Summary
BK polyomavirus (BKPyV or BKV) is a non-enveloped, circular double-stranded DNA virus that may exceed 80% seroprevalence in adults. BKV infection typically occurs during childhood, and the majority of adults are latently infected. While BKV infection is rarely associated with clinical disease in most individuals, in immunosuppressed individuals, reactivation may cause kidney (BK-associated nephropathy) or bladder (hemorrhagic cystitis and ureteral stenosis) injury. No antiviral therapies have been approved for the treatment of BKV infection. Reducing immunosuppression is the most effective therapy, although this is not feasible in many patients. Thus, a robust understanding of viral pathogenesis and viral diversity remains important for the development of future therapeutic strategies. Studies of BKV diversity are quite sparse compared to other common viral infections; thus, much of our understanding of BVK variability and evolution relies heavily analogous studies of other viruses such as HIV or viral hepatitis. We provide a comprehensive review of BKV diversity at the population and individual level with careful consideration of how viral variability may impact viral replication, pathogenesis, tropism, and protein function. We also discuss a number of outstanding questions related to BK virus diversity that should be explored rigorously in future studies.
Keywords: BK polyomavirus, BK virus, diversity, variation, evolution
1 |. INTRODUCTION
BK polyomavirus (BKPyV or BKV) was first isolated from the urine of a kidney transplant recipient in 1971. BKV belongs to the Polyomaviridae family of non-enveloped, circular double-stranded DNA viruses that is ~5000 nucleotides in length.1,2 Human polyomaviruses are thought to be transmitted through direct person-to-person contact and via contaminated surfaces, food, and water, although confirmation of these transmission routes is difficult due to the asymptomatic nature of primary infections and/or clinically non-specific (eg, flu-like) presentations.3 BKV infection typically occurs during childhood, and the majority of adults are latently infected, with the virus remaining dormant in the epithelial cells of the kidney and bladder in immunocompetent persons. Depending upon the study population, the seroprevalence of BKV may exceed 80% in adults.4 While BKV infection in most individuals is rarely associated with clinical disease, in immunosuppressed individuals, reactivation may cause kidney (BK-associated nephropathy) or bladder (hemorrhagic cystitis and ureteral stenosis) injury. More rarely, BKV is associated with progressive multifocal leukoencephalopathy meningitis and encephalitis, retinitis, pneumonitis, prostate cancer, and HIV-associated salivary gland disease renal carcinoma.5
BKV encodes multiple regulatory regions and structural proteins (VP1, VP2, and VP3). VP1 is the major capsid protein of virions and accounts for ~80% of their protein content. The non-coding control region (NCCR) of ~400 nucleotides contains the origin of replication and regulatory sequences for early and late transcription.5 A number of transcription factor binding sites exist within the NCCR and serve as determinants of host cell tropism. An excellent review of BKV biology has been published previously by Helle et al.6 No antiviral therapies have been approved for the treatment of BKV infection. Therefore, lowering of immunosuppression is the most effective therapy, although this is not feasible in many patients. Thus, a robust understanding of viral pathogenesis and viral diversity remains important for the development of future therapeutic strategies.
2 |. VIRUS DIVERSITY IS CLINICALLY RELEVANT
Studies of BKV diversity are quite sparse compared to other common viral infections such as HIV, hepatitis B virus (HBV), or hepatitis C virus (HCV) (see Figure S1 for the annual number of publications from 1987 to 2019). However, understanding how viral diversity impacts these exemplar infections is informative for designing future studies of BKV diversity. For example, viral variation is a defining feature of HIV disease. Within an individual, HIV exists as a population of related yet distinct viral variants termed the viral quasispecies that permits rapid, adaptive responses to immunologic selection pressures, antiviral therapies, and/or the cellular microenvironment, thereby contributing to viral fitness.7 Minor viral mutations can significantly alter key biological properties, including cellular tropism, co-receptor utilization, drug resistance, neutralization properties, and transactivation. The HIV long-terminal repeats (LTRs) contain cis-acting sequences that are recognized by viral and cellular proteins and regulate viral gene expression. The LTR exhibits diversity at the population and individual levels, and minor changes within an LTR can have significant effects on replication kinetics, cell tropism, transmission, and disease progression.8–24 Similarly, it is well established that variability within the HIV env and pol genes contribute to cell tropism, pathogenesis, replicative fitness, and/or drug resistance.
Viral diversity occurs during HCV infection, and the HCV quasispecies is a key determinant of replication capacity or fitness, cell tropism, immunologic escape, and antiviral drug resistance.25–39 Importantly, resistance associated mutations are relatively common during HCV infection and compromise treatment outcomes, while numerous studies have identified key positions within the HCV polymerase that reduce or enhance viral replication in vitro.40–53 Such polymorphisms can lead to rapid resistance and impact HCV replication and viral fitness in the absence of antiviral agents.
Analysis of complete HBV genomes has demonstrated the existence of multiple genotypes that influence chronicity, disease severity, and antiviral response rates.54,55 Moreover, the HBV mutation rate is relatively high due to the low fidelity of the viral reverse transcriptase,56 and HBV also exists as a viral quasispecies within an infected individual. This heterogeneity facilitates rapid, adaptive changes in response to immune selection pressure and antiviral therapy and has consequences for viral persistence and HBV-associated disease.57 Mutations within all HBV open reading frames may impact HBsAg expression, viral detection, vaccine escape, and/or HBV replication.
Given the central role of diversity to many common viral pathogens, robust evaluation of BKV diversity is essential for understanding the varied course of clinical disease.58,59 A number of investigators have suggested that cell determinants of BKV infection, such as cellular receptors and cell type-specific transcription can regulate viral infection and expression and contribute to differences in cell tropism between different BKV isolates.58 As reviewed by Moens and Van Ghelue,58 multiple amino acid substitutions have been described in the VP1, large T antigen, and agnoprotein; however, biological characterization of these mutations has rarely been performed. Similarly, the landscape of the NCCR, as well as polymorphisms within viral coding regions, may impact cell permissiveness of BKV isolates. BK virus diversity will almost certainly facilitate drug resistance if BK virus-specific therapies are developed in the future.
