Abstract
In 2012, a novel human coronavirus emerged and was tentatively named “Middle East respiratory syndrome coronavirus” (MERS-CoV). The high mortality rate of MERS-CoV focused attention on the ecology of the virus. It has been found that MERS-CoV belongs to the group C lineage of the genus Betacoronavirus. Coronavirus surveillance studies in different populations of bats have suggested that they are probable reservoirs for this novel virus, and phylogenetic analysis of both the spike (S1) and RNA-dependent RNA polymerase proteins of MERS-CoV have revealed that it is related to bat viruses. Recently, the MERS-CoV and its neutralizing antibodies were detected in dromedary camels. Despite the limited number of reported cases of person-to-person transmission, the rapid evolution of the virus poses a continuous threat to humans worldwide. This paper reviews the current state of knowledge regarding the virology, clinical spectrum, evolution, diagnosis and treatment of MERS-CoV infections.
Keywords: Arabian Peninsula, Severe Acute Respiratory Syndrome, Camel Milk, Dromedary Camel, Severe Acute Respiratory Syndrome Coronavirus
Introduction
Coronaviruses (CoV) are able to infect humans, birds and many animal species [68]. In humans, coronavirus infections result mostly in mild respiratory, enteric and neurological diseases [65, 66]. In June 2012, a novel coronavirus was detected in a Saudi patient who had experienced pneumonia and renal failure. Virus genome sequencing demonstrated that the virus belonged to lineage C of the genus Betacoronavirus and was phylogenetically related to the bat coronaviruses HKU4 and HKU5, which had previously been found in lesser bamboo bats and Japanese Pipistrelle bats in Hong Kong.
In September 2012, another patient from Qatar with a similar clinical picture also proved positive by means of real-time reverse transcription polymerase chain reaction (rRT-PCR) assay [5]. This finding was followed by the detection of a closely related virus in archival samples from two Jordanian patients who died in April 2012 after unexplained respiratory illness [24].
The outbreaks of severe acute respiratory syndrome coronavirus (SARS-CoV) infection in the year 2003 and the Middle East respiratory syndrome coronavirus (MERS-CoV) infection in 2012 showed that these viruses can cause fatal human disease (death rates were ~11 % and ~43 %, respectively) [6, 9]. Before the discovery of MERS-CoV, five coronaviruses were known to infect human populations. HCoV-229E and HCoV-OC43 were identified in the early 1960s as the causative agents of respiratory infection [29, 64] while SARS-CoV, HCoV-NL63 and HCoV-HKU1 were identified in 2003, 2004 and 2005, respectively [53].
Most of the emerging infectious pathogens originated from animals. Betacoronavirus A, OC43, possibly jumped from a bovine source in the 1890s [69] while betacoronavirus B, the SARS virus, jumped from bats to civets, then to humans [37]. Like its counterparts, the emergence of MERS-CoV may constitute another model of interspecies transmission that has yet to be characterized in detail. The current article provides a review of recent findings and speculations regarding MERS-CoV.
Taxonomy
Coronaviruses belong to the order Nidovirales, family Coronaviridae and subfamily Coronavirinae, which consists of four genera: Alphacoronavirus, Betacoronavirus, Deltacoronavirus and Gammacoronavirus. Their taxonomy is based on antigenic relationships [44, 49] and has subsequently been largely supported by gene sequencing [35]. Human coronaviruses belong to two genera: Alphacoronavirus and Betacoronavirus. HCoV-229E and HCoV-NL63 belong to the genus Alphacoronavirus. The genus Betacoronavirus consists of four lineages, A-D. SARS-CoV belongs to lineage B, but HCoV-OC43 and HCoV-HKU1 belong to lineage A. Bat coronaviruses (BtCoV) belong to lineages C and D of the betacoronaviruses. The sequencing data has suggested that the sixth discovered human coronavirus, MERS-CoV, belongs to lineage C betacoronaviruses. Based on the RdRp, a bat-derived isolate, BtCoV/8-724/Pip_pyg/ROU/2009, shows 98 % identity to MERS-CoV [1]. Comparison of the S1 and RdRp gene sequences of different coronaviruses from different species with those from MERS-CoV, including available bat coronaviruses, has been conducted (Table 1). Unfortunately, no available spike protein sequence of BtCoV/8-724/Pip_pyg/ROU/2009 was available in the GenBank database and therefore this was not included in the comparison. In phylogenetic trees, MERS-CoV clearly clustered with lineage C of the genus Betacoronavirus (Fig. 1), relatively close to BtCoV/133/2005 and BtCoV HKU4-2, which had the highest S1 amino acid sequence identity to MERS-CoV (60 %); however, the amino acid sequence identity to MERS-CoV was higher (up to 94 %) when the RdRp of BtCoV HKU5 and BtCoV HKU5-3 were compared. Interestingly, Beta CoV/KW2E-F93/Nyc_spec/ GHA/2010 showed 40 % and 92 % S1 and RdRp amino acid sequence identity to MERS-CoV, respectively (Table 1). This finding suggests that genotyping based on the polymerase gene should be followed by confirmation based on S1 genotyping (Table 1, Fig. 1). The phylogenetic relationship of the Saudi Arabian MERS-CoV sequences revealed that they are subdivided into two main clades: clade A, comprising EMC/2012 and Jordan-N3 and clade B, comprising Munich/AbuDhabi, England-Qatar, Riyadh-3, Bisha-1, Riyadh-1 cluster, Hafr-AlBatin-1, Riyadh-2, Buraidah-1 and England2-HPA [15].
Table 1.
Multisequence alignment of the S1 protein and RdRp of MERS-CoV with those of different coronaviruses
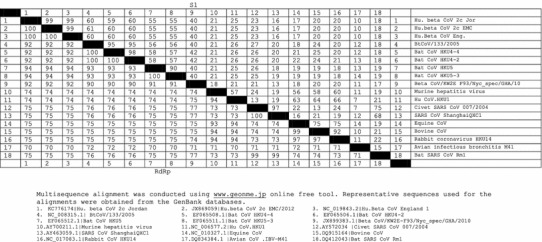
Fig. 1.
