ABSTRACT
In essentially all eukaryotes, proteins can be modified by the attachment of small ubiquitin-related modifier (SUMO) proteins to lysine side chains to produce branched proteins. This process of ‘SUMOylation’ plays essential roles in plant and animal development by altering protein function in spatially and temporally controlled ways. In this Primer, we explain the process of SUMOylation and summarize how SUMOylation regulates a number of signal transduction pathways. Next, we discuss multiple roles of SUMOylation in the epigenetic control of transcription. In addition, we evaluate the role of SUMOylation in the etiology of neurodegenerative disorders, focusing on Parkinson's disease and cerebral ischemia. Finally, we discuss the possibility that SUMOylation may stimulate survival and neurogenesis of neuronal stem cells.
KEY WORDS: SUMO, Signal transduction, Epigenetics, Neurodegenerative disorder, Ubiquitin-like protein, Post-translational protein modification
Summary: In this Primer, we review the roles of SUMO, a ubiquitin-like protein, in developmental processes such as signaling, epigenetic regulation, neurogenesis and neurodegeneration.
Introduction
Post-translational protein modification (PTM) is a dynamic process in which chemical groups of varying complexity are covalently attached to proteins (Mann and Jensen, 2003). Such modifications often lead to changes in protein shape, stability and affinity, and thus play an essential role in regulating and diversifying protein function. Many PTMs result from the attachment of small chemical groups (such as phosphate, methyl or acetyl groups), but PTMs can also involve the conjugation of large biomolecules, such as lipids, carbohydrates, polyADP-ribose or proteins. For example, ubiquitin-like proteins (UBLs) can become attached to target proteins via an amide linkage between the UBL C-terminal carboxyl group and a target protein lysine side chain to produce branched proteins (van der Veen and Ploegh, 2012). One such protein, small ubiquitin-related modifier (SUMO), is a highly conserved ∼90 amino acid UBL (Bayer et al., 1998; Mahajan et al., 1997; Matunis et al., 1996). SUMO conjugation (also termed ‘SUMOylation’) is reversible and is used to regulate many processes with central roles in development including signal transduction, protein subcellular localization, protein aggregation and the epigenetic control of transcription.
SUMO family proteins are found in essentially all eukaryotes. Whereas Drosophila melanogaster and Saccharomyces cerevisiae have only one SUMO gene, vertebrates and flowering plants have at least four (Augustine and Vierstra, 2018; Matunis and Rodriguez, 2016). The list of developmental roles for SUMOylation in both animals and plants is growing. For example, misregulation of SUMOylation is associated with various developmental defects, such as an inability to form regular seeds in plants, severe syncytial cleavage defects, embryonic patterning defects and haltere-to-wing transformation in fruit flies, and neural tube and heart defects in frogs (Alkuraya et al., 2006; Augustine and Vierstra, 2018; Bertke et al., 2019; Nie et al., 2009; Smith et al., 2011). In humans, the activity of proteins important in craniofacial developmental is regulated by SUMOylation (reviewed by Pauws and Stanier, 2017). Although not the focus of this Primer, the role of SUMOylation has also been extensively studied in the context of cancer and the response to DNA damage (reviewed by Seeler and Dejean, 2017).
In this Primer, we (1) discuss the basic SUMO-conjugation pathway, (2) provide examples of the ways in which SUMOylation regulates protein-protein interactions in signaling pathways that are essential in development, (3) discuss roles for SUMOylation in the epigenetic regulation of gene activity and (4) discuss the involvement of SUMOylation in debilitating neurodegenerative diseases. In addition, SUMOylation has many additional roles that are beyond the scope of this Primer. These include interactions between SUMO and regulatory factors, such as Bicoid, Medea, Spalt and Notch (Cao and Courey, 2017; Epps and Tanda, 1998; Miles et al., 2008; Sánchez et al., 2010; Zhu et al., 2017), and several roles for SUMOylation in development and function of the innate immune system (Bhaskar et al., 2002; Koltun et al., 2017; Liu et al., 2013).
The SUMO-conjugation pathway
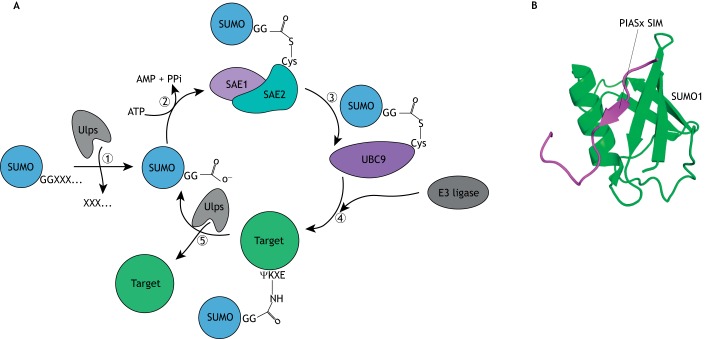
The attachment of SUMO to lysine residues in target proteins (Box 1), like conjugation of all ubiquitin family proteins, is a multi-step process (Fig. 1) (Hannoun et al., 2010; Kim et al., 2002; Matunis and Rodriguez, 2016). After translation, SUMO is initially inactive and before SUMOylation can occur it undergoes a maturation step, which involves the removal of a C-terminal sequence following a diglycine motif (Gly-Gly) by a member of the ubiquitin-like protease/sentrin-specific protease (Ulp/SENP) family (Fig. 1A, step 1) (Nayak and Müller, 2014). In the subsequent SUMO activation step, ATP is consumed to adenylate an active site cysteine residue in the SUMO-activating enzyme [a heterodimer of SAE1 and SAE2 (also known as UBA2)] (Fig. 1A, step 2). The adenylate group is then displaced by the C-terminal carboxyl group of mature SUMO, forming a thioester linkage between SUMO and the activating enzyme. Following the activation step, SUMO is transferred to a cysteine residue in UBC9 (UBE2I), the SUMO-conjugating enzyme (Fig. 1A, step 3). UBC9 then promotes the formation of an isopeptide bond between SUMO and a target protein by transferring SUMO to the ɛ-amino group of a lysine side chain forming an amide linkage (Fig. 1A, step 4).
Box 1. Diverse SUMO targets.
Eukaryotic cells contain hundreds or thousands of SUMOylated proteins. Identification of the set of SUMOylated proteins (the SUMO-ome) in a developing organism or tissue can provide information about the many roles of SUMOylation in development (Hendriks et al., 2018; Hendriks and Vertegaal, 2016; Lumpkin et al., 2017; Nie et al., 2009; Pirone et al., 2017; Tammsalu et al., 2015). This generally involves affinity purification of SUMOylated proteins followed by multi-dimensional liquid chromatography/mass spectroscopy to identify the purified proteins. Higher confidence and more useful information can be obtained by using the power of mass spectroscopy to simultaneously map SUMO acceptor lysines while identifying SUMOylated proteins (Hendriks et al., 2018; Hendriks and Vertegaal, 2016; Lumpkin et al., 2017; Tammsalu et al., 2015). Such approaches have led to evidence for roles of SUMOylation in such developmental processes as signal transduction, epigenetic transcriptional control and neurogenesis (Anderson et al., 2017; Nie et al., 2009).
Fig. 1.
The SUMOylation pathway and SUMO-interacting motifs. (A) (1) SUMO is cleaved to remove the entire C-terminal extension following the diglycine (GG) motif, by a SUMO-specific protease in the Ulp/SENP (Ulps) family, to generate mature SUMO. (2) Mature SUMO is attached to an active site cysteine residue in the SUMO-activating enzyme (a heterodimer of SAE1 and SAE2) in a process that requires ATP. (3) SUMO is transferred from the activating enzyme to a cysteine residue in UBC9, the SUMO-conjugating enzyme. (4) SUMO is transferred to a lysine side chain in a target protein. The target lysines are often embedded in a sequence resembling the four amino acid consensus sequence shown (ΨKXE, where Ψ is any hydrophobic amino acid and X is any amino acid). Target selection sometimes involves the action of an E3 SUMO ligase. (5) SUMOylation is a reversible process and its removal from target proteins is catalyzed by Ulp/SENP family SUMO proteases. (B) Many of the biological effects of SUMOylation are mediated by SUMO-interacting motifs (SIMs). These motifs generally consist of a short stretch of hydrophobic amino acids that bind to SUMO in a groove between a β-strand of the SUMO β-sheet and an α-helix. In doing so, the SIM forms a β-strand that adds to the SUMO β-sheet in either a parallel or an anti-parallel orientation. In the example shown here, the SIM from a PIAS family SUMO ligase, PIASx, interacts in parallel with the second SUMO β-strand (Song et al., 2004). The image was generated using PDB ID 2asq.