3 |. BKV SUBTYPES/SUBGROUPS
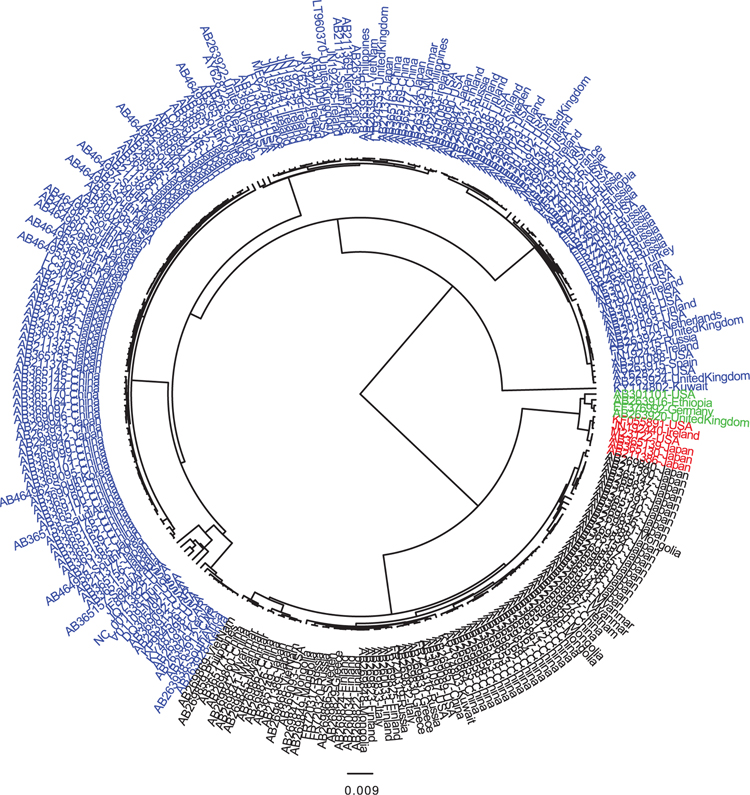
Prior to 1993, BKV strains worldwide were classified using a serological method.60 In 1993, Jin et al PCR amplified and sequenced a 287 base pair (bp) region of the VP1 gene - designated the 287-bp typing region - that provided evidence of distinct BKV genotypes.61 Based on a relatively small number of full-length BKV genomes currently available, it is now appreciated that BKV consists of antigenically distinct serotypes and multiple subtypes (I-IV)62,63 as shown in Figure 1. Notably, BK isolates from distinct individuals are rarely identical, suggesting adaptation of BK viral sequences to the unique immunologic environment of their host. Subtype I is the most common worldwide, followed by subtype IV. Subtype I can be divided further into subgroups Ia, Ib, and Ic based on nucleotide similarity. Subtype II is rare and has limited sequence data available for analysis. Additionally, subtypes II, III, and IV, along with subgroups Ib1 and Ib1, function as five fully distinct serotypes.64 Unfortunately, BKV genotyping studies have not been conducted in most countries, and it is unclear if the published reports are representative of the BK subtypes circulating in that region. Thus, the true geographic distribution of these subtypes is not fully appreciated and requires further investigation. For example, in the United States, BKV seropositivity rates are >85% in adults,65 and all subtypes have been detected although not in equal proportions, as shown in Table 1.63,66–70 Further study in larger cohorts and distinct at-risk populations should provide a more accurate assessment of the circulating BKV subtypes and their relative proportion.
FIGURE 1.
Full-length BK genomes were downloaded from GenBank. For individuals with multiple sequences available, a single representative sequence was chosen for inclusion. 258 sequences were evaluated using a Bayesian inference approach as implemented in the BEAST v1.10.1 software and are labeled using their GenBank accession number and country of origin. Blue sequences belong to BK subtype I, green sequences to subtypes II/III, and red sequences to subtype IV
TABLE 1.
The distribution of BKV subtypes in the United States
It is important to note that BKV diversity is influenced by a variety of selection pressures including the adaptive immune response, co-infections, and immunosuppressive and/or antiviral agents. Thus, BKV sequences within unrelated individuals (ie, no evidence of epidemiologic linkage) are rarely identical. For example, Krumbholz et al evaluated within subtype/subgroup and intersubtype/subgroup variability of 92 unique full-length BKV sequences.71 Between subtype I subgroups and between subtypes (I–IV), the mean genetic distances ranged from 1.06% to 5.5%, while intrasubtype/intrasubgroup genetic distances were lower at 0.18% to 0.47%. These data demonstrate that careful consider of the viral quasispecies may impact a variety of clinically relevant outcomes such as diagnosis of infection, viral replication, immune response, disease outcome, and treatment response.
It has been suggested that a particular BKV subtype may have greater potential to cause clinical disease compared to other subtypes.72 Bauer et al found that a single amino acid substitution within VP1 can affect binding of a mouse polyomavirus to its receptor and shorten the time to lethality by more than 3-fold,73 although detailed studies of BK virus interactions with its receptor(s) remain uncommon. A recent study suggests that BKV genotypes may differ in cellular entry tropism and pathogenic potential.64 Using reporter pseudoviruses, Pastrana et al demonstrated that BKV I, II, III, and IV are distinct serotypes and that BKV subgenotypes Ib1 and Ib2 also represent distinct serotypes.64 These differences map to a hypervariable region including residues 61 to 82 of the VP1. Another study found that distinct BKV isolates representing subtypes I, II, III, and IV replicated at different rates in the same cell type.74 Sequence variability within the transcription factor binding sites in the canonical NCCR exist and may contribute to virus reactivation, replication, and pathogenesis.59,75–81 Polyomavirus NCCRs differ in length, sequence, and organization, and basal NCCR activity depends on the activation state of the host cell type.59 A number of BKV NCCR variants have been reported, although few have been evaluated for their ability to alter replication. Olsen et al replaced the NCCR of the Dunlop isolate of BKV with several NCCRs from urine or kidney allograph biopsies and noted considerable variation in replication efficiency in vitro, suggesting that NCCR variability does impact replication rate.77
Studies have also demonstrated functional differences amongst BK genotypes/subgroups. For example, subtype I isolates grow more efficiently in human renal epithelial cells compared to subtype IV isolates.82 In renal transplant recipients, urine viral loads were higher for individuals infected with BK genotype Ia compared to Ib1.83 Genotype Ia was also associated with positive immune selection pressure at VP1 loops, which was absent in genotype Ib1 sequences. However, this has not been investigated further with other BK genotypes/subgroups or in other geographic regions. Virulence determinants also reside within VP1; therefore, mutations within this region may alter receptor specificity or other phenotypic properties of BKV.73,84 For example, of 500+ complete VP1 genes currently available in GenBank, a number of unique amino acid sequences are presented as shown in Figure 2. Whether these distinct viral sequences represent VP1s with divergent functions/genotypes remains to be evaluated in vitro. An interlaboratory collaboration in France reported that polymorphisms within regions targeted by PCR primers during BK viral load assays were a major source of variability.85 Thus, accurate measures of BK viremia may be challenging until BKV diversity is carefully considered during primer design and evaluation.
