Viral primary infections and reactivations are common complications in patients after solid organ transplantation (SOT) and hematopoietic stem cell transplantation (HSCT) and are associated with high morbidity and mortality. Among these patients, viral infections are frequently associated with viremia. Beyond the usual well-known viruses that are part of the routine clinical management of transplant recipients, numerous other viral signatures or genomes can be identified in the blood of these patients.
KEYWORDS: blood, blood donors, bone marrow transplantation, diagnostics, solid organ transplantation, transplantation, virus
SUMMARY
Viral primary infections and reactivations are common complications in patients after solid organ transplantation (SOT) and hematopoietic stem cell transplantation (HSCT) and are associated with high morbidity and mortality. Among these patients, viral infections are frequently associated with viremia. Beyond the usual well-known viruses that are part of the routine clinical management of transplant recipients, numerous other viral signatures or genomes can be identified in the blood of these patients. The identification of novel viral species and variants by metagenomic next-generation sequencing has opened up a new field of investigation and new paradigms. Thus, there is a need to thoroughly describe the state of knowledge in this field with a review of all viral infections that should be scrutinized in high-risk populations. Here, we review the eukaryotic DNA and RNA viruses identified in blood, plasma, or serum samples of pediatric and adult SOT/HSCT recipients and the prevalence of their detection, with a particular focus on recently identified viruses and those for which their potential association with disease remains to be investigated, such as members of the Polyomaviridae, Anelloviridae, Flaviviridae, and Astroviridae families. Current knowledge of the clinical significance of these viral infections with associated viremia among transplant recipients is also discussed. To ensure a comprehensive description in these two populations, individuals described as healthy (mostly blood donors) are considered for comparative purposes. The list of viruses that should be on the clinicians’ radar is certainly incomplete and will expand, but the challenge is to identify those of possible clinical significance.
INTRODUCTION
Humans are infected by numerous DNA and RNA viruses during their lifetime. Some of these viral infections can be completely eliminated after an acute phase, but others may lead to latent phases (at risk of reactivations later in life) or to chronic and persistent infections, according to the type of virus. The associated clinical manifestations range from asymptomatic infections to fatal disease. These clinical features are determined not only by the intrinsic virulence and tropism of the virus but also, and very importantly, by the interactions with the host immune system and the state of its immunity.
The human virome refers to the viral component of the human microbiome found on the body surface, in blood (plasma, serum, or blood cells), or inside any bodily fluid or tissue, including eukaryotic and prokaryotic DNA and RNA viruses and endogenous viral elements integrated into host chromosomes, with or without clinical manifestations (1–4). It is characterized by dynamic intrapersonal changes that evolve during early life and later by intrapersonal mid- and long-term stability (5–8), with a specific composition according to the different body sites (2, 5, 7, 9–20). These viruses potentially interact with other members of the microbiome as part of “transkingdom interactions,” as well as the host immune system, leading to the dynamic building of an individual immunophenotype and virotype (4, 21).
Until the last decade, most data regarding the blood virome composition originated from the detection of DNA and RNA virus genomes using specific molecular assays. Metagenomic next-generation sequencing (mNGS) broadened the vision of the blood virome due to its unbiased and exhaustive approach. It also raised new issues, particularly regarding the risk of contamination, spurious bioinformatic findings, or the difficulties fulfilling Koch’s postulates (22, 23). Despite these limitations, mNGS opened the door to the identification of unexpected infections and new commensal viruses, leading to novel paradigms regarding viral infections and/or reactivations. In this regard, viral infections with detectable viremia deserve a systematic and comprehensive description, especially in highly immunocompromised populations. Two populations are of special interest. The first population is solid organ transplant (SOT) and hematopoietic stem cell transplant (HSCT) recipients, which represent the most immunocompromised patients in whom unrecognized infections can lead to serious complications. Viral primary infections and reactivations, which are frequently associated with viremia, are among the most common complications after transplantation and are associated with high morbidity and mortality (24–26). The graft itself and the multiple blood product transfusions represent potential sources of virus transmission. Recent studies have described the blood virome of SOT and HSCT recipients and the clinical impact of some DNA virus infections with detectable viremia (27–29). The second population concerns individuals described as healthy, such as blood donors, in whom the detection of viral genomes in blood without any symptoms is not a rare event (30). In their case, a description of the blood virome is of special importance, as these individuals can be considered a control population, including in the context of optimizing blood product safety (31).
The identification and understanding of the pathogenic role posttransplantation of cytomegalovirus (CMV) and BK virus, among other viruses, over the past decades have proven to be of paramount importance to transplant recipients and have contributed to the reduction of posttransplantation morbidity and mortality, as well as improvements in their clinical management. The identification of multiple novel viral species and variants due to mNGS has opened up a new field of investigation and new paradigms that deserve clinicians’ attention. There is a pressing need to precisely describe the state of knowledge in this field, with a systematic review of the landscape of all viral infections that should be under scrutiny in high-risk populations. This article reviews the eukaryotic DNA and RNA viruses identified in human blood, plasma, or serum samples of SOT and HSCT recipients, with a special focus on recently identified viruses and those for which their potential association with disease remains to be investigated. Clinicians involved in the clinical management of SOT and HSCT recipients need to be aware of these viral species and their potential clinical impacts. To provide a comprehensive description of these viral infections with detectable blood viremia in these two populations, we also compare these populations with nontransplanted individuals, which consist mostly of blood donors and individuals described as healthy.
DNA VIRUSES
Double-Stranded DNA Viruses
Herpesviridae.
Primary infection and reactivation of viruses belonging to the Herpesviridae family, such as herpes simplex virus 1 (HSV1) and HSV2, varicella-zoster virus (VSV), Epstein-Barr virus (EBV), and cytomegalovirus (CMV), can be associated with severe complications among SOT and HSCT recipients (32–38). For several decades now, specific monitoring, preventive, and therapeutic strategies have been implemented in the clinical management of this patient population (32–38). Apart from the above-mentioned viral species that are not discussed here, other viruses of the Herpesviridae family deserve the clinician’s attention.
(i) Human herpesvirus 6A and 6B.
(a) Overview.
According to the 10th Report of the International Committee on Taxonomy of Viruses (ICTV) (https://talk.ictvonline.org/ictv-reports/ictv_online_report/) in 2018, human herpesvirus 6 (HHV-6) belongs to the Betaherpesvirinae subfamily and comprises two species, HHV-6A and HHV-6B. HHV-6 tropism encompasses various cells, such as CD4+ and CD8+ T lymphocytes, monocytes, macrophages, bone marrow progenitors, and central nervous system cells (astrocytes, oligodendrocytes, microglial cells, neurons) (39, 40). Following primary infection, a specific immune response is mounted with the production of IgM and IgG (the latter persists throughout life), and cellular immunity is also thought to play a major role with specific CD4+ and CD8+ T cell responses (39). The seroprevalence rates in the general adult population are estimated to be >95% in developed countries (of note, current serological assays do not distinguish HHV-6A and HHV-6B) (39, 41).
HHV-6 persists in the host and is capable of reactivation, but the mechanisms of HHV-6 latency still remain unclear. The existence of covalently closed circular episomes associated with cellular nuclear proteins, as has been observed with other herpesviruses, has not been demonstrated for HHV-6A or HHV-6B (39, 40). The HHV-6A/6B genome can, however, integrate into host chromosomes of somatic cells and/or gametes, specifically into the telomeres, due to the presence of telomeric repeats at the end of its genome (40). The presence of chromosomally integrated HHV-6 (ciHHV-6) may result from de novo infection or from transmission to an offspring of integrated HHV-6 in germ cells. Integration is suggested by some authors to be the main latency mechanism for HHV-6, and studies show that ciHHV-6 may lead to reactivation (39, 40). It is estimated that 0.2% to 1% of the general population in developed countries have ciHHV-6 (39). The prevalence of ciHHV-6 is estimated to be about 1% in umbilical cord blood and healthy blood donors from the United States and the United Kingdom, 1.28% to 5% in liver transplant recipients from the United States and the United Kingdom, 0.74% in an Italian cohort of SOT recipients, 1.92% to 2.13% in kidney transplant recipients from the United Kingdom and the United States, and 1.4% to 1.86% of HSCT recipients from Italy and the United States (42, 43).
Primary infections occur mostly through saliva at a young age. The clinical manifestations, if any, consist of an acute febrile illness (exanthema subitum) and, in a small percentage of cases, seizures or gastrointestinal or respiratory manifestations (41, 44). Most cases of primary infections are caused by HHV-6B in Europe, the United States, and Japan (44). The epidemiology of HHV-6A is less known but was found to be predominant in Africa (44).
Among transplant recipients, primary infection concerns mostly pediatric recipients, particularly those less than 3 years old (45). Primary infections are rare in adult recipients and may result from donor graft transmission (46–48). Among adult transplant recipients, most HHV-6 infections are due to reactivation in the context of immunosuppression and typically occur 2 to 4 weeks after transplantation (41). HHV-6B reactivation seems to be more frequent than HHV-6A reactivation. HHV-6A is rarely identified and accounts for up to 3% of reactivation after transplantation (49). Reactivation occurs in approximately 30% of SOT recipients (50) and 45% of HSCT recipients (49) but varies widely due to the variability of diagnostic assays and the diagnostic challenge of distinguishing active infection from ciHHV-6. Associated manifestations include fever, rash, myelosuppression, and neurological manifestations, including encephalitis, for which a causal role of HHV-6 has been demonstrated (44, 49). HHV-6 has also been associated with pneumonitis and hepatitis, but the causal role remains unclear. Among SOT recipients, HHV-6 reactivation has also been associated with allograft rejection, an increased risk of CMV disease, and fungal and opportunistic infections (50, 51). Among HSCT recipients, it has been associated with delayed engraftment, acute graft-versus-host disease (GvHD), CMV reactivation, and increased mortality after HSCT (52–54).
The diagnosis of HHV-6 active infection relies on the detection of the HHV-6 genome in blood samples but is challenging, since genome detection can be the result of HHV-6 viremia during primary infection or reactivation, as well as the presence of ciHHV-6. Distinguishing ciHHV-6 from primary and reactivated infections is of particular importance in the clinical setting of transplanted patients, as HHV-6 replication can be associated with the above-mentioned clinical manifestations and patients will require antiviral therapy. Among transplant recipients, current guidelines do not recommend routine testing for HHV-6 viremia but rather testing in the case of clinical manifestations using quantitative real-time PCR in blood samples (50, 54). Diverse techniques can be used to distinguish ciHHV-6 from active replication (39, 50, 51, 54). ciHHV-6 is suggested by persistently elevated viral loads in whole blood (approximately 6 log10 copies/ml) with minimal fluctuation over time and by 100- to 1,000-fold lower viral loads in serum or plasma. ciHHV-6 can be excluded by a negative PCR result from a blood sample pretransplantation. This condition is, in contrast, confirmed by a positive HHV-6 PCR in hair follicles or nails. The gold standard test for confirming ciHHV-6 is still fluorescence in situ hybridization (FISH), which is labor-intensive. Droplet digital PCR is a convenient and accurate method and is more frequently used for ciHHV-6 investigations.
(b) Detection in blood samples of nontransplanted individuals.
Few data exist on the prevalence of HHV-6 DNA detection among nonimmunocompromised individuals. Among 176 Japanese pediatric patients visiting medical facilities for various reasons, HHV-6 DNA was detected in four (2.2%) whole-blood samples (55). The prevalence of HHV-6 DNA detection among blood donors was estimated to be 3.5% in a Greek cohort of 401 subjects and 6.1% in a cohort of 198 subjects from Burkina Faso (56, 57). Metagenomic analysis of whole-blood samples of more than 8,000 individuals considered healthy revealed the identification of HHV-6A and HHV-6B in 1.5% and 5% of individuals, respectively (30). HHV-6A and HHV-6B sequences were also detected using mNGS in red blood cells and fresh frozen plasma products (58).
(c) Detection in blood samples of SOT recipients.
In a Finnish study of 84 pediatric liver, kidney, or heart transplant recipients, the screening of HHV-6 using real-time PCR in plasma samples revealed that 19/22 (86%) seronegative patients developed primary infection and 29/62 (47%) seropositive patients presented reactivation (45); 35% of patients presented symptoms such as fever, rash, diarrhea, or seizures (45). In a Canadian study of 154 pediatric liver transplant recipients, HHV-6 DNA was detected using real-time PCR in whole-blood samples of 25 (16.2%) patients and also in plasma samples from 4 (16%) of those 25 patients (59). Seven of 25 patients (28%) had symptoms consistent with HHV-6 infection. The screening of 495 whole-blood samples of 34 adult liver or kidney transplant recipients revealed that 11% of samples were positive for HHV-6 (60). Of note, HHV-6 infection was associated with graft rejection (60).
(d) Detection in blood samples of HSCT recipients.
Most published data concern HHV-6 among HSCT recipients. The first detection of HHV-6 DNA usually occurs in the early period post-HSCT. HHV-6B has been identified with incidences ranging from 29% to 67% in plasma samples of pediatric and adult HSCT patients, at a median time of 3 weeks (53). In a French study of 235 allogeneic HSCT (allo-HSCT) patients, 82% of HHV-6 reactivation episodes occurred during the first 100 days posttransplantation (61). Among 49 adult Japanese HSCT recipients, screening for HHV-6 in plasma at 30 days posttransplantation was positive in 53.1% of patients (62). In a U.S. cohort of 315 allo-HSCT recipients, HHV-6 DNA was detected in plasma samples of 111 (35%) patients at a median of 20 days post-HSCT (52). The screening of plasma samples of 404 U.S. adult allo-HSCT recipients with real-time PCR revealed that HHV-6B and HHV-6A were detected in 46% and 0.2% of patients, respectively, with median viral loads of 7.41E2 copies/ml (range, 2.29E2 to 3.24E3 copies/ml) for HHV-6B (27). When screening with multiplex PCR for herpesviruses, polyomaviruses, adenovirus, and parvovirus B19 in whole blood, HHV-6 was the most frequently detected in two Japanese studies of pediatric and adult patients. HHV-6 was detected in 41.7% (10/24) of autologous HSCT (auto-HSCT) recipients and in 60% (63/105) of allo-HSCT patients (63, 64). In a French study of 78 allo- and auto-HSCT pediatric and adult patients, peripheral blood mononuclear cell (PBMC) samples were screened for HHV-6 and revealed that 3/66 (4.5%) patients were positive before HSCT (65). After transplantation, HHV-6 was detected at least once in 31/78 (39.7%) patients, with a median time of 19 days (65). HHV-6 and human herpesvirus 7 (HHV-7) genomes were simultaneously detected in 24 (30.8%) patients. Among the allo-HSCT recipients included in this study, CMV prophylaxis with acyclovir did not influence the rate of virus detection (65).
(ii) Human herpesvirus 7.
(a) Overview.
According to the 10th Report of the ICTV in 2018 (https://talk.ictvonline.org/ictv-reports/ictv_online_report/), human herpesvirus 7 (HHV-7) belongs to the Betaherpesvirinae subfamily. HHV-7 has a tropism for CD4+ T cells (66) and is presumed to be transmitted via saliva. Primary infection occurs mainly during childhood and is responsible for mild disease, and most adults are seropositive for HHV-7 (67). CD4+ T cells are the site of HHV-7 latency, during which the virus might persist as episomes; some data revealed that HHV-7 can integrate into the host’s chromosomes, similar to HHV-6 (67, 68).
As with HHV-6, most HHV-7 infections among transplant recipients are presumed to be the consequence of reactivation and typically may occur within 2 to 4 weeks after transplantation in up to approximately 40% of SOT and HSCT recipients, with a limited duration of detectable HHV-7 DNA in blood samples and low viral loads (49, 50, 67). Overt manifestations of HHV-7 infection seem to be uncommon among adult SOT recipients and may consist of nonspecific febrile syndrome and myelosuppression (50). HHV-7 diseases and viremia are also uncommon among pediatric SOT recipients (50, 69), while viremia and associated clinical manifestations are poorly studied among HSCT recipients. HHV-7 reactivation has been associated with central nervous system (CNS) disease, hepatitis, pneumonitis, myelosuppression, acute GvHD, and potentially an increased risk of CMV reactivation (49, 67, 70, 71).
Routine screening for HHV-7 viremia is not recommended in asymptomatic patients nor is prophylaxis or preemptive therapy in transplant recipients (50).
As with other Herpesviridae such as EBV, CMV, and HHV-6, the prevalence rates of HHV-7 DNA detection in blood samples should be interpreted cautiously, considering their latent phase in lymphocytes and the use of potentially active antiviral prophylaxis and treatments in transplant recipients, such as (val)ganciclovir.
(b) Detection in blood samples of nontransplanted individuals.
Metagenomic analysis of the whole-blood virome of more than 8,000 individuals considered healthy showed that HHV-7 sequences were the most frequently detected, with a prevalence estimated at 20.37% (30). HHV-7 sequences were also detected in fresh frozen plasma products by use of metagenomics (58). Among 176 Japanese children visiting medical facilities for various reasons, two (1.1%) had detectable HHV-7 DNA in blood samples (55). A higher prevalence has been reported when screening the PBMCs of 31 French adult PBMC donors, with 80.7% positive for HHV-7 (65), and these results are in line with the tropism and latency of HHV-7 in lymphocytes.
(c) Detection in blood samples of SOT recipients.
The prevalence of HHV-7 detection in SOT recipients varies among studies, according to the various populations tested, PCR assays, and whole blood versus plasma screening. The screening of HHV-7 in whole-blood samples of 34 pediatric kidney and liver transplant recipients revealed that it was more frequently detected than EBV, CMV, and HHV-6, in particular when analyzing the leukocyte fraction compared to plasma (60). Codetection with EBV was frequent in whole-blood samples (62%) (60). Notably, 85% of infections were determined to be either exclusively latent or lytic (60). Interestingly, coinfection with EBV and HHV-7 was associated with a 5.6-fold increased risk of events of kidney rejection (60). In a Canadian study, the prevalence of HHV-7 detection in the plasma or whole blood of 204 pediatric liver transplant recipients was estimated at 5.4% (59).
(d) Detection in blood samples of HSCT recipients.
In a French study of 78 allo- and auto-HSCT pediatric and adult patients, HHV-7 DNA was detected in PBMC samples in 29/66 (43.9%) patients before HSCT (65) and at least once in 43/78 (55.1%) patients posttransplantation, with a median time of 21.5 days (65). Interestingly, the rates of HHV-7 DNA detection and the viral loads were comparable before and after HSCT, in comparison to HHV-6, which reactivated more frequently after transplantation (65). The prevalence of HHV-7 detection in plasma was estimated at 20.8% in a Czech cohort of 125 pediatric allo-HSCT recipients (72) and 18.3% in a Japanese cohort of adult HSCT recipients (62). The prevalence of HHV-7 detection in whole-blood samples varied from 3.1%, among a Canadian cohort of 163 pediatric allo-HSCT recipients, with a median time of detection of 21 days after HSCT (71), to 4.2% and 8.6% among 24 auto-HSCT and 105 allo-HSCT Japanese pediatric and adult recipients, respectively (63, 64).
Human Polyomaviridae.
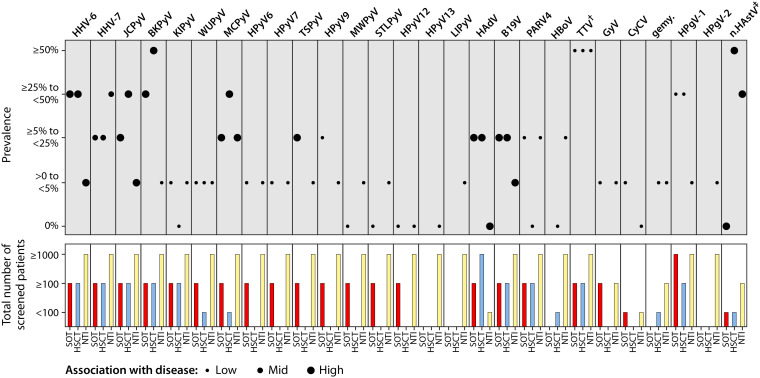
Polyomaviridae are nonenveloped DNA viruses phylogenetically classified into four genera based on the amino acid sequence of the large tumor antigen (LTAg) (73, 74). There are 14 known human polyomaviruses (HPyV), and the estimated prevalence for each one is displayed in Table 1. According to the current state of knowledge for some human polyomaviruses, infection mostly occurs early in life (75). Furthermore, the detection of antibodies against more than one polyomavirus is frequent (76). It increases with age after childhood with variable kinetics and a peak prevalence according to the species, possibly suggesting different routes of transmission and mechanisms of persistence (77–79). While considering that replication of viruses such as JC polyomavirus (JCPyV) and BK polyomavirus (BKPyV) are known to be associated with potentially dramatic consequences in immunocompromised patients and deserve specific monitoring and clinical management (75, 80–82), their detection in blood needs to be better described. Regarding the detection of other human polyomaviruses, this section will outline the current state of knowledge among transplant recipients.
TABLE 1.
Seroprevalence of human Polyomaviridae in pediatric and adult populationsa
| Genus | Species | Species abbreviation | Seroprevalence (%) |
|
|---|---|---|---|---|
| Pediatric populations | Adult populations | |||
| Alphapolyomarivus | Merkel cell polyomavirus | HPyV5 | 23–34 | 25–81.9 |
| Trichodysplasia spinulosa-associated polyomavirus | HPyV8 | Unknown | 79.6–80.9 | |
| Human polyomavirus 9 | HPyV9 | 20 | 17.6–47 | |
| Human polyomavirus 12 | HPyV12 | 34–80 | 4–97.3 | |
| New Jersey polyomavirus | HPyV13 | 7.5–33 | 5.1–57.5 | |
| Betapolyomavirus | BK polyomavirus | HPyV1 | 73 | 82–98.9 |
| JC polyomavirus | HPyV2 | 21 | 35–63.2 | |
| Karolinska Institute polyomavirus | HPyV3 | 56 | 55–91.6 | |
| Washington University polyomavirus | HPyV4 | 54 | 69–98.9 | |
| Deltapolyomavirus | Human polyomavirus 6 | HPyV6 | Unknown | 69–83.8 |
| Human polyomavirus 7 | HPyV7 | Unknown | 35–71.7 | |
| Malawi polyomavirus | HPyV10 | Unknown | 99.1–99.5 | |
| Saint Louis polyomavirus | HPyV11 | 34–61 | 64.8–93.3 | |
| Lyon IARC polyomavirus | HPyV14b | Unknown | 5.9 | |
(i) JC polyomavirus.
(a) Overview.
There is only one JC polyomavirus (JCPyV) serotype, with seven genotypes that have different epidemiologies according to geographical location (83). JCPyV transmission probably occurs via contact with urine (84). Clinical manifestations of primary infection remain unclear. JCPyV infection seems to occur early in life and with a lower prevalence than that of BK polyomavirus (BKPyV), i.e., from 16% in children from 1 to 5 years of age to 34% in adults from 21 to 50 years and 51% in adults over 70 years (78). The overall seroprevalence in European and U.S. individuals is estimated to be 39% to 70% (78, 83, 85, 86). After primary infection, JCPyV enters a phase of latency, with the main reported site being the reno-urinary tract (87). Progressive multifocal leukoencephalopathy (PML), a rare and late complication in transplant recipients, has been associated with JCPyV reactivation in immunocompromised patients (e.g., HIV-infected patients, patients treated with rituximab, multiple sclerosis patients treated with natalizumab, and SOT and HSCT recipients). JCPyV reactivation has also been associated with other central nervous system disorders (e.g., JCPyV-associated encephalopathy) and JCPyV-associated nephropathy (JCPyVAN) in kidney transplant recipients, the latter being less frequent than BKPyV-associated nephropathy (BKPyVAN) (83, 88). The cellular immune response is thought to play a major role in controlling JCPyV replication, particularly in diseases such as PML (83). A review of the literature identified 44 cases of PML in SOT recipients reported from 1958 to 2010, and another review identified 43 cases in allo- and auto-HSCT recipients (89, 90). In those patients, JCPyV screening in blood samples is barely reported. Among HIV-infected patients with PML, JCPyV viremia is not more frequent than in matched HIV-infected control patients (83). Among the few studies regarding JCPyVAN, JCPyV viremia was reported in some patients with low viral loads of approximately 1E3 copies/ml and transient viremia, contrasting with the high viral loads (>1E6 copies/ml) in urine samples (88, 91, 92).
(b) Detection in blood samples of nontransplanted individuals.
In comparison to the closely related and also clinically important BKPyV, JCPyV is less frequently detected in blood samples of healthy and immunocompromised individuals. This difference could be explained by different patterns of replication, sites of latency, and different ways of containment by the immune system. The JCPyV genome was not detected in plasma samples of a large cohort of 400 Swiss blood donors, comprising 58% JCPyV-seropositive individuals and 19% individuals with detectable replication in urine (86). Serum samples of 1,016 Dutch blood donors were screened with three different multiplex PCR assays to detect the 14 known human polyomaviruses: JCPyV was detected in five (0.5%) donors with low viral loads (range, 0.9E1 to 3.7E1 copies/ml), and all were JCPyV seropositive (93). However, the low viral load results should be interpreted with caution and may reflect assay variability. Some studies have investigated the detection of JCPyV in the PBMCs of immunocompetent adults, with a prevalence ranging from 0.9% to 83%, possibly reflecting methodological differences among studies (94–96).
(c) Detection in blood samples of SOT recipients.
Among adult kidney transplant recipients, the JCPyV genome was detectable in the plasma or blood samples of 0.9% to 25% of patients of several cohorts from the United States (n = 20), Australia (n = 167), United Kingdom (n = 112), and Brazil (n = 12) (97–100). JCPyV viremia reported among pediatric kidney transplant recipients varied from 12.1% (16/132 patients) to 20.8% (22/106 patients) (101, 102), with the differences between these two cohorts potentially attributable to different immunosuppressive treatments and patient age. Among these children, JCPyV viremia was more frequent and associated with higher viral loads among patients with viruria (102). Detection rates of JCPyV in plasma were stable during the first 3 years posttransplantation, with immunosuppression being a risk factor, followed by a drop in prevalence thereafter (102), thus highlighting the role of the immune system, especially the altered cellular immunity occurring after transplantation, in the containment of JCPyV primary infection or reactivation (103).
(d) Detection in blood samples of HSCT recipients.
JCPyV reactivation is not rare among HSCT recipients, as revealed in a study by Wittmann et al. (104). In this cohort of 164 adult allo-HSCT recipients from Israel, the retrospective screening of JCPyV in whole-blood samples collected as part of routine investigations during a mean follow-up period of 7 months (standard deviation [SD], 6.7 months) revealed that 40 (24.4%) patients experienced either transient (n = 20) or persistent (n = 20) reactivation, which was defined as two or more consecutive positive samples (104). Viral loads were higher in patients with JCPyV DNA detected in multiple consecutive samples, with median viral loads up to 3.88E3 copies/ml for those with more than 10 positive samples (range, 1.13E2 to 2.50E6 copies/ml). Among patients experiencing transient reactivation, the median time to reactivation was 119 days (range, 7 to 908 days) after transplantation, 50% of patients had either acute or chronic GvHD, and 60% died within a median period of 19.5 days (range, 2 to 643 days) after JCPyV reactivation. Among patients experiencing persistent reactivation, the median time to reactivation was shorter (49 days; range, 7 to 959 days), 85% of patients were under immunosuppressive therapy for acute and/or chronic GvHD, and 75% died within a median period of 126 days (range, 11 to 435 days) after JCPyV reactivation. Neurological complications (including 7 suspected cases of PML, of which 2 were confirmed) occurred in 60% of patients with persistent reactivation. The concomitant use of more than three immunosuppressive treatments was associated with an increased risk of persistent viremia (104). The results of this study highlight the impact of strong immunosuppression on JCPyV reactivation and suggest that JCPyV persistent reactivation and/or high viral loads might be associated with adverse outcomes.
In another study of 30 adult allo-HSCT recipients from Israel, the prevalence of JCPyV DNA detection in plasma samples decreased with time after transplantation and was 4/22 (18%), 0/21 (0%), and 1/22 (5%) of patients at 3, 6, and 12 to 18 months after transplantation, respectively (105). The overall mean viral load was 5.65E2 copies/ml (range, 2.70E2 to 2.9E3 copies/ml). JCPyV was also detected in PBMCs of 2/22 (9%), 4/21 (19%), and 0/22 patients at 3, 6, and 12 to 18 months, respectively. JCPyV viremia was not associated with any clinical manifestation or GvHD, and no patient developed PML in this study (105).
(ii) BK polyomavirus.
(a) Overview.
BK polyomavirus (BKPyV) is classified into four different subtypes (I to IV), correlated with a genotypic classification based on the epitope region of the VP1 gene (106); subtype I is predominant in Europe, Africa, and Asia (107). BKPyV is presumed to be transmitted through oral and respiratory routes (103, 108). The clinical manifestations of primary infection remain unclear and may consist of nonspecific febrile episodes (103). BKPyV infection appears to occur early in life, as revealed by European and U.S. seroepidemiological studies, which also found that prevalence increases with age, ranging from 38% in children of 1 to 5 years to 87% in adults of 21 to 50 years, with an estimated overall seroprevalence of 82 to 98.9% (Table 1) (78, 85, 86, 93). After primary infection, BKPyV persists in a latent phase in the reno-urinary tract; data are conflicting regarding latency in PBMCs and brain (87, 103, 108). As with JCPyV, the cellular immune response plays an important role in controlling BKPyV replication. Among HIV-infected patients, BKPyV reactivation is frequent and in most cases asymptomatic (103). In transplant recipients, BKPyV reactivation is associated with BKPyVAN in SOT recipients, ureteric stenosis specifically in renal transplant recipients, and hemorrhagic cystitis in HSCT recipients (103). Up to approximately 10% of kidney transplant recipients develop BKPyVAN and potentially further graft failure (109, 110). The detection of the BKPyV genome in blood or plasma samples is the cornerstone of BKPyVAN investigations and diagnosis (80, 110–113). BKPyV genome detection in blood or plasma samples has rarely been described among HSCT recipients, as the diagnosis of BKPyV hemorrhagic cystitis relies mostly on its detection in urine samples (81). Nevertheless, recent data showed that BKPyV viremia precedes hemorrhagic cystitis (with a median time of 17 days in a cohort of pediatric patients [114]). Plasmatic viral loads of >1E4 copies/ml are associated with an increased risk of hemorrhagic cystitis among pediatric and adult allo-HSCT recipients (114–116).
(b) Detection in blood samples of nontransplanted individuals.
The BKPyV genome was not detected in plasma samples of a large cohort of 400 Swiss blood donors, comprising 82% individuals that were BKPyV seropositive (86), whereas it has been reported in 15% of serum samples of a Brazilian cohort of 20 blood donors (100). Screening of serum samples of 1,016 Dutch blood donors using three different multiplex PCR assays to detect the 14 known human polyomaviruses led to the detection of BKPyV DNA in one (0.1%) donor who was also BKPyV seropositive (93).
(c) Detection in blood samples of SOT recipients.
The incidence of viremia among kidney transplant recipients varies widely among studies, according to several factors, i.e., time after transplantation, immunosuppressive regimens, and type of sample (blood or urine) initially screened. The incidence in adults initially screened in plasma ranged from 9.2% to 84% up to 1 year after transplantation (97, 100, 109, 110, 112, 117–121). In a recent study of 99 kidney transplant recipients from Finland, BKPyV DNA was detected at least in one plasma sample of 84% of patients by use of a multiplex PCR assay for the detection of 13 human polyomaviruses, with a median viral load of 3.6E4 copies/ml (range, 1E2 to 1.5E9 copies/ml) (121). The highest incidences of BKPyV DNA detection and viral loads above 1E4 copies/ml are most commonly observed within 6 months posttransplantation, with a median detection time of 2 to 4 months (98, 112, 119, 122). A recent study investigated the impact of pretransplantation seroreactivity of donor-recipient pairs on posttransplantation BKPyV viremia and BKPyVAN development (123), considering that BKPyV infection might originate from the kidney allograft (124) and that BKPyV seroreactivity correlates with BKPyV replication after transplantation (125). The pretransplantation donor BKPyV IgG level was associated with the recipient’s BKPyV viremia and BKPyVAN development and was the strongest pretransplantation factor for viremia and BKPyVAN in multivariate analysis; the recipient’s BKPyV seroreactivity was inversely associated with BKPyV viremia and BKPyVAN, but these results were nonsignificant (123). These results suggest that pretransplantation BKPyV seroreactivity of donor-recipient pairs might be useful for BKPyV infection risk assessment.
(d) Detection in blood samples of HSCT recipients.
BKPyV viremia occurs in approximately 33% to 55% of pediatric and adult HSCT patients, including in studies with a follow-up of up to 1 year posttransplantation (27, 114, 126, 127). In a cohort of 273 pediatric allo-HSCT recipients, the cumulative incidence for BKPyV viremia (above 1E3 copies/ml) was 15% during a median follow-up period of 58 months. BKPyV viremia did not predict GvHD, overall survival, or nonrelapse mortality (128). The higher incidence of BKPyV viremia among patients having undergone double umbilical transplantation than among unrelated peripheral blood stem cell-transplanted patients (58% versus 32% and 97% versus 23.5%, respectively) highlights the impact of the source of the graft and the immune system reconstitution after HSCT for the control of viral infections (127). In a study by Hill et al. (27) that investigated coinfections of HSCT patients with double-stranded DNA (dsDNA) viruses, plasma samples collected weekly over 100 days posttransplantation from pediatric and adult allo-HSCT recipients were screened by real-time PCR for dsDNA viruses, including BKPyV, EBV, HHV-6A/B, and adenovirus. BKPyV was detected in 54% of adult allo-HSCT recipients, either alone or with other coinfecting viruses, mainly CMV and HHV-6 (27). The burden of these coinfections, expressed as the cumulative area under the curve of the viral load, was associated with an increase in overall mortality with a dose-dependent relationship (27), but the respective impact of each virus remains undetermined.
