Abstract
Karyotyping, the practice of visually examining and recording chromosomal abnormalities, is commonly used to diagnose diseases of genetic origin, including cancers. Karyotypes are recorded as text written in the International System for Human Cytogenetic Nomenclature (ISCN). Downstream analysis of karyotypes is conducted manually, due to the visual nature of analysis and the linguistic structure of the ISCN. The ISCN has not been computer-readable and, as such, prevents the full potential of these genomic data from being realized. In response, we developed CytoGPS, a platform to analyze large volumes of cytogenetic data using a Loss-Gain-Fusion model that converts the human-readable ISCN karyotypes into a machine-readable binary format. As proof of principle, we applied CytoGPS to cytogenetic data from the Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer, a National Cancer Institute hosted database of over 69,000 karyotypes of human cancers. Using the Jaccard coefficient to determine similarity between karyotypes structured as binary vectors, we were able to identify novel patterns from 4,696 Mitelman CML karyotypes, such as the co-occurrence of trisomy 19 and 21. The CytoGPS platform unlocks the potential for large-scale, comparative analysis of cytogenetic data. This methodological platform is freely available at CytoGPS.org.
Keywords: CytoGPS, Karyotypes, Cytogenetics, Chronic myeloid leukemia, Data science, Bioinformatics
Introduction
Precision medicine leverages an individual’s genotype to optimize their likelihood of achieving and maintaining health through prevention and targeted therapy. Karyotyping, the practice of visually examining and recording chromosomal abnormalities, is one of the earliest and most common genotyping techniques [1]. The utility of karyotyping for diagnosis and prognosis has been demonstrated in numerous genetic and neoplastic diseases. The results often have important clinical implications for patient care, and are regularly and widely used for clinical decision making [1]. Conventional karyotypic analysis is a component of the standard-of-care for virtually all hematologic malignancies [1]. Because cytogenetic data have been used clinically for decades, many institutions have databases that contain tens of thousands of karyotypes. For example, the National Cancer Institute (NCI) hosts the Mitelman Database of Chromosome Aberrations and Gene Fusions, which contains more than 69,000 karyotypes collected since 1971 [2]. These karyotypes represent a wealth of historic data that is waiting be unlocked for biomedical research.
Karyotypes are stored as text using a standard human-interpretable notation, the International System for Human Cytogenetic Nomenclature (ISCN). ISCN codes are complex, variable, and have not been translated into a machine-readable format able to identify clinically relevant patterns. Further, several versions of the ISCN standard have been introduced over the decades. Following the release of the earliest version of ISCN in 1971, the standard has been revised nine times, most recently in 2018 [3]. These revisions often contain significant changes from previous standards, compounding the difficulty of translation across and between cytogenetic data sets. Thus, to date little of the information embedded in karyotypic databases has been mined for research purposes.
Current use of karyotype data for research is limited to those patterns that are visually apparent to cytogeneticists. Although the ISCN is meant to be readable by humans, it is often difficult to interpret due to the volume, complexity, and variability of the information contained therein. To the knowledge of the authors, no commercial or open source software converts ISCN-based karyotypes into a computational model. The closest is CyDAS [4], which has two significant drawbacks: 1) it employs regular expressions to parse karyotypes, and therefore cannot support automatic diagnosis and recovery from syntactic failures, and 2) CyDAS cannot parse some special karyotypes, such as derivative chromosomes. Due to the lack/limitation of existing software, potential clinically relevant patterns in long and complex karyotypes remain hidden and thus, unused. To leverage this wealth of existing karyotypic data in its entirety, we developed a computational tool, the CytoGenetic Pattern Sleuth (CytoGPS), which is designed to translate raw karyotypes into a computable form [5]. This form represents chromosomal abnormalities as loss, gain, or fusion (LGF) events at the resolution of cytogenetic bands. Karyotypes transformed by CytoGPS can be analyzed in aggregate with a diverse array of approaches.
