Abstract
In advanced prostate cancer, resistance to androgen deprivation therapy is achieved through numerous mechanisms, including loss of the androgen receptor (AR) allowing for AR-independent growth. Therapeutic options are limited for AR-independent castration-resistant prostate cancer (CRPC), and defining mechanisms critical for survival is of utmost importance for targeting this lethal disease. Our studies focus on identifying telomere maintenance mechanism (TMM) hallmarks adopted by CRPC to promote survival. TMMs are responsible for telomere elongation to instill replicative immortality and prevent senescence, with the two TMM pathways available being telomerase and alternative lengthening of telomeres (ALT). Here, we show that AR-independent CRPC demonstrates an atypical ALT-like phenotype with variable telomerase expression and activity, whereas AR-dependent models lack discernible ALT hallmarks. In addition, AR-independent CRPC cells exhibited elevated levels of SLX4IP, a protein implicated in promoting ALT. SLX4IP overexpression in AR-dependent C4-2B cells promoted an ALT-like phenotype and telomere maintenance. SLX4IP knockdown in AR-independent DU145 and PC-3 cells led to ALT-like hallmark reduction, telomere shortening, and induction of senescence. In PC-3 xenografts, this effect translated to reduced tumor volume. Using an in vitro model of AR-independent progression, loss of AR in AR-dependent C4-2B cells promoted an atypical ALT-like phenotype in an SLX4IP-dependent manner. Insufficient SLX4IP expression diminished ALT-like hallmarks and resulted in accelerated telomere loss and senescence.
Implications:
This study demonstrates a unique reliance of AR-independent CRPC on SLX4IP-mediated ALT-like hallmarks and loss of these hallmarks induces telomere shortening and senescence, thereby impairing replicative immortality.
Introduction
Prostate cancer relies on androgen receptor (AR) signaling, and advanced disease is treated with androgen deprivation therapy, despite the nearly unavoidable progression to castration-resistant prostate cancer (CRPC; refs. 1-3). Castration resistance is acquired through a number of AR-dependent mechanisms, such as AR mutations, amplifications, and splice variants (4). Resistance is also attained following AR loss and activation of AR-independent pathways (5-9). Accordingly, second-generation antiandrogens have been developed for CRPC (10, 11), however, they provide little benefit in CRPC patients with AR loss (6, 12). Moreover, AR-dependent CRPC treatment with second-generation agents induces AR loss (6). Because of the lack of efficacious therapeutics and poor prognosis of AR-independent CRPC, it is crucial to identify regulatory mechanisms required for AR-independent CRPC survival.
Our studies have focused on defining telomere maintenance mechanism (TMM) phenotypes in AR-dependent and -independent CRPC. Cancer cells acquire replicative immortality by activating TMMs, which allows for maintained telomere length over successive cell divisions (13-16). As replicative polymerases are unable to fully replicate the ends of linear DNA (17), telomeric DNA of somatic cells progressively shortens with each replication cycle until a minimum threshold limit is reached provoking senescence to prevent deleterious genomic consequences (18). Conversely, malignancies employ TMMs, through activation of telomerase or the alternative lengthening of telomeres (ALT) pathway, to evade this terminal fate and instill replicative immortality (13-16). Telomerase is a reverse transcriptase capable of catalyzing the addition of telomeric repeats to chromosomal ends (13) and is the primary contributor to telomere elongation in 85% to 90% of cancers (19). ALT activation accounts for the remaining 10% to 15% of cancers (15) and involves telomere-directed homologous recombination (HR) to promote elongation (14). The ALT mechanism remains challenging to characterize due to the inability to measure ALT activity directly, unlike the telomerase pathway (19, 20). As such, the identification of ALT relies upon surrogate measures, including ALT-associated promyelocytic leukemia protein (PML) bodies (APB) found at sites of telomeric HR (21), extratelomeric circular C-rich DNA byproducts of ALT called C-circles (22), and significant telomere length heterogeneity (14). Historically, ALT was predominantly associated with cancers with undetectable telomerase activity (15), but emerging data indicate TMM identity is not universally binary; instead, both telomerase and ALT are inherently programmed in malignancy and coexist to varying degrees, further confounding TMM characterization (23, 24). Despite the complicated mechanism and identification of ALT, recent evidence has begun to illuminate the underestimation of ALT’s contribution to telomere maintenance, including prostate cancer.
Both in vitro and in vivo, telomere elongation in primary prostate cancer has been wholly attributed to telomerase (25, 26). This is not surprising as androgens in primary disease models promote expression of the telomerase reverse transcriptase component, TERT (26). Historically, CRPC, regardless of AR expression, was assumed to rely upon telomerase for telomere elongation (25), which is unsurprising due to the challenges surrounding ALT identification, presence of detectable telomerase activity, and underestimation of TMM coexistence. However, a case study comparing primary and castration-resistant disease identified ALT hallmarks in a distant metastatic site, which were lacking in the primary tumor (27). This finding represents one of the few documented occurrences of the understudied ALT phenotype in prostate cancer and, more importantly, highlights a link between TMM coexistence and disease progression. Thus, understanding the acquisition of ALT hallmarks in prostate cancer progression is critical for understanding the purpose of TMM coexistence.
Additional examples of TMM coexistence have emerged in cases beyond prostate cancer progression, underscoring the complexity of malignant TMM programming (28-30). Moreover, cancer cells initiate a shift from telomerase to ALT as a mechanism of resistance to telomerase inhibition highlighting the necessity of both TMM programs, although both mechanisms may not be engaged equally or simultaneously (31-34). Further supporting the potential of TMM coexistence, telomerase reconstitution or fusion of telomerase- and ALT-positive cells led to a blended TMM phenotypic presentation (35, 36). Despite the identification of TMM coexistence, key regulators have yet to be well characterized. Recently, SLX4IP was implicated as a regulator of TMM plasticity. SLX4IP interacts with SLX4, a nuclease scaffold essential for ALT-mediated telomere maintenance (37, 38). SLX4 shuttles its associated nucleases and SLX4IP to participate in the resolution of HR junctions at the telomere following its recruitment by TRF2, an essential telomere end-binding protein (37, 39). Knowledge regarding the function of SLX4IP beyond this interaction is limited, but emerging data demonstrate SLX4IP differentially regulates TMMs based on cancer type. Specifically, SLX4IP knockout promotes ALT hallmarks in osteosarcoma models (40), but in breast cancer models, SLX4IP knockdown leads to the loss of ALT hallmarks and induction of telomerase (41). Despite this disease-dependent difference, it is evident that SLX4IP is involved in TMM plasticity, although its involvement in TMM coexistence linked to prostate cancer disease progression remains unexplored.
To understand the acquisition of ALT hallmarks and TMM coexistence in CRPC, we have characterized the TMM phenotype of five diverse CRPC in vitro models. Surprisingly, highly variable telomerase expression and activity were noted among CRPC models. CRPC cell lines utilizing AR-independent modes of resistance exhibit an atypical ALT phenotype coupled with elevated SLX4IP expression, whereas AR-dependent models demonstrate a lack of ALT hallmarks. In addition, we show that overexpression of SLX4IP promotes unique ALT-like telomeric PML localization events closely mimicking traditional APBs. In contrast, we show SLX4IP knockdown leads to loss of ALT-like PML localization events in AR-independent CRPC cell lines. Consequently, these cells undergo a notable reduction in telomere length coupled with the induction of senescence-associated markers, which was partially rescued following telomerase overexpression. The observed SLX4IP-mediated promotion of ALT-like PML localization events and telomere maintenance were further confirmed after inducing AR loss in AR-dependent CRPC cells via androgen deprivation. Herein, we identify a unique relationship between SLX4IP and ALT-like hallmarks that is necessary for the maintenance of telomere length and, therefore, replicative immortality of AR-independent CRPC in vitro.
Materials and Methods
Cell culture
Cell lines were obtained from the ATCC and maintained at 37°C in 5% CO2. U2OS, HCT116, WI-38 VA13, and HEK293T cells were cultured in DMEM (Gibco) with 10% [volume/volume (v/v)] FBS (Sigma), 1% of penicillin 10,000 U/mL, streptomycin 10,000 μg/mL, and 25 μg/mL Amphotericin B Mixture (Antibiotic-Antimycotic, Gibco). C4-2B, CWR-R1, 22Rv1, DU145, and PC-3 cells were cultured in RPMI1640 (Gibco) with 10% FBS and 1% antibiotic-antimycotic. LAPC4 cells were cultured in Iscove’s modified Dulbecco’s Medium (Sigma) with 10% (v/v) FBS (Sigma) and 1 nmol/L Dihydrotestosterone (Sigma). For androgen-deprived conditions, C4-2B cells were cultured in RPMI1640 with no Phenol Red (Gibco) with 10% Charcoal-Stripped FBS (Sigma) and 1% antibiotic-antimycotic. Parental and genetically modified cell lines were subjected to Mycoplasma testing via MycoAlert PLUS Detection Kit (July 2018, Lonza) and short tandem repeat validation [April 2019, Case Western Reserve University (CWRU) Genomics Core, Cleveland, OH]. To determine population doubling (PD) times, cells were plated and trypsinized at 72 and 120 hours. Using trypan blue exclusion, viable cell number was determined by using the Countess II (Thermo Fisher Scientific). All experiments, including designated early-passage timepoints, were completed with cells from PD 2 through 8. Late-passage timepoints were completed with cells from the latest PD included in each telomere length experiment or after.
Stable cell line generation
To generate cell lines stably overexpressing 3X FLAG-tagged-SLX4IP, a gBlock was designed to incorporate a 3X FLAG tag. The 3X FLAG-SLX4IP DNA sequence (NCBI nucleotide, NM_001009608.3) was flanked with a SalI endonuclease restriction site on the C-terminus and a BamHI restriction site on the N-terminus. The SLX4IP open reading frame was engineered for bacterial codon optimization using the gBlock Gene Fragment design tool (Integrated DNA Technologies). The gBlock was cloned into pBABE-puro, a gift from Hartmut Land, Jay Morgenstern, and Bob Weinberg (Addgene, #1764; ref. 42). Retrovirus was produced using Lipofectamine 2000 (Thermo Fisher Scientific) to cotransfect pBABE-puro or pBABE-puro.SLX4IP.3XFLAG with pCMV-VSV-G (at a ratio of 6:1, Clontech) into GP2-293T cells (Clontech). Virus was harvested and cleared via centrifugation and/or sterile filtration at 48 hours posttransfection and infections were carried out in the presence of 10 μg/mL of Polybrene (Sigma) followed by selection in 1 (C4-2B) or 1.5 μg/mL (DU145 and PC-3) of Puromycin (Sigma). To generate cell lines stably overexpressing telomerase, an identical strategy was used as outlined above using pBABE-puro.UThTERT+U3-hTR-500, a gift from Kathleen Collins (Addgene#27665; ref. 43).
