Abstract
DNA is both a fundamental building block of life and a fascinating natural polymer. The advent of single-molecule manipulation tools made it possible to exert controlled force on individual DNA molecules and measure their mechanical response. Such investigations elucidated the elastic properties of DNA and revealed its distinctive structural configurations across force regimes. In the meantime, a detailed understanding of DNA mechanics laid the groundwork for single-molecule studies of DNA-binding proteins and DNA-processing enzymes that bend, stretch, and twist DNA. These studies shed new light on the metabolism and transactions of nucleic acids, which constitute a major part of the cell’s operating system. Furthermore, the marriage of single-molecule fluorescence visualization and force manipulation has enabled researchers to directly correlate the applied tension to changes in the DNA structure and the behavior of DNA-templated complexes. Overall, experimental exploitation of DNA mechanics has been and will continue to be a unique and powerful strategy for understanding how molecular machineries recognize and modify the physical state of DNA to accomplish their biological functions.
INTRODUCTION
The DNA double helix is arguably the most celebrated discovery in biology during the past century1–3. In the ensuing decades, individual DNA molecules were directly visualized under electron and fluorescence microscopes4–7. During the past 25 years, the development of single-molecule manipulation methods has led to elucidation of the mechanical properties of DNA reviewed in8, 9]. Single-molecule methods circumvent the need for synchronization and allow for real-time observation. These tools, initially used to observe and manipulate DNA, have also been employed to dissect the molecular mechanism of DNA-binding proteins and DNA-processing enzymes reviewed in10, 11]. In this perspective, we will first review how single-molecule force spectroscopy has enabled detailed investigation of the elastic properties of DNA. We will then discuss how the knowledge of DNA mechanics has facilitated the studies of DNA-based biological processes such as DNA packaging and replication. Finally, we will discuss how recent development in combined fluorescence and force microscopy has allowed direct correlation between DNA structure and mechanics.
DNA MECHANICS
DNA has several unique properties as a polymer: its extensive hydrogen bonding and base stacking render it highly stiff; the negative charge on every backbone phosphate also makes it one of the most charged polymers known in nature. Characterization of the elastic properties of DNA was greatly facilitated by the development of single-molecule force manipulation methods, which exert controlled force to a biomolecule and precisely measure its mechanical response12. The viscoelastic properties of DNA have been investigated by magnetic tweezers13, 14, micro fibers15, hydrodynamic flow16, and optical tweezers (Fig. 1A)17. The force-extension (F-x) behavior of double-stranded DNA (dsDNA) can be well described by a worm-like-chain (WLC) model (Fig. 1B):
where F is the applied force, x is the DNA extension, kB is the Boltzmann constant, T is the absolute temperature, and L0 is the DNA contour length (maximum end-to-end distance)18. The WLC model employs a coarse-grained treatment on DNA—ignoring local variations in sequence and helical structure—and characterizes the flexibility of the polymer with a single parameter: the persistence length (Lp). Lp can be intuitively understood as the length scale over which the direction of the polymer chain persists under thermal fluctuations. In a typical physiological buffer, Lp for dsDNA is ~50 nm or ~150 base pairs (bp), and has been shown to depend on the ionic strength and valency19, 20. However, DNA exhibits surprisingly high bendability at short length scales (<100 bp)21. This extreme flexibility can be described by modified WLC models that allow for the formation of transient kinks or bubbles22, 23. Moreover, a “twistable” WLC model was proposed24 to account for the response of the helical structure of DNA to torsional tensions25, 26.
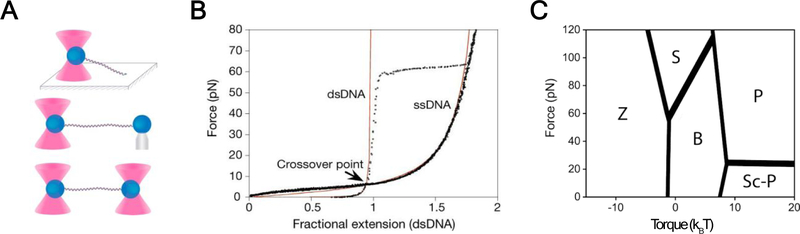
Figure 1.
Single-molecule manipulation to elucidate DNA mechanics (A) Force measurements of single DNA molecules are enabled via multiple optical-trapping geometries, in which one end of the DNA is tethered to an optically trapped micron-sized bead, and the other end is tethered to a microscope slide surface (Top), a micropipette-suctioned bead (Middle), or another optically trapped bead (Bottom). Figure reprinted with permission from27. Copyright (2014) American Chemical Society. (B) Force-extension behavior of ssDNA and dsDNA. The red lines indicate the WLC model prediction. At forces above the crossover point (~6 pN), ssDNA is longer than dsDNA. Figure reproduced with permission from28. (C) Force-torque phase diagram of dsDNA. Note that along the borders between regions, adjacent phases coexist in equilibrium. Z: Z-DNA; S: S-DNA; B: B-DNA; P: P-DNA (extended and overtwisted); Sc-P: plectonemically supercoiled DNA. Figure adapted with permission from29. Copyright (2001) American Physical Society.
A flexible polymer tends to coil randomly in solution, resulting in an end-to-end distance much shorter than its contour length. Thus, pulling a flexible chain into an extended one is entropically unfavorable. A tension of ~6 pN—a typical amount of force exerted by biomolecular motors—is needed to stretch dsDNA to 95% of its contour length (Fig. 1B). In comparison, single-stranded DNA (ssDNA) has a much shorter persistence length (on the order of 1 nm) and is more contractile, adopting more compact conformations than dsDNA at low forces30. At higher forces, the extension of ssDNA exceeds that of dsDNA and reaches nearly twice as long as dsDNA at full extension (Fig. 1B).
DNA predominantly exists in a right-handed B-form in aqueous solution, but can adopt a shorter and wider A-form structure under dehydrating conditions. Moreover, DNA with specific sequences can assume a left-handed Z form31. Besides these three well-known biologically active forms, single-molecule pulling experiments have revealed additional forms of DNA under specific force regimes. In particular, when the applied tension was increased to ~65 pN, B-DNA was found to transition to an extended form that is ~70% longer15, 17. There has been considerable debate regarding whether this overstretched form of dsDNA represents a new base-paired structure (coined the term “S-DNA”), or comprises two denatured ssDNA32, 33. Recent studies showed that both mechanisms can be simultaneously at work: force-induced denaturation is favored when AT content in the DNA is high; while for GC-rich sequences, the DNA undergoes a reversible overstretching transition into an elongated, underwound, yet base-paired form, supporting the existence of S-DNA34, 35.
Besides tension-induced structural transitions, torque can also cause DNA to adopt distinct conformations36. For torsionally constrained DNA (via multiple attachment points to the bead at each terminus), an overwound structure with the backbones tightly wrapped around and the base pairs flipped out, known as P-DNA, was observed when the DNA overstretched at forces >110 pN37. The twist elasticity of DNA has been extensively reviewed elsewhere38, 39. Through a combination of experimental manipulation and statistical-mechanical modeling, the force-torque phase diagrams for DNA have been depicted (Fig. 1C)29, 40.
