Abstract
Background
In December, 2019, a novel zoonotic severe acute respiratory syndrome-related coronavirus emerged in China. The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) became pandemic within weeks and the number of human infections and severe cases is increasing. We aimed to investigate the susceptibilty of potential animal hosts and the risk of anthropozoonotic spill-over infections.
Methods
We intranasally inoculated nine fruit bats (Rousettus aegyptiacus), ferrets (Mustela putorius), pigs (Sus scrofa domesticus), and 17 chickens (Gallus gallus domesticus) with 105 TCID50 of a SARS-CoV-2 isolate per animal. Direct contact animals (n=3) were included 24 h after inoculation to test viral transmission. Animals were monitored for clinical signs and for virus shedding by nucleic acid extraction from nasal washes and rectal swabs (ferrets), oral swabs and pooled faeces samples (fruit bats), nasal and rectal swabs (pigs), or oropharyngeal and cloacal swabs (chickens) on days 2, 4, 8, 12, 16, and 21 after infection by quantitative RT-PCR (RT-qPCR). On days 4, 8, and 12, two inoculated animals (or three in the case of chickens) of each species were euthanised, and all remaining animals, including the contacts, were euthanised at day 21. All animals were subjected to autopsy and various tissues were collected for virus detection by RT-qPCR, histopathology immunohistochemistry, and in situ hybridisation. Presence of SARS-CoV-2 reactive antibodies was tested by indirect immunofluorescence assay and virus neutralisation test in samples collected before inoculation and at autopsy.
Findings
Pigs and chickens were not susceptible to SARS-CoV-2. All swabs, organ samples, and contact animals were negative for viral RNA, and none of the pigs or chickens seroconverted. Seven (78%) of nine fruit bats had a transient infection, with virus detectable by RT-qPCR, immunohistochemistry, and in situ hybridisation in the nasal cavity, associated with rhinitis. Viral RNA was also identified in the trachea, lung, and lung-associated lymphatic tissue in two animals euthanised at day 4. One of three contact bats became infected. More efficient virus replication but no clinical signs were observed in ferrets, with transmission to all three direct contact animals. Mild rhinitis was associated with viral antigen detection in the respiratory and olfactory epithelium. Prominent viral RNA loads of 0–104 viral genome copies per mL were detected in the upper respiratory tract of fruit bats and ferrets, and both species developed SARS-CoV-2-reactive antibodies reaching neutralising titres of up to 1/1024 after 21 days.
Interpretation
Pigs and chickens could not be infected intranasally by SARS-CoV-2, whereas fruit bats showed characteristics of a reservoir host. Virus replication in ferrets resembled a subclinical human infection with efficient spread. Ferrets might serve as a useful model for further studies—eg, testing vaccines or antivirals.
Funding
German Federal Ministry of Food and Agriculture.
Introduction
Coronaviruses are enveloped viruses with a large, single-stranded RNA genome of positive polarity.1 Although numerous coronaviruses have been identified in animals or humans,2 two β-coronaviruses that emerged in the past 20 years are remarkable: severe acute respiratory syndrome coronavirus (SARS-CoV);3 and Middle East respiratory syndrome coronavirus (MERS-CoV).4 Both viruses presumably originated from bats,5 but have adapted to further animals such as palm civets6 or dromedary camels,4 from which sporadic or sustained spill-over infections occurred, resulting in abundant (in the case of SARS-CoV)7 or limited human-to-human infection chains (in the case of MERS-CoV).8
In December, 2019, another SARS-CoV-related zoonotic β-coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), started spreading pandemically from Wuhan, China. Similar to findings for SARS-CoV and MERS-CoV, β-coronaviruses that are closely related to SARS-CoV-2 were found in bats.9, 10
Because of the zoonotic origin of SARS-CoV-2 from the likely bat reservoir, several questions concerning the susceptibility of animals arise. First, how susceptible are putative reservoir hosts such as bats? Second, what is the risk of possible spill-over infections from humans to farmed animals? And finally, what animal would serve as suitable animal models of human infection to study antivirals and vaccine prototypes? Viral receptor structure might be used as an important predictive factor of susceptibility. SARS-CoV and SARS-CoV-2 have been shown to use the same receptor molecule, angiotensin-converting enzyme 2 (ACE2),11 for contact with the receptor-binding-domain of the spike protein. Based on findings from molecular studies, the ACE2 proteins of non-human primates, pigs, cats, and ferrets closely resemble the human ACE2 receptor. Therefore, these species might be susceptible to SARS-CoV-2 infection, as has been shown for SARS-CoV.12, 13 Bats, as a major reservoir host of β-coronaviruses and especially SARS-CoV-related viruses,14 need to be further studied to better understand the viral replication, shedding, transmission, or persistence in a putative reservoir host species.
Research in context.