To generate cell lines with stable knockdown of SLX4IP, two short hairpin RNAs (shRNA) targeting SLX4IP in the pLKO.1-puro lentiviral expression plasmid were purchased from Sigma (clone IDs: NM_001009608.1-426s1c1 and NM_001009608.1-247s1c1). Lentivirus was produced by using Lipofectamine 2000 (Thermo Fisher Scientific) to cotransfect HEK293T cells with pLKO.1-puro Non-Mammalian shRNA control (Sigma, #SCH002), pLKO.1-puro. shSLX4IP.1, or pLKO.1-puro.shSLX4IP.2 with pMD2.G, pRRE, and pRSV-Rev, which were gifts from Didier Trono (at a ratio of 4:1:1:1, Addgene, #12259, #12251, #12253; ref. 44). Viral infections and selection were carried out as described above.
Mouse xenograft studies and IHC
In vivo experiments were performed with approval from the Institutional Animal Care and Use Committee at CWRU (Protocol 2017–0126, Cleveland, OH), which is certified by the American Association of Accreditation for Laboratory Animal Care. Six-week-old male nude mice (NCR nu/nu), bred on-site at The Athymic Animal & Preclinical Therapeutics Facility at CWRU, were inoculated with a 1:1 mix of Matrigel (Gibco, 354234) and 1 × 106 cells subcutaneously. Mouse weights and tumor volumes were determined three times weekly. Tumor volumes were calculated using the hemiellipsoid volume formula following Vernier caliper measurements of tumor diameters in three dimensions. At 30 days postinoculation, tumors were excised and processed for IHC and telomerase activity. IHC and hematoxylin and eosin staining were carried out as published previously (45). Primary antibody dilutions used are as follows: SLX4IP (1:200, Sigma, HPA046372), CDKN1A or p21 (1:500, Cell Signaling Technology, 2947), MKI67 or Ki-67 (1:1,000, Sigma, HPA001164), and active caspase-3 (1:1,000, Millipore, AB3623). Three fields of view for each slide were imaged using an Olympus BX43 Upright Microscope and cells were manually counted and scored using ImageJ software (46).
RNA isolation and analysis
RNA was isolated using TRizol Reagent (Invitrogen), reverse transcribed using the High-Capacity RNA-to-cDNA Kit (Thermo Fisher Scientific), and quantified via qRT-PCR on an Applied Biosystems StepOnePlus Real-time PCR System as described previously (47). Established primer pairs for TERT and GAPDH were used (47).
Western blotting
Western blotting was completed as described previously (47). Briefly, 50 μg of lysate was resolved on a 4% to 20% Mini-PROTEAN TGX Gel (Bio-Rad) and transferred to Nitrocellulose Membrane (Millipore) using the Trans-Blot Turbo (Bio-Rad). Blots were blocked in 5% nonfat milk diluted in TBST and incubated overnight at 4°C with primary antibody. Blots were incubated for 1 hour at room temperature with either horseradish peroxidase–conjugated (Santa Cruz Biotechnology), IRDye 800CW (LI-COR), or IRDye 680RD (LI-COR) secondary antibodies diluted to 1:10,000. Development with ProtoGlow ECL Reagent (National Diagnostics) was used when necessary. Blots were imaged on an Odyssey Fc Imaging System (LI-COR), quantified using Image Studio 5.2 software, and normalized to GAPDH. Primary antibody dilutions used are as follows: 1:20,000 GAPDH (Cell Signaling Technology, 2118), 1:5,000 SLX4IP (Sigma, HPA046372), 1:5,000 FLAG (Sigma, F1804), 1:10,000 AR (Santa Cruz Biotechnology, sc-816), 1:3,000 ENO2 (Cell Signaling Technology, 9536), 1:5,000 p21 (Cell Signaling Technology, 2947), and 1:2,000 ATRX (Cell Signaling Technology, 14820).
Telomere-repeat amplification protocol
The real-time Q-TRAP assay was performed as published previously (48). Tumor lysates were prepared following homogenization with Kontes PELLET PESTLE Grinders (Kimble) under similar lysis conditions. Reactions were performed on an Applied Biosystems StepOnePlus Real-time PCR System. Collected Ct values were then converted to relative telomerase activity units using the telomerase-positive standard curve generated as described previously. Technique validation experiments were carried out with telomerase-positive HEK293T and HCT116 cells and telomerase-negative U2OS and WI-38 VA13 cells, with a standard curve generated using HCT116 cells (Supplementary Fig. S1A and S1B). Additional negative controls were generated by incubating telomerase-positive samples with 1 μg of RNase at 37° C for 20 minutes.
Immunofluorescence-FISH analysis
A previously published immunofluorescence-FISH (IF-FISH) protocol was carried out on cells seeded on sterile glass coverslips and tumor sections (47). Coverslips were incubated with PML antibody (Santa Cruz Biotechnology, sc-5621) diluted 1:200 for 1 hour at room temperature. Alexa-488–conjugated anti-rabbit secondary antibody (The Jackson Laboratory) was diluted 1:100 and added to coverslips for overnight incubation at 4°C. Hybridization with 333 ng/mL of a TelC-Cy5–labeled peptide nucleic acid (PNA) oligonucleotide telomere probe (N-CCTAACCTAACCTAA-C, PNA BIO) in PNA buffer (10 mmol/L Tris, pH 7.5 and 70% formamide) was carried out at 95°C for 5 minutes followed by overnight incubation at room temperature. Coverslips were stained with 2 μg/mL of DAPI for 10 minutes and mounted with Fluoromount-G (Thermo Fisher Scientific). Stained slides were imaged following blinding using the Leica HyVolution SP8 gated STED Microscope and LAS X software. Cell count and regions of interest (ROI) representing potential colocalization events were identified in ImageJ (46). These ROIs were analyzed via Coloc 2 using the Manders’ correlation algorithm (49), where a correlation coefficient of ≥0.25 was further analyzed. This coefficient represents the total PML signal relative to the intersection of the PML and telomeric signal in a given ROI. Following this analysis, ROIs were categorized as true APBs (coefficients of >0.85) versus ALT-like PML localization events (coefficients of 0.25–0.85). Each cell was categorized on the basis of highest ROI correlation coefficient found within the cell. Colocalization events within U2OS control cells were included for comparison.
C-circle isolation and analysis
A previously published C-circle dot blot assay was performed with minor modifications (50). Genomic DNA was harvested using the Genelute Mammalian Genomic DNA Miniprep Kit (Sigma) according to the manufacturer’s instructions. DNA (160 ng) was incubated under conditions described in the protocol published previously. Reactions were blotted onto Hybond Membranes (Thermo Fisher Scientific) and UV cross-linked (Spectrolinker XL-1000, Spectronics). Membranes were hybridized with 30 μg/mL of Digoxigenin (DIG) conjugated telomere probe (CCCTAACCCTAACCCTAACCCTAA-DIG, Integrated DNA Technologies) in Digoxigenin Easy Hyb Buffer (Roche) overnight at 37°C. Saline-sodium citrate (SSC, 0.5×) buffer with 0.1% SDS was used to remove excess probe. Membranes were blocked in Digoxigenin Blocking Buffer (Roche) for 30 minutes followed by incubation with 1:3,000 dilution of Digoxigenin-AP, Fab Fragments (Roche) for 3 hours at room temperature. Digoxigenin Wash Buffer (Roche) was used to remove excess antibody and amplified C-circles were detected with CDP-Star Kit (Roche). Membranes were imaged with the Odyssey Fc Imager (LI-COR) and signal intensity ratios were calculated as described previously using Image Studio 5.2 Software. Technique validation experiments were carried out with 160 ng of DNA from telomerase-positive HEK293T, HCT116, and A549 cells and a standard curve of DNA from ALT-positive U2OS and WI-38 VA13 cells (Supplementary Fig. S1C).
Telomere restriction fragment analysis
Genomic DNA was harvested using the GenElute Mammalian Genomic DNA Miniprep Kit (Sigma) according to the manufacturer’s instructions and telomere restriction fragment (TRF) analysis was carried out as published previously (47). Membranes were hybridized as described in the above C-circle analysis protocol at 42°C. SSC (2×) buffer with 0.1% SDS was used to remove excess probe. Membranes were blocked in Digoxigenin Blocking Buffer (Roche) for 30 minutes followed by incubation with 1:10,000 dilution of Digoxigenin-AP, Fab Fragments (Roche) for 3 hours at room temperature. Additional membrane processing and imaging steps were carried out as described above in the C-circle analysis protocol.
Senescence-associated β-galactosidase activity staining
Cells were seeded on sterile glass coverslips and grown to 80% confluency. Senescence β-galactosidase Staining Kit was used following the manufacturer’s protocol (Cell Signaling Technology). Slides were imaged following blinding using an Olympus BX43 Upright Microscope and cells were manually counted and scored using ImageJ software (46).
Statistical analysis
Statistical analyses were performed using two-tailed Student t test (in vitro and in vivo data comparing two groups) or one-way ANOVA with multiple comparisons (in vitro data comparing at least three groups) in GraphPad Prism 8 software. Patient expression data were analyzed through the Prostate Cancer Transcriptome Atlas (PCTA) on the basis of disease course using rank-sums test between metastatic CRPC (mCRPC) and primary subsets (www.thepcta.org; ref. 51). P values less than 0.05 were considered statistically significant. All in vitro data are represented as mean values from three independent experiments performed in triplicate, unless otherwise noted.
Results
AR-independent CRPC in vitro models exhibit an atypical ALT-like phenotype
Because of the understudied nature of ALT’s contribution to telomere maintenance in CRPC, we sought to determine whether TMM phenotype correlated with AR dependence. Five in vitro CRPC models were interrogated for telomerase expression, activity, ALT hallmarks, and telomere length distribution. C4-2B, CWR-R1, and 22Rv1 cell lines retained AR expression (AR-dependent; refs. 52, 53), whereas DU145 and PC-3 cells exhibited AR loss (AR-independent; refs. 54, 55). Despite being castration sensitive, LAPC4 cells represent a characterized telomerase-positive AR-dependent model capable of acquiring ALT hallmarks (56), and were included as controls. Relative mRNA expression of TERT and telomerase activity across cell lines were analyzed alongside telomerase-positive HCT116 and ALT-positive U2OS controls (Fig. 1A and B; Supplementary Fig. S1D). Interestingly, significant variability was noted among CRPC cell lines regardless of AR status.
