Abstract
Naturally occurring tRNA mutants are known that suppress +1 frameshift mutations by means of an extended anticodon loop, and a few have been used in protein mutagenesis. In an effort to expand the number of possible ways to uniquely and efficiently encode unnatural amino acids, we have devised a general strategy to select tRNAs with the ability to suppress four-base codons from a library of tRNAs with randomized 8 or 9 nt anticodon loops. Our selectants included both known and novel suppressible four-base codons and resulted in a set of very efficient, non-cross-reactive tRNA/four-base codon pairs for AGGA, UAGA, CCCU and CUAG. The most efficient four-base codon suppressors had Watson-Crick complementary anticodons, and the sequences of the anticodon loops outside of the anticodons varied with the anticodon. Additionally, four-base codon reporter libraries were used to identify “shifty” sites at which +1 frameshifting is most favorable in the absence of suppressor tRNAs in Escherichia coli. We intend to use these tRNAs to explore the limits of unnatural polypeptide biosynthesis, both in vitro and eventually in vivo. In addition, this selection strategy is being extended to identify novel five- and six-base codon suppressors.
Keywords: four-base codon, suppressors, genetic code, shifty codons
Introduction
Pioneering work by Crick, Brenner and co-workers in the early 1960s established the triplet nature of the genetic code (Crick et al., 1961), and 40 years later only rare exceptions are known to the universal correspondence of mRNA codon to amino acid (Fox, 1987). However, it has also become clear that frame maintenance during translation is not absolute Atkins et al 1991, Kurland 1992, Parker 1989. In addition to the inherent accuracy limits of the ribosome in maintaining frame, shifts can be promoted by mutant tRNAs and can occur with high frequency at “programmed” sites in mRNA. Frameshifts can be stimulated by such elements as upstream rRNA-binding motifs, stem-loop structures and underused codons. Indeed, these programmed shifts can be critical to protein production, e.g. the gag-pol polyprotein of Rous Sarcoma virus, the Ty1 and Ty3 elements of yeast, the dnaX gene of DNA Pol III tau and gamma units, and the self-regulating RF2 protein in Escherichia coli (Gesteland et al., 1992).
As early as 1968 it was recognized that frameshift mutations could be externally suppressed (Riyasaty & Atkins, 1968). Shortly thereafter, Yourno & Tanemura (1970) showed that protein from a suppressed frameshift was of wild-type sequence, and Riddle & Carbon (1973) showed that the external frameshift suppressor sufD in Salmonella is tRNAGly with a four-base anticodon CCCC instead of the wild-type CCC. (Note that we adopt the convention herein of writing both codon and anticodon sequences in the 5′ to 3′ direction.) These experiments conclusively demonstrated that certain tRNAs with extended anticodons can read non-triplet codons. Most of the known four-base codon suppressors have subsequently been isolated from Salmonella or yeast and are largely single-base insertion mutants of tRNAPro, tRNAGly or tRNALys reading CCCX, GGGX or AAAX, where it is not always known which X can be read. The known four-base codon suppressors tend to act with efficiencies from very poor to approaching 20 % (Atkins et al., 1991).
Less well known are +1 suppressors with more extended anticodon loops. For example, the Salmonella sufT621 derivative of tRNAArg causes insertion of arginine in response to CCGU. A synthetic mutant of this tRNA, which bears a spontaneous mutation in the DHU loop of unknown importance, has a nine-base anticodon loop that causes +1 frameshifting (Tuohy et al., 1992). The only other known tRNAs with nine-base anticodon loops mediating +1 frameshifting are the related tRNAPro-derived yeast suppressors SUF7, SUF8, SUF9 and SUF11, which lack a 31:39 anticodon stem pair (Atkins et al., 1991).
The specificity of pairing is not entirely predictable for extended-anticodon tRNAs. For example, when the suppression of GGGN codons by tRNAs bearing NCCC anticodons (SUF16 derivatives) was analyzed, it was found that the only non-functional N:N pair was G:G. This result indicates that Watson-Crick pairing at the fourth codon nucleotide is even less strictly enforced than at the third “wobble” position for normal codons. However, the identity of the N:N pair does affect suppression efficiency, and canonical pairings in the fourth position gave the strongest suppression (Gaber & Culbertson, 1984). Additionally, it has been found that sufJ, a modified tRNAThr with anticodon UGGU, is capable of decoding ACCC and ACCU in addition to ACCA (ACCG was not tested; Bossi & Roth, 1981).
In an engineered system, Curran & Yarus (1987) tested the ability of derivatives of the glutamine-inserting amber suppressor Su7 with NCUA anticodons to suppress UAGN codons. While the canonical pairs tended to suppress well, some non-canonical pairs were also efficient. For example, the best pair arose from a CCUA anticodon and a UAGA codon; also, the GCUA tRNA preferred UAGA and UAGG to its canonical UAGC. The ability to read a four-base codon versus a three-base codon for these extended-anticodon tRNAs was also assessed, by using tRNAs that decode four-base codons whose first three bases encode a stop codon. In this case there is no competitive translation with a canonical tRNA. In general, four-base decoding predominated when a fourth Watson-Crick pair was possible and was less likely to predominate otherwise. The authors proposed that this is a consequence of mRNA-induced stabilization of a tRNA conformation that favors four-base decoding, possible only when a fourth canonical base-pair is present (Curran & Yarus, 1987).
In contrast, Qian et al. (1998) have proposed that, at least in some cases, extended-anticodon tRNAs merely render the tRNA less capable of three-base decoding and in the process make room for other tRNAs to promiscuously interact with the codon by a slippage mechanism. For example in Salmonella, mutant tRNAPro with a GGGG anticodon is methylated in such a way (underlined G37) that pairing with the mRNA cytosine is obstructed. Apparently, the tRNAPro with cmo5UGG anticodon, which typically reads CCA, CCG and CCU, can read CCC in the absence of its normal cognate tRNAGGG, but favors slippage since there are only two canonical base-pairs.
Four-base codon suppression has begun to be used for in vitro protein engineering experiments, initially by Ma et al., to insert alanine at UAGG and AGGU with complementary anticodons engineered into tRNAAla Kramer et al 1998, Ma et al 1993. Sisido’s group has used extended-anticodon tRNAs to insert unnatural amino acids into proteins in vitro, employing known suppressible four-base codons such as AGGU and CGGG Hohsaka et al 1996, Hohsaka et al 1999b, Murakami et al 1998. Significantly, Hohsaka et al. (1999a), used both of these codons together in a single transcript to insert two different unnatural amino acid residues into the same protein site-specifically.
In an effort to develop improved tRNA/four-base codon pairs for protein mutagenesis, Moore et al. examined the ability of Su6 (amber tRNALeu) derivatives with NCUA anticodons to suppress UAGN codons and also monitored decoding in the −1 and 0 frames. In an RF1-deficient E. coli strain, UCUA suppressed the UAGA codon with up to 26 % efficiency with little decoding in the 0 or −1 frames (Moore et al., 2000). This pair was also fairly efficient in the Curran & Yarus study, and differences between the results of independent experiments are likely due, in part, to the extent of aminoacylation and maturation with different tRNA scaffolds. It is clear that decoding four-base codons with complementary anticodons is generally favorable, but the rules that govern the comparative suppression efficiencies for different four-base codons remain unclear.
Likewise, it is not exhaustively known what governs the likelihood of frameshift in the absence of suppressor tRNAs. Atkins and co-workers established the generality of “leakiness” in frame maintenance by measuring the weak activity from insertion and deletion mutants of the gene for β-galactosidase induced by the acridine derivative ICR-191D. Sixteen mutants varied in activity by 100-fold, with the strongest 0.06 % of wild-type (Atkins et al., 1972). This, and studies of immunoprecipitated truncation products of β-galactosidase (Manley, 1978) place the average frequency of frameshift in the range of 5×10−4 per translocation event. However, factors such as ribosomal mutation (rpsL and ram), streptomycin treatment, amino acid starvation and “hungry” codons greatly increase the frequency of frame errors Atkins et al 1972, Weiss and Gallant 1983, Weiss et al 1988a. For example, in E. coli, tandem repeat of AGG, a rare arginine codon read by a minor tRNA, results in +1 frameshift up to 50 % of the time (Spanjaard & van Duin, 1988).
