Abstract
Background
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CO-V-2), was first reported in Wuhan, Hubei province, China has now rapidly spread over 50 countries. For the prevention and control of infection, Taiwan Centers for Disease Control initiated testing of SARS-CoV-2 on January 24th 2020 for persons suspected with this disease. Until February 28th, 43 flu-like symptomatic patients were screened in China Medical University Hospital.
Methods
Two patients were confirmed positive for SARS-CoV-2 infection by rRT-PCR as COVID-19 patients A and B. Causative pathogens for included patients were detected using FilmArray™ Respiratory Panel. We retrospectively analyzed the clinical presentations, laboratory data, radiologic findings, and travel and exposure contact histories, of the COVID-19 patients in comparison to those with other respiratory infections.
Results
Through contact with Taiwan No. 19 case patient on 27th January, COVID-19 patients A and B were infected. Both patients had no identified comorbidities and developed mild illness with temporal fever, persistent cough, and lung interstitial infiltrates. Owing to the persistence of positive SARS-CoV-2 in respiratory specimen, the two COVID-19 patients are still in the isolation rooms despite recovery until 10th of March. The results of FilmArrayTM Respiratory Panel revealed 22 of the 41 non-COVID-19 patients were infected by particular pathogens. In general, seasonal respiratory pathogens are more prevalent than SARS-CoV-2 in symptomatic patients in non- COVID-19 endemic area during the flu season. Since all patients shared similar clinical and laboratory findings, expanded surveillance of detailed exposure history for suspected patients and application of rapid detection tools are highly recommended.
Keywords: COVID-19, SARS-CO-V-2, FilmArray™ Respiratory Panel
Introduction
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CO-V-2), which is a member of Betacoronavirus. The outbreak of COVID-19 was first reported in Wuhan, Hubei province, China on December 2019, has now rapidly spread over 50 countries.1 , 2 Clinical spectrum of this disease varied from mild to severe. Fever (88%), cough (67%), and fatigue (34%) were the most common symptoms presented by COVID-19 patients3 which were similar to those with infections caused by other respiratory viruses, such as Influenza A/B, respiratory syncytial virus, and rhinovirus.4 , 5
For the prevention and control of COVID-19, Taiwan Centers for Disease Control (CDC) initiated testing of SARS-CoV-2 on January 24th 2020 for persons who had a travel history to China and presented fever or any respiratory symptoms within 14 days. Until February 28th, 2105 cases were screened and 34 of them were diagnosed of COVID-19. During this period, 43 suspected patients were admitted to a medical center located in central Taiwan, and two of them were positive for SARS-CoV-2. Through multiplex PCR analysis with FilmArray™ Respiratory Panel, the non-COVID-19 patients were later diagnosed of infections with other respiratory pathogens. Undoubtedly, early screening and diagnosis is crucial for the treatment and control of COVID-19. To establish the diagnostic protocol for this disease, in this report, we comparatively analyzed the clinical presentations, laboratory data, radiologic findings, and travel and exposure contact histories, of the COVID-19 patients with those with other respiratory infections.
Materials and methods
Patients and clinical data collection
During January 20th to February 19th 2020, a total of 43 patients who were admitted to China Medical University Hospital (CMUH) met the screening criteria of COVID-19 reported by Taiwan CDC. In the negative pressure isolation room of CMUH emergency room, demographic and travel history data of the patients were collected, and portable chest X-ray and naso-oropharyngeal sampling for viral identification were also completed. For cases reporting of COVID-19 infection, blood tests were performed after their admission to the negative pressure isolation wards.