Evidence before this study
We searched PubMed and bioRxiv for articles using the search terms “SARS-CoV-2”, or “COVID-19”, and “animal model”, or “ferret”, or “bat”, or “pig”, or “chicken”, for articles published in English between inception and April 10, 2020. Little information is available on whether severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can infect animals and whether some species have the potential of becoming epidemiological animal reservoirs or could represent suitable animal models for testing vaccines and antiviral drugs. Infection of ferrets and cats by severe acute respiratory syndrome coronavirus (SARS-CoV) has been shown. Field infections of pigs were also reported, whereas poultry did not appear to be affected. For SARS-CoV, non-human primate and ferret models were used. Two studies on ferrets, one on chickens and pigs, and two on non-human primates indicated similar results for SARS-CoV-2. However, data on the susceptibility of bat species, as well as detailed analyses including viral loads and histopathology of SARS-CoV-2 in ferrets and their contact animals do not exist. Furthermore, the results of the study of the inoculation of pigs and chickens require confirmation and extension because the pig results contrast with receptor binding predictions and chickens represent the most important livestock in Asia.
Added value of this study
We found that neither pigs nor chickens showed any signs of infection and none of the contact animals became infected. This finding is of particular importance for risk analysis in these farmed animals, which are kept in large numbers and in contact with humans. Moreover, the virus replicated in the upper respiratory tract of fruit bats, and was transmitted to contact animals. This finding indicates that fruit bats, which are kept and bred in captivity, can serve as a reservoir host model, but also emphasises the risk to free-living bats (eg, in ecological bat protection programmes). Finally, ferret infections resulted in a high replication rate of SARS-CoV-2 in the nasal cavity, as confirmed by immunohistochemistry and in situ hybridisation. The transmission to contacts was highly efficient and high virus titres were detected in the ferrets' nasal cavities. We showed that only minor viral adaptions occurred during infection of ferrets with a human SARS-CoV-2 isolate. Our results suggest that the ferret is a highly suitable model for testing vaccines and antiviral treatment for their effect on viral excretion and transmission.
Implications of all the available evidence
Our results support previous findings indicating a negligible risk of anthropozoonotic transmission to pigs and chickens, but a substantial risk for bats and ferrets. Fruit bats show a different pattern of infection than ferrets, but both can serve as model animals. Because of distinct differences, for example in the immune system, between humans and bats, bats are not considered suitable models for testing preventive or therapeutic measures but might represent an appropriate model for a potential reservoir host. However, infection in ferrets, next to that in non-human primates, most closely resembles human infection and therefore ferrets could be used as an animal model for testing vaccines and antivirals.
We aimed to investigate virus replication and shedding, the clinical course, pathohistological changes, and transmission of SARS-CoV-2 in four animal species.
Methods
Animals and study design
We used 12 Egyptian fruit bats (Rousettus aegyptiacus, seven [58%] female, five [42%] male, born between 2015 and 2019 in the Friedrich-Loeffler-Institut [FLI] breeding colony), 12 female ferrets (Mustela putorius, aged 9–12 months, originating from the FLI breeding colony), 12 male pigs (Sus scrofa domesticus, German landrace breed, aged 9 weeks, raised by a commercial breeding farm with a high veterinary hygiene standard [Bundeshybridzuchtprogramm, Dahlenburg, Germany]), and 20 chickens (Gallus gallus domesticus, white leghorn, aged 5 weeks, 12 [60%] male, 8 [40%] female, hatched from specific-pathogen-free eggs [VALO BioMedia, Osterholz-Scharmbeck, Germany]). The number of animals included in the study was determined using a statistical experiment plan (appendix p 4). Fruit bats were kept in groups of four and pigs in groups of six. Ferrets were kept together in one cage, chickens were kept in free-run conditions with nests and perches. All animals were offered water ad libitum and were fed and checked for clinical scores daily and by video supervision during the 21-day study period by animal caretakers and study researchers. All animals tested negative for SARS-CoV-2 genome and antibodies by indirect immunofluorescence assay (IIFA) and virus neutralisation test (VNT) before the experiment. The ACE2 receptor of pigs and ferrets, but not chickens, resembles the human receptor. However, we included chickens in our experiments because of their importance as livestock in Asia. Egyptian fruit bats were chosen as a model bat species because of their availability due to having our own breeding colony.
Fruit bats 1–9, ferrets 1–9, and pigs 1–9 were infected intranasally, whereas chickens 1–17 received oculo-oronasal 105 TCID50 SARS-CoV-2 2019_nCoV Muc-IMB-1 per animal. This dose was chosen on the basis of the virus titre of our stock and the applicable volumes to the respective animals, and was considered to be potentially relevant in the context of anthropozoonotic transmission. The inoculum was administered to both nostrils using a pipette (fruit bats, ferrets, and chickens) or an intranasal spraying device (pigs; Teleflex Medical, Fellbach, Germany).