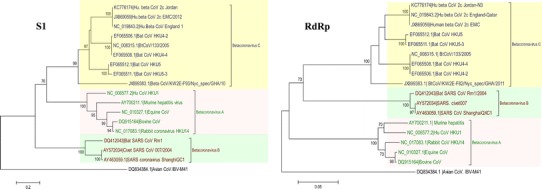
Phylogenetic analysis of the S1 and RNA-dependent RNA polymerase of MERS-CoV in comparison to those of selected coronaviruses from the GenBank database. The betacoronavirus strains were selected to be representative of different lineages and were rooted to IBV-M41 Gammacoronavirus. The robustness of individual nodes of the tree was assessed using 1000 bootstrap replicates and bootstrap values 70 % and higher are indicated at key nodes
Nomenclature
Since its initial discovery, MERS-CoV isolates have appeared under various names including human coronavirus-Erasmus Medical Center (hCoV-EMC), human betacoronavirus 2c England-Qatar, human betacoronavirus 2C Jordan-N3, betacoronavirus England 1 and novel coronavirus (NCoV) [19]. Currently, the Coronavirus Study Group (CSG) of the International Committee on Taxonomy of Viruses has decided to refer to the new coronavirus as Middle East respiratory syndrome coronavirus (MERS-CoV) [19].
Viral genome and proteins
The genome structure of MERS-CoV (30,119 nt) is similar to that of other coronaviruses. RNAs are capped and polyadenylated with the 5′ two-thirds of the genome encoding the non-structural proteins (NSPs) involved in viral replication and the remaining 3′ third of the genome encoding the structural genes in addition to four accessory genes interspersed within the structural gene region [67]. At the 5′ end of the genome there is a leader sequence (67 nt), which is followed by an untranslated region (UTR). At the 3′ end of the RNA genome there is another UTR, followed by a poly(A) sequence of variable length. Transcription-regulatory sequences (TRSs: 5′ AACGAA 3′) are found at the 3′ end of the leader sequence and at different positions upstream of genes in the genomic 3′-proximal domain of MERS-CoV [67]. The MERS-CoV genome contains at least 10 predicted open reading frames (ORFs): ORF1a, ORF1b, S, 3, 4a, 4b, 5, E, M and N [67], with sixteen predicted nonstructural proteins being encoded by ORF1a/b. Moreover, several unique group-specific ORFs that are not essential for virus replication are encoded by MERS-CoV [60]. The functions of these group-specific ORFs are unknown; however, by analogy to other coronaviruses, they may encode structural proteins or interferon antagonist genes [63]. Open reading frames ORF2, -6, -7 and -8a are translated from subgenomic mRNAs predicted to encode the four canonical structural genes: a 180/90-kDa spike glycoprotein (S), a ∼23-kDa membrane glycoprotein (M), a small envelope protein (E) and a ∼50-kDa nucleocapsid protein (N), respectively [67].
The spike (S) glycoprotein of coronaviruses is a heavily glycosylated, petal-shaped, large protein on the surface of the virion. The S glycoprotein is cleaved by a cellular protease into S1 and S2 subunits, which remain noncovalently on the viral envelop prior to fusion activation [26, 35]. S proteins of the alphacoronavirus group are not cleaved [18]; however, cleavage of MERS-CoV-S protein by trypsin, TMPRSS2 or -4 or cathepsin L is required to activate the membrane fusion activity of S, leading to virus entry and syncytium formation [54]. The S1 protein forms the globular portion of the spikes and possesses sequences that are responsible for receptor binding on the surface of host cells. S1 sequences are variable, with various degrees of deletion and substitutions among different coronavirus strains or isolates. Mutations in S1 sequences have been associated with altered antigenicity and pathogenicity of the virus [4, 39]. Meanwhile, S2 sequences constitute the stalk of the spikes and are less variable [17]. Recently, analysis of the MERS-CoV genomes has revealed the expected accumulation of genetic diversity, including changes in the S protein [15].
MERS-CoV receptor
Dipeptidyl peptidase 4 (DPP4) (or so-called CD26) is a 766-aa type II transmembrane glycoprotein that has been shown to be the functional receptor for MERS-CoV in permissive cell lines [56]. Moreover, non-permissive cell lines become susceptible to virus infection upon the exogenous expression of this receptor. DPP4 is also expressed on the cell surface in the kidneys, lungs, small intestine, liver, parotid gland, spleen, testes, prostate and activated leukocytes [43, 46, 56], which explains the capacity of MERS-CoV to infect cells of these organs [56]. Interestingly, MERS-CoV can bind to bat DPP4 [56].
Clinical features
Globally, a total of 179 cases with 76 fatalities had been reported up to 21 January 2014. The distribution of the cases is as follows: two from Jordan, nine from Qatar, twelve from the United Arab Emirates, three from Tunisia, three from Oman, two from Kuwait, three from the UK, two from France and one from Italy, with the rest (n:142) coming from Saudi Arabia. Of the Saudi Arabian cases, three were from Jeddah, sixty one from Riyadh, one from Al-Qasim, one from Bisha, seven from Asir, five from Al-Taif, eight from Hafr El-Batin, seven from Medina, two from Al-Jouf, one from Wadi Al-Dawaser and forty six from the Eastern district [24, 52, 61, 75]. All index cases in Europe had a history of travel to the Middle East. The age range was from 2 to 94 years and the recorded case fatality was higher in males than in females: 52 and 24 % respectively [50]. In confirmed cases of MERS-CoV, individuals developed acute, serious respiratory tract disease. Of the first 47 cases in Saudi Arabia, 89 % required management in intensive care and 72 % required mechanical ventilation [2]. Typical symptoms have included fever, cough and breathing difficulties, myalgia, nausea, vomiting and diarrhoea. An earlier fatal case that was transferred from the UAE to Germany showed symptoms of rapid onset of non-productive cough, then pneumonia and thrombocytopaenia until day 14, when the patient suffered from renal insufficiency, requiring dialysis [23]. Renal insufficiency or failure has indeed been noted for many MERS patients; however, it is unclear whether this is due to the infection or is a side-effect of anti-microbial agents and other medications used in treatment, or indeed whether it possibly influences disease progression, or some combination of these factors. A cluster of cases in a hospital in Saudi Arabia was centred on a dialysis unit, with 13 of 25 probable and confirmed cases involving end-stage renal disease [3]. The number of asymptomatic cases in which only mild symptoms were experienced has increased recently. Fifteen mild cases have been identified through contact tracing in late June and July 2013, including four health-care workers in Saudi Arabia [71, 76]. Two out of three Tunisian patients have also suffered from a mild version of the disease, with complete recovery [75]. To date, 46 subclincal cases of MERS-CoV were reported worldwide [75]. Severe cases of MERS-CoV infection have tended to be recorded in older patients with underlying conditions, while milder cases have been recorded in younger persons who experienced contact infection. Underlying comorbidity included diabetes, chronic renal disease, chronic heart disease, hypertension, chronic pulmonary disease [2], malignancy [30] and receiving a renal transplant [28]. Co-infection with other pathogens, including influenza A virus, parainfluenza virus, herpes simplex virus and pneumococcus, has been reported [74]. It can be speculated that co-infections serve to aggravate the clinical consequences of MERS infection.
