Abstract
Infectious diseases prevalent in humans and animals are caused by pathogens that once emerged from other animal hosts. In addition to these established infections, new infectious diseases periodically emerge. In extreme cases they may cause pandemics such as COVID-19; in other cases, dead-end infections or smaller epidemics result. Established diseases may also re-emerge, for example by extending geographically or by becoming more transmissible or more pathogenic. Disease emergence reflects dynamic balances and imbalances, within complex globally distributed ecosystems comprising humans, animals, pathogens, and the environment. Understanding these variables is a necessary step in controlling future devastating disease emergences.
Keywords: infection, epidemic, epizootic, pandemic, One Health, zoonosis, virology, disease ecology, medical history, COVID-19
Understanding the dynamic balance and interplay between complex and global ecosystems comprising humans, animals, pathogens, and the environment provides perspectives on how we got to the COVID-19 pandemic.
Introduction
Unimagined just a few short months ago, the ongoing COVID-19 pandemic has upended our entire planet, quickly challenging past assumptions and future certainties. It possesses simultaneously three characteristics that have allowed it to render an historic assault on the human species, triggering a virtual global “lockdown” as the only weapon against uncontrolled spread. It combines the characteristics of being a virus that to our knowledge has never before infected humans in a sustained manner, together with its extraordinary efficiency in transmitting from person to person and its relatively high level of morbidity and mortality, especially among seniors and those with underlying co-morbidities. It indeed is the perfect storm of an emerging infectious disease.
Yet, pandemics such as COVID-19 are not entirely new phenomena. Newly emerging (and re-emerging) infectious diseases have been threatening humans since the neolithic revolution, 12,000 years ago, when human hunter-gatherers settled into villages to domesticate animals and cultivate crops (Dobson and Carper, 1996; Morens et al., 2020b; Morens et al., 2008a). These beginnings of domestication were the earliest steps in man’s systematic, widespread manipulation of nature. Ancient emerging zoonotic diseases (see Box 1 ) with deadly consequences include smallpox, falciparum malaria, measles, and bubonic/pneumonic plague. Some, e.g., the Justinian plague (541 AD) and the Black Death (1348 AD), killed substantial proportions of humans in the “known” world, i.e., the world known to those whose recordings of it survive, predominantly in Asia, the Middle East, and Europe.
Box 1. Terms Related to Emerging Infectious Diseases.
Antigenic immunodominance: Ability of a protein epitope to elicit an immune response greater than the response to one or more adjacent epitopes
Cell tropism: Ability of a pathogen to infect a particular cell type
Endemic: Noun and adjective denoting prevalence of human infection
Enzootic: Noun and adjective denoting prevalence of animal infection
Epidemic, Pandemic: Noun and adjective denoting highly incident disease (epidemic) or spread that is global or covers very large geographic areas (pandemic)
Epizootic, Panzootic: Noun and adjective analogous to epidemic and pandemic, but with respect to animal diseases
Fomite: An inanimate object that transmits infection, e.g., a towel or doorknob
Host-Switching, Spillover: Process by which a pathogen adapted to one host species becomes adapted to another host species
Disease emergence: Appearance of a disease in a new host
Zoonosis: A human infection caused by an animal pathogen that may be either a dead-end infection or that may initiate person-to-person spread
Only a century ago, the 1918 influenza pandemic killed 50 million or more people, apparently the deadliest event in recorded human history (Morens and Taubenberger, 2020). The HIV/AIDS pandemic, recognized in 1981, has so far killed at least 37 million. And the past decade has witnessed unprecedented pandemic explosions: H1N1 “swine” influenza (2009), chikungunya (2014), and Zika (2015), as well as pandemic-like emergences of Ebola fever over large parts of Africa (2014 to the present).
Since there are four endemic coronaviruses that circulate globally in humans, coronaviruses must have emerged and spread pandemically in the era prior to the recognition of viruses as human pathogens. The severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV) emerged from an animal host, likely a civet cat, in 2002–2003, to cause a near-pandemic before disappearing in response to public health control measures. The related Middle East respiratory syndrome (MERS) coronavirus (MERS-CoV) emerged into humans from dromedary camels in 2012, but has since been transmitted inefficiently among humans (Cui et al., 2019). COVID-19, recognized in late 2019, is but the latest example of an unexpected, novel, and devastating pandemic disease. One can conclude from this recent experience that we have entered a pandemic era (Morens et al., 2020a; Morens et al., 2020b). The causes of this new and dangerous situation are multifaceted, complex, and deserving of serious examination.
Infectious Diseases that Have Emerged in the Past
In thinking about these recent infectious disease emergences, it is necessary to first consider currently existing infectious diseases that newly emerged in the past and then over time became endemic (prevalent in humans) or enzootic (prevalent in animals) (Fauci and Morens, 2012; Morens et al., 2020b; Morens and Fauci, 2012; Morens et al., 2004). Such existing diseases may provide important clues about the mechanisms of disease emergence and persistence and why thus far we have been largely unable to prevent and control many of them.
The fact that many past emerging infectious microbes and viruses (hereafter grouped together as “microbes”) have adapted to stable co-existence with humans is evidenced by the presence of endogenous retroviruses in human DNA (Johnson, 2019) and by latently infecting herpesviruses such as herpes simplex (HSV), cytomegalovirus (CMV), Epstein-Barr virus (EBV), and varicella-zoster virus (VZV). VZV, for example, is a highly cytolytic, highly contagious, and potentially fatal virus that has adapted to long-term survival in human populations via a complex survival mechanism. Unlike other highly contagious human-adapted respiratory viruses such as measles—whose survival requires very large populations in order to avoid exhausting susceptible persons—VZV establishes latent non-cytolytic infections in human ganglia, periodically reactivating into an infectious/cytolytic form (zoster) that can be transmitted—even in populations without circulating varicella (“chicken pox”)—to new birth cohorts of susceptible persons to be manifested as highly contagious varicella.
Human infectious agents such as retroviruses, herpesviruses, and many others tell us that long-ago emergences of certain diseases can result in long-term microbial survival by co-opting certain of our genetic, cellular, and immune mechanisms to ensure their continuing transmission. In the terminology of British biologist Richard Dawkins, evolution occurs at the level of gene competition and we, phenotypic humans, are merely genetic “survival machines” in the competition between microbes and humans (Dawkins, 1976). It may be a matter of perspective who is in the evolutionary driver’s seat. This perspective has implications for how we think about and react to emerging infectious disease threats.
From the human point of view, the fact that modern endemic diseases emerged and became established, at some unobserved time in the past (Table 1 ) (Morens et al., 2004), and that some of these diseases survived by adopting complicated long-term survival strategies, provides a compelling rationale for dual strategies for immediate and long-term control. First, in the immediate sense, it is important to mitigate spread of infection, illness, and death. Second, it is critical to prevent the persistence of microbes that may lead to additional emergences that are cumulatively as deadly, or more so, than the original emergences (Dobson and Carper, 1996). That viral genetic descendants of the 1918 influenza pandemic virus are still causing seasonal outbreaks throughout the world, and still killing cumulatively millions of people a century later (Morens and Taubenberger, 2020), is a powerful reminder that single disease emergences can have consequences beyond immediate morbidity and mortality. In the ancient ongoing struggle between microbes and man, genetically more adaptable microbes have the upper hand in consistently surprising us and often catching us unprepared.
Table 1.
Emerging Infectious Diseases in History
| Year | Name | Deaths | Comments |
|---|---|---|---|
| 430 BCE | “Plague of Athens” | ∼100,000 | First identified trans-regional pandemic |
| 541 | Justinian plague (Yersinia pestis) | 30–50 million | Pandemic; killed half of world population |
| 1340s | “Black Death” (Yersinia pestis) | ∼50 million | Pandemic; killed at least a quarter of world population |
| 1494 | Syphilis (Treponema pallidum) | >50,000 | Pandemic brought to Europe from the Americas |
| c. 1500 | Tuberculosis | High millions | Ancient disease; became pandemic in Middle Ages |
| 1520 | Hueyzahuatl (Variola major) | 3.5 million | Pandemic brought to New World by Europeans |
| 1793–1798 | “The American plague” | ∼25,000 | Yellow fever terrorized colonial America |
| 1832 | 2nd cholera pandemic (Paris) | 18,402 | Spread from India to Europe/Western Hemisphere |
| 1918 | “Spanish” influenza | ∼50 million | Led to additional pandemics in 1957, 1968, 2009 |
| 1976–2020 | Ebola | 15,258 | First recognized in 1976; 29 regional epidemics to 2020 |
| 1981 | Acute hemorrhagic conjunctivitis | rare deaths | First recognized in 1969; pandemic in 1981 |
| 1981 | HIV/AIDS | ∼37 million | First recognized 1981; ongoing pandemic |
| 2002 | SARS | 813 | Near-pandemic |
| 2009 | H1N1 “swine flu” | 284,000 | 5th influenza pandemic of century |
| 2014 | Chikungunya | uncommon | Pandemic, mosquito-borne |
| 2015 | Zika | ∼1,000?∗ | Pandemic, mosquito-borne |
Selected important emerging and re-emerging infectious diseases of the past and present, 430 BCE–2020 CE. Mortality estimates are in most cases imprecise; see text.
Zika mortality has not been fully established. Most deaths are fetal or related to outcomes of severe congenital infections.
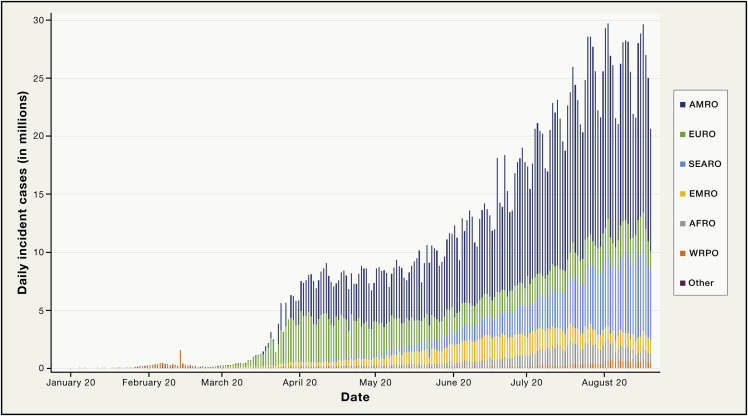
The latest example of this, the COVID-19 pandemic, which emerged in December 2019, is still exploding globally (Figure 1 ). At time of writing, over 22 million cases have been detected, with over 800,000 deaths recorded (World Health Organization); however, these are undoubtedly significant undercounts, reflecting early and still problematic access to diagnostic testing coupled with incomplete diagnoses of fatal cases. As COVID-19 is caused by a novel virus (SARS-CoV-2) producing a spectrum of disease whose clinical, pathologic, and epidemiologic patterns have never before been observed, we are gaining insights only incrementally. At some time in the future we will be better able to compare and contrast COVID-19 to other important emerging diseases; however, at this time we are still just entering a steep learning curve that will surely keep surprising us as we struggle to control what is already among the deadliest pandemics of the past century.