The best-known (+1 and −1) frameshifting events, both programmed and non-programmed, occur because of single-base repeats constituting a “slippery run” of mRNA. For example, the +1 frameshift that avoids the UGA stop codon at the programmed site in RF2 occurs at the sequence CUUU, mediated by the CUU-decoding tRNALeu (Weiss et al., 1987). While it is easy to understand how a long repeat is a frame-maintenance conundrum for the ribosome, it is equally striking that high levels of shifting are observed only in the presence of other “stimulatory” elements (Atkins et al., 1990), such as a Shine-Delgarno-like sequence upstream and a UGA stop codon downstream of the RF2 shift site Weiss et al 1987, Weiss et al 1988b; an RNA-pseudoknot downstream of the shift site for the F2 protein of coronavirus IBV (Brierley et al., 1989); and the rare AGG codon adjacent downstream to the CUU slip site in yeast Ty elements (Belcourt & Farabaugh, 1990).
We were interested in employing a library approach to examine all possible four-base codons for their propensity to shift and, moreover, to exhaustively identify tRNAs that will efficiently suppress four-base codons. To this end, we have installed the sequence NNNN into various codons of the gene for β-lactamase and selected with ampicillin in the presence and absence of derivatives of tRNA2 Ser with randomized, extended (eight or nine nucleotide) anticodon loops. We have successfully identified at least four non-interacting sets of efficient four-base codon-tRNA pairs, and we have defined a strikingly simple pattern that underlies “shiftiness” in four-base codons. These pairs will be used in our ongoing efforts to engineer bacteria capable of inserting unnatural amino acid residues into proteins.
Results
Library construction and characterization
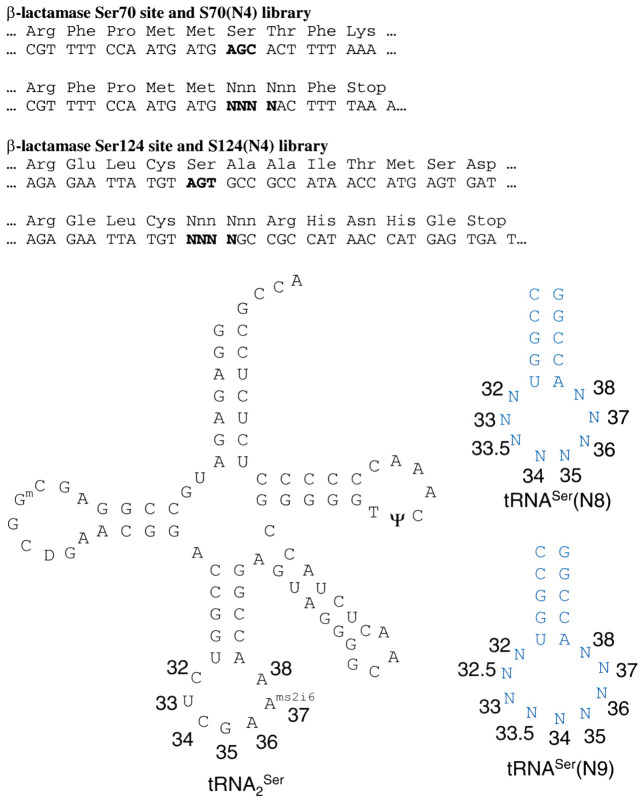
Since the basis of four-base codon suppression is poorly understood, we adopted a combinatorial approach to examine the suppression of all possible four-base codons by all possible appropriately sized tRNA anticodon loops. Two types of libraries were generated: four-base codon reporters and tRNA suppressors (Figure 1). The reporter system involved replacement of a conserved (S70) or permissive (S124) codon in the gene for β-lactamase with four random nucleotides. These libraries, which each contain 44=256 unique members, bear an insertion mutation at the site of interest, producing an inactive enzyme as a result of the frame alteration that renders transformed bacteria incapable of survival on ampicillin. To our surprise, however, approximately 0.5 % of the members of these reporter libraries survived on moderate to high concentrations of ampicillin (100-2000 μg ml−1). Invariably, sequencing of these clones showed that the β-lactamase genes contained only three nucleotides at the sites of interest, corresponding to Ser and Cys at the conserved S70 site and corresponding to all amino acids except Cys, Met, Pro, Trp and Tyr at the permissive S124 site (although this is probably largely due to undersampling of three-base sequence space, since the amount of contamination was small). Presumably, these “deletion” mutants, which occur at rates much higher than the 10−6 upper-limit estimated for deletion in vivo (Cupples et al., 1990), are the result of inefficiency in the coupling and capping reactions employed during synthesis of the oligonucleotides used to generate the libraries.
Figure 1.
Schematic of β-lactamase libraries and tRNASer libraries. Insertion of NNNN at Ser70 or Ser124 results in abortive translation unless the frameshift is suppressed.
To remove this contamination, cells transformed with the reporter libraries were grown in small batches and analyzed on agar plates for moderate levels of ampicillin resistance (100 μg ml−1). By pooling the cultures that contained no ampicillin resistance at this level, uncontaminated, oversampled reporter libraries were obtained (about 5000 clones for the S124(N4) library and 10,000 clones for the S70(N4) library). Sequencing of 48-96 clones from each reporter library showed that the libraries were random at each nucleotide and sufficiently unbiased that unique four-base codons were pulled out only once or twice among these clones.
Because the β-lactamase reporter libraries were constructed from pBR322-dervived vectors containing ColE1 origins, tRNA suppressor libraries were constructed with pACYC184-derived vectors with p15a origins to permit co-maintenance within a single bacterium. These libraries consisted of derivatives of tRNA2 Ser with the anticodon loop (7 nt) replaced with eight or nine random nucleotides, transcribed under the control of the strong lpp promoter and efficient rrnC terminator. This tRNA was chosen because seryl-tRNA synthetase (SerRS) does not recognize the anticodon loop of tRNASer. Major recognition elements are, instead, in the long variable arm, acceptor stem, and the D and TΨC loops, where the SerRS contacts the tRNA Biou et al 1994, Price et al 1993. The choice of a tRNA scaffold and cognate synthetase that is permissive to anticodon loop structure ensures that suppression efficiency is related to codon-dependent effects rather than the effects of variable aminoacylation with different anticodon sequences. For each of these libraries, 24 clones were sequenced and found to be unique and unbiased.
Selected four-base codons at S124 and S70
The libraries were crossed by the transformation of the appropriate tRNA library into competent cells of the reporter strains, followed by selection on media containing various concentrations of ampicillin. Survival on higher concentrations of ampicillin requires that more β-lactamase be present in the cell, which is related to the extent to which the tRNA in the selectant is able to mediate frameshift at the site of mutation in the reporter. Thus, survival at the highest levels of ampicillin indicates the presence of tRNAs that read the co-transformed four-base codons most efficiently.
The library of 8 nt anticodon loop tRNAs was first crossed with the S124 and S70 reporter libraries, choosing two sites to examine the effects of mRNA context. In addition to different general contexts in the gene, these sites also have different local contexts with different nucleotides 5′ and 3′ to the four-base codon (Figure 1). At the permissive S124 site, four-base codons selected at moderate ampicillin concentrations (200-300 μg ml−1) included a subset of AGGN, CCCN, CGGN, GGGN and UAGN, for which four-base codon suppressors are known, as well as CUAG, UCAA, CCUU, CUCU, GGAC and UGCG (Table 1). Only one of these, AGGA, could be suppressed at 1000 μg ml−1 ampicillin. At the S70 site, however, AGGG, CUCU and UAGA were also suppressed at 1000 μg ml−1, and more codons were represented at the 300 μg ml−1 level than for the S124 site. Suppression of the S70 site, which is the catalytic Ser of TEM-1 β-lactamase and requires Ser or Cys for activity, confirms that these four-base codons are suppressed by our modified seryl-tRNA.
Table 1.
Selected four-base codons at S124 and S70 at high and moderate ampicillin concentrations
| S124 |
S70 |
||
|---|---|---|---|
| 1000 μg ml−1 | 300 μg ml−1 | 1000 μg ml−1 | 300 μg ml−1 |
| AGGA | AGGA | AGGA | AGGA |
| AGGG | AGGG | AGGC | |
| AGGC | CUCU | AGGG | |
| CCCU | UAGA | AGGU | |
| UAGA | CCAU | ||
| CGGG | CCCC | ||
| UCAA | CCCU | ||
| CGGC | |||
| CUAC | |||
| CUAG | |||
| CUCU | |||
| UAGA | |||
Since there was no apparent pattern to the sequences of the suppressed four-base codons, we devised an experiment to examine whether or not any four-base codon could be suppressed, at least at low levels. Six clones from the S70(N4) reporter library were selected at random and crossed against the tRNASer(N8) library. From these clones (Table 2), tRNAs that efficiently suppress UUCU, GGAU and CGGA to at least 200 μg ml−1 ampicillin could be found. AAUG and ACGC suppressors were generally much weaker, between 5 and 10 μg ml−1 ampicillin. A suppressor for UGAA, based on the UGA stop codon, could not be found, however, even at 5 μg ml−1 ampicillin.
