Abstract
Targeted molecular diagnostic tests and accurate immunoassays have transformed the landscape of clinical virology, calling into question the usefulness of traditional viral culture. Here we present a case where viral culture, followed by metagenomic sequencing, was central to the diagnosis of an unexpected viral infection, with significant clinical and public health implications.
Keywords: metagenomic sequencing, poxvirus, vaccinia, vaccine-transmitted, viral culture
Targeted molecular diagnostics and immunoassays have transformed the landscape of clinical virology, calling into question the usefulness of viral culture. This case demonstrates how viral culture, followed by metagenomic sequencing, was central to the diagnosis of an unexpected viral infection.
Recent advances in molecular diagnostic technologies have revolutionized the detection of viral infections and spawned debates over the continued utility of viral culture in clinical microbiology [1, 2]. Viral culture is relatively slow, labor intense, and requires highly specialized expertise. Additionally, viruses with significant biosafety concerns, such as hemorrhagic fever viruses, orthopoxviruses, and the novel coronavirus (SARS-CoV-2), may grow in routine viral culture [3, 4], which poses potential risk to laboratorians. As a result, many clinical laboratories have discontinued its use in favor of more rapid and sensitive nucleic acid amplification tests (NAATs). It should be noted, however, that the NAAT assays commonly used in clinical practice are designed to detect only a limited number of the most common viruses and may be unable to detect genetically diverse viral strains. Thus, culture retains important advantages over targeted molecular testing. Culture allows for the isolation of a wide variety of viruses, including potential new or emerging pathogens, not expected by clinicians as long as viral replication can be confirmed by detection of cytopathic effect (CPE) or viral antigens. Strain variability typically has less of an impact on virus replication in culture than it does for targeted gene- or protein-based assays. Additionally, culture provides information on virus viability and is useful for phenotypic antiviral susceptibility testing [2]. Maintaining culture capabilities, at least as a reference method, still has benefits for patient care. Here we present an example of how viral culture was essential in the diagnosis and subsequent management of an unexpected infectious disease.
CASE PRESENTATION
Associated Regional and University Pathologists, Inc. (ARUP Laboratories), received a skin swab specimen collected from a “buttock lesion” for viral culture, along with a request to “rule out hand, foot, and mouth disease.” The Supplementary Data contains additional details on the laboratory methods and reagents used in this case. For culture, an aliquot of the specimen transport media was inoculated onto primary rhesus monkey kidney (pRhMK) cells, as well as Buffalo green monkey kidney, human lung carcinoma (A549), and human embryonic lung fibroblast (MRC-5) cell lines. CPE resembling that produced by herpes simplex virus (HSV) was observed in the pRhMK and MRC-5 monolayers on day 4 of incubation. However, testing of MRC-5 cells for HSV-1 and HSV-2 antigens using commercially available direct fluorescent antibody (DFA) reagents was negative.
To exclude the possibility of nonspecific cytotoxicity, supernatant from all culture vials was filtered and the eluate passed to new cell lines. CPE was again observed only in the pRhMK and MRC-5 vials on day 4 postfiltration. Testing of MRC-5 and pRhMK displaying CPE for enterovirus antigens (on day 4 and day 5, respectively) using commercially available DFA reagent was negative. Two days later, repeat staining for enterovirus, HSV-1, and HSV-2 using different commercially available DFA kits to rule out potential false-negative results was also negative. Staining for additional common viruses capable of causing skin lesions and growth in cell culture was also performed. In addition to the above viruses, cells displaying CPE tested negative for varicella-zoster virus, cytomegalovirus, adenovirus, and rubeola virus antigens. Molluscum contagiosum virus, a poxvirus, has been reported to produce HSV-like CPE in standard viral cell culture [5]. Therefore, given the negative routine culture workup, molluscum contagiosum was considered to be high on the virologic differential diagnosis. Because molluscum contagiosum infection has a common clinical presentation, laboratory confirmation is usually not pursued.
