Summary
Recombinant HIV-1 envelope glycoproteins (Env) of ever increasing sophistication have been evaluated as vaccine candidates for over 30 years. Structurally defined mimics of native trimeric Env (e.g., SOSIP trimers) present multiple epitopes for broadly neutralizing Abs (bNAbs) and their germline precursors, but elicitation of bNAbs remains elusive. Here, we argue that the interactions of Env with the immune system render it exceptional among viral vaccine antigens and hinder its immunogenicity in absolute and comparative terms. In other words, Env binds to CD4 on key immune cells and transduces signals that may compromise their function. Moreover, the extensive array of oligomannose glycans on Env shields peptidic B-cell epitopes, impedes the presentation of T-helper cell epitopes, and attracts mannose binding proteins, which could affect the Ab response. We suggest lines of research for assessing how to overcome obstacles the exceptional features of Env impose to the creation of a successful HIV-1 vaccine.
Keywords: HIV-1, envelope glycoprotein, neutralizing Abs, vaccine, adaptive immunity, innate immunity, HIV-1 Env trimers, mannose glycans, CD4, nanoparticles
Introduction
The recent abandonment, due to futility, of the latest HIV-1 vaccine efficacy trial, HVTN-702, increases the emphasis on immunization strategies for inducing broadly neutralizing Abs (bNAbs) (Cohen, 2020). The HIV-1 Envelope glycoproteins (Env) are the basis of many vaccine development strategies, particularly those for eliciting bNAbs. That statement has been true for around 35 years, and yet we still lack Env-based immunogens that elicit Abs with the breadth and potency sufficient to counter global HIV-1 diversity (Escolano et al., 2017; Kwong and Mascola, 2018). Our collective failure is certainly not for want of effort. Sophisticated and, at times, very well-funded immunogen design programs harness knowledge of Env and bNAb structure, and of the ontogeny of bNAbs and their co-evolution with Env in vivo (Bonsignori et al., 2017; McGuire, 2019; Sok and Burton, 2018; Ward and Wilson, 2017).
Many factors render bNAb induction difficult, including the multiple defenses HIV-1 has evolved to counter humoral immunity. For example, relatively conserved Env regions that would bind Abs are well protected by a dense array of glycans, while other vulnerable sites become exposed only transiently when receptor-triggered conformational changes drive Env-mediated fusion of the virus and host cell membranes (Crispin et al, 2018; Klasse, 2012, West et al., 2014). The tolerance of Env to sequence changes without loss of function facilitates rapid escape from bNAbs that emerge in or are administered to HIV-1-infected people clinically (Escolano et al., 2017; Kwong and Mascola, 2018; Schommers et al., 2020; Sok and Burton, 2018; West et al., 2014). During infection, bNAbs evolve in a minority of individuals, usually only after extensive and prolonged somatic hypermutation (SHM) of precursor Abs (Abbott et al., 2018; Kepler and Wiehe, 2017; Klein et al., 2013a). But are there other dimensions to the problems of eliciting bNAbs by Env-based immunogens? Here, we argue that HIV-1 Env is an inherently poor immunogen because it has exceptional properties. We discuss research strategies that could help overcome these hurdles to a more effective HIV-1-vaccine design.
HIV-1 Env structure and function
The native, functional Env complex is a trimeric spike structure, and typically only ~15 are present on the HIV-1 virion surface (Klein and Bjorkman, 2010). The spikes interact with cell-surface receptors (CD4 and then CCR5 or CXCR4) in a process that drives virus-cell membrane fusion in the first stage of the replication cycle (Klasse, 2012; Weiss, 2013). By binding to a sufficient number of spikes before or, more rarely, during the fusion process, NAbs protect target cells from HIV-1 infection (Klasse, 2007, 2014; Regoes and Magnus, 2015). The native trimer comprises three non-covalently associated gp120 and gp41 subunits: gp120 engages the entry receptors; gp41 anchors the spike into the virus membrane and contains the fusion peptide, which merges the viral and cell membranes (Julien et al., 2013; Lee et al., 2016; Lyumkis et al., 2013; Pancera et al., 2014). Almost half the mass of the trimer consists of N-linked glycans; there are ~90 individual glycan sites per trimer, most of them on gp120. Recombinant gp140 mimics of the viral spike (native-like trimers such as the SOSIP design), engineered for increased stability and rendered soluble by eliminating the membrane-anchoring domain of gp41, are widely used in immunogen-design programs (for example: Dey et al., 2018; Escolano et al., 2017; Sanders and Moore, 2017; Sok and Burton, 2018). Some earlier constructs such as gp120 monomers and non-native gp140 pseudo-trimers are, however, still being evaluated clinically (Bekker et al., 2020; Excler and Kim, 2019). The points made below apply to most of these different Env-immunogen designs (Figure 1). The various forms of Env, including membrane-anchored SOSIP trimers, generally induce similar binding Ab titers in various species. But native-like SOSIP trimers elicit the strongest autologous neutralization titers when administered as soluble proteins or DNA vectors in rabbits and macaques or on virus-like particles in guinea pigs (Aldon et al., 2018; Crooks et al., 2007; Pauthner et al., 2017; Sanders and Moore, 2017; Sanders et al., 2015).
Figure 1. HIV-1 Env is a highly glycosylated, metastable trimer of heterodimers.
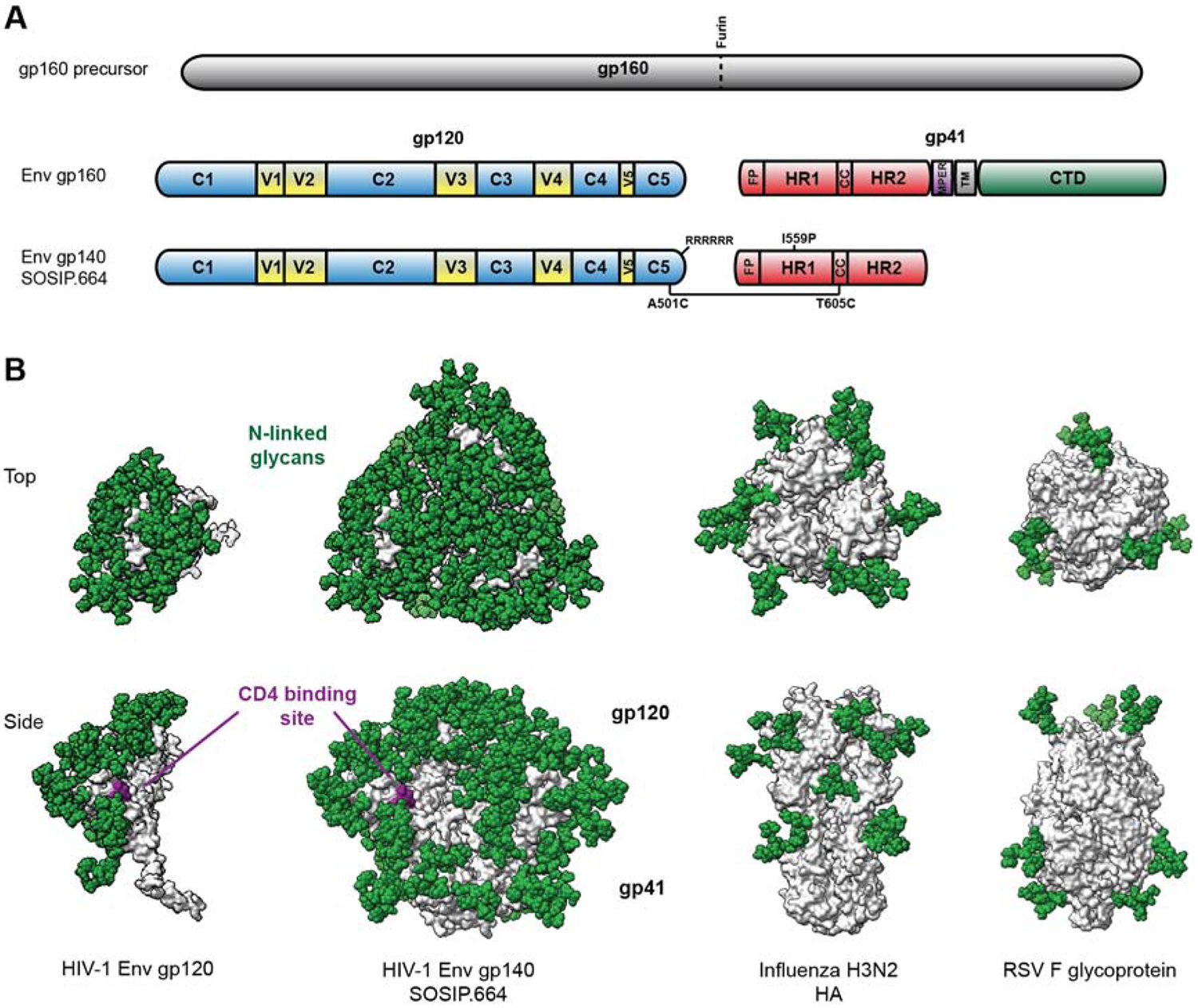
(A) The linear polypeptide map highlights the gp120 and gp41 subunits, constant and variable regions (C and V), and the engineered sequence changes conferring stability to recombinant HIV-1 Env SOSIP.664 trimers. The site on the precursor for cleavage by furin in the Golgi is marked; RRRRRR is a motif for enhance cleavage; FP=fusion peptide; HR=heptad repeat region; CC=disulfide bonded loop in gp41; MPER=membrane-proximate external region; TM=transmembrane region; CTD=cytoplasmic domain. (B) Structural comparison of HIV-1 Env (a gp120 subunit and the SOSIP.664 trimer), the influenza hemagglutinin (HA) and the RSV F glycoprotein. Peptidic surfaces are shown in grey and idealized oligomannose (Man-9) N-linked glycans as green spheres. The CD4bs of HIV-1 Env is highlighted in purple. The top view (upper row) is defined as looking down towards the viral membrane. The models are based on PDB 6mco, 4o5n and 6q0s.
It is certainly possible to raise high titers of Abs when HIV-1 Env is co-administered with a strong adjuvant. These Abs tend to recognize simple, peptidic epitopes and they lack useful, and often any, neutralizing activity. It is also plausible that inducing such non-neutralizing Abs (non-NAbs) or narrowly acting NAbs distracts the immune system from responding to the more complex, less immunogenic, but crucial epitopes for bNAbs (Havenar-Daughton et al., 2018; Haynes et al., 2012; Klein et al., 2013b; Ringe et al., 2019). A key aspect of bNAb epitopes is that they often comprise both peptidic and glycan components; all the known ones on gp120 include, or even consist entirely of, glycans (Crispin et al., 2018; Ward and Wilson, 2017). Since glycans are largely seen as self by the immune system, their immunogenicity requires the breaking of tolerance (Kelsoe and Haynes, 2018). In short, the high titers of non-NAbs or NAbs that act only against highly sensitive (Tier-1) viruses, which are common endpoints in animal studies and clinical trials, are not useful for assessing Env immunogenicity. Even for non-NAb or ELISA-based antigen-binding Ab endpoints, however, the evidence suggests that HIV-1 Env is a relatively poor immunogen, which does not induce sustained Ab titers (Fouts et al., 2015; Fouts et al., 2003; Klasse et al., 2012; Lewis, 2010; Lewis et al., 2014; Robb et al., 2012). Thus, Ab responses to many vaccines persist in humans for decades, often at protective levels (Amanna and Slifka, 2010). In contrast, anti-Env half-lives are orders of magnitude shorter and are, of course, generally non-protective (Lewis, 2010; Lewis et al., 2014). Why is there such a difference?