Table 2.
Selection of tRNA suppressors of randomly chosen codons
| Codon | Maximum suppression level | tRNA sequences |
|---|---|---|
| UGAA | >5 | None detected |
| AAUG | 5-10 | CUCGUUAC |
| CUCGUUAA | ||
| ACGC | 5-10 | CUGCGUAU |
| CUACGUAU | ||
| GGACGUAA | ||
| UUCU | >200 | UUAGAAAG |
| AUAGAAAG | ||
| ACGGAAAC | ||
| UUAGAAAC | ||
| CUAGAAAC | ||
| ACAGAAAC | ||
| CGGGAAAA | ||
| GGAU | >200 | GUGUCCAU |
| CAGUCCAU | ||
| UGAUCCAU | ||
| CCAUCCAG | ||
| UAGUCCAC | ||
| UCAUCCAC | ||
| CCAUCCAC | ||
| UAAUCCAC | ||
| AAGUCCAA | ||
| CGGA | >200 | CGGCCCGG |
| UUUCCGUA | ||
| CCUCCGUA | ||
| GUUCCGGC | ||
| CUUCCGCU | ||
| ACUCCGCG | ||
| GUUCCGGC | ||
| ACUCCGUA | ||
| AAUCCGCU | ||
| ACUCCGUA | ||
| ACUCCGUU |
Suppression levels are in ampicillin concentration, μg ml−1.
The procedure for removal of the three-base contamination at the catalytic S70 site cannot remove those clones that contain a random three-base codon that corresponds to a missense mutation (i.e. anything other than Ser or Cys). As a result, upon crossing this library with the tRNA library, a number of clones were isolated that contained missense three-base codons that were presumably being suppressed by the Ser-inserting tRNA library member. Interestingly, the most efficiently suppressed, AGG and CGG at 1000 μg ml−1, correspond to the first three nucleotides of some of the most efficiently suppressed four-base codons (Table 3). It was not determined whether the tRNA suppressors contain 8-nt anticodon loops or 7 nt (i.e. normal) loops as a result of the same type of “deletion” seen in the reporter libraries.
Table 3.
Codons for which Ser missense suppressors were slected at S70
| [Amp] | Codon | Acid |
|---|---|---|
| 1000 | AGG | Arg |
| CGC | Arg | |
| 300 | ACA | Thr |
| AGG | Arg | |
| AUA | Ile | |
| CCA | Pro | |
| CCU | Pro | |
| CGA | Arg | |
| CGG | Arg | |
| UUG | Leu | |
| 100 | ACG | Thr |
| AUA | Ile | |
| CCU | Pro | |
| CGA | Arg | |
| CGG | Arg | |
| CUC | Leu | |
| GCC | Ala | |
| GGC | Gly | |
| GUG | Val | |
| UUA | Tyr |
Ampicillin concentrations are in μg ml−1.
Sequence variation of tRNA suppressors of four-base codons
In order to examine the diversity of tRNA anticodon loops that could suppress each selected codon, the reporter plasmid was isolated and the tRNASer(N8) library was re-crossed against individual reporter sequences at S124. These were then selected at a variety of levels of ampicillin for each reporter, and the tRNA sequences were examined. In each case, suppression at the highest levels was mediated by a tRNA bearing the complementary anticodon (Table 4). Interestingly, the sequences external to the anticodon (i.e. the two nucleotides on either side of the anticodon) converged on different sequences depending upon the anticodon. To examine the effect of sequence variation in the anticodon loop outside of the anticodon per se, tRNAs from the tRNASer(N8) library were selected at different levels of ampicillin (20 μg ml−1 to 1500 μg ml−1) against a single reporter codon at S124 (Table 5). For this experiment, we selected the most efficiently suppressed codon (AGGA) to examine the widest array of suppression efficiency. At 1500 μg ml−1 ampicillin, the sequences converged on MUUCCUAM, where M=A or C. At 300 μg ml−1, the convergence was reduced to NBUCCURN, where B=C or G or U and R=A or G. At 20 mg/ml, the consensus was reduced to NNNCCUDN, where D=A or G or U, with loss of canonical recognition of the fourth base of the codon but maintenance of the other three bases. This allows us to rank the different anticodon positions in importance for suppression efficiency: Watson-Crick complementarity to the first three bases of the codon is most critical, followed by complementarity at the fourth base, the presence of a purine at position 37 (3′ to the anticodon) and the presence of a U at position 33 (5′ to the anticodon).
Table 4.
Selectants from re-cross of individual four-base codons against tRNASer(N8)
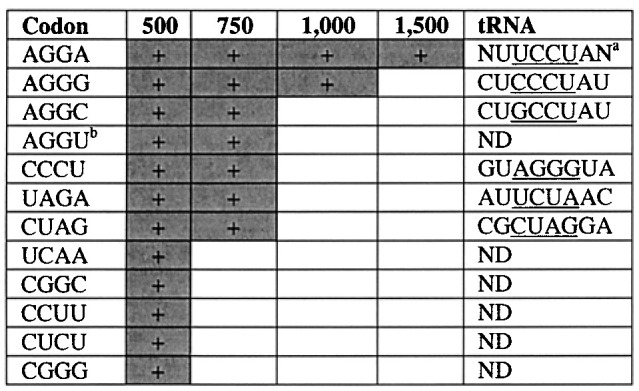
Ampicillin survival levels are in μg ml−1 for codons at S124. ND=not determined.
a Even at the highest levels of ampicillin selection, three tRNAs were found for the AGGA anticodon: A…A, C…A, C…C.
b Determined at S70.
Table 5.
Selected eight-nt anticodon loop tRNAs to suppress AGGA at moderate and low ampicillin levels
| 300 μg ml−1 | 20 μg ml−1 | |||
|---|---|---|---|---|
| ACUCCUAA | CUUCCUAC | AAUCCUAG | CAUCCUAU | UAUCCUGU |
| ACUCCUAC | GGUCCUAU | AAUCCUAU | CGGCCUAU | UGUCCUAG |
| AGUCCUAC | GUUCCUAA | ACUCCUAU | CGUCCUGU | UGUCCUAU |
| AGUCCUGC | UCUCCUAA | ACGCCUAU | CGUCCUUU | UGUCCUGC |
| AUUCCUAG | UCUCCUAC | ACUCCUAU | CUACCUAU | UUUCCUAU |
| CCUCCUAA | UUUCCUAC | AGGCCUGG | CUUCCUAA | UUACCUAA |
| CCUCCUAC | UUUCCUAG | AGUCCUAC | GGUCCUGA | UUUCCUAG |
| CGUCCUAU | UUUCCUAU | AGUCCUUG | GUUCCUAA | UUUCCUAU |
| CUUCCUAA | AUUCCUAG | GUUCCUAU | UUUCCUGU | |
| CAACCUAA | GUUCCUGU | |||
| CAUCCUAA | UAUCCUAA | |||
Sequences were found one to six times each. Only the anticodon loop sequence is shown.
Analogously, to examine how efficiently AGGA could be suppressed by 9-nt anticodon loop tRNAs, the AGGA reporter at S124 was crossed against tRNASer(N9) and selected at 20 and 300 μg ml−1 ampicillin. At 300 μg ml−1 ampicillin, the consensus sequence was BNKUCCUAH, where K=G or U and H=A or C or U (Table 6). However, at 20 μg ml−1, the consensus sequence degenerated to NNNUCCUAN. In a single case out of 38 clones, CUUCCUAUU, the extra nucleotide in the loop was on the 3′ side of the anticodon (UCCU). At both levels of ampicillin selection, tRNAs with 8 nt anticodon loops were among the selectants, presumably as a result of “deletion” from inefficient capping during oligonucleotide synthesis seen in the reporter libraries. These deletions dominate the selection at higher ampicillin levels, indicating that suppression of four-base codons (or at least AGGA) is more robust with tRNAs bearing 8 nt anticodon loops rather than 9-nt loops, overall.
Table 6.
