Abstract
Sugarcane Bacilliform Guadeloupe A Virus (SCBGAV, genus Badnavirus, family Caulimoviridae) is an emerging, deleterious pathogen of sugarcane which presents a substantial barrier to producing high sugarcane earnings. Sugarcane bacilliform viruses (SCBVs) are one of the main species that infect sugarcane. During the last 30 years, significant genetic changes in SCBV strains have been observed with a high risk of disease incidence associated with crop damage. SCBV infection may lead to significant losses in biomass production in susceptible sugarcane cultivars. The circular, double-stranded (ds) DNA genome of SCBGAV (7.4 Kb) is composed of three open reading frames (ORFs) on the positive strand that replicate by a reverse transcriptase. SCBGAV can infect sugarcane in a semipersistent manner via the insect vectors sugarcane mealybug species. In the current study, we used miRNA target prediction algorithms to identify and comprehensively analyze the genome-wide sugarcane (Saccharum officinarum L.)-encoded microRNA (miRNA) targets against the SCBGAV. Mature miRNA target sequences were retrieved from the miRBase (miRNA database) and were further analyzed for hybridization to the SCBGAV genome. Multiple computational approaches—including miRNA-target seed pairing, multiple target positions, minimum free energy, target site accessibility, maximum complementarity, pattern recognition and minimum folding energy for attachments—were considered by all algorithms. Among them, sof-miR396 was identified as the top effective candidate, capable of targeting the vital ORF3 of the SCBGAV genome. miRanda, RNA22 and RNAhybrid algorithms predicted hybridization of sof-miR396 at common locus position 3394. The predicted sugarcane miRNAs against viral mRNA targets possess antiviral activities, leading to translational inhibition by mRNA cleavage. Interaction network of sugarcane-encoded miRNAs with SCBGAV genes, created using Circos, allow analyze new targets. The finding of the present study acts as a first step towards the creation of SCBGAV-resistant sugarcane through the expression of the identified miRNAs.
Keywords: Computational algorithms, R language, miRNA, Saccharum officinarum, Sugarcane Bacilliform Guadeloupe A Virus, Target prediction, Gene silencing, miRanda, Virus-host interaction, RNA interference
Introduction
Sugarcane bacilliform virus (SCBV, genus: Badnavirus, family Caulimoviridae) is a circular, non-enveloped bacilliform, monopartite DNA virus that harbors about 7.5 kb double-stranded (ds) DNA with three open reading frames (ORFs) (Sun et al., 2016). SCBV was first reported in the sugarcane cultivar (B34104) in the South American continent (Autrey et al., 1992) and disseminated into many sugarcane (Saccharum officinarum L.)-growing regions around the globe (Autrey et al., 1992). SCBV is implicated in yield losses in sugarcane cultivars. SCBV infection may lead to significant losses in biomass production in susceptible sugarcane cultivars (Lockhart & Autrey, 2000). SCBV infect sugarcane cultivars utilized for sugar and bioenergy production and constitute a major threat to the global exchange of sugarcane germplasm (Borah et al., 2013). Variable follicle symptoms were observed in some sugarcane varieties showing severe chlorotic mottling, stunted growth and pronounced freckle (Lockhart, Ireyt & Comstock, 1996; Viswanathan, Alexander & Garg, 1996). SCBV has wide host range. It comprises members of plant families Poaceae (sugarcane, johnsongrass, rice, sorghum, Panicum maximum, Rottboellia exaltata and Brachiaraia sp.) and Musaceae (bananas) (Borah et al., 2013; Lockhart & Autrey, 2000; Lockhart, Ireyt & Comstock, 1996). SCBVs are highly endemic in S. officinarum germplasm. These viruses can also infect commercial sugarcane (Saccharum interspecific hybrids), S. barberi, S. robustum, S. spontaneum and S. sinensis (Lockhart, Ireyt & Comstock, 1996). SCBV is transmitted naturally by sap-sucking insect vector mealybug species (Saccharicoccus sacchari and Dysmicoccus boninsis) through vegetative cuttings. Agrobacterium-mediated inoculation is used to disseminate SCBV infection experimentally (Bouhida, Lockhart & Olszewski, 1993; Lockhart, Ireyt & Comstock, 1996; Viswanathan, Alexander & Garg, 1996). Previous studies have revealed that SCBV is a heterogeneous badnavirus species. SCBV exist in nature as a complex of viruses or distinct strains and genetic variants. Currently, four distinct SCBV species have been classified by the International Committee on Taxonomy of Viruses (ICTV). These are Sugarcane Bacilliform Guadeloupe A Virus (SCBGAV), Sugarcane Bacilliform Guadeloupe D Virus (SCBGDV) Sugarcane bacilliform MO virus (SCBMOV-MOR) and Sugarcane bacilliform IM virus (SCBIMV-QLD) originating from Guadeloupe, Morocco and Australia, respectively (Adams & Carstens, 2012; Adams et al., 2016; Bouhida, Lockhart & Olszewski, 1993; Geijskes et al., 2002; Muller et al., 2011).
SCBGAV was first identified as a sugarcane R570 hybrid variety in Guadeloupe in 2011 (Muller et al., 2011). SCBGAV was classified as a new species in the genus badnavirus under the SCBV-A group as assigned by ICTV (Adams et al., 2016). The disease symptoms appear in the form of chlorosis, leaf freckle and mottling. Some infected plants were also observed to exhibit no symptoms. Recently in China, SCBV-infection resulted in reduced sucrose content, juice, stalk weigh, purity and gravity in sugarcane plants (Ahmad et al., 2019).
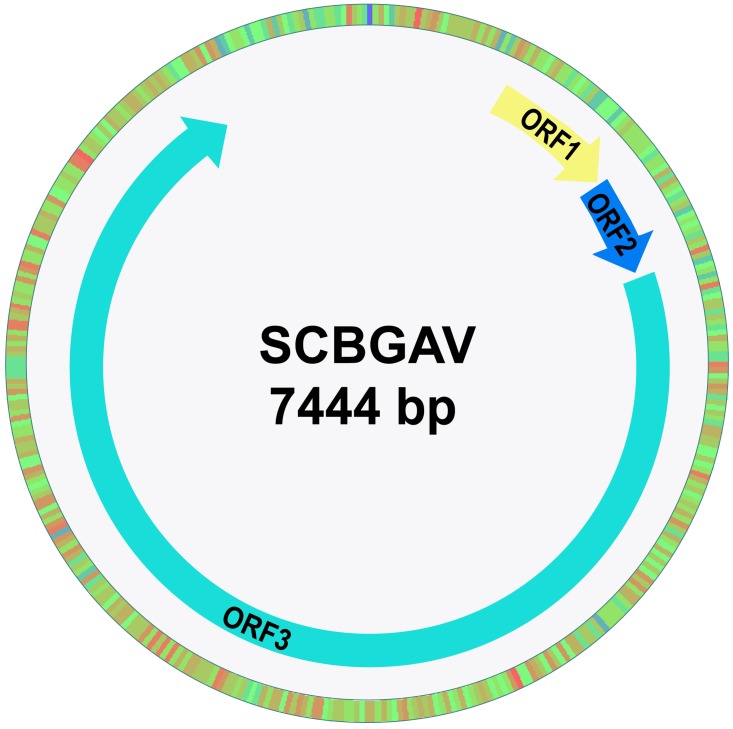
SCBGAV is a mealybug-transmitted monopartite badnavirus that infects sugarcane and is composed of a 7,444 bp circular, dsDNA genome containing three ORFs on the positive strand that replicate by a virus-encoded reverse transcriptase (RT). The two small proteins of sizes (176–185 and 122–135 amino acids), were encoded by the ORF1 and ORF2 respectively. The ORF3 was a key component of SCBGAV genome, encoded by a polyprotein (1,786–1,933 amino acids) (Muller et al., 2011).