The well-characterized elasticity combined with its ease of construction makes DNA a popular choice as molecular handles41 for single-molecule studies of protein/RNA folding42, 43 and biomolecular motors44. More recently, researchers have utilized DNA origami technology to construct bundled DNA beams that are much more rigid than conventional dsDNA for ultra-high-resolution measurements45, 46
EXPLOIT DNA MECHANICS TO STUDY BIOLOGY
DNA is under constant tension inside the cell: it is wrapped around histones47, unwound by helicases48, twisted and untwisted by RNA polymerases49 and topoisomerases50, to name a few examples. The elucidation of the mechanical properties of DNA has greatly facilitated the study of molecular machines that act on DNA, yielding mechanistic insights into their force-generation mechanisms and the coordination of their components51. Below we use two example systems to demonstrate how the knowledge of DNA elasticity has inspired creative assays for studying biological processes.
Viral DNA packaging
The exact derivation of DNA extension as a function of force makes it possible to convert movement in distance into changes in base pairs, thereby providing exquisite insight into the operating mechanism of protein machineries that translocate on DNA, such as RNA polymerases52 and viral DNA packaging motors53. In the latter case, dsDNA genomes are pumped into preformed protein capsids during viral assembly. The ring-shaped packaging motors are among the most powerful molecular machines found in nature. To compact the stiff, highly charged dsDNA to near-crystalline densities into a small capsid, the packaging motor needs to generate a large amount of force—as high as 60 pN—in order to overcome major energetic barriers. The packaging motor of bacteriophage φ29, one of the best-characterized molecular machines thus far, is a homopentameric ring ATPase that packs a 19.3-kbp genome into a capsid 50 nm in height and 40 nm in diameter54. Using a dual-trap optical tweezers instrument, the Bustamante group was able to detect discrete DNA translocation cycles of the ring motor (Fig. 2A). Under low external forces (<10 pN), DNA is translocated in 10-bp cycles, each consisting of an ATP-binding dwell phase and a DNA-translocating burst phase (Fig. 2B). Under high external forces (30–40 pN), each 10-bp burst is decelerated and can be shown to be composed of four 2.5-bp steps. Each step is powered by the release of an inorganic phosphate molecule produced from ATP hydrolysis. Subsequent experiments using nucleotide analogs and ATPase mutants mapped specific chemical transitions (such as ATP hydrolysis and ADP release) onto the dwell-burst pathway, revealing intricate, clocklike coordination among the five ring subunits (Fig. 2C)55–57.
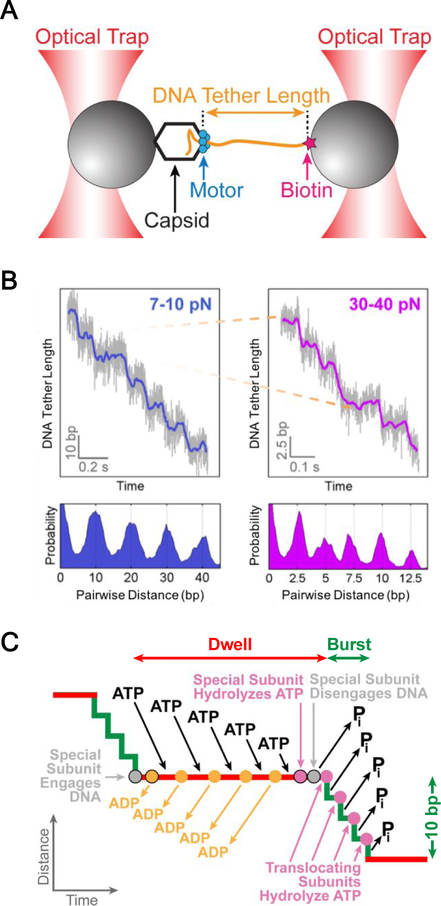
Figure 2.
Optical-trapping assay to study viral DNA packaging (A) Schematic of a dual-trap optical tweezers assay to study DNA translocation by the bacteriophage φ29 packaging motor. Figure reproduced with permission from58. (B) (Top) Representative packaging traces collected at low (Left) and high force (Right). At high force, the 10-bp bursts seen at low force are decelerated enough to reveal 2.5-bp steps. (Bottom) Pairwise distance analysis for the corresponding traces. Figure reproduced with permission from56. (C) Mechanochemical model for the φ29 packaging motor showing DNA translocation cycles with a dwell-burst structure. Figure adapted with permission from58.
The non-integer 2.5-bp step size is surprising. This result argues against any mechanism in which every motor subunit makes identical chemical contacts with the DNA during translocation. It further suggests that only four of the five subunits participate in DNA translocation per cycle. Indeed, it was shown that the translocating subunits make nonspecific contacts with the DNA during the burst phase59, and that the ring symmetry is broken through specific electrostatic interaction with DNA backbone phosphates, bestowing a special regulatory role upon the contacting subunit56, 57.
It was proposed60 and experimentally observed61 that DNA inside the capsid is organized into a spool, which may require rotation of the DNA in order to relieve the torsional strain. In addition, the small difference between the 10.0-bp burst size of the motor and the 10.4-bp helical pitch of B-form DNA entails that the DNA may need to rotate relative to the motor to make crucial electrostatic contacts at the beginning of each cycle. To directly probe DNA rotation during translocation, a third “rotor bead” was introduced to the standard two-bead optical tweezers assay, which allowed angular changes of the DNA around its helical axis to be monitored concomitantly with its linear translocation58. A similar setup was previously used to measure the twist elasticity of DNA62. The DNA was shown to rotate on average by ~1.5 degrees per base pair in a left-handed direction. Thus, the packaging motor can simultaneously generate force and torque, both of which can reach values high enough to denature DNA. The quantitation of DNA rotation also suggests that the same subunit makes the specific DNA interactions cycle after cycle, thus significantly constraining the possible models for the identity of the special subunit. After a 10-bp burst, the DNA backbone winds by 346 degrees. Thus a 14-degree rotation is required to re-align the DNA with the subunit that makes contacts in the previous cycle, thereby yielding an average rotation density of 1.4 degree/bp.
DNA packaging has long been anticipated to slow down as DNA fills up the capsid due to the mounting internal pressure working against the motor. This was directly proven by the single-molecule packaging assay showing that the velocity of the φ29 packaging motor drops from an initial value of >100 bp/s to essentially zero when the entire genome length is internalized63. Subsequent high-resolution measurements revealed that the internal pressure affects multiple aspects of the mechanochemical cycle of the motor, including slowing ATP binding during the dwell phase and DNA translocation during the burst phase58. These results led to an estimation of the final internal pressure to be ~20 atm—a remarkably large number that is consistent with predictions from analytical modeling and numerical simulation studies64–66. Furthermore, the φ29 packaging motor was observed to take smaller bursts per cycle (10 bp at low filling vs. 9 bp at high filling) and smaller elementary steps (2.5 bp at low filling vs. 2.3 bp at high filling). An accompanying change in the DNA rotation density was also observed (~1.5 degree/bp at low filling vs. ~5 degree/bp at high filling). These concerted adjustments ensure that the distinct functions and coordination of the ring subunits are preserved even in the face of drastically different operating conditions58. In addition, a recent simulation study suggested that, instead of being a passive substrate, the DNA itself is an active component of the packaging machinery, driving its own translocation by undergoing cyclic conformational distortions inside the viral portal channel67. Overall, this model system showcases how knowledge in DNA mechanics allows for a detailed mechanistic dissection of the 3D trajectories of molecular machines that track on DNA.