Selected nine-nt anticodon loop tRNAs to suppress AGGA at moderate and low ampicillin levels
| 300 μg ml−1 | 20 μg ml−1 | ||
|---|---|---|---|
| CAUUCCUAU | AAUUCCUAC | CCUUCCUAA | GAUUCCUAU |
| CCUUCCUAU | ACGUCCUAC | CCUUCCUAU | GCUUCCUAU |
| CUGUCCUAA | ACUUCCUAC | CUGUCCUAC | UCGUCCUAG |
| GCUUCCUAU | ACUUCCUAU | CUGUCCUAU | UCUUCCUAA |
| GGUUCCUAU | AGUUCCUAC | CUUUCCUAA | UCUUCCUAG |
| UUGUCCUAC | AUUUCCUAA | CUUUCCUAU | UUGUCCUAC |
| AUUUCCUAC | GACUCCUAC | UUUUCCUAC | |
| CACUCCUAC | GAGUCCUAC | UUUUCCUAU | |
| CCAUCCUAU | GAUUCCUAC | CUUCCUAUU | |
Sequences were found one to four times each. Only the anticodon loop sequence is shown.
Specificity of tRNA suppressors of four-base codons
The most robust suppressors of each of the most favorable codons were isolated and re-crossed against the S70(N4) reporter library with selection at very low ampicillin (10 μg ml−1). Here, S70 was selected, since the site is slightly easier to suppress, and we were interested in examining extremely low levels of suppression, if they existed. Selectants were then transferred to increasingly higher concentrations of ampicillin and sequenced to identify the extent to which each tRNA is capable of suppressing other four-base codons. In each case, suppression of the canonical four-base codon was highly favored, with suppression of related codons with non-canonical pairing at the fourth codon position possible at fivefold lower ampicillin at best (Table 7). The tRNA that efficiently reads AGGA (1500 μg ml−1 ampicillin), for example, reads AGGU to only 100 μg ml−1, and AGGG and AGGC to only 50 μg ml−1. In contrast, suppression of no other codon could be detected for the tRNA that suppresses CUAG.
Table 7.
Codons selected by most efficient four-base codon suppressors
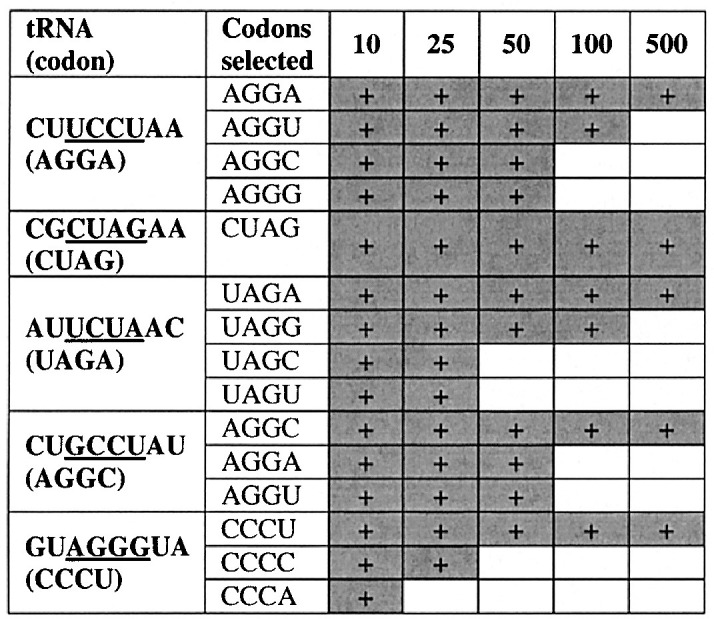
Sequences for the anticodon loop are shown. Codon in parentheses is that which elicited the tRNA in selection against a single reporter (Table 3). Suppression levels are indicated for each tRNA/four-base codon pair in μg ml−1 ampicillin.
Efficiency of frameshift mediated by four-base codon suppressors
The level of readthrough of the four-base codons and the efficiency of suppression of these codons were quantified by determining the amount of β-lactamase per cell with the chromogenic substrate nitrocefin. Convenient arbitrary units were defined to normalize the activity for the number of cells assayed and to subtract out the effects of the slow background hydrolysis of nitrocefin (see Materials and Methods). Spontaneous frameshifting at of the four-base codons at S124 was no more than twice the background hydrolysis by TOP10 E. coli (0.8–1.7 units versus 0.9 unit), which was comparable to readthrough of the amber stop codon (1.2 units, Figure 2). Suppression of the four-base codons was in the range of 17 to 240 units, or 2.5–35 % of wild-type (AGT or β-lactamase on pBR322). However, in this assay, the very-efficient supD suppressor of the amber stop codon exhibited virtually the same β-lactamase activity as observed in cells with AGT (Ser) at the S124 site. This may indicate that it is possible to saturate the amount of β-lactamase that is exported to the periplasm in this system. Therefore, while the activities allow us to compare frameshift efficiencies, we cannot strictly correlate these activity data with suppression efficiencies as compared to translation of wild-type β-lactamase.
Figure 2.
Suppression efficiency of four-base codon/tRNA pairs measured by nitrocefin turnover. See Materials and Methods for definition of units. Readthrough of UAG, AGGA, CCCU, CUAG and UAGA was 0.8–1.7 units, while TOP10 cells alone exhibit about 0.9 unit. Suppression of four-base codons resulted in 17–240 units of activity, compared to 970 for suppression of UAG by supD. Results are the average of three to five independent trials per data point.
Identification of “shifty” codons
It was found empirically during the process of removing three-base contamination from the libraries that no four-base codon was read through by the natural translational machinery at levels sufficient to permit survival at 100 μg ml−1 ampicillin. However, at lower levels of ampicillin, some four-base codons can be read at low levels, or are inherently shifty (Table 8). Strikingly, each of these four-base codons has a tandem repeat in the central two nucleotides (a purine repeat in one case). It is also noteworthy that some of the best suppressed codons (e.g. AGGC, CGGN and CGAU) but not all of them (e.g. AGGA) are among this shifty set. For some codons (e.g. AGGN, AAAN, and GGGN), only a subset of the family is present, indicating that slipping at a particular three-base codon is strongly influenced by the base that follows. Some of these, such as AGGA, are probably only slightly less shifty (5 μg ml−1), but it becomes difficult to examine libraries at this low level of selection due to background survival of satellites on ampicillin.
Table 8.
“Shifty” four-base codons detected at the S124 site
| 25 μg ml−1 | 10 μg ml−1 |
|---|---|
| AAAA | AAAA |
| AGGC | AGGC |
| CGGC | CGAU |
| CUUA | CGGA |
| CUUU | CGGC |
| GGGA | CGGG |
| UUUC | CGGU |
| UUUU | CUUA |
| CUUC | |
| CUUU | |
| GGGA | |
| UUUC | |
| UUUU |
Selection levels are given in ampicillin concentration.
Discussion
Suppression of four-base codons
When the first frameshift suppressors were isolated, it was thought that only repeating codons like GGGG could be suppressed. However, a number of exceptions, like ACCN and UAGN, were later found or engineered (Atkins et al., 1991). Among those efficient suppressors identified in this study, it is clear that there is no sequence pattern that generally underlies efficient four-base suppression. For example, CUAG is completely non-repetitive, and GGGG, AAAA and UUUU are not among the best suppressors. However, the most efficiently suppressed four-base codons correspond to three-base codons (plus one 3′ nucleotide) with low usage and low tRNA abundance in E. coli (Table 9). For example, AGG is the least used codon in the E. coli genome (Inokuchi & Yamao, 1995). Nevertheless, the best suppressors do not simply correspond to the least represented tRNAs or codons, since we found no robust suppressors of highly underrepresented codons AGAN, AUAN or CGAN. Also, among randomly selected codons that vary in usage from overrepresented (AAUG and UUCU) to moderate representation (ACGC) to underrepresented (GGAU and CGGA), and that lack any evident sequence pattern, weak to moderate suppressors could be found (Table 9). Of course, the procedure employed here selects for growth on ampicillin and therefore requires both an efficient four-base codon/tRNA interaction and a lack of toxicity to the cell. We cannot rule out the possibility of efficient but toxic suppressors of highly represented four-base codons.
Table 9.