FIGURE 2.
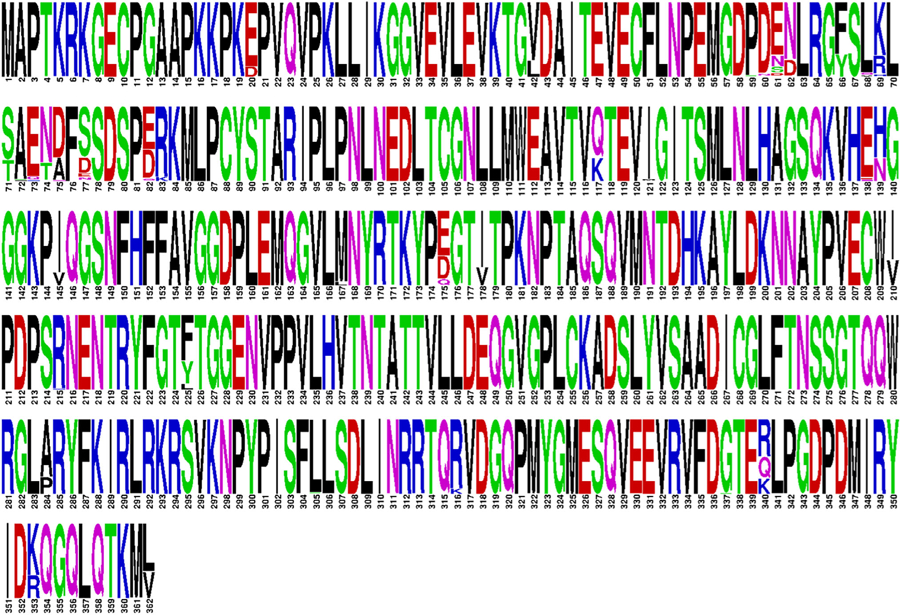
532 complete VP1 genes were downloaded from GenBank and aligned. Duplicate sequences were removed, and the resulting dataset is shown as a WebLogo with the height of each amino acid representing its relative proportion at that position of VP1
4 |. INTRAPATIENT BKV DIVERSITY
It was recently reported that BKV has the highest mutation rate for double-stranded DNA viruses - one that is similar to single-stranded RNA viruses.86 Yet, only a very limited number of studies have examined BKV diversity within an individual. Chen et al67 studied one patient with HIV, one with Capillary Leak Syndrome, and one healthy control from the United States/West Africa. Takasaka et al87 studies six renal transplant recipients from Japan. Kapusinszky et al70 studied one pediatric renal transplant patient with nephropathy from the United States. Data from these three studies were reconstructed using a Bayesian inference approach and colored by their patient of origin as shown in Figure 3. Chen et al found that full-length viral sequences from BKV cases formed monophyletic clusters of related yet distinct viral variants - termed the viral quasispecies and that the NCCR has unique nucleotide differences within each variant.67 Similarly, Takasaka et al evaluated full-length genomes from renal transplant patients with the four of six having multiple viral variants detected.87 Analysis of VP1 sequences found BK viral variants in 25 of 25 individuals evaluated.68 Compared to healthy controls, BK viremia/nephropathy was characterized by altered sequence complexity and immune selection pressure. Quasispecies variability also exists within the NCCR.76 The quasispecies evolves over time and may impact replication.87–89 A recent study reported that intrapatient variation also modified the specific receptor glycans that were engaged by BKV during host cell entry.89 Moreover, next generation sequencing of full-length BKV genomes identified an average of 110 polymorphisms per sample.86 Amongst all amino acid positions, 37.9% were polymorphic in the agnoprotein, 12.4% in VP1, 10.0% in VP2, 11.2% in VP3, 8.2% in the LTA, and 8.7% in STA.
FIGURE 3.
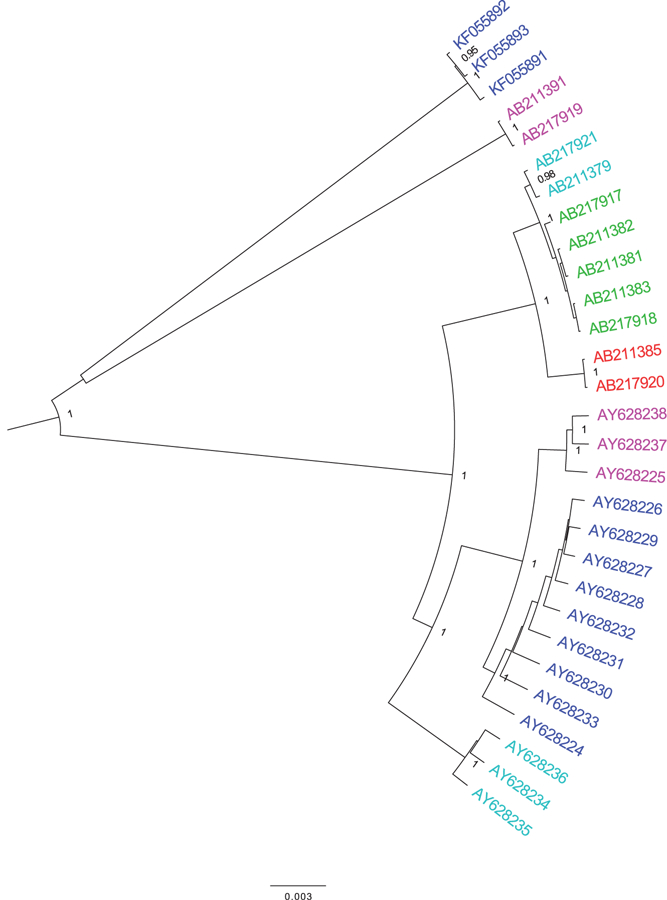
Full-length genomes from three studies of BK quasispecies diversity were downloaded from GenBank. Chen et al.67 studied one patient with HIV, one with Capillary Leak Syndrome, and one healthy control from the United States/West Africa. Takasaka et al.87 studied six renal transplant recipients from Japan. Kapusinszky et al.70 studied one pediatric renal transplant patient with nephropathy from the United States. Sequences were evaluated using a Bayesian inference approach labeled using their GenBank accession number and country of origin. Sequences are colored by their patient of origin
Infection with multiple BK viruses is also common, occurring in 12 of 25 individuals in one study.68 Similarly, Jin et al found dual infections with more than one subtype in HIV-infected patients, children, and pregnant women but not in bone marrow recipients.90 Recombination represents another important driver of viral evolution and has been observed within the BKV VP1.68 However, large, population-based studies of BKV dual infection and recombination have not been performed to date. Moreover, complementation can rescue non-functional BK variants91 highlighting the potential importance of recombination. BKV may exhibit distinct subtypes in the urine vs plasma suggesting tissue-specific selection pressures and viral compartmentalization.92
5 |. FUTURE DIRECTIONS
Multiple nucleotides and amino acid residues are known to impact BK virus replication, pathogenesis, tropism, and protein function. These include amino acids that regulate viral entry, replication, and/or assembly, multiple structural protein modifications, nuclear localization signals, amino acids responsible for transformation, transcription factor binding sites, T cell epitopes, and viral motifs that permit interactions with cellular proteins.5,6,66,93–103 BKV also contains a microRNA (BKPyV-3p-miRNA) that targets large T-antigen transcripts, thereby downregulating viral replication.79,104–110 Collectively, these data imply that a number of outstanding questions related to BK virus diversity remain to be explored in depth, including:
Is BKV subtype/subgroup associated with specific disease presentations such as hemorrhagic cystitis after hematopoietic cell transplantation and/or BKV-associated nephropathy after hematopoietic or solid organ transplantation? This question is best answered in large cohort studies in which multiple BKV subtypes are circulating and disease outcomes are carefully monitored. Over-representation of a particular BKV subtype/subgroup in certain at-risk and/or disease-specific populations would then provide preliminary evidence of distinct pathogenesis associated with different BKV subtypes/subgroups.
