Abstract
Background and Objectives
Peritoneal metastases (PM) from primary colorectal cancer (pCRC) are associated with poor outcomes; however, molecular differences are not well defined.
Methods
We compared unpaired tumor profiles of patients with pCRC and PM from Caris Life Sciences. Testing included next-generation sequencing of 592 genes, microsatellite instability (MSI) and tumor mutational burden (TMB). Mutations were test-defined as pathogenic (PATH).
Results
Six hundred seventeen pCRC and 348 PM patients had similar gender (55% male) and age (median 59). PATHs were similar between PM and pCRC in KRAS, BRAF, SMAD2, SMAD4, and PTEN. pCRC PATHs were increased in APC (76% vs 48%, P < .01), ARID1A (29% vs 12%, P < .05), TP53 (72% vs 53%, P < .01), PIK3CA (22% vs 15%, P < .05), and FBXW7 (13% vs 7%, P < .01) compared with PM. Mucinous PM had more PATHs in GNAS (19% vs 8%, P = .032) while nonmucinous PM had more PATHs in BRAF (13% vs 8%, P = .027). Right-sided PM had decreased PATHs in APC (39% vs 68%, P < .0001), ARID1A (7% vs 38%, P < .004), and TP53 (48% vs 65%, P = .033) while there were no difference for left-sided PM. Nine percent of pCRC and 6% of PM were MSI-high (P = NS). There was no difference in TMB-high, TMB-intermediate, or TMB-low between PM and pCRC.
Conclusions
PM have similar rates of KRAS mutation with increased PATHs in GNAS (mucinous) and BRAF (nonmucinous) compared to pCRC. No differences in MSI or TMB were identified between PM and pCRC tumors. These findings inform future study into the molecular profile of PM.
Keywords: BRAF, carcinomatosis, colon, GNAS, KRAS, molecular profile
1 |. INTRODUCTION
Colorectal cancer (CRC) is the fourth most common malignancy and the second leading cause of cancer‐related death in the United States. CRC with locoregional lymph node spread has a 5‐year overall survival (OS) of 70% while spread to distant organs carries a significantly worse prognosis with a 5‐year OS of 12%.1 Metastasis to the liver is the most frequent site of distant spread2 while the peritoneal surface is the second most common site of metastasis, involving approximately 10% of patients with CRC at the time of initial presentation and the sole site of recurrence in up to 25% of patients with CRC.3,4 Peritoneal metastasis is associated with a poor prognosis with a survival ranging from 5 to 7 months with systemic chemotherapy alone.5,6 When compared with other sites of metastasis, PM is associated with a significantly shorter progression‐free survival (PFS) and OS.7
Molecular profiling has been used to identify predictive and prognostic biomarkers and identify novel therapeutic targets for patients with metastatic CRC.8 Approximately, 40% of CRC are characterized by mutations in the KRAS gene.9 RAS mutations have been implicated in CRC carcinogenesis, response to chemotherapy, and adversely affect the survival of patients with CRC,10 and are predictive of resistance to EGFR‐directed therapy (cetuximab and panitumumab).11,12 In addition, the presence of microsatellite instability (MSI), programmed death‐ligand 1 (PD‐L1), expression and tumor mutation burden (TMB) are predictive of potential tumor response to immune checkpoint inhibitor therapy.13
Advances in molecular therapeutics have significantly improved survival for CRC over the last two decades. Recent efforts have moved toward relying on the biology of each tumor revealed by multiplatform tumor profiling testing to tailor therapies. While the role of KRAS and BRAF mutations have been examined in metastatic CRC to the liver,14,15 the impact of molecular therapeutics for peritoneal‐only CRC metastasis has not been thoroughly examined. It is recognized that peritoneal metastases are poorly responsive to treatment with traditional 5‐FU systemic chemotherapy while more contemporary chemotherapy containing oxaliplatin or irinotecan offer improved survival.16 Molecular profiling of peritoneal metastases may aid in the identification of biomarkers that are predictive of response or selection of more appropriate treatment.7 We hypothesized that peritoneal metastases have a unique molecular profile, which is distinct from the primary colorectal cancer (pCRC) tumor and sought to elucidate the genomic alterations of peritoneal metastases by comparing the molecular profile of peritoneal metastases from CRC to unmatched pCRC patient samples.
2 |. METHODS
Primary and metastatic colorectal adenocarcinoma tumor samples were submitted to Caris Life Sciences, a CLIA, ISO15189, and CAP certified/accredited laboratory (Dallas, TX) for evaluation. Peritoneal CRC metastases were compared with primary CRC samples while other metastatic sites, nonmetastatic CRC, and appendiceal origin were excluded. These tumors were initially certified for adequate sample size and histological subtype by board‐certified pathologists. Corresponding formalin‐fixed paraffin‐embedded (FFPE) samples were then processed for analysis using one or more of the following profiling platforms: next-generation sequencing (NGS), immunohistochemistry (IHC), or fusion analysis.17 TMB, MSI, and copy number variation (CNV) analyses were performed using data from the NGS panel. Clinical information including tumor stage, nodal stage, and treatment information was unavailable.