Codon usage and tRNA abundance for codons suppressible with 8 nt anticodon loop tRNAs and “shifty” codons
| Codon (acid) | Codon usage (0.1 %)a | tRNA abundance (relative)b | S124 suppression with tRNA | S124 readthrough (without tRNA) |
|---|---|---|---|---|
| AGG (Arg) | 0.9 | ND | + | + |
| CCA (Pro) | 7.4 | ND | + | |
| CCC (Pro) | 3.4 | ND | + | |
| CGG (Arg) | 3.6 | Minor | + | + |
| CUA (Leu) | 2.5 | Minor | + | |
| CUC (Leu) | 9.3 | 0.30 | + | |
| UAG (amber) | ND | N/A | + | |
| UCA (Ser) | 5.6 | 0.25 | + | |
| AAA (Lys) | 38.4 | 1.00 | + | |
| CGA (Arg) | 2.6 | 0.90 | + | |
| CUU (Leu) | 9.9 | 0.30 | + | |
| GGG (Gly) | 8.6 | 0.10 | + | |
| UUU (Phe) | 16.9 | 0.35 | + | |
| AAU (Asn) | 14.8 | 0.60 | weak | |
| ACG (Thr) | 11.4 | ND | weak | |
| GGA (Gly) | 5.5 | 0.20 | weak | |
| UUC (Phe) | 19.1 | 0.35 | weak |
ND, no data. N/A, not applicable. Minor, abundance too low for accurate measurement (Inokuchi & Yamao, 1995).
Usage in E. coli. If all sense codons were used equally, usage would be 16.4.
Relative to tRNA3Leu with codon usage 56.3.
The identity of the fourth base has a profound effect on the ability of the four-base codon to be suppressed. For the AGGN family, the series appears to be AGGA>AGGG>AGGC≈AGGU for canonical suppressors. However, Hohsaka et al. (1996), saw little difference in the in vitro suppression levels among these four codons using a yeast tRNAPhe scaffold, perhaps due to the extreme overexpression conditions he employed in contrast to the moderate, natural β-lactamase promoter employed here. Likewise, only UAGA of UAGN, and CGGC and CGGG of CGGN, are among the most strongly suppressed codons.
Some of the four-base codons that we identified were already known, including AGGN, used in protein engineering Hohsaka et al 1996, Ma et al 1993, CCCU, which is SUF8 (Cummins et al., 1985) and UAGA Curran and Yarus 1987, Moore et al 2000. However, CUAG is apparently a completely novel efficiently suppressed four-base codon, consistent with the fact that CTAG is underrepresented in the bacterial genome (Burge et al., 1992). These suppressors acted with efficiencies of 2.5–35 % of the maximum activity detectable with a nitrocefin-based assay (AGU or supD/UAG), which is consistent with activities from the most efficient natural and engineered suppressors known. Also, fairly efficient suppressors for the four-base codons CUAC, CCAU, UCAA, CCUU and CUCU were found Table 1, Table 4. Examination of the anticodon loop sequences shows that the best tRNAs for each codon have a canonical anticodon, but the rest of the loop does not necessarily converge on the sequence consensus observed in E. coli tRNAs. For example, the tRNA that suppresses CUAG has G33 and G37 instead of U33 and A37, and the tRNA that suppresses CCCU has a U37. (See Figure 1 for numbering of nucleotides in the 8 nt and 9 nt anticodon loops.) Presumably, these mutations adjust the loop conformation to allow maximally favorable interaction between the codon and anticodon, which is quite possibly different from the most favorable conformation for a three-base anticodon. These data support a model in which the extended-codon tRNA directly reads all four bases of the codon (as opposed to a slippage model), since (1) the most efficient suppressors invariably had Watson-Crick complementarity at all four bases of the codon, (2) the suppression at S70 required insertion of a serine or cysteine residue, demonstrating that the modified tRNASer was delivering the amino acid, and (3) there is no special pattern underlying the suppressed codons, as might be expected if slippage were occurring.
Diversity among tRNAs that suppress four-base codons can also be assessed by comparing the tRNA sequences selected with a single reporter codon. This approach gives us some idea about the tolerance of the E. coli translational apparatus to variation in the tRNAs for suppression of four-base codons (presumably combined with tRNA maturation and aminoacylation differences, which are probably minimal here). Figure 3 shows the consensus sequences for tRNAs able to suppress AGGA at various levels of ampicillin compared with the known E. coli tRNAs. The best suppressors of this codon maintain the universal U33 and highly favored A37 seen in E. coli tRNAs. At the 300 μg ml−1 ampicillin level, U33 or C33 and A37 are favored, but G is present in some clones at both sites. When much weaker suppressors were selected (20 μg ml−1 ampicillin), no base is conserved at position 33 and, though A37 is favored, G37 and U37 are seen. This calls into question the necessity of the U33 to make the sharp turn seen in the X-ray crystal structure of tRNAPhe Holbrook et al 1978, Sussman and Kim 1976. Even the 33.5 position corresponding to AGGA is not entirely conserved in these weak suppressors, though U33.5 is still strongly favored. Some 5 % of the clones selected at 300 μg ml−1 and 19 % at 20 μg ml−1 have the possibility of an A:U or U:A pair at the 32:38 site, which would formally extend the anticodon stem by one pair and leave a 6 nt loop. Other work has suggested that it is unlikely that a 6 nt loop can present a four-base anticodon (Atkins et al., 1991), and five of the seven clones of this type retain complementary UCCU anticodons, perhaps suggesting why only the weaker A:U pair is seen at this site and not the C:G pair. Nevertheless, the possibility of 6 nt anticodon loop tRNAs decoding four-base codons cannot be ruled out.
Figure 3.
Nucleotide representation at anticodon loop sites in E. coli tRNAs and moderate and weak tRNA suppressors of AGGA with 8 nt and 9 nt anticodon loops. Suppression is at S124 and listed in ampicillin concentration of μg ml−1.
When this selection scheme was applied to 9 nt anticodon loop tRNAs that suppress AGGA (Figure 3), a UCCU anticodon was found in each tRNA and, in every clone but one, the extra nucleotide is added 5′ to the anticodon (as was always the case with the 8 nt anticodon loop tRNAs). At 300 μg ml−1 ampicillin, U38 is favored, while either pyrimidine is favored at 20 μg ml−1. This differs both from the E. coli preference for A38 and lack of preference in 8 nt tRNAs. U33 is strongly preferred at both ampicillin concentrations, much more so than in the 8 nt anticodon loops. Also, while there is degeneracy at the 32 position, the stronger suppressors do not have A32, possibly to avoid 32:38 pairing with the favored U38. At 300 μg ml−1 ampicillin, three of the nine clones have the possibility of a weak G32:U38 wobble pair (resulting in a 7 nt anticodon loop and extended anticodon stem), but none can canonically pair. However, among the weaker suppressors, two of 37 clones can have a U:A or A:U pair and five can have a G:C pair. Particularly in the latter case, it is possible that the 7 nt anticodon loop with an extended anticodon stem is the active conformation suppressing the four-base codon. Indeed, recent work by Auffinger & Westhof (1999) suggests structural conservation at the 32:38 position in all tRNAs and excludes G:C pairs, perhaps lending credence to the view that this pair is part of the stem, not the loop, in these cases.
In addition to exhibiting high suppression efficiency, any useful four-base codon suppressor must also not cross-react with other codons (three or four-base) to ensure protein sequence integrity. It is helpful in this regard that all of the useful four-base codons are based on rare natural codons. This greatly reduces the potential need to silently mutate related codons in the gene of interest (i.e. if one employed AGGA suppression in a gene containing AGG codons) to avoid missense or frameshift suppression at other sites. Moreover, that these codons are extremely rare and that the suppressors are not toxic to E. coli suggests that there is not a notable problem with suppression of three or four-base codons by these tRNAs genome-wide. Equally important is the fact that the suppressor tRNAs do not interact with other codons. Selection with the best suppressor tRNAs from the library of all four-base codons resulted in, at worst, limited sets of highly related codons. For example, no other codon was selected even at very low ampicillin concentration with the tRNA that suppressed CUAG (Table 7). In the cases of UAGA and AGGA, suppression of other UAGN or AGGN codons at more moderate levels of ampicillin (though still at least fivefold lower than in the canonical case) probably places a practical limit on the use of similar four-base codons in a single transcript. For example, AGGA and AGGU are probably sufficiently cross-reactive that they should be avoided in a single transcript, whereas AGGA, CUAG and UAGA and CCCU are likely to be simultaneously useful.