Are there specific viral mutations associated with altered BKV DNA levels? Based on our current understanding of viral pathogens, it is quite reasonable to suspect that BKV polymorphisms impact the overall function of viral proteins and/or their interactions with the host’s immune system. This hypothesis can be evaluated by rigorous phylogenetic and signature pattern analysis of viral sequences from individuals with high BKV viremia or viruria vs individuals with low BKV viremia or viruria, although potential confounders such as age, race, gender, BKV subtype/subgroup, use of immunosuppressive agents, and/or the presence of other co-infections must be considered.
Are there specific viral mutations associated with the presence of BKV in the plasma? While detection of BKD in the plasma is less common than detection in the urine, it is important to evaluate whether the viruses circulating in both compartments are identical or represent viruses that are uniquely adapted for replication within a particular cell type or tissue.
Similarly, are there specific viral mutations that are associated with the presence/absence of BKV aggregates in the urine - referred to as Haufen particles - which are sensitive and specific for biopsyproven BK nephropathy111,112?
Is BKV diversity correlated with absolute lymphocyte count? There are data suggesting that BKV DNA levels are elevated when CD4 T cell counts are less than 200 cells/mm3.113–115 However, the impact of immunosuppression - and distinct types of immunosuppression - on BKV diversity has rarely been evaluated.84
Does diversity within BKV proteins predict ELISPOT response to BKV infection and/or reactivation? BKV-specific immune responses may control BKV replication and reactivation; thus, viral diversity within key immunologic epitopes may also impact viral replication and reactivation.66,102,116,117 This is of significant clinical interest and requires robust evaluation of BKV-specific immune responses and the presence/absence of BKV epitope diversity in well-characterized clinical cohorts.
Does BKV diversity change in response to immunosuppression and is this dependent upon the specific immunosuppressive strategy utilized?
How does BKV diversity impact the development of a successful immunization strategy?
Does BKV diversity impact the detection of seropositive transplant donors and recipients? With other DNA viruses such as CMV and EBV, risk of post-transplant infection is related to donor and recipient serostatus mismatching. Similar data on the risk of BK virus infection related to donor or recipient serostatus are less available. As BK serological assays become more widely used in clinical practice, it will be important to understand if BK diversity will influence these measurements and the associated clinical relevance.
6 |. CONCLUSIONS
Understanding viral diversity has led to improved clinical outcomes for patients with HIV, HBV, and HCV. Our knowledge of the associations between BK virus diversity and clinical disease is much less compared to these other well-studied viral infections. The significance of learning about viral diversity will be greater as new therapeutic strategies for BK emerge. For example, novel treatments, such as the infusion of third party or donor-derived virus-specific T cells (VSTs), have shown promise in the treatment of refractory viral infections in immunosuppressed patients. However, manufacturing VSTs is a time and resource intensive process, and not all centers can provide such therapies to all of their patients in need. Therefore, understanding viral diversity may provide for the identification of the patients at highest risk of disease, allowing for the rational targeting of therapy to the patients who are likely to derive the most benefit.
Supplementary Material
ACKNOWLEDGEMENTS
Dr. Laskin is supported by the National Institute of Diabetes and Digestive and Kidney Diseases through grant K23 DK101600. Dr. Davies is supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development under award number R01 HD093773.
Funding information
Eunice Kennedy Shriver National Institute of Child Health and Human Development, Grant/Award Numbers: HD093773, R01; National Institute of Diabetes and Digestive and Kidney Diseases, Grant/Award Number: K23 DK101600
Footnotes
CONFLICT OF INTEREST
The authors have no conflicts of interest to report.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of this article.