SARS-CoV-2 detection by real-time reverse transcription polymerase chain reaction (rRT-PCR)
The loads of SARS-CoV-2 in the naso-oropharyngeal specimens were performed, using rRT-PCR, upon and 24 h after admission to identify the COVID-19-infected patients. For the purpose of COVID-19 de-isolation, sputum and pharyngeal specimens were examined for SARS-CoV-2 rRT-PCR every 3–7 days based clinical conditions. The specimens collected from each of the admitted patients was incubated with Vero cells in Dulbecco's Modified Eagle Medium at 37 °C for 1 h. Total RNA was extracted from the culture supernatant using MagNA Pure 96 system (Roche, Manheim, Germany) and subjected to the detection of SARS-CoV-2 using rRT-PCR. Based on the US CDC guideline, the primers and probes we used for the detection of PdPR, RdRP, and E_Sarbeco genes were RdRP_SARSr-F2-GTG ARA TGG TCA TGT GTG GCG G, RdRP_SARSr-R1-CAR ATG TTA AAS ACA CTA TTA GCA TA, RdRP_SARSr-P1-FAM-CCA GGT GGW ACR TCA TCM GGT GAT GC-BBQ, E_Sarbeco_F1-ACA GGT ACG TTA ATA GTT AAT AGC GT, E_Sarbeco_R2-ATA TTG CAG CAG TAC GCA CAC A, and E_Sarbeco_P1-FAM-ACA CTA GCC ATC CTT ACT GCG CTT CG-BBQ.6
Respiratory pathogens detection by FilmArray™ Respiratory Panel
The nasopharyngeal specimens collected from the non-COVID-19 patients were subjected to the detection of respiratory pathogens using FilmArray™ Respiratory Panel (BioFire Diagnostics, bioMérieux, Utah, USA). A total of 20 respiratory pathogens, including the viral pathogens: Influenza A (untyped, A/H1, A/H1-2009, A/H3), Influenza B, Adenovirus, Coronaviruses (HKU1, NL63, 229E, OC43), Human Metapneumovirus, Human Rhinovirus/Enterovirus, Parainfluenza (types 1, 2, 3, 4), and Respiratory Syncytial Virus; and the bacterial pathogens: Bordetella pertussis, Chlamydophila pneumoniae, and Mycoplasma pneumoniae, could be simultaneously detected by nested real-time PCR in the FilmArray™ instrument in 1 h with high sensitivity and specificity.7, 8, 9
Statistical analysis
The categorical variable was summarized as n/N (%), where N is the total number of patients with available data and continuous variable was summarized as mean (SD) or median (IQR). We assessed differences between COVID-19 and non-COVID-19 using χ2 test or Fisher's exact test for categorical variables and Mann–Whitney U test for continuous variable. The MedCalc Statistical Software version 17.4 was applied for all analyses.
Results
A total of 43 patients suspected with COVID-19 were admitted to CMUH during January 20th to February 19th 2020. Two of them were later confirmed to be positive for SARS-CoV-2 infection by rRT-PCR. As shown in Table 1 , the ages of patients ranged from 3 to 68 years (mean age of 34.0 years). The male-to-female ratio was 0.65. Of all patients, 23% (n = 10) came back from Guangdong province, 9% (n = 4) from Wuhan city, and 2% (n = 1) from Hubei province areas other than Wuhan city, and 7% (n = 3) went to other country but transited from Hong Kong airport. Of the 10 patients who had contact history, 4 had close contact with people who visited Guangdong or Hubei province, and 6 had connected to COVID-19 patients. The two COVID-19 patients confirmed in this study had no travel or contact history with people who had been to China but had a family dinner on 27th January with the patient who was the first COVID-19 mortality case (Taiwan NO. 19) confirmed on 15th February. About 44% of the non-COVID19 patients had at least one comorbidity; however, the two confirmed cases, COVID-19 patients A and B, presented no comorbid conditions.
Table 1.