Here, we describe the development of the CytoGPS model and, as proof of principle, apply it to all 4,969 karyotypes in the Mitelman database with a diagnosis of chronic myelogenous leukemia (CML). CML is a cytogenetically well-understood disease, defined by presence of the Philadelphia translocation, t(9;22)(q34;q11.2) [6]. We expected the database to include some cases where this translocation would not appear, for two reasons. First, some of the data extend back to 1971, before the t(9;22)(q34;q11.2) became a diagnostic standard of CML, and might be classified differently using current criteria. Second, in up to 10% of CML cases, the t(9;22)(q34;q11.2) cannot be detected on routine karyotyping either because 3 or more chromosomes are involved in the translocation, or it is a so-called “cryptic” translocation, beyond the limits of resolution of routine karyotyping [7]. Regardless, we demonstrate that CytoGPS can find both known subtypes defined by the most important secondary cytogenetic abnormalities in CML as well as novel abnormalities.
Materials and Methods
Dataset
We first applied CytoGPS to 66,362 cancer-related karyotypes in ISCN notation obtained from the Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer (http://cgap.nci.nih.gov/Chromosomes/Mitelman). A subsequent search of this database for cases with the diagnosis of CML identified 4,969 karyotypes. All Mitelman karyotypes were subjected to analysis by CytoGPS.
CytoGPS Algorithm
CytoGPS converts ISCN karyotypes to binary vectors through a novel parsing and mapping algorithm. CytoGPS is implemented using ANTLR (ANother Tool for Language Recognition) [8], a lexer and parser generator aimed at building and walking parse trees. CytoGPS breaks the karyotype text into individual elements based on the ISCN grammar. Multiple clones in the same karyotype (separated by slashes) are returned as separate binary vectors, in the same order; a note in the output file identifies secondary clones. Karyotypes are first processed with syntactic parsing to identify the text boundaries of individual chromosomal aberrations in a string. CytoGPS can parse both left-to-right and right-to-left, allowing parsing of complex chromosomal abnormalities, such as derivative chromosomes, and of karyotypes that use the terms stemline (sl), sideline (sdl), and idem. For example, the karyotype:
which contains a complex derivative chromosome 1, would be parsed from right to left, beginning with the inv(3)(q23q26). Parsed individual aberrations are classified using a rule-based mapping language represented within the LGF biological model. We developed a rule for each aberration in the ISCN based on the biological events that occur during that cytogenetic aberration. In order to ensure that any parsed and mapped karyotype is represented in its entirety, we designed the mapping function to fail when it encounters a single error within a case. Thus, a karyotype with five aberrations, four of which can be mapped but one that fails mapping, will be discarded as having failed completely. This stringent design allows for careful analysis of karyotypes because we are only observing complete karyotypes.
Statistical Methods
All statistical analyses were performed using version 3.6.0 of the R Statistical Programming Environment. We used the Jaccard coefficient to measure similarity between karyotypes. The Jaccard coefficient is defined as J = N11/(N11 + N10 + N01), where each Nij is the number of entries equal to i in the first LGF karyotype and j in the second karyotype [9]. We chose the Jaccard coefficient because karyotype vectors are sparsely populated, and the Jaccard coefficient does not give any weight to “normal” N00 matches. We performed clustering by applying partitioning around medoids (PAM) [10] to distances constructed from the Jaccard coefficient. The number of clusters was determined using the R packages PCDimension (version 1.1.12) and Thresher (version 1.1.2) [11, 12]. To visualize clusters, we used a nonlinear dimension reduction technique, t-distributed Stochastic Neighbor Embedding (t-SNE) [13]. To summarize the associations between cytogenetic abnormalities and individual clusters, we computed the most frequent aberrations by cluster.
Results
CytoGPS Validation
We first applied CytoGPS to 66,362 cancer-related karyotypes in ISCN notation from the Mitelman database. We evaluated parser and mapping success or failure on a sampling of cases evaluated visually by expert cytogeneticists (NAH, LVA). CytoGPS successfully parsed and mapped 70% of the entries into the LGF model. Evaluation of parsing or mapping success identified four kinds of failures (Table 1). First, uncertain karyotypes comprised 20% of the total karyotypes (13,400) and represented 66% of the failures. In ISCN, question marks indicate abnormal chromosomes or chromosome structures that cannot be fully ascertained by the cytogeneticist [14]. Second, mapping failures represented 8.2% of the total (5,482), or 27% of failures. These were syntactically valid ISCN karyotypes that could not be mapped using the LGF model because they contained either 1) a unique abnormality, such as a five-way translocation event, that was so rare it lacked a mapper rule, or 2) clearly incorrect content, such as chromosome 70. Third, syntax failures comprised 1.6% of the total (1,055), representing, 5.2% of the failures. These usually occurred when the entry contained characters such as dashes or parentheses in locations not permitted by any ISCN standard. Finally, the “other” failure category consisted of 0.4% of the total (280), making up 1.4% of the failures. This category contained all other failures, including large derivative chromosomes not translated by the mapping function. The uncertain karyotype failures are an intentional cause of parser failure designed to preserve only complete karyotypes for analysis. Removing these from the total failure, parsing accuracy is high, with a 93% success rate. Consequently, the vast majority of failures (27%) were produced by the mapping function.