Sugarcane (Saccharum officinarum L.) has inherited an active immunity that is composed of microRNAs (miRNAs) to combat infection. The miRNAs are a class of 21–23 nucleotide-long, endogenous, noncoding RNAs that govern gene regulation and expression at the post-transcriptional level. In plants, this mechanism occurs through the process of mRNA cleavage or translational repression. They are synthesized after the processing of hairpin miRNA precursors using an RNase-III-like enzyme (Dicer) to control gene expression (Brodersen & Voinnet, 2006).
RNA silencing, through miRNAs present in the host plant, thus imparts natural immunity and resistance to the host against foreign genetic elements including plant viruses (Carbonell & Daròs, 2019; Qu, Ye & Fang, 2007).
Viral miRNA degradation is the simplest molecular approach to combat viral infection in plants. Successful demonstration about changes in the nucleotide transcripts of mature miRNAs does not affect their biogenesis and the application of modified miRNAs to target Potyviruses in Arabidopsis was reported first time (Niu et al., 2006). Several studies have been successful in generating resistance against plant viruses using amiRNA-technology in crop plants such as tomato (Chen et al., 2019; Zhang et al., 2011) and cotton (Akmal, Baig & Khan, 2017; Ali et al., 2013). Most recently, amiRNA-mediated gene silencing has been demonstrated against Potexvirus and Tobamovirus successfully (Petchthai, Le Yee & Wong, 2018).
miRNA: mRNA target identification, validation and interactions are the basis for discerning the important functions of miRNAs in the greater perspective of regulatory network leading to different biotic developments. Computational algorithms for identification and analysis of miRNA targets enable the process of narrowing down possible target binding sites for experimental validation at transgenic level. In-silico strategies model how miRNAs target specific mRNAs and a number of algorithms are accessible, each with a diverse approach to miRNA target prediction and identification. Although, it might be valuable to have access to a variety of tools and algorithms with different capabilities, the user is challenged with a significant option in determining which algorithm/tool to apply (Peterson et al., 2014).
The development of artificial microRNAs (amiRNAs)-mediated gene silencing method offers a highly precise, targeted approach that gives several advantages over RNAi-mediated gene silencing strategy. Design, construction and validation of the amiRNAs depend upon the assembly of an endogenous precursor miRNA, which is substituted with a selected miRNA nucleotide transcript complementary to the target sequence (Niu et al., 2006; Ossowski, Schwab & Weigel, 2008).
The objective of the present research work is to predict sugarcane-encoded miRNAs that have the potential to develop resistance against badnaviruses, especially the SCBGAV. The major research objective was designed to identify the potential host-derived miRNAs in the SCBGAV genome and to screen the most promising miRNAs to understand the complex host-virus interactions. The novel amiRNA silencing method has been adopted for the first time to explore as a source of creating resistance to a monopartite badnavirus. The predicted miRNAs may provide help for the generation of SCBGAV-resistant sugarcane plants through genetic engineering in the future.
Materials & Methods
Retrieval of mature miRNAs of sugarcane
The sugarcane (Saccharum-officinarum L) and (Saccharum spp.) plant miRNA sequences were accessed from the microRNA database, miRBase (http://www.mirbase.org/cgi-bin/browse.pl/) (Griffiths-Jones et al., 2006; Griffiths-Jones et al., 2007; Kozomara, Birgaoanu & Griffiths-Jones, 2018; Kozomara & Griffiths-Jones, 2013). A total of 28 miRNAs were retrieved (Table S1).
SCBGAV genome retrieval and annotation
The nucleotide sequence of SCBGAV dsDNA genome (accession number NC_038382.1) composed of 7,444 bp was retrieved from the NCBI database. pDRAW32 plasmid map software (v 1.1.129) was used for annotation of genome features of SCBGAV (Fig. 1).
Figure 1. Genome organization of Sugarcane Bacilliform Guadeloupe A Virus.
Three ORFs are shown with arrows. The double stranded DNA genome of SCBGAV is composed of 7,444 bp.
miRNA target prediction in SCBGAV genome
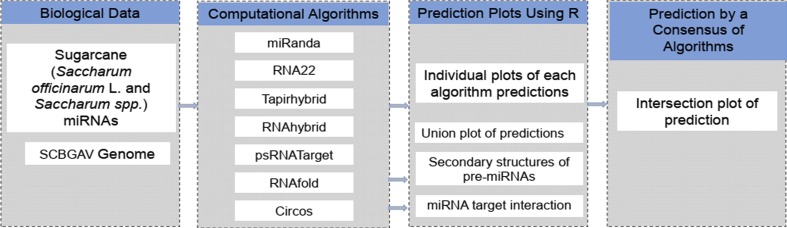
Four computational algorithms (Table 1)—named miRanda, RNA22, RNAhybrid and psRNATarget—were selected for the screening of mature miRNAs of sugarcane against the SCBGAV genome to identify the miRNAs target positions (Table 1). Nucleotide sequences of the SCBGAV genome and sugarcane miRNAs were recorded in FASTA format to process further using computational algorithms with defined parameters. A detailed workflow pipeline was designed to identify miRNA binding sites within the SCBGAV genome (Fig. 2).
Table 1. miRNA target prediction algorithms.
Characteristic features considered by miRNA target prediction algorithms under study.
| Algorithms | Main parameters | Availability |
|---|---|---|
| miRanda | Seed pairing, target site accessibility, multiple target sites and translation inhibition | Web server and source code (http://cbio.mskcc.org/miRNA2003/miranda.html) |
| RNA22 | Seed pairing, target site accessibility and multiple target sites | Only Web server (https://cm.jefferson.edu/rna22/Interactive/) |
| RNAhybrid | Seed pairing, target site accessibility, multiple target sites and translation inhibition | Web server and source code (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid) |
| psRNATarget | Target site accessibility, multiple target sites and translation inhibition | Only Web server (http://plantgrn.noble.org/psRNATarget/) |
| Tapirhybrid | Seed pairing, target site accessibility and multiple target sites | Web server and source code (http://bioinformatics.psb.ugent.be/webtools/tapir/) |
| Targetfinder | Seed pairing | Only source code |
| Target-align | Multiple target sites | Web server and source code (Xie & Zhang, 2010) |
Figure 2. A schematic pipeline depicting the strategy of miRNA prediction from SCBGAV genome.
A schematic representation of miRNA target prediction in SCBGAV genome pipeline. Biological data set contains the kinds of data obtained for this study from NCBI (badnavirus genome) and miRBase (miRNAs). Computational algorithm group enlists three kinds of tools for the prediction of miRNA, secondary structure and miRNA-target interaction. R language was used to make plots and select data using in-house scripts/codes.
miRanda
miRanda is a computational algorithm (John et al., 2004) used to predict and identify the potential plant genomic miRNA targets (Archak & Nagaraju, 2007). It was designed to consider the following algorithm properties: sequence complementarity, RNA-RNA duplex dimerization, free energy and cross-species evolutionary conservation of molecular target sites. These properties yield output to weigh all matches and mismatch scores for gap penalties and for base pairs. It also promotes the prediction of multiple miRNA target sites including the ones with imperfect binding in the seed region within the 3′ UTR of the target site thereby enhancing the specificity (Betel et al., 2008; Witkos, Koscianska & Krzyzosiak, 2011). miRanda software was downloaded from the source website (http://cbio.mskcc.org/miRNA2003/miranda.html), and the SCBGAV genome was assessed to determine whether there were any possible targets for sugarcane miRNAs. The miRanda algorithm was run after defining standard settings. These include: Gap Open Penalty = −9.0, Score Threshold = 130, Minimum free energy (MFE) threshold (= −15.00 kcal/mol). MFE is an important statistical parameter to screen potential targets.
RNA22
RNA22 is a novel diverse web server (http://cm.jefferson.edu/rna22v1.0/), designed to implement a pattern-based approach to detect potential miRNA target sites (Miranda et al., 2006). The minimum folding energy (MFE) is a key parameter to screen possible miRNA without a cross-species conservation filter (Loher & Rigoutsos, 2012). Specificity and sensitivity values were selected at 61% and 63%, respectively. The value of the seed size was selected as 7 with unpaired base 1 permitted inside the seed region. There was no limit set to the maximum number of G:U wobbles in the seed region. The paired bases with minimum number and MFE were selected at 12 and −13.50 kcal/mol, respectively.