In the case of ubiquitylation, there are hundreds of E3 ubiquitin ligases that catalyze the transfer of ubiquitin from the conjugating enzyme to the final target and these ligases are essential for ubiquitylation (Hershko and Ciechanover, 1998; Zheng and Shabek, 2017). SUMOylation involves a much smaller number of E3 ligases and, in vitro, selective SUMOylation can occur without E3 ligases. Nevertheless, a number of SUMO ligases [such as the Siz/PIAS proteins, promyelocytic leukemia (PML) protein, and Ran BP2] have been discovered, and appear to be required for many instances of target protein selection in vivo (Castro et al., 2012; Constanzo et al., 2016; Gali et al., 2012; Gareau and Lima, 2010; Higuchi et al., 2019; Moreno-Ayala et al., 2015; Rabellino et al., 2017).
SUMO acceptor lysine residues are sometimes embedded in the motif: ΨKXE, where Ψ is any hydrophobic amino acid and X is any amino acid (Rodriguez et al., 2001; Sampson et al., 2001). However, recent mass spectrometry studies have indicated that only a minority of SUMOylated lysines reside in such a motif (Hendriks et al., 2018; Lumpkin et al., 2017).
SUMOylation is a reversible process owing to the presence of numerous SUMO-deconjugating enzymes, which hydrolyze the isopeptide linkage between SUMO and a target protein (Fig. 1A, step 5). Many SUMO-deconjugating enzymes are related to cysteine proteases, including the members of the Ulp family (the same proteins involved in SUMO maturation, see above) (Nayak and Müller, 2014). To account for the fact that these enzymes are involved in both deconjugation and maturation, we sometimes refer to them as SUMO proteases rather than as SUMO deconjugases. As SUMO proteases have roles that both favor and disfavor the conjugated state, it is not always clear if a phenotype associated with altered levels of one of these enzymes results from increased or decreased levels of SUMOylation.
Mechanisms of SUMO action
SUMO often modulates target protein function by directing non-covalent interactions with other proteins through SUMO-interacting motifs (SIMs) (Hecker et al., 2006; Kerscher, 2007; Song et al., 2004). SIMs typically consist of a stretch of about four hydrophobic amino acid residues sometimes preceded or followed by a stretch of negatively charged residues. SIMs often bind in a groove between the α-helix and the second β-strand of SUMO and form an extended intermolecular β-sheet (Fig. 1B). SUMO-targeted ubiquitin ligases (STUbLs) are an interesting class of SIM-containing proteins that are directed to a SUMOylated protein by a SUMO/SIM interaction where they direct ubiquitylation (Sriramachandran and Dohmen, 2014).
Often, a single SUMO moiety is attached to an acceptor lysine in the target protein. However, poly-SUMOylation (the attachment of SUMO to additional molecules of SUMO, forming a chain of SUMO polypeptides attached to a single lysine in the target) does occur, often leading to distinct functional consequences. For example, activation of the STUbL RNF4 occurs upon RNF4 dimerization. RNF4 dimerization only occurs when RNF4 binds to poly-SUMO chains in its substrates, which leads to ubiquitin/26S proteasome-dependent degradation of both the substrate and RNF4 itself (Rojas-Fernandez et al., 2014).
Although there is no single mechanistic role for SUMOylation in development – any more than there is a single mechanistic role for any small PTM (e.g. phosphorylation) – SUMOylation frequently functions by controlling the subcellular localization of target proteins. For example, studies of the STUbL Degringolade (Dgrn), a Drosophila homolog of RNF4, have shown that Dgrn is maternally contributed to the Drosophila embryo and is essential for many developmental processes, such as the early mitotic cycles in the syncytial embryo (Abed et al., 2011, 2018). Dgrn recognizes the SUMOylated form of the co-repressor Groucho (Gro) and leads to sequestration of Gro in a compartment where it is unable to repress transcription.
SUMOylation targets are rarely quantitatively SUMOylated. In fact, in the case of most targets, only a small percentage of a SUMO target protein is SUMOylated at any given time. This is paradoxical because, as will become clear, SUMOylation often modulates the activity of nearly the entire population of a protein target. This phenomenon has been termed the ‘SUMO enigma’ (Hay, 2005). One possible explanation is the idea of a ‘hit and run’ phenomenon, in which transient SUMOylation can have some kind of lasting effect on protein structure even after a protein has been de-SUMOylated (e.g. by forcing the adoption of a metastable conformation or by triggering other PTMs).
Regulation of signal transduction by SUMO
The ability of cells to send and receive signals is crucial for the development of multicellular organisms. Cross-talk between the SUMO conjugation pathway and many different signal transduction pathways modulates this signaling in a context-dependent manner. Here, we discuss the Ras/MAPK, JNK and Hedgehog pathways, which provide three of the best-studied examples of such cross-talk.
Regulation of Ras/MAPK signaling by SUMO
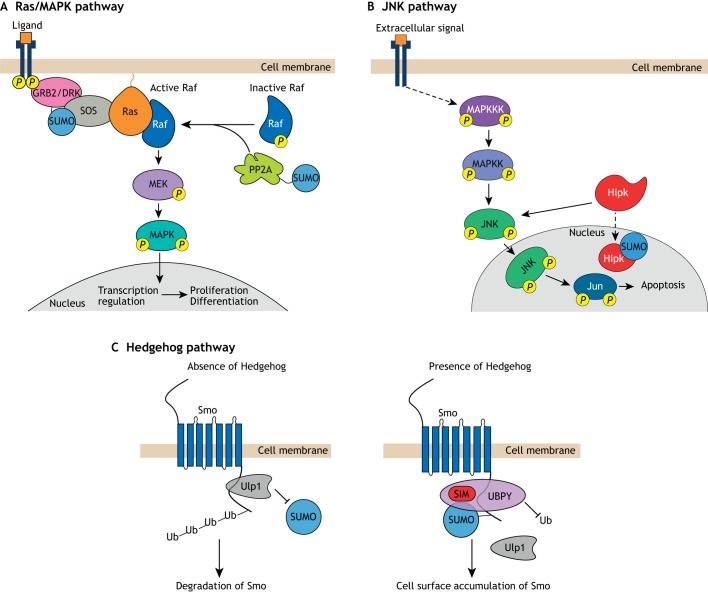
The Ras/mitogen-activated protein kinase (Ras/MAPK) pathway is highly conserved and essential for cell proliferation and differentiation in all eukaryotes (Fig. 2A) (Shilo, 2014). The Ras/MAPK pathway is activated when a ligand, such as insulin, epidermal growth factor or fibroblast growth factor, binds to a cognate receptor tyrosine kinase (RTK) (Shilo, 2014; Sopko and Perrimon, 2013). Upon ligand binding, RTKs undergo autophosphorylation, leading to the recruitment of the Son of Sevenless (SOS) GTP exchange factor through the growth factor receptor bound protein 2/Downstream of Receptor Kinase (GRB2/DRK) adaptor protein. SOS activates Ras, a small GTPase, by promoting the release of GDP and the binding of GTP to Ras. This exchange leads to the activation of a protein phosphorylation cascade involving the sequential action of three Ser/Thr kinases: Raf, MAPK/Erk kinase (MEK) and MAPK. Raf activation depends, in part, on the removal of an inhibitory phosphate group from Raf by protein phosphatases (Abraham et al., 2000; Ory et al., 2003). Activated MAPK then relocalizes to the nucleus where it regulates gene expression by phosphorylating transcription factors (Fig. 2A).
Fig. 2.