Table 1.
Parsing and mapping failures classified by failure type.
| Failure Group | Number of Karyotypes |
Percent of total karyotypes |
Percent of failures |
|---|---|---|---|
| Uncertain karyotype with ‘?’ | 13,400 | 20.2% | 66.3% |
| Mapping | 5,482 | 8.3% | 27.1% |
| Syntax | 1,055 | 1.6% | 5.2% |
| Other | 280 | 0.4% | 1.4% |
Chronic Myeloid Leukemia
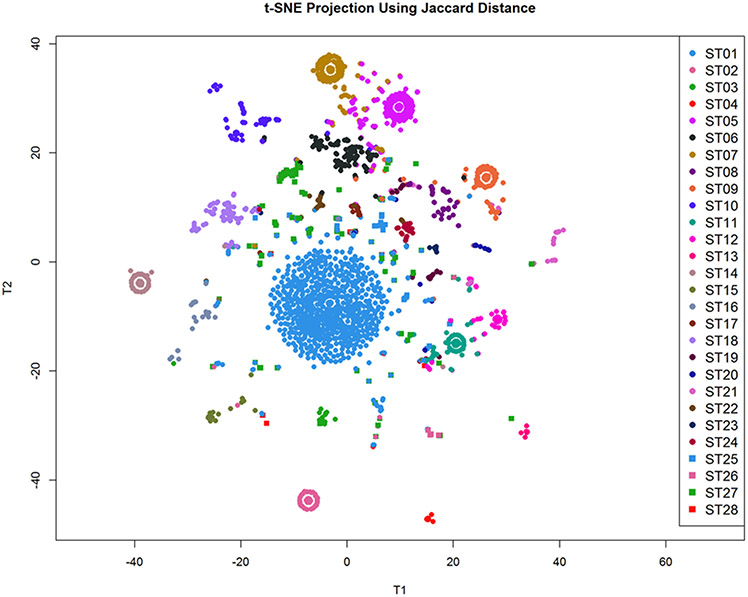
Next, we applied CytoGPS to the 4,969 karyotypes with a diagnosis of CML. Each karyotype was processed through the CytoGPS system, parsed and mapped into the binary LGF model. We then calculated the Jaccard coefficient between all pairs of binary LGF karyotypes and constructed a Jaccard distance matrix (1 – J). Using the PCDimension and Thresher packages, we determined that there were 28 clusters of LGF karyotypes. We then clustered the karyotypes using PAM and visualized them using t-SNE (Figure 1). This visualization supports the finding of 28 distinct clusters, many of which are readily identified as eye-shaped nuclei surrounding a center, all with an identical karyotype. Karyotypes containing a small number of additional abnormalities visually fan out from the center.
Figure 1: T-SNE projection of the CML LGF karyotype data using the Jaccard distance demonstrates approximately 28 distinguishable clusters.
Each cluster is represented by an ‘eye like’ structure, and is defined largely by a single abnormality or a set of abnormalities. Karyotypes that contain additional abnormalities fan out from center.