Comparative immunogenicity studies
The few immunogenicity studies that directly compare Ab (and sometimes cellular) responses to HIV-1 Env and a different antigen can be highly informative, although there are often interpretative caveats. Do the two immunogens have comparable physical properties? For example, can soluble HIV-1 Env be fairly compared with a Hepatitis B Surface Antigen (HBsAg) multivalent nanoparticle (NP)? Are de novo and possible recall responses being unreasonably treated as equivalent? Are the immunogen doses optimal? Are mice appropriate for HIV-1 Env immunogenicity studies? Nonetheless, in most reports that we have identified, HIV-1 Env is substantially less immunogenic than its comparators, and the anti-Env Ab responses often have atypical properties.
We are aware of only a single comparative human study, a Phase 1 trial with HIV-1 gp120 and the rabies virus G-protein, both immunogens delivered either mucosally or systemically by a canarypox vector-prime followed by soluble protein boosting. The resulting IgG and IgA responses to gp120 were less frequent than to the rabies-G and canarypox-vector proteins in the systemic-delivery groups, although with no significant differences after mucosal immunization (Wright et al., 2004). The paper contained no information on the Ab titers or decay rates to the various antigens, a gap that could perhaps be filled retroactively if samples are still in storage. A much earlier experiment in baboons compared HIV-1 gp120 monomers with the particulate HBsAg vaccine in Alum adjuvant. Initially, HBsAg induced much stronger Ab responses than gp120 and although the titers to the two immunogens did eventually converge, anti-gp120 Abs then declined more rapidly than anti-HBsAg Abs (Anderson et al., 1989). Thus, gp120 may elicit adequate memory B-cell and short-lived plasma-cell responses, but fewer long-lived plasma-cells. Although the particulate presentation of HBsAg complicates interpretation, that study provided an early clue that gp120 might be a poor immunogen compared with those the vaccine field had previously encountered successfully (Anderson et al., 1989). In contrast, when adjuvanted HIV-1 non-native gp140s and influenza HA1 proteins were given to different legs of the same rhesus macaques, ELISA binding Ab titers and decay kinetics were similar. The authors concluded that the transience of both Ab responses resulted from inherent property of soluble protein immunogens (Sundling et al., 2013). The seasonal influenza vaccine, which is based on inactivated virions, also induces short-lived responses in humans with significantly reduced protection 91–180 days after immunization (Young et al., 2018). Nonetheless, a vector-prime, inactivated virion-boost influenza vaccine regimen can elicit autologous ID90 NAb titers > 1 × 104 in mice, and can even broaden the response in animals, including macaques, by partly directing it to cross-reactive epitopes on the HA trimer stem and away from the immunodominant strain-specific epitopes on its head (Wei et al., 2010). These findings suggest that HA immunogenicity is intrinsically stronger and more amenable to improvement than HIV-1 Env. Indirect comparisons support this supposition: HIV-1 Env SOSIP trimer immunogens induce transient autologous Tier-2 NAb ID50 titers in the range 102–103 when administered as a bolus in a strong adjuvant (3M052 in polylactic glycolic acid, PLGA, nanoparticles) and somewhat higher, ~ 103, when given gradually by an osmotic-pump (Pauthner et al., 2017). These peak titers are often below the threshold for protection against autologous SHIV challenge (~ 3 × 102), and even when initially above that mark, they drop below it within weeks (Pauthner et al., 2019).
Ab responses to Env tend to be qualitatively atypical. When DNA vaccines encoding either HIV-1 gp120 or influenza HA were administered to mice, the Ab responses to the two proteins differed IgG-subclass and cytokine profiles. Thus, the mice responded to HA with Th1-associated high IgG2a/IgG1 ratios but to gp120 with Th2-associated low IgG2a/IgG1 ratios. This finding was attributed to the gp120-specific induction of IL-10 (Daly et al., 2005), although IL-10 has since emerged as distinct from Th2-associated cytokines, being instead both the inducer and product of Type 1 regulatory (Tr1) cells (Martinez-Sanchez et al., 2018). In other mouse experiments, Addavax-adjuvanted pseudo-trimeric HIV-1 gp120 constructs derived from three strains were compared with particulate HBsAg (Yu et al., 2016; Yuan et al., 2018). The major differences between the immunogens were that, compared with HBsAg, gp120 induced weak germinal center reactions, slow Ab recall responses, poor B-cell memory and low endpoint titers for all IgG subclasses, but strong MHC class II expression on B cells and high frequencies of Tfh-helper cells. Programmed death (PD)−1+ T cells were also more frequent in the gp120-immunized mice, which was proposed to contribute to the slow Ab-recall response. The IgG2a/IgG1 ratios for gp120 were low, but significantly higher than for HBsAg (Yu et al., 2016; Yuan et al., 2018). Also in mice, soluble HIV-1 gp140 behaved unusually in that it induced much lower IgG2a/IgG1 ratios than HIV-1 Gag, Influenza-HA and RSV-F proteins (Hess et al., 2019). Furthermore, the IgG subclass-skewing effect of HIV-1 Env was imparted to the anti-Gag IgG response against an Env-Gag fusion protein; the anti-Gag IgG2a/IgG1 ratio then resembled that of the response to Env but not to Gag alone. The unusual IgG isotype profile was attributed to an interaction between HIV-1 Env and mannose C-type lectin receptors (MCLRs) (Hess et al., 2019). Along similar thematic lines, when mice received monomeric HIV-1 gp120 or Gag proteins in the presence or absence of the mannose-binding protein griffithsin, IgG1-dominated anti-gp120 responses were strongly increased by griffithsin but anti-Gag IgG1 responses were not. The formation of gp120-griffithsin complexes probably accounted for an increase in the anti-griffithsin response when gp120, but not Gag, was a co-immunogen (Banerjee et al., 2012).
The above differences in murine IgG subclass profiles reflect Th1 vs. Th2 polarization that may not have any bearing on how the primate immune system responds to HIV-1 Env (Gor et al., 2003; Martinez-Sanchez et al., 2018). They are, however, indicative of unusual, Env-dependent variations in the interplay between innate and adaptive immunity that could be relevant across species. Nonetheless, any problems with HIV-1 Env immunogenicity in primates are not rooted in IgG subclass profiles per se. The primate immune response to Env in both virus-infected and Env-immunized subjects is generally strongly dominated by IgG1, which is not associated with Th1 vs. Th2 polarization in humans (Banerjee et al., 2010; Klasse and Blomberg, 1987; Wahren et al., 1988). Most bNAbs are also of the IgG1 subclass (https://www.hiv.lanl.gov/content/immunology). Indeed, a study of bNAb development using plasma from >4,000 HIV-1-infected patients found that an IgG1-dominated response to HIV-1 antigens and IgG1 Ab binding to the BG505 SOSIP trimer were the two best predictors of HIV-1 neutralization breadth (Kadelka et al., 2018).
Unusually modest benefits to nanoparticle presentation of HIV-1 Env
One method to improve the immunogenicity of vaccine antigens in general is to present them on NPs of, typically, 25–50 nm diameter (Figure 2). Thus, multivalent antigen display may drive stronger, longer-lasting Ab responses through efficient cross-linking of B-cell receptors (BCR) and improved antigen trafficking, endocytic uptake and thereby presentation (Brinkkemper and Sliepen, 2019; Tokatlian et al., 2019). The licensed HPV and HBsAg vaccines involve NPs, as do impressive programs to create influenza and RSV vaccines (Darricarrere et al., 2018; Hsia et al., 2016; Kanekiyo et al., 2019; Marcandalli et al., 2019). Multiple studies in animals show that NP presentation substantially improves the quantity and quality of Ab responses, compared with the delivery of the same antigens as soluble proteins (Brinkkemper and Sliepen, 2019). Thus, NP display of RSV antigens enhanced NAb titers over 10-fold (Marcandalli et al., 2019). Ongoing or new studies may show even greater increases. NP presentation also allows the creation of multivalent, antigenically mosaic immunogens, which can improve NAb breadth by increasing the avidity of interactions specifically with the most cross-reactive BCRs, as shown for influenza HA (Kanekiyo et al., 2019).
Figure 2. Icosahedral nanoparticle display of HIV-1 Env SOSIP.664 trimers.
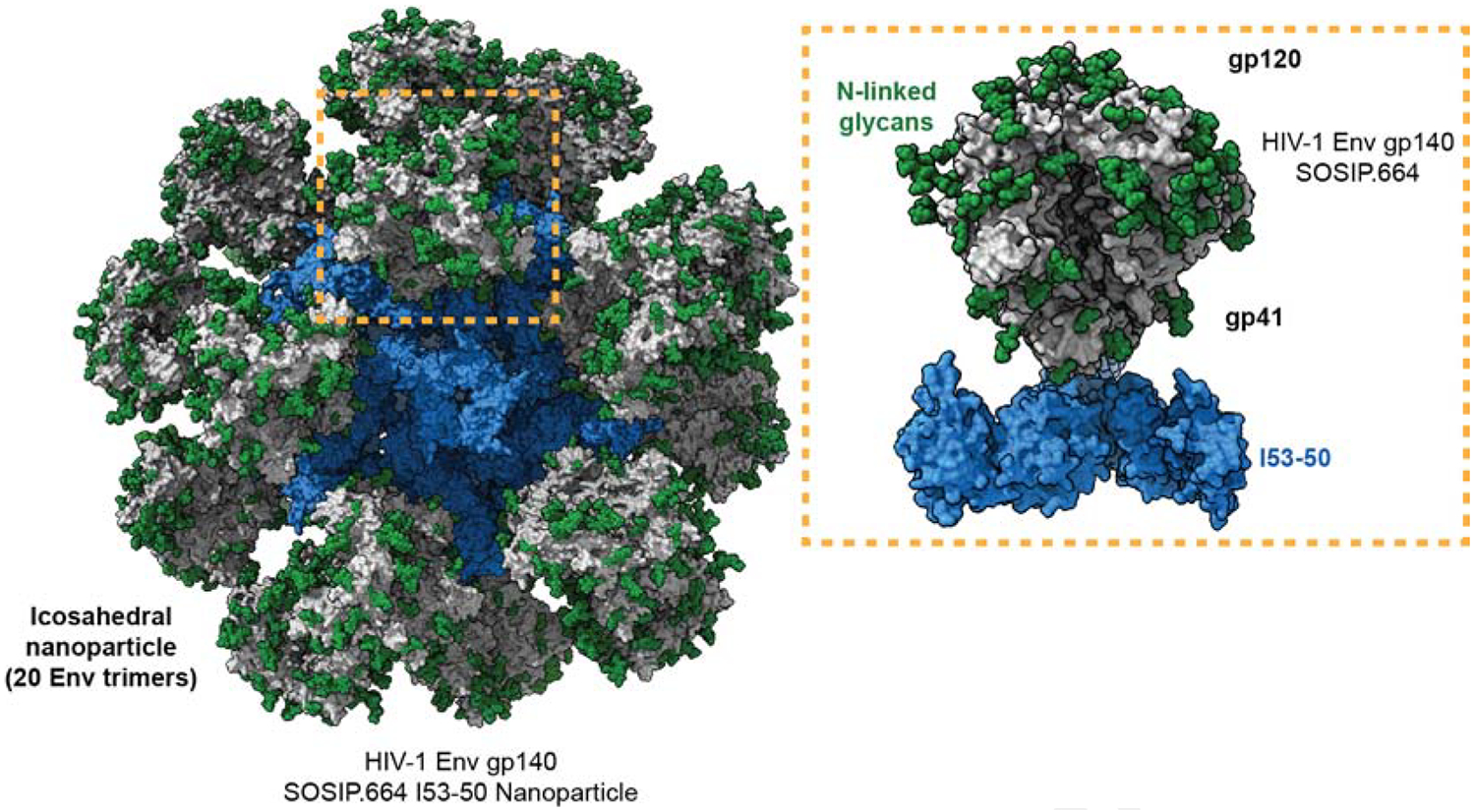
The entire nanoparticle with 20 HIV-1 Env SOSIP trimers distributed in icosahedral symmetry is shown to the left with idealized N-linked glycans in green, peptidic Env surfaces in grey, and the non-Env components in blue. To the right a magnified trimer with its non-Env scaffold component is shown with the same color code (Brouwer et al., 2019). The model is based on PDB 6p6f and EMD-20261.
