Abstract
In order to further the present knowledge of the emerging severe acute respiratory syndrome-associated coronavirus (SARS-CoV), 486 different specimens from 54 patients with a clinical diagnosis of SARS were investigated for the presence of viral RNA, and 314 plasma specimens of 73 patients were examined for IgM and IgG antibodies specific against SARS-CoV using an indirect ELISA. Viral RNA was detectable in 28 of the 54 patients tested. Cumulative data showed that 67 of the 73 SARS patients demonstrated seroconversion by week 5 of illness. In contrast, only 1 of 278 healthy subjects enrolled in the study was found to be positive for the IgG antibody. Coexistence of viral RNA in plasma and specific antibodies was simultaneously observed over three consecutive weeks in two critical cases. In three convalescent patients in particular, cultivable SARS-CoV was detected in stool or urine specimens for longer than 4 weeks (29–36 days). These findings suggest that SARS-CoV may remain viable in the excretions of convalescent patients.
Keywords: Severe Acute Respiratory Syndrome, Urine Specimen, Stool Specimen, Severe Acute Respiratory Syndrome, Severe Acute Respiratory Syndrome Patient
Introduction
Severe acute respiratory syndrome (SARS) is a newly emerging human infectious disease that has resulted globally in 774 deaths from 8,096 probable cases to date [1, 2]. A novel coronavirus has been identified as the etiological agent of SARS and been designated as the SARS-associated coronavirus (SARS-CoV) [3–7]. SARS-CoV contains a single-stranded plus-sense RNA genome approximately 30 kb in length, which typically has five major open reading frames (ORFs) that encode the replicase (R) polyproteins, and the spike (S), the envelope (E), the membrane (M), and the nucleocapsid (N) proteins. SARS-CoV can infect African green monkey kidney (Vero E6) cells in vitro and experimentally cause a similar disease in cynomolgus macaques (macaca fascicularis) [8].
It has been reported that around 90% (83.3–100%) of patients with probable SARS generated antibodies against SARS-CoV [9–11], while the figure was negligible in a healthy population [12]. For early laboratory diagnostics, assays based on conventional nested and/or real-time reverse transcription-polymerase chain reactions (RT-PCR) have been developed to detect viral RNA in the clinical specimens of SARS patients [3, 13, 14–16]. Even though the viral RNA and the antibody profiles in SARS patients have been well studied, the simultaneous detection of both profiles in sera or plasma has rarely been documented. In particular, there is a paucity of data concerning viral vitality in the excretions of SARS patients in the convalescent phase.
In the study presented here viral RNA was detected in different types of clinical specimens. Dynamic profiles of SARS-CoV RNA in plasma and antibodies against SARS-CoV were simultaneously investigated in patients with probable SARS. The viral RNA-positive stool and urine samples from some patients were studied further for viral vitality. The results obtained further the available knowledge regarding the laboratory diagnostics and prophylactic control of this emerging infection.
Materials and methods
Participants and specimens
A total of 486 clinical specimens from 54 patients with a clinical diagnosis of SARS were tested for the presence of both viral RNA and specific antibodies against SARS-CoV. The specimens included 213 plasma samples (isolated from citrate sodium-anticoagulated peripheral blood), 115 blood cell samples (remaining part of the blood from the plasma), 81 sputum/throat swab samples, 47 stool specimens, and 30 urine specimens. When possible, specimens were collected weekly. Moreover, 101 plasma specimens from an additional 19 patients were tested solely for the presence of SARS-CoV antibody. The patients were all hospitalized in our unit from March to June 2003, they all had a clinical diagnosis of SARS, and all of them received ribavirin/steroid combination therapy. In addition, plasma samples from 278 healthy subjects were analyzed for the presence of antibodies specific against SARS-CoV. All specimens were stored at −70°C until tested.
