Targeting a key enzyme in SARS-CoV-2
Scientists across the world are working to understand severe acute respiratory syndrome–coronavirus 2 (SARS-CoV-2), the virus that causes coronavirus disease 2019 (COVID-19). Zhang et al. determined the x-ray crystal structure of a key protein in the virus' life cycle: the main protease. This enzyme cuts the polyproteins translated from viral RNA to yield functional viral proteins. The authors also developed a lead compound into a potent inhibitor and obtained a structure with the inhibitor bound, work that may provide a basis for development of anticoronaviral drugs.
Optimized inhibitor of a key enzyme of the novel coronavirus exhibits pronounced lung tropism.
Abstract
The coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome–coronavirus 2 (SARS-CoV-2) is a global health emergency. An attractive drug target among coronaviruses is the main protease (Mpro, also called 3CLpro) because of its essential role in processing the polyproteins that are translated from the viral RNA. We report the x-ray structures of the unliganded SARS-CoV-2 Mpro and its complex with an α-ketoamide inhibitor. This was derived from a previously designed inhibitor but with the P3-P2 amide bond incorporated into a pyridone ring to enhance the half-life of the compound in plasma. On the basis of the unliganded structure, we developed the lead compound into a potent inhibitor of the SARS-CoV-2 Mpro. The pharmacokinetic characterization of the optimized inhibitor reveals a pronounced lung tropism and suitability for administration by the inhalative route.
In December 2019, a new coronavirus caused an outbreak of pulmonary disease in the city of Wuhan, the capital of Hubei province in China, and has since spread globally (1, 2). The virus has been named severe acute respiratory syndrome–coronavirus 2 (SARS-CoV-2) (3) because the RNA genome is about 82% identical to that of the SARS coronavirus (SARS-CoV); both viruses belong to clade b of the genus Betacoronavirus (1, 2). The disease caused by SARS-CoV-2 is called coronavirus disease 2019 (COVID-19). Whereas at the beginning of the outbreak, cases were connected to the Huanan seafood and animal market in Wuhan, efficient human-to-human transmission led to exponential growth in the number of cases. On 11 March 2020, the World Health Organization (WHO) declared the outbreak a pandemic. As of 9 April, there were >1,500,000 cumulative cases globally, with a ~5.9% case fatality rate.
One of the best-characterized drug targets among coronaviruses is the main protease (Mpro, also called 3CLpro) (4). Along with the papain-like protease(s), this enzyme is essential for processing the polyproteins that are translated from the viral RNA (5). The Mpro operates at no fewer than 11 cleavage sites on the large polyprotein 1ab (replicase 1ab, ~790 kDa); the recognition sequence at most sites is Leu-Gln↓(Ser, Ala, Gly) (↓ marks the cleavage site). Inhibiting the activity of this enzyme would block viral replication. Because no human proteases with a similar cleavage specificity are known, such inhibitors are unlikely to be toxic.
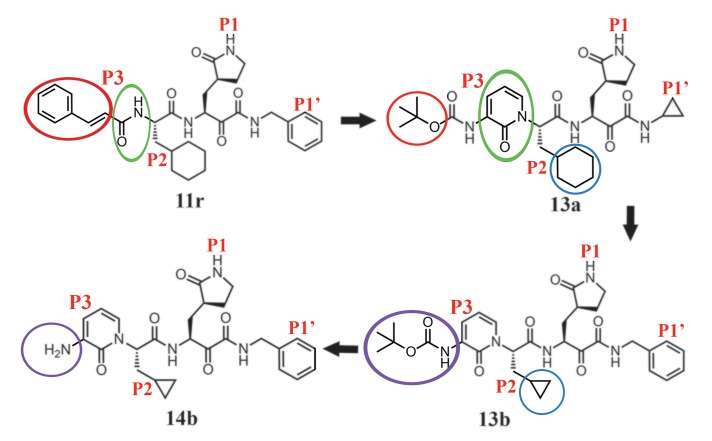
Previously, we designed and synthesized peptidomimetic α-ketoamides as broad-spectrum inhibitors of the main proteases of betacoronaviruses and alphacoronaviruses as well as the 3C proteases of enteroviruses (6). The best of these compounds (11r; Fig. 1) showed an half-maximal effective concentration (EC50) of 400 pM against Middle East respiratory syndrome–coronavirus (MERS-CoV) in Huh7 cells as well as low-μM EC50 values against SARS-CoV and a whole range of enteroviruses in various cell lines, although the antiviral activity seemed to depend to a great extent on the cell type used in the experiments (6). To improve the half-life of the compound in plasma, we modified 11r by hiding the P3-P2 amide bond within a pyridone ring (Fig. 1, green ovals) in the expectation that this might prevent cellular proteases from accessing this bond and cleaving it. Further, to increase the solubility of the compound in plasma and to reduce its binding to plasma proteins, we replaced the hydrophobic cinnamoyl moiety by the somewhat less hydrophobic Boc group (Fig. 1, red ovals) to give 13a (see scheme S1 for synthesis).