There are, then, substantial grounds to pursue the NP approach with HIV-1 Env. The greatest benefits of NP presentation may be for immunogens with such low affinities for BCRs that they are completely unable to activate B-cells when delivered as soluble antigens (i.e., KD values in the μM range). The avidity benefits of NP presentation could bring the functional KD value into a range allowing B-cell activation. HIV-1 Env trimers with complete glycan shields might be one example of such an inert immunogen (Wagh et al., 2018). However, only modest, if any, immunogenic improvements occurred when early-generation HIV-1 Env was tested on NPs (Bale et al., 2017; Ingale et al., 2016; Martinez-Murillo et al., 2017; Tokatlian et al., 2018). Design limitations such as NP instability in vivo and poor epitope display on non-native Env may have compromised these studies. It is now evident that presenting more modern native-like trimers on NPs of various designs also confers at most modest (i.e., ~3-fold) increases in NAb titers, and does so only when the immunogenic epitopes are located close to the apex of the trimer (Brinkkemper and Sliepen, 2019; Brouwer et al., 2019; Brouwer and Sanders, 2019; Ringe et al., 2020; Tokatlian et al., 2019). The relevance of epitope location may reflect varied NAb epitope accessibility on the NP surface and occlusion by mannose binding lectins (MBL), as discussed further below (Brouwer et al., 2019; Ringe et al., 2020).
We are unaware of comparative experiments with HIV-1 Env NPs and those based on other viral antigens. Indirect comparisons are inevitably inexact, but the available data do suggest that NP presentation of HIV-1 Env is substantially less beneficial than in other NP-vaccine systems (Brinkkemper and Sliepen, 2019). Two unusual structure-function facets of HIV-1 Env merit discussion: The capacity of the gp120 subunits to bind to the cell surface CD4 and transduce signals that may affect immune cell function; and the extremely high content of mannose-rich glycans on the surface of Env. Neither feature is shared by other vaccine-relevant antigens, but both may influence the induction and maturation of anti-Env Abs in general or indeed of bNAbs.
Do CD4 binding and Env conformational changes influence immunogenicity?
The ability of gp120 to signal by ligating CD4 in vitro has long been observed in multiple cell systems. Generally, when amounts of gp120 that occur in infection are added to cultures of CD4+ cells, the consequences are almost invariably inimical to normal cell function (Klasse and Moore, 2004; Schwartz et al., 1994; Weinhold et al., 1989; Weissman et al., 1997). The purity of the gp120 proteins, particularly commercial products, is sometimes questionable, and any LPS or other contaminants could interfere with the cell function. Nonetheless, gp120 binding to CD4 in vitro can affect the normal properties of various cells important to immune responses in vivo (e.g., CD4+ T-helper cells, dendritic cells, monocytes and macrophages). One example is that HIV-1 gp120 monomers, in vitro, impaired the maturation of human plasmacytoid dendritic cells and hence the activation of co-cultured B cells through cell surface interactions with both CD4 and MCLRs (Chung et al., 2012). In another report, gp120 was found to suppress B-cell proliferation by binding to α4β7-integrin receptors on the B-cell surface and thereby increasing the expression of inhibitory cytokines and receptors (Jelicic et al., 2013).
It is dubious whether such events play much role in the pathogenesis of HIV-1 infection because gp120 concentrations in vivo are both low and vastly exceeded by the quantities of circulating anti-gp120 Abs that block gp120-CD4 binding (Callahan and Norcross, 1989; Klasse and Moore, 2004; Moore et al., 1994). However, as gp120 vaccines are administered to naïve volunteers in amounts up to 500 μg/dose, the local concentrations at or near injection sites (e.g., in draining lymph nodes) will be high, at least in the short term. Suppressive signals might be transduced during the critical early phases of an immune response, and CD4 binding could also reduce the concentration of free Env immunogen available for eliciting immune responses. Similar considerations apply to native-like trimer immunogens, although the latter are usually engineered to bind CD4 with lower affinity than gp120 monomers (Chuang et al., 2017; de Taeye et al., 2015; Joyce et al., 2017; Torrents de la Pena et al., 2017; Zhang et al., 2018).
Are Ab responses to Env compromised by the binding of the immunogen to CD4 on immune system cells in vivo? A definitive answer still remains to be obtained. This topic can only be addressed in species that express CD4 capable of binding HIV-1 Env with high affinity, which do not include rabbits, guinea pigs or mice. A mutation, W427S, in the CD4 binding site (CD4bs) of gp120 was reported to enhance effector memory Tfh-cell responses, reduce non-specific Tfh-cell responses, and increase the gp120-specific Ab titers in wild-type BALB/c mice (Yu et al., 2014). As HIV-1 gp120 does not bind to murine CD4, the explanation for these observations is obscure. Unmodified HIV-1 Env also binds inefficiently to macaque CD4, limiting the power of any conclusions drawn so far from this species. In a macaque study involving non-native HIV-1 gp140 proteins with standard and disabled CD4-binding capacities, there were similar binding-Ab and non-NAb responses to the two immunogens (Sundling et al., 2013). However, the standard gp140 did not incorporate sequence changes that were later shown to be required for high affinity HIV-1 Env binding to macaque CD4 (Li et al., 2016). A more definitive macaque experiment could now be performed to compare the immunogenicity of CD4-optimized and CD4-disabled native-like trimers, or even gp120 monomers with the same sequence changes. Alternatively, SIV Env variants could be used.
In a complementary strategy, a CD4-optimized or -disabled Env mutant could be injected, alone or together with a different protein (e.g., HIV-1 Gag), into macaque lymphoid tissue or delivered more conventionally. The goal would be to assess whether CD4-binding or another property of gp120 interferes with the development or maintenance of the immune response to gp120 or the co-administered immunogen. Does CD4 engagement in lymphoid tissues trigger apoptosis or otherwise disrupt the cytokine milieu within germinal centers? Are memory responses affected? Is there an impact on the rate of decline of post-peak Ab titers? The answers are achievable and could strongly improve immunogen design.
Even in mice, where high affinity CD4 binding by Env does not occur, there is evidence that Env interferes with the immunogenicity of co-immunized antigens. For example, in mice given DNA plasmids expressing HIV-1 Gag protein with or without gp120, Ab and cellular responses to Gag were suppressed by gp120. Curiously, the inhibitory effect of HIV-1 gp120 was not mimicked by the corresponding glycoproteins from SIV or EIAV, or by influenza HA (Toapanta et al., 2007).
CD4-binding will also occlude bNAb epitopes overlapping the CD4bs. Again, the impact can only be determined by comparative studies in species with high CD4-Env affinity. A different but related question is whether reducing CD4-binding or CD4-induced conformational changes qualitatively improves the immunogenicity of Env by preventing responses to previously cryptic and potentially distractive non-NAb epitopes. Multiple SOSIP trimer variants have been designed and tested to explore this hypothesis, but although some reductions in non-NAb responses have been observed, they have not been accompanied by major increases in autologous or heterologous NAb titers (Chuang et al., 2017; de Taeye et al., 2015; Henderson et al., 2020; Joyce et al., 2017; Kulp et al., 2017; Ringe et al., 2017; Torrents de la Pena et al. 2017; Zhang et al., 2018). Additional research in this area is ongoing, including attempts to better suppress the immunogenicity of non-NAb neo-epitopes at the base of soluble trimers (Kulp et al., 2017; Ringe et al., 2020).
One of the mechanisms by which HIV-1 Env reduces the antigenicity of the CD4bs is entropic or conformational masking (Kwong et al., 2002). Whether the conformational flexibility inherent in HIV-1 Env glycoproteins also affects their immunogenicity by, for example, masking potential bNAb epitopes has been an open question for many years. This general topic was indirectly revisited recently by the use of biophysical assays to study conformational transitions within SOSIP trimers in vitro (Lu et al., 2019; Stadtmueller et al., 2018). However, different spectroscopy techniques, DEER (double electron-electron resonance) and smFRET (single-molecule fluorescence resonance energy transfer), yielded discordant outcomes (Lu et al., 2019; Stadtmueller et al., 2018). The interpretation of the data obtained by smFRET, and hence the conclusions drawn in one of the studies, have also been questioned (Lu et al., 2019; Moore, 2019; Pan et al., 2020). Furthermore, it is unknown to what extent conformational transitions that take place over the microsecond to millisecond range affect the induction of Ab responses in vivo in the hours to days after immunization (Moore, 2019). An answer might be obtained if a native-like trimer immunogen could be designed to adopt only one, optimal conformation, as judged by multiple and reliable in vitro assays. No such immunogen has yet emerged.
Glycans mould the response to both B-cell and T-helper cell epitopes
Compared with other virus-derived immunogens, HIV-1 Env has an exceptionally high content of glycans. As noted, there can be over 30 individual glycan moieties on each protomer of a trimeric spike or recombinant mimic. In comparison, an influenza HA protomer typically contains about 3 glycans while RSV F has 5 or 6 (Figure 1). The glycan shield on HIV-1 Env plays a thoroughly studied and well-understood role in resistance to, and escape from, NAbs; equally fundamental is the inclusion of glycan moieties in all known bNAb epitopes on gp120 and some on gp41 (Crispin et al., 2018; Ward and Wilson, 2017; Zhou et al., 2017). In short, effective bNAb responses must accommodate the presence of glycans.
Less well explored is how glycans affect T-helper cell epitopes (THCEs) (Wolfert and Boons, 2013). Extensive SHM, driven by T-cell help, is critical to the emergence of many bNAbs. Much more research in this area seems justified, particularly using modern trimeric Env immunogens, which were not available when most of the experiments reviewed below were performed 10–20 years ago. Moreover, knowledge of Env structure is much more detailed and sophisticated now than then, perhaps allowing the structure and function of glycans and THCEs to be better understood than in most of the studies outlined below.
Although some THCEs are present in HIV-1 Env sequences, their availability for presentation by MHC class II (MHC-II) to T-helper cells may be highly restricted. Thus, glycans close to proteolytic sites can sterically block the access of antigen-processing proteases in the lysosomal compartment and hence prevent the liberation of 10–25–residue, THCE-containing Env peptides that would otherwise be loaded into the MHC-II peptide binding cleft. But not all N-and O-linked glycosides will survive lysosomal degradation, and some resulting glycol-peptides can be presented by MHC-II. Nonetheless, there are so many bulky glycans on HIV-1 Env that too much carbohydrate may survive processing to allow peptide binding into the MHC-II cleft (Purcell et al., 2008; Werdelin et al., 2002).
Early studies of glycans and THCEs generally involved immunizing mice with gp120 monomers or gp140 pseudo-trimers, experimental systems that complicate extrapolations to the use of more modern native-like trimers in primates. It is also fair to assume that the structural constraints on the presentation of THCEs will be stronger for a fully assembled, covalently stabilized trimer than for a monomeric gp120 subunit (Landry, 2000). Reports from 2001–2002 were among the first to analyze how Env glycosylation and gp120 structure might restrict the presentation of THCEs (Sarkar et al., 2002a, b; Surman et al., 2001). The first study was based on peptide assays of T-helper cell responses in mice immunized with DNA expressing a non-native gp140 (Surman et al., 2001). The identified THCE sequences were then plotted on the only available Env structure, that of a gp120 core (Kwong et al., 1998). Three “T-help hotspots” were identified in the C2, V3 and V4/C4 regions of gp120, and another near gp41-residue 650. It was noted that N-glycans flank most clusters of human and murine THCEs (Surman et al., 2001). In a conceptually related approach, macaques were infected with a live, attenuated SIV vaccine (hence at least some of the Env was fully native). The THCEs were identified by screening 20mer peptides and then plotted on the linear sequence of SIV Env, but without placing the information in a tertiary-structural context. Here, the most common epitopes included gp120 residues 341–360 (C3) and 421–470 (V4–V5), in linear regions with no or few glycans. Other sites were flagged in gp41 (Sarkar et al., 2002a, b). The most detailed of several later papers based on mice immunized with gp120 or gp140, delivered as DNA plasmids or soluble proteins, again depicted the THCE data on the gp120-core crystal structure (Brown et al., 2003).