Figure 1.
Global Daily Incident Cases of COVID-19 by World Health Organization Region as of August 18, 2020
The data (World Health Organization) show that beginning in March 2020, the pandemic exploded in Europe and the Americas, particularly in the United States, was blunted in these two regions between March and May 2020, and then began to explode anew in the Americas and to a lesser extent in Europe beginning in late May. Since May 2020, the pandemic has been increasing significantly in the SEARO as well as the AFRO regions. WRPO, Western Pacific; AFRO, Africa; EMRO, Eastern Mediterranean; SEARO, Southeast Asia; EURO, Europe; AMRO, Americas.
Definitions of Emerging Infectious Diseases
The once-emerging/now prevalent diseases mentioned above, e.g., many viruses causing upper respiratory, enteric, or dermal/mucosal infections, are not considered to be truly emerging even when they vary seasonally or geographically; however, upon this background of existing diseases, new diseases still continue to emerge.
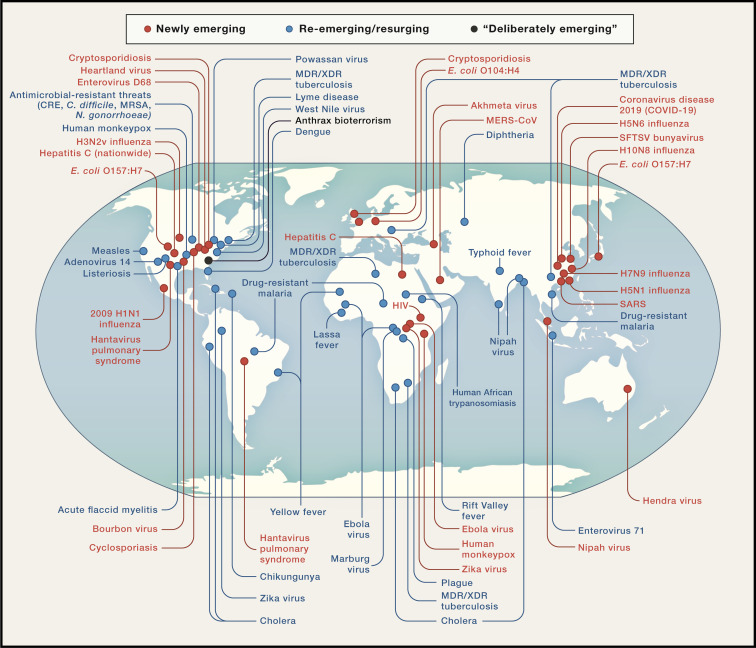
Emerging diseases have been categorized as newly emerging, re-emerging, or “deliberately emerging,” that is, associated with bioterrorism (Table 2 ; Figure 2 ) (Morens and Fauci, 2012; Morens et al., 2004, 2008a). To these we add “accidentally emerging” human-generated diseases, such as repeated emergences of vaccine-derived polioviruses (VDPVs) resulting from naturally occurring back-mutations of live virus vaccines, as well as a live human-engineered vaccine that escaped to cause a new epizootic disease: naturally transmitted vaccinia (Lum et al., 1967). Although these four categories are distinct, they are also interrelated: newly emerging diseases can persist and then re-emerge and can also become agents of deliberate or accidental release. An example crossing the latter two categories is the 1979 Sverdlovsk (now Yekaterinaberg) anthrax accident, in which an unintentional explosion in a Russian bioweapons factory released anthrax into the air, resulting in at least 100 human deaths (Meselson et al., 1994). Such deliberately emerging and accidentally emerging diseases represent a special case in which emergence/epidemicity is best prevented by global biosafety cooperation.
Table 2.
Major Categories of Emerging Infectious Diseases
| Newly emerging infectious diseases | Diseases recognized in humans for the first time, e.g., HIV/AIDS (1981), Nipah virus (1999), SARS (2002), MERS (2012), COVID-19 (2019) |
| Re-emerging infectious diseases | Diseases that have historically infected humans but continue to re-appear either in new locations (e.g., West Nile in the United States and Russia in 1999) or in resistant forms (e.g., methicillin-resistant Staphylococcus aureus) |
| Deliberately emerging infectious diseases | Diseases associated with intent to harm, including mass bioterrorism |
| Accidentally emerging infectious diseases | Diseases created by humans that are released unintentionally, e.g., epizootic vaccinia and transmissible vaccine-derived polioviruses |
Not included are currently established endemic diseases that are presumed to have been newly emerging at some time in the past and then went on to develop long-term persistence in human or animal populations (see text).
Figure 2.
Recent Emerging Infectious Diseases
The global extent of newly emerging, re-emerging, and “deliberately emerging” infectious disease from 1981 to the present (2020).
Among possible ways to achieve such cooperation are by strengthening the United Nations and its agencies, particularly the World Health Organization and the Office International des Épizooties (OIE; World Organisation for Animal Health); by supporting collaborative multinational research in prevention of disease emergence; by studying high-consequence pathogens under appropriate safety and containment conditions; by demanding renewed international intergovernmental efforts at the global level to collaborate on research related to the risks of global pathogen emergence risks and how to prevent them; and by preventing bioweapons development.
A fifth related category, significant because it emphasizes the importance of responding to disease emergence threats with countermeasures, is that of diseases that are “de-emerging,” i.e., those that have been eliminated or even eradicated or that are in the process of elimination and/or eradication (Table 3 ) (Dowdle and Cochi, 2011; Hopkins, 2013; Tomori, 2011). Smallpox and the veterinary disease rinderpest were declared eradicated in 1980 and 2011, respectively. SARS, which emerged in 2002–2003 and spread globally to 29 countries, infecting 8,096 people and killing 813, was controlled and ultimately eliminated from human spread by effective public health efforts (Cui et al., 2019). By some definitions, SARS was thereby eradicated, although it presumably remains in enzootic circulation and could re-emerge from nature, as Ebola viruses have been doing for the past 44 years (Baseler et al., 2017).
Table 3.
Selected Human Infectious Diseases
 |
Variables that relate to their potential for eradication. (Top) Selected infectious diseases that have either been eradicated, are now being targeted for eradication, or are being significantly controlled by public health and medical or veterinary actions. (Bottom) Human infectious diseases that are currently considered non-eradicable but for which some important disease aspects could potentially be eliminated with existing tools (e.g., eliminating human rabies without or before eradicating rabies in wild animals). The information is based on published data reflecting 2008 determinations (https://www.cartercenter.org), supplemented by additional widely available publications. Columns show disease features that favor eradicability (blue circles), are of uncertain relevance to eradicability (yellow), or are expected to make eradication more difficult or impossible (red). Some of these features (e.g., ease of detecting disease and immunity, seasonality in tropical versus temperate climates) are subjective and situationally variable; see text and references for the individual diseases.
∗Sterile immunity for life refers to the ability of a natural infection or vaccine to induce a type and degree of immunity that prevents infection/reinfection and eliminates carriage and transmissibility to others and to animal and environmental reservoirs.
Other diseases nearing eradication include dracunculiasis, lymphatic filariasis, measles, polio, and rubella (Table 3). Such successes in eradication/control reflect the availability of improved tools and strategies for prevention and control, as well as international public and private efforts to reduce their substantial mortality and morbidity. Successes in eradication and control of infectious diseases remind us that we are not helpless in the face of emerging diseases. Eradicating/controlling existing diseases and preventing/controlling newly emerging diseases are related efforts demanding the same scientific, public health, and civic/political focus that will be required to successfully address this formidable challenge.
Variables in Disease Emergence: The Agent, Host, and Environment
Microbes that cause human diseases by definition have existed in some other environmental niche before emerging to infect humans and other animals. While some such organisms have long been human pathogens that mutated into new forms—e.g., re-emergences of antibiotic-resistant bacteria such as methicillin-resistant Staphylococcus aureus (MRSA)—most are, and historically have been, zoonotic (Woolhouse and Gowtage-Sequeria, 2005; Woolhouse et al., 2005). Such zoonotic microbial emergences are often associated with mutational mechanisms allowing host-switching from animals to humans, as discussed below.
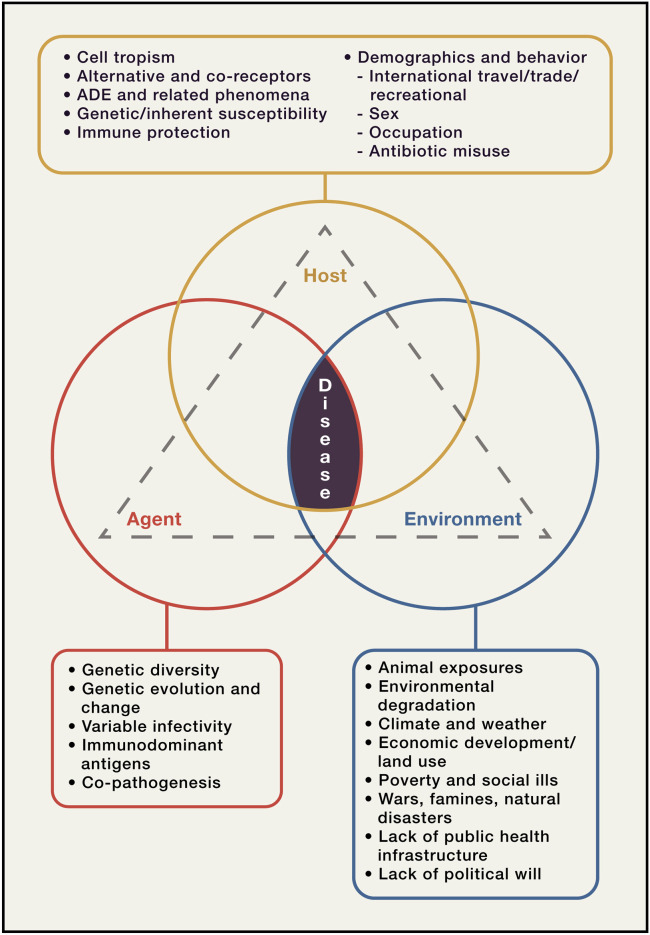
The triad of causations of emerging and other diseases, as conceptualized for over a century, represents interactions between infectious agents, their hosts, and the environment (Figure 3 ). This conceptualization acknowledges the reality that, while infectious diseases themselves are necessarily “caused” by microbial agents, emergences that produce epidemics and pandemics are also significantly determined by co-factors related to the host and to host-environmental interactions (Centers for Disease Control and Prevention, 2011).