Fig. 1. Chemical structures of α-ketoamide inhibitors 11r, 13a, 13b, and 14b.
Colored ovals and circles highlight the modifications from one development step to the next (see text).
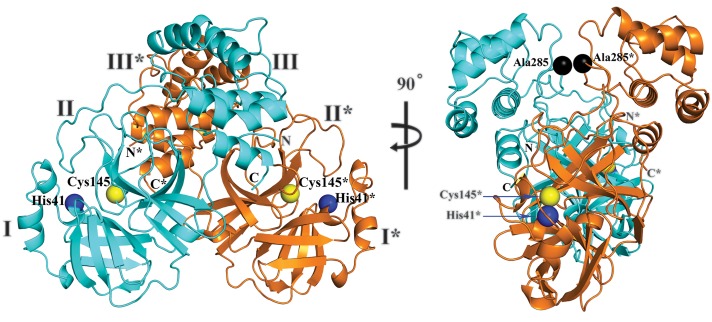
To examine whether the introduced pyridone ring is compatible with the three-dimensional structure of the target, we determined the crystal structure, at 1.75 Å resolution, of the Mpro of SARS-CoV-2 (Fig. 2). The three-dimensional structure is highly similar to that of the SARS-CoV Mpro, as expected from the 96% sequence identity (see fig. S8); the root mean square deviation between the two free-enzyme structures is 0.53 Å for all Cα positions [comparison between SARS-CoV-2 Mpro structure and SARS-CoV Mpro, PDB entry 2BX4 (7)]. The chymotrypsin-like and picornavirus 3C protease–like domains I and II (residues 10 to 99 and 100 to 182, respectively) are six-stranded antiparallel β barrels that harbor the substrate-binding site between them. Domain III (residues 198 to 303), a globular cluster of five helices, is involved in regulating the dimerization of the Mpro, mainly through a salt-bridge interaction between Glu290 of one protomer and Arg4 of the other (8). The tight dimer formed by SARS-CoV-2 Mpro has a contact interface of ~1394 Å2, predominantly between domain II of molecule A and the NH2-terminal residues (“N-finger”) of molecule B, with the two molecules oriented perpendicular to one another (Fig. 2). Dimerization of the enzyme is necessary for catalytic activity, because the N-finger of each of the two protomers interacts with Glu166 of the other protomer and thereby helps shape the S1 pocket of the substrate-binding site (9). To reach this interaction site, the N-finger is squeezed in between domains II and III of the parent monomer and domain II of the other monomer.
Fig. 2. Three-dimensional structure of SARS-CoV-2 Mpro in two different views.
One protomer of the dimer is shown in light blue, the other one in orange. Domains are labeled by Roman numerals. Amino acid residues of the catalytic site are indicated as yellow spheres for Cys145 and blue spheres for His41. Asterisks mark residues from protomer B (orange). Black spheres indicate the positions of Ala285 for each of the two domains III (see text). Chain termini are labeled N and C for molecule A (light blue) and N* and C* for molecule B (orange).
Interestingly, in the SARS-CoV but not in the SARS-CoV-2 Mpro dimer, there is a polar interaction between the two domains III involving a 2.60-Å hydrogen bond between the side-chain hydroxyl groups of residue Thr285 of each protomer, supported by a hydrophobic contact between the side chain of Ile286 and Thr285 Cγ2. In SARS-CoV-2, the threonine is replaced by alanine (indicated by the black spheres in Fig. 2) and the isoleucine by leucine (fig. S8). It was previously shown that replacing Ser284, Thr285, and Ile286 by alanine residues in SARS-CoV Mpro leads to enhancement of the catalytic activity of the protease by a factor of 3.6, concomitant with a slightly closer packing of the two domains III of the dimer against one another (10). This was accompanied by changes in enzyme dynamics that transmit the effect of the mutation to the catalytic center. Indeed, the Thr285 → Ala replacement observed in the SARS-CoV-2 Mpro also allows the two domains III to approach each other more closely (the distance between the Cα atoms of residues 285 in molecules A and B is 6.77 Å in SARS-CoV Mpro and 5.21 Å in SARS-CoV-2 Mpro, and the distance between the centers of mass of the two domains III shrinks from 33.4 Å to 32.1 Å). However, the catalytic efficiency of SARS-CoV-2 Mpro is only slightly higher, if at all [turnover number (kcat)/Michaelis constant (Km) = 3426.1 ± 416.9 s–1 M–1] than that of SARS-CoV Mpro (kcat/Km = 3011.3 ± 294.6 s–1 M–1). Further, the estimated dissociation constant of dimerization is the same (~2.5 μM) for the two enzymes, as determined by analytical ultracentrifugation (fig. S10).