DNA replication
Many DNA-based cellular processes involve interconversion between dsDNA and ssDNA. DNA duplex is unzipped into two separate strands by a multitude of helicases or degraded into ssDNA by exonucleases. On the other hand, DNA polymerases copy ssDNA templates into duplexes. The differential elasticity between ssDNA and dsDNA has been exploited to follow the progression of these biochemical reactions in real time without having to fluorescently label the enzymes. This was first used to study bacteriophage T7 DNA polymerase by optical tweezers28 and magnetic tweezers68. Interestingly, it was found that at high forces (>40 pN), the nucleolytic activity of the polymerase is greatly stimulated, effectively converting the polymerase into an exonuclease28. Moreover, monitoring the enzymatic behavior as a function of applied tension unveiled intermediate states in the kinetic pathway that are related to the proofreading activity of the polymerase69.
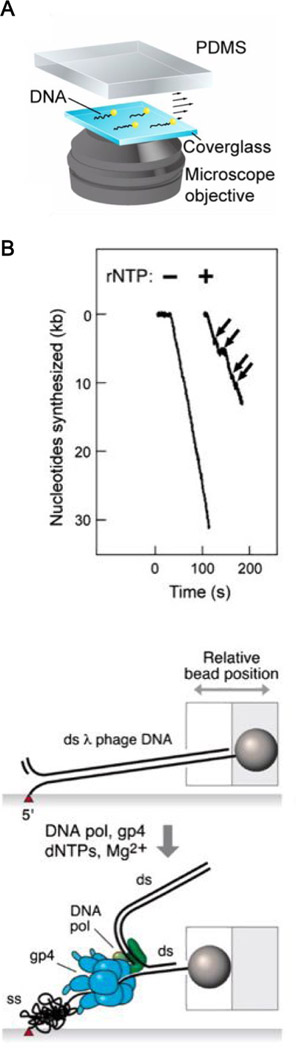
van Oijen and coworkers developed a single-molecule DNA flow-stretching assay that can monitor many molecules at the same time, thus affording a higher throughput than the optical tweezers assay. In this setup, bacteriophage λ genomic DNA is anchored to a coverslip surface on one end and conjugated to a micron-sized bead on the other end. The DNA is then stretched by a hydrodynamic flow (Fig. 3A). This setup was first utilized to study the activity of phage λ exonuclease, which converts dsDNA into ssDNA70. Later, this setup was applied to the phage T7 DNA replication system71. Here a bead is attached to the parental end of a forked DNA substrate. At a typical stretching force of ~2 pN, ssDNA is much shorter than dsDNA (Fig. 1B). Leading-strand synthesis results in a shortening of the tether length due to accumulation of the lagging-strand ssDNA (Fig. 3B). In the presence of lagging-strand synthesis, gradual shortening of the DNA followed by sudden lengthening was observed, which was interpreted as formation and subsequent fast release of replication loops in the lagging strand. These looping events were also observed in single-molecule FRET72 and magnetic tweezers assays73. Further analysis of the loop sizes and lag times between loops suggested that the initiation of primer synthesis (“signaling” mechanism) and the encounter with a downstream Okazaki fragment (“collision” mechanism) can both serve as a trigger for loop release. Such a dual-trigger mechanism ensures timely reset of the enzymatic apparatus at the replication fork after the completion of each round of Okazaki fragment synthesis74. Thus, this simple and elegant assay, chiefly utilizing DNA elasticity, yielded key insights into the coordination between continuous leading-strand synthesis and discontinuous lagging-strand synthesis.
Figure 3.
Flow-stretching assay to study DNA replication (A) Schematic of a DNA flow-stretching assay. Individual DNA molecules are tethered to the surface of a flow cell on one end and conjugated to a bead on the other end. Figure reproduced with permission from75. (B) (Top) Representative single-molecule trajectories with and without ribonucleotides required for lagging-strand priming. Arrows indicate pausing events. (Bottom) Schematic depicting that leading-strand synthesis causes conversion of the 5’ tail of the tethered strand from dsDNA to ssDNA, resulting in a decrease in the tether length. Figure reproduced with permission from76, originally adapted with permission from71.
The same group later developed a more sophisticated assay to simultaneously follow T7 leading-strand synthesis and lagging-strand loop formation by attaching two beads to two arms of the forked DNA77. This assay revealed that the looping events in the lagging strand contain both ssDNA loops formed during priming (priming loop, more frequent) and ss-ds ones that support Okazaki fragment synthesis (replication loop, less frequent). This study also showed that lagging-strand polymerases are often released from the replisome to complete Okazaki fragment synthesis behind the fork. These results depict a highly plastic replisome that can access multiple reaction pathways to achieve efficient and robust replication.
The DNA flow-stretching assay has also been applied to the E. coli replisome to investigate the processivity of the DNA polymerase III holoenzyme and its regulation by the DnaB helicase and DnaG primase78. More recently, it was demonstrated in a eukaryotic system from S. cerevisiae75, which utilizes dedicated leading- and lagging-strand polymerases and more regulatory factors. Meanwhile, a number of other single-molecule assays based on fluorescence imaging of stretched DNA (for example, rolling circle and DNA curtains) have also been developed to study various aspects of DNA replication, such as initiation, lesion bypass, and polymerase exchange79–82.
CORRELATIVE INTERROGATION OF DNA STRUCTURE AND MECHANICS
The combination of single-molecule fluorescence spectroscopy and force spectroscopy allows multiplexed measurements of DNA conformation and protein-DNA interaction83–87. Such interrogation has enabled direct correlation between the force applied to DNA and the structural transitions that it undergoes, and has yielded key information on the stoichiometry, dynamics, and force dependence of DNA-templated molecular assemblies.
Force-induced conversion between dsDNA and ssDNA
Using a single-molecule instrument that combines confocal fluorescence microscopy, dual-trap optical tweezers, and automated microfluidics, Wuite, Peterman and coworkers directly visualized the structural transitions of DNA during overstretching33. They used the intercalating dye YOYO to stain dsDNA that was linked to optically trapped beads via the 3’ end of each strand (Fig. 4A). While the dsDNA molecule was stained along its full length under low forces, unstained segments appeared and grew when the tension was raised above 65 pN (Fig. 4B). These segments preferentially initiated from free DNA ends and internal nicks. To test whether these segments correspond to ssDNA regions, the authors then used fluorescently labeled ssDNA-binding protein (SSB), which wraps around ssDNA88. SSB foci were observed at edges of YOYO-labeled dsDNA segments and became brighter as the force increased. These results were interpreted as SSB binding to relaxed ssDNA unpeeled from free ends and nicks. To study overstretching of DNA without preferred nucleation sites, the authors also designed DNA substrates that were linked to beads via both ends of each strand to prevent peeling from free ends (Fig. 4C). Notably, these torsionally constrained constructs displayed an overstretching plateau at much higher forces (~110 pN), in agreement with an earlier report89. Single-stranded regions were generated above 110 pN, as indicated by the binding of RPA, another ssDNA-binding protein that can bind to ssDNA under tension (unlike SSB). This observation can be interpreted as the formation of single-stranded “melting bubbles”. Overall, these results demonstrated force-induced melting of dsDNA into ssDNA during overstretching regardless of the DNA attachment geometry.