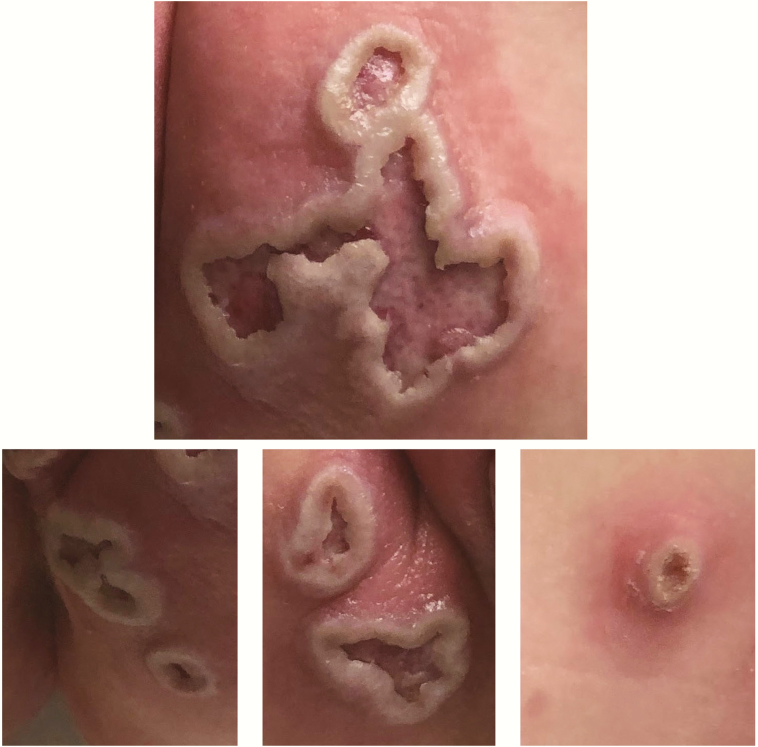
At this point, the medical providers were contacted to discuss the case and obtain additional patient history. The patient was a 10-month-old female infant who presented to a hospital in another state for evaluation of fever and new skin lesions (Figure 1). On exam, the diaper area had 10 circular to irregularly shaped ulcers, 1–2.5 cm in size, with undermined borders, all painful to touch. The parents reported “blisters” that developed 4–5 days prior and ulcerated in the following days. Scattered red macules were noted in perioral and acral locations. Overall, the skin lesions were not consistent with molluscum contagiosum.
Figure 1.
Skin lesions. Ulcerative lesions in the buttock area ranging in size from 1 to 2.5 cm. Consent to use these images for publication was obtained from the patient’s family.
The patient had received no medical care before this illness, and she was behind in her vaccines. She had not traveled. There were no animal exposures or recent changes in topical hygiene products. The patient’s mother had a history of moderate to severe atopic dermatitis and reported a recent illness and flare of her skin disease. On exam, she had numerous circular collarettes of scale over her forearms and arms from a vesicular eruption that began the week prior but was resolving without intervention. The patient’s father, an active duty military member, had a 1-cm vesicular, crusted plaque above his right upper lip, which he attributed to a cold sore. Neither parent, nor other close contacts, had received recent live-virus vaccinations.
ADDITIONAL DIAGNOSTIC TESTING
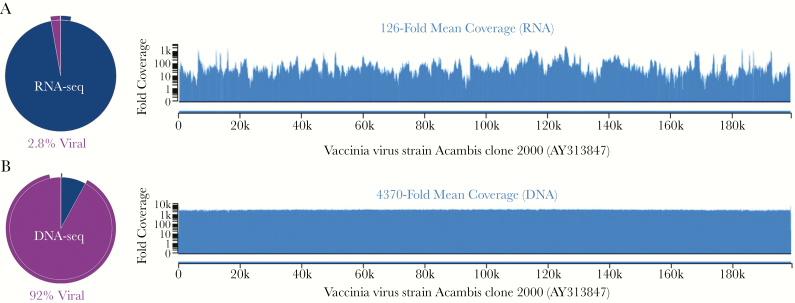
In an attempt to identify the unknown virus, metagenomic next-generation RNA and DNA sequencing (mNGS) and electron microscopy (EM) were performed using culture material inactivated in a biosafety cabinet. Briefly, RNA and DNA sequencing generated >1.2 × 107 single-end 150-bp sequencing reads per library (IDbyDNA, Salt Lake City, UT, USA). Sequencing data analyzed with the v2 Explify Platform (IDbyDNA) identified vaccinia virus. mNGS generated a full-length viral genome with >4300-fold coverage (Figure 2). Vaccinia virus Acambis clone 2000 was the most similar virus strain matching the patient’s isolate in public databases (ACAM2000, GenBank accession number AY313847), containing only a single nucleotide polymorphism (49676G>A, numbering from AY313847). This vaccinia virus strain is used in 1 of the current live virus smallpox vaccines available in the United States (Sanofi Pasteur Biologics, Cambridge, MA, USA). In addition, low levels of Bovine polyomavirus 1 (most similar to KU200259, 71.6% genome coverage) and Macacine betaherpesvirus 3 (Rhesus cytomegalovirus, most similar to KX689268, 22.7% genome coverage) were detected, most likely representing contamination of the bovine serum and primate cells used as part of viral culture [6].