(iii) Karolinska Institute polyomavirus.
(a) Overview.
Karolinska Institute polyomavirus (KIPyV) was discovered in 2007 using random PCR with respiratory samples of symptomatic patients (129), but its mode of transmission has not been established. Respiratory tract infections have been associated with the detection of KIPyV in respiratory samples of pediatric and adult immunocompetent patients (130–132) and transplant recipients (75, 98, 133, 134), but evidence of proven pathology is still lacking. The seroprevalence rates are detailed in Table 1.
(b) Detection in blood samples of nontransplanted individuals.
Among two cohorts of French blood donors (n = 640; n = 130) and one cohort of Italian bone marrow donors (n = 26) screened for KIPyV in blood samples, KIPyV was detected in 0.5% to 7.6% (135–137). In a study in which KIPyV was screened in blood samples of 100 pregnant Hungarian women, all results were negative (138). The screening of KIPyV by real-time PCR among 100 Australian blood donors yielded no positive results (139). In a study by Csoma et al. (140), screening with PCR of 200 plasma samples from blood donors revealed all samples to be negative. The screening of serum samples of 1,016 Dutch blood donors using three different multiplex PCR assays to detect the 14 known human polyomaviruses led to the detection of KIPyV in only one (0.1%) donor who was also KIPyV seropositive (93).
(c) Detection in blood samples of SOT and HSCT recipients.
Among adult kidney transplant recipients from two Hungarian cohorts, KIPyV was detected by PCR in plasma samples of 7/195 (3.6%) and 6/76 (7.9%) patients (133, 140). KIPyV DNA was not detected in blood samples of several cohorts of SOT and HSCT recipients from Italy (n = 167; n = 100) (98, 141), Australia (n = 100) (139), Italy (26 adult HSCT recipients) (137), and Finland (53 pediatric HSCT recipients) (142). Finally, in the study in which KIPyV was discovered, the screening of serum, whole blood, and frozen leukocytes of 17 HSCT recipients and 192 blood donors by a classic PCR obtained negative results (129).
(iv) Washington University polyomavirus.
(a) Overview.
Washington University polyomavirus (WUPyV) was discovered in 2007 using mNGS on a nasopharyngeal aspirate sample from a child presenting with pneumonia (143). The mode of transmission of WUPyV has not been established. As with KIPyV, respiratory tract infections have been associated with the detection of WUPyV in respiratory samples of pediatric and adult immunocompetent patients (130–132) and transplant recipients (75, 98, 133, 134), but evidence of proven pathology is still lacking. The seroprevalence rates are detailed in Table 1.
(b) Detection in blood samples of nontransplanted individuals.
WUPyV has been detected by real-time PCR in blood samples from French blood donors at a low prevalence of 0.8% (1/130 subjects) (136). Among 26 Italian bone marrow donors, WUPyV was not detected using real-time PCR on plasma samples (n = 26) (137). Results from WUPyV screening of blood from 100 pregnant women were negative (138), similar to plasma samples of 640 French blood donors (135). Among 1,016 Dutch blood donors, the screening of sera using multiplex PCR assays to detect human polyomaviruses revealed positive results for WUPyV DNA in two (0.2%) donors, both being WUPyV seropositive (93).
(c) Detection in blood samples of SOT recipients.
Among two Hungarian cohorts of 77 and 195 kidney transplant recipients and one Australian cohort of 167 kidney transplant recipients, WUPyV DNA was detected in blood samples of 5.3% (4/77), 2.6% (5/195), and 1.2% (2/167) of patients, respectively (98, 133, 140).
(d) Detection in blood samples of HSCT recipients.
In an Italian study of 100 SOT and HSCT recipients, PCR screening of WUPyV in blood samples revealed negative results (139), similar to studies of 26 Italian adult HSCT recipients and 53 pediatric HSCT recipients from Finland (137, 142). Among HSCT recipients, the absence of detection of KIPyV and WUPyV might be explained by several factors: the small number of screened patients (26 adult HSCT patients [137] and 53 pediatric HSCT patients [142]), a true low prevalence of infection or associated viremia, timing of reactivation after transplantation, and technical aspects, such as the type of PCR assay used (144).
(v) Merkel cell polyomavirus.
(a) Overview.
Merkel cell carcinoma (MCC) is a malignant skin cancer associated with clonal integration of Merkel cell polyomavirus (MCPyV) DNA in MCC cells (145), for which transplant recipients have an increased risk (146). The route of transmission and clinical manifestations of primary infection remain unknown. The skin is a major site of infection, but the specific type of host cell remains to be determined (147). MCPyV is the only known oncovirus among human polyomaviruses. High seroprevalence rates, as reported in a study of the general U.S. population, reveal that MCPyV infection is frequent (Table 1) (78, 85, 148).
(b) Detection in blood samples of nontransplanted individuals.
MCPyV DNA was detected in 3.8% (39/1,016) of serum samples from Dutch blood donors by using a multiplex PCR assay, of which 30/39 (77%) were MCPyV seropositive (93). Studies using metagenomics revealed that the MCPyV genome could be detected in red blood cells and fresh frozen plasma units of Swiss blood donors (58) and in plasma pools of febrile Kenyan children (149). Metagenomic analysis of whole-blood samples of more than 8,000 individuals considered healthy revealed that MCPyV sequences were detected in 0.59% of subjects (30). MCPyV DNA was also detected in blood samples using real-time PCR assays in 2.6% (5/190) and 10% (4/40) of Italian and Korean blood donors, respectively (150, 151), in up to 10% of elderly patients from Finland (152), in 5% of an Italian cohort of hepatitis C virus (HCV)-infected patients (153), and in 39% of blood samples of untreated, HIV-infected Japanese patients (154). Furthermore, MCPyV DNA could be detected in buffy coat samples of a cohort of Italian blood donors (at very low copy numbers, ranging from 10 to 100 copies/100,000 cells) (155).
(c) Detection in blood samples of SOT recipients.
MCPyV DNA was detected in blood samples of 1.8% to 16.7% of adult kidney transplant recipients, without any associated overt disease reported so far (97, 98, 121, 151). In a recent study of 30 Korean kidney transplant recipients, MCPyV DNA was detected in whole-blood samples with the highest prevalence reported so far among SOT patients (16.7%) (median viral loads of 5.7E3 copies/ml detected late after transplantation [mean time, 35 months posttransplantation]) (151). In the same study, the prevalence among 30 blood donors was 10% (151). In another recent study from Finland, the detection rate of MCPyV DNA in plasma samples of 99 kidney transplant recipients using a multiplex PCR assay for the detection of 13 human polyomaviruses was 8.1%, with a median viral load of 5.9E2 copies/ml (range, 1.2E2 to 1.4E3 copies/ml); MCPyV DNA was detected 1 month before transplantation (1 case), on the day of transplantation (2 cases), and from 1 to 8 months after transplantation (all 96 remaining cases) (121). MCPyV and BKPyV codetection was reported in 6/8 MCPyV-positive cases (121).
(d) Detection in blood samples of HSCT recipients.
MCPyV DNA was detected in 45% of 40 adult allo-HSCT recipients by mNGS performed on plasma samples collected 30 days posttransplantation (28).
(vi) Human polyomavirus 6 and 7.
(a) Overview.
Human polyomavirus 6 (HPyV6) and HPyV7 were first discovered in 2010 by Schowalter et al. (156) in skin swab specimens by using rolling circle amplification, and the route of transmission remains unknown. They have been detected mostly in skin specimens of nontransplanted individuals and transplanted recipients with or without dermatological diseases, but the association with clinical manifestation is not established (157–161).
(b) Detection in blood samples of nontransplanted individuals.
HPyV6 DNA was detected in one subject using multiplex PCR assays for human polyomavirus screening in serum samples of 1,016 Dutch blood donors; the subject was also HPyV6 seropositive (93), but HPyV7 was not detected in any sample (93).
(c) Detection in blood samples of SOT recipients.
HPyV6 DNA has been detected in 1/167 (0.6%) kidney transplant recipients by use of a real-time PCR assay on whole-blood specimens (98), but HPyV7 was not detected in blood samples of this cohort. HPyV6 and HPyV7 were not detected in any blood samples of 161 patients (comprising 125 immunocompromised and 36 immunocompetent patients) undergoing routine BKPyV, CMV, or adenovirus testing (162). HPyV7 was detected by real-time PCR in the PBMCs and skin biopsy specimens of two lung transplant recipients presenting with a rash (161).
(d) Detection in blood samples of HSCT recipients.
The detection of HPyV6 and HPyV7 in blood samples of HSCT recipients has not been studied so far.
(vii) Trichodysplasia spinulosa-associated polyomavirus.
(a) Overview.
Trichodysplasia spinulosa-associated polyomavirus (TSPyV) was first identified in 2010 by use of rolling circle amplification performed on typical cutaneous spicules of a heart transplant patient presenting with trichodysplasia spinulosa (163), a skin disease occurring in severely immunocompromised hosts that is characterized by follicular distention and keratotic spine formation, predominantly affecting the face (164). Trichodysplasia spinulosa may be associated with TSPyV primary infection, rather than reactivation, as demonstrated by serology. The disease is associated with viremia that may precede the development of skin lesions (165–168). High seroprevalence rates in children may indicate early infection during childhood (Table 1).
(b) Detection in blood samples of nontransplanted individuals.
TSPyV DNA was detected using a multiplex PCR assay in 0.5% (5/1,016) of serum samples from Dutch blood donors, of which 4/5 (80%) were TSPyV seropositive (93). TSPyV was not identified in screenings of Finnish and Brazilian cohorts of individuals described as healthy (serum samples of a cohort of 394 elderly subjects, blood samples of a cohort of 71 adults, and serum samples of another cohort of 229 children and adults) (152, 169, 170).
(c) Detection in blood samples of SOT recipients.
TSPyV DNA detection rates in blood samples of kidney transplant recipients without associated skin disease vary widely from one study to another and range from 1/167 (0.6%) patients with persistent infection and high blood viral loads (mean viral load, 1.94E5 copies/ml [98]) and 1/99 (1.1%) patients with a viral load below the limit of quantification (121) to 19/71 (26.8%) patients with a median viral load of 1E5 copies/ml (range, 8E2 to 1.2E6 copies/ml) (169). These differences in prevalence may be due to different geographical locations (Brazil [169] and Australia [98]) and real-time PCR assays targeting two different regions (a region between the large and small T antigen genes [169] and a region in the VP1 gene [98], respectively).
(d) Detection in blood samples of HSCT recipients.
The detection of TSPyV in blood samples of HSCT recipients has not been studied so far.
(viii) Human polyomavirus 9.
(a) Overview.
Human polyomavirus 9 (HPyV9) was initially identified in a serum sample of a kidney transplant recipient as part of a large study for HPyV screening in clinical samples (171). The tropism, routes of transmission, and clinical manifestations of HPyV9 infection remain unknown. HPyV9 seroprevalence rates increase with age, as revealed by Italian and Greek studies, with rates ranging from 10.4% in children of 1 to 4 years to 41% among adults of 60 to 69 years and 69.9% in adults over 80 years (76, 172, 173) (Table 1).
(b) Detection in blood samples of nontransplanted individuals.
HPyV9 DNA was detected in 0.4% (4/1,016) of serum samples from Dutch blood donors by use of a multiplex PCR assay; all were HPyV9 seronegative (93). In a cohort of 87 blood donors, screening by real-time PCR was negative in all patients (174), but HPyV9 DNA was detected in plasma samples of 6/100 (6%) pregnant and 2/100 (2%) nonpregnant women (138). In a recent study, HPyV9 DNA was amplified from PBMCs of a patient with HIV infection by rolling circle amplification; further screening of PBMCs of 40 healthy subjects and 9 HIV-infected patients was negative (175). However, as suggested by the authors, the small amount of material available for the experiments may explain these negative results (175).
(c) Detection in blood samples of SOT recipients.
Among a cohort of kidney transplant recipients from the Netherlands, 33% of patients were seropositive at the time of transplantation, which increased to 46% at 12 months posttransplantation (174). The question of viral clearance associated with the appearance of antibodies has still not been specifically addressed, but viremia and seroreactivity were not correlated in this Dutch study. Among a cohort of 101 kidney transplant and kidney-pancreas transplant recipients, the cumulative incidence of HPyV9 DNA detection in serum was 20% (174). None of the patients had a positive sample by real-time PCR before transplantation, but 3% of patients had consecutive positive samples after transplantation, with an incidence peak at 3 months posttransplantation (174). Furthermore, HPyV9 DNA was detected significantly more frequently in BKPyV DNA-positive samples than in negative samples (174). Among two Australian patient cohorts comprising a cohort of 112 kidney transplant recipients (98) and a cohort of immunocompetent and immunocompromised patients (162), HPyV9 DNA was not detected by real-time PCR in any blood or other samples. These discrepancies with the results of van der Meijden et al. (174) might be explained by a different local epidemiology or by different real-time PCR sensitivities or specificities. In another recent study from Finland, the detection rate of HPyV9 DNA in plasma samples of 99 kidney transplant recipients using a multiplex PCR assay for the detection of 13 human polyomaviruses was 1%, with a viral load of 1E4 copies/ml at 5 months posttransplantation (121).
(d) Detection in blood samples of HSCT recipients.
The detection of HPyV9 in blood samples of HSCT recipients has not been studied so far.
(ix) Human polyomaviruses 10 to 14: human polyomavirus 10 (Malawi polyomavirus), human polyomavirus 11 (Saint Louis polyomavirus), human polyomavirus 12, human polyomavirus 13 (New Jersey polyomavirus), and human polyomavirus 14 (Lyon IARC polyomavirus).
(a) Overview.
Since 2008, five human polyomavirus species have been identified. Human polyomavirus 10 (Malawi polyomavirus [MWPyV]) was first identified in 2008 by shotgun pyrosequencing of purified virus-like particles recovered from a stool sample of a healthy and asymptomatic child living in Malawi (176). In that study, the use of classic PCR and sequencing of reads closely related to MWPyV allowed the first identification of human polyomavirus 11 (Saint Louis polyomavirus [STLPyV]) in a fecal sample of an immunocompetent child in Malawi (177). Human polyomavirus 12 (HPyV12) was first identified by Korup et al. in Germany using a generic polyomavirus PCR targeting the VP1 gene region in liver specimens of deceased individuals who donated organs for transplantation, leading to the further design of a real-time PCR and detection in other tissues (intestinal and stool samples) (178). In 2014, New Jersey polyomavirus (human polyomavirus 13 [HPyV13]) was first identified using mNGS in a muscle biopsy specimen from a pancreas transplant recipient (179). The HPyV13 genome was detected in sera collected 10 months later (viral load, 1.6E5 copies/ml) but not in a pretransplant serum sample, thus revealing posttransplant persistent infection (179). Finally, a 14th novel putative human polyomavirus (Lion IARC polyomavirus [LIPyV]) was isolated and characterized in 2017, but it has not yet been classified by the ICTV (https://talk.ictvonline.org/ictv-reports/ictv_online_report/) (180).
Regarding these human polyomaviruses, data regarding their detection in blood samples are scarce and their potential associations with diseases (including gastrointestinal diseases) remain to be determined.
(b) Detection in blood samples of nontransplanted individuals.
Among blood donors, serum samples of a large cohort of 1,016 Dutch blood donors have been screened using multiplex real-time PCR for human polyomaviruses (93). The genomes of MWPyV, STLPyV, HPyV13, and LIPyV were detected in one (0.1%) seropositive, one (0.1%) seronegative, one (0.1%) seronegative, and one (0.1%) seronegative individual, respectively (93). The detection of HPyV12 DNA in 10/1,016 (1%) subjects was not confirmed by sequencing (93). Among children and adults described as healthy, MWPyV was not detected using real-time PCR assays in blood samples of an Italian cohort of 200 children or in 200 serum samples of Chinese adults (181, 182).
(c) Detection in blood samples of SOT recipients.
Among SOT recipients, the screening for MWPyV and STLPyV DNA in 261 plasma samples from an adult cohort of kidney transplant recipients in Saint Louis, MO (USA), revealed negative results (177). MWPyV, STLPyV, and HPyV12 DNA was not detected in whole-blood samples of an Australian cohort of 167 adult kidney transplant recipients (98). The screening of 45 serum samples of liver transplant recipients for HPyV-12 by real-time PCR gave negative results (178).
(d) Detection in blood samples of HSCT recipients.
There is currently no published study regarding the screening of human polyomaviruses 10 to 14 in blood samples of HSCT recipients.
Adenoviridae.
(i) Human adenovirus.
(a) Overview.
Human adenoviruses (HAdVs) are classified into seven species (A to G) and, further, into 67 types that are characterized by different cell tropisms and associated diseases (183). HAdV-F and -G are mostly associated with gastroenteritis, HAdV-B, -C, and -E with pneumonia, HAdV-C with hepatitis, HAdV-A, -B, and -D with meningoencephalitis, HAdV-B with cystitis, and HAdV-B and -D with keratoconjunctivitis (183). HAdVs circulate worldwide, but predominant species and types vary according to geographic regions (184–188), with, for instance, HAdV-B being the predominant species in the Japanese population (184, 185, 187).
In immunocompetent individuals, HAdV is transmitted mainly via inhalation of aerosolized droplets, direct conjunctival inoculation, or the fecal-oral route; most infections occur at a young age and are mild and self-limited (183). The innate and adaptive immune responses contribute to viral clearance, with the development of a T-cell response that is cross-reactive with different HAdV species (183). HAdV can persist in a latent state in various cells, including tonsillar and intestinal lymphocytes and lung epithelial cells (183).
In transplant recipients, HAdV infections can arise mainly from reactivation of persistent HAdV but also from de novo infection or from the transplanted organ (183, 189, 190). Infections are more frequent among pediatric transplant recipients, ranging from 3% to 60% and 4% to 20% in pediatric and adult SOT recipients, respectively, and 6% to 40% and 3% to 15% in pediatric and adult allo-HSCT recipients (183, 191). HAdV infection can be asymptomatic in transplant patients but can also encompass diverse clinical manifestations, such as fever, pneumonia, hepatitis, nephritis, hemorrhagic cystitis, enteritis, and disseminated disease (defined in HSCT recipients as multiple organ involvement with ≥2 positive PCR results in blood or other clinical samples) (183, 189). In SOT recipients, the transplanted organ is often the site of HAdV disease.
Most infections occur 2 to 3 months after transplantation (183). Several factors have been associated with HAdV reactivation in HSCT patients: a young age, the use of HLA-mismatched grafts, T-cell-depleted grafts, anti-thymocyte globulin, a delayed immune reconstitution, and acute GvHD (192–196). HSCT recipients with detectable viremia within 100 days after transplantation are at high risk of developing disseminated disease. HAdV-associated diseases are more severe and mortality rates are higher in pediatric than adult recipients, in particular in the presence of HAdV viremia (183, 191, 197). HAdV infection and disease in pediatric patients are associated with lower overall survival, higher nonrelapse mortality (128), and up to 15% to 50% of deaths after HSCT (195, 198). A sustained or elevated plasmatic viral load is associated with disseminated disease and clinical severity, longer hospitalization, and mortality in adult HSCT recipients (186, 187, 192, 194, 199, 200). Monitoring by real-time PCR with blood samples in asymptomatic patients is not recommended in adult and pediatric SOT and adult HSCT recipients but is recommended in high-risk pediatric HSCT recipients (191, 197). The blood viral load threshold at which preemptive antiviral treatment should be initiated is not established and ranges from 1E2 to 1E6 copies/ml (183).
(b) Detection in blood samples of nontransplanted individuals.
Among immunocompetent individuals, HAdV viremia can be detected in the context of acute infection. HAdV DNA detection in blood samples of asymptomatic individuals is poorly studied. In a Japanese study, the screening of blood samples of 50 healthy adults by real-time PCR for HAdV-B and -C revealed negative results (187).
(c) Detection in blood samples of SOT recipients.
In a multicenter study comparing CMV prophylaxis with valganciclovir to that with oral ganciclovir in 57 centers worldwide, 263 adult SOT recipients (comprising liver, heart, kidney, and kidney-pancreas recipients) were screened for HAdV using real-time PCR on whole-blood samples collected at or around days 7, 28, 56, and 100, as well as at 6 and 12 months posttransplantation (201). HAdV DNA was detected in blood samples of 19/263 (7.2%) patients, including 10/121 (8.3%) liver transplant recipients, 6/92 (6.5%) kidney transplant recipients, and 3/45 (6.7%) heart transplant recipients. Of 19 positive patients, 11 (58%) were asymptomatic at the time of viremia and only 1 (5%) presented acute rejection. CMV disease developed in 4/19 (21%) HAdV-positive patients (201).
In a U.S. cohort of 75 pediatric kidney transplant recipients screened monthly for HAdV in plasma samples over a 2-year period, HAdV was detected in 14.7% of patients, with a median time of 173 days to first detection (202). These patients were more likely to develop CMV viremia but did not have more adverse clinical outcomes (202).
(d) Detection in blood samples of HSCT recipients.
In a cohort of 215 adult T-cell-depleted graft HSCT recipients, 8% of patients had detectable viremia at a median time of 57 days (interquartile range [IQR], 2.5 to 4 days). Of these, 33% developed adenovirus disease with viremia preceding the development at a median time of 11 days (range, 3 to 37 days) (200). In this cohort, codetection of HAdV with other dsDNA viruses was frequent, with two viruses or more detected in 44% of patients. CMV, EBV, and HHV-6 were detected concomitantly with HAdV in 67%, 22%, and 17% of patients, respectively (200). When specifically considering pediatric allo-HSCT, the prevalence of HAdV viremia varied from 15% among 291 U.K. allo-HSCT recipients tested in whole-blood samples, to 37.5% among Japanese auto- and allo-HSCT recipients tested in blood samples, to 42.3% among 26 U.K. allo-HSCT recipients tested in whole-blood samples (187, 192, 194). Among adults, the incidence has been reported to be lower, ranging from 7.4% among 27 Swedish allo-HSCT recipients to 19.7% among 76 U.K. allo-HSCT recipients (195, 203). Coinfection with HAdV-B and -C has been reported in pediatric and adult cases (187). In a recent large multicenter European study of 1,736 pediatric and 2,540 adult transplant recipients, the cumulative incidences of HAdV viremia above 1E3 copies/ml during a 6-month follow-up period after allo-HSCT were 14% and 1.5% in pediatric and adult recipients, respectively (median time to first detection, 21 and 61 days for pediatric and adult patients, respectively) (196). In another cohort of 48 adult and pediatric allo-HSCT recipients followed up over 16 weeks, 11% of all recipients had detectable HAdV DNA in plasma, and of these, 57.8% had asymptomatic viremia (188).
Parvoviridae.
Parvoviridae are small nonenveloped viruses which are classified based on phylogenetic analysis of the amino acid sequence of the large nonstructural protein NS1. According to the ICTV classification (https://talk.ictvonline.org/ictv-reports/ictv_online_report/), they are divided into two subfamilies, Parvovirinae and Densovirinae, which infect vertebrates and invertebrates, respectively (204). Aside from the well-known parvovirus B19, other members of the Parvoviridae family deserve further discussion.
(i) Parvovirus B19.
(a) Overview.
Parvovirus B19 (B19V) was discovered in 1975 by Cossart et al. in serum samples of individuals screened for HBV infection (205). According to the ICTV classification (https://talk.ictvonline.org/ictv-reports/ictv_online_report/), B19V is a member of the Parvovirinae family (genus Erythroparvovirus) and comprises three genotypes, G1 to G3, that share the same pathogenic and antigenic characteristics. Genotype 1 is the most common genotype currently circulating worldwide; prior to the 1960s, genotype 2 was as common or more common than genotype 1 and has rarely been found in blood or to be associated with acute disease since then (204, 206). B19V productive infection is restricted to bone marrow erythroid progenitor cells in various stages of erythroid differentiation and leads to cell death (apoptosis), which results in aplastic crisis (204). B19V tropism also extends to nonerythroid-lineage tissues, but there is no clear evidence that infection is productive (204). B19V is transmitted mainly by the respiratory route, and the mechanisms by which the virus reaches the bone marrow are currently unknown. B19V can also be transmitted via blood products or vertically from the mother to the fetus (204). A B19V-specific cellular immunity develops and is directed against the capsid proteins VP1 and VP2 and the nonstructural protein NS1, with an important role of the CD8 T cell response in the control of acute infection. The humoral response is characterized by the development of IgM and IgG directed against the VP1 and VP2 proteins and the lifelong persistence of IgG (204).
The seroprevalence of B19V increases with age, with rates ranging from approximately 20% in children of 1 to 9 years old and 50% to 70% in children of 10 to 17 years old, as reported by an Israeli and a German study (207, 208); seroprevalence rates estimated in adult population-based studies range from 62.3% (1,323 Dutch individuals) to 72.1% (6,583 German individuals) (208, 209). Among the elderly, the prevalence rates rise to 79.1% among German individuals of 65 to 69 years old and to 77.7% among Japanese individuals of 70 years and older (208, 210). The reported seroprevalence rates among blood donors worldwide range from 24.6% (448 Chinese donors) (211), 27.6% (500 Iranian donors) (212), 33.6% (8,244 Chinese donors included in a meta-analysis) (213), 34.1% (800 Indian donors) (214), and 48.6% (360 donors from the Democratic Republic of the Congo) (215) to 53% (480 Brazilian donors) (216).
In nonimmunocompromised patients, most infections are asymptomatic. Clinical manifestations of acute infection range from hydrops fetalis in the fetus, typical erythema infectiosum in healthy children, and arthralgia in healthy adults to transient aplastic crisis in patients with a high rate of red blood cell turnover (e.g., sickle cell disease patients). Persistent infection among immunocompromised patients can lead to pure red cell aplasia (204). The diagnosis of acute infection should rely on the combination of real-time PCR and serological assays, considering the low sensitivity of some serological assays (217). Primary infection is associated with B19V viremia with high viral loads (up to 1E9 copies/ml) that decreases (with viral loads around 1E4 copies/ml) in parallel with IgG development (218, 219). Nevertheless, B19V DNA can be detectable at lower viral loads in blood samples after IgG development and resolution of symptoms for prolonged periods of time (weeks to years), even in nonimmunocompromised patients. Such prolonged viremia has been described among asymptomatic blood donors with viral loads ranging from <1E3 copies/ml up to 1E6 copies/ml (212, 218–225). Some data suggest that this prolonged detection of B19V DNA at low viral loads (mostly <1E3 copies/ml) might reflect the presence of naked DNA (detected by the use of more sensitive assays) rather than virions and that infections of blood product recipients are currently rare events with the use of current inactivation and other blood safety procedures (221, 226).
Clinical manifestations associated with B19V infection may be atypical in immunocompromised patients. Regarding SOT recipients, fever, arthralgia, and rash are observed in approximately 25%, 7%, and 6% of patients (227, 228). The manifestations mediated by the immune system such as arthralgia and rash can be blunted or absent in these patients. Anemia is, however, almost a constant finding, and leukopenia and thrombocytopenia can also be observed (227, 228). Graft loss or dysfunction and organ-invasive diseases (e.g., hepatitis, pneumonitis, and carditis) have been associated with B19V infection, but the causality remains unclear (227, 228). The diagnosis of B19V infection is confirmed by PCR using blood samples at onset of disease in most patients, as reported in a review of the literature, including 74 SOT patients (53 kidney, 12 heart/lung, 9 liver) with positive PCR in 87% of patients (228).
Regarding HSCT recipients, rash, cytopenia, and arthralgia were observed in 50%, 50%, and 11%, respectively, of 28 allo-HSCT recipients infected with B19V that were identified by literature review (229). Most patients developed a rash early after HSCT (≤3 months), whereas anemia occurred later after HSCT (>3 months). The median time of onset of manifestations was 2.7 months (range, 0.1 to 96 months) (229). Organ diseases involving heart, liver, or lung were also described in this study, as well as in other studies of pediatric and adult patients; for SOT recipients, causality remains unclear (230–232). In a review by Katoh et al., the diagnosis of B19V infection was confirmed by PCR using blood or other samples (bone marrow specimens, skin specimens, heart, liver, or lung) in 26/28 (92.9%) cases and viremia was present in all patients (229). The detection of B19V in blood samples at the onset of disease was also a constant finding in another review that included 24 HSCT recipients (20 allogeneic and 4 autologous transplant recipients) (228).
(b) Detection in blood samples of nontransplanted individuals.
Several reports concerning blood donors have reported prevalence rates of B19V DNA detection in blood samples ranging from 0.008% to 1.9%. Among 3,957 Chinese blood donors, B19V DNA was detected by real-time PCR in plasma samples of 23 (0.58%) subjects, with viral loads ranging from 2.48E2 to 6.38E4 copies/ml (211). A meta-analysis including 48,923 Chinese blood donors reported B19V DNA detection in 0.7% of subjects (95% confidence interval [CI], 0.2% to 2.4%) (213). In a cohort of Australian blood donors, B19V genome was detected in the plasma samples of 10/4,232 (0.24%) patients, with a mean viral load of 3.17E3 IU/ml (standard deviation, 1.77E6 IU/ml) (224). A large German study investigated B19V infection in plasma samples of 139,155 blood donations from 2006 to 2008 and revealed positive cases in only 11 (0.008%) donations (233). Among Brazilian blood donors, B19V was detected in plasma samples of 1/477 (0.21%) donors (234). In another Brazilian study, B19V DNA was detected in the plasma samples of 9/480 (1.9%) individuals with viral loads ranging from 2.7E3 to 6.7E5 copies/ml, and IgG was concomitantly detected in 7/9 (77.8%) individuals (216). No related posttransfusion event was reported in these two studies (216, 234). Similar results were reported in an Iranian study that included 500 donors, with B19V detected in plasma samples of 1.2% of individuals, with viral loads of >1E6 copies/ml, and IgG was concurrently detected in 4/6 (66.7%) cases (212). Finally, a Japanese population-based epidemiological survey of B19V infection in which B19V was screened by real-time PCR in serum samples of 2,081 individuals revealed that 0.2% of subjects were positive (210).
(c) Detection in blood samples of SOT recipients.
The prevalence of B19V infection after transplantation is difficult to estimate, considering different diagnostic procedures (serology versus PCR), patient selection bias, and diverse durations of follow-up. B19V infection appears to be uncommon or is rarely reported among adult and pediatric transplant recipients (228–230).
Among a cohort of 137 Italian SOT recipients (58 liver, 59 kidney, and 20 heart transplant) retrospectively screened for B19V by a competitive PCR enzyme immunoassay on serum samples collected up to 6 months after transplantation, 2 patients (1.5%) were found to be positive for B19V, including one patient with a febrile episode (235). In a German study including 268 SOT recipients (147 kidney, 91 liver, 30 heart transplant), the screening of B19V using real-time PCR in serum and plasma samples collected before transplantation and up to 6 months after transplantation revealed positive results with low viral loads (<600 to 1,100 genome equivalent [geq]/ml) (limit of quantification of the assay, 600 geq/ml or 215 IU/ml) in 9 patients (3.4%), with positive results before transplantation in some patients (236). As reported in most of the studies reviewed here, genotype 1 was the most frequent (88.9%). All except two heart transplant recipients were IgG positive before transplantation. The presence of B19V viremia was not associated with anemia (236). In the same study, the authors examined B19V DNA detection in healthy adults, which was similar in terms of prevalence (3/120 [2.5%] individuals) and viral loads to that of the SOT recipients (236). The prevalence of B19V DNA detection was reported at 1.8% in a U.K. cohort of 110 adult transplant recipients screened for B19V in plasma samples by nested PCR (237). In one of these cases, B19V might have been transmitted via kidney transplantation. In the other case, B19V infection has been suggested as a cause of graft failure. Epidemics of B19V infection have also been reported among SOT recipients, as described in a study by Ki et al. that retrospectively screened the sera of 167 adult kidney transplant recipients, by qualitative and quantitative real-time PCR, every 2 months over a follow-up period of 6 months after transplantation (238). Fifty-two of 167 patients (31.1%) had at least one sample positive, including 20 patients with B19V-positive samples during 2 months or more (238).
The prevalence of B19V infection is higher among SOT recipients presenting with anemia. In a group of 48 Italian adult renal transplant recipients with anemia, B19V infection was confirmed by positive nested PCR in serum samples of 11 patients (23%) and occurred in 73% of patients within 3 months after transplantation (239). In another group of 19 U.S. adult liver transplant recipients with anemia, the screening of serum samples for B19V by classic PCR revealed that 6 patients (31.6%) were viremic (240). The viral load of B19V among recipients with anemia has been reported at levels up to 1E9 copies/ml (241–243). Of note, qualitative PCR assays are often used, and viral loads are therefore not described.