To test viral transmission by direct contact, three naive contact animals (bats, ferrets, and pigs 10–12 and chickens 18–20) were added 24 h after inoculation. Animals were monitored for body temperature (pigs, fruit bats, and ferrets) and bodyweight (fruit bats and ferrets) throughout the experiment. Viral shedding was tested using nasal washes and rectal swabs (ferrets), oral swabs and pooled faeces samples (fruit bats), nasal and rectal swabs (pigs), or oropharyngeal and cloacal swabs (chicken) on days 2, 4, 8, 12, 16, and 21 after infection. On days 4, 8, and 12, two inoculated animals (or three in the case of chickens) of each species were euthanised. All remaining animals, including the contacts, were euthanised at day 21 (figure 1 ). All animals were subjected to autopsy. For virus detection and histopathology, nasal conchae, trachea, lung, tracheobronchial lymph node (not for chickens), heart, liver, spleen, duodenum, colon or cecum, pancreas, kidney, adrenal gland, skeletal muscle, inguinal skin, and brain tissues were collected.
Figure 1.
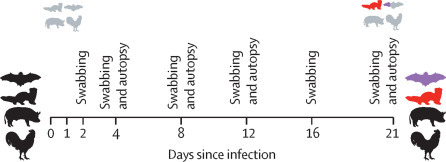
Outline of the in vivo experiments
On days 4, 8, and 12, two fruit bats, ferrets, and domestic pigs were euthanised. The same schedule was applied to three chickens at each timepoint. All remaining animals, including the contacts, were euthanised on day 21. Black animals (n=9 for bats, ferrets, and pigs; n=17 for chickens) were inoculated intranasally (or oculo-oronasally for chickens) with 105 TCID50. Grey animals (n=3 for each species) indicates direct contact animals included 1 day after inoculation. On the right-hand side, black and grey animals were not susceptible; red animals became infected and showed strong viral shedding; and purple animals were infected but displayed only little virus shedding.
The animal experiments were assessed and approved by the ethics committee of the State Office of Agriculture, Food Safety, and Fisheries in Mecklenburg–Western Pomerania (LALLF M-V: LVL MV/TSD/7221.3-2-010/18-12). All procedures were carried out in approved biosafety level 3 facilities.
RNA extraction, detection of SARS-CoV-2 genome, and reactive antibodies and pathology
Total RNA was extracted from oral, nasal, rectal, faecal, and tissue samples and from nasal washes using the NucleoMagVet kit (MachereyNagel, Düren, Germany) according to the manufacturer's instructions in an elution volume of 100 μL. SARS-CoV-2 RNA was detected by the E-gene Sarbeco 6-carboxyfluorescein quantitative RT-PCR (RT-qPCR) as described by Corman and colleagues,15 detection limit 1 genome copy per μL RNA eluate. Serum samples were tested for the presence of SARS-CoV-2 reactive antibodies by IIFA and VNT, with a cutoff of less than 1/16.
At autopsies, tissues were fixed in 10% formalin, embedded in paraffin, cut at 3 μm sections, and stained with haematoxylin and eosin. Immunohistochemistry was done using an anti-SARS nucleocapsid antibody (Novus Biologicals NB100-56576, Colorado, USA) according to the standardised avidin-biotin-peroxidase complex method, producing a red labelling and haematoxylin counterstain. To confirm immunohistochemistry, RNA in situ hybridisation (RNAScope, Advanced Cell Diagnostics, Newark, CA, USA) was done on selected tissues using SARS-CoV-2 spike protein probes.
Further details on virus, cells, virus titration, RNA extraction, RT-qPCR, next-generation sequencing, antibody detection, histopathology, immunohistochemistry, and in situ hybridisation, as well as porcine cell and embryonated chicken egg experiments are given in the appendix (pp 1–5).
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
No clinical signs, fever, bodyweight loss, or mortality were observed in any of the 12 bats. Oral shedding was observed in all nine infected bats from days 2–12, with one of the three remaining infected bats still virus-positive at day 12. Oral shedding was also detected once at day 2 in contact bat 11 and until day 8 in contact bat 10 (figure 2A ). The virus was isolated from one oral swab on day 2 (101·75 TCID50 per mL; fruit bat 8). Faecal shedding was observed in all three bat cages at days 2 and 4, with quantification cycle (Cq) ranging from 29·54 to 36·43 (appendix p 10). SARS-CoV-2 genome (Cq 23·16–38·97; 1·96 × 104 to 1·32 × 101 genome copies per μL) was detected in the nasal epithelium in seven (78%) of nine infected bats euthanised at days 4, 8, and 21, with two giving negative results at days 8 and 12 (fruit bats 4 and 6). The nasal epithelium of one contact animal contained viral RNA on day 21 (Cq value 32·89; 3·12 genome copies per μL; fruit bat 10). At day 4, genome was also detected in respiratory tissues (trachea [2/2], lung [1/2], and lung-associated lymphatic tissue [2/2]) and at lower levels in heart, skin, duodenum, and adrenal gland tissues (fruit bat 2 at day 4), and in duodenum, skin, and adrenal gland tissues at day 8 (either fruit bat 3 or 4; figure 2C; appendix p 6). Virus only could be cultivated from the trachea (102·25 TCID50 per mL) and the nasal epithelium (101·75 TCID50 per mL) of fruit bat 2 at day 4.
Figure 2.