Tapirhybrid
Tapirhybrid is a novel web server (http://bioinformatics.psb.ugent.be/webtools/tapir) designed for the prediction of plant microRNA targets, including microRNA target mimics, using a very fast and precise algorithm (Bonnet et al., 2010). It considers many important parameters including seed pairing, multiple target sites and target site accessibility. It is one of the most highly recommended tools for plant microRNA prediction due to its great accuracy (Srivastava et al., 2014). It was designed to run using the RNAhybrid algorithm for microRNA target prediction and produces miRNA target scores (on the basis of the number of mismatches), the number of GU pairs, gaps and mismatches in the seed region. It is regarded as a high precision algorithm to enhance the accuracy of the predicted results. The setting for input parameters were standardized as score < =8 and MFE_ratio > =0.5.
RNAhybrid
RNAhybrid is a novel algorithm used to predict hybridization of miRNA and mRNA that is based on MFE (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid). Site complementarity, target-site abundance and MFE are the unique parameters of RNAhybrid algorithm. MFE was set a threshold of −20 kcl/mol. RNAhybrid can also assign a p-value for the miRNA: miRNA interaction based on the number of binding sites within the 3′ UTR sequence, which is a measure of target-site abundance. The other filters remained fixed as default parameters (Krüger & Rehmsmeier, 2006).
psRNATarget
psRNATarget is a new web server (http://plantgrn.noble.org/psRNATarget/) that was developed to predict small RNA (sRNA) in plants. It was used to analyze complementary matching between target mRNA sequence and sRNA sequence based on a scoring schema (Dai, Zhuang & Zhao, 2018). Evaluation of target-site accessibility on the basis of UPE (unpaired energy) is another important feature of the psRNATarget algorithm. The standard parameter set for our analysis was as follows: penalty for (extending gap = 0.5, opening gap = 2, G.U pair = 1, other mismatches = 1), HSP size = 19, seed region = 2–7 nucleotides. The minimum expectation score was 7.0.
RNAfold algorithm
RNAfold algorithm was used to generate the stable secondary structure of pre-miRNAs from the precursors of the mature miRNAs (Lorenz et al., 2011). The parameters set for secondary structure prediction were: minimum free energy and partition function; avoid isolated base pairs and dangling energy on both sides of helix in any case; RNA parameters; rescale energy parameters at a given temperature 37C; interactive RNA secondary structure plot; RNA secondary structure plots with reliability annotation (partition function folding) and mountain plot (Turner & Mathews, 2009). Further, these predicted secondary structures were manually curated using rules reported by Zhang et al. (2005).
Circos algorithm
A Circos plot was developed between sugarcane-encoded miRNA and SCBGAV genes by applying the Circos algorithm (Krzywinski et al., 2009).
Statistical analysis
Predicted miRNA data obtained by applying four diverse algorithms were further analyzed using R-Language (v3.1.1, software version 3.5.1) (Gandrud, 2016). The graphical representation of the predicted miRNAs was processed using in house-scripts (readxl and ggplot2 packages).
Results
miRNA target prediction in genome of SCBGAV
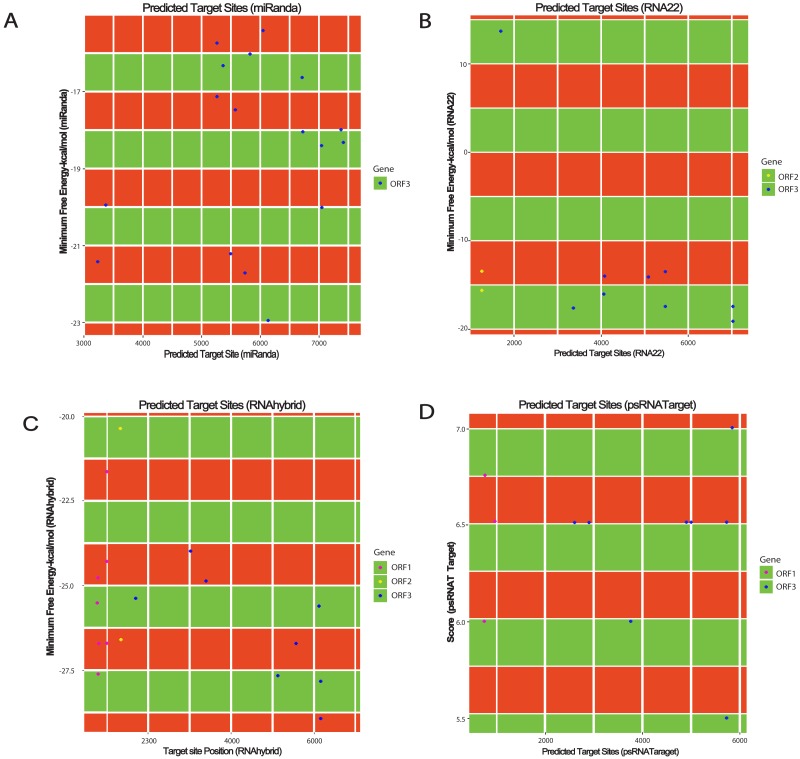
In order to predict sugarcane miRNA targets in the SCBGAV genome, a combination of above mentioned computational algorithms were employed to magnify high accuracy of miRNA target prediction. This strategy also filters out the false-positive results. miRanda (John et al., 2004) was selected to implement various miRNA target prediction parameters. These are target site conservation and prediction of several miRNA target binding sites. Afterwards, RNA22 was widely used for pattern-recognition in the target sequence (Miranda et al., 2006). RNAhybrid is a unique algorithm which determines miRNA and mRNA hybridization using following unique parameters (Krüger & Rehmsmeier, 2006). This algorithm is applied to calculate (MFE) minimum free energy and mode of target inhibition suggested as per the Brodersen et al. (2008) conclusion. psRNATarget is the most widely used plant miRNA target prediction tool which deals with a large set of data. It was designed to predict plant sRNA targets by analyzing complementary matching between sRNA and target mRNA sequence and by evaluating target site accessibility (Dai, Zhuang & Zhao, 2018). No sugarcane miRNAs were predicted in the SCBGAV genome using Tapirhybrid algorithm. Figure 3 represents all the genomic positions targeted by sugarcane miRNAs by the various algorithms applied (Tables S2, S3 and File S1).
Figure 3. miRNA target prediction results of Sugarcane Bacilliform Guadeloupe A Virus.
(A) Target prediction results from miRanda, (B) target prediction by RNA22 (C) target prediction by RNAhybrid and (D) target prediction by psRNATarget.
Prediction of sugarcane-encoded miRNA targets in the SCBGAV ORF1
There is little information available about the function of this protein. ORF1 (538–1,071 nucleotides) encodes a hypothetical protein represented as P1 (177 amino acids). RNAhybrid predicted binding of sof-miR159e at locus 733, ssp-miR169 at locus 752, ssp-miR437 (a, c) at locus 978, and ssp-miR444 (a, b, c-3p) at locus 774 (Fig. 3C). In addition, psRNATarget predicted hybridization of ssp-miR166 at locus 981, ssp-miR444a at locus 775, and ssp-miR444b at position 756 (Fig. 3D).