Figure 2.
Identification and characterization of vaccinia virus from tissue culture by shotgun metagenomic RNA and DNA sequencing. A, Shotgun RNA sequencing revealed 2.8% of sequencing reads to be of viral origin (pie chart), resulting in a mean 126-fold coverage of the vaccinia virus genome. B, Ninety-two percent of shotgun DNA sequencing reads were of viral origin, producing >4000-fold mean coverage and allowing for construction of a high-quality viral consensus genome for the patient strain covering 199 077 nucleotides (99.92% of the reference genome). Vaccinia virus strain Acambis clone 2000, used in the second-generation smallpox vaccine (ACAM2000, GenBank accession number AY313847), was the most similar strain in public databases. Only a single nucleotide polymorphism (49676G>A, numbering from AY313847) relative to ACAM2000 was identified.
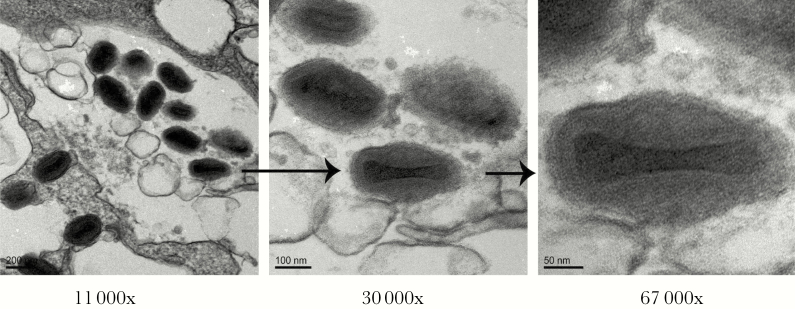
Electron micrographs were performed as previously described [7]. These images were also consistent with a poxvirus infection, as evidenced by the presence of a large (~350 × 270 nm) brick-shaped virion (Figure 3). EM does not differentiate members of the poxvirus family [8].
Figure 3.
Transmission electron micrograph of MRC-5 cells infected with vaccinia virus. Representative images of vaccinia virus are shown at different magnifications (11 000×, 30 000×, and 67 000×). The large (~200 × 300 nm) brick-shaped virions with biconcave (dumbbell-shaped) electron-dense cores containing the viral genome are characteristic of poxviruses.
DISCUSSION
Routine smallpox vaccination of the American public ended in 1972 after the disease was eradicated from the United States. However, the government has maintained a vaccination program for selected health care workers, first responders, and military personnel out of concerns that the orthopoxvirus variola virus (smallpox) could be used as an agent of bioterrorism [9, 10]. Two smallpox vaccines are currently approved by the US Food and Drug Administration. Both contain live vaccinia virus, a closely related orthopoxvirus that provides cross-protection against variola and other orthopoxviruses. The newest product (JYNNEOS, Bavarian Nordic) is a live, nonreplicating virus vaccine [11]. The JYNNEOS vaccine was approved by the Food and Drug Administration in September of 2019, and therefore was not available at the time of this event. In contrast, ACAM2000 contains replication-competent virus that can be shed from the intradermal vaccination site for up to 3 weeks, and this vaccine was in use during the time this patient developed infection [12–14].
Transmission of vaccinia to household-, sexual-, and sports-related contacts of recent vaccinees has been reported [15–19]. Tertiary transmission from contacts of vaccinees, including to an infant, has also been previously described [20]. At the time of this report, the child’s parents had not undergone testing for vaccinia exposure, and a follow-up epidemiologic investigation failed to identify another potential source of exposure. As neither parent had been recently vaccinated, we consider occult transmission from a recent vaccinee to the mother, and then from mother to child, to be the most likely etiology of the child’s skin lesions in this case. We postulate that the mother’s rash may have been a mild form of eczema vaccinatum, a rare complication of vaccination most commonly seen in vaccinees or their close contacts with a history of atopic dermatitis or eczema [21]. Given the distribution of the child’s lesions, we suspect that transmission occurred during diaper care and that the sores on the baby’s hands and mouth were a result of autoinoculation. As many military members in the father’s workplace receive ACAM2000 vaccine, it is theoretically possible that contact transmission resulted in unrecognized secondary vaccinia infection in the father, with or without tertiary infection in the mother, subsequently followed by infection in this infant. Secondary transmission to the child through fomites in the household is also possible, although less likely based on the location of the lesions and given a lack of identifiable family contact with known recent vaccinees.
