Abstract
Eukaryotic cells must accurately monitor the integrity of the mitochondrial network to overcome environmental insults and respond to physiological cues. The mitochondrial unfolded protein response (UPRmt ) is a mitochondrial-to-nuclear signaling pathway that maintains mitochondrial proteostasis, mediates signaling between tissues, and regulates organismal aging. Aberrant UPRmt signaling is associated with a wide spectrum of disorders, including congenital diseases as well as cancers and neurodegenerative diseases. Here, we review recent research into the mechanisms underlying UPRmt signaling in Caenorhabditis elegans and discuss emerging connections between the UPRmt signaling and a translational regulation program called the integrated stress response (ISR). Further study of the UPRmt will potentially enable development of new therapeutic strategies for inherited metabolic disorders and diseases of aging.
Keywords: Mitochondria, Mitochondrial Unfolded Protein Response, Integrated Stress Response, Stress Signaling
Physiological roles of mitochondrial stress signaling
Endosymbiosis of an aerobic bacterium enabled archaea-eukaryotic cells to produce energy from atmospheric oxygen. This bacterium transitioned into the mitochondria, a network comprised of a double-membraned organelle that retained a diminutive genome encoding oxidative phosphorylation (OXPHOS) proteins, tRNAs, and rRNAs [1]. In addition to producing ATP through respiration, mitochondria also exert control over cellular metabolism [2], signaling cascades both within cells and between tissues [3–5], and the regulation of organismal lifespan [6].
The import, folding, and quality control of the mitochondrial proteome is regulated by a transcriptional program called the mitochondrial unfolded protein response (UPRmt) [7–12]. The UPRmt is classically understood as a transcriptional response that increases the expression of mitochondrial chaperones in response to protein misfolding within mitochondria [13–14]. However, contemporary evidence has revealed that the UPRmt protects cells from a broader range of mitochondrial stresses, including OXPHOS dysfunction, perturbed protein import arising from mitochondrial protein misfolding, ATP depletion, or dissipation of mitochondrial inner membrane potential [15], and bacterial pathogens that target mitochondria [16]. Alongside this expanded list of stressors, the UPRmt has been implicated in biological processes beyond simply determining the abundance of mitochondrial chaperones. These processes include regulating development [17], innate immune signaling [16], aging [18], cardioprotection [19], and preserving cellular and organismal fitness when challenged with high levels of amyloid β, the primary component of the plaques observed in the brains of patients with Alzheimer’s disease[20]. Although acute activation of the UPRmt enables adaptation to environmental stressors and physiological cues, chronic UPRmt signaling is potentially maladaptive and represents a therapeutic target for a broad spectrum of disorders. UPRmt signaling drives the propagation of mutant mitochondrial genomes that culminate in congenital metabolic disorders [21–23]. Moreover, dysregulated UPRmt signaling contributes to the pathogenesis of diseases of aging including cancers [24–25] and neurodegenerative disorders [5, 26]. Here, we provide an update on the UPRmt [27], particularly emphasizing links between UPRmt signaling and a broader stress response program involved in translational regulation called the integrated stress response (ISR).
Characterization of the UPRmt in Caenorhabditis elegans
Although the UPRmt was initially characterized in mammalian cells [13–14], the discovery that this pathway was intact in C. elegans enabled researchers to utilize RNAi to rapidly screen for genes involved in sensing and responding to mitochondrial dysfunction [28]. These efforts revealed an essential role for the mitochondrial protease CLPP in triggering the UPRmt and culminated in the identification of proteins involved in the nuclear response to mitochondrial dysfunction: UBL-5, DVE-1, and ATFS-1 [29–30].
Transcription of genes induced during the UPRmt depends on atfs-1, a gene encoding a bZIP domain as well as a N-terminal mitochondrial targeting sequence (MTS) and a nuclear localization sequence. Under basal conditions, ATFS-1 is imported into mitochondria and subsequently degraded by the Lon protease (Fig 1A). When mitochondrial protein import capacity is reduced during stress ATFS-1 instead traffics to the nucleus to drive transcription of genes involved in mitochondrial proteostasis (Fig 1A). Thus, ATFS-1 directly coordinates the import capacity of the mitochondrial network with the induction of a mitochondrial quality control program [20–21].
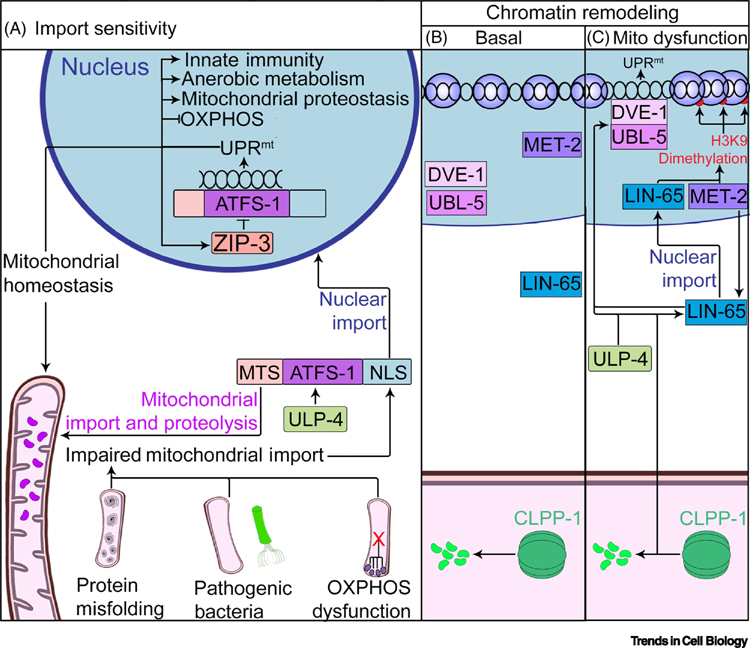
Figure 1:
UPRmt Signaling in Caenorhabditis elegans.
In C. elegans, UPRmt activation is sensitive to the protein import capacity of the mitochondrial network and requires chromatin remodelling. (A) ATFS-1 harbors a mitochondrial targeting sequence (MTS) and a nuclear localization sequence (NLS). The MTS promotes the import and degradation of ATFS-1within healthy mitochondria. However, if mitochondrial import is impaired, which can be caused by OXPHOS dysfunction, or misfolded protein accumulation within the mitochondrial matrix, the NLS of ATFS-1 directs it to the nucleus to activate the UPRmt. This step requires ATFS-1 desumoylation by ULP-4, which enhances its transcriptional activity. The bZIP protein ZIP-3 also serves as a negative regulator of ATFS-1 activation, functioning in a negative feedback loop. (B) Under basal conditions, LIN-65 resides in the cytosol, DVE-1 and UBL-5 exhibit a diffuse nuclear localization, and the methyltransferase MET-2 is inactive. (C) Mitochondrial stress stimulates MET-2 activity which promotes nuclear import of LIN-65, and together these proteins catalyze H3K9 dimethylation and chromatin condensation, in a manner dependent on CLPP-1. Chromatin condensation, CLPP-1, and SUMO cleavage by ULP-4 activity drive redistribution of DVE-1/UBL-5 to the promoters of UPRmt responsive genes.