REFERENCES
- 1.Gardner SD, Field AM, Coleman DV, Hulme B. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet. 1971;1(7712):1253–1257. [DOI] [PubMed] [Google Scholar]
- 2.Moens U, Calvignac-Spencer S, Lauber C, et al. ICTV virus taxonomy profile: Polyomaviridae. J Gen Virol. 2017;98:1159–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirsch HH, Babel N, Comoli P, et al. European perspective on human polyomavirus infection, replication and disease in solid organ transplantation. Clin Microbiol Infect. 2014;20(Supplement 7):74–88. [DOI] [PubMed] [Google Scholar]
- 4.Kean JM, Rao S, Wang M, Garcea R. Seroepidemiology of human polyomaviruses. PLoS Pathog. 2009;5(3):e1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moens U, Krumbholz A, Ehlers B, et al. Biology, evolution, and medical importance of polyomaviruses: an update. Infect Genet Evol. 2017;54:18–38. [DOI] [PubMed] [Google Scholar]
- 6.Helle F, Brochot E, Handala L, et al. Biology of the BKPyV: an update. Viruses. 2017;9(11):E327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackard JT, Cohen DE, Mayer K. Human immunodeficiency virus superinfection and recombination: current state of knowledge and potential clinical consequences. Clin Infect Dis. 2002;34(8):1108–1114. [DOI] [PubMed] [Google Scholar]
- 8.Kondo M, Shima T, Nishizawa M, et al. Identification of attenuated variants of HIV-1 circulating recombinant form 01_AE that are associated with slow disease progression due to gross genetic alterations in the nef/long terminal repeat sequences. J Infect Dis. 2005;192(1): 56–61. [DOI] [PubMed] [Google Scholar]
- 9.McNearney T, Hornickova Z, Templeton A, et al. Nef and LTR sequence variation from sequentially derived human immunodeficiency virus type 1 isolates. Virology. 1995;208:388–398. [DOI] [PubMed] [Google Scholar]
- 10.Ramírez de Arellano E, Martín C, Soriano V, Alcamí J, Holguín A. Genetic analysis of the long terminal repeat (LTR) promoter region in HIV-1-infected individuals with different rates of disease progression. Virus Genes. 2007;34(2):111–116. [DOI] [PubMed] [Google Scholar]
- 11.Nonnemacher MR, Irish BP, Liu Y, Mauger D, Wigdahl B. Specific sequence configurations of HIV-1 LTR G/C box array result in altered recruitment of Sp isoforms and correlate with disease progression. J Neuroimmunol. 2004;157(1–2):39–47. [DOI] [PubMed] [Google Scholar]
- 12.Hogan TH, Stauff DL, Krebs FC, Gartner S, Quiterio SJ, Wigdahl B. Structural and functional evolution of human immunodeficiency virus type 1 long terminal repeat CCAAT/enhancer binding protein sites and their use as molecular markers for central nervous system disease progression. J Neurovirol. 2003;9(1):55–68. [DOI] [PubMed] [Google Scholar]
- 13.Fang G, Burger H, Chappey C, et al. Analysis of transition from long-term nonprogressive to progressive infection identifies sequences that may attenuate HIV type 1. AIDS Res Hum Retroviruses. 2001;17 (15):1395–1404. [DOI] [PubMed] [Google Scholar]
- 14.Hiebenthal-Millow K, Greenough TC, Bretttler DB, et al. Alterations in HIV-1 LTR promoter activity during AIDS progression. Virology. 2003;317(1):109–118. [DOI] [PubMed] [Google Scholar]
- 15.Ait-Khaled M, McLaughlin JE, Johnson MA, Emery V. Distinct HIV-1 long terminal repeat quasispecies present in nervous tissues compared to that in lung, blood and lymphoid tissues of an AIDS patient. AIDS. 1995;9(7):675–683. [DOI] [PubMed] [Google Scholar]
- 16.Ross HL, Nonnemacher MR, Hogan TH, et al. Interaction between CCAAT/enhancer binding protein and cyclic AMP response element binding protein 1 regulates human immunodeficiency virus type 1 transcription in cells of the monocyte/macrophage lineage. J Virol. 2001;75(4):1842–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naghavi M, Salminen M, Sonnerborg A, Vahlne A. DNA sequence of the long terminal repeat of human immunodeficiency virus type 1 subtype a through G. AIDS Res Hum Retroviruses. 1999;15(5):485–488. [DOI] [PubMed] [Google Scholar]
- 18.Naghavi M, Schwartz S, Sonnerborg A, Vahlne A. Long terminal repeat promoter/enhancer activity of different subtypes of HIV type 1, AIDS Res Hum Retroviruses. 1999;15(14):1293–1303. [DOI] [PubMed] [Google Scholar]
- 19.Oskarsson T, Hreggvidsdóttir HS, Agnarsdóttir G, et al. Duplicated sequence motif in the long terminal repeat of maedivisna virus extends cell tropism and is associated with neurovirulence. J Virol. 2007;81(8):4052–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quinones-Mateu ME, Mas A, de Lera TL, et al. LTR and tat variability of HIV-1 isolates from patients with divergent rates of disease progression. Virus Res. 1998;57:11–20. [DOI] [PubMed] [Google Scholar]
- 21.Visco-Comandini U, Yun Z, Vahlne A, Sonnerborg A. No association of HIV type 1 long terminal repeat sequence pattern with long-term nonprogression and in vivo viral replication levels in European subjects. AIDS Res Hum Retroviruses. 1999;15(7):609–617. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L, Huang Y, Yuan H, Chen B, Ip J, Ho D. Genotypic and phenotypic characterization of long terminal repeat sequences from long-term survivors of human immunodeficiency virus type 1 infection. J Virol. 1997;71(7):5608–5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Estable M, Bell B, Merzouki A, Montaner J, O’Shaughnessy M, Sadowski I. Human immunodeficiency virus type 1 long terminal repeat variants from 42 patients representing all stages of infection display a wide range of sequence polymorphism and transcription activity. J Virol. 1996;70(6):4053–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirchhoff F, Greenough TC, Hamacher M, Sullivan JL, Desrosiers R. Activity of human immunodeficiency virus type 1 promoter/TAR regions and tat1 genes derived from individuals with different rates of disease progression. Virology. 1997;232:319–331. [DOI] [PubMed] [Google Scholar]
- 25.Simmonds P. Genetic diversity and evolution of hepatitis C virus–15 years on. J Gen Virol. 2004;85(11):3173–3188. [DOI] [PubMed] [Google Scholar]
- 26.Brambilla S, Bellati G, Asti M, et al. Dynamics of hypervariable region 1 variation in hepatitis C virus infection and correlation with clinical and virological features of liver disease. Hepatology. 1998;27(6): 1678–1686. [DOI] [PubMed] [Google Scholar]
- 27.Hayashi J, Furusyo N, Ariyama I, Sawayama Y, Etoh Y, Kashiwagi S. A relationship between the evolution of hepatitis C virus variants, liver damage, and hepatocellular carcinoma in patients with hepatitis C viremia. J Infect Dis. 2000;181:1523–1527. [DOI] [PubMed] [Google Scholar]
