, Wit FW, Bakker M, Jurriaans S, Prins JM, Berkhout B, Cell-associated HIV-1 RNA predicts viral rebound and disease progression after discontinuation of temporary early ART, JCI Insight 5(6) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Kearney MF, Wiegand A, Shao W, Coffin JM, Mellors JW, Lederman M, Gandhi RT, Keele BF, Li JZ, Origin of Rebound Plasma HIV Includes Cells with Identical Proviruses That Are Transcriptionally Active before Stopping of Antiretroviral Therapy, J Virol 90(3) (2016) 1369–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].De Scheerder MA, Vrancken B, Dellicour S, Schlub T, Lee E, Shao W, Rutsaert S, Verhofstede C, Kerre T, Malfait T, Hemelsoet D, Coppens M, Dhondt A, De Looze D, Vermassen F, Lemey P, Palmer S, Vandekerckhove L, HIV Rebound Is Predominantly Fueled by Genetically Identical Viral Expansions from Diverse Reservoirs, Cell Host Microbe (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Wiegand A, Spindler J, Hong FF, Shao W, Cyktor JC, Cillo AR, Halvas EK, Coffin JM, Mellors JW, Kearney MF, Single-cell analysis of HIV-1 transcriptional activity reveals expression of proviruses in expanded clones during ART, Proc Natl Acad Sci U S A 114(18) (2017) E3659–E3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Pinzone MR, VanBelzen DJ, Weissman S, Bertuccio MP, Cannon L, Venanzi-Rullo E, Migueles S, Jones RB, Mota T, Joseph SB, Groen K, Pasternak AO, Hwang WT, Sherman B, Vourekas A, Nunnari G, O’Doherty U, Longitudinal HIV sequencing reveals reservoir expression leading to decay which is obscured by clonal expansion, Nature communications 10(1) (2019) 728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Prator CA, Thanh C, Kumar S, Pan T, Peluso MJ, Bosch R, Jones N, Milush JM, Bakkour S, Stone M, Busch MP, Deeks SG, Hunt PW, Henrich TJ, Circulating CD30+CD4+ T Cells Increase Before Human Immunodeficiency Virus Rebound After Analytical Antiretroviral Treatment Interruption, J Infect Dis 221(7) (2020) 1146–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Fennessey CM, Pinkevych M, Immonen TT, Reynaldi A, Venturi V, Nadella P, Reid C, Newman L, Lipkey L, Oswald K, Bosche WJ, Trivett MT, Ohlen C, Ott DE, Estes JD, Del Prete GQ, Lifson JD, Davenport MP, Keele BF, Genetically-barcoded SIV facilitates enumeration of rebound variants and estimation of reactivation rates in nonhuman primates following interruption of suppressive antiretroviral therapy, PLoS Pathog 13(5) (2017) e1006359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Ho Y-C, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DIS, Lai J, Blankson JN, Siliciano JD, Siliciano RF, Replication-Competent Noninduced Proviruses in the Latent Reservoir Increase Barrier to HIV-1 Cure, Cell 155(3) (2013) 540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Anderson EM, Simonetti FR, Gorelick RJ, Hill S, Gouzoulis MA, Bell J, Rehm C, Perez L, Boritz E, Wu X, Wells D, Hughes SH, Rao V, Coffin JM, Kearney MF, Maldarelli F, Dynamic Shifts in the HIV Proviral Landscape During Long Term Combination Antiretroviral Therapy: Implications for Persistence and Control of HIV Infections, Viruses 12(2) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Rothenberger MK, Keele BF, Wietgrefe SW, Fletcher CV, Beilman GJ, Chipman JG, Khoruts A, Estes JD, Anderson J, Callisto SP, Schmidt TE, Thorkelson A, Reilly C, Perkey K, Reimann TG, Utay NS, Nganou Makamdop K, Stevenson M, Douek DC, Haase AT, Schacker TW, Large number of rebounding/founder HIV variants emerge from multifocal infection in lymphatic tissues after treatment interruption, Proc Natl Acad Sci U S A (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Lu CL, Pai JA, Nogueira L, Mendoza P, Gruell H, Oliveira TY, Barton J, Lorenzi JCC, Cohen YZ, Cohn LB, Klein F, Caskey M, Nussenzweig MC, Jankovic M, Relationship between intact HIV-1 proviruses in circulating CD4(+) T cells and rebound viruses emerging during treatment interruption, Proc Natl Acad Sci U S A 115(48) (2018) E11341–E11348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Kwon KJ, Timmons AE, Sengupta S, Simonetti FR, Zhang H, Hoh R, Deeks SG, Siliciano JD, Siliciano RF, Different human resting memory CD4(+) T cell subsets show similar low inducibility of latent HIV-1 proviruses, Sci Transl Med 12(528) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Einkauf KB, Lee GQ, Gao C, Sharaf R, Sun X, Hua S, Chen SM, Jiang C, Lian X, Chowdhury FZ, Rosenberg ES, Chun TW, Li JZ, Yu XG, Lichterfeld M, Intact HIV-1 proviruses accumulate at distinct chromosomal positions during prolonged antiretroviral therapy, J Clin Invest 129(3) (2019) 988–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Rato S, Rausell A, Munoz M, Telenti A, Ciuffi A, Single-cell analysis identifies cellular markers of the HIV permissive cell, PLoS Pathog 13(10) (2017) e1006678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Golumbeanu M, Cristinelli S, Rato S, Munoz M, Cavassini M, Beerenwinkel N, Ciuffi A, Single-Cell RNA-Seq Reveals Transcriptional Heterogeneity in Latent and Reactivated HIV-Infected Cells, Cell reports 23(4) (2018) 942–950. [DOI] [PubMed] [Google Scholar]