Suppression of four-base codons at the S124 and S70 sites of β-lactamase reveals that the S70 site is slightly easier to suppress, since in the AGGG, CUCU and UAGA codons suppressed at the 1000 μg ml−1 level. These codons were selected at the 200 or 300 μg ml−1 level at S124, but not 1000 μg ml−1. Since all of the codons selected at the 300 μg ml−1 level at S124 are present at that level or above at S70, it is likely that these codons can generally be suppressed at any site. The local context of the codon probably affects overall suppression efficiency, a concept that has been demonstrated many times for frameshift and stop-codon suppression Ayer and Yarus 1986, Belcourt and Farabaugh 1990, Bossi and Roth 1980, Tate et al 1996. However, there are no conspicuous sequences flanking the S70 site, such as repeated rare codons, upstream Shine-Delgarno sequence or stable downstream mRNA structure. Interestingly, amber stop codons (UAG) are often suppressed most efficiently when followed by an A, as the S70 site is here, though the reason for this is not clear Ayer and Yarus 1986, Bossi and Roth 1980.
Tolerance for errors in translation
It is clearly critical for the survival of any organism to maintain a great deal of fidelity both in the replication of hereditary information and in the transduction of that information to generate proteins. However, the actual amount of tolerance for errors is difficult to appreciate. It is estimated that DNA replication fidelity is between 10−8 and 10−11 per base in vivo, but transcriptional and translational fidelity is closer to 10−4 per codon (Parker, 1989). Manley (1978) demonstrated that a measured 5×10−4 frameshift error rate corresponds to approximately 30 % of incipient translations of a given protein resulting in some sort of abortive synthesis, due in large part to the relative prevalence of stop codons in the non-coding frames. Especially for genes that are transcribed at very low levels, this appears to be a significant energetic waste that is tolerated by the cell, and one must wonder if it is not being exploited as a subtle, stochastic mechanism of regulation or is an evolutionary artifact.
Equally striking is the ease with which the normal translational apparatus can be thwarted with no apparent detriment to the cell. For example, we found efficient missense suppressors of a variety of codons corresponding to a wide variety of amino acids. While it is true that these tended to correspond to underrepresented codons (Table 9), even the least used codon must appear in the largely transcribed E. coli genome hundreds to thousands of times. Hong et al. (1998) found that a glutaminyl-tRNA synthetase mutant that inserted glutamic acid for glutamine less than 5 % of the time resulted in a 40 % increase in doubling time of the bacteria. However, none of the missense suppressors found here had demonstrably toxic effects, and were selected at levels comparable to about 15 % suppression efficiency as measured with the nitrocefin assay for our four-base suppressors. Moreover, at many of these same rare codons, very efficient suppressors of four-base codons could be maintained without deleterious effects on cell growth, and in at least one case (UUCU) a moderately efficient suppressor was found for an overrepresented codon. It is conceivable that this is an evolutionary requirement, wherein only those organisms that could tolerate a reasonable level of translational errata were sufficiently free to adapt to environmental stresses.
Another more general limitation on the fidelity of protein production is that some sites in mRNA are “shiftier” than others. These tend to be highly repetitive sequences; specifically, sequences with tandem repeats in the second and third position of the codon. Some of these correspond to codons used widely throughout the genome (AAA accounts for 3.84 % of codons in E. coli). A number of these sequences have been exploited by nature at programmed frameshift sites. For example, E. coli RF2 is produced when a UGA stop codon is averted by slippage on a CUUU site, which was seen here. In the yeast Ty1 element, slippage occurs on CUUA to generate the TYB protein, a pol analog (Farabaugh, 1996). In the case of the RF2 site, engineered alteration of the CUU to UUA, CUA, GUA, GUG or AUA caused drastic decreases in shifting, but alteration to GUU, UUU, CCC or CCU retained higher levels. While only UUUU was found in our selection, the other sites except for CCUU clearly retain the internal tandem-repeat motif Curran 1993, Weiss et al 1987.
If one extends Curran’s proposal that shiftiness is related to the ability of the cognate tRNA on the zero frame to re-pair in the +1 frame, and one assumes that re-pairing is only favorable for Watson-Crick or wobble pairs at the first and second position of the +1 codon, the following list of four-base codons would be expected to shift: AGGN, AAAN, AAGN, UUUN, CUUN, CCCN, CCUN and GGGN. The selected codons differ from this list in three ways: (i) the identities of N are anomalous; (ii) AAGN, CCCN and CCUN were not found; and (iii) CGGN and CGAU were found. The potential advantage of additionally re-pairing the third base would explain AAAA, UUUU and CUUU but not AGGC, GGGA, UUUC or the other CUUN, which largely do not even follow wobble rules. It is unlikely that AAGN, CCUN and CCCN were simply not in the library, since the sequence space is small (256) and more than 70 clones were sequenced including duplicates of most sequences. Probably, CCUN and CCCN did not appear due to lack of tolerance for Pro at S124, as CCN codes for Pro, and perhaps the central wobble re-pair in AAGN is especially unfavorable. Shiftiness at CGGN and CGAU is not explained by this hypothesis, especially since no re-pairing is possible with CGAU, but it is of note that both of these codons are highly underrepresented in E. coli. It seems likely that the full explanation of inherent shiftiness at a given codon is a combination of the ability of the tRNA to re-pair in the new frame, the abundance of the tRNA and the context of the codon generally.
We believe that an extended repertoire of sites for efficient suppression will be a great aid to the site-specific insertion of multiple unnatural amino acids, both in vitro and eventually in vivo with our developing technology for use in E. coli. We are currently selecting for unnatural amino acid specificity from three different aminoacyl-tRNA synthetases Liu and Schultz 1999, Pastrnak et al 2000, Wang et al 2000, and this raises the eventual need for multiple, non-interacting sites for the insertion of unnatural amino acids. Preliminary experiments suggest that efficient suppression of even more extended codons (five-base and perhaps six-base) is possible with tRNAs with even more extended anticodon loops, and we are currently examining this in depth (J.C.A. and P.G.S., unpublished results). This approach is complementary to our approach in collaboration with Romesberg et al., of generating additional codons by designing novel base-pairs (Ogawa et al., 2000), and has the present advantage of being immediately useful in vitro for a wide range of experiments to probe the limits of biosynthesis of unnatural polypeptides.
Materials and methods
Strains, plasmids and materials
Subcloning was carried out in E. coli strains DH10B (Gibco Life Technologies) and TOP10 (Invitrogen), and selections were carried out in TOP10 (which is rpsL). PCR reactions were carried out according to standard protocols with Taq or Pfu (Promega) polymerases. Standard protocols were employed for subcloning with restriction enzymes (NEB) and T4 DNA ligase (NEB or Roche).
Construction of four-base codon reporter libraries
A derivative of pBR322 was silently mutagenized with a PCR oligonucleotide-directed approach to contain BstEII and PstI sites flanking the Ser124 site in the β-lactamase gene. A synthetic linker was inserted between the BstEII and PstI sites to inactivate the β-lactamase and generate pBRBstPstXmaKO. A cassette from the β-lactamase gene with BstEII and PstI ends containing four random nucleotides in place of the Ser124 codon (Figure 1) was generated by extension of two overlapping synthetic oligonucleotides with phage T7 DNA polymerase (NEB) and subcloned into pBRBstPstXmaKO. Randomized sites in the oligonucleotides were synthesized (Operon) with a pre-mix of phosphoramidites and were outside the complementary overlap region. Similarly, a derivative of pBR322 was silently mutagenized to contain XhoI and XbaI sites flanking Ser70 of β-lactamase, and a linker was inserted to inactivate the β-lactamase gene and generate pBRS70KO. A library cassette was generated and subcloned as above with synthetic oligonucleotides synthesized on a Perceptive Biosystems Expedite synthesizer, using a pre-mix of phosphoramidites (Glen Research) to replace the codon for Ser70 with four random nucleotides.
Construction of tRNA anticodon-loop libraries
Vector pACGFP, derived from pAC123 (Liu et al., 1997), contains a linker derived from pGFPuv (Clontech) between unique EcoRI and PstI restriction sites flanked by the strong lpp promoter and rrnC terminator. A cassette that corresponds to the sequence for tRNA2 Ser with eight or nine random nucleotides in place of the 7 nt anticodon loop was made by extension of synthetic oligonucleotides (Genosys) with the Klenow fragment of E. coli DNA polymerase I (NEB) and inserted into pACGFP between EcoRI and PstI sites.
Selection and library crossing
Electrocompetent TOP10 cells were transformed with β-lactamase four-base codon reporter libraries to make reporter strains. Since it was found that the β-lactamase reporter libraries were contaminated with genes bearing randomized three-base codons at the sites of interest instead of randomized four-base codons (see Results), libraries were diluted and inoculated into 96-well plates with approximately 100 cells per well. Wells containing cultures that were able to grow on agar plates with 100 μg ml−1 of ampicillin were discarded. Clean cultures were pooled and amplified, DNA was prepared and digested with an enzyme selective for the linker to remove background, and the libraries were retransformed into TOP10 cells.