Concerning pediatric SOT recipients, a Swiss study investigated the prevalence of B19V infection among 54 pediatric liver transplant recipients who were followed up over a 6-month period after transplantation (242). B19V DNA was detected in blood samples of 5/54 patients (9.2%) with viral loads of 1E3 geq/ml in most samples. In all cases, viremia was observed within the first 3 months after transplantation. B19V viremia was more frequent in tacrolimus-based regimens than in cyclosporine-based regimens (242).
(d) Detection in blood samples of HSCT recipients.
B19V detection has been reported at similar prevalence rates in HSCT recipients. B19V DNA was detected in a Swedish cohort of 77 adult allo-HSCT recipients by quantitative real-time PCR in serum samples of 2 patients (2.6%), one at 3 months and one at 1 year after transplantation, without any clinical manifestations despite peak viral loads of 2E5 and 4.7E8 copies/ml, respectively (244). In the same study, 1/20 (5%) pediatric allo-HSCT recipients was found to be B19V viremic at 3 weeks after transplantation, with a peak viral load of 1E5 copies/ml (244). In a Finnish study, B19V DNA was screened by classic PCR using serum samples of 201 allo-HSCT recipients, who were all found to be negative during the first 6 months after HSCT; of 119 patients with available samples for screening during a 26-month period after HSCT, B19V was detected in 3 patients (2.5%) (245).
Among a cohort of 171 Turkish pediatric allo-HSCT recipients screened weekly and retrospectively for B19V, using real-time PCR in plasma samples during a follow-up period of 2 years, 12 patients (7%) were found to be positive (246). B19V infection occurred in the first 100 days after HSCT in 91.7% of patients, and B19V viremia was cleared after 1 month in 41.7% of patients (246). In a Japanese study investigating viral infections among 24 pediatric and adult auto-HSCT recipients by screening whole-blood samples with a real-time multiplex assay targeting 13 viruses, B19V was identified in one patient (4.2%) (64). Another study by the same group performed the same investigations among 105 pediatric and adult allo-HSCT recipients in which B19V was not detected (63). A Finnish study revealed high prevalence rates of B19V DNA detection in serum samples of pediatric allo-HSCT, at 30.2% (16/53) of patients (247). The mean viral loads were 4.6E3 copies/ml before transplantation (9/53 patients) and 9.9E7 copies/ml, 1.1E10 copies/ml, and 1.6E2 copies/ml at 1 (6/53), 2 (4/53), and 3 months (1/25) after HSCT, respectively (247). Interestingly, no clinical manifestations were associated with B19V viremia. The authors did not report the occurrence of any outbreak during the period of the study.
The transmission of B19V might rarely occur via HSCT, as suggested in case reports of auto- and allo-HSCT (248, 249).
(ii) Human parvovirus 4.
(a) Overview.
According to the ICTV classification (https://talk.ictvonline.org/ictv-reports/ictv_online_report/), human parvovirus 4 (PARV4) is a member of the Parvoviridae family (genus Tetraparvovirus) and comprises three genotypes, G1 to G3, with genotype G1 circulating predominantly in Europe and North America, G2 in Asia and Brazil, and G3 in Africa (250). PARV4 was first identified in 2005 in plasma samples of individuals with symptoms of acute viral infection following high-risk behavior for HIV infection (251). The transmission route remains unclear. Higher seroprevalence rates among intravenous drug users and polytransfused patients might suggest that PARV4 is a blood-borne virus (250, 252, 253), even though the high seroprevalence rates in other populations suggest other routes of transmission (254, 255). The clinical manifestations associated with PARV4 infection remain poorly understood (250). Some data suggest that PARV4 has the potential for latency and reactivation (250).
The seroprevalence in several cohorts from Africa was estimated at 37% among 167 blood donors in Burkina Faso, 25% among 238 individuals in the general population in Cameroon, 35% among 221 military members of the Democratic Republic of the Congo, and 20% among 170 blood donors in South Africa (255). In another cohort of pediatric and adult individuals from the South African general population in which 69% were HIV-infected patients, 48% (238/492) of adults and 21% (50/234) of children were PARV4 IgG seropositive. The prevalence was higher in HIV-positive (52%) than HIV-negative (24%) individuals (256). The higher prevalence among adults than in children was confirmed in another cohort of 157 adult and pediatric outpatients in South Africa, with prevalence rates of 49% and 33% in adults and children, respectively (254). In Europe, the reported seroprevalence rates include 0.9% among 228 Danish women (257), 4.76% among 608 U.K. blood donors (258), 11% among 99 HIV-infected men who have sex with men, and 95% of 94 intravenous drug-using HIV-infected patients from Switzerland (259).
(b) Detection in blood samples of nontransplanted individuals.
Among 289 adults from China, PARV4 DNA was detected in 20.4% of subjects (260). PARV4 DNA was detected in the plasma of 32 (8.9%) of 361 children from Ghana (median age, 15 months), with a median viral load of 4.53E2 copies/ml (range, 2E2 to 3E4 copies/ml) (261). The detection of PARV4 DNA in blood samples taken several months apart in the same subjects raises the question of persistent infection or reactivation (262). Among blood donors, the variation of reported PARV4 DNA detection rates using real-time or nested PCR assays in blood samples included 1% in an Italian cohort (n = 100) (263), 2% in a French cohort (n = 304) (264), 3.95% in a Thai cohort (n = 176) (252), and 16.6% in an Iranian cohort (n = 120) (265). Screening of PARV4 DNA with real-time PCR in 29 IgG4-seropositive blood donor sera from the United Kingdom yielded only negative results, possibly due to the low number of included patients (258). PARV4 DNA was also detected in manufacturing plasma pools, with a prevalence rate ranging from 4% (14/351 pools) (266) to 26.2% (51/195 pools) and viral loads ranging from 2.8E3 copies/ml to 2.4E7 copies/ml (267). The lower prevalence rates in pooled plasma than in individual samples in some studies need to be interpreted when considering the dilution of individual samples (266). The high prevalence rate and viral loads of PARV4 in blood products and the lack of data regarding potential associated clinical manifestations might raise concerns regarding transfusion safety.
(c) Detection in blood samples of SOT recipients.
Data are scarce among transplant recipients, but PARV4 DNA was detected in 13.5% of a French cohort of 104 adult lung transplant recipients (268). In an Italian study in which PARV4 was screened by nested PCR in serum samples of 126 kidney transplant recipients, collected at 6 and 12 months posttransplantation, results were positive in 1.6% of patients (263).
(d) Detection in blood samples of HSCT recipients.
In the same Italian study, the screening of PARV4 by nested PCR in serum samples of 107 allo-HSCT recipients revealed only negative results (263). Among 53 pediatric allo-HSCT recipients from Finland, none of the sera tested by real-time PCR were positive for PARV4 DNA (247).
(iii) Human bocavirus.
(a) Overview.
Human bocavirus (HBoV) belongs to the Parvovirinae subfamily of the Parvoviridae family. The subfamily is divided into eight genera comprising the genus Erythroparvovirus, which includes parvovirus B19, and the genus Bocaparvovirus (204, 269). HBoV comprises two species (primate bocaparvovirus 1 and 2) and four variants (HBoV1, 2a to c, 3, and 4) (204, 269). HBoV1 was first identified in 2005 by random PCR performed on nasopharyngeal aspirates of children with respiratory tract infections (270); during 2009 to 2010, three other HBoVs (HBoV2 to -4) were identified in stool samples (271–273). The mechanisms of persistence, latency, reactivation, and reinfection are poorly understood. HBoV circulates worldwide, but in contrast to HBoV2 to -4, for which the pathogenic role has been poorly described, HBoV1 has been associated with respiratory tract disease. High viral loads of HBoV1 in respiratory samples are often associated with symptoms, but the pathogenic contribution of HBoV1, especially in coinfections, remains unclear (204, 274–277).
Most seroepidemiological investigations have been conducted among patients with respiratory infections (275, 278–280) and are therefore not representative of the seroprevalence in the general population. Among asymptomatic patients, most seroepidemiological studies were conducted in China (281–283). In a study of 884 healthy pediatric and adult subjects, the seroprevalence was 69.2% for HBoV1 and 64.4% for HBoV2, with the highest prevalence rates in children aged 3 to 5 years (281). Of note, the cross-reactivity of HBoV1 and HBoV2 may have contributed to an overestimation of prevalence (281). Another study reported prevalence rates for HBoV ranging from 59.1% to 78.7% in sera from 400 and 141 adults collected in 1996 to 1997 and 2005, respectively (282). Among 6,096 samples from plasma donors, the seroprevalence of HBoV1 was 13% in samples with undetectable viremia and 20.2% in those with detectable viremia (283).
(b) Detection in blood samples of nontransplanted individuals.
The detection of HBoV DNA in blood during acute respiratory disease has been reported in nonimmunocompromised patients and pediatric HSCT recipients (284–287). A Chinese study screened 1,626 blood samples from asymptomatic children and adults, and 3.8% were positive for HBoV (288). HBoV DNA detection in blood donor samples has been reported in several studies. In a Chinese study, the screening of 6,096 plasma samples of blood donors revealed 9.06% samples to be positive, with high viral loads ranging from 5E2 to 3E6 copies/ml (283). In an Italian study, the screening of 272 serum samples revealed detection of HBoV1 or HBoV2 DNA in 5.5% of samples, with viral loads ranging from 1.8E2 to 3.3E3 copies/ml (289). The real-time PCR screening of 300 whole-blood samples from blood donors in Saudi Arabia gave positive results in 7% of individuals, with low viral loads (<1E3 copies/ml) (290). In a French study, 167 manufacturing plasma pools from blood donors proved to be HBoV DNA negative when screened, possibly reflecting dilution below the detection limits for individual low viral load samples (266).
(c) Detection in blood samples of SOT recipients.
There is currently no published study regarding the screening of HBoV in blood samples of SOT recipients.
(d) Detection in blood samples of HSCT recipients.
An Australian study screened whole-blood samples collected from 31 immunocompromised children with acute leukemia or post-stem cell transplant recipients without any symptoms by real-time PCR for HBoV and found positive results in 2.6% specimens, with one patient accounting for 50% of the positive samples (291). HBoV1 to -4 infection was investigated in 19 Brazilian adult allo-HSCT recipients by real-time PCR performed on 130 sera. HBoV1 to -4 DNA was detected in 9.2% of samples, with median viral loads of 3.6E5 copies/ml (range, 4.6E4 to 1.9E6 copies/ml), and with a first detection time ranging from 8 days before to 216 days after transplantation; most patients were either asymptomatic or had gastrointestinal symptoms (292). In a longitudinal study from Finland of 53 pediatric HSCT recipients, no patient had detectable HBoV1 DNA detected in serum samples (247).
Single-Stranded DNA Viruses
Anelloviridae.
(i) Torque teno virus, torque teno mini virus, and torque teno midi virus.
(a) Overview.
Anelloviridae are nonenveloped viruses containing a circular, negative, single-stranded DNA (ssDNA) genome (293), and the family is classified into several genera: Alphatorquevirus, comprising torque teno virus (TTV); Betatorquevirus, comprising torque teno mini virus (TTMV); and Gammatorquevirus, comprising torque teno midi virus (TTMDV) (293). According to the ICTV classification (https://talk.ictvonline.org/ictv-reports/ictv_online_report/), the cutoff value for species distinction is 35% of nucleotide sequence identity. The Alphatorquevirus genus includes 29 species (TTV 1 to 29), the Betatorquevirus genus includes 12 species (TTMV 1 to 12), and the Gammatorquevirus genus includes two species (TTMDV 1 and 2). TTV is the most well-described anellovirus, and the 29 species are classified into five genogroups with at least 50% nucleotide sequence divergence and further into genotypes with differences of up to 30% in genomic sequence (293). In humans, more than 1E10 TTV virions are produced daily, with 90% of virions circulating in the plasma replaced every day (294). Different genotypes can coinfect the same individual (295). The genotypic variability of TTV detected among different tissues of the same individual may reflect a compartmentalization of TTV infection (296, 297). This genotypic diversity has contributed to difficulties in the development of serological assays and thus the precise estimation of seroprevalence. The seroprevalence of TTV varies according to geographical location and is globally estimated to be higher than 50% in the general population (293). A European seroprevalence study performed among 178 adults and 108 children in 2013 using an assay targeting five TTV strains belonging to four genogroups revealed that seroprevalence rates reach 43% in healthy children aged 2 to 4 years and 42 % in healthy adults (298).
Anelloviruses are presumed to be transmitted by blood transfusion, but their detection in various biological samples (e.g., nasal secretion, saliva, feces) suggests combined modes of transmission such as fecal-oral, respiratory, sexual, and vertical transmission or through transplantation (299). The association between anellovirus infection and specific diseases remains unproven. According to the current state of knowledge, considering the fact that most humans are either chronically infected or continuously reinfected, the likelihood that anelloviruses cause diseases is low, and this raises the possibility that anelloviruses are commensal with humans (299).
Despite no currently existing culture model for TTV, several data suggest viral replication in PBMCs and lymphocytes (300–302). TTV DNA has been detected in blood, plasma, cord blood, and PMBCs (58, 297, 300, 303–305). In PBMCs and other blood cells, the TTV genome has been detected in monocytes and natural killer cells, as well as in B and T lymphocytes (297, 300, 303, 306). TTV has also been detected in bone marrow, spleen (296, 307), and other diverse clinical specimens (296, 308–317).
(b) Detection in blood samples of nontransplanted individuals.
The prevalence and relative abundance of TTV in studies using mNGS on blood samples are difficult to estimate considering the heterogeneity of mNGS techniques and the reporting methods in publications. However, Anelloviridae are considered major members of the human blood virome, and some studies have reported them as the most abundant virus in blood samples (28, 29, 305). Recent metagenomics studies demonstrated a high prevalence of Anelloviridae genome detection in almost all analyzed blood pools (58, 305). Studies in which the prevalence of TTV DNA detection in blood was determined using real-time PCR assays revealed a higher prevalence among polytransfused patients and intravenous drug users than in individuals described as healthy. The high prevalence of TTV reported among polytransfused beta-thalassemic patients might underline infection through blood transfusion with diverse genotypes (318, 319). Among individuals described as healthy, the prevalence of TTV DNA detection in blood varies according to the geographical location, the different genotypes coinfecting the same individual, and the PCR assays used (304, 318–322). The results of screening of TTV DNA in healthy Swiss adults were shown to be positive in 51/74 (69%) individuals, with a median plasmatic viral load of 1.7E2 copies/ml (range, 0 to 5.4E4 copies/ml) (320). TTV DNA was detected using specific conventional PCR in serum samples of 23/39 (59%) healthy Japanese adults (304) and in 47/91 (51.6%) serum samples of blood donors from Turkey using heminested PCR (319). Among a Romanian cohort of 701 individuals described as healthy, the screening of TTMV, TTMDV, and TTV DNA by heminested PCR in whole-blood samples was positive in 225 (40.1%), 325 (54.4%), and 410 (68.2%) individuals, respectively; codetection was frequent (321). A Japanese study in which specific real-time PCR assays were performed on serum samples of 305 healthy individuals (100 adults, 205 children, and 44 infants of ≤1 year old) revealed that TTMDV, TTMV, and TTV DNA was detected in 75.2%, 84.8%, and 93.3% of subjects, respectively (322).
(c) Detection in blood samples of SOT recipients.
Regarding SOT recipients, a U.S. metagenomics study revealed that Anelloviridae represent 68% of the total DNA viral sequences detected in the blood of heart and lung transplant recipients (29). In this study, the relative abundance of Anelloviridae changed with time after transplantation, with a rapid increase during the first 6 months posttransplantation in parallel with a decrease in the relative abundance of the Herpesviridae and Adenoviridae. Thereafter, an opposite trend was observed, depending on antiviral and immunosuppressive drug administration (29).
As demonstrated in other studies, the type of immunosuppressive regimen and the administration of anti-T lymphocyte drugs after transplantation are associated with TTV viremia and viral loads (323–326). In an Italian study performed in 280 adult liver or kidney transplant recipients whose plasma samples were screened using real-time PCR for TTV during 1 year of follow-up, 92% of patients had detectable TTV DNA in plasma samples before transplantation (325). TTV viremia evolved according to the immunosuppressive regimen, with an increase in viral loads to a peak of 1E6.9 copies/ml at day 90 in kidney transplant recipients and 1E6.5 copies/ml at day 80 in liver transplant recipients (325). In contrast, TTV DNA levels were not correlated with immunosuppressive trough levels in a German cohort of 34 lung transplant recipients (327). Of note, all patients had detectable TTV DNA in plasma samples collected after transplantation (327). In a recent Spanish study of 63 adult liver transplant recipients, TTV DNA was detectable in the plasma samples of 93.7% of patients before transplantation (mean viral load, 1E4.14 copies/ml; IQR, 1E3.42 to 1E5.08 copies/ml) and in all patients posttransplantation (328). TTV DNA increased after liver transplantation, with a peak 3 months posttransplantation (mean viral load, 1E7.01 copies/ml; IQR, 1E6.05 to 1E8.08 copies/ml) and decreased after 6 months (328). These kinetics demonstrate an increase of TTV viral load reaching a peak (up >1E9.5 copies/ml) at 3 months posttransplantation, and similar increases have also been reported among other cohorts of lung transplant recipients (327, 329).
The potential role of TTV as a marker of immunosuppression or as a predictor of clinical outcomes among adult SOT recipients has recently been investigated in several studies (325, 327–330). A study by Ruiz et al. of TTV in adult liver transplant recipients suggested such a potential role by revealing that TTV viral loads were significantly lower during episodes of acute cellular rejection (328). Another study among adult lung transplant recipients reported that among patients with biopsy-proven rejection, a 10-fold decrease in TTV plasmatic viral load had a sensitivity of 0.74 and a specificity of 0.99 for biopsy-proven rejection (327). In the study by Ruiz et al., the area under the curve of TTV viral load (calculated using TTV mean viral loads at several time points during 1 year of follow-up) was significantly higher among adult patients receiving triple immunosuppressive therapy than in those receiving double immunosuppressive therapy (328). Furthermore, TTV viral loads were significantly higher during CMV infections (328). In an Italian study of a cohort of 280 adult liver and kidney transplant recipients, the plasmatic TTV viral load measured from days 0 to 10 posttransplantation was significantly higher in patients with CMV DNA-positive samples than in those with CMV-negative samples during the first 4 months after transplantation. A TTV viral load above 1E3.45 copies/ml within the first 10 days posttransplant was associated with a higher risk of CMV reactivation (325). In two prospective studies from Austria of two cohorts of 143 adult lung transplant recipients and 169 kidney transplant recipients, a TTV viral load above 3E9 copies/ml and TTV viral load kinetics were associated with the occurrence of infections posttransplantation (including bacterial, fungal, and viral infections) (329, 330). Of the 110/143 adult lung transplant recipients for which the pretransplantation plasma samples were available, 91/110 (82.3%) tested positive for TTV DNA by using real-time PCR, and TTV DNA was detected in all patients after transplantation (329). The same study revealed that the highest TTV viral load during the first 3 months posttransplantation was associated with the risk of infection, with a 5-fold increased risk for each 10-fold increase in viral load (329). Among 169 adult kidney transplant recipients, 80% of patients had detectable TTV DNA in plasma samples screened with real-time PCR, with a median viral load of 1.9E4 copies/ml (IQR, 2.4E3 to 8E4 copies/ml) (330). As reported in other studies described above, the TTV viremia kinetics peaked at 3 months after transplantation, with a median viral load of 4.3E8 copies/ml (IQR, 3.6E7 to 2.9E9 copies/ml), followed by a reduction to a mean viral load of 4.2E6 copies/ml at 9 months posttransplantation (330). The mean TTV viral load at stabilization of viremia (after 3 months posttransplantation) was associated with the development of further infectious complications (330). Taken together, these results suggest that the TTV viral load might be used as a predictor for clinical outcomes after transplantation and might provide relevant information for the management of the immunosuppressive regimen among adult SOT patients.
(d) Detection in blood samples of HSCT recipients.
TTV infection is highly prevalent among patients with hematological disease before HSCT. Plasmatic TTV DNA was detected in 44/55 (80%) patients in a Spanish cohort (331) and 40/50 (80%) patients in an Austrian cohort with a median plasmatic viral load of 1E5.37 copies/ml (IQR, 1E3.51 to 1E6.44 copies/ml) (332). Albert et al. (331) showed that TTV plasmatic viral loads before conditioning and at the time of allo-HSCT were comparable, irrespective of the type of hematological disease. In a study by Masouridi-Levrat et al. (320), the TTV plasmatic viral load at the time of transplantation was higher among patients suffering from acute lymphocytic leukemia and non-Hodgkin’s lymphoma, irrespective of the conditioning regimen. Finally, recent results from a German cohort of 123 adult allo-HSCT recipients showed that TTV was detected in whole-blood samples of 62 (50.4%) patients before transplantation and that the TTV viral load was significantly higher in patients with an underlying lymphatic malignancy (20/26, 76.9%) than in those with a myeloid malignancy (42/97, 43.3%) (333).
TTV viremia has been shown to be sustained and to occur almost universally after allo-HSCT. This has been demonstrated in several studies, including an Austrian study that included 50 patients for which TTV screening was performed by real-time PCR in plasma samples every month for 1 year after transplantation (332). In this study, the peak TTV viral load was reached at day 79 (IQR, 50 to 117 days) after transplantation and with peak viral loads of approximately 8E10 copies/ml. In a German study that included 23 allo-HSCT recipients screened for TTV with real-time PCR in whole-blood samples at days 30, 100, and 200 after transplantation, all patients had TTV DNA detected after transplantation. In a group of high-risk transplant recipients who developed acute GvHD requiring escalation of immunosuppression and/or persisting or symptomatic non-TTV viral infections such as EBV or CMV reactivations, the median peak viral load was 1.4E10 copies/ml (range, 1.0E7 to 1.87E12 copies/ml) at day 100 after transplantation (334). TTV DNA was also detected in the plasma samples of all 33 allo-HSCT recipients after transplantation and continued to be detected in all available samples until 210 days after transplantation (335).
In a Swiss study including 40 adult HSCT recipients for whom plasma samples were analyzed by mNGS at day 30 after transplantation, sequences of anelloviruses were identified in 16/40 (40%) patients: TTV sequences were identified in 15 patients, TTMV sequences were identified in four patients, TTMDV sequences were identified in two patients, and codetections were observed in 5 patients (336).
Similarly, as in SOT recipients, TTV viremia evolves over time after allo-HSCT. In parallel with the absolute lymphocyte count, there is a decrease in TTV plasmatic DNA load after conditioning (7, 333, 335, 337). In several studies, the TTV peak viral load was reported at 80 to 100 days after HSCT, with viral loads up to 1E12 copies/ml, followed by a stable plateau over 1 year after transplantation (331–335, 338). The TTV viral load increase paralleled the increase of the absolute lymphocyte count (331, 333, 338), and TTV viremia was correlated with the absolute lymphocyte count after engraftment (332). Furthermore, the type of malignancy, conditioning regimen, the use of antithymocyte globulin, and acute GvHD requiring systemic treatment and particularly the use of corticosteroids are associated with TTV viremia dynamics and viral loads (332, 335). All these data on TTV viremia dynamics after transplantation could support the fact that lymphocytes are an important site of replication for TTV.
A German study recently investigated the role of TTV as a potential marker for the degree of immunosuppression among HSCT recipients (333). In a cohort of 123 adult allo-HSCT recipients, higher TTV viral loads in whole-blood samples after HSCT was significantly associated with complete remission before allo-HSCT. Nevertheless, TTV viral load was not associated with viral reactivations (CMV, EBV, and adenovirus), different T and B cell counts, acute GvHD, relapse, or death (333). Another German study of 50 allo-HSCT recipients failed to demonstrate that TTV viral loads could predict immune-related complications, such GvHD, disease relapse, and CMV or EBV reactivation (332). In a study by Albert et al., the median area under the curve of TTV DNA levels between days 90 and 210 was significantly higher in patients who received corticosteroids within this time frame for treatment of acute or chronic GvHD than in controls (335). Finally, in a recent study of 168 allo-HSCT recipients, higher TTV viral loads at 100 days after transplantation have been associated with worse overall survival and a higher risk of acute GvHD and infections (339). In summary, despite the fact that TTV might mirror the degree of immunosuppression, further data are needed to assess the role of TTV as a potential marker of immunosuppression among HSCT recipients.
(ii) Gyrovirus.
(a) Overview.
The genus Gyrovirus (GyV) has recently been reassigned to the Anelloviridae family (340) and comprises eight species, including species identified in avian and human samples. The human GyV genome was detected in an Italian study in 3/50 (6%) kidney transplant recipients by using real-time PCR (with primers targeting the VP1 gene) with plasma samples (341).
In the absence of any proof of replication in cell cultures, any animal model, and any available serological assay, the tropism, pathogenic mechanisms, and seroprevalence of GyV still remain unknown. The detection of GyV DNA in human clinical samples such as stools (in which avian and human species have been detected) may reflect poultry product consumption without human infection (342). Furthermore, data have shown that several viral genomes detected in human samples are in fact linked to laboratory reagents and, despite the report by Asplund et al. (23) of no such detection, this hypothesis should be taken into consideration.
(b) Detection in blood samples of nontransplanted individuals.
In the same study in which GyV was first identified, the screening of plasma samples of 50 blood donors and 50 liver transplant recipients was negative (341).
In another study, human GyV DNA was detected with a low prevalence in 3/352 (0.85%) plasma samples of blood donors in France by using an in-house real-time PCR assay (with primers targeting the VP2 gene) (343). Biagini et al. repeated the screening of the same samples with the PCR assay used in the study of Maggi et al. (341) and obtained negative results (343), thus underlining the importance of the performance and targets of the PCR assays used in these studies. Human GyV was not detected using real-time PCR in purified CD34+ hematopoietic stem cells or in umbilical cord blood (PCR with primers targeting the VP1 gene) (344).
(c) Detection in blood samples of SOT and HSCT recipients.
The detection of GyV in blood samples of SOT and HSCT recipients has not been studied so far.
Circoviridae.
(i) Cyclovirus.
(a) Overview.
Cycloviruses (CyCVs) are classified in a clade close to the circovirus clade. CyCVs are nonenveloped viruses with a circular single-stranded DNA genome. CyCV-VS5700009 was identified using mNGS for the screening of cerebrospinal fluid (CSF) samples of adult patients from Malawi with unexplained paraplegia (345). Despite the identification of CyCV DNA in human clinical samples, there is no proof of replication in human cells. There is currently no available serological assay. The transmission route, tropism, pathogenic mechanisms, and potential association with human diseases still need to be clarified. Recent data suggest that CyCV detected in human samples might originate from parasitic arthropods on the skin, even if certain CyCVs, such as CyCV-VN, for instance, belong to a lineage exclusively detected in mammals that is distinct from arthropod lineages (346).
(b) Detection in blood samples of nontransplanted individuals.
After the identification of CyCV-VS5700009, further screening using real-time PCR revealed 8/54 (15%) positive serum samples and 4/40 (10%) positive CSF samples (345). However, these results were challenged by data demonstrating potential false-positive results with the same real-time PCR assay (347). The screening of another species, cyclovirus Vietnam (CyCV-VN), using real-time PCR with serum samples of 79 healthy adult individuals, produced negative results (348). In a study investigating the etiology of community-acquired sepsis with mNGS in 384 sera of pediatric and adult patients from Vietnam, sequences of CyCV-VN were identified in one (0.3%) sample (349). In the context of a virus discovery study, mNGS was performed on pooled plasma samples (from 352 individual samples) of blood donors from seven African countries (Mauritania [n = 44], Mali [n = 52], Niger [n = 28], Cameroon [n = 68], Burundi [n = 40], Madagascar [n = 48], Democratic Republic of the Congo [n = 72]) and led to the detection of sequences of CyCV-VN. The analysis by seminested PCR of unpooled plasma samples revealed a prevalence of 38.1% (16/42 samples) and of 43.1% when 50 additional samples were screened (350), with all positive samples coming from patients from Madagascar (350). These results could be explained by the circulation of different CyCV-VN variants across these African regions.
(c) Detection in blood samples of SOT recipients.
CyCV-VN was identified by real-time PCR in serum samples of 1/32 (3%) adult kidney transplant recipients (348).
(d) Detection in blood samples of HSCT recipients.
There is currently no study that investigated the detection of CyCV in blood samples of HSCT recipients.
Genomoviridae.
(i) Gemycircularvirus.
(a) Overview.
Gemycircularvirus (Gemini-like myco-infecting circular virus) is the sole genus of the Mycodnaviridae family of small nonenveloped viruses with a genome consisting of circular ssDNA and are thus classified as circular Rep-encoding ssDNA (CRESS DNA) viruses (351). These viruses are widespread in the environment, and the only reported hosts are fungi. The detection of the gemicircularvirus genome in human clinical samples raises several hypotheses, e.g., infection of some commensal or pathogenic fungi in human hosts, true human infection, or skin or reagent contamination. The transmission route, tropism, pathogenic mechanisms, and potential association with human diseases are still not established.
(b) Detection in blood samples of nontransplanted individuals.
Gemycircularvirus is identified at a low prevalence in blood samples. A German group identified gemycircularvirus DNA using rolling circle amplification in serum and brain samples of a patient with multiple sclerosis (352). mNGS analysis of European and U.S. plasma pools of blood donors allowed the detection of gemycircularvirus sequences in one pool (353). Screening of blood samples of 256 blood donors in France obtained negative results (354). In an Italian study in which the screening of plasma samples was performed using real-time PCR assays targeting nine gemycircularvirus species, gemycircularvirus DNA was identified in 1/290 (0.3%) healthy individuals (355). In a recent study investigating the etiology of community-acquired sepsis by mNGS in 384 sera from pediatric and adult patients from Vietnam, sequences of gemycircularvirus were identified and confirmed by specific PCR in 3 (0.4%) samples (349).
(c) Detection in blood samples of SOT recipients.
The detection of gemycircularvirus in blood samples of SOT recipients has not been studied.
(d) Detection in blood samples of HSCT recipients.
Among 65 Italian HSCT recipients, the screening of plasma samples using real-time PCR assays targeting nine gemycircularvirus species gave positive results in one patient (1.5%) (355).
RNA VIRUSES
Positive ssRNA viruses
Flaviviridae.
(i) Human pegivirus 1.
(a) Overview.
According to the 10th Report of the ICTV in 2018 (https://talk.ictvonline.org/ictv-reports/ictv_online_report/), Pegivirus genera are members of the Flaviviridae family and are classified as comprising pegivirus species A to K. The novel human pegivirus 2 was independently identified in 2015 and has provisionally been named human hepegivirus 1 (HHpgV-1) by Kapoor et al. (356) or human pegivirus 2 (HPgV-2) by Berg et al. (357). An update of the taxonomy of pegiviruses was proposed in 2016 by Smith et al. (358), and pegivirus C now corresponds to pegivirus 1 and pegivirus H corresponds to pegivirus 2. HPgV-1 was independently discovered by two groups in 1995 and 1996 (359, 360). This enveloped virus contains a positive-sense RNA genome (361), and seven genotypes have been described with a varied geographical distribution (362–364). The tropism of HPgV-1 was initially thought to be hepatic, as the virus had been detected in patients with non-A/B/C hepatitis, but further studies found no evidence of replication in the liver and instead demonstrated a tropism for lymphocytes (361). Coinfection with HCV has been reported in some studies (365–367).
Seroprevalence is assessed by the detection of antibodies targeting the envelope protein E2. In contrast to hepatitis C virus (HCV; another member of the Flaviviridae) for which chronic infection is associated with concomitant circulating antibodies, the development of antibodies directed against the E2 glycoprotein of HPgV-1 is associated with viral clearance (361, 368, 369). In a large U.S. cohort of healthy individuals, 84/1,316 (6.4%) subjects were seropositive for HPgV-1 (370). This seroprevalence rate was 14% in another cohort of blood donors, 32% among hemophiliacs, and 54% among HCV chronically infected patients and intravenous drug users (369). In a recent meta-analysis investigating the prevalence of HPgV-1 DNA detection in blood donors worldwide, the authors also analyzed HPgV-1 pooled seroprevalence in 19 studies including 8,925 individuals, revealing a prevalence of 9% (95% CI, 7.2% to 11.4%) (371).
HPgV-1 is considered to be responsible for persistent infection in immunocompetent patients, with HPgV-1 RNA detectable for several weeks to years, as shown in some longitudinal follow-up studies among healthy individuals (372, 373). HPgV-1 positive-sense RNA and negative-sense replication intermediates have been detected in bone marrow and the spleen, while only positive-sense RNA has been detected in PBMCs (361). Concerning PBMCs, HPgV-1 RNA has been detected in CD4/8 T lymphocytes and B lymphocytes, as well as natural killer cells and monocytes isolated from healthy individuals (374). The interactions between HPgV-1 and the immune system are only partially understood (361). So far, HPgV-1 infection has not been associated with any overt disease, although an association with non-Hodgkin’s lymphoma has been described (370, 375). HPgV-1 might be a commensal virus of humans (361). Some data from HIV-infected patients even raised the possibility of a potential benefit of HPgV infection. Coinfection of HIV and HPgV-1 has been reported in several studies in 8% to 23% of HIV-infected patients (362, 376–379). A recent study performed a retrospective analysis of data of 20 years of follow-up of HIV-infected patients and revealed that before the era of highly active antiretroviral therapy (HAART), patients coinfected with HIV and HPgV-1 (HPgV-1 viremic patients) had a significantly higher CD4 count, lower HIV viremia, and lower mortality due to AIDS-related diseases (infections and not lymphoma) than anti-E2-positive patients and nonviremic HPgV-1 patients (380). Several mechanisms of interaction between HPgV-1 and HIV have been studied to explain this effect (381). This advantage observed in the pre-HAART era disappeared with the systematic use of HAART (380).