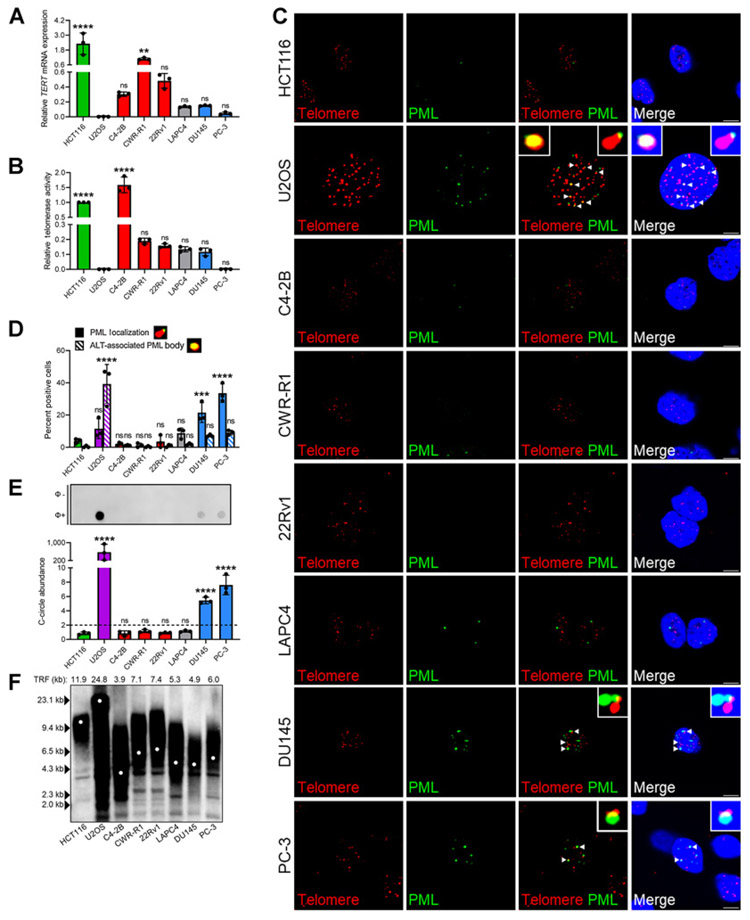
Figure 1.
Identification of atypical ALT phenotype in AR-independent CRPC in vitro. Relative TERT mRNA expression (A) and telomerase activity (B) across CRPC cell lines. C4-2B, CWR-R1, and 22Rv1 retain AR expression (red) and DU145 and PC-3 exhibit AR loss (blue). ALT-positive U2OS (purple), telomerase-positive HCT116 (green), and telomerase-positive LAPC4 cells retaining AR expression (gray) were included as controls. All groups were compared with U2OS control. C, Representative IF-FISH images demonstrating the presence (arrowheads) or absence of PML (green) at telomeres (red) with zoom insets provided. Scale bar, 5 μm. D, Quantification of C for percent positive cells with at least one APB. Cells lacking APBs were quantified for ALT-like PML localization events. Representative foci are included defining these events. All groups were compared with HCT116 control. E, Representative dot blot demonstrating the presence of telomeric C-circles. Φ+ lane indicates reactions with Φ29 polymerase and Φ− lane indicates control reaction. Quantification of C-circle abundance, defined as the signal ratio of Φ+ to Φ− reaction is shown below. Dotted line at y = 2 indicates threshold suggesting ALT activity. All groups were compared with HCT116 control. F, TRF analysis demonstrating average telomere length across cell lines. Data represented as mean ± SD; n = 3 (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). ns, not significant.
With the exception of C4-2B cells, CRPC cell lines exhibited limited telomerase activity. Based on this result, we reasoned ALT-like processes might be engaged to assist with telomere maintenance in CRPC cells with limited telomerase activity. As ALT activity cannot be directly measured, we evaluated surrogate measures of ALT activity, including APB abundance, C-circles, and telomere length heterogeneity (14, 21, 22). APBs were visualized via IF-FISH for ALT-associated PML protein localizing at the telomere, which is highly indicative of ALT-mediated HR events (21). Traditionally, APBs are instances of nearly complete overlap of PML foci with ultra-bright telomeric foci illustrated in the ALT-positive U2OS control (Fig. 1C and D; ref. 21). However, we identified atypical telomeric PML localization events exhibiting only partial overlap, representing an ALT-like hallmark (Fig. 1C and D). AR-independent DU145 and PC-3 cell lines had a modest proportion of APB-positive cells at approximately 7% and 9%, respectively, compared with ALT-positive control, with approximately 39%; however, AR-dependent cell lines had significantly fewer APB-positive cells ranging from approximately 0.3% to 1.8% (Fig. 1C and D). In addition, DU145 and PC-3 cell lines had a significantly greater proportion of cells with ALT-like PML localization events at approximately 22% and 34%, respectively, whereas AR-dependent cell lines exhibited these events infrequently, ranging from approximately 1.2% to 8.7%. To complement this finding, telomeric C-circles, which are byproducts of ALT-related events, were evaluated via C-circle amplification (CCA; ref. 50). A signal ratio greater than or equal to two when comparing the CCA reaction with control is highly indicative of ALT (50), and only AR-independent cell lines reached this threshold, albeit at a significantly lower level than U2OS control (Fig. 1E). Finally, telomere length was evaluated to determine whether CRPC cell lines contain long, heterogeneous telomeres, a known characteristic of traditional ALT-positive cells (14). Interestingly, DU145 and PC-3 telomeres did not mimic the telomere length distribution present in the U2OS control, and only exhibited an average telomere length of approximately 4.9 and 6 kb, respectively (Fig. 1F). Moreover, they did not differ dramatically from the telomere length distribution patterns of their AR-dependent counterparts.
Taken together, these data indicate CRPC cell lines exhibit variable telomerase expression and activity. Those that were AR-dependent did not contain an appreciable amount of ALT-like hallmarks, supporting telomerase as the primary TMM. Conversely, in DU145 and PC-3 cells lacking AR expression, relatively low telomerase activity was accompanied by an atypical ALT-like phenotype, including ALT-like telomeric PML localization events and C-circles, but lacking long, heterogeneous telomeres.
SLX4IP expression correlates with AR independence and ALT-like hallmarks
ALT hallmarks, including those in CRPC (27), often correlate with loss of chromatin remodeler, ATRX. While ATRX deletions are potential ALT-initiating events, these genomic alterations are not universally causative of ALT hallmarks across in vitro models (56, 57). Mutational analysis of ATRX (58-60) and protein expression across CRPC cell lines revealed DU145 and PC-3 cells, which display an atypical ALT-like phenotype, retain wild-type ATRX expression (Fig. 2A). Thus, ATRX loss may be responsible for ALT initiation in some cases, but is noncontributory to the ALT-like hallmarks that we observed. As such, additional ALT regulators were investigated.
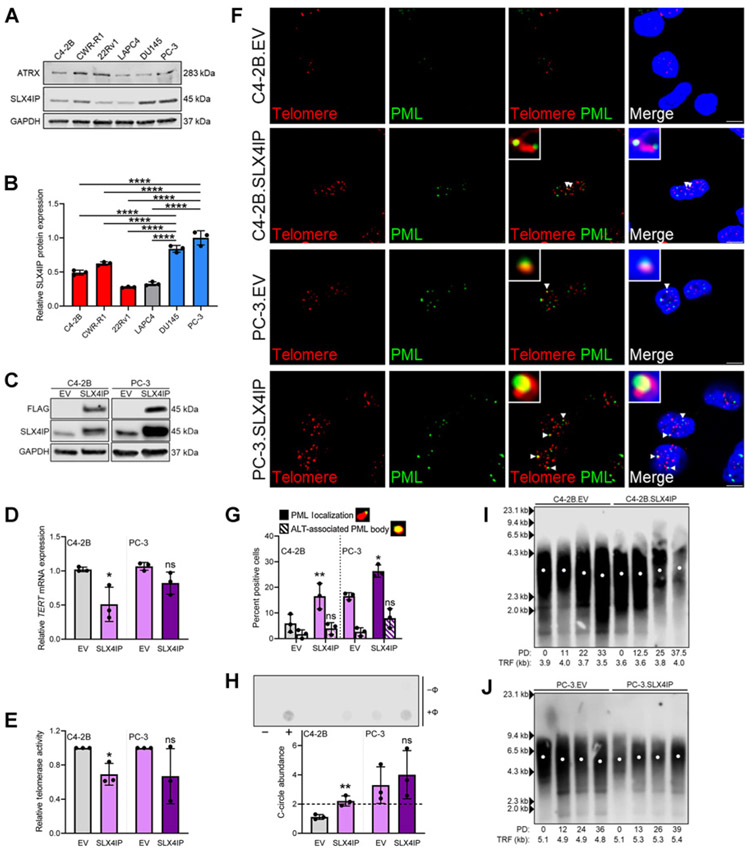
Figure 2.
SLX4IP promotes ALT-like PML localization events in CRPC in vitro. A, ATRX and SLX4IP protein expression across CRPC cell lines. B, Quantification of SLX4IP expression in A. C, Confirmation of stable SLX4IP overexpression in C4-2B and PC-3 cells. EV, empty vector. Relative TERT mRNA expression (D) and telomerase activity (E) following stable SLX4IP overexpression in C4-2B and PC-3 cells. F, Representative IF-FISH images demonstrating the presence (arrowheads) or absence of PML (green) at telomeres (red), with zoom insets provided. Scale bar, 5 μm. G, Quantification of F for percent positive cells with at least one APB. Cells lacking typical APBs were quantified for ALT-like PML localization events. Representative foci are included defining these events. H, Representative dot blot demonstrating the presence of telomeric C-circles in C4-2B and PC-3 cells with SLX4IP overexpression. Φ+ lane indicates reactions with Φ29 polymerase and Φ− lane indicates control reaction. Telomerase-positive HCT116 (−) and ALT-positive WI-38 VA-13 (+) cells were included as controls. Quantification of C-circle abundance, defined as the signal ratio of Φ+ to Φ− reaction is shown below. Dotted line at y = 2 indicates threshold signal ratio suggesting ALT activity. TRF analysis demonstrating telomere length changes over 45 days in C4-2B (I) and PC-3 (J) cells overexpressing SLX4IP versus control. PD, calculated PDs. Data represented as mean ± SD; n = 3 (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). ns, not significant.
The relatively uncharacterized protein, SLX4IP, was identified as a promoter of ALT-mediated telomere maintenance in breast cancer models (41); however, its role in CRPC remains ambiguous. Inspection of SLX4IP expression data from the PCTA (51) revealed significantly higher expression in patients with mCRPC, which includes both AR-dependent and -independent cases, when compared with those with primary disease (Supplementary Fig. S2). Interestingly, relative SLX4IP protein expression in DU145 and PC-3 cells, which exhibit ALT-like hallmarks and AR independence, was significantly higher than in AR-dependent cell lines that lack ALT-like hallmarks (Fig. 2A and B). The correlation among elevated SLX4IP expression, ALT-like hallmarks, and known role of SLX4IP in telomere maintenance supports SLX4IP as a potential regulator of the observed atypical ALT phenotype in AR-independent CRPC in vitro (37, 38, 41).