To examine leakiness in-frame maintenance, about 106 cells of the reporter strain were plated on LB agar containing 10 or 25 μg ml−1 ampicillin. All the selectants were pooled from the plates, diluted and re-plated under the same selective conditions to isolate about 100 well-spaced colonies. Colonies were amplified in small cultures of rich medium and the cells were washed with water and subjected to PCR to amplify the region of interest in β-lactamase. These reactions were treated with ExoI and shrimp alkaline phosphatase (USB), heated to denature the enzymes and sequenced.
To select the best suppressors of four-base codons, the tRNASer-derived libraries were transformed into electrocompetent cells of the reporter library strains. These libraries were plated on selective media containing various concentrations of ampicillin using a vast excess of cells. For example, 109 cells were used for the cross at 300 μg ml−1 ampicillin of the tRNASer(N8) library against the S124(N4) reporter library, which theoretically has 2×107 unique combinations. To sequence the β-lactamase reporters in the selectants, colonies were amplified in liquid culture and DNA prepared and digested with an enzyme specific to the tRNA-bearing plasmid. This DNA was retransformed into TOP10 cells and sequenced as above. Sequencing of the tRNAs from selectants was done analogously with destruction of the reporter vector.
Assay for suppression efficiency
TOP10 cells bearing a specific four-base codon at S124 were isolated, and chemically competent cells of these strains were prepared by the method of Inoue et al. (1990). The corresponding tRNA-bearing plasmids were isolated and transformed into the strains bearing the specific members of the S124(N4) library. Cells bearing the specific members of the S124(N4) library and controls, specifically, S124 mutants UAG, AGGA, CCCU, CUAG, UAGA and wild-type pBR322, were assayed for β-lactamase activity in the presence and absence of the corresponding tRNAs. Cells were grown in 2YT to A 600 between 0.5 and 1.5 (mid to late-log phase), washed with 50 mM sodium phosphate buffer (pH 7) containing 10 % (w/v) DMSO, and assayed with 1 mM nitrocefin (Oxoid). To measure the low read-through activities of the TAG and four-base codons alone, 5 ml of cells from culture was concentrated into 1 ml assays and observed at 486 nm at 20, 30 and 40 minutes. For higher activities from suppression with tRNAs, 50–250 μl of culture was added directly to the assays and the A 486 nm was observed every 30 seconds to one minute for 15 to 30 minutes. Since the concentration of nitrocefin is far above the K M of 110 μM (Sigal et al., 1984) and far less than 10 % of the substrate was consumed, the rate of substrate turnover is proportional to the concentration of β-lactamase contributed by the cells, and therefore the amount of enzyme from coding, readthrough or suppression. All assays were carried out at room temperature. Arbitrary units were defined as:Units=500,000ΔA486/minutevol×A600 where vol is the culture volume (in μl) in the 1 ml assay, and the rate of change in absorbance at 486 nm was adjusted for the background rate of hydrolysis of nitrocefin, which was 100 times slower than the slowest rate measured from TOP10 cells alone.
Acknowledgements
This work was supported by the Department of the Army, the Office of Naval Research and the Skaggs Research Institute. T.J.M. and J.C.A. are NSF pre-doctoral fellows.
Footnotes
Edited by M. Gottesman
References
- Atkins J.F., Elseviers D., Gorini L. Low activity of galactosidase in frameshift mutants of Escherichia coli. Proc. Natl Acad. Sci. USA. 1972;69:1192–1195. doi: 10.1073/pnas.69.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins J.F., Weiss R.B., Gesteland R.F. Ribosome gymnastics — degree of difficulty 9.5, style 10.0. Cell. 1990;62:413–423. doi: 10.1016/0092-8674(90)90007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins J.F., Weiss R.B., Thompson S., Gesteland R.F. Towards a genetic dissection of the basis of triplet decoding, and its natural subversion: programmed reading frame shifts and hops. Annu. Rev. Genet. 1991;25:201–228. doi: 10.1146/annurev.ge.25.120191.001221. [DOI] [PubMed] [Google Scholar]
- Auffinger P., Westhof E. Singly and bifurcated hydrogen-bonded base-pairs in tRNA anticodon hairpins and ribozymes. J. Mol. Biol. 1999;292:467–483. doi: 10.1006/jmbi.1999.3080. [DOI] [PubMed] [Google Scholar]
- Ayer D., Yarus M. The context effect does not require a fourth base-pair. Science. 1986;231:393–395. doi: 10.1126/science.3510456. [DOI] [PubMed] [Google Scholar]
- Belcourt M.F., Farabaugh P.J. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell. 1990;62:339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biou V., Yaremchuk A., Tukalo M., Cusack S. The 2.9 Å crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNA(Ser) Science. 1994;263:1404–1410. doi: 10.1126/science.8128220. [DOI] [PubMed] [Google Scholar]
- Bossi L., Roth J.R. The influence of codon context on genetic code translation. Nature. 1980;286:123–127. doi: 10.1038/286123a0. [DOI] [PubMed] [Google Scholar]
- Bossi L., Roth J.R. Four-base codons ACCA, ACCU and ACCC are recognized by frameshift suppressor sufJ. Cell. 1981;25:489–496. doi: 10.1016/0092-8674(81)90067-2. [DOI] [PubMed] [Google Scholar]
- Brierley I., Digard P., Inglis S.C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C., Campbell A.M., Karlin S. Over- and under-representation of short oligonucleotides in DNA sequences. Proc. Natl Acad. Sci. USA. 1992;89:1358–1362. doi: 10.1073/pnas.89.4.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F.H.C., Barnett L., Brenner S., Watts-Tobin R.J. General nature of the genetic code for proteins. Nature. 1961;192:1227–1232. doi: 10.1038/1921227a0. [DOI] [PubMed] [Google Scholar]
- Cummins C.M., Culbertson M.R., Knapp G. Frameshift suppressor mutations outside the anticodon in yeast proline tRNAs containing an intervening sequence. Mol. Cell. Biol. 1985;5:1760–1771. doi: 10.1128/mcb.5.7.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupples C.G., Cabrera M., Cruz C., Miller J.H. A set of lacZ mutations in Escherichia coli that allow rapid detection of specific frameshift mutations. Genetics. 1990;125:275–280. doi: 10.1093/genetics/125.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran J.F. Analysis of effects of tRNA: message stability on frameshift frequency at the Escherichia coli RF2 programmed frameshift site. Nucl. Acids Res. 1993;21:1837–1843. doi: 10.1093/nar/21.8.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran J.F., Yarus M. Reading frame selection and transfer RNA anticodon loop stacking. Science. 1987;238:1545–1550. doi: 10.1126/science.3685992. [DOI] [PubMed] [Google Scholar]
- Farabaugh P.J. Programmed translational frameshifting. Microbiol. Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox T.D. Natural variation in the genetic code. Annu. Rev. Genet. 1987;21:67–91. doi: 10.1146/annurev.ge.21.120187.000435. [DOI] [PubMed] [Google Scholar]
- Gaber R.F., Culbertson M.R. Codon recognition during frameshift suppression in Saccharomyces cerevisiae. Mol. Cell. Biol. 1984;4:2052–2061. doi: 10.1128/mcb.4.10.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteland R.F., Weiss R.B., Atkins J.F. Recoding: reprogrammed genetic decoding. Science. 1992;257:1640–1641. doi: 10.1126/science.1529352. [DOI] [PubMed] [Google Scholar]
- Hohsaka T., Ashizuka Y., Murakami H., Sisido M. Incorporation of nonnatural amino acids into streptavidin through in vitro frame-shift suppression. J. Am. Chem. Soc. 1996;118:9778–9779. [Google Scholar]