2.1 |. Next‐generation sequencing
NGS was performed for 592 genes using genomic DNA isolates from FFPE tumor samples. The Agilent SureSelect XT system was used to enrich for the 592 whole‐gene targets and with subsequent sequencing performed via the Illumina NextSeq® platform. All variants disclosed herein were detected with more than 99% confidence based on allele frequency and amplicon coverage, with a more than 500X average sequencing depth of coverage and an analytic sensitivity of 5% for variant frequency. Results were test‐defined as pathogenic, presumed pathogenic, variant of unknown significance (VUS), or wild‐type. Both pathogenic and presumed pathogenic were considered as pathogenic mutations (PATH) and were examined against wild‐type, while unclassified, equivocal, and VUS results were excluded.
2.2 |. Immunohistochemistry
IHC analysis was performed to determine the expression of 27 proteins on FFPE tumor samples using automated staining techniques. The primary antibody clones used were as follows: ALK, Androgen Receptor, cMET (SP44), EGFR (2‐18C9), ER (SP1), ERCC1 (8F1), HER2/Neu (4B5), MGMT (MT23.3), MLH1 (M1), MSH2 (G2191129), MSH6 (44), PD‐1, PD‐L1 (SP142), PGP (C494), PMS2 (EPR3927), PR (1E2/100), PTEN (6H2.1), RRM1 (polyclonal), SPARC monoclonal (122511), SPARC (polyclonal), TLE3 (M‐201), TOPO1 (1D6), TOP2A (3F6), TrkA/B/C, TS (TS106/4H4B1), and TUBB3 (neuronal class III beta‐tubulin polyclonal). The results were evaluated independently by board‐certified pathologists and were categorized as positive or negative based on previously established thresholds.
2.3 |. Tumor mutation burden
TMB was calculated based on the total number of nonsynonymous somatic mutations per megabase (MMb) identified through NGS and/or Sanger sequencing. All single‐nucleotide polymorphisms (SNPs) recognized in dbSNP (version 137) or in the 1000 Genomes Project database were excluded from the analyses. The threshold of ≥17 MMb was used to define TMB‐high, and TMB‐low was defined as ≤6 MMb, while all values in between were TMB‐intermediate. In addition, TMB thresholds were analyzed at all values ranging from ≥5 to ≥20 MMb and compared for significant differences.
2.4 |. Microsatellite instability
MSI status was determined using NGS as previously described.18 This methodology analyzed 7317 target microsatellite loci altered by somatic insertion or deletion. The number of microsatellite loci that were altered were counted for each sample and only insertions or deletions that increased or decreased the number of repeats were considered. MSI‐NGS results were compared with results from over 2000 matching clinical cases analyzed with traditional polymerase chain reaction‐based methods. MSI status was test‐defined as high with ≥46 altered loci for tumors of colorectal origin, which generated a sensitivity of 95.8% and a specificity of 99.4%. Results were recorded as MSI‐high or MSI‐low/microsatellite stable (MSS).
2.5 |. Statistics
Population proportions of biomarker positive pCRC and PM samples were compared as whole populations and by primary tumor site location (right‐sided [RS], left‐sided [LS], rectal) and evaluated for significant differences using a one‐tailed χ2 distribution analysis. Biomarkers were considered positive (n Biomarker Positive/n Total Viable Samples) or negative (n Biomarker Negative/n Total Viable Samples), and all unequivocal, unclassified, and indeterminate results were excluded from the analysis. P values of <.05 were considered significant. As all the data contained within this dataset were deidentified, this study was considered exempt from the institutional review board.
3 |. RESULTS
3.1 |. Tumor characteristics
The Caris Life Sciences (Dallas, TX) tumor database was queried for pCRC tumors and PM originating from known primary CRC from 2008 to 2017, which resulted in a dataset of 617 pCRC tumors and 348 PM with a median age of 59 (range 16–93), where the majority of patients were male (n = 526, 55%). These tumors were further categorized by sidedness (right, left, or rectum), histological subtype (mucinous, signet ring cell, or goblet cell) and tumor grade (low, intermediate, high) based on deidentified clinical data extracted from pathology reports. Table 1 gives an overview of patient demographics and tumor characteristics.
TABLE 1.
Patient demographics and tumor characteristics of pCRC and PM specimens
| Variable | pCRC N = 617 |
PM N = 348 |
|---|---|---|
| Gender | ||
| Male | 339 | 190 |
| Female | 281 | 158 |
| Age, y | 59 (16–91) | 59 (20–93) |
| Primary site | ||
| Right colon | 189 | 45 |
| Left colon | 232 | 29 |
| Rectum | 147 | 22 |
| NOS | 49 | 252 |
| Histology | ||
| Mucinous | 74 | 126 |
| Signet ring cell | 14 | 36 |
| Goblet cell | 1 | 1 |
| Grade | ||
| Low | 110 | 34 |
| Moderate | 303 | 44 |
| High | 49 | 11 |
Note: Data presented as N (range).
Abbreviations: pCRC, primary colorectal tumor; PM, peritoneal metastases; NOS, not otherwise specified.