To investigate the ability of SLX4IP to promote ALT-like hallmarks in CRPC cell lines, a 3X FLAG-tagged SLX4IP construct was stably overexpressed via retroviral transduction in C4-2B (C4-2B.SLX4IP) and PC-3 cells (PC-3.SLX4IP; Fig. 2C). In C4-2B.SLX4IP cells, a significant reduction in TERT expression and telomerase activity was concomitant with SLX4IP overexpression (Fig. 2D and E; Supplementary Fig. S2). This effect was diminished in PC-3 cells that have limited telomerase expression and activity at baseline (Fig. 2D and E; Supplementary Fig. S2). As predicted, SLX4IP overexpression coincided with a significant increase in the percentage of cells with ALT-like PML localization events in both C4-2B.SLX4IP and PC-3.SLX4IP cell populations (Fig. 2F and G). Although modest increases in the percentage of APB-positive cells were noted with SLX4IP overexpression, the changes were deemed not statistically significant. Finally, SLX4IP overexpression resulted in a significant elevation in C-circle signal ratio in C4-2B cells (Fig. 2H). A slight, but statistically insignificant, elevation was noted in C-circle abundance in PC-3.SLX4IP cells. Together, these data indicate SLX4IP overexpression is capable of promoting ALT-like hallmarks to varying degrees but is most consistently correlated with elevations in ALT-like PML localization events.
These data support the role of SLX4IP in promoting ALT-like hallmarks, but it is unclear whether SLX4IP expression correlates with telomere length changes. After defining PD time (Supplementary Fig. S2), TRF analysis revealed control C4-2B and PC-3 cells exhibited minimal telomere length changes over 45 days (Fig. 2I and J). If SLX4IP-mediated ALT-like hallmarks are not promoting telomere maintenance it would be expected that, at least in the case of C4-2B. SLX4IP cells, telomeres would shorten because of the observed significant reduction in telomerase expression and activity (Fig. 2D and E). However, slight telomere lengthening was noted in both C4-2B.SLX4IP and PC-3.SLX4IP cells, supporting the role of SLX4IP-mediated ALT-like PML localization events in promoting telomere maintenance (Fig. 2I and J; Supplementary Fig. S2). Despite this observation, changes in telomere length distribution were not present, consistent with the atypical ALT-like presentation described above.
Here, we establish elevated SLX4IP expression promotes ALT-like PML localization events and C-circles in C4-2B cells and elevates ALT-like hallmarks from baseline in PC-3 cells. Moreover, these phenotypic changes are coupled to telomere maintenance, suggesting SLX4IP-mediated ALT-like PML localization promotes functional telomeric elongation events.
SLX4IP is essential for ALT-like hallmarks and telomere maintenance in AR-independent CRPC
To identify the effects of SLX4IP loss on the atypical ALT phenotype of AR-independent CRPC in vitro, stable cell lines with SLX4IP knockdown (KD.1 and KD.2) and nontargeting control (NS) derived from PC-3 cells were generated using lentiviral transduction (Fig. 3A and B). An approximate 50% knockdown of SLX4IP in PC-3 cells led to a reduction in both percentage of APB-positive cells and percentage of cells with ALT-like telomeric PML localization events (Fig. 3C and D), while having no significant effect on C-circle abundance (Fig. 3E). With this specific loss of ALT-like PML localization events, we reasoned SLX4IP knockdown may trigger a compensatory induction of telomerase to provide adequate telomere maintenance (61). However, TERT expression and telomerase activity were not induced, but marginally diminished, in PC-3.KD.1 and KD.2 cells (Fig. 3F and G; Supplementary Fig. S3). To solidify the role of SLX4IP-mediated ALT-like PML localization events in telomere maintenance, telomere length changes were investigated following SLX4IP knockdown.
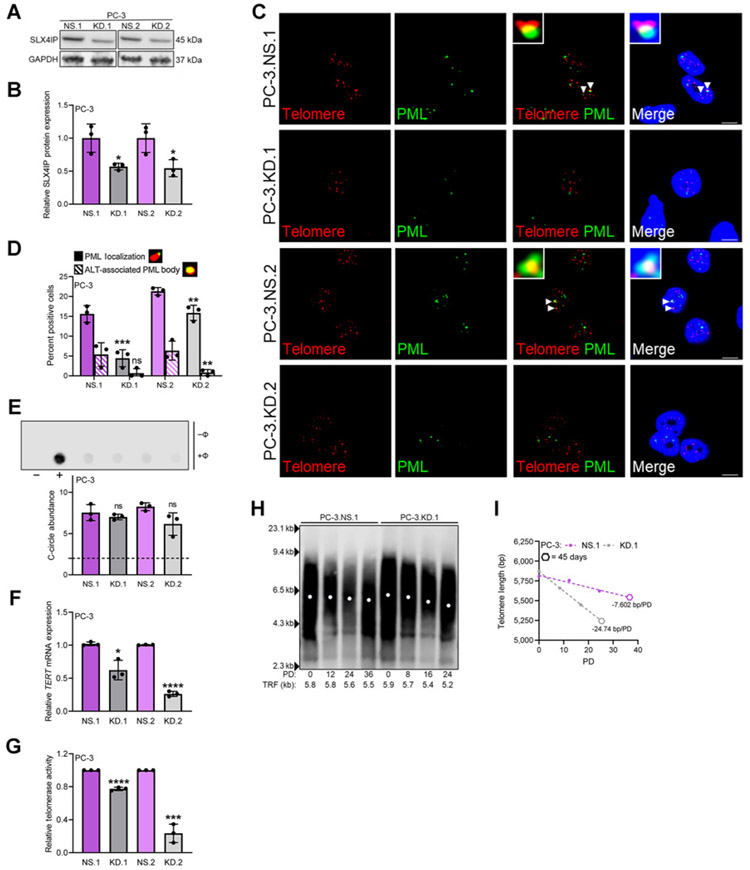
Figure 3.
SLX4IP knockdown is accompanied by disappearance of ALT-like PML localization events and accelerated telomere shortening. A, Confirmation of stable SLX4IP knockdown using two shRNAs (KD.1 and KD.2) in PC-3 cells with scrambled shRNA control (NS) at the protein level. B, Quantification of A. C, Representative IF-FISH images demonstrating the presence (arrowheads) or absence of PML (green) at telomeres (red), with zoom insets provided. Scale bar, 5 μm. D, Quantification of C for percent positive cells with at least one APB. Cells lacking typical APBs were quantified for ALT-like PML localization events. Representative foci are included defining these events. E, Representative dot blot demonstrating the presence of telomeric C-circles. Φ+ lane indicates reactions with Φ29 polymerase and Φ− lane indicates control reaction. Telomerase-positive HCT116 (−) and ALT-positive U2OS (+) cells were included as controls. Quantification of C-circle abundance, defined as the signal ratio of Φ+ to Φ− reaction is shown below. Dotted line at y = 2 indicates threshold signal ratio suggesting ALT activity. Relative TERT mRNA expression (F) and telomerase activity (G) following stable SLX4IP knockdown in PC-3 cells. H, TRF analysis demonstrating telomere length changes over 45 days in PC-3 cells with SLX4IP knockdown (KD.1) versus control (NS.1). PD, Calculated PDs. I, Telomere length changes over time measured via PDs from H. Simple linear regression models are depicted by dotted lines and slopes reported as bp/PD. Hexagon data points represent 45-day timepoints. Data represented as mean + SD; n = 3 (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). ns, not significant.
If the observed SLX4IP-mediated atypical ALT phenotype is involved in telomere maintenance, then a reduction in SLX4IP expression without compensatory telomerase induction should lead to accelerated telomere shortening (34). To test this hypothesis, telomere length changes were evaluated over 45 days coupled with PD time determination. Calculation of PD time revealed a dramatic reduction in cell number over time in PC-3.KD.1 and KD.2 cells suggesting an impairment in proliferative capacity, a phenotype observed with impaired telomere maintenance (refs. 18, 62; Supplementary Fig. S3). In addition, PC-3.KD.1 and KD.2 cells demonstrated an average telomere loss of approximately 700–800 bp over 24 PDs, whereas control lines exhibited minimal telomere length changes in the equivalent number of PDs (Fig. 3H; Supplementary Fig. S3). This translates to more than triple the rate of telomere loss in control lines (Fig. 3I; Supplementary Fig. S3). Because cells unable to sufficiently maintain telomere length are programmed to senesce to prevent loss of genomic information as they approach a minimum threshold length (18, 62), we reasoned that accelerated telomeric shortening in PC-3.KD.1 and KD.2 would lead to premature senescence. As expected, a considerable increase in the proportion of senescence-associated β-galactosidase (SA β-gal) staining (63) was observed in late-passage cells with SLX4IP knockdown compared with late-passage controls (Fig. 4A and B). Elevations in relative p21 protein expression, a senescence-associated marker (64), further corroborated the senescent phenotype (Fig. 4C and D). Moreover, SLX4IP-mediated senescence is most likely due to inadequate telomere length maintenance, as no significant differences in β-gal–positive cells (Fig. 4A and B) or p21 protein expression (Fig. 4C and D) were observed in early-passage PC-3.NS.1, NS.2, KD.1, and KD.2 cells.
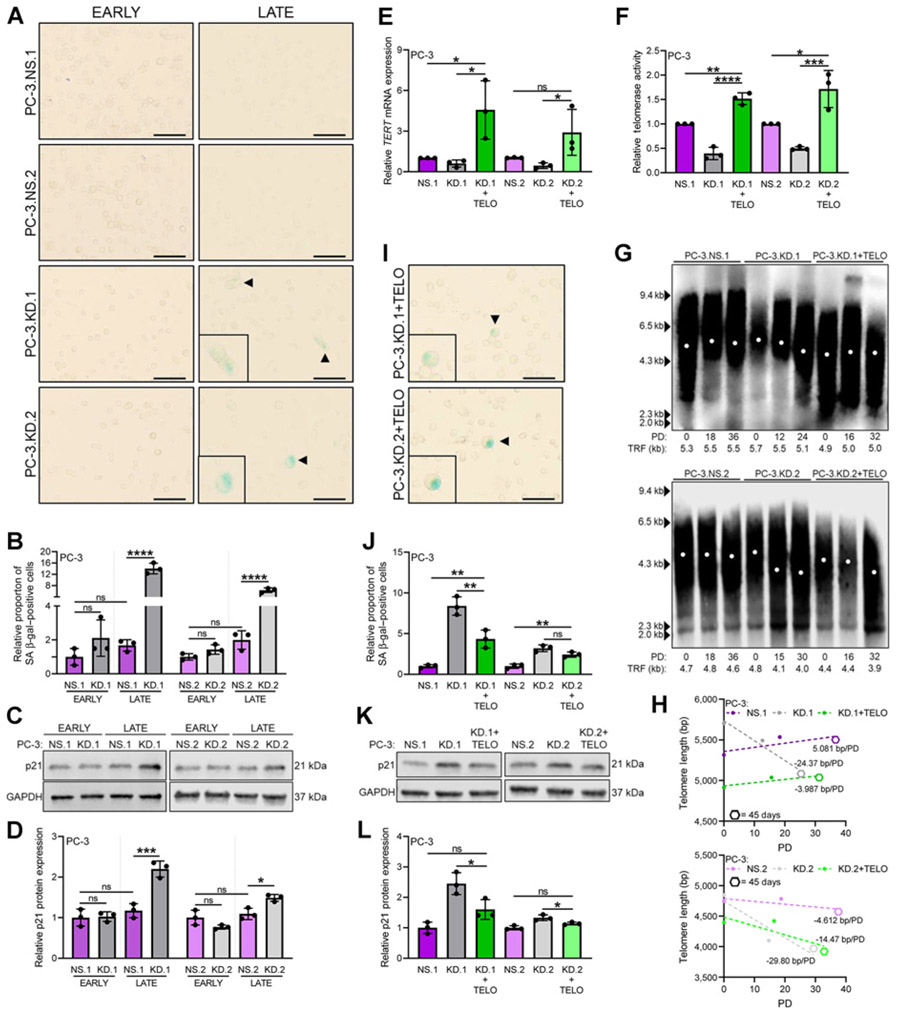
Figure 4.