RNA extraction
RNA extraction was performed in the biosafety level 3 (P3) laboratory at the Beijing 302 Hospital. RNA was extracted directly from plasma and urine samples. Sputum samples were shaken for 30 min with an equal volume of 1.0% acetylcysteine and 0.9% sodium chloride, and the supernatant was isolated by centrifugation (10,000g × 3 min). Throat swabs and stool samples were suspended with phosphate-buffered saline (PBS) containing 10 U/ml RNasin (Promega, Madison, WI, USA), shaken for 10 min, and the supernatant was isolated by centrifugation as mentioned above. Blood cells (from 1.8 ml of blood) were treated with an equal volume of Trizol (Invitrogen, Carlsbad, CA, USA), followed by isolating the aqueous phase by centrifugation (12,000g × 15 min). RNA was extracted using the QIAmp Viral RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
Nested RT-PCR
Two groups of nested primers were used for amplifying the viral RNA. One set of BNI primers recommended by the Word Health Organization was used to target the R polyprotein encoding region (ORF1b) of SARS-CoV [17]. For this, the outer sense/antisense primers were 5′-ATGAATTACCAAGTCAATGGTTAC-3′/5′-CATAACCAGTCGGTACAGCTA-3′, and the inner sense/antisense primers were 5′-GAAGCTATTCGTCACGTTCG-3′/CTGTAGAAAATCCTAGCTGGAG-3′, resulting in a 109-bp-long fragment complementary to nucleotide 18182-18290 of the BJ01 strain viral genome (GenBank accession no. AY278488). Another set of primers was used to target the N protein-encoding region of SARS-CoV designed using sequence analysis software, MacVector (Accelrys, San Diego, CA, USA). For this, the outer sense/antisense primers were 5′-TCACTCAAGCATTTGGGAGACG-3′/5′-TCCTTTTTAGGCTCTGTTGGTGGG-3′, and the inner sense/antisense primers were 5′-TGCACAATTTGCTCCAAGTGC-3′/5′TGCTTGTTCAGCAGTATGACGTTG-3′, resulting in a 156-bp-long fragment complementary to nucleotide 29015-29170 of the BJ01 strain viral genome. The one-step RT-PCR Kit (Qiagen) was used for reverse transcription and the first round of PCR amplification with outer primers. Thermal cycling consisted of 50°C for 30 min; 95°C for 15 min; 10 cycles at 95°C for 15 s, at 57.5°C for 15 s (decreasing by 1.5°C every other cycle), and at 72°C for 30 s; 40 cycles at 95°C for 15 s, at 56°C for 15 s, and at 72°C for 30 s. Afterwards, 2 μl of the product was used as a template for the second round of PCR amplification in 100-μl volume with inner primers and Taq DNA polymerase (MBI Fermentas, Hanover, MD, USA). Thermal cycling consisted of 30 cycles at 95°C for 10 s, at 56°C for 10 s, and at 72°C for 20 s. All reactions were carefully carried out to avoid contamination.
Positive and negative controls were included in each reaction run. RT-PCR products were analyzed by electrophoresis in 2.5% agarose gel containing 0.5 μg/ml of ethidium bromide. To confirm the results, DNA in the electrophoretic bands of the right size was recovered using the QIAquick Gel Extraction kit (Qiagen) and cloned into a pGEM-T vector system (Promega) for sequence analysis. DNA sequences were obtained with the use of an automated ABI377 sequencer (Applied Biosystems, Foster City, CA, USA). Analysis and comparison of nucleotides were carried out with the DNAstar computer software (DNAstar, Madison, WI, USA).
Antibody detection
Plasma IgM and IgG antibodies specific against SARS-CoV were detected using two commercially available ELISA kits (Jibiai Biotech, Beijing, China) according to the manufacturer’s instructions. Indirect assays were employed using both ELISA kits with culture-obtained SARS-CoV antigen for the capture of antibodies.
Virus passage and isolation
These experiments were carried out in the P3 laboratory at Beijing Genomics Institute, Chinese Academy of Sciences. Stool was suspended in PBS. The supernatant was isolated by centrifugation, sterilized by passing through a 0.22 μm Millipore filter, and spread onto a monolayer of Vero E6 cells in a 24-well plate. Urine was clarified by centrifugation, sterilized, and spread onto the cells in the same way. As a control, PBS was used instead of a clinical specimen in triplicate wells. After incubation at 37°C for 2 h, the infection solution was replaced with fresh Dulbecco minimum essential medium with 5% fetal calf serum (Gibco BRL, Grand Island, NY, USA). The cultures were examined for cytopathic effect (CPE) each day. At day 7, a blind passage was performed. The cells were collected by vigorously blowing the monolayer cells with the medium. The cell suspension was treated with three rounds of freeze-thaw conditions, clarified by centrifugation, and spread onto fresh Vero E6 cells. The blind passage was performed once each week for 3 weeks, and discarded if no CPE was observed. If CPE appeared, cells were collected for electron microscopy observation and RT-PCR identification. The isolation of SARS-CoV was confirmed by sequence analysis of the RT-PCR products.