Regulation of signal transduction by SUMO. (A) Regulation of the Ras/MAPK pathway by SUMO. Upon ligand binding, a receptor tyrosine kinase dimerizes leading to auto-phosphorylation on tyrosine residues. Phosphorylation of the receptor activates a set of proteins that results in the phosphorylation of the downstream kinase MAPK. MAPK translocates into the cell nucleus to activate various transcription factors. This pathway is regulated by SUMO at multiple points. For example, in mammals SUMOylation of the adaptor protein GRB2 increases its affinity for the guanine nucleotide exchange factor SOS, and in Drosophila SUMOylation of PP2A may stimulate the removal of an inhibitory phosphate group from Raf. (B) Regulation of the Drosophila JNK pathway by Hipk and SUMOylation. When extracellular signals are sensed, a phosphorylation cascade results in phosphorylation of JNK proteins and their translocation into the nucleus to activate apoptotic genes through phosphorylation of transcription factors such as Jun. This process is enhanced by cytoplasmic Hipk; SUMOylation of Hipk results in its sequestration in the nucleus, thus reducing apoptosis. (C) Regulation of Drosophila Hedgehog signaling by SUMO. In the absence of Hedgehog signaling (left), the SUMO deconjugase Ulp1 binds Smo and removes SUMO from Smo preventing it from accumulating in a SUMOylated form. Smo protein is then subject to poly-ubiquitylation (Ub), which targets it to the proteasome for degradation. In the presence of Hedgehog signaling (right), Ulp1 dissociates from Smo, which therefore accumulates in a SUMOylated form. This leads to the recruitment of UBPY (also known as Usp8) through an interaction between its SIM and SUMO. UBPY is a deubiquitylase and therefore prevents accumulation of ubiquitylated Smo, thus preventing Smo degradation and allowing its accumulation on the cell surface.
SUMOylation regulates Ras/MAPK signaling at various points in the pathway. In vitro, mammalian GRB2 is SUMOylated and this SUMOylation enhances the oncogenic potential of a mouse colon cancer cell line (Qu et al., 2014). Knockdown of GRB2 in these cells significantly reduces Ras/MAPK signaling, cell motility and the size of tumors that form after subcutaneous injection of the cells into immunocompromised mice. All these effects are rescued by the expression of wild-type GRB2, but not by the expression of a mutant form of GRB2 in which the major SUMO acceptor lysine is changed to arginine. SUMOyation of GRB2 enhances Ras/MAPK signaling by increasing the affinity of GRB2 for SOS.
There is also evidence that the Ras protein itself is a direct target of SUMOylation in mammalian cells (Choi et al., 2018a,b). Mammalian K-Ras is SUMOylated on lysine 42 and mutation of this lysine impairs signaling, as evidenced by reduced phosphorylation of MEK and MAPK. In addition, the mutation of this lysine impairs Ras-dependent cell migration in vivo, a phenotype that is also observed in the presence of a UBC9 inhibitor.
Evidence that SUMOylation may be relevant to Ras pathway function in development comes from studies of patterning of the follicle cell epithelium during Drosophila oogenesis. Ras/MAPK signaling is required cell-autonomously for the dorsal follicle cell fate (Schweitzer and Shilo, 1997), and epistasis studies indicate that SUMO is needed for this function (Schnorr et al., 2001). Although the mechanism by which SUMO modulates Ras signaling in Drosophila is not known, a proteomic study (Nie et al., 2009) shows that protein phosphatase 2A (PP2A), which is known to remove an inhibitory phosphate group from Raf (Abraham et al., 2000; Ory et al., 2003), is a SUMOylation target.
Regulation of JNK signaling by SUMO
Jun N-terminal kinases (JNKs) belong to the MAPK superfamily and regulate many development processes including apoptosis and wound healing (Bosch et al., 2005; Dhanasekaran and Reddy, 2008; Gazel et al., 2006). A variety of findings suggest that JNK may be regulated by mechanisms involving SUMOylation.
During Drosophila development, RNAi depletion of SUMO in the wing disc increases apoptosis, and reducing JNK signaling rescues this apoptotic phenotype (Huang et al., 2011). In the wing disc, lysine 25 of homeodomain-interacting protein kinase (Hipk) is targeted for SUMOylation to promote Hipk nuclear localization. The sequestration of Hipk in the nucleus inhibits it from phosphorylating JNK in the cytoplasm, thereby preventing JNK-induced apoptosis (Fig. 2B). This mechanism may be at least partially conserved in mammals: in mice, SUMOylation of HIPK2 (on lysine 25) prevents JNK-induced apoptosis. Concordantly, SUMO deconjugation allows JNK to drive apoptosis, presumably through JNK phosphorylation by cytoplasmic HIPK2 (Hofmann et al., 2005).
JNK signaling in the human inflammatory response that occurs in the liver is also regulated by SUMOylation (Schneider Aguirre and Karpen, 2013). JNK-mediated phosphorylation of the retinoid X receptor (RXR), a member of the nuclear receptor transcription factor superfamily, is part of this response. This phosphorylation, which reduces RXR activity thereby downregulating the expression of RXR targets, is partly dependent upon SUMOylation of RXR at lysine 108. Accordingly, both the inhibition of the SUMOylation pathway and the mutagenesis of RXR lysine 108 increase expression levels of RXR targets. Indeed, stimulation with TNFα, an inflammatory cytokine that signals through the TNF receptor, increases SUMOylation of RXR and this effect is blocked by the pharmacological inhibition of JNK (Schneider Aguirre and Karpen, 2013). It is not yet clear how SUMOylation of RXR might prevent activation of RXR targets.
Regulation of Hedgehog signaling by SUMO
The Hedgehog signaling pathway is essential in the regulation of animal development, and misregulation of Hedgehog signaling is implicated in human developmental diseases (Briscoe and Thérond, 2013). Briefly, binding of extracellular Hedgehog (Hh) to the transmembrane protein Patched (Ptc) allows accumulation and activation of the transmembrane protein Smoothened (Smo). In the absence of Smo activity, Gli/Ci family transcription factors are proteolytically processed into forms that act as repressors. However, once active Smo accumulates as a result of Hedgehog signaling, it inhibits Gli/Ci processing, thereby allowing these factors to activate Hedgehog target genes.
Studies in Drosophila reveal a key role for SUMO in activation of Hedgehog signaling (Ma et al., 2016; Zhang et al., 2017). Knockdown of components of the SUMOylation pathway, including Ubc9 (Lwr in Drosophila), SUMO (Smt3) and the SUMO ligase dPIAS [Protein Inhibitor of Activated STAT; also known as Su(var)2-10], have been found to enhance wing developmental defects resulting from a dominant-negative form of Smo. Furthermore, knockdown of these gene products blocks accumulation of Smo on the cell surface and leads to reduced expression of Hedgehog target genes.
A combined in vivo/in vitro analysis in Drosophila and S2 cells suggests a novel mechanism for SUMO action in Hedgehog signaling (Fig. 2C). Induction of Hedgehog signaling results in the dissociation of the SUMO deconjugase Ulp1 from Smo, leading to the accumulation of a form of Smo SUMOylated on lysine 851 (Ma et al., 2016). It appears that SUMOylation of Smo stabilizes Smo by recruiting a SIM-containing deubiquitylating enzyme, thus preventing degradation of Smo via the ubiquitin-proteasome pathway. The role of SUMO in Hedgehog signaling appears to be conserved in mammals (Cox et al., 2010; Han et al., 2012; Ma et al., 2016).
SUMOylation in epigenetic transcriptional control
Epigenetic traits are phenotypes that can be stably inherited, but that do not result from alterations in DNA sequence (Bonasio et al., 2010; Deans and Maggert, 2015; Henikoff and Greally, 2016). Instead, they commonly result from DNA modifications (e.g. cytosine methylation) or histone modifications (e.g. methylation of lysine residues in histone N-terminal tails), which are then transmitted from the parental chromosome to the daughter chromosomes during DNA replication. Epigenetic mechanisms play crucial roles in both plant and animal development by allowing transcriptional states to be maintained during cell division and even trans-generationally through the gametes.
A number of studies suggest that the regulatory factors responsible for chromatin remodeling and modification are subject to SUMOylation, and that this modification alters their activities. Three instances of such regulation are discussed below.
Regulation of Polycomb group function by SUMO
One of the well-studied systems for epigenetic regulation involves the action of the Trithorax group (TrxG) and Polycomb group (PcG) genes (Schuettengruber et al., 2017). These two groups of genes work antagonistically: TrxG genes encode proteins involved in the maintenance of the transcriptionally active state, whereas PcG genes encode proteins required for the maintenance of the transcriptionally silent state.