Figure 3.
Infectious Agents, Hosts, and the Environment: Determinants of Disease Emergence and Persistence
Diseases, including emerging diseases, result from interactions between infectious agents, hosts, and the environment. Adapted from Fauci and Morens (2012); Morens and Fauci (2012); Morens et al. (2004), (2008a).
The Role of the Infectious Agent in the Emergence of Infectious Diseases
Considerations of the emergences of infectious diseases begin with the infectious agent itself. Although many established diseases, such as tuberculosis, malaria, and cholera, are bacterial or protozoal, and yet others are caused by fungi (e.g., cryptococci) or agents such as Rickettsia or prions, the majority of important newly emerging and re-emerging diseases in the past century have been viral. This review therefore emphasizes viruses, including SARS-CoV, and SARS-CoV-2, influenza, arboviruses, and hemorrhagic fever viruses among others.
Genetic instability of microorganisms is an inherent property allowing rapid microbial evolution to adapt to ever-changing ecologic niches. This is particularly true of RNA viruses such as influenza viruses, flaviviruses, enteroviruses, and coronaviruses, which have inherently deficient or absent polymerase error-correction mechanisms and are transmitted as quasispecies or swarms of many, often hundreds or thousands of, genetic variants.
Emergences of viral diseases begin with the genetic plasticity of the infectious agent, which may repeatedly encounter ecologic niches into which it can evolve and adapt under facilitative circumstances, e.g., those provided by the hosts in the context of the host environment. For viruses transmitted by person-to-person mechanisms, transmission by quasispecies may increase the likelihood that one or more viral variants within the quasispecies will be infectious for cells of a new host, leading to infection, viral amplification, and expansion of a new and different quasispecies, facilitating onward transmission (see below).
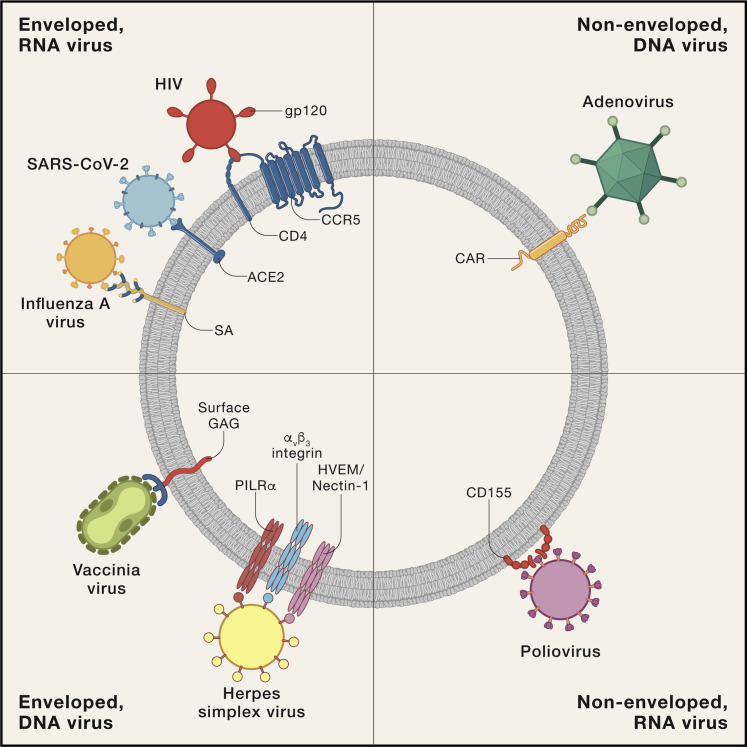
Other determinants of emerging infectious agents include cell tropism, ability to circumvent innate immune responses, and antigenic immunodominance, among others. Many viruses enter cells via one or more cell receptors (Figure 4 ) (Dai et al., 2020; Jayawardena et al., 2020); some infect different cells via different receptors, while some cell receptors may be entry points for multiple different types of viruses. The situation is extraordinarily complex from the point of view of both virus and host, with a bewildering array of receptors, alternate receptors, and co-receptors and of countless viruses able to utilize them, reflecting that “viruses have deep evolutionary roots in the cellular world” (Baranowski et al., 2001). This is exemplified by the SARS-like bat β-coronaviruses, or sarbecoviruses, whose receptor binding domains appear to be hyper-evolving by sampling a variety of mammalian receptors (Hu et al., 2017).
Figure 4.
Variable Mechanisms of Viral Entry into Host Cells
Examples of cell receptors for various DNA and RNA viruses. The cartoon image shows a spherical cell with different receptors for different categories of selected viruses. Viruses and cells are not reflective of relative sizes. The figure is suggested by the text and images of Jayawardena et al. (2020).
Many viruses enter cells via phagocytic or pinocytic endocytosis, the latter including clathrin-mediated or claveolin-mediated endocytosis, yet other viruses enter cells via fusion or direct penetration (Dai et al., 2020). SARS-CoV and SARS-CoV-2 are β-coronaviruses that enter human cells via angiotensin-converting enzyme-2 (ACE-2) receptors, whose non-human counterpart receptors are ubiquitous on cells of other species (Cui et al., 2019; Hasan et al., 2020). This means that coronaviruses of many other mammalian species may essentially be pre-adapted to human infectivity. Evidence suggests that there are many bat coronaviruses pre-adapted to emerge, and possibly to emerge pandemically (Andersen et al., 2020; Hasan et al., 2020; Hu et al., 2017; Menachery et al., 2016; Wang et al., 2018; Zhou et al., 2020a).
Influenza A viruses (IAVs) infect cells via binding to terminal sialic acids found on lumenal respiratory epithelial cells of avian as well as mammalian and human hosts and contain a neuraminidase that cleaves these same receptors to allow viral release, which facilitates onward viral transmission (Morens and Taubenberger, 2020; Taubenberger et al., 2019). Since sialic acids are found on a wide variety of mammalian and non-mammalian cells, it is not surprising that many viruses attach to these receptors, although viral affinities to receptors are complicated. For example, in vitro lectin assays suggest variable affinities of influenza for different types of terminal sialic acids, e.g., those with α-2,3 (ostensibly avian) and those with α-2,6 (ostensibly mammalian) linkages. However, autopsy studies have confirmed fatal human infections caused by IAVs that prefer either receptor, reflecting the complexity of virus-host interactions across the animal kingdom.
Yet another aspect of infecting viruses is that some (e.g., HIV, influenza) express immunodominant epitopes on external proteins that dominate the elicitation of immune responses, resulting in less robust immune responses to other, often adjacent, epitopes. This may have the effect of blunting, or limiting the breadth of, an otherwise optimal host immune response. Almost all viruses have proteins that inhibit innate protective host responses, such as the host interferon response (Blanco-Melo et al., 2020; Mesev et al., 2019). Some viruses are able to infect FcR bearing cells via antibody-dependent infection enhancement or ADE—a mechanism by which virus-IgG complexes are “tied down” by FcRs to the cell surface, facilitating cell entry via another receptor (Morens, 1994; Sullivan, 2001). During the 2002–2003 SARS epidemic, it was found that both post-infectious and vaccine-elicited antibody caused in vitro ADE with SARS-CoV (Jaume et al., 2012; Wang et al., 2016, 2014; Yip et al., 2014). It is not known whether this phenomenon, studied mostly in vitro, has implications for natural human coronavirus infections or vaccinations (Wan et al., 2020); however, it does represent a potential safety concern associated with vaccine development for SARS-CoV and SARS-CoV-2.
In considering SARS-CoV-2 vaccine development and antibody treatment/prophylaxis, it is also of some concern that natural infection with the feline coronavirus (FECV) initiates a non-fatal infection that sometimes leads to development of viral variants (known as feline infectious peritonitis viruses [FIPVs]) that are macrophage-tropic and that can bind to host anti-spike protein antibody (antibody against the external viral protein that attaches to ACE-2 receptors and elicits protective immunity) to allow viruses to enter macrophages via FcRs (ADE), leading to a distinct and universally fatal disease known as feline infectious peritonitis, or FIP (Vennema et al., 1990; Weiss and Scott, 1981). Post-infectious gene editing by the host has also been proposed as a mechanism for development of subacute sclerosing pan-encephalitis (SSPE), a fatal human disease associated with persistent natural measles virus infection complicated by generation of host-edited mutant viruses (Cattaneo et al., 1986). This represents yet another variation on mechanisms of viral emergence (Baranowski et al., 2001; Cattaneo et al., 1986; Novella et al., 2011). Fortunately, viruses such as those that cause SSPE, derived from in-host gene editing, are not necessarily transmissible.
In experimental studies, early FIPV-associated feline deaths result from both FIPV spike protein vaccination and passive transfusion with anti-FIPV antibody (Vennema et al., 1990; Weiss and Scott, 1981). Neither FECV nor FIPV are phylogenetically close to SARS-CoV or SARS-CoV-2, both being α-coronaviruses utilizing aminopeptidase N or other protein or glycan receptors rather than the ACE-2 receptors that bind SARS β-coronaviruses. But evidence for ADE with multiple different α- and β-coronaviruses suggests that as we proceed to develop SARS-CoV-2 vaccines and therapeutic antibodies, much remains to be learned about this complex viral family.
Also of importance to the infectivity of newly emerging infectious diseases are viral genetic properties associated with pathogenicity and co-pathogenicity, exemplified most clearly with pandemic IAVs. The 1918 H1N1 pandemic virus, which killed an estimated 50 million people (equivalent to 200 million when adjusted to the 2020 population) was particularly lethal because of at least two inherent properties: (1) an avian-descended H1 hemagglutinin (HA) that is unusually cytopathic and immunopathogenic compared to the HAs of most other IAVs and (2) a marked co-pathogenic ability—the viral genetic basis of which remains poorly understood—to precipitate fatal bacterial bronchopneumonias in association with pneumopathogenic bacteria carried silently in the human upper respiratory tract (Morens et al., 2008b; Morens and Taubenberger, 2020; Taubenberger et al., 2019). We now know that not only avian H1s but also 4 of the other 15 avian HAs found within the wild waterfowl and shore birds (Anseriformes and Charidriiformes) reservoir have similar pathogenic properties, and thus they represent future threats for highly fatal pandemic emergences (Morens and Taubenberger, 2020; Taubenberger et al., 2019). A perhaps even more shocking example of pandemic emergence associated with enhanced pathogenicity is that of Zika, a flavivirus known for decades. Zika had never caused a human epidemic; however, in 2015, it suddenly spread pandemically around the global tropical belt, causing millions of infections and severe fetal losses and birth defects (Fauci and Morens, 2016). The apparent cause of the pandemic was a mutation resulting in the change of a single amino acid in the external viral glycoprotein (Shan et al., 2020).