Interestingly, the MTS of ATFS-1 is relatively weak [31], and this aspect is essential for the nuclear activity of this protein. When the endogenous MTS of ATFS-1 is replaced with the more robust MTS derived from an ATP synthase subunit, worms fail to induce the UPRmt in response to proteotoxic stress. The relative inefficiency of this MTS likely renders ATFS-1 sensitive to mild perturbations to mitochondrial import [32].
ATFS-1 activity within the nucleus is antagonized by a bZIP protein called ZIP-3. ZIP-3 is typically degraded by the proteasome in a manner dependent on the ubiquitin ligase WWP-1. Inhibition of ZIP-3 results in constitutive UPRmt activation under unstressed conditions, while stabilizing ZIP-3 by preventing its ubiquitination hampers induction of the UPRmt during mitochondrial stress. As ATFS-1 promotes the transcription of ZIP-3, this data suggests that ZIP-3 functions in a negative feedback loop that terminates or limits UPRmt activation (Fig 1A) [33].
Harboring a homeobox domain that enables binding to the promoter of hsp-60, DVE-1 has recently been placed downstream of chromatin remodeling driven by mitochondrial dysfunction. This reorganization of chromatin depends on the histone methyltransferase MET-2 and LIN-65. Inhibition of either component diminishes histone H3K9 methylation and blocks UPRmt induction downstream of CLPP-1. Genetic and localization analyses revealed that MET-2 is required for nuclear accumulation of LIN-65. In turn, LIN-65 promotes the redistribution and transcriptional activity of DVE-1 (Fig 1B) [19, 34]. Two histone H3K27 demethylases called JMJD-1.2 and JMJD-3.1 were also identified as essential for UPRmt induction. Notably, overexpression of either of these demethylases elicits a transcriptional response similar to that observed during OXPHOS dysfunction [35]. These data collectively highlight chromatin remodeling as an essential precursor to the DVE-1 mediated transcriptional response to mitochondrial stress.
Although ATFS-1 and DVE-1 appear to function independently, both are regulated by covalent attachment of the small ubiquitin like modifier (SUMO) protein. A peptidase involved in cleaving SUMO moieties, ULP-4, was previously identified as required for the UPRmt [36]. ATFS-1 and DVE-1 are SUMOlyated at K326 and K327, respectively. This modification blocks nuclear import of DVE-1 and accelerates proteolysis of ATFS-1. Mutations that abolish SUMOlyation of these transcription factors enable UPRmt activation even in the absence of ulp-4 [37].
Though work on the UPRmt has focused on the expression levels of mitochondrial chaperones, RNAseq has revealed that these nuclear factors regulate a much broader transcriptional profile. These include enzymes required for anaerobic metabolism [38], proteins involved in cytosolic protein quality control, anti-microbial peptides and innate immune factors that promote clearance of pathogens [15, 33, 35–36]. The UPRmt thus not only safeguards the proteostatic capacity of mitochondria, but also enables cell- and organism-wide adaptation to mitochondrial stress.
Highlighting this concept, research into how mitochondrial dysfunction enhances longevity in C. elegans has revealed that UPRmt signaling proceeds between tissues, not just within the cells experiencing mitochondrial stress. Modest respiratory chain dysfunction early in life leads to delayed development and extended lifespan in C. elegans [39–40]. This phenotype can be reproduced by intestinal or neuronal knockdown of OXPHOS components. Strikingly, neuronal knockdown of OXPHOS components can trigger the UPRmt in the intestine [41]. This cell non-autonomous induction of the UPRmt depends on secretion of the Wnt ligand egl-20 from serotonin-producing neurons, recycling of Wnt cargo receptors by the retromer complex, and neuronal expression of an ortholog of the follicle-stimulating hormone receptor [4, 42–43]. Tissue-specific mitochondrial dysfunction can also trigger a mitochondrial stress response in distal tissues via secretion of Fibroblast Growth Factor 21 (FGF21) in mammals [44–45]. Further study of cell non-autonomous UPRmt signaling in C. elegans will likely shed further light on organismal responses to tissue-specific insults.
The UPRmt in mammals
Considerable evidence from mammalian systems has highlighted the integrated stress response (ISR) as a central element of the UPRmt [46–49]. The ISR is an adaptive translation program regulated by four distinct kinases that are activated by different stressors and that converge on phosphorylation of a single serine residue of the α subunit of the eukaryotic translation initiation factor 2 (eIF2) complex. Phosphorylation of S51 on eIF2α results in reduced global translation alongside the selective synthesis of proteins involved in environmental adaptation. This triaging enables cells to instead focus on survival and quality control processes, or, if faced with overwhelming stress, initiate apoptosis [50]. The ISR is also activated by mitochondrial stress in C. elegans, and this pathway enables worms to survive respiratory dysfunction. However, the ISR is dispensable for induction of the UPRmt in worms [51]. In mammals, however, the ISR acts as an essential precursor to UPRmt activation. Sensing, surviving, and adapting to mitochondrial dysfunction depends on attenuated protein synthesis, privileged translation of transcription factors, and the activity of the eIF2α kinases.
Translational attenuation
Translation initiation depends on the loading of eIF2 with GTP by eIF2B [52–54]. However, phosphorylation of eIF2α converts eIF2 from a substrate into an inhibitor of eIF2B [32–33], leading to reduced global translation (see Box 1, Fig 2A) [37–41]. Although the mechanisms underlying eIF2α phosphorylation have largely been characterized within the context of extreme stress, phosphorylated eIF2α has been observed in most cell types, even in the absence of stress (50). Moreover, mitochondrial dysfunction causes only a mild increase in the abundance of phosphorylated eIF2α (52–54), suggesting that a modest inhibition of protein synthesis may suffice to preserve mitochondrial function, potentially by reducing the load on the mitochondrial-resident chaperones and proteases during stress.
Box 1: Translational control by eIF2α phosphorylation.
During cap-dependent translation, eIF2 assembles into the Ternary Complex (TC) with GTP and methionylated initiator tRNA (Met-tRNAi). The TC then docks onto the 40S ribosome, initiating binding of capped mRNAs and scanning for a start codon. Base-pairing between the Met-tRNAi and an AUG stimulates GTP hydrolysis and dissociation of eIF2, followed by recruitment of the 60S ribosome and initiation of translation. Subsequently, the guanine exchange factor eIF2B catalyzes exchange of GDP for GTP on eIF2, recycling eIF2 for subsequent rounds of translation initiation (Fig 2A) [52–54].