(b) Detection in blood samples of nontransplanted individuals.
The prevalence of plasmatic HPgV-1 RNA detection has been reported to range from 0.5% to 13.7% among blood donors in diverse studies from Germany (n = 90), China (n = 506; n = 694), the United States (n = 1,316), and Qatar (n = 612) (365, 370, 382–384). In a recent study, the prevalence of HPgV-1 RNA detection by a nested PCR assay on plasma samples of 431 blood donors of the Brazilian Amazon was 9.5% (385). A recent meta-analysis of 102 studies, including 97,348 blood donors from China, reported a pooled prevalence of HPgV-1 RNA detection in whole-blood and plasma samples of 3.25% (95% CI, 2.35% to 4.26%) (386). Two metagenomics studies identified HPgV-1 and Anelloviridae sequences in pooled blood product samples from French and Swiss blood donors in most of the analyzed pools (58, 305). A recent meta-analysis investigated the prevalence of HPgV-1 in blood samples of blood donors through the review of 79 studies from several countries in North America, South America, Europe, Asia, Africa, and Oceania (371). Among a total of 35,468 blood donors, HPgV-1 DNA detection in blood samples was 3.1% (95% CI, 2.4% to 4.1%) (371). The authors assessed the pooled HPgV-1 prevalence rates according to continents: 1.7% (95% CI, 1.1% to 2.6%) in North America, 9.1% (95% CI, 6.4% to 12.7%) in South America, 2.3% (95% CI, 2% to 2.8%) in Europe, and 2.4% (95% CI, 1.4% to 4%) in Asia. Only two studies each were available for Africa and Australia, with reported prevalence rates of 8.8% to 18.9% for South Africa and 3.3% to 4% for Australia, respectively (371).
(c) Detection in blood samples of SOT recipients.
Among SOT recipients, the prevalence of HPgV-1 RNA detection in kidney transplant recipients ranges from 16.7% to 52.9% (387–391). Plasmatic viral loads range from 1E2 to 1E7 copies/ml. In an adult European cohort of 51 heart and 72 liver recipients, 36% and 41% of patients, respectively, had detectable HPgV-1 RNA in sera (392). In a recent Japanese study, the screening of whole-blood samples by reverse transcription-PCR (RT-PCR) among 313 liver transplant recipients was positive in 14.1% patients (393), but the authors did not identify any association of HPgV-1 RNA detection with clinical outcomes such as acute and chronic rejection and graft loss (393). Data are conflicting concerning the role of blood product transfusion in the transmission of HPgV-1 posttransplantation (384, 391, 392, 394). So far, no association has been demonstrated regarding HPgV-1 infection and clinical outcomes after transplantation, such as graft rejection or survival (392, 393).
(d) Detection in blood samples of HSCT recipients.
Among adult allo-HSCT recipients, HPgV-1 RNA plasmatic detection is frequent and occurred in 45/108 (41.7%) and 35/188 (18.6%) recipients in two Chinese cohorts (365, 384) and in 14/40 (35%) recipients in a Swiss cohort (28). In the latter study, HPgV-1 and anelloviruses were the most frequently identified viruses by mNGS (28). The HPgV-positive group of patients comprised higher proportions of patients with myeloid malignancies and a lower proportion of patients with lymphoid malignancies than the HPgV-1-negative patients (28). As in immunocompetent individuals, HPgV-1 is responsible for persistent infections, with detectable viremia for months after transplantation (395). The tropism of HPgV-1 for PBMCs and the high prevalence of HPgV-1 infection among blood donors raises the question of HPgV-1 transmission with HSCT, but data are conflicting. In a study by Ljungman et al. (396), all HPgV-1 RNA-negative patients before transplantation remained negative posttransplantation. In a study by Akiyama et al. (397), 11% of HPgV-1 RNA-negative patients before transplantation had de novo infections, irrespective of blood transfusions. The prevalence of HPgV-1 RNA detection in blood samples (without specification of blood sample type) was significantly higher among a cohort of 188 HSCT patients than among a cohort of 694 blood donors (18.6% and 2.3%, respectively) (384). In this study, the number of transfused blood products was significantly higher in patients for whom HPgV-1 RNA screening was initially negative and became positive in a second sample during follow-up (6 patients), in comparison to 40 patients for whom the test remained negative (40 patients) (384).
The impact of HPgV-1 in HSCT recipients requires further study. So far, HPgV-1 infection has not been significantly associated with clinical outcomes, such as GvHD or survival (28, 395, 396). Nevertheless, higher rates of progression-free survival and GvHD-free, relapse-free survival, as well as lower acute GvHD of grades 2 to 4 and relapse, were reported among HPgV-1-infected patients than among HPgV-1-negative patients, but these results were not significant (28).
(ii) Human pegivirus 2.
(a) Overview.
Human pegivirus 2 (HPgV-2) was identified by mNGS in the plasma of an HCV-infected patient (357). Codetection of antibodies to E2 and RNA is infrequent for HPgV-1 (5.88%) in comparison to HPgV-2, for which antibodies to E2 are detected in most HPgV-2 viremic individuals (92.86%), as observed in HCV chronically infected patients (357, 398).
As with HPgV-1, HPgV-2 transmission has been suggested via transfusion of blood products (356). So far, no other route of transmission has been studied and no study has evaluated the impact of HPgV2 infection.
(b) Detection in blood samples of nontransplanted individuals.
After the first identification of HPgV-2, the same group performed further real-time PCR screening of plasma samples in a U.S. cohort of 2,176 individuals that comprised patients infected with HIV, HBV, or HCV, as well as blood donors; plasma samples of 11/2,440 (0.45%) individuals were positive, with viral loads ranging from 1E2.5 to 1E6.6 copies/ml, and all blood donors (n = 476) had undetectable HPgV-2 RNA (357). More specifically, 19/452 (4.2%) blood donors, 71/742 (9.5%) HCV chronically infected patients, and 58/494 (11.7%) HIV-infected patients were HPgV-2 RNA positive (357). In a recent Chinese study, 6/4,017 (0.15%) blood donors were positive for anti-HPgV-2, and all were negative for HPgV-2 RNA (399). In the same study, 30/2,440 (1.23%) and 7/2,440 (0.29%) of HCV chronically infected patients were HPgV-2 seropositive and had detectable plasmatic RNA, respectively (399). Anti-HPgV-2 and HPgV-2 RNA detection was more prevalent among HCV-infected patients (399), and this potential association was reported by other groups (400, 401). Among 384 serum samples of pediatric and adult patients with community-acquired sepsis from Vietnam analyzed by mNGS, HPgV-2-specific sequences were identified in one (0.3%) sample (349).
(c) Detection in blood samples of SOT and HSCT recipients.
The detection of HPgV-2 in blood samples of SOT and HSCT recipients has not been studied so far.
Astroviridae.
(i) Novel human astrovirus.
(a) Overview.
According to the ICTV classification (https://talk.ictvonline.org/ictv-reports/ictv_online_report/), human astroviruses (HAstVs) are nonenveloped, single-stranded, positive-sense RNA viruses belonging to the Astroviridae family and are further divided into the classic HAstV (Mamastrovirus 1 species) and the novel HAstVs (MLB and VA), which are particularly diverse (402). MLB HAstVs (Mamastrovirus 6) are classified into three types or clades (MLB1, MLB2, and MLB3). VA HAstVs are composed of Mamastrovirus 8 species including VA2 (HMO-B) and VA4, Mamastrovirus 9 species comprising VA1 (HMO-C) and VA3 (HMO-A), and the VA5 clade (402). Unlike with classic HAstV, the route of transmission, pathogenesis, and associated clinical manifestations of novel human HAstV are not fully established (402). Some novel HAstVs have been associated with encephalitis and meningitis in immunocompromised patients and have been detected in respiratory samples of patients with respiratory manifestations (402, 403).
The seroprevalence of novel HAstV is poorly described, with one study reporting a seroprevalence rate of 65% for novel HAstV-VA1 in a U.S. adult cohort and rates ranging from 20% to 36% in a U.S. cohort of pediatric patients with ages ranging from 6 to 12 months to 5 to 10 years old, respectively (404).
(b) Detection in blood samples of nontransplanted individuals.
Novel HAstV identification in blood samples results mainly from the screening of respiratory, stool, and CSF samples by mNGS or real-time RT-PCR and further screening of available blood samples of positive cases. Among blood donors, a Swiss study by Lau et al. identified specific sequences of novel HAstV-MLB2 in a fresh frozen plasma unit by using mNGS, with results confirmed by real-time RT-PCR (58). Among a cohort of U.S. children with fever of unknown origin and afebrile controls, the screening of 176 nasopharyngeal swabs and plasma samples by mNGS allowed the detection of novel HAstV-MLB2 sequences in nasopharyngeal and plasma samples in one case (405). As part of a study that screened respiratory samples for novel HAstV by several specific real-time RT-PCRs, a case of novel HAstV-MLB2 disseminated infection was identified in several clinical specimens, including a serum sample with a viral load of 2.66E3 copies/ml from an immunocompetent pediatric patient with fever, respiratory, and digestive manifestations (406). Another case of novel HAstV-MLB2 disseminated infection in a pediatric patient with neutropenia, fever, respiratory, and digestive manifestations was identified by mNGS. Results were confirmed by real-time RT-PCR with a viral load of 4.5E5 copies/ml (407). In the same U.S. study, the plasma samples of 90 children with fever and 98 afebrile controls were screened with an astrovirus consensus RT-PCR and were all negative (407).
(c) Detection in blood samples of SOT and HSCT recipients.
In a Swiss study, two cases of novel HAstV-MLB2 disseminated infection in adult patients with meningitis were identified by mNGS, with the patients comprising one nonimmunocompromised and one HSCT recipient patient (408). The same group performed a retrospective screen for classic and novel HAstVs in stool and CSF samples from pediatric and adult patients at a tertiary hospital, with a specially designed real-time RT-PCR (rRT-PCR) panel for novel HAstV-MLB1 to -MLB3 and HAstV-VA1 to -VA4 detection. Results showed that 20/654 stool samples were positive for novel HAstV, of which 40% were from SOT and HSCT recipients, but none of the available serum or plasma samples of these patients were positive (409). Novel HAstV-VA1 RNA was detected by mNGS and real-time RT-PCR in CSF and serum samples of an 18-month-old HSCT recipient from the United Kingdom with encephalitis (410).
SYNTHESIS, CRITICAL APPRAISAL, AND PERSPECTIVE
Synthesis
Beyond the usual well-known culprits that are part of the routine clinical management of SOT and HSCT recipients, such as the herpesviruses (i.e., HSV, VZV, EBV, and CMV), BKPyV and JCPyV, adenovirus, and parvovirus B19, numerous other viral signatures or genomes can be identified in the blood of these immunocompromised patients. The clinical significance of infections with the viruses described in this review are well described for some of them, while others need to be further investigated. We aimed to provide a systematic and detailed description of the current state of knowledge regarding the DNA and RNA viruses of the blood virome that clinicians should keep in mind, with a review of the prevalence of their detection in blood samples among pediatric and adult SOT and HSCT recipients, as well as nontransplanted individuals, which were mostly blood donors and patients described as healthy. We also aimed to review current knowledge of the clinical significance of these DNA and RNA virus infections with associated viremia among transplant recipients. The detection of bacteriophages in blood samples was not addressed in this review. mNGS contributed to the detection of numerous sequences of diverse bacteriophages in various clinical samples, including blood samples. The significance of their presence (corresponding to infection or experimental contamination from the environment or reagents [23]) and their clinical significance have not been established. This topic deserves further study and discussion in a separate specialized article.
From a general perspective, infectious complications are one of the most common complications after transplantation and are associated with significant morbidity and mortality (25, 411, 412). Among allo-HSCT recipients, a large Swiss multicenter study reported that viral infections were the most common infections after allo-HSCT, representing 74.2% of all infectious complications, followed by bacterial (22.4%) and fungal (3.5%) infections (411). In this study, the underlying disease, donor type, CMV serological constellation, and GvHD were associated with the incidence rate of infections; furthermore, there was an increased risk for 1-year nonrelapse mortality associated with all pathogens (411). Other risks factors, such as HLA compatibility, the conditioning regimen, the type of graft, acute and chronic GvHD, and T-cell depletion, have been associated with an increased risk of infectious complications in general (25, 26, 411, 413). Among SOT recipients, a study from the Swiss Transplant Cohort Study has revealed that bacterial infections were the most common infectious complications, representing 62.5% of all infections, followed by viral (29.5%) and fungal (7.5%) infections (412); the viral infections mostly concerned kidney, liver, and lung transplant recipients and were mostly caused by herpesviruses (412).
Despite a tight follow-up and preemptive and therapeutic strategies, viral infections are still associated with high morbidity and mortality among transplant recipients and deserve the clinician’s attention (25, 26, 411–413).
The list of viruses that clinicians should be aware of is subject to constant expansion and updating, but the challenge now is to identify those of possible clinical significance. Although the detection of DNA and RNA viruses in blood samples among transplanted patients is still rarely reported for some viruses described in this review, possibly reflecting the lack of data from systematic screening or true low prevalence, such results should heighten the attention of the clinician who is in charge of SOT and HSCT recipients. In particular, those viruses for which recent data have provided evidence of associated disease and for which specific diagnostic assays might be available (e.g., novel HAstV) deserve more studies.
According to the results of this review, DNA and RNA viruses could be classified into several groups, according to the prevalence of their genome detection in blood and their pathogenicity and associated clinical manifestations (Fig. 1). For some viruses (such as HHV-6, B19V, and novel HAstV), infection with detectable viremia is known to be associated with specific diseases and clinical outcomes. Other viruses (such as TTV and HPgV-1) have limited virulence and are detected in blood at high replication levels. Those might be considered “commensals,” and despite their lack of associated overt clinical manifestations, they do interact with the immune system. Their clinical impact remains to be determined. Other viruses detected in blood samples of immunocompromised patients (such as the recently discovered novel human polyomaviruses HBoV1 and PARV4) have pathogenicities that are still unknown or debated and/or are rarely reported as a cause of disease. Finally, for some viruses (such as gemycircularvirus or GyV), data are still controversial regarding the association of their detection in blood samples and human infection. The detection of viral sequences in the blood or viremia is not synonymous with disease; thus, further studies are needed to identify viruses that belong to the human blood virome and to identify those with clinical significance to improve diagnosis and management of transplant recipients.
FIG 1.
Schematic representation of the prevalence of the detection of each viral species considered in this review, in whole-blood, plasma, or serum samples of pediatric and adult SOT recipients, HSCT recipients, and nontransplanted individuals. The prevalence rates of the detection of each viral species for each population screened are classified into five categories according to the mean prevalence rates reported in the publications reviewed and are reported on the ordinate axis. The sizes of the dots represent the degrees of evidence of association of viral infections with diseases, according to the publications reviewed. Histograms represent the total number of screened patients reported in the publications reviewed. Symbols: †, according to data discussed in this review, TTV has not been reported as associated with diseases despite being suggested as a potential predictor for clinical outcomes after transplantation; ‡, the novel HAstV prevalence represented here might be overestimated, considering the screening strategies in some of the studies reviewed, in which the screening of blood samples was performed only in patients with other types of clinical samples positive for novel HAstV. Abbreviations: SOT, solid organ transplant; HSCT, hematopoietic stem cell transplant; NTI, nontransplanted individuals; HHV-6, human herpesvirus 6; HHV-7, human herpesvirus 7; JCPyV, JC polyomavirus; BKPyV, BK polyomavirus; KIPyV, Karolinska Institute polyomavirus; WUPyV, Washington University polyomavirus; MCPyV, Merkel cell polyomavirus; HPyV6, human polyomavirus 6; HPyV7, human polyomavirus 7; TSPyV, trichodysplasia spinulosa-associated polyomavirus; HPyV9, human polyomavirus 9; MWPyV, Malawi polyomavirus; STLPyV, Saint Louis polyomavirus; HPyV12, human polyomavirus 12; HPyV13, human polyomavirus 13; LIPyV, Lyon IARC polyomavirus; HAdV, human adenovirus; B19V, parvovirus B19; PARV4, human parvovirus 4; HBoV, human bocavirus; TTV, torque teno virus; GyV, gyrovirus; CyCV, cyclovirus; gemy., gemycircularvirus; HPgV-1, human pegivirus-1; HPgV-2, human pegivirus-2; n. HAstV, novel human astrovirus.
Critical Appraisal
This review reveals heterogeneous results across studies regarding the detection of DNA and RNA viruses in blood samples of transplant recipients and patients described as healthy. This heterogeneity might be explained by several factors.
Epidemiological aspects.
The prevalence of infection or the seroprevalence can vary according to the local epidemiology of some viral species, genotypes, or serotypes and the specificity of the diagnostic assays used. The unbiased mNGS approach can overcome this issue and provide a broader vision of the epidemiology of some viral infections. The current state of knowledge varies widely according to viral species, with some recently identified and studied in few or small cohorts and others (i.e., BKPyV, HAdV, and B19V) for which investigations have been widely performed for decades.
Clinical factors.
Complex alterations of immunity among SOT and HSCT recipients are associated with diverse risks of viral infections, with diverse kinetics and magnitudes of viral replication. Thus, among SOT recipients, the type of transplanted organ, antirejection treatment, or antiviral prophylaxis plays a major role. Among HSCT recipients, whether the transplant is allogeneic or autologous, the type of conditioning regimen, and GvHD prophylaxis and treatment, as well as blood product transfusion, also play a major role. The differences in clinical management (in particular regarding antiviral prophylactic and therapeutic strategies) across studies from different centers that were published over the past decades should also be considered. Furthermore, the timing of sampling is a key factor for the estimation of the prevalence or incidence of viral infections in transplant recipients, since the risk of viral primary infections and reactivations changes over time after transplantation. Among transplant recipients, particularly HSCT patients, blood transfusions may represent a mode of transmission of some viruses that are not screened for in blood product safety tests, despite inactivation procedures (414). As reviewed here, data are, however, conflicting. Among blood donors, the heterogeneity of results may also highlight the heterogeneity of some selected populations, comprising patients with diverse conditions and degrees of immunocompetence.
Dynamic changes of the blood virome composition.
Acute or chronic infection and reactivation can be associated with persistent or transient viremia. The study designs and virological outcomes (e.g., point prevalence or cumulative incidence) contribute to the differences in prevalence or incidence rates of infection. Longitudinal studies using (RT-)PCR assays or mNGS provide a dynamic description of viral infections and are particularly important in transplant recipients.
Technical aspects.
The type of blood sample (whole blood, plasma, serum, peripheral blood cells) has an impact on the detection of viral genomes and the magnitude of viral loads. This is particularly relevant for herpesviruses such as EBV, CMV, and HHV-6. Another important point is the diversity of the diagnostic assays performed in the studies reviewed here. Diverse (RT-)PCR assays have been used in studies published over the past 15 to 20 years, including classic (RT-)PCR, nested (RT-)PCR, real-time (RT-)PCR, single or multiplex (RT-)PCR assays, commercial kits assays, and in-house assays. All these assays have their own performances, with differing limits of detection and quantification and diverse target sequences. mNGS is also characterized by methodological heterogeneity, with diverse preanalytic methods, mNGS protocols with the use of various mNGS platforms, single- or paired-end protocols, various bioinformatic pipelines and databases, and de novo analysis (415). Importantly, recent data have also highlighted the issue of laboratory reagent contamination, which is at the heart of concerns in virus discovery studies (23).
Perspectives
Most studies reviewed here aimed to investigate one or only a few viruses in a specific population and to explore the impact of viremia on clinical outcomes. Further studies investigating viral codetections, their kinetics in blood after transplantation, and their potential association with clinical outcomes are needed. The burden of these coinfections needs to be better assessed in studies that consider viruses that are currently not part of routine investigations of transplant recipients. Some studies recently addressed this question by investigating the role of dsDNA viral infections among HSCT recipients. The detection of multiple dsDNA viruses (including EBV, CMV, HHV-6, HAdV, and BKPyV) in blood samples after allo-HSCT was associated with increased overall mortality in a dose-dependent relationship (27, 416). The kinetics of dsDNA detection after allo-HSCT with high viral loads has been shown to predict mortality (417). Nevertheless, so far no study has investigated the overall impact of DNA and RNA viruses together. Finally, as recently investigated for TTV, other viruses may represent potential biomarkers that might contribute to the adjustment of clinical management after transplantation. This could lead to the adaptation of therapeutic strategies, including antiviral and immunosuppressive strategies.
In view of the increasing number of viruses identified and the potential associated clinical manifestations, comprehension of the human blood virome is a growing challenge that affects SOT and HSCT recipients and will help to improve diagnosis and management of transplant recipients.
ACKNOWLEDGMENTS
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
We have no conflicts of interest to declare.
Marie-Céline Zanella contributed to the conceptualization and methodology of the review, to the writing of the original draft, and to the reviewing and editing of the review. Samuel Cordey contributed to the supervision and to the writing and editing of the review. Laurent Kaiser contributed to the conceptualization, supervision, and writing and editing of the review.
We thank Florian Laubscher and Patrick Lane of ScEYEnce Studios for their contribution in designing the figure. We thank Manuel Schibler for his constructive comments and Rosemary Sudan and Erik Boehm for editorial assistance.
Biographies

Marie-Céline Zanella, M.D., obtained her pharmacy (2006) and medical (2011) degrees at the University of Geneva. She is also an ECFMG-certified physician. She completed her residency in internal medicine and has had a clinical fellowship in infectious diseases at the Geneva University Hospitals since 2016. She is involved in clinical research in the field of virology of transplantation and diagnostics.

Samuel Cordey, Ph.D., is a board-certified specialist in clinical microbiology and was in charge of the Swiss National Reference Centre for Influenza between 2014 and 2018. He obtained his Ph.D. in life sciences in 2006 at the University of Geneva. He then joined the Laboratory of Virology of the Geneva University Hospitals (2007), where he divides his time between translational research and routine diagnostics, including emerging viruses. His present research interests are related to the investigation of diseases of unrecognized viral etiology by using molecular and high-throughput sequencing, their associated epidemiology (among others, human astroviruses), and the development of diagnostic methods.

Laurent Kaiser obtained his M.D. degree at the University of Geneva in 1987. He completed his training in infectious diseases and clinical microbiology at the Geneva University Hospitals and the University of Virginia, Charlottesville, Virginia, USA. He is currently Head of the Division of Infectious Diseases and the Laboratory of Virology at the Geneva University Hospitals, Director of the Geneva Center for Viral Emerging Diseases and the Swiss National Reference Centres for Influenza and Emerging Viruses, and Full Professor at the University of Geneva. He is an expert in respiratory and emerging viruses and is involved in high-profile health emergency epidemics, such as epidemics of Ebola virus and other emerging viruses. His research areas are clinical virology, particularly respiratory viruses, viral infections in transplant recipients, and emerging viruses. He is a member of the American Society of Microbiology and the European Society for Clinical Virology.
REFERENCES
- 1.Rascovan N, Duraisamy R, Desnues C. 2016. Metagenomics and the human virome in asymptomatic individuals. Annu Rev Microbiol 70:125–141. doi: 10.1146/annurev-micro-102215-095431. [DOI] [PubMed] [Google Scholar]
- 2.Popgeorgiev N, Temmam S, Raoult D, Desnues C. 2013. Describing the silent human virome with an emphasis on giant viruses. Intervirology 56:395–412. doi: 10.1159/000354561. [DOI] [PubMed] [Google Scholar]
- 3.Foxman EF, Iwasaki A. 2011. Genome-virome interactions: examining the role of common viral infections in complex disease. Nat Rev Microbiol 9:254–264. doi: 10.1038/nrmicro2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vu DL, Kaiser L. 2017. The concept of commensal viruses almost 20 years later: redefining borders in clinical virology. Clin Microbiol Infect 23:688–690. doi: 10.1016/j.cmi.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lim ES, Zhou Y, Zhao G, Bauer IK, Droit L, Ndao IM, Warner BB, Tarr PI, Wang D, Holtz LR. 2015. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med 21:1228–1234. doi: 10.1038/nm.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Minot S, Bryson A, Chehoud C, Wu GD, Lewis JD, Bushman FD. 2013. Rapid evolution of the human gut virome. Proc Natl Acad Sci U S A 110:12450–12455. doi: 10.1073/pnas.1300833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abeles SR, Robles-Sikisaka R, Ly M, Lum AG, Salzman J, Boehm TK, Pride DT. 2014. Human oral viruses are personal, persistent and gender-consistent. ISME J 8:1753–1767. doi: 10.1038/ismej.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broecker F, Russo G, Klumpp J, Moelling K. 2017. Stable core virome despite variable microbiome after fecal transfer. Gut Microbes 8:214–220. doi: 10.1080/19490976.2016.1265196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foulongne V, Sauvage V, Hebert C, Dereure O, Cheval J, Gouilh MA, Pariente K, Segondy M, Burguiere A, Manuguerra JC, Caro V, Eloit M. 2012. Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS One 7:e38499. doi: 10.1371/journal.pone.0038499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zoll J, Rahamat-Langendoen J, Ahout I, de Jonge MI, Jans J, Huijnen MA, Ferwerda G, Warris A, Melchers WJ. 2015. Direct multiplexed whole genome sequencing of respiratory tract samples reveals full viral genomic information. J Clin Virol 66:6–11. doi: 10.1016/j.jcv.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wylie KM. 2017. The virome of the human respiratory tract. Clin Chest Med 38:11–19. doi: 10.1016/j.ccm.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rani A, Ranjan R, McGee HS, Metwally A, Hajjiri Z, Brennan DC, Finn PW, Perkins DL. 2016. A diverse virome in kidney transplant patients contains multiple viral subtypes with distinct polymorphisms. Sci Rep 6:33327. doi: 10.1038/srep33327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stone L. 2018. Microbiota: bacteriophage diversity in the urinary microbiome. Nat Rev Urol 15:135. doi: 10.1038/nrurol.2018.23. [DOI] [PubMed] [Google Scholar]