SARS-CoV-2 viral genome loads during the study period
Oral swabs of fruits bats (A), nasal washes of ferrets (B), tissues collected from fruit bats (C), and tissues collected from ferrets (D) that were experimentally infected with SARS-CoV-2 and the contact animals. Genome copies per μL RNA eluate were calculated on the basis of a quantified standard RNA. Each extracted sample was eluted in 100 μL. Limit of detection was 1 genome copy per μL RNA. Contact fruit bats were infected but displayed negligible shedding of the virus, whereas contact ferrets became infected and showed strong viral shedding. Viral genome was undetectable in fruit bats 6 (day 12), 11 and 12 (day 21), and ferrets 5 (day 12) and 8 (day 21). SARS-CoV-2=severe acute respiratory syndrome coronavirus 2.
SARS-CoV-2-reactive antibodies were observed in all inoculated bats by IIFA starting from day 8 and in one contact bat (bat 10) on day 21 with titres of 1/16. Only a slight increase in antibody levels could be observed between day 8 and day 21 (with varying titres between 1/16 and 1/64). Neutralising antibodies could be detected in the same fruit bats with titres up to 1/64 (table ).
Table.
Serological evidence of SARS-CoV-2 infection in fruit bats and ferrets
| Indirect immunofluorescence assay | Virus neutralisation test | |
|---|---|---|
| Fruit bats | ||
| Fruit bat 1, day 4 | <1/16 | <1/16 |
| Fruit bat 2, day 4 | <1/16 | <1/16 |
| Fruit bat 3, day 8 | 1/16 | 1/32 |
| Fruit bat 4, day 8 | 1/16 | 1/32 |
| Fruit bat 5, day 12 | 1/16 | 1/32 |
| Fruit bat 6, day 12 | 1/32 | 1/16 |
| Fruit bat 7, day 21 | 1/64 | 1/64 |
| Fruit bat 8, day 21 | 1/32 | 1/32 |
| Fruit bat 9, day 21 | 1/64 | 1/32 |
| Fruit bat 10, day 21 | 1/16 | 1/16 |
| Fruit bat 11, day 21 | <1/16 | <1/16 |
| Fruit bat 12, day 21 | <1/16 | <1/16 |
| Ferrets | ||
| Ferret 1, day 4 | <1/16 | <1/16 |
| Ferret 2, day 4 | <1/16 | <1/16 |
| Ferret 3, day 8 | 1/128 | <1/16 |
| Ferret 4, day 8 | 1/512 | <1/16 |
| Ferret 5, day 12 | 1/64 | <1/16 |
| Ferret 6, day 12 | 1/4096 | <1/16 |
| Ferret 7, day 21 | 1/4096 | 1/128 |
| Ferret 8, day 21 | 1/8192 | 1/1024 |
| Ferret 9, day 21 | 1/4096 | 1/1024 |
| Ferret 10, day 21 | <1/16 | <1/16 |
| Ferret 11, day 21 | <1/16 | <1/16 |
| Ferret 12, day 21 | 1/8192 | 1/256 |
Serum samples taken before inoculation were below the cutoff (<1/16) for indirect immunofluorescence assay and virus neutralisation test. These data were obtained from serum samples collected at autopsy. SARS-CoV-2=severe acute respiratory syndrome coronavirus 2.
Autopsy revealed no gross pathological lesions. Fruit bat 10 was pregnant. Histopathology revealed minimal to mild rhinitis at day 4 (fruit bats 1 and 2), with epithelial necrosis, oedema, infiltrating lymphocytes and neutrophils, and intraluminal cellular debris (figure 3A ). Viral antigen detection, restricted to foci in the nasal respiratory epithelium and single cells of the non-respiratory epithelium (fruit bats 1 and 2; figure 3B, C), were confirmed by in situ hybridisation. Viral antigen was absent at later timepoints, but moderate rhinitis was detected at day 8 (fruit bats 3 and 4), day 12 (fruit bat 6), and to a milder extent at day 21 (fruit bats 7 and 11), indicating the presence of previous replication sites. Despite the detection of viral RNA by RT-qPCR, no viral antigen was detectable in the lung tissue. However, three of the inoculated (fruit bats 1, 4, and 5) as well as one contact animal (fruit bat 10) showed inflammatory infiltrates in the lung (appendix pp 6–8, 11). Slightly increased numbers of alveolar macrophages were found at all timepoints (appendix p 8). Gram stain did not detect intralesional bacteria. None of the other organs were found positive for viral antigen and no further relevant morphological changes were detected.
Figure 3.