3.2 |. NGS results
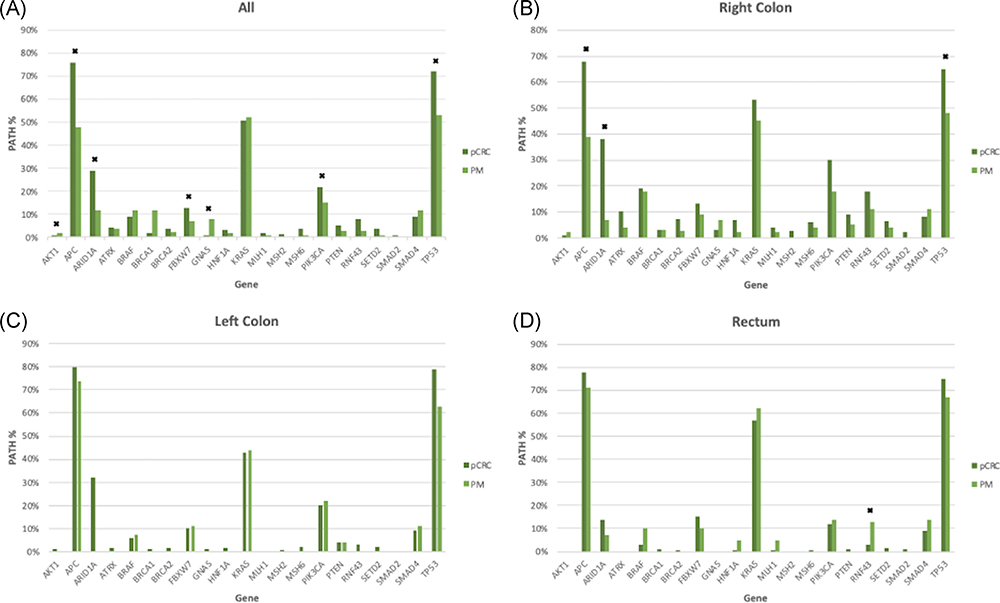
No difference was seen in the rate of PATHs between pCRC and PM for KRAS, BRAF (V600E [n = 87], and nonclassical [n = 8]), SMAD2, SMAD4, or, PTEN; however, pCRC tumor samples showed an increased rate of PATHs in APC (76% vs 48%, P < .0001), TP53 (72% vs 53%, P < .0001), ARID1A (29% vs 12%, P = .018), PIK3CA (22% vs 15%, P = .016), and FBXW7 (13% vs 7%, P < .008). In contrast, PATHs in GNAS (8% vs 1%, P < .0001) and AKT1 (2% vs 1%, P = .044) were higher in PM than pCRC tumor samples. Genes with significant differences in rate of PATHs between pCRC and PM as well as other selected CRC genes are depicted in Figure 1A.
FIGURE 1.
Comparison of pathogenic gene mutation rates between primary CRC (pCRC) and peritoneal metastases of all samples (A), right colon origin (B), left colon origin (C), and rectum origin (D). * significant values (P < .05). pCRC, primary colorectal cancer
In comparing rates of PATHs by sidedness, RS pCRC vs RS PM results were similar to whole population comparison with the rate of PATHs being higher in RS pCRC for ARID1A (38% vs 7%, P = .004), APC (68% vs 39%, P < .0001), and TP53 (65% vs 48%, P 0< =.033; Figure 1B). There were no differences in PATHS between PM and LS pCRC samples (Figure 1C). Lastly, when comparing rectal tumors samples, the occurrence of PATHs were higher in rectal PM compared with rectal pCRC in RNF43 (13% vs 3%, P = .038; Figure 1D).
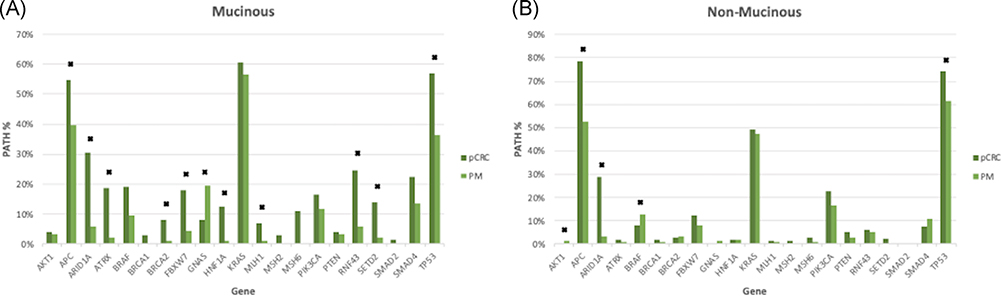
PATHs were compared based on mucinous or nonmucinous histology. Mucinous PMs were compared with mucinous pCRC and had a higher rate of GNAS (11% vs 8%, P = .032) and lower rate of APC (15% vs 55%, P = .044), TP53 (36% vs 57%, P = .006), and ARID1A (25% vs 30%, P = .004) as well as several others (Figure 2A). Nonmucinous PMs had a higher rate of BRAF (13% vs 8%, P = .027) and lower rate of APC, TP53, and ARID1A (all P < .05) than nonmucinous pCRC (Figure 2B). The comparison of mucinous and nonmucinous PM and pCRC PATHs is shown in Table S1.
FIGURE 2.
Comparison of mucinous pathogenic gene mutation rates between pCRC and PM of mucinous (A) and nonmucinous (B) origin. * significant values (P < .05). pCRC, primary colorectal cancer; PM, peritoneal metastases
3.3 |. IHC results
PM presented with significant differences in protein expression compared with pCRC for 6 of the 27 analyzed IHCs, including higher rates of protein expression in TOPO1 (62% vs 52%, P < .01), ERCC1 (27% vs 18%, P < .01), and MLH1 (96% vs 92%, P < .05) with lower expression rates in PD‐1 (36% vs 65%, P < .01), TOP2A, (76% vs 100%, P < .01), and PTEN (64% vs 72%, P < .05). When comparing PM and pCRC by sidedness, no significant differences in rates of protein expression were seen in RS samples; however, as compared with LS pCRC, LS PM showed a higher rate of protein expression in TOPO1 (69% vs 43%, P < .01) and PD‐L1 (9% vs 2%, P < .05) and a much lower rate of expression of MGMT (58% vs 100%, P < .05). When comparing rectal tumor samples, rectal PM showed a significant difference only in PTEN (60% vs 81%, P < .05) expression rate compared with rectal pCRC.