- 14.Santiago-Rodriguez TM, Ly M, Bonilla N, Pride DT. 2015. The human urine virome in association with urinary tract infections. Front Microbiol 6:14. doi: 10.3389/fmicb.2015.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller-Ensminger T, Garretto A, Brenner J, Thomas-White K, Zambom A, Wolfe AJ, Putonti C. 2018. Bacteriophages of the urinary microbiome. J Bacteriol 200:e00738-17. doi: 10.1128/JB.00738-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manrique P, Bolduc B, Walk ST, van der Oost J, de Vos WM, Young MJ. 2016. Healthy human gut phageome. Proc Natl Acad Sci U S A 113:10400–10405. doi: 10.1073/pnas.1601060113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carding SR, Davis N, Hoyles L. 2017. Review article: the human intestinal virome in health and disease. Aliment Pharmacol Ther 46:800–815. doi: 10.1111/apt.14280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Columpsi P, Sacchi P, Zuccaro V, Cima S, Sarda C, Mariani M, Gori A, Bruno R. 2016. Beyond the gut bacterial microbiota: the gut virome. J Med Virol 88:1467–1472. doi: 10.1002/jmv.24508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell AB, Oliver BG, Glanville AR. 2016. Translational aspects of the human respiratory virome. Am J Respir Crit Care Med 194:1458–1464. doi: 10.1164/rccm.201606-1278CI. [DOI] [PubMed] [Google Scholar]
- 20.Wylie KM, Mihindukulasuriya KA, Zhou Y, Sodergren E, Storch GA, Weinstock GM. 2014. Metagenomic analysis of double-stranded DNA viruses in healthy adults. BMC Biol 12:71. doi: 10.1186/s12915-014-0071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cadwell K. 2015. The virome in host health and disease. Immunity 42:805–813. doi: 10.1016/j.immuni.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes EC. 2019. Reagent contamination in viromics: all that glitters is not gold. Clin Microbiol Infect 25:1167–1168. doi: 10.1016/j.cmi.2019.06.019. [DOI] [PubMed] [Google Scholar]
- 23.Asplund M, Kjartansdottir KR, Mollerup S, Vinner L, Fridholm H, Herrera JAR, Friis-Nielsen J, Hansen TA, Jensen RH, Nielsen IB, Richter SR, Rey-Iglesia A, Matey-Hernandez ML, Alquezar-Planas DE, Olsen PVS, Sicheritz-Ponten T, Willerslev E, Lund O, Brunak S, Mourier T, Nielsen LP, Izarzugaza JMG, Hansen AJ. 2019. Contaminating viral sequences in high-throughput sequencing viromics: a linkage study of 700 sequencing libraries. Clin Microbiol Infect 25:1277–1285. doi: 10.1016/j.cmi.2019.04.028. [DOI] [PubMed] [Google Scholar]
- 24.Timsit JF, Sonneville R, Kalil AC, Bassetti M, Ferrer R, Jaber S, Lanternier F, Luyt CE, Machado F, Mikulska M, Papazian L, Pene F, Poulakou G, Viscoli C, Wolff M, Zafrani L, Van Delden C. 2019. Diagnostic and therapeutic approach to infectious diseases in solid organ transplant recipients. Intensive Care Med 45:573–591. doi: 10.1007/s00134-019-05597-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Srinivasan A, Wang C, Srivastava DK, Burnette K, Shenep JL, Leung W, Hayden RT. 2013. Timeline, epidemiology, and risk factors for bacterial, fungal, and viral infections in children and adolescents after allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transplant 19:94–101. doi: 10.1016/j.bbmt.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin-Pena A, Aguilar-Guisado M, Espigado I, Parody R, Miguel Cisneros J. 2011. Prospective study of infectious complications in allogeneic hematopoietic stem cell transplant recipients. Clin Transplant 25:468–474. doi: 10.1111/j.1399-0012.2010.01286.x. [DOI] [PubMed] [Google Scholar]
- 27.Hill JA, Mayer BT, Xie H, Leisenring WM, Huang ML, Stevens-Ayers T, Milano F, Delaney C, Sorror ML, Sandmaier BM, Nichols G, Zerr DM, Jerome KR, Schiffer JT, Boeckh M. 2017. The cumulative burden of double-stranded DNA virus detection after allogeneic HCT is associated with increased mortality. Blood 129:2316–2325. doi: 10.1182/blood-2016-10-748426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vu DL, Cordey S, Simonetta F, Brito F, Docquier M, Turin L, van Delden C, Boely E, Dantin C, Pradier A, Roosnek E, Chalandon Y, Zdobnov EM, Masouridi-Levrat S, Kaiser L. 2019. Human pegivirus persistence in human blood virome after allogeneic haematopoietic stem-cell transplantation. Clin Microbiol Infect 25:225–232. doi: 10.1016/j.cmi.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 29.De Vlaminck I, Khush KK, Strehl C, Kohli B, Luikart H, Neff NF, Okamoto J, Snyder TM, Cornfield DN, Nicolls MR, Weill D, Bernstein D, Valantine HA, Quake SR. 2013. Temporal response of the human virome to immunosuppression and antiviral therapy. Cell 155:1178–1187. doi: 10.1016/j.cell.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moustafa A, Xie C, Kirkness E, Biggs W, Wong E, Turpaz Y, Bloom K, Delwart E, Nelson KE, Venter JC, Telenti A. 2017. The blood DNA virome in 8,000 humans. PLoS Pathog 13:e1006292. doi: 10.1371/journal.ppat.1006292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sauvage V, Gomez J, Boizeau L, Laperche S. 2017. The potential of viral metagenomics in blood transfusion safety. Transfus Clin Biol 24:218–222. doi: 10.1016/j.tracli.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 32.Wilck MB, Zuckerman RA, AST Infectious Diseases Community of Practice . 2013. Herpes simplex virus in solid organ transplantation. Am J Transplant 13(Suppl 4):121–127. doi: 10.1111/ajt.12105. [DOI] [PubMed] [Google Scholar]
- 33.Pergam SA, Limaye AP, AST Infectious Diseases Community of Practice . 2013. Varicella zoster virus in solid organ transplantation. Am J Transplant 13(Suppl 4):138–146. doi: 10.1111/ajt.12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allen UD, Preiksaitis JK, AST Infectious Diseases Community of Practice . 2013. Epstein-Barr virus and posttransplant lymphoproliferative disorder in solid organ transplantation. Am J Transplant 13(Suppl 4):107–120. doi: 10.1111/ajt.12104. [DOI] [PubMed] [Google Scholar]
- 35.Razonable RR, Humar A, AST Infectious Diseases Community of Practice . 2013. Cytomegalovirus in solid organ transplantation. Am J Transplant 13(Suppl 4):93–106. doi: 10.1111/ajt.12103. [DOI] [PubMed] [Google Scholar]
- 36.Styczynski J, Reusser P, Einsele H, de la Camara R, Cordonnier C, Ward KN, Ljungman P, Engelhard D, Second European Conference on Infections in Leukemia . 2009. Management of HSV, VZV and EBV infections in patients with hematological malignancies and after SCT: guidelines from the Second European Conference on Infections in Leukemia. Bone Marrow Transplant 43:757–770. doi: 10.1038/bmt.2008.386. [DOI] [PubMed] [Google Scholar]
- 37.Styczynski J, van der Velden W, Fox CP, Engelhard D, de la Camara R, Cordonnier C, Ljungman P, Sixth European Conference on Infections in Leukemia, a joint venture of the Infectious Diseases Working Party of the European Society of Blood and Marrow Transplantation (EBMT-IDWP), the Infectious Diseases Group of the European Organization for Research and Treatment of Cancer (EORTC-IDG), the International Immunocompromised Host Society (ICHS), and the European Leukemia Network . 2016. Management of Epstein-Barr virus infections and post-transplant lymphoproliferative disorders in patients after allogeneic hematopoietic stem cell transplantation: Sixth European Conference on Infections in Leukemia (ECIL-6) guidelines. Haematologica 101:803–811. doi: 10.3324/haematol.2016.144428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ljungman P, de la Camara R, Robin C, Crocchiolo R, Einsele H, Hill JA, Hubacek P, Navarro D, Cordonnier C, Ward KN, European Conference on Infections in Leukaemia Group . 2019. Guidelines for the management of cytomegalovirus infection in patients with haematological malignancies and after stem cell transplantation from the 2017 European Conference on Infections in Leukaemia (ECIL 7). Lancet Infect Dis 19:e260–e272. doi: 10.1016/S1473-3099(19)30107-0. [DOI] [PubMed] [Google Scholar]
- 39.Agut H, Bonnafous P, Gautheret-Dejean A. 2015. Laboratory and clinical aspects of human herpesvirus 6 infections. Clin Microbiol Rev 28:313–335. doi: 10.1128/CMR.00122-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaufer BB, Flamand L. 2014. Chromosomally integrated HHV-6: impact on virus, cell and organismal biology. Curr Opin Virol 9:111–118. doi: 10.1016/j.coviro.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 41.De Bolle L, Naesens L, De Clercq E. 2005. Update on human herpesvirus 6 biology, clinical features, and therapy. Clin Microbiol Rev 18:217–245. doi: 10.1128/CMR.18.1.217-245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pellett PE, Ablashi DV, Ambros PF, Agut H, Caserta MT, Descamps V, Flamand L, Gautheret-Dejean A, Hall CB, Kamble RT, Kuehl U, Lassner D, Lautenschlager I, Loomis KS, Luppi M, Lusso P, Medveczky PG, Montoya JG, Mori Y, Ogata M, Pritchett JC, Rogez S, Seto E, Ward KN, Yoshikawa T, Razonable RR. 2012. Chromosomally integrated human herpesvirus 6: questions and answers. Rev Med Virol 22:144–155. doi: 10.1002/rmv.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hill JA, Magaret AS, Hall-Sedlak R, Mikhaylova A, Huang ML, Sandmaier BM, Hansen JA, Jerome KR, Zerr DM, Boeckh M. 2017. Outcomes of hematopoietic cell transplantation using donors or recipients with inherited chromosomally integrated HHV-6. Blood 130:1062–1069. doi: 10.1182/blood-2017-03-775759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ablashi D, Agut H, Alvarez-Lafuente R, Clark DA, Dewhurst S, DiLuca D, Flamand L, Frenkel N, Gallo R, Gompels UA, Hollsberg P, Jacobson S, Luppi M, Lusso P, Malnati M, Medveczky P, Mori Y, Pellett PE, Pritchett JC, Yamanishi K, Yoshikawa T. 2014. Classification of HHV-6A and HHV-6B as distinct viruses. Arch Virol 159:863–870. doi: 10.1007/s00705-013-1902-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ylinen E, Lehtinen S, Jahnukainen T, Karlsson T, Loginov R, Mannonen L, Lautenschlager I, Jalanko H. 2017. Human herpes virus 6 infection in pediatric organ transplant patients. Pediatr Transplant 21. doi: 10.1111/petr.12905. [DOI] [PubMed] [Google Scholar]
- 46.Kamble RT, Clark DA, Leong HN, Heslop HE, Brenner MK, Carrum G. 2007. Transmission of integrated human herpesvirus-6 in allogeneic hematopoietic stem cell transplantation. Bone Marrow Transplant 40:563–566. doi: 10.1038/sj.bmt.1705780. [DOI] [PubMed] [Google Scholar]
- 47.Clark DA, Nacheva EP, Leong HN, Brazma D, Li YT, Tsao EH, Buyck HC, Atkinson CE, Lawson HM, Potter MN, Griffiths PD. 2006. Transmission of integrated human herpesvirus 6 through stem cell transplantation: implications for laboratory diagnosis. J Infect Dis 193:912–916. doi: 10.1086/500838. [DOI] [PubMed] [Google Scholar]
- 48.Yamada Y, Osumi T, Imadome KI, Takahashi E, Ohye T, Yoshikawa T, Tomizawa D, Kato M, Matsumoto K. 2017. Transmission of chromosomally integrated human herpesvirus 6 via cord blood transplantation. Transpl Infect Dis 19:e12636. doi: 10.1111/tid.12636. [DOI] [PubMed] [Google Scholar]
- 49.Hill JA, Zerr DM. 2014. Roseoloviruses in transplant recipients: clinical consequences and prospects for treatment and prevention trials. Curr Opin Virol 9:53–60. doi: 10.1016/j.coviro.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pellett Madan R, Hand J, AST Infectious Diseases Community of Practice . 2019. Human herpesvirus 6, 7, and 8 in solid organ transplantation: guidelines from the American Society of Transplantation Infectious Diseases Community of Practice. Clin Transplant 33:e13518. doi: 10.1111/ctr.13518. [DOI] [PubMed] [Google Scholar]
- 51.Le J, Gantt S, AST Infectious Diseases Community of Practice . 2013. Human herpesvirus 6, 7 and 8 in solid organ transplantation. Am J Transplant 13(Suppl 4):128–137. doi: 10.1111/ajt.12106. [DOI] [PubMed] [Google Scholar]
- 52.Zerr DM, Boeckh M, Delaney C, Martin PJ, Xie H, Adler AL, Huang ML, Corey L, Leisenring WM. 2012. HHV-6 reactivation and associated sequelae after hematopoietic cell transplantation. Biol Blood Marrow Transplant 18:1700–1708. doi: 10.1016/j.bbmt.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Pagter PJ, Schuurman R, Meijer E, van Baarle D, Sanders EA, Boelens JJ. 2008. Human herpesvirus type 6 reactivation after haematopoietic stem cell transplantation. J Clin Virol 43:361–366. doi: 10.1016/j.jcv.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 54.Ward KN, Hill JA, Hubacek P, de la Camara R, Crocchiolo R, Einsele H, Navarro D, Robin C, Cordonnier C, Ljungman P, European Conference on Infections in Leukaemia (ECIL) . 2019. Guidelines from the 2017 European Conference on Infections in Leukaemia for management of HHV-6 infection in patients with hematologic malignancies and after hematopoietic stem cell transplantation. Haematologica 104:2155–2163. doi: 10.3324/haematol.2019.223073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hara S, Kimura H, Hoshino Y, Tanaka N, Nishikawa K, Ihira M, Yoshikawa T, Morishima T. 2002. Detection of herpesvirus DNA in the serum of immunocompetent children. Microbiol Immunol 46:177–180. doi: 10.1111/j.1348-0421.2002.tb02683.x. [DOI] [PubMed] [Google Scholar]
- 56.Traore L, Tao I, Bisseye C, Diarra B, Compaore TR, Nebie Y, Assih M, Ouedraogo A, Zohoncon T, Djigma F, Ouermi D, Barro N, Sanou M, Ouedraogo RT, Simpore J. 2016. Molecular diagnostic of cytomegalovirus, Epstein Barr virus and herpes virus 6 infections among blood donors by multiplex real-time PCR in Ouagadougou, Burkina Faso. Pan Afr Med J 24:298. doi: 10.11604/pamj.2016.24.298.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rouka E, Kyriakou D. 2015. Molecular epidemiology of human herpesviruses types 1–6 and 8 among Greek blood donors. Transfus Med 25:276–279. doi: 10.1111/tme.12202. [DOI] [PubMed] [Google Scholar]
- 58.Lau P, Cordey S, Brito F, Tirefort D, Petty TJ, Turin L, Guichebaron A, Docquier M, Zdobnov EM, Waldvogel-Abramowski S, Lecompte T, Kaiser L, Preynat-Seauve O. 2017. Metagenomics analysis of red blood cell and fresh-frozen plasma units. Transfusion 57:1787–1800. doi: 10.1111/trf.14148. [DOI] [PubMed] [Google Scholar]
- 59.Al Fawaz T, Ng V, Richardson SE, Barton M, Allen U. 2014. Clinical consequences of human herpesvirus-6 DNAemia in peripheral blood in pediatric liver transplant recipients. Pediatr Transplant 18:47–51. doi: 10.1111/petr.12176. [DOI] [PubMed] [Google Scholar]
- 60.Sánchez-Ponce Y, Varela-Fascinetto G, Romo-Vázquez J, López-Martínez B, Sánchez-Huerta J, Parra-Ortega I, Fuentes-Pananá E, Morales-Sánchez A. 2018. Simultaneous detection of beta and gamma human herpesviruses by multiplex qPCR reveals simple infection and coinfection episodes increasing risk for graft rejection in solid organ transplantation. Viruses 10:730. doi: 10.3390/v10120730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dulery R, Salleron J, Dewilde A, Rossignol J, Boyle EM, Gay J, de Berranger E, Coiteux V, Jouet JP, Duhamel A, Yakoub-Agha I. 2012. Early human herpesvirus type 6 reactivation after allogeneic stem cell transplantation: a large-scale clinical study. Biol Blood Marrow Transplant 18:1080–1089. doi: 10.1016/j.bbmt.2011.12.579. [DOI] [PubMed] [Google Scholar]
- 62.Gotoh M, Yoshizawa S, Katagiri S, Suguro T, Asano M, Kitahara T, Akahane D, Okabe S, Tauchi T, Ito Y, Ohyashiki K. 2014. Human herpesvirus 6 reactivation on the 30th day after allogeneic hematopoietic stem cell transplantation can predict grade 2–4 acute graft-versus-host disease. Transpl Infect Dis 16:440–449. doi: 10.1111/tid.12229. [DOI] [PubMed] [Google Scholar]
- 63.Inazawa N, Hori T, Hatakeyama N, Yamamoto M, Yoto Y, Nojima M, Suzuki N, Shimizu N, Tsutsumi H. 2015. Large-scale multiplex polymerase chain reaction assay for diagnosis of viral reactivations after allogeneic hematopoietic stem cell transplantation. J Med Virol 87:1427–1435. doi: 10.1002/jmv.24161. [DOI] [PubMed] [Google Scholar]
- 64.Inazawa N, Hori T, Nojima M, Saito M, Igarashi K, Yamamoto M, Shimizu N, Yoto Y, Tsutsumi H. 2017. Virus reactivations after autologous hematopoietic stem cell transplantation detected by multiplex PCR assay. J Med Virol 89:358–362. doi: 10.1002/jmv.24621. [DOI] [PubMed] [Google Scholar]
- 65.Boutolleau D, Fernandez C, Andre E, Imbert-Marcille BM, Milpied N, Agut H, Gautheret-Dejean A. 2003. Human herpesvirus (HHV)-6 and HHV-7: two closely related viruses with different infection profiles in stem cell transplantation recipients. J Infect Dis 187:179–186. doi: 10.1086/367677. [DOI] [PubMed] [Google Scholar]
- 66.Lusso P, Secchiero P, Crowley RW, Garzino-Demo A, Berneman ZN, Gallo RC. 1994. CD4 is a critical component of the receptor for human herpesvirus 7: interference with human immunodeficiency virus. Proc Natl Acad Sci U S A 91:3872–3876. doi: 10.1073/pnas.91.9.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Agut H, Bonnafous P, Gautheret-Dejean A. 2016. Human herpesviruses 6A, 6B, and 7. Microbiol Spectr 4:10.1128/microbiolspec.DMIH2-0007-2015. doi: 10.1128/microbiolspec.DMIH2-0007-2015. [DOI] [PubMed] [Google Scholar]
- 68.Prusty BK, Gulve N, Rasa S, Murovska M, Hernandez PC, Ablashi DV. 2017. Possible chromosomal and germline integration of human herpesvirus 7. J Gen Virol 98:266–274. doi: 10.1099/jgv.0.000692. [DOI] [PubMed] [Google Scholar]
- 69.Feldstein AE, Razonable RR, Boyce TG, Freese DK, El-Youssef M, Perrault J, Paya CV, Ishitani MB. 2003. Prevalence and clinical significance of human herpesviruses 6 and 7 active infection in pediatric liver transplant patients. Pediatr Transplant 7:125–129. doi: 10.1034/j.1399-3046.2003.00028.x. [DOI] [PubMed] [Google Scholar]
- 70.Fule Robles JD, Cheuk DK, Ha SY, Chiang AK, Chan GC. 2014. Human herpesvirus types 6 and 7 infection in pediatric hematopoietic stem cell transplant recipients. Ann Transplant 19:269–276. doi: 10.12659/AOT.889995. [DOI] [PubMed] [Google Scholar]
- 71.Khanani M, Al-Ahmari A, Tellier R, Allen U, Richardson S, Doyle JJ, Gassas A. 2007. Human herpesvirus 7 in pediatric hematopoietic stem cell transplantation. Pediatr Blood Cancer 48:567–570. doi: 10.1002/pbc.20829. [DOI] [PubMed] [Google Scholar]
- 72.Hubacek P, Sedlacek P, Keslova P, Formankova R, Stary J, Kulich M, Cinek O. 2008. Incidence of HHV7 in donors and recipients of allogeneic hematopoietic stem cell transplantation. Pediatr Blood Cancer 50:935–936. doi: 10.1002/pbc.21436. [DOI] [PubMed] [Google Scholar]
- 73.Moens U, Calvignac-Spencer S, Lauber C, Ramqvist T, Feltkamp MCW, Daugherty MD, Verschoor EJ, Ehlers B, ICTV Report Consortium . 2017. ICTV virus taxonomy profile: Polyomaviridae. J Gen Virol 98:1159–1160. doi: 10.1099/jgv.0.000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Polyomaviridae Study Group of the International Committee On Taxonomy Of Viruses, Calvignac-Spencer S, Feltkamp MC, Daugherty MD, Moens U, Ramqvist T, Johne R, Ehlers B. 2016. A taxonomy update for the family Polyomaviridae. Arch Virol 161:1739–1750. doi: 10.1007/s00705-016-2794-y. [DOI] [PubMed] [Google Scholar]
- 75.Hirsch HH, Babel N, Comoli P, Friman V, Ginevri F, Jardine A, Lautenschlager I, Legendre C, Midtvedt K, Munoz P, Randhawa P, Rinaldo CH, Wieszek A, ESCMID Study Group of Infection in Compromised Hosts . 2014. European perspective on human polyomavirus infection, replication and disease in solid organ transplantation. Clin Microbiol Infect 20(Suppl 7):74–88. doi: 10.1111/1469-0691.12538. [DOI] [PubMed] [Google Scholar]
- 76.Karachaliou M, Waterboer T, Casabonne D, Chalkiadaki G, Roumeliotaki T, Michel A, Stiakaki E, Chatzi L, Pawlita M, Kogevinas M, de Sanjose S. 2016. The natural history of human polyomaviruses and herpesviruses in early life—the Rhea Birth Cohort in Greece. Am J Epidemiol 183:671–679. doi: 10.1093/aje/kwv281. [DOI] [PubMed] [Google Scholar]
- 77.Gaboriaud P, Ferté M, Arnold F, Leblond V, Nicol J, Debare H, Le Meur M, Martini F, Tognon M, Touzé A. 2018. Age-specific seroprevalence of human polyomavirus 12 and Saint Louis and New Jersey polyomaviruses. Emerg Microbes Infect 7:22. doi: 10.1038/s41426-018-0026-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kean JM, Rao S, Wang M, Garcea RL. 2009. Seroepidemiology of human polyomaviruses. PLoS Pathog 5:e1000363. doi: 10.1371/journal.ppat.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Viscidi RP, Rollison DE, Sondak VK, Silver B, Messina JL, Giuliano AR, Fulp W, Ajidahun A, Rivanera D. 2011. Age-specific seroprevalence of Merkel cell polyomavirus, BK virus, and JC virus. Clin Vaccine Immunol 18:1737–1743. doi: 10.1128/CVI.05175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hirsch HH, Randhawa P, AST Infectious Diseases Community of Practice . 2013. BK polyomavirus in solid organ transplantation. Am J Transplant 13(Suppl 4):179–188. doi: 10.1111/ajt.12110. [DOI] [PubMed] [Google Scholar]
- 81.Cesaro S, Dalianis T, Hanssen Rinaldo C, Koskenvuo M, Pegoraro A, Einsele H, Cordonnier C, Hirsch HH, ECIL-6 Group . 2018. ECIL guidelines for the prevention, diagnosis and treatment of BK polyomavirus-associated haemorrhagic cystitis in haematopoietic stem cell transplant recipients. J Antimicrob Chemother 73:12–21. doi: 10.1093/jac/dkx324. [DOI] [PubMed] [Google Scholar]
- 82.Yuan C, Deberardinis C, Patel R, Shroff SM, Messina SA, Goldstein S, Mori S. 2018. Progressive multifocal leukoencephalopathy after allogeneic stem cell transplantation: case report and review of the literature. Transpl Infect Dis 20:e12879. doi: 10.1111/tid.12879. [DOI] [PubMed] [Google Scholar]
- 83.Hirsch HH, Kardas P, Kranz D, Leboeuf C. 2013. The human JC polyomavirus (JCPyV): virological background and clinical implications. APMIS 121:685–727. doi: 10.1111/apm.12128. [DOI] [PubMed] [Google Scholar]
- 84.Berger JR, Miller CS, Mootoor Y, Avdiushko SA, Kryscio RJ, Zhu H. 2006. JC virus detection in bodily fluids: clues to transmission. Clin Infect Dis 43:e9. doi: 10.1086/504947. [DOI] [PubMed] [Google Scholar]
- 85.Gossai A, Waterboer T, Nelson HH, Michel A, Willhauck-Fleckenstein M, Farzan SF, Hoen AG, Christensen BC, Kelsey KT, Marsit CJ, Pawlita M, Karagas MR. 2016. Seroepidemiology of human polyomaviruses in a US population. Am J Epidemiol 183:61–69. doi: 10.1093/aje/kwv155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Egli A, Infanti L, Dumoulin A, Buser A, Samaridis J, Stebler C, Gosert R, Hirsch HH. 2009. Prevalence of polyomavirus BK and JC infection and replication in 400 healthy blood donors. J Infect Dis 199:837–846. doi: 10.1086/597126. [DOI] [PubMed] [Google Scholar]
- 87.Chesters PM, Heritage J, McCance DJ. 1983. Persistence of DNA sequences of BK virus and JC virus in normal human tissues and in diseased tissues. J Infect Dis 147:676–684. doi: 10.1093/infdis/147.4.676. [DOI] [PubMed] [Google Scholar]
- 88.Drachenberg CB, Hirsch HH, Papadimitriou JC, Gosert R, Wali RK, Munivenkatappa R, Nogueira J, Cangro CB, Haririan A, Mendley S, Ramos E. 2007. Polyomavirus BK versus JC replication and nephropathy in renal transplant recipients: a prospective evaluation. Transplantation 84:323–330. doi: 10.1097/01.tp.0000269706.59977.a5. [DOI] [PubMed] [Google Scholar]
- 89.Adrianzen Herrera D, Ayyappan S, Jasra S, Kornblum N, Derman O, Shastri A, Mantzaris I, Verma A, Braunschweig I, Janakiram M. 2019. Characteristics and outcomes of progressive multifocal leukoencephalopathy in hematologic malignancies and stem cell transplant—a case series. Leuk Lymphoma 60:395–401. doi: 10.1080/10428194.2018.1474523. [DOI] [PubMed] [Google Scholar]
- 90.Mateen FJ, Muralidharan R, Carone M, van de Beek D, Harrison DM, Aksamit AJ, Gould MS, Clifford DB, Nath A. 2011. Progressive multifocal leukoencephalopathy in transplant recipients. Ann Neurol 70:305–322. doi: 10.1002/ana.22408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Janphram C, Worawichawong S, Disthabanchong S, Sumethkul V, Rotjanapan P. 2017. Absence of JC polyomavirus (JCPyV) viremia in early post-transplant JCPyV nephropathy: a case report. Transpl Infect Dis 19:e12761. doi: 10.1111/tid.12761. [DOI] [PubMed] [Google Scholar]
- 92.Lautenschlager I, Jahnukainen T, Kardas P, Lohi J, Auvinen E, Mannonen L, Dumoulin A, Hirsch HH, Jalanko H. 2014. A case of primary JC polyomavirus infection-associated nephropathy. Am J Transplant 14:2887–2892. doi: 10.1111/ajt.12945. [DOI] [PubMed] [Google Scholar]
- 93.Kamminga S, van der Meijden E, de Brouwer C, Feltkamp M, Zaaijer H. 2019. Prevalence of DNA of fourteen human polyomaviruses determined in blood donors. Transfusion 59:3689–3697. doi: 10.1111/trf.15557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.De Santis R, Azzi A. 1996. Duplex polymerase chain reaction for the simultaneous detection of the human polyomavirus BK and JC DNA. Mol Cell Probes 10:325–330. doi: 10.1006/mcpr.1996.0044. [DOI] [PubMed] [Google Scholar]
- 95.Dolei A, Pietropaolo V, Gomes E, Di Taranto C, Ziccheddu M, Spanu MA, Lavorino C, Manca M, Degener AM. 2000. Polyomavirus persistence in lymphocytes: prevalence in lymphocytes from blood donors and healthy personnel of a blood transfusion centre. J Gen Virol 81:1967–1973. doi: 10.1099/0022-1317-81-8-1967. [DOI] [PubMed] [Google Scholar]
- 96.Dorries K, Vogel E, Gunther S, Czub S. 1994. Infection of human polyomaviruses JC and BK in peripheral blood leukocytes from immunocompetent individuals. Virology 198:59–70. doi: 10.1006/viro.1994.1008. [DOI] [PubMed] [Google Scholar]
- 97.Husseiny MI, Anastasi B, Singer J, Lacey SF. 2010. A comparative study of Merkel cell, BK and JC polyomavirus infections in renal transplant recipients and healthy subjects. J Clin Virol 49:137–140. doi: 10.1016/j.jcv.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bialasiewicz S, Rockett RJ, Barraclough KA, Leary D, Dudley KJ, Isbel NM, Sloots TP. 2016. Detection of recently discovered human polyomaviruses in a longitudinal kidney transplant cohort. Am J Transplant 16:2734–2740. doi: 10.1111/ajt.13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Saundh BK, Baker R, Harris M, Hale A. 2016. A prospective study of renal transplant recipients reveals an absence of primary JC polyomavirus infections. J Clin Virol 77:101–105. doi: 10.1016/j.jcv.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 100.Castro T, Fink MCD, Figueiredo M, Braz-Silva PH, Pannuti CM, Ortega KL, Gallottini M. 2017. Polyomavirus BK and JC in individuals with chronic kidney failure, kidney transplantation, and healthy controls. J Clin Virol 89:5–9. doi: 10.1016/j.jcv.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 101.Ettenger R, Chin H, Kesler K, Bridges N, Grimm P, Reed EF, Sarwal M, Sibley R, Tsai E, Warshaw B, Kirk AD. 2017. Relationship among viremia/viral infection, alloimmunity, and nutritional parameters in the first year after pediatric kidney transplantation. Am J Transplant 17:1549–1562. doi: 10.1111/ajt.14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hocker B, Tabatabai J, Schneble L, Oh J, Thiel F, Pape L, Rusai K, Topaloglu R, Kranz B, Klaus G, Printza N, Yavascan O, Fichtner A, Krupka K, Bruckner T, Waldherr R, Pawlita M, Schnitzler P, Hirsch HH, Tonshoff B. 2018. JC polyomavirus replication and associated disease in pediatric renal transplantation: an international CERTAIN Registry study. Pediatr Nephrol 33:2343–2352. doi: 10.1007/s00467-018-4029-9. [DOI] [PubMed] [Google Scholar]
- 103.Hirsch HH, Steiger J. 2003. Polyomavirus BK. Lancet Infect Dis 3:611–623. doi: 10.1016/S1473-3099(03)00770-9. [DOI] [PubMed] [Google Scholar]
- 104.Wittmann T, Horowitz N, Benyamini N, Henig I, Zuckerman T, Rowe JM, Kra-Oz Z, Szwarcwort Cohen M, Oren I, Avivi I. 2015. JC polyomavirus reactivation is common following allogeneic stem cell transplantation and its preemptive detection may prevent lethal complications. Bone Marrow Transplant 50:984–991. doi: 10.1038/bmt.2015.68. [DOI] [PubMed] [Google Scholar]
- 105.Tan CS, Broge TA Jr, Ngo L, Gheuens S, Viscidi R, Bord E, Rosenblatt J, Wong M, Avigan D, Koralnik IJ. 2014. Immune reconstitution after allogeneic hematopoietic stem cell transplantation is associated with selective control of JC virus reactivation. Biol Blood Marrow Transplant 20:992–999. doi: 10.1016/j.bbmt.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jin L, Gibson PE, Knowles WA, Clewley JP. 1993. BK virus antigenic variants: sequence analysis within the capsid VP1 epitope. J Med Virol 39:50–56. doi: 10.1002/jmv.1890390110. [DOI] [PubMed] [Google Scholar]
- 107.Zheng HY, Nishimoto Y, Chen Q, Hasegawa M, Zhong S, Ikegaya H, Ohno N, Sugimoto C, Takasaka T, Kitamura T, Yogo Y. 2007. Relationships between BK virus lineages and human populations. Microbes Infect 9:204–213. doi: 10.1016/j.micinf.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 108.Reploeg MD, Storch GA, Clifford DB. 2001. Bk virus: a clinical review. Clin Infect Dis 33:191–202. doi: 10.1086/321813. [DOI] [PubMed] [Google Scholar]
- 109.Huang G, Chen LZ, Qiu J, Wang CX, Fei JG, Deng SX, Li J, Chen GD, Zhang L, Fu Q, Zeng WT, Zhao DQ. 2010. Prospective study of polyomavirus BK replication and nephropathy in renal transplant recipients in China: a single-center analysis of incidence, reduction in immunosuppression and clinical course. Clin Transplant 24:599–609. doi: 10.1111/j.1399-0012.2009.01141.x. [DOI] [PubMed] [Google Scholar]
- 110.Hirsch HH, Knowles W, Dickenmann M, Passweg J, Klimkait T, Mihatsch MJ, Steiger J. 2002. Prospective study of polyomavirus type BK replication and nephropathy in renal-transplant recipients. N Engl J Med 347:488–496. doi: 10.1056/NEJMoa020439. [DOI] [PubMed] [Google Scholar]
- 111.Nickeleit V, Klimkait T, Binet IF, Dalquen P, Del Zenero V, Thiel G, Mihatsch MJ, Hirsch HH. 2000. Testing for polyomavirus type BK DNA in plasma to identify renal-allograft recipients with viral nephropathy. N Engl J Med 342:1309–1315. doi: 10.1056/NEJM200005043421802. [DOI] [PubMed] [Google Scholar]
- 112.Almeras C, Vetromile F, Garrigue V, Szwarc I, Foulongne V, Mourad G. 2011. Monthly screening for BK viremia is an effective strategy to prevent BK virus nephropathy in renal transplant recipients. Transpl Infect Dis 13:101–108. doi: 10.1111/j.1399-3062.2011.00619.x. [DOI] [PubMed] [Google Scholar]
- 113.Viscount HB, Eid AJ, Espy MJ, Griffin MD, Thomsen KM, Harmsen WS, Razonable RR, Smith TF. 2007. Polyomavirus polymerase chain reaction as a surrogate marker of polyomavirus-associated nephropathy. Transplantation 84:340–345. doi: 10.1097/01.tp.0000275205.41078.51. [DOI] [PubMed] [Google Scholar]
- 114.Cesaro S, Tridello G, Pillon M, Calore E, Abate D, Tumino M, Carucci N, Varotto S, Cannata E, Pegoraro A, Barzon L, Palu G, Messina C. 2015. A prospective study on the predictive value of plasma BK virus-DNA load for hemorrhagic cystitis in pediatric patients after stem cell transplantation. J Pediatric Infect Dis Soc 4:134–142. doi: 10.1093/jpids/piu043. [DOI] [PubMed] [Google Scholar]