Results
One hundred and nine specimens from 28 of 54 SARS patients were positive for SARS-CoV RNA by nested RT-PCR, resulting in a 51.9% detection rate. The amplified fragments were 109 and 156 bp in length and were confirmed to be parts of the SARS-CoV polymerase gene and N gene, respectively, by sequence analysis (Fig. 1). Of 108 positive samples, 55 were double positive for both R and N gene fragment amplification. Individual single-positive amplification of R or N gene fragments was observed in 28 and 25 samples, respectively. Viral RNA was detectable in 42.1% (8/19) and 61.3% (19/31) of patients tested at week 1 and week 2 of illness, respectively. The detection profile of viral RNA from different specimens is shown in Fig. 2. Specimens obtained from the respiratory tract were more likely than other specimens to yield viral RNA in the early stage of illness, with seven of eight tested patients (7 of 12 tested specimens) positive at week 1. During the convalescent phase of the infection (i.e., >4 weeks after disease onset), viral RNA was detected in different specimens of 10 of 32 (31.3%) patients.
Fig. 1.
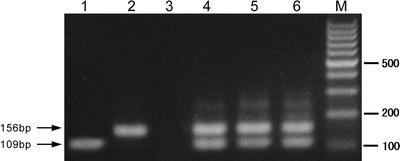
Electrophoretic analysis of RT-PCR products for SARS-CoV RNA amplified from clinical specimens. Lanes1−6 were loaded with RT-PCR products; laneM was loaded with 100 bp DNA marker; 109bp and 156bp-long bands correspond to the amplified R (ORF 1b) and N gene fragments of SARS-CoV, respectively
Fig. 2.
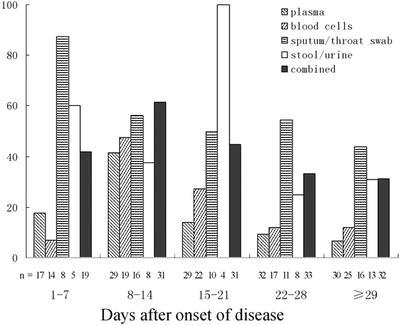
Percent of positive results for SARS-CoV RNA detected by RT-PCR in different clinical specimens of 54 patients with a clinical diagnosis of SARS. Combined represents the percentage of all patients from whom any of the specimens was positive for SARS-CoV RNA. n represents the number of patients in whom SARS-CoV was detected
Figure 3 shows the dynamic seroconversion in cumulative percentages of patients with probable SARS. From week 1 to week 5 of illness, the rates of IgM-positive antibody detection were 0% (0/22), 44.0% (22/50), 76.5% (52/68), 81.9% (59/72), and 68.5% (50/73) per week, respectively, while the rates for IgG antibody were 0% (0/22), 38.0% (19/50), 72.1% (49/68), 84.7% (61/72), and 90.4% (66/73), respectively. The combined (IgM and/or IgG) seroconversion rates were 0% (0/22), 54.0% (27/50), 82.4% (56/68), 88.9% (64/72), and 91.8% (67/73), respectively. In contrast, only 1 of 278 (0.36%) healthy subjects was positive for the SARS-CoV-specific IgG antibody.
Fig. 3.
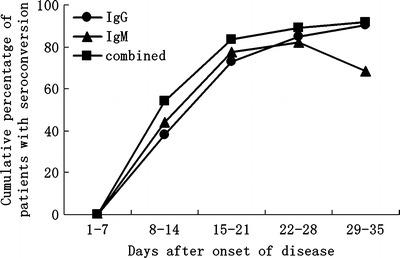
Cumulative percentage of seroconversion among 73 patients with a clinical diagnosis of SARS. Combined represents the cumulative percentage of patients demonstrating IgM and/or IgG seroconversion
Because of the difficulties associated with clinical sample collection, only 148 plasma samples could be collected from 25 SARS patients during three consecutive weeks and tested for both SARS-CoV RNA and antibodies. Among these patients, 21 had detectable antibodies, and 12 had one or more specimens positive for viral RNA (Table 1). Two patients (no. 1 and 10) were persistently positive for viral RNA in plasma up to 5–7 weeks after disease onset. It is notable that the coexistence of specific antibodies and viral RNA was observed for as long as 3 weeks in these two patients.