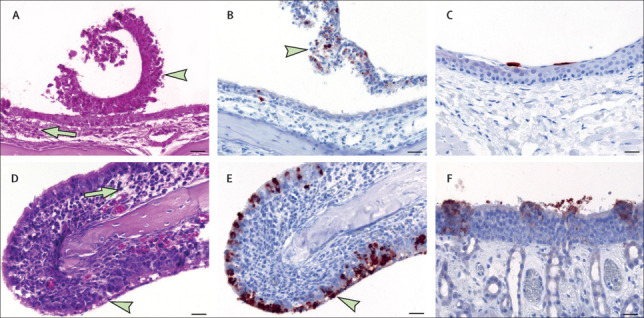
SARS-CoV-2-associated rhinitis and antigen detection at day 4
(A) Rhinitis in a bat, with intraluminal debris (arrowhead), slight mucosal oedema, and minimal inflammation (arrow). (B) Nasal respiratory epithelium in a bat, showing intralesional viral antigen mainly within intraluminal debris. (C) Non-respiratory epithelium in a bat, with single antigen positive cells and no inflammation. (D) Rhinitis in a ferret, with degeneration and necrosis of the respiratory epithelium (arrowhead), slight mucosal oedema, and numerous infiltrates (arrow). (E) Nasal respiratory epithelium in a ferret, showing intralesional, abundant viral antigen. (F) Olfactory epithelium in a ferret, showing multifocal, intralesional viral antigen. Parts A and D show histopathology, haematoxylin and eosin stain; bar 20 μm. Parts B, C, E, and F show immunohistochemistry, Avidin-Biotin Complex method, aminoethyl carbazole chromogen (red-brown), Mayer's haematoxylin counter stain (blue); bar 20 μm. SARS-CoV-2=severe acute respiratory syndrome coronavirus 2.
None of the 12 ferrets showed clinical signs or loss of bodyweight during the study period, and body temperatures remained normal. Viral shedding was detected in nasal washes in eight of nine inoculated ferrets between days 2 and 8, with cycle quantification values ranging from 21·77 to 36·35 (8·44 × 103 to 0·34 genome copies per μL). Virus isolation was successful from nasal washes collected on days 2 (ferrets 2, 3, and 4; 102·5–102·875 TCID50 per mL) and 4 (ferret 4; 102·75 TCID50 per mL). All three contact ferrets were infected by direct contact with the other inoculated ferrets. The first RT-qPCR-positive nasal wash sample in a contact ferret was observed on day 8 (ferret 12). Ferret 12 showed viral shedding on days 8 (cycle quantification value 37·03) and 12 (28·59). Ferret 11 tested positive in nasal washes between days 12, 16, and 21 (37·39, 26·15, and 36·93) and ferret 10 on days 16 (28·04) and 21 (30·00; figure 2B). Analysis of the rectal swabs showed minor amounts of viral RNA in four ferrets at singular timepoints, with cycle quantification values between 33·97 and 38·45 (appendix p 10).
At least one of the two ferrets that were euthanised at day 4 (ferrets 1 and 2) was RT-qPCR positive in various tissues (nasal conchae, lung, muscle, skin, trachea, lung lymph node, and colon), with the highest viral genome load in the nasal conchae (cycle quantification values 24·31 and 26·21; 1·93 × 103 and 5·26 × 102 genome copies per μL). The two ferrets euthanised at day 8 (ferrets 3 and 4) were positive in the nasal conchae (34·77 and 21·57; 1·61 × 101 and 1·21 × 104 genome copies per μL). On day 12, one of two ferrets was also positive in the nasal conchae (ferret 6; cycle quantification value 29·26). The last three inoculated ferrets were euthanised at day 21. These animals showed only very weak RT-qPCR positivity in the cerebrum (ferret 7; 37·78) and in the colon (ferret 9; 37·47). The three contact ferrets that were euthanised on day 21 were all positive in the nasal conchae (cycle quantification values 26·29–36·51). Additionally, RT-qPCR positive samples were collected from muscle, lung, cerebrum, cerebellum, and trachea tissue, which were all positive in ferrets 10 and 11, whereas lung lymph node, skin, and adrenal gland tissues were only positive in one animal (figure 2D; appendix p 6). Virus could be cultivated only from the nasal conchae of ferrets 1, 2, and 10 (100·8125–103·875 TCID50 per mL), from the lung and trachea tissues of ferret 1 (101·625 TCID50 per mL).
SARS-CoV-2 antibodies were detected by IIFA from day 8 in all inoculated ferrets with varying titres (1/64–1/8192; table). One (33%) of three contact animals also showed high antibody titres (ferret 12; highest reactive serum dilution 1/8192), whereas the others remained negative. Neutralising antibodies were observed in the three (33%) inoculated ferrets (ferrets 7, 8, and 9) that were euthanised on day 21 and one (33%) contact animal (ferret 12) by VNT (table).
In ferrets, no gross lesions were detected by autopsy. At day 4, viral antigen was associated with rhinitis, showing epithelial degeneration and necrosis, intraluminal cellular debris, and mild inflammation (figure 3D–F). A more severe rhinitis developed at days 8 and 12. At day 21, rhinitis was only slightly detectable (ferret 7) or absent (ferrets 8 and 9). We also observed an antigen-associated rhinitis in the contact ferrets (ferrets 10 and 11). Viral antigen was detected in the nasal cavity at days 4 (ferrets 1 and 2), 8 (ferret 3), and 21 (contact ferrets 10 and 11) in the nasal respiratory and olfactory epithelium as well as in the olfactory epithelium of the vomero-nasal organ (ferret 11; appendix p 12). Selected immunohistochemistry results were confirmed by in situ hybridisation (appendix p 13). No viral antigen was identified in the lung tissue. Three infected ferrets (ferrets 1–3) at days 4 and 8 and all contact animals showed inflammatory infiltrates in the lung (appendix pp 6–8, 11). Slightly increased numbers of alveolar macrophages were found at all timepoints (appendix p 8). Gram stain did not detect intralesional bacteria. None of the other organs was found positive for viral antigen, and no further relevant morphological alterations were detected.