- 115.Erard V, Kim HW, Corey L, Limaye A, Huang ML, Myerson D, Davis C, Boeckh M. 2005. BK DNA viral load in plasma: evidence for an association with hemorrhagic cystitis in allogeneic hematopoietic cell transplant recipients. Blood 106:1130–1132. doi: 10.1182/blood-2004-12-4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Laskin BL, Denburg M, Furth S, Diorio D, Goebel J, Davies SM, Jodele S. 2013. BK viremia precedes hemorrhagic cystitis in children undergoing allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transplant 19:1175–1182. doi: 10.1016/j.bbmt.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Costa C, Bergallo M, Astegiano S, Terlizzi ME, Sidoti F, Segoloni GP, Cavallo R. 2008. Monitoring of BK virus replication in the first year following renal transplantation. Nephrol Dial Transplant 23:3333–3336. doi: 10.1093/ndt/gfn289. [DOI] [PubMed] [Google Scholar]
- 118.Koukoulaki M, Grispou E, Pistolas D, Balaska K, Apostolou T, Anagnostopoulou M, Tseleni-Kotsovili A, Hadjiconstantinou V, Paniara O, Saroglou G, Legakis N, Drakopoulos S. 2009. Prospective monitoring of BK virus replication in renal transplant recipients. Transpl Infect Dis 11:1–10. doi: 10.1111/j.1399-3062.2008.00342.x. [DOI] [PubMed] [Google Scholar]
- 119.Bagchi S, Gopalakrishnan V, Srivastava SK, Upadhayay A, Singh G, Bhowmik D, Mahajan S, Dinda A, Agarwal SK. 2017. BK polyomavirus infection after renal transplantation: surveillance in a resource-challenged setting. Transpl Infect Dis 19:e12770. doi: 10.1111/tid.12770. [DOI] [PubMed] [Google Scholar]
- 120.Sood P, Senanayake S, Sujeet K, Medipalli R, Zhu YR, Johnson CP, Hariharan S. 2012. Management and outcome of BK viremia in renal transplant recipients: a prospective single-center study. Transplantation 94:814–821. doi: 10.1097/TP.0b013e31826690c6. [DOI] [PubMed] [Google Scholar]
- 121.Wang Y, Strassl R, Helantera I, Aberle SW, Bond G, Hedman K, Weseslindtner L. 2019. Multiplex analysis of human polyomavirus diversity in kidney transplant recipients with BK virus replication. J Clin Virol 120:6–11. doi: 10.1016/j.jcv.2019.08.012. [DOI] [PubMed] [Google Scholar]
- 122.Schachtner T, Babel N, Reinke P. 2015. Different risk factor profiles distinguish early-onset from late-onset BKV-replication. Transpl Int 28:1081–1091. doi: 10.1111/tri.12601. [DOI] [PubMed] [Google Scholar]
- 123.Wunderink HF, van der Meijden E, van der Blij-de Brouwer CS, Mallat MJK, Haasnoot GW, van Zwet EW, Claas ECJ, de Fijter JW, Kroes ACM, Arnold F, Touzé A, Claas FHJ, Rotmans JI, Feltkamp MCW. 2017. Pretransplantation donor-recipient pair seroreactivity against BK polyomavirus predicts viremia and nephropathy after kidney transplantation. Am J Transplant 17:161–172. doi: 10.1111/ajt.13880. [DOI] [PubMed] [Google Scholar]
- 124.Urbano PRP, Nali L, Oliveira RDR, Sumita LM, Fink M, Pierrotti LC, Bicalho CDS, David-Neto E, Pannuti CS, Romano CM. 2019. Variable sources of BK virus in renal allograft recipients. J Med Virol 91:1136–1141. doi: 10.1002/jmv.25409. [DOI] [PubMed] [Google Scholar]
- 125.Bohl DL, Brennan DC, Ryschkewitsch C, Gaudreault-Keener M, Major EO, Storch GA. 2008. BK virus antibody titers and intensity of infections after renal transplantation. J Clin Virol 43:184–189. doi: 10.1016/j.jcv.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Erard V, Storer B, Corey L, Nollkamper J, Huang ML, Limaye A, Boeckh M. 2004. BK virus infection in hematopoietic stem cell transplant recipients: frequency, risk factors, and association with postengraftment hemorrhagic cystitis. Clin Infect Dis 39:1861–1865. doi: 10.1086/426140. [DOI] [PubMed] [Google Scholar]
- 127.Illiaquer M, Imbert-Marcille BM, Guillaume T, Planche L, Rimbert M, Bressollette-Bodin C, Le Bourgeois A, Peterlin P, Garnier A, Le Houerou C, Moreau P, Mohty M, Chevallier P. 2017. Impact of stem cell graft on early viral infections and immune reconstitution after allogeneic transplantation in adults. J Clin Virol 93:30–36. doi: 10.1016/j.jcv.2017.05.019. [DOI] [PubMed] [Google Scholar]
- 128.Admiraal R, de Koning CCH, Lindemans CA, Bierings MB, Wensing AMJ, Versluys AB, Wolfs TFW, Nierkens S, Boelens JJ. 2017. Viral reactivations and associated outcomes in the context of immune reconstitution after pediatric hematopoietic cell transplantation. J Allergy Clin Immunol 140:1643–1650.e9. doi: 10.1016/j.jaci.2016.12.992. [DOI] [PubMed] [Google Scholar]
- 129.Allander T, Andreasson K, Gupta S, Bjerkner A, Bogdanovic G, Persson MA, Dalianis T, Ramqvist T, Andersson B. 2007. Identification of a third human polyomavirus. JVI 81:4130–4136. doi: 10.1128/JVI.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Huijskens EG, van Erkel AJ, Peeters MF, Rossen JW. 2010. Human polyomavirus KI and WU in adults with community acquired pneumonia in The Netherlands, 2008–2009. J Clin Virol 49:306–307. doi: 10.1016/j.jcv.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hansen-Estruch C, Coleman KK, Thoon KC, Low JG, Anderson BD, Gray GC. 2018. Prevalence of respiratory polyomaviruses among pediatric patients with respiratory symptoms in Singapore. Front Pediatr 6:228. doi: 10.3389/fped.2018.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rao S, Lucero MG, Nohynek H, Tallo V, Lupisan SP, Garcea RL, Simoes EAF, ARIVAC Consortium . 2016. WU and KI polyomavirus infections in Filipino children with lower respiratory tract disease. J Clin Virol 82:112–118. doi: 10.1016/j.jcv.2016.07.013. [DOI] [PubMed] [Google Scholar]
- 133.Csoma E, Meszaros B, Asztalos L, Gergely L. 2015. WU and KI polyomaviruses in respiratory, blood and urine samples from renal transplant patients. J Clin Virol 64:28–33. doi: 10.1016/j.jcv.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 134.Rao S, Garcea RL, Robinson CC, Simoes EA. 2011. WU and KI polyomavirus infections in pediatric hematology/oncology patients with acute respiratory tract illness. J Clin Virol 52:28–32. doi: 10.1016/j.jcv.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Touinssi M, Galicher V, de Micco P, Biagini P. 2013. Molecular epidemiology of KI and WU polyomaviruses in healthy blood donors, south-eastern France. J Med Virol 85:1444–1446. doi: 10.1002/jmv.23602. [DOI] [PubMed] [Google Scholar]
- 136.Babakir-Mina M, Ciccozzi M, Farchi F, Bergallo M, Cavallo R, Adorno G, Perno CF, Ciotti M. 2010. KI and WU polyomaviruses and CD4+ cell counts in HIV-1-infected patients, Italy. Emerg Infect Dis 16:1482–1485. doi: 10.3201/eid1609.100211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Porrovecchio R, Babakir-Mina M, Rapanotti MC, Arcese W, Perno CF, Ciotti M. 2013. Monitoring of KI and WU polyomaviruses in hematopoietic stem cell transplant patients. J Med Virol 85:1122–1124. doi: 10.1002/jmv.23565. [DOI] [PubMed] [Google Scholar]
- 138.Csoma E, Sapy T, Meszaros B, Gergely L. 2012. Novel human polyomaviruses in pregnancy: higher prevalence of BKPyV, but no WUPyV, KIPyV and HPyV9. J Clin Virol 55:262–265. doi: 10.1016/j.jcv.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 139.Barzon L, Squarzon L, Militello V, Trevisan M, Palu G. 2009. Human KI and WU polyomavirus infection in immunocompromised subjects. J Clin Virol 45:370. doi: 10.1016/j.jcv.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 140.Csoma E, Meszaros B, Asztalos L, Konya J, Gergely L. 2011. Prevalence of WU and KI polyomaviruses in plasma, urine, and respiratory samples from renal transplant patients. J Med Virol 83:1275–1278. doi: 10.1002/jmv.22083. [DOI] [PubMed] [Google Scholar]
- 141.Bialasiewicz S, Whiley DM, Lambert SB, Nissen MD, Sloots TP. 2009. Detection of BK, JC, WU, or KI polyomaviruses in faecal, urine, blood, cerebrospinal fluid and respiratory samples. J Clin Virol 45:249–254. doi: 10.1016/j.jcv.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 142.Rahiala J, Koskenvuo M, Sadeghi M, Waris M, Vuorinen T, Lappalainen M, Saarinen-Pihkala U, Allander T, Soderlund-Venermo M, Hedman K, Ruuskanen O, Vettenranta K. 2016. Polyomaviruses BK, JC, KI, WU, MC, and TS in children with allogeneic hematopoietic stem cell transplantation. Pediatr Transplant 20:424–431. doi: 10.1111/petr.12659. [DOI] [PubMed] [Google Scholar]
- 143.Gaynor AM, Nissen MD, Whiley DM, Mackay IM, Lambert SB, Wu G, Brennan DC, Storch GA, Sloots TP, Wang D. 2007. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog 3:e64. doi: 10.1371/journal.ppat.0030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bergallo M, Terlizzi ME, Astegiano S, Ciotti M, Babakir-Mina M, Perno CF, Cavallo R, Costa C. 2009. Real time PCR TaqMan assays for detection of polyomaviruses KIV and WUV in clinical samples. J Virol Methods 162:69–74. doi: 10.1016/j.jviromet.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Feng H, Shuda M, Chang Y, Moore PS. 2008. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Koljonen V, Sahi H, Bohling T, Makisalo H. 2016. Post-transplant Merkel cell carcinoma. Acta Derm Venereol 96:442–447. doi: 10.2340/00015555-2284. [DOI] [PubMed] [Google Scholar]
- 147.Harms PW, Harms KL, Moore PS, DeCaprio JA, Nghiem P, Wong MKK, Brownell I, International Workshop on Merkel Cell Carcinoma Research (WMCC) Working Group . 2018. The biology and treatment of Merkel cell carcinoma: current understanding and research priorities. Nat Rev Clin Oncol 15:763–776. doi: 10.1038/s41571-018-0103-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Carter JJ, Paulson KG, Wipf GC, Miranda D, Madeleine MM, Johnson LG, Lemos BD, Lee S, Warcola AH, Iyer JG, Nghiem P, Galloway DA. 2009. Association of Merkel cell polyomavirus-specific antibodies with Merkel cell carcinoma. J Natl Cancer Inst 101:1510–1522. doi: 10.1093/jnci/djp332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ngoi CN, Siqueira J, Li L, Deng X, Mugo P, Graham SM, Price MA, Sanders EJ, Delwart E. 2016. The plasma virome of febrile adult Kenyans shows frequent parvovirus B19 infections and a novel arbovirus (Kadipiro virus). J Gen Virol 97:3359–3367. doi: 10.1099/jgv.0.000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mazzoni E, Rotondo JC, Marracino L, Selvatici R, Bononi I, Torreggiani E, Touze A, Martini F, Tognon MG. 2017. Detection of Merkel cell polyomavirus DNA in serum samples of healthy blood donors. Front Oncol 7:294. doi: 10.3389/fonc.2017.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jin HT, Park SJ, Choi EK, Kim YS. 2019. The frequency of Merkel cell polyomavirus in whole blood from immunocompetent and immunosuppressed patients with kidney disease and healthy donors. Microb Pathog 131:75–80. doi: 10.1016/j.micpath.2019.03.020. [DOI] [PubMed] [Google Scholar]
- 152.Sadeghi M, Aronen M, Chen T, Jartti L, Jartti T, Ruuskanen O, Soderlund-Venermo M, Hedman K. 2012. Merkel cell polyomavirus and trichodysplasia spinulosa-associated polyomavirus DNAs and antibodies in blood among the elderly. BMC Infect Dis 12:383. doi: 10.1186/1471-2334-12-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Comar M, Zanotta N, Del Savio R, Vascotto F, Calabrese N, Zorat F, Pozzato G. 2014. No evidence of polyomavirus and EBV infections in Italian patients with mixed cryoglobulinemia infected chronically with HCV. J Med Virol 86:666–671. doi: 10.1002/jmv.23867. [DOI] [PubMed] [Google Scholar]
- 154.Fukumoto H, Sato Y, Hasegawa H, Katano H. 2013. Frequent detection of Merkel cell polyomavirus DNA in sera of HIV-1-positive patients. Virol J 10:84. doi: 10.1186/1743-422X-10-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pancaldi C, Corazzari V, Maniero S, Mazzoni E, Comar M, Martini F, Tognon M. 2011. Merkel cell polyomavirus DNA sequences in the buffy coats of healthy blood donors. Blood 117:7099–7101. doi: 10.1182/blood-2010-09-310557. [DOI] [PubMed] [Google Scholar]
- 156.Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. 2010. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe 7:509–515. doi: 10.1016/j.chom.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fava P, Merlino C, Novelli M, Ponti R, Galliano I, Montanari P, Tovo PA, Fierro MT, Bergallo M. 2016. HPyV6, HPyV7 and TSPyV DNA sequences detection in skin disease patients and healthy subjects. J Eur Acad Dermatol Venereol 30:624–627. doi: 10.1111/jdv.13094. [DOI] [PubMed] [Google Scholar]
- 158.Hampras SS, Locke FL, Chavez JC, Patel NS, Giuliano AR, Miller K, Gheit T, Tommasino M, Rollison DE. 2018. Prevalence of cutaneous viral infections in incident cutaneous squamous cell carcinoma detected among chronic lymphocytic leukemia and hematopoietic stem cell transplant patients. Leuk Lymphoma 59:911–917. doi: 10.1080/10428194.2017.1342822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hashida Y, Higuchi T, Matsuzaki S, Nakajima K, Sano S, Daibata M. 2018. Prevalence and genetic variability of human polyomaviruses 6 and 7 in healthy skin among asymptomatic individuals. J Infect Dis 217:483–493. doi: 10.1093/infdis/jix516. [DOI] [PubMed] [Google Scholar]
- 160.Wieland U, Silling S, Hellmich M, Potthoff A, Pfister H, Kreuter A. 2014. Human polyomaviruses 6, 7, 9, 10 and trichodysplasia spinulosa-associated polyomavirus in HIV-infected men. J Gen Virol 95:928–932. doi: 10.1099/vir.0.061259-0. [DOI] [PubMed] [Google Scholar]
- 161.Ho J, Jedrych JJ, Feng H, Natalie AA, Grandinetti L, Mirvish E, Crespo MM, Yadav D, Fasanella KE, Proksell S, Kuan SF, Pastrana DV, Buck CB, Shuda Y, Moore PS, Chang Y. 2015. Human polyomavirus 7-associated pruritic rash and viremia in transplant recipients. J Infect Dis 211:1560–1565. doi: 10.1093/infdis/jiu524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rockett RJ, Sloots TP, Bowes S, O’Neill N, Ye S, Robson J, Whiley DM, Lambert SB, Wang D, Nissen MD, Bialasiewicz S. 2013. Detection of novel polyomaviruses, TSPyV, HPyV6, HPyV7, HPyV9 and MWPyV in feces, urine, blood, respiratory swabs and cerebrospinal fluid. PLoS One 8:e62764. doi: 10.1371/journal.pone.0062764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.van der Meijden E, Janssens RW, Lauber C, Bouwes Bavinck JN, Gorbalenya AE, Feltkamp MC. 2010. Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient. PLoS Pathog 6:e1001024. doi: 10.1371/journal.ppat.1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kazem S, van der Meijden E, Feltkamp MC. 2013. The trichodysplasia spinulosa-associated polyomavirus: virological background and clinical implications. APMIS 121:770–782. doi: 10.1111/apm.12092. [DOI] [PubMed] [Google Scholar]
- 165.van der Meijden E, Horvath B, Nijland M, de Vries K, Racz E, Diercks GF, de Weerd AE, Clahsen-van Groningen MC, van der Blij-de Brouwer CS, van der Zon AJ, Kroes AC, Hedman K, van Kampen JJ, Riezebos-Brilman A, Feltkamp MC. 2017. Primary polyomavirus infection, not reactivation, as the cause of trichodysplasia spinulosa in immunocompromised patients. J Infect Dis 215:1080–1084. doi: 10.1093/infdis/jiw403. [DOI] [PubMed] [Google Scholar]
- 166.Borgogna C, Albertini S, Zavattaro E, Veronese F, Peruzzi L, Meijden E, Feltkamp MCW, Tosoni A, Volpe A, Boldorini R, Gariglio M. 2019. Primary trichodysplasia spinulosa polyomavirus infection in a kidney transplant child displaying virus-infected decoy cells in the urine. J Med Virol 91:1896–1900. doi: 10.1002/jmv.25519. [DOI] [PubMed] [Google Scholar]
- 167.Chamseddin BH, Tran B, Lee EE, Pastrana DV, Buck CB, Wang RC, Kirkorian AY. 2019. Trichodysplasia spinulosa in a child: identification of trichodysplasia spinulosa-associated polyomavirus in skin, serum, and urine. Pediatr Dermatol 36:723–724. doi: 10.1111/pde.13857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Pierrotti LC, Urbano PRP, Nali L, Romano CM, Bicalho CDS, Arnone M, Valente NS, Pannuti CS, David-Neto E, Azevedo LS. 2019. Viremia and viruria of trichodysplasia spinulosa-associated polyomavirus before the development of clinical disease in a kidney transplant recipient. Transpl Infect Dis 21:e13133. doi: 10.1111/tid.13133. [DOI] [PubMed] [Google Scholar]
- 169.Urbano PR, Nali LH, Bicalho CS, Pierrotti LC, David-Neto E, Pannuti CS, Romano CM. 2016. New findings about trichodysplasia spinulosa-associated polyomavirus (TSPyV)—novel qPCR detects TSPyV-DNA in blood samples. Diagn Microbiol Infect Dis 84:123–124. doi: 10.1016/j.diagmicrobio.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 170.Sadeghi M, Aaltonen LM, Hedman L, Chen T, Soderlund-Venermo M, Hedman K. 2014. Detection of TS polyomavirus DNA in tonsillar tissues of children and adults: evidence for site of viral latency. J Clin Virol 59:55–58. doi: 10.1016/j.jcv.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 171.Scuda N, Hofmann J, Calvignac-Spencer S, Ruprecht K, Liman P, Kuhn J, Hengel H, Ehlers B. 2011. A novel human polyomavirus closely related to the African green monkey-derived lymphotropic polyomavirus. J Virol 85:4586–4590. doi: 10.1128/JVI.02602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Nicol JT, Robinot R, Carpentier A, Carandina G, Mazzoni E, Tognon M, Touze A, Coursaget P. 2013. Age-specific seroprevalences of Merkel cell polyomavirus, human polyomaviruses 6, 7, and 9, and trichodysplasia spinulosa-associated polyomavirus. Clin Vaccine Immunol 20:363–368. doi: 10.1128/CVI.00438-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Nicol JT, Touze A, Robinot R, Arnold F, Mazzoni E, Tognon M, Coursaget P. 2012. Seroprevalence and cross-reactivity of human polyomavirus 9. Emerg Infect Dis 18:1329–1332. doi: 10.3201/eid1808.111625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.van der Meijden E, Wunderink HF, van der Blij-de Brouwer CS, Zaaijer HL, Rotmans JI, Bavinck JN, Feltkamp MC. 2014. Human polyomavirus 9 infection in kidney transplant patients. Emerg Infect Dis 20:991–999. doi: 10.3201/eid2006.140055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lednicky JA, Butel JS, Luetke MC, Loeb JC. 2014. Complete genomic sequence of a new human polyomavirus 9 strain with an altered noncoding control region. Virus Genes 49:490–492. doi: 10.1007/s11262-014-1119-z. [DOI] [PubMed] [Google Scholar]
- 176.Siebrasse EA, Reyes A, Lim ES, Zhao G, Mkakosya RS, Manary MJ, Gordon JI, Wang D. 2012. Identification of MW polyomavirus, a novel polyomavirus in human stool. J Virol 86:10321–10326. doi: 10.1128/JVI.01210-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lim ES, Reyes A, Antonio M, Saha D, Ikumapayi UN, Adeyemi M, Stine OC, Skelton R, Brennan DC, Mkakosya RS, Manary MJ, Gordon JI, Wang D. 2013. Discovery of STL polyomavirus, a polyomavirus of ancestral recombinant origin that encodes a unique T antigen by alternative splicing. Virology 436:295–303. doi: 10.1016/j.virol.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Korup S, Rietscher J, Calvignac-Spencer S, Trusch F, Hofmann J, Moens U, Sauer I, Voigt S, Schmuck R, Ehlers B. 2013. Identification of a novel human polyomavirus in organs of the gastrointestinal tract. PLoS One 8:e58021. doi: 10.1371/journal.pone.0058021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Mishra N, Pereira M, Rhodes RH, An P, Pipas JM, Jain K, Kapoor A, Briese T, Faust PL, Lipkin WI. 2014. Identification of a novel polyomavirus in a pancreatic transplant recipient with retinal blindness and vasculitic myopathy. J Infect Dis 210:1595–1599. doi: 10.1093/infdis/jiu250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Gheit T, Dutta S, Oliver J, Robitaille A, Hampras S, Combes JD, McKay-Chopin S, Le Calvez-Kelm F, Fenske N, Cherpelis B, Giuliano AR, Franceschi S, McKay J, Rollison DE, Tommasino M. 2017. Isolation and characterization of a novel putative human polyomavirus. Virology 506:45–54. doi: 10.1016/j.virol.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ma FL, Li DD, Wei TL, Li JS, Zheng LS. 2017. Quantitative detection of human Malawi polyomavirus in nasopharyngeal aspirates, sera, and feces in Beijing, China, using real-time TaqMan-based PCR. Virol J 14:152. doi: 10.1186/s12985-017-0817-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Papa N, Zanotta N, Knowles A, Orzan E, Comar M. 2016. Detection of Malawi polyomavirus sequences in secondary lymphoid tissues from Italian healthy children: a transient site of infection. Virol J 13:97. doi: 10.1186/s12985-016-0553-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lion T. 2014. Adenovirus infections in immunocompetent and immunocompromised patients. Clin Microbiol Rev 27:441–462. doi: 10.1128/CMR.00116-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hatakeyama N, Suzuki N, Kudoh T, Hori T, Mizue N, Tsutsumi H. 2003. Successful cidofovir treatment of adenovirus-associated hemorrhagic cystitis and renal dysfunction after allogenic bone marrow transplant. Pediatr Infect Dis J 22:928–929. doi: 10.1097/01.inf.0000091399.29505.21. [DOI] [PubMed] [Google Scholar]
- 185.Akiyama H, Kurosu T, Sakashita C, Inoue T, Mori S, Ohashi K, Tanikawa S, Sakamaki H, Onozawa Y, Chen Q, Zheng H, Kitamura T. 2001. Adenovirus is a key pathogen in hemorrhagic cystitis associated with bone marrow transplantation. Clin Infect Dis 32:1325–1330. doi: 10.1086/319992. [DOI] [PubMed] [Google Scholar]
- 186.Lion T, Baumgartinger R, Watzinger F, Matthes-Martin S, Suda M, Preuner S, Futterknecht B, Lawitschka A, Peters C, Pötschger U, Gadner H. 2003. Molecular monitoring of adenovirus in peripheral blood after allogeneic bone marrow transplantation permits early diagnosis of disseminated disease. Blood 102:1114–1120. doi: 10.1182/blood-2002-07-2152. [DOI] [PubMed] [Google Scholar]
- 187.Takayama R, Hatakeyama N, Suzuki N, Yamamoto M, Hayashi T, Ikeda Y, Ikeda H, Nagano H, Ishida T, Tsutsumi H. 2007. Quantification of adenovirus species B and C viremia by real-time PCR in adults and children undergoing stem cell transplantation. J Med Virol 79:278–284. doi: 10.1002/jmv.20796. [DOI] [PubMed] [Google Scholar]
- 188.Grimley MS, Chemaly RF, Englund JA, Kurtzberg J, Chittick G, Brundage TM, Bae A, Morrison ME, Prasad VK. 2017. Brincidofovir for asymptomatic adenovirus viremia in pediatric and adult allogeneic hematopoietic cell transplant recipients: a randomized placebo-controlled phase II trial. Biol Blood Marrow Transplant 23:512–521. doi: 10.1016/j.bbmt.2016.12.621. [DOI] [PubMed] [Google Scholar]
- 189.Florescu DF, Hoffman JA, AST Infectious Diseases Community of Practice . 2013. Adenovirus in solid organ transplantation. Am J Transplant 13(Suppl 4):206–211. doi: 10.1111/ajt.12112. [DOI] [PubMed] [Google Scholar]
- 190.Veltrop-Duits LA, van Vreeswijk T, Heemskerk B, Thijssen JC, El Seady R, Jol-van der Zijde EM, Claas EC, Lankester AC, van Tol MJ, Schilham MW. 2011. High titers of pre-existing adenovirus serotype-specific neutralizing antibodies in the host predict viral reactivation after allogeneic stem cell transplantation in children. Clin Infect Dis 52:1405–1413. doi: 10.1093/cid/cir231. [DOI] [PubMed] [Google Scholar]
- 191.Florescu DF, Schaenman JM, AST Infectious Diseases Community of Practice . 2019. Adenovirus in solid organ transplant recipients: guidelines from the American Society of Transplantation Infectious Diseases Community of Practice. Clin Transplant 33:e13527. doi: 10.1111/ctr.13527. [DOI] [PubMed] [Google Scholar]
- 192.Hiwarkar P, Gaspar HB, Gilmour K, Jagani M, Chiesa R, Bennett-Rees N, Breuer J, Rao K, Cale C, Goulden N, Davies G, Amrolia P, Veys P, Qasim W. 2013. Impact of viral reactivations in the era of pre-emptive antiviral drug therapy following allogeneic haematopoietic SCT in paediatric recipients. Bone Marrow Transplant 48:803–808. doi: 10.1038/bmt.2012.221. [DOI] [PubMed] [Google Scholar]
- 193.van Tol MJ, Kroes AC, Schinkel J, Dinkelaar W, Claas EC, Jol-van der Zijde CM, Vossen JM. 2005. Adenovirus infection in paediatric stem cell transplant recipients: increased risk in young children with a delayed immune recovery. Bone Marrow Transplant 36:39–50. doi: 10.1038/sj.bmt.1705003. [DOI] [PubMed] [Google Scholar]
- 194.Walls T, Hawrami K, Ushiro-Lumb I, Shingadia D, Saha V, Shankar AG. 2005. Adenovirus infection after pediatric bone marrow transplantation: is treatment always necessary? Clin Infect Dis 40:1244–1249. doi: 10.1086/429235. [DOI] [PubMed] [Google Scholar]
- 195.Chakrabarti S, Mautner V, Osman H, Collingham KE, Fegan CD, Klapper PE, Moss PA, Milligan DW. 2002. Adenovirus infections following allogeneic stem cell transplantation: incidence and outcome in relation to graft manipulation, immunosuppression, and immune recovery. Blood 100:1619–1627. doi: 10.1182/blood-2002-02-0377. [DOI] [PubMed] [Google Scholar]
- 196.Sedlacek P, Petterson T, Robin M, Sivaprakasam P, Vainorius E, Brundage T, Chandak A, Mozaffari E, Nichols G, Voigt S. 2019. Incidence of adenovirus infection in hematopoietic stem cell transplantation recipients: findings from the AdVance Study. Biol Blood Marrow Transplant 25:810–818. doi: 10.1016/j.bbmt.2018.12.753. [DOI] [PubMed] [Google Scholar]
- 197.Matthes-Martin S, Feuchtinger T, Shaw PJ, Engelhard D, Hirsch HH, Cordonnier C, Ljungman P, Fourth European Conference on Infections in Leukemia . 2012. European guidelines for diagnosis and treatment of adenovirus infection in leukemia and stem cell transplantation: summary of ECIL-4 (2011). Transpl Infect Dis 14:555–563. doi: 10.1111/tid.12022. [DOI] [PubMed] [Google Scholar]
- 198.Howard DS, Phillips IG, Reece DE, Munn RK, Henslee-Downey J, Pittard M, Barker M, Pomeroy C. 1999. Adenovirus infections in hematopoietic stem cell transplant recipients. Clin Infect Dis 29:1494–1501. doi: 10.1086/313514. [DOI] [PubMed] [Google Scholar]
- 199.Erard V, Huang ML, Ferrenberg J, Nguy L, Stevens-Ayers TL, Hackman RC, Corey L, Boeckh M. 2007. Quantitative real-time polymerase chain reaction for detection of adenovirus after T cell-replete hematopoietic cell transplantation: viral load as a marker for invasive disease. Clin Infect Dis 45:958–965. doi: 10.1086/521851. [DOI] [PubMed] [Google Scholar]
- 200.Lee YJ, Huang YT, Kim SJ, Maloy M, Tamari R, Giralt SA, Papadopoulos EB, Jakubowski AA, Papanicolaou GA. 2016. Adenovirus viremia in adult CD34(+) selected hematopoietic cell transplant recipients: low incidence and high clinical impact. Biol Blood Marrow Transplant 22:174–178. doi: 10.1016/j.bbmt.2015.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Humar A, Kumar D, Mazzulli T, Razonable RR, Moussa G, Paya CV, Covington E, Alecock E, Pescovitz MD, PV16000 Study Group . 2005. A surveillance study of adenovirus infection in adult solid organ transplant recipients. Am J Transplant 5:2555–2559. doi: 10.1111/j.1600-6143.2005.01033.x. [DOI] [PubMed] [Google Scholar]
- 202.Engen RM, Huang ML, Park GE, Smith JM, Limaye AP. 2018. Prospective assessment of adenovirus infection in pediatric kidney transplant recipients. Transplantation 102:1165–1171. doi: 10.1097/TP.0000000000002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Gustafson I, Lindblom A, Yun Z, Omar H, Engstrom L, Lewensohn-Fuchs I, Ljungman P, Broliden K. 2008. Quantification of adenovirus DNA in unrelated donor hematopoietic stem cell transplant recipients. J Clin Virol 43:79–85. doi: 10.1016/j.jcv.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 204.Qiu J, Soderlund-Venermo M, Young NS. 2017. Human parvoviruses. Clin Microbiol Rev 30:43–113. doi: 10.1128/CMR.00040-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Cossart YE, Field AM, Cant B, Widdows D. 1975. Parvovirus-like particles in human sera. Lancet 1:72–73. doi: 10.1016/S0140-6736(75)91074-0. [DOI] [PubMed] [Google Scholar]
- 206.Norja P, Hokynar K, Aaltonen LM, Chen R, Ranki A, Partio EK, Kiviluoto O, Davidkin I, Leivo T, Eis-Hubinger AM, Schneider B, Fischer HP, Tolba R, Vapalahti O, Vaheri A, Soderlund-Venermo M, Hedman K. 2006. Bioportfolio: lifelong persistence of variant and prototypic erythrovirus DNA genomes in human tissue. Proc Natl Acad Sci U S A 103:7450–7453. doi: 10.1073/pnas.0602259103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Miron D, Horovitz Y, Luder A, Ohnona FS, Schlesinger Y. 2010. Age-related immunoglobulin G seroprevalence of human parvovirus B-19 in Israeli children. Isr Med Assoc J 12:277–279. [PubMed] [Google Scholar]
- 208.Rohrer C, Gartner B, Sauerbrei A, Bohm S, Hottentrager B, Raab U, Thierfelder W, Wutzler P, Modrow S. 2008. Seroprevalence of parvovirus B19 in the German population. Epidemiol Infect 136:1564–1575. doi: 10.1017/S0950268807009958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.van Rijckevorsel GG, Sonder GJ, Schim van der Loeff MF, van den Hoek JA. 2009. Population-based study on the seroprevalence of parvovirus B19 in Amsterdam. J Med Virol 81:1305–1309. doi: 10.1002/jmv.21528. [DOI] [PubMed] [Google Scholar]
- 210.Ihara T, Furusyo N, Hayashi T, Toyoda K, Murata M, Hayashi J. 2013. A population-based epidemiological survey of human parvovirus B19 infection: a project of the Kyushu and Okinawa Population Study (KOPS). Arch Virol 158:2465–2472. doi: 10.1007/s00705-013-1746-z. [DOI] [PubMed] [Google Scholar]
- 211.Ke L, He M, Li C, Liu Y, Gao L, Yao F, Li J, Bi X, Lv Y, Wang J, Hirsch ML, Li W. 2011. The prevalence of human parvovirus B19 DNA and antibodies in blood donors from four Chinese blood centers. Transfusion 51:1909–1918. doi: 10.1111/j.1537-2995.2011.03067.x. [DOI] [PubMed] [Google Scholar]
- 212.Zadsar M, Aghakhani A, Banifazl M, Kazemimanesh M, Tabatabaei Yazdi SM, Mamishi S, Bavand A, Sadat Larijani M, Ramezani A. 2018. Seroprevalence, molecular epidemiology and quantitation of parvovirus B19 DNA levels in Iranian blood donors. J Med Virol 90:1318–1322. doi: 10.1002/jmv.25195. [DOI] [PubMed] [Google Scholar]
- 213.Li X, Lin Z, Liu J, Tang Y, Yuan X, Li N, Lin Z, Chen Y, Liu A. 2020. Overall prevalence of human parvovirus B19 among blood donors in mainland China: a PRISMA-compliant meta-analysis. Medicine (Baltimore, MD) 99:e19832. doi: 10.1097/MD.0000000000019832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Raturi G, Kaur P, Kaur G. 2018. Seroprevalence of human parvovirus B19 amongst North Indian blood donors—do current donor testing guidelines need a relook? Transfus Apher Sci 57:646–650. doi: 10.1016/j.transci.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 215.Chabo Byaene A, Kabututu ZP, Abou Rayia DM, El-Sokkary M. 2020. The seroprevalence of parvovirus B19 antibody in blood donors at the National Blood Transfusion Center in Kinshasa. J Med Virol 92:288–294. doi: 10.1002/jmv.25613. [DOI] [PubMed] [Google Scholar]