During stress, eIF2α is phosphorylated at serine 51, transforming eIF2 into an inhibitor of eIF2B. When phosphorylated, eIF2 binds to a distinct site on eIF2B, restricting the nucleotide-binding domain of eIF2 from accessing the catalytic GEF domain of eIF2B. This change in association also prevents eIF2B from catalyzing nucleotide exchange even on non-phosphorylated eIF2 complexes [55–56], leading to a depletion of eIF2-GTP and inhibition of cap-dependent translation.
Paradoxically, this depletion of ternary complexes enables translation of transcripts harboring upstream open reading frames (uORFs) that are translated under standard conditions and that inhibit translation of downstream coding sequences [60]. Reduced ternary complex availability decreases the likelihood that a ribosome will acquire eIF2-GTP prior to reaching the uORF start codon. Consequently, this uORF is bypassed and the downstream coding sequence is translated (Fig 2B–C) [53, 61–62]. Transcripts that are selectively translated during the ISR often encode proteins involved in stress-response pathways, including the transcription factors ATF4, ATF5, and CHOP [61–64].
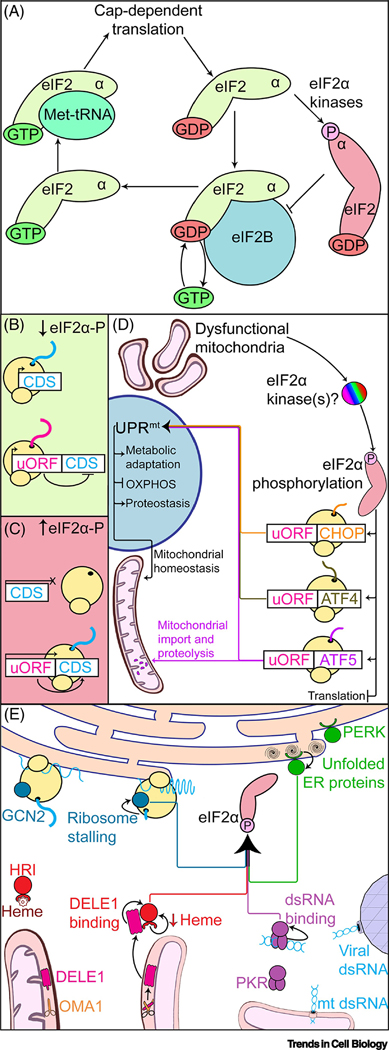
Figure 2.
The Integrated Stress Response (ISR) Is Required for UPRmt Induction in Mammals. (A) GTP-laden eIF2 binds to methionylated initiator tRNAs (Met-tRNA) to initiate cap-dependent translation. Translation initiation stimulates GTP hydrolysis on eIF2, causing the complex to dissociate. eIF2B then catalyzes exchange of GDP for GTP on eIF2, enabling further rounds of translation initiation. Phosphorylation of eIF2α at S51 (eIF2α-P) converts eIF2 into an inhibitor of eIF2B, which suppresses cap-dependent translation. (B) When eIF2α-P levels are low, monocistronic transcripts are translated, as are the uORFs of polycistronic transcripts. Translation of downstream coding sequences (CDS) is inhibited by uORF translation. (C) High eIF2α-P levels reduce cap-dependent translation. Ribosomes scan past uORFs and instead initiate translation at the downstream CDS. (D) Mitochondrial dysfunction stimulates phosphorylation of eIF2α, though it is unclear which kinases are responsible for the diverse stressors that perturb mitochondrial function. eIF2α phosphorylation enables translation of the transcription factors CHOP, ATF4, and ATF5. ATF5 activity is negatively regulated by mitochondrial import into healthy mitochondria. CHOP, ATF4, and ATF5 stimulate transcription of genes that enable recovery from mitochondrial insults. (E) eIF2α is phosphorylated by four different kinases that are activated by different stimuli. Gcn2 associates with ribosomes and is activated by stalled translation. PERK is an ER membrane protein activated by accumulation of unfolded proteins within the ER lumen. HRI is activated in the absence of heme or binding to DELE1, a mitochondrial protein exported to the cytosol upon cleavage by the stress-activated protease OMA1. PKR activity is stimulated upon binding dsRNA, including those generated in the mitochondrial matrix during transcription.
Translation attenuation has been shown to promote mitochondrial activity and shield cells from mitochondrial dysfunction. Translation of TIM17A, an essential component of the TIM23 import channel, is suppressed when eIF2α is phosphorylated in response to ER dysfunction or arsenite exposure. Reduced translation of TIM17A is accompanied by accelerated turnover of this protein, ultimately resulting in diminished presequence-dependent import. This reduction in mitochondrial protein import is likely protective, as knockdown of TIM17A increased resistance of C. elegans and HEK293T cells to the oxidative stressor paraquat [57].
Translational attenuation has emerged as a potential treatment for mitochondrial diseases. This potential strategy was first established in a study on a Saccharomyces cerevisiae model of a neurodegenerative disorder caused by a dominant-negative mutation in the mitochondrial adenine translocase AAC2. Yeast cells expressing this allele exhibited mitochondrial depolarization and reduced lifespan. Strikingly, these phenotypes were suppressed by knocking out genes required for high translational activity or by treating cells with low concentrations of cycloheximide, an inhibitor of cytosolic translation [58]. This phenomenon appears conserved, as cycloheximide treatment also improved respiration and suppressed autophagy in human fibroblast and podocyte models of respiratory chain dysfunction [59]. Reducing translation may thus represent a potential therapeutic intervention for a broad spectrum of mitochondrial disorders.
Privileged translation during the ISR
Although eIF2α phosphorylation leads to reduced global protein synthesis, some mRNAs with upstream open reading frames (uORFs) are selectively translated during the ISR [60]. Ribosomes typically translate the first start codon they encounter. Translation of a uORF generally suppresses that of downstream coding sequences (Fig 2B). Phosphorylation of eIF2α reduces the availability of ternary complexes and decreases the likelihood that a ribosome will reacquire eIF2-GTP prior to reaching a uORF start codon. Consequently, these uORFs are bypassed during the ISR, while downstream coding sequences are subjected to privileged translation (Fig 2C) [53, 61–62].
The transcription factors ATF4, ATF5, and CHOP (C/EBP Homology Protein) all harbor uORFs and require eIF2α phosphorylation for their translation [61–64]. Each of these transcription factors has been characterized within the context of ER stress responses [61, 65–68]. However, they have also emerged as essential mediators of a response to mitochondrial dysfunction with similarities to the UPRmt in C. elegans (Fig 2D). CHOP was the first factor implicated in mediating the UPRmt. Treatment of rat liver cells with mitochondrial-specific stressors resulted in increased expression of mitochondrial chaperones and proteases without affecting expression levels of chaperones localized to the endoplasmic reticulum (ER) or cytosol [13–14]. Elevated mitochondrial chaperone expression coincided with induction of CHOP. Moreover, CHOP-binding elements were identified in the promoters of these mitochondrial chaperones. However, other conserved sequences were also observed in these promoters, suggesting that CHOP does not act alone in inducing the UPRmt [14, 69].