Identification of miRNA targets in the SCBGAV ORF2
The DNA binding protein, represented as P2, is encoded by the ORF2. ORF2 has the least number of predicted targets by sugarcane miRNAs; only three miRNA of sugarcane (sof- miR167 (a, b) and ssp-miR528) were targeted at common locus 1,301, indicated by RNA22 (Fig. 3B). Similarly, RNAhybrid predicted bindings of sof-miR167 (a, b) at locus 1,304 and ssp-miR1432 at locus 1301 (Fig. 3C).
miRNA target prediction in the ORF3
The polyprotein is encoded by the ORF3 which contains several functional units. These proteins are coat, movement, aspartic protease, RT, and ribonuclease H (Bouhida, Lockhart & Olszewski, 1993; Geijskes et al., 2002; King et al., 2011). The RT/RNase H coding region is a commonly used taxonomic marker for badnaviruses. It is considered sufficient to address the sequence diversity present within Badnavirus genomes (Bousalem, Douzery & Seal, 2008). For the ORF3 of SCBGAV, seven different kinds of miRNAs were predicted by miRanda: (sof-miR159 (a, b, c, d, and e), sof-miR168a, sof-miR396, ssp-miR166, ssp-miR827, ssp-miR1128 and ssp-miR1432) (Fig. 3A). RNA22 predicted hybridization of the following miRNAs: sof-miR168 (a, b), sof-miR396, sof-miR408 (a, b, c and d) and ssp-miR444 (a, b) (Fig. 3B). Suitable miRNAs for targeting ORF3 were hypothesized by RNAhybrid to interact with the ORF3 of SCBGAV. These are sof-miR159 (a, b, d), sof-miR168a, sof-miR396, sof-miR408 (a, b, c, d and e), ssp-miR166, ssp-miR827, and ssp-miR1128 (Fig. 3C). Moreover, sof-miR159 (a, b, c, d), sof-miR167 (a, b), sof-miR396, ssp-miR528, ssp-miR444b and ssp-miR444c-3p were identified by psRNATarget (Fig. 3D).
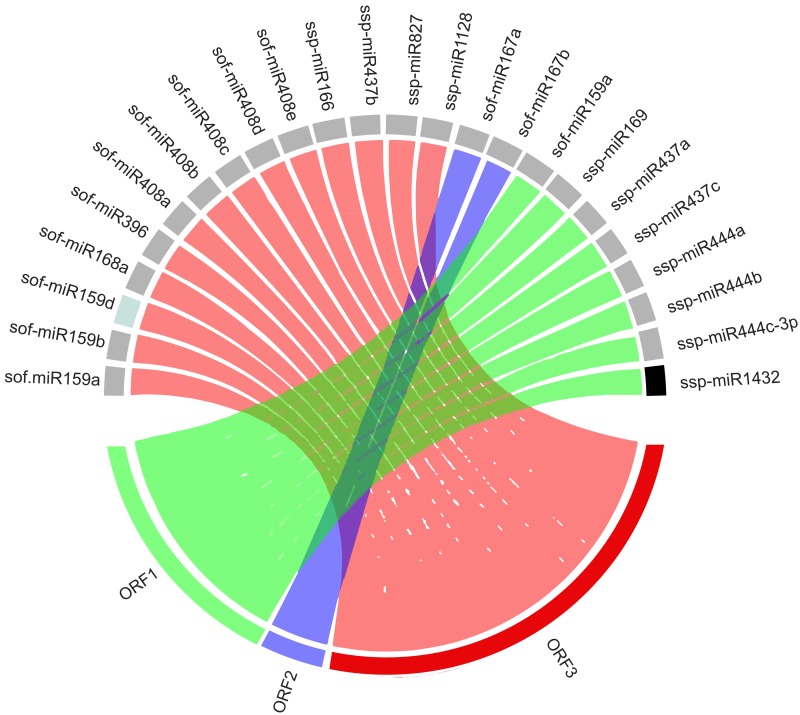
Visualization of miRNA target interaction
A Circos plot was used to study the visualization of sugarcane miRNAs with SCBGAV-targets (ORFs) that display particular evidence for potential targets. We have for the first time reported sugarcane-encoded miRNAs and their targets simultaneously constructed in this manner (Fig. 4).
Figure 4. Circos plot showing network interaction between sugarcane-encoded miRNAs and their SCBGAV targets.
Sugarcane miRNAs (predicted by a consensus of algorithms) for SCBGAV silencing
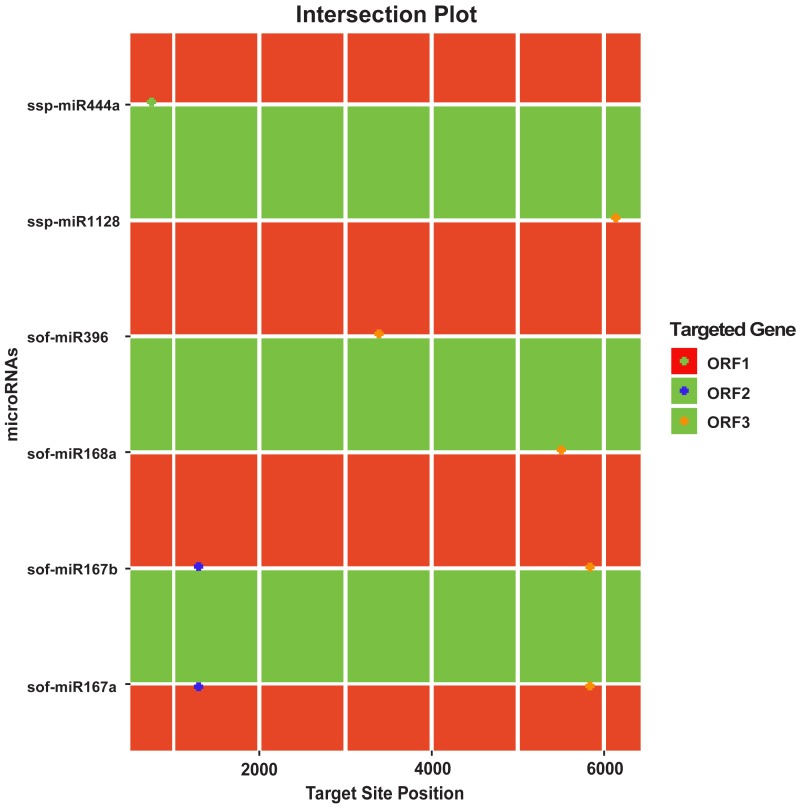
Among all the predicted miRNAs for SCBGAV silencing, only four miRNAs (sof-miR167 (a, b), sof-miR396 and ssp-miR528) were predicted by all of the algorithms used (Fig. 5, Table 2 and Table S4). We have verified the efficacy of these miRNA targets against SCBGAV following suppression of RNAi-mediated badnavirus control through translational inhibition or cleavage of viral mRNA. Moreover, six miRNAs (sof-miR167 (a, b) at locus 5846 and 1310, sof-miR168a at locus 5506, sof-miR396 at locus 3394, ssp-miR444a at locus 775 and ssp-miR1128 at locus 6148) were predicted at the common locus by at least two of the algorithms used (Fig. 6). Out of 28 sugarcane miRNAs, only one miRNA (sof-miR396) was predicted by at least three algorithms used to have binding site at the same locus position 3394 (Table 3).
Figure 5. Venn diagram representing common sugarcane miRNAs predicted by all algorithms.
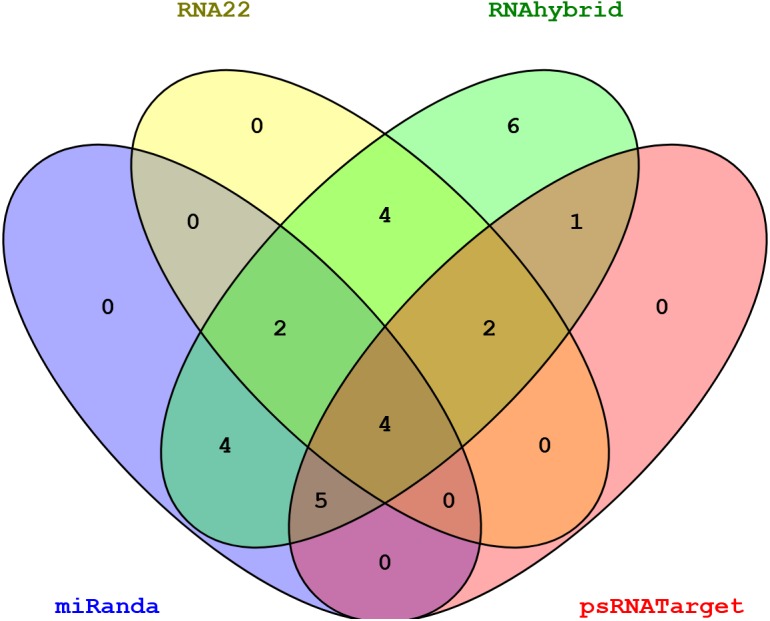
Table 2. miRNA-target pairs were selected from miRanda analysis.