A hypothesis that local polypeptide instability in model antigens would promote THCE processing and presentation was formulated, supported in silico and then corroborated in vivo in gp120-immunized mice and HIV-1-infected humans. The dominant THCEs were located in the gp120 outer domain (Dai et al., 2001; Landry, 2000; Mirano-Bascos et al., 2008). Furthermore, the influence of glycans on THCE presentation is not always negative: mutation of glycan sites in gp160 can suppress the priming of T cells that recognize glycan-proximate epitopes (Sjolander et al., 1996). Indeed, although eliminating either the N448 or the N230 glycan did not affect endocytic uptake or lysosomal trafficking, it interfered with the presentation of N-terminally located THCEs. Mutating N448 reduced proteolytic processing, but a change at N230 acted by an unidentified mechanism (Li et al., 2009).
Disulfide bonds have a complex influence on THCE presentation (Li et al., 2002). There are typically eight intramolecular disulfide bonds in a monomeric HIV-1 gp120 subunit, and another within gp41 (Checkley et al., 2011). Additional inter- and intra-protomeric disulfide bonds are deliberately engineered into recombinant native-like trimers of various designs, to confer additional stability (Chuang et al., 2017; de Taeye et al., 2015; Joyce et al., 2017; Kwon et al., 2015; Torrents de la Pena et al., 2017; Zhang et al., 2018). A correlation between local polypeptide flexibility and THCE localization suggested that eliminating some disulfide bonds from gp120 monomers would promote antigen processing and THCE presentation (Landry, 2000). But when the 298–331, 378–418 and 378–445 gp120 disulfide bonds were individually knocked out, T-cell helper responses in BALB/c mice were actually reduced, although Ab responses were enhanced. Specifically, the 378–418 mutation redirected the Ab specificity towards the CD4bs (Mirano-Bascos et al., 2010). However, in CBA mice, in which MHC-II have well-defined binding properties, disrupting a single gp120 disulfide-bond redirected responses to cryptic CD4+ T-cell epitopes (Li et al., 2014). Thus, manipulating THCE epitopes in an immunogen as complex as a gp120 monomer can have unpredictable and undesired outcomes. By extension, liberating THCE peptides from native-like trimers by proteolytic cleavage, for MHC-II loading, must be even more complicated. Stabilizing these trimers to optimize the presentation of bNAb epitopes and to reduce the exposure of non-NAb sites may compromise the presentation of THCEs, particularly when additional disulfide bonds are introduced. In short, there could be a direct conflict between optimizing the liberation of THCEs and presenting bNAb epitopes on the same engineered trimer. But inducing HIV-1-specific CD4+ T-helper cells carries a serious risk, as their hyper-susceptibility to HIV-1 infection could be disastrous in the vaccine context (Douek et al., 2002). On this argument, any promotion of endogenous THCEs in Env may be best avoided, even to the extent of eliminating the few existing ones by targeted mutagenesis. Research here may be fruitful.
An alternative approach is to provide exogenous THCEs unrelated to HIV-1 sequences as C-terminal extensions to Env flanked by cathepsin cleavage sites, or intrastructurally as free peptides incorporated within NPs (Elsayed et al., 2018; Lake and Mitchison, 1976). Thus, there were some initial increases in Ab responses when the PADRE THCE was genetically fused to gp120 monomers or, more recently, native-like trimers (Grundner et al., 2004; Ringe et al., 2020). THCE peptides can also be attached to, or incorporated within, NPs (Bale et al., 2016; Damm et al., 2019; Elsayed et al., 2018; Hsia et al., 2016). Various other immune-stimulatory molecules such as BAFF, APRIL and CD40L have been genetically fused to Env proteins but without major immunogenicity benefits (Gupta et al., 2015; Liu et al., 2020; Melchers et al., 2012). Vaccines such as tetanus toxoid, against which most vaccinated people have already developed T-helper cell responses, can also be exploited (Damm et al., 2019; Elsayed et al., 2018). These approaches to improving Env immunogenicity all merit further exploration with the current generation of trimers.
The infant macaque model may also lend itself to assessing the various influences on immunogenicity outlined above: bNAbs can develop more rapidly after SIV/SHIV infection of young animals than adults, and with less hypermutation, in a kappa-chain maturation-dependent manner (Simonich et al., 2019; Simonich et al., 2016). Although the frequencies of Tfh cells and germinal-center B cells were greater in SHIV-infected infant macaques than adults, however, autologous Tier 2 NAbs developed with similar kinetics (Nelson et al., 2019). Ab responses in infants are generally weak; although the priming of memory B cells is effective before and at birth, affinity maturation is constricted until after 4–6 months of age (Siegrist and Aspinall, 2009). Hence, the immaturity of the SHM machinery in infants may open other routes to bNAb development that merit more exploration.
Mannose moieties are not just components of shielding glycans and bNAb epitopes
The glycans on HIV-1 Env are unusually rich in terminal mannose residues, even more so for native-like trimers than gp120 monomers (Cao et al., 2018; Crispin et al., 2018). This mannose density arises from steric hindrance, particularly on native-like trimers, to glycan-processing enzymes as Env migrates along the secretory pathway (Checkley et al., 2011). Hence, discrete areas of oligomannose glycans extend over the trimer surface (Crispin et al., 2018). These mannose-rich patches are targets for MBLs and MCLRs, components of the innate immune system contributing to complement activity and the presentation of mannose-rich antigens. It seems plausible that other viral vaccine antigens with far less mannose may not proceed along the routes available to HIV-1 Env.
Although MBLs bind to soluble HIV-1 Env, they do so more avidly when Env is presented on NPs (Ezekowitz et al., 1989; Ringe et al., 2020; Tokatlian et al., 2019). This is relevant in vivo because SOSIP trimer ferritin-NPs are trafficked to follicular dendritic cells and germinal centers in mice via an MBL-dependent antigen trafficking pathway that is much less available to the corresponding soluble trimers (Tokatlian et al., 2019). In vitro, MBL binding to SOSIP trimers on iron oxide-NPs can occlude NAb epitopes that are located away from the trimer apex region (Ringe et al., 2020) (Figure 3). That observation may help explain why presenting Env trimers on NPs is of comparatively limited benefit (Brinkkemper and Sliepen, 2019).
Figure 3. Presumed MBL interacting surface on HIV-1 Env.
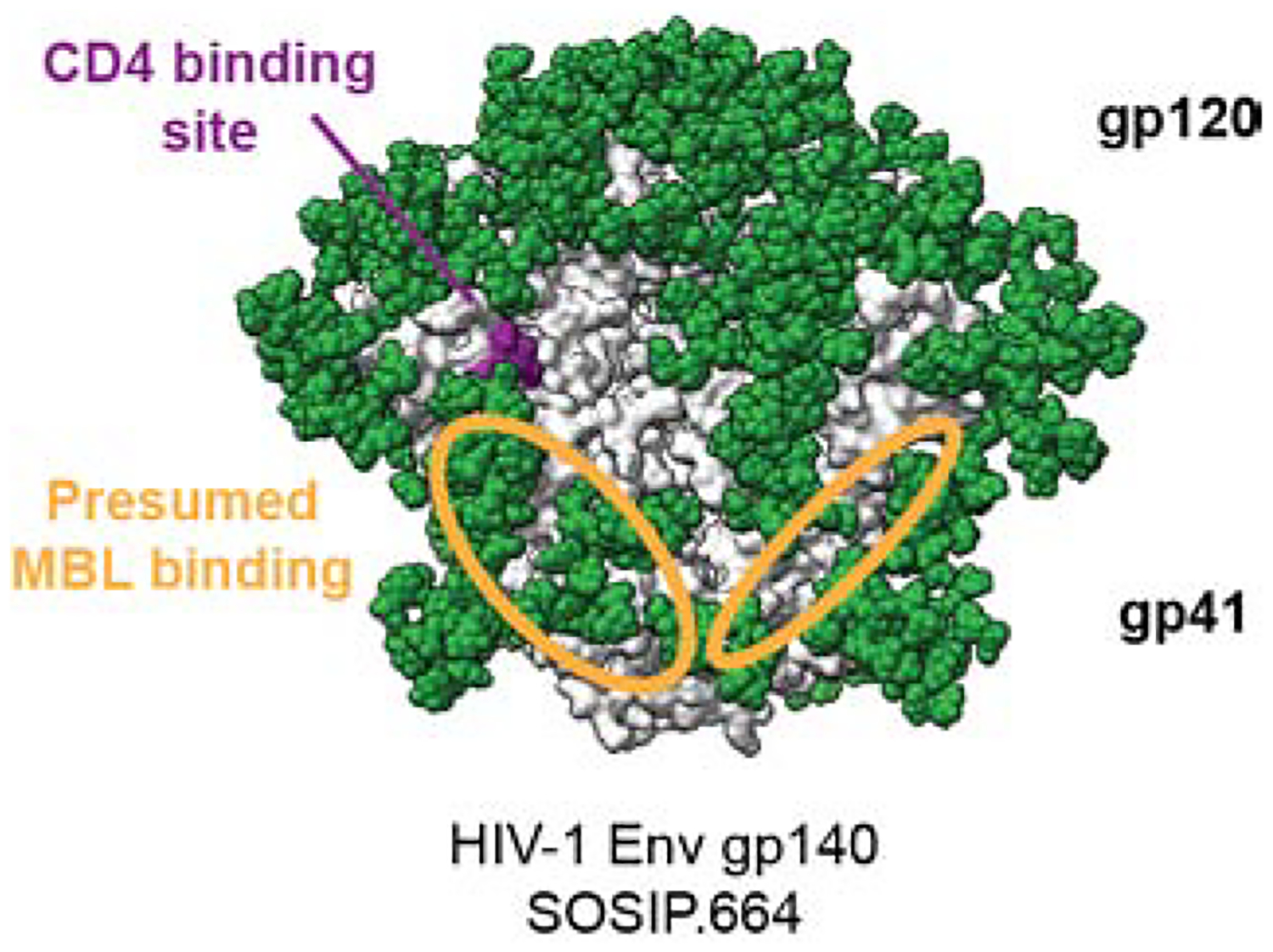
The approximate areas of MBL interaction with terminal mannose residues on an HIV-1 Env SOSIP.664 trimer are indicated on two protomers as orange ellipses; the third is hidden from view. The area extends over the gp120-gp41 interface from the CD4bs site towards the trimer base. The model is based on data obtained from a competition ELISA with MBL and bNAbs (Ringe et al., 2020).
Earlier studies with gp120 also indicated the relevance of interactions between mannose moieties and MCLRs in vitro (Shan et al., 2007) and MBL or MCLR in vivo (Banerjee et al., 2009, 2010, 2012). Thus, depleting mannose moieties on monomeric HIV-1 gp120 skewed the IgG isotype response of immunized mice away from IgG1 and towards IgG2a and IgG3, and increased the overall anti-gp120 titers (Banerjee et al., 2009). As noted above, it has been suggested that MCLRs play a role in the atypical IgG subclass profile induced in mice by HIV-1 gp140s (Hess et al., 2019).