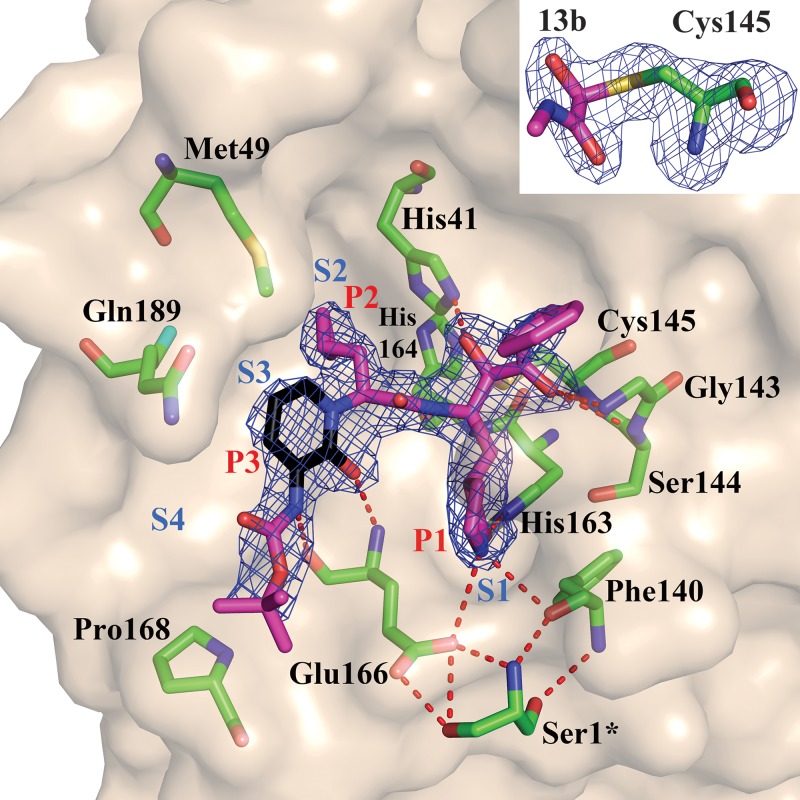
We used this crystal structure to dock the α-ketoamide 13a; this suggested that the pyridone ring might have some steric clash with the side chain of Gln189. However, in our previous work (6), we had found Gln189 to be quite flexible, and therefore we went ahead with 13a as a lead. The plasma half-life of this compound in mice was increased by a factor of ~3 relative to 11r (from 0.3 hours to 1.0 hours), the in vitro kinetic plasma solubility was improved by a factor of ~19 (from 6 μM for 11r to 112 μM for 13a), and the thermodynamic solubility increased by a factor of ~13 (from 41 μM to 530 μM). Binding to mouse plasma protein was reduced from 99% to 97% [many drugs have plasma protein binding of >90% (11)]. However, relative to 11r (IC50 = 0.18 ± 0.02 μM), the structural modification led to some loss of inhibitory activity against the main protease of SARS-CoV-2 (IC50 = 2.39 ± 0.63 μM) as well as the 3C proteases (3Cpro) of enteroviruses. 11r was designed for broad-spectrum activity, with the P2 cyclohexyl moiety intended to fill a pocket in the enterovirus 3Cpro. The S2 pocket of the betacoronavirus Mpro (Fig. 3) features substantial plasticity, enabling it to adapt to the shape of smaller inhibitor moieties (6). To enhance the antiviral activity against betacoronaviruses of clade b (SARS-CoV-2 and SARS-CoV), we sacrificed the goal of broad-spectrum activity and replaced the P2 cyclohexyl moiety of 13a by the smaller cyclopropyl in 13b (Fig. 1, blue circles). Here, we present x-ray crystal structures in two different crystal forms, at 1.95 and 2.20 Å resolution, of the complex between α-ketoamide 13b and the Mpro of SARS-CoV-2. One structure is in space group C2 (Fig. 3), where both protomers of the Mpro dimer are bound by crystal symmetry to have identical conformations; the other is in space group P212121, where the two protomers are independent of each other and free to adopt different conformations. Indeed, we find that in the latter crystal structure, the key residue Glu166 adopts an inactive conformation in protomer B (as evidenced by its distance from His172 and the lack of H-bonding interaction with the P1 moiety of the inhibitor), even though compound 13b is bound in the same mode as in molecule A. This phenomenon has also been observed with the SARS-CoV Mpro (12) and is consistent with the half-site activity described for this enzyme (13). In all copies of the inhibited SARS-CoV-2 Mpro, the inhibitor binds to the shallow substrate-binding site at the surface of each protomer, between domains I and II (Fig. 3).