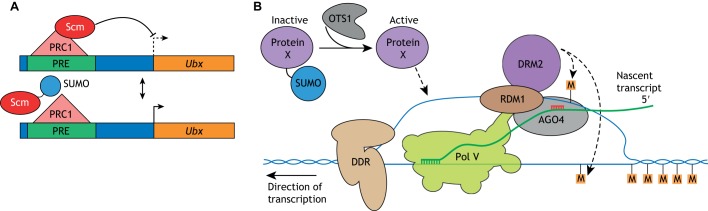
One important PcG gene encodes the SUMOylation target Sex-combs on midleg (Scm) (Schwartz and Pirrotta, 2013). PcG-mediated silencing in Drosophila requires the recruitment of Scm to Polycomb response elements (PREs) in target genes (Smith et al., 2011; Wang et al., 2010). Knockdown of SUMO or mutagenesis of the SUMO acceptor lysines in Scm in vitro increases recruitment of Scm to a PRE in the Ultrabithorax (Ubx) locus and reduces Ubx expression. Conversely, knockdown of the SUMO deconjugase Ulp1 leads to decreased Scm recruitment and increased Ubx expression (Smith et al., 2011). Thus, SUMO appears to negatively regulate Scm function (Fig. 3A).
Fig. 3.
Transcriptional gene silencing in Drosophila and Arabidopsis. (A) Polycomb group silencing and SUMO. Polycomb repressive complex 1 (PRC1) is recruited to Polycomb response elements (PREs) where it works with Scm to repress the transcription of Ubx. The SUMOylation of Scm impairs its recruitment to the PRE, and thus SUMOylation of Scm is required to allow Ubx expression under some conditions. (B) Transcriptional gene silencing and SUMOylation. In plants, AGO4 (in a complex with a siRNA shown in red), together with the aid of RDM1, recruits a DNA methylase (DRM2) to transcriptionally engaged RNA polymerase V (Pol V), which leads to RNA-dependent DNA methylation (RdRM) and transcriptional gene silencing (TGS). This silencing depends on the DNA damage response (DDR) chromatin-remodeling complex. SUMO appears to regulate this process, because mutations in the SUMO deconjugase OTS1 and the SUMO ligases SIZ1 and MMS21 compromise silencing. This suggests the existence of a SUMO-conjugated protein (Protein X) that promotes TGS when it is in its deconjugated form. M, cytosine methylation.
Evidence that regulation of PcG function by SUMO occurs in vivo is provided by experiments in which SUMO was knocked down by RNAi in the haltere disc of Drosophila (Smith et al., 2011). As the cell culture experiments discussed above indicate that SUMO antagonizes Scm function, SUMO knockdown should lead to increased Scm function and therefore to Ubx repression. In accordance with this expectation, SUMO knockdown in the developing haltere results in a partial haltere-to-wing transformation, analogous to the Ubx loss-of-function phenotype (Lewis, 1963).
Although these results above indicate that SUMO can antagonize PcG-mediated silencing, in other contexts SUMO may enhance such silencing (Gill, 2010; Kang et al., 2010; Zhang et al., 2004). In mouse embryos, loss of function of the SUMO protease SENP2 leads to myocardial defects (Kang et al., 2010), which may result from de-SUMOylation of PC2 [a component of Polycomb repressive complex 1 (PRC1) in humans; also known as CBX4]. PRC1 is normally recruited to PcG target genes, where it may lead to their compaction and silencing. PC2 SUMOylation appears to be required for this recruitment and, therefore, reducing the activity of the SUMO pathway leads to derepression of PC2 targets in the myocardium.
An indication for the involvement of SUMOylation in PcG function may come from studies investigating the effect of SUMOylation on nuclear architecture. Like human PC2, the PC2 Drosophila homolog Polycomb (Pc) is also SUMOylated (Gonzalez et al., 2014) and is normally localized in nuclear foci called Pc bodies. Reduced SUMO pathway activity results in fewer, but more concentrated, foci, whereas reduced levels of a SUMO-deconjugating enzyme results in more diffuse Pc localization. The role of these Pc bodies in PcG function is not clear, but it is possible that they serve to mediate long-range chromatin interactions required for silencing.
SUMOylation may also positively regulate homeotic gene transcription during Drosophila development through effects on TrxG function. This is illustrated by the regulation of Ubx by the product of the tonalli (tna) TrxG gene, a possible E3 SUMO ligase. TnaA interacts with Osa, a subunit of the Drosophila SWI/SNF chromatin remodeling complexes and a possible SUMOylation target (Monribot-Villanueva et al., 2013; Nie et al., 2009). Although TnaA is not required for normal Ubx expression, it may nonetheless play a role in fine-tuning this expression as ectopic Ubx expression resulting from Pc mutations is significantly decreased in tna mutants (Rosales-Vega et al., 2018).
Regulation of transcriptional gene silencing in plants by SUMO
A second link between SUMOylation and epigenetic regulation is provided by studies of RNA-dependent DNA methylation, which directs transcriptional gene silencing (TGS) in flowering plants (Gallego-Bartolome et al., 2019; Matzke and Mosher, 2014). In Arabidopsis, small interfering RNAs (siRNAs) from heterochromatic regions of the genome are loaded onto ARGONAUTE 4 (AGO4). The siRNA then guides AGO4 to nascent transcripts associated with transcriptionally engaged RNA polymerase V (a Pol II-like RNA polymerase unique to flowering plants). This results in the recruitment of the DNA methyltransferase DRM2 to produce 5-methylcytosine residues in the DNA leading to heterochromatic silencing. The action of Pol V in TGS requires assistance from the DRD1/DMS3/RDM1 (DDR) chromatin remodeling complex (Fig. 3B).
A screen for Arabidopsis genes required for silencing of a TGS-sensitive reporter gene has uncovered a role for the SUMO protease OVERLY TOLERANT TO SALT 1 (OTS1) (Liu et al., 2017). In this study, mutation of the catalytic cysteine residue in OTS1 leads to reduced TGS, supporting the idea that the SUMO-deconjugation function of OTS1 plays a direct role in silencing.
Additional SUMO pathway components (including four other SUMO proteases and two SUMO ligases) also play roles in TGS. Mutations in the SUMO ligases have similar effects on TGS as SUMO protease mutations: both lead to a partial loss of silencing (Liu et al., 2017). As SUMO proteases are required for SUMO maturation in addition to SUMO deconjugation, the effects of mutating the SUMO proteases and the ligases may both stem from a positive role for SUMO in TGS. Alternatively, it is possible that the dynamic cycling of a SUMO-conjugation target between conjugated and unconjugated states is required for TGS. Further illumination of the role of SUMO conjugation in TGS will require identification of the relevant SUMO-conjugation targets.
SUMO as an enforcer of distinct chromatin states in mammalian cells
SUMOylation may play a role in maintaining cellular identity during mammalian development (Cossec et al., 2018). Analysis of the genome-wide distribution of SUMO by chromatin immunoprecipitation using an anti-SUMO antibody, reveals distinct ‘SUMO chromatin landscapes’ when comparing embryonic stem cells (ESCs) with mouse embryonic fibroblasts (MEFs). In MEFs, SUMO primarily associates with active enhancers, whereas in ESCs it primarily associates with heterochromatin enriched for trimethylated histone H3 lysine 9, which is associated with transcriptionally silent heterochromatin and loss of pluripotency.
Evidence that these different SUMO landscapes are of functional significance comes from studies looking at the role of the SUMOylation pathway in reprogramming. Reduced SUMO pathway activity in MEFs enhances the formation of induced pluripotent stem cells following co-expression of the canonical reprogramming transcription factors [Oct4 (Pou5f1), Klf4, Sox2 and Myc]. This reprogramming is apparently the result of chromatin closing at MEF enhancers, accompanied by the loss of the reprogramming factors from these sites and their relocalization to pluripotency enhancers (Cossec et al., 2018).
Whereas SUMO appears to be required for the maintenance of the differentiated state in MEFs, it seems to be required to stabilize the pluripotent state in ESCs. Specifically, reduced SUMO pathway activity in ESCs leads to loss of H3 lysine 9 trimethylation and may reflect a general ability of SUMO to prevent the differentiation of undifferentiated cells. Indeed, SUMO pathway impairment also enhances the retinoic acid induced differentiation of cancer cells (Cossec et al., 2018).
Although the chromosomal SUMOylation targets that are required for the enforcement of chromosomal states have not been identified, the histones themselves are a possible target. A number of studies have demonstrated that histone SUMOylation (Galisson et al., 2011; Hendriks et al., 2018; Lamoliatte et al., 2017; Nie et al., 2009; Shiio and Eisenman, 2003), and histone H4 SUMOylation has links to gene silencing via the recruitment of co-repressors, such as histone deacetylase and heterochromatin protein 1 (Shiio and Eisenman, 2003). As discussed above, PcG and TrxG proteins are also regulated by SUMOylation and could play a role in the enforcement of chromosomal states (Monribot-Villanueva et al., 2017).