Pathogenic variability undoubtedly applies to many other virus types, exemplified, for example, by comparing the highly pathogenic Ebola Zaire strain to the closely related but lowly pathogenic Ebola Reston strain (Baseler et al., 2017). Although not yet adequately studied, pathogenic variability might also be a property of animal coronaviruses. That all three recently emerging human coronaviruses (the agents of SARS, MERS, and COVID-19) exact a high degree of human morbidity and mortality suggests that enzootic coronaviruses as a whole may be inherently pathogenic for humans. On the one hand, preliminary data suggest that SARS-CoV-2 may elicit an unbalanced innate immune response associated with decreased expression of interferons I and III and with increased inflammatory cytokine production (Mesev et al., 2019), consistent with preliminary COVID-19 findings (Vanderheiden et al., 2020). On the other hand, preliminary data suggest that viral-bacterial co-pathogenesis may be of somewhat less concern with SARS-CoV-2 than with influenza, measles, and other pathogenic respiratory viruses, although comprehensive clinical/autopsy series have not yet been published.
With regard to coronavirus disease severity, it is worth considering the conventional wisdom (not always correct) that viruses that kill their hosts limit their own ability to be transmitted and that, if transmitted person to person, they would be expected to be selected for attenuation of pathogenicity over time. It is conceivable that the four endemic coronaviruses of humans—the β-coronaviruses OC43 and HKU1 and the α-coronaviruses 229E and NL63—emerged long ago as zoonotic and perhaps highly pathogenic viruses that evolved into attenuated forms over time (Cui et al., 2019). Such natural attenuation of pathogenicity has not yet been observed with the agents of SARS, MERS, or COVID-19, although with a SARS-CoV-2 case-fatality in the range of 1%, and with evidence for significant asymptomatic and presymptomatic transmission in a largely susceptible population, evidence for selection pressures for attenuation may not be detectable in the short term.
Moreover, viral evolution toward lower pathogenicity does not apply to all infectious diseases. For many other organisms such as cholera (expressing a bacteriophage toxin causing diarrhea) or rotavirus infection (with an NSP4 diarrhea-causing toxin), or for cough-inducing tuberculosis and many respiratory viruses, damage to the gastrointestinal tract and lungs, respectively, facilitates transmission, since diarrhea and coughing expel infectious pathogens into the environment, increasing the chance of infecting additional hosts. Disease severity thus reflects a balancing act between killing or incapacitating hosts, on the one hand, and optimizing microbial transmission, and therefore survival, on the other. Similar principles may also apply to non-viral diseases with environmental modes of transmission: for example, with enzootic anthrax, host-killing may be an important transmission mechanism, as rotting carcasses leave anthrax spores in the ground to reignite future infections (Turner et al., 2014). Agent-host interaction variables are often exceedingly complex.
The Role of the Host in the Emergence of Infectious Diseases
Host variables that underlie the emergence of infectious diseases include those variables specific to individuals within the host population and those variables that relate to the host population as a whole (Morens and Fauci, 2012; Morens et al., 2004, 2008a). Since a virus replicates within the cells of the host, and since viruses usually infect hosts via specific receptors on the cells of various tissues and organs, the new host must express cellular receptors or other cell-surface properties to which the virus can bind and initiate viral internalization.
Major portals of host entry for infectious agents include those that are visibly external to the environment such as the skin or that can be reached directly from the environment such as the respiratory and gastrointestinal tracts, as well as organs reached systemically such as the liver, heart, and other internal organs. Human beings have many different organ systems, each with many different cell types, and with each cell having arrays of different receptors; therefore, it is not surprising that switching of a pathogen from an animal host to humans results in very different clinical and epidemiologic outcomes, including different disease manifestations and transmission mechanisms. These factors ultimately relate to the potential for establishment of infection in the new host as well as the likelihood of sustained transmission within the new host population and, as such, have a bearing on whether host-switching succeeds or fails.
SARS-CoV and SARS-CoV-2 enter cells via ACE-2 receptors (Wang et al., 2020), found on lung alveolar epithelial cells, gastrointestinal enterocytes, arterial and venous endothelial cells, and arterial smooth muscle cells, among other cell types (Hamming et al., 2004; Wang et al., 2020), which explains the excretion of SARS-CoV-2 and potential transmission via the respiratory and enteric routes. With regard to the latter, although SARS-CoV-2 infects cells of the gastrointestinal tract, fecal transmission has not to date been implicated in significant person-to-person viral spread. Different viruses utilize widely different strategies for binding to, penetrating, and entering cells, e.g., polioviruses, HIV, influenza viruses, coronaviruses, and others (Figure 4) (Bowers et al., 2017; Cicala et al., 2011; Jayawardena et al., 2020; Laureti et al., 2018).
Some viruses enter cells via binding to two different proximate receptors, a primary and secondary receptor, e.g., receptors for binding and for fusion (for example, numerous flaviviruses [Laureti et al., 2018]). As noted, mechanisms of viral entry into cells are exceedingly variable and complex (Jayawardena et al., 2020). Viruses may also infect macrophages and macrophage-like cells, as is the case with mosquito-borne and tick-borne flaviviruses. These viruses are injected into perivascular dermal tissue by their respective vectors and are taken up by dendritic cells and carried to regional lymph nodes, where they initiate systemic infection. ADE, discussed above, and other ADE-like phenomena, may also facilitate cellular infection (Morens, 1994; Sullivan, 2001).
Tissue/cell tropism also has a bearing on the types of immune responses that are elicited. For example, in a systemic infection like measles, high-level viremia is associated with infection of multiple organs, tissues, and cell types; the resulting broad systemic B and T cell responses lead to lifelong protection from reinfection. In contrast, influenza A viruses and respiratory syncytial virus (RSV), among many other respiratory viruses that infect surface epithelial cells, do not cause viremia and infect only surface respiratory epithelial cells. As a result, infectious virions do not have intimate interactions with the systemic immune system. The major site of influenza virus-immune system interaction is in the semi-organized tear duct-, nasal-, and mucosal-associated lymphoid tissues (TALT, NALT, and MALT), as well as the post-natally generated inducible bronchus-associated iBALT (Moyron-Quiroz et al., 2007), leading to tissue compartmentalization of the immune response, perhaps in part explaining the weakly protective immune responses of naturally acquired or vaccine-associated influenza and RSV.
Preliminary evidence from clinical and pathological studies of both SARS-CoV and SARS-CoV-2, which indicate viral infection of multiple tissues, is consistent with elicitation of robust and hopefully long-lasting protective immunity, providing a potential for control of COVID-19 with vaccines. More ominously, expression of ACE-2 receptors on endothelial and numerous other cells, and autopsy evidence of significant SARS-C0V-2 endothelial infection (Fox et al., 2020), are consistent with systemic viral infection causing both pulmonary and extra-pulmonary pathology, including widespread microthrombus formation, among other outcomes.
Some emerging viruses encounter pre-existing partial population immunity, e.g., pandemic influenza viruses. Most notably, in the influenza pandemics of 1968 and 2009, caused by an H3N2 and an H1N1 virus, respectively, segments of the population had pre-existing immunity that interfered with early viral spread and possibly with viral evolution. Although insufficient to prevent emergence, such population immunity did protect certain segments of the population (Morens and Taubenberger, 2020; Taubenberger et al., 2019). In fact, in most influenza pandemics, influenza-specific case-fatality in the elderly, which increases regularly over about age 60 with seasonal influenza, is nevertheless apparently blunted by poorly characterized immune effects of prior influenza exposures, emphasizing the complexity of viral infectivity and host resistance factors (Morens and Taubenberger, 2011). In contrast, there are suggestions that decreases in natural infections with, or vaccinations against, pathogens may facilitate the emergence of related organisms, e.g., increased incidence of human monkeypox after the cessation of smallpox vaccination that followed eradication of the monkeypox-related smallpox virus (Lloyd-Smith, 2013), or the long-held theory that existing or newly evolving enteroviruses will emerge, or are already emerging, to fill an “ecologic niche” created by the near-eradication of the three polioviruses (Rieder et al., 2001).
In the case of COVID-19, some evidence suggests the absence of pre-existing population immunity, afforded by exposure to the four endemic coronaviruses, sufficient to prevent infection (Corman et al., 2018). Although these endemic viruses share few epitopes that cross-react significantly with SARS-CoV-2 in serologic studies, it has been speculated that endemic coronavirus cross-protection may nevertheless prevent or at least limit the severity of disease in some, especially in young persons (Nickbakhsh et al., 2020).
There is preliminary but growing evidence that infectious disease severity or even susceptibility may in some cases be related to host genetic variables associated with the innate immune response, as is the case with epidemiologic information concerning severe disease caused by H5N1 poultry-associated influenza (Morens and Taubenberger, 2015). Several host genes have been provisionally linked to susceptibility to such severe viral disease outcomes (Nguyen et al., 2020; Tang et al., 2008), including the interferon-related transmembrane protein 3 (IFITM3) SNP rs22522-C allele (Everitt et al., 2012; Zheng et al., 2017), which has been proposed (based on very preliminary data) to be involved in severity of SARS-CoV-2 disease (Zhang et al., 2020). In addition, differential interactions with IFITMs such as IFITM3 include inhibiting the human endemic α-coronavirus while enhancing entry of SARS-CoV and MERS-CoV β-coronaviruses (Huang et al., 2011; Zhao et al., 2018).
With respect to COVID-19, several studies have associated blood group A in the ABO system with disease severity, although the mechanisms of this effect are not yet clear. ABO system associations with infectious diseases have also been shown for infections with noroviruses, H. pylori, and falciparum malaria; however, any such associations might also be indirect markers for unrelated genes. Newer data are beginning to define human immonotype risks for more severe disease (Mathew et al., 2020), as well as posssible genetic signatures of severe diseases (Gussow et al., 2020). At this time, data establishing specific genetic susceptibilities remain inconclusive for most diseases, including coronavirus diseases. This is a research area likely to be important in the future, since identification of susceptibilities for human disease severity has great implications for prevention, diagnosis, and treatment.
Among the most important host factors for infection and for disease emergences/re-emergences are those associated with human behaviors, e.g., population growth, crowding, human movement, and many others, including behaviors that either perturb the environment or result in new human-created ecologic niches (Figure 3). Regarding human movement, both the 1347–1348 Black Death (bubonic/pneumonic plague) and the 1832 cholera pandemic (which traveled from India to Europe and then to the Western Hemisphere) were spread along major trade and travel routes. In 1831–1832, 45 years before a coherent “germ theory” would be articulated, it was clear that as cholera spread slowly westward, it moved no faster than coaches and ships traveled: it slowed down in the winter as travel slowed down, and it picked up again in the summer as travel increased.