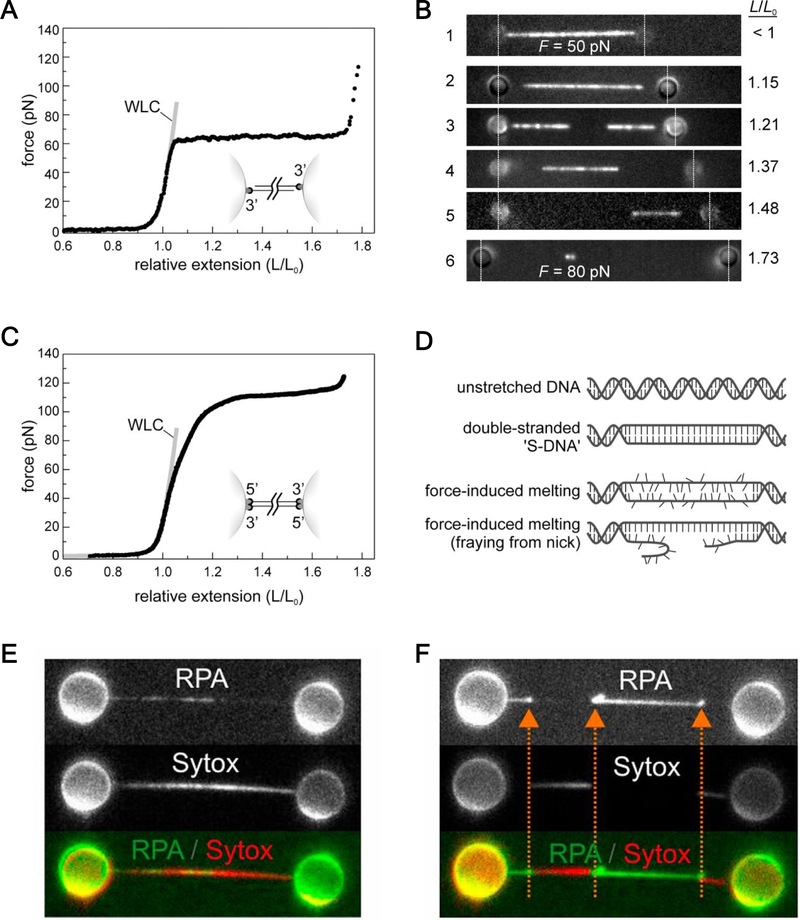
Figure 4.
DNA structural transitions revealed by combined fluorescence-force microscopy (A) Representative force-extension curve for dsDNA tethered to optically trapped beads via the 3’ end of each strand. (B) Fluorescence images of a dsDNA molecule tethered by the geometry depicted in (A) and stained with the intercalating dye YOYO. At tensions resulting in tether lengths greater than the contour length (L0), unstained regions emerged, signifying the overstretching transition. (C) Representative force-extension curve for dsDNA tethered to beads via both ends of each strand. Here the overstretching transition occurs at ~110 pN. (D) Cartoon representations of various structural forms when dsDNA is overstretched. (E) Fluorescence images of an overstretched DNA that is topologically closed but torsionally relaxed. At low ionic strength, melting bubbles are visualized with fluorescent RPA. dsDNA regions are indicated by the intercalating dye Sytox. (F) Fluorescence images of an overstretched and nicked DNA. Here strand unpeeling is favored, as indicated by the orange arrows. Panels A-D are reproduced with permission from33. Panels E and F are reproduced with permission from90.
In a follow-up study90, the same group focused on the competition between three processes during overstretching: strand unpeeling, localized base-pair breaking (melting bubbles), and S-DNA formation (strand unwinding with base-pairing maintained) (Fig. 4D). The authors found that all three mechanisms are at work. In topologically closed but torsionally relaxed DNA where free ends and nicks are lacking, melting bubbles form preferentially at AT-rich regions at low ionic strength (Fig. 4E). The same type of construct was also used by another group to show that strand unpeeling is not a requirement for the overstretching transition at 65 pN91. High ionic strength, by contrast, stabilizes the double helix and inhibits melting-bubble formation. Instead, a different structural form results that cannot by stained by either RPA or Sytox (a dsDNA-intercalating dye). The authors interpreted this form, likely base-paired, as evidence for S-DNA. Increasing ionic strength also suppresses unpeeling and promotes S-DNA formation for topologically open DNA. Thus, the balance between these different processes during overstretching is dependent on DNA sequence, topology, and salt concentration (Fig. 4F). An accompanying paper employed magnetic tweezers to study the same transitions and reached essentially the same conclusion92. By investigating the temperature effect, the latter study calculated the entropic contribution from each of the three processes. The unpeeling and bubble-melting transitions are hysteretic with a positive entropy change of 17 cal/(K·mol), similar to thermal melting, whereas the B-to-S transition is nonhysteretic with a small negative entropy change of −2 cal/(K·mol). Moreover, a recent study adopted concurrent fluorescence polarization imaging and force manipulation to show that base pairs in S-DNA are substantially more tilted than those in B-DNA93.
In these types of studies, it is worth keeping in mind the potential perturbation of DNA structure introduced by the intercalating dyes and ssDNA-binding proteins94, 95. Thus, a low concentration of these reagents is recommended whenever possible. It has also been reported that the fluorescence intensity of intercalating dyes can be used as a sensor for the local tension in dsDNA96.
Formation of nucleoprotein filaments
Many essential genomic transactions, such as DNA replication, repair, and recombination, involve the generation of ssDNA. Single-molecule studies of these reactions are aided by various experimental strategies to produce long ssDNA templates, such as force-induced duplex melting97, 98. Using combined fluorescence and force spectroscopy, Ha and coworkers showed that SSB can rapidly diffuse along ssDNA, which facilitates its redistribution99. The force dependence of SSB movement suggests that it migrates on DNA via intersegment transfer100.
Besides SSB and RPA, there are other proteins that bind to ssDNA—notably RecA for prokaryotes and Rad51 for eukaryotes. These ATPases form nucleoprotein filaments on ssDNA in an ATP-dependent manner and play critical roles in homology search and pairing during homologous recombination101. In an earlier study, the Bustamante group interrogated the mechanical properties of RecA-ssDNA and RecA-dsDNA filaments at various nucleotide states102. It was found that RecA significantly stiffens and elongates DNA (both RecA-ssDNA and RecA-dsDNA filaments are ~1.5 times longer than bare B-form DNA). Combining fluorescence imaging and flow stretching, the Kowalczykowski group monitored the nucleation and bidirectional growth of RecA-dsDNA filaments103 and examined the inhibitory effect of SSB on RecA-ssDNA filament assembly104. The same group also investigated the process of homology search by RecA-ssDNA filaments on dsDNA105. They found that the number of available DNA conformations, which decreases as the end-to-end distance of the target dsDNA is increased, exerts a major influence on the rate of homologous pairing. Using a dual-molecule manipulation setup combining optical tweezers and magnetic tweezers, the Dekker group also studied the process of homology recognition106. By controlling the supercoiling state of the target dsDNA, the authors found that homologous pairing is strongly enhanced by underwinding of the target DNA, which facilitates transient engagement of the incoming duplex by a secondary DNA-binding site in the RecA filament. Both of these studies pointed to a mechanism that harnesses nonspecific and weak interactions between the RecA filament and target dsDNA for rapid homology search.