Demographic characteristics, travel and contact history, and clinical features of the patients suspected with COVID-19 in this study.
| COVID-19 (n = 2) | Non-COVID-19 (n = 41) | All Patients (n = 43) | P value | |
|---|---|---|---|---|
| Demographic | ||||
| Age (range), year | 45.0 (39–51) | 34.0 (3–68) | 34.0 (3–68) | 0.6237 |
| Male | 1 (50%) | 16 (39%) | 17 (40%) | |
| Female | 1 (50%) | 25 (61%) | 26 (60%) | |
| Co-morbidities | ||||
| Diabetes mellitus | 4 (10%) | 4 (9%) | 1 | |
| Hypertension | 2 (5%) | 2 (5%) | 1 | |
| Coronary artery disease | 1 (2%) | 1 (2%) | 1 | 1 |
| Airway disease | 4 (10%) | 4 (9%) | 1 | |
| Liver cirrhosis | 1 (2%) | 1 (2%) | 1 | |
| Autoimmune disease | 4 (10%) | 4 (9%) | 1 | |
| Malignancy | 2 (5%) | 2 (5%) | 1 | |
| Travel/Contact history | ||||
| Travel to | ||||
| Wuhan | 4 (10%) | 4 (9%) | ||
| Hubei | 1 (2%) | 1 (2%) | ||
| Guangdong | 10 (24%) | 10 (23%) | ||
| Other province of China | 15 (37%) | 15 (35%) | ||
| Transit in Hong Kong | 3 (7%) | 3 (7%) | ||
| Contact persons traveling to | ||||
| Wuhan | 2 (5%) | 2 (5%) | ||
| Hubei | 1 (2%) | 1 (2%) | ||
| Guangdong | 1 (2%) | 1 (2%) | ||
| Contact with confirmed COVID-19 patient | 2 (100%) | 4 (10%) | 6 (14%) | |
| Symptoms | ||||
| Fever | 2 (100%) | 22 (54%) | 24 (56%) | 0.495 |
| Nonproductive cough | 1 (50%) | 22 (54%) | 23 (53%) | 1 |
| Productive cough | 1 (50%) | 8 (20%) | 9 (21%) | 0.3787 |
| Dyspnea | 1 (50%) | 3 (7%) | 4 (9%) | 0.1794 |
| Chest tightness | 3 (7%) | 3 (7%) | 1 | |
| Rhinorrhea | 1 (50%) | 15 (37%) | 16 (37%) | 1 |
| Stuff nose | 5 (12%) | 5 (12%) | 1 | |
| Sore throat | 1 (50%) | 8 (20%) | 9 (21%) | 0.3787 |
| Headache | 5 (12%) | 5 (12%) | 1 | |
| Myalgia | 6 (15%) | 6 (14%) | 1 | |
| Abdominal pain | 1 (50%) | 2 (5%) | 2 (5%) | 1 |
| Diarrhea | 1 (50%) | 4 (10%) | 4 (9%) | 1 |
| Nausea/Vomiting | 3 (7%) | 3 (7%) | 1 | |
Clinical course of COVID-19 patient A and B
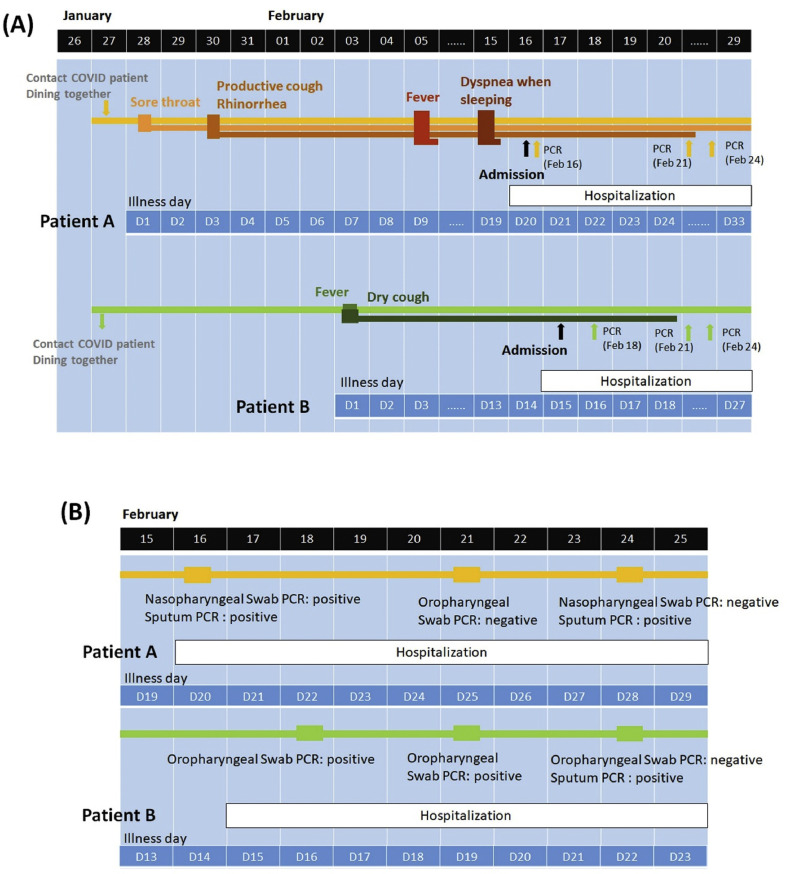
The first symptom of COVID-19 patient A was sore throat, which developed on the day (28th January, illness day 1; Fig. 1 A) next to his contact with Taiwan No. 19 case patient. Productive cough and rhinorrhea appeared on illness day 3. Fever up to 39.1 °C and dyspnea developed on illness day 9 and 19, respectively. On 16th February (illness day 20), this patient was admitted to CMUH and diagnosed of COVID-19, as confirmed by positive rRT-PCR tests of