Locus position was selected by consensus analysis of at least two algorithms.
| SugarcanemicroRNAs | miRNA-target pair | Locusposition | MFE*(kcal/mol) |
|---|---|---|---|
| sof-miR167(a, b) | Query: 3′ gucUAGUACGACCGUCGAAGu 5′
Ref: 5′ aaaATCAAGTT-GCAGCCTCa 3′ |
5846–5865 | −26.60 |
| sof-mi396 | Query: 3′ guCAAGUUC-UUUCGACACCUu 5′
Ref: 5′ agGATTAGGTGATGCTGTGGAg 3′ |
3394–3415 | −24.90 |
| ssp-miR528 | Query: 3′ gaggAGACGUACGGGGAAGGu 5′
Ref: 5′ gcgaTCCGC-CCCCCCTTCC- 3′ |
7426–7444 | −28.0 |
Notes.
MFE (Minimum Free Energy)* and mode of target Inhibition** was determined by RNAhybrid.
Figure 6. Intersection plot showing sugarcane miRNAs predicted from at least two algorithms.
Table 3. Sugarcane miRNAs and their target positions in the SCBGAV genome identified by all four algorithms.
| miRNA name | Position miRanda | Position RNA22 | Position RNAhybrid | Position psRNATarget | MFE* miRanda | MFE** RNA22 | MFE RNAhybrid | Expectation psRNATarget |
|---|---|---|---|---|---|---|---|---|
| sof-miR156 | 7104 | −23.2 | ||||||
| sof-miR159a | 5282 | 1659 | 3779 | −17.18 | −25.4 | 6 | ||
| sof-miR159a(1) | 6739 | −18.11 | ||||||
| sof-miR159b | 5282 | 1659 | 3779 | −17.18 | −25.4 | 6 | ||
| sof-miR159b(1) | 6739 | −18.11 | ||||||
| sof-miR159c | 6739 | 6896 | 3779 | −16.70 | -28 | 6 | ||
| sof-miR159d | 5282 | 1659 | 3779 | −17.18 | −25.4 | 6 | ||
| sof-miR159d(1) | 6739 | −18.11 | ||||||
| sof-miR159e | 5282 | 733 | −15.81 | −25.5 | ||||
| sof-miR167a | 5846 | 1310 | 1304 | 5846 | −16.08 | −15.80 | −26.6 | 7 |
| sof-miR167b | 5846 | 1310 | 1304 | 5846 | −16.08 | −15.80 | −26.6 | 7 |
| sof-miR168a | 5506 | 5506 | 6084 | −21.24 | −17.70 | −25.6 | ||
| sof-miR168a(1) | 7050 | 7050 | −18.45 | −17.10 | ||||
| sof-miR168a(2) | 5137 | −14.30 | ||||||
| sof-miR168b | 7050 | 7050 | 6937 | −20.05 | −19.40 | −25.6 | ||
| sof-miR168b(1) | 5506 | −13.70 | ||||||
| sof-miR396 | 3394 | 3394 | 3394 | 5732 | −19.99 | −17.80 | −24.9 | 5.5 |
| sof-miR396(1) | 4115 | −14.20 | ||||||
| sof-miR408a | 1735 | 5136 | −13.70 | −27.6 | ||||
| sof-miR408b | 1735 | 5136 | −13.70 | −27.6 | ||||
| sof-miR408c | 1735 | 5136 | −13.70 | −27.6 | ||||
| sof-miR408d | 1735 | 5136 | −13.70 | −27.6 | ||||
| sof-miR408e | 6152 | −28.9 | ||||||
| ssp-miR166 | 3249 | 5572 | 981 | −21.45 | −26.7 | 6.5 | ||
| ssp-miR166(1) | 5589 | −17.54 | ||||||
| ssp-miR166(2) | 5761 | −21.74 | ||||||
| ssp-miR169 | 752 | −24.8 | ||||||
| ssp-miR437a | 978 | −21.7 | ||||||
| ssp-miR437b | 6829 | −21.8 | ||||||
| ssp-miR437c | 978 | −24.3 | ||||||
| ssp-miR528 | 7426 | 1310 | 7407 | 2897 | −18.37 | −13.70 | -28 | 6.5 |
| ssp-miR528(1) | 7395 | −18.05 | ||||||
| ssp-miR827 | 5378 | 3010 | −16.40 | -24 | ||||
| ssp-miR444a | 4103 | 774 | 775 | −16.30 | −27.6 | 6.75 | ||
| ssp-miR444b | 4103 | 774 | 756 | −16.30 | −27.6 | 6.0 | ||
| ssp-miR444b(1) | 2615 | 6.5 | ||||||
| ssp-miR444b(2) | 5009 | 6.5 | ||||||
| ssp-miR444b(3) | 5731 | 6.5 | ||||||
| ssp-miR444c-3p | 774 | 4940 | −26.7 | 6.5 | ||||
| ssp-miR1128 | 6141 | 6148 | −22.96 | −27.8 | ||||
| ssp-miR1432 | 6070 | 1301 | −15.48 | −20.4 |
Notes.
*MFE: Minimum free energy measured in /Kcal/mol where *MFE represents minimum folding energy measured in Kcal/mol.
Prediction of secondary structures
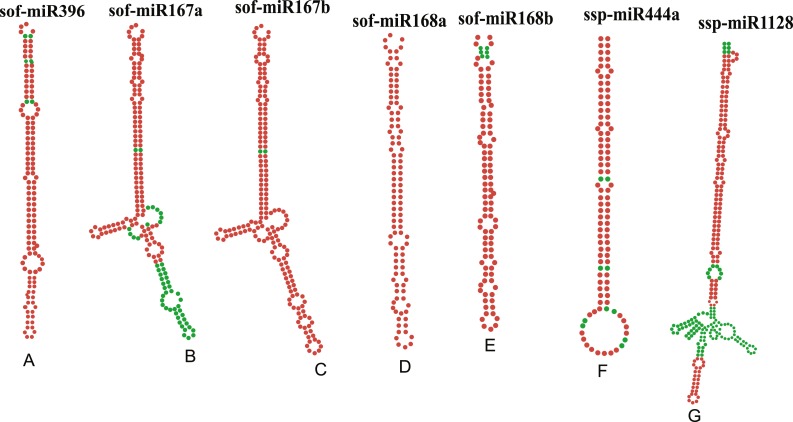
Stable secondary structures of the consensus sugarcane miRNA precursor sequences was obtained using RNAfold algorithm. The precursor sequences were manually curated. Top seven stable secondary structures (based on MFEI value) of consensus sugarcane miRNAs are shown (Fig. 7). We have characterized the salient features of precursor miRNA, such as MFE (minimum free energy) to determine the secondary structure of nucleic acids. In our studies, MFE range from −56.1 to −101.7 kcal/mol, GC content (42–65%) and MFEI ranges from −0.7 to −1.17 (Table 4).
Figure 7. Stable secondary structures of top seven pre-miRNAs, precursors of the mature sugarcane miRNAs found in the study as the miRNAs detected by a consensus of algorithms.
Names with minimum free energy (MFE) are represented: (A) sof-MIR396 (−67.40 Kcal/mol) (B) sof-MIR167a (−82.70 Kcal/mol) (C) sof-MIR167b (−86.00 Kcal/mol) (D) sof-MIR168a (−66.20 Kcal/mol), (E) sof-MIR168b (−56.10 Kcal/mol), (F) ssp-MIR444a (−57.70 Kcal/mol), (G) ssp-MIR1128 (−101.70).