The full consequences of HIV-1 Env interactions with MBL or MCLRs are far from clear, but an atypically high mannose content may be yet another complex influence on the interplay between Env and the humoral immune system that is not shared with most other pathogens. Such complexities may even extend to the adjuvant field. The different antigen-trafficking and -presentation properties of mannose-rich vs. more typical antigens (soluble or NP) could affect the responses to TLR-activators and other adjuvant components. Whether these influences matter in practice can be explored by preparing mannose-depleted Env through enzymatic digestion. Conversely, mannose–enriched glycoproteins can be expressed in mutant producer cells (Crispin et al., 2018). It may also be possible, although challenging, to knock-out MBL binding sites on HIV-1 Env by targeted mutagenesis. MBL-knock-out mice represent another resource, although there are other limitations to this species for the induction of NAbs.
What adjuvants best support the induction of bNAbs?
Adjuvants are crucial for providing supporting stimuli, including T-cell help, to HIV-1 Env. Various adjuvants have been tested with early forms of Env in multiple species, including macaques (Francica et al., 2015). A particularly strong adjuvant effect occurred with cholera toxin, which acts in a Th17-dependent manner, in that it increased responses to gp120 1,000-fold when both molecules were administered to mice as DNA (Bagley et al., 2003). It is of course important to verify that any chosen adjuvant does not compromise the structural integrity of the Env immunogen, particularly for native-like trimers (Ozorowski et al., 2018).
To date, only few adjuvants have been tested for their ability to support the immunogenicity of native-like SOSIP trimers in animals. An ISCOM-based adjuvant, “ISCOMATRIX”, worked well in rabbits and macaques (Pauthner et al., 2017; Sanders et al., 2015). “Adjuplex” and 3M052-PLGA adjuvants are also effective with native-like trimers in macaques, much more so than Alum (Zhou et al., 2017; Arunachalam, 2020). Substantial immunogenicity increases were found when SOSIP trimers were stably attached to Alum particles to allow gradual and sustained release of the antigen in vivo (Moyer et al., 2020). Slow delivery by osmotic pumps was also beneficial, probably by mimicking more conventional delivery at gradually increasing doses (Cirelli et al., 2019; Pauthner et al., 2017; Tam et al., 2016). At least one adjuvant, GLA-LSQ, which was predicted based on its performance with other antigens to be strong, however, fared very poorly in conjunction with soluble and NP-presented SOSIP trimers in rabbits and macaques (Brouwer et al., 2019; Ringe et al., 2020). A human trial of the prototypic native-like trimer, BG505 SOSIP.664, with multiple different adjuvants is now ongoing, in concert with similarly designed experiments in small animals (Clinicaltrials.gov: NCT04177355). Another human trial of the same trimer with the AS01b adjuvant is also underway (NCT03699241), as is one with a similar trimer delivered in Alum (NCT03783130). Animal studies have shown that the kinetics of non-NAb and autologous Tier-2 NAb responses induced by the same SOSIP trimer can differ, the former Abs arising earlier (Klasse et al., 2016; Ringe et al., 2017; Sanders et al., 2015). The above human trials may therefore reveal that eliciting Abs against glycan-affected NAb/bNAb epitopes on native-like trimers requires a different adjuvant from those that work well for the simpler antigens and epitopes present on earlier HIV-1 proteins and other vaccine antigens. Whatever the outcome, the clinical data should be very informative about how to immunize humans (and animals) with native-like trimers. What is the best adjuvant? If autologous Nab responses occur, how do they compare with those associated with protection of macaques immunized with the same trimers and then SHIV-challenged? As noted earlier, for trimer-only immunization the protective ID50 are ~3 × 102 (Pauthner et al., 2019; Arunachalam et al., 2020).
Given the importance of SHM for inducing bNAbs, it will be critical that the adjuvant fully supports this process, including the interactions between dendritic cells and lymphocytes. Dendritic cells are key players in the interface between innate and adaptive immunity, which is central to the actions of adjuvants, not least for boosting Ab responses (Boscardin et al., 2006; Pulendran and Ahmed, 2006). Dermally derived CD14+ dendritic cells can be triggered by TLR ligands to express B cell stimulatory cytokines that, in turn, stimulate naïve B cells. Thus, in vitro, activation of these dendritic cells by a combination of ligands for TLR-3, -4 and -7/8 induced naïve B cells to proliferate and differentiate into CD27+ CD38+ B cells that secrete high levels of IgG and IgA (Matthews et al., 2012; Matthews et al., 2013).
In both mice and humans, a CpG-based, TLR-9-activating adjuvant supported the production of high levels of IgG Abs to a malaria antigen, but a more sophisticated analysis then revealed that SHM of these Abs was unexpectedly weak (Akkaya et al., 2018). All Ab endpoints are not equally useful when assessing immunogenicity. The reported antagonizing of Ab affinity maturation by TLR-9 activation, as in in the above malaria-vaccine study, could be a substantial hindrance to eliciting bNAbs by HIV-1 Env immunogens. Another concern about TLR-9 as an adjuvant target for Env vaccines arises from in vitro studies in which gp120 inhibited the CpG-induced maturation of plasmacytoid dendritic cells in a CD4-and MCLR-dependent manner, and hence their ability to stimulate B cells (Chung et al., 2012). Similarly, inhibitory effects of gp120 were seen in a co-cultures of CD14+ dermal dendritic cells and B cells when TLR-9 was activated by CpG, but not when TLR-3, -4 or -7/8 were activated (Matthews et al., 2012; Matthews et al., 2013).
Conclusion
Despite over 30 years of study in multiple species, including many thousands of immunized humans, several aspects of how Ab responses to HIV-1 Env are generated remain unknown. Comparative immunogenicity experiments can be particularly informative about the exceptional properties of HIV-1 Env, but they are rarely performed and have not always been thoroughly analyzed. Those lacunae are understandable for human trials, given their complexities and expense, but here there is a role for the macaque models that may be predictive for humans.
Env immunogen design has improved over the past decade, largely through high-resolution structures of native-like trimers. These methods will remain necessary for further advances in HIV-1 vaccine development, but they may not be sufficient. It is entirely possible that no single, fixed-composition Env, whether a native-like trimer or not, will ever be able to mimic the complicated bNAb-evolution processes in infected people. The sophisticated techniques now available to study bNAb ontogeny reveal how difficult it is for the humoral immune response to overcome the multiple defenses that have evolved to hinder the antigenicity and immunogenicity of the native trimer (Klein et al., 2013b; Kwong and Mascola 2018; West et al., 2014). Immunological approaches such as, but not limited to, germline-targeting may breach some of these barriers (McGuire, 2019; Medina-Ramirez et al., 2017; Escolano et al, 2017). More attention could also be paid to human genetics. What factors underlie the well-documented variation of at least 100-fold in the magnitude of the Ab responses to Env variants within vaccine cohorts, whether animal or human (Corey et al., 2015; Gilbert et al., 2005)? In appropriately designed comparative immunogenicity studies (see above), would poor responders to HIV-1 Env immunogens also respond weakly to other viral protein immunogens, and vice versa? Can advanced genetic approaches such as systems vaccinology provide answers (Cortese et al., 2020; Pulendran, 2019; Querec et al., 2009)?
In conclusion, we suggest that all Env glycoprotein-based strategies would benefit from an improved knowledge of how these elaborate, and highly unusual, molecular constructs interact with the innate and adaptive immune systems. The immunology of Env vaccine development may turn out to be as critical as structure-guided design to the eventual success of the overall concept.
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health grant P01 AI110657 and Bill and Melinda Gates Foundation grants OPP1132237 and INV-002022. R.W.S is a recipient of a Vici grant from the Netherlands Organization for Scientific Research (NWO).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abbott RK, Lee JH, Menis S, Skog P, Rossi M, Ota T, Kulp DW, Bhullar D, Kalyuzhniy O, Havenar-Daughton C, et al. (2018). Precursor frequency and affinity determine B cell competitive fitness in germinal centers, tested with germline-targeting HIV vaccine immunogens. Immunity 48, 133–146 e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkaya M, Akkaya B, Kim AS, Miozzo P, Sohn H, Pena M, Roesler AS, Theall BP, Henke T, Kabat J, et al. (2018). Toll-like receptor 9 antagonizes Ab affinity maturation. Nat Immunol 19, 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldon Y, McKay PF, Allen J, Ozorowski G, Felfodine Levai R, Tolazzi M, Rogers P, He L, de Val N, Fabian K, et al. (2018). Rational Design of DNA-Expressed Stabilized Native-Like HIV-1 Envelope Trimers. Cell Rep 24, 3324–3338 e3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanna IJ, and Slifka MK (2010). Mechanisms that determine plasma cell lifespan and the duration of humoral immunity. Immunol Rev 236, 125–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunachalam PS, P. T, Joag C, Bollimpelli V, Scott SV, D. MK, Wimmers F, Burton SL, Labranche CC, Petitdemange C, Gangadhara S, et al. (2020). Vaccine that induces tissue-resident T cells durably prevents HIV with lower neutralizing antibody titers. Nat Med (submitted). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KP, Lucas C, Hanson CV, Londe HF, Izu A, Gregory T, Ammann A, Berman PW, and Eichberg JW (1989). Effect of dose and immunization schedule on immune response of baboons to recombinant glycoprotein 120 of HIV-1. J Infect Dis 160, 960–969. [DOI] [PubMed] [Google Scholar]
- Bagley KC, Shata MT, Onyabe DY, DeVico AL, Fouts TR, Lewis GK, and Hone DM (2003). Immunogenicity of DNA vaccines that direct the coincident expression of the 120 kDa glycoprotein of human immunodeficiency virus and the catalytic domain of cholera toxin. Vaccine 21, 3335–3341. [DOI] [PubMed] [Google Scholar]