- 28.Curran R, Jameson C, Craggs J, et al. Evolutionary trends of the first hypervariable region of the hepatitis C virus E2 protein in individuals with differing liver disease severity. J Gen Virol. 2002;83:11–23. [DOI] [PubMed] [Google Scholar]
- 29.Martell M, Esteban J, Quer J, et al. Dynamic behavior of hepatitis C virus quasispecies in patients undergoing orthotopic liver transplantation. J Virol. 1994;68(5):3425–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gretch D, Polyak S, Wilson J, Carithers R, Perkins J, Corey L. Tracking hepatitis C virus quasispecies major and minor variants in symptomatic and asymptomatic liver transplant recipients. J Virol. 1996; 70:7622–7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sullivan D, Wilson J, Carithers R, Perkins J, Gretch D. Multigene tracking of hepatitis C virus quasispecies after liver transplantation: correlation of genetic diversification in the envelope region with asymptomatic or mild disease patterns. J Virol. 1998;72(12):10036–10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ray S, Wang Y, Laeyendecker O, Ticehurst J, Villano S, Thomas D. Acute hepatitis C virus structural gene sequences as predictors of persistent viremia: hypervariable region 1 as a decoy. J Virol. 1999; 73(4):2938–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farci P, Shimoda A, Coiana A, et al. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000; 288:339–344. [DOI] [PubMed] [Google Scholar]
- 34.Manzin A, Solforosi L, Petrelli E, et al. Evolution of hypervariable region 1 of hepatitis C virus in primary infection. J Virol. 1998;72(7): 6271–6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Donlin MJ, Cannon NA, Aurora R, et al. Contribution of genome-wide HCV genetic differences to outcome of interferon-based therapy in Caucasian American and African American patients. PLoS One. 2010;5(2):e9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Donlin MJ, Cannon NA, Yao E, et al. Pretreatment sequence diversity differences in the full-length hepatitis C virus open reading frame correlate with early response to therapy. J Virol. 2007;81(15): 8211–8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pawlotsky J. Hepatitis C virus resistance to antiviral therapy. Hepatology. 2000;32(5):889–896. [DOI] [PubMed] [Google Scholar]
- 38.Layden-Almer JE, Kuiken C, Ribeiro RM, et al. Hepatitis C virus genotype 1a NS5A pretreatment sequence variation and viral kinetics in African American and white patients. J Infect Dis. 2005;192(6): 1078–1087. [DOI] [PubMed] [Google Scholar]
- 39.Cannon NA, Donlin MJ, Fan X, Aurora R. JE; T, group V-CS. Hepatitis C virus diversity and evolution in the full open-reading frame during antiviral therapy. PLoS One. 2008;3(5):e2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheney IW, Naim S, Lai VC, et al. Mutations in NS5B polymerase of hepatitis C virus: impacts on in vitro enzymatic activity and viral RNA replication in the subgenomic replicon cell culture. Virology. 2002;297(2):298–306. [DOI] [PubMed] [Google Scholar]
- 41.Clemente-Casares P, Lopez-Jimenez AJ, Bellon-Echeverria I, et al. De novo polymerase activity and oligomerization of hepatitis C virus RNA-dependent RNA-polymerases from genotypes 1 to 5. PLoS One. 2011;6(4):e18515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ishii K, Tanaka Y, Yap CC, Aizaki H, Matsuura Y, Miyamura T. Expression of hepatitis C virus NS5B protein: characterization of its RNA polymerase activity and RNA binding. Hepatology. 1999;29(4): 1227–1235. [DOI] [PubMed] [Google Scholar]
- 43.Labonte P, Axelrod V, Agarwal A, Aulabaugh A, Amin A, Mak P. Modulation of hepatitis C virus RNA-dependent RNA polymerase activity by structure-based site-directed mutagenesis. J Biol Chem. 2002;277(41):38838–38846. [DOI] [PubMed] [Google Scholar]
- 44.Lohmann V, Korner F, Herian U, Bartenschlager R. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J Virol. 1997;71(11):8416–8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mishima K, Sakamoto N, Sekine-Osajima Y, et al. Cell culture and in vivo analyses of cytopathic hepatitis C virus mutants. Virology. 2010;405(2):361–369. [DOI] [PubMed] [Google Scholar]
- 46.Moradpour D, Brass V, Bieck E, et al. Membrane association of the RNA-dependent RNA polymerase is essential for hepatitis C virus RNA replication. J Virol. 2004;78(23):13278–13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murayama A, Date T, Morikawa K, et al. The NS3 helicase and NS5B-to-3’X regions are important for efficient hepatitis C virus strain JFH-1 replication in Huh7 cells. J Virol. 2007;81(15):8030–8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murayama A, Weng L, Date T, et al. RNA polymerase activity and specific RNA structure are required for efficient HCV replication in cultured cells. PLoS Pathog. 2010;6(4):e1000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmitt M, Scrima N, Radujkovic D, et al. A comprehensive structure-function comparison of hepatitis C virus strain JFH1 and J6 polymerases reveals a key residue stimulating replication in cell culture across genotypes. J Virol. 2011;85(6):2565–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cento V, Chevaliez S, Perno C. Resistance to direct-acting antiviral agents: clinical utility and significance. Curr Opin HIV AIDS. 2015;10 (5):381–389. [DOI] [PubMed] [Google Scholar]
- 51.Lontok E, Harrington P, Howe A, et al. Hepatitis C virus drug resistance-associated substitutions: state of the art summary. Hepatology. 2015;62(5):1623–1632. [DOI] [PubMed] [Google Scholar]
- 52.Sorbo MC, Cento V, Di Maio VC, et al. Hepatitis C virus drug resistance associated substitutions and their clinical relevance: update 2018. Drug Resist Updat. 2018;37:17–39. [DOI] [PubMed] [Google Scholar]
- 53.Gaudieri S, Rauch A, Pfafferott K, et al. Hepatitis C virus drug resistance and immune-driven adaptations: relevance to new antiviral therapy. Hepatology. 2009;49(4):1069–1082. [DOI] [PubMed] [Google Scholar]
- 54.Schaefer S. Hepatitis B virus: significance of genotypes. J Viral Hepat. 2005;12(2):111–124. [DOI] [PubMed] [Google Scholar]
- 55.Tanwar S, Dusheiko G. Is there any value to hepatitis B virus genotype analysis? Curr Gastroenterol Rep. 2012;14(1):37–46. [DOI] [PubMed] [Google Scholar]
- 56.Okamoto H, Imai M, Kametani M, Nakamura T, Mayumi M. Genomic heterogeneity of hepatitis B virus in a 54-year-old woman who contracted the infection through materno-fetal transmission. Jpn J Exp Med. 1987;57(4):231–236. [PubMed] [Google Scholar]
- 57.Pawlotsky J. The concept of hepatitis B virus mutant escape. J Clin Virol. 2005;34(suppl 1):S125–S129. [DOI] [PubMed] [Google Scholar]
- 58.Moens U, Ghelue M. Polymorphism in the genome of non-passaged human polyomavirus BK: implications for cell tropism and the pathological role of the virus. Virology. 2005;331:209–231. [DOI] [PubMed] [Google Scholar]