- 216.Slavov SN, Rodrigues ES, Sauvage V, Caro V, Diefenbach CF, Zimmermann AM, Covas DT, Laperche S, Kashima S. 2019. Parvovirus B19 seroprevalence, viral load, and genotype characterization in volunteer blood donors from southern Brazil. J Med Virol 91:1224–1231. doi: 10.1002/jmv.25453. [DOI] [PubMed] [Google Scholar]
- 217.Bredl S, Plentz A, Wenzel JJ, Pfister H, Most J, Modrow S. 2011. False-negative serology in patients with acute parvovirus B19 infection. J Clin Virol 51:115–120. doi: 10.1016/j.jcv.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 218.Lindblom A, Isa A, Norbeck O, Wolf S, Johansson B, Broliden K, Tolfvenstam T. 2005. Slow clearance of human parvovirus B19 viremia following acute infection. Clin Infect Dis 41:1201–1203. doi: 10.1086/444503. [DOI] [PubMed] [Google Scholar]
- 219.Manaresi E, Gallinella G, Zuffi E, Bonvicini F, Zerbini M, Musiani M. 2002. Diagnosis and quantitative evaluation of parvovirus B19 infections by real-time PCR in the clinical laboratory. J Med Virol 67:275–281. doi: 10.1002/jmv.2218. [DOI] [PubMed] [Google Scholar]
- 220.Juhl D, Gorg S, Hennig H. 2014. Persistence of parvovirus B19 (B19V) DNA and humoral immune response in B19V-infected blood donors. Vox Sang 107:226–232. doi: 10.1111/vox.12162. [DOI] [PubMed] [Google Scholar]
- 221.Molenaar-de Backer MW, Russcher A, Kroes AC, Koppelman MH, Lanfermeijer M, Zaaijer HL. 2016. Detection of parvovirus B19 DNA in blood: viruses or DNA remnants? J Clin Virol 84:19–23. doi: 10.1016/j.jcv.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 222.Matsukura H, Shibata S, Tani Y, Shibata H, Furuta RA. 2008. Persistent infection by human parvovirus B19 in qualified blood donors. Transfusion 48:1036–1037. doi: 10.1111/j.1537-2995.2008.01704.x. [DOI] [PubMed] [Google Scholar]
- 223.Lefrere JJ, Servant-Delmas A, Candotti D, Mariotti M, Thomas I, Brossard Y, Lefrere F, Girot R, Allain JP, Laperche S. 2005. Persistent B19 infection in immunocompetent individuals: implications for transfusion safety. Blood 106:2890–2895. doi: 10.1182/blood-2005-03-1053. [DOI] [PubMed] [Google Scholar]
- 224.Styles CE, Hoad VC, Gorman E, Roulis E, Flower R, Faddy HM. 2019. Modeling the parvovirus B19 blood safety risk in Australia. Transfusion 59:295–302. doi: 10.1111/trf.14965. [DOI] [PubMed] [Google Scholar]
- 225.Kooistra K, Mesman HJ, de Waal M, Koppelman MH, Zaaijer HL. 2011. Epidemiology of high-level parvovirus B19 viraemia among Dutch blood donors, 2003–2009. Vox Sang 100:261–266. doi: 10.1111/j.1423-0410.2010.01423.x. [DOI] [PubMed] [Google Scholar]
- 226.Juhl D, Hennig H. 2018. Parvovirus B19: what is the relevance in transfusion medicine? Front Med (Lausanne) 5:4. doi: 10.3389/fmed.2018.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Eid AJ, Ardura MI, AST Infectious Diseases Community of Practice . 2019. Human parvovirus B19 in solid organ transplantation: guidelines from the American Society of Transplantation Infectious Diseases Community of Practice. Clin Transplant 33:e13535. doi: 10.1111/ctr.13535. [DOI] [PubMed] [Google Scholar]
- 228.Eid AJ, Brown RA, Patel R, Razonable RR. 2006. Parvovirus B19 infection after transplantation: a review of 98 cases. Clin Infect Dis 43:40–48. doi: 10.1086/504812. [DOI] [PubMed] [Google Scholar]
- 229.Katoh D, Ochi Y, Hiramoto N, Morita M, Yabushita T, Shimomura Y, Yamashita D, Ono Y, Yoshioka S, Yonetani N, Matsushita A, Imai Y, Hashimoto H, Ishikawa T. 2020. Parvovirus B19 infection in adult patients after allogeneic stem cell transplantation: our experience of five cases and literature review. Bone Marrow Transplant 55:653–656. doi: 10.1038/s41409-019-0533-1. [DOI] [PubMed] [Google Scholar]
- 230.Broliden K. 2001. Parvovirus B19 infection in pediatric solid-organ and bone marrow transplantation. Pediatr Transplant 5:320–330. doi: 10.1034/j.1399-3046.2001.00035.x. [DOI] [PubMed] [Google Scholar]
- 231.Beske F, Modrow S, Sorensen J, Schmidt H, Kriener S, Allwinn R, Klingebiel T, Schwabe D, Lehrnbecher T. 2007. Parvovirus B19 pneumonia in a child undergoing allogeneic hematopoietic stem cell transplantation. Bone Marrow Transplant 40:89–91. doi: 10.1038/sj.bmt.1705693. [DOI] [PubMed] [Google Scholar]
- 232.Klumpen HJ, Petersen EJ, Verdonck LF. 2004. Severe multiorgan failure after parvovirus B19 infection in an allogeneic stem cell transplant recipient. Bone Marrow Transplant 34:469–470. doi: 10.1038/sj.bmt.1704612. [DOI] [PubMed] [Google Scholar]
- 233.Eis-Hubinger AM, Reber U, Edelmann A, Kalus U, Hofmann J. 2014. Parvovirus B19 genotype 2 in blood donations. Transfusion 54:1682–1684. doi: 10.1111/trf.12591. [DOI] [PubMed] [Google Scholar]
- 234.Slavov SN, Goncalves de Noronha LA, Gonzaga FAC, Pimentel BMS, Kashima S, Haddad R. 2019. Low human parvovirus B19 (B19V) DNA prevalence in blood donors from Central-West Brazil. J Med Microbiol 68:622–626. doi: 10.1099/jmm.0.000957. [DOI] [PubMed] [Google Scholar]
- 235.Gallinella G, Manaresi E, Venturoli S, Grazi GL, Musiani M, Zerbini M. 1999. Occurrence and clinical role of active parvovirus B19 infection in transplant recipients. Eur J Clin Microbiol Infect Dis 18:811–813. doi: 10.1007/s100960050406. [DOI] [PubMed] [Google Scholar]
- 236.Plentz A, Wurdinger M, Kudlich M, Modrow S. 2013. Low-level DNAemia of parvovirus B19 (genotypes 1–3) in adult transplant recipients is not associated with anaemia. J Clin Virol 58:443–448. doi: 10.1016/j.jcv.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 237.Zolnourian ZR, Curran MD, Rima BK, Coyle PV, O'Neill HJ, Middleton D. 2000. Parvovirus B19 in kidney transplant patients. Transplantation 69:2198–2202. doi: 10.1097/00007890-200005270-00043. [DOI] [PubMed] [Google Scholar]
- 238.Ki CS, Kim IS, Kim JW, Lee NY, Kim SH, Lee KW, Kim SJ, Joh JW, Huh WS, Oh HY. 2005. Incidence and clinical significance of human parvovirus B19 infection in kidney transplant recipients. Clin Transplant 19:751–755. doi: 10.1111/j.1399-0012.2005.00415.x. [DOI] [PubMed] [Google Scholar]
- 239.Cavallo R, Merlino C, Re D, Bollero C, Bergallo M, Lembo D, Musso T, Leonardi G, Segoloni GP, Ponzi AN. 2003. B19 virus infection in renal transplant recipients. J Clin Virol 26:361–368. doi: 10.1016/S1386-6532(02)00104-X. [DOI] [PubMed] [Google Scholar]
- 240.Ndimbie OK, Frezza E, Jordan JA, Koch W, van Thiel DH. 1996. Parvovirus B19 in anemic liver transplant recipients. Clin Diagn Lab Immunol 3:756–760. doi: 10.1128/CDLI.3.6.756-760.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Liang TB, Li DL, Yu J, Bai XL, Liang L, Xu SG, Wang WL, Shen Y, Zhang M, Zheng SS. 2007. Pure red cell aplasia due to parvovirus B19 infection after liver transplantation: a case report and review of the literature. World J Gastroenterol 13:2007–2010. doi: 10.3748/wjg.v13.i13.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Wurdinger M, Modrow S, Plentz A. 2017. Impact of parvovirus B19 viremia in liver transplanted children on anemia: a retrospective study. Viruses 9:149. doi: 10.3390/v9060149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Pinto NC, Newman C, Gomez CA, Khush KK, Moayedi Y, Lee R, Teuteberg JJ, Montoya JG. 2019. Parvovirus B19-induced severe anemia in heart transplant recipients: case report and review of the literature. Clin Transplant 33:e13498. doi: 10.1111/ctr.13498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Ohrmalm L, Wong M, Aust C, Ljungman P, Norbeck O, Broliden K, Tolfvenstam T. 2012. Viral findings in adult hematological patients with neutropenia. PLoS One 7:e36543. doi: 10.1371/journal.pone.0036543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Soderlund M, Ruutu P, Ruutu T, Asikainen K, Franssila R, Hedman K. 1997. Primary and secondary infections by human parvovirus B19 following bone marrow transplantation: characterization by PCR and B-cell molecular immunology. Scand J Infect Dis 29:129–135. doi: 10.3109/00365549709035872. [DOI] [PubMed] [Google Scholar]
- 246.Atay D, Akcay A, Erbey F, Ozturk G. 2018. The impact of alternative donor types on viral infections in pediatric hematopoietic stem cell transplantation. Pediatr Transplant 22:e13109. doi: 10.1111/petr.13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Rahiala J, Koskenvuo M, Norja P, Meriluoto M, Toppinen M, Lahtinen A, Väisänen E, Waris M, Vuorinen T, Saarinen-Pihkala U, Lappalainen M, Allander T, Ruuskanen O, Hedman K, Söderlund-Venermo M, Vettenranta K. 2013. Human parvoviruses B19, PARV4 and bocavirus in pediatric patients with allogeneic hematopoietic SCT. Bone Marrow Transplant 48:1308–1312. doi: 10.1038/bmt.2013.63. [DOI] [PubMed] [Google Scholar]
- 248.Arnold DM, Neame PB, Meyer RM, Soamboonsrup P, Luinstra KE, O'Hoski P, Garner J, Foley R. 2005. Autologous peripheral blood progenitor cells are a potential source of parvovirus B19 infection. Transfusion 45:394–398. doi: 10.1111/j.1537-2995.2005.04267.x. [DOI] [PubMed] [Google Scholar]
- 249.Wasak-Szulkowska E, Grabarczyk P, Rzepecki P. 2008. Pure red cell aplasia due to parvovirus B19 infection transmitted probably through hematopoietic stem cell transplantation. Transpl Infect Dis 10:201–205. doi: 10.1111/j.1399-3062.2007.00266.x. [DOI] [PubMed] [Google Scholar]
- 250.Matthews PC, Sharp C, Simmonds P, Klenerman P. 2017. Human parvovirus 4 ‘PARV4’ remains elusive despite a decade of study. F1000Res 6:82. doi: 10.12688/f1000research.9828.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Jones MS, Kapoor A, Lukashov VV, Simmonds P, Hecht F, Delwart E. 2005. New DNA viruses identified in patients with acute viral infection syndrome. J Virol 79:8230–8236. doi: 10.1128/JVI.79.13.8230-8236.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Lurcharchaiwong W, Chieochansin T, Payungporn S, Theamboonlers A, Poovorawan Y. 2008. Parvovirus 4 (PARV4) in serum of intravenous drug users and blood donors. Infection 36:488–491. doi: 10.1007/s15010-008-7336-4. [DOI] [PubMed] [Google Scholar]
- 253.Simmonds P, Manning A, Kenneil R, Carnie FW, Bell JE. 2007. Parenteral transmission of the novel human parvovirus PARV4. Emerg Infect Dis 13:1386–1388. doi: 10.3201/eid1309.070428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Matthews PC, Sharp CP, Malik A, Gregory WF, Adland E, Jooste P, Goulder PJ, Simmonds P, Klenerman P. 2015. Human parvovirus 4 infection among mothers and children in South Africa. Emerg Infect Dis 21:713–715. doi: 10.3201/eid2104.141545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Sharp CP, Vermeulen M, Nébié Y, Djoko CF, LeBreton M, Tamoufe U, Rimoin AW, Kayembe PK, Carr JK, Servant-Delmas A, Laperche S, Harrison GLA, Pybus OG, Delwart E, Wolfe ND, Saville A, Lefrère JJ, Simmonds P. 2010. Changing epidemiology of human parvovirus 4 infection in sub-Saharan Africa. Emerg Infect Dis 16:1605–1607. doi: 10.3201/eid1610.101001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Sharp CP, Gregory WF, Hattingh L, Malik A, Adland E, Daniels S, van Zyl A, Carlson JM, Wareing S, Ogwu A, Shapiro R, Riddell L, Chen F, Ndung'u T, Goulder PJR, Klenerman P, Simmonds P, Jooste P, Matthews PC. 2017. PARV4 prevalence, phylogeny, immunology and coinfection with HIV, HBV and HCV in a multicentre African cohort. Wellcome Open Res 2:26. doi: 10.12688/wellcomeopenres.11135.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.von Linstow ML, Rosenfeldt V, Lindberg E, Jensen L, Hedman L, Li X, Vaisanen E, Hedman K, Norja P. 2015. Absence of novel human parvovirus (PARV4) in Danish mothers and children. J Clin Virol 65:23–25. doi: 10.1016/j.jcv.2015.01.021. [DOI] [PubMed] [Google Scholar]
- 258.Maple PA, Beard S, Parry RP, Brown KE. 2013. Testing UK blood donors for exposure to human parvovirus 4 using a time-resolved fluorescence immunoassay to screen sera and Western blot to confirm reactive samples. Transfusion 53:2575–2584. doi: 10.1111/trf.12278. [DOI] [PubMed] [Google Scholar]
- 259.Simmons R, Sharp C, McClure CP, Rohrbach J, Kovari H, Frangou E, Simmonds P, Irving W, Rauch A, Bowness P, Klenerman P, Swiss HIV Cohort Study . 2012. Parvovirus 4 infection and clinical outcome in high-risk populations. J Infect Dis 205:1816–1820. doi: 10.1093/infdis/jis291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Yu X, Zhang J, Hong L, Wang J, Yuan Z, Zhang X, Ghildyal R. 2012. High prevalence of human parvovirus 4 infection in HBV and HCV infected individuals in Shanghai. PLoS One 7:e29474. doi: 10.1371/journal.pone.0029474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.May J, Drexler JF, Reber U, Sarpong N, Adjei O, Panning M, Drosten C, Eis-Hübinger AM. 2012. Human parvovirus 4 viremia in young children, Ghana. Emerg Infect Dis 18:1690–1692. doi: 10.3201/eid1810.111836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Chen MY, Hung CC, Lee KL. 2015. Detection of human parvovirus 4 viremia in the follow-up blood samples from seropositive individuals suggests the existence of persistent viral replication or reactivation of latent viral infection. Virol J 12:94. doi: 10.1186/s12985-015-0326-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Vallerini D, Barozzi P, Quadrelli C, Bosco R, Potenza L, Riva G, Gregorini G, Sandrini S, Tironi A, Montagnani G, De Palma M, Torelli G, Delwart E, Luppi M. 2008. Parvoviruses in blood donors and transplant patients, Italy. Emerg Infect Dis 14:185–186. doi: 10.3201/eid1401.070610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Touinssi M, Brisbarre N, Picard C, Frassati C, Dussol B, Uch R, Gallian P, Cantaloube JF, Micco P, Biagini P. 2010. Parvovirus 4 in blood donors, France. Emerg Infect Dis 16:165–166. doi: 10.3201/eid1601.090517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Asiyabi S, Nejati A, Shoja Z, Shahmahmoodi S, Jalilvand S, Farahmand M, Gorzin AA, Najafi A, Haji Mollahoseini M, Marashi SM. 2016. First report of human parvovirus 4 detection in Iran. J Med Virol 88:1314–1318. doi: 10.1002/jmv.24485. [DOI] [PubMed] [Google Scholar]
- 266.Fryer JF, Delwart E, Hecht FM, Bernardin F, Jones MS, Shah N, Baylis SA. 2007. Frequent detection of the parvoviruses, PARV4 and PARV5, in plasma from blood donors and symptomatic individuals. Transfusion 47:1054–1061. doi: 10.1111/j.1537-2995.2007.01235.x. [DOI] [PubMed] [Google Scholar]
- 267.Ma YY, Guo Y, Zhao X, Wang Z, Lv MM, Yan QP, Zhang JG. 2012. Human parvovirus PARV4 in plasma pools of Chinese origin. Vox Sang 103:183–185. doi: 10.1111/j.1423-0410.2012.01605.x. [DOI] [PubMed] [Google Scholar]
- 268.Touinssi M, Reynaud-Gaubert M, Gomez C, Thomas P, Dussol B, Berland Y, Basire A, Picard C, Cantaloube JF, de Micco P, Biagini P. 2011. Parvovirus 4 in French in-patients: a study of hemodialysis and lung transplant cohorts. J Med Virol 83:717–720. doi: 10.1002/jmv.22003. [DOI] [PubMed] [Google Scholar]
- 269.Cotmore SF, Agbandje-McKenna M, Chiorini JA, Mukha DV, Pintel DJ, Qiu J, Soderlund-Venermo M, Tattersall P, Tijssen P, Gatherer D, Davison AJ. 2014. The family Parvoviridae. Arch Virol 159:1239–1247. doi: 10.1007/s00705-013-1914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung-Lindell A, Andersson B. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci U S A 102:12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Arthur JL, Higgins GD, Davidson GP, Givney RC, Ratcliff RM. 2009. A novel bocavirus associated with acute gastroenteritis in Australian children. PLoS Pathog 5:e1000391. doi: 10.1371/journal.ppat.1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Kapoor A, Slikas E, Simmonds P, Chieochansin T, Naeem A, Shaukat S, Alam MM, Sharif S, Angez M, Zaidi S, Delwart E. 2009. A newly identified bocavirus species in human stool. J Infect Dis 199:196–200. doi: 10.1086/595831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Kapoor A, Simmonds P, Slikas E, Li L, Bodhidatta L, Sethabutr O, Triki H, Bahri O, Oderinde BS, Baba MM, Bukbuk DN, Besser J, Bartkus J, Delwart E. 2010. Human bocaviruses are highly diverse, dispersed, recombination prone, and prevalent in enteric infections. J Infect Dis 201:1633–1643. doi: 10.1086/652416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Blessing K, Neske F, Herre U, Kreth HW, Weissbrich B. 2009. Prolonged detection of human bocavirus DNA in nasopharyngeal aspirates of children with respiratory tract disease. Pediatr Infect Dis J 28:1018–1019. doi: 10.1097/INF.0b013e3181a854ae. [DOI] [PubMed] [Google Scholar]
- 275.Kantola K, Hedman L, Allander T, Jartti T, Lehtinen P, Ruuskanen O, Hedman K, Soderlund-Venermo M. 2008. Serodiagnosis of human bocavirus infection. Clin Infect Dis 46:540–546. doi: 10.1086/526532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Allander T. 2008. Human bocavirus. J Clin Virol 41:29–33. doi: 10.1016/j.jcv.2007.10.026. [DOI] [PubMed] [Google Scholar]
- 277.Pinana JL, Madrid S, Perez A, Hernandez-Boluda JC, Gimenez E, Terol MJ, Calabuig M, Navarro D, Solano C. 2018. Epidemiologic and clinical characteristics of coronavirus and bocavirus respiratory infections after allogeneic stem cell transplantation: a prospective single-center study. Biol Blood Marrow Transplant 24:563–570. doi: 10.1016/j.bbmt.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Korppi M, Jartti T, Hedman K, Soderlund-Venermo M. 2010. Serologic diagnosis of human bocavirus infection in children. Pediatr Infect Dis J 29:387. doi: 10.1097/INF.0b013e3181ce8e81. [DOI] [PubMed] [Google Scholar]
- 279.Endo R, Ishiguro N, Kikuta H, Teramoto S, Shirkoohi R, Ma X, Ebihara T, Ishiko H, Ariga T. 2007. Seroepidemiology of human bocavirus in Hokkaido prefecture, Japan. J Clin Microbiol 45:3218–3223. doi: 10.1128/JCM.02140-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Kantola K, Hedman L, Arthur J, Alibeto A, Delwart E, Jartti T, Ruuskanen O, Hedman K, Soderlund-Venermo M. 2011. Seroepidemiology of human bocaviruses 1–4. J Infect Dis 204:1403–1412. doi: 10.1093/infdis/jir525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Hao Y, Gao J, Zhang X, Liu N, Li J, Zheng L, Duan Z. 2015. Seroepidemiology of human bocaviruses 1 and 2 in China. PLoS One 10:e0122751. doi: 10.1371/journal.pone.0122751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Zhao LQ, Qian Y, Zhu RN, Deng J, Wang F, Dong HJ, Sun Y, Li Y. 2009. Human bocavirus infections are common in Beijing population indicated by sero-antibody prevalence analysis. Chin Med J (Engl) 122:1289–1292. [PubMed] [Google Scholar]
- 283.Li H, He M, Zeng P, Gao Z, Bian G, Yang C, Li W. 2015. The genomic and seroprevalence of human bocavirus in healthy Chinese plasma donors and plasma derivatives. Transfusion 55:154–163. doi: 10.1111/trf.12785. [DOI] [PubMed] [Google Scholar]
- 284.Schenk T, Maier B, Hufnagel M, Strahm B, Kontny U, Neumann-Haefelin D, Falcone V. 2011. Persistence of human bocavirus DNA in immunocompromised children. Pediatr Infect Dis J 30:82–84. doi: 10.1097/INF.0b013e3181f12fcf. [DOI] [PubMed] [Google Scholar]
- 285.Schenk T, Strahm B, Kontny U, Hufnagel M, Neumann-Haefelin D, Falcone V. 2007. Disseminated bocavirus infection after stem cell transplant. Emerg Infect Dis 13:1425–1427. doi: 10.3201/eid1309.070318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Allander T, Jartti T, Gupta S, Niesters HGM, Lehtinen P, Osterback R, Vuorinen T, Waris M, Bjerkner A, Tiveljung-Lindell A, van den Hoogen BG, Hyypiä T, Ruuskanen O. 2007. Human bocavirus and acute wheezing in children. Clin Infect Dis 44:904–910. doi: 10.1086/512196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Karalar L, Lindner J, Schimanski S, Kertai M, Segerer H, Modrow S. 2010. Prevalence and clinical aspects of human bocavirus infection in children. Clin Microbiol Infect 16:633–639. doi: 10.1111/j.1469-0691.2009.02889.x. [DOI] [PubMed] [Google Scholar]
- 288.Tong R, Shen L, Yin W, Zhou W, Lu J, Zheng M, Bi S, Lou Y, Tan W. 2013. Prevalence of human parvovirus B19, bocavirus, and PARV4 in blood samples from the general population of China and lack of a correlation between parvovirus and hepatitis B co-infection. PLoS One 8:e64391. doi: 10.1371/journal.pone.0064391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Bonvicini F, Manaresi E, Gentilomi GA, Di Furio F, Zerbini M, Musiani M, Gallinella G. 2011. Evidence of human bocavirus viremia in healthy blood donors. Diagn Microbiol Infect Dis 71:460–462. doi: 10.1016/j.diagmicrobio.2011.08.019. [DOI] [PubMed] [Google Scholar]
- 290.Abdel-Moneim AS, Mahfouz ME, Zytouni DM. 2018. Detection of human bocavirus in Saudi healthy blood donors. PLoS One 13:e0193594. doi: 10.1371/journal.pone.0193594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Tozer SJ, Lambert SB, Whiley DM, Bialasiewicz S, Lyon MJ, Nissen MD, Sloots TP. 2009. Detection of human bocavirus in respiratory, fecal, and blood samples by real-time PCR. J Med Virol 81:488–493. doi: 10.1002/jmv.21409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Bogolepov NK, Velikanov II, Sapel’nikova LI. 1976. Effect of Kislovodsk health resort treatment on cardiocerebral disorders in middle-aged and old patients with arteriosclerosis. Vrach Delo 1976:35–38. (In Russian.) [PubMed] [Google Scholar]
- 293.Spandole S, Cimponeriu D, Berca LM, Mihăescu G. 2015. Human anelloviruses: an update of molecular, epidemiological and clinical aspects. Arch Virol 160:893–908. doi: 10.1007/s00705-015-2363-9. [DOI] [PubMed] [Google Scholar]
- 294.Maggi F, Pistello M, Vatteroni M, Presciuttini S, Marchi S, Isola P, Fornai C, Fagnani S, Andreoli E, Antonelli G, Bendinelli M. 2001. Dynamics of persistent TT virus infection, as determined in patients treated with alpha interferon for concomitant hepatitis C virus infection. J Virol 75:11999–12004. doi: 10.1128/JVI.75.24.11999-12004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Ball JK, Curran R, Berridge S, Grabowska AM, Jameson CL, Thomson BJ, Irving WL, Sharp PM. 1999. TT virus sequence heterogeneity in vivo: evidence for co-infection with multiple genetic types. J Gen Virol 80:1759–1768. doi: 10.1099/0022-1317-80-7-1759. [DOI] [PubMed] [Google Scholar]
- 296.Okamoto H, Nishizawa T, Takahashi M, Asabe S, Tsuda F, Yoshikawa A. 2001. Heterogeneous distribution of TT virus of distinct genotypes in multiple tissues from infected humans. Virology 288:358–368. doi: 10.1006/viro.2001.1097. [DOI] [PubMed] [Google Scholar]
- 297.Okamoto H, Kato N, Iizuka H, Tsuda F, Miyakawa Y, Mayumi M. 1999. Distinct genotypes of a nonenveloped DNA virus associated with posttransfusion non-A to G hepatitis (TT virus) in plasma and peripheral blood mononuclear cells. J Med Virol 57:252–258. doi:. [DOI] [PubMed] [Google Scholar]
- 298.Chen T, Vaisanen E, Mattila PS, Hedman K, Soderlund-Venermo M. 2013. Antigenic diversity and seroprevalences of Torque teno viruses in children and adults by ORF2-based immunoassays. J Gen Virol 94:409–417. doi: 10.1099/vir.0.046862-0. [DOI] [PubMed] [Google Scholar]
- 299.Kaczorowska J, van der Hoek L. 2020. Human anelloviruses: diverse, omnipresent and commensal members of the virome. FEMS Microbiol Rev 44:305–313. doi: 10.1093/femsre/fuaa007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Maggi F, Fornai C, Zaccaro L, Morrica A, Vatteroni ML, Isola P, Marchi S, Ricchiuti A, Pistello M, Bendinelli M. 2001. TT virus (TTV) loads associated with different peripheral blood cell types and evidence for TTV replication in activated mononuclear cells. J Med Virol 64:190–194. doi: 10.1002/jmv.1035. [DOI] [PubMed] [Google Scholar]
- 301.Desai M, Pal R, Deshmukh R, Banker D. 2005. Replication of TT virus in hepatocyte and leucocyte cell lines. J Med Virol 77:136–143. doi: 10.1002/jmv.20426. [DOI] [PubMed] [Google Scholar]
- 302.Mariscal LF, López-Alcorocho JM, Rodríguez-Iñigo E, Ortiz-Movilla N, de Lucas S, Bartolomé J, Carreño V. 2002. TT virus replicates in stimulated but not in nonstimulated peripheral blood mononuclear cells. Virology 301:121–129. doi: 10.1006/viro.2002.1545. [DOI] [PubMed] [Google Scholar]
- 303.Poovorawan Y, Theamboonlers A, Vimolket T, Jantaradsamee P, Kaew-In N, Hirsch P. 1999. Detection of TTV in peripheral blood mononuclear cells of intravenous drug users. Tohoku J Exp Med 188:47–54. doi: 10.1620/tjem.188.47. [DOI] [PubMed] [Google Scholar]
- 304.Matsubara H, Michitaka K, Horiike N, Kihana T, Yano M, Mori T, Onji M. 2001. Existence of TT virus DNA and TTV-like mini virus DNA in infant cord blood: mother-to-neonatal transmission. Hepatol Res 21:280–287. doi: 10.1016/S1386-6346(01)00115-2. [DOI] [PubMed] [Google Scholar]
- 305.Sauvage V, Laperche S, Cheval J, Muth E, Dubois M, Boizeau L, Hebert C, Lionnet F, Lefrere JJ, Eloit M. 2016. Viral metagenomics applied to blood donors and recipients at high risk for blood-borne infections. Blood Transfus 14:400–407. doi: 10.2450/2016.0160-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Takahashi M, Asabe S, Gotanda Y, Kishimoto J, Tsuda F, Okamoto H. 2002. TT virus is distributed in various leukocyte subpopulations at distinct levels, with the highest viral load in granulocytes. Biochem Biophys Res Commun 290:242–248. doi: 10.1006/bbrc.2001.6183. [DOI] [PubMed] [Google Scholar]
- 307.Kikuchi K, Miyakawa H, Abe K, Kako M, Katayama K, Fukushi S, Mishiro S. 2000. Indirect evidence of TTV replication in bone marrow cells, but not in hepatocytes, of a subacute hepatitis/aplastic anemia patient. J Med Virol 61:165–170. doi:. [DOI] [PubMed] [Google Scholar]
- 308.Naganuma M, Tominaga N, Miyamura T, Soda A, Moriuchi M, Moriuchi H. 2008. TT virus prevalence, viral loads and genotypic variability in saliva from healthy Japanese children. Acta Paediatr 97:1686–1690. doi: 10.1111/j.1651-2227.2008.00962.x. [DOI] [PubMed] [Google Scholar]
- 309.Deng X, Terunuma H, Handema R, Sakamoto M, Kitamura T, Ito M, Akahane Y. 2000. Higher prevalence and viral load of TT virus in saliva than in the corresponding serum: another possible transmission route and replication site of TT virus. J Med Virol 62:531–537. doi:. [DOI] [PubMed] [Google Scholar]
- 310.Prasetyo AA, Desyardi MN, Tanamas J, Suradi, Reviono, Harsini, Kageyama S, Chikumi H, Shimizu E. 2015. Respiratory viruses and torque teno virus in adults with acute respiratory infections. Intervirology 58:57–68. doi: 10.1159/000369211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Pifferi M, Maggi F, Andreoli E, Lanini L, Marco ED, Fornai C, Vatteroni ML, Pistello M, Ragazzo V, Macchia P, Boner A, Bendinelli M. 2005. Associations between nasal torquetenovirus load and spirometric indices in children with asthma. J Infect Dis 192:1141–1148. doi: 10.1086/444389. [DOI] [PubMed] [Google Scholar]
- 312.Lewandowska DW, Schreiber PW, Schuurmans MM, Ruehe B, Zagordi O, Bayard C, Greiner M, Geissberger FD, Capaul R, Zbinden A, Boni J, Benden C, Mueller NJ, Trkola A, Huber M. 2017. Metagenomic sequencing complements routine diagnostics in identifying viral pathogens in lung transplant recipients with unknown etiology of respiratory infection. PLoS One 12:e0177340. doi: 10.1371/journal.pone.0177340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Matsubara H, Michitaka K, Horiike N, Yano M, Akbar SM, Torisu M, Onji M. 2000. Existence of TT virus DNA in extracellular body fluids from normal healthy Japanese subjects. Intervirology 43:16–19. doi: 10.1159/000025018. [DOI] [PubMed] [Google Scholar]
- 314.Ross RS, Viazov S, Runde V, Schaefer UW, Roggendorf M. 1999. Detection of TT virus DNA in specimens other than blood. J Clin Virol 13:181–184. doi: 10.1016/S1386-6532(99)00015-3. [DOI] [PubMed] [Google Scholar]
- 315.Saláková M, Němeček V, Tachezy R. 2009. TTV and HPV co-infection in cervical smears of patients with cervical lesions. BMC Infect Dis 9:118. doi: 10.1186/1471-2334-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Calcaterra S, Zaniratti MS, Serraino D, Peroni M, Abbate I, Cappiello G, Piselli P, Pavia C, Rezza G, Ippolito G, Capobianchi MR. 2001. Cervicovaginal shedding of TT virus in HIV-infected women. J Hum Virol 4:343–345. [PubMed] [Google Scholar]
- 317.Inami T, Konomi N, Arakawa Y, Abe K. 2000. High prevalence of TT virus DNA in human saliva and semen. J Clin Microbiol 38:2407–2408. doi: 10.1128/.38.6.2407-2408.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Hu YW, Al-Moslih MI, Al Ali MT, Khameneh SR, Perkins H, Diaz-Mitoma F, Roy JN, Uzicanin S, Brown EG. 2005. Molecular detection method for all known genotypes of TT virus (TTV) and TTV-like viruses in thalassemia patients and healthy individuals. J Clin Microbiol 43:3747–3754. doi: 10.1128/JCM.43.8.3747-3754.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Erensoy S, Sayıner AA, Türkoğlu S, Canatan D, Akarca US, Sertöz R, Özacar T, Batur Y, Badur S, Bilgiç A. 2002. TT virus infection and genotype distribution in blood donors and a group of patients from Turkey. Infection 30:299–302. doi: 10.1007/s15010-002-2185-z. [DOI] [PubMed] [Google Scholar]
- 320.Masouridi-Levrat S, Pradier A, Simonetta F, Kaiser L, Chalandon Y, Roosnek E. 2016. Torque teno virus in patients undergoing allogeneic hematopoietic stem cell transplantation for hematological malignancies. Bone Marrow Transplant 51:440–442. doi: 10.1038/bmt.2015.262. [DOI] [PubMed] [Google Scholar]
- 321.Spandole-Dinu S, Cimponeriu DG, Crăciun A-M, Radu I, Nica S, Toma M, Alexiu OA, Iorga CS, Berca L-M, Nica R. 2018. Prevalence of human anelloviruses in Romanian healthy subjects and patients with common pathologies. BMC Infect Dis 18:334. doi: 10.1186/s12879-018-3248-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Ninomiya M, Takahashi M, Nishizawa T, Shimosegawa T, Okamoto H. 2008. Development of PCR assays with nested primers specific for differential detection of three human anelloviruses and early acquisition of dual or triple infection during infancy. J Clin Microbiol 46:507–514. doi: 10.1128/JCM.01703-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Focosi D, Macera L, Boggi U, Nelli LC, Maggi F. 2015. Short-term kinetics of torque teno virus viraemia after induction immunosuppression confirm T lymphocytes as the main replication-competent cells. J Gen Virol 96:115–117. doi: 10.1099/vir.0.070094-0. [DOI] [PubMed] [Google Scholar]
- 324.Focosi D, Macera L, Pistello M, Maggi F. 2014. Torque teno virus viremia correlates with intensity of maintenance immunosuppression in adult orthotopic liver transplant. J Infect Dis 210:667–668. doi: 10.1093/infdis/jiu209. [DOI] [PubMed] [Google Scholar]