SLX4IP knockdown triggers accumulation of senescence-associated markers. A, Representative bright-field images demonstrating SA β-gal staining (arrowheads) for senescence in early- and late-passage PC-3 cells with stable knockdown of SLX4IP. Scale bar, 20 μm. B, Quantification of relative proportion of SA β-gal–positive cells in A. C, p21 expression following knockdown of SLX4IP in early- and late-passage PC-3 cells. D, Quantification of relative p21 expression in C. Confirmation of stable overexpression of telomerase in PC-3.KD.1 and KD.2 with SLX4IP knockdown via relative TERT mRNA expression (E) and telomerase activity (F). G, TRF analysis demonstrating telomere length changes over 45 days in PC-3 cells with SLX4IP knockdown (KD.1 and KD.2) and telomerase overexpression (+TELO). PD, calculated PDs. H, Telomere length changes over time measured via PDs from G. Simple linear regression models are depicted by dotted lines and slopes reported as bp/PD. Hexagon data points represent 45-day timepoints. I, Representative bright-field images demonstrating SA β-gal staining (arrowheads) for senescence in late-passage PC-3.KD.1 and KD.2 cells with telomerase overexpression. Scale bar, 20 μm. J, Quantification of relative proportion of SA β-gal–positive cells in I compared with controls. K, p21 expression in late-passage PC-3.KD.1 and KD.2 cells with telomerase overexpression. L, Quantification of relative p21 expression in K compared with controls. Data represented as mean ± SD; n = 3 (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). ns, not significant.
In contrast to PC-3 cells, SLX4IP knockdown in DU145 cells revealed a compensatory increase in telomerase expression and activity coupled with a reduction in cells with ALT-like telomeric PML localization (Supplementary Fig. S4). Although these cells have the ability to induce telomerase expression and promote functional assembly of the enzyme, a significant impairment in PD time and modest telomere shortening were observed (Supplementary Fig. S5). Moreover, SLX4IP knockdown led to a prominent increase in both β-gal positively stained cells and relative p21 protein expression in late-passage DU145-derived cell lines that were not present in early-passage cells (Supplementary Fig. S5). Altogether, these data demonstrate diminished SLX4IP expression is associated with the induction of senescence in late-passage PC-3 and DU145 cells.
The inability of telomerase induction to fully rescue DU145.KD.1 and KD.2 cell lines from telomere shortening and senescent-associated markers raises the question of whether stable telomerase overexpression in PC-3.KD.1 and KD.2 cells, which do not induce telomerase, can promote sufficient telomere maintenance and prevent senescence. To answer this question, PC-3.KD.1 and KD.2 cells with stable telomerase overexpression (PC-3.KD.1+TELO and PC-3.KD.2+TELO) were generated via retroviral transduction and followed over 45 days. Following confirmation of telomerase overexpression (Fig. 4E and F; Supplementary Fig. S5) and PD determination (Supplementary Fig. S5), TRF analysis revealed minimal telomere length changes in PC-3.KD.1+TELO cells over 32 PDs versus approximately 600 bp loss in PC-3.KD.1 cells over 24 PDs (Fig 4G). Telomere length reductions of approximately 500 bp in PC-3.KD.2+TELO cells over 32 PDs versus approximately 800 bp in only 30 PDs in PC-3.KD.2 cells were also noted (Fig 4G). These data translate to a deceleration in the rate of telomere loss to only approximately 4 and approximately 14 bp per PD in PC-3.KD.1+TELO and PC-3. KD.2+TELO cells, respectively (Fig. 4H). Telomere length preservation via telomerase was further reflected by a significant reduction in the proportion of SA β-gal–positive cells and p21 expression in PC-3.KD.1+TELO cells and p21 expression in PC-3.KD.2+TELO cells when compared with cells with SLX4IP knockdown alone (Fig. 4I-L). Together, these data support that the senescent phenotype observed following SLX4IP knockdown is very likely through a telomere-based mechanism.
Loss of SLX4IP-mediated ALT-like PML localization events revealed senescence-associated markers in both PC-3 and DU145 cells demonstrating AR independence. Remarkably, the compensatory induction of telomerase observed in DU145 cells was unable to completely prevent telomere shortening and senescence. This effect was recapitulated following telomerase reconstitution in PC-3 cells with SLX4IP knockdown. These data support ALT-like PML localization is necessary for adequate telomere maintenance in AR-independent CRPC in vitro models. In addition, these data identify SLX4IP as an essential regulator of the atypical ALT phenotype in this AR-independent environment.
SLX4IP-mediated ALT-like hallmark depletion and induction of senescence is maintained in vivo
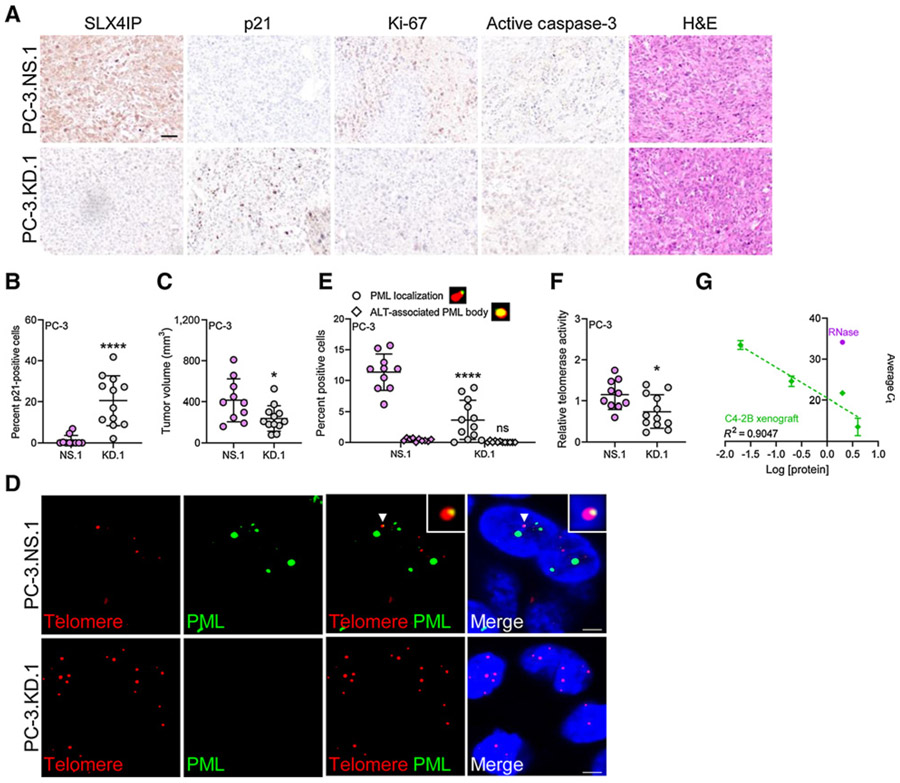
To ascertain whether loss of ALT-like PML localization events via SLX4IP knockdown is maintained in vivo, male nude mice were inoculated subcutaneously with PC-3.NS.1 or PC-3.KD.1 cells and tumor volumes were subsequently monitored over 30 days. IHC revealed SLX4IP knockdown led to a significant increase in the percentage of p21 positively stained cells (Fig. 5A and B). Consequently, this effect translated to a significant reduction in average tumor volume at time of sacrifice (Fig. 5C). As no considerable difference in Ki-67 or active caspase-3 staining was observed between groups, reductions in tumor volume were likely due to the senescent phenotype associated with SLX4IP knockdown (Fig. 5A). Indeed, SLX4IP knockdown resulted in a reduction of ALT-like PML localization events (Fig. 5D and E). Analogous to our in vitro studies, evaluation of telomerase activity among xenografts did not reveal a compensatory increase in telomerase activity following SLX4IP knockdown (Fig. 5F and G). Together, these in vivo results further support SLX4IP as a mediator of ALT-like PML localization in AR-independent CRPC.
Figure 5.
SLX4IP knockdown in AR-independent CRPC leads to ALT-like hallmark loss, reduced tumor volume, and induction of senescent-associated markers in vivo. A, Representative IHC 40× images demonstrating SLX4IP, p21, Ki-67, active caspase-3, and hematoxylin and eosin (H&E) staining from PC-3.NS.1 and KD.1 xenograft sections. Scale bar, 60 μm. B, Quantification of p21 staining in A for percent p21-positive cells. C, Average tumor volume of PC-3.NS.1 (n = 12) and KD.1 (n = 10) on day 30. D, Representative IF-FISH images demonstrating the presence (arrowheads) or absence of PML (green) at telomeres (red), with zoom insets provided. Scale bar, 5 μm. E, Quantification of D for percent positive cells with at least one APB. Cells lacking typical APBs were quantified for ALT-like PML localization events. Representative foci are included defining these events. F, Relative telomerase activity following SLX4IP knockdown in PC-3 xenografts. G, Representative standard curve of telomerase-positive C4-2B xenograft using Q-TRAP protocol for F. RNase-treated C4-2B sample included as negative control. Data represented as mean ± SD (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). ns, not significant.
Androgen-deprived conditions promote ALT-like PML localization in an SLX4IP-dependent manner
Androgen deprivation therapy triggers AR independence in patients (2, 65-67). Similarly, AR-dependent cell lines undergo AR loss when placed in growth media lacking androgens (68-70). Because we identified a unique relationship between ALT-like hallmarks and AR-independent CRPC, we reasoned AR-dependent CRPC cells lacking ALT-like hallmarks grown in androgen-deprived conditions may limit telomerase and gain ALT-like hallmarks as they transition to AR independence. Moreover, this promotion of ALT-like hallmarks, specifically ALT-like PML localization, is expected to be reliant on SLX4IP. Therefore, C4-2B cells with stable knockdown of SLX4IP (KD.1 and KD.2) and NS were generated (Fig. 6A and B) and placed in charcoal-stripped media (+CSS) to mimic androgen deprivation and induce AR loss (68-71).