Fig. 3. Compound 13b in the substrate-binding cleft located between domains I and II of the Mpro in the monoclinic crystal form (space group C2).
Fobs – Fcalc density is shown for the inhibitor (contouring level 3σ). Carbon atoms of the inhibitor are magenta, except in the pyridone ring, which is black; oxygen atoms are red, nitrogens blue, and sulfur yellow. Light blue symbols Sn (n = 1, 2, 3…) indicate the canonical binding pockets for moieties Pn (n = 1, 2, 3…) (red symbols) of the peptidomimetic inhibitor. Hydrogen bonds are indicated by dashed red lines. Note the interaction between Ser1*, the N-terminal residue of molecule B, and Glu166 of molecule A, which is essential for keeping the S1 pocket in the correct shape and the enzyme in the active conformation. Inset: Thiohemiketal formed by the nucleophilic attack of the catalytic cysteine onto the α-carbon of the inhibitor. The stereochemistry of the α-carbon is S. Fobs − Fcalc density (contoured at 3σ) is shown in blue. See fig. S9 for more details.
Through the nucleophilic attack of the catalytic Cys145 onto the α-keto group of the inhibitor, a thiohemiketal is formed in a reversible reaction. This is clearly reflected in the electron density (Fig. 3, inset); the stereochemistry of this chiral moiety is S in all copies of compound 13b in these structures. The oxyanion (or hydroxyl) group of this thiohemiketal is stabilized by a hydrogen bond from His41, whereas the amide oxygen of 13b accepts a hydrogen bond from the main-chain amides of Gly143, Cys145, and partly Ser144, which form the canonical “oxyanion hole” of the cysteine protease. It is an advantage of the α-ketoamides that their warhead can interact with the catalytic center of the target proteases through two hydrogen-bonding interactions (6) rather than only one, as with other warheads such as aldehydes (14) or Michael acceptors (15).
The P1 γ-lactam moiety, designed as a glutamine surrogate (15, 16), is deeply embedded in the S1 pocket of the protease, where the lactam nitrogen donates a three-center (bifurcated) hydrogen bond to the main-chain oxygen of Phe140 (3.20/3.10/3.28 Å; values for the structure in space group C2/space group P212121 molecule A/space group P212121 molecule B) and to the Glu166 carboxylate [3.35/3.33/(3.55) Å], and the carbonyl oxygen accepts a 2.57/2.51/2.81 Å hydrogen bond from the imidazole of His163. The P2 cyclopropyl methyl moiety fits snugly into the S2 subsite, which has shrunk by 28 Å3 relative to the complex between compound 13a with P2 = cyclohexyl methyl and the SARS-CoV Mpro (17). The pyridone in the P3-P2 position of the inhibitor occupies the space normally filled by the substrate’s main chain; its carbonyl oxygen accepts a 2.89/2.99/3.00 Å hydrogen bond from the main-chain amide of residue Glu166. Further, the P3 amide donates a 2.83/2.96/2.87 Å hydrogen bond to the main-chain oxygen of Glu166. Embedded within the pyridone, the P2 nitrogen can no longer donate a hydrogen bond to the protein (the H-bond prevented from forming would connect the P2 nitrogen and the side-chain oxygen of Gln189; these two atoms are highlighted in fig. S9). However, our previous crystal structures showed that the P2 main-chain amide of the linear α-ketoamides does not make a hydrogen bond with the protein in all cases, so this interaction does not seem to be critical (6). The protecting Boc group on P3 does not occupy the canonical S4 site of the protease [in contrast to the protecting groups of other inhibitors in complex with the SARS-CoV Mpro (18)] but is located near Pro168 (3.81/4.17/3.65 Å) (Fig. 3); as a result of this interaction, the latter residue moves outward by more than 2 Å (relative to the structure of the free enzyme). This contact explains why removing the Boc group as in compound 14b (Fig. 1, purple ovals) weakens the inhibitory potency of this compound by a factor of ~2. Interestingly, there is a space between the pyridone ring of 13b, the main chain of residue Thr190, and the side chain of Gln189 (smallest distance: 3.6 Å), which is filled by a dimethyl sulfoxide (DMSO) molecule in the C2 crystal structure and a water molecule in the P212121 structure. This suggests that P3 moieties more bulky than pyridone may be accepted here.