Table 4. Predicted characterized features of secondary structures of precursor miRNAs.
| miRNA ID | miRNA sequence | Length miRNA | Length precursor | MFE1 (Kcal/mol) | AMFE2 | MFEI3 | (G + C)% |
|---|---|---|---|---|---|---|---|
| sof-miR396 | UUCCACAGCUUUCUUGAACUG | 21 | 134 | −67.40 | −50.29 | −1.17 | 42.85 |
| sof-miR167a | UGAAGCUGCCAGCAUGAUCUG | 21 | 188 | −82.70 | −43.98 | −0.83 | 52.38 |
| sof-miR167b | UGAAGCUGCCAGCAUGAUCUG | 21 | 188 | −86.00 | −45.74 | −0.87 | 52.38 |
| sof-miR168a | UCGCUUGGUGCAGAUCGGGAC | 21 | 104 | −66.20 | −63.65 | −1.02 | 61.90 |
| sof-miR168b | UCGCUUGGGCAGAUCGGGAC | 20 | 103 | −56.10 | −54.46 | −0.83 | 65.00 |
| ssp-miR444a | UGCAGUUGUUGCCUCAAGCUU | 21 | 105 | −57.70 | −54.95 | −1.14 | 47.61 |
| ssp-miR1128 | UACUACUCCCUCCGUCCCAAA | 21 | 275 | −101.70 | −36.98 | −0.70 | 52.38 |
Notes.
1MFE represents minimum free energy. 2AMFE is an adjusted minimum free energy calculated by MFE/length of precursor*100. MFEI defines as minimum free energy index which was calculated by AMFE/(G + C) %.
Discussion
For possible miRNA target prediction in the genome of SCBGAV, a combination of the aforementioned computational tools was used in order to filter out the false-positive results and to increase the accuracy of miRNA target prediction. miRanda was implemented to validate various parameters, from target site conservation to whole genome prediction of miRNA target genes (John et al., 2004; Riffo-Campos, Riquelme & Brebi-Mieville, 2016). Then, RNAhybrid and psRNATarget were used, both of which are strongly prescribed for plant miRNA target identification (Dai, Zhuang & Zhao, 2018; Srivastava et al., 2014; Zhang & Verbeek, 2010). RNA22 is a novel algorithm that applies an in-silico strategy that is highly divergent in comparison to other algorithms. Pattern-based recognition is the key feature of this algorithm (Min & Yoon, 2010; Miranda et al., 2006).
The results from this study suggest that SCBGAV is susceptible to targeting by consensus sugarcane-encoded miRNAs. The genome components of SCBGAV (ORF1, ORF2 and ORF3) seemed to be principally prone sites for sugarcane-miRNA regulation. While in vivo demonstration requires validating functional efficacy, the degree of complementarity between target mRNA and miRNA concludes the fate of the predicted sites. A full complementarity binding between target mRNA and miRNA sequence results in endonucleolytic cleavage and disruption. Contrary to this, partial target-site complementarity characteristically down-regulates target-gene expression by suppressing translation of target mRNA.
The present study identifies suitable sugarcane-encoded miRNAs to exhibit a stronger degree of target-site complementarities within the ORF1, ORF2 and ORF3 of SCBGAV. These predicted miRNAs may be utilized to develop effective amiRNA constructs, which could be used to enhance the sugarcane immunity to SCBGAV. Pairing multiple miRNAs with a single mRNA induces effective RNA silencing (Doench & Sharp, 2004). In the current study, we have predicted several miRNA targets which were associated with SCBGAV (ORF1, ORF2, and ORF3) genes at multiple loci. A deeper understanding of these vital ORFs involved in SCBGAV epidemic via miRNA-mediated control of gene expression would significantly assist in the development of molecular approaches to combat the dissemination of SCBGAV. The miRNA-target pairs ensuring MFE exceeding the threshold standards were predicted.
Our study is unique from previous studies in that it provides an in silico approach for comprehensive analysis of host-derived miRNAs using seven computational algorithms and advanced statistical approaches. Previous studies that have identified plant-encoded miRNA Targets in plant viruses have been limited by small numbers of miRNAs screened, small sample sizes, lack of independent validation sets, and the use of inappropriate statistical methods to present miRNA interaction data. Notably, we have validated the efficacy of the consensus seven sugarcane-encoded miRNA targets (detected two algorithms at the same locus) against SCBGAV following suppression of RNAi-mediated viral combat. These are achieved by translational inhibition or cleavage of viral miRNA.
Applying a combination of four diverse computational algorithms (miRanda, RNA22, Tapirhybrid and psRNATarget), the predicted consensus results are highly reliable and robust for assessing interaction between sugarcane miRNAs and viral genome (Oliveira et al., 2017). This miRNA-viral gene interaction was further verified by RNAhybrid (standard MFE predicted) and Circos algorithms (network visualization). False-positive miRNA-target interactions predicted by computational algorithms depend upon the mechanism of the miRNA-target recognition (Pinzón et al., 2017). In addition, MFE is one of the key factors to affect the miRNA-targets interactions for results sorting. A lower MFE value has high potential to develop miRNA-target complex (Hammell et al., 2008; Kertesz et al., 2007). We have applied a stringent cut-off −15 kcal/mol for miRanda and −20 kcal/mol for RNAhybrid to narrow down miRNA candidates.
In order to control false-positive prediction, union and intersection approach was applied. Union approach based on grouping of one or more algorithmic tools which results increase of number of true targets as well the number of false targets. It decreases specificity at the cost of increasing sensitivity. Using intersection approach relies on the mixing two or more tools which improves the specificity at the cost of decreasing sensitivity (Witkos, Koscianska & Krzyzosiak, 2011). Our analysis indicated that both the strategies showed the highest performance as shown in (Figs. 5 and 6). These miRNAs were identified and considered as the most desirable sugarcane miRNAs against the genome of SCBGAV. These predicted miRNAs were identified after setting the parameters of minimum free energy, seed pairing, target site accessibility, folding energy and pattern recognition, thus integrating all aspects of miRNA target prediction. Therefore, they are the most suitable selections for gene silencing.
Similar studies have been published previously using multiple computational algorithms to identify a best fit miRNA for a desirable target to understand plant-virus interaction. First time, genome-wide identification and comprehensive analysis was carried out to predict maize (Zea-mays)-encoded miRNA targets against maize chlorotic mottle virus (MCMV) (Iqbal et al., 2017). We have adopted the same novel computational strategy to predict the sugarcane-encode miRNA targets in the SCBGAV genome to combat badnaviruses in sugarcane.
As the amiRNAs have high specificity to the designed target gene, detrimental off-target effects can be minimized, permitting their silencing expression to be stably transmitted to future generations (Ossowski, Schwab & Weigel, 2008; Zhao et al., 2009). Furthermore, the small size of amiRNA permits for the insertion of multiple and distinct amiRNAs within a single gene expression cassette, which can then be transformed to develop transgenic plant resistant to multiple viruses simultaneously (Niu et al., 2006; Park, Zhai & Lee, 2009; Schwab et al., 2010). We have designed future work to validate this promising amiRNA-based strategy can in fact be used to develop durable SCBGAV- resistance in transgenic sugarcane.
Conclusions
The current study details an organized computational approach for the identification of host-derived miRNAs aimed at silencing viral genes affecting the sugarcane host plant by amiRNA-mediated interference targeting different genes of the SCBGAV. This study offers an enhanced means to computationally investigate the best-candidate miRNAs against badnaviruses, prior to cloning. As our approach allows a narrow-range of match-mismatch in microRNA-mRNA attachment, it screens most of the falsely predicted attachments. These predicted miRNAs were identified after setting the parameters of MFE, seed pairing, target site accessibility, folding energy and pattern recognition, thus using key features of miRNA target prediction. Most importantly, we identified six miRNAs (sof-miR167 (a, b), sof-miR168a, sof-miR396, ssp-miR444a and ssp-miR1128) that could target the SCBGAV genome. Among them, sof-miR396 was predicted as the top-ranking effective candidate, capable of targeting the vital ORF3. To further refine our results, we also identified key interaction of miRNA networks and target genes by constructing Circos graph of consensus miRNAs and their targets. This short-listed miRNA is the best candidates to be utilized in sugarcane plant transformation for the development of SCBGAV-resistant sugarcane cultivars. These results offer an evidence of idea for the construction of innovative amiRNA-based therapeutics against emerging SCBGAV badnaviruses.