Table 1.
Serial detection of SARS-CoV RNA and antibodies against SARS-CoV in the plasma of 25 hospitalized patients with a clinical diagnosis of SARS
| Pt. no. | Age (year) | Sex | Course (day) | Manifestation or result | Day after disease onset (IgM/IgG/RNA) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1–7 | 8–14 | 15–21 | 22–28 | 29–35 | 36–42 | 43–49 | 50–56 | |||||
| 1 | 20 | M | 77 | Critical | ND | ND | −/−/+ | −/−/+ | +/+/+ | +/+/+ | +/+/+ | +/+/− |
| 2 | 27 | M | 34 | General | −/−/− | +/+/− | +/+/− | +/+/− | +/+/− | |||
| 3 | 42 | F | 73 | Critical | ND | −/−/+ | −/+/− | +/+/− | +/+/− | +/+/− | +/+/− | −/+/− |
| 4 | 59 | M | 73 | Critical | ND | +/+/+ | +/+/− | +/+/− | +/+/− | +/+/− | +/+/− | −/+/− |
| 5 | 39 | M | 41 | General | −/−/− | +/+/+ | −/+/− | −/+/− | −/+/− | −/+/− | ||
| 6 | 33 | M | 59 | General | ND | −/+/+ | +/+/− | +/+/− | +/+/− | +/+/− | +/+/− | +/+/− |
| 7 | 51 | M | 59 | Dead | ND | −/+/+ | +/+/− | +/+/− | +/+/− | −/+/− | −/+/− | −/+/− |
| 8 | 26 | M | 41 | General | ND | +/+/− | +/+/− | +/+/− | +/+/− | +/+/− | ||
| 9 | 32 | M | 30 | General | −/−/− | −/−/− | −/−/− | −/−/− | −/−/− | |||
| 10 | 31 | F | 47 | Critical | −/−/− | −/−/+ | +/+/+ | +/+/+ | −/+/+ | −/+/− | −/+/− | |
| 11 | 19 | F | 59 | General | ND | −/−/+ | −/−/+ | +/+/− | +/+/− | +/+/− | +/+/− | +/+/− |
| 12 | 31 | M | 50 | Critical | ND | −/+/− | +/+/− | +/+/− | +/+/− | +/+/− | −/+/− | −/+/− |
| 13 | 42 | M | 49 | Critical | ND | −/−/− | −/−/− | +/+/− | +/+/− | +/+/− | +/+/− | |
| 14 | 13 | F | 34 | General | −/−/− | −/−/− | −/−/− | −/−/− | −/−/− | |||
| 15 | 45 | F | 31 | General | −/−/− | −/−/− | +/−/− | +/+/− | +/+/− | |||
| 16 | 46 | F | 36 | Critical | −/−/− | −/−/+ | −/−/+ | −/−/+ | −/+/− | +/+/− | ||
| 17 | 33 | M | 41 | Critical | ND | −/+/− | ND | +/+/− | +/+/− | +/+/− | ||
| 18 | 38 | F | 36 | General | −/−/− | −/−/− | +/−/− | +/−/− | +/−/− | −/−/− | ||
| 19 | 21 | M | 67 | Critical | −/−/− | −/−/− | +/+/− | +/+/− | +/+/− | +/+/− | +/+/− | ND |
| 20 | 20 | M | 38 | General | −/−/− | −/−/− | −/−/− | −/−/− | −/−/− | −/−/− | ||
| 21 | 48 | M | 38 | Critical | −/−/+ | +/−/+ | +/+/− | +/+/− | +/+/− | +/+/− | ||
| 22 | 9 | M | 36 | General | −/−/− | −/−/+ | −/−/− | −/−/+ | −/−/− | −/−/− | ||
| 23 | 45 | M | 30 | General | ND | −/−/− | −/+/− | −/+/− | ND | |||
| 24 | 32 | M | 47 | General | −/−/+ | +/+/+ | +/+/− | +/+/− | +/+/− | +/+/− | ND | |
| 25 | 18 | M | 46 | General | ND | +/−/− | +/−/− | +/+/− | +/+/− | +/+/− | +/+/− | |
M, male; F, female; +, positive; −, negative; ND, not detected
Twenty-one stool and urine specimens were employed for in vitro isolation of SARS-CoV. Sputum specimens were not employed because the use of acetylcysteine in the viral RNA extraction process may have destroyed the protein structure of the virus. Plasma and blood specimens were not employed because of the low viral load in these samples. Nested RT-PCR showed that 16 of the 21 stool and urine specimens were positive for viral RNA (10 for both the R and N gene fragments; 6 for either the R or the N gene fragment), and five were negative for viral RNA. SARS-CoV was isolated from all ten of the specimens that were positive for both the R and N gene fragments of viral RNA after 2–3 generations of passage in culture and the emergence of CPE and the virus in Vero E6 cells (Fig. 4); identity was confirmed by RT-PCR followed by sequence analysis. In contrast, the virus was not isolated from any of the other 11 specimens (Table 2).