The 1889 influenza pandemic traveled westward from Asia to Europe along railroad lines and then was exported globally along shipping routes. The 1957 influenza pandemic was spread by ships, but 11 years later the 1968 influenza pandemic was spread along air routes, the first example of global pandemic spread by airplanes. In 1981, acute hemorrhagic conjunctivitis was spread between international air hubs in the tropics and some temperate zones (e.g., to Florida and North Carolina). In 2002–2003, SARS was exported by air from Hong Kong to the Western Hemisphere and Europe. In 2019–2020, SARS-CoV-2 was spread globally from China in a similar manner. These many ancient and modern examples reflect the extraordinary importance of human population growth and movement in spreading diseases: the more populous and crowded we as a species become, and the more we travel, the more we provide opportunities for emerging diseases.
The Role of the Environment in the Emergence of Infectious Diseases
Many other human activities related to the environment have important consequences for disease emergence (Allen et al., 2017; Dobson and Carper, 1996; Fauci and Morens, 2012; Morens et al., 2020b; Morens and Fauci, 2012; Morens et al., 2004, 2008a, 2019; Morens and Taubenberger, 2020). Human water storage practices in Northern Africa, beginning about 5,000 years ago, led to the emergence of a new, exclusively human-adapted mosquito, Aedes aegypti, which created a secondary ecologic niche for the emergence of yellow fever virus and, centuries later, dengue, chikungunya, and Zika, all of which then went on to spread pandemically. Depots of used rubber tires create ideal breeding sites for a related human-adapted mosquito, Aedes albopictus, which in recent decades has spread globally across the tropical and subtropical zones, transmitting many of these same arboviral diseases widely, if less efficiently. Predictably, viruses and viral vectors have adapted to environmental influences: a single locus mutation in the Aedes aegypti-adapted Indian Ocean strain of chikungunya virus has newly adapted it, without loss of fitness for aegypti, to Aedes albopictus, widely prevalent in the region (Tsetsarkin and Weaver, 2011).
Land-management practices have been associated with re-emergences of Eastern equine encephalitis (Morens et al., 2019); deforestation with emergences of Zika and Hendra viruses; road-building and environmental degradation with the spread of Bolivian hemorrhagic fever and HIV (infections spread by truckers and truck stop prostitution); and poverty, crowding, and poor sanitation with re-emergences of many diseases such as tuberculosis and cholera. For centuries, wars have precipitated the re-emergences of many diseases (Dobson and Carper, 1996; Fauci and Morens, 2012; Morens and Fauci, 2012; Morens et al., 2004, 2008a, 2020b; Morens and Taubenberger, 2020), e.g., the Serbian typhus epidemic during World War I, which killed 150,000 people, mostly civilians. In the 1700s, when typhus had not been identified as a specific disease, textbooks listed two separate conditions under the nosologic terms “war typhus” and “jail typhus,” reflecting human activities that provoked long-ago disease emergences. A classic epidemiology text published over a century ago, still studied today, is titled Epidemics Resulting from Wars (Prinzing, 1916).
It is suspected that SARS-CoV-2 emerged in 2019, as did SARS-CoV in 2002, and as did H5N1 and H7N9 poultry-associated influenza, in 1997 and 2013, respectively, from wet markets in China (Morens et al., 2020a; Morens et al., 2020b). These four diseases may thus represent four deadly emergences, within an 18-year span, from one cultural practice in one region of the world. These and many other examples (Dobson and Carper, 1996; Fauci and Morens, 2012; Lu et al., 2020; Morens and Fauci, 2012; Morens et al., 2004, 2008a, 2019, 2020b; Morens and Taubenberger, 2020; Zhang and Holmes, 2020; Zhou et al., 2020b) constitute a powerful argument that human activities and practices have become the key determinant of disease emergence.
Emergence of Diseases Leading to Epidemicity and Endemicity
Whatever human behaviors lead to the emergence of infectious diseases, the newly emerged organism cannot survive continually in humans without adapting to one of several direct or indirect mechanisms of person-to-person spread (Table 4 ). Many emerging organisms, such as the hantaviral agents of hantavirus pulmonary syndrome and Korean hemorrhagic fever, or the arenaviruses causing Argentine, Bolivian, and Lassa hemorrhagic fevers, result from dead-end exposures to reservoir rodents and rarely spread from person to person. Preventing and controlling these types of emergences thus focus on the enzootic reservoir.
Table 4.
Mechanisms of Infection Transmission from Person to Person and from Animal to Person
| Respiratory including Environmental | Gastrointestinal including Environmental | Inoculation |
|
|---|---|---|---|
| Direct | Vectorborne | ||
| Influenza∗ | Cholera | Anthrax2 | Chikungunya∗ |
| Human coronaviruses | Noroviruses | Dracunculiasis3 | Dengue∗ |
| Measles | Rotaviruses | Gonorrhea | Lyme disease∗ |
| Rhinoviruses | Salmonellosis | Hepatitis B and C | Malaria∗ |
| SARS,∗ COVID-19,∗ MERS1,∗ | Typhoid fever | HIV∗ | Yellow fever |
| Some human enteroviruses∗ | Some human enteroviruses | Syphilis | Zika∗ |
Selected endemic and emerging infectious diseases transmitted to humans via multiple mechanisms. Many diseases are transmitted by more than one mechanism. For example, most of the respiratory-transmitted diseases are also transmitted by hands and on fomites; most of the gastrointestinal diseases are uncommonly transmitted directly and more commonly transmitted by water, food, and on fomites. Some enteroviruses are predominantly transmitted by the fecal-oral (gastrointestinal) route, whereas others are predominantly transmitted by the respiratory route. Anthrax is transmitted by respiratory, gastrointestinal, and inoculation routes. Understanding mechanisms of transmission is important not only because it helps control those diseases that emerge but also because it provides opportunities to control multiple diseases transmitted by the same mechanisms.
New/important emergences and re-emergences in the past 4 decades.
MERS is largely a zoonotic disease, although person-to-person spread can occur with relative inefficiency.
Anthrax is acquired by inoculation, inhalation, or ingestion of spores, but the cutaneous form of anthrax can be spread person to person.
Dracunculiasis is environmentally acquired as part of a complex life cycle including human expulsion of pathogens into the water and water-borne transmission to other humans.
Person-to-person transmissibility, a necessary step in establishing epidemicity and pandemicity after host-switching, can occur via one or more of four basic mechanisms (Table 4): respiratory;gastrointestinal; environmental spread via an intermediate environmental state such as waterborne, foodborne, and fomite-mediated; and environmental spread via inoculation, including vectorborne. It is noteworthy that these mechanisms reflect not only human societal practices such as sanitary practices, embracing physical closeness, and sexual activities but also ways in which people interact with the environment such as storage and drinking of water from different sources, food procurement, and food preparation practice. Even so, many variables interact to produce different clinical and epidemiologic aspects of disease transmitted from person to person. For example, while both Ebola and SARS-CoV-2 are potentially fatal diseases transmitted person to person, SARS-CoV-2 is, like most respiratory diseases, associated with asymptomatic infection in many and is often transmitted to others by an asymptomatic or pre-symptomatic infected person. Ebola, on the other hand, has a lower frequency of asymptomatic infection and is of low transmissibility right up to the time of illness onset. Moreover, the mechanisms of person-to-person transmission are distinct for these two diseases. SARS-CoV-2 is transmitted via hands and fomites, respiratory droplets and aerosol, including transmission by superspreading events where large numbers of individuals are infected by a single person, almost invariably in closed crowded settings. Ebola, on the other hand, is transmitted via virus-contaminated bodily secretions, and infection is usually acquired by persons touching contaminated fluids or fomites, for example in providing nursing care, burial services, handling towels, bed linens, utensils, etc. (Baseler et al., 2017). Although general principles of infection transmission are understood, specific diseases may vary in clinical and epidemiologic features that bear upon the type and intensity of transmission.
In this context, ongoing research into the origin of SARS-CoV-2 seeks to learn where, how, and why the virus emerged as a human pandemic disease (Boni et al., 2020; Latinne et al., 2020). SARS-CoV-2 clusters phylogenetically within an extensive but still not fully characterized universe of wild bat β-coronaviruses found in many species over much of the globe (Anthony et al., 2017; Cui et al., 2019; Hu et al., 2017; Letko et al., 2020; Lu et al., 2020; Morens et al., 2020a; Zhang and Holmes, 2020; Zhou et al., 2020b). The discovery that its closest identified viral relatives are enzootic in horseshoe (Rhinolophus) bats (Zhou et al., 2020a, 2020b) indicates that SARS-CoV-2 probably emerged from an as-yet-unidentified bat reservoir either directly or after infection of an intermediate host such as a pangolin (Boni et al., 2020; Letko et al., 2020; Li et al., 2020; Zhou et al., 2020a, 2020b). As was true of SARS-CoV 18 years ago, the specific determinants of SARS-CoV-2 emergence remain obscure. Gaining a better understanding of the enormous reservoir of bat coronaviruses has been an urgent priority since the 2002 SARS epidemic, and remains so today. Considerable surveillance and phylogenetic and experimental work remains to be done. In 2020, it is among our most urgent research priorities (Latinne et al., 2020).
One of the most important unanswered questions we face in the ongoing COVID-19 pandemic relates to the evolutionary potential of human-adapted SARS-CoV-2. Will it, similar to human IAVs in recent centuries, evolve to persist as a permanent human pathogen by mutating to escape the population herd immunity it creates? And if it does persist, will it attenuate over time, as the four endemic coronaviruses may have done centuries ago? Or, on the other hand, will it increase in pathogenicity as the pandemic H3N2 IAV has done over the past 52 years? Because SARS-CoV-2 lacks a segmented genome, we are spared at least one genetic trick (gene reassortment) underlying IAV pandemic emergences; however, like human IAVs, SARS-CoV-2 can be expected to evolve by mutation as it spreads through human populations, and it has the additional capacity of evolving by genetic recombination. Enzootic evidence supports a very high degree of recombination of SARS-like CoVs in nature. Will these capacities allow SARS-CoV-2 to escape from population immunity elicited by natural infection or future vaccination? Only additional time, and much important research, will begin to answer these questions.