Similar to RecA, Rad51 binding also extends the DNA by about 50%107. The disassembly of Rad51 nucleoprotein filaments occurs at the filament terminus one monomer at a time upon ATP hydrolysis, and is sensitive to tension, stalling at forces above 50 pN108. By investigating the competition between RPA and Rad51 for DNA binding, Greene and coworkers showed that free RPA in solution inhibits Rad51 filament nucleation, but the elongation of Rad51 filaments on RPA-coated ssDNA is not significantly impacted109. Interestingly, it was recently reported that RecA/Rad51 polymerizes faster on S-DNA than on B-DNA110, implying potential physiological functions of S-DNA.
OUTLOOK
Single-molecule force-manipulation techniques enable precise measurements made at scales highly relevant to the description of DNA mechanics (picoNewtons and nanometers). Once its elastic properties were well understood, DNA became an indispensable tool for studying the mechanism of DNA-based biological processes. The integration of fluorescence detection modality into single-molecule force spectroscopy allows the conformational and mechanical dynamics of DNA or DNA-templated complexes to be simultaneously monitored. These instruments are increasingly becoming commercially available111, providing non-specialists with access to this type of investigation. At the same time, physicists and biophysicists are constantly making technological innovations to further enhance the multiplexity and throughput of single-molecule assays112, 113. Another critical element for achieving the full power of this type of studies is progress made in the in vitro reconstitution of complex biochemical systems—such as the eukaryotic replisome114–116—which allows the function of each component to be unambiguously dissected. Finally, characterization of the physical properties of chromatin and whole chromosomes will contribute to the understanding of genome organization and transcriptional regulation inside the cell117. It is anticipated that continued exploration of the mechanical nature of the double helix will lead to a deeper appreciation of how this fundamental molecule orchestrates the operation of living systems.
ACKNOWLEDGEMENTS
M.R.W. is an Anderson Cancer Center Postdoctoral Fellow at the Rockefeller University. S.L. is supported by the Robertson Foundation and the National Institutes of Health (DP2HG010510).
Footnotes
Publisher's Disclaimer: This document is confidential and is proprietary to the American Chemical Society and its authors. Do not copy or disclose without written permission. If you have received this item in error, notify the sender and delete all copies.
REFERENCES
- [1]. Watson JD, and Crick FH (1953) Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid, Nature 171, 737–738. [DOI] [PubMed] [Google Scholar]
- [2]. Wilkins MH, Stokes AR, and Wilson HR (1953) Molecular structure of deoxypentose nucleic acids, Nature 171, 738–740. [DOI] [PubMed] [Google Scholar]
- [3]. Franklin RE, and Gosling RG (1953) Molecular configuration in sodium thymonucleate, Nature 171, 740–741. [DOI] [PubMed] [Google Scholar]
- [4]. Griffith J, Huberman JA, and Kornberg A (1971) Electron microscopy of DNA polymerase bound to DNA, Journal of moìecuìar biology 55, 209–214. [DOI] [PubMed] [Google Scholar]
- [5]. Morikawa K, and Yanagida M (1981) Visualization of individual DNA molecules in solution by light microscopy: DAPI staining method, Journal of biochemistry 89, 693–696. [DOI] [PubMed] [Google Scholar]
- [6]. Houseal TW, Bustamante C, Stump RF, and Maestre MF (1989) Real-time imaging of single DNA molecules with fluorescence microscopy, Biophysical journal 56, 507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7]. Gurrieri S, Smith SB, Wells KS, Johnson ID, and Bustamante C (1996) Real-time imaging of the reorientation mechanisms of YOYO-labelled DNA molecules during 90 degrees and 120 degrees pulsed field gel electrophoresis, Nucleic acids research 24, 4759–4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8]. Bustamante C, Bryant Z, and Smith SB (2003) Ten years of tension: single-molecule DNA mechanics, Nature 421, 423–427. [DOI] [PubMed] [Google Scholar]
- [9]. Bryant Z, Oberstrass FC, and Basu A (2012) Recent developments in single-molecule DNA mechanics, Current opinion in structural biology 22, 304–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10]. Ha T (2016) Probing Nature’s Nanomachines One Molecule at a Time, Biophysical journal 110, 1004–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11]. Bustamante C (2017) Molecular machines one molecule at a time, Protein science: a publication of the Protein Society 26, 1245–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12]. Neuman KC, and Nagy A (2008) Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy, Nature methods 5, 491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13]. Smith SB, Finzi L, and Bustamante C (1992) Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads, Science 258, 1122–1126. [DOI] [PubMed] [Google Scholar]
- [14]. Strick TR, Allemand JF, Bensimon D, Bensimon A, and Croquette V (1996) The elasticity of a single supercoiled DNA molecule, Science 271, 1835–1837. [DOI] [PubMed] [Google Scholar]
- [15]. Cluzel P, Lebrun A, Heller C, Lavery R, Viovy JL, Chatenay D, and Caron F (1996) DNA: an extensible molecule, Science 271, 792–794. [DOI] [PubMed] [Google Scholar]
- [16]. Perkins TT, Smith DE, Larson RG, and Chu S (1995) Stretching of a single tethered polymer in a uniform flow, Science 268, 83–87. [DOI] [PubMed] [Google Scholar]
- [17]. Smith SB, Cui Y, and Bustamante C (1996) Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules, Science 271, 795–799. [DOI] [PubMed] [Google Scholar]
- [18]. Bustamante C, Marko JF, Siggia ED, and Smith S (1994) Entropic elasticity of lambda-phage DNA, Science 265, 1599–1600. [DOI] [PubMed] [Google Scholar]
- [19]. Wang MD, Yin H, Landick R, Gelles J, and Block SM (1997) Stretching DNA with optical tweezers, Biophysical journal 72, 1335–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20]. Baumann CG, Smith SB, Bloomfield VA, and Bustamante C (1997) Ionic effects on the elasticity of single DNA molecules, Proceedings of the National Academy of Sciences of the United States of America 94, 6185–6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21]. Vafabakhsh R, and Ha T (2012) Extreme bendability of DNA less than 100 base pairs long revealed by single-molecule cyclization, Science 337, 1097–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22]. Yan J, and Marko JF (2004) Localized single-stranded bubble mechanism for cyclization of short double helix DNA, Physical review letters 93, 108108. [DOI] [PubMed] [Google Scholar]
- [23]. Wiggins PA, Phillips R, and Nelson PC (2005) Exact theory of kinkable elastic polymers, Physical review. E, Statistical, nonlinear, and soft matter physics 71, 021909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24]. Gross P, Laurens N, Oddershede LB, Bockelmann U, Peterman EJ, and Wuite GJ (2011) Quantifying how DNA stretches, melts and changes twist under tension, Nat Phys 7, 731–736. [Google Scholar]