Fig. 4.

Observation of cytopathic effect (CPE) and SARS-CoV particles in Vero E6 cells infected with SARS-CoV isolated from the stool sample of a convalescent-phase patient with SARS. a–b Light microscopy images of Vero E6 cells without/with CPE. c Electron microscopy image of Vero E6 cells with CPE showing the SARS-CoV particles
Table 2.
Isolation of SARS-CoV in urine and stool samples by culture with Vero E6 cells
| Pt. no.a | Sample no. | Sampling day after disease onset | RT-PCR results (R gene/N gene) | Virus isolation |
|---|---|---|---|---|
| 5 | urine 94 | 8 d | +/+ | + |
| stool 158 | 8 d | +/+ | + | |
| stool 131 | 11 d | +/+ | + | |
| 6 | urine 107 | 36 d | +/+ | + |
| 11 | stool 159 | 29 d | +/+ | + |
| 24 | stool 163 | 23 d | +/+ | + |
| stool 124 | 26 d | +/+ | + | |
| urine 95 | 21 d | +/+ | + | |
| stool 154 | 24 d | +/+ | + | |
| stool 141 | 33 d | +/+ | + | |
| 16 | stool 156 | 10 d | −/+ | − |
| urine 111 | 6 d | +/− | − | |
| 15 | stool 126 | 21 d | −/+ | − |
| 21 | stool 160 | 11 d | +/− | − |
| stool 145 | 33 d | +/− | − | |
| urine 96 | 11 d | +/− | − | |
| 9 | stool 142 | 3 d | −/− | − |
| 20 | stool 120 | 18 d | −/− | − |
| 2 | stool 144 | 12 d | −/− | − |
| 12 | urine 92 | 22 d | −/− | − |
| 22 | urine 89 | 14 d | −/− | − |
+, positive; −, negative
aPatient numbers correspond with those listed in Table 1
Discussion
As the area most affected by the SARS epidemic in the world to date, Beijing claimed 2,521 cases and 191 deaths from early March to late May 2003 [18]. Of these cases, 132 of the SARS patients were hospitalized and treated in our unit at the Beijing 302 Hospital, including the first cluster of case-patients in the area [19]. The criteria for laboratory confirmation of SARS-CoV infection are based on the following methods: SARS-CoV RNA detection by RT-PCR, serological detection of SARS-CoV-related antibody, and isolation of SARS-CoV by cell culture [11, 20].
Several assays have been developed to detect viral RNA using conventional nested RT-PCR and real-time RT-PCR. Though real-time RT-PCR assays have a quantitative advantage and are reported to have higher sensitivity/specificity in some cases, the requirements of expensive reagents and special instruments are obstacles preventing their widespread use in ordinary laboratories. During the crucial time span of the SARS epidemic, a real-time RT-PCR assay was still being developed. Therefore, conventional nested RT-PCR still served as a practicable assay at the time. The method using BNI outer/inner primers was reported to have a lower cost but similar sensitivity and specificity compared to three in-house real-time RT-PCR assays [21]. We observed that simultaneous employment of the primers targeting both R (ORF 1b) and N regions of SARS-CoV could increase the method’s detection sensitivity. This finding is identical to that contained in a previous report by Bressler et al. [16]. These authors suggested that the N gene is present in all of the sub-genomic RNAs of SARS-CoV instead of in only one of them (ORF 1b of the R gene). However, we believe the major reason might be that most of the SARS-CoV is not intact in clinical specimens. Thus, amplification of different regions of SARS-CoV will increase the chance of detecting the disintegrated virus genome.