nasopharyngeal swab and sputum. Despite of relatively mild symptoms, this patient developed a prolonged and insidious disease course and had gastrointestinal symptoms, presented as upper abdominal dull pain and diarrhea during hospitalization. COVID-19 patient B who also contacted with Taiwan No. 19 case patient on 27th January developed fever up to 38 °C on 6 days later (3rd February, illness day 1; Fig. 1A) and had dry cough as the primary manifestation that persisted until illness day 19. During the third sampling on 24th February, the loads of SARS-CoV-2 in the naso-oropharyngeal specimens from COVID-19 patients A and B were both at an undetectable level. However, their sputum was still tested positive for SARS-CoV-2. Patient A's sputum remained positive rRT-PCR on illness day 28, and then turned negative on illness day 35. The oropharyngeal specimens rRT-PCR of patient B altered to detectable again on illness day 29 and 33, and sputum rRT-PCR also remained positive at least until illness day 33.
Figure 1.
Clinical symptoms (A) and SARS-CoV-2 rRT-PCR tests (B) of COVID-19 patient A and B. Upon exposure to Taiwan No. 19 case patient on 27th January, COVID-19 patient A and B developed symptoms on 28th January and 3rd February, respectively. Naso-oropharyngeal or sputum specimens collected from both of them were tested positive for SARS-CoV-2 on three samplings during hospitalization.
Clinical features and laboratory data of COVID-19 patients in comparison with non-COVID-19 group
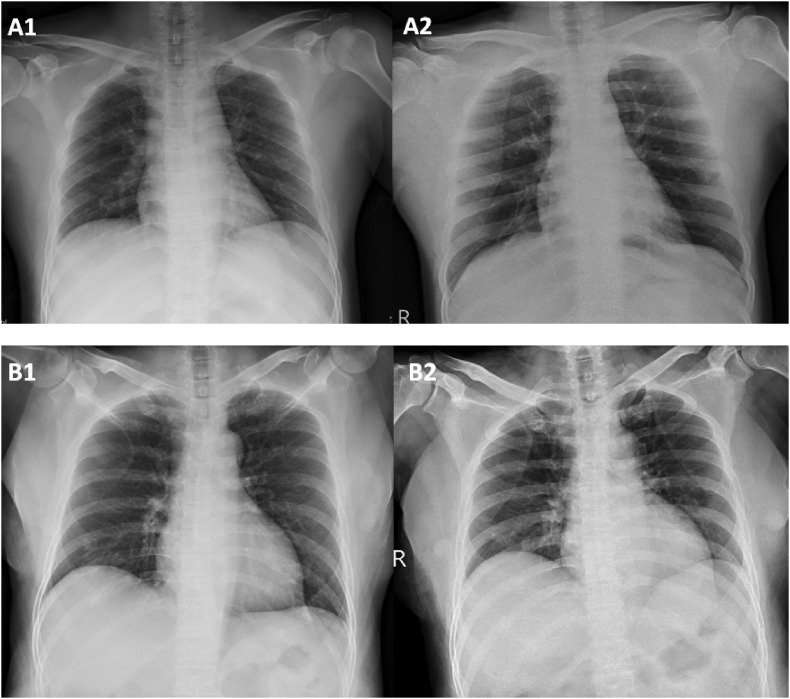
The most common symptoms reported by both groups included fever (56%) and nonproductive cough (53%), followed by rhinorrhea (37%), sore throat (21%), productive cough (21%), and dyspnea (9%) (Table 1). Screening of chest radiographs were abnormal for 55.8% of the 43 patients, mostly interstitial infiltrates. The initial chest radiograph of COVID-19 patient A on hospital admission (illness day 20) was normal without active lesions (Fig. 2 A; left panel). However, on day 6 of admission (illness day 25), interstitial infiltrates were noted on his right upper lung (Fig. 2A; right panel). Left lower lung interstitial infiltrates were revealed by the chest radiograph of COVID-19 patient B on admission (illness day 15; Fig. 2B; left panel) and persisted for 4 days (Fig. 2B; right panel).
Figure 2.