- [25]. Strick TR, Allemand JF, Bensimon D, and Croquette V (1998) Behavior of supercoiled DNA, Biophysical journal 74, 2016–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26]. Gore J, Bryant Z, Nollmann M, Le MU, Cozzarelli NR, and Bustamante C (2006) DNA overwinds when stretched, Nature 442, 836–839. [DOI] [PubMed] [Google Scholar]
- [27]. Heller I, Hoekstra TP, King GA, Peterman EJ, and Wuite GJ (2014) Optical tweezers analysis of DNA-protein complexes, Chemical reviews 114, 3087–3119. [DOI] [PubMed] [Google Scholar]
- [28]. Wuite GJ, Smith SB, Young M, Keller D, and Bustamante C (2000) Single-molecule studies of the effect of template tension on T7 DNA polymerase activity, Nature 404, 103–106. [DOI] [PubMed] [Google Scholar]
- [29]. Sarkar A, Leger JF, Chatenay D, and Marko JF (2001) Structural transitions in DNA driven by external force and torque, Physical review. E, Statistical, nonlinear, and soft matter physics 63, 051903. [DOI] [PubMed] [Google Scholar]
- [30]. Camunas-Soler J, Ribezzi-Crivellari M, and Ritort F (2016) Elastic Properties of Nucleic Acids by Single-Molecule Force Spectroscopy, Annual review of biophysics 45, 65–84. [DOI] [PubMed] [Google Scholar]
- [31]. Calladine CR, Drew HR, Luisi BF, and Travers AA (2004) Understanding DNA: The molecule and how it works, Elsevier Academic Press. [Google Scholar]
- [32]. Williams MC, Rouzina I, and Bloomfield VA (2002) Thermodynamics of DNA interactions from single molecule stretching experiments, Accounts of chemical research 35, 159–166. [DOI] [PubMed] [Google Scholar]
- [33]. van Mameren J, Gross P, Farge G, Hooijman P, Modesti M, Falkenberg M, Wuite GJ, and Peterman EJ (2009) Unraveling the structure of DNA during overstretching by using multicolor, single-molecule fluorescence imaging, Proceedings of the National Academy of Sciences of the United States of America 106, 18231–18236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34]. Fu H, Chen H, Marko JF, and Yan J (2010) Two distinct overstretched DNA states, Nucleic acids research 38, 5594–5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35]. Bosaeus N, El-Sagheer AH, Brown T, Smith SB, Akerman B, Bustamante C, and Norden B (2012) Tension induces a base-paired overstretched DNA conformation, Proceedings of the National Academy of Sciences of the United States of America 109, 15179–15184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36]. Allemand JF, Bensimon D, Lavery R, and Croquette V (1998) Stretched and overwound DNA forms a Pauling-like structure with exposed bases, Proceedings of the National Academy of Sciences of the United States of America 95, 14152–14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37]. King GA, Peterman EJ, and Wuite GJ (2016) Unravelling the structural plasticity of stretched DNA under torsional constraint, Nature communications 7, 11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38]. Forth S, Sheinin MY, Inman J, and Wang MD (2013) Torque measurement at the single-molecule level, Annual review of biophysics 42, 583–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39]. Lipfert J, van Oene MM, Lee M, Pedaci F, and Dekker NH (2015) Torque spectroscopy for the study of rotary motion in biological systems, Chemical reviews 115, 1449–1474. [DOI] [PubMed] [Google Scholar]
- [40]. Sheinin MY, Forth S, Marko JF, and Wang MD (2011) Underwound DNA under tension: structure, elasticity, and sequence-dependent behaviors, Physical review letters 107, 108102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41]. Hao Y, Canavan C, Taylor SS, and Maillard RA (2017) Integrated Method to Attach DNA Handles and Functionally Select Proteins to Study Folding and Protein-Ligand Interactions with Optical Tweezers, Scientific reports 7, 10843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42]. Liphardt J, Onoa B, Smith SB, Tinoco I Jr., and Bustamante C (2001) Reversible unfolding of single RNA molecules by mechanical force, Science 292, 733–737. [DOI] [PubMed] [Google Scholar]
- [43]. Liu K, Maciuba K, and Kaiser CM (2019) The Ribosome Cooperates with a Chaperone to Guide Multi-domain Protein Folding, Molecular cell 74, 310–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44]. Zananiri R, Malik O, Rudnizky S, Gaydar V, Kreiserman R, Henn A, and Kaplan A (2019) Synergy between RecBCD subunits is essential for efficient DNA unwinding, eLife 8, e40836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45]. Pfitzner E, Wachauf C, Kilchherr F, Pelz B, Shih WM, Rief M, and Dietz H (2013) Rigid DNA beams for high-resolution single-molecule mechanics, Angewandte Chemie 52, 7766–7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46]. Kilchherr F, Wachauf C, Pelz B, Rief M, Zacharias M, and Dietz H (2016) Single-molecule dissection of stacking forces in DNA, Science 353, aaf5508. [DOI] [PubMed] [Google Scholar]
- [47]. Hall MA, Shundrovsky A, Bai L, Fulbright RM, Lis JT, and Wang MD (2009) High-resolution dynamic mapping of histone-DNA interactions in a nucleosome, Nature structural & molecular biology 16, 124–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48]. Burnham DR, Kose HB, Hoyle RB, and Yardimci H (2019) The mechanism of DNA unwinding by the eukaryotic replicative helicase, Nature communications 10, 2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49]. Ma J, Bai L, and Wang MD (2013) Transcription under torsion, Science 340, 1580–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50]. Stone MD, Bryant Z, Crisona NJ, Smith SB, Vologodskii A, Bustamante C, and Cozzarelli NR (2003) Chirality sensing by Escherichia coli topoisomerase IV and the mechanism of type II topoisomerases, Proceedings of the National Academy of Sciences of the United States of America 100, 8654–8659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51]. Bustamante C, Cheng W, and Mejia YX (2011) Revisiting the central dogma one molecule at a time, Cell 144, 480–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52]. Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, and Block SM (2005) Direct observation of base-pair stepping by RNA polymerase, Nature 438, 460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53]. Moffitt JR, Chemla YR, Aathavan K, Grimes S, Jardine PJ, Anderson DL, and Bustamante C (2009) Intersubunit coordination in a homomeric ring ATPase, Nature 457, 446–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54]. Mao H, Saha M, Reyes-Aldrete E, Sherman MB, Woodson M, Atz R, Grimes S, Jardine PJ, and Morais MC (2016) Structural and Molecular Basis for Coordination in a Viral DNA Packaging Motor, Cell reports 14, 2017–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55]. Chemla YR, Aathavan K, Michaelis J, Grimes S, Jardine PJ, Anderson DL, and Bustamante C (2005) Mechanism of force generation of a viral DNA packaging motor, Cell 122, 683–692. [DOI] [PubMed] [Google Scholar]