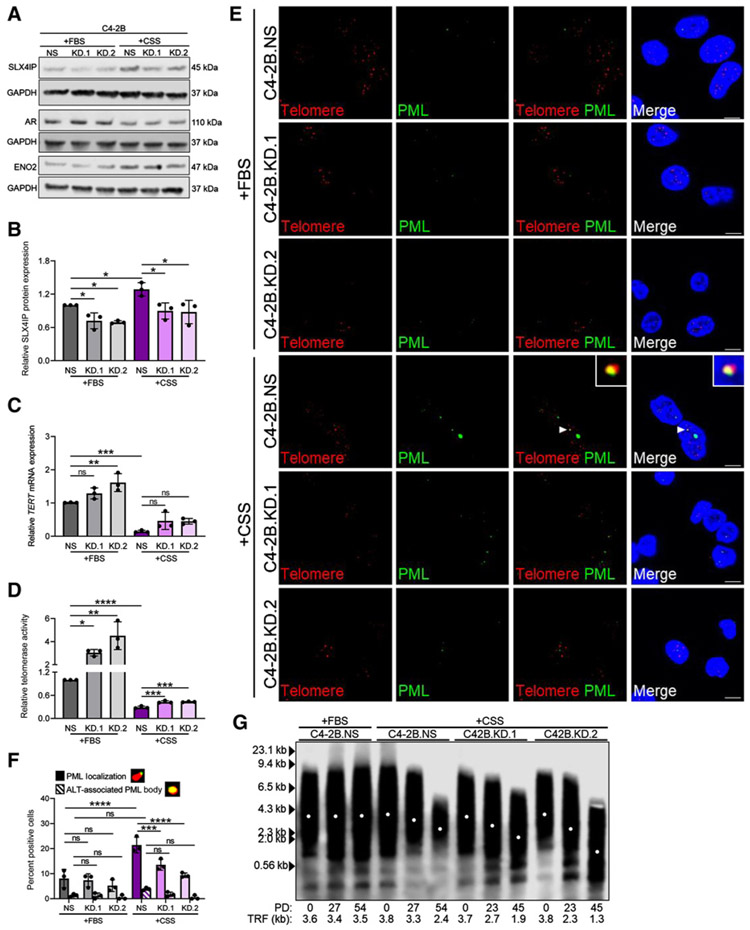
Figure 6.
Androgen deprivation triggers SLX4IP-dependent ALT-like PML localization events. A, Confirmation of stable SLX4IP knockdown using two shRNAs (KD.1 and KD.2) in C4-2B cells with scrambled shRNA control (NS) and confirmation of AR-independent transition via AR and ENO2 expression following growth in androgen-deprived conditions (+CSS) versus normal growth media (+FBS). B, Quantification of SLX4IP expression in A. Relative TERT mRNA expression (C) and telomerase activity (D) following SLX4IP knockdown in C4-2B cells grown in +FBS or +CSS. E, Representative IF-FISH images demonstrating the presence (arrowheads) or absence of PML (green) at telomeres (red), with zoom insets provided. Scale bar, 5 μm. F, Quantification of E for percent positive cells with at least one APB. Cells lacking typical APBs were quantified for ALT-like PML localization events. Representative foci are included defining these events. G, TRF analysis demonstrating telomere length changes in C42B cells with SLX4IP knockdown (KD.1 and KD.2) grown in +CSS versus C4-2B.NS grown in +CSS or +FBS. PD, calculated PDs. Data represented as mean ± SD; n = 3 (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). ns, not significant.
Analysis of SLX4IP expression revealed a modest, yet significant increase in SLX4IP expression when comparing C4-2B.NS cells cultured in +CSS, hypothesized to adopt an ALT-like phenotype, with their normal growth media (+FBS) counterparts (Fig. 6A and B). Reductions in AR expression and upregulation of ENO2 expression, markers of AR independence, across groups confirmed the transition was initiated under androgen-deprived conditions (Fig. 6A; Supplementary Fig. S6; refs. 71, 72). Notably, a difference in AR and ENO2 expression was not observed between cell lines grown in +CSS, indicating SLX4IP itself is not responsible for inducing AR independence.
As expected, relatively high telomerase expression and activity were noted in cells grown in +FBS, however, both were significantly reduced with +CSS (Fig. 6C and D; Supplementary Fig. S6). Moreover, few APB-positive cells and ALT-like PML localization events were identified in C4-2B.NS, KD.1, and KD.2 cells grown in +FBS. Following +CSS growth, C4-2B.NS cells exhibited a significant increase in the percentage of cells with ALT-like PML localization events, but not APBs (Fig. 6E and F). This significant increase was diminished with SLX4IP knockdown. Together, these data support C4-2B cells shift from high telomerase expression and activity to low telomerase expression and activity with ALT-like PML localization events following androgen deprivation. Without sufficient SLX4IP expression, this phenotypic shift is blunted.
Reductions in telomerase with a blunted atypical ALT phenotype make telomere shortening an expected fate. Following PD time determination (Supplementary Fig. S6), cell lines grown in each condition were followed for 45 days to characterize telomere length changes. As expected, C4-2B cells grown in +FBS regardless of SLX4IP expression exhibited minimal telomere length changes consistent with their telomerase-positive status (Supplementary Fig. S6). C4-2B-derived cell lines grown in +CSS demonstrated telomere shortening; however, this phenotype was markedly amplified in the case of SLX4IP knockdown (Fig 6G; Supplementary Fig. S6). Specifically, C4-2B.KD.1 and KD.2 cells exhibited a dramatic loss of approximately 1,800 and approximately 2,500 bp in 45 PDs compared with the approximately 1,400 bp loss in 54 PDs in C4-2B.NS cells. Insufficient telomere maintenance was unsurprising because C4-2B–derived cell lines demonstrate limited telomerase expression and activity following growth in +CSS, regardless of SLX4IP expression. However, the accelerated loss of telomere length in C4-2B.KD.1 and KD.2 cells supports the role of ALT-like PML localization events in promoting some degree of telomere maintenance in the C4-2B.NS cells following androgen deprivation, albeit not to the same degree as telomerase in +FBS.
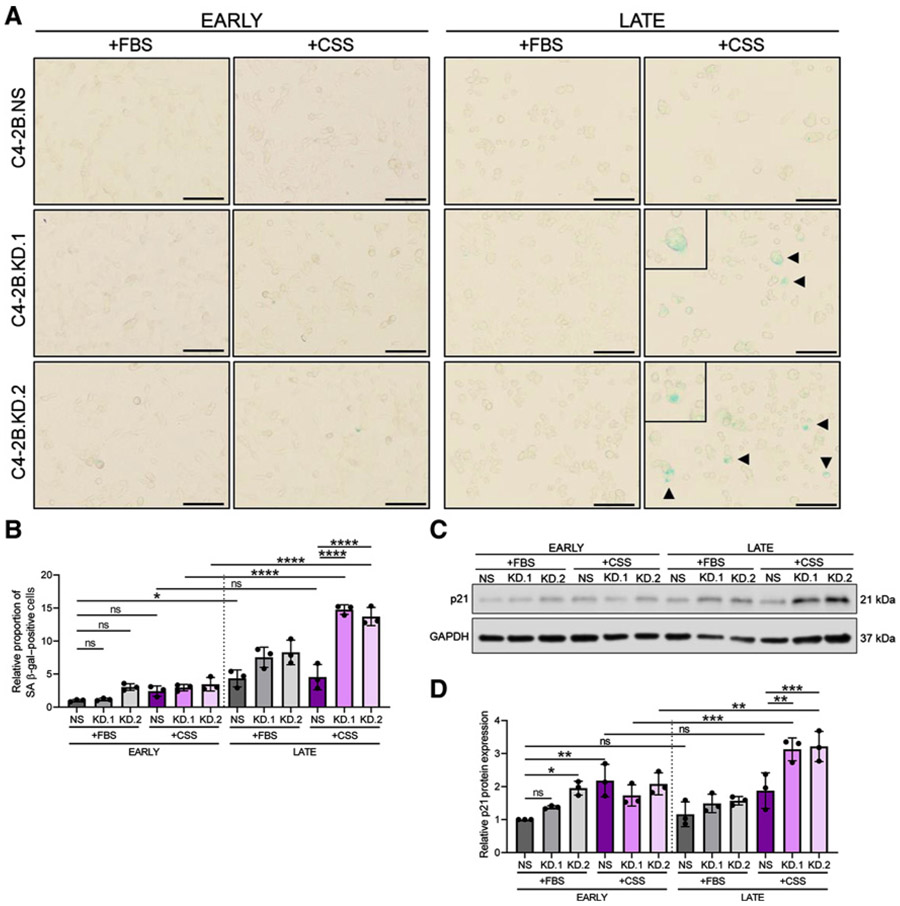
After identification of telomere shortening, senescent-associated markers were evaluated. Early-passage comparisons revealed SLX4IP knockdown and growth in +CSS independently trigger immediate β-gal positivity (Fig 7A and B) and elevations in relative p21 protein expression (Fig 7C and D) to some degree. This pattern remained consistent from early- to late-passage cells with the exception of C4-2B. KD.1 and KD.2 cells grown in +CSS, which exhibited dramatic telomere shortening. β-gal staining of these late-passage C4-2B.KD.1 and KD.2 cells revealed a significant increase in β-gal–positive cells compared with C4-2B.NS cells grown in +CSS and compared with C4-2B.KD.1 and KD.2 cells grown in +FBS (Fig. 7A and B). Consequently, these late-passage β-gal–positive populations can likely be attributed to the impaired telomere maintenance highlighted above. Corroborating this pattern, relative p21 protein expression mimicked our previous finding and was significantly elevated in late-passage SLX4IP-knockdown cells cultured in +CSS (Fig. 7C and D). Therefore, with androgen deprivation, AR-dependent CRPC limits telomerase expression and activity and gains ALT-like PML localization events in an SLX4IP-dependent manner. Without sufficient SLX4IP expression and promotion of these ALT-like hallmarks, telomere shortening and senescence are induced in this in vitro model of disease progression following androgen deprivation.
Figure 7.
SLX4IP knockdown triggers accumulation of senescence-associated markers following androgen deprivation. A, Representative bright-field images demonstrating SA β-gal staining (arrowheads) for senescence in early- and late-passage C4-2B cells with stable knockdown of SLX4IP grown in +CSS and +FBS. Scale bar, 20 μm. B, Quantification of relative proportion of SA β-gal–positive cells in A. C, Relative p21 expression in early- and late-passage C4-2B cells with stable knockdown of SLX4IP grown in +CSS and +FBS. D, Quantification of relative p21 expression in C. Data represented as mean ± SD; n = 3 (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ***, P < 0.0001). ns, not significant.