SUMOylation in neurodegenerative disease
Numerous studies have demonstrated roles for SUMOylation in neurodegenerative diseases, such as Parkinson's disease (Eckermann, 2013), Alzheimer's disease (Knock et al., 2018), and Huntington's disease (Anderson et al., 2017). A common feature of many of these pathologies is the aggregation of neuronal proteins and, in many cases, SUMO has been found to either enhance or protect against this aggregation. The following section focuses on the role of SUMOylation in Parkinson's disease, because studies of this neurodegenerative disorder are the most revealing regarding the mechanism of SUMO action. We then conclude by briefly reviewing evidence that SUMOylation may have a role in resistance to cerebral ischemia, as well as the survival and differentiation of neuronal stem cells.
SUMOylation of α-synuclein in Parkinson's disease
One of the best-studied SUMO targets in neurodegeneration is α-synuclein, which may have a causative role in Parkinson's disease, a widespread neurodegenerative disorder characterized by tremors and slowed movement (Baba et al., 1998; Spillantini et al., 1997; Wakabayashi et al., 2007). Parkinson's disease results from the degeneration of dopaminergic neurons in the midbrain and is characterized by the appearance of protein inclusions called Lewy bodies in the cytosol of neurons, of which α-synuclein aggregates are a major component.
α-Synuclein is thought to be involved in membrane trafficking and could have a role in normal neuronal development (George et al., 1995; Sulzer and Edwards, 2019). It is extensively SUMOylated (Anderson et al., 2017), although the exact function of α-synuclein SUMOylation is not clear. One study suggests that SUMOylation may enhance the formation of α-synuclein aggregates and thereby contribute to Parkinson's disease onset (Rott et al., 2017). This study showed that SUMOylation of α-synuclein is stimulated by the SUMO ligase PIAS2 (protein inhibitor of activated STAT 2) and overexpression of PIAS2 results in increased intracellular concentrations of α-synuclein. Conversely, α-synuclein levels can be reduced using ginkgolic acid, which blocks SUMOylation by inhibiting the SUMO-activating enzyme. Inhibiting the 26S proteasome prevents the ginkgolic acid-induced loss of α-synuclein, suggesting that SUMOylation stabilizes α-synuclein by preventing its ubiquitin-dependent proteasomal degradation.
In addition, the study showed that α-synuclein mutants that are associated with disease exhibit increased aggregation and proteinase K resistance, and that mutant α-synuclein is more extensively SUMOylated than wild-type α-synuclein. Together, these results indicate that such mutations may lead to disease by promoting SUMOylation (Rott et al., 2017). Accordingly, Parkinson's disease brains contain elevated levels of SUMOylated α-synuclein as well as elevated overall levels of SUMOylated proteins and PIAS2. These findings support the hypothesis that SUMOylation contributes to the onset of Parkinson's disease by preventing proteasomal degradation of α-synuclein, which in turn leads to over-accumulation and aggregation of α-synuclein. They also indicate that SUMOylation might actively encourage α-synuclein aggregation.
Although it is generally agreed that SUMOylation has a role to play in Parkinson's disease through conjugation to α-synuclein, the mechanism remains controversial (Anderson et al., 2017). For example, an early study failed to show an effect of SUMOylation on α-synuclein ubiquitylation (Kim et al., 2011). Furthermore, another finding suggests that SUMOylation of α-synuclein prevents – rather than stimulates – aggregation, thereby lessening its toxicity (Krumova et al., 2011), consistent with a mechanism by which SUMOylation could play a general role in protecting cells against misfolded proteins (Guo et al., 2014). In this pathway, the SUMO ligase PML binds misfolded proteins and directs their SUMOylation, leading to their ubiquitylation and proteasomal degradation by the SUMO-targeted ubiquitin ligase RNF4. In conclusion, the role of SUMOylation in neurodegenerative disease is complex. Even in the case of a single disease such as Parkinson's, it appears that – depending on context – SUMOylation may have either neuroprotective or neurodegenerative effects.
SUMOylation in cerebral ischemia
The neurodegenerative disorder cerebral ischemia results from neuronal death caused by hypoxic stress. Studies of cerebral ischemia provide evidence that SUMOylation directly regulates neurogenesis and has a role in the recovery from neuronal damage. In mice and humans, cerebral ischemia leads to significant increases in SUMOylated proteins in the area surrounding the cerebral lesion (Bernstock et al., 2018; Yang et al., 2008).
Mouse neural stem cells (NSCs) have been used to explore the possibility of a causal link between SUMOylation and resistance to ischemia (Bernstock et al., 2019). UBC9 overexpression in NSCs leads to a global increase in protein SUMOylation and to changes in the gene expression profile in vitro, such as downregulation of genes related to apoptosis, and upregulation of neural differentiation genes and genes required for neuronal survival after oxygen deprivation. Perhaps most excitingly, UBC9 overexpression increases survival and wound integration of NSCs transplanted into sites of ischemic damage in mouse brains. These findings suggest that manipulation of SUMO pathway activity may lead to improved outcomes of stem-cell therapies.
Conclusions
This Primer has touched on some of the essential roles of SUMOylation in plant and animal development. These include roles in multiple signaling pathways, epigenetic regulation, and neuronal development and degeneration. There is no simple relationship between developmental complexity and the number of proteins encoded in an organism's genome (Szathmáry et al., 2001). By turning linear proteins into branched proteins, SUMOylation can increase protein functional diversity without a commensurate increase in the coding capacity of the genome. This could provide part of the explanation for increased developmental complexity in the absence of increased genomic complexity.
Acknowledgements
We would like to thank anonymous reviewers and Reviews Editor Alex Eve for many excellent comments and suggestions that significantly improved this Primer.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
O.M. received support from a National Institute of General Medical Sciences MARC fellowship (T34GM008563). A.J.C. received support from the National Institutes of Health (GM63596). Deposited in PMC for release after 12 months.
References
- Abed M., Barry K. C., Kenyagin D., Koltun B., Phippen T. M., Delrow J. J., Parkhurst S. M. and Orian A. (2011). Degringolade, a SUMO-targeted ubiquitin ligase, inhibits hairy/groucho-mediated repression. EMBO J. 30, 1289-1301. 10.1038/emboj.2011.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abed M., Bitman-Lotan E. and Orian A. (2018). The biology of SUMO-targeted ubiquitin ligases in Drosophila development, immunity, and cancer. J. Dev. Biol. 6, E2 10.3390/jdb6010002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham D., Podar K., Pacher M., Kubicek M., Welzel N., Hemmings B. A., Dilworth S. M., Mischak H., Kolch W. and Baccarini M. (2000). Raf-1-associated protein phosphatase 2A as a positive regulator of kinase activation. J. Biol. Chem. 275, 22300-22304. 10.1074/jbc.M003259200 [DOI] [PubMed] [Google Scholar]
- Alkuraya F. S., Saadi I., Lund J. J., Turbe-Doan A., Morton C. C. and Maas R. L. (2006). SUMO1 haploinsufficiency leads to cleft lip and palate. Science 313, 1751 10.1126/science.1128406 [DOI] [PubMed] [Google Scholar]
- Anderson D. B., Zanella C. A., Henley J. M. and Cimarosti H. (2017). Sumoylation: implications for neurodegenerative diseases. Adv. Exp. Med. Biol. 963, 261-281. 10.1007/978-3-319-50044-7_16 [DOI] [PubMed] [Google Scholar]