Chest radiographs of COVID-19 patient A (A1 and A2) and patient B (B1 and B2). No active lung lesion was noted in patient A on admission (16th February; illness day 20) (A1), while right upper lung interstitial infiltrates were presented on day 6 on admission (21st February; illness day 25) (A2). Left lower lung interstitial infiltrates were noted in patient B on admission (17th February; illness day 15) (B1) and persisted to day 5 of admission (21st February; illness day 19) (B2).
Both the COVID-19 patients had normal white blood cell (WBC) counts without lymphocytopenia and the neutrophil to lymphocyte ratio in the normal range (Table 2 ). Of the non-COVID-19 patients, 83% had normal WBC counts and 29% had lymphocytopenia. The average neutrophil to lymphocyte ratio was 3.3. No statistically significant differences were found between the COVID-19 and non-COVID-19 groups in levels of creatinine, aspartate aminotransferase (AST), alanine aminotransferase (ALT), bilirubin, C-reactive protein (CRP), creatine kinase (CK), and lactate dehydrogenase (LDH) (Table 2).
Table 2.
Laboratory data of the patients suspected with COVID-19 in this study.
| COVID-19 (n = 2) | Non-COVID-19 (n = 41) | P value | |
|---|---|---|---|
| WBC, 103/uL (3600–11200) | 6300 (5900–6700) | 7700 (6700–10325) | 0.1492 |
| <3600 | 0 | 2 (4.9%) | 0.7868 |
| 3600-11200 | 2 (100%) | 33 (80.5%) | |
| >11,200 | 0 | 6 (14.6%) | |
| Neutrophil, 103/uL | 3934 (3198–4670) | 5350 (4183–7510) | 0.2045 |
| Neutrophil, % | 62.0 (54.2–69.7) | 70.4 (62.7–76.5) | 0.3561 |
| Lymphocytes, 103/uL | 1829 (1534–2124) | 1565 (972–1975) | 0.4889 |
| Lymphocyte, % | 29.5 (22.9–36.0) | 21.0 (12.2–27.4) | 0.2258 |
| <1000 (lymphopenia) | 0 | 12 (29.3%) | 1 |
| >1000 | 2 (100%) | 29 (70.7%) | |
| Neutrophil to lymphocytes ratio | 2.3 (1.5–3.0) | 3.1 (2.3–6.5) | 0.2259 |
| Creatinine, mg/dL (0.7–1.3) | 0.81 (0.64–0.97) | 0.68 (0.60–0.80) | 0.3753 |
| Aspartate aminotransferase, IU/L (13–39) | 18.0 (17.0–19.0) | 19.0 (14.3–24.0) | 0.716 |
| Alanine aminotransferase, IU/L (5–40) | 30.0 (18.0–42.0) | 14.0 (11.0–24.5) | 0.2133 |
| Bilirubin, mg/dL (0.2–1.3) | 0.36 (0.35–0.37) | 0.54 (0.42–0.71) | 0.1174 |
| C-reactive protein, mg/dL (≤1) | 0.23 (0.20–0.26) | 0.42 (0.08–1.45) | 0.4782 |
| ≤1 | 2 (100%) | 28 (70.0%) | 1 |
| >1 | 0 | 12 (30.0%) | |
| Creatine kinase, IU/L (30–223) | 92.0 (53.0–131.0) | 74.0 (50.3–114.0) | 0.6281 |
| Lactate dehydrogenase, U/L (140–271) | 190 (184.0–196.0) | 178.0 (147.5–192.8) | 0.4313 |