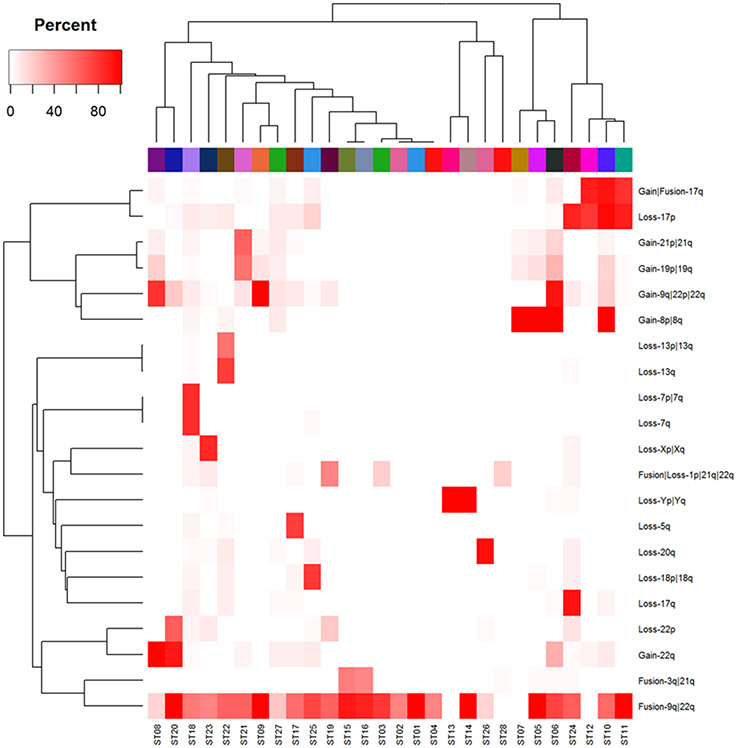
The informative LGF features that helped separate sample clusters were themselves clustered (using PAM and the Jaccard distance) into 21 groups, each of which had a clear interpretation as a unique cytogenetic abnormality. We then computed the percentage of karyotypes in each sample-cluster that exhibited each of the clustered abnormalities (Figure 2). Using these percentages, we were able to describe the set of abnormalities that characterized each sample cluster (Table 2). We defined the characteristic abnormalities per cluster as abnormalities that occurred in 50% or more of the karyotypes within a cluster. For this reason, multiple clusters could have the same defining cytogenetic abnormalities while having different low level cytogenetic abnormalities that did not reach the 50% population threshold. This methodology recovered cytogenetic subgroups associated with each of the most common secondary abnormalities in CML: gain of chromosome 8 (+8), monosomies of chromosomes Y and 7 (−Y and −7), deletion of a portion of the long arm of chromosome 7 (del(7q)), balanced translocation of chromosome 3 band and chromosome 21 band (t(3;21)(q26;q22), an extra copy of the Philadelphia chromosome (der(22)t(9;22)(q34;q11.2)), and an isochromosome 17 formed by the centromeric fusion of two long arms of chromosome 17 (i(17q)) [15]. We detected subtypes with loss of one chromosome (−5, −13, −17, −18, or −X) in addition to the characteristic t(9;22). Our clustering analysis also detected more complex relationships, such as the co-occurrence of trisomy 19 and 21 in the same patient samples [16]. These findings indicate that CytoGPS can detect known as well as novel or complex cytogenetic subgroups in CML.
Figure 2: Heat map of recurrent aberrations by cluster.
This heat map shows the relationship between the most common cytogenetic abnormalities (y-axis) by each individual cluster (x-axis). This illustrates that different clusters are cytogenetically defined by certain commonly recurring abnormalities.
Table 2: Frequent Aberrations by Cluster.
This table shows each cluster, the number of karyotypes in that cluster and the defining cytogenetic abnormalities of each cluster. Some clusters have identical defining cytogenetic abnormalities while having different low level cytogenetic abnormalities that did not reach the 50% population threshold.