Nested RT-PCR may give occasional false-positive reactions with stool specimens [15]. We tested 36 stool specimens and 24 sputum specimens from healthy subjects using the assay. False-positive reactions were observed in four of the stool specimens. Therefore, all positive results from the stool specimens of the SARS patients were confirmed by sequence analysis of the RT-PCR products. In addition, 64 of the 108 positive samples were amplified and sequenced for the S gene of SARS-CoV, and we confirmed that all of these samples contained S gene fragments of SARS-CoV [22]. Employing various specimens may increase the detection rate for viral RNA. In the present study, viral RNA was detectable in 42.1% (8/19) and 61.3% (19/31) of patients with probable SARS during week 1 and week 2 of illness, respectively. In contrast, specific antibodies against SARS-CoV were detectable in 0% (0/22) and 50% (27/54) of patients in the same respective weeks. Therefore, viral RNA detection is particularly valuable for early-stage diagnostics. However, false-positive results have to be carefully excluded, especially for stool specimens. Employment of nasopharyngeal aspirate collected by professional physicians and use of qualified real-time RT-PCR assays may increase the method’s sensitivity/specificity [13]. From week 2 following disease onset, serology is the gold standard for identifying true-positive cases, since SARS-CoV is new to humans and we commonly lack immunity against it.
In our study, 6 of 73 patients with a clinical diagnosis of SARS were negative for seroconversion, and two of them were children (patients no. 14 and 22 in Table 1). It is interesting, however, that viral RNA was detectable in both of these patients (i.e., in 1 blood cell sample of patient 14 and in 2 plasma samples of patient 22). Other investigators have reported that about half of their pediatric patients with a clinical diagnosis of SARS failed to develop detectable antibodies specific against SARS-CoV [23]. Weak humoral immune response in pediatric SARS patients may be responsible for this unusual phenomenon. Since our observation of four other adult patients who did not seroconvert was terminated after 4–6 weeks of illness, and seroconversion can still occur during weeks 7 and 8 of illness [24], one possible explanation for the negative antibody detection results may be a delayed antibody response. It is also possible that the antibody response of some patients is below the sensitivity of the ELISA test. However, considering that the SARS diagnosis in our patients was based on clinical manifestations and characteristics on chest radiograph during an emergency situation, clinical misdiagnosis of the patients who were negative for SARS-CoV RNA cannot be excluded.
Of 278 healthy subjects tested, only one individual was positive for IgG antibody in plasma, suggesting that silent infection with SARS-CoV is unlikely. In general, seroconversion occurs in parallel with eradication of the virus in plasma or serum. In our study, however, persistent coexistence of SARS-CoV RNA and antibodies against SARS-CoV was observed in the plasma of two patients whose clinical status was critical. Other investigators also reported the long-term persistence of SARS-CoV RNA in the serum of SARS-CoV-specific IgG seroconverters [25], and we previously documented the presence of SARS-CoV quasispecies in individual patients [26]. Therefore, it remains to be clarified whether persistent coexistence of viral RNA and specific antibodies is correlated with dynamic genetic variation of the virus.
Through in vitro passage of Vero E6 cells, SARS-CoV was isolated from seven stool and three urine specimens that were positive for both the N and R regions of viral RNA using nested RT-PCR. These included two stool specimens and one urine specimen collected more than 4 weeks after disease onset. To our knowledge, the isolation of viable SARS-CoV from the urine of a convalescent patient has not been reported previously. Our isolation of living SARS-CoV from the stool specimens of convalescent SARS patients is discrepant with one report [15] but in accordance with the findings of another investigation [27]. The discrepancy may be due to differences in the isolation protocols. Our results suggest that viable SARS-CoV might exist in the excretions of convalescent-phase patients. Since it is not yet known precisely how long SARS-CoV can remain active in vivo, measures should be taken to prevent the spread of this infectious agent via the excretions of convalescent individuals.