Detection of respiratory pathogens for non-COVID-19 patients
FilmArray™ Respiratory Panel, a multiplex PCR assay that comprehensively detects 20 respiratory viruses and bacteria,8 was used for the identification of respiratory pathogens for non-COVID-19 patients. Coronavirus229E/OC43 was identified in 3 patients (7%), Influenza A/B in 4 patients (9%), Human Rhinovirus/Enterovirus in 5 patients (12%), Adenovirus in 3 patients (7%), other sporadic viral pathogens in 4 patients (9%), and Mycoplasma pneumonia in 3 patients (7%) (Table 3 ). Three of them (7%) had more than one pathogen detected. Of the 4 Influenza A or B patients, all had fever (100%), 3 had rhinorrhea (75%), 2 had nonproductive cough (50%), 1 had productive cough (25%), and 2 suffered from myalgia (50%). Among the 3 patients with Coronavirus (229E/OC43) infection, only 1 had fever (33%), and the others had variable upper respiratory tract and gastrointestinal (GI) tract symptoms, including diarrhea and abdominal pain. No pathogens were found by FilmArray™ for two of non-COVID-19 patients, and both had alveolar infiltrates on the chest radiograph. All the 43 patients’ respiratory specimens were tested twice for COVID-19 by rt-PCR within the first 2 days of admission.
Table 3.
Other respiratory pathogens detected in the non-COVID-19 patients by FilmArray Respiratory Panel.
| Respiratory pathogen | Non-COVID-19 (n = 38) |
|---|---|
| CoV 229E | 1 (2%) |
| Coronavirus OC43 | 2 (5%) |
| Influenza A | 3 (7%) |
| Influenza B | 1 (2%) |
| Parainfluenza 2 | 1 (2%) |
| Parainfluenza 3 | 1 (2%) |
| Human Rhinovirus/Enterovirus | 2 (5%) |
| Adenovirus | 3 (7%) |
| RSV | 2 (5%) |
| Mycoplasma pneumoniae | 2 (5%) |
| No pathogen | 21 (55%) |
| One pathogen | 14 (37%) |
| More than one pathogen | 3 (8%) |
Discussion
In comparison with non-COVID-19 patients, no specific symptoms or laboratory data were noted in the early phase of COVID-19 illness. Therefore, the contact and travel history are the main screening criteria for SARS-CoV-2. Rapid detection tools such as FilmArrayTM Respiratory Panel can aid in early diagnosis of causative pathogens. As demonstrated in this report, the development of mild COVID-19 illness was insidiously initiated and persisted for a long period. The prolonged duration of SARS-CoV-2 detection in naso-oropharyngeal specimens of the COVID-19 patients with mild illness is of concern in epidemic areas.