- [56]. Chistol G, Liu S, Hetherington CL, Moffitt JR, Grimes S, Jardine PJ, and Bustamante C (2012) High degree of coordination and division of labor among subunits in a homomeric ring ATPase, Cell 151, 1017–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57]. Tafoya S, Liu S, Castillo JP, Atz R, Morais MC, Grimes S, Jardine PJ, and Bustamante C (2018) Molecular switch-like regulation enables global subunit coordination in a viral ring ATPase, Proceedings of the National Academy of Sciences of the United States of America 115, 7961–7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58]. Liu S, Chistol G, Hetherington CL, Tafoya S, Aathavan K, Schnitzbauer J, Grimes S, Jardine PJ, and Bustamante C (2014) A viral packaging motor varies its DNA rotation and step size to preserve subunit coordination as the capsid fills, Cell 157, 702–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59]. Aathavan K, Politzer AT, Kaplan A, Moffitt JR, Chemla YR, Grimes S, Jardine PJ, Anderson DL, and Bustamante C (2009) Substrate interactions and promiscuity in a viral DNA packaging motor, Nature 461, 669–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60]. Hendrix RW (1978) Symmetry mismatch and DNA packaging in large bacteriophages, Proceedings of the National Academy of Sciences of the United States of America 75, 4779–4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61]. Xu J, Wang D, Gui M, and Xiang Y (2019) Structural assembly of the tailed bacteriophage varphi29, Nature communications 10, 2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62]. Bryant Z, Stone MD, Gore J, Smith SB, Cozzarelli NR, and Bustamante C (2003) Structural transitions and elasticity from torque measurements on DNA, Nature 424, 338–341. [DOI] [PubMed] [Google Scholar]
- [63]. Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, and Bustamante C (2001) The bacteriophage straight phi29 portal motor can package DNA against a large internal force, Nature 413, 748–752. [DOI] [PubMed] [Google Scholar]
- [64]. Kindt J, Tzlil S, Ben-Shaul A, and Gelbart WM (2001) DNA packaging and ejection forces in bacteriophage, Proceedings of the National Academy of Sciences of the United States of America 98, 13671–13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65]. Purohit PK, Inamdar MM, Grayson PD, Squires TM, Kondev J, and Phillips R (2005) Forces during bacteriophage DNA packaging and ejection, Biophysical journal 88, 851–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66]. Spakowitz AJ, and Wang ZG (2005) DNA packaging in bacteriophage: is twist important?, Biophysical journal 88, 3912–3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67]. Sharp KA, Lu X-J, Cingolani G, and Harvey SC (2019) DNA conformational changes play a force-generating role during bacteriophage genome packaging, Biophysical journal 116, 2172–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68]. Maier B, Bensimon D, and Croquette V (2000) Replication by a single DNA polymerase of a stretched single-stranded DNA, Proceedings of the National Academy of Sciences of the United States of America 97, 12002–12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69]. Ibarra B, Chemla YR, Plyasunov S, Smith SB, Lazaro JM, Salas M, and Bustamante C (2009) Proofreading dynamics of a processive DNA polymerase, The EMBO journal 28, 2794–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70]. van Oijen AM, Blainey PC, Crampton DJ, Richardson CC, Ellenberger T, and Xie XS (2003) Single-molecule kinetics of lambda exonuclease reveal base dependence and dynamic disorder, Science 301, 1235–1238. [DOI] [PubMed] [Google Scholar]
- [71]. Lee JB, Hite RK, Hamdan SM, Xie XS, Richardson CC, and van Oijen AM (2006) DNA primase acts as a molecular brake in DNA replication, Nature 439, 621–624. [DOI] [PubMed] [Google Scholar]
- [72]. Pandey M, Syed S, Donmez I, Patel G, Ha T, and Patel SS (2009) Coordinating DNA replication by means of priming loop and differential synthesis rate, Nature 462, 940–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73]. Manosas M, Spiering MM, Zhuang Z, Benkovic SJ, and Croquette V (2009) Coupling DNA unwinding activity with primer synthesis in the bacteriophage T4 primosome, Nature chemical biology 5, 904–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74]. Hamdan SM, Loparo JJ, Takahashi M, Richardson CC, and van Oijen AM (2009) Dynamics of DNA replication loops reveal temporal control of lagging-strand synthesis, Nature 457, 336–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75]. Lewis JS, Spenkelink LM, Schauer GD, Hill FR, Georgescu RE, O’Donnell ME, and van Oijen AM (2017) Single-molecule visualization of Saccharomyces cerevisiae leading-strand synthesis reveals dynamic interaction between MTC and the replisome, Proceedings of the National Academy of Sciences of the United States of America 114, 10630–10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76]. van Oijen AM (2007) Honey, I shrunk the DNA: DNA length as a probe for nucleic-acid enzyme activity, Biopolymers 85, 144–153. [DOI] [PubMed] [Google Scholar]
- [77]. Duderstadt KE, Geertsema HJ, Stratmann SA, Punter CM, Kulczyk AW, Richardson CC, and van Oijen AM (2016) Simultaneous Real-Time Imaging of Leading and Lagging Strand Synthesis Reveals the Coordination Dynamics of Single Replisomes, Molecular cell 64, 1035–1047. [DOI] [PubMed] [Google Scholar]
- [78]. Tanner NA, Hamdan SM, Jergic S, Loscha KV, Schaeffer PM, Dixon NE, and van Oijen AM (2008) Single-molecule studies of fork dynamics in Escherichia coli DNA replication, Nature structural & molecular biology 15, 170–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79]. Loparo JJ, Kulczyk AW, Richardson CC, and van Oijen AM (2011) Simultaneous single-molecule measurements of phage T7 replisome composition and function reveal the mechanism of polymerase exchange, Proceedings of the National Academy of Sciences of the United States of America 108, 3584–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80]. Duzdevich D, Warner MD, Ticau S, Ivica NA, Bell SP, and Greene EC (2015) The dynamics of eukaryotic replication initiation: origin specificity, licensing, and firing at the single-molecule level, Molecular cell 58, 483–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81]. Graham JE, Marians KJ, and Kowalczykowski SC (2017) Independent and Stochastic Action of DNA Polymerases in the Replisome, Cell 169, 1201–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82]. Sparks JL, Chistol G, Gao AO, Raschle M, Larsen NB, Mann M, Duxin JP, and Walter JC (2019) The CMG Helicase Bypasses DNA-Protein Cross-Links to Facilitate Their Repair, Cell 176, 167–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83]. Harada Y, Funatsu T, Murakami K, Nonoyama Y, Ishihama A, and Yanagida T (1999) Single-molecule imaging of RNA polymerase-DNA interactions in real time, Biophysical journal 76, 709–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84]. Lang MJ, Fordyce PM, and Block SM (2003) Combined optical trapping and single-molecule fluorescence, Journal of biology 2, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85]. Candelli A, Wuite GJ, and Peterman EJ (2011) Combining optical trapping, fluorescence microscopy and micro-fluidics for single molecule studies of DNA-protein interactions, Physical chemistry chemical physics: PCCP 13, 7263–7272. [DOI] [PubMed] [Google Scholar]
- [86]. Comstock MJ, Ha T, and Chemla YR (2011) Ultrahigh-resolution optical trap with single-fluorophore sensitivity, Nature methods 8, 335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87]. Graves ET, Duboc C, Fan J, Stransky F, Leroux-Coyau M, and Strick TR (2015) A dynamic DNA-repair complex observed by correlative single-molecule nanomanipulation and fluorescence, Nature structural & molecular biology 22, 452–457. [DOI] [PubMed] [Google Scholar]