The results of this retrospective study depict the dynamic profiles of viral persistence and seroconversion in SARS patients and reveal the novel finding that viable SARS-CoV can be shed in the urine and stool of some convalescent-phase patients. The results further the current knowledge of this emerging infectious disease and offer implications for clinical diagnostics and infection control.
Acknowledgments
We acknowledge the research funding from Beijing Natural Science Foundation (Number: 03F017) and Sino-UK Collaboration Foundation for SARS Immunopathogenesis Study (Number: H030230100130). We would like to thank Dr. P. Kretchmer for his assistance in editing this manuscript
References
- 1.World Health Organization (2004) Summary of probable SARS with onset of illness from 1 November 2002 to 31 July 2003. Available at: http://www.who.int/csr/sars/country/table2004_04_21/en/. Cited 8 May 2004
- 2.Tambyah PA. SARS: responding to an unknown virus. Eur J Clin Microbiol Infect Dis. 2004;23:589–595. doi: 10.1007/s10096-004-1175-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA, Berger A, Burguiere AM, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra JC, Muller S, Rickerts V, Sturmer M, Vieth S, Klenk HD, Osterhaus AD, Schmitz H, Doerr HW. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 4.Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey CD, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ, SARS Working Group A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 5.Marra MA, Jones SJ, Astell CR, Holt RA, Brooks-Wilson A, Butterfield YS, Khattra J, et al. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 6.Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W, Nicholls J, Yee WK, Yan WW, Cheung MT, Cheng VC, Chan KH, Tsang DN, Yung RW, Ng TK, Yuen KY, SARS Study Group Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Penaranda S, Bankamp B, Maher K, Chen MH, Tong S, Tamin A, Lowe L, Frace M, DeRisi JL, Chen Q, Wang D, Erdman DD, Peret TC, Burns C, Ksíazek TG, Rollin PE, Sanchez A, Liffick S, Holloway B, Limor J, McCaustland K, Olsen-Rasmussen M, Fouchier R, Gunther S, Osterhaus AD, Drosten C, Pallansch MA, Anderson LJ, Bellini WJ. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 8.Fouchier RA, Kuiken T, Schutten M, van Amerongen G, van Doornum GJ, van den Hoogen BG, Peiris M, Lim W, Stohr K, Osterhaus AD. Aetiology: Koch’s postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peiris JS, Chu CM, Cheng VC, Chan KS, Hung IF, Poon LL, Law KI, Tang BS, Hon TY, Chan CS, Chan KH, Ng JS, Zheng BJ, Ng WL, Lai RW, Guan Y, Yuen KY, HKU/UCH SARS Study Group Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li G, Chen X, Xu A. Profile of specific antibodies to the SARS-associated coronavirus. N Engl J Med. 2003;349:508–509. doi: 10.1056/NEJM200307313490520. [DOI] [PubMed] [Google Scholar]
- 11.Chen X, Zhou B, Li M, Liang X, Wang H, Yang G, Wang H, Le X. Serology of severe acute respiratory syndrome: implications for surveillance and outcome. J Infect Dis. 2004;189:1158–1163. doi: 10.1086/380397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leung GM, Chung PH, Tsang T, Lim W, Chan SK, Chau P, Donnelly CA, Ghani AC, Fraser C, Riley S, Ferguson NM, Anderson RM, Law YL, Mok T, Ng T, Fu A, Leung PY, Peiris JS, Lam TH, Hedley AJ. SARS-CoV antibody prevalence in all Hong Kong patient contacts. Emerg Infect Dis. 2004;10:1653–1656. doi: 10.3201/eid1009.040155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poon LL, Chan KH, Wong OK, Yam WC, Yuen KY, Guan Y, Lo YM, Peiris JS. Early diagnosis of SARS coronavirus infection by real time RT-PCR. J Clin Virol. 2003;28:233–238. doi: 10.1016/j.jcv.2003.