Clinical manifestations of viral respiratory infections range from asymptomatic or flu-like symptoms such as fever, nonproductive or productive cough, myalgia, rhinorrhea, and sore throat, to acute respiratory distress syndrome (ARDS) or multiple organs failure.10 Various viral pathogens cause community-acquired infections during the cold season in Taiwan. Of which, influenza A/B, human Metapneumovirus, adenovirus, parainfluenza virus 1/2/3, coronavirus 229E/NL63/OC43, rhinovirus, and respiratory syncytial virus, are the most common.11 The lack of specificity for clinical symptoms, laboratory data, and chest radiographs of COVID-19, as demonstrated in this report and previous studies,3 , 4 , 12 makes differential diagnosis of this disease from other viral respiratory infections difficult. Rapid detection tools of common respiratory virus and bacteria, such as FilmArray™ Panel used here, help clinicians to determine causative pathogens of pneumonia in patients who are suspected with COVID-19 but have negative rRT-PCR result for SARS-CoV-2. One of the huge challenges for preventing and controlling of this pandemic-prone disease13 , 14 is the transmission of SARS-CoV-2 from COVID-19 patients who were asymptomatic or presented flu-like symptoms.15 , 16 Under this circumstances, quarantine and active surveillance of suspected patients and the application of rapid detection tools for etiology confirmation is strongly recommended.
The confirmed COVID-19 cases in Taiwan belonged to two groups: international migration and locally transmitted. Both COVID-19 patients in this report had no travel history. Upon exposure to Taiwan No. 19 case patient during a family dinner, they were infected. There were two additional family clusters of infections and a cluster of 8 patients infected in a single hospital in Taiwan until 6th March 2020. The reproductive number of COVID-19, ranged from 1.4 to 6.49,17 was averagely higher than severe acute respiratory symptoms (SARS) in 2003, and Middle East Respiratory Syndrome (MERS) in 2012.18 Community spread of COVID-19 has occurred in several countries in different continents. China, Hong Kong, Macao, Korea, Italy, and Iran are reported epidemic areas. To prevent the COVID-19 pandemic, we need to expand the exposure risk management to persons who have travel history to epidemic areas, sick or cluster contacts, and to patients who have pneumonia or ARDS without clear explanation.
COVID-19 is highly contagious. However, 75–80% of patients have mild illness. Patients who have two or more comorbidities, such as COPD, diabetes, and malignancy, are prone to become severely ill or die.5 Although mild illness without lymphocytopenia was developed in both the COVID-19 patients A and B, who had no comorbid conditions, the loads of SARS-CoV-2 persisted for a long duration (Fig. 1B). Some COVID-19 patients who were considered recovered after symptoms resolved still carried detectable levels of SARS-CoV-2 for 5–13 days.19 The naso-oropharyngeal specimens, collected from the third sampling of COVID-19 patients A and B, showed negative for SARS-CoV-2. However, their sputum specimens collected on the same day were tested positive. Until 6th March, the two COVID-19 patients are still in the isolation room, pending for the latest sputum rRT-PCR results. Despite of mild upper and lower respiratory tract symptoms, COVID-19 patient A suffered from diarrhea throughout the illness course. Consistent with the recent study that gastrointestinal symptoms, including diarrhea, vomiting, and abdominal pain, were developed in some COVID-19 patients.20 Fecal specimens of these patients were tested positive for SARS-CoV-2. Potential fecal-oral transmission in patients with COVID-19 is of concern.
Seasonal viral respiratory pathogens prevalent in Taiwan include Rhinoviruses/enteroviruses, respiratory syncytial virus and human metapneumovirus,11 which were similar in this study. From the database of Taiwan's CDC (https://nidss.cdc.gov.tw/ch/), severe influenza infection, in February 2020, had 67 cases, which was a remarkable decrease from, in February 2019, 256 cases. This could be due to the national reinforcement of infection control measures and immigration policy for coping with COVID-19 since the end of January to February 2020.
In conclusion, seasonal respiratory pathogens are more prevalent than SARS-CoV-2 in the non-endemic patients suspected with COVID-19. All the patients shared similar clinical features and laboratory data. The scope of quarantine and active surveillance of suspected patients should be expanded. The application of rapid detection tools, such as FilmArray™ Respiratory Panel, is highly recommended for the determination of causative pathogens. The long duration of SARS-CoV-2 shedding and the transmission in patients asymptomatic or with mild illness drastically increase the pandemic potential of COVID-19. Owing to the early wave of SARS-CoV-2 transmission in Taiwan, our study was limited by the small number of cases for screening and included 2 COVID-19 patients with mild illness. Further studies of SARS-CoV-2 pathogenesis are warranted to illuminate the full spectrum of COVID-19 and the treatment strategy.