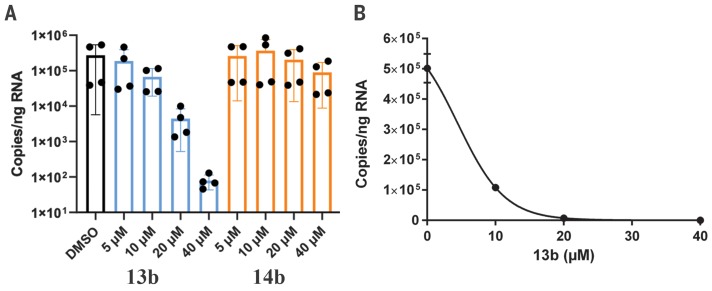
Compound 13b inhibits the purified recombinant SARS-CoV-2 Mpro with IC50 = 0.67 ± 0.18 μM. The corresponding IC50 values for inhibition of the SARS-CoV Mpro and the MERS-CoV Mpro are 0.90 ± 0.29 μM and 0.58 ± 0.22 μM, respectively. In a SARS-CoV replicon (19), RNA replication is inhibited with EC50 = 1.75 ± 0.25 μM. In human Calu-3 cells infected with SARS-CoV-2, an EC50 of 4 to 5 μM was observed, whereas compound 14b lacking the Boc group was almost inactive (Fig. 4). This suggests that the hydrophobic and bulky Boc group is necessary to cross the cellular membrane and that an even more hydrophobic moiety might be advantageous here, although this may again lead to increased plasma protein binding, as observed for the cinnamoyl-containing 11r.
Fig. 4. Compound 13b inhibits SARS-CoV-2 replication in human Calu-3 lung cells.
(A) Calu-3 cells were infected with SARS-CoV-2 using a multiplicity of infection (MOI) of 0.05. Varying amounts (5, 10, 20, or 40 μM) of 13b (blue bars) or 14b (orange bars) were added. DMSO was used as vehicle control (black bar). Total RNA was isolated from cell lysates, and viral RNA content was analyzed by quantitative polymerase chain reaction. Data are means ± SD of two biological experiments with two technical replicates each. (B) For the estimation of the EC50 value of compound 13b against SARS-CoV-2, a dose-response curve was prepared (GraphPad).
To assess the absorption-distribution-metabolism-excretion (ADME) properties of the pyridone-containing α-ketoamides, we first investigated compound 13a. Metabolic stability in mouse and human microsomes was good, with intrinsic clearance rates Clint_mouse = 32.0 μl min–1 (mg protein)–1 and Clint_human = 21.0 μl min–1 (mg protein)–1. This means that after 30 min, ~80% and 60% (for mouse and human, respectively) of residual compound remained metabolically stable. Pharmacokinetic studies in CD-1 mice using the subcutaneous route at 20 mg/kg showed that 13a stayed in plasma for up to 4 hours but was excreted via urine for up to 24 hours. The maximum plasma concentration (Cmax) was determined at 334.5 ng ml−1 and the mean residence time was ~1.6 hours. Although 13a seemed to be cleared very rapidly from plasma, at 24 hours it was found at 135 ng/g tissue in the lung and at 52.7 ng ml−1 in bronchio-alveolar lavage fluid (BALF), which suggests that it was mainly distributed to tissue. Next, we investigated 13b for its pharmacokinetic properties in CD-1 mice using the subcutaneous route as well, but at 3 mg kg−1. The ADME parameters of 13b were similar to those of 13a; in addition, binding to human plasma proteins was found to be 90%. The Cmax of 13b was determined at 126.2 ng ml−1. This is around 37% of the Cmax detected for 13a, although the 13b dosage was lower by a factor of ~7. The mean residence time for 13b was extended to 2.7 hours and the plasma half-life in mice was 1.8 hours. In addition, 13b showed a less rapid clearance relative to 13a (table S3). During the pharmacokinetic study with 13b, we monitored its lung tissue levels. After 4 hours, 13b was still found at ~13 ng g−1 in lung tissue. This lung tropism of 13a and 13b is beneficial given that COVID-19 affects the lungs. In addition to subcutaneous administration, 13b was nebulized using an inhalation device at 3 mg kg−1. After 24 hours, 13b was found at 33 ng g−1 in lung tissue. Inhalation was tolerated well and mice did not show any adverse effects, which suggests that direct administration of the compound to the lungs would be possible. Given these favorable pharmacokinetic results, our study provides a useful framework for the development of the pyridone-containing inhibitors toward anticoronaviral drugs.