3.4 |. TMB results
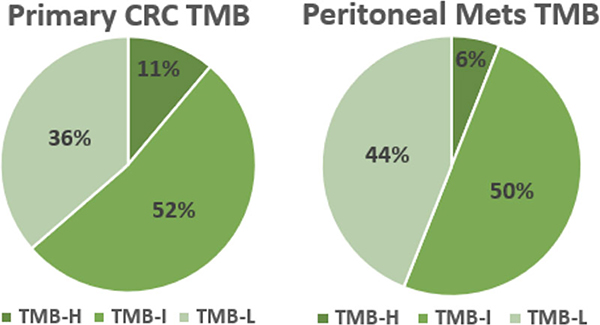
At the time of sample acquisition, test‐determined thresholds for TMB defined TMB‐high as ≥17 mutations per megabase (MMb), TMB‐intermediate as 7 to 16 MMb, and TMB‐low as ≤6 MMb. By these thresholds, there were no significant differences between any of the abovementioned values for pCRC vs PM both as whole populations or based on sidedness (Figure 3).
FIGURE 3.
Comparison of tumor mutation burden (TMB) between primary CRC (pCRC) and peritoneal metastases. TMB low (TMB‐L) was defined as ≤6 mutations/megabase (MMb), TMB‐intermediate (TMB‐I) as 7 to 16 MMb and TMB‐high (TMB‐H) as ≥17 MMb. There was no significant difference in TMB between pCRC and PM. pCRC, primary colorectal cancer; PM, peritoneal metastases
TMB was evaluated as continuous values ranging from ≥5 to ≥20 MMb to identify potential differences between pCRC and PM tumor samples. Significant differences were found at values of ≥5, 7, 8, 9, 12, 13, 14, 15, 16, and 18 MMb with the largest difference occurring at ≥8 MMb. Primary CRC tumor samples with ≥8 MMb occurred 10% more frequently than PM tumors samples (53% vs 43%, P < .05). No significant differences in TMB values were found when evaluated by sidedness. Table S2 provides additional results from the TMB analysis.
3.5 |. MSI results
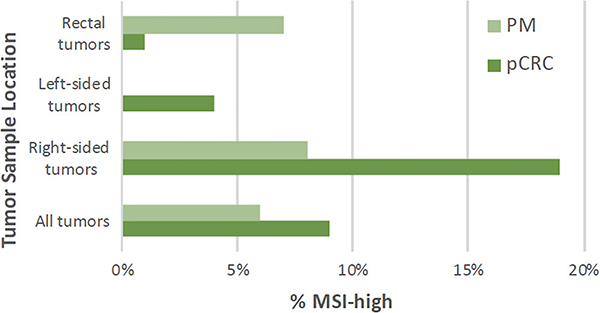
MSI status was available for 601 pCRC and 224 PM tumor samples. Nine percent of pCRC and 6% of PM tumor samples were MSI‐high. RS pCRC tumors were more frequently MSI‐high while PM from rectal tumor samples were more frequently MSI‐high. No PM tumor samples from LS pCRC were MSI‐high. There was no significant difference in MSI‐high status when comparing pCRC and PM samples as whole populations or by sidedness (Table 2, Figure 4).
TABLE 2.
MSI-status compared as whole population and by tumor sidedness
| All |
Right colon |
Left colon |
Rectum |
|||||
|---|---|---|---|---|---|---|---|---|
| MSI | pCRC | PM | pCRC | PM | pCRC | PM | pCRC | PM |
| Samples | 601 | 224 | 183 | 36 | 225 | 22 | 145 | 14 |
| MSI-high | 55 (9) | 13 (6) | 35 (19) | 3 (8) | 9 (4) | 0 | 2 (1) | 1 (7) |
| Difference | −3% | −11% | −4% | −6% | ||||
| P | .12 | .12 | na | .15 | ||||
Abbreviations: MSI, microsatellite instability; pCRC, primary colorectal tumor; PM, peritoneal metastases.
FIGURE 4.
Comparison of microsatellite instability (MSI) rates between primary CRC (pCRC) and peritoneal metastases between all tumors and based on tumor sidedness. There was no significant difference in MSI-high status between pCRC and PM. pCRC, primary colorectal cancer; PM, peritoneal metastases
3.6 |. Paired sample results
Paired samples from both the pCRC and PM were available for 14 patients, all of which were mucinous histology. There were similar rates of PATHs for APC (67% vs 69%), KRAS (58% vs 58%), and TP53 (50% vs 67%), none of which were significant. Rates of GNAS were also similar between pCRC and PM (8% each).