Fortunately, no additional cases of transmission events were identified, and the patient improved with supportive care. Vaccinia immune globulin and antivirals with antivaccinia activity (eg, tecovirimat, cidofovir, and brincidofovir) were considered for treatment of the skin lesions [22] but ultimately were not administered. Although rare, spread to close contacts is considered a significant adverse event of vaccination that requires reporting to the Vaccine Adverse Events Reporting System (VAERS) [10, 23]. Serious cases should also be reported to the state health department and/or the Centers for Disease Control and Prevention (CDC), as was done in this case [23].
Clinical laboratories should be aware that vaccinia virus, as well as other poxviruses such as monkeypox and variola, are capable of growth in routine clinical cell culture. Diploid cells infected with vaccinia virus are described as having a characteristic granular appearance, often with focal areas of degenerated cells and cytoplasmic bridges. This description, however, is relatively nonspecific. In general, recognition of CPE not typical for common viruses should prompt clinical laboratorians to consider the potential presence of a poxvirus or other emerging pathogen that may require enhanced biocontainment measures. Vaccinia virus is a biosafety level 2 (BSL2) organism, which is the standard level of containment for clinical virology work. However, transmission to a laboratory worker manipulating live vaccinia virus has been reported [24]. The Advisory Committee on Immunization Practices (ACIP) recommends smallpox vaccination at least every 10 years for laboratory workers who handle cultures or animals infected with non–highly attenuated vaccinia or other orthopoxviruses [9], but this is not routine for the clinical virology workforce.
Laboratory confirmation of orthopoxvirus infection, including vaccinia, is not commercially available. For cases where orthopoxviruses are suspected, viral culture should not be performed and specimens should be referred to state or local health departments for testing via the Laboratory Response Network (LRN) or CDC [25]. In this case, however, vaccinia infection was not initially considered given the lack of clear exposure in the patients and CPE most closely resembling HSV in the clinical laboratory. Identification of the virus was ultimately accomplished using a combination of traditional and advanced diagnostic methods.
In summary, this case illustrates the utility of viral culture combined with the power of mNGS for identifying viruses to the strain level in specimens from patients with a disease of unknown etiology. Although metagenomic sequencing could have theoretically been applied directly to the initial skin swab, its performance relative to conventional tests needs to be demonstrated, and costs have limited broader adoption for upfront testing. As sequencing costs come down and bioinformatic tools for sequence analysis are simplified, the debate over the role of viral culture in clinical virology will likely need to be revisited yet again.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
The authors would like to thank Dr. Margaret Ryan with the Defense Health Agency for her assistance in investigating this case and the patient’s family for their willingness to share the case history and images of the skin lesions.
Financial support. S.S. and D.V. were supported by 1R01 CA81133 from the National Institutes of Health.
Potential conflicts of interest. R.S. is a co-founder and Chief Medical Officer of IDbyDNA. H.X. is an employee of IDbyDNA. The University of Utah owns Intellectual Property in the mNGS platform used in this study. The other authors have no conflicts to declare. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Author contributions. R.M.M. drafted the manuscript and coordinated communication among authors. K.B. provided patient medical history and case presentation. J.L. supervised routine diagnostic virology testing. H.X. performed metagenomic sequencing. R.S. analyzed metagenomic sequencing. D.V. performed transmission electron micrograph. All authors critically revised the manuscript. S.S. and K.E.H. supervised the study.
Statement of data availability. The vaccinia virus metagenomic sequence data from this study have been submitted to the National Center for Biotechnology Information (NCBI) sequence read archive (SRA). The consensus sequence of the vaccinia virus strain has been deposited under the NCBI accession number MT227314.
References
- 1. Hodinka RL. Point: is the era of viral culture over in the clinical microbiology laboratory? J Clin Microbiol 2013; 51:2–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Leland DS, Ginocchio CC. Role of cell culture for virus detection in the age of technology. Clin Microbiol Rev 2007; 20:49–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burd EM. Ebola virus: a clear and present danger. J Clin Microbiol 2015; 53:4–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. CDC grows SARS-CoV-2, the virus that causes COVID-19, in cell culture Available at: https://www.cdc.gov/coronavirus/2019-ncov/about/grows-virus-cell-culture.html. Published 15 February 2020. Accessed 7 March 2020.