Acknowledgments
We thank Y. Kusov and G. Hansen, as well as A. Aljnabi, for determining the inhibitory activities of compounds in a SARS-CoV replicon and against recombinant MERS-CoV Mpro, respectively; T. Biet for recording 13C NMR spectra; A. Ahlers, J. Schreiber, and L. Litz for excellent technical assistance; K. Chen for continuous organizational support; and the staff at beamline 14.2 of BESSY II, Berlin, Germany, for help with diffraction data collection. Funding: We thank the German Center for Infection Research (DZIF) for financial support (projects TTU01, grant 8011801806, and TTU09, grant 8004709710). Author contributions: Conceptualization: L.Z., D.L., R.H.; investigation: L.Z., D.L., X.S., U.C., L.S., S.B., K.R., R.H.; contribution of research materials: C.D.; writing (original draft preparation): R.H., D.L., K.R.; writing (review and editing): L.Z., D.L., U.C., L.S., K.R., R.H.; visualization: L.Z., L.S.; supervision: R.H.; funding acquisition: R.H., S.B., K.R. Competing interests: The University of Lübeck has filed a patent application covering compounds 13a and 13b as well as related compounds with a pyridone structure in the P3-P2 position, with L.Z., D.L., and R.H. as inventors. Data and materials availability: Crystallographic coordinates and structure factors are available from the PDB under accession codes 6Y2E (unliganded Mpro), 6Y2F (complex with 13b in space group C2), and 6Y2G (complex with 13b in space group P212121). The plasmid encoding the SARS-CoV-2 Mpro is freely available. The available amounts of inhibitors are limited. This work is licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/. This license does not apply to figures/photos/artwork or other content included in the article that is credited to a third party; obtain authorization from the rights holder before using such material.
Supplementary Materials
science.sciencemag.org/content/368/6489/409/suppl/DC1
Materials and Methods
Supplementary Text
Scheme S1
Figs. S1 to S10
Tables S1 to S3
References and Notes
- 1.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L., A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E. C., Zhang Y.-Z., A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020). 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorbalenya A. E., Baker S. C., Baric R. S., de Groot R. J., Drosten C., Gulyaeva A. A., Haagmans B. L., Lauber C., Leontovich A. M., Neuman B. W., Penzar D., Perlman S., Poon L. L. M., Samborskiy D., Sidorov I. A., Sola I., Ziebuhr J., Severe acute respiratory syndrome-related coronavirus: The species and its viruses – a statement of the Coronavirus Study Group. Nat. Microbiol. 5, 536–544 (2020). 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anand K., Ziebuhr J., Wadhwani P., Mesters J. R., Hilgenfeld R., Coronavirus main proteinase (3CLpro) structure: Basis for design of anti-SARS drugs. Science 300, 1763–1767 (2003). 10.1126/science.1085658 [DOI] [PubMed] [Google Scholar]
- 5.Hilgenfeld R., From SARS to MERS: Crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 281, 4085–4096 (2014). 10.1111/febs.12936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L., Lin D., Kusov Y., Nian Y., Ma Q., Wang J., von Brunn A., Leyssen P., Lanko K., Neyts J., de Wilde A., Snijder E. J., Liu H., Hilgenfeld R., α-Ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: Structure-based design, synthesis, and activity assessment. J. Med. Chem. acs.jmedchem.9b01828 (2020). 10.1021/acs.jmedchem.9b01828 [DOI] [PubMed] [Google Scholar]
- 7.Tan J., Verschueren K. H. G., Anand K., Shen J., Yang M., Xu Y., Rao Z., Bigalke J., Heisen B., Mesters J. R., Chen K., Shen X., Jiang H., Hilgenfeld R., pH-dependent conformational flexibility of the SARS-CoV main proteinase (Mpro) dimer: Molecular dynamics simulations and multiple X-ray structure analyses. J. Mol. Biol. 354, 25–40 (2005). 10.1016/j.jmb.2005.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi J., Song J., The catalysis of the SARS 3C-like protease is under extensive regulation by its extra domain. FEBS J. 273, 1035–1045 (2006). 10.1111/j.1742-4658.2006.05130.