Virus morbidity
Coronaviruses spread very rapidly and effectively among the susceptible hosts, and MERS-CoV morbidity is assumed to be higher than recorded. It is possible that MERS-CoV induces mild to moderate infections in healthy people; however, severe disease occurs in immunocompromised patients. This assumption can be discounted or confirmed once virological and/or serological diagnostic tests specific for MERS-CoV are adopted routinely. The first cases of MERS-CoV infection outside the Arabian Peninsula have usually been associated with a history of travel to those countries where there is no evidence of contact with laboratory-confirmed infected patients. The only logical explanation for the widespread occurrence of the MERS-CoV virus in these countries is that MERS-CoV can induce mild to moderate disease across a widespread epidemiological base that does not necessitate hospitalization, as speculated above. Interestingly, in terms of testing for the disease, in the case of one patient, false negative result was recorded from nasopharyngeal swab, even though positive result was obtained in a pulmonary lavage sample from the same patient [58]. Lower respiratory tract samples are therefore to be recommended whenever possible and nasopharyngeal samples should only be used when lower respiratory tract samples are not available.
Virus tracking and natural reservoirs
The rapid identification of the causative agent represents the basis for an effective response for disease control. The fact that respiratory diseases rarely result from a single entity makes the rapid identification of novel causative agents complicated; however, in the case of MERS-CoV infection, confusion persisted until September 2012, when the virus in a Saudi patient was identified as a previously unknown coronavirus [77].
The role of bats
Bats have been found to be the reservoir hosts of dozens of viruses, including severe acute respiratory syndrome coronaviruses (SARS-CoVs), henipaviruses, lyssaviruses, filoviruses, adenoviruses, picornaviruses and herpesviruses [1, 7, 12, 27, 36, 38, 40, 41, 62, 70]. Such viruses have existed in bats for a long time, and the question, therefore, is why they suddenly jumped to human hosts. The virus might have been transmitted to humans in ancient times but has only been discovered recently. It is more likely, however, that the jump to human hosts can be attributed to changes in human lifestyle. Thus, one potential future plan to restrict infection would be to limit direct and indirect contact between bats and humans. In the meantime, understanding these routes of transmission may help us to prevent future outbreaks of known and/or currently unknown diseases that leap from bats to humans directly or via intermediate animal amplifiers. Although the natural reservoir of MERS-CoV has not yet been identified, its high sequence similarity to bat viruses is suggestive of an origin in bats. A MERS-CoV sequence identical to that of the virus isolated from an index-case patient in Bisha 2c EMC/2012 was detected in a faecal pellet from Taphozous perforatus. The bat sample was collected <12 km from the home of the index case in an abandoned date palm orchard and <1 km from his place of employment. The research team observed roosting bats and guano in abandoned wells and ruins in the nearby areas [45]. Interestingly, Al-Ahsa is ranked among the most important date-palm-production cities in Saudi Arabia. It is likely that food or water of farm animals in areas with palm orchards may be contaminated with bat guano, saliva, and/or urine, and this may constitute an indirect mean of transmission to people. MERS-CoV-related viruses have not yet been detected in fruit bats. Meanwhile, drinking of raw sap is not known in the Arabian Peninsula, so the scenario of drinking raw date palm sap that has been identified as a risk factor for human Nipah virus infection [42, 55] is an unlikely possibility.
Possible animal reservoirs
In contrast to SARS, the cultural and feeding traditions in the Arabian Peninsula minimize the potential for an effective amplifying host. Normally, well-cooked meat, mainly from sheep, goats and camels, is used, thus excluding a wide variety of meat-borne pathogens. Animal slaughtering is routinely conducted under acceptable hygienic conditions, and meat is sold commercially in a chilled state. Although, there is no current evidence for the presence of MERS-CoV in camel milk, non-heat-treated raw camel milk is commonly consumed, especially in the Bedouin culture, and this may constitute a source of infection to humans. Recently, MERS-CoV neutralizing antibodies were detected in dromedary camels from Oman (100 %), the Canary Islands (14 %), and Egypt (94-98 %), which suggests the widespread infection of camelids with MERS-CoV or a closely related virus [51, 58]. This finding raises a concern about the role of camels in the transmission of the disease to humans. Consumption of raw milk and direct contact with camels could, therefore, be routes of virus transmission. More recently, the presence of MERS-CoV has been confirmed in three camels in a herd of 14 animals. Animals exhibited either no symptoms or only mild symptoms when samples were taken and remained so during the following 40 days [72].
In a few cases, in Saudi Arabia and the Arabian Peninsula, native citizens consume the meat of lizards of the genus Uromastyx after hunting, and this may constitute a form of animal contact that is unique to the Arabian Peninsula. Although these lizards are almost exclusively herbivorous, analysis of faeces from Uromastyx has revealed the presence of sheep droppings, date kernels and feathers [16]. Interestingly, the remains of invertebrates, vertebrates and stones have been detected in the faeces of Uromastyx aegyptia microlepis, which suggests an element of scavenging in the feeding behaviour of this type of lizard [8]. Potentially, therefore, Uromastyx could play a role in the ecology of MERS-CoV. In addition, baboons are present in some mountain areas in the Arabian Peninsula, and therefore, people living in these areas may have direct or indirect contact to baboons. The Eastern areas of Saudi Arabia, including the Al-Ahsa district, where a large proportion of cases have been reported, do not contain baboons, however. Of course, the presence of unknown amplifying hosts is a distinct possibility, and there is an urgent need for widespread seroprevalence and virological screening for MERS-CoV in different fauna present in the Arabian Peninsula in order to discover the amplifying host(s).