Complete virus genome sequencing revealed two non-synonymous single nucleotide exchanges after the ferret passage. In Orf1a of the polyprotein coding region, we detected a C12723A substitution, resulting in a Thr to Ile amino acid substitution. In the surface glycoprotein coding gene (S gene), an A23038C substitution was found, which resulted in an Asn to Thr amino acid substitution. The C12723A substitution was only detected in inoculated ferret 4, whereas the A23038C substitution within the surface glycoprotein coding gene was found in inoculated (ferret 4) and in contact animals (ferrets 10 and 11).
No clinical signs, including increased body temperatures, were observed in any of the 12 pigs or 20 chickens. All collected samples were negative for SARS-CoV-2 genome. SARS-CoV-2-reactive antibodies were not detected. No lesions were detected at autopsy, histopathology was not done, and embryonated chicken eggs that were inoculated with SARS-CoV-2 were non-permissive (data not shown). Three porcine cell lines (PK-15, SK-6, and ST) inoculated with SARS-CoV-2 developed no cytopathic effect, but two (SK-6 and ST) showed virus replication (appendix pp 9, 14).
Discussion
Our study focused on four animal species, which are potentially relevant as models in research (fruit bats and ferrets) or could pose a risk as a viral reservoir following anthropozoonotic spill-over infections into animals that are used in food production (pigs and chickens).
Neither pigs nor chickens were susceptible to SARS-CoV-2 by intranasal or oculo-oronasal infection. All swabs, organ samples, and contact animals were negative for SARS-CoV-2 RNA and did not seroconvert. Our findings on the non-permissiveness of chickens to SARS-CoV-2 infection were similar to those of previous reports showing the lack of susceptibility of chickens to SARS-CoV16 and support other study findings.17 We showed that this finding extends to embryonated chicken eggs, which are a classic substrate for isolation and propagation of a plethora of zoonotic viruses. These data are also in agreement with findings on the chicken ACE2 receptor,18 which contains alterations in three of five critical residues (Lys31Glu, Glu35Arg, and Met82Arg). By contrast, similar predictions suggested that pigs and ferrets were likely to be susceptible to SARS-CoV-2 because of these animals' matching ACE2 receptor-binding sites.18 However, our study, as well as the report by Shi and colleagues,17 found no susceptibility of pigs by the intranasal inoculation route. Nevertheless, we showed permissiveness of two out of three porcine cell lines tested. The young age of the pigs might have had an influence as an age dependency has been found in other animals—eg, monkeys.19 To further exclude an anthropozoonotic spill-over infection into farm animals, further experiments should focus on Bovidae or other animals, which are predicted to be susceptible according to cell culture data.20
We found that intranasal inoculation of fruit bats resulted in a transient infection in the respiratory tract and virus shedding in fruit bats. SARS-CoV-2 genomes could be detected by RT-qPCR in nasal conchae, trachea, lung, tracheal lymph node, skin, and duodenum tissue. Infectious virus was isolated from nasal conchae and trachea. Oral viral RNA shedding was detectable up to day 12 after infection, but infectious virus could only be isolated at day 2. Immunohistochemistry and in situ hybridisation found that most inoculated fruit bats had viral genome in their nasal cavity at day 4. Rhinitis was associated with the presence of viral antigen, mainly in the respiratory epithelium. Despite the absence of viral antigen at later timepoints, rhinitis was still identifiable, indicating previous replication sites. Some infected animals and one contact fruit bat presented with mild inflammation in the lung, which should be addressed in future studies because no viral antigen or bacteria were detectable. Starting from day 8, all inoculated bats developed a weak immune response. The virus was transmitted to one out of the three contact fruit bats. The affected animal was at an early pregnancy. Several studies show an increased virus detection rate in bats during the reproductive phase, probably due to the associated immunosuppression.21 The other two contact animals were seronegative. In fruit bat 11, the transmission of viral RNA possibly did not result in sufficient local replication, which could explain the single positive oral swab result and the lack of antibody production. β-coronaviruses were shown to infect a variety of bat species with few clinical signs, even during active virus shedding.22 Moreover, low antibody titres are typical for bats.23 Although Egyptian fruit bats express ACE2 in the intestine and respiratory tract, a study found little evidence of virus replication and seroconversion after infection with SARS-like coronaviruses; however, serum samples of some of these bats, collected before the infection, were already reactive with SARS spike or nucleocapsid proteins.24 Our data suggest that intranasal infection of Rousettus aegyptiacus could reflect reservoir host status and therefore represent a useful model, although this species is certainly not the original reservoir of SARS-CoV-2 because these bats are not present in China, the epicentre of the pandemic. Furthermore, we showed that bat-to-bat transmission is possible. Consequently, although our findings for Rousettus aegyptiacus might not apply to all bat species, as over 1200 of them exist, our results indicate bats are at risk of being infected anthropozoonotically by SARS-CoV-2. Therefore, during the pandemic, all contact with bats (eg, during research programmes or ecological analyses), should be avoided.