- 59.Ajuh ET, Wu Z, Kraus E, et al. Novel human polyomavirus noncoding control regions differ in bidirectional gene expression according to host cell, large T-antigen expression, and clinically occurring rearrangements. J Virol. 2018;92(7):e02231–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Knowles WA, Gibson PE, Gardner S. Serological typing scheme for BK-like isolates of human polyomavirus. J Med Virol. 1989;28(2): 118–123. [DOI] [PubMed] [Google Scholar]
- 61.Jin L, Gibson PE, Booth JC, Clewley J. Genomic typing of BK virus in clinical specimens by direct sequencing of polymerase chain reaction products. J Med Virol. 1993;41(1):11–17. [DOI] [PubMed] [Google Scholar]
- 62.Luo C, Bueno M, Kant J, Martinson J, Randhawa P. Genotyping schemes for polyomavirus BK, using gene-specific phylogenetic trees and single nucleotide polymorphism analysis. J Virol. 2009;83 (5):2285–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sharma PM, Gupta G, Vats A, Shapiro R, Randhawa P. Phylogenetic analysis of polyomavirus BK sequences. J Virol. 2006;80:8869–8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pastrana DV, Ray U, Magaldi TG, Schowalter RM, Çuburu N, Buck C. BK polyomavirus genotypes represent distinct serotypes with distinct entry tropism. J Virol. 2013;87(18):10105–10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gossai A, Waterboer T, Nelson HH, et al. Seroepidemiology of human polyomaviruses in a US population. Am J Epidemiol. 2016; 183(1):61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sahoo MK, Tan SK, Chen SF, et al. Limited variation in BK virus T-cell epitopes revealed by next-generation sequencing. J Clin Microbiol. 2015;53(10):3226–3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen Y, Sharp PM, Fowkes M, Kocher O, Joseph JT, Koralnik I. Analysis of 15 novel full-length BK virus sequences from three individuals: evidence of a high intra-strain genetic diversity. J Gen Virol. 2004;85:2651–2663. [DOI] [PubMed] [Google Scholar]
- 68.Luo C, Hirsch HH, Kant J, Randhawa P. VP-1 quasispecies in human infection with polyomavirus BK. J Med Virol. 2012;84(1):152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhong S, Randhawa PS, Ikegaya H, et al. Distribution patterns of BK polyomavirus (BKV) subtypes and subgroups in American, European and Asian populations suggest co-migration of BKV and the human race. J Gen Virol. 2009;90:144–152. [DOI] [PubMed] [Google Scholar]
- 70.Kapusinszky B, Chen SF, Sahoo MK, et al. BK polyomavirus subtype III in a pediatric renal transplant patient with nephropathy. J Clin Microbiol. 2013;51(12):4255–4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krumbholz A, Bininda-Emonds OR, Wutzler P, Zell R. Evolution of four BK virus subtypes. Infect Genet Evol. 2008;8(5):632–643. [DOI] [PubMed] [Google Scholar]
- 72.Yogo Y, Sugimoto C, Zhong S, Homma Y. Evolution of the BK polyomavirus: epidemiological, anthropological and clinical implications. Rev Med Virol. 2009;19(4):185–199. [DOI] [PubMed] [Google Scholar]
- 73.Bauer PH, Bronson RT, Fung SC, et al. Genetic and structural analysis of a virulence determinant in polyomavirus VP1. J Virol. 1995;69 (12):7925–7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tremolada S, Delbue S, Larocca S, et al. Polymorphisms of the BK virus subtypes and their influence on viral in vitro growth efficiency. Virus Res. 2010;149:190–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Carr MJ, McCormack GP, Mutton KJ, Crowley B. Unique BK virus non-coding control region (NCCR) variants in hematopoietic stem cell transplant recipients with and without hemorrhagic cystitis. J Med Virol. 2006;78:4. [DOI] [PubMed] [Google Scholar]
- 76.Perets TT, Silberstein I, Rubinov J, Sarid R, Mendelson E, Shulman L. High frequency and diversity of rearrangements in polyomavirus bk noncoding regulatory regions cloned from urine and plasma of Israeli renal transplant patients and evidence for a new genetic subtype. J Clin Microbiol. 2009;47(5):1402–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Olsen GH, Hirsch HH, Rinaldo C. Functional analysis of polyomavirus BK non-coding control region quasispecies from kidney transplant recipients. J Med Virol. 2009;81(11):1959–1967. [DOI] [PubMed] [Google Scholar]
- 78.Gosert R, Rinaldo CH, Funk GA, et al. Polyomavirus BK with rearranged noncoding control region emerge in vivo in renal transplant patients and increase viral replication and cytopathology. J Exp Med. 2008;205(4):841–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Virtanen E, Seppälä H, Helanterä I, et al. BK polyomavirus microRNA expression and sequence variation in polyomavirus-associated nephropathy. J Clin Virol. 2018;102:70–76. [DOI] [PubMed] [Google Scholar]
- 80.Bethge T, Hachemi HA, Manzetti J, Gosert R, Schaffner W, Hirsch H. Sp1 sites in the noncoding control region of BK polyomavirus are key regulators of bidirectional viral early and late gene expression. J Virol. 2015;89(6):3396–3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bethge T, Ajuh E, Hirsch H. Imperfect symmetry of Sp1 and core promoter sequences regulates early and late virus gene expression of the bidirectional BK polyomavirus noncoding control region. J Virol. 2016;90(22):10083–10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nukuzuma S, Takasaka T, Zheng HY, et al. Subtype I BK polyomavirus strains grow more efficiently in human renal epithelial cells than subtype IV strains. J Gen Virol. 2006;87:1893–1901. [DOI] [PubMed] [Google Scholar]
- 83.Varella RB, Zalona AC, Diaz NC, Zalis MG, Santoro-Lopes G. BK polyomavirus gentoypes Ia and Ib1 exhibit different biological properties in renal transplant patients. Virus Res. 2018;243: 65–68. [DOI] [PubMed] [Google Scholar]
- 84.Karalic D, Lazarevic I, Banko A, Cupic M, Jevtovic D, Jovanovic T. Molecular characterization of BK virus in patients infected with human immunodeficiency virus. Med Microbiol Immunol. 2016;205: 185–193. [DOI] [PubMed] [Google Scholar]
- 85.Solis M, Meddeb M, Sueur C, et al. Sequence variation in amplification target genes and standards influences interlaboratory comparison of BK virus DNA load measurement. J Clin Microbiol. 2015;53:3842–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Domingo-Calap P, Schubert B, Joly M, et al. An unusually high substitution rate in transplant-associated BK polyomavirus in vivo is further concentrated in HLA-C-bound viral peptides. PLoS Pathog. 2018;14:e1007368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Takasaka T, Goya N, Ishida H, et al. Stability of the BK polyomavirus genome in renal-transplant patients without nephropathy. J Gen Virol. 2006;87:303–306. [DOI] [PubMed] [Google Scholar]