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anand K., Palm G. J., Mesters J. R., Siddell S. G., Ziebuhr J., Hilgenfeld R., Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 21, 3213–3224 (2002). 10.1093/emboj/cdf327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim L., Shi J., Mu Y., Song J., Dynamically-driven enhancement of the catalytic machinery of the SARS 3C-like protease by the S284-T285-I286/A mutations on the extra domain. PLOS ONE 9, e101941 (2014). 10.1371/journal.pone.0101941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kratochwil N. A., Huber W., Müller F., Kansy M., Gerber P. R., Predicting plasma protein binding of drugs: A new approach. Biochem. Pharmacol. 64, 1355–1374 (2002). 10.1016/S0006-2952(02)01074-2 [DOI] [PubMed] [Google Scholar]
- 12.Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., Gao G. F., Anand K., Bartlam M., Hilgenfeld R., Rao Z., The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. U.S.A. 100, 13190–13195 (2003). 10.1073/pnas.1835675100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen H., Wei P., Huang C., Tan L., Liu Y., Lai L., Only one protomer is active in the dimer of SARS 3C-like proteinase. J. Biol. Chem. 281, 13894–13898 (2006). 10.1074/jbc.M510745200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu L., George S., Schmidt M. F., Al-Gharabli S. I., Rademann J., Hilgenfeld R., Peptide aldehyde inhibitors challenge the substrate specificity of the SARS-coronavirus main protease. Antiviral Res. 92, 204–212 (2011). 10.1016/j.antiviral.2011.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan J., George S., Kusov Y., Perbandt M., Anemüller S., Mesters J. R., Norder H., Coutard B., Lacroix C., Leyssen P., Neyts J., Hilgenfeld R., 3C protease of enterovirus 68: Structure-based design of Michael acceptor inhibitors and their broad-spectrum antiviral effects against picornaviruses. J. Virol. 87, 4339–4351 (2013). 10.1128/JVI.01123-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dragovich P. S., Zhou R., Skalitzky D. J., Fuhrman S. A., Patick A. K., Ford C. E., Meador J. W. 3rd, Worland S. T., Solid-phase synthesis of irreversible human rhinovirus 3C protease inhibitors. Part 1: Optimization of tripeptides incorporating N-terminal amides. Bioorg. Med. Chem. 7, 589–598 (1999). 10.1016/S0968-0896(99)00005-X [DOI] [PubMed] [Google Scholar]
- 17.L. Zhang, D. Lin, R. Hilgenfeld, Crystal structure of the complex resulting from the reaction between the SARS-CoV main protease and tert-butyl (1-((S)-3-cyclohexyl-1-(((S)-4-(cyclopropylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl) amino)-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate, PDB ID 6Y7M (2020). 10.2210/pdb6Y7M/pdb [DOI]
- 18.L. Zhu, R. Hilgenfeld, Crystal structure of SARS coronavirus main protease complexed with an alpha, beta-unsaturated ethyl ester inhibitor SG85, PDB ID 3TNT (2012). 10.2210/pdb3TNT/pdb [DOI]
- 19.Kusov Y., Tan J., Alvarez E., Enjuanes L., Hilgenfeld R., A G-quadruplex-binding macrodomain within the “SARS-unique domain” is essential for the activity of the SARS-coronavirus replication-transcription complex. Virology 484, 313–322 (2015). 10.1016/j.virol.2015.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xue X., Yang H., Shen W., Zhao Q., Li J., Yang K., Chen C., Jin Y., Bartlam M., Rao Z., Production of authentic SARS-CoV Mpro with enhanced activity: Application as a novel tag-cleavage endopeptidase for protein overproduction. J. Mol. Biol. 366, 965–975 (2007). 10.1016/j.jmb.2006.11.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mueller U., Darowski N., Fuchs M. R., Förster R., Hellmig M., Paithankar K. S., Pühringer S., Steffien M., Zocher G., Weiss M. S., Facilities for macromolecular crystallography at the Helmholtz-Zentrum Berlin. J. Synchrotron Radiat. 19, 442–449 (2012). 10.1107/S0909049512006395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krug M., Weiss M. S., Heinemann U., Mueller U., XDSAPP: A graphical user interface for the convenient processing of diffraction data using XDS. J. Appl. Crystallogr. 45, 568–572 (2012). 10.1107/S0021889812011715 [DOI] [Google Scholar]
- 23.Evans P., Scaling and assessment of data quality. Acta Crystallogr. D 62, 72–82 (2006). 10.1107/S0907444905036693 [DOI] [PubMed] [Google Scholar]