SARS-CoV-2 replicated efficiently in ferrets. Almost all intranasally infected ferrets shed virus between days 2 and 8. Viral genome was detected by RT-qPCR in nasal washes and infectious virus was isolated from two animals at days 2 and 4. Only one ferret was RT-qPCR negative at all sampling points and developed only a weak IIFA titre. All other inoculated ferrets showed increasing SARS-CoV-2-reactive antibodies starting from day 8. The measured antibody concentrations were generally much higher in ferrets than in bats, indicating a more prominent virus replication in the infected animals. For IIFA, this finding might also be explained by the use of different secondary antibodies. Neutralising antibodies were only detected at day 21, but also with high titres in ferrets, whereas we detected neutralising antibodies in bats from day 8 at low titres. This finding might indicate a reservoir host infection, which deserves more detailed analysis in future studies.
SARS-CoV-2 was efficiently transmitted to three ferrets by direct contact. In those animals, viral RNA was present in nasal washes starting from day 8 and detected mostly in the nasal conchae, but also in lung, trachea, lung lymph node or cerebrum, and cerebellum tissue. The absence of seroconversion at day 21 in two ferrets was most likely due to the late transmission. Viral antigen within the upper respiratory tract was confirmed by strong positive immunohistochemistry and in situ hybridisation in the nasal cavity. In the case of SARS-CoV, the virus was found to replicate in the upper and lower respiratory tract, and the animals developed no or mild clinical disease, characterised by nasal discharge, sneezing, and fever.25 We used high-throughput sequencing to analyse the complete genome of the virus in the inoculum and in samples from the inoculated ferrets and found two non-synonymous single nucleotide substitutions after the ferret passage, showing adaptations to this animal model.
Our results are in line with those of two reports17, 26 that showed productive SARS-CoV-2 infection in ferrets with no or mild clinical signs. Kim and colleagues26 described increased body temperatures in ferrets, but in our study all body temperatures were within a normal physiological range. Moreover, limited histopathology and tissue tropism data were available in both those studies. Our study adds important detailed histopathology substantiating the nasal cavity as the main SARS-CoV-2 replication site, having measured viral antigen in the respiratory and olfactory epithelium and observed signs of rhinitis. At later timepoints, rhinitis was still present despite the absence of viral antigen. Studies27, 28 have described the expression of two host factors, ACE2 and TMPRSS2 proteases, which facilitate SARS-CoV-2 binding, replication, and accumulation in the olfactory epithelium in humans. These studies suggest that the partial loss of sense of smell reported in some patients might be caused by direct damage of the olfactory receptor neurons or another, yet unidentified factor,27, 28 which could also be true for ferrets. Several animals showed pulmonary inflammation, and neither viral antigen nor bacteria were identified. The pathogenesis and significance of these observations should be addressed in future studies. Testing a broader tissue spectrum, including salivary glands, the lower urinary tract, full gastrointestinal tract, and the cerebrospinal fluid, will help to increase understanding of the source of viral RNA in secretions, excretions, and in the brain. Generally, RT-qPCR detected viral genome in a substantially broader spectrum of tissues compared with antigen detected by immunohistochemistry, mainly because of the higher sensitivity of RT-qPCR.
In summary, farmed animals such as chickens and pigs were resistant against intranasal SARS-CoV-2 inoculation under our experimental conditions. The small number as well as the young age of the animals were limitations and do not allow us to fully exclude the possibility of transmission among farm animals, which is relevant for risk assessment and epidemiology of the infection. By contrast, we showed that ferrets and fruits bats could be productively infected. SARS-CoV-2 infection in ferrets, in particular, which resembled a mild infection in humans, might serve as a useful animal model for testing prototypic COVID-19 vaccines and antivirals.
Data sharing
Sequence data are available under study accession number PRJEB37671.
Acknowledgments
Acknowledgments
The authors are very grateful to Roman Wölfel (German Armed Forces Institute of Microbiology) for providing the SARS-CoV-2 isolate used in this study. We thank Bernd Köllner for generating the anti-bat monoclonal antibody. We also acknowledge Mareen Lange, Christian Korthase, Silvia Schuparis, Gabriele Czerwinski, and Patrick Zitzow for their excellent technical assistance and Frank Klipp, Doreen Fiedler, Harald Manthei, René Siewert, Christian Lipinski, Ralf Henkel, and Domenique Lux for their excellent support in the animal experiments.