- Augustine R. C. and Vierstra R. D. (2018). SUMOylation: re-wiring the plant nucleus during stress and development. Curr. Opin. Plant Biol. 45, 143-154. 10.1016/j.pbi.2018.06.006 [DOI] [PubMed] [Google Scholar]
- Baba M., Nakajo S., Tu P. H., Tomita T., Nakaya K., Lee V. M., Trojanowski J. Q. and Iwatsubo T. (1998). Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson's disease and dementia with Lewy bodies. Am. J. Pathol. 152, 879-884. [PMC free article] [PubMed] [Google Scholar]
- Bayer P., Arndt A., Metzger S., Mahajan R., Melchior F., Jaenicke R. and Becker J. (1998). Structure determination of the small ubiquitin-related modifier SUMO-1. J. Mol. Biol. 280, 275-286. 10.1006/jmbi.1998.1839 [DOI] [PubMed] [Google Scholar]
- Bernstock J. D., Ye D. G., Griffin A., Lee Y.-J., Lynch J., Latour L. L., Friedman G. K., Maric D. and Hallenbeck J. M. (2018). Cerebral ischemia increases small ubiquitin-like modifier conjugation within human penumbral tissue: radiological-pathological correlation. Front. Neurol. 8, 738 10.3389/fneur.2017.00738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstock J. D., Peruzzotti-Jametti L., Leonardi T., Vicario N., Ye D., Lee Y. J., Maric D., Johnson K. R., Mou Y., Van Den Bosch A. et al. (2019). SUMOylation promotes survival and integration of neural stem cell grafts in ischemic stroke. EBioMedicine 42, 214-224. 10.1016/j.ebiom.2019.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertke M. M., Dubiak K. M., Cronin L., Zeng E. and Huber P. W. (2019). A deficiency in SUMOylation activity disrupts multiple pathways leading to neural tube and heart defects in Xenopus embryos. BMC Genomics 20, 386 10.1186/s12864-019-5773-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar V., Smith M. and Courey A. J. (2002). Conjugation of Smt3 to dorsal may potentiate the Drosophila immune response. Mol. Cell. Biol. 22, 492-504. 10.1128/MCB.22.2.492-504.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R., Tu S. and Reinberg D. (2010). Molecular signals of epigenetic states. Science 330, 612-616. 10.1126/science.1191078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M., Serras F., Martín-Blanco E. and Baguñà J. (2005). JNK signaling pathway required for wound healing in regenerating Drosophila wing imaginal discs. Dev. Biol. 280, 73-86. 10.1016/j.ydbio.2005.01.002 [DOI] [PubMed] [Google Scholar]
- Briscoe J. and Thérond P. P. (2013). The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 14, 416-429. 10.1038/nrm3598 [DOI] [PubMed] [Google Scholar]
- Cao J. and Courey A. J. (2017). SUMO in Drosophila development. Adv. Exp. Med. Biol. 963, 249-257. 10.1007/978-3-319-50044-7_15 [DOI] [PubMed] [Google Scholar]
- Castro P. H., Tavares R. M., Bejarano E. R. and Azevedo H. (2012). SUMO, a heavyweight player in plant abiotic stress responses. Cell. Mol. Life Sci. 69, 3269-3283. 10.1007/s00018-012-1094-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B. H., Chen C., Philips M. and Dai W. (2018a). RAS GTPases are modified by SUMOylation. Oncotarget 9, 4440-4450. 10.18632/oncotarget.23269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B. H., Philips M. R., Chen Y., Lu L. and Dai W. (2018b). K-Ras Lys-42 is crucial for its signaling, cell migration, and invasion. J. Biol. Chem. 293, 17574-17581. 10.1074/jbc.RA118.003723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constanzo J. D., Deng M., Rindhe S., Tang K.-J., Zhang C.-C. and Scaglioni P. P. (2016). Pias1 is essential for erythroid and vascular development in the mouse embryo. Dev. Biol. 415, 98-110. 10.1016/j.ydbio.2016.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossec J. C., Theurillat I., Chica C., Bua Aguin S., Gaume X., Andrieux A., Iturbide A., Jouvion G., Li H., Bossis G. et al. (2018). SUMO safeguards somatic and pluripotent cell identities by enforcing distinct chromatin states. Cell Stem Cell 23, 742-757.e8. 10.1016/j.stem.2018.10.001 [DOI] [PubMed] [Google Scholar]
- Cox B., Briscoe J. and Ulloa F. (2010). SUMOylation by Pias1 regulates the activity of the Hedgehog dependent Gli transcription factors. PLoS ONE 5, e11996 10.1371/journal.pone.0011996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans C. and Maggert K. A. (2015). What do you mean, ‘epigenetic’? Genetics 199, 887-896. 10.1534/genetics.114.173492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanasekaran D. N. and Reddy E. P. (2008). JNK signaling in apoptosis. Oncogene 27, 6245-6251. 10.1038/onc.2008.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckermann K. (2013). SUMO and Parkinson's disease. Neuromolecular Med. 15, 737-759. 10.1007/s12017-013-8259-5 [DOI] [PubMed] [Google Scholar]
- Epps J. L. and Tanda S. (1998). The Drosophila semushi mutation blocks nuclear import of bicoid during embryogenesis. Curr. Biol. 8, 1277-1280. 10.1016/S0960-9822(07)00538-6 [DOI] [PubMed] [Google Scholar]
- Gali H., Juhasz S., Morocz M., Hajdu I., Fatyol K., Szukacsov V., Burkovics P. and Haracska L. (2012). Role of SUMO modification of human PCNA at stalled replication fork. Nucleic Acids Res. 40, 6049-6059. 10.1093/nar/gks256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galisson F., Mahrouche L., Courcelles M., Bonneil E., Meloche S., Chelbi-Alix M. K. and Thibault P. (2011). A novel proteomics approach to identify SUMOylated proteins and their modification sites in human cells. Mol. Cell. Proteomics 10, M110.004796 10.1074/mcp.M110.004796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolome J., Liu W., Kuo P. H., Feng S., Ghoshal B., Gardiner J., Zhao J. M., Park S. Y., Chory J. and Jacobsen S. E. (2019). Co-targeting RNA polymerases IV and V promotes efficient de novo DNA methylation in Arabidopsis. Cell 176, 1068-1082.e19. 10.1016/j.cell.2019.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareau J. R. and Lima C. D. (2010). The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11, 861-871. 10.1038/nrm3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazel A., Banno T., Walsh R. and Blumenberg M. (2006). Inhibition of JNK promotes differentiation of epidermal keratinocytes. J. Biol. Chem. 281, 20530-20541. 10.1074/jbc.M602712200 [DOI] [PubMed] [Google Scholar]
- George J. M., Jin H., Woods W. S. and Clayton D. F. (1995). Characterization of a novel protein regulated during the critical period for song learning in the zebra finch. Neuron 15, 361-372. 10.1016/0896-6273(95)90040-3 [DOI] [PubMed] [Google Scholar]
- Gill G. (2010). SUMO weighs in on polycomb-dependent gene repression. Mol. Cell 38, 157-159. 10.1016/j.molcel.2010.04.006 [DOI] [PubMed] [Google Scholar]
- Gonzalez I., Mateos-Langerak J., Thomas A., Cheutin T. and Cavalli G. (2014). Identification of regulators of the three-dimensional polycomb organization by a microscopy-based genome-wide RNAi screen. Mol. Cell 54, 485-499. 10.1016/j.molcel.2014.03.004 [DOI] [PubMed] [Google Scholar]
- Guo L., Giasson B. I., Glavis-Bloom A., Brewer M. D., Shorter J., Gitler A. D. and Yang X. (2014). A cellular system that degrades misfolded proteins and protects against neurodegeneration. Mol. Cell 55, 15-30. 10.1016/j.molcel.2014.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L., Pan Y. and Wang B. (2012). Small ubiquitin-like Modifier (SUMO) modification inhibits GLI2 protein transcriptional activity in vitro and in vivo. J. Biol. Chem. 287, 20483-20489. 10.1074/jbc.M112.359299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannoun Z., Greenhough S., Jaffray E., Hay R. T. and Hay D. C. (2010). Post-translational modification by SUMO. Toxicology 278, 288-293. 10.1016/j.tox.2010.07.013 [DOI] [PubMed] [Google Scholar]
- Hay R. T. (2005). SUMO: a history of modification. Mol. Cell 18, 1-12. 10.1016/j.molcel.2005.03.012 [DOI] [PubMed] [Google Scholar]
- Hecker C.-M., Rabiller M., Haglund K., Bayer P. and Dikic I. (2006). Specification of SUMO1- and SUMO2-interacting motifs. J. Biol. Chem. 281, 16117-16127. 10.1074/jbc.M512757200 [DOI] [PubMed] [Google Scholar]