Supplemental Information
Sugarcane-encoded miRNAs predicted from all four algorithms. Genome positions and the output parameters including minimum free energy, folding energy and expectation have been included.
Consensus sugarcane-encoded miRNA were selected from all four algorithms.
Acknowledgments
We are highly thankful to our lab colleagues for their assistance in data analysis. We thank to Dr. Zhiqiang Xia (ITBB) for assistance in constructing Circos plot. We are thankful to our senior lab colleagues Xiao-Yan Feng and Lin-Bo Shen for reading the revised manuscript as well as the editor and two referees for their valuable comments on this manuscript. We are highly thankful to a qualified native English speaking editor at the PeerJ Experts for editing proper English language.
Funding Statement
This study is supported by the National Natural Science Foundation of China (No. 31771865), the Sugar Crop Research System (CARS-170301) and the “Talented Young Scientist Program” (TYSP-4th Batch 2017-2018) of China Project ID (Pakistan-18-004). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Muhammad Aleem Ashraf, Email: muhammad.aleemashraf@iub.edu.pk.
Shuzhen Zhang, Email: zhangshuzhen@itbb.org.cn.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Fakiha Ashraf and Xiaowen Hu analyzed the data, performed the experiments, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Muhammad Aleem Ashraf analyzed the data, conceived and designed the experiments, performed the experiments, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Shuzhen Zhang conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Data Availability
The following information was supplied regarding data availability:
Saccharum officinarum L. miRNA data is available at miRBase: MI0001754–MI0001769, which can be accessed by species name.
Additional data is available at NCBI: NC_038382.1.
Tools used to analyze the data are available at the Jefferson Computational Medicine Center RNA22 (https://cm.jefferson.edu/rna22/), BiBiServ (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid), and psRNATarget (http://plantgrn.noble.org/psRNATarget/).
References
- Adams & Carstens (2012).Adams MJ, Carstens E. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2012) Archives of Virology. 2012;157:1411–1422. doi: 10.1007/s00705-012-1299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams et al. (2016).Adams MJ, Lefkowitz EJ, King AM, Harrach B, Harrison RL, Knowles NJ, Kropinski AM, Krupovic M, Kuhn JH, Mushegian AR. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2016) Archives of Virology. 2016;161:2921–2949. doi: 10.1007/s00705-016-2977-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad et al. (2019).Ahmad K, Sun S-R, Chen J-L, Huang M-T, Fu H-Y, Gao S-J. Presence of diverse sugarcane bacilliform viruses infecting sugarcane in China revealed by pairwise sequence comparisons and phylogenetic analysis. The Plant Pathology Journal. 2019;35:41–50. doi: 10.5423/PPJ.OA.08.2018.0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akmal, Baig & Khan (2017).Akmal M, Baig MS, Khan JA. Suppression of cotton leaf curl disease symptoms in Gossypium hirsutum through over expression of host-encoded miRNAs. Journal of Biotechnology. 2017;263:21–29. doi: 10.1016/j.jbiotec.2017.10.003. [DOI] [PubMed] [Google Scholar]
- Ali et al. (2013).Ali I, Amin I, Briddon RW, Mansoor S. Artificial microRNA-mediated resistance against the monopartite begomovirus Cotton leaf curl Burewala virus. Virology Journal. 2013;10:231. doi: 10.1186/1743-422X-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archak & Nagaraju (2007).Archak S, Nagaraju J. Computational prediction of rice (Oryza sativa) miRNA targets. Genomics, Proteomics & Bioinformatics. 2007;5:196–206. doi: 10.1016/S1672-0229(08)60007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autrey et al. (1992).Autrey L, Boolell S, Jones E, Lociçhart B, Nadie A. Distribution of sugarcane bacilliform virus in various geographical regions. ISSCT. Proceedings XXI Congress. 1992:5–14. [Google Scholar]
- Betel et al. (2008).Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA. org resource: targets and expression. Nucleic Acids Research. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet et al. (2010).Bonnet E, He Y, Billiau K, Van de Peer Y. TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics. 2010;26:1566–1568. doi: 10.1093/bioinformatics/btq233. [DOI] [PubMed] [Google Scholar]
- Borah et al. (2013).Borah BK, Sharma S, Kant R, Johnson AA, Saigopal DVR, Dasgupta I. Bacilliform DNA-containing plant viruses in the tropics: commonalities within a genetically diverse group. Molecular Plant Pathology. 2013;14:759–771. doi: 10.1111/mpp.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhida, Lockhart & Olszewski (1993).Bouhida M, Lockhart B, Olszewski NE. An analysis of the complete sequence of a sugarcane bacilliform virus genome infectious to banana and rice. Journal of General Virology. 1993;74:15–22. doi: 10.1099/0022-1317-74-1-15. [DOI] [PubMed] [Google Scholar]
- Bousalem, Douzery & Seal (2008).Bousalem M, Douzery EJ, Seal S. Taxonomy, molecular phylogeny and evolution of plant reverse transcribing viruses (family Caulimoviridae) inferred from full-length genome and reverse transcriptase sequences. Archives of Virology. 2008;153:1085. doi: 10.1007/s00705-008-0095-9. [DOI] [PubMed] [Google Scholar]
- Brodersen et al. (2008).Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- Brodersen & Voinnet (2006).Brodersen P, Voinnet O. The diversity of RNA silencing pathways in plants. Trends in Genetics. 2006;22:268–280. doi: 10.1016/j.tig.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Carbonell & Daròs (2019).Carbonell A, Daròs J-A. Design, synthesis, and functional analysis of highly specific artificial small RNAs with antiviral activity in plants. Antiviral Resistance in Plants. 2019;2028:231–246. doi: 10.1007/978-1-4939-9635-3_13. [DOI] [PubMed] [Google Scholar]
- Chen et al. (2019).Chen L, Meng J, He XL, Zhang M, Luan YS. Solanum lycopersicum microRNA1916 targets multiple target genes and negatively regulates the immune response in tomato. Plant, Cell & Environment. 2019;42:1393–1407. doi: 10.1111/pce.13468. [DOI] [PubMed] [Google Scholar]
- Dai, Zhuang & Zhao (2018).Dai X, Zhuang Z, Zhao PX. psRNATarget: a plant small RNA target analysis server (2017 release) Nucleic Acids Research. 2018;46:W49–W54. doi: 10.1093/nar/gky316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench & Sharp (2004).Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes & Development. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandrud (2016).Gandrud C. Reproducible research with R and R studio. Chapman and Hall/CRC Press; 2016. [Google Scholar]
- Geijskes et al. (2002).Geijskes RJ, Braithwaite KS, Dale J, Harding R, Smith GR. Sequence analysis of an Australian isolate of sugarcane bacilliform badnavirus. Archives of Virology. 2002;147:2393–2404. doi: 10.1007/s00705-002-0879-2. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones et al. (2006).Griffiths-Jones S, Grocock RJ, Van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Research. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones et al. (2007).Griffiths-Jones S, Saini HK, Van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Research. 2007;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell et al. (2008).Hammell M, Long D, Zhang L, Lee A, Carmack CS, Han M, Ding Y, Ambros V. mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein–enriched transcripts. Nature Methods. 2008;5:813–819. doi: 10.1038/nmeth.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal et al. (2017).Iqbal MS, Jabbar B, Sharif MN, Ali Q, Husnain T, Nasir IA. In silico MCMV silencing concludes potential host-derived miRNAs in maize. Frontiers in Plant Science. 2017;8:372. doi: 10.3389/fpls.2017.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John et al. (2004).John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLOS Biology. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz et al. (2007).Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nature Genetics. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- King et al. (2011).King AM, Lefkowitz E, Adams MJ, Carstens EB. ElsevierVirus taxonomy: ninth report of the International committee on taxonomy of viruses. 2011