| Cluster | Number of Karyotypes |
Defining Cytogenetic Aberrations >50% of karyotypes have event |
|---|---|---|
| 1 | 861 | t(9;22)(q34;q11.2) |
| 2 | 178 | t(9;22)(q34;q11.2) |
| 3 | 58 | t(9;22)(q34;q11.2) |
| 4 | 70 | t(9;22)(q34;q11.2) |
| 5 | 364 | t(9;22)(q34;q11.2),+8 |
| 6 | 243 | t(9;22)(q34;q11.2),+8,add(9q),+22 |
| 7 | 296 | +8 |
| 8 | 149 | +9q,+22 |
| 9 | 243 | t(9;22)(q34;q11.2),add(9q),+22 |
| 10 | 239 | t(9;22)(q34;q11.2),+8,i(17)(q10) |
| 11 | 220 | t(9;22)(q34;q11.2),i(17)(q10) |
| 12 | 159 | i(17)(q10) |
| 13 | 68 | −Y |
| 14 | 159 | t(9;22)(q34;q11.2),−Y |
| 15 | 96 | t(9;22)(q34;q11.2),t(3;21)(q26;q22) |
| 16 | 139 | t(9;22)(q34;q11.2),t(3;21)(q26;q22) |
| 17 | 43 | t(9;22)(q34;q11.2),−5 |
| 18 | 225 | t(9;22)(q34;q11.2),−7 |
| 19 | 64 | t(9;22)(q34;q11.2),t(1;21;22) |
| 20 | 55 | t(9;22)(q34;q11.2),i(22)(q10) |
| 21 | 110 | t(9;22)(q34;q11.2),+19,+21 |
| 22 | 53 | t(9;22)(q34;q11.2),−13 |
| 23 | 32 | t(9;22)(q34;q11.2),−X |
| 24 | 71 | t(9;22)(q34;q11.2),−17 |
| 25 | 49 | t(9;22)(q34;q11.2),−18 |
| 26 | 47 | t(9;22)(q34;q11.2), del(20q) |
| 27 | 223 | NA |
| 28 | 24 | NA |
Discussion
Most of the specific abnormalities that define each of these clusters have been reported in the literature; thus providing verification and validation of CytoGPS as a new, viable methodology. Isochromosome 17 has been noted as relevant in CML cases particularly due to the resulting loss of the TP53 gene [15]. Trisomy 8 is another common cytogenetic event in CML; it is the most common secondary event after the Philadelphia chromosome [17]. This finding is supported by our analysis, both by the number of cases with trisomy 8 and the abundance of trisomy 8 as a defining characteristic in multiple clusters. Rarer events such as fusions associated with 3q (the defining characteristic of Cluster 1) have also been noted in the literature [18].
Many of these clusters can be “paired” based on the presence or absence of the t(9;22) translocation. Clusters 5 and 7 are both defined as having trisomy 8 yet Cluster 5 has the t(9;22) translocation while Cluster 7 does not. Both clusters have a comparable number of cases despite the ascertainment bias that CML is defined by the t(9;22) translocation and thus we would expect a much larger number of cases with the translocation than without. This mirrored cluster structure is due to two major reasons: 1) the age of the samples and 2) cryptic translocations. The first reason, the age of the samples, relates to the timing of CML’s disease definition. The Mitelman database goes back to 1971, pre-dating the disease definition of CML by the t(9;22) translocation. Many of the CML samples that were analyzed in the study thus pre-date this disease definition. The second reason that many of these samples do not appear to have the t(9;22) translocation relates to cryptic translocations. A cryptic translocation is defined as the event where a translocation has occurred, but it is too small to be detectable by conventional cytogenetics [19]. Such events would appear only from a more molecular assay such as Fluorescence in situ Hybridization (FISH) [20]. That is, many of these cases that do not appear to have the t(9;22) translocation probably have a cryptic translocation, thus, it went undetected and was not displayed in the ISCN karyotype.
Another interesting finding is the relationship of common cytogenetic abnormalities to each other. As seen in Figure 2, the majority of gain events split from the majority of loss events at the top of the dendrogram. This indicates that when a patient has a gain event they are more likely to have additional gains rather than losses. This phenomenon has been observed in other hematologic malignancies such as chronic lymphocytic leukemia (CLL) [21]. This further strengthens the argument that CytoGPS is able to uncover the underlying biology contained in karyotype data. Further studies will determine the clinical relevance of these findings.
The methods described here can be applied to any collection of ISCN karyotypes. We expect leukemias and lymphomas, which predominate in many institutional cytogenetics databases, to be the first diseases to the studied in detail. The free version of CytoGPS can be accessed via the web browser CytoGPS.org [5]. CytoGPS exports the binary vectors of each clone-karyotype both as Excel spreadsheets and as JSON files, which can be read into a wide variety of statistical software. As shown in this paper, these can be used for unsupervised clustering analyses, but both the binary vectors and the cluster assignments can also be used in supervised analyses to find markers or predictors of clinically important variable such as response to therapy.