- 5. Bell CA, Eberly AP, Takata G, et al. Specimens from a vesicular lesion caused by molluscum contagiosum virus produced a cytopathic effect in cell culture that mimicked that produced by herpes simplex virus. J Clin Microbiol 2006; 44:283–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schuurman R, van Steenis B, Sol C. Bovine polyomavirus, a frequent contaminant of calf serum. Biologicals 1991; 19:265–70. [DOI] [PubMed] [Google Scholar]
- 7. Verma D, Thompson J, Swaminathan S. Spironolactone blocks Epstein-Barr virus production by inhibiting EBV SM protein function. Proc Natl Acad Sci U S A 2016; 113:3609–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pauli G, Blümel J, Burger R, et al. Orthopox viruses: infections in humans. Transfus Med Hemother 2010; 37:351–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Petersen BW, Harms TJ, Reynolds MG, Harrison LH. Use of vaccinia virus smallpox vaccine in laboratory and healthcare workers at risk for occupational exposure to orthopoxviruses - recommendations of the Advisory Committee on Immunization Practices (ACIP), 2015. MMWR Morb Mortal Wkly Rep 2016; 65: 257–62. [DOI] [PubMed] [Google Scholar]
- 10.Defense Health Agency Immunization Healthcare Division. What you need to know about smallpox vaccine. Available at: https://health.mil/Military-Health-Topics/Health-Readiness/Immunization-Healthcare/Vaccine-Preventable-Diseases/Smallpox . Published 25 July 2019. Accessed 3 December 2019. [Google Scholar]
- 11. Pittman PR, Hahn M, Lee HS, et al. Phase 3 efficacy trial of modified vaccinia ankara as a vaccine against smallpox. N Engl J Med 2019; 381:1897–908. [DOI] [PubMed] [Google Scholar]
- 12. Lane JM, Fulginiti VA. Transmission of vaccinia virus and rationale for measures for prevention. Clin Infect Dis 2003; 37:281–4. [DOI] [PubMed] [Google Scholar]
- 13. ACAM2000 Product Insert. Gaithersburg, MD: Emergent BioSolutions Inc;2007. [Google Scholar]
- 14. Nalca A, Zumbrun EE. ACAM2000: the new smallpox vaccine for United States Strategic National Stockpile. Drug Des Devel Ther 2010; 4:71–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Centers for Disease Control and Prevention. Vulvar vaccinia infection after sexual contact with a military smallpox vaccinee — Alaska, 2006. MMWR Morb Mortal Wkly Rep 2007; 56(17):417–9. [PubMed] [Google Scholar]
- 16. Centers for Disease Control and Prevention. Secondary and tertiary transmission of vaccinia virus after sexual contact with a smallpox vaccinee - San Diego, California, 2012. MMWR Morb Mortal Wkly Rep 2013; 62:145–7. [PMC free article] [PubMed] [Google Scholar]
- 17. Young GE, Hidalgo CM, Sullivan-Frohm A, et al. Secondary and tertiary transmission of vaccinia virus from US military service member. Emerg Infect Dis 2011; 17:718–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hughes CM, Blythe D, Li Y, et al. Vaccinia virus infections in martial arts gym, Maryland, USA, 2008. Emerg Infect Dis 2011; 17:730–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Centers for Disease Control and Prevention. Household transmission of vaccinia virus from contact with a military smallpox vaccinee—Illinois and Indiana, 2007. MMWR Morb Mortal Wkly Rep 2007; 56:478–81. [PubMed] [Google Scholar]
- 20. Garde V, Harper D, Fairchok MP. Tertiary contact vaccinia in a breastfeeding infant. JAMA 2004; 291:725–7. [DOI] [PubMed] [Google Scholar]
- 21. Reed JL, Scott DE, Bray M. Eczema vaccinatum. Clin Infect Dis 2012; 54:832–40. [DOI] [PubMed] [Google Scholar]
- 22. Medical management of adverse reactions Available at: https://www.cdc.gov/smallpox/clinicians/vaccine-medical-management6.html. Published 5 December 2016. Accessed 31 December 2019.
- 23. Cono J, Casey CG, Bell DM; Centers for Disease Control and Prevention. Smallpox vaccination and adverse reactions guidance for clinicians. MMWR Recomm Rep 2003; 52:1–28. [PubMed] [Google Scholar]
- 24. Davies E, Peake L, Woolard D, et al. Laboratory-acquired vaccinia virus infection—Virginia, 2008. MMWR Morb Mortal Wkly Rep 2009; 58:797–800. [PubMed] [Google Scholar]
- 25. Smallpox: laboratory personnel Available at: https://www.cdc.gov/smallpox/lab-personnel/index.html. Published 8 October 2019. Accessed 3 December 2019.
- 26. Reynolds ES. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol 1963; 17:208–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.