Experimental animal model
MERS-CoV has failed to replicate in Syrian hamsters following intratracheal inoculation and inoculation via aerosol [21], and the best-known current experimental model for MERS-CoV infection is the rhesus macaque [22, 47]. Experimental infection using a combination of intra-tracheal, nasal, oral and ocular routes (7 × 106 TCID50) resulted in a transient lower respiratory tract infection after 24 h [47]. Symptoms included elevated temperature, reduced appetite, increased respiration, piloerection, cough and a hunched posture. Histopathological assays showed multifocal to coalescent bright red lesions that developed to dark reddish purple areas of pulmonary inflammation in the lower respiratory tract [22, 47]. These were accompanied by fibrous adhesions, and edematous, atelectatic and consolidation areas in the lungs. MERS-CoV replicated mainly in the alveolar pneumocytes and resulted in interstitial pneumonia. The severity of the disease observed in humans may be explained by the ability of MERS-CoV to replicate in the lower respiratory tract [22]. Blood tests showed an early elevated number of white cells, with other values being within normal ranges, suggesting an asystemic but organ-specific infection. These clinical findings were corroborated by virological tests in which the virus was isolated from lung tissues and quantified by means of RT-PCR [47]. The rhesus macaque model therefore offers a powerful tool to study the full replication cycle of the virus as well as to develop efficacious antiviral drugs and vaccines.
Culture systems
MERS-CoV can replicate in different mammalian cell lines [10]. In humans, it can replicate in the respiratory tract (lung adenocarcinoma cell line A549, embryonic fibroblast cell line HFL and polarized airway epithelium cell line Calu-3), kidney (embryonic kidney cell line; HEK), liver cells (hepatocellular carcinoma cell line; Huh-7), and the intestinal tract (colorectal adenocarcinoma cell line; Caco-2) [10]. The range of human tissue tropism is broader than that for all other HCoVs, including SARS coronavirus, HKU1, 229E, OC43 and NL63. MERS-CoV can also infect cell lines originating from primates, pigs, bats, civet cats and rabbits [10].
Diagnosis
Viral loads were highest in samples from the lower respiratory tract (up to 1.2 × 106 copies per mL), followed by oronasal swabs (5370 copies per mL). Low concentrations of the virus were detected in urine and faecal samples, but there was no evidence of the virus in blood, suggesting a low infection risk during non-respiratory-care procedures [23]. A diagnostic rRT-PCR assay is now available for the detection of MERS-CoV [13, 14]. The rRT-PCR assays have been rigorously validated and are highly specific and sensitive. rRT-PCR assays for both upE and 1A can be used in combination as detection and confirmatory assays. Genotype sequencing-based assays for both the RNA-dependent RNA polymerase gene (RdRp) and the N gene have also been developed. Primers and probe sequences are listed in Table 2. A simplified and biologically safe protocol for the detection of antibody response by immunofluorescence microscopy has been developed using convalescent patient serum. A highly specific assay for the detection of antibodies against MERS-CoV using protein microarray technology has also been developed [57]. Serology provides valuable information on the rates of infection among populations and risk groups. A two-stage approach for detecting antibodies to recombinant MERS-CoV nucleocapsid (N) using ELISA, followed by a confirmatory test using either a whole-virus indirect fluorescent antibody (IFA) test or a microneutralization test, has been developed by CDC [73]. A safe and convenient pseudovirus-based inhibition assay has been developed to detect MERS-CoV neutralizing antibodies and to screen for viral-entry inhibitors [78].
Table 2.
Oligonucleotides sequence of the primers and probes used for MERS-CoV
| RT-PCR assay | Primers | Reference |
|---|---|---|
| upE gene assay | upE-Fwd-GCAACGCGCGATTCAGTT | [13] |
| upE-Rev-GCCTCTACACGGGACCCATA | ||
| FAM-CTCTTCACATAATCGCCCCGAGCTCG-TAMRA | ||
| ORF1b assay | ORF1b-Fwd-TTCGATGTTGAGGGTGCTCAT | [13] |
| ORF1b-Rev-TCACACCAGTTGAAAATCCTAATTG | ||
| FAM-CCCGTAATGCATGTGGCACCAATGT-TAMRA | ||
| ORF 1a assay | EMC-Orf1a-Fwd CCACTACTCCCATTTCGTCAG | [14] |
| EMC-Orf1a-Rev CAGTATGTGTAGTGCGCATATAAGCA | ||
| EMCOrf1a-Prb FAM-TTGCAAATTGGCTTGCCCCCACT-TAMRA | ||
| NSeq assaya | EMC-SeqN-Fwd CCTTCGGTACAGTGGAGCCA | [14] |
| EMC-SeqN-Rev GATGGGGTTGCCAAACACAAAC | ||
| EMC-SeqN-Fnest TGACCCAAAGAATCCCAACTAC | ||
| RdRpSeq assayb | RdRpSeq-Fwd TGCTATWAGTGCTAAGAATAGRGC | [14] |
| RdRpSeq-Rev GCATWGCNCWGTCACACTTAGG | ||
| RdRpSeq-Rnest CACTTAGGRTARTCCCAWCCCA |
The second round was only done if no product was visible by agarose gel electrophoresis after the first round
aFragment size: first round, 306-312 bp (including primers); second round, 279-285 bp (including primers)
bFragment size: first round, 242 bp (including primers); second round, 228 bp (including primers)
Antivirals
The treatment of MERS-CoV-infected human airway epithelium cultures with IFN of type I or III has been found to efficiently reduce viral replication. This finding supports the recommendation of IFN as a promising treatment for MERS-CoV [33]. Interestingly, MERS-CoV was found to be 50-100 times more sensitive to interferon-alpha (IFN-α) treatment than SARS-CoV. Cyclosporin A is another compound that inhibited the replication of MERS-CoV in cell cultures [20]. An in vitro assay showed that the p38 mitogen-activated protein kinase (MAPK) inhibitor SB203580 possesses a promising antiviral effect against MERS-HCoV [32]. SB203580 also showed effective antiviral effects against encephalomyocarditis virus [31], respiratory syncytial virus [59] and HIV [34]. A combination of ribavirin and IFN-α2b should be further evaluated for the possible management of MERS-CoV, since treatment of MERS-CoV-infected rhesus macaques with a combination of ribavirin and IFN-α2b reduced virus replication and improved clinical outcomes [25]. IFN-β1b with mycophenolic acid was also found to be a successful combination that possessed antiviral activity against MERS-CoV [11]. An important question is whether MERS-CoV-infected patients who were on diabetes treatment with DPP4 inhibitors could eliminate the virus due to the inhibition of MERS-CoV S1 protein binding to the DPP4 receptor, which consequently prevents viral entry into the target cells. Monoclonal antibodies (mAbs) against human CD26 derived from clone 2F9 had neutralizing activity against MERS-CoV. The potential values of these mAbs in MERS therapy, especially of the humanized mAb YS110 used in a phase 1 cancer treatment study, should therefore be subjected to further evaluation [48].