- [164].Bradley T, Ferrari G, Haynes BF, Margolis DM, Browne EP, Single-Cell Analysis of Quiescent HIV Infection Reveals Host Transcriptional Profiles that Regulate Proviral Latency, Cell reports 25(1) (2018) 107–117 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Kazmierski J, Postmus D, Wyler E, Fischer C, Jansen J, Meixenberger K, Vitcetz SN, Sohn M, Sauer S, Bannert N, Landthaler M, Goffinet C, Single Cell RNA-Sequencing-based Analysis of CD4+ T-Cell Subset-Specific Susceptibility to Transcriptional Modulation by HIV-1 Latency-Reversing Agents, bioRxiv (2020) 2020.05.04.075119. [Google Scholar]
- [166].Cohn LB, da Silva IT, Valieris R, Huang AS, Lorenzi JCC, Cohen YZ, Pai JA, Butler AL, Caskey M, Jankovic M, Nussenzweig MC, Clonal CD4(+) T cells in the HIV-1 latent reservoir display a distinct gene profile upon reactivation, Nat Med 24(5) (2018) 604–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Liu R, Yeh YJ, Varabyou A, Collora JA, Sherrill-Mix S, Talbot CC Jr., Mehta S, Albrecht K, Hao H, Zhang H, Pollack RA, Beg SA, Calvi RM, Hu J, Durand CM, Ambinder RF, Hoh R, Deeks SG, Chiarella J, Spudich S, Douek DC, Bushman FD, Pertea M, Ho YC, Single-cell transcriptional landscapes reveal HIV-1-driven aberrant host gene transcription as a potential therapeutic target, Sci Transl Med 12(543) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Cavrois M, Banerjee T, Mukherjee G, Raman N, Hussien R, Rodriguez BA, Vasquez J, Spitzer MH, Lazarus NH, Jones JJ, Ochsenbauer C, McCune JM, Butcher EC, Arvin AM, Sen N, Greene WC, Roan NR, Mass Cytometric Analysis of HIV Entry, Replication, and Remodeling in Tissue CD4+ T Cells, Cell reports 20(4) (2017) 984–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Neidleman J, Luo X, Frouard J, Xie G, Hsiao F, Ma T, Morcilla V, Lee A, Telwatte S, Thomas R, Tamaki W, Wheeler B, Hoh R, Somsouk M, Vohra P, Milush J, James KS, Archin NM, Hunt PW, Deeks SG, Yukl SA, Palmer S, Greene WC, Roan NR, The Atlas of the In Vivo HIV CD4 T Cell Reservoir, bioRxiv (2020) 2020.06.27.175745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Baxter AE, Niessl J, Fromentin R, Richard J, Porichis F, Charlebois R, Massanella M, Brassard N, Alsahafi N, Delgado GG, Routy JP, Walker BD, Finzi A, Chomont N, Kaufmann DE, Single-Cell Characterization of Viral Translation-Competent Reservoirs in HIV-Infected Individuals, Cell Host Microbe 20(3) (2016) 368–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Grau-Exposito J, Luque-Ballesteros L, Navarro J, Curran A, Burgos J, Ribera E, Torrella A, Planas B, Badia R, Martin-Castillo M, Fernandez-Sojo J, Genesca M, Falco V, Buzon MJ, Latency reversal agents affect differently the latent reservoir present in distinct CD4+ T subpopulations, PLoS Pathog 15(8) (2019) e1007991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Pardons M, Fromentin R, Pagliuzza A, Routy JP, Chomont N, Latency-Reversing Agents Induce Differential Responses in Distinct Memory CD4 T Cell Subsets in Individuals on Antiretroviral Therapy, Cell reports 29(9) (2019) 2783–2795 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Deleage C, Schuetz A, Alvord WG, Johnston L, Hao XP, Morcock DR, Rerknimitr R, Fletcher JL, Puttamaswin S, Phanuphak N, Dewar R, McCune JM, Sereti I, Robb M, Kim JH, Schacker TW, Hunt P, Lifson JD, Ananworanich J, Estes JD, Impact of early cART in the gut during acute HIV infection, JCI Insight 1(10) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Santangelo PJ, Rogers KA, Zurla C, Blanchard EL, Gumber S, Strait K, Connor-Stroud F, Schuster DM, Amancha PK, Hong JJ, Byrareddy SN, Hoxie JA, Vidakovic B, Ansari AA, Hunter E, Villinger F, Whole-body immunoPET reveals active SIV dynamics in viremic and antiretroviral therapy-treated macaques, Nature methods 12(5) (2015) 427–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Beckford Vera D, Schulte B, Henrich T, Flavell R, Seo Y, Abdelhafez Y, Badawi R, Cherry S, VanBrocklin H, First-in-human total-body PET imaging of HIV with 89Zr-VRC01 on the EXPLORER, Journal of Nuclear Medicine 61(supplement 1) (2020) 545. [Google Scholar]