4 |. DISCUSSION
Approximately, 50% to 60% of patients with CRC develop the metastatic disease with the liver as the most common site of metastasis19 while approximately 17% with metastatic CRC develop peritoneal carcinomatosis.7,20 PMs are associated with a poor outcome, however, and a 30% reduction in OS when compared with those with other sites of metastasis.7 The outcome for patients who present with ascites or malignant obstruction from PM is even worse with an expected median survival of less than 4 months.21,22 The current goal of treatment for PM is palliative and consists of systemic chemotherapy or supportive care with surgery reserved for obstruction.23 Cytoreduction and hyperthermic intraperitoneal chemotherapy (HIPEC) has been utilized for appropriately selected patients with reports of improved OS over systemic chemotherapy alone.24,25 However, recent randomized evidence suggests limited benefit when HIPEC is performed with oxaliplatin over cytoreduction alone. Recurrence after resection of peritoneal metastases is high, however, and dependent upon several factors including the burden of disease, primary disease location and histology.26,27 Given the heterogenous nature and lack of standardized treatment approach, identification of unique molecular alterations that may establish profiles or biomarkers that are predictive of peritoneal metastases and in turn, lead to clinically meaningful treatment strategies is important.
Results from the current study identified a similar incidence of pathogenic gene mutations in traditional CRC‐associated genes including APC, TP53, and KRAS with some unique differences observed between pCRC and PM tumor samples. These results are in concordance with the findings of El‐Deiry et al28 which demonstrated the presence of mutated APC, TP53 and KRAS in 68%, 58% and 44% of pCRC tumors, respectively. RAS mutations are early events in the development of CRC and a correlation exists between the mutation status of the primary tumor and site of metastasis.29 RAS mutations have a negative effect in metastatic CRC and KRAS mutation specifically is associated with a worse prognosis, earlier onset of metastasis, and more aggressive metastatic behavior.30,31 In the current study, KRAS mutation was observed in approximately 50% of pCRC and PM tumor samples. TP53, APC, ARID1A, FBXW7, and PIK3CA mutations were more frequently present in pCRC and less commonly observed in PM.
BRAF PATHs (92% classic V600E) were present in 9% of pCRC tumors and 12% of PM. Approximately, 5% to 9% of CRC are characterized by mutation in the BRAF gene, and while more rare, are associated with more aggressive behavior and a worse prognosis.32 Tran et al33 similarly identified that BRAF mutations are associated with a higher percentage of nodal and peritoneal metastasis compared with BRAF wild‐type tumors. Furthermore, results of the Medical Research Council COIN trial demonstrated no benefit of addition of cetuximab to standard chemotherapy in first‐line treatment of BRAF‐mutated metastatic CRC.34 Interestingly, we found nonmucinous PMs harbored more BRAF aberrations, which is counterintuitive to the previous association of mucinous CRC with BRAF V600E. This is hypothesis‐generating and could be explained by a discordance between primary samples and metastases, possibly indicating a more aggressive nature of BRAF‐altered nonmucinous lesions with a propensity for PM.
The rate of GNAS mutation was higher in mucinous PM in the current study. GNAS mutation has been previously described in mucinous neoplasms of the pancreas and peritoneal carcinomatosis from appendiceal adenocarcinomas.35,36 While the clinical significance of GNAS mutation in primary CRC is vaguely understood, a recent review by Khan et al,37 demonstrated that GNAS mutation was more common in mucinous CRC adenocarcinoma. In that series, GNAS mutation was present in 6.9% of mucinous adenocarcinomas and was a poor prognostic factor on univariate analysis. In contradistinction, GNAS mutation was present in less than 1% of nonmucinous adenocarcinomas. In the current study, GNAS was present in 8% of mucinous pCRC and 0% of nonmucinous pCRC. Conversely, GNAS mutation was observed in 19% of mucinous PM and 1% of nonmucinous PM. GNAS mutation has prognostic significance and is associated with a shorter PFS in patients receiving low‐dose capecitabine and bevacizumab in relapsed appendiceal malignancies.38 The results of the current analysis are important when considering the refractory nature of PM to systemic chemotherapy and represents a potential area for further study.
TOPO1 and ERCC1 protein expression rates were higher in PM than pCRC tumors. Rectal cancers have been shown to have higher expression of TOPO1 than RS or LS colon cancers.39 TOPO1 expression potentially identifies a population that may benefit from irinotecan therapy. The UK Medical Research Council FOCUS trial randomized 1688 patients with metastatic CRC to single‐agent FU or FU plus irinotecan or oxaliplatin. Patients with high TOPO1 expression (HR 0.60, median benefit 5.3 months) had better OS with first‐line combination chemotherapy with irinotecan compared to those with moderate or low TOPO1 expression (HR 0.92 and 1.09, respectively, P = .005).40 ERCC1 expression has also been identified as a predictive biomarker for peritoneal carcinomatosis from colorectal, gastric, and ovarian malignancies.28,41 ERCC1 protein expression predicts potential resistance to oxaliplatin chemotherapy. Li et al,42 examined ERCC1 expression in 255 patients who received either fluoropyrimidine‐based chemotherapy (N = 95) or oxaliplatin‐based chemotherapy (N = 160). ERCC1 positive patients who received oxaliplatin had lower 5‐year DFS and OS than those with ERCC1 negative tumors (54% vs 72%, P = .009% and 60% vs 78%, P = .002, respectively), suggesting resistance to oxaliplatin. While these results are interesting, the predictive ability of protein expression for determining chemosensitivities remains controversial.