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poon LL, Chan KH, Wong OK, Cheung TK, Ng I, Zheng B, Seto WH, Yuen KY, Guan Y, Peiris JS. Detection of SARS coronavirus in patients with severe acute respiratory syndrome by conventional and real-time quantitative reverse transcription-PCR assays. Clin Chem. 2004;50:67–72. doi: 10.1373/clinchem.2003.023663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan KH, Poon LL, Cheng VC, Guan Y, Hung IF, Kong J, Yam LY, Seto WH, Yuen KY, Peiris JS. Detection of SARS coronavirus in patients with suspected SARS. Emerg Infect Dis. 2004;10:294–299. doi: 10.3201/eid1002.030610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bressler AM, Nolte FS. Preclinical evaluation of two real-time, reverse transcription-PCR assays for detection of the severe acute respiratory syndrome coronavirus. J Clin Microbiol. 2004;42:987–991. doi: 10.1128/JCM.42.3.987-991.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.World Health Organization (2003) PCR primers for SARS developed by WHO network laboratories [monograph on the Internet]. Available at: http://www.who.int/csr/sars/primers/en/. Cited 20 April 2003
- 18.Liang WN, Mi J, Information Branch Joint Leadership Group of SARS Prevention and Control in Beijing Epidemiological features of severe acute respiratory syndrome in Beijing. Zhonghua Liu Xing Bing Xue Za Zhi. 2003;24:1096–1099. [PubMed] [Google Scholar]
- 19.Zhou XZ, Zhao M, Wang FS, Jiang TJ, Li YG, Nie WM, Zhou ZP, Wang Y, He J, Fan R, Zhao JM, Zhang XW. Epidemiologic features, clinical diagnosis and therapy of first cluster of patients with severe acute respiratory syndrome in Beijing area. Zhonghua Yi Xue Za Zhi. 2003;83:1018–1022. [PubMed] [Google Scholar]
- 20.Berger A, Drosten C, Doerr HW, Sturmer M, Preiser W. Severe acute respiratory syndrome (SARS)-paradigm of an emerging viral infection. J Clin Virol. 2004;29:13–22. doi: 10.1016/j.jcv.2003.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahony JB, Petrich A, Louie L, Song X, Chong S, Smieja M, Chernesky M, Loeb M, Richardson S, Ontario Laboratory Working Group for the Rapid Diagnosis of Emerging Infections Performance and cost evaluation of one commercial and six in-house conventional and real-time reverse transcription-PCR assays for detection of severe acute respiratory syndrome coronavirus. J Clin Microbiol. 2004;42:1471–1476. doi: 10.1128/JCM.42.4.1471-1476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu D, Zhang Z, Chu F, Li Y, Jin L, Zhang L, Gao GF, Wang FS. Genetic variation of SARS coronavirus in Beijing hospital. Emerg Infect Dis. 2004;10:789–794. doi: 10.3201/eid1005.030875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li ZZ, Shen KL, Wei XM, Wang HL, Lu J, Tian H, Sun GQ, Zeng JJ, Hu YH, Zhao SY, Yin J, Feng XL, Jiang ZF, Yang YH. Clinical analysis of pediatric SARS cases in Beijing. Zhonghua Er Ke Za Zhi. 2003;41:574–577. [PubMed] [Google Scholar]
- 24.Leong HN, Chan KP, Khan AS, Oon L, Se-Thoe SY, Bai XL, Yeo D, Leo YS, Ang B, Ksiazek TG, Ling AE. Virus-specific RNA and antibody from convalescent-phase SARS patients discharged from hospital. Emerg Infect Dis. 2004;10:1745–1750. doi: 10.3201/eid1010.040026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsueh PR, Hsiao CH, Yeh SH, Wang WK, Chen PJ, Wang JT, Chang SC, Kao CL, Yang PC, SARS Research Group of National Taiwan University College of Medicine and National Taiwan University Hospital Microbiologic characteristics, serologic responses, and clinical manifestations in severe acute respiratory syndrome, Taiwan. Emerg Infect Dis. 2003;9:1163–1167. doi: 10.3201/eid0909.030367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu D, Zhang Z, Wang FS. SARS-associated coronavirus quasispeies in individual patients. New Engl J Med. 2004;350:1366–1367. doi: 10.1056/NEJMc032421. [DOI] [PubMed] [Google Scholar]
- 27.Tian W, Deng YJ, Wu QF, Wang XM, Yang L, Zhang F, Liu CH, Bao QY, Wang J, Yang H, Liu M. Isolation and identification of SARS-CoV from stool of SARS patients in different phase of disease. World Chin J Digestol. 2004;12:1246–1248. [Google Scholar]