Discussion
Currently, there exists a lack of targeted therapeutic options for patients with AR-independent CRPC, which encompasses a group of heterogeneous diseases, including therapeutically acquired neuroendocrine prostate cancer (6) and double-negative prostate cancer (9). Here, we report that SLX4IP-dependent ALT-like PML localization events coexist with telomerase to promote telomere maintenance and perpetuate replication in in vitro models of this highly resistant CRPC cohort. Whether these ALT-like hallmarks are universally required for telomere maintenance across all models of AR independence and whether this phenotype emerges as a consequence of androgen deprivation in patient tissues remain to be evaluated. Several in vitro models undergo AR-independent reprogramming following acquisition of resistance to second-generation antiandrogens indicated for CRPC treatment (73). Investigation of SLX4IP-mediated telomere maintenance in these models of therapy-acquired AR independence would elucidate whether this is consistent across AR-independent CRPC heterogeneity, complementing our findings.
The inability of AR-independent CRPC to overcome even an approximately 50% loss of SLX4IP expression, and subsequent senescence, provides an elegant therapeutic target, but the knowledge void around SLX4IP-mediated ALT-like hallmarks must first be addressed. Our studies demonstrated a strong relationship between SLX4IP, ALT-like PML localization events, and telomere maintenance, yet it is unclear as to why other ALT hallmarks remain unchanged. The incomplete ALT phenotype observed in our studies may be due to the retention of functional ATRX (57). ATRX alterations have been identified in a small subset of mCRPC cases, including the CRPC case demonstrating ALT hallmarks, but these mutations are rare (27, 58, 59, 74). Elevations in SLX4IP and ATRX loss may be required for CRPC to recapitulate a traditional ALT phenotype. Coincidentally, several studies have demonstrated that ATRX loss is unable to initiate the traditional ALT phenotype; instead, an incomplete atypical ALT phenotypic change has been observed, similar to our findings (40, 75, 76). For example, ATRX knockout in mouse embryonic cells in vivo did not cause an increase in telomere length or heterogeneity (75) and ATRX knockout in telomerase-positive glioma cell lines did not increase telomeric C-circle abundance (76). Contrary to this hypothesis, investigation of osteosarcoma tumors revealed limited telomerase activity and significant C-circle abundance despite retention of ATRX expression (40), supporting the possibility of an ALT-like phenotype on a background of functional ATRX. Although nontraditional, the ALT-like phenotype described in our studies linked to SLX4IP expression warrants further characterization to determine whether ALT-like processes are driving this unique presentation and whether elevations in SLX4IP expression are stepping stones to bona fide ALT.
The coexistence of an atypical ALT-like phenotype on a background of variable telomerase activity described in our studies challenges the traditional dichotomous relationship of ALT and telomerase. Several groups have demonstrated androgens transcriptionally regulate TERT in primary prostate cancer (26, 77, 78). Following progression to CRPC, AR mutants and splice variants are capable of differentially regulating telomerase expression (78). Furthermore, these AR mutants and splice variants coexist in vitro and in vivo making telomerase expression the collective output of AR perturbations present (4). Despite this complexity, AR-mediated regulation of telomerase supports our data demonstrating the reliance of AR-independent CRPC in vitro models on SLX4IP-mediated ALT-like hallmarks. It is possible that either AR suppresses SLX4IP transcriptionally or TERT, shown to have noncanonical extratelomeric functions, interferes with SLX4IP (79). Following progression to AR-independent CRPC, we have shown SLX4IP is free to promote TMM coexistence and ALT-like hallmarks to compensate for limited telomerase activity. Interestingly, late-passage AR-independent DU145 cells demonstrated rebound telomerase expression and activity following ALT-like hallmark reduction despite maintained AR loss, whereas PC-3 cells did not. This compensatory effect was described in other malignant investigations (41), but why this varies between two AR-independent CRPC cell lines remain undetermined. Nonetheless, the observed induction of telomerase was incapable of completely filling the TMM deficiency in the timeframe studied and both cell lines underwent telomere shortening and senescence. Because of this attempted resistance to SLX4IP-mediated telomere shortening, defining mechanisms regulating both SLX4IP and compensatory telomerase in AR-independent CRPC will be crucial for describing the purpose of TMM coexistence associated with disease progression.
In summary, AR-independent CRPC exhibits a unique ALT-like phenotype with variable telomerase activity, while CRPC retaining AR lacks ALT hallmarks, relying on telomerase in vitro. ALT-like hallmarks correlate with elevated SLX4IP expression and introduction of exogenous SLX4IP promotes ALT-like PML localization events and telomere maintenance. In addition, loss of SLX4IP in AR-independent CRPC renders an environment lacking ALT-like PML localization events triggering telomere shortening and senescence. This phenotype was recapitulated in vivo resulting in reduced tumor volume. Triggering AR-independent progression, AR-dependent CRPC cells grown in androgen-deprived conditions limit telomerase and exhibit ALT-like PML localization events in an SLX4IP-dependent manner. These ALT-like hallmarks promoted a greater degree of telomere length preservation compared with those cell lines lacking sufficient SLX4IP expression. Together, these data reveal SLX4IP-mediated telomere maintenance as a unique mechanism necessary for the replicative immortality of AR-independent CRPC in vitro.
Supplementary Material
Acknowledgments
The authors would like to thank all members of the Taylor and Grabowska laboratories for helpful comments and suggestions related to this investigation. The Grabowska laboratory was supported by start-up funds (to M.M. Grabowska) provided by the Case Research Institute, a joint venture between University Hospitals and Case Western Reserve University. The Taylor laboratory was supported by the NIH (R01 GM133841 and R01 CA240993 to D.J. Taylor). We would also like to acknowledge the Molecular Therapeutics Training Program (T32 GM008056 to T.L. Mangosh).
Footnotes
Authors’ Disclosures
T.L. Mangosh reports grants from Case Western Reserve University Molecular Therapeutics Training Program during the conduct of the study. M.M. Grabowska reports personal fees from Synchronicity Pharma outside the submitted work. D.J. Taylor reports grants from NIGMS and NCI during the conduct of the study, personal fees and other from Rappta Therapeutics outside the submitted work, as well as has an ownership interest in, provides consultation for, and has license agreements with Rappta Therapeutics. No disclosures were reported by the other author.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
References
- 1.Salonen AJ, Taari K, Ala-Opas M, Viitanen J, Lundstedt S, Tammela TLJ. The FinnProstate study VII: intermittent versus continuous androgen deprivation in patients with advanced prostate cancer. J Urol 2012;187:2074–81. [DOI] [PubMed] [Google Scholar]
- 2.Beltran H, Tomlins S, Aparicio A, Arora V, Rickman D, Ayala G, et al. Aggressive variants of castration-resistant prostate cancer. Clin Cancer Res 2014;20:2846–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohler JL, Antonarakis ES, Armstrong AJ, D’Amico AV, Davis BJ, Dorff T, et al. Prostate cancer, version 2.2019. JNCCN J Natl Compr Cancer Netw 2019;17:479–505. [DOI] [PubMed] [Google Scholar]
- 4.Ho Y, Dehm SM. Androgen receptor rearrangement and splicing variants in resistance to endocrine therapies in prostate cancer. Endocrinology 2017;158:1533–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 2016;22:298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aggarwal R, Huang J, Alumkal JJ, Zhang L, Feng FY, Thomas G V, et al. Clinical and genomic characterization of treatment-emergent small-cell neuroendocrine prostate cancer: a multi-institutional prospective study. J Clin Oncol 2018;36:2492–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anthony di Sant’agnese P, de Mesy Jensen KL. Neuroendocrine differentiation in prostatic carcinoma. Hum Pathol 1987;39:135–48. [DOI] [PubMed] [Google Scholar]
- 8.Mosquera JM, Beltran H, Park K, MacDonald TY, Robinson BD, Tagawa ST, et al. Concurrent AURKA and MYCN gene amplifications are harbingers of lethal treatment- related neuroendocrine prostate cancer. Neoplasia 2013;15:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bluemn EG, Coleman IM, Lucas JM, Coleman RT, Hernandez-Lopez S, Tharakan R, et al. Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell 2017;32:474–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med 2012;367:1187–97. [DOI] [PubMed] [Google Scholar]
- 11.Fizazi K, Scher HI, Molina A, Logothetis CJ, Chi KN, Jones RJ, et al. Abiraterone acetate for treatment of metastatic castration-resistant prostate cancer: final overall survival analysis of the COU-AA-301 randomised, double-blind, placebo-controlled phase 3 study. Lancet Oncol 2012;13:983–92. [DOI] [PubMed] [Google Scholar]
- 12.Abida W, Cyrta J, Heller G, Prandi D, Armenia J, Coleman I, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A 2019;116:11428–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in tetrahymena extracts. Cell 1985;43:405–13. [DOI] [PubMed] [Google Scholar]
- 14.Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J 1995;14:4240–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dilley RL, Greenberg RA. ALTernative telomere maintenance and cancer. Trends Cancer 2015;1:145–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PLC, et al. Specific association of human telomerase activity with immortal cells and cancer. Science 1994;266:2011–5. [DOI] [PubMed] [Google Scholar]
- 17.De Lange T How telomeres solve the end-protection problem. Science 2009;326:948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayflick L The illusion of cell immortality. Br J Cancer 2000;83:841–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer Part A 1997;33:787–91. [DOI] [PubMed] [Google Scholar]
- 20.Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet 2010;11:319–30. [DOI] [PubMed] [Google Scholar]
- 21.Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR, Reddel RR. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res 1999;59:4175–9. [PubMed] [Google Scholar]
- 22.Cesare AJ, Griffith JD. Telomeric DNA in ALT Cells is characterized by free telomeric circles and heterogeneous t-loops. Mol Cell Biol 2004;24:9948–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Vitis M, Berardinelli F, Sgura A. Telomere length maintenance in cancer: at the crossroad between telomerase and alternative lengthening of telomeres (ALT). Int J Mol Sci 2018;19:606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heaphy CM, Subhawong AP, Hong SM, Goggins MG, Montgomery EA, Gabrielson E, et al. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am J Pathol 2011;179:1608–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sommerfeld HJ, Meeker AK, Piatyszek MA, Bova GS, Shay JW, Coffey DS. Telomerase activity: a prevalent marker of malignant human prostate tissue. Cancer Res 1996;56:218–22. [PubMed] [Google Scholar]