The use of mutational or neoantigen burden has been investigated as a predictive biomarker. TMB measures the total number of nonsynonymous somatic mutations identified per megabase of the genomic area. TMB high tumors may be more responsive to immune checkpoint inhibitors, such as pembrolizumab or nivolumab.43 The clinical significance of high TMB in metastatic CRC and peritoneal metastasis remains unknown. No difference in TMB high status, as defined by Caris at the time of testing, was identified between pCRC (11%) and PM (6%) tumor samples in the current study. Very high TMB has been correlated and strongly associated with MSI‐high status.44 RS colon cancers are more frequently MSI-high/mismatch repair‐deficient (MMRd) and display a higher mutation burden.39 Recently, nivolumab, a PD‐1 immune checkpoint inhibitor, was found to provide a durable response in patients with previously treated metastatic DNA MMRd/MSI high CRC and represents an additional treatment option for select patients.45
Mucinous histology and primary tumor sidedness are important prognostic and predictive factors in the treatment of metastatic CRC. Mucinous and signet ring cell histology were represented in a large proportion of peritoneal metastases in this study. Mucinous adenocarcinomas are more common in RS colon cancers and both mucinous and signet ring cell adenocarcinoma are associated with a high rate of peritoneal metastases, ranging from 22% to 45%.26,46,47 Regarding sidedness, RS colon cancers are associated with a worse outcome than LS colon or rectal cancers. Retrospective analysis of six randomized trials (CRYSTAL, FIRE‐3, CALGB 80405, PRIME, PEAK, and 20050181) demonstrated that RS KRAS wild‐type metastatic colon cancers had a worse PFS, OS, and objective response rate than LS primary colon cancers.48 In that study, bevacizumab resulted in better outcomes than EGFR antibody therapy among patients with RS cancers, whereas EGFR antibody therapy performed better than bevacizumab for KRAS wild‐type LS cancers. The reason for this clinical difference in outcomes is poorly understood, however, and likely differences in molecular biology are to explain. Common initial tumor‐initiating events involving APC, KRAS, and TP53 genes are observed irrespective of sidedness for RS, LS, and rectal cancers but different mutational behavior characterized by significant somatic and proteomic differences at each location have been identified.49 RS cancers have higher rates of microsatellite instability, mutational burden, and BRAF and PIK3CA mutation rates than LS colon and rectal cancers, whereas rectal cancers have higher rates of TOPO1 expression and Her2/neu amplification.39 Given these observations and the results of the current study, molecular analysis of peritoneal metastases is warranted to better understand tumor biology and potentially guide appropriate treatment.
Limitations to the current study include a large number of deidentified samples from a heterogeneous patient population with varying stages of disease and treatment status. Clinical outcome information such as demographic, primary tumor location, clinical or pathologic stage, synchronous or metachronous metastases, neoadjuvant or adjuvant treatment or survival information was not available for analysis. While peritoneal metastàses may develop from a variety of locations, we specifically examined the colorectal origin and excluded appendiceal origin as it is recognized that gene expression from CRC is quite distinct from appendiceal cancer.50 Molecular profiling was limited to those genes that represent the most common driver mutations and not whole exome testing or variants of unknown significance, which may limit applicability and accuracy. Lastly, most of the samples were not paired and represented unmatched whole populations of PM to unmatched whole populations of pCRC cancers. Paired samples were available for only a small number of patients and did not reveal any differences in mutations (unpublished data).
5 |. CONCLUSION
Compared with pCRC, PM have unique molecular differences. PM from CRC have similar rates of KRAS mutation but a lower rate of mutation in TP53, APC, and PIK3CA. We found GNAS mutations are more commonly associated with mucinous PMs while BRAF mutations are associated with nonmucinous PMs. There was no difference in MSI or test‐specified TMB between PM and pCRC tumors. Similarly, protein expression of TOPO1 and ERCC1 in PM are increased, although the clinical significance of this finding is unknown. Nuances to the molecular profile of CRC PMs are hypothesis‐generating and should be expanded upon by coupling with clinical data.
Supplementary Material
ACKNOWLEDGMENTS
Support was provided solely from institutional and/or departmental sources. This study was collaborative with Caris Life Sciences.
Footnotes
CONFLICT OF INTERESTS
Dr Xiu and Dr Korn are employed by Caris Life Sciences. Dr Marshall is a consultant for Caris Life Sciences. The other authors declare that there are no conflict of interests. The content is solely the responsibility of the authors.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from Caris Life Sciences (Phoenix, AZ). Restrictions apply to the availability of these data, which were used under license for this study. Data are available with the permission of Caris Life Sciences.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- 2.Fong Y, Cohen AM, Fortner JG, et al. Liver resection for colorectal metastases. J Clin Oncol. 1997;15:938–946. [DOI] [PubMed] [Google Scholar]
- 3.Dawson LE, Russell AH, Tong D, Wisbeck WM. Adenocarcinoma of the sigmoid colon: sites of initial dissemination and clinical patterns of recurrence following surgery alone. J Surg Oncol. 1983;22:95–99. [DOI] [PubMed] [Google Scholar]