- [88]. Suksombat S, Khafizov R, Kozlov AG, Lohman TM, and Chemla YR (2015) Structural dynamics of E. coli single-stranded DNA binding protein reveal DNA wrapping and unwrapping pathways, eLife 4, e08193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89]. Leger JF, Romano G, Sarkar A, Robert J, Bourdieu L, Chatenay D, and Marko JF (1999) Structural transitions of a twisted and stretched DNA molecule, Physical review letters 83, 1066–1069. [Google Scholar]
- [90]. King GA, Gross P, Bockelmann U, Modesti M, Wuite GJ, and Peterman EJ (2013) Revealing the competition between peeled ssDNA, melting bubbles, and S-DNA during DNA overstretching using fluorescence microscopy, Proceedings of the National Academy of Sciences of the United States of America 110, 3859–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91]. Paik DH, and Perkins TT (2011) Overstretching DNA at 65 pN does not require peeling from free ends or nicks, Journal of the American Chemical Society 133, 3219–3221. [DOI] [PubMed] [Google Scholar]
- [92]. Zhang X, Chen H, Le S, Rouzina I, Doyle PS, and Yan J (2013) Revealing the competition between peeled ssDNA, melting bubbles, and S-DNA during DNA overstretching by single-molecule calorimetry, Proceedings of the National Academy of Sciences of the United States of America 110, 3865–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93]. Backer AS, Biebricher AS, King GA, Wuite GJL, Heller I, and Peterman EJG (2019) Single-molecule polarization microscopy of DNA intercalators sheds light on the structure of S-DNA, Science advances 5, eaav1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94]. Shokri L, Rouzina I, and Williams MC (2009) Interaction of bacteriophage T4 and T7 single-stranded DNA-binding proteins with DNA, Physical biology 6, 025002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95]. Biebricher AS, Heller I, Roijmans RF, Hoekstra TP, Peterman EJ, and Wuite GJ (2015) The impact of DNA intercalators on DNA and DNA-processing enzymes elucidated through force-dependent binding kinetics, Nature communications 6, 7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96]. King GA, Biebricher AS, Heller I, Peterman EJG, and Wuite GJL (2018) Quantifying Local Molecular Tension Using Intercalated DNA Fluorescence, Nano letters 18, 2274–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97]. Candelli A, Hoekstra TP, Farge G, Gross P, Peterman EJ, and Wuite GJ (2013) A toolbox for generating single-stranded DNA in optical tweezers experiments, Biopolymers 99, 611–620. [DOI] [PubMed] [Google Scholar]
- [98]. Lee KS, Balci H, Jia H, Lohman TM, and Ha T (2013) Direct imaging of single UvrD helicase dynamics on long single-stranded DNA, Nature communications 4, 1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99]. Zhou R, Kozlov AG, Roy R, Zhang J, Korolev S, Lohman TM, and Ha T (2011) SSB functions as a sliding platform that migrates on DNA via reptation, Cell 146, 222–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100]. Lee KS, Marciel AB, Kozlov AG, Schroeder CM, Lohman TM, and Ha T (2014)Ultrafast redistribution of E. coli SSB along long single-stranded DNA via intersegment transfer, Journal of molecular biology 426, 2413–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101]. Bell JC, and Kowalczykowski SC (2016) Mechanics and Single-Molecule Interrogation ofDNA Recombination, Annual review of biochemistry 85, 193–226. [DOI] [PubMed] [Google Scholar]
- [102]. Hegner M, Smith SB, and Bustamante C (1999) Polymerization and mechanical properties of single RecA-DNA filaments, Proceedings of the National Academy of Sciences of the United States of America 96, 10109–10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103]. Galletto R, Amitani I, Baskin RJ, and Kowalczykowski SC (2006) Direct observation of individual RecA filaments assembling on single DNA molecules, Nature 443, 875–878. [DOI] [PubMed] [Google Scholar]
- [104]. Bell JC, Plank JL, Dombrowski CC, and Kowalczykowski SC (2012) Direct imaging of RecA nucleation and growth on single molecules of SSB-coated ssDNA, Nature 491, 274–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105]. Forget AL, and Kowalczykowski SC (2012) Single-molecule imaging of DNA pairing by RecA reveals a three-dimensional homology search, Nature 482, 423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106]. De Vlaminck I, van Loenhout MT, Zweifel L, den Blanken J, Hooning K, Hage S, Kerssemakers J, and Dekker C (2012) Mechanism of homology recognition in DNA recombination from dual-molecule experiments, Molecular cell 46, 616–624. [DOI] [PubMed] [Google Scholar]
- [107]. Hilario J, Amitani I, Baskin RJ, and Kowalczykowski SC (2009) Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules, Proceedings of the National Academy of Sciences of the United States of America 106, 361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108]. van Mameren J, Modesti M, Kanaar R, Wyman C, Peterman EJ, and Wuite GJ (2009) Counting RAD51 proteins disassembling from nucleoprotein filaments under tension, Nature 457, 745–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109]. Ma CJ, Gibb B, Kwon Y, Sung P, and Greene EC (2017) Protein dynamics of human RPA and RAD51 on ssDNA during assembly and disassembly of the RAD51 filament, Nucleic acids research 45, 749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110]. Zhao XC, Fu H, Song L, Yang YJ, Zhou EC, Liu GX, Chen XF, Li Z, Wu WQ, and Zhang XH (2019) S-DNA and RecA/RAD51-Mediated Strand Exchange in Vitro, Biochemistry 58, 2009–2016. [DOI] [PubMed] [Google Scholar]
- [111]. van Mameren J, Wuite GJL, and Heller I (2018) Introduction to Optical Tweezers:Background, System Designs, and Commercial Solutions, Methods in moìecuìar biology 1665, 3–23. [DOI] [PubMed] [Google Scholar]
- [112]. Brouwer I, Sitters G, Candelli A, Heerema SJ, Heller I, de Melo AJ, Zhang H, Normanno D, Modesti M, Peterman EJ, and Wuite GJ (2016) Sliding sleeves of XRCC4-XLF bridge DNA and connect fragments of broken DNA, Nature 535, 566–569. [DOI] [PubMed] [Google Scholar]
- [113]. Nathwani B, Shih WM, and Wong WP (2018) Force Spectroscopy and Beyond: Innovations and Opportunities, Biophysical journal 115, 2279–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114]. Georgescu RE, Schauer GD, Yao NY, Langston LD, Yurieva O, Zhang D, Finkelstein J, and O’Donnell ME (2015) Reconstitution of a eukaryotic replisome reveals suppression mechanisms that define leading/lagging strand operation, eLife 4, e04988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115]. Yeeles JT, Deegan TD, Janska A, Early A, and Diffley JF (2015) Regulated eukaryotic DNA replication origin firing with purified proteins, Nature 519, 431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116]. Wasserman MR, Schauer GD, O’Donnell ME, and Liu S (2019) Replication Fork Activation Is Enabled by a Single-stranded DNA Gate in CMG Helicase, Cell, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117]. Stephens AD, Banigan EJ, and Marko JF (2019) Chromatin’s physical properties shape the nucleus and its functions, Current opinion in cell biology 58, 76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]