- Hohsaka T., Ashizuka Y., Sasaki H., Murakami H., Sisido M. Incorporation of two different nonnatural amino acids independently into a single protein through extension of the genetic code. J. Am. Chem. Soc. 1999;121:12194–12195. [Google Scholar]
- Hohsaka T., Kajihara D., Ashizuka Y., Murakami H., Sisido M. Efficient incorporation of nonnatural amino acids with large aromatic groups into streptavidin in in vitro protein synthesizing systems. J. Am. Chem. Soc. 1999;121:34–40. [Google Scholar]
- Holbrook S.R., Sussman J.L., Warrant R.W., Kim S.H. Crystal structure of yeast phenylalanine transfer RNA. II. Structural features and functional implications. J. Mol. Biol. 1978;123:631–660. doi: 10.1016/0022-2836(78)90210-3. [DOI] [PubMed] [Google Scholar]
- Hong K.W., Ibba M., Soll D. Retracing the evolution of amino acid specificity in glutaminyl-tRNA synthetase. FEBS Letters. 1998;434:149–154. doi: 10.1016/s0014-5793(98)00968-5. [DOI] [PubMed] [Google Scholar]
- Inokuchi H., Yamao F. Structure and expression of prokaryotic tRNA genes. In: Soll D., RajBhandary U.L., editors. tRNA: Structure, Biosynthesis and Function. ASM Press; Washington, DC: 1995. pp. 17–30. [Google Scholar]
- Inoue H., Nojima H., Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- Kramer G., Kudlicki W., Hardesty B. In vitro engineering using synthetic tRNAs with altered anticodons including four-nucleotide anticodons. Methods Mol. Biol. 1998;77:105–116. doi: 10.1385/0-89603-397-X:105. [DOI] [PubMed] [Google Scholar]
- Kurland C.G. Translational accuracy and the fitness of bacteria. Annu. Rev. Genet. 1992;26:29–50. doi: 10.1146/annurev.ge.26.120192.000333. [DOI] [PubMed] [Google Scholar]
- Liu D.R., Schultz P.G. Progress toward the evolution of an organism with an expanded genetic code. Proc. Natl Acad. Sci. USA. 1999;96:4780–4785. doi: 10.1073/pnas.96.9.4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.R., Magliery T.J., Pastrnak M., Schultz P.G. Engineering a tRNA and aminoacyl-tRNA synthetase for the site-specific incorporation of unnatural amino acids into proteins in vivo. Proc. Natl Acad. Sci. USA. 1997;94:10092–10097. doi: 10.1073/pnas.94.19.10092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Kudlicki W., Odom O.W., Kramer G., Hardesty B. In vitro protein engineering using synthetic tRNA(Ala) with different anticodons. Biochemistry. 1993;32:7939–7945. doi: 10.1021/bi00082a015. [DOI] [PubMed] [Google Scholar]
- Manley J.L. Synthesis and degradation of termination and premature-termination fragments of beta-galactosidase in vitro and in vivo. J. Mol. Biol. 1978;125:407–432. doi: 10.1016/0022-2836(78)90308-x. [DOI] [PubMed] [Google Scholar]
- Moore B., Persson B.C., Nelson C.C., Gesteland R.F., Atkins J.F. Quadruplet codons: implications for code expansion and the specification of translation step size. J. Mol. Biol. 2000;298:195–209. doi: 10.1006/jmbi.2000.3658. [DOI] [PubMed] [Google Scholar]
- Murakami H., Hohsaka T., Ashizuka Y., Sisido M. Site-directed incorporation of p-nitrophenylalanine into streptavidin and site-to-site photoinduced electron transfer from a pyrenyl group to a nitrophenyl group on the protein framework. J. Am. Chem. Soc. 1998;120:7520–7529. [Google Scholar]
- Ogawa A.K., Wu Y., McMinn D.L., Liu L., Schultz P.G., Romesberg F.E. Efforts toward the expansion of the genetic alphabet: information storage and replication with unnatural hydrophobic base-pairs. J. Am. Chem. Soc. 2000;122:3274–3287. [Google Scholar]
- Parker J. Errors and alternatives in reading the universal genetic code. Microbiol. Rev. 1989;53:273–298. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastrnak M., Magliery T.J., Schultz P.G. A new orthogonal suppressor tRNA/aminoacyl-tRNA synthetase pair for evolving an organism with an expanded genetic code. Helv. Chim. Acta. 2000;83:2277–2286. [Google Scholar]
- Price S., Cusack S., Borel F., Berthet-Colominas C., Leberman R. Crystallization of the seryl-tRNA synthetase: tRNAS(ser) complex of Escherichia coli. FEBS Letters. 1993;324:167–170. doi: 10.1016/0014-5793(93)81386-e. [DOI] [PubMed] [Google Scholar]
- Qian Q., Li J., Zhao H., Hagervall T.G., Farabaugh P.J., Bjork G.R. A new model for phenotypic suppression of frameshift mutations by mutant tRNAs. Mol. Cell. 1998;1:471–482. doi: 10.1016/s1097-2765(00)80048-9. [DOI] [PubMed] [Google Scholar]
- Riddle D.L., Carbon J. Frameshift suppression: a nucleotide addition in the anticodon of a glycine transfer RNA. Nature New Biol. 1973;242:230–234. doi: 10.1038/newbio242230a0. [DOI] [PubMed] [Google Scholar]
- Riyasaty S., Atkins J.F. External suppression of a frameshift mutant in Salmonella. J. Mol. Biol. 1968;34:541–557. doi: 10.1016/0022-2836(68)90179-4. [DOI] [PubMed] [Google Scholar]
- Sigal I.S., DeGrado W.F., Thomas B.J., Petteway S.R., Jr Purification and properties of thiol beta-lactamase. A mutant of pBR322 beta-lactamase in which the active site serine has been replaced with cysteine. J. Biol. Chem. 1984;259:5327–5332. [PubMed] [Google Scholar]
- Spanjaard R.A., van Duin J. Translation of the sequence AGG-AGG yields 50 % ribosomal frameshift. Proc. Natl Acad. Sci. USA. 1988;85:7967–7971. doi: 10.1073/pnas.85.21.7967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman J.L., Kim S. Three-dimensional structure of a transfer rna in two crystal forms. Science. 1976;192:853–858. doi: 10.1126/science.775636. [DOI] [PubMed] [Google Scholar]
- Tate W.P., Poole E.S., Dalphin M.E., Major L.L., Crawford D.J., Mannering S.A. The translational stop signal: codon with a context, or extended factor recognition element? Biochimie. 1996;78:945–952. doi: 10.1016/s0300-9084(97)86716-8. [DOI] [PubMed] [Google Scholar]
- Tuohy T.M., Thompson S., Gesteland R.F., Atkins J.F. Seven, eight and nine-membered anticodon loop mutants of tRNA(2Arg) which cause +1 frameshifting. Tolerance of DHU arm and other secondary mutations. J. Mol. Biol. 1992;228:1042–1054. doi: 10.1016/0022-2836(92)90313-9. [DOI] [PubMed] [Google Scholar]
- Wang L., Magliery T.J., Liu D.R., Schultz P.G. A new functional suppressor tRNA/aminoacyl-tRNA synthetase pair for the in vivo incorporation of unnatural amino acids into proteins. J. Am. Chem. Soc. 2000;122:5010–5011. [Google Scholar]
- Weiss R., Gallant J. Mechanism of ribosome frameshifting during translation of the genetic code. Nature. 1983;302:389–393. doi: 10.1038/302389a0. [DOI] [PubMed] [Google Scholar]
- Weiss R.B., Dunn D.M., Atkins J.F., Gesteland R.F. Slippery runs, shifty stops, backward steps, and forward hops: −2, −1, +1, +2, +5, and +6 ribosomal frameshifting. Cold Spring Harbor Symp. Quant. Biol. 1987;52:687–693. doi: 10.1101/sqb.1987.052.01.078. [DOI] [PubMed] [Google Scholar]
- Weiss R., Lindsley D., Falahee B., Gallant J. On the mechanism of ribosomal frameshifting at hungry codons. J. Mol. Biol. 1988;203:403–410. doi: 10.1016/0022-2836(88)90008-3. [DOI] [PubMed] [Google Scholar]
- Weiss R.B., Dunn D.M., Dahlberg A.E., Atkins J.F., Gesteland R.F. Reading frame switch caused by base-pair formation between the 3′ end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J. 1988;7:1503–1507. doi: 10.1002/j.1460-2075.1988.tb02969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yourno J., Tanemura S. Restoration of in-phase translation by an unlinked suppressor of a frameshift mutation in Salmonella typhimurium. Nature. 1970;5231:422–426. doi: 10.1038/225422a0. [DOI] [PubMed] [Google Scholar]
Further reading
- Winey M., Mendenhall M.D., Cummins C.M., Culbertson M.R., Knapp G. Splicing of a yeast proline tRNA containing a novel suppressor mutation in the anticodon stem. J. Mol. Biol. 1986;192:49–63. doi: 10.1016/0022-2836(86)90463-8. [DOI] [PubMed] [Google Scholar]