- Hendriks I. A. and Vertegaal A. C. O. (2016). A comprehensive compilation of SUMO proteomics. Nat. Rev. Mol. Cell Biol. 17, 581-595. 10.1038/nrm.2016.81 [DOI] [PubMed] [Google Scholar]
- Hendriks I. A., Lyon D., Su D., Skotte N. H., Daniel J. A., Jensen L. J. and Nielsen M. L. (2018). Site-specific characterization of endogenous SUMOylation across species and organs. Nat. Commun. 9, 2456 10.1038/s41467-018-04957-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. and Greally J. M. (2016). Epigenetics, cellular memory and gene regulation. Curr. Biol. 26, R644-R648. 10.1016/j.cub.2016.06.011 [DOI] [PubMed] [Google Scholar]
- Hershko A. and Ciechanover A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425-479. 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- Higuchi C., Yamamoto M., Shin S. W., Miyamoto K. and Matsumoto K. (2019). Perturbation of maternal PIASy abundance disrupts zygotic genome activation and embryonic development via SUMOylation pathway. Biol. Open 8, bio048652 10.1242/bio.048652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann T. G., Jaffray E., Stollberg N., Hay R. T. and Will H. (2005). Regulation of homeodomain-interacting protein kinase 2 (HIPK2) effector function through dynamic small ubiquitin-related modifier-1 (SUMO-1) modification. J. Biol. Chem. 280, 29224-29232. 10.1074/jbc.M503921200 [DOI] [PubMed] [Google Scholar]
- Huang H., Du G., Chen H., Liang X., Li C., Zhu N., Xue L., Ma J. and Jiao R. (2011). Drosophila Smt3 negatively regulates JNK signaling through sequestering Hipk in the nucleus. Development 138, 2477-2485. 10.1242/dev.061770 [DOI] [PubMed] [Google Scholar]
- Kang X., Qi Y., Zuo Y., Wang Q., Zou Y., Schwartz R. J., Cheng J. and Yeh E. T. H. (2010). SUMO-specific protease 2 is essential for suppression of polycomb group protein-mediated gene silencing during embryonic development. Mol. Cell 38, 191-201. 10.1016/j.molcel.2010.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O. (2007). SUMO junction—what's your function? New insights through SUMO-interacting motifs. EMBO Rep. 8, 550-555. 10.1038/sj.embor.7400980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. I. L., Baek S. H. and Chung C. H. (2002). Versatile protein tag, SUMO: its enzymology and biological function. J. Cell. Physiol. 191, 257-268. 10.1002/jcp.10100 [DOI] [PubMed] [Google Scholar]
- Kim Y. M., Jang W. H., Quezado M. M., Oh Y., Chung K. C., Junn E. and Mouradian M. M. (2011). Proteasome inhibition induces alpha-synuclein SUMOylation and aggregate formation. J. Neurol. Sci. 307, 157-161. 10.1016/j.jns.2011.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knock E., Matsuzaki S., Takamura H., Satoh K., Rooke G., Han K., Zhang H., Staniszewski A., Katayama T., Arancio O. et al. (2018). SUMO1 impact on Alzheimer disease pathology in an amyloid-depositing mouse model. Neurobiol. Dis. 110, 154-165. 10.1016/j.nbd.2017.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltun B., Shackelford E., Bonnay F., Matt N., Reichhart J. M. and Orian A. (2017). The SUMO-targeted ubiquitin ligase, Dgrn, is essential for Drosophila innate immunity. Int. J. Dev. Biol. 61, 319-327. 10.1387/ijdb.160250ao [DOI] [PubMed] [Google Scholar]
- Krumova P., Meulmeester E., Garrido M., Tirard M., Hsiao H.-H., Bossis G., Urlaub H., Zweckstetter M., Kügler S., Melchior F. et al. (2011). Sumoylation inhibits alpha-synuclein aggregation and toxicity. J. Cell Biol. 194, 49-60. 10.1083/jcb.201010117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamoliatte F., McManus F. P., Maarifi G., Chelbi-Alix M. K. and Thibault P. (2017). Uncovering the SUMOylation and ubiquitylation crosstalk in human cells using sequential peptide immunopurification. Nat. Commun. 8, 14109 10.1038/ncomms14109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E. B. (1963). Genes and developmental pathways. Am. Zool. 3, 33-56. 10.1093/icb/3.1.33 [DOI] [Google Scholar]
- Liu X., Chen W., Wang Q., Li L. and Wang C. (2013). Negative regulation of TLR inflammatory signaling by the SUMO-deconjugating enzyme SENP6. PLoS Pathog. 9, e1003480 10.1371/journal.ppat.1003480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Yan X., Kong X., Zhao Y., Gong Z., Jin J. B. and Guo Y. (2017). Transcriptional gene silencing maintained by OTS1 SUMO protease requires a DNA-dependent polymerase V-dependent pathway. Plant Physiol. 173, 655-667. 10.1104/pp.16.01365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumpkin R. J., Gu H., Zhu Y., Leonard M., Ahmad A. S., Clauser K. R., Meyer J. G., Bennett E. J. and Komives E. A. (2017). Site-specific identification and quantitation of endogenous SUMO modifications under native conditions. Nat. Commun. 8, 1171 10.1038/s41467-017-01271-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma G., Li S., Han Y., Li S., Yue T., Wang B. and Jiang J. (2016). Regulation of smoothened trafficking and Hedgehog signaling by the SUMO pathway. Dev. Cell 39, 438-451. 10.1016/j.devcel.2016.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan R., Delphin C., Guan T., Gerace L. and Melchior F. (1997). A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88, 97-107. 10.1016/S0092-8674(00)81862-0 [DOI] [PubMed] [Google Scholar]
- Mann M. and Jensen O. N. (2003). Proteomic analysis of post-translational modifications. Nat. Biotechnol. 21, 255-261. 10.1038/nbt0303-255 [DOI] [PubMed] [Google Scholar]
- Matunis M. J. and Rodriguez M. S. (2016). Concepts and methodologies to study protein SUMOylation: an overview. Methods Mol. Biol. 1475, 3-22. 10.1007/978-1-4939-6358-4_1 [DOI] [PubMed] [Google Scholar]
- Matunis M. J., Coutavas E. and Blobel G. (1996). A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135, 1457-1470. 10.1083/jcb.135.6.1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M. A. and Mosher R. A. (2014). RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15, 394-408. 10.1038/nrg3683 [DOI] [PubMed] [Google Scholar]
- Miles W. O., Jaffray E., Campbell S. G., Takeda S., Bayston L. J., Basu S. P., Li M., Raftery L. A., Ashe M. P., Hay R. T. et al. (2008). Medea SUMOylation restricts the signaling range of the Dpp morphogen in the Drosophila embryo. Genes Dev. 22, 2578-2590. 10.1101/gad.494808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monribot-Villanueva J., Juárez-Uribe R. A., Palomera-Sánchez Z., Gutiérrez-Aguiar L., Zurita M., Kennison J. A. and Vázquez M. (2013). TnaA, an SP-RING protein, interacts with Osa, a subunit of the chromatin remodeling complex BRAHMA and with the SUMOylation pathway in Drosophila melanogaster. PLoS ONE 8, e62251 10.1371/journal.pone.0062251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monribot-Villanueva J., Zurita M. and Vázquez M. (2017). Developmental transcriptional regulation by SUMOylation, an evolving field. Genesis 55, e23009 10.1002/dvg.23009 [DOI] [PubMed] [Google Scholar]
- Moreno-Ayala R., Schnabel D., Salas-Vidal E. and Lomelí H. (2015). PIAS-like protein Zimp7 is required for the restriction of the zebrafish organizer and mesoderm development. Dev. Biol. 403, 89-100. 10.1016/j.ydbio.2015.04.013 [DOI] [PubMed] [Google Scholar]
- Nayak A. and Müller S. (2014). SUMO-specific proteases/isopeptidases: SENPs and beyond. Genome Biol. 15, 422 10.1186/s13059-014-0422-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie M., Xie Y., Loo J. A. and Courey A. J. (2009). Genetic and proteomic evidence for roles of Drosophila SUMO in cell cycle control, Ras signaling, and early pattern formation. PLoS ONE 4, e5905 10.1371/journal.pone.0005905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ory S., Zhou M., Conrads T. P., Veenstra T. D. and Morrison D. K. (2003). Protein phosphatase 2A positively regulates Ras signaling by dephosphorylating KSR1 and Raf-1 on critical 14-3-3 binding sites. Curr. Biol. 13, 1356-1364. 10.1016/S0960-9822(03)00535-9 [DOI] [PubMed] [Google Scholar]