The Enigma of Host-Switching
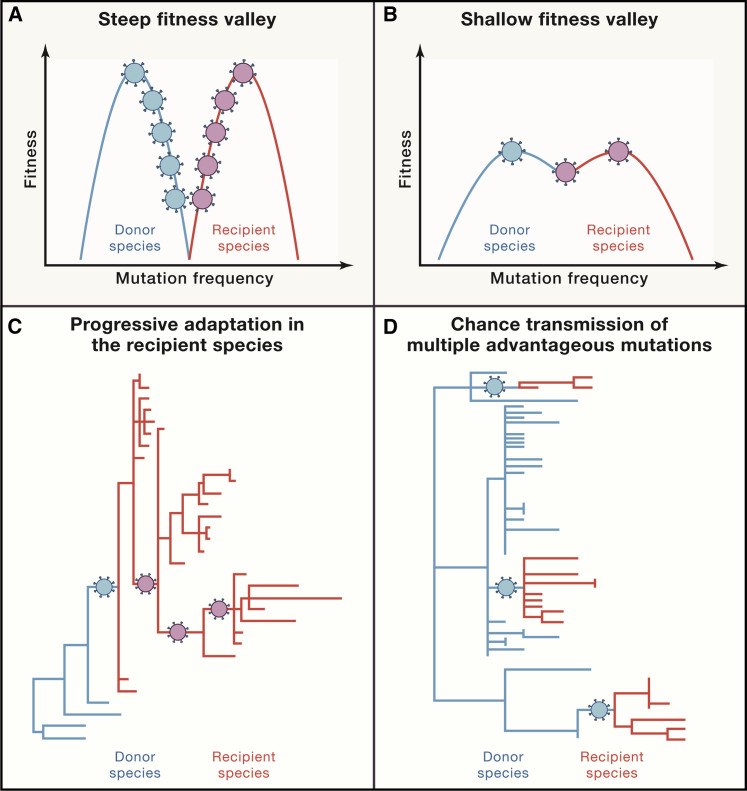
Perhaps the single biggest mystery of emerging diseases is how microorganisms, including animal-adapted microorganisms, switch species to infect humans. Host-switching from animal to human has long been conceptualized as the crossing of a fitness valley, in which a virus adapted to transmitting between members of host species A must somehow simultaneously develop the capacity to productively infect cells of new host species B (Figure 5 ) (Dolan et al., 2018; Geoghegan and Holmes, 2018; Kuiken et al., 2006; Parrish et al., 2008). In this paradigm, the depth of the fitness valley reflects host-to-host barrier challenges that have to be overcome. Our understanding of mechanisms of viral evolution and host-switching is nevertheless incomplete, in part because of inability to reconcile experimental within-host viral evolutionary findings and natural-world findings that examine viral evolution during outbreaks, including new disease emergences (Geoghegan and Holmes, 2018). Among many complicating factors, rapid intense person-to-person transmission of new viruses in large human populations often produces complex genetic diversity, confounding attempts to link viral variation to viral phenotypic changes and selection pressures eliciting them.
Figure 5.
Proposed Molecular Mechanisms of Host-Switching
Proposed mechanisms of cross-species host-switching of infectious agents (after Kuiken et al., 2006). Steep (A) and shallow (B) fitness valleys between donor and recipient host species reflect adaptational barriers that need to be crossed. (A) and (C) show that a greater number of sequential adaptational mutations are needed in (A) to cross the fitness valley and then adapt to the new host, as compared to the situation shown in (B) and (D), where greater donor-host similarities facilitate switching. (C) and (D) represent the associated phylogenetic trees: in (C), the donor host and recipient host viruses most go through significant adaptational steps, including those associated with initial transmission in the new host. In (D), the new host receives an infectious agent that is partially pre-adapted; successful emergence requires fewer adaptational mutations.
The situation is particularly complicated for the most important category of newly emerging disease agents: the RNA viruses, which include SARS-CoV, SARS-CoV-2, MERS, Ebola, and influenza, as well as dengue, Zika, and other arboviruses. These viruses evolve as, and are transmitted as, complex quasispecies, or viral swarms, which contain many viral variants of differing degrees of relatedness. It is unclear whether and/or to what extent transmission/host-switching reflects Darwinian evolution of novel virus variants, as opposed to evolution based on whole-quasispecies fitness (Geoghegan and Holmes, 2018). In the latter conceptualization, viral quasispecies evolve together as a diverse array of optimally fit, less optimally fit, and least optimally fit variants, trading off perfect host fitness for adaptational flexibility.
While it has long been assumed that the major determinants of host-switching are the evolutionary closeness of hosts A and B and the diversity of their transmitting quasispecies, recent research suggests that pathogen opportunity may be the major determinant of host-switching (Anishchenko et al., 2006; Araujo et al., 2015). In essence, even a virus poorly fit to a potential host can adapt to infect that host if given enough chances. The implications are profound. If host-switching is opportunity driven, e.g., for SARS- CoV and CoV-2, Nipah, and Hendra, then prevention and control will have to focus not only on the infectious agents themselves but also on human behaviors, such as the animal-human interface, represented by shopping in live animal markets, preparation and consumption of bush meat, intensive farming/animal husbandry, environmental degradation, and other human behaviors (Allen et al., 2017; Carroll et al., 2018).
Looked at another way, infectious diseases may be emerging into humans with greater frequency than appreciated; however, historically they have not usually achieved sustained-enough transmission that would lead to detection of the emergence. For example, before the recognized emergence of Zika, low human seroprevalence rates had been detected in enzootic areas for decades, but without detection of human outbreaks. Before the emergence of SARS-CoV-2, antibodies to it or to closely related sarbecoviruses were detected in humans exposed to bat coronaviruses (Wang et al., 2018). MERS has emerged from dromedary camels to humans on multiple occasions; however, despite limited chains of subsequent human-to-human spread, it has not become an established human disease. H5N1 avian influenza has infected millions of humans; however, only a small percentage manifest a recognized disease (often severe or fatal), and human-to-human transmission has been rarely reported (Morens and Taubenberger, 2015). These and many other examples suggest that disease emergence into humans may be common; however, sustained transmission between humans has been a rare occurrence resulting from incompletely understood factors. But in looking at the recent spate of deadly emergences noted above, we must now ask whether human behaviors that perturb the human-microbial status quo have reached a tipping point that forecasts the inevitability of an acceleration of disease emergences.
Such enigmas need to be better understood if we are to control emergences of infectious diseases (Dobson et al., 2020). Better understanding of the obscure mechanisms of emergences might allow us to anticipate emergence risks by (1) surveilling and characterizing taxonomic groups of potentially pre-emergent viruses, including viruses that may be the nearest to emerging, e.g., coronaviruses, henipaviruses, flaviviruses, arenaviruses, and filoviruses; (2) conducting intensive research on suspected high-risk viral categories to identify, in experimental animals, conserved epitopes for vaccine development and targets for antiviral therapies; (3) characterizing mechanisms of potential human transmission in experimental animal studies; (4) developing control mechanisms in areas such as animal husbandry, wildlife interaction, and rodent, vector, and mosquito/tick control; (5) devising environmental, land management, wildlife conservation/control programs; and (6) utilizing new virologic, informatic, and technological approaches to understand viral evolution and even predict emergence potential (Allen et al., 2017).
There are many examples where disease emergences reflect our increasing inability to live in harmony with nature. Nipah virus emergence followed agricultural burning of forests, which led to displacement of infected bats; bats then went to roost in trees that shaded intensively farmed pigs that were crowded into small areas, which led to infection of pigs via bat droppings, which in turn led to human outbreaks in pig farmers (Morens et al., 2004). Fish meal farming by fishermen in waters around the globe damages ecosystems by over-fishing and deprives local residents of food sources, leading to poverty and human movement, which exacerbate the potential for disease emergence. In Asia, the fish meal is used for breeding and over-feeding of farmed animals, often in intensive farming practices, which increase the likelihood of zoonotic diseases. Yellow fever, dengue, chikungunya, and Zika are all associated with urban crowding, poor sanitation, and water storage. Over many centuries, urbanization and crowding has led to rodent infestation and to rodent-borne diseases such as plague, murine typhus, and rat-bite fever. The ongoing COVID-19 pandemic reminds us that overcrowding in dwellings and places of human congregation (sports venues, bars, restaurants, beaches, airports), as well as human geographic movement, catalyzes disease spread.
Living in greater harmony with nature will require changes in human behavior as well as other radical changes that may take decades to achieve: rebuilding the infrastructures of human existence, from cities to homes to workplaces, to water and sewer systems, to recreational and gatherings venues. In such a transformation we will need to prioritize changes in those human behaviors that constitute risks for the emergence of infectious diseases. Chief among them are reducing crowding at home, work, and in public places as well as minimizing environmental perturbations such as deforestation, intense urbanization, and intensive animal farming. Equally important are ending global poverty, improving sanitation and hygiene, and reducing unsafe exposure to animals, so that humans and potential human pathogens have limited opportunities for contact. It is a useful “thought experiment” to note that until recent decades and centuries, many deadly pandemic diseases either did not exist or were not significant problems. Cholera, for example, was not known in the West until the late 1700s and became pandemic only because of human crowding and international travel, which allowed new access of the bacteria in regional Asian ecosystems to the unsanitary water and sewer systems that characterized cities throughout the Western world. This realization leads us to suspect that some, and probably very many, of the living improvements achieved over recent centuries come at a high cost that we pay in deadly disease emergences. Since we cannot return to ancient times, can we at least use lessons from those times to bend modernity in a safer direction? These are questions to be answered by all societies and their leaders, philosophers, builders, and thinkers and those involved in appreciating and influencing the environmental determinants of human health.
Summary and Conclusions
SARS-CoV-2 is a deadly addition to the long list of microbial threats to the human species. It forces us to adapt, react, and reconsider the nature of our relationship to the natural world. Emerging and re-emerging infectious diseases are epiphenomena of human existence and our interactions with each other, and with nature. As human societies grow in size and complexity, we create an endless variety of opportunities for genetically unstable infectious agents to emerge into the unfilled ecologic niches we continue to create. There is nothing new about this situation, except that we now live in a human-dominated world in which our increasingly extreme alterations of the environment induce increasingly extreme backlashes from nature.
Science will surely bring us many life-saving drugs, vaccines, and diagnostics; however, there is no reason to think that these alone can overcome the threat of ever more frequent and deadly emergences of infectious diseases. Evidence suggests that SARS, MERS, and COVID-19 are only the latest examples of a deadly barrage of coming coronavirus and other emergences. The COVID-19 pandemic is yet another reminder, added to the rapidly growing archive of historical reminders, that in a human-dominated world, in which our human activities represent aggressive, damaging, and unbalanced interactions with nature, we will increasingly provoke new disease emergences. We remain at risk for the foreseeable future. COVID-19 is among the most vivid wake-up calls in over a century. It should force us to begin to think in earnest and collectively about living in more thoughtful and creative harmony with nature, even as we plan for nature’s inevitable, and always unexpected, surprises.
Acknowledgments
We thank John and Aaron Weddle for graphics assistance.