Global transcriptomic analyses have validated the presence of CHOP-binding elements in many of the promoters of genes induced by mitochondrial dysfunction, while also revealing abundant ATF4-binding motifs [47–48]. HeLa cells lacking a functional copy of ATF4 failed to upregulate several mitochondrial enzymes and exhibited a reduction in ATP-dependent respiration. Gene expression analysis of human and mouse tissues also revealed a tight correlation between ATF4 induction and UPRmt- responsive genes [48].
A targeted screening approach revealed a role for ATF5 in mounting a transcriptional response to mitochondrial dysfunction. The ability of worms lacking ATFS-1 to induce hsp60pr::GFP expression in response to mitochondrial dysfunction was rescued by ATF5 expression, but not ATF4. Notably, knockdown of ATF5 in HEK293 cells dampened induction of UPRmt responsive genes and reduced proliferation of cells subjected to mitochondrial stress. When overexpressed in HeLa cells, ATF5 colocalizes with mitochondria and the nucleus, while endogenously expressed ATF5 from mouse liver extracts cofractionates with mitochondrial membranes. Collectively, these results suggest that the transcriptional activity of ATF5 is regulated by mitochondrial import, analogous to ATFS-1 [46].
Although each of these transcription factors has been implicated in promoting transcription of genes required to overcome mitochondrial dysfunction, it is unclear if they function individually or in concert. Research on the role of these transcription factors in responding to ER and cytosolic stress demonstrates that they regulate the expression of one another. Genetic evidence indicates that during ER stress, ATF4 operates upstream of CHOP and ATF5, while expression of ATF5 depends on both CHOP and ATF4 [65–67].
Interestingly, recent evidence indicates that activation of these transcription factors proceeds in a distinct sequence during mitochondrial stress. Elevated transcript levels of ATF4 and ATF5 are detected in the skeletal muscle of 22-month old mice harboring a mutation in the mitochondrial DNA helicase that culminates in late-onset respiratory chain dysfunction [22]. Increased ATF5 mRNA can be detected up to 6 months beforehand, before any apparent induction of ATF4 can be detected [44]. Transcript levels of the FGF21, a prominent hormone involved in glucose uptake secreted by the liver when the ISR is triggered [45], also rise alongside ATF5. Knockout of FGF21 suppresses induction of ATF4, but not ATF5, and rescues neuronal respiratory chain dysfunction in these mutant mice [44]. These data suggest ATF5 acts upstream of ATF4 during mitochondrial stress, a reverse of what is observed during ER stress. Clarifying the sequence in which these proteins act will be essential to better understanding the transcriptional response to mitochondrial dysfunction.
The eIF2α kinases
Four different kinases are capable of phosphorylating eIF2α to promote the ISR [70]. These kinases share a conserved catalytic core flanked by different accessory domains that enable activation of each kinase in response to divergent environmental and physiological cues (Fig 2E) [71]. Notably, each of these kinases has been implicated in sensing and responding to cues related to mitochondrial dysfunction.
PERK
PKR-like ER kinase (PERK) localizes to the ER and is activated when unfolded proteins accumulate within this organelle (see Box 2) [72–73]. Despite being restricted to the ER, PERK has also been implicated in modulating mitochondrial function, particularly during stress induced by reactive oxygen species (ROS). HEK293 cells treated with menadione, a compound that stimulates ROS production within mitochondria, showed an increase in eIF2α phosphorylation, but this phosphorylation was blocked when cells were cotreated with a chemical inhibitor of PERK [49]. MEFs subjected to photooxidative stress exhibited PERK-dependent release of cytochrome C, dissipation of mitochondrial membrane potential, and an increase in the number of contacts between the mitochondria and ER. These latter two phenotypes can be rescued with a kinase dead allele of PERK, suggesting an eIF2α-independent role for PERK in mediating signaling between the ER and mitochondria [74].
Box 2: The eIF2α kinases.
The ISR is regulated by four different kinases that have modest steady-state activity and are activated in response to distinct environmental and physiological cues (Fig 2E). The ISR kinases are PERK, PKR, HRI, and GCN2.
PERK is a type I integral membrane protein that localizes to the ER. The kinase domain of PERK faces the cytosol, while its regulatory domain resides within the lumen of the ER. Binding of unfolded proteins to the luminal domain stimulates PERK activity, ultimately resulting in phosphorylation of eIF2α [72–73]. PERK has been implicated in activating the UPRmt [49, 74] during oxidative stress and in adapting mitochondrial function to ER stress [49,74–75].
GCN2 phosphorylates eIF2α during amino acid limitation, enabling cells to survive extended periods of starvation. Historically, Gcn2 was thought to become activated through interactions with uncharged tRNAs, which accumulate during starvation [50, 76]. However, emerging evidence suggests that interactions with tRNAs may not represent the primary mechanism underlying Gcn2 activation. Gcn2 was found to phosphorylate eIF2α in a mouse model of neurodegeneration caused by ribosome stalling, despite the apparent absence of an increase in uncharged tRNA levels [77]. Furthermore, a recent study demonstrated that Gcn2 is far more potently activated in vitro by ribosomes than by tRNAs. This activation stems from interactions between Gcn2 and the ribosomal P-stalk, a complex that promotes translation elongation [78]. Collectively, these results suggest that Gcn2 is activated by P-stalks derived from stalled ribosomes. GCN2 is activated during mitochondrial dysfunction in worms and mammals [51, 79–82].
PKR plays an essential role in the response to viral infection. Binding of PKR to viral double-stranded RNAs stimulate autophosphorylation and activation of PKR, resulting in phosphorylation of eIF2α, attenuated production of proteins required for viral replication, and induction of apoptosis [84–85]. PKR has also been implicated in driving cell cycle progression and responding to metabolic, oxidative, and ER stress [86–88]. In these circumstances, PKR activation can be stimulated through interactions with either endogenous mRNAs or with an accessory protein called PACT [86–90]. The most common binding partner for PKR in uninfected cells is dsRNA derived from mitochondria [91]. PKR has also been implicated in UPRmt activation in a murine IBD model [95].
HRI plays a central role in erythropoiesis by coordinating translational activity with the availability of heme, the prosthetic group of hemoglobin [96]. Heme biosynthesis proceeds between the cytosol and mitochondria, culminating with the incorporation of iron into a heme precursor within the mitochondrial matrix [97]. Heme binds to and inhibits the kinase activity of HRI, terminating ISR induction and enabling translation of globin chains [96, 98–99]. Protein misfolding in the cytosol can also activate HRI [100]. HRI also regulates mitochondrial activity during erythropoiesis [101]. However, HRI can also be activated by binding to DELE1 even in the presence of heme, a mitochondrial protein that translocates to the cytosol when cleaved by the stress-activated protease OMA1 [102–103].