- Bale JB, Gonen S, Liu Y, Sheffler W, Ellis D, Thomas C, Cascio D, Yeates TO, Gonen T, King NP, et al. (2016). Accurate design of megadalton-scale two-component icosahedral protein complexes. Science 353, 389–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale S, Goebrecht G, Stano A, Wilson R, Ota T, Tran K, Ingale J, Zwick MB, and Wyatt RT (2017). Covalent Linkage of HIV-1 Trimers to Synthetic Liposomes Elicits Improved B Cell and Ab Responses. J Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee K, Andjelic S, Klasse PJ, Kang Y, Sanders RW, Michael E, Durso RJ, Ketas TJ, Olson WC, and Moore JP (2009). Enzymatic removal of mannose moieties can increase the immune response to HIV-1 gp120 in vivo. Virology 389, 108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee K, Klasse PJ, Sanders RW, Pereyra F, Michael E, Lu M, Walker BD, and Moore JP (2010). IgG subclass profiles in infected HIV type 1 controllers and chronic progressors and in uninfected recipients of Env vaccines. AIDS Res Hum Retroviruses 26, 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee K, Michael E, Eggink D, van Montfort T, Lasnik AB, Palmer KE, Sanders RW, Moore JP, and Klasse PJ (2012). Occluding the mannose moieties on human immunodeficiency virus type 1 gp120 with griffithsin improves the Ab responses to both proteins in mice. AIDS Res Hum Retroviruses 28, 206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker LG, Tatoud R, Dabis F, Feinberg M, Kaleebu P, Marovich M, Ndung’u T, Russell N, Johnson J, Luba M, et al. (2020). The complex challenges of HIV vaccine development require renewed and expanded global commitment. Lancet 395, 384–388. [DOI] [PubMed] [Google Scholar]
- Bonsignori M, Liao HX, Gao F, Williams WB, Alam SM, Montefiori DC, and Haynes BF (2017). Ab-virus co-evolution in HIV infection: paths for HIV vaccine development. Immunol Rev 275, 145–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscardin SB, Hafalla JC, Masilamani RF, Kamphorst AO, Zebroski HA, Rai U, Morrot A, Zavala F, Steinman RM, Nussenzweig RS, et al. (2006). Antigen targeting to dendritic cells elicits long-lived T cell help for Ab responses. J Exp Med 203, 599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkkemper M, and Sliepen K (2019). Nanoparticle vaccines for inducing HIV-1 neutralizing Abs. Vaccines (Basel) 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer PJM, Antanasijevic A, Berndsen Z, Yasmeen A, Fiala B, Bijl TPL, Bontjer I, Bale JB, Sheffler W, Allen JD, et al. (2019). Enhancing and shaping the immunogenicity of native-like HIV-1 envelope trimers with a two-component protein nanoparticle. Nat Commun 10, 4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer PJM, and Sanders RW (2019). Presentation of HIV-1 envelope glycoprotein trimers on diverse nanoparticle platforms. Curr Opin HIV AIDS 14, 302–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SA, Stambas J, Zhan X, Slobod KS, Coleclough C, Zirkel A, Surman S, White SW, Doherty PC, and Hurwitz JL (2003). Clustering of Th cell epitopes on exposed regions of HIV envelope despite defects in Ab activity. J Immunol 171, 4140–4148. [DOI] [PubMed] [Google Scholar]
- Callahan LN, and Norcross MA (1989). Inhibition of soluble CD4 therapy by Abs to HIV. Lancet 2, 734–735. [DOI] [PubMed] [Google Scholar]
- Cao L, Pauthner M, Andrabi R, Rantalainen K, Berndsen Z, Diedrich JK, Menis S, Sok D, Bastidas R, Park SR, et al. (2018). Differential processing of HIV envelope glycans on the virus and soluble recombinant trimer. Nat Commun 9, 3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checkley MA, Luttge BG, and Freed EO (2011). HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J Mol Biol 410, 582–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang GY, Geng H, Pancera M, Xu K, Cheng C, Acharya P, Chambers M, Druz A, Tsybovsky Y, Wanninger TG, et al. (2017). Structure-Based Design of a Soluble Prefusion-Closed HIV-1 Env Trimer with Reduced CD4 Affinity and Improved Immunogenicity. J Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung NP, Matthews K, Klasse PJ, Sanders RW, and Moore JP (2012). HIV-1 gp120 impairs the induction of B cell responses by TLR9-activated plasmacytoid dendritic cells. J Immunol 189, 5257–5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli KM, Carnathan DG, Nogal B, Martin JT, Rodriguez OL, Upadhyay AA, Enemuo CA, Gebru EH, Choe Y, Viviano F, et al. (2019). Slow Delivery Immunization Enhances HIV Neutralizing Ab and Germinal Center Responses via Modulation of Immunodominance. Cell 177, 1153–1171 e1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J (2020). Combo of two HIV vaccines fails its big test. Science 367, 611–612. [DOI] [PubMed] [Google Scholar]
- Corey L, Gilbert PB, Tomaras GD, Haynes BF, Pantaleo G, and Fauci AS (2015). Immune correlates of vaccine protection against HIV-1 acquisition. Sci Transl Med 7, 310rv317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese M, Sherman AC, Rouphael NG, and Pulendran B (2020). Systems Biological Analysis of Immune Response to Influenza Vaccination. Cold Spring Harb Perspect Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispin M, Ward AB, and Wilson IA (2018). Structure and immune recognition of the HIV glycan shield. Annu Rev Biophys. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks ET, Moore PL, Franti M, Cayanan CS, Zhu P, Jiang P, de Vries RP, Wiley C, Zharkikh I, Schulke N, et al. (2007). A comparative immunogenicity study of HIV-1 virus-like particles bearing various forms of envelope proteins, particles bearing no envelope and soluble monomeric gp120. Virology 366, 245–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai G, Steede NK, and Landry SJ (2001). Allocation of helper T-cell epitope immunodominance according to three-dimensional structure in the human immunodeficiency virus type I envelope glycoprotein gp120. J Biol Chem 276, 41913–41920. [DOI] [PubMed] [Google Scholar]
- Daly LM, Johnson PA, Donnelly G, Nicolson C, Robertson J, and Mills KH (2005). Innate IL-10 promotes the induction of Th2 responses with plasmid DNA expressing HIV gp120. Vaccine 23, 963–974. [DOI] [PubMed] [Google Scholar]
- Damm D, Rojas-Sanchez L, Theobald H, Sokolova V, Wyatt RT, Uberla K, Epple M, and Temchura V (2019). Calcium Phosphate Nanoparticle-Based Vaccines as a Platform for Improvement of HIV-1 Env Ab Responses by Intrastructural Help. Nanomaterials (Basel) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darricarrere N, Pougatcheva S, Duan X, Rudicell RS, Chou TH, DiNapoli J, Ross TM, Alefantis T, Vogel TU, Kleanthous H, et al. (2018). Development of a Pan-H1 Influenza Vaccine. J Virol 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Taeye SW, Ozorowski G, Torrents de la Pena A, Guttman M, Julien JP, van den Kerkhof TL, Burger JA, Pritchard LK, Pugach P, Yasmeen A, et al. (2015). Immunogenicity of Stabilized HIV-1 Envelope Trimers with Reduced Exposure of Non-neutralizing Epitopes. Cell 163, 1702–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey AK, Cupo A, Ozorowski G, Sharma VK, Behrens AJ, Go EP, Ketas TJ, Yasmeen A, Klasse PJ, Sayeed E, et al. (2018). cGMP production and analysis of BG505 SOSIP.664, an extensively glycosylated, trimeric HIV-1 envelope glycoprotein vaccine candidate. Biotechnol Bioeng 115, 885–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, et al. (2002). HIV preferentially infects HIV-specific CD4+ T cells. Nature 417, 95–98. [DOI] [PubMed] [Google Scholar]
- Elsayed H, Nabi G, McKinstry WJ, Khoo KK, Mak J, Salazar AM, Tenbusch M, Temchura V, and Uberla K (2018). Intrastructural Help: Harnessing T Helper Cells Induced by Licensed Vaccines for Improvement of HIV Env Ab Responses to Virus-Like Particle Vaccines. J Virol 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escolano A, Dosenovic P, and Nussenzweig MC (2017). Progress toward active or passive HIV-1 vaccination. J Exp Med 214, 3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excler JL, and Kim JH (2019). Novel prime-boost vaccine strategies against HIV-1. Expert Rev Vaccines 18, 765–779. [DOI] [PubMed] [Google Scholar]
- Ezekowitz RA, Kuhlman M, Groopman JE, and Byrn RA (1989). A human serum mannose-binding protein inhibits in vitro infection by the human immunodeficiency virus. J Exp Med 169, 185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts TR, Bagley K, Prado IJ, Bobb KL, Schwartz JA, Xu R, Zagursky RJ, Egan MA, Eldridge JH, LaBranche CC, et al. (2015). Balance of cellular and humoral immunity determines the level of protection by HIV vaccines in rhesus macaque models of HIV infection. Proc Natl Acad Sci U S A 112, E992–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts TR, DeVico AL, Onyabe DY, Shata MT, Bagley KC, Lewis GK, and Hone DM (2003). Progress toward the development of a bacterial vaccine vector that induces high-titer long-lived broadly neutralizing Abs against HIV-1. FEMS Immunol Med Microbiol 37, 129–134. [DOI] [PubMed] [Google Scholar]
- Francica JR, Sheng Z, Zhang Z, Nishimura Y, Shingai M, Ramesh A, Keele BF, Schmidt SD, Flynn BJ, Darko S, et al. (2015). Analysis of immunoglobulin transcripts and hypermutation following SHIV(AD8) infection and protein-plus-adjuvant immunization. Nat Commun 6, 6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert PB, Peterson ML, Follmann D, Hudgens MG, Francis DP, Gurwith M, Heyward WL, Jobes DV, Popovic V, Self SG, et al. (2005). Correlation between immunologic responses to a recombinant glycoprotein 120 vaccine and incidence of HIV-1 infection in a phase 3 HIV-1 preventive vaccine trial. J Infect Dis 191, 666–677. [DOI] [PubMed] [Google Scholar]
- Gor DO, Rose NR, and Greenspan NS (2003). TH1-TH2: a procrustean paradigm. Nat Immunol 4, 503–505. [DOI] [PubMed] [Google Scholar]
- Grundner C, Pancera M, Kang JM, Koch M, Sodroski J, and Wyatt R (2004). Factors limiting the immunogenicity of HIV-1 gp120 envelope glycoproteins. Virology 330, 233–248. [DOI] [PubMed] [Google Scholar]
- Gupta S, Clark ES, Termini JM, Boucher J, Kanagavelu S, LeBranche CC, Abraham S, Montefiori DC, Khan WN, and Stone GW (2015). DNA vaccine molecular adjuvants SP-D-BAFF and SP-D-APRIL enhance anti-gp120 immune response and increase HIV-1 neutralizing Ab titers. J Virol 89, 4158–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenar-Daughton C, Abbott RK, Schief WR, and Crotty S (2018). When designing vaccines, consider the starting material: the human B cell repertoire. Curr Opin Immunol 53, 209–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Kelsoe G, Harrison SC, and Kepler TB (2012). B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat Biotechnol 30, 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R, Lu M, Zhou Y, Mu Z, Parks R, Han Q, Hsu AL, Carter E, Blanchard SC, Edwards RJ, et al. (2020). Disruption of the HIV-1 Envelope allosteric network blocks CD4-induced rearrangements. Nat Commun 11, 520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess R, Storcksdieck Genannt Bonsmann M, Lapuente D, Maaske A, Kirschning C, Ruland J, Lepenies B, Hannaman D, Tenbusch M, and Uberla K (2019). Glycosylation of HIV Env Impacts IgG Subtype Responses to Vaccination. Viruses 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia Y, Bale JB, Gonen S, Shi D, Sheffler W, Fong KK, Nattermann U, Xu C, Huang PS, Ravichandran R, et al. (2016). Design of a hyperstable 60-subunit protein dodecahedron. [corrected]. Nature 535, 136–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingale J, Stano A, Guenaga J, Sharma SK, Nemazee D, Zwick MB, and Wyatt RT (2016). High-Density Array of Well-Ordered HIV-1 Spikes on Synthetic Liposomal Nanoparticles Efficiently Activate B Cells. Cell Rep 15, 1986–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelicic K, Cimbro R, Nawaz F, Huang da W, Zheng X, Yang J, Lempicki RA, Pascuccio M, Van Ryk D, Schwing C, et al. (2013). The HIV-1 envelope protein gp120 impairs B cell proliferation by inducing TGF-beta1 production and FcRL4 expression. Nat Immunol 14, 1256–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce MG, Georgiev IS, Yang Y, Druz A, Geng H, Chuang GY, Kwon YD, Pancera M, Rawi R, Sastry M, et al. (2017). Soluble Prefusion Closed DS-SOSIP.664-Env Trimers of Diverse HIV-1 Strains. Cell Rep 21, 2992–3002. [DOI] [PubMed] [Google Scholar]
- Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, et al. (2013). Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342, 1477–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadelka C, Liechti T, Ebner H, Schanz M, Rusert P, Friedrich N, Stiegeler E, Braun DL, Huber M, Scherrer AU, et al. (2018). Distinct, IgG1-driven Ab response landscapes demarcate individuals with broadly HIV-1 neutralizing activity. J Exp Med 215, 1589–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekiyo M, Joyce MG, Gillespie RA, Gallagher JR, Andrews SF, Yassine HM, Wheatley AK, Fisher BE, Ambrozak DR, Creanga A, et al. (2019). Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat Immunol 20, 362–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsoe G, and Haynes BF (2018). What Are the Primary Limitations in B-Cell Affinity Maturation, and How Much Affinity Maturation Can We Drive with Vaccination? Breaking through Immunity’s Glass Ceiling. Cold Spring Harb Perspect Biol 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepler TB, and Wiehe K (2017). Genetic and structural analyses of affinity maturation in the humoral response to HIV-1. Immunol Rev 275, 129–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasse J, and Blomberg J (1987). Patterns of Abs to human immunodeficiency virus proteins in different subclasses of IgG. J Infect Dis 156, 1026–1030. [DOI] [PubMed] [Google Scholar]
- Klasse PJ (2007). Modeling how many envelope glycoprotein trimers per virion participate in human immunodeficiency virus infectivity and its neutralization by Ab. Virology 369, 245–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasse PJ (2012). The molecular basis of HIV entry. Cell Microbiol 14, 1183–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasse PJ (2014). Neutralization of virus infectivity by Abs: old problems in new perspectives. Adv Biol 2014, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasse PJ, LaBranche CC, Ketas TJ, Ozorowski G, Cupo A, Pugach P, Ringe RP, Golabek M, van Gils MJ, Guttman M, et al. (2016). Sequential and simultaneous immunization of rabbits with HIV-1 envelope glycoprotein SOSIP.664 trimers from Clades A, B and C. PLoS Pathog 12, e1005864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasse PJ, and Moore JP (2004). Is there enough gp120 in the body fluids of HIV-1-infected individuals to have biologically significant effects? Virology 323, 1–8. [DOI] [PubMed] [Google Scholar]
- Klasse PJ, Sanders RW, Cerutti A, and Moore JP (2012). How can HIV-type-1-Env immunogenicity be improved to facilitate Ab-based vaccine development? AIDS Res Hum Retroviruses 28, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev IS, Pancera M, Zhou T, Incesu RB, Fu BZ, et al. (2013a). Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell 153, 126–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Mouquet H, Dosenovic P, Scheid JF, Scharf L, and Nussenzweig MC (2013b). Abs in HIV-1 vaccine development and therapy. Science 341, 1199–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein JS, and Bjorkman PJ (2010). Few and far between: how HIV may be evading Ab avidity. PLoS Pathog 6, e1000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp DW, Steichen JM, Pauthner M, Hu X, Schiffner T, Liguori A, Cottrell CA, Havenar-Daughton C, Ozorowski G, Georgeson E, et al. (2017). Structure-based design of native-like HIV-1 envelope trimers to silence non-neutralizing epitopes and eliminate CD4 binding. Nat Commun 8, 1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YD, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, Joyce MG, Guttman M, Ma X, Narpala S, et al. (2015). Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat Struct Mol Biol 22, 522–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, et al. (2002). HIV-1 evades Ab-mediated neutralization through conformational masking of receptor-binding sites. Nature 420, 678–682. [DOI] [PubMed] [Google Scholar]
- Kwong PD, and Mascola JR (2018). HIV-1 vaccines based on Ab identification, B cell ontogeny, and epitope structure. Immunity 48, 855–871. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, and Hendrickson WA (1998). Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human Ab. Nature 393, 648–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBranche CC, Henderson R, Hsu A, Behrens S, Chen X, Zhou T, Wiehe K, Saunders KO, Alam SM, Bonsignori M, et al. (2019). Neutralization-guided design of HIV-1 envelope trimers with high affinity for the unmutated common ancestor of CH235 lineage CD4bs broadly neutralizing Abs. PLoS Pathog 15, e1008026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake P, and Mitchison NA (1976). Associative control of the immune response to cell surface antigens. Immunol Commun 5, 795–805. [DOI] [PubMed] [Google Scholar]
- Landry SJ (2000). Helper T-cell epitope immunodominance associated with structurally stable segments of hen egg lysozyme and HIV gp120. J Theor Biol 203, 189–201. [DOI] [PubMed] [Google Scholar]
- Lee JH, Andrabi R, Su CY, Yasmeen A, Julien JP, Kong L, Wu NC, McBride R, Sok D, Pauthner M, et al. (2017). A broadly neutralizing Ab targets the dynamic HIV envelope trimer apex via a long, rigidified, and anionic beta-hairpin structure. Immunity 46, 690–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Ozorowski G, and Ward AB (2016). Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 351, 1043–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis GK (2010). Challenges of Ab-mediated protection against HIV-1. Expert Rev Vaccines 9, 683–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis GK, DeVico AL, and Gallo RC (2014). Ab persistence and T-cell balance: two key factors confronting HIV vaccine development. Proc Natl Acad Sci U S A 111, 15614–15621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang S, Kong R, Ding W, Lee FH, Parker Z, Kim E, Learn GH, Hahn P, Policicchio B, et al. (2016). Envelope residue 375 substitutions in simian-human immunodeficiency viruses enhance CD4 binding and replication in rhesus macaques. Proc Natl Acad Sci U S A 113, E3413–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Xu CF, Blais S, Wan Q, Zhang HT, Landry SJ, and Hioe CE (2009). Proximal glycans outside of the epitopes regulate the presentation of HIV-1 envelope gp120 helper epitopes. J Immunol 182, 6369–6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Haque MA, and Blum JS (2002). Role of disulfide bonds in regulating antigen processing and epitope selection. J Immunol 169, 2444–2450. [DOI] [PubMed] [Google Scholar]
- Li T, Steede NK, Nguyen HN, Freytag LC, McLachlan JB, Mettu RR, Robinson JE, and Landry SJ (2014). Comprehensive analysis of contributions from protein conformational stability and major histocompatibility complex class II-peptide binding affinity to CD4+ epitope immunogenicity in HIV-1 envelope glycoprotein. J Virol 88, 9605–9615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Clayton K, Gao W, Li Y, Zealey C, Budylowski P, Schwartz J, Yue FY, Bie Y, Rini J, et al. (2020). Trimeric HIV-1 gp140 fused with APRIL, BAFF, and CD40L on the mucosal gp140-specific Ab responses in mice. Vaccine 38, 2149–2159. [DOI] [PubMed] [Google Scholar]
- Lu M, Ma X, Castillo-Menendez LR, Gorman J, Alsahafi N, Ermel U, Terry DS, Chambers M, Peng D, Zhang B, et al. (2019). Associating HIV-1 envelope glycoprotein structures with states on the virus observed by smFRET. Nature 568, 415–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, et al. (2013). Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342, 1484–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcandalli J, Fiala B, Ols S, Perotti M, de van der Schueren W, Snijder J, Hodge E, Benhaim M, Ravichandran R, Carter L, et al. (2019). Induction of potent neutralizing Ab responses by a designed protein nanoparticle vaccine for respiratory syncytial virus. Cell 176, 1420–1431 e1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Murillo P, Tran K, Guenaga J, Lindgren G, Adori M, Feng Y, Phad GE, Vazquez Bernat N, Bale S, Ingale J, et al. (2017). Particulate Array of Well-Ordered HIV Clade C Env Trimers Elicits Neutralizing Abs that Display a Unique V2 Cap Approach. Immunity 46, 804–817 e807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Sanchez ME, Huerta L, Alvarez-Buylla ER, and Villarreal Lujan C (2018). Role of Cytokine Combinations on CD4+ T Cell Differentiation, Partial Polarization, and Plasticity: Continuous Network Modeling Approach. Front Physiol 9, 877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews K, Chung NP, Klasse PJ, Moore JP, and Sanders RW (2012). Potent induction of Ab-secreting B cells by human dermal-derived CD14+ dendritic cells triggered by dual TLR ligation. J Immunol 189, 5729–5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews K, Chung NP, Klasse PJ, Moutaftsi M, Carter D, Salazar AM, Reed SG, Sanders RW, and Moore JP (2013). Clinical adjuvant combinations stimulate potent B-cell responses in vitro by activating dermal dendritic cells. PLoS One 8, e63785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire AT (2019). Targeting broadly neutralizing Ab precursors: a naive approach to vaccine design. Curr Opin HIV AIDS 14, 294–301. [DOI] [PubMed] [Google Scholar]
- Medina-Ramirez M, Garces F, Escolano A, Skog P, de Taeye SW, Del Moral-Sanchez I, McGuire AT, Yasmeen A, Behrens AJ, Ozorowski G, et al. (2017). Design and crystal structure of a native-like HIV-1 envelope trimer that engages multiple broadly neutralizing Ab precursors in vivo. J Exp Med 214, 2573–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchers M, Bontjer I, Tong T, Chung NP, Klasse PJ, Eggink D, Montefiori DC, Gentile M, Cerutti A, Olson WC, et al. (2012). Targeting HIV-1 envelope glycoprotein trimers to B cells by using APRIL improves Ab responses. J Virol 86, 2488–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirano-Bascos D, Steede NK, Robinson JE, and Landry SJ (2010). Influence of disulfide-stabilized structure on the specificity of helper T-cell and Ab responses to HIV envelope glycoprotein gp120. J Virol 84, 3303–3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirano-Bascos D, Tary-Lehmann M, and Landry SJ (2008). Antigen structure influences helper T-cell epitope dominance in the human immune response to HIV envelope glycoprotein gp120. Eur J Immunol 38, 1231–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP (2019). Nature https://www.nature.com/articles/s41586-019-1101-y#article-comments.