- 325.Maggi F, Focosi D, Statzu M, Bianco G, Costa C, Macera L, Spezia PG, Medici C, Albert E, Navarro D, Scagnolari C, Pistello M, Cavallo R, Antonelli G. 2018. Early post-transplant torquetenovirus viremia predicts cytomegalovirus reactivations in solid organ transplant recipients. Sci Rep 8:15490. doi: 10.1038/s41598-018-33909-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Handala L, Descamps V, Morel V, Castelain S, Francois C, Duverlie G, Helle F, Brochot E. 2019. No correlation between torque teno virus viral load and BK virus replication after kidney transplantation. J Clin Virol 116:4–6. doi: 10.1016/j.jcv.2019.03.018. [DOI] [PubMed] [Google Scholar]
- 327.Frye BC, Bierbaum S, Falcone V, Kohler TC, Gasplmayr M, Hettich I, Durk T, Idzko M, Zissel G, Hengel H, Muller-Quernheim J. 2019. Kinetics of torque teno virus-DNA plasma load predict rejection in lung transplant recipients. Transplantation 103:815–822. doi: 10.1097/TP.0000000000002436. [DOI] [PubMed] [Google Scholar]
- 328.Ruiz P, Martinez-Picola M, Santana M, Munoz J, Perez-Del-Pulgar S, Koutsoudakis G, Sastre L, Colmenero J, Crespo G, Navasa M. 2019. Torque teno virus is associated with the state of immune suppression early after liver transplantation. Liver Transpl 25:302–310. doi: 10.1002/lt.25374. [DOI] [PubMed] [Google Scholar]
- 329.Jaksch P, Kundi M, Görzer I, Muraközy G, Lambers C, Benazzo A, Hoetzenecker K, Klepetko W, Puchhammer-Stöckl E. 2018. Torque teno virus as a novel biomarker targeting the efficacy of immunosuppression after lung transplantation. J Infect Dis 218:1922–1928. doi: 10.1093/infdis/jiy452. [DOI] [PubMed] [Google Scholar]
- 330.Strassl R, Schiemann M, Doberer K, Görzer I, Puchhammer-Stöckl E, Eskandary F, Kikic Ž, Gualdoni GA, Vossen MG, Rasoul-Rockenschaub S, Herkner H, Böhmig GA, Bond G. 2018. Quantification of torque teno virus viremia as a prospective biomarker for infectious disease in kidney allograft recipients. J Infect Dis 218:1191–1199. doi: 10.1093/infdis/jiy306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Albert E, Solano C, Pascual T, Torres I, Macera L, Focosi D, Maggi F, Gimenez E, Amat P, Navarro D. 2017. Dynamics of torque teno virus plasma DNAemia in allogeneic stem cell transplant recipients. J Clin Virol 94:22–28. doi: 10.1016/j.jcv.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 332.Wohlfarth P, Leiner M, Schoergenhofer C, Hopfinger G, Goerzer I, Puchhammer-Stoeckl E, Rabitsch W. 2018. Torquetenovirus dynamics and immune marker properties in patients following allogeneic hematopoietic stem cell transplantation: a prospective longitudinal study. Biol Blood Marrow Transplant 24:194–199. doi: 10.1016/j.bbmt.2017.09.020. [DOI] [PubMed] [Google Scholar]
- 333.Schmitz J, Kobbe G, Kondakci M, Schuler E, Magorsch M, Adams O. 2019. The value of torque teno virus (TTV) as a marker for the degree of immunosuppression in adult patients after hematopoietic stem cell transplantation (HSCT). Biol Blood Marrow Transplant 26:643–650. doi: 10.1016/j.bbmt.2019.11.002. [DOI] [PubMed] [Google Scholar]
- 334.Gilles R, Herling M, Holtick U, Heger E, Awerkiew S, Fish I, Holler K, Sierra S, Knops E, Kaiser R, Scheid C, Di Cristanziano V. 2017. Dynamics of torque teno virus viremia could predict risk of complications after allogeneic hematopoietic stem cell transplantation. Med Microbiol Immunol 206:355–362. doi: 10.1007/s00430-017-0511-4. [DOI] [PubMed] [Google Scholar]
- 335.Albert E, Solano C, Gimenez E, Focosi D, Perez A, Macera L, Pinana JL, Mateo EM, Boluda JCH, Maggi F, Navarro D. 2019. Kinetics of alphatorquevirus plasma DNAemia at late times after allogeneic hematopoietic stem cell transplantation. Med Microbiol Immunol 208:253–258. doi: 10.1007/s00430-019-00586-w. [DOI] [PubMed] [Google Scholar]
- 336.Vu D, Simonetta F, Cordey S, Turin L, Docquier M, Brito F, Zdobnov E, Masouridi-Levrat S, Pradier A, Dantin C, Boely E, Van Delden C, Chalandon Y, Roosnek E, Kaiser L. 2017. Pegivirus in hematopoietic stem cell transplant recipients: a longitudinal prevalence study and association with immune reconstitution and clinical outcomes:O019. Bone Marrow Transpl 52:S21–S22. [Google Scholar]
- 337.Maggi F, Focosi D, Albani M, Lanini L, Vatteroni ML, Petrini M, Ceccherini-Nelli L, Pistello M, Bendinelli M. 2010. Role of hematopoietic cells in the maintenance of chronic human torquetenovirus plasma viremia. J Virol 84:6891–6893. doi: 10.1128/JVI.00273-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Focosi D, Maggi F, Albani M, Macera L, Ricci V, Gragnani S, Di Beo S, Ghimenti M, Antonelli G, Bendinelli M, Pistello M, Ceccherini-Nelli L, Petrini M. 2010. Torquetenovirus viremia kinetics after autologous stem cell transplantation are predictable and may serve as a surrogate marker of functional immune reconstitution. J Clin Virol 47:189–192. doi: 10.1016/j.jcv.2009.11.027. [DOI] [PubMed] [Google Scholar]
- 339.Pradier A, Masouridi-Levrat S, Bosshard C, Dantin C, Vu D-L, Zanella M-C, Boely E, Tapparel C, Kaiser L, Chalandon Y, Simonetta F, Roosnek E. 2020. Torque teno virus as a potential biomarker for complications and survival after allogeneic hematopoietic stem cell transplantation. Front Immunol 11:998. doi: 10.3389/fimmu.2020.00998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Rosario K, Breitbart M, Harrach B, Segales J, Delwart E, Biagini P, Varsani A. 2017. Revisiting the taxonomy of the family Circoviridae: establishment of the genus Cyclovirus and removal of the genus Gyrovirus. Arch Virol 162:1447–1463. doi: 10.1007/s00705-017-3247-y. [DOI] [PubMed] [Google Scholar]
- 341.Maggi F, Macera L, Focosi D, Vatteroni ML, Boggi U, Antonelli G, Eloit M, Pistello M. 2012. Human gyrovirus DNA in human blood, Italy. Emerg Infect Dis 18:956–959. doi: 10.3201/eid1806.120179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Chu DK, Poon LL, Chiu SS, Chan KH, Ng EM, Bauer I, Cheung TK, Ng IH, Guan Y, Wang D, Peiris JS. 2012. Characterization of a novel gyrovirus in human stool and chicken meat. J Clin Virol 55:209–213. doi: 10.1016/j.jcv.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Biagini P, Bedarida S, Touinssi M, Galicher V, de Micco P, Philippe de Micco . 2013. Human gyrovirus in healthy blood donors, France. Emerg Infect Dis 19:1014–1015. doi: 10.3201/eid1906.130228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Macera L, Focosi D, Giannelli R, Bulleri M, Zucca A, Scatena F, Pistello M, Ceccherini Nelli L, Maggi F. 2013. Human gyrovirus is not found in human CD34+ hematopoietic stem cells from peripheral blood or umbilical cord. J Clin Virol 57:182–183. doi: 10.1016/j.jcv.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 345.Smits SL, Zijlstra EE, van Hellemond JJ, Schapendonk CM, Bodewes R, Schurch AC, Haagmans BL, Osterhaus AD. 2013. Novel cyclovirus in human cerebrospinal fluid, Malawi, 2010–2011. Emerg Infect Dis 19:1511–1513. doi: 10.3201/eid1909.130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Dennis TPW, Flynn PJ, de Souza WM, Singer JB, Moreau CS, Wilson SJ, Gifford RJ. 2018. Insights into circovirus host range from the genomic fossil record. J Virol 92:e00145-18. doi: 10.1128/JVI.00145-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Chan MC, Kwok SW, Chan PK. 2015. False-positive PCR detection of cyclovirus Malawi strain VS5700009 in human cerebrospinal fluid. J Clin Virol 68:76–78. doi: 10.1016/j.jcv.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 348.Macera L, Focosi D, Vatteroni ML, Manzin A, Antonelli G, Pistello M, Maggi F. 2016. Cyclovirus Vietnam DNA in immunodeficient patients. J Clin Virol 81:12–15. doi: 10.1016/j.jcv.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 349.Anh NT, Hong NTT, Nhu LNT, Thanh TT, Lau CY, Limmathurotsakul D, Deng X, Rahman M, Chau NVV, van Doorn HR, Thwaites G, Delwart E, Tan LV. 2019. Viruses in Vietnamese patients presenting with community-acquired sepsis of unknown cause. J Clin Microbiol 57:e00386-19. doi: 10.1128/JCM.00386-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Sauvage V, Gomez J, Barray A, Vandenbogaert M, Boizeau L, Tagny CT, Rakoto O, Bizimana P, Guitteye H, Cire BB, Soumana H, Tchomba JS, Caro V, Laperche S. 2018. High prevalence of cyclovirus Vietnam (CyCV-VN) in plasma samples from Madagascan healthy blood donors. Infect Genet Evol 66:9–12. doi: 10.1016/j.meegid.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 351.Krupovic M, Ghabrial SA, Jiang D, Varsani A. 2016. Genomoviridae: a new family of widespread single-stranded DNA viruses. Arch Virol 161:2633–2643. doi: 10.1007/s00705-016-2943-3. [DOI] [PubMed] [Google Scholar]
- 352.Lamberto I, Gunst K, Muller H, Zur Hausen H, de Villiers EM. 2014. Mycovirus-like DNA virus sequences from cattle serum and human brain and serum samples from multiple sclerosis patients. Genome Announc 2:e00848-14. doi: 10.1128/genomeA.00848-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Zhang W, Li L, Deng X, Blumel J, Nubling CM, Hunfeld A, Baylis SA, Delwart E. 2016. Viral nucleic acids in human plasma pools. Transfusion 56:2248–2255. doi: 10.1111/trf.13692. [DOI] [PubMed] [Google Scholar]
- 354.Uch R, Fournier PE, Robert C, Blanc-Tailleur C, Galicher V, Barre R, Jordier F, de Micco P, Raoult D, Biagini P. 2015. Divergent gemycircularvirus in HIV-positive blood, France. Emerg Infect Dis 21:2096–2098. doi: 10.3201/eid2111.150486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Macera L, Spezia PG, Medici C, Falasca F, Sciandra I, Antonelli G, Focosi D, Pistello M, Maggi F. 2019. Low prevalence of gemycircularvirus DNA in immunocompetent and immunocompromised subjects. New Microbiol 42:118–120. [PubMed] [Google Scholar]
- 356.Kapoor A, Kumar A, Simmonds P, Bhuva N, Singh Chauhan L, Lee B, Sall AA, Jin Z, Morse SS, Shaz B, Burbelo PD, Lipkin WI. 2015. Virome analysis of transfusion recipients reveals a novel human virus that shares genomic features with hepaciviruses and pegiviruses. mBio 6:e01466-15. doi: 10.1128/mBio.01466-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Berg MG, Lee D, Coller K, Frankel M, Aronsohn A, Cheng K, Forberg K, Marcinkus M, Naccache SN, Dawson G, Brennan C, Jensen DM, Hackett J Jr, Chiu CY. 2015. Discovery of a novel human pegivirus in blood associated with hepatitis C virus co-infection. PLoS Pathog 11:e1005325. doi: 10.1371/journal.ppat.1005325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Smith DB, Becher P, Bukh J, Gould EA, Meyers G, Monath T, Muerhoff AS, Pletnev A, Rico-Hesse R, Stapleton JT, Simmonds P. 2016. Proposed update to the taxonomy of the genera Hepacivirus and Pegivirus within the Flaviviridae family. J Gen Virol 97:2894–2907. doi: 10.1099/jgv.0.000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Linnen J, Wages J Jr, Zhang-Keck ZY, Fry KE, Krawczynski KZ, Alter H, Koonin E, Gallagher M, Alter M, Hadziyannis S, Karayiannis P, Fung K, Nakatsuji Y, Shih JW, Young L, Piatak M Jr, Hoover C, Fernandez J, Chen S, Zou JC, Morris T, Hyams KC, Ismay S, Lifson JD, Hess G, Foung SK, Thomas H, Bradley D, Margolis H, Kim JP. 1996. Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science 271:505–508. doi: 10.1126/science.271.5248.505. [DOI] [PubMed] [Google Scholar]
- 360.Simons JN, Leary TP, Dawson GJ, Pilot-Matias TJ, Muerhoff AS, Schlauder GG, Desai SM, Mushahwar IK. 1995. Isolation of novel virus-like sequences associated with human hepatitis. Nat Med 1:564–569. doi: 10.1038/nm0695-564. [DOI] [PubMed] [Google Scholar]
- 361.Chivero ET, Stapleton JT. 2015. Tropism of human pegivirus (formerly known as GB virus C/hepatitis G virus) and host immunomodulation: insights into a highly successful viral infection. J Gen Virol 96:1521–1532. doi: 10.1099/vir.0.000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Reshetnyak VI, Karlovich TI, Ilchenko LU. 2008. Hepatitis G virus. World J Gastroenterol 14:4725–4734. doi: 10.3748/wjg.14.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Ghai RR, Sibley SD, Lauck M, Dinis JM, Bailey AL, Chapman CA, Omeja P, Friedrich TC, O'Connor DH, Goldberg TL. 2013. Deep sequencing identifies two genotypes and high viral genetic diversity of human pegivirus (GB virus C) in rural Ugandan patients. J Gen Virol 94:2670–2678. doi: 10.1099/vir.0.055509-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Luk KC, Berg MG, Naccache SN, Kabre B, Federman S, Mbanya D, Kaptue L, Chiu CY, Brennan CA, Hackett J Jr. 2015. Utility of metagenomic next-generation sequencing for characterization of HIV and human pegivirus diversity. PLoS One 10:e0141723. doi: 10.1371/journal.pone.0141723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Li G, Ma HH, Lau GK, Leung YK, Yao CL, Chong YT, Tang WH, Yao JL. 2002. Prevalence of hepatitis G virus infection and homology of different viral strains in Southern China. World J Gastroenterol 8:1081–1087. doi: 10.3748/wjg.v8.i6.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Nicolosi Guidicelli S, Lopez-Guillermo A, Falcone U, Conconi A, Christinat A, Rodriguez-Abreu D, Grisanti S, Lobetti-Bodoni C, Piffaretti JC, Johnson PW, Mombelli G, Cerny A, Montserrat E, Cavalli F, Zucca E. 2012. Hepatitis C virus and GBV-C virus prevalence among patients with B-cell lymphoma in different European regions: a case-control study of the International Extranodal Lymphoma Study Group. Hematol Oncol 30:137–142. doi: 10.1002/hon.1015. [DOI] [PubMed] [Google Scholar]
- 367.Komatsu H, Fujisawa T, Inui A, Sogo T, Morinishi Y, Miyagawa Y, Inui M. 1999. GBV-C/HGV infection in children with chronic hepatitis C. J Med Virol 59:154–159. doi:. [DOI] [PubMed] [Google Scholar]
- 368.Gutierrez RA, Dawson GJ, Knigge MF, Melvin SL, Heynen CA, Kyrk CR, Young CE, Carrick RJ, Schlauder GG, Surowy TK, Dille BJ, Coleman PF, Thiele DL, Lentino JR, Pachucki C, Mushahwar IK. 1997. Seroprevalence of GB virus C and persistence of RNA and antibody. J Med Virol 53:167–173. doi:. [DOI] [PubMed] [Google Scholar]
- 369.Tacke M, Schmolke S, Schlueter V, Sauleda S, Esteban JI, Tanaka E, Kiyosawa K, Alter HJ, Schmitt U, Hess G, Ofenloch-Haehnle B, Engel AM. 1997. Humoral immune response to the E2 protein of hepatitis G virus is associated with long-term recovery from infection and reveals a high frequency of hepatitis G virus exposure among healthy blood donors. Hepatology 26:1626–1633. doi: 10.1002/hep.510260635. [DOI] [PubMed] [Google Scholar]
- 370.Chang CM, Stapleton JT, Klinzman D, McLinden JH, Purdue MP, Katki HA, Engels EA. 2014. GBV-C infection and risk of NHL among U.S. adults. Cancer Res 74:5553–5560. doi: 10.1158/0008-5472.CAN-14-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Yang N, Dai R, Zhang X. 2020. Global prevalence of human pegivirus-1 in healthy volunteer blood donors: a systematic review and meta-analysis. Vox Sang 115:107–119. doi: 10.1111/vox.12876. [DOI] [PubMed] [Google Scholar]
- 372.Dawson GJ, Schlauder GG, Pilot-Matias TJ, Thiele D, Leary TP, Murphy P, Rosenblatt JE, Simons JN, Martinson FE, Gutierrez RA, Lentino JR, Pachucki C, Muerhoff AS, Widell A, Tegtmeier G, Desai S, Mushahwar IK. 1996. Prevalence studies of GB virus-C infection using reverse transcriptase-polymerase chain reaction. J Med Virol 50:97–103. doi:. [DOI] [PubMed] [Google Scholar]
- 373.Lefrère J-J, Loiseau P, Maury J, Lasserre J, Mariotti M, Ravera N, Lerable J, Lefèvre G, Morand-Joubert L, Girot R. 1997. Natural history of GBV-C/hepatitis G virus infection through the follow-up of GBV-C/hepatitis G virus-infected blood donors and recipients studied by RNA polymerase chain reaction and anti-E2 serology. Blood 90:3776–3780. doi: 10.1182/blood.V90.9.3776. [DOI] [PubMed] [Google Scholar]
- 374.Chivero ET, Bhattarai N, Rydze RT, Winters MA, Holodniy M, Stapleton JT. 2014. Human pegivirus RNA is found in multiple blood mononuclear cells in vivo and serum-derived viral RNA-containing particles are infectious in vitro. J Gen Virol 95:1307–1319. doi: 10.1099/vir.0.063016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Krajden M, Yu A, Braybrook H, Lai AS, Mak A, Chow R, Cook D, Tellier R, Petric M, Gascoyne RD, Connors JM, Brooks-Wilson AR, Gallagher RP, Spinelli JJ. 2010. GBV-C/hepatitis G virus infection and non-Hodgkin lymphoma: a case control study. Int J Cancer 126:2885–2892. doi: 10.1002/ijc.25035. [DOI] [PubMed] [Google Scholar]
- 376.Li L, Deng X, Linsuwanon P, Bangsberg D, Bwana MB, Hunt P, Martin JN, Deeks SG, Delwart E. 2013. AIDS alters the commensal plasma virome. J Virol 87:10912–10915. doi: 10.1128/JVI.01839-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Miao Z, Gao L, Song Y, Yang M, Zhang M, Lou J, Zhao Y, Wang X, Feng Y, Dong X, Xia X. 2017. Prevalence and clinical impact of human pegivirus-1 infection in HIV-1-infected individuals in Yunnan, China. Viruses 9:28. doi: 10.3390/v9020028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Bonsall D, Gregory WF, Ip CL, Donfield S, Iles J, Ansari MA, Piazza P, Trebes A, Brown A, Frater J, Pybus OG, Goulder P, Klenerman P, Bowden R, Gomperts ED, Barnes E, Kapoor A, Sharp CP, Simmonds P. 2016. Evaluation of viremia frequencies of a novel human pegivirus by using bioinformatic screening and PCR. Emerg Infect Dis 22:671–678. doi: 10.3201/eid2204.151812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Baggio-Zappia GL, Hernandes Granato CF. 2009. HIV-GB virus C co-infection: an overview. Clin Chem Lab Med 47:12–19. doi: 10.1515/CCLM.2009.001. [DOI] [PubMed] [Google Scholar]
- 380.Ernst D, Greer M, Akmatova R, Pischke S, Wedemeyer H, Heiken H, Tillmann HL, Schmidt RE, Stoll M. 2014. Impact of GB virus C viraemia on clinical outcome in HIV-1-infected patients: a 20-year follow-up study. HIV Med 15:245–250. doi: 10.1111/hiv.12094. [DOI] [PubMed] [Google Scholar]
- 381.Giret MT, Kallas EG. 2012. GBV-C: state of the art and future prospects. Curr HIV/AIDS Rep 9:26–33. doi: 10.1007/s11904-011-0109-1. [DOI] [PubMed] [Google Scholar]
- 382.Stark K, Bienzle U, Hess G, Engel AM, Hegenscheid B, Schluter V. 1996. Detection of the hepatitis G virus genome among injecting drug users, homosexual and bisexual men, and blood donors. J Infect Dis 174:1320–1323. doi: 10.1093/infdis/174.6.1320. [DOI] [PubMed] [Google Scholar]
- 383.AbuOdeh RO, Al-Absi E, Ali NH, Khalili M, Al-Mawlawi N, Hadwan TA, Althani AA, Nasrallah GK. 2015. Detection and phylogenetic analysis of human pegivirus (GBV-C) among blood donors and patients infected with hepatitis B virus (HBV) in Qatar. J Med Virol 87:2074–2081. doi: 10.1002/jmv.24289. [DOI] [PubMed] [Google Scholar]
- 384.Li Z, Li Y, Liang Y, Hu L, Chen S. 2019. Prevalence and risk factors of human pegivirus type 1 infection in hematopoietic stem cell transplantation patients. Int J Infect Dis 85:111–113. doi: 10.1016/j.ijid.2019.05.032. [DOI] [PubMed] [Google Scholar]
- 385.Slavov SN, Maraninchi Silveira R, Hespanhol MR, Sauvage V, Rodrigues ES, Fontanari Krause L, Bittencourt HT, Caro V, Laperche S, Covas DT, Kashima S. 2019. Human pegivirus-1 (HPgV-1) RNA prevalence and genotypes in volunteer blood donors from the Brazilian Amazon. Transfus Clin Biol 26:234–239. doi: 10.1016/j.tracli.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 386.Wang T, Chen J, Zhang Q, Huang X, Xie N, Zhang J, Cai T, Zhang Y, Xiong H. 2019. Prevalence of hepatitis G virus infection among 67,348 blood donors in mainland China. BMC Public Health 19:685. doi: 10.1186/s12889-019-6948-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Ramos Filho R, Carneiro MA, Teles SA, Dias MA, Cardoso DD, Lampe E, Yoshida CF, Martins RM. 2004. GB virus C/hepatitis G virus infection in dialysis patients and kidney transplant recipients in Central Brazil. Mem Inst Oswaldo Cruz 99:639–643. doi: 10.1590/S0074-02762004000600019. [DOI] [PubMed] [Google Scholar]
- 388.Erensoy S, Zeytinoglu A, Goksel S, Ozacar T, Ozkahya M, Ok E, Tuumlrkoglu S, Bilgic A. 2002. GB virus C/hepatitis G virus infection among renal transplant recipients in Izmir, Turkey: molecular analysis of phylogenetic groups. Int J Infect Dis 6:242–243. doi: 10.1016/S1201-9712(02)90121-9. [DOI] [PubMed] [Google Scholar]
- 389.Abraham P, John GT, Raghuraman S, Radhakrishnan S, Thomas PP, Jacob CK, Sridharan G. 2003. GB virus C/hepatitis G virus and TT virus infections among high risk renal transplant recipients in India. J Clin Virol 28:59–69. doi: 10.1016/S1386-6532(02)00239-1. [DOI] [PubMed] [Google Scholar]
- 390.De Filippi F, Lampertico P, Soffredini R, Rumi MG, Lunghi G, Aroldi A, Tarantino A, Ponticelli C, Colombo M. 2001. High prevalence, low pathogenicity of hepatitis G virus in kidney transplant recipients. Dig Liver Dis 33:477–479. doi: 10.1016/S1590-8658(01)80025-6. [DOI] [PubMed] [Google Scholar]
- 391.Isaacson AH, Bhardwaj B, Qian K, Davis GL, Kato T, Mizokami M, Lau JY. 1999. Hepatitis G virus infection in renal transplant recipients. J Viral Hepat 6:151–160. doi: 10.1046/j.1365-2893.1999.00149.x. [DOI] [PubMed] [Google Scholar]
- 392.Kallinowski B, Seipp S, Dengler T, Klar E, Theilmann L, Stremmel W. 2002. Clinical impact of hepatitis G virus infection in heart and liver transplant recipients. Transplant Proc 34:2288–2291. doi: 10.1016/S0041-1345(02)03239-6. [DOI] [PubMed] [Google Scholar]
- 393.Izumi T, Sakata K, Okuzaki D, Inokuchi S, Tamura T, Motooka D, Nakamura S, Ono C, Shimokawa M, Matsuura Y, Mori M, Fukuhara T, Yoshizumi T. 2019. Characterization of human pegivirus infection in liver transplantation recipients. J Med Virol 91:2093–2100. doi: 10.1002/jmv.25555. [DOI] [PubMed] [Google Scholar]
- 394.Elkayam O, Hassoba HM, Ferrell LD, Garcia-Kennedy R, Gish RG, Wright TL, Laffler T, Traylor D, Hunt G, Rosenthal P. 1999. GB virus C (GBV-C/HGV) and E2 antibodies in children preliver and postliver transplant. Pediatr Res 45:795–798. doi: 10.1203/00006450-199906000-00002. [DOI] [PubMed] [Google Scholar]
- 395.Corbi C, Traineau R, Esperou H, Ravera N, Portelette E, Benbunan M, Gluckman E, Loiseau P. 1997. Prevalence and clinical features of hepatitis G virus infection in bone marrow allograft recipients. Bone Marrow Transplant 20:965–968. doi: 10.1038/sj.bmt.1701005. [DOI] [PubMed] [Google Scholar]
- 396.Ljungman P, Halasz R, Hagglund H, Sonnerborg A, Ringden O. 1998. Detection of hepatitis G virus/GB virus C after allogeneic bone marrow transplantation. Bone Marrow Transplant 22:499–501. doi: 10.1038/sj.bmt.1701366. [DOI] [PubMed] [Google Scholar]
- 397.Akiyama H, Nakamura N, Tanikawa S, Sakamaki H, Onozawa Y, Shibayama T, Tanaka S, Tsuda F, Okamoto H, Miyakawa Y, Mayumi M. 1998. Incidence and influence of GB virus C and hepatitis C virus infection in patients undergoing bone marrow transplantation. Bone Marrow Transplant 21:1131–1135. doi: 10.1038/sj.bmt.1701239. [DOI] [PubMed] [Google Scholar]
- 398.Coller KE, Berg MG, Frankel M, Forberg K, Surani R, Chiu CY, Hackett J Jr, Dawson GJ. 2016. Antibodies to the novel human pegivirus 2 are associated with active and resolved infections. J Clin Microbiol 54:2023–2030. doi: 10.1128/JCM.00515-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Wang H, Wan Z, Xu R, Guan Y, Zhu N, Li J, Xie Z, Lu A, Zhang F, Fu Y, Tang S. 2018. A novel human pegivirus, HPgV-2 (HHpgV-1), is tightly associated with hepatitis C virus (HCV) infection and HCV/human immunodeficiency virus type 1 coinfection. Clin Infect Dis 66:29–35. doi: 10.1093/cid/cix748. [DOI] [PubMed] [Google Scholar]
- 400.Forberg K, Rodgers MA, Dawson GJ, Sauleda S, Olivo A, Vallari A, Bes M, Piron M, Cloherty GA, Berg MG. 2020. Human pegivirus 2 exhibits minimal geographic and temporal genetic diversity. Virology 539:69–79. doi: 10.1016/j.virol.2019.10.012. [DOI] [PubMed] [Google Scholar]
- 401.Shui J, Liu W, Liang Y, Zhang J, Wan Z, Wang H, Qu X, Tang S. 2019. Infection of human pegivirus 2 (HPgV-2) is associated with hepatitis C virus but not hepatitis B virus infection in people who inject drugs. J Gen Virol 100:968–974. doi: 10.1099/jgv.0.001266. [DOI] [PubMed] [Google Scholar]
- 402.Vu DL, Bosch A, Pinto RM, Guix S. 2017. Epidemiology of classic and novel human astrovirus: gastroenteritis and beyond. Viruses 9:33. doi: 10.3390/v9020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Vu DL, Cordey S, Brito F, Kaiser L. 2016. Novel human astroviruses: novel human diseases? J Clin Virol 82:56–63. doi: 10.1016/j.jcv.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 404.Burbelo PD, Ragheb JA, Kapoor A, Zhang Y. 2013. The serological evidence in humans supports a negligible risk of zoonotic infection from porcine circovirus type 2. Biologicals 41:430–434. doi: 10.1016/j.biologicals.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Wylie KM, Mihindukulasuriya KA, Sodergren E, Weinstock GM, Storch GA. 2012. Sequence analysis of the human virome in febrile and afebrile children. PLoS One 7:e27735. doi: 10.1371/journal.pone.0027735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Cordey S, Zanella MC, Wagner N, Turin L, Kaiser L. 2018. Novel human astroviruses in pediatric respiratory samples: a one-year survey in a Swiss tertiary care hospital. J Med Virol 90:1775–1778. doi: 10.1002/jmv.25246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Holtz LR, Wylie KM, Sodergren E, Jiang Y, Franz CJ, Weinstock GM, Storch GA, Wang D. 2011. Astrovirus MLB2 viremia in febrile child. Emerg Infect Dis 17:2050–2052. doi: 10.3201/eid1711.110496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Cordey S, Vu D-L, Schibler M, L'Huillier AG, Brito F, Docquier M, Posfay-Barbe KM, Petty TJ, Turin L, Zdobnov EM, Kaiser L. 2016. Astrovirus MLB2, a new gastroenteric virus associated with meningitis and disseminated infection. Emerg Infect Dis 22:846–853. doi: 10.3201/eid2205.151807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Cordey S, Vu DL, Zanella MC, Turin L, Mamin A, Kaiser L. 2017. Novel and classical human astroviruses in stool and cerebrospinal fluid: comprehensive screening in a tertiary care hospital, Switzerland. Emerg Microbes Infect 6:e84. doi: 10.1038/emi.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Brown JR, Morfopoulou S, Hubb J, Emmett WA, Ip W, Shah D, Brooks T, Paine SM, Anderson G, Virasami A, Tong CY, Clark DA, Plagnol V, Jacques TS, Qasim W, Hubank M, Breuer J. 2015. Astrovirus VA1/HMO-C: an increasingly recognized neurotropic pathogen in immunocompromised patients. Clin Infect Dis 60:881–888. doi: 10.1093/cid/ciu940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Vu DL, Dayer JA, Masouridi-Levrat S, Combescure C, Boely E, Khanna N, Mueller NJ, Kleber M, Medinger M, Halter J, Passweg J, Muller AM, Schanz U, Chalandon Y, Neofytos D, van Delden C, Kaiser L, Swiss Transplant Cohort Study . 2020. Microbiologically documented infections after adult allogeneic hematopoietic cell transplantation: a 5-year analysis within the Swiss Transplant Cohort study. Transpl Infect Dis 2020:e13289. doi: 10.1111/tid.13289. [DOI] [PubMed] [Google Scholar]
- 412.van Delden C, Stampf S, Hirsch HH, Manuel O, Meylan P, Cusini A, Hirzel C, Khanna N, Weisser M, Garzoni C, Boggian K, Berger C, Nadal D, Koller M, Saccilotto R, Mueller NJ, Swiss Transplant Cohort Study . 2020. Burden and timeline of infectious diseases in the first year after solid organ transplantation in the Swiss Transplant Cohort study. Clin Infect Dis 2020:ciz1113. doi: 10.1093/cid/ciz1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Miller HK, Braun TM, Stillwell T, Harris AC, Choi S, Connelly J, Couriel D, Goldstein S, Kitko CL, Magenau J, Pawarode A, Reddy P, Riwes M, Yanik GA, Levine JE. 2017. Infectious risk after allogeneic hematopoietic cell transplantation complicated by acute graft-versus-host disease. Biol Blood Marrow Transplant 23:522–528. doi: 10.1016/j.bbmt.2016.12.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Waldvogel-Abramowski S, Taleb S, Alessandrini M, Preynat-Seauve O. 2019. Viral metagenomics of blood donors and blood-derived products using next-generation sequencing. Transfus Med Hemother 46:87–93. doi: 10.1159/000499088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Junier T, Huber M, Schmutz S, Kufner V, Zagordi O, Neuenschwander S, Ramette A, Kubacki J, Bachofen C, Qi W, Laubscher F, Cordey S, Kaiser L, Beuret C, Barbie V, Fellay J, Lebrand A. 2019. Viral metagenomics in the clinical realm: lessons learned from a Swiss-wide ring trial. Genes (Basel) 10:655. doi: 10.3390/genes10090655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Huang YT, Kim SJ, Lee YJ, Burack D, Nichols P, Maloy M, Perales MA, Giralt SA, Jakubowski AA, Papanicolaou GA. 2017. Co-infections by double-stranded DNA (dsDNA) viruses after ex vivo T-cell depleted, CD34+ selected hematopoietic cell transplantation (HCT). Biol Blood Marrow Transplant 23:1759–1766. doi: 10.1016/j.bbmt.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Hill JA, Mayer BT, Xie H, Leisenring WM, Huang ML, Stevens-Ayers T, Milano F, Delaney C, Jerome KR, Zerr DM, Nichols G, Boeckh M, Schiffer JT. 2018. Kinetics of double-stranded DNA viremia after allogeneic hematopoietic cell transplantation. Clin Infect Dis 66:368–375. doi: 10.1093/cid/cix804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Moens U, Ludvigsen M, Van Ghelue M. 2011. Human polyomaviruses in skin diseases. Patholog Res Int 2011:123491. doi: 10.4061/2011/123491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Sroller V, Hamšíková E, Ludvíková V, Vochozková P, Kojzarová M, Fraiberk M, Saláková M, Morávková A, Forstová J, Němečková S. 2014. Seroprevalence rates of BKV, JCV, and MCPyV polyomaviruses in the general Czech Republic population. J Med Virol 86:1560–1568. doi: 10.1002/jmv.23841. [DOI] [PubMed] [Google Scholar]
- 420.Nguyen NL, Le BM, Wang D. 2009. Serologic evidence of frequent human infection with WU and KI polyomaviruses. Emerg Infect Dis 15:1199–1205. doi: 10.3201/eid1508.090270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Trusch F, Klein M, Finsterbusch T, Kuhn J, Hofmann J, Ehlers B. 2012. Seroprevalence of human polyomavirus 9 and cross-reactivity to African green monkey-derived lymphotropic polyomavirus. J Gen Virol 93:698–705. doi: 10.1099/vir.0.039156-0. [DOI] [PubMed] [Google Scholar]
- 422.Kamminga S, van der Meijden E, Feltkamp MCW, Zaaijer HL. 2018. Seroprevalence of fourteen human polyomaviruses determined in blood donors. PLoS One 13:e0206273. doi: 10.1371/journal.pone.0206273. [DOI] [PMC free article] [PubMed] [Google Scholar]