- 26.Guo C, Armbruster BN, Price DT, Counter CM. In vivo regulation of hTERT expression and telomerase activity by androgen. J Urol 2003;170:615–8. [DOI] [PubMed] [Google Scholar]
- 27.Haffner MC, Mosbruger T, Esopi DM, Fedor H, Heaphy CM, Walker DA, et al. Tracking the clonal origin of lethal prostate cancer. J Clin Invest 2013;123:4918–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hakin-Smith V, Jellinek DA, Levy D, Carroll T, Teo M, Timperley WR, et al. Alternative lengthening of telomeres and survival in patients with glioblastoma multiforme. Lancet 2003;361:836–8. [DOI] [PubMed] [Google Scholar]
- 29.Omori Y, Nakayama F, Li D, Kanemitsu K, Semba S, Ito A, et al. Alternative lengthening of telomeres frequently occurs in mismatch repair system-deficient gastric carcinoma. Cancer Sci 2009;100:413–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu B, Peng M, Song Q. The co-expression of telomerase and ALT pathway in human breast cancer tissues. Tumor Biol 2014;35:4087–93. [DOI] [PubMed] [Google Scholar]
- 31.Hu J, Hwang SS, Liesa M, Gan B, Sahin E, Jaskelioff M, et al. Antitelomerase therapy provokes ALT and mitochondrial adaptive mechanisms in cancer. Cell 2012;148:651–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bechter OE, Zou Y, Walker W, Wright WE, Shay JW. Telomeric recombination in mismatch repair deficient human colon cancer cells after telomerase inhibition. Cancer Res 2004;64:3444–51. [DOI] [PubMed] [Google Scholar]
- 33.Queisser A, Heeg S, Thaler M, von Werder A, Opitz OG. Inhibition of telomerase induces alternative lengthening of telomeres during human esophageal carcinogenesis. Cancer Genet 2013;206:374–86. [DOI] [PubMed] [Google Scholar]
- 34.Xue Y, Li L, Zhang D, Wu K, Chen Y, Zeng J, et al. Twisted epithelial-to-mesenchymal transition promotes progression of surviving bladder cancer T24 cells with hTERT-dysfunction. PLoS One 2011;6:e27748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cerone MA. Telomere maintenance by telomerase and by recombination can coexist in human cells. Hum Mol Genet 2001;10:1945–52. [DOI] [PubMed] [Google Scholar]
- 36.Perrem K, Colgin LM, Neumann AA, Yeager TR, Reddel RR. Coexistence of alternative lengthening of telomeres and telomerase in hTERT-transfected GM847 cells. Mol Cell Biol 2001;21:3862–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wan B, Yin J, Horvath K, Sarkar J, Chen Y, Wu J, et al. SLX4 assembles a telomere maintenance toolkit by bridging multiple endonucleases with telomeres. Cell Rep 2013;4:861–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H, Chen Z, Ye Y, Ye Z, Cao D, Xiong Y, et al. SLX4IP acts with SLX4 and XPF-ERCC1 to promote interstrand crosslink repair. Nucleic Acids Res 2019;47:10181–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson JSJ, Tejera AM, Castor D, Toth R, Blasco MA, Rouse J. Localization-dependent and -independent roles of SLX4 in regulating telomeres. Cell Rep 2013;4:853–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panier S, Maric M, Hewitt G, Mason-Osann E, Gali H, Dai A, et al. SLX4IP antagonizes promiscuous BLM activity during ALT maintenance. Mol Cell 2019;76:27–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson NJ, Morrison-Smith CD, Gooding AJ, Schiemann BJ, Jackson MW, Taylor DJ, et al. SLX4IP and telomere dynamics dictate breast cancer metastasis and therapeutic responsiveness. Life Sci Alliance 2020;3:e201900427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morgenstern JP, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res 1990;18:3587–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong JMY, Collins K. Telomerase RNA level limits telomere maintenance in X-linked dyskeratosis congenita. Genes Dev 2006;20:2848–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, et al. A third-generation lentivirus vector with a conditional packaging system. J Virol 1998;72:8463–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Popovics P, Awadallah WN, Kohrt SE, Case TC, Miller NL, Ricke EA, et al. Prostatic osteopontin expression is associated with symptomatic benign prostatic hyperplasia. Prostate 2020;80:731–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012;9:676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng X, Hernandez-Sanchez W, Xu M, Whited TL, Baus D, Zhang J, et al. Administration of a nucleoside analog promotes cancer cell death in a telomerase-dependent manner. Cell Rep 2018;23:3031–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herbert BS, Hochreiter AE, Wright WE, Shay JW. Nonradioactive detection of telomerase activity using the telomeric repeat amplification protocol. Nat Protoc 2006;1:1583–90. [DOI] [PubMed] [Google Scholar]
- 49.Manders EMM, Verbeek FJ, Aten JA. Measurement of co-localization of objects in dual-colour confocal images. J Microsc 1993;169:375–82. [DOI] [PubMed] [Google Scholar]
- 50.Henson JD, Lau LM, Koch S, Martin La Rotta N, Dagg RA, Reddel RR. The C-circle assay for alternative-lengthening-of-telomeres activity. Methods 2017;114:74–84. [DOI] [PubMed] [Google Scholar]
- 51.You S, Knudsen BS, Erho N, Alshalalfa M, Takhar M, Ashab HAD, et al. Integrated classification of prostate cancer reveals a novel luminal subtype with poor outcome. Cancer Res 2016;76:4948–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thalmann GN, Anezinis PE, Chang SM, Zhau HE, Kim EE, Hopwood VL, et al. Androgen-independent cancer progression and bone metastasis in the LNCaP model of human prostate cancer. Cancer Res 1994;54:2577–81. [PubMed] [Google Scholar]
- 53.Sramkoski RM, Pretlow TG, Giaconia JM, Pretlow TP, Schwartz S, Sy MS, et al. A new human prostate carcinoma cell line, 22Rv1. Vitr Cell Dev Biol Anim 1999;35:407–9. [DOI] [PubMed] [Google Scholar]
- 54.Stone KR, Mickey DD, Wunderli H, Mickey GH, Paulson DF. Isolation of a human prostate carcinoma cell line (DU 145). Int J Cancer 1978;21:274–81. [DOI] [PubMed] [Google Scholar]
- 55.Kaighn ME, Narayan KS, Ohnuki Y, Lechner JF, Jones LW. Establishment and characterization of a human prostatic carcinoma cell line (PC-3). Invest Urol 1979;17:16–23. [PubMed] [Google Scholar]
- 56.Graham MK, Kim J, Da J, Brosnan-Cashman JA, Rizzo A, Del Valle JAB, et al. Functional loss of ATRX and TERC activates alternative lengthening of telomeres (ALT) in LAPC4 prostate cancer cells. Mol Cancer Res 2019;17:2480–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amorim JP, Santos G, Vinagre J, Soares P. The role of ATRX in the alternative lengthening of telomeres (ALT) phenotype. Genes 2016;7:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghandi M, Huang FW, Jané-Valbuena J, Kryukov G V, Lo CC, McDonald ER, et al. Next-generation characterization of the cancer cell line encyclopedia. Nature 2019;569:503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Plantinga MJ, Pascarelli KM, Merkel AS, Lazar AJ, Von Mehren M, Lev D, et al. Telomerase suppresses formation of ALT-associated single-stranded telomeric C-circles. Mol Cancer Res 2013;11:557–67. [DOI] [PubMed] [Google Scholar]
- 62.D’Adda Di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature 2003;426:194–8. [DOI] [PubMed] [Google Scholar]
- 63.Debacq-Chainiaux F, Erusalimsky JD, Campisi J, Toussaint O. Protocols to detect senescence-associated beta-galactosidase (SA-βgal) activity, a biomarker of senescent cells in culture and in vivo. Nat Protoc 2009;4:1798–806. [DOI] [PubMed] [Google Scholar]
- 64.Brown JP, Wei W, Sedivy JM. Bypass of senescence after disruption of p21 (CIP1)/(WAF1) gene in normal diploid human fibroblasts. Science 1997;277:831–4. [DOI] [PubMed] [Google Scholar]
- 65.Lipianskaya J, Cohen A, Chen CJ, Hsia E, Squires J, Li Z, et al. Androgen-deprivation therapy-induced aggressive prostate cancer with neuroendocrine differentiation. Asian J Androl 2014;16:541–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Beltran H, Tagawa ST, Park K, MacDonald T, Milowsky MI, Mosquera JM, et al. Challenges in recognizing treatment-related neuroendocrine prostate cancer. J Clin Oncol 2012;30:e386–9. [DOI] [PubMed] [Google Scholar]
- 67.Komiya A, Yasuda K, Watanabe A, Fujiuchi Y, Tsuzuki T, Fuse H. The prognostic significance of loss of the androgen receptor and neuroendocrine differentiation in prostate biopsy specimens among castration-resistant prostate cancer patients. Mol Clin Oncol 2013;1:257–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen R, Dorai T, Szaboles M, Katz AE, Olsson CA, Buttyan R. Transdifferentiation of cultured human prostate cancer cells to a neuroendocrine cell phenotype in a hormone-depleted medium. Urol Oncol 1997;3:67–75. [DOI] [PubMed] [Google Scholar]
- 69.Burchardt T, Burchardt M, Chen MW, Cao Y, De La Taille A, Shabsigh A, et al. Transdifferentiation of prostate cancer cells to a neuroendocrine cell phenotype in vitro and in vivo. J Urol 1999;162:1800–5. [PubMed] [Google Scholar]
- 70.Pernicová Z, Slabáková E, Fedr R, Šimečková Š, Jaroš J, Suchánková T, et al. The role of high cell density in the promotion of neuroendocrine transdifferentiation of prostate cancer cells. Mol Cancer 2014;13:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fiandalo MV, Wilton JH, Mantione KM, Wrzosek C, Attwood KM, Wu Y, et al. Serum-free complete medium, an alternative medium to mimic androgen deprivation in human prostate cancer cell line models. Prostate 2018;78:213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ather MH, Abbas F, Faruqui N, Israr M, Pervez S. Correlation of three immunohistochemically detected markers of neuroendocrine differentiation with clinical predictors of disease progression in prostate cancer. BMC Urol 2008;8:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kregel S, Chen JL, Tom W, Krishnan V, Kach J, Brechka H, et al. Acquired resistance to the second-generation androgen receptor antagonist enzalutamide in castration-resistant prostate cancer. Oncotarget 2016;7:26259–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012. 487:239–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Clynes D, Jelinska C, Xella B, Ayyub H, Taylor S, Mitson M, et al. ATRX dysfunction induces replication defects in primary mouse cells. PLoS One 2014;9:e92915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brosnan-Cashman JA, Yuan M, Graham MK, Rizzo AJ, Myers KM, Davis C, et al. ATRX loss induces multiple hallmarks of the alternative lengthening of telomeres (ALT) phenotype in human glioma cell lines in a cell line-specific manner. PLoS One 2018;13:e0204159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu S, Qi Y, Ge Y, Duplessis T, Rowan BG, Ip C, et al. Telomerase as an important target of androgen signaling blockade for prostate cancer treatment. Mol Cancer Ther 2010;9:2016–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moehren U, Papaioannou M, Reeb CA, Grasselli A, Nanni S, Asim M, et al. Wild-type but not mutant androgen receptor inhibits expression of the hTERT telomerase subunit: a novel role of AR mutation for prostate cancer development. FASEB J 2008;22:1258–67. [DOI] [PubMed] [Google Scholar]
- 79.Li Y, Tergaonkar V. Noncanonical functions of telomerase: implications in telomerase-targeted cancer therapies. Cancer Res 2014;74:1639–44. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.