Another recent study demonstrated that metabolic adaptation of U2OS cells to media without glucose depends on signaling by PERK. Metabolites required for secretory protein glycosylation become quickly depleted after cells are shifted to glucose, triggering protein misfolding within the ER and activation of PERK. PERK subsequently phosphorylates eIF2α, triggering translation of ATF4, which activates transcription of Supercomplex Assembly Factor 1 (SCAF1). SCAF1 promotes the assembly of respiratory chain supercomplexes, enabling cells to respire and proliferate after being shifted from glucose to galactose [75]. This work suggests that PERK optimizes the capacity of the mitochondria to meet the metabolic demands of the ER.
GCN2
Gcn2 associates with ribosomes and phosphorylates eIF2α during amino acid limitation. Historically, Gcn2 was thought to become activated through interactions with uncharged tRNAs [50, 76]. However, recent biochemical, structural, and murine genetic evidence suggests that ribosomal stalling mediates Gcn2 activation (See Box 2) [77–78].
The C. elegans ortholog of Gcn2 plays a protective role during mitochondrial dysfunction. A genetic screen for kinases and phosphatases that modulate induction of hsp-60pr::gfp during mitochondrial stress revealed that knockdown of gcn-2 enhances expression of this transgene, while knockdown of the ortholog of PERK had no effect. Moreover, gcn-2 knockdown in worms subjected to mitochondrial stress resulted in developmental delay, reduced oxygen consumption, diminished motility, and caused mitochondrial fragmentation [51]. In contrast to mammalian systems wherein ATF4 and ATF5 act downstream of ISR induction [47–48], gcn-2 and atfs-1 function independent of one another, as worms harboring mutations in both genes exhibit more severe developmental defects than when only one of the genes is mutated [51].
Gcn2 has also been identified as a potential mediator of the mitochondrial stress responses in mammals. The ISR is induced in HeLa cells treated with doxycycline, an inhibitor of mitochondrial translation. Phosphorylation of eIF2α in doxycycline-treated cells can be suppressed with siRNA against Gcn2, but not with siRNA against any of the other eIF2α kinases [79]. Elevated Gcn2 activity has also been observed in gastrointestinal cancer cells treated with the mitochondrial ATPase inhibitor oligomycin [80]. Treatment of these cells with oligomycin stimulated a metabolic shift from oxidative phosphorylation to glycolysis, increased resistance to the chemotherapeutic cisplatin, and enhanced proliferation when implanted into nude mice [80–82]. Notably, knockdown of Gcn2, but not PERK, suppressed phosphorylation of eIF2α and rendered these cancer cells sensitive to cisplatin, even after an oligomycin pretreatment [82]. Given that mitochondrial stress is associated with tumor progression, metastasis, and adverse patient outcomes, these results highlight a potential role for Gcn2 in cancer pathogenesis [24, 83].
PKR
Protein Kinase R (PKR) is canonically activated by binding to double-stranded RNA, enabling cells to sense and respond to viral infection [84–85]. However, PKR also contributes to stress responses in uninfected cells. In these scenarios, PKR activation proceeds through interactions with endogenous RNAs or proteins (Box 2) [86–90].
Surprisingly, a recent report demonstrated that PKR can frequently be found in complex with mitochondrial RNA [91]. The entire mitochondrial genome is transcribed as a polycistronic transcript that is subsequently processed into individual mRNAs. Transcription can proceed from both the heavy and light strand of the mitochondrial genome, but those synthesized from the light strand are generally rapidly degraded [92]. However, Kim and coworkers demonstrate that mtRNAs derived from the light strand can evade destruction and base-pair with mitochondrial transcripts from the heavy strand. Knockdown of the mitochondrial RNA polymerase suppressed PKR activation by okadaic acid or staurosporine, and treatment with the latter compound resulted in accumulation of mtRNAs in the cytosol [91]. Coupled with recent reports on the roles of cytosolic mtDNA in driving inflammation and apoptosis, these results highlight an emerging connection between the release of nucleic acids from the mitochondria and induction of stress signaling pathways [93–94].
PKR signaling has been implicated in mediating the mitochondrial unfolded protein response during intestinal inflammation both in cultured cells and in vivo. Overexpression of a terminally misfolded mitochondrial protein in a mouse intestinal epithelial cell line resulted in eIF2α phosphorylation and increased expression of Hsp60. These phenotypes could be blocked by knocking down PKR with siRNA or with a chemical inhibitor of PKR. Moreover, Hsp60 induction was also observed in the intestine of a mouse model of inflammatory bowel disease (IBD), and elevated Hsp60 levels and PKR activity was also observed in intestinal epithelial cells isolated from human patients with IBD. Hsp60 induction and intestinal inflammation were both suppressed in PKR −/− mice, suggesting that PKR-driven mitochondrial stress signaling contributes to the pathology of IBD [95].
HRI
The heme regulated eIF2α kinase (HRI) phosphorylates eIF2α in erythroblasts in the absence of heme, an essential hemoglobin prosthetic group that harbors an iron ion. Binding of heme inhibits the kinase activity of HRI, enabling translation to proceed again (Box 2) [96–98]. Mice lacking HRI do not exhibit any gross defects under basal conditions, although reticulocytes derived from such animals show attenuated eIF2α phosphorylation and elevated translation rates. However, HRI −/− mice fed an iron-deficient diet exhibit an accumulation of protein aggregates within reticulocytes, elevated apoptosis of late-stage erythroblasts, and anemia. Thus, HRI regulates erythropoiesis by coordinating translation of globin chains with the availability of cofactors required for hemoglobin folding and assembly [96, 99]. Interestingly, a recent report demonstrated that HRI can also be activated by an accumulation of misfolded proteins in the cytosol and that HRI activation stimulates expression of cytosolic chaperones and proteases, suggesting that HRI controls a cytosolic UPR [100–101].
In addition to controlling cytosolic translation, a recent report uncovered a role for HRI in regulating translation within the mitochondria during erythropoiesis. Ribosome profiling experiments revealed enhanced translation of components of the mitochondrial ribosome and respiratory chain in erythroid cells derived from iron-starved HRI −/− mice. Protein translation was elevated in these cells even in the presence of cycloheximide, but could be blocked with both cycloheximide and chloramphenicol, which inhibit cytoplasmic and mitochondrial translation, respectively. Despite increased translation of respiratory chain components within the mitochondria, HRI −/− erythroid cells exhibited reduced respiratory activity and an increase in mitochondrial mass when deprived of iron. HRI thus regulates mitochondrial biogenesis, translation, and activity during erythroblast differentiation [101].