- 24.Evans P. R., An introduction to data reduction: Space-group determination, scaling and intensity statistics. Acta Crystallogr. D 67, 282–292 (2011). 10.1107/S090744491003982X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S., Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011). 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vagin A., Teplyakov A., Molecular replacement with MOLREP. Acta Crystallogr. D 66, 22–25 (2010). 10.1107/S0907444909042589 [DOI] [PubMed] [Google Scholar]
- 27.Lebedev A. A., Young P., Isupov M. N., Moroz O. V., Vagin A. A., Murshudov G. N., JLigand: A graphical tool for the CCP4 template-restraint library. Acta Crystallogr. D 68, 431–440 (2012). 10.1107/S090744491200251X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010). 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A., REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 (2011). 10.1107/S0907444911001314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y., Kati W., Chen C. M., Tripathi R., Molla A., Kohlbrenner W., Use of a fluorescence plate reader for measuring kinetic parameters with inner filter effect correction. Anal. Biochem. 267, 331–335 (1999). 10.1006/abio.1998.3014 [DOI] [PubMed] [Google Scholar]
- 31.P. H. Brown, A. Balbo, P. Schuck, Characterizing protein-protein interactions by sedimentation velocity analytical ultracentrifugation. Curr. Protoc. Immunol. Chapter 18, Unit 18.15 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Schuck P., Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 (2000). 10.1016/S0006-3495(00)76713-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.M. T. Laue, B. D. Shah, T. M. Rigdeway, S. L. Pelletier, in Analytical Ultracentrifugation in Biochemistry and Polymer Science, S. Harding, A. Rowe, J. Horton, Eds. (Royal Society of Chemistry, 1992), pp. 90–125. [Google Scholar]
- 34.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 35.Tian Q., Nayyar N. K., Babu S., Chen L., Tao J., Lee S., Tibbetts A., Moran T., Liou J., Guo M., Kennedy T. P., An efficient synthesis of a key intermediate for the preparation of the rhinovirus protease inhibitor AG7088 via asymmetric dianionic cyanomethylation of N-Boc-L-(+)-glutamic acid dimethyl ester. Tetrahedron Lett. 42, 6807–6809 (2001). 10.1016/S0040-4039(01)01416-2 [DOI] [Google Scholar]
- 36.Corman V. M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D. K. W., Bleicker T., Brünink S., Schneider J., Schmidt M. L., Mulders D. G. J. C., Haagmans B. L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J. L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M. P. G., Drosten C., Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 25, (2020). 10.2807/1560-7917.ES.2020.25.3.2000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y., Huo M., Zhou J., Xie S., PKSolver: An add-in program for pharmacokinetic and pharmacodynamic data analysis in Microsoft Excel. Comput. Methods Programs Biomed. 99, 306–314 (2010). 10.1016/j.cmpb.2010.01.007 [DOI] [PubMed] [Google Scholar]
- 38.Hsu W. C., Chang H. C., Chou C. Y., Tsai P. J., Lin P. I., Chang G. G., Critical assessment of important regions in the subunit association and catalytic action of the severe acute respiratory syndrome coronavirus main protease. J. Biol. Chem. 280, 22741–22748 (2005). 10.1074/jbc.M502556200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bräutigam C. A., Calculations and publication-quality illustrations for analytical ultracentrifugation data. Methods Enzymol. 562, 109–133 (2015). 10.1016/bs.mie.2015.05.001 [DOI] [PubMed] [Google Scholar]
- 40.Weiss M. S., Hilgenfeld R., On the use of the merging R factor as a quality indicator for X-ray data. J. Appl. Crystallogr. 30, 203–205 (1997). 10.1107/S0021889897003907 [DOI] [Google Scholar]
- 41.Karplus P. A., Diederichs K., Linking crystallographic model and data quality. Science 336, 1030–1033 (2012). 10.1126/science.1218231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010). 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
science.sciencemag.org/content/368/6489/409/suppl/DC1
Materials and Methods
Supplementary Text
Scheme S1
Figs. S1 to S10
Tables S1 to S3