Conclusion and recommendations
Lessons learnt from the SARS outbreak were able to be applied when MERS-CoV emerged in the Middle East during 2012-2013. As a result of rapid data sharing, a reliable and powerful diagnostic of MERS-CoV became available worldwide within just a few months of the virus’s first isolation and identification. Like SARS CoV, MERS-CoV is closely related to bat viruses with an as yet unknown reservoir. The high prevalence of camels that are seropositive for MERS-CoV, or a closely related virus, and the isolation of MERS-CoV from camels are suggestive of a possible amplifying host, and therefore, further studies to monitor virus infections in camels are highly recommended in order to assess their potential role in MERS-CoV virus transmission. The transmission of the MERS-CoV to humans in general needs further study to clarify the role of the amplifying host and whether the virus is transmitted from bats to camels and then to humans, or whether other amplifying hosts are involved. The factors associated with the increased virulence and mortality rate in men compared to women should also be investigated. However, in the Arab countries, people usually try to avoid hospitalisation, which is more frequent for adults than children, and for females more than males. Determinants of host susceptibility to MERS-CoV should be carefully and intensively studied, and whether this depends on the presence of specific receptors or whether there are other host metabolic determinants. An important question that requires large and comprehensive serological surveillance is whether MERS-CoV was introduced to and circulated in the general population in the countries of the Middle East, or possibly other countries worldwide, prior to the first reported cases. Many reliable assays have been developed that allow serological screening and surveillance should be conducted on a wide scale among humans and different animal species to determine the exact rate of spread of MERS-CoV.
Conflict of interest
The author has no competing interest.
References
- 1.Annan A, Baldwin HJ, Corman VM, Klose SM, Owusu M, Nkrumah EE, Badu EK, Anti P, Agbenyega O, Meyer B, Oppong S, Sarkodie YA, Kalko EK, Lina PH, Godlevska EV, Reusken C, Seebens A, Gloza-Rausch F, Vallo P, Tschapka M, Drosten C, Drexler JF. Human Betacoronavirus 2c EMC/2012-related Viruses in Bats, Ghana and Europe. Emerg Infect Dis. 2013;3:456–459. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, Al-Rabiah FA, Al-Hajjar S, Al-Barrak A, Flemban H, Al-Nassir WN, Balkhy HH, Al-Hakeem RF, Makhdoom HQ, Zumla AI, Memish ZA. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis. 2013;13:752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Assiri A, McGeer A, Perl TM, Price CS, Al Rabeeah AA, Cummings DA, Alabdullatif ZN, Assad M, Almulhim A, Makhdoom H, Madani H, Alhakeem R, Al-Tawfiq JA, Cotten M, Watson SJ, Kellam P, Zumla AI, Memish ZA, KSA MERS-CoV Investigation Team, Hospital outbreak of Middle East respiratory syndrome coronavirus. N Engl J Med. 2013;369:407–416. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballesteros ML, Sanchez CM, Enjuanes L. Two amino acid changes at the N-terminus of transmissible gastroenteritis coronavirus spike protein result in the loss of enteric tropism. Virology. 1997;227:378–388. doi: 10.1006/viro.1996.8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bermingham A, Chand MA, Brown CS, Aarons E, Tong C, Langrish C, Hoschler K, Brown K, Galiano M, Myers R, Pebody RG, Green HK, Boddington NL, Gopal R, Price N, Newsholme W, Drosten C, Fouchier RA, Zambon M (2012) Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill 17. pii: 20290 [PubMed]
- 6.Butler D. Receptor for new coronavirus found. Nature. 2013;495:149–150. doi: 10.1038/495149a. [DOI] [PubMed] [Google Scholar]
- 7.Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev. 2006;19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castilla AM, Richer R, Herrel A, Conkey AAT, Tribuna J, Al-Thani M. First evidence of scavenging behaviour in the herbivorous lizard Uromastyx aegyptia microlepis. J Arid Environ. 2011;75:671–673. doi: 10.1016/j.jaridenv.2011.02.005. [DOI] [Google Scholar]
- 9.Chan-Yeung M, Xu RH. SARS: epidemiology. Respirol. 2003;8:S9–S14. doi: 10.1046/j.1440-1843.2003.00518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan JF, Chan KH, Choi GK, To KK, Tse H, Cai JP, Yeung ML, Cheng VC, Chen H, Che XY, Lau SK, Woo PC, Yuen KY. Differential cell line susceptibility to the emerging novel human betacoronavirus 2c EMC/2012: implications for disease pathogenesis and clinical manifestation. J Infect Dis. 2013;207:1743–1752. doi: 10.1093/infdis/jit123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan JF, Chan KH, Kao RY, To KK, Zheng BJ, Li CP, Li PT, Dai J, Mok FK, Chen H, Hayden FG, Yuen KY (2013) Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J Infect 13. pii: S0163-4453 [DOI] [PMC free article] [PubMed]
- 12.Chu DK, Poon LL, Guan Y, Peiris JS. Novel astroviruses in insectivorous bats. J Virol. 2008;82:9107–9114. doi: 10.1128/JVI.00857-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corman VM, Eckerle I, Bleicker T, Zaki A, Landt O, Eschbach-Bludau M, van Boheemen S, Gopal R, Ballhause M, Bestebroer TM, Muth D, Müller MA, Drexler JF, Zambon M, Osterhaus AD, Fouchier RM, Drosten C (2012) Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill 17. pii: 20285 [DOI] [PubMed]