Two recent papers revealed a direct mechanism linking mitochondrial dysfunction to HRI activation. Both groups performed independent genome-wide screens to identify factors required for ISR induction in cultured cells exposed to mitochondrial toxins, as monitored by translation of a fluorescent protein fused to the uORFs of either CHOP or ATF4. In both studies, ISR induction required HRI, the mitochondrial protease OMA1, and a relatively unstudied mitochondrial protein called DELE1. Genetic and biochemical analyses revealed that mitochondrial stress triggers cleavage of DELE1 by OMA1. Cleaved DELE1 traffics to the cytosol where it binds to and activates HRI (Figure 2E). Of note, mitochondrial dysfunction stimulates HRI even in the presence of heme. Importantly, expression of the cleaved form of DELE1 was sufficient to stimulate HRI-dependent eIF2α phosphorylation independent of mitochondrial stress. Combined, both papers demonstrate a direct signal transduction pathway through which mitochondrial stress leads to phosphorylation of eIF2α in the cytosol to activate the ISR [102–103].
Concluding Remarks
Research in C. elegans has established that UPRmt signaling contributes to an expansive set of physiological functions. This work has also demonstrated that mitochondrial import capacity and chromatin remodeling represent central regulators of UPRmt. The essentiality of the ISR stands as a notable divergence between UPRmt signaling in C. elegans and mammals. ISR induction improves mitochondrial function in worms but is not essential for UPRmt activation. By contrast, the ISR is inextricable from the UPRmt in mammals. Translational attenuation driven by the ISR protects cells from mitochondrial dysfunction. Moreover, the ISR stimulates uORF-regulated translation of CHOP, ATF4, and ATF5. These proteins subsequently activate a transcriptional program that rewires cellular metabolism and enhances mitochondrial proteostasis.
Despite these advances, our understanding of the relationship between mitochondrial stress signaling and the ISR remains limited (see Outstanding Questions). Though HRI can trigger ISR signaling during mitochondrial dysfunction, each of the other eIF2α kinases have also been implicated in maintaining mitochondrial homeostasis. It will be essential to determine which mitochondrial insults each of these kinases responds to. Downstream of these kinases, it is unclear whether CHOP, ATF4, and ATF5 are subject to post-translational regulation akin to how ATFS-1 activity is regulated by mitochondrial import. Global translational attenuation has been proposed to enhance proteostasis within the cytosol, ER, and mitochondria. However, it has not been explicitly addressed whether global translational attenuation enables mitochondrial recovery during the ISR. We anticipate that further exploring the connections between mitochondrial homeostasis and translational regulation will enable the development of new therapeutic strategies to target the slew of pathologies associated with mitochondrial dysfunction.
Outstanding Questions.
The ISR and UPRmt operate independently to preserve mitochondrial network integrity in C. elegans, while the UPRmt and ISR are intertwined in mammals. What is the evolutionary advantage of integrating the UPRmt into the ISR?
Do the eIF2α kinases enhance UPRmt induction in response to discrete mitochondrial insults, or is there overlap in conditions that lead to their activation?
Does the integrity of the mitochondrial network influence the activity of CHOP, ATF4, or ATF5 following ISR-dependent synthesis?
Do CHOP, ATF4, and ATF5 act alone or in concert?
What is the role of the ATFS-1 and ATF5 that accumulates within mitochondria?
Does translational attenuation represent an essential component of the UPRmt, and if so, how does reduced translation impact mitochondrial function?
Highlights.
The UPRmt in C. elegans is a transcriptional program that requires chromatin remodeling and the transcription factor ATFS-1 which, is regulated by mitochondrial protein import efficiency.
The UPRmt in C. elegans rewires cellular metabolism, contributes to innate immunity, mediates signaling between tissues, and regulates development and organismal aging. In mammals, the UPRmt is embedded within the integrated stress response (ISR), a program that enables selective translation of transcripts harboring upstream open reading frames (uORFs).
In mammals, UPRmt induction is mediated by the transcription factors CHOP, ATF4, and a functional ortholog of ATFS-1, ATF5.
The kinases responsible for activating the ISR have all been implicated in maintaining mitochondrial homeostasis or responding to mitochondrial dysfunction.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sloan DB, et al. (2018). Cytonuclear integration and co-evolution. Nature Reviews Genetics 19, 635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spinelli JB, and Haigis MC (2018). The multifaceted contributions of mitochondria to cellular metabolism. Nature Cell Biology 20, 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chandel NS (2015). Evolution of Mitochondria as Signaling Organelles. Cell Metab 22, 204–206. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Q, et al. (2018). The Mitochondrial Unfolded Protein Response Is Mediated Cell-Non-autonomously by Retromer-Dependent Wnt Signaling. Cell 174, 870–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilkins HM, et al. (2017). Mitochondria-Derived Damage-Associated Molecular Patterns in Neurodegeneration. Front Immunol 8, 508–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dall KB, and Færgeman NJ (2019). Metabolic regulation of lifespan from a C. elegans perspective. Genes Nutr 14, 25–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pickles S, et al. (2018). Mitophagy and Quality Control Mechanisms in Mitochondrial Maintenance. Current Biology 28, R170–R185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfanner N, et al. (2019). Mitochondrial proteins: from biogenesis to functional networks. Nature Reviews Molecular Cell Biology 20, 267–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng MY, et al. (1989). Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature 337, 620–625. [DOI] [PubMed] [Google Scholar]
- 10.Leustek T, et al. (1989). A member of the Hsp70 family is localized in mitochondria and resembles Escherichia coli DnaK. Proceedings of the National Academy of Sciences of the United States of America 86, 7805–7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang PJ, et al. (1990). Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature 348, 137–143. [DOI] [PubMed] [Google Scholar]
- 12.Bolliger L, et al. . (1994). A mitochondrial homolog of bacterial GrpE interacts with mitochondrial hsp70 and is essential for viability. The EMBO Journal 13, 1998–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinus RD, et al. (1996). Selective induction of mitochondrial chaperones in response to loss of the mitochondrial genome. European journal of biochemistry 240, 98–103. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Q, et al. (2002). A mitochondrial specific stress response in mammalian cells. The EMBO journal 21, 4411–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nargund AM, et al. (2012). Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 337, 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pellegrino MW, et al. (2014). Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature 516, 414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohrin M, et al. (2015). Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science (New York, NY) 347, 1374–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Houtkooper RH, et al. (2013). Mitonuclear protein imbalance as a conserved longevity mechanism. Nature 497, 451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang YT, et al. (2019). Cardioprotection by the mitochondrial unfolded protein response requires ATF5. Am J Physiol Heart Circ Physiol 317, H472–h478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sorrentino V, et al. (2017). Enhancing mitochondrial proteostasis reduces amyloid-beta proteotoxicity. Nature 552, 187–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin YF, et al. (2016). Maintenance and propagation of a deleterious mitochondrial genome by the mitochondrial unfolded protein response. Nature 533, 416–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khan NA, et al. (2017). mTORC1 Regulates Mitochondrial Integrated Stress Response and Mitochondrial Myopathy Progression. Cell Metab 26, 419–428.e415. [DOI] [PubMed] [Google Scholar]