- 4.Russell AH, Tong D, Dawson LE, et al. Adenocarcinoma of the retroperitoneal ascending and descending colon: sites of initial dissemination and clinical patterns of recurrence following surgery alone. Int J Radiat Oncol Biol Phys. 1983;9:361–365. [DOI] [PubMed] [Google Scholar]
- 5.Chu DZJ, Lang NP, Thompson C, Osteen PK, Westbrook KC. Peritoneal carcinomatosis in nongynecologic malignancy. A prospective study of prognostic factors. Cancer. 1989;63:364–367. [DOI] [PubMed] [Google Scholar]
- 6.Koppe MJ, Boerman OC, Oyen WJ, Bleichrodt RP. Peritoneal carcinomatosis of colorectal origin: incidence and current treatment strategies. Ann Surg. 2006;243:212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franko J, Shi Q, Goldman CD, et al. Treatment of colorectal peritoneal carcinomatosis with systemic chemotherapy: a pooled analysis of north central cancer treatment group phase III trials N9741 and N9841. J Clin Oncol. 2012;30:263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmoll HJ, Van Cutsem E, Stein A, et al. ESMO consensus guidelines for management of patients with colon and rectal cancer. a personalized approach to clinical decision making. Ann Oncol. 2012;23:2479–2516. [DOI] [PubMed] [Google Scholar]
- 9.Roth AD, Tejpar S, Delorenzi M, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC‐3, EORTC 40993, SAKK 60‐00 trial. J Clin Oncol. 2010;28:466–474. [DOI] [PubMed] [Google Scholar]
- 10.Mise Y, Zimmitti G, Shindoh J, et al. RAS mutations predict radiologic and pathologic response in patients treated with chemotherapy before resection of colorectal liver metastases. Ann Surg Oncol. 2015;22:834–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lièvre A, Bachet JB, Le Corre D, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. [DOI] [PubMed] [Google Scholar]
- 12.Karapetis CS, Khambata‐Ford S, Jonker DJ, et al. K‐RAS mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. [DOI] [PubMed] [Google Scholar]
- 13.Gibney GT, Weiner LM, Atkins MB. Predictive biomarkers for checkpoint inhibitor‐based immunotherapy. Lancet Oncol. 2016;17: e542–e551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brudvik KW, Jones RP, Giuliante F, et al. RAS mutation clinical risk score to predict survival after resection of colorectal liver metastases. Ann Surg. 2019;269:120–126. [DOI] [PubMed] [Google Scholar]
- 15.Karagkounis G, Torbenson MS, Daniel HD, et al. Incidence and prognostic impact of KRAS and BRAF mutation in patients undergoing liver surgery for colorectal metastases. Cancer. 2013;119:4137–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franko J, Ibrahim Z, Gusani NJ, Holtzman MP, Bartlett DL, Zeh HJ. Cytoreductive surgery and hyperthermic intraperitoneal chemoperfusion versus systemic chemotherapy alone for colorectal peritoneal carcinomatosis. Cancer. 2010;116:3756–3762. [DOI] [PubMed] [Google Scholar]
- 17.Kelly KR, Friedberg JW, Park SI, et al. Phase I study of the investigational aurora A kinase inhibitor alisertib plus rituximab or rituximab/vincristine in relapsed/refractory aggressive B‐cell lymphoma. Clin Cancer Res. 2018;24:6150–6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vanderwalde A, Spetzler D, Xiao N, Gatalica Z, Marshall J. Microsatellite instability status determined by next‐generation sequencing and compared with PD‐L1 and tumor mutational burden in 11,348 patients. Cancer Med. 2018;7:746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoo PS, Lopez‐Soler RI, Longo WE, Cha CH. Liver resection for metastatic colorectal cancer in the age of neoadjuvant chemotherapy and bevacizumab. Clin Colorectal Cancer. 2006;6:202–207. [DOI] [PubMed] [Google Scholar]
- 20.Franko J, Shi Q, Meyers JP, et al. Prognosis of patients with peritoneal metastatic colorectal cancer given systemic therapy: an analysis of individual patient data from prospective randomised trials from the Analysis and Research in Cancers of the Digestive System (ARCAD) database. Lancet Oncol. 2016;17:1709–1719. [DOI] [PubMed] [Google Scholar]
- 21.Blair SL, Chu DZ, Schwarz RE. Outcome of palliative operations for malignant bowel obstruction in patients with peritoneal carcinomatosis from nongynecological cancer. Ann Surg Oncol. 2001;8:632–637. [DOI] [PubMed] [Google Scholar]
- 22.Helyer LK, Law CHL, Butler M, Last LD, Smith AJ, Wright FC. Surgery as a bridge to palliative chemotherapy in patients with malignant bowel obstruction from colorectal cancer. Ann Surg Oncol. 2007;14:1264–1271. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi H, Okabayashi K, Tsuruta M, Hasegawa H, Yahagi M, Kitagawa Y. Self‐expanding metallic stents versus surgical intervention as palliative therapy for obstructive colorectal cancer: A meta-analysis. World J Surg. 2015;39:2037–2044. [DOI] [PubMed] [Google Scholar]
- 24.Verwaal VJ, van Ruth S, de Bree E, et al. Randomized trial of cytoreduction and hyperthermic intraperitoneal chemotherapy versus systemic chemotherapy and palliative surgery in patients with peritoneal carcinomatosis of colorectal cancer. J Clin Oncol. 2003;21:3737–3743. [DOI] [PubMed] [Google Scholar]
- 25.Verwaal VJ, Bruin S, Boot H, van Slooten G, van Tinteren H. 8‐year follow‐up of randomized trial: cytoreduction and hyperthermic intraperitoneal chemotherapy versus systemic chemotherapy in patients with peritoneal carcinomatosis of colorectal cancer. Ann Surg Oncol. 2008;15:2426–2432. [DOI] [PubMed] [Google Scholar]
- 26.Hugen N, van de Velde CJ, de Wilt JH, Nagtegaal ID. Metastatic pattern in colorectal cancer is strongly influenced by histological subtype. Ann Oncol. 2014;25:651–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Oudheusden TR, Nienhuijs SW, Luyer MD, et al. Incidence and treatment of recurrent disease after cytoreductive surgery and intraperitoneal chemotherapy for peritoneally metastasized colorectal cancer: a systematic review. Eur J Surg Oncol. 2015;41:1269–1277. [DOI] [PubMed] [Google Scholar]