- 14.Corman VM, Müller MA, Costabel U, Timm J, Binger T, Meyer B, Kreher P, Lattwein E, Eschbach-Bludau M, Nitsche A, Bleicker T, Landt O, Schweiger B, Drexler JF, Osterhaus AD, Haagmans BL, Dittmer U, Bonin F, Wolff T, Drosten C (2012) Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Euro Surveill 17. pii: 20334 [DOI] [PubMed]
- 15.Cotten M, Watson SJ, Kellam P, Al-Rabeeah AA, Makhdoom HQ, Assiri A, Al-Tawfiq JA, Alhakeem RF, Madani H, Alrabiah FA, Hajjar SA, Al-Nassir WN, Albarrak A, Flemban H, Balkhy HH, Alsubaie S, Palser AL, Gall A, Bashford-Rogers R, Rambaut A, Zumla AI, Memish ZA. Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: a descriptive genomic study. Lancet. 2013;382:1993–2002. doi: 10.1016/S0140-6736(13)61887-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cunningham PL. Notes on the diet, survival rate, and burrow specifics of Uromastyx aegyptius microlepis from the United Arab Emirates. Asiatic Herpetol Res. 2001;9:30–33. [Google Scholar]
- 17.de Groot RJ, Luytjes W, Horzinek MC, van der Zeijst BA, Spaan WJ, Lenstra JA. Evidence for a coiled-coil structure in the spike proteins of coronaviruses. J Mol Biol. 1987;196:963–966. doi: 10.1016/0022-2836(87)90422-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Groot RJ, Van Leen RW, Dalderup MJ, Vennema H, Horzinek MC, Spaan WJ. Stably expressed FIPV peplomer protein induces cell fusion and elicits neutralizing antibodies in mice. Virology. 1989;171:493–502. doi: 10.1016/0042-6822(89)90619-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Groot RJ, Baker SC, Baric RS, Brown CS, Drosten C, Enjuanes L, Fouchier RA, Galiano M, Gorbalenya AE, Memish Z, Perlman S, Poon LL, Snijder EJ, Stephens GM, Woo PC, Zaki AM, Zambon M, Ziebuhr J. Middle East respiratory syndrome coronavirus (MERS-CoV); announcement of the coronavirus study group. J Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Wilde AH, Ray VS, Oudshoorn D, Bestebroer TM, van Nieuwkoop S, Limpens RW, Posthuma CC, van der Meer Y, Bárcena M, Haagmans BL, Snijder EJ, van den Hoogen BG. Human coronavirus-EMC replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-alpha treatment. J Gen Virol. 2013;94:1749–1760. doi: 10.1099/vir.0.052910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Wit E, Prescott J, Baseler L, Bushmaker T, Thomas T, Lackemeyer MG, Martellaro C, Milne-Price S, Haddock E, Haagmans BL, Feldmann H, Munster VJ. The Middle East respiratory syndrome coronavirus (MERS-CoV) does not replicate in Syrian hamsters. PLoS One. 2013;8:e69127. doi: 10.1371/journal.pone.0069127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Wit E, Rasmussen AL, Falzarano D, Bushmaker T, Feldmann F, Brining DL, Fischer ER, Martellaro C, Okumura A, Chang J, Scott D, Benecke AG, Katze MG, Feldmann H, Munster VJ. Middle East respiratory syndrome coronavirus (MERS-CoV) causes transient lower respiratory tract infection in rhesus macaques. Proc Natl Acad Sci USA. 2013;110:16598–16603. doi: 10.1073/pnas.1310744110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drosten C, Seilmaier M, Corman VM, Hartmann W, Scheible G, Sack S, Guggemos W, Kallies R, Muth D, Junglen S, Müller MA, Haas W, Guberina H, Röhnisch T, Schmid-Wendtner M, Aldabbagh S, Dittmer U, Gold H, Graf P, Bonin F, Rambaut A, Wendtner CM. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect. 2013;13:745–751. doi: 10.1016/S1473-3099(13)70154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.ECDC (2013) European Centre for Disease Prevention and Control. Rapid Risk Assessment: Severe respiratory disease associated with Middle East respiratory syndrome coronavirus (MERS-CoV), 6 November 2013
- 25.Falzarano D, de Wit E, Rasmussen AL, Feldmann F, Okumura A, Scott DP, Brining D, Bushmaker T, Martellaro C, Baseler L, Benecke AG, Katze MG, Munster VJ, Feldmann H. Treatment with interferon-α2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nat Med. 2013;19:1313–1317. doi: 10.1038/nm.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frana MF, Behnke JN, Sturman LS, Holmes KV. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: host-dependent differences in proteolytic cleavage and cell fusion. J Virol. 1985;56:912–920. doi: 10.1128/jvi.56.3.912-920.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freuling CM, Beer M, Conraths FJ, Finke S, Hoffmann B, Keller B, Kliemt J, Mettenleiter TC, Mühlbach E, Teifke JP, Wohlsein P, Müller T. Novel lyssavirus in Natterer’s bat, Germany. Emerg Infect Dis. 2011;17:1519–1522. doi: 10.3201/eid1708.110201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guery B, Poissy J, el Mansouf L, Séjourné C, Ettahar N, Lemaire X, Vuotto F, Goffard A, Behillil S, Enouf V, Caro V, Mailles A, Che D, Manuguerra JC, Mathieu D, Fontanet A, van der Werf S, group M-Cs Clinical features and viral diagnosis of two cases of infection with Middle East respiratory syndrome coronavirus: a report of nosocomial transmission. Lancet. 2013;381:2265–2272. doi: 10.1016/S0140-6736(13)60982-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamre D, Procknow JJ. A new virus isolated from human respiratory tract. Proc Soc Exp Biol Med. 1966;121:190–193. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- 30.Health Protection Agency (HPA) UK Novel Coronavirus Investigation team Evidence of person-to-person transmission within a family cluster of novel coronavirus infections, United Kingdom, February 2013. Euro Surveill. 2013;18:20427. doi: 10.2807/ese.18.11.20427-en. [DOI] [PubMed] [Google Scholar]