Acknowledgments
Funding
This work was supported by the National Library of Medicine (NLM) grant number T15 LM011270, the National Cancer Institute grant number R03 CA235101, and by Pelotonia Intramural Research Funds from the James Cancer Center, Columbus Ohio
References
- 1.Shuman S, Structure, mechanism, and evolution of the mRNA capping apparatus. Prog Nucleic Acid Res Mol Biol, 2001. 66: p. 1–40. [DOI] [PubMed] [Google Scholar]
- 2.Heim S and Mitelman F, Cancer cytogenetics: chromosomal and molecular genetic aberrations of tumor cells. 2015: John Wiley & Sons. [Google Scholar]
- 3.Stevens-Kroef M, et al. , Cytogenetic Nomenclature and Reporting. Methods Mol Biol, 2017. 1541: p. 303–309. [DOI] [PubMed] [Google Scholar]
- 4.Hiller B, et al. , CyDAS: a cytogenetic data analysis system. Bioinformatics, 2005. 21(7): p. 1282–3. [DOI] [PubMed] [Google Scholar]
- 5.Abrams ZB, et al. , CytoGPS: a web-enabled karyotype analysis tool for cytogenetics. Bioinformatics, 2019. 35(24): p. 5365–5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rowley JD, A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature, 1973. 243(5405): p. 290–3. [DOI] [PubMed] [Google Scholar]
- 7.Vardiman J, et al. , Chronic myeloid leukaemia, BCR-ABL1–positive, in WHO Classification of Tumors of Haematopoietic and Lymphoid Tissue, Swerdlow SH, et al. , Editors. 2017, IARC Press: Lyon, France. p. 30–36. [Google Scholar]
- 8.Parr T, ANTLR: Another tool for language recognition. 2006. [Google Scholar]
- 9.Jaccard P, The distribution of the flora in the alpine zone. 1. New phytologist, 1912. 11(2): p. 37–50. [Google Scholar]
- 10.J. KP and Rousseeuw L, Finding Groups in Data: An Introduction to Cluster Analysis. 1990, Hoboken, NJ: John Wiley & Sons. [Google Scholar]
- 11.Wang M, et al. , Thresher: determining the number of clusters while removing outliers. BMC Bioinformatics, 2018. 19(1): p. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang M, Kornblau SM, and Coombes KR, Decomposing the Apoptosis Pathway Into Biologically Interpretable Principal Components. Cancer Inform, 2018. 17: p. 1176935118771082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Maaten L and Hinton G, Visualizing data using t-SNE. Journal of machine learning research, 2008. 9(November): p. 2579–2605. [Google Scholar]
- 14.Shaffer LG, McGowan-Jordan J, and Schmid M, ISCN 2013: an international system for human cytogenetic nomenclature (2013). 2013: Karger Medical and Scientific Publishers. [Google Scholar]
- 15.Meggendorfer M, et al. , SETBP1 mutations occur in 9% of MDS/MPN and in 4% of MPN cases and are strongly associated with atypical CML, monosomy 7, isochromosome i(17)(q10), ASXL1 and CBL mutations. Leukemia, 2013. 27(9): p. 1852–60. [DOI] [PubMed] [Google Scholar]
- 16.Johansson B, Fioretos T, and Mitelman F, Cytogenetic and molecular genetic evolution of chronic myeloid leukemia. Acta Haematol, 2002. 107(2): p. 76–94. [DOI] [PubMed] [Google Scholar]
- 17.Bakshi SR, et al. , Trisomy 8 in leukemia: A GCRI experience. Indian J Hum Genet, 2012. 18(1): p. 106–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Togasaki E, et al. , Frequent somatic mutations in epigenetic regulators in newly diagnosed chronic myeloid leukemia. Blood Cancer J, 2017. 7(4): p. e559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilch ES and Morton CC, Historical and Clinical Perspectives on Chromosomal Translocations. Adv Exp Med Biol, 2018. 1044: p. 1–14. [DOI] [PubMed] [Google Scholar]
- 20.Bayani J and Squire JA, Fluorescence in situ Hybridization (FISH). Curr Protoc Cell Biol, 2004. Chapter 22: p. Unit 22.4. [DOI] [PubMed] [Google Scholar]
- 21.Baliakas P, et al. , Additional trisomies amongst patients with chronic lymphocytic leukemia carrying trisomy 12: the accompanying chromosome makes a difference. haematologica, 2016. 101(7): p. e299–e302. [DOI] [PMC free article] [PubMed] [Google Scholar]