Funding
This study was supported by a grant, CMUH DMR-108-189, from China Medical University Hospital, Taichung, Taiwan.
Contributor Information
Wen-Hsin Hsih, Email: kelly9502016@gmail.com.
Meng-Yu Cheng, Email: d23214@mail.cmuh.org.tw.
Yi-Chyi Lai, Email: yclai@csmu.edu.tw.
Min-Chi Lu, Email: luminchi@outlook.com.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X. Clinical characteristics of 2019 novel coronavirus disease in China. N Engl J Med. 2020 https://DOI:10.1056/NEJMoa2002032 [Google Scholar]
- 4.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. Jama. 2020 doi: 10.1001/jama.2020.1585. https://doi:10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. https://doi:10.2807/1560-7917.ES.2020.25.3.2000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang H.S., Tsai C.L., Chang J., Hsu T.C., Lin S., Lee C.C. Multiplex PCR system for the rapid diagnosis of respiratory virus infection: systematic review and meta-analysis. Clin Microbiol Infect. 2018;24(10):1055–1063. doi: 10.1016/j.cmi.2017.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poritz M.A., Blaschke A.J., Byington C.L., Meyers L., Nilsson K., Jones D.E. FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection. PloS One. 2011;6(10) doi: 10.1371/journal.pone.0026047. https://doi:10.1371/journal.pone.0026047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee S.H., Ruan S.Y., Pan S.C., Lee T.F., Chien J.Y., Hsueh P.R. Performance of a multiplex PCR pneumonia panel for the identification of respiratory pathogens and the main determinants of resistance from the lower respiratory tract specimens of adult patients in intensive care units. J Microbiol Immunol Infect. 2019;52(6):920–928. doi: 10.1016/j.jmii.2019.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hui D.S. Review of clinical symptoms and spectrum in humans with influenza A/H5N1 infection. Respirology. 2008;13(Suppl 1):S10–S13. doi: 10.1111/j.1440-1843.2008.01247.x. [DOI] [PubMed] [Google Scholar]
- 11.Litwin C.M., Bosley J.G. Seasonality and prevalence of respiratory pathogens detected by multiplex PCR at a tertiary care medical center. Arch Virol. 2014;159(1):65–72. doi: 10.1007/s00705-013-1794-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee P.I., Hsueh P.R. Emerging threats from zoonotic coronaviruses-from SARS and MERS to 2019-nCoV. J Microbiol Immunol Infect. 2020;53:365–367. doi: 10.1016/j.jmii.2020.02.001. https://doi:10.1016/j.jmii.2020.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105924. https://doi:10.1016/j.ijantimicag.2020.105924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lipsitch M., Swerdlow D.L., Finelli L. Defining the epidemiology of Covid-19—studies needed. N Engl J Med. 2020 doi: 10.1056/NEJMp2002125. https://doi:10.1056/NEJMp2002125 [DOI] [PubMed] [Google Scholar]
- 16.Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382(10):970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y., Gayle A.A., Wilder-Smith A., Rocklöv J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J Trav Med. 2020 doi: 10.1093/jtm/taaa021. https://doi:10.1093/jtm/taaa021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hon K.L. Severe respiratory syndromes: travel history matters. Trav Med Infect Dis. 2013;11(5):285–287. doi: 10.1016/j.tmaid.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lan L., Xu D., Ye G., Xia C., Wang S., Li Y. Positive RT-PCR test results in patients recovered from COVID-19. Jama. 2020 doi: 10.1001/jama.2020.2783. https://doi:10.1001/jama.2020.2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu J., Han B., Wang J. COVID-19: gastrointestinal manifestations and potential fecal-oral transmission. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.02.054. https://doi:10.1053/j.gastro.2020.02.054 [DOI] [PMC free article] [PubMed] [Google Scholar]