- Pauws E. and Stanier P. (2017). Sumoylation in craniofacial disorders. Adv. Exp. Med. Biol. 963, 323-335. 10.1007/978-3-319-50044-7_19 [DOI] [PubMed] [Google Scholar]
- Pirone L., Xolalpa W., Sigurðsson J. O., Ramirez J., Pérez C., González M., de Sabando A. R., Elortza F., Rodriguez M. S., Mayor U. et al. (2017). A comprehensive platform for the analysis of ubiquitin-like protein modifications using in vivo biotinylation. Sci. Rep. 7, 40756 10.1038/srep40756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y., Chen Q., Lai X., Zhu C., Chen C., Zhao X., Deng R., Xu M., Yuan H., Wang Y. et al. (2014). SUMOylation of Grb2 enhances the ERK activity by increasing its binding with Sos1. Mol. Cancer 13, 95 10.1186/1476-4598-13-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabellino A., Andreani C. and Scaglioni P. P. (2017). The role of PIAS SUMO E3-ligases in cancer. Cancer Res. 77, 1542-1547. 10.1158/0008-5472.CAN-16-2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M. S., Dargemont C. and Hay R. T. (2001). SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276, 12654-12659. 10.1074/jbc.M009476200 [DOI] [PubMed] [Google Scholar]
- Rojas-Fernandez A., Plechanovová A., Hattersley N., Jaffray E., Tatham M. H. and Hay R. T. (2014). SUMO chain-induced dimerization activates RNF4. Mol. Cell 53, 880-892. 10.1016/j.molcel.2014.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales-Vega M., Hernández-Becerril A., Murillo-Maldonado J. M., Zurita M. and Vázquez M. (2018). The role of the trithorax group TnaA isoforms in Hox gene expression, and in Drosophila late development. PLoS ONE 13, e0206587 10.1371/journal.pone.0206587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rott R., Szargel R., Shani V., Hamza H., Savyon M., Abd Elghani F., Bandopadhyay R. and Engelender S. (2017). SUMOylation and ubiquitination reciprocally regulate alpha-synuclein degradation and pathological aggregation. Proc. Natl. Acad. Sci. USA 114, 13176-13181. 10.1073/pnas.1704351114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson D. A., Wang M. and Matunis M. J. (2001). The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 276, 21664-21669. 10.1074/jbc.M100006200 [DOI] [PubMed] [Google Scholar]
- Sánchez J., Talamillo A., Lopitz-Otsoa F., Pérez C., Hjerpe R., Sutherland J. D., Herboso L., Rodríguez M. S. and Barrio R. (2010). Sumoylation modulates the activity of Spalt-like proteins during wing development in Drosophila. J. Biol. Chem. 285, 25841-25849. 10.1074/jbc.M110.124024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider Aguirre R. and Karpen S. J. (2013). Inflammatory mediators increase SUMOylation of retinoid X receptor alpha in a c-Jun N-terminal kinase–dependent manner in human hepatocellular carcinoma cells. Mol. Pharmacol. 84, 218-226. 10.1124/mol.113.085555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorr J. D., Holdcraft R., Chevalier B. and Berg C. A. (2001). Ras1 interacts with multiple new signaling and cytoskeletal loci in Drosophila eggshell patterning and morphogenesis. Genetics 159, 609-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B., Bourbon H.-M., Di Croce L. and Cavalli G. (2017). Genome regulation by polycomb and trithorax: 70 Years and counting. Cell 171, 34-57. 10.1016/j.cell.2017.08.002 [DOI] [PubMed] [Google Scholar]
- Schwartz Y. B. and Pirrotta V. (2013). A new world of polycombs: unexpected partnerships and emerging functions. Nat. Rev. Genet. 14, 853-864. 10.1038/nrg3603 [DOI] [PubMed] [Google Scholar]
- Schweitzer R. and Shilo B. Z. (1997). A thousand and one roles for the Drosophila EGF receptor. Trends Genet. 13, 191-196. 10.1016/S0168-9525(97)01091-3 [DOI] [PubMed] [Google Scholar]
- Seeler J.-S. and Dejean A. (2017). SUMO and the robustness of cancer. Nat. Rev. Cancer 17, 184-197. 10.1038/nrc.2016.143 [DOI] [PubMed] [Google Scholar]
- Shiio Y. and Eisenman R. N. (2003). Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. USA 100, 13225-13230. 10.1073/pnas.1735528100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilo B.-Z. (2014). The regulation and functions of MAPK pathways in Drosophila. Methods 68, 151-159. 10.1016/j.ymeth.2014.01.020 [DOI] [PubMed] [Google Scholar]
- Smith M., Mallin D. R., Simon J. A. and Courey A. J. (2011). Small ubiquitin-like modifier (SUMO) conjugation impedes transcriptional silencing by the polycomb group repressor sex comb on midleg. J. Biol. Chem. 286, 11391-11400. 10.1074/jbc.M110.214569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Durrin L. K., Wilkinson T. A., Krontiris T. G. and Chen Y. (2004). Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. USA 101, 14373-14378. 10.1073/pnas.0403498101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R. and Perrimon N. (2013). Receptor tyrosine kinases in Drosophila development. Cold Spring Harb. Perspect. Biol. 5, a009050 10.1101/cshperspect.a009050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillantini M. G., Schmidt M. L., Lee V. M.-Y., Trojanowski J. Q., Jakes R. and Goedert M. (1997). Alpha-synuclein in Lewy bodies. Nature 388, 839-840. 10.1038/42166 [DOI] [PubMed] [Google Scholar]
- Sriramachandran A. M. and Dohmen R. J. (2014). SUMO-targeted ubiquitin ligases. Biochim. Biophys. Acta 1843, 75-85. 10.1016/j.bbamcr.2013.08.022 [DOI] [PubMed] [Google Scholar]
- Sulzer D. and Edwards R. H. (2019). The physiological role of alpha-synuclein and its relationship to Parkinson's disease. J. Neurochem. 150, 475-486. 10.1111/jnc.14810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szathmáry E., Jordán F. and Pál C. (2001). Molecular biology and evolution. Can genes explain biological complexity? Science 292, 1315-1316. 10.1126/science.1060852 [DOI] [PubMed] [Google Scholar]
- Tammsalu T., Matic I., Jaffray E. G., Ibrahim A. F. M., Tatham M. H. and Hay R. T. (2015). Proteome-wide identification of SUMO modification sites by mass spectrometry. Nat. Protoc. 10, 1374-1388. 10.1038/nprot.2015.095 [DOI] [PubMed] [Google Scholar]
- van der Veen A. G. and Ploegh H. L. (2012). Ubiquitin-like proteins. Annu. Rev. Biochem. 81, 323-357. 10.1146/annurev-biochem-093010-153308 [DOI] [PubMed] [Google Scholar]
- Wakabayashi K., Tanji K., Mori F. and Takahashi H. (2007). The lewy body in Parkinson's disease: molecules implicated in the formation and degradation of alpha-synuclein aggregates. Neuropathology 27, 494-506. 10.1111/j.1440-1789.2007.00803.x [DOI] [PubMed] [Google Scholar]
- Wang L., Jahren N., Miller E. L., Ketel C. S., Mallin D. R. and Simon J. A. (2010). Comparative analysis of chromatin binding by sex comb on midleg (SCM) and other polycomb group repressors at a Drosophila Hox gene. Mol. Cell. Biol. 30, 2584-2593. 10.1128/MCB.01451-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Sheng H., Warner D. S. and Paschen W. (2008). Transient global cerebral ischemia induces a massive increase in protein sumoylation. J. Cereb. Blood Flow Metab. 28, 269-279. 10.1038/sj.jcbfm.9600523 [DOI] [PubMed] [Google Scholar]
- Zhang H., Smolen G. A., Palmer R., Christoforou A., van den Heuvel S. and Haber D. A. (2004). SUMO modification is required for in vivo Hox gene regulation by the Caenorhabditis elegans polycomb group protein SOP-2. Nat. Genet. 36, 507-511. 10.1038/ng1336 [DOI] [PubMed] [Google Scholar]
- Zhang J., Liu Y., Jiang K. and Jia J. (2017). SUMO regulates the activity of smoothened and Costal-2 in Drosophila Hedgehog signaling. Sci. Rep. 7, 42749 10.1038/srep42749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N. and Shabek N. (2017). Ubiquitin ligases: structure, function, and regulation. Annu. Rev. Biochem. 86, 129-157. 10.1146/annurev-biochem-060815-014922 [DOI] [PubMed] [Google Scholar]
- Zhu X., Ding S., Qiu C., Shi Y., Song L., Wang Y., Wang Y., Li J., Wang Y., Sun Y. et al. (2017). SUMOylation negatively regulates angiogenesis by targeting endothelial NOTCH signaling. Circ. Res. 121, 636-649. 10.1161/CIRCRESAHA.117.310696 [DOI] [PMC free article] [PubMed] [Google Scholar]