- 31.Hirasawa K, Kim A, Han HS, Han J, Jun HS, Yoon JW. Effect of p38 mitogen-activated protein kinase on the replication of encephalomyocarditis virus. J Virol. 2003;77:5649–5656. doi: 10.1128/JVI.77.10.5649-5656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Josset L, Menachery VD, Gralinski LE, Agnihothram S, Sova P, Carter VS, Yount BL, Graham RL, Baric RS, Katze MG. Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. MBio. 2013;4:e00165-00113. doi: 10.1128/mBio.00165-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kindler E, Jónsdóttir HR, Muth D, Hamming OJ, Hartmann R, Rodriguez R, Geffers R, Fouchier RA, Drosten C, Müller MA, Dijkman R, Thiel V. Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential. MBio. 2013;4:e00611–e00612. doi: 10.1128/mBio.00611-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar S, Orsini MJ, Lee JC, McDonnell PC, Debouck C, Young PR. Activation of the HIV-1 long terminal repeat by cytokines and environmental stress requires an active CSBP/p38 MAP kinase. J Biol Chem. 1996;271:30864–30869. doi: 10.1074/jbc.271.48.30864. [DOI] [PubMed] [Google Scholar]
- 35.Lai MMC, Perlman S, Anderson LJ (2007) Coronaviridae. In: Knipe DM, Howley PM, Griffin DE, Martin MA, Lamb RA, Roizman B, Straus SE (eds) Fields of virology, 5th edn. Lippincott Williams & Wilkins, USA, pp 1305–1336
- 36.Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, Wong BH, Wong SS, Leung SY, Chan KH, Yuen KY. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lau SK, Li KS, Huang Y, Shek CT, Tse H, Wang M, Choi GK, Xu H, Lam CS, Guo R, Chan KH, Zheng BJ, Woo PC, Yuen KY. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. J Virol. 2010;84:2808–2819. doi: 10.1128/JVI.02219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lau SK, Woo PC, Lai KK, Huang Y, Yip CC, Shek CT, Lee P, Lam CS, Chan KH, Yuen KY. Complete genome analysis of three novel picornaviruses from diverse bat species. J Virol. 2011;85:8819–8828. doi: 10.1128/JVI.02364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leparc-Goffart I, Hingley ST, Chua MM, Jiang X, Lavi E, Weiss SR. Altered pathogenesis of a mutant of the murine coronavirus MHV-A59 is associated with a Q159L amino acid substitution in the spike protein. Virology. 1997;239:1–10. doi: 10.1006/viro.1997.8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, Délicat A, Paweska JT, Gonzalez JP, Swanepoel R. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- 41.Li Y, Ge X, Zhang H, Zhou P, Zhu Y, Zhang Y, Yuan J, Wang LF, Shi Z. Host range, prevalence, and genetic diversity of adenoviruses in bats. J Virol. 2010;84:3889–3897. doi: 10.1128/JVI.02497-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luby SP, Rahman M, Hossain MJ, Blum LS, Husain MM, Gurley E. Foodborne transmission of Nipah virus, Bangladesh. Emerg Infect Dis. 2006;12:1888–1894. doi: 10.3201/eid1212.060732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mamgain S, Mathur S, Kothiyal P. Immunomodulatory activity of DPP4. J Pharmacol Clin Toxicol. 2013;1:1006. [Google Scholar]
- 44.McIntosh K, Kapikian AZ, Hardison KA, Hartley JW, Chanock RM. Antigenic relationships among the coronaviruses of man and between human and animal coronaviruses. J Immunol. 1969;102:1109–1118. [PubMed] [Google Scholar]
- 45.Memish ZA, Mishra N, Olival KJ, Fagbo SF, Kapoor V, Epstein JH, Alhakeem R, Durosinloun A, Al Asmari M, Islam A, Kapoor A, Briese T, Daszak P, Al Rabeeah AA, Lipkin WI. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg Infect Dis. 2013;19:1819–1823. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mentlein R. Dipeptidyl-peptidase IV (CD26)—role in the inactivation of regulatory peptides. Regul Pept. 1999;85:9–24. doi: 10.1016/S0167-0115(99)00089-0. [DOI] [PubMed] [Google Scholar]
- 47.Munster VJ, de Wit E, Feldmann H. Pneumonia from human coronavirus in a macaque model. N Engl J Med. 2013;368:1560–1562. doi: 10.1056/NEJMc1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohnuma K, Haagmans BL, Hatano R, Raj VS, Mou H, Iwata S, Dang NH, Jan Bosch B, Morimoto C. Inhibition of Middle East respiratory syndrome coronavirus infection by anti-CD26 monoclonal antibody. J Virol. 2013;87:13892. doi: 10.1128/JVI.02448-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pensaert MB, Debouck P, Reynolds DJ. An immunoelectron microscopic and immunofluorescent study on the antigenic relationship between the coronavirus-like agent, CV 777, and several coronaviruses. Arch Virol. 1981;68:45–52. doi: 10.1007/BF01315166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Penttinen P, Kaasik-Aaslav K, Friaux A, Donachie A, Sudre B, Amato-Gauci A, Memish Z, Coulombier D (2013) Euro Surveill 18. pii: 20596 [DOI] [PubMed]
- 51.Perera RA, Wang P, Gomaa MR, El-Shesheny R, Kandeil A, Bagato O, Siu LY, Shehata MM, Kayed AS, Moatasim Y, Li M, Poon LL, Guan Y, Webby RJ, Ali MA, Peiris JS, Kayali G (2013) Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt. Euro Surveill 18. pii: 20574 [DOI] [PubMed]
- 52.ProMed (2013) ProMed. http://healthmap.org/promed/
- 53.Pyrc K, Berkhout B, van der Hoek L. The novel human coronaviruses NL63 and HKU1. J Virol. 2007;81:3051–3057. doi: 10.1128/JVI.01466-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qian Z, Dominguez SR, Holmes KV. Role of the spike glycoprotein of human Middle East respiratory syndrome coronavirus (MERS-CoV) in virus entry and syncytia formation. PLoS One. 2013;8:e76469. doi: 10.1371/journal.pone.0076469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rahman MA, Hossain MJ, Sultana S, Homaira N, Khan SU, Rahman M. Date palm sap linked to Nipah virus outbreak in Bangladesh, 2008. Vector Borne Zoonotic Dis. 2012;12:65–72. doi: 10.1089/vbz.2011.0656. [DOI] [PubMed] [Google Scholar]