Contributors
KS, MR, AG, JSc, DHo, AB-B, TH, and CG did the animal experiments. KS, MR, AG, and JSc did molecular, serological and classical virological analyses. AB and JSe did animal necropsies. AB did histopathology, immunohistochemistry, and in situ hybridisation analysis. DHö, CW, and KS added sequencing and quantification data. KS, AG, DHo, TH, TCM, ABB, and MB designed the study. KS, MR, AG, AB, and MB wrote the manuscript. All authors critically evaluated and approved the manuscript.
Declaration of interests
We declare no competing interests.
Supplementary Material
References
- 1.Masters PS, Perlmen S. Coronaviridae. In: Knipe DM, Howley PM, editors. Fields virology. Lippincott Williams & Wilkins; Philadelphia: 2013. pp. 825–858. [Google Scholar]
- 2.Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. In: Maier H, Bickerton E, Britton P, editors. Coronaviruses. Methods in molecular biology. vol 1282. Humana Press; New York, NY: 2015. pp. 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsang KW, Ho PL, Ooi GC. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1977–1985. doi: 10.1056/NEJMoa030666. [DOI] [PubMed] [Google Scholar]
- 4.Haagmans BL, Al Dhahiry SHS, Reusken CBEM. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis. 2014;14:140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li W, Shi Z, Yu M. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 6.Song HD, Tu CC, Zhang GW. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc Natl Acad Sci USA. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherry JD, Krogstad P. SARS: the first pandemic of the 21st century. Pediatr Res. 2004;56:1–5. doi: 10.1203/01.PDR.0000129184.87042.FC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drosten C, Meyer B, Müller MA. Transmission of MERS-coronavirus in household contacts. N Engl J Med. 2014;371:828–835. doi: 10.1056/NEJMoa1405858. [DOI] [PubMed] [Google Scholar]
- 9.Zhou P, Yang XL, Wang XG. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu F, Zhao S, Yu B. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann M, Kleine-Weber H, Schroeder S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271. doi: 10.1016/j.cell.2020.02.052. 80.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gretebeck LM, Subbarao K. Animal models for SARS and MERS coronaviruses. Curr Opin Virol. 2015;13:123–129. doi: 10.1016/j.coviro.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen W, Yan M, Yang L. SARS-associated coronavirus transmitted from human to pig. Emerg Infect Dis. 2005;11:446–448. doi: 10.3201/eid1103.040824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang LF, Shi Z, Zhang S, Field H, Daszak P, Eaton BT. Review of bats and SARS. Emerg Infect Dis. 2006;12:1834–1840. doi: 10.3201/eid1212.060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corman VM, Landt O, Kaiser M. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:23–30. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swayne DE, Suarez DL, Spackman E. Domestic poultry and SARS coronavirus, southern China. Emerg Infect Dis. 2004;10:914–916. doi: 10.3201/eid1005.030827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi J, Wen Z, Zhong G. Susceptibility of ferrets, cats, dogs, and different domestic animals to SARS-coronavirus-2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94:e00127–e00220. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu P, Qi F, Xu Y. Age-related rhesus macaque models of COVID-19. Animal Model Exp Med. 2020;3:93–97. doi: 10.1002/ame2.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luan J, Jin X, Lu Y, Zhang L. SARS-CoV-2 spike protein favors ACE2 from Bovidae and Cricetidae. J Med Virol. 2020 doi: 10.1002/jmv.25817. published online April 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drexler JF, Corman VM, Wegner T. Amplification of emerging viruses in a bat colony. Emerg Infect Dis. 2011;17:449–456. doi: 10.3201/eid1703.100526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi Z, Hu Z. A review of studies on animal reservoirs of the SARS coronavirus. Virus Res. 2008;133:74–87. doi: 10.1016/j.virusres.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schountz T, Baker ML, Butler J, Munster V. Immunological control of viral infections in bats and the emergence of viruses highly pathogenic to humans. Front Immunol. 2017;8 doi: 10.3389/fimmu.2017.01098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Doremalen N, Schäfer A, Menachery VD. SARS-like coronavirus WIV1-CoV does not replicate in Egyptian fruit bats (Rousettus aegyptiacus) Viruses. 2018;10:e727. doi: 10.3390/v10120727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Enkirch T, von Messling V. Ferret models of viral pathogenesis. Virology. 2015;479–480:259–270. doi: 10.1016/j.virol.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim YI, Kim SG, Kim SM. Infection and rapid transmission of SARS-CoV-2 in Ferrets. Cell Host Microbe. 2020;27:704. doi: 10.1016/j.chom.2020.03.023. 09.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brann DH, Tsukahara T, Weinreb C. Non-neural expression of SARS-CoV-2 entry genes in the olfactory epithelium suggests mechanisms underlying anosmia in COVID-19 patients. bioRxix. 2020 doi: 10.1101/2020.03.25.009084. published online March 28. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Butowt R, Bilinska K. SARS-CoV-2: Olfaction, brain infection, and the urgent need for clinical samples allowing earlier virus detection. ACS Chem Neurosci. 2020;11:1200–1203. doi: 10.1021/acschemneuro.0c00172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data are available under study accession number PRJEB37671.