- Moore JP, Cao Y, Ho DD, and Koup RA (1994). Development of the anti-gp120 Ab response during seroconversion to human immunodeficiency virus type 1. J Virol 68, 5142–5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer TJ, Kato Y, Abraham W, Chang JYH, Kulp DW, Watson N, Turner HL, Menis S, Abbott RK, Bhiman JN, et al. (2020). Engineered immunogen binding to alum adjuvant enhances humoral immunity. Nat Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson AN, Goswami R, Dennis M, Tu J, Mangan RJ, Saha PT, Cain DW, Curtis AD, Shen X, Shaw GM, et al. (2019). Simian-Human Immunodeficiency Virus SHIV.CH505-Infected Infant and Adult Rhesus Macaques Exhibit Similar Env-Specific Ab Kinetics, despite Distinct T-Follicular Helper and Germinal Center B Cell Landscapes. J Virol 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozorowski G, Cupo A, Golabek M, LoPiccolo M, Ketas TA, Cavallary M, Cottrell CA, Klasse PJ, Ward AB, and Moore JP (2018). Effects of Adjuvants on HIV-1 Envelope Glycoprotein SOSIP Trimers In Vitro. J Virol 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Peng H, Chen B, and Harrison SC (2020). Cryo-EM Structure of Full-length HIV-1 Env Bound With the Fab of Ab PG16. J Mol Biol 432, 1158–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, et al. (2014). Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauthner M, Havenar-Daughton C, Sok D, Nkolola JP, Bastidas R, Boopathy AV, Carnathan DG, Chandrashekar A, Cirelli KM, Cottrell CA, et al. (2017). Elicitation of Robust Tier 2 Neutralizing Ab Responses in Nonhuman Primates by HIV Envelope Trimer Immunization Using Optimized Approaches. Immunity 46, 1073–1088 e1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauthner MG, Nkolola JP, Havenar-Daughton C, Murrell B, Reiss SM, Bastidas R, Prevost J, Nedellec R, von Bredow B, Abbink P, et al. (2019). Vaccine-Induced Protection from Homologous Tier 2 SHIV Challenge in Nonhuman Primates Depends on Serum-Neutralizing Ab Titers. Immunity 50, 241–252 e246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B (2019). Immunology taught by vaccines. Science 366, 1074–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B, and Ahmed R (2006). Translating innate immunity into immunological memory: implications for vaccine development. Cell 124, 849–863. [DOI] [PubMed] [Google Scholar]
- Purcell AW, van Driel IR, and Gleeson PA (2008). Impact of glycans on T-cell tolerance to glycosylated self-antigens. Immunol Cell Biol 86, 574–579. [DOI] [PubMed] [Google Scholar]
- Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, Pirani A, Gernert K, Deng J, Marzolf B, et al. (2009). Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol 10, 116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regoes RR, and Magnus C (2015). The role of chance in primate lentiviral infectivity: from protomer to host organism. Prog Mol Biol Transl Sci 129, 327–351. [DOI] [PubMed] [Google Scholar]
- Ringe RP, Cruz Portillo VM, Dosenovic P, Ketas TJ, Ozorowski G, Nogal B, Perez L, LaBranche CC, Lim J, Francomano E, et al. (2020). Neutralizing Ab induction by HIV-1 Envelope glycoprotein SOSIP trimers on iron oxide nanoparticles may be impaired by mannose binding lectin. J Virol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringe RP, Ozorowski G, Rantalainen K, Struwe WB, Matthews K, Torres JL, Yasmeen A, Cottrell CA, Ketas TJ, LaBranche CC, et al. (2017). Reducing V3 antigenicity and immunogenicity on soluble, native-like HIV-1 Env SOSIP trimers. J Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringe RP, Pugach P, Cottrell CA, LaBranche CC, Seabright GE, Ketas TJ, Ozorowski G, Kumar S, Schorcht A, van Gils MJ, et al. (2019). Closing and Opening Holes in the Glycan Shield of HIV-1 Envelope Glycoprotein SOSIP Trimers Can Redirect the Neutralizing Ab Response to the Newly Unmasked Epitopes. J Virol 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb ML, Rerks-Ngarm S, Nitayaphan S, Pitisuttithum P, Kaewkungwal J, Kunasol P, Khamboonruang C, Thongcharoen P, Morgan P, Benenson M, et al. (2012). Risk behaviour and time as covariates for efficacy of the HIV vaccine regimen ALVAC-HIV (vCP1521) and AIDSVAX B/E: a post-hoc analysis of the Thai phase 3 efficacy trial RV 144. Lancet Infect Dis 12, 531–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, et al. (2013). A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing Abs. PLoS Pathog 9, e1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, and Moore JP (2017). Native-like Env trimers as a platform for HIV-1 vaccine design. Immunol Rev 275, 161–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, et al. (2015). HIV-1 VACCINES. HIV-1 neutralizing Abs induced by native-like envelope trimers. Science 349, aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Kalia V, Murphey-Corb M, and Montelaro RC (2002a). Characterization of CD4+ T helper cell fine specificity to the envelope glycoproteins of simian immunodeficiency virus. J Med Primatol 31, 194–204. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Kalia V, Murphey-Corb M, and Montelaro RC (2002b). Detailed analysis of CD4+ Th responses to envelope and Gag proteins of simian immunodeficiency virus reveals an exclusion of broadly reactive Th epitopes from the glycosylated regions of envelope. J Immunol 168, 4001–4011. [DOI] [PubMed] [Google Scholar]
- Schommers P, Gruell H, Abernathy ME, Tran MK, Dingens AS, Gristick HB, Barnes CO, Schoofs T, Schlotz M, Vanshylla K, et al. (2020). Restriction of HIV-1 Escape by a Highly Broad and Potent Neutralizing Ab. Cell 180, 471–489 e422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz O, Alizon M, Heard JM, and Danos O (1994). Impairment of T cell receptor-dependent stimulation in CD4+ lymphocytes after contact with membrane-bound HIV-1 envelope glycoprotein. Virology 198, 360–365. [DOI] [PubMed] [Google Scholar]
- Shan M, Klasse PJ, Banerjee K, Dey AK, Iyer SP, Dionisio R, Charles D, Campbell-Gardener L, Olson WC, Sanders RW, et al. (2007). HIV-1 gp120 mannoses induce immunosuppressive responses from dendritic cells. PLoS Pathog 3, e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist CA, and Aspinall R (2009). B-cell responses to vaccination at the extremes of age. Nat Rev Immunol 9, 185–194. [DOI] [PubMed] [Google Scholar]
- Simonich CA, Doepker L, Ralph D, Williams JA, Dhar A, Yaffe Z, Gentles L, Small CT, Oliver B, Vigdorovich V, et al. (2019). Kappa chain maturation helps drive rapid development of an infant HIV-1 broadly neutralizing Ab lineage. Nat Commun 10, 2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonich CA, Williams KL, Verkerke HP, Williams JA, Nduati R, Lee KK, and Overbaugh J (2016). HIV-1 Neutralizing Abs with Limited Hypermutation from an Infant. Cell 166, 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjolander S, Bolmstedt A, Akerblom L, Horal P, Olofsson S, Morein B, and Sjolander A (1996). N-linked glycans in the CD4-binding domain of human immunodeficiency virus type 1 envelope glycoprotein gp160 are essential for the in vivo priming of T cells recognizing an epitope located in their vicinity. Virology 215, 124–133. [DOI] [PubMed] [Google Scholar]
- Sok D, and Burton DR (2018). Recent progress in broadly neutralizing Abs to HIV. Nat Immunol 19, 1179–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtmueller BM, Bridges MD, Dam KM, Lerch MT, Huey-Tubman KE, Hubbell WL, and Bjorkman PJ (2018). DEER Spectroscopy Measurements Reveal Multiple Conformations of HIV-1 SOSIP Envelopes that Show Similarities with Envelopes on Native Virions. Immunity 49, 235–246 e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundling C, Martinez P, Soldemo M, Spangberg M, Bengtsson KL, Stertman L, Forsell MN, and Karlsson Hedestam GB (2013). Immunization of macaques with soluble HIV type 1 and influenza virus envelope glycoproteins results in a similarly rapid contraction of peripheral B-cell responses after boosting. J Infect Dis 207, 426–431. [DOI] [PubMed] [Google Scholar]
- Surman S, Lockey TD, Slobod KS, Jones B, Riberdy JM, White SW, Doherty PC, and Hurwitz JL (2001). Localization of CD4+ T cell epitope hotspots to exposed strands of HIV envelope glycoprotein suggests structural influences on antigen processing. Proc Natl Acad Sci U S A 98, 4587–4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam HH, Melo MB, Kang M, Pelet JM, Ruda VM, Foley MH, Hu JK, Kumari S, Crampton J, Baldeon AD, et al. (2016). Sustained antigen availability during germinal center initiation enhances Ab responses to vaccination. Proc Natl Acad Sci U S A 113, E6639–E6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toapanta FR, Craigo JK, Montelaro RC, and Ross TM (2007). Reduction of anti-HIV-1 Gag immune responses during co-immunization: immune interference by the HIV-1 envelope. Curr HIV Res 5, 199–209. [DOI] [PubMed] [Google Scholar]
- Tokatlian T, Kulp DW, Mutafyan AA, Jones CA, Menis S, Georgeson E, Kubitz M, Zhang MH, Melo MB, Silva M, et al. (2018). Enhancing Humoral Responses Against HIV Envelope Trimers via Nanoparticle Delivery with Stabilized Synthetic Liposomes. Sci Rep 8, 16527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokatlian T, Read BJ, Jones CA, Kulp DW, Menis S, Chang JYH, Steichen JM, Kumari S, Allen JD, Dane EL, et al. (2019). Innate immune recognition of glycans targets HIV nanoparticle immunogens to germinal centers. Science 363, 649–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrents de la Pena A, Julien JP, de Taeye SW, Garces F, Guttman M, Ozorowski G, Pritchard LK, Behrens AJ, Go EP, Burger JA, et al. (2017). Improving the immunogenicity of native-like HIV-1 envelope trimers by hyperstabilization. Cell Rep 20, 1805–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagh K, Kreider EF, Li Y, Barbian HJ, Learn GH, Giorgi E, Hraber PT, Decker TG, Smith AG, Gondim MV, et al. (2018). Completeness of HIV-1 Envelope Glycan hield at Transmission Determines Neutralization Breadth. Cell Rep 25, 893–908 e897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahren B, Chiodi F, Ljunggren K, Putney S, Kurth R, Gallo RC, and Fenyo EM (1988). B and T cell reactivities after immunization of macaques with HIV subcomponents. AIDS Res Hum Retroviruses 4, 199–210. [DOI] [PubMed] [Google Scholar]
- Ward AB, and Wilson IA (2017). The HIV-1 envelope glycoprotein structure: nailing down a moving target. Immunol Rev 275, 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CJ, Boyington JC, McTamney PM, Kong WP, Pearce MB, Xu L, Andersen H, Rao S, Tumpey TM, Yang ZY, et al. (2010). Induction of broadly neutralizing H1N1 influenza Abs by vaccination. Science 329, 1060–1064. [DOI] [PubMed] [Google Scholar]
- Weinhold KJ, Lyerly HK, Stanley SD, Austin AA, Matthews TJ, and Bolognesi DP (1989). HIV-1 GP120-mediated immune suppression and lymphocyte destruction in the absence of viral infection. J Immunol 142, 3091–3097. [PubMed] [Google Scholar]
- Weiss RA (2013). Thirty years on: HIV receptor gymnastics and the prevention of infection. BMC Biol 11, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman D, Rabin RL, Arthos J, Rubbert A, Dybul M, Swofford R, Venkatesan S, Farber JM, and Fauci AS (1997). Macrophage-tropic HIV and SIV envelope proteins induce a signal through the CCR5 chemokine receptor. Nature 389, 981–985. [DOI] [PubMed] [Google Scholar]
- Werdelin O, Meldal M, and Jensen T (2002). Processing of glycans on glycoprotein and glycopeptide antigens in antigen-presenting cells. Proc Natl Acad Sci U S A 99, 9611–9613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP Jr., Scharf L, Scheid JF, Klein F, Bjorkman PJ, and Nussenzweig MC (2014). Structural insights on the role of Abs in HIV-1 vaccine and therapy. Cell 156, 633–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfert MA, and Boons GJ (2013). Adaptive immune activation: glycosylation does matter. Nat Chem Biol 9, 776–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PF, Mestecky J, McElrath MJ, Keefer MC, Gorse GJ, Goepfert PA, Moldoveanu Z, Schwartz D, Spearman PW, El Habib R, et al. (2004). Comparison of systemic and mucosal delivery of 2 canarypox virus vaccines expressing either HIV-1 genes or the gene for rabies virus G protein. J Infect Dis 189, 1221–1231. [DOI] [PubMed] [Google Scholar]
- Young B, Sadarangani S, Jiang L, Wilder-Smith A, and Chen MI (2018). Duration of Influenza Vaccine Effectiveness: A Systematic Review, Meta-analysis, and Meta-regression of Test-Negative Design Case-Control Studies. J Infect Dis 217, 731–741. [DOI] [PubMed] [Google Scholar]
- Yu HT, Tian D, Wang JY, Guo CX, Li Y, Wang X, Li D, Zhang FM, Zhuang M, and Ling H (2014). An HIV-1 envelope immunogen with W427S mutation in CD4 binding site induced more T follicular helper memory cells and reduced non-specific Ab responses. PLoS One 9, e115047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HT, Wang JY, Tian D, Wang MX, Li Y, Yuan L, Chen WJ, Li D, Zhuang M, and Ling H (2016). Comparison of the patterns of Ab recall responses to HIV-1 gp120 and hepatitis B surface antigen in immunized mice. Vaccine 34, 6276–6284. [DOI] [PubMed] [Google Scholar]
- Yuan L, Chen WJ, Wang JY, Li Y, Tian D, Wang MX, Yu HT, Xu YC, Li D, Zhuang M, et al. (2018). Divergent Primary Immune Responses Induced by Human Immunodeficiency Virus-1 gp120 and Hepatitis B Surface Antigen Determine Ab Recall Responses. Virol Sin 33, 502–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Gorman J, Geng H, Liu Q, Lin Y, Tsybovsky Y, Go EP, Dey B, Andine T, Kwon A, et al. (2018). Interdomain Stabilization Impairs CD4 Binding and Improves Immunogenicity of the HIV-1 Envelope Trimer. Cell Host Microbe 23, 832–844 e836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Doria-Rose NA, Cheng C, Stewart-Jones GBE, Chuang GY, Chambers M, Druz A, Geng H, McKee K, Kwon YD, et al. (2017). Quantification of the impact of the HIV-1-glycan shield on Ab elicitation. Cell Rep 19, 719–732. [DOI] [PMC free article] [PubMed] [Google Scholar]