- Kozomara, Birgaoanu & Griffiths-Jones (2018).Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Research. 2018;47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara & Griffiths-Jones (2013).Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research. 2013;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger & Rehmsmeier (2006).Krüger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Research. 2006;34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski et al. (2009).Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Research. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart & Autrey (2000).Lockhart B, Autrey LJC. A guide to sugarcane diseases. Editions Quae, CIRAD; France: 2000. Sugarcane bacilliform virus; pp. 266–272. [Google Scholar]
- Lockhart, Ireyt & Comstock (1996).Lockhart B, Ireyt MJ, Comstock JC. Sugarcane germplasm conservation and exchange. Australian Centre for International Agricultural Research; Brisbane, Canberra: 1996. Sugarcane Bacilliform Virus, sugarcane mild mosaic virus and sugarcane yellow leaf syndrome; pp. 108–112. [Google Scholar]
- Loher & Rigoutsos (2012).Loher P, Rigoutsos I. Interactive exploration of RNA22 microRNA target predictions. Bioinformatics. 2012;28:3322–3323. doi: 10.1093/bioinformatics/bts615. [DOI] [PubMed] [Google Scholar]
- Lorenz et al. (2011).Lorenz R, Bernhart SH, Zu Siederdissen CH, Tafer H, Flamm C, Stadler PF, Hofacker IL. ViennaRNA package 2.0. Algorithms for Molecular Biology. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min & Yoon (2010).Min H, Yoon S. Got target?: computational methods for microRNA target prediction and their extension. Experimental & Molecular Medicine. 2010;42:233–244. doi: 10.3858/emm.2010.42.4.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda et al. (2006).Miranda KC, Huynh T, Tay Y, Ang Y-S, Tam W-L, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Muller et al. (2011).Muller E, Dupuy V, Blondin L, Bauffe F, Daugrois J-H, Nathalie L, Iskra-Caruana M-L. High molecular variability of sugarcane bacilliform viruses in Guadeloupe implying the existence of at least three new species. Virus Research. 2011;160:414–419. doi: 10.1016/j.virusres.2011.06.016. [DOI] [PubMed] [Google Scholar]
- Niu et al. (2006).Niu Q-W, Lin S-S, Reyes JL, Chen K-C, Wu H-W, Yeh S-D, Chua N-H. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nature Biotechnology. 2006;24:1420–1428. doi: 10.1038/nbt1255. [DOI] [PubMed] [Google Scholar]
- Oliveira et al. (2017).Oliveira AC, Bovolenta LA, Nachtigall PG, Herkenhoff ME, Lemke N, Pinhal D. Combining results from distinct microRNA target prediction tools enhances the performance of analyses. Frontiers in Genetics. 2017;8:59. doi: 10.3389/fgene.2017.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski, Schwab & Weigel (2008).Ossowski S, Schwab R, Weigel D. Gene silencing in plants using artificial microRNAs and other small RNAs. The Plant Journal. 2008;53:674–690. doi: 10.1111/j.1365-313X.2007.03328.x. [DOI] [PubMed] [Google Scholar]
- Park, Zhai & Lee (2009).Park W, Zhai J, Lee J-Y. Highly efficient gene silencing using perfect complementary artificial miRNA targeting AP1 or heteromeric artificial miRNA targeting AP1 and CAL genes. Plant Cell Reports. 2009;28:469–480. doi: 10.1007/s00299-008-0651-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petchthai, Le Yee & Wong (2018).Petchthai U, Le Yee CS, Wong S-M. Resistance to CymMV and ORSV in artificial microRNA transgenic Nicotiana benthamiana plants. Scientific Reports. 2018;8:9958. doi: 10.1038/s41598-018-28388-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson et al. (2014).Peterson SM, Thompson JA, Ufkin ML, Sathyanarayana P, Liaw L, Congdon CB. Common features of microRNA target prediction tools. Frontiers in Genetics. 2014;5:23. doi: 10.3389/fgene.2014.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinzón et al. (2017).Pinzón N, Li B, Martinez L, Sergeeva A, Presumey J, Apparailly F, Seitz H. microRNA target prediction programs predict many false positives. Genome Research. 2017;27:234–245. doi: 10.1101/gr.205146.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, Ye & Fang (2007).Qu J, Ye J, Fang R. Artificial microRNA-mediated virus resistance in plants. Journal of Virology. 2007;81:6690–6699. doi: 10.1128/JVI.02457-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riffo-Campos, Riquelme & Brebi-Mieville (2016).Riffo-Campos Á, Riquelme I, Brebi-Mieville P. Tools for sequence-based miRNA target prediction: what to choose? International Journal of Molecular Sciences. 2016;17:1987. doi: 10.3390/ijms17121987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab et al. (2010).Schwab R, Ossowski S, Warthmann N, Weigel D. Directed gene silencing with artificial microRNAs. Plant MicroRNAs. 2010;592:71–88. doi: 10.1007/978-1-60327-005-2_6. [DOI] [PubMed] [Google Scholar]
- Srivastava et al. (2014).Srivastava PK, Moturu TR, Pandey P, Baldwin IT, Pandey SP. A comparison of performance of plant miRNA target prediction tools and the characterization of features for genome-wide target prediction. BMC Genomics. 2014;15:348. doi: 10.1186/1471-2164-15-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun et al. (2016).Sun S-R, Damaj MB, Alabi OJ, Wu X-B, Mirkov TE, Fu H-Y, Chen R-K, Gao S-J. Molecular characterization of two divergent variants of sugarcane bacilliform viruses infecting sugarcane in China. European Journal of Plant Pathology. 2016;145:375–384. doi: 10.1007/s10658-015-0851-0. [DOI] [Google Scholar]
- Turner & Mathews (2009).Turner DH, Mathews DH. NNDB: the nearest neighbor parameter database for predicting stability of nucleic acid secondary structure. Nucleic Acids Research. 2009;38:D280–D282. doi: 10.1093/nar/gkp892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan, Alexander & Garg (1996).Viswanathan R, Alexander K, Garg I. Detection of sugarcane bacilliform virus in sugarcane germplasm. Acta Virologica. 1996;40:5–8. [PubMed] [Google Scholar]
- Witkos, Koscianska & Krzyzosiak (2011).Witkos MT, Koscianska E, Krzyzosiak JW. Practical aspects of microRNA target prediction. Current Molecular Medicine. 2011;11:93–109. doi: 10.2174/156652411794859250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie & Zhang (2010).Xie F, Zhang B. Target-align: a tool for plant microRNA target identification. Bioinformatics. 2010;26(23):3002–3003. doi: 10.1093/bioinformatics/btq568. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2011).Zhang X, Li H, Zhang J, Zhang C, Gong P, Ziaf K, Xiao F, Ye Z. Expression of artificial microRNAs in tomato confers efficient and stable virus resistance in a cell-autonomous manner. Transgenic Research. 2011;20:569–581. doi: 10.1007/s11248-010-9440-3. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2005).Zhang BH, Pan XP, Wang QL, George PC, Anderson TA. Identification and characterization of new plant microRNAs using EST analysis. Cell Research. 2005;15:336–360. doi: 10.1038/sj.cr.7290302. [DOI] [PubMed] [Google Scholar]
- Zhang & Verbeek (2010).Zhang Y, Verbeek FJ. Comparison and integration of target prediction algorithms for microRNA studies. Journal of Integrative Bioinformatics. 2010;7:169–181. doi: 10.2390/biecoll-jib-2010-127. [DOI] [PubMed] [Google Scholar]
- Zhao et al. (2009).Zhao T, Wang W, Bai X, Qi Y. Gene silencing by artificial microRNAs in Chlamydomonas. The Plant Journal. 2009;58:157–164. doi: 10.1111/j.1365-313X.2008.03758.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sugarcane-encoded miRNAs predicted from all four algorithms. Genome positions and the output parameters including minimum free energy, folding energy and expectation have been included.
Consensus sugarcane-encoded miRNA were selected from all four algorithms.
Data Availability Statement
The following information was supplied regarding data availability:
Saccharum officinarum L. miRNA data is available at miRBase: MI0001754–MI0001769, which can be accessed by species name.
Additional data is available at NCBI: NC_038382.1.
Tools used to analyze the data are available at the Jefferson Computational Medicine Center RNA22 (https://cm.jefferson.edu/rna22/), BiBiServ (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid), and psRNATarget (http://plantgrn.noble.org/psRNATarget/).