References
- Allen T., Murray K.A., Zambrana-Torrelio C., Morse S.S., Rondinini C., Di Marco M., Breit N., Olival K.J., Daszak P. Global hotspots and correlates of emerging zoonotic diseases. Nat. Commun. 2017;8:1124. doi: 10.1038/s41467-017-00923-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anishchenko M., Bowen R.A., Paessler S., Austgen L., Greene I.P., Weaver S.C. Venezuelan encephalitis emergence mediated by a phylogenetically predicted viral mutation. Proc. Natl. Acad. Sci. USA. 2006;103:4994–4999. doi: 10.1073/pnas.0509961103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony S.J., Johnson C.K., Greig D.J., Kramer S., Che X., Wells H., Hicks A.L., Joly D.O., Wolfe N.D., Daszak P., et al. PREDICT Consortium Global patterns in coronavirus diversity. Virus Evol. 2017;3:vex012. doi: 10.1093/ve/vex012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo S.B., Braga M.P., Brooks D.R., Agosta S.J., Hoberg E.P., von Hartenthal F.W., Boeger W.A. Understanding Host-Switching by Ecological Fitting. PLoS ONE. 2015;10:e0139225. doi: 10.1371/journal.pone.0139225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranowski E., Ruiz-Jarabo C.M., Domingo E. Evolution of cell recognition by viruses. Science. 2001;292:1102–1105. doi: 10.1126/science.1058613. [DOI] [PubMed] [Google Scholar]
- Baseler L., Chertow D.S., Johnson K.M., Feldmann H., Morens D.M. The Pathogenesis of Ebola Virus Disease. Annu. Rev. Pathol. 2017;12:387–418. doi: 10.1146/annurev-pathol-052016-100506. [DOI] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Moller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni M.F., Lemey P., Jiang X., Lam T.T., Perry B.W., Castoe T.A., Rambaut A., Robertson D.L. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat. Microbiol. 2020 doi: 10.1038/s41564-020-0771-4. Published online July 28, 2020. [DOI] [PubMed] [Google Scholar]
- Bowers J.R., Readler J.M., Sharma P., Excoffon K.J.D.A. Poliovirus Receptor: More than a simple viral receptor. Virus Res. 2017;242:1–6. doi: 10.1016/j.virusres.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D., Daszak P., Wolfe N.D., Gao G.F., Morel C.M., Morzaria S., Pablos-Méndez A., Tomori O., Mazet J.A.K. The Global Virome Project. Science. 2018;359:872–874. doi: 10.1126/science.aap7463. [DOI] [PubMed] [Google Scholar]
- Cattaneo R., Schmid A., Rebmann G., Baczko K., Ter Meulen V., Bellini W.J., Rozenblatt S., Billeter M.A. Accumulated measles virus mutations in a case of subacute sclerosing panencephalitis: interrupted matrix protein reading frame and transcription alteration. Virology. 1986;154:97–107. doi: 10.1016/0042-6822(86)90433-2. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention . Principles of Epidemiology in Public Health Practice: An Introduction to Applied Epidemiology and Biostatistics. Third Edition. Centers for Disease Control and Prevention; 2011. Lesson 1: Introduction to Epidemiology.https://www.cdc.gov/csels/dsepd/ss1978/index.html [Google Scholar]
- Cicala C., Arthos J., Fauci A.S. HIV-1 envelope, integrins and co-receptor use in mucosal transmission of HIV. J. Transl. Med. 2011;9(Suppl 1):S2. doi: 10.1186/1479-5876-9-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Muth D., Niemeyer D., Drosten C. Hosts and Sources of Endemic Human Coronaviruses. Adv. Virus Res. 2018;100:163–188. doi: 10.1016/bs.aivir.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Zhang X., Ostrikov K., Abrahamyan L. Host receptors: the key to establishing cells with broad viral tropism for vaccine production. Crit. Rev. Microbiol. 2020;46:147–168. doi: 10.1080/1040841X.2020.1735992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawkins R. Oxford University Press; Oxxford, England: 1976. The Selfish Gene. [Google Scholar]
- Dobson A.P., Carper E.R. Infectious diseases and human population history. BioScience. 1996;46:115–126. [Google Scholar]
- Dobson A.P., Pimm S.L., Hannah L., Kaufman L., Ahumada J.A., Ando A.W., Bernstein A., Busch J., Daszak P., Engelmann J., et al. Ecology and economics for pandemic prevention. Science. 2020;369:379–381. doi: 10.1126/science.abc3189. [DOI] [PubMed] [Google Scholar]
- Dolan P.T., Whitfield Z.J., Andino R. Mapping the Evolutionary Potential of RNA Viruses. Cell Host Microbe. 2018;23:435–446. doi: 10.1016/j.chom.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdle W.R., Cochi S.L. The principles and feasibility of disease eradication. Vaccine. 2011;29(Suppl 4):D70–D73. doi: 10.1016/j.vaccine.2011.04.006. [DOI] [PubMed] [Google Scholar]
- Everitt A.R., Clare S., Pertel T., John S.P., Wash R.S., Smith S.E., Chin C.R., Feeley E.M., Sims J.S., Adams D.J., et al. GenISIS Investigators. MOSAIC Investigators IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484:519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauci A.S., Morens D.M. The perpetual challenge of infectious diseases. N. Engl. J. Med. 2012;366:454–461. doi: 10.1056/NEJMra1108296. [DOI] [PubMed] [Google Scholar]
- Fauci A.S., Morens D.M. Zika Virus in the Americas--Yet Another Arbovirus Threat. N. Engl. J. Med. 2016;374:601–604. doi: 10.1056/NEJMp1600297. [DOI] [PubMed] [Google Scholar]
- Fox S.E., Akmatbekov A., Harbert J.L., Li G., Quincy Brown J., Vander Heide R.S. Pulmonary and cardiac pathology in African American patients with COVID-19: an autopsy series from New Orleans. Lancet Respir. Med. 2020;8:681–686. doi: 10.1016/S2213-2600(20)30243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan J.L., Holmes E.C. Evolutionary Virology at 40. Genetics. 2018;210:1151–1162. doi: 10.1534/genetics.118.301556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gussow A.B., Auslander N., Faure G., Wolf Y.I., Zhang F., Koonin E.V. Genomic determinants of pathogenicity in SARS-CoV-2 and other human coronaviruses. bioRxiv. 2020 doi: 10.1101/2020.04.05.026450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A., Paray B.A., Hussain A., Qadir F.A., Attar F., Aziz F.M., Sharifi M., Derakhshankhah H., Rasti B., Mehrabi M., et al. A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1754293. Published online April 22, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins D.R. Disease eradication. N. Engl. J. Med. 2013;368:54–63. doi: 10.1056/NEJMra1200391. [DOI] [PubMed] [Google Scholar]
- Hu B., Zeng L.P., Yang X.L., Ge X.Y., Zhang W., Li B., Xie J.Z., Shen X.R., Zhang Y.Z., Wang N., et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13:e1006698. doi: 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang I.C., Bailey C.C., Weyer J.L., Radoshitzky S.R., Becker M.M., Chiang J.J., Brass A.L., Ahmed A.A., Chi X., Dong L., et al. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7:e1001258. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaume M., Yip M.S., Kam Y.W., Cheung C.Y., Kien F., Roberts A., Li P.H., Dutry I., Escriou N., Daeron M., et al. SARS CoV subunit vaccine: antibody-mediated neutralisation and enhancement. Hong Kong Med. J. 2012;18(Suppl 2):31–36. [PubMed] [Google Scholar]
- Jayawardena N., Poirier J.T., Burga L.N., Bostina M. Virus-Receptor Interactions and Virus Neutralization: Insights for Oncolytic Virus Development. Oncolytic Virother. 2020;9:1–15. doi: 10.2147/OV.S186337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W.E. Origins and evolutionary consequences of ancient endogenous retroviruses. Nat. Rev. Microbiol. 2019;17:355–370. doi: 10.1038/s41579-019-0189-2. [DOI] [PubMed] [Google Scholar]
- Kuiken T., Holmes E.C., McCauley J., Rimmelzwaan G.F., Williams C.S., Grenfell B.T. Host species barriers to influenza virus infections. Science. 2006;312:394–397. doi: 10.1126/science.1122818. [DOI] [PubMed] [Google Scholar]
- Latinne A., Hu B., Olival K.J., Zhu G., Zhang L., Li H., Chmura A.A., Field H.E., Zambrana-Torrelio C., Epstein J.H., et al. Origin and cross-species transmission of bat coronaviruses in China. bioRxiv. 2020 doi: 10.1101/2020.05.31.116061. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Laureti M., Narayanan D., Rodriguez-Andres J., Fazakerley J.K., Kedzierski L. Flavivirus Receptors: Diversity, Identity, and Cell Entry. Front. Immunol. 2018;9:2180. doi: 10.3389/fimmu.2018.02180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Seifert S.N., Olival K.J., Plowright R.K., Munster V.J. Bat-borne virus diversity, spillover and emergence. Nat. Rev. Microbiol. 2020;18:461–471. doi: 10.1038/s41579-020-0394-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Giorgi E.E., Marichann M.H., Foley B., Xiao C., Kong X.P., Chen Y., Korber B., Gao F. Emergence of SARS-CoV-2 through Recombination and Strong Purifying Selection. bioRxiv. 2020 doi: 10.1126/sciadv.abb9153. 2020.03.20.000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Smith J.O. Vacated niches, competitive release and the community ecology of pathogen eradication. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013;368:20120150. doi: 10.1098/rstb.2012.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum G.S., Soriano F., Trejos A., Llerena J. Vaccinia epidemic and epizootic in El Salvador. Am. J. Trop. Med. Hyg. 1967;16:332–338. doi: 10.4269/ajtmh.1967.16.332. [DOI] [PubMed] [Google Scholar]
- Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D’Andrea K., et al. UPenn COVID Processing Unit Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020 doi: 10.1126/science.eabc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Sims A.C., Debbink K., Agnihothram S.S., Gralinski L.E., Graham R.L., Scobey T., Plante J.A., Royal S.R., et al. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. USA. 2016;113:3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson M., Guillemin J., Hugh-Jones M., Langmuir A., Popova I., Shelokov A., Yampolskaya O. The Sverdlovsk anthrax outbreak of 1979. Science. 1994;266:1202–1208. doi: 10.1126/science.7973702. [DOI] [PubMed] [Google Scholar]
- Mesev E.V., LeDesma R.A., Ploss A. Decoding type I and III interferon signalling during viral infection. Nat. Microbiol. 2019;4:914–924. doi: 10.1038/s41564-019-0421-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens D.M. Antibody-dependent enhancement of infection and the pathogenesis of viral disease. Clin. Infect. Dis. 1994;19:500–512. doi: 10.1093/clinids/19.3.500. [DOI] [PubMed] [Google Scholar]