- 23.Nicolas E, et al. (2019). Disease-Associated Genetic Variation in Human Mitochondrial Protein Import. American Journal of Human Genetics 104, 784–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kenny TC, et al. (2019). Mitohormesis Primes Tumor Invasion and Metastasis. Cell reports 27, 2292–2303.e2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roth KG, et al. (2019). The Mitochondrion as an Emerging Therapeutic Target in Cancer. Trends in Molecular Medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez BA, et al. (2017). Dysregulation of the Mitochondrial Unfolded Protein Response Induces Non-Apoptotic Dopaminergic Neurodegeneration in C. elegans Models of Parkinson’s Disease. J Neurosci 37, 11085–11100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haynes CM, et al. (2013). Evaluating and responding to mitochondrial dysfunction: the mitochondrial unfolded-protein response and beyond. Trends in cell biology 23, 311–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoneda T, et al. (2004). Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. Journal of Cell Science 117, 4055–4066. [DOI] [PubMed] [Google Scholar]
- 29.Haynes CM, et al. (2007). ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Developmental Cell 13, 467–480. [DOI] [PubMed] [Google Scholar]
- 30.Haynes CM, et al. (2010). The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Molecular Cell 37, 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melber A, and Haynes CM (2018). UPR(mt) regulation and output: a stress response mediated by mitochondrial-nuclear communication. Cell Res 28, 281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rolland SG, et al. (2019). Compromised Mitochondrial Protein Import Acts as a Signal for UPR(mt). Cell reports 28, 1659–1669.e1655. [DOI] [PubMed] [Google Scholar]
- 33.Deng P, et al. (2019). Mitochondrial UPR repression during Pseudomonas aeruginosa infection requires the bZIP protein ZIP-3. Proceedings of the National Academy of Sciences of the United States of America 116, 6146–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian Y, et al. (2016). Mitochondrial Stress Induces Chromatin Reorganization to Promote Longevity and UPR(mt). Cell 165, 1197–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merkwirth C, et al. (2016). Two Conserved Histone Demethylases Regulate Mitochondrial Stress-Induced Longevity. Cell 165, 1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y, et al. (2014). Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature 508, 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao K, et al. (2019). SUMO peptidase ULP-4 regulates mitochondrial UPR-mediated innate immunity and lifespan extension. eLife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nargund AM, et al. (2015). Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPR(mt). Molecular Cell 58, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dillin A, et al. (2002). Rates of behavior and aging specified by mitochondrial function during development. Science (New York, NY) 298, 2398–2401. [DOI] [PubMed] [Google Scholar]
- 40.Moehle EA, et al. (2019). Mitochondrial proteostasis in the context of cellular and organismal health and aging. The Journal of biological chemistry 294, 5396–5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Durieux J, et al. (2011). The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell 144, 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berendzen KM, et al. (2016). Neuroendocrine Coordination of Mitochondrial Stress Signaling and Proteostasis. Cell 166, 1553–1563.e1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim S, and Sieburth D (2019). FSHR-1/GPCR Regulates the Mitochondrial Unfolded Protein Response in Caenorhabditis elegans. Genetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forsstrom S, et al. (2019). Fibroblast Growth Factor 21 Drives Dynamics of Local and Systemic Stress Responses in Mitochondrial Myopathy with mtDNA Deletions. Cell Metab 30, 1040–1054.e1047. [DOI] [PubMed] [Google Scholar]
- 45.Potthoff MJ (2017). FGF21 and metabolic disease in 2016: A new frontier in FGF21 biology. Nat Rev Endocrinol 13, 74–76. [DOI] [PubMed] [Google Scholar]
- 46.Fiorese CJ, et al. (2016). The Transcription Factor ATF5 Mediates a Mammalian Mitochondrial UPR. Current biology : CB 26, 2037–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Munch C, and Harper JW (2016). Mitochondrial unfolded protein response controls matrix pre-RNA processing and translation. Nature 534, 710–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quiros PM, et al. (2017). Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. The Journal of cell biology 216, 2027–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Samluk L, et al. (2019). Cytosolic translational responses differ under conditions of severe short-term and long-term mitochondrial stress. Molecular biology of the cell 30, 1864–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pakos-Zebrucka K, et al. (2016). The integrated stress response. EMBO Rep 17, 1374–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baker BM, et al. (2012). Protective coupling of mitochondrial function and protein synthesis via the eIF2alpha kinase GCN-2. PLoS Genet 8, e1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hinnebusch AG, et al. (2016). Translational control by 5’-untranslated regions of eukaryotic mRNAs. Science (New York, NY) 352, 1413–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Young SK, and Wek RC (2016). Upstream Open Reading Frames Differentially Regulate Gene-specific Translation in the Integrated Stress Response. The Journal of biological chemistry 291, 16927–16935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang H, et al. (2019). Function and Evolution of Upstream ORFs in Eukaryotes. Trends in biochemical sciences 44, 782–794. [DOI] [PubMed] [Google Scholar]
- 55.Kashiwagi K, et al. (2019). Structural basis for eIF2B inhibition in integrated stress response. Science (New York, NY) 364, 495–499. [DOI] [PubMed] [Google Scholar]
- 56.Kenner LR, et al. (2019). eIF2B-catalyzed nucleotide exchange and phosphoregulation by the integrated stress response. Science (New York, NY) 364, 491–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rainbolt TK, et al. (2013). Stress-regulated translational attenuation adapts mitochondrial protein import through Tim17A degradation. Cell Metab 18, 908–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang X, et al. (2008). Reduced cytosolic protein synthesis suppresses mitochondrial degeneration. Nature cell biology 10, 1090–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peng M, et al. (2015). Inhibiting cytosolic translation and autophagy improves health in mitochondrial disease. Human molecular genetics 24, 4829–4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andreev DE, et al. (2015). Translation of 5’ leaders is pervasive in genes resistant to eIF2 repression. eLife 4, e03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vattem KM, and Wek RC (2004). Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America 101, 11269–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Palam LR, et al. . (2011). Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. The Journal of biological chemistry 286, 10939–10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jousse C, et al. (2001). Inhibition of CHOP translation by a peptide encoded by an open reading frame localized in the chop 5’UTR. Nucleic acids research 29, 4341–4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Watatani Y, et al. (2008). Stress-induced translation of ATF5 mRNA is regulated by the 5’-untranslated region. The Journal of biological chemistry 283, 2543–2553. [DOI] [PubMed] [Google Scholar]