- 28.El‐Deiry WS, Vijayvergia N, Xiu J, et al. Molecular profiling of 6,892 colorectal cancer samples suggests different possible treatment options specific to metastatic sites. Cancer Biol Ther. 2015;16:1726–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Artale S, Sartore‐Bianchi A, Veronese SM, et al. Mutations of KRAS and BRAF in primary and matched metastatic sites of colorectal cancer. J Clin Oncol. 2008;26:4217–4219. [DOI] [PubMed] [Google Scholar]
- 30.Richman SD, Seymour MT, Chambers P, et al. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan:results from the MRC FOCUS trial. J Clin Oncol. 2009;27:5931–5937. [DOI] [PubMed] [Google Scholar]
- 31.Kemeny NE, Chou JF, Capanu M, et al. KRAS mutation influences recurrence patterns in patients undergoing hepatic resection of colorectal metastases. Cancer. 2014;120:3965–3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Cutsem E, Köhne CH, Láng I, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first‐line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29:2011–2019. [DOI] [PubMed] [Google Scholar]
- 33.Tran B, Kopetz S, Tie J, et al. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer. 2011;117:4623–4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maughan TS, Adams RA, Smith CG, et al. Addition of cetuximab to oxaliplatin‐based first‐line combination chemotherapy for treatment of advanced colorectal cancer: results of the randomised phase 3 MRC COIN trial. Lancet. 2011;377:2103–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Furukawa T, Kuboki Y, Tanji E, et al. Whole‐exome sequencing uncovers frequent GNAS mutations in intraductal papillary mucinous neoplasms of the pancreas. Sci Rep. 2011;1:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishikawa G, Sekine S, Ogawa R, et al. Frequent GNAS mutations in low‐grade appendiceal mucinous neoplasms. Br J Cancer. 2013;108:951–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khan M, Loree JM, Advani SM, et al. Prognostic implications of mucinous differentiation in metastatic colorectal carcinoma can be explained by distinct molecular and clinicopathologic characteristics. Clin Colorectal Cancer. 2018;17:e699–e709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pietrantonio F, Berenato R, Maggi C, et al. GNAS mutations as prognostic biomarker in patients with relapsed peritoneal pseudomyxoma receiving metronomic capecitabine and bevacizumab: a clinical and translational study. J Transl Med. 2016;14:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salem ME, Weinberg BA, Xiu J, et al. Comparative molecular analyses of left‐sided colon, right‐sided colon, and rectal cancers. Oncotarget. 2017;8:86356–86368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braun MS, Richman SD, Quirke P, et al. Predictive biomarkers of chemotherapy efficacy in colorectal cancer: results from the UK MRC FOCUS trial. J Clin Oncol. 2008;26:2690–2698. [DOI] [PubMed] [Google Scholar]
- 41.Arienti C, Tesei A, Verdecchia G, et al. Peritoneal carcinomatosis from ovarian cancer: chemosensitivity test and tissue markers as predictors of response to chemotherapy. J Transl Med. 2011;9:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li P, Fang YJ, Li F, Ou QJ, Chen G, Ma G. ERCC1, defective mismatch repair status as predictive biomarkers of survival for stage III colon cancer patients receiving oxaliplatin‐based adjuvant chemotherapy. Br J Cancer. 2013;108:1238–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Champiat S, Ferté C, Lebel‐Binay S, Eggermont A, Soria JC. Exomics and immunogenics: bridging mutational load and immune checkpoints efficacy. Oncoimmunology. 2014;3:e27817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salem ME, Puccini A, Grothey A, et al. Landscape of tumor mutation load, mismatch repair deficiency, and PD‐L1 expression in a large patient cohort of gastrointestinal cancers. Mol Cancer Res. 2018;16:805–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Overman MJ, McDermott R, Leach JL, et al. Nivolumab in patients with metastatic DNA mismatch repair‐deficient or microsatellite instability‐high colorectal cancer (CheckMate 142): an open‐label, multicentre, phase 2 study. Lancet Oncol. 2017;18:1182–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Catalano V, Loupakis F, Graziano F, et al. Mucinous histology predicts for poor response rate and overall survival of patients with colorectal cancer and treated with first‐line oxaliplatin‐ and/or irinotecan‐based chemotherapy. Br J Cancer. 2009;100:881–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeh CY, Wang JY, Chen JS, et al. Clinical significance of signet ring cell rectal carcinoma. Int J Colorectal Dis. 2004;19:102–107. [DOI] [PubMed] [Google Scholar]
- 48.Arnold D, Lueza B, Douillard JY, et al. Prognostic and predictive value of primary tumour side in patients with RAS wild‐type metastatic colorectal cancer treated with chemotherapy and EGFR directed antibodies in six randomized trials. Ann Oncol. 2017;28:1713–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Imperial R, Ahmed Z, Toor OM, et al. Comparative proteogenomic analysis of right‐sided colon cancer, left‐sided colon cancer and rectal cancer reveals distinct mutational profiles. Mol Cancer. 2018;17:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Levine EA, Blazer DG 3rd, Kim MK, et al. Gene expression profiling of peritoneal metastases from appendiceal and colon cancer demonstrates unique biologic signatures and predicts patient outcomes. J Am Coll Surg. 2012;214:599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.