- Morens D.M., Fauci A.S. Emerging infectious diseases in 2012: 20 years after the institute of medicine report. MBio. 2012;3 doi: 10.1128/mBio.00494-12. e00494-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens D.M., Taubenberger J.K. Pandemic influenza: certain uncertainties. Rev. Med. Virol. 2011;21:262–284. doi: 10.1002/rmv.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens D.M., Taubenberger J.K. How low is the risk of influenza A(H5N1) infection? J. Infect. Dis. 2015;211:1364–1366. doi: 10.1093/infdis/jiu530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens D.M., Taubenberger J.K. In: Preparing for Pandemics in the Modern World. Blackburn C.C., editor. Texas A & M University Press; 2020. The 1918 Influenza Pandemic: A Still-Mysterious Litmus Test for Pandemic Prevention and Control. [Google Scholar]
- Morens D.M., Folkers G.K., Fauci A.S. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430:242–249. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens D.M., Folkers G.K., Fauci A.S. Emerging infections: a perpetual challenge. Lancet Infect. Dis. 2008;8:710–719. doi: 10.1016/S1473-3099(08)70256-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens D.M., Taubenberger J.K., Fauci A.S. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J. Infect. Dis. 2008;198:962–970. doi: 10.1086/591708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens D.M., Folkers G.K., Fauci A.S. Eastern Equine Encephalitis Virus - Another Emergent Arbovirus in the United States. N. Engl. J. Med. 2019;381:1989–1992. doi: 10.1056/NEJMp1914328. [DOI] [PubMed] [Google Scholar]
- Morens D.M., Breman J.G., Calisher C.H., Doherty P.C., Hahn B.H., Keusch G.T., Kramer L.D., LeDuc J.W., Monath T.P., Taubenberger J.K. The Origin of COVID-19 and Why It Matters. Am. J. Trop. Med. Hyg. 2020 doi: 10.4269/ajtmh.20-0849. Published online July 22, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens D.M., Daszak P., Taubenberger J.K. Escaping Pandora’s Box - Another Novel Coronavirus. N. Engl. J. Med. 2020;382:1293–1295. doi: 10.1056/NEJMp2002106. [DOI] [PubMed] [Google Scholar]
- Moyron-Quiroz J., Rangel-Moreno J., Carragher D.M., Randall T.D. The function of local lymphoid tissues in pulmonary immune responses. Adv. Exp. Med. Biol. 2007;590:55–68. doi: 10.1007/978-0-387-34814-8_4. [DOI] [PubMed] [Google Scholar]
- Nguyen A., David J.K., Maden S.K., Wood M.A., Weeder B.R., Nellore A., Thompson R.F. Human Leukocyte Antigen Susceptibility Map for Severe Acute Respiratory Syndrome Coronavirus 2. J. Virol. 2020;94 doi: 10.1128/JVI.00510-20. e00510-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickbakhsh S., Ho A., Marques D.F.P., McMenamin J., Gunson R.N., Murcia P.R. Epidemiology of Seasonal Coronaviruses: Establishing the Context for the Emergence of Coronavirus Disease 2019. J. Infect. Dis. 2020;222:17–25. doi: 10.1093/infdis/jiaa185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novella I.S., Presloid J.B., Smith S.D., Wilke C.O. Specific and nonspecific host adaptation during arboviral experimental evolution. J. Mol. Microbiol. Biotechnol. 2011;21:71–81. doi: 10.1159/000332752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish C.R., Holmes E.C., Morens D.M., Park E.C., Burke D.S., Calisher C.H., Laughlin C.A., Saif L.J., Daszak P. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol. Mol. Biol. Rev. 2008;72:457–470. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinzing F. Clarendon Press; 1916. Epidemics Resulting From Wars. [Google Scholar]
- Rieder E., Gorbalenya A.E., Xiao C., He Y., Baker T.S., Kuhn R.J., Rossmann M.G., Wimmer E. Will the polio niche remain vacant? Dev. Biol. (Basel) 2001;105:111–122. discussion 149–150. [PubMed] [Google Scholar]
- Shan C., Xia H., Haller S.L., Azar S.R., Liu Y., Liu J., Muruato A.E., Chen R., Rossi S.L., Wakamiya M., et al. A Zika virus envelope mutation preceding the 2015 epidemic enhances virulence and fitness for transmission. Proc. Natl. Acad. Sci. USA. 2020 doi: 10.1073/pnas.2005722117. Published online August 3, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan N.J. Antibody-mediated enhancement of viral disease. Curr. Top. Microbiol. Immunol. 2001;260:145–169. doi: 10.1007/978-3-662-05783-4_8. [DOI] [PubMed] [Google Scholar]
- Tang F., Liu W., Zhang F., Xin Z.T., Wei M.T., Zhang P.H., Yang H., Ly H., Cao W.C. IL-12 RB1 genetic variants contribute to human susceptibility to severe acute respiratory syndrome infection among Chinese. PLoS ONE. 2008;3:e2183. doi: 10.1371/journal.pone.0002183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger J.K., Kash J.C., Morens D.M. The 1918 influenza pandemic: 100 years of questions answered and unanswered. Sci. Transl. Med. 2019;11:eaau5485. doi: 10.1126/scitranslmed.aau5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomori O. From smallpox eradication to the future of global health: innovations, application and lessons for future eradication and control initiatives. Vaccine. 2011;29(Suppl 4):D145–D148. doi: 10.1016/j.vaccine.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Tsetsarkin K.A., Weaver S.C. Sequential adaptive mutations enhance efficient vector switching by Chikungunya virus and its epidemic emergence. PLoS Pathog. 2011;7:e1002412. doi: 10.1371/journal.ppat.1002412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner W.C., Kausrud K.L., Krishnappa Y.S., Cromsigt J.P., Ganz H.H., Mapaure I., Cloete C.C., Havarua Z., Küsters M., Getz W.M., Stenseth N.C. Fatal attraction: vegetation responses to nutrient inputs attract herbivores to infectious anthrax carcass sites. Proc. Biol. Sci. 2014;281 doi: 10.1098/rspb.2014.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderheiden A., Ralfs P., Chirkova T., Upadhyay A.A., Zimmerman M.G., Bedoya S., Aoued H., Tharp G.M., Pellegrini K.L., Manfredi C., et al. Type I and Type III IFN Restrict SARS-CoV-2 Infection of Human Airway Epithelial Cultures. J Virol. 2020 doi: 10.1128/JVI.00985-20. Published online July 22, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., de Groot R.J., Harbour D.A., Dalderup M., Gruffydd-Jones T., Horzinek M.C., Spaan W.J. Early death after feline infectious peritonitis virus challenge due to recombinant vaccinia virus immunization. J. Virol. 1990;64:1407–1409. doi: 10.1128/jvi.64.3.1407-1409.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Sun S., Tai W., Chen J., Geng Q., He L., Chen Y., Wu J., Shi Z., et al. Molecular Mechanism for Antibody-Dependent Enhancement of Coronavirus Entry. J. Virol. 2020;94:e02015-19. doi: 10.1128/JVI.02015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.F., Tseng S.P., Yen C.H., Yang J.Y., Tsao C.H., Shen C.W., Chen K.H., Liu F.T., Liu W.T., Chen Y.M., Huang J.C. Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem. Biophys. Res. Commun. 2014;451:208–214. doi: 10.1016/j.bbrc.2014.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang L., Kuwahara K., Li L., Liu Z., Li T., Zhu H., Liu J., Xu Y., Xie J., et al. Immunodominant SARS Coronavirus Epitopes in Humans Elicited both Enhancing and Neutralizing Effects on Infection in Non-human Primates. ACS Infect. Dis. 2016;2:361–376. doi: 10.1021/acsinfecdis.6b00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Li S.Y., Yang X.L., Huang H.M., Zhang Y.J., Guo H., Luo C.M., Miller M., Zhu G., Chmura A.A., et al. Serological Evidence of Bat SARS-Related Coronavirus Infection in Humans, China. Virol. Sin. 2018;33:104–107. doi: 10.1007/s12250-018-0012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R.C., Scott F.W. Antibody-mediated enhancement of disease in feline infectious peritonitis: comparisons with dengue hemorrhagic fever. Comp. Immunol. Microbiol. Infect. Dis. 1981;4:175–189. doi: 10.1016/0147-9571(81)90003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse M.E., Gowtage-Sequeria S. Host range and emerging and reemerging pathogens. Emerg. Infect. Dis. 2005;11:1842–1847. doi: 10.3201/eid1112.050997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse M.E., Haydon D.T., Antia R. Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol. Evol. 2005;20:238–244. doi: 10.1016/j.tree.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization WHO Coronavirus Diease (COVID-19) Dashboard. https://covid19.who.int/.
- Yip M.S., Leung N.H., Cheung C.Y., Li P.H., Lee H.H., Daëron M., Peiris J.S., Bruzzone R., Jaume M. Antibody-dependent infection of human macrophages by severe acute respiratory syndrome coronavirus. Virol. J. 2014;11:82. doi: 10.1186/1743-422X-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.Z., Holmes E.C. A Genomic Perspective on the Origin and Emergence of SARS-CoV-2. Cell. 2020;181:223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Qin L., Zhao Y., Zhang P., Xu B., Li K., Liang L., Zhang C., Dai Y., Feng Y., et al. Interferon-Induced Transmembrane Protein 3 Genetic Variant rs12252-C Associated With Disease Severity in Coronavirus Disease 2019. J. Infect. Dis. 2020;222:34–37. doi: 10.1093/infdis/jiaa224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Sehgal M., Hou Z., Cheng J., Shu S., Wu S., Guo F., Le Marchand S.J., Lin H., Chang J., Guo J.T. Identification of Residues Controlling Restriction versus Enhancing Activities of IFITM Proteins on Entry of Human Coronaviruses. J. Virol. 2018;92:e01535-17. doi: 10.1128/JVI.01535-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X.-y., Bian P.-y., Ye C.-t., Ye W., Ma H.-w., Tang K., Zhang C.-m., Lei Y.-f., Wei X., Wang P.-z., et al. Interferon-Induced Transmembrane Protein 3 Inhibits Hantaan Virus Infection, and Its Single Nucleotide Polymorphism rs12252 Influences the Severity of Hemorrhagic Fever with Renal Syndrome. Front. Immunol. 2017;7:535. doi: 10.3389/fimmu.2016.00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Chen X., Hu T., Li J., Song H., Liu Y., Wang P., Liu D., Yang J., Holmes E.C., et al. A Novel Bat Coronavirus Closely Related to SARS-CoV-2 Contains Natural Insertions at the S1/S2 Cleavage Site of the Spike Protein. Curr Biol. 2020;30:2196–2203.3. doi: 10.1016/j.cub.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]