- 56.Raj VS, Mou H, Smits SL, Dekkers DH, Müller MA, Dijkman R, Muth D, Demmers JA, Zaki A, Fouchier RA, Thiel V, Drosten C, Rottier PJ, Osterhaus AD, Bosch BJ, Haagmans BL. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reusken C, Mou H, Godeke GJ, van der Hoek L, Meyer B, Müller MA, Haagmans B, de Sousa R, Schuurman N, Dittmer U, Rottier P, Osterhaus A, Drosten C, Bosch BJ, Koopmans M. Specific serology for emerging human coronaviruses by protein microarray. Euro Surveill. 2013;10:20441. doi: 10.2807/1560-7917.es2013.18.14.20441. [DOI] [PubMed] [Google Scholar]
- 58.Reusken CB, Haagmans BL, Müller MA, Gutierrez C, Godeke GJ, Meyer B, Muth D, Raj VS, Vries LS, Corman VM, Drexler JF, Smits SL, El Tahir YE, De Sousa R, van Beek J, Nowotny N, van Maanen K, Hidalgo-Hermoso E, Bosch BJ, Rottier P, Osterhaus A, Gortázar-Schmidt C, Drosten C, Koopmans MP. Respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis. 2013;3:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rixon HW, Brown G, Murray JT, Sugrue RJ. The respiratory syncytial virus small hydrophobic protein is phosphorylated via a mitogen-activated protein kinase p38-dependent tyrosine kinase activity during virus infection. J Gen Virol. 2005;86:375–384. doi: 10.1099/vir.0.80563-0. [DOI] [PubMed] [Google Scholar]
- 60.Scobey T, Yount BL, Sims AC, Donaldson EF, Agnihothram SS, Menachery VD, Graham RL, Swanstrom J, Bove PF, Kim JD, Grego S, Randell SH, Baric RS. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc Natl Acad Sci USA. 2013;110:16157–16162. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.SMH (2013) Saudi ministry of health: novel coronavirus. http://www.moh.gov.sa/en/CoronaNew/PressReleases/Pages/default.aspx
- 62.To KK, Hung IF, Chan JF, Yuen KY. From SARS coronavirus to novel animal and human coronaviruses. J Thorac Dis. 2013;5:S103–S108. doi: 10.3978/j.issn.2072-1439.2013.06.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Totura AL, Baric RS. SARS coronavirus pathogenesis: Host innate immune responses and viral antagonism of interferon. Curr Opin Virol. 2012;2:264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tyrrell DA, Bynoe ML. Cultivation of viruses from a high proportion of patients with colds. Lancet. 1966;1:76–77. doi: 10.1016/S0140-6736(66)92364-6. [DOI] [PubMed] [Google Scholar]
- 65.Vabret A, Dina J, Gouarin S, Petitjean J, Corbet S, Freymuth F. Detection of the new human coronavirus HKU1: A report of 6 cases. Clin Infect Dis. 2006;42:634–639. doi: 10.1086/500136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vabret A, Dina J, Gouarin S, Tripey V, Brouard J, Freymuth F. Human (non-severe acute respiratory syndrome) coronavirus infections in hospitalised children in France. J Paed Child Health. 2008;44:176–181. doi: 10.1111/j.1440-1754.2007.01246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, Osterhaus AD, Haagmans BL, Gorbalenya AE, Snijder EJ, Fouchier RA (2012) Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio 3. pii: e00473-00412 [DOI] [PMC free article] [PubMed]
- 68.van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, Osterhaus ADME, Haagmans BL, Gorbalenya AE, Snijder EJ, Fouchier RAM (2012) Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. Mbio 3. pii: e00473-00412 [DOI] [PMC free article] [PubMed]
- 69.Vijgen L, Keyaerts E, Moës E, Thoelen I, Wollants E, Lemey P, Vandamme AM, Van Ranst M. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J Virol. 2005;3:1595–1604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Watanabe S, Maeda K, Suzuki K, Ueda N, Iha K, Taniguchi S, Shimoda H, Kato K, Yoshikawa Y, Morikawa S, Kurane I, Akashi H, Mizutani T. Novel betaherpesvirus in bats. Emerg Infect Dis. 2010;16:986–988. doi: 10.3201/eid1606.091567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.WHO (2013) World Health Organization. Middle East respiratory syndrome coronavirus (MERS-CoV)—updates 23 September 2012 to 28 August 2013. WHO 28 August 2013
- 72.WHO (2013) World Health Organization. Middle East respiratory syndrome coronavirus (MERS-CoV)—update http://www.who.int/csr/don/2013_11_29/en/index.html. WHO [DOI] [PMC free article] [PubMed]
- 73.WHO (2013) World Health Organization. Laboratory testing for Middle East respiratory syndrome coronavirus interim recommendations
- 74.WHO . WHO guidelines for investigation of cases of human infection with Middle East respiratory syndrome coronavirus (MERS-CoV) UK: WHO; 2013. pp. 1–22. [Google Scholar]
- 75.WHO (2013) World Health Organization. Global alert and response: coronavirus infections. http://www.who.int/csr/don/archive/disease/coronavirus_infections/en/index.html
- 76.WHO (2013) World Health Organization. Middle East Respiratory Syndrome Coronavirus (MERS-CoV) - summary and literature update as of 9 July 2013. WHO 9 July 2013
- 77.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 78.Zhao G, Du L, Ma C, Li Y, Li L, Poon VK, Wang L, Yu F, Zheng BJ, Jiang S, Zhou Y. A safe and convenient pseudovirus-based inhibition assay to detect neutralizing antibodies and screen for viral entry inhibitors against the novel human coronavirus MERS-CoV. Virol J. 2013;10:266. doi: 10.1186/1743-422X-10-266. [DOI] [PMC free article] [PubMed] [Google Scholar]