- 88.Sugimoto C, Hara K, Taguchi F, Yogo Y. Growth efficiency of naturally occurring BK virus variants in vivo and in vitro. J Virol. 1989;63 (7):3195–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Peretti A, Geoghegan EM, Pastrana DV, et al. Characterization of BK polyomaviruses from kidney transplant recipients suggests a role for APOBEC3 in driving in-host virus evolution. Cell Host Microbe. 2018; 23(5):628–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jin L, Pietropaolo V, Booth JC, Ward KH, Brown D. Prevalence and distribution of BK virus subtypes in healthy people and immunocompromised patients detected by PCR restriction enzyme analysis. Clin Diagn Virol. 1995;3(3):285–295. [DOI] [PubMed] [Google Scholar]
- 91.Myhre MR, Olsen GH, Gosert R, Hirsch HH, Rinaldo C. Clinical polyomavirus BK variants with agnogene deletion are non-functional but rescued by trans-complementation. Virology. 2010; 398(1):12–20. [DOI] [PubMed] [Google Scholar]
- 92.Ledesma J, Bouza E, González-Nicolás MA. García de Viedma D, Rodríguez-Sánchez B, Muñoz P. BK polyomavirus genotyping at inter- and intra-patient level in Spain. J Med Virol. 2013;85(8):1402–1408. [DOI] [PubMed] [Google Scholar]
- 93.Harris KF, Christensen JB, Imperiale M. BK virus large T antigen: interactions with the retinoblastoma family of tumor suppressor proteins and effects on cellular growth control. J Virol. 1996;70:2378–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Harris KF, Christensen JB, Radany EH, Imperiale MJ. Novel mechanisms of E2F induction by BK virus large-T antigen: requirement of both the pRb-binding and the J domains. Mol Cell Biol. 1998;18:1746–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dugan AS, Gasparovic ML, Tsomaia N, et al. Identification of amino acid residues in BK virus VP1 that are critical for viability and growth. J Virol. 2007;81(21):11798–11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Neu U, Allen SA, Blaum BS, et al. A structure-guided mutation in the major capsid protein retargets BK polyomavirus. PLoS Pathog. 2013; 9(10):e1003688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen PL, Hsu PH, Fang CY, et al. Phosphorylation of Ser-80 of VP1 and Ser-254 of VP2 is essential for human BK virus propagation in tissue culture. J Gen Virol. 2011;92:2637–2645. [DOI] [PubMed] [Google Scholar]
- 98.Johannessen M, Myhre MR, Dragset M, Tümmler C, Moens U. Phosphorylation of human polyomavirus BK agnoprotein at Ser-11 is mediated by PKC and has an important regulative function. Virology. 2008;379(1):97–109. [DOI] [PubMed] [Google Scholar]
- 99.DeCaprio JA, Garcea R. A cornucopia of human polyomaviruses. Nat Rev Microbiol. 2013;11:264–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nakshatri H, Pater MM, Pater A. Functional role of BK virus tumor antigens in transformation. J Virol. 1988;62:4613–4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fang CY, Chen HY, Wang M, et al. Global analysis of modifications of the human BK virus structural proteins by LC-MS/MS. Virology. 2010;402(1):164–176. [DOI] [PubMed] [Google Scholar]
- 102.Mani J, Schmitt M. Cellular immunotherapy for patients with reactivation of JC and BK polyomaviruses after transplantation. Cytotherapy. 2014;16(10):1325–1335. [DOI] [PubMed] [Google Scholar]
- 103.Li J, Melenhorst J, Hensel N, et al. T-cell responses to peptide fragments of the BK virus T antigen: implications for cross-reactivity of immune response to JC virus. J Gen Virol. 2006;87: 2951–2960. [DOI] [PubMed] [Google Scholar]
- 104.Seo GJ, Fink LH, O’Hara B, Atwood WJ, Sullivan C. Evolutionarily conserved function of a viral microRNA. J Virol. 2008;82:9823–9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Broekema NM, Imperiale M. miRNA regulation of BK polyomavirus replication during early infection. Proc Natl Acad Sci U S A. 2013; 110:8200–8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lagatie O, Tritsmans L, Stuyver L. The miRNA world of polyomaviruses. Virol J. 2013;10:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cullen B. Viruses and microRNAs. Nat Genet. 2006;38:S25–S30. [DOI] [PubMed] [Google Scholar]
- 108.Bauman Y, Mandelboim O. MicroRNA based immunoevasion mechanism of human polyomaviruses. RNA Biol. 2011;8: 591–594. [DOI] [PubMed] [Google Scholar]
- 109.Bauman Y, Nachmani D, Vitenshtein A, et al. An identical miRNA of the human JC and BK polyoma viruses targets the stress-induced ligand ULBP3 to escape immune elimination. Cell Host Microbe. 2011;9:93–102. [DOI] [PubMed] [Google Scholar]
- 110.Bauman Y, Drayman N, Ben-Nun-Shaul O, et al. Downregulation of the stress-induced ligand ULBP1 following SV40 infection confers viral evasion from NK cell cytotoxicity. Oncogene. 2016;7:15369–15381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Laskin BL, Singh HK, Beier UH, et al. The noninvasive urinary polyomavirus Haufen test predicts BK virus nephropathy in children after hematopoietic cell transplantation: a pilot study. Transplantation. 2016;1000(10):e81–e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Singh HK, Andreoni KA, Madden V, et al. Presence of urinary Haufen accurately predicts polyomavirus nephropathy. J Am Soc Nephrol. 2009;20(2):416–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hu C, Huang Y, Su J, Wang M, Zhou Q, Zhu B. The prevalence and isolated subtypes of BK polyomavirus reactivation among patients infected with human immunodeficiency virus-1 in southeastern China. Arch Virol. 2018;163:1463–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jagannath S, Sachithanandham J, Ramalingam VV, et al. BK virus characterisation among HIV-1-infected individuals and its association with immunosuppression. Indian J Med Microbiol. 2018;36(2): 172–177. [DOI] [PubMed] [Google Scholar]
- 115.Behzad-Behbahani A, Klapper PE, Vallely PJ, Cleator GM, Khoo S. Detection of BK virus and JC virus DNA in urine samples from immunocompromised (HIV-infected) and immunocompetent (HIV-non-infected) patients using polymerase chain reaction and microplate hybridisation. J Clin Virol. 2004;29:224–229. [DOI] [PubMed] [Google Scholar]
- 116.Binggeli S, Egli A, Schaub S, et al. Polymovirus BK-specific cellular immune response to VP1 and large T-antigen in kidney transplant recipients. Am J Transplant. 2007;7:1131–1139. [DOI] [PubMed] [Google Scholar]
- 117.Cioni M, Leboeuf C, Comoli P, Ginevri F, Hirsch H. Characterization of immunodominant BK polyomavirus 9mer epitope T cell responses. Am J Transplant. 2016;16:1193–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.