- 65.Zhou D, et al. (2008). Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. The Journal of biological chemistry 283, 7064–7073. [DOI] [PubMed] [Google Scholar]
- 66.Teske BF, et al. (2013). CHOP induces activating transcription factor 5 (ATF5) to trigger apoptosis in response to perturbations in protein homeostasis. Molecular biology of the cell 24, 2477–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Juliana CA, et al. (2017). ATF5 regulates beta-cell survival during stress. Proceedings of the National Academy of Sciences of the United States of America 114, 1341–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang Y, et al. (2017). Transcription Factor C/EBP Homologous Protein in Health and Diseases. Front Immunol 8, 1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aldridge JE, et al. (2007). Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements. PLoS One 2, e874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Taniuchi S, et al. (2016). Integrated stress response of vertebrates is regulated by four eIF2alpha kinases. Scientific reports 6, 32886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Donnelly N, et al. (2013). The eIF2alpha kinases: their structures and functions. Cell Mol Life Sci 70, 3493–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harding HP, et al. (1999). Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397, 271–274. [DOI] [PubMed] [Google Scholar]
- 73.Marciniak SJ, et al. (2006). Activation-dependent substrate recruitment by the eukaryotic translation initiation factor 2 kinase PERK. The Journal of cell biology 172, 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Verfaillie T, et al. (2012). PERK is required at the ER-mitochondrial contact sites to convey apoptosis after ROS-based ER stress. Cell Death Differ 19, 1880–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Balsa E, et al. (2019). ER and Nutrient Stress Promote Assembly of Respiratory Chain Supercomplexes through the PERK-eIF2alpha Axis. Molecular cell 74, 877–890.e876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vazquez de Aldana CR, et al. (1994). Multicopy tRNA genes functionally suppress mutations in yeast eIF-2 alpha kinase GCN2: evidence for separate pathways coupling GCN4 expression to unchanged tRNA. Molecular and cellular biology 14, 7920–7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ishimura R, et al. (2016). Activation of GCN2 kinase by ribosome stalling links translation elongation with translation initiation. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Inglis AJ, et al. (2019). Activation of GCN2 by the ribosomal P-stalk. Proceedings of the National Academy of Sciences of the United States of America 116, 4946–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Michel S, et al. (2015). Inhibition of mitochondrial genome expression triggers the activation of CHOP-10 by a cell signaling dependent on the integrated stress response but not the mitochondrial unfolded protein response. Mitochondrion 21, 58–68. [DOI] [PubMed] [Google Scholar]
- 80.Martinez-Reyes I, et al. (2012). AMPK and GCN2-ATF4 signal the repression of mitochondria in colon cancer cells. Biochem J 444, 249–259. [DOI] [PubMed] [Google Scholar]
- 81.Sanchez-Arago M, et al. (2010). Selection of cancer cells with repressed mitochondria triggers colon cancer progression. Carcinogenesis 31, 567–576. [DOI] [PubMed] [Google Scholar]
- 82.Wang SF, et al. (2016). Mitochondrial dysfunction enhances cisplatin resistance in human gastric cancer cells via the ROS-activated GCN2-eIF2alpha-ATF4-xCT pathway. Oncotarget 7, 74132–74151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Deng P, and Haynes CM (2017). Mitochondrial dysfunction in cancer: Potential roles of ATF5 and the mitochondrial UPR. Semin Cancer Biol 47, 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dey M, et al. (2005). Mechanistic link between PKR dimerization, autophosphorylation, and eIF2alpha substrate recognition. Cell 122, 901–913. [DOI] [PubMed] [Google Scholar]
- 85.Gal-Ben-Ari S, et al. (2018). PKR: A Kinase to Remember. Front Mol Neurosci 11, 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Patel CV, et al. (2000). PACT, a stress-modulated cellular activator of interferon-induced double-stranded RNA-activated protein kinase, PKR. The Journal of biological chemistry 275, 37993–37998. [DOI] [PubMed] [Google Scholar]
- 87.Nakamura T, et al. (2010). Double-stranded RNA-dependent protein kinase links pathogen sensing with stress and metabolic homeostasis. Cell 140, 338–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim Y, et al. (2014). PKR is activated by cellular dsRNAs during mitosis and acts as a mitotic regulator. Genes Dev 28, 1310–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li S, et al. (2006). Molecular basis for PKR activation by PACT or dsRNA. Proceedings of the National Academy of Sciences of the United States of America 103, 10005–10010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Youssef OA, et al. (2015). Potential role for snoRNAs in PKR activation during metabolic stress. Proceedings of the National Academy of Sciences of the United States of America 112, 5023–5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim Y, et al. (2018). PKR Senses Nuclear and Mitochondrial Signals by Interacting with Endogenous Double-Stranded RNAs. Molecular cell 71, 1051–1063.e1056. [DOI] [PubMed] [Google Scholar]
- 92.D’Souza AR, and Minczuk M (2018). Mitochondrial transcription and translation: overview. Essays Biochem 62, 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sliter DA, et al. (2018). Parkin and PINK1 mitigate STING-induced inflammation. Nature 561, 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McArthur K, et al. (2018). BAK/BAX macropores facilitate mitochondrial herniation and mtDNA efflux during apoptosis. Science (New York, NY) 359. [DOI] [PubMed] [Google Scholar]
- 95.Rath E, et al. (2012). Induction of dsRNA-activated protein kinase links mitochondrial unfolded protein response to the pathogenesis of intestinal inflammation. Gut 61, 1269–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen JJ, and Zhang S (2019). Heme-regulated eIF2alpha kinase in erythropoiesis and hemoglobinopathies. Blood 134, 1697–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Severance S, and Hamza I (2009). Trafficking of heme and porphyrins in metazoa. Chem Rev 109, 4596–4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rafie-Kolpin M, et al. (2000). Two heme-binding domains of heme-regulated eukaryotic initiation factor-2alpha kinase. N terminus and kinase insertion. The Journal of biological chemistry 275, 5171–5178. [DOI] [PubMed] [Google Scholar]
- 99.Han AP, et al. (2001). Heme-regulated eIF2alpha kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. The EMBO journal 20, 6909–6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Abdel-Nour M, et al. (2019). The heme-regulated inhibitor is a cytosolic sensor of protein misfolding that controls innate immune signaling. Science (New York, NY) 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang S, et al. (2019). HRI coordinates translation necessary for protein homeostasis and mitochondrial function in erythropoiesis. eLife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fessler E, et al. (2020). Haploid genetic stress profiling identifies DELE1 as the spearhead of a pathway that relays mitochondrial perturbation to the cytosol. Nature Published online March 4 2020 10.1038/s41586-020-2076-4 [DOI] [Google Scholar]
- 103.Guo X, et al. (2020) Mitochondrial dysfunction triggers the integrated stress response through OMA1, DELE1 and HRI. Nature Published online March 4, 2020 10.1038/s41586-020-2078